Abstract
Withania somnifera (L.) Dunal is a valued medicinal plant with pharmaceutical applications. The present study was undertaken to analyze the salicylic acid induced leaf transcriptome of W. somnifera. A total of 45.6 million reads were generated and the de novo assembly yielded 73,523 transcript contig with average transcript contig length of 1620 bp. A total of 71,062 transcripts were annotated and 53,424 of them were assigned GO terms. Mapping of transcript contigs to biological pathways revealed presence of 182 pathways. Seventeen genes representing 12 pathogenesis-related (PR) families were mined from the transcriptome data and their pattern of expression post 17 and 36 hours of salicylic acid treatment was documented. The analysis revealed significant up-regulation of all families of PR genes by 36 hours post treatment except WsPR10. The relative fold expression of transcripts ranged from 1 fold to 6,532 fold. The two families of peroxidases including the lignin-forming anionic peroxidase (WsL-PRX) and suberization-associated anionic peroxidase (WsS-PRX) recorded maximum expression of 377 fold and 6532 fold respectively, while the expression of WsPR10 was down-regulated by 14 fold. Additionally, the most stable reference gene for normalization of qRT-PCR data was also identified. The effect of SA on the accumulation of major secondary metabolites of W. somnifera including withanoside V, withaferin A and withanolide A was also analyzed and an increase in content of all the three metabolites were detected. This is the first report on expression patterns of PR genes during salicylic acid signaling in W. somnifera.
Introduction
Withania somnifera (L.) Dunal belonging to family Solanceae is commonly known as Ashwagandha or Indian ginseng and is a valued medicinal plant with pharmaceutical and nutraceutical applications. It is widely used in traditional medical systems of India and Africa as an adaptogens or vitalizers. The phytochemical analysis of root and leaf tissues of this species has been extensively studied [1], [2] and the major metabolites reported include alkaloids (isopelletierine, anaferine), steroidal lactones (withanolides, withaferins), saponins (sitoindoside VII and VIII) and withanolides. These chemical components possess anti-inflammatory, anti-stress, antitumor, antioxidant, anti-aging, immunomodulatory properties, hemopoetic effect, rejuvenating effect and provide cardiovascular protection [3], [4], [5], [6]. In a recent report, the leaf and root transcriptome of W. somnifera was analyzed to elucidate the withanolide biosynthetic pathway [7].
Molecular signaling during plant defense response is widely documented and involves three major pathways including salicylic acid (SA) dependent pathway predominant during interactions with biotrophic pathogens and jasmonic acid (JA) and ethylene (ET) dependent pathways effective during necrotrophy and herbivory. Extensive cross talk between the pathways has been reported [8], [9], [10]. Other phytohormones like abscisic acid, gibberellins, auxins, cytokinins, and brassinosteroids are also documented to regulate plant immune response [11], [12] indicating that plant growth and defense are tightly linked [13].
SA is the key hormone during biotic defense response and levels of SA and its glycosylated conjugate (SAG) in tissues are known to drastically accumulate both locally and systemically after pathogen infection [14]. Additionally, blockade of SA, impairs deployment of systemic acquired resistance (SAR) [15]. The best characterized SA induced genes (SAIGs) include the pathogenesis – related (PR) gene families coding for proteins with antimicrobial activity [16], [17]. Studies at molecular level have indicated that the SAIGs are activated by transcriptional control rather than by increase in the mRNA levels [18]. An extensive reprogramming from primary to secondary pathways with down-regulation of non-essential cellular activities is also reported during SA signaling [19].
Exogenously application of SA was reported to enhance disease resistance and induce PR gene expression in a wide variety of plant species like sunflower, wheat, Musa sp. and pepper [20], [21], [22], [23]. Further, the expression of PR genes during host-pathogen interaction has been extensively documented in solanaceous species like tomato [24], [25], tobacco [26], potato [27], [28], [29] and Capsicum [30]. The accumulation of PR proteins/up-regulation of PR genes during host – pathogen interaction in woody perennials was reviewed and the predominantly reported PRs in trees included PR-1, PR-2, PR-3, PR-5, PR-9, PR-10 and PR-12 [31].
Transcriptome analysis to comprehend gene expression during pathogen infection was recently reported from several species including Musa sp. [32], [33], wheat [34], potato [35], Arabidopsis [36], peach [37], Lactuca sativa [38] and Citrus sp. [39], [40]. However, to our knowledge the transcriptome induced by SA in plants has not been reported. In the present study, the leaf transcriptome of W. somnifera during exogenous application of SA was characterized. The RNA-Seq approach employed in the present study to analyze the global expression of transcripts during SA signaling is the first report on understanding the SAIGs in this species.
Materials and Methods
Plant material
Withania somnifera seeds were germinated in vitro and axillary shoots from one month old plants were used as explants for micropropagation. Multiple shoot induction was done in MS media supplemented with 0.5 mg/L BA and cultures were incubated in 25±2°C, 40–50% relative humidity with photoperiod of 16 h light and 8 h dark conditions. The proliferated multiple shoots were maintained by regular sub-culturing every 4–5 weeks.
Exogenous application of salicylic acid (SA)
Initially, the optimization of SA concentration for exogenous application was conducted by incubating the leaf discs from in vitro raised plantlets to different concentration of SA (5 mM, 10 mM and 20 mM) at room temperature. A control of sterile water treatment was included to document the elicitation of SA. Observations were made for appearance of yellowing and necrotic symptoms in both SA and water treated (control) leaf discs.
In subsequent experiments, 5 mM SA was spayed uniformly on the plantlets aseptically at every 12 hours for a total time period of 36 hours. The control treatment included exogenous application of sterile water on the plantlets. Two hours after the final treatment, plantlets were washed thoroughly with sterile water and leaves were excised and immersed in RNA stabilization reagent, RNAlater (Qiagen, Hilden, Germany) for RNA isolation and transcriptome analysis.
The experiment on reference gene selection was conducted on leaves harvested from water treated (control) and SA spayed (treated) plantlets harvested 17 hours post application. Expression profiling of PR genes and estimation of secondary metabolites were conducted on SA sprayed leaves harvested from 17 and 36 hour post SA treatment against water treated control leaves harvested at 36 hour.
RNA extraction, library construction and sequencing
Total RNA from 36 hour post SA treated leaves was isolated using Plant tissue total RNA extraction spin kit (Chromous Biotech Pvt Ltd, India) and RNA integrity was confirmed using the 2100 Bioanalyzer (Agilent Technologies Inc., Santa Clara, CA). Subsequently, TruSeq RNA Sample Preparation Kit (Illumina Inc., San Diego, CA, USA) was used for purification and fragmentation of RNA, cDNA synthesis, end repair and adapter ligation followed by enrichment with PCR to create a cDNA library suitable for cluster generation following manufacturer's protocol. The QC of the amplified library was determined using high sensitivity bioanalyzer chip (Agilent Technologies Inc., Santa Clara, CA). The sequencing of the cDNA library was performed on Illumina Genome Analyzer IIx sequencer and 72 base paired end sequencing was conducted.
De novo assembly and sequence annotation
The raw reads generated were filtered for weak and low signals (mean quality score > = 20) followed by adaptor trimming using Trimmomatic read trimming tool for Illumina NGS data [41]. The high quality (HQ) reads were then assembled de novo into contigs with De-brujin graph based assembler Velvet 1.2.07 (http://www.ebi.ac.uk/~zerbino/velvet/) [42] on different kmers. The contig assembly was followed by transcriptome assembly with default parameters using Oases transcriptome Assembly pipeline 0.2.08 (http://www.ebi.ac.uk/zerbino/oases/) [43]. The de novo assembly validation was conducted using CLC Genomics Workbench (CLC Bio, Aarhus, Denmark). The functional annotation was performed by aligning the transcript contigs to non-redundant (Nr) database of NCBI using BLASTx for green plants (http://www.ncbi.nlm.nih.gov) with cut off E value 1e -06 to identify transcripts with significant similarity.
Analysis and validation of transcript contig assembly
Gene ontology (GO) mapping of transcript contigs were performed to classify their functions and categorize them into biological, molecular and cellular functions [44]. The Accession IDs derived from BLASTx were directly searched in GO database. GO terms for annotated transcript contigs were retrieved using different databases including UniProtKB (http://www.uniprot.org/help/uniprotkb), TAIR (www.arabidopsis.org/) and Sol Genomics Network (SGN) (http://solgenomics.net/). The E-value distribution and sequence similarity distribution was determined to evaluate the success of the alignment for a given sequence database and the overall performance of the alignments, respectively. The experimental evidence for existence of the protein was determined through the Evidence Code (EC) distribution of the annotated transcript contigs. The annotation distribution graph was prepared to determine the number of GO terms assigned per sequence.
Functional characterization and pathway analysis
The ortholog assignment and mapping of the transcript contigs to biological pathways were performed according to the Kyoto Encyclopedia of Genes and Genomes (KEGG) automatic annotation server (KAAS) [45]. All transcript contigs were compared against the KEGG database using BLASTx, with default threshold bit –score value of 60.
Identification of simple sequence repeats (SSRs)
All transcript contigs in the draft assembly were analyzed for presence of SSRs using MISA standalone SSR tool (http://pgrc.ipk-gatersleben.de/misa). SSR motifs from di- to hexa-nucleotide were identified with the criteria of atleast 6 repeats for di- and five repeats for tri-, tetra-, penta- and hexa- nucleotide.
Discovery of miRNAs
The potential conserved miRNAs in the transcriptome data was identified by mapping the transcript contigs against known plant hairpin (5,077) and mature (5,855) miRNA sequences deposited in miRBase version 19 (http://www.mirbase.org/) using CLC Genomic Work bench [46].
Selection of reference genes for normalization of qRT-PCR data
Expression profiling of transcripts in biological systems using qRT-PCR obligates the use of a stable reference or house-keeping gene for normalization of data. In the present study, a set of six commonly used reference genes, viz., 60 S ribosomal protein L2 (WsRPL), actin (WsAct), glyceraldehyde-3-phosphate dehydrogenase (WsGAPDH), α-tubulin (WsTUB), ADP-ribosylation factor (WsARF) and histone H2B (WsH2B) were mined from the transcriptome data and analyzed for its suitability as a reference gene for the given tissue and experimental condition. Primer pairs were designed using Beacon Designer (http://www.premierbiosoft.com/molecular_beacons), analyzed using BLASTn and Primer-BLAST (www.ncbi.nlm.nih.gov/tools/primer-blast) to ensure specificity and were subsequently used in qRT-PCR analysis (table 1).
Table 1. Details of primer pairs and amplicon size of reference genes used for normalization of qRT-PCR data in W. somnifera.
| Gene Name and ID | Sequence of Primer pairs | Amplicon length (bp) | Tm (°C) |
| Forward Primer Sequence 5′-3′ | |||
| Reverse Primer Sequence 5′-3′ | |||
| 60 S ribosomal protein L2 (WsRPL) | GAGGACGTACTGAGAAACCTATG TACTAGCATGACCAATGTGTTGA | 156 | 79.4 |
| Actin (WsAct) | AGATATTCAGCCTCTTGTCTGTG ATTGAGCCTCATCACCAACATA | 170 | 81.1 |
| Glyceraldehyde-3-phosphate dehydrogenase (WsGAPDH) | ATGCTCAAGTATGATTCCACTCA GAAGACACCAGAAGATTCAACAAC | 174 | 78.2 |
| α-Tubulin (WsTUB) | AAATGCTTGCTGGGAACTTTAC TCCTGTCCTCACTTCATCAATG | 193 | 82.2 |
| ADP-ribosylation factor (WsARF) | GAGATTGTTACCACTATTCCTACCA CACGATCACGATCATTACTATCAAC | 178 | 78.6 |
| Histone H2B (WsH2B) | TTCTAGCAAGTCAATGGGTATCAT CTTTAGTTCCTTCAGAAACAGCAT | 188 | 78.1 |
Leaves were harvested from water treated (control) and SA treated plantlets (as described earlier) after 17 hours post treatment and stored at −80°C until further use. Total RNA was isolated individually from all tissues using Plant tissue total RNA extraction spin kit (Chromous Biotech Pvt Ltd, India) using manufacturer's protocol. The qRT-PCR reactions were performed in fast optical reaction tube (Microamp- Applied Biosystems, USA) and a 20 µl reaction included 50 ng of cDNA, 10.0 µl SYBR Green JumpStart Taq ReadyMix (Sigma, St. Louis, MO, USA), 150 nM each of forward and reverse primer and 0.8 µl of 20 mg/ml BSA. All reactions were conducted as three independent technical replicates in ABI PRISM 7500 Step One plus Sequence Detection System (Applied Biosystems, USA) using the following program - one cycle of 95°C for 10 min; 40 cycles of 95°C for 15 sec and 60°C for 1 min. The melting curve was determined for each primer pair to confirm the specificity of the amplified product.
Statistical analysis for stability of gene expression
The expression level and stability of the six selected endogenous genes were evaluated with statistical programs including geNorm [47] and Normfinder [48] downloaded from GenEX standard software (http://GenEx.gene-quantification.info) and BestKeeper, an Excel-based tool [49] downloaded from (http://www.gene-quantification.de/bestkeeper.html). Expression levels were assessed based on the number of amplification cycles needed to reach a fixed threshold (Cycle threshold - Ct) in the exponential phase of PCR.
Ct values were imported to GenEX software and analyzed using geNorm and Normfinder tools following the developer's instruction. In geNorm analysis, the data in logarithmic value was automatically converted to linear scale and the gene pair with lowest M value (average expression stability value) was considered to have most stable expression [47]. NormFinder uses data in the logarithmic scale and takes into consideration information of groupings of samples and predicts the optimal reference based on the variability values [48]. In BestKeeper, the average Ct values were used to analyze the stability value of the candidate genes. BestKeeper creates a pair-wise correlation coefficient between each gene and generates the BestKeeper index as well as calculates coefficient of variance (CV) and the standard deviation (SD) of the Ct values using the whole data set [49]. Genes that exhibited the lowest coefficient of variance and standard deviation (CV±SD) were identified as the most stable gene. Genes that show a SD greater than 1 were considered unstable [50].
Data mining for PR genes and expression profiling during SA signaling
The annotated transcriptome data was manually mined for pathogenesis –related genes [16] and the identity was confirmed by BLASTx analysis (table 2). The transcript contigs from each PR gene families were individually aligned using Clustal W2 (http://www.ebi.ac.uk/Tools/msa/clustalw2/) and distinct genes having pair-wise score less than 70 from each family were selected for expression profiling. Primer pairs for 17 genes representing 12 PR families were designed and synthesized (IDT Technologies, Canada) for qRT-PCR (table 2).
Table 2. Details of primer pairs and amplicon size of pathogenesis - related genes used for expression profiling during SA signaling.
| S. No. | PR Family | Name | Sequence similarity | Primer Pairs | Amplicon size (bp) | Tm (°C) |
| 1 | PR-1 | Pathogenesis- related protein PR-1 (WsPR1) | Capsicum annum (AY560589.1) | TGATGAGAAGCAATGGTATGACTAT CGATCAGACATCAGTTGGAAGT | 183 | 79.8 |
| 2 | PR-2 | β-1,3-glucanase (WsB13G) | Solanum tuberosum (JX838875.1) | ACATTGCTTCGTCTATCAAAGTTTC CACCATGAGGTAAGAACCAGTT | 158 | 78.5 |
| 3 | PR-3 | Class I chitinase (WsCHTN1) | Solanum tuberosum (U02605.1) | CTCAATCACCAAAGCCATCTTG AATTCCGCAGTACCTTCTGTAAA | 194 | 81.9 |
| 4 | Class II chitinase (WsCHTN2) | Solanum tuberosum (U49969.1) | CACAAGACAACAAGCCATCATG TAGAATCCAATTCGATCATCCACTT | 175 | 80 | |
| 5 | Class IV chitinase (WsCHTN4) | Nicotiana tabacum (AB267862.1) | CTTCAAGCAATAATGGAGGTTCAG CTCACGCTTAGAATCATCAGTAGA | 185 | 76.1 | |
| 6 | PR-5 | Thaumatin-like protein (WsTHAU) | Solanum lycopersicum (XM_004230458.1) | ACGTCTTTGACACCGATGAATA ACATAGTCAGTAGAAGAGCAAGTG | 166 | 78.1 |
| 7 | PR-6 | Cystatin-like protein (WsCYST) | Solanum tuberosum (DQ191655.1) | GTTGAAGATGGTCCTACCTTTACT CCTCAGCATTAGCATGAACAATC | 196 | 79.4 |
| 8 | Serine protease inhibitor like protein (WsSPI) | Solanum lycopersicum (XP_004234308.1) | ATGCCCGTCAAATTCATTAAGTTT TCCTCCAGTCTCCAACAATCTA | 171 | 76.5 | |
| 9 | PR-8 | Class III chitinase (WsCHTN3) | Capsicum annum (AB267862.1) | GAACTTGGATCACCACTTCATTAC GTAGTGAACTGACATGGAGGATT | 197 | 77.1 |
| 10 | PR-9 | Lignin-forming anionic peroxidase (WsL-PRX) | Solanum lycopersicum (XM_004250354.1) | TCCACATTCTATGATCGCACTT AACGCAGTCTTCTCACTAACAA | 194 | 78.3 |
| 11 | Suberization-associated anionic peroxidase (WsS-PRX) | Solanum tuberosum (AAA33837.1) | GTGCAAAGAGAAATTCAGACAAGT AGAATACCTCCATCACAACCATC | 175 | 80.5 | |
| 12 | PR-10 | PR-10 type pathogenesis-related protein (WsPR10) | Nicotiana tabacum (AB518291.1) | AGTTGCTCATATAGAAGTCAAGTGT TCCATCATAGTTCAATCTCCATTCA | 169 | 76.2 |
| 13 | PR-11 | Chitinase, class V (WsCHTN5) | Nicotiana tabacum (X77110.1) | TGCGAACAATCATGGTCTTAGA TCCTGAGTAACAATAATCTCCAACA | 163 | 78.4 |
| 14 | PR-12 | Defensin (WsDFSN) | Capsicum annum (X95730.1) | TGCTGGTTTTTGCTACTGAGGCA CAGAAGCAACGGCGACGGAATC | 151 | 81.4 |
| 15 | PR-14 | Nonspecific lipid transfer protein 2 (WsLTPa) | Capsicum annum (AY496100.1) | GTTGTGGTGGAGTTAAGAATTTGAT GGGCTGATCTTGTAAGGAATGT | 169 | 80.3 |
| 16 | Non-specific lipid-transfer protein-like protein At2g13820-like (WsLTPb) | Solanum lycopersicum (XP_004229337.1) | GCACTTCAACTCAATGTTACACTT CAACAGAAGGAATGGGACTATTTG | 193 | 79 | |
| 17 | PR-16 | Germin-like protein subfamily 1 member 20 (WsGER1-20) | Solanum tuberosum (AFW90592.1) | TAATGGCTGTGGTGACTTCAATA GGCATATTCAAACCCGATTTAAAGA | 169 | 78.4 |
Total RNA was isolated from leaves harvested from 17 and 36 hours SA treated plantlets while leaves harvested from 36 hour post water treated plantlets were used as control. qRT-PCR reactions were conducted as described earlier for reference gene selection. The melting curve was determined for each primer pair to confirm the specificity of the amplified product and all reactions were conducted in three independent technical replicates. The qRT-PCR data was analyzed using the ΔΔCT method described by Livak and Schmittgen [51]. While calculating the ΔΔCT, undetermined Ct values were imputed to 40 [52] and fold decrease was calculated as the reciprocal of the fold change [53].
Statistical analysis of data
The fold expression of transcripts between control and SA treated cDNA pools were statistically analyzed by T-Test using SPSS software (version 20.0) and difference between treatments were considered statistically significant when P<0.01.
Quantification of secondary metabolites during SA signaling
Leaves were harvested from water treated (control) and SA treated plantlets (as described earlier) after 17 and 36 hours post treatment and air dried. The samples were subsequently ground into fine powder and used for secondary metabolite analysis.
Extraction of metabolites
Three hundred mg of each leaf samples were extracted directly with chloroform hexane using a tissue homogenizer (Kinematica Polytron Homogenizer PT 6100). All solvents used for the extraction were of HPLC grade (Qualigen fine chemicals, India). The solvent portion was collected by filtration and the process was repeated until the chloroform layer was almost colourless. The combined extracts were filtered and the filtrate was concentrated under reduced pressure using rotovap (Laborota 4000, Heidolph, Germany) followed by high vacuum drying (EZ-2, Genevac, USA) to remove traces of solvent. Subsequently, the samples were lyophilized and stored at −20°C for analysis of metabolites. The dried samples were later dissolved (3 mg/ml) in methanol and filtered through 0.45 µm filter and degassed for one minute. The external standards used in HPLC analysis included withanosides-V, withaferin-A and withanolide-A (Natural Remedies Pvt. Ltd., Bangalore, India). The stock solutions of external standards were prepared in methanol at the concentration of 1 mg/mL.
HPLC analysis of secondary metabolites
The estimation of the three metabolites were performed on a Waters liquid chromatograph equipped with a Waters 600 controller, a Waters Delta 600 solvent delivery system, a Rheodyne 7125 sample injector fitted with a 20 µL loop, and a Waters 2996 Photodiode Array Detector, with Waters Empowered 2.154 software. A Supelco 516 C18 (4.6 mm×25 cm) reverse phase analytical column equipped with a Waters µBondapak C18 10 µm precolumn was used for estimation. The wavelength scan range of the PDA was set to 190–350 nm and the presence of withanosides-V, withaferin-A and withanolide-A was detected at 227 nm. A. The isocratic mobile phase consisted of 60% acetonitrile containing 0.1% acetic acid (solvent A) and 40% water containing 0.1% acetic acid (solvent-B) at a flow rate of 1.0 mL min−1. The metabolites were estimated in comparison to the external standards and the results were presented as µg mg−1 of dry weight of leaf tissue.
Results
Optimization of SA treatment
Leaf discs subjected to different concentrations of SA showed varying levels of necrotic symptoms. Yellowing of leaf discs was observed in all treatments by one hour of SA application and complete necrosis was observed in 20 mM SA treatment. Hence, the optimal concentration was selected as 5 mM (Figure S1), which was used for further experiments.
Subsequently, plantlets were sprayed with 5 mM SA in intervals as described earlier and a control with plantlets sprayed with sterile water was maintained, to document the effect of SA. The SA treated plantlets showed initial symptoms of yellowing by 17th hours and by 36th hours the symptoms were prominent (Figure 1). No symptoms were observed in water treated control plantlets.
Figure 1. Effect of salicylic acid on in vitro grown plantlets of W. somnifera.

0 = Plantlets immediately after application of SA; 17 h = Plantlets after 17 hours post SA treatment; 36 h = Plantlets after 36 hours post SA treatment.
Transcriptome sequencing and Read Statistics
Total RNA was isolated from leaves harvested from 36 hours post SA treated plantlets. The RNA was quantified and 10 µg of total RNA at a concentration of 400 ng/µl with OD260/280 = 1.8, RNA 28 S:18 S≥1.0 and RNA Integrity Number (RIN) of 7.0 was used for cDNA library construction. The cDNA library was sequenced using Illumina Genome Analyzer IIx Sequencer. The raw paired - end - sequence data was deposited in NCBIs Short Read Archive with the accession number SRA107547. A total of 45.6 million, 72 base paired – end reads (3.28 Gb) was generated. The raw reads were subjected to quality control and the total number of HQ reads was 87.26% (39.8 million reads).
De novo assembly and functional annotation
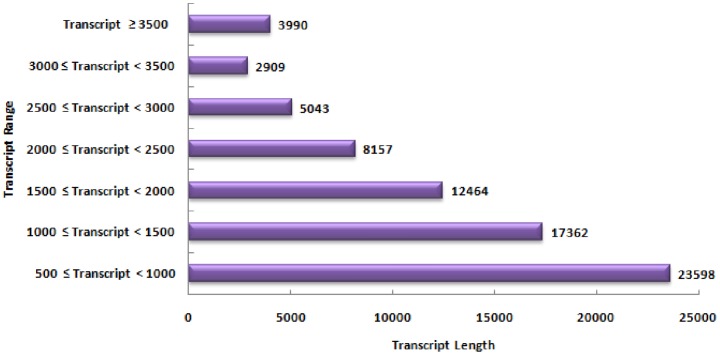
The de novo assembly generated 73,523 transcript contigs with average transcript contig length of 1620 bp and the maximum length of contig transcript was 9489 bp. The total number of bases in transcript contigs was 119,136,311 bases (1.19 Gb). The distribution of transcript contig length is shown in figure 2. N50 (the smallest contig size in which half the assembly is represented) is the statistics used to assess the quality of sequence assembly and higher values suggest better assembly. In the present study the N50 was determined to be 1,978 bp. The assembled transcript contigs were annotated using BLASTx against Nr database for Viridiplantae (table S1). A total of 71,062 (96.65%) transcript were annotated while 2,461 transcripts had no significant BLAST hits. Maximum percent of W. somnifera sequences showed significant similarity with Vitus vinifera, followed by Populus trichocarpa and Ricinus communis. The E- value distribution and sequence similarity distribution are provided as Figure S2 and S3. The sequence similarity distribution revealed that about 80% of transcript contigs had positive alignment length which ranged from 50–90 percent.
Figure 2. Distribution of transcript contig length in RNA-Seq data of W. somnifera.
Gene Ontology (GO) Classification
The annotated transcript contigs were mapped on GO database and 53,424 (75%) sequences were assigned GO terms while 17,683 were not assigned any functional categories. The GO terms for the annotated transcript contigs were retrieved from different databases and maximum terms could be derived from UniProtKB followed by TAIR (Figure S4). The evidence code distribution for sequences and their translated products showed an over-representation of electronic annotations (IEA), although other non-automatic codes, such as inferred from direct assay (IDA), inferred from sequence or structural similarity (ISS) and inferred by mutant phenotype (IMP) were also well represented (Figure S5A and S5B). The annotation score distribution revealed that maximum number of transcript contigs annotated with 55–70 percent similarity (Figure S6).
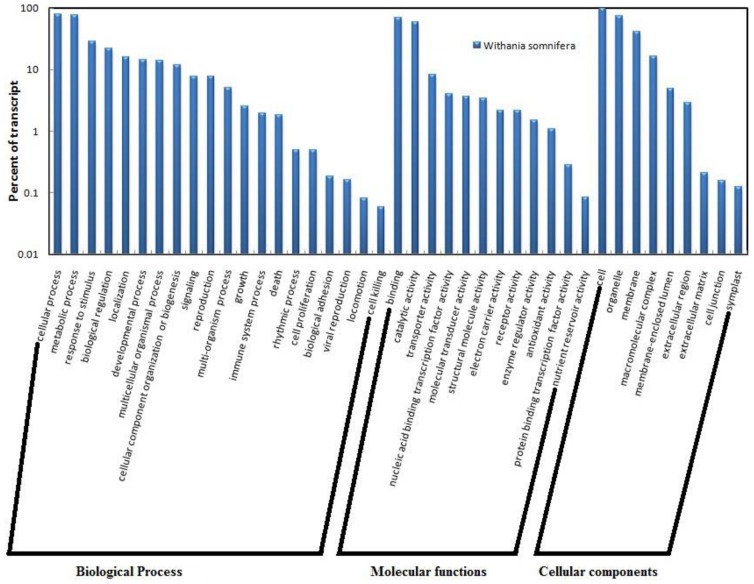
The GO terms were grouped into different levels for all the three ontology domains i.e., biological processes, molecular functions and cellular components (figure S7). The number of transcript contigs categorized under biological function was 37,831 while 44,216 transcript contigs grouped under molecular functions. The cellular components clustered 38,312 transcript contigs. Each transcript contigs could be multi-functional and hence can lie in more than one GO term. The “metabolic process” and “cellular process” constituted the main biological processes in SA treated leaves of W. somnifera with 70% transcript contigs grouping to these ontologies. The main “metabolic function” included primary metabolic processes, biosynthetic processes, nitrogen metabolism while the “cellular process” included response to stimulus, cellular development process, cellular localization, cell communication and cell adhesion. “Catalytic activity” including transferase, hydrolase, oxidoreductase, ligase, lyase and isomerase activity were the main molecular functions in SA treated leaf tissues of this species. The ‘binding activity’ including protein, nucleotide, lipid and cofactor binding accounted for about 55% of the molecular functions. The cellular component represented by 38,312 transcript contigs mainly included genes involved in cell function (99%) followed by transcripts related to ‘organelle’ functioning (7%) (figure 3).
Figure 3. Gene Ontology classification of transcript contigs grouped under biological processes, molecular functions and cellular components.
Pathway annotation using KEGG
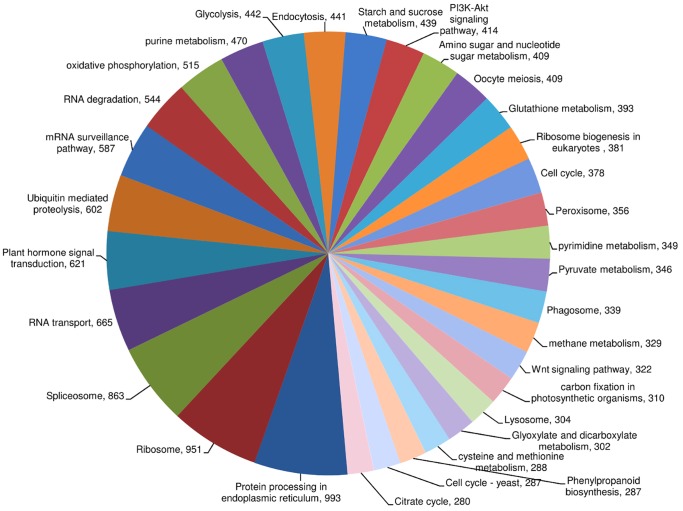
Ortholog assignment and mapping of transcript contigs to biological pathways were performed using KEGG (table S2). The annotated transcript contigs were assigned to 182 pathways and the major representation of transcript contigs was from protein processing in endoplasmic reticulum [PATH: ko04141; 993 transcript contigs] followed by ribosome [PATH: ko03010; 951 transcript contigs], spliceosome [PATH: ko03040; 863 transcript contigs], RNA transport [PATH: ko03013; 665 transcript contigs] and plant hormone signal transduction [PATH: ko04075; 621 transcript contigs] (Figure 4).
Figure 4. Distribution of transcript contigs to different biological pathways.
Values indicate number of transcript contig representation in each pathway. Pathways with number of transcript contigs below 280 are not shown in the chart.
Identification of SSRs
The leaf transcriptome data of W. somnifera generated a total of 4,250 SSRs with maximum representation of tri-nucleotide SSRs (2457) followed by di-nucleotide (1576), hexa-nulceotide (116) and tetra-nucleotide (86). Minimum number of SSRs (15) was registered under the category of penta-nucleotide.
Identification of miRNAs
Hairpin and mature miRNAs in W. somnifera leaf transcriptome was identified by searching the public miRNA database. A total of 911 miRNAs were identified including 51 hairpin and 860 mature plant miRNAs. The mature miRNAs were distributed across 101 families and included isoforms found in various plant species. The largest family was miR169 with 18 members followed by miR171 (14 members), miR166 (12 members) and miR160 (9 members). Further, the family of miR393 and miR395 constituted eight members each.
Selection of reference gene for normalization of qRT-PCR data
Gene expression stabilities of six genes including WsRPL, WsAct, WsGAPDH, WsTUB, WsARF and WsH2B were analyzed for their suitability in normalization of qRT-PCR data. The melt curve analysis of the six reference genes is provided in figure S8. The identification of the most stable reference gene was statistically derived using three independent programs. In geNorm analysis, WsTUB and WsRPL produced the lowest M value (0.21) while WsH2B had the highest M value (1.01) indicating that WsTUB and WsRPL had the most stable expression and WsH2B was the least stable. Normfinder analysis revealed WsRPL (0.11) as the best reference gene with lowest variability value followed by WsTUB. WsH2B (2.12) was predicted as the least stable. Similarly, in BestKeeper analysis WsTUB had a CV±SD value of 1.15±0.36, revealing highest stability followed by WsRPL (1.25±0.35). WsARF and WsH2B documented the least stability with values of 3.9±1.13 and 3.16±0.94 respectively. All the three programs revealed WsTUB as the most suitable reference genes for quantitative gene expression studies in W. somnifera during SA signaling. Hence, WsTUB was used for data normalization in subsequent experiments conducted on expression profiling of PR genes.
Expression profiling of PR genes during SA signaling
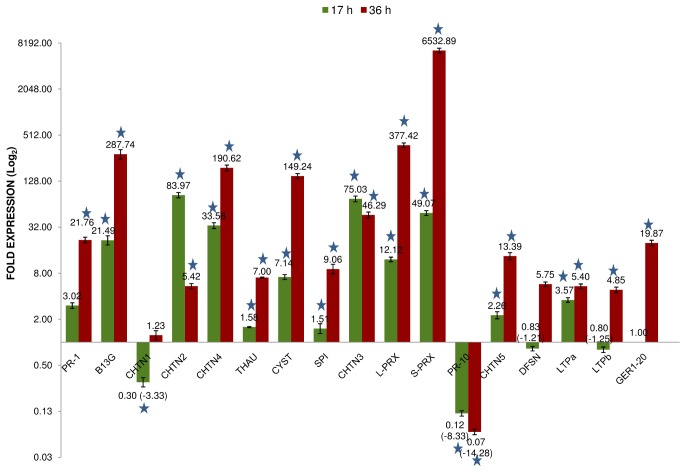
The effect of SA on the temporal expression of 17 selected PR genes in leaves of W. somnifera was investigated across two time points (17 and 36 hours). The melt curve analysis of all the primer pairs revealed single product and absence of non-specific bands (figure S9). The expression of thirteen PR genes belonging to 10 families including PR1, chitinases (PR3, PR8, PR11), peroxidases (PR9), glucanase (PR2), thaumatin – like (PR5), cystatin (PR6), serine protease inhibitor (PR6), one member of lipid transfer protein (PR14) and germin-like (PR16) were up-regulated by 1.4 fold to 83 fold after 17 hours of SA treatment. The class II chitinase (WsCHTN2) documented maximum up-regulation by 83 fold followed by the class III chitinase (WsCHTN3) belonging to PR8 with 75 fold relative increase in expression when compared with its expression in water treated control leaf tissues. The expression of four genes including class I chitinase (WsCHTN1), WsPR10, defensin (WsDFSN) and one member of LTP (WsLTPb) were down-regulated post 17 hour SA treatment (figure 5). The down-regulation of WsPR10 and WsCHTN1 by 8 and 3 fold respectively was significant in comparison to other down-regulated transcripts.
Figure 5. Expression profiling of Pathogenesis-related gene families in W. somnifera.
Results are mean of triplicate data from independent replications and the error bars represent standard deviation. The gene IDs are given in Table-2 and the number on each bar represent relative fold expression. Asterisk indicate statistical significance of P<0.01. 17 h = Expression of PR genes in 17 hours post SA treated leaf tissues; 36 h = Expression of PR genes in 36 hours post SA treated leaf tissues.
The fold expression of all transcripts except WsPR10 was up-regulated by 36 hours of SA treatment and the expression levels ranged from 1 fold to 6532 fold. Very high levels of expression was recorded for peroxidases with 377 and 6532 fold for WsL-PRX and WsS-PRX followed by glucanase with 287 fold, class IV chitinase (WsCHTN4) by 190 fold and cystatin (WsCYST) by 149 fold. The expression of class I chitinase (WsCHTN1), defensin (WsDFSN) and LTP (WsLTPb) which were slightly down-regulated after 17 hours of SA treatment, showed up-regulation by 1 to 5 fold after 36 hours of SA treatment. However, WsPR10 continued to show down-regulation by 14 fold after 36 hours of treatment (Figure 5).
Quantification of secondary metabolites during SA signaling
The content of three major secondary metabolites including withanoside V, withaferin A and withanolide A in the leaf tissues of water treated control and 17 and 36 hour post SA treatment was estimated. An increase in the content of all the three metabolites was recorded. The maximum effect of SA was documented on withanoside V, with an increase in the content by 0.857 µg mg−1 in comparison to negligible levels in control. Withaferin A recorded an increase from 0.499 µg mg−1 in water treated control to 1.26 µg mg−1 in 36 hours SA treated leaf tissues. The withanolide A content was marginally increased by SA application (Table 3; figure 6).
Table 3. Estimation of secondary metabolites in control and salicylic acid treated leaf tissues of W. somnifera.
| Standards | Leaf tissue samples | ||
| Control | SA-17 | SA-36 | |
| Withanoside V | ND | 0.4802±0.0500 | 0.8571±0.1010 |
| Withaferin A | 0.4985±0.0600 | 0.9755±0.1002 | 1.2598±0.1025 |
| Withanolide A | 0.0597±0.0164 | 0.0963±7.3000e-3 | 0.1087±0.0219 |
Mean values ±SD of µg mg−1 of dry weight of leaf sample; ND = Not Detected; Control = leaf samples from water treated plantlets; SA-17 = leaf samples from 17 hours post SA treated plantlets; SA-36 = leaf samples from 36 hours post SA treated plantlets.
Figure 6. Quantitative variation in secondary metabolite content by exogenous application of salicylic acid.
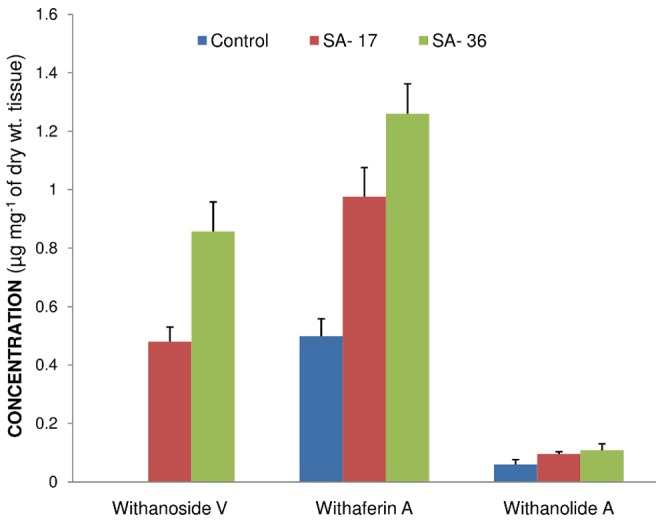
Control = Metabolite content in water treated leaf tissues; SA-17 = Metabolite content in 17 hours post SA treated leaf tissues; SA-36 = Metabolite content in 36 hours post SA treated leaf tissues. Presence of withanoside V was not detected in water treated (control) leaf tissues.
Discussion
Comparison of leaf transcriptome data in W. somnifera
The first report on de novo assembly, functional annotation and pathway analysis in W. somnifera leaf and root transcriptome was recently reported to identify putative genes involved in the withanolides biosynthesis [7]. The present study targeted the analysis of transcripts expressed in salicylic acid treated leaf tissues. The sequencing platforms used in both the studies were different wherein pyrosequencing was employed in the earlier study while Illumina platform was used in the present analysis. The number of transcript contigs annotated from the present assembly was 71,062, which was significantly higher than the transcripts annotated in the earlier study (38,961) using Nr database. The maximum number of transcripts annotated in the earlier report using four databases was 54,639 [7]. The probable reasons for over representation of transcripts in the present study could be attributed to the different assembly and annotation pipelines used across the two reports.
The functional classification and pathway assignment was performed using KEGG and a total of 124 pathways were reported from leaf and root transcriptomes by Gupta and co workers [7] while in the present study, 182 pathways were identified. Significant differences in the number of transcript contigs annotated under different secondary metabolite pathways were documented. In the present study, a total of 1354 transcript contigs was categorized under secondary metabolite pathways while 1068 unigenes were annotated from the previous study. A comparison of the number of transcript contigs/unigenes represented under the different secondary metabolite pathways across both the studies is given in table S3. This difference in transcript representation may be attributed to the total number of annotated transcripts, which significantly differed in both the studies.
Changes in ploidy level are known to significantly shape the plant genomes [54] and impact genetic and epigenetic aspects including gene expression [55], [56]. In Arabidopsis thaliana, altered gene expression was reported between autotetraploid and diploid ecotypes [57]. Similarly, in buffalo grass (Buchloe dactyloides), a significant difference in the number of transcripts assembled from the transcriptome data was documented between the tetraploid and hexaploid cultivar [58] suggesting that ploidy levels can impact the number of annotated transcripts during de novo assembly. In W. somnifera, considerable morphogenetic diversity is documented in Indian populations and intraspecific diploid (2n = 24), tetraploid (2n = 48) and hexaploid (2n = 72) cytotypes are reported [59]. The autotetraploid cytotype of this species was reported to yield higher root alkaloid in comparison to their diploid counterparts [60]. Hence, the difference in the number of annotated transcript contigs/unigenes between the two studies in Withania can also be attributed to a probable difference in ploidy level of the genotypes sequenced in the two studies. The ploidy level of the cytotype used in the present study was not determined while the same was not reported in the earlier study [7].
N50 is a statistical approach to assess the quality of the sequence assembly and higher N50 value indicate better assembly. The N50 of the present assembly was 1,978 bp suggesting optimum assembly and was higher than several plant transcriptome assemblies published earlier including Coctus pictus [61], Cicer arietinum [62], Daucus carota [63], Cajanus cajan [64] and Euphorbia fischeriana [65].
Discovery of EST-SSRs from transcriptome sequencing has significantly facilitated a cost effective method for identification of genic SSRs [66]. These SSRs can be used in genetic diversity analysis and for linkage/QTL/association mapping studies tagging traits of interest [67]. The higher rates of cross species transferability of EST-SSRs [68], [69] due to synteny across species has been demonstrated in rice [70], bread wheat [71], Capsicum [68], [72], sugarcane [73] and cotton [74]. In the present study, a total of 4250 SSRs were identified in comparison to the previous study [7] were a total of 2553 SSRs were reported in leaf transcriptome. Both studies registered higher number of tri- nucleotide repeats and minimum representation of penta- nucleotide SSRs. The abundance of tri-nucleotide repeats have been earlier reported from Catharanthus roseus [75], Ipomoea batatas [76] and Leymus chinensis [77].
In Withania somnifera, marker studies for diversity analysis are limited to RAPD, AFLP and ISSR [78], [79], [80]. Hence, the identification of EST-SSRs from the present study and the earlier report [7] will facilitate future studies on molecular markers in this species.
Elicitor induced production of withanolides under culture conditions
The production of secondary metabolites under in vitro condition are reported to be enhanced by exogenous application of elicitors (biotic and abiotic) in culture media [81] and methyl jasmonate and salicylic acid are widely reported to induce production of secondary metabolites under culture conditions [82], [83]. In W. somnifera, the exogenous application of salacin was reported to induce the production of withaferin A in suspension cultures [84] while chitosan, methyl jasmonate and SA induced the production of withanolides in adventitious root and hairy root culture [83], [85], [86], [87]. Similarly, the present study recorded increase in production of three major metabolites of W. somnifera including withanoside V, withaferin A and withanolide A in leaf tissues, subsequent to exogenous application of SA.
Selection of reference gene for qRT-PCR
Reliable quantification of gene expression levels by qRT-PCR requires the standardization and fine-tuning of several parameters, such as amount of initial sample, RNA recovery and integrity, enzymatic efficiencies of cDNA synthesis, PCR amplification and overall transcriptional activity of the tissues or cells analyzed [88]. Among various methods, internal control genes (reference genes) are most commonly used to normalize qRT-PCR data and reduce possible errors generated during quantification of gene expression [88], [89]. Nevertheless, this method relies on the choice of appropriate house-keeping genes, which ideally has stable expression under different experimental conditions and in different tissue types. In W. somnifera, there are no reports on selection of endogenous reference gene for normalization of qRT-PCR data under any experimental conditions or tissue types. In the earlier reports, actin was used as the reference gene for data normalization [90], [91]. However, in the present study actin was not identified as a stable gene for data normalization while WsTUB was documented to be the most suitable reference gene for quantitative gene expression studies. In members solanaceae family like potato, tobacco, tomato and Capsicum annuum, several house-keeping genes were screened to identify the most stable reference gene for a given experimental condition. In potato, ef1α and ribosomal protein (L2) was reported as the most stable gene during biotic and abiotic stress treatments and actin and tubulin where found to be least stable [92]. Similarly, in tobacco ef1α and L25 was reported as most stable for qRT-PCR studies for developmentally distinct tissues and abiotic stresses [93]. However, in Capsicum annuum, beta tubulin and ubiquitin-conjugating protein showed high stability in sample pools with abiotic stress and hormonal treatments [94]. In tomato, the most stable reference gene for analyzing the gene expression during the interaction with the endophyte Fusarium oxysporum was TUB and PP2ACS for roots and EF1 and PP2ACS for cotyledons [95]. These studies highlight that use of universal reference gene for qRT-PCR may not be ideal for data normalization. Hence, screening of stable reference genes for a given tissue type and experimental condition is a pre-requisite for data validation. The present study in Withania is the first report on identification of stable reference gene, which can support future gene expression studies in this important medicinal plant.
Expression of SAIGs during SA signaling
Salicylic acid is a phenolic compound which plays a central role in plant defense signaling network [96]. It is important for basal defense, protein- mediated defense and systemic acquired resistance [97], [98], [99], [100]. SA-mediated immune response is integral part of both PAMP-triggered and effector-triggered immunity [100] and also a prerequisite for activation of SAR [101]. Earlier studies have indicated that pathogen infection leads to SA accumulation both in locally infected tissues and distal uninfected tissues that develop SAR [102], [103] and concurrently results in up-regulation of PR genes [104]. Studies have revealed that SA also plays an important role in controlling the cellular redox balance at the onset of SAR [105], [106]. The SA associated gene expression has been grouped into three categories in Arabidopsis, type I including genes encoding enzymes that are directly involved in SA biosynthesis, type II including proteins that do not act directly on SA biosynthesis but mutations in these genes lead to compromised SA accumulation and disease susceptibility and type III including genes which act downstream of SA accumulation like NPR1, a major signal transducer of SA and PR genes [107], [108]. Exogenous application of SA can mimic the endogenous increase that occurs during pathogen infection and elicit SAR.
The PR proteins/genes which are considered signatures of the SA signaling are PR1, PR2 and PR5 [109], [110]. SA signaling mutants and transgenics expressing bacterial salicylate hydroxylase with reduced SA accumulation have impaired ability of SAR and reduced expression of PR1, PR2 and PR5 [111], [112], [113]. In Arabidopsis, several mutants with impaired disease response have been developed to understand the signaling pathways operational during pathogenesis. Mutant phenotypes with increased SA levels including constitutive immunity (cim) [114], constitutive expression of PR proteins (cpr) [115], [116] and defense no cell death (dnd1) [117] recorded higher expression levels of PR genes. Similarly, mutants with impaired SA accumulation like pad4 [118] and SA induction–deficient (sid) [119] documented low expression of PR1 and increased disease symptoms, reiterating the predominant role of SA in disease resistance and induction of PR genes/proteins. In the present study the temporal expression of 17 genes representing 12 pathogenesis-related (PR) families were analyzed during SA signaling.
Plant chitinases classified under PR protein families PR3, PR4, PR8 and PR11 [120], [121] include one of the most characterized families of PR proteins which catalyze the hydrolysis of chitin present in fungal cell wall and exoskeleton of insects. The induction of different classes of chitinases during exogenous application of SA was reported in Pinus elliottii [122]; cucumber [123]; cotton [124]; Castanea sativa [125]; tobacco [126]; sweet cherry [127], [128], grape berries [129], sorghum [130], Casuarina equisetifolia [131], Malus hupehensis [22] and tomato [132]. In Vitis vinifera, two classes of chitinases (Class I and Class III) were analyzed for their expression during SA mediated SAR and results revealed that the class III chitinase expressed in distal leaves, suggesting it as a reliable indicator of SAR [133]. Similarly, in the present study, a significant up-regulation of three chitinases including WsCHTN2, WsCHTN3 and WsCHTN4 and moderate induction of WsCHTN5 was documented. However, the present study also documented the down-regulation of a class I chitinase (WsCHTN1) after 17 hours of SA treatment. In concurrence to the present result, study in Vitis vinifera revealed no significant change in expression of class I chitinase when challenged with SA [133].
Beta-1,3 glucanase classified under PR-2 play a direct role in fungal defense by hydrolyzing the fungal cell wall and an indirect role by generating oligosaccharide elicitors [134]. In the present study, this gene (WsB13G) was up-regulated by 287 fold after 36 hours of SA treatment. Similarly, earlier studies where exogenous application of SA induced expression of PR-2 are reported from tomato [132], Eucalyptus grandis [135], Casuarina equisetifolia [136], cotton [124], sweet cherry [127], [128], grape berries [129] and tobacco [126]. However, down-regulation of PR-2 is also reported in sorghum [130] and wheat [34] during SA signaling and pathogen infection.
PR-1 is induced by pathogens and salicylic acid and is commonly identified as a marker for SAR. Their antifungal activity suggests involvement in plant defense, but their mode of action or relationship to other proteins is unknown. In W. somnifera, WsPR1 was up-regulated by 21-fold subsequent to 36 hours of SA treatment. Similarly, induction of PR-1 was reported during SA treatment from Arabidopsis [36], [137], tomato [138] and tobacco [126], [139]. In Malus hupehensis seedlings, application of SA enhanced the expression of MhPR1, MhPR5 and MhPR8 after 48 hours of post –induction [22].
Peroxidases (PR-9) are heme-containing oxido-reductases and its activities have been correlated with plant resistance. They are involved in the oxidation of phenolic residues to cell wall polymers in pathogen-infected tissues. In the present study, two peroxidases viz., lignin-forming and suberization-associated anionic peroxidase was induced by 377-fold and 6532-fold on SA application. Similarly, up-regulation of peroxidases is reported from sorghum [130], sunflower [141], Vigna unguiculata [140] and Polygonum minus [142]. In woody perennials like Pyrus bretschneideri cv. aYali, exogenous application of SA induced the accumulation of several PR proteins including chitinase, glucanase and peroxidase. Further, the SA treated leaves showed reduced symptoms of ring rot disease caused by Physalospora piricola [143]. The increased peroxidase activity could be due to the increased lignin biosynthesis, which forms the basal defense response in plants [144].
PR-10 proteins including pollen allergens are present as multigene family in seed plants and are developmentally and environmentally regulated. PR-10 proteins are reported from numerous dicots, including parsley, pea, potato, bean, soybean, celery and alfalfa and monocots like asparagus, rice, lily and sorghum [145]. PR-10 from W. somnifera (WsPR10) was down-regulated by 8-fold and 14-fold after 17 and 36 hours of SA application respectively. Similarly, in western white pine, the wound inducible PmPR10 transcript was partially suppressed by SA [145]. The expression of root specific PR-10 induced by drought and salt (RSOsPR10) was strongly inhibited by SA treatment in rice roots [146]. However, up-regulation of PR-10 has been reported from Arabidopsis [137], sorghum [130], soybean [147], asparagus [148], Medicago sativa [149], bean [150], rice [151], [152], [153] and Lithospermum erythrorhizon [154] during SA signaling and pathogenesis. This revealed that different signal transduction pathways might be involved in activation of different classes of PR10 to different environmental stresses [145].
The other PR genes which were up-regulated after 36 hours of SA treatment in Withania included thaumatin-like WsTHAU (PR5), WsCYST and WsSPI, (PR6), WsDEFN (PR12), lipid transfer proteins (PR14) and germin-like WsGER1 (PR16). Concurrently, the up-regulation of thaumatin-like PR-5 during SA treatment was reported from Malus hupehensis [22], sorghum [130], wheat [155], Arachis diogoi [156] and Eucalyptus grandis [135].
The synthetic analogs of SA including Benzothidiazole, benzo (1,2,3) thiadiazole-7-carbothioic acid S-methyl ester (BTH), 2,6-dichloroisonicotinic acid (INA) and 2,6-dichloroisonicotinic acid (DCINA) are chemical inducers of SAR and are commercially used to induce resistance to pathogenic infection in crops [157]. These functional analogs are also known to induce PR proteins/genes [26], [158], [159]. In sugar beet, BTH induced accumulation of chitinase and glucanase [160] while in soybean, the chemical up-regulated expression of PR-1, PR-3a, PR-3b, PR-9, and PR- 10 [157]. In banana, exogenous application of BTH caused prolonged expression of chitinase and reduced the symptoms of anthracnose disease [161]. Similarly, exogenous application of aspirin and BTH induced the expression of PR10 in Lithospermum erythrorhizon [154]. In sunflower, acetylsalicylic acid (asprin) induced the expression of four PR proteins including PR1, PR2, PR3 and PR5 which comprised 80% of the intercellular fluid proteins of induced leaf discs [20]. In barley seedlings, exogenous application of DCINA induced disease resistance against Erysiphe graminis f. sp. hordei and the acquired resistance was associated with increased accumulation of PR transcripts including PR1, chitinase and peroxidase [162]. Similarly, up-regulation of CaPR1, CaPR4, CaPR9 and CaCHI2 was reported in pepper during elicitation by BTH [23], while in maize, PR1 and PR5 were induced by BTH and INA. [163].
Recently, with the introduction of cost-effective NGS platforms for transcriptome sequencing, studies on understanding the global gene expression patterns during pathogenesis are being undertaken in several plant species. Host transcriptome analysis during interaction with pathogens are reported in banana - Fusarium oxysporum [32], Musa acuminate - Mycosphaerella musicola [33], wheat - Fusarium graminearum [34], potato- Phytophthora infestans [35], peach - Xanthomonas arboricola pv. Pruni [37] and Lactuca sativa - Botrytis cinerea [38]. These studies have also highlighted the up-regulation of PR gene families during pathogenesis.
The results on expression patterns of different PR genes during SA treatment documented in the present study and earlier studies from other plant species indicate that most of the pathways mediated by SA are analogous, but gene expression patterns can be species/genotype specific.
The present study is an attempt to characterize the SA mediated transcriptome in W. somnifera, a non-model medicinal species. The data generated in this study can support future studies in understanding the transcriptional regulation and networking of different pathways during pathogen defense response in Withania and other allied species from the Solanaceae family.
Supporting Information
Effect on different concentration of salicylic acid on leaf discs of W. somnifera.
(TIF)
E-value distribution of transcript contigs from RNA-Seq data of W. somnifera.
(TIF)
Sequence similarity distribution of transcript contigs from RNA-Seq data of W. somnifera.
(TIF)
GO mapping of transcript contigs from RNA-Seq data of W. somnifera to different databases.
(TIF)
Evidence code distribution of transcript contigs (A) and annotated transcript contigs (B) from RNA-Seq data of W. somnifera.
(TIF)
Annotation score distribution of transcript contigs from RNA-Seq data of W. somnifera.
(TIF)
GO-level-wise sequence distribution of transcript contigs for (a) Biological processes (b) Molecular functions (c) Cellular components. The initial levels denote the general function of transcript contigs and with progression of levels the function of the transcript contigs becomes more specific. Each transcript contigs can be multi-functional and hence, can lie in more than one ontology domain.
(TIF)
Melt curve analysis of reference genes used for normalization of qRT-PCR data.
(TIF)
Melt curve analysis of PR genes used for expression profiling.
(TIF)
Annotation of W. somnifera leaf transcriptome using Nr database.
(XLSX)
Classification of transcript contigs to biological pathways in W. somnifera using KEGG database.
(XLS)
Comparison of number of transcript contigs represented under different secondary metabolite pathways in two independent studies conducted on W. somnifera.
(DOC)
Acknowledgments
The authors acknowledge Dr. R. Viswanathan, Principal Scientist and Head (Plant Protection), Sugarcane Breeding Institute, Coimbatore, India for providing facilities to conduct qRT-PCR. The authors thank the Department of Biotechnology, Ministry of Science and Technology, Government of India, for the financial support.
Funding Statement
The authors thank the Department of Biotechnology, Ministry of Science and Technology, Government of India, for the financial support (Project ID: BT/PR/12597/PBD/16/867/2009). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Rastogi RP, Mehrotra BN (1998) Compendium of Indian Medicinal Plants. Vol.6. Central Drug Research Institute & NISCAIR. New Delhi.
- 2. Bharti SK, Bhatia A, Tewari SK, Sidhu OP, Roy R (2011) Application of HR-MAS NMR spectroscopy for studying chemotype variations of Withania somnifera (L.) Dunal. Magnetic Resonance in Chemistry 49: 659–667. [DOI] [PubMed] [Google Scholar]
- 3. Mishra LC, Singh BB, Dagenais S (2000) Scientific basis for the therapeutic use of Withania somnifera (Ashwagandha): A review. Alternative Medicine Review 5: 334–346. [PubMed] [Google Scholar]
- 4. Verma SK, Kumar A (2011) Therapeutic uses of Withania somnifera (ashwagandha) with a note on Withanolides and its pharmacological actions. Asian Journal of Pharmaceutical and Clinical Research 4: 1–4. [Google Scholar]
- 5. Siriwardhane AS, Dharmadasa RM, Samarasinghe K (2013) Distribution of Withaferin A, an anticancer potential agent, in different parts of two varieties of Withania somnifera (L.) Dunal. grown in Sri Lanka. Pakistan Journal of Biological Sciences 16: 141–144. [DOI] [PubMed] [Google Scholar]
- 6. Khedgikar V, Kushwaha P, Gautam J, Verma A, Changkija B, et al. (2013) Withaferin A: a proteasomal inhibitor promotes healing after injury and exerts anabolic effect on osteoporotic bone. Cell Death and Disease 4: e778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Gupta P, Goel R, Pathak S, Srivastava A, Singh SP, et al. (2013b) De novo assembly, functional annotation and comparative analysis of Withania somnifera leaf and root transcriptomes to identify putative genes involved in the withanolides biosynthesis. PLoS ONE 8: e62714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Glazebrook J (2005) Contrasting mechanisms of defense against biotrophic and necrotrophic pathogens. Annu Rev Phytopathol 43: 205–227. [DOI] [PubMed] [Google Scholar]
- 9. De Vos M, Van Oosten VR, Van Poecke RM, Van Pelt JA, Pozo MJ, et al. (2005) Signal signature and transcriptome changes of Arabidopsis during pathogen and insect attack. Mol Plant Microbe Interact 18: 923–937. [DOI] [PubMed] [Google Scholar]
- 10. Spoel SH, Johnson JS, Dong X (2007) Regulation of tradeoffs between plant defenses against pathogens with different lifestyles. Proc Natl Acad Sci USA 104: 18842–18847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Robert-Seilaniantz A, Grant M, Jones JDG (2011) Hormone crosstalk in plant disease and defense: More than just jasmonate-salicylate antagonism. Annu Rev Phytopathol 49: 317–343. [DOI] [PubMed] [Google Scholar]
- 12. Pieterse CMJ, Van der Does D, Zamioudis C, Leon-Reyes A, Van Wees SCM (2012) Hormonal modulation of plant immunity. Annu Rev Cell Dev Biol 28: 489–521. [DOI] [PubMed] [Google Scholar]
- 13. Van der Does D, Leon-Reyes A, Koornneef A, Van Verk MC, Rodenburg N, et al. (2013) Salicylic acid suppresses jasmonic acid signaling downstream of SCFCOI1-JAZ by targeting GCC promoter motifs via transcription factor ORA59. Plant Cell 25: 744–761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Lee J, Nam J, Park HC, Na G, Miura K, et al. (2007) Salicylic acid-mediated innate immunity in Arabidopsis is regulated by SIZ1 SUMO E3 ligase. Plant J 49: 79–90. [DOI] [PubMed] [Google Scholar]
- 15. Wildermuth MC, Dewdney J, Wu G, Ausubel FM (2001) Isochorismate synthase 1002 is required to synthesize salicylic acid for plant defence. Nature 414: 562–565. [DOI] [PubMed] [Google Scholar]
- 16. van Loon LC, Rep M, Pieterse CMJ (2006) Significance of inducible defense-related proteins in infected plants. Annu Rev Phytopathol 44: 135–162. [DOI] [PubMed] [Google Scholar]
- 17. Reymond P, Farmer EE (1998) Jasmonate and salicylate as global signals for defense gene expression. Curr Opin Plant Biol 1: 404–411. [DOI] [PubMed] [Google Scholar]
- 18. Blanco F, Salinas P, Cecchini N, Jordana X, Van Hummelen P, et al. (2009) Early genomic responses to salicylic acid in Arabidopsis . Plant Mol Biol 70: 79–102. [DOI] [PubMed] [Google Scholar]
- 19. Somssich IE, Hahlbrock K (1998) Pathogen defence in plants-a paradigm of biological complexity. Trends Plant Sci 3: 86–90. [Google Scholar]
- 20. Jung JL, Fritig B, Hahne G (1993) Sunflower (Helianthus annuus L.) Pathogenesis-related proteins (induction by aspirin (acetylsalicylic Acid) and characterization). Plant Physiol 101: 873–880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Görlach J, Volrath S, Knauf-Beiter G, Hengy G, Beckhove U, et al. (1996) Benzothiadiazole, a novel class of inducers of systemic acquired resistance, activates gene expression and disease resistance in wheat. Plant Cell 8: 629–643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Zhang J, Du X, Wang Q, Chen X, Lv D, et al. (2010) Expression of pathogenesis related genes in response to salicylic acid, methyl jasmonate and 1 aminocyclopropane-1-carboxylic acid in Malus hupehensis (Pamp.) Rehd. BMC Research Notes 3: 208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Lee B, Park YS, Yi HS, Ryu CM (2013) Systemic induction of the small antibacterial compound in the leaf exudate during benzothiadiazole-elicited systemic acquired resistance in pepper. Plant Pathol J 29: 1–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Niderman T, Genetet I, BruyèreT, Gees R, Stintzi A, et al. (1995) Pathogenesis related PR-1 proteins are antifungal. Plant Physiol 108: 17–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Taheri P, Tarighi S (2012) The role of pathogenesis-related proteins in the tomato-Rhizoctonia solani interaction. Journal of Botany Doi.org/doi:10.1155/2012/137037
- 26. Ward ER, Payne GP, Moyer MB, Williams SC, Dincher SS, et al. (1991) Differential regulation of β-l, 3-glucanase messenger RNAs in response to pathogen infection. Plant Physiol 96: 390–397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Kombrink E, Schroder M, Hahlbrock K (1988) Several “pathogenesis-related” proteins in potato are 1,3-,β glucanases and chitinases. Proc Natl Acad Sci 85: 782–786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Zhu B, Chen TH, Li PH (1995) Expression of three osmotin-like protein genes in response to osmotic stress and fungal infection in potato. Plant Mol Biol 28: 17–26. [DOI] [PubMed] [Google Scholar]
- 29. Orłowska E, Fiil A, Kirk HG, Llorente B, Cvitanich C (2012) Differential gene induction in resistant and susceptible potato cultivars at early stages of infection by Phytophthora infestans . Plant Cell Rep 31: 187–203. [DOI] [PubMed] [Google Scholar]
- 30. Soh HC, Park AR, Park S, Back K, Yoon JB, et al. (2012) Comparative analysis of pathogenesis-related protein 10 (PR10) genes between fungal resistant and susceptible peppers. European Journal of Plant Pathology 132: 37–48. [Google Scholar]
- 31. Veluthakkal R, Ghosh Dasgupta M (2010) Pathogenesis-related genes and proteins in forest tree species. Trees 24: 993–1006. [Google Scholar]
- 32. Wang Z, Zhang JB, Jia CH, Liu JH, Li YQ, et al. (2012) De Novo characterization of the banana root transcriptome and analysis of gene expression under Fusarium oxysporum f. sp. Cubense tropical race 4 infection. BMC Genomics 13: 650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Passos MA, de Cruz VO, Emediato FL, de Teixeira CC, Azevedo VC, et al. (2013) Analysis of the leaf transcriptome of Musa acuminata during interaction with Mycosphaerella musicola: gene assembly, annotation and marker development. BMC Genomics 14: 78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Xiao J, Jin X, Jia X, Wang H, Cao A, et al. (2013) Transcriptome-based discovery of pathways and genes related to resistance against Fusarium head blight in wheat landrace Wangshuibai. BMC Genomics 14: 197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Gao L, Tu ZJ, Millett BP, Bradeen JM (2013) Insights into organ-specific pathogen defense responses in plants: RNA-seq analysis of potato tuber-Phytophthora infestans interactions. BMC Genomics 14: 340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Zhu QH, Stephen S, Kazan K, Jin G, Fan L, et al. (2013) Characterization of the defense transcriptome responsive to Fusarium oxysporum infection in Arabidopsis using RNA-seq. Gene 512: 259–266. [DOI] [PubMed] [Google Scholar]
- 37. Socquet-Juglard D, Kamber T, Pothier JF, Christen D, Gessler C, et al. (2013) Comparative RNA-Seq analysis of early-infected peach leaves by the invasive phytopathogen Xanthomonas arboricola pv. pruni. PLoS ONE 8: e54196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. De Cremer K, Mathys J, Vos C, Froenicke L, Michelmore RW, et al. (2013) RNAseq-based transcriptome analysis of Lactuca sativa infected by the fungal necrotroph Botrytis cinerea . Plant Cell Environ 36: 1992–2007. [DOI] [PubMed] [Google Scholar]
- 39. Martinelli F, Uratsu SL, Albrecht U, Reagan RL, Phu ML, et al. (2012) Transcriptome profiling of citrus fruit response to huanglongbing disease. PLoS ONE 7: e38039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Rodrigues CM, de Souza AA, Takita MA, Kishi LT, Machado MA (2013) RNA-Seq analysis of Citrus reticulata in the early stages of Xylella fastidiosa infection reveals auxin-related genes as a defense response. BMC Genomics 14: 676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Lohse M, Bolger AM, Nagel A, Fernie AR, Lunn JE, et al. (2012) RobiNA: a user-friendly, integrated software solution for RNA-Seq-based transcriptomics. Nucleic Acids Res 40: W622–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Zerbino DR, Birney E (2008) Velvet: Algorithms for de novo short read assembly using de Bruijn graphs. Genome Res 18: 821–829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Schulz MH, Zerbino DR, Vingron M, Birney E (2012) Oases: robust de novo RNA-seq assembly across the dynamic range of expression levels. Bioinformatics 28: 1086–1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, et al. (2000) Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet 25: 25–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Moriya Y, Itoh M, Okuda S, Yoshizawa AC, Kanehisa M (2007) KAAS: an automatic genome annotation and pathway reconstruction server. Nucl Acids Res 35 (suppl 2)W182–W185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Kozomara A, Griffiths-Jones S (2011) miRBase: integrating microRNA annotation and deep sequencing data. Nucl Acids Res 39 (suppl 1)D152–D157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Vandesompele J, De Preter K, Pattyn F, Poppe B, Van Roy N, et al. (2002) Accurate normalisation of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol 3: 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Andersen CL, Jensen JL, Orntoft TF (2004) Normalization of real-time quantitative reverse transcription-PCR data: a model-based variance estimation approach to identify genes suited for normalization, applied to bladder and colon cancer data sets. Cancer Res 64: 5245–5250. [DOI] [PubMed] [Google Scholar]
- 49. Pfaffl MW, Tichopad A, Prgomet C, Neuvians TP (2004) Determination of stable housekeeping genes, differentially regulated target genes and sample integrity: BestKeeper – Excel-based tool using pair-wise correlations. Biotechnol Lett 26: 509–515. [DOI] [PubMed] [Google Scholar]
- 50. Migocka M, Papierniak A (2010) Identification of suitable reference genes for studying gene expression in cucumber plants subjected to abiotic stress and growth regulators. Mol Breeding 28: 343–357. [Google Scholar]
- 51. Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCT method. Methods 25: 402–408. [DOI] [PubMed] [Google Scholar]
- 52. Goswami RS, Atenafu EG, Xuan Y, Waldron L, Reis PP, et al. (2013) A microRNA signature obtained from the comparison of aggressive and indolent non-Hodgkin lymphomas: Potential prognostic value in mantle cell lymphoma J Clin Oncol. 31: 2903–2911. [DOI] [PubMed] [Google Scholar]
- 53.Peirson SN, Butler JN, Foster RG (2003) Experimental validation of novel and conventional approaches to quantitative real-time PCR data analysis. Nucleic Acids Research 31: : 14 e73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. De Bodt S, Maere S, Van de Peer Y (2005) Genome duplication and the origin of angiosperms.Trends Ecol Evol. 20: 591–597. [DOI] [PubMed] [Google Scholar]
- 55. Osborn TC, Pires JC, Birchler JA, Auger DL, Chen ZJ, et al. (2003) Understanding mechanisms of novel gene expression in polyploids. Trends Genet 19: 141–147. [DOI] [PubMed] [Google Scholar]
- 56. Comai L (2005) The advantages and disadvantages of being polyploid. Nat Rev Genet 6: 836–846. [DOI] [PubMed] [Google Scholar]
- 57. Yu Z, Habererb G, Matthesa M, Ratteic T, Mayerb KFX, et al. (2013) Impact of natural genetic variation on the transcriptome of autotetraploid Arabidopsis thaliana . Proc Natl Acad Sci USA 107: 17809–17814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Wachholtz M, Heng-Moss T, Twigg P, Baird L, Lu G, et al. (2013) Transcriptome analysis of two buffalograss cultivars. BMC Genomics 14: 613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Singhal VK, Kumar P (2008) Cytomixis during microsporogenesis in the diploid and tetraploid cytotypes of Withania somnifera (L.) Dunal, (Solanaceae). Comparative Cytogenetics 2: 85–92. [Google Scholar]
- 60. Vidya KM, Hegde L, Saraswathi G, Vijay Kumar BM (2013) Evaluation of diploids and autotetraploid genotypes of ashwagandha (Withania somnifera Dunal.) for growth, yield and quality characters. The Asian J Hort 8: 97–100. [Google Scholar]
- 61. Annadurai RS, Jayakumar V, Mugasimangalam RC, Katta MAVSK, Anand S, et al. (2012) Next generation sequencing and de novo transcriptome analysis of Costus pictus D. Don, a non-model plant with potent anti-diabetic properties. BMC Genomics 13: 663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Garg R, Patel RK, Tyagi AK, Jain M (2011) De novo assembly of chickpea transcriptome using short reads for gene discovery and marker identification. DNA Res 18: 53–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Iorizzo M, Senalik DA, Grzebelus D, Bowman M, Cavagnaro PF, et al. (2011) De novo assembly and characterization of the carrot transcriptome reveals novel genes, new markers, and genetic diversity. BMC Genomics 12: 389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Kudapa H, Bharti AK, Cannon SB, Farmer AD, Mulaosmanovic B, et al. (2012) A comprehensive transcriptome assembly of pigeonpea (Cajanus cajan L.) using sanger and second-generation sequencing platforms. Mol Plant 5: 1020–1028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Barrero RA, Chapman B, Yang Y, Moolhuijzen P, Keeble-Gagnère G, et al. (2011) De novo assembly of Euphorbia fischeriana root transcriptome identifies prostratin pathway related genes. BMC Genomics 12: 600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Temnykh S, DeClerck G, Lukashova A, Lipovich L, Cartinhour S, et al. (2001) Computational and experimental analysis of microsatellites in rice (Oryza sativa L.): frequency, length variation, transposon associations and genetic marker potential. Genome Res 11: 1441–1452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Thiel T, Michalek W, Varshney RK, Graner A (2003) Exploiting EST 941 databases for the development and characterization of gene derived SSR-markers in barley (Hordeum vulgare L.). Theor Appl Genet 106: 411–422. [DOI] [PubMed] [Google Scholar]
- 68. Portis E, Nagy I, Sasva Z, Stagelri A, Barchi L, et al. (2007) The design of Capsicum spp. SSR assays via analysis of in silico DNA sequence, and their potential utility for genetic mapping. Plant Sci 172: 640–648. [Google Scholar]
- 69. Varshney RK, Sigmund R, Boerner A, Korzun V, Stein N, et al. (2005) Interspecific transferability and comparative mapping of barley EST-SSR markers in wheat, rye, and rice. Plant Sci 168: 195–202. [Google Scholar]
- 70. Kantety RV, La Rota M, Matthews DE, Sorrells ME (2002) Data mining for simple sequence repeats in expressed sequence tags from barley, maize, rice, sorghum and wheat. Plant Mol Biol 48: 501–510. [DOI] [PubMed] [Google Scholar]
- 71. Gupta PK, Rustgi S, Sharma S, Singh R, Kumar N, et al. (2003) Transferable EST-SSR markers for the study of polymorphism and genetic diversity in bread wheat. Mol Genet Genome 270: 315–323. [DOI] [PubMed] [Google Scholar]
- 72. Minamiyama Y, Tsuro M, Hirai M (2006) An SSR-based linkage map of Capsicum annuum . Mol Breed 18: 157–169. [Google Scholar]
- 73. Cordeiro GM, Casu R, McIntyre CL, Manners JM, Henry RJ (2001) Microsatellite markers from sugarcane (Saccharum spp.) ESTs cross transferable to Erianthus and Sorghum . Plant Sci 160: 1115–1123. [DOI] [PubMed] [Google Scholar]
- 74. Park YH, Alabady MS, Ulloa M, Sickler B, Wilkins TA, et al. (2005) Genetic mapping of new cotton fiber loci using EST derived microsatellites in an interspecific recombinant inbred line cotton population. Mol Genet Genome 274: 428–441. [DOI] [PubMed] [Google Scholar]
- 75.Kumar S, Shah N, Garg V, Bhatia S (2014) Large scale in-silico identification and characterization of simple sequence repeats (SSRs) from de novo assembled transcriptome of Catharanthus roseus (L.) G. Don. Plant Cell Rep doi:10.1007/s00299-014-1569-8 [DOI] [PubMed]
- 76. Xie F, Burklew CE, Yang Y, Liu M, Xiao P, et al. (2012) De novo sequencing and a comprehensive analysis of purple sweet potato (Ipomoea batatas L.) transcriptome. Planta 236: 101–113. [DOI] [PubMed] [Google Scholar]
- 77. Chen S, Huang X, Yan X, Liang Y, Wang Y, et al. (2013) Transcriptome Analysis in Sheepgrass (Leymus chinensis): A Dominant Perennial Grass of the Eurasian Steppe. PLoS ONE 8(7): e67974 doi:10.1371/journal.pone.0067974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Mir BA, Koul S, Kumar A, Kaul MK, Soodan AS, et al. (2011) Assessment and characterization of genetic diversity in Withania somnifera (L.) Dunal using RAPD and AFLP markers. African Journal of Biotechnology 10: 14746–14756. [Google Scholar]
- 79. Tripathi N, Saini N, Kumar S, Tiwari S (2012) Assessment of genetic diversity among Withania somnifera collected from central India using RAPD and ISSR analysis. J Med Arom Plant Sci Biotechnol 6: 133–139. [Google Scholar]
- 80. Udayakumar R, Kasthurirengan S, Mariashibu TS, Sudhakar B, Ganapathi A, et al. (2013) Analysis of genetic variation among populations of Withania somnifera (L.) in South India based on RAPD Markers. European Journal of Medicinal Plants 3: 266–280. [Google Scholar]
- 81. Radman R, Saez T, Bucke C, Keshavarz T (2003) Elicitation of plants and microbial cell systems. Biotechnol Appl Biochem 37: 91–102. [DOI] [PubMed] [Google Scholar]
- 82. Ketchum REB, Gibson DM, Croteau RB, Schuler ML (1999) The kinetics of taxoid accumulation in cell suspension cultures of Taxus following elicitation with methyl jasmonate. Biotechnol Bioeng 62: 97–105. [DOI] [PubMed] [Google Scholar]
- 83. Sivanandhan G, Arun M, Mayavan S, Rajesh M, Jeyaraj M, et al. (2012b) Optimization of elicitation conditions with methyl jasmonate and salicylic acid to improve the productivity of withanolides in the adventitious root culture of Withania somnifera (L.) Dunal. Appl Biochem Biotechnol 168: 681–696. [DOI] [PubMed] [Google Scholar]
- 84. Ciddi V (2006) Withaferin A from cell cultures of Withania somnifera . Indian J Pharm Sci 68: 490–492. [Google Scholar]
- 85. Sivanandhan G, Arun M, Mayavan S, Rajesh M, Mariashibu TS, et al. (2012a) Chitosan enhances withanolides production in adventitious root cultures of Withania somnifera (L.) Dunal. Ind Crop Prod 37: 124–129. [Google Scholar]
- 86. Sivanandhan G, Kapil Dev G, Jeyaraj M, Rajesh M, Arjunan A, et al. (2013) Increased production of withanolide A, withanone, and withaferin A in hairy root cultures of Withania somnifera (L.) Dunal elicited with methyl jasmonate and salicylic acid. Plant Cell Tiss Organ Cult 114: 121–129. [Google Scholar]
- 87. Doma M, Abhayankar G, Reddy VD, Kavi Kishor PB (2012) Carbohydrate and elicitor enhanced withanolide (withaferin A and withanolide A) accumulation in hairy root cultures of Withania somnifera (L.). Indian Journal of Experimental Biology 50: 484–490. [PubMed] [Google Scholar]
- 88. Ginzinger DG (2002) Gene quantification using real-time quantitative PCR: an emerging technology hits the mainstream. Exp Hematol 30: 503–512. [DOI] [PubMed] [Google Scholar]
- 89. Huggett J, Dheda K, Bustin S, Zumla A (2005) Real-time RT-PCR normalization; strategies and considerations. Genes Immun 6: 279–284. [DOI] [PubMed] [Google Scholar]
- 90. Gupta P, Akhtar N, Tewari SK, Sangwan RS, Trivedi PK (2011) Differential expression of farnesyl diphosphate synthase gene from Withania somnifera in different chemotypes and in response to elicitors. Plant Growth Regulation 65: 93–100. [Google Scholar]
- 91. Gupta P, Agarwal AV, Akhtar N, Sangwan RS, Singh SP, et al. (2013a) Cloning and characterization of 2-C-methyl-D-erythritol-4-phosphate pathway genes for isoprenoid biosynthesis from Indian ginseng, Withania somnifera . Protoplasma 250: 285–295. [DOI] [PubMed] [Google Scholar]
- 92. Nicot N, Hausman J, Hoffmann L, Evers D (2005) Housekeeping gene selection for real time RT-PCR normalization in potato during biotic and abiotic stress. J Exp Bot 56: 2907–2914. [DOI] [PubMed] [Google Scholar]
- 93. Schmidt GW, Delaney SK (2010) Stable internal reference genes for normalization of real-time RT-PCR in tobacco (Nicotiana tabacum) during development and abiotic stress. Mol Genet Genomics 283: 233–241. [DOI] [PubMed] [Google Scholar]
- 94. Wan H, Yuan W, Ruan M, Ye Q, Wang R, et al. (2011) Identification of reference genes for reverse transcription quantitative real-time PCR normalization in pepper (Capsicum annuum L.). Biochem Biophys Res Commun 416: 24–30. [DOI] [PubMed] [Google Scholar]
- 95. Aimé S, Alabouvette C, Steinberg C, Olivain C (2013) The endophytic strain Fusarium oxysporum Fo47: A good candidate for priming the defense responses in tomato roots. Mol Plant-Microbe Interact 26: 918–926. [DOI] [PubMed] [Google Scholar]
- 96. Vlot AC, Dempsey DA, Klessig DF (2009) Salicylic acid, a multifaceted hormone to combat disease. Annu Rev Phytopathol 47: 177–206. [DOI] [PubMed] [Google Scholar]
- 97. Blanco-Ulate B, Vincenti E, Powell ALT, Cantu D (2013) Tomato transcriptome and mutant analyses suggest a role for plant stress hormones in the interaction between fruit and Botrytis cinerea . Front Plant Sci 4: 142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Hammond-Kosack KE, Jones JD (1996) Resistance gene-dependent plant defense responses. Plant Cell 8: 1773–1791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Ryals J, Neuenschwander UH, Willits MG, Molina A, Steiner H-Y, et al. (1996) Systemic acquired resistance. Plant Cell 8: 1809–1819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Tsuda K, Sato M, Glazebrook J, Cohen JD, Katagiri F (2008) Interplay between MAMP948 triggered and SA-mediated defense responses. Plant J 53: 763–775. [DOI] [PubMed] [Google Scholar]
- 101. Durrant WE, Dong X (2004) Systemic acquired resistance. Annu Rev Phytopathol 42: 185–209. [DOI] [PubMed] [Google Scholar]
- 102. Malamy J, Carr JP, Klessig DF, Raskin I (1990) Salicylic acid: a likely endogenous signal in the resistance response of tobacco to viral infection. Science 250: 1002–1004. [DOI] [PubMed] [Google Scholar]
- 103. Métraux J-P, Signer H, Ryals J, Ward E, Wyss-Benz M, et al. (1990) Increase in salicylic acid at the onset of systemic acquired resistance. Science 250: 1004–1006. [DOI] [PubMed] [Google Scholar]
- 104. An C, Mou Z (2011) Salicylic Acid and its Function in Plant Immunity Journal of Integrative Plant Biology. 53: 412–428. [DOI] [PubMed] [Google Scholar]
- 105. Mateo A, Funck D, Muhlenbock P, Kular B, Mullineaux PM, et al. (2006) Controlled levels of salicylic acid are required for optimal photosynthesis and redox homeostasis. J Exp Bot 57: 1795–1807. [DOI] [PubMed] [Google Scholar]
- 106.Holuigue L, Salinas P, Blanco F, Garreto'n V (2007) Salicylic acid and reactive oxygen species in the activation of stress defense genes. In: Hayat S, Ahmad A, editors. Salicylic acid—a plant hormone. Springer, Dordrecht. pp. 197–246.
- 107. Lu H (2009) Dissection of salicylic acid-mediated defense signaling networks. Plant Signal Behav 4: 713–717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Ng G, Seabolt S, Zhang C, Salimian S, Watkins TA, et al. (2011) Genetic dissection of salicylic acid-mediated defense signaling networks in Arabidopsis . Genetics 189: 851–859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Kunkel BN, Brooks DM (2002) Cross talk between signaling pathways in pathogen defense. Curr Opin Plant Biol 5: 325–331. [DOI] [PubMed] [Google Scholar]
- 110. Delaure SL, Hemelrijck WV, Bolle MFCD, Cammue BPA, De Coninck BMA (2008) Building up plant defenses by breaking down proteins. Plant Sci 174: 375–385. [Google Scholar]
- 111. Cao H, Bowling SA, Gordon AS, Dong X (1994) Characterization of an Arabidopsis mutant that is nonresponsive to inducers of systemic acquired resistance. Plant Cell 6: 1583–1592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112. Delaney TP, Friedrich LE, Ryals J (1995) Arabidopsis signal transduction mutant defective in chemically and biologically induced disease resistance. Proc Natl Acad Sci USA 92: 6602–6606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113. Shah J, Tsui F, Klessig DF (1997) Characterization of a salicylic acid-insensitive mutant (sai1) of Arabidopsis thaliana, identified in a selective screen utilizing the SA- inducible expression of the tms2 gene. Mol Plant Microbe Interact 10: 69–78. [DOI] [PubMed] [Google Scholar]
- 114. Maleck K, Neuenschwander U, Cade RM, Dietrich RA, Dangl JL, et al. (2002) Isolation and characterization of broad-spectrum disease-resistant Arabidopsis mutants. Genetics 160: 1661–1671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115. Bowling SA, Guo A, Cao H, Gordon S, Klessig DF, et al. (1994) A mutation in Arabidopsis that leads to constitutive expression of systemic acquired resistance. Plant Cell 6: 1845–1857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116. Clarke JD, Liu Y, Klessig DF, Dong X (1998) Uncoupling PR gene expression from NPR1 and bacterial resistance: Characterization of the dominant Arabidopsis cpr6-1 mutant. Plant Cell 10: 557–569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117. Yu IC, Parker J, Bent AF (1998) Gene-for-gene disease response resistance without the hypersensitive response in Arabidopsis dnd1 mutant. Proc Natl Acad Sci USA 95: 7819–7824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118. Zhou N, Tootle TL, Tsui F, Klessig DF, Glazebrook J (1998) PAD4 functions upstream from salicylic acid to control defense responses in Arabidopsis . Plant Cell 10: 1021–1030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119. Nawrath C, Métraux JP (1999) Salicylic acid induction–deficient mutants of Arabidopsis express PR-2 and PR-5 and accumulate high levels of camalexin after pathogen inoculation. The Plant Cell 11: 1393–1404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120. van Loon LC, van Strien EA (1999) The families of pathogenesis-related proteins, their activities, and comparative analysis of PR-1 type proteins. Physiol Mol Plant Pathol 55: 85–97. [Google Scholar]
- 121.Neuhaus JM (1999) Plant Chitinases (PR-3,PR-4,PR-8, PR-11). In: Datta SK, Muthukrishnan, S. CRC Press, Boca Raton. pp. 77–105
- 122. Davis JM, Wu H, Cooke JE, Reed JM, Luce KS, et al. (2002) Pathogen challenge, salicylic acid, and jasmonic acid regulate expression of chitinase gene homologs in pine. Mol Plant Microbe Interact 15: 380–387. [DOI] [PubMed] [Google Scholar]
- 123. Lawton KA, Beck J, Potter S, Ward E, Ryals J (1994) Regulation of cucumber class III chitinase gene expression. Mol Plant Microbe Interact 7: 48–57. [DOI] [PubMed] [Google Scholar]
- 124. Zhang LY, Zheng XH, Tang HL, Zhu JW, Yang JM (2003) Increase of b-1,3-glucanse and chitinase activities in cotton callus cells treated by salicylic acid and toxin of Verticillium dahlia . Acta Botanica Sinica 45: 802–808. [Google Scholar]
- 125. Harfouche AL, Rugini E, Mencarelli F, Botondi R, Muleo R (2008) 721 Salicylic acid induces H2O2 production and endochitinase gene expression but not ethylene biosynthesis in Castanea sativa in vitro model system. J Plant Physiol 165: 734–744. [DOI] [PubMed] [Google Scholar]
- 126. Vidal S, debLeon IP, Denecke J, Tapio Palva E (1997) Salicylic acid and the plant pathogen Erwinia carotovora induce defence genes via antagonistic pathways. Plant Journal 11: 115–123. [Google Scholar]
- 127. Yao H J, Tian SP (2005) Effects of pre- and post-harvest application of salicylic acid or methyl jasmonate on inducing disease resistance of cherry fruit in storage. Postharvest Biology and Technology 35: 253–262. [Google Scholar]
- 128. Xu X, Tian S (2008) Salicylic acid alleviated pathogen-induced oxidative stress in harvested sweet cherry fruit. Postharvest Biology and Technology 49: 379–385. [Google Scholar]
- 129. Derckel JP, Audran JC, Haye B, Lambert B, Legendre L (1998) Characterization, induction by wounding and salicylic acid, and activity against Botrytis cinerea of chitinases and β-1,3 glucanases of ripening grape berries. Physiologia Plantarum 104: 56–64. [Google Scholar]
- 130. Salzman RA, Brady JA, Finlayson SA, Buchanan CD, Summaer EJ, et al. (2005) Transcriptional profiling of sorghum induced by methyl jasmonate, salicylic acid, and aminocyclopropane carboxylic acid reveals cooperative regulation and novel gene responses. Plant Physiol 138: 352–368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131. Veluthakkal R, Ghosh Dasgupta M (2012) Isolation and characterization of pathogen defence related class I chitinase from the actinorhizal tree Casuarina equisetifolia . Forest Pathology 42: 467–480. [Google Scholar]
- 132. Ding CK, Wang CY, Gross KC, Smith DL (2002) Jasmonate and salicylate induce the expression of pathogenesis-related-protein genes and increase resistance to chilling injury in tomato fruit. Planta 214: 895–901. [DOI] [PubMed] [Google Scholar]
- 133.Busam C, Kassemeyer HH, Matern U (1997) Differential Expression of Chitinases in Vitis vinifera L. Responding to Systemic Acquired Resistance Activators or Fungal Challenge. Plant Physiol 115: : 1029–1038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134. Klarzynski O, Plesse B, Joubert JM, Yvin JC, Kopp M, et al. (2000) Linear b-1, 3-glucans are elicitors of defense responses in tobacco. Plant physiol 124: 1027–1037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135. Naidoo R, Ferreira L, Beger DK, Myburg AA, Naidoo S (2013) The identification and differential expression of Eucalyptus grandis pathogenesis- related genes in response to salicylic acid and methyl jasmonate. Front Plant Sci 4: 43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Ghosh Dasgupta M, Veluthakkal R, Karpaga Raja Sundari B (2013) Characterization of genes expressed in Casuarina equisetifolia in response to elicitation by cell wall components of Trichosporium vesiculosum. Silvae Genetica (In Press).
- 137. Datta R, Sinha R, Chattopadhyay S (2013) Changes in leaf proteome profile of Arabidopsis thaliana in response to salicylic acid. J Biosci 38: 317–328. [DOI] [PubMed] [Google Scholar]
- 138. Wang YY, Li BQ, Qin GZ, Li L, Tian S (2011) Defense response of tomato fruit at different maturity stages to salicylic acid and ethephon. Scientia Horticulturae 129: 183–188. [Google Scholar]
- 139. Xu Y, Chang PFL, Liu D, Narasimhan ML, Raghothama KG, et al. (1994) Plant defense genes are synergistically induced by ethylene and methyl jasmonate. Plant Cell 6: 1077–1085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Fernandes CF, Moraes VC, Vasconcelosb IM, Silveirab JA, Oliveira JT (2006) Induction of an anionic peroxidase in cowpea leaves by exogenous salicylic acid. J Plant Physiol 163: : 1040–1048. [DOI] [PubMed] [Google Scholar]
- 141. Cag S, Cevahir-Oz G, Sarsag M, Goren-Saglam N (2009) Effect of salicylic acid on pigment, protein content and peroxidase activity in excised sunflower cotyledons. Pak J Bot 41: 2297–2303. [Google Scholar]
- 142. Ee SF, Oh JM, Mohd Noor N, Kwon TR, Mohamed-Hussein ZA, et al. (2013) Transcriptome profiling of genes induced by salicylic acid and methyl jasmonate in Polygonum minus . Mol Biol Rep 40: 2231–2241. [DOI] [PubMed] [Google Scholar]
- 143. Gao LJ, Zhang YX (2013) Effects of Salicylic Acid on the Expression of SOD, PPO Isozymes and NPR1 in Pear. Acta Horticulturae Sinica 40: 41–48. [Google Scholar]
- 144. Hatfield R, Vermerris W (2001) Lignin formation in plants. The dilemma of linkage specificity. Plant Physiol 126: 1351e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145. Liu JJ, Ekramoddoullah AKM, Yu X (2003) Differential expression of multiple PR10 proteins in western white pine following wounding, fungal infection and cold-hardening. Physiol Plantarum 119: 544–553. [Google Scholar]
- 146. Takeuchi K, Gyohda A, Tominaga M, Kawakatsu M, Hatakeyama A, et al. (2011) RSOsPR10 Expression in response to environmental stresses is regulated antagonistically by jasmonate/ethylene and salicylic acid signaling pathways in rice roots. Plant Cell Physiol 52: 1686–1696. [DOI] [PubMed] [Google Scholar]
- 147. Crowell DN, John MF, Russell D, Amasino RM (1992) Characterization of a stress-induced, developmentally regulated gene family from soybean. Plant Mol Biol 18: 459–466. [DOI] [PubMed] [Google Scholar]
- 148. Warner SA, Gill A, Draper J (1994) The developmental expression of the asparagus intracellular PR protein (AoPR1) gene correlates with sites of phenylpropanoid biosynthesis. Plant J 6: 31–43. [DOI] [PubMed] [Google Scholar]
- 149. Breda C, Sallaud C, el-Turk J, Buffard D, de Kozak I, et al. (1996) Defense reaction in Medicago sativa: a gene encoding a class 10 PR protein is expressed in vascular bundles, Mol. Plant Microbe Interact 9: 713–719. [DOI] [PubMed] [Google Scholar]
- 150. Walter MH, Liu JW, Wunn J, Hess D (1996) Bean ribonuclease-like pathogenesis-related protein genes (Ypr10) display complex patterns of developmental dark-induced and exogenous-stimulus-dependent expression. Eur J Biochem 239: 281–293. [DOI] [PubMed] [Google Scholar]
- 151. Jwa NS, Agrawal GK, Rakwal R, Park CH, Agrawal VP (2001) Molecular cloning and characterization of a novel jasmonate inducible pathogenesis-related class 10 protein gene, JIOsPR10, from rice (Oryza sativa L.) seedling leaves. Biochem Biophys Res Commun 286: 973–983. [DOI] [PubMed] [Google Scholar]
- 152. Hwang SH, Lee IA, Yie SW, Hwang DJ (2008) Identification of an OsPR10a promoter region responsive to salicylic acid. Planta 227: 1141–1150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153. McGee JD, Hamer JE, Hodges TK (2001) Characterization of a PR-10 pathogenesis-related gene family induced in rice during infection with Magnaporthe grisea . Mol Plant Microbe Interact 14: 877–886. [DOI] [PubMed] [Google Scholar]
- 154. Hwang HJ, Kima H, Yu HJ, Oh MH, Lee I, et al. (2003) Gene encoding pathogenesis related 10 protein of Lithospermum erythrorhizon is responsive to exogenous stimuli related to the plant defense system. Plant Science 165: 1297–1302. [Google Scholar]
- 155. Jayaraj J, Muthukrishnan S, Liang GH, Velazhahan R (2004) Jasmonic acid and salicylic acid induce accumulation of β-1,3-glucanase and thaumatin-like proteins in wheat and enhance resistance against Stagonospora nodorum . Biol Plant 48: 425–430. [Google Scholar]
- 156. Singh NK, Kumar KRR, Kumar D, Shukla P, Kirti PB (2013) Characterization of a pathogen induced thaumatin-like protein gene AdTLP from Arachis diogoi, a wild peanut. PLoS ONE 8(12): e83963 doi:10.1371/journal.pone.0083963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157. Han Q, Feng H, Zhao H, Huang L, Wang X, et al. (2013) Effect of a benzothiadiazole on inducing resistance of soybean to Phytophthora sojae . Protoplasma 250: 471–481. [DOI] [PubMed] [Google Scholar]
- 158. Bovie C, Ongena M, Thonart P, Dommes J (2004) Cloning and expression analysis of cDNAs corresponding to genes activated in cucumber showing systemic acquired resistance after BTH treatment. BMC Plant Biol 26: 4–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159. Dao TTH, Puig RC, Kima HK, Erkelens C, Lefeber AWM, et al. (2009) Effect of benzothiadiazole on the metabolome of Arabidopsis thaliana . Plant Physiol Biochem 47: 146–152. [DOI] [PubMed] [Google Scholar]
- 160. Burketova L, Sindelarova M, Sindelarov L (1999) Benzothiadiazole as an inducer of β-1,3- glucanase and chitinase isozymes in sugar beet. Biol Plant 42: 423–430. [Google Scholar]
- 161. Ma BC, Tang WL, Ma LY, Li LL, Zhang LB, et al. (2009) The role of chitinase gene expression in the defense of harvested banana against anthracnose disease. J Amer Soc Hort Sci 134: 379–386. [Google Scholar]
- 162. Kogel KH, Beckhove U, Dreschers J, M¨unch S, Romm'e Y (1994) Acquired resistance in barley: the resistance mechanism induced by 2,6-dichloroisonicotinic acid is a phenocopy of a genetically based mechanism governing race-specific powdery mildew resistance. Plant Physiol 106: 1269–1277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163. Morris SW, Vernooij B, Titatarn S, Starrett M, Thomas S, et al. (1998) Induced resistance responses in maize. Mol Plant Microbe Interact 11: 643–658. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Effect on different concentration of salicylic acid on leaf discs of W. somnifera.
(TIF)
E-value distribution of transcript contigs from RNA-Seq data of W. somnifera.
(TIF)
Sequence similarity distribution of transcript contigs from RNA-Seq data of W. somnifera.
(TIF)
GO mapping of transcript contigs from RNA-Seq data of W. somnifera to different databases.
(TIF)
Evidence code distribution of transcript contigs (A) and annotated transcript contigs (B) from RNA-Seq data of W. somnifera.
(TIF)
Annotation score distribution of transcript contigs from RNA-Seq data of W. somnifera.
(TIF)
GO-level-wise sequence distribution of transcript contigs for (a) Biological processes (b) Molecular functions (c) Cellular components. The initial levels denote the general function of transcript contigs and with progression of levels the function of the transcript contigs becomes more specific. Each transcript contigs can be multi-functional and hence, can lie in more than one ontology domain.
(TIF)
Melt curve analysis of reference genes used for normalization of qRT-PCR data.
(TIF)
Melt curve analysis of PR genes used for expression profiling.
(TIF)
Annotation of W. somnifera leaf transcriptome using Nr database.
(XLSX)
Classification of transcript contigs to biological pathways in W. somnifera using KEGG database.
(XLS)
Comparison of number of transcript contigs represented under different secondary metabolite pathways in two independent studies conducted on W. somnifera.
(DOC)