Abstract
TMEM106B is a major risk factor for frontotemporal lobar degeneration with TDP-43 pathology. TMEM106B localizes to lysosomes, but its function remains unclear. We show that TMEM106B knockdown in primary neurons affects lysosomal trafficking and blunts dendritic arborization. We identify microtubule-associated protein 6 (MAP6) as novel interacting protein for TMEM106B. MAP6 over-expression inhibits dendritic branching similar to TMEM106B knockdown. MAP6 knockdown fully rescues the dendritic phenotype of TMEM106B knockdown, supporting a functional interaction between TMEM106B and MAP6. Live imaging reveals that TMEM106B knockdown and MAP6 overexpression strongly increase retrograde transport of lysosomes in dendrites. Downregulation of MAP6 in TMEM106B knockdown neurons restores the balance of anterograde and retrograde lysosomal transport and thereby prevents loss of dendrites. To strengthen the link, we enhanced anterograde lysosomal transport by expressing dominant-negative Rab7-interacting lysosomal protein (RILP), which also rescues the dendrite loss in TMEM106B knockdown neurons. Thus, TMEM106B/MAP6 interaction is crucial for controlling dendritic trafficking of lysosomes, presumably by acting as a molecular brake for retrograde transport. Lysosomal misrouting may promote neurodegeneration in patients with TMEM106B risk variants.
Keywords: dendrites, frontotemporal lobar degeneration, lysosomes, microtubule-associated protein 6, TMEM106B
See also: S Debaisieux & G Schiavo (March 2014)
Introduction
Frontotemporal lobar degeneration (FTLD) is the third most common neurodegenerative disease after Alzheimer's disease and Parkinson's disease (reviewed in Rademakers et al, 2012). The clinical presentation is diverse and the symptoms include dementia, behavioral changes, as well as speech and language impairment. Additional symptoms of upper or lower motoneuron disease are common and indicate a partial overlap with amyotrophic lateral sclerosis (ALS). Moreover, FTLD is heterogeneous in terms of pathology and genetics. The majority of cases show neuronal cytoplasmic aggregates of the nuclear DNA/RNA-binding protein TDP-43 (Neumann et al, 2006). Pathogenic mutations in TARDBP, the gene coding for TDP-43 are rare and predominantly cause ALS (Sreedharan et al, 2008). Familial forms of FTLD with TDP-43 pathology are mainly caused by hexanucleotide repeat expansion in C9ORF72 (DeJesus-Hernandez et al, 2011; Renton et al, 2011) and dominant loss-of-function mutations in the growth factor progranulin (GRN) (Cruts et al, 2006).
A multicentric genome-wide association study identified TMEM106B as a risk factor for FTLD with TDP-43 pathology (Van Deerlin et al, 2010). Three SNPs in linkage disequilibrium (LD) (rs1020004, rs6966915, and rs1990622) have been identified in coding and non-coding regions of TMEM106B with genome-wide significance. These SNPs conferred the strongest risk in patients also carrying a GRN mutation suggesting a functional interaction between TMEM106B and GRN. The genetic association of TMEM106B variants with FTLD-TDP was replicated with high confidence (Cruchaga et al, 2011; Finch et al, 2011; Van der Zee et al, 2011). Homozygocity of the protective minor allele of the non-coding SNP rs1990622 (7 kb downstream of the gene) is observed only in 2.6% of patients and 19.1% of controls. This protective allele is associated with higher GRN levels in plasma (Finch et al, 2011). The TMEM106B coding variant T185S, although not significantly associated with the risk of FTLD-TDP, but in perfect LD with rs1990622, seems to slightly increase TMEM106B protein levels (Nicholson et al, 2013). Although TMEM106B variants are not associated with ALS per se, the risk allele is linked to cognitive impairment in these patients (Vass et al, 2011). The minor allele of rs1990622 protects against hippocampal sclerosis and TDP-43 pathology in Alzheimer patients suggesting a more general role in neurodegeneration independent of GRN (Rutherford et al, 2012).
TMEM106B is a type 2 transmembrane protein and primarily localizes to late endosomes and lysosomes (Brady et al, 2012; Chen-Plotkin et al, 2012; Lang et al, 2012). TMEM106B overexpression leads to aberrant vacuole formation and enlarged lysosomes that are less acidic, which may be accompanied by slightly enhanced GRN levels and thus argues against a gain-of-function role of TMEM106B risk variants in FTLD (Brady et al, 2012; Chen-Plotkin et al, 2012). Lysosomal and autophagosomal inhibitors increase TMEM106B expression (Brady et al, 2012; Lang et al, 2012). Thus, the increased TMEM106B expression observed in GRN mutation carriers may be due to impaired lysosomal/autophagosomal function, which has been described in GRN knockout mice (Ahmed et al, 2010; Chen-Plotkin et al, 2012; Wils et al, 2012). Interestingly, homozygous GRN mutations cause neuronal ceroid lipofuscinosis, a lysosomal storage disorder (Smith et al, 2012). Pathogenic mutations in CHMP2B and VCP further support a strong endo-lysosomal dysfunction component in FTLD, as they affect membrane fusion and vesicle sorting within the endo-lysosomal and autophagosomal system (Filimonenko et al, 2007; Ju et al, 2009; Urwin et al, 2010; Ritz et al, 2011).
Despite its lysosomal localization, none of the previous publications reported altered lysosomal function or morphology or any effect on GRN expression upon TMEM106B knockdown (Brady et al, 2012; Chen-Plotkin et al, 2012; Lang et al, 2012). The physiological role of TMEM106B is therefore unknown. Here, we combine loss-of-function experiments and proteomics to identify the microtubule-associated protein 6 (MAP6) as novel interacting protein for TMEM106B. We show that interaction of TMEM106B and MAP6 regulates dendritic trafficking of lysosomes and affects dendrite morphology.
Results
TMEM106B knockdown causes lysosomal clustering in HeLa cells
TMEM106B shows a predominantly lysosomal localization, but its function in lysosomes is unknown. Transfection of HeLa cells with siRNAs against TMEM106B strongly reduced TMEM106B protein expression without affecting the pH-dependent proteolytic maturation of the lysosomal protease Cathepsin B (Fig 1A). However, immunostaining of lysosomes against the marker protein LAMP2 revealed a striking change in lysosomal localization upon TMEM106B knockdown (Fig 1B). Lysosomes were distributed throughout the cytoplasm in control-transfected cells, but appeared tightly clustered near the nucleus in cells devoid of TMEM106B, although the cell size was unaltered. Automated quantitative image analysis measuring the average distance of lysosomes to the nucleus and the lysosomal distribution using the Clark aggregation index (Clark & Evans, 1954) confirmed the shift of lysosomes towards the nucleus and the more compact subcellular distribution of lysosomes upon TMEM106B knockdown (Fig 1C). Overexpressing siRNA-resistant human TMEM106B (HA-hT106b*) in cells transfected with TMEM106B siRNA rescued normal lysosomal localization (compare HA-T106b*-expressing cell to the neighboring cells in Supplementary Fig S1) thus confirming siRNA specificity. These data suggest that TMEM106B controls lysosomal localization and trafficking, which may be particularly important in highly polarized cells such as neurons.
Figure 1.

- HeLa cells were transfected with control siRNA (siCtrl) or siRNA against TMEM106B (siTMEM). Immunoblot detects TMEM106B at 51 kDa. Immature and processed mature Cathepsin B is detected at 40 kDa and 25 kDa, respectively. β-actin levels confirm equal loading.
- siRNA-transfected HeLa cells were immunostained with antibodies against LAMP2 (green) and β-catenin (red). Scale bar represents 25 μm.
- Quantification of lysosomal positioning and clustering upon TMEM106B knockdown in HeLa cells. Positioning of lysosomes was determined by measuring the mean distance of the lysosomes to the nucleus and clustering was quantified by using the Clark aggregation index (Clark & Evans, 1954). In three independent experiments, a total of 510 control cells and 422 TMEM106B knockdown cells were analyzed. Mann–Whitney U-test, *** denotes P < 0.001.
TMEM106B localizes to late-endosomes and lysosomes throughout the somatodendritic compartment in primary neurons
Since accumulation of lysosomes in the soma and proximal dendrites is a common finding in brain tissue of FTLD patients particularly with GRN mutations (Chen-Plotkin et al, 2012; Busch et al, 2013), we analyzed the localization of TMEM106B in rat primary hippocampal neurons. Consistent with previous reports (Chen-Plotkin et al, 2012; Lang et al, 2012), endogenous TMEM106B colocalized with LAMP1-positive late-endosomal/lysosomal vesicles in the cell body and in dendrites (Fig 2A). Quantitative correlation analysis confirms good colocalization of TMEM106B with LAMP1 (Pearson's coefficient 0.66 ± 0.02), but not synaptic vesicles (labeled with SV2) and early or recycling endosomes (labeled with transferrin receptor TfR) (Supplementary Fig S2A and B).
Figure 2.
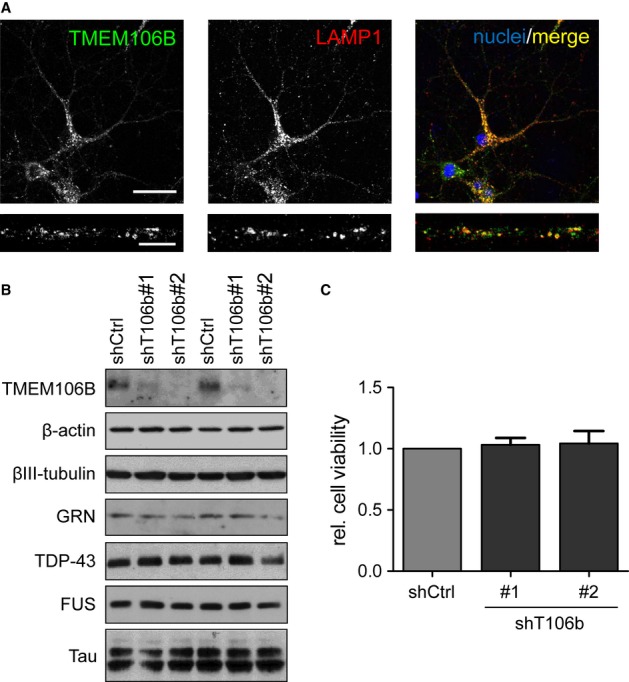
- Primary rat hippocampal neurons (DIV12) immunostained with antibodies against TMEM106B (green) and LAMP1 (red). Merged image indicates widespread co-localization in a single confocal plane. Scale bar represents 50 μm (overview) or 10 μm (dendrite segment).
- Primary rat cortical neurons (DIV7+6) were transduced with lentiviruses expressing either TMEM106B shRNA #1 or #2 or (shT106b) a control shRNA (shCtrl). Immunoblots with the indicated antibodies.
- Primary rat cortical neurons (DIV7+6) were transduced with the indicated lentiviruses. Cell viability upon TMEM106B knockdown was measured using an XTT assay. TMEM106B knockdown had no statistically significant effect (one-way ANOVA). n = 3 independent experiments, mean ± s.e.m.
To investigate the role of TMEM106B in neurons, we took advantage of lentiviral knockdown using two specific shRNAs targeting TMEM106B (shT106b#1 and #2). Cortical neurons were transduced at day 7 in vitro (DIV7) with individual shRNAs for 5 days (DIV7+5). Both shRNAs strongly reduced TMEM106B expression compared to the control shRNA (shCtrl), without affecting the expression of β-actin, βIII-tubulin or the FTLD-associated proteins GRN, TDP-43, FUS and Tau (Fig 2B). We detected no overt toxicity upon TMEM106B knockdown under these conditions using an XTT-based viability assay (Fig 2C) indicating that loss of TMEM106B alone does not cause neurodegeneration.
To analyze the morphology of individual cells by immunofluorescence, we transfected hippocampal neurons with shRNA constructs targeting TMEM106B. The punctate somatodendritic TMEM106B staining disappeared almost completely in neurons transfected with both TMEM106B-specific short hairpin constructs (Supplementary Fig S3A). While the distribution of lysosomes showed only a subtle trend towards clustering in the soma of TMEM106B shRNA-transfected neurons (Supplementary Fig S3B), the dendritic arborization appeared less complex suggesting TMEM106B may affect the function or distribution of lysosomes particularly in dendrites. Together, these data indicate that in neurons TMEM106B is localized to lysosomes throughout the somatodendritic compartment without grossly affecting their somatic distribution and neuronal viability.
TMEM106B is essential for dendrite branching and maintenance
We observed a blunted dendritic arborization in TMEM106B shRNA-transfected neurons compared to shCtrl-transfected cells (Supplementary Fig S3A). To quantitatively assess these changes, we cotransfected hippocampal neurons at DIV7 or 14 with the shRNA constructs together with a GFP-expressing plasmid to outline cell morphology. Five days after transfection the most noticeable effect was significantly reduced complexity of the dendritic arbor (Fig 3A). We quantified this phenotype by Sholl analysis, which measures the number of dendrites crossing concentric circles around the cell body (Sholl, 1953). Despite the nearly normal number of primary dendrites in TMEM106B knockdown neurons, we observed a striking reduction in dendritic branching compared to control transfected neurons. This effect was similar for neurons transfected at DIV7 and 14 (Fig 3B), indicating that TMEM106B is also required for maintenance of already established dendritic arborization in mature neurons. While overall dendritic branching was less complex in TMEM10B knockdown neurons, we found that the remaining principal dendrite was longer in the shRNA-treated cells (Supplementary Fig S4A). To exclude off-target effects, we used lentiviral co-expression of shRNA-resistant rat TMEM106B (T106b*) in knockdown neurons to restore basal TMEM106B levels (Fig 3C), while avoiding TMEM106B aggregation seen at higher expression levels (Brady et al, 2012; Chen-Plotkin et al, 2012). Expression of shRNA-resistant T106b* in TMEM106B shRNA-transfected hippocampal neurons restored dendritic arborization (Fig 3D and E) almost to control levels, while the expression of T106b* together with the control shRNA had no effect on dendritic arborization.
Figure 3.

TMEM106B knockdown impairs dendrite branching and maintenance.
A, B Primary rat hippocampal neurons (DIV7+5 and DIV14+5) were cotransfected with the indicated shRNAs and GFP to visualize cell morphology. The dendritic arborization was quantified manually and blinded to the experimental condition by Sholl analysis using MetaMorph software. Neurons transfected with either TMEM106B shRNA#1 or #2 are significantly different from control shRNA transfected cells (at DIV7 + 5: shT106b#1: from 25 to 62.5 μm radius P < 0.001, 100 and 112.5 μm P < 0.001; shT106b#2: from 25 to 75 μm P < 0.001. Analysis at DIV14+5: shT106b#1: from 37.5 to 62.5 μm P < 0.001, at 75 μm P < 0.05; shT106b#2: 37.5 to 62.5 μm P < 0.001, at 25 and 87.5 μm P < 0.05). n > 38 neurons per condition, three independent experiments, mean ± s.e.m. Two-way ANOVA. Scale bar represents 100 μm.
C Primary rat cortical neurons (DIV7+6) were cotransduced with a shRNA-expressing lentivirus (shT106b #2) and a lentivirus expressing shRNA-resistant TMEM106B (T106b*) or GFP as control to titrate TMEM106B expression. Immunoblots with the indicated antibodies.
D, E Primary rat hippocampal neurons were virally infected (DIV6) with either mCherry (RFP) or shRNA-resistant TMEM106B mutant (T106b*) and cotransfected (DIV7) with either a control shRNA or the indicated TMEM106B shRNA and GFP to outline neuron morphology for additional 5 days. Sholl analysis as above. shRNA-resistant TMEM106B overexpression fully rescued the phenotype of the less potent TMEM106B shRNA #1 (shCtrl+RFP versus shT106b+RFP: from 25 to 50 μm P < 0.001, shT106b+RFP versus shT106b+T106b*: 25 and 62.5 μm P < 0.05, 37.5 and 50 μm P < 0.001) and partially rescued the more potent shRNA #2 (shCtrl+RFP versus shT106b+RFP: from 12.5 to 50 μm P < 0.001, shT106b+RFP versus shT106b+T106b*: from 12.5 to 25 μm P < 0.001, at 37.5 μm P < 0.05). n > 38 neurons per condition, three independent experiments, mean ± s.e.m., two-way ANOVA. Scale bar represents 100 μm.
To further characterize whether impairment of lysosomal function inhibits dendritic branching we transfected hippocampal neurons (DIV7+5) with wild-type Rab7a, constitutively active Rab7a (Q67L) or dominant-negative Rab7a (T22N), which blocks cargo transport from early to late endosomes and lysosomes and inhibits lysosomal biogenesis (Mukhopadhyay et al, 1997; Press et al, 1998; Bucci et al, 2000). Neurons transfected with wild-type or constitutively active Rab7a appeared normal. In contrast, dendrites in neurons expressing the dominant-negative Rab7a showed a less complex branching pattern reminiscent of TMEM106B knockdown suggesting that proper lysosomal function is required for dendrite development (Supplementary Fig S4B and C, compare to Fig 3A and B).
Moreover, compared to the abundant mushroom-shaped spines in control neurons, the dendritic protrusions in TMEM106B knockdown cells appeared less dense and thinner (Supplementary Fig S4D). Reduction of the pre-and post-synaptic marker proteins (synaptophysin and PSD-95) in cortical neurons transduced with TMEM106B knockdown virus corroborate synapse loss at a biochemical level (Supplementary Fig S4E and F).
We detected some endogenous TMEM106B also in the axonal compartment in developing neurons at DIV4 (Supplementary Fig S5A), which allows better morphological analysis of axons than in mature neurons. Interestingly, developmental TMEM106B knockdown (DIV0+4) increased axon length by 40% (Supplementary Fig S5B and C), confirming that TMEM106B knockdown has no general toxic effect, but differentially affects axons, dendrites and synapses. Due to its predominant somatodendritic localization, we focused our further analysis on the dendritic phenotype.
Taken together, the late endosomal/lysosomal protein TMEM106B is required for proper maintenance of all except the primary dendrites possibly through a specific effect on lysosomes in the dendritic compartment.
TMEM106B interacts with the microtubule-binding protein MAP6 in the brain
In order to tie TMEM106B function to known cellular pathways, we performed immunoprecipitation experiments from P15 rat brain to identify interacting proteins using a proteomics approach. In three independent experiments with a total of five replicates the only common protein specifically identified in TMEM106B immunoprecipitates under these conditions was the microtubule-associated protein 6 (MAP6), also known as stable tubule-only polypeptide (STOP) (Bosc et al, 1996). Apart from microtubules, MAP6 is known to interact with the actin cytoskeleton and the Golgi apparatus (Baratier et al, 2006; Gory-Faure et al, 2006). LC-MS/MS analysis identified a total of 11 peptides unique for MAP6 (23% sequence coverage) in TMEM106B immunoprecipitates, but none in controls (Supplementary Fig S6A). To confirm the proteomics data, we repeated the experiment and analyzed TMEM106B and MAP6 immunoprecipitates by immunoblotting. Coimmunoprecipitation in both directions corroborates the specific interaction of TMEM106B and MAP6 (Fig 4A and B).
Figure 4.
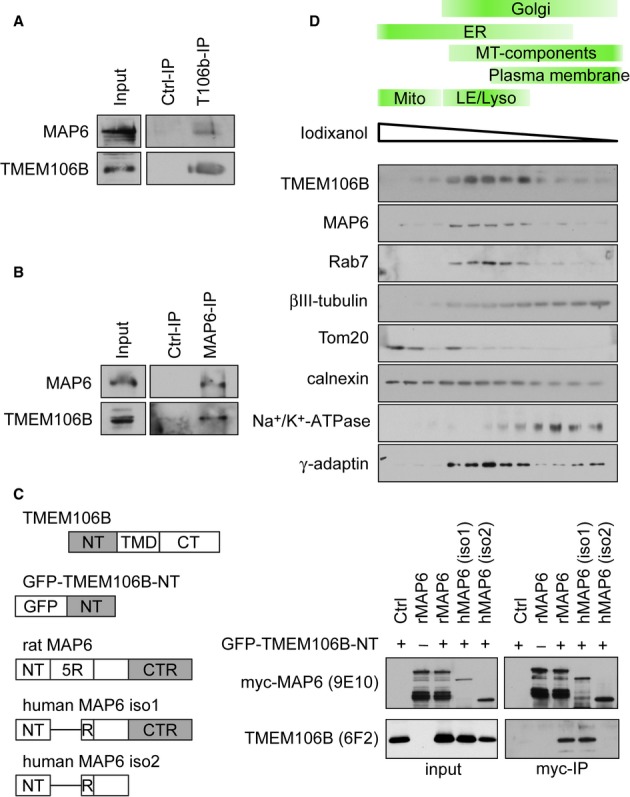
TMEM106B interacts with MAP6 in rat brain.
A, B TMEM106B (A) and MAP6 (B) were immunoprecipitated from P15 rat brain. Immunoblots with the indicated antibodies.
C Interacting domains of TMEM106B and MAP6. The cytoplasmic N-terminal domain (NT) of TMEM106B fused to GFP. Transmembrane domain (TMD) and C-terminal domain (CT) were removed to avoid aggregation. MAP6 contains a central repeat domain, consisting of five repeats (R) of a 46-amino-acid motif in rat but only one repeat in humans. The shorter isoform 2 lacks the C-terminal repeat domain (CTR) consisting of up to 28 imperfect repeats. The right panel shows coimmunoprecipitation of TMEM106B and MAP6 variants. GFP-TMEM106B-NT and the indicated myc-tagged MAP6 variants were co-expressed in HEK293FT cells. MAP6 variants were immunoprecipitated with myc-beads. Immunoblots with the indicated antibodies.
D Subcellular compartments from rat brain were fractionated using discontinuous iodixanol density gradient (2.5–30%). Immunoblots with the indicated antibodies.
To determine the interacting domains we transfected variants of MAP6 and TMEM106B in HEK293 cells. We expressed only the cytoplasmic N-terminus of TMEM106B tagged with GFP to avoid aggregation of the full-length construct. We could robustly detect GFP-N-TMEM106B in immunoprecipitates of rat MAP6 and the long neuron-enriched isoform 1 of human MAP6, but not in the C-terminally truncated isoform 2, indicating that the C-terminal domain of MAP6 interacts with the TMEM106B N-terminus (Fig 4C).
Consistent with previous observations (Baratier et al, 2006; Gory-Faure et al, 2006), double immunofluorescence shows predominant microtubule-like staining of MAP6 in dendrites and axons and vesicular staining in the soma with some overlap with TMEM106B-positive vesicles (Supplementary Fig S6B).
To further support the biological relevance of the interaction between TMEM106B and MAP6, we performed subcellular fractionation of rat brain on a discontinuous iodixanol density gradient. While ßIII-tubulin was broadly distributed over the whole gradient, TMEM106B and MAP6 levels peaked in endosomal/lysosomal fractions (marker protein Rab7) supporting close contact of the two proteins in the same cellular compartment (Fig 4D).
Together, our data establish the physical interaction of TMEM106B and MAP6 and imply a role of TMEM106B in microtubule-dependent processes, which are central to neurite development and maintenance (Hoogenraad & Bradke, 2009).
MAP6 knockdown rescues TMEM106B knockdown
Since TMEM106B knockdown strongly reduced dendritic branching, we asked whether MAP6 and the interaction between TMEM106B and MAP6 in particular affects dendrite morphology. Interestingly, MAP6 overexpression in hippocampal neurons (DIV7+5) strongly reduced dendrite branching and thus phenocopies the effect of TMEM106B knockdown (Fig 5A and B). We generated a shRNA construct against MAP6 to perform loss-of-function studies (Supplementary Fig S6C). MAP6 knockdown enhanced dendritic complexity and particularly the distal branching (Supplementary Fig S7A and B). This effect was fully rescued by overexpressing shRNA-resistant human MAP6 indicating a specific effect of the MAP6 shRNA.
Figure 5.

MAP6 knockdown and nocodazole treatment rescue the branching effect of TMEM106B knockdown.
A, B Primary rat hippocampal neurons (DIV7+5) were cotransfected with either a MAP6 overexpression construct or an empty vector control and GFP. Quantification of dendritic arborization by Sholl analysis. Neurons overexpressing MAP6 are significantly less branched than controls (n > 40 per condition; three independent experiments, mean ± s.e.m., at 12.5, 25, 75 and 87.5 μm radius P < 0.05, from 37.5 to 62.5 μm P < 0.001). Scale bar represents 100 μm.
C, D Primary rat hippocampal neurons (DIV7+5) were cotransfected with combinations of control shRNA (shCtrl), TMEM106B shRNA (shT106b) and MAP6 shRNA (shMAP6) together with GFP. Sholl analysis as above. MAP6 knockdown restores branching in TMEM106B knockdown neurons (n > 40 per condition; 3 independent experiments, mean ± s.e.m., two-way ANOVA: shCtrl versus shT106b#2 + shCtrl: from 25 to 62.5 μm radius P < 0.001. shT106b#2 + shCtrl versus shT106b#2 + shMAP6: from 25 to 100 μm P < 0.001, at 112.5 μm P < 0.05. shCtrl versus shMAP6 + shCtrl: at 87.5 μm P < 0.05, from 100 to 112.5 μm P < 0.001). Scale bar represents 100 μm.
E, F Primary rat hippocampal neurons (DIV7+5) were cotransfected with either shCtrl or shT106b#2 and GFP. 10 nM nocodazole was added to neurons in the treatment group freshly every 36 h for a total of 5 days. Nocodazole-treated neurons branched significantly more than untreated T106b#2-transfected neurons (n > 40 cells per condition; three independent experiments, mean ± SEM, two-way ANOVA: shCtrl versus shT106b#2: from 12.5 to 50 μm P < 0.001, 62.5 μm P < 0.05. shT106b#2 versus shT106b#2 + Noco: from 12.5 to 25 μm P < 0.001, at 37.5 μm P < 0.01). Scale bar represent 100 μm.
We therefore speculated that MAP6 knockdown might alleviate the impact of TMEM106B knockdown. MAP6 knockdown alone enhanced branching preferentially in the distal part of the dendrite (Fig 5 C and D, but see also supplemental Fig S7A and B). Strikingly, combined knockdown of MAP6 and TMEM106B fully restored also proximal dendritic branching compared to TMEM106B knockdown alone suggesting the two interacting proteins act in a common pathway (Fig 5C and D).
Since MAP6 knockdown is predicted to enhance microtubule dynamics (Bosc et al, 1996), we also tested if pharmacological destabilization of microtubules associated with enhanced dynamics could rescue the TMEM106B knockdown phenotype. Prolonged treatment with low concentrations of nocodazole (10 nM, added fresh every 36 h) for 5 days after transfection partially rescued the blunted dendritic morphology in TMEM106B shRNA transfected neurons (Fig 5E and F). The slight difference in the rescue capacity of MAP6 knockdown and nocodazole treatment might be due to additional effects apart from changing lysosomal transport and microtubule dynamics.
We conclude that the dendrite loss by TMEM106B knockdown can be rescued by additionally reducing MAP6 expression or by increasing microtubule dynamics pharmacologically, which strongly suggests that the functional interaction of TMEM106B and MAP6 is microtubule-dependent.
TMEM106B and MAP6 regulate retrograde lysosomal trafficking in dendrites
Vesicular trafficking in dendrites and axons strongly relies on microtubules and microtubule-associated proteins (Dehmelt & Halpain, 2005; Kapitein & Hoogenraad, 2011). To test whether the interaction between TMEM106B and MAP6 affects lysosomal trafficking in dendrites we performed live imaging of late endosomes and lysosomes labeled with Rab7a-GFP. We analyzed TMEM106B knockdown neurons at a time point before dendritic loss becomes apparent to exclude secondary effects (DIV6+3). Accelerated movies of lysosomal trafficking revealed a population of highly mobile lysosomes in the dendrites of TMEM106B knockdown neurons that was not present in controls (Supplementary Movies S1 and S2). Quantitative analysis of kymographs visualizing the trafficking of dendritic lysosomes showed a more than two-fold increase in the number of mobile vesicles in the dendrites of TMEM106B knockdown neurons (Fig 6A and B). This effect was predominantly due to enhanced retrograde motility while the number of anterogradely transported vesicles and vesicles without net-movement (during the 5 min recording time) remained unaffected (Fig 6B). This imbalance towards retrograde transport implies a progressive net loss of lipid membranes from distal dendrites which may be the cause of progressive dendrite loss. Reintroduction of TMEM106B by viral overexpression of shRNA-resistant T106b* fully restored the balance of anterograde and retrograde movement and thereby excludes off-target effects of the TMEM10B shRNA (Supplementary Fig S7C and D). Both the total distance a mobile vesicle traveled and the average velocity in the 5 min interval were increased upon TMEM106B knockdown (Supplementary Fig S8A and B). This effect was primarily due to increased speed and distance of retrogradely transported lysosomes. The overall density of dendritic Rab7a-positive vesicles was not affected by TMEM106B knockdown (Supplementary Fig S8C).
Figure 6.

- Primary hippocampal neurons (DIV6+3) were transfected with Rab7a-GFP and either TMEM106B shRNA #2 or a Ctrl shRNA and live imaged every second for 5 min to visualize late-endosomal/lysosomal trafficking in dendrites. Dendrite segments and kymographs of dendritic movement of Rab7a-GFP labeled vesicles. Scale bar represents 60 s (vertical) and 20 μm (horizontal).
- Quantitative analysis of vesicle movement from 5 min kymographs. Vesicles were manually classified according to their movement.
- Primary hippocampal neurons were transfected with either rat MAP6 or empty vector and Rab7a-GFP. Scale bar represents 60 s and 20 μm.
- Analysis of Rab7a-positive vesicle movement as above.
Data information: In (B) and (D), between five and 11 neurons per condition were analyzed per experiment in at least three independent experiments, mean ± s.e.m., unpaired t-test: * denotes P < 0.05, ** denotes P < 0.01.
To exclude unspecific effects on dendritic trafficking in general, we analyzed the transport of mitochondria using dsRed fused to a mitochondrial targeting sequence. Importantly, TMEM106B knockdown had no effect on mitochondrial density and motility in dendrites supporting a specific effect of TMEM106B on lysosomal trafficking (Supplementary Fig S8D and E).
As a further control for the specificity of TMEM106B knockdown on dendritic trafficking of lysosomes, we analyzed the movement of Rab7a-GFP labeled vesicles in axons. Hippocampal neurons were nucleofected directly before plating and analyzed by live imaging at DIV4. We found no apparent difference in the number or the direction of moving lysosomes (Supplementary Fig S5D and E).
Since overexpression of MAP6 reduced dendritic arborization similar to knockdown of TMEM106B, we asked whether lysosomal trafficking is similarly affected under both conditions. Indeed, overexpression of MAP6 accelerated retrograde transport of Rab7a vesicles comparable to TMEM106B knockdown without affecting anterograde transport (Fig 6C and D) and overall vesicle density (Supplementary Fig S8F). Interestingly, live imaging of co-expressed MAP6-GFP and LAMP1-RFP revealed that some moving lysosomes are labeled with MAP6-GFP, indicating that excess MAP6-GFP that can no longer bind to microtubules may still bind to TMEM106B on lysosomes (Supplementary Fig S9). This implies a dominant-negative effect of MAP6 overexpression. Thus, TMEM106B and MAP6 both regulate retrograde transport of lysosomes along dendritic microtubules.
MAP6 knockdown and nocodazole treatment rescue TMEM106B knockdown phenotype by rebalancing lysosomal transport in dendrites
MAP6 knockdown or treatment with low doses of nocodazole rescued the branching deficit in TMEM106B knockdown neurons (see Fig 5). To test whether MAP6 knockdown could also correct the enhanced retrograde transport of lysosomes, we cotransfected TMEM106B shRNA #2 together with MAP6 shRNA and performed live cell imaging of Rab7a-GFP labeled vesicles. Surprisingly, MAP6 knockdown alone as well as double knockdown of TMEM106B and MAP6 (Fig 7A and C) enhanced lysosome motility. However, retrograde and anterograde transport were both increased to an equal level, suggesting balanced lysosomal transport is important for dendrite development and maintenance.
Figure 7.

MAP6 knockdown and nocodazole treatment rebalance lysosomal trafficking in TMEM106B knockdown neurons.
A Primary hippocampal neurons were transfected with either shCtrl, shT106b#2 + shCtrl, shCtrl+shMAP6 or shT106b#2 + shMAP6 and Rab7a-GFP to visualize late endosomal/lysosomal trafficking. Dendrite segment and kymographs of dendritic movement of Rab7a-GFP-labeled vesicles. Scale bars represent 60 s and 20 μm.
B Primary hippocampal neurons were transfected with either shCtrl or shT106b#2 and Rab7a-GFP. Fresh nocodazole (10 nM) was added to neurons of the treatment group every 36 h.
C, D Quantitative analysis of vesicle movement from 5 min kymographs. Vesicles were manually classified according to their movement. Between six and 10 neurons per condition were analyzed per experiment in at least three independent experiments, mean ± s.e.m., unpaired t-test: * denotes P < 0.05, *** denotes P < 0.001.
Similar results were obtained for the rescue with low-dose nocodazole treatment. While untreated TMEM106B knockdown neurons showed an increased number of retrogradely moving vesicles, neurons which were additionally treated with 10 nM nocodazole for 5 days showed enhanced movement in both directions (Fig 7B and D). Thus, genetic or pharmacologic re-balancing of dendritic transport of lysosomes rescues dendrite loss in TMEM106B knockdown neurons.
Enhancing anterograde transport of lysosomes promotes dendritic branching and rescues the TMEM106B phenotype
To further confirm that TMEM106B and MAP6 affect dendritic branching through their effect on lysosomal trafficking we aimed to manipulate lysosomal transport independent of TMEM106B and MAP6. Rab7-interacting lysosomal protein (RILP) is known to recruit dynein-dynactin motor complexes to lysosomes and thus promotes trafficking towards the minus end of microtubules (Cantalupo et al, 2001; Jordens et al, 2001). To investigate how RILP affects lysosomes in neurons, we co-expressed the dominant-negative C-terminal fragment of RILP together with Rab7-GFP and quantified lysosomal trafficking in dendrites. dnRILP expression specifically enhanced anterograde movement, without affecting retrograde motility (Fig 9A and B). Moreover, prolonged expression of dnRILP resulted in enhanced dendritic complexity indicating that anterograde lysosomal transport is required for dendritic branching (Fig 8C and D).
Figure 9.

- Interaction of TMEM106B and MAP6 inhibits retrograde transport of lysosomes in dendrites.
- TMEM106B knockdown and MAP6 overexpression specifically enhance retrograde transport of lysosomes.
- MAP6 knockdown moderately enhances trafficking of lysosomes in both direction and rescues TMEM106B knockdown.
Figure 8.
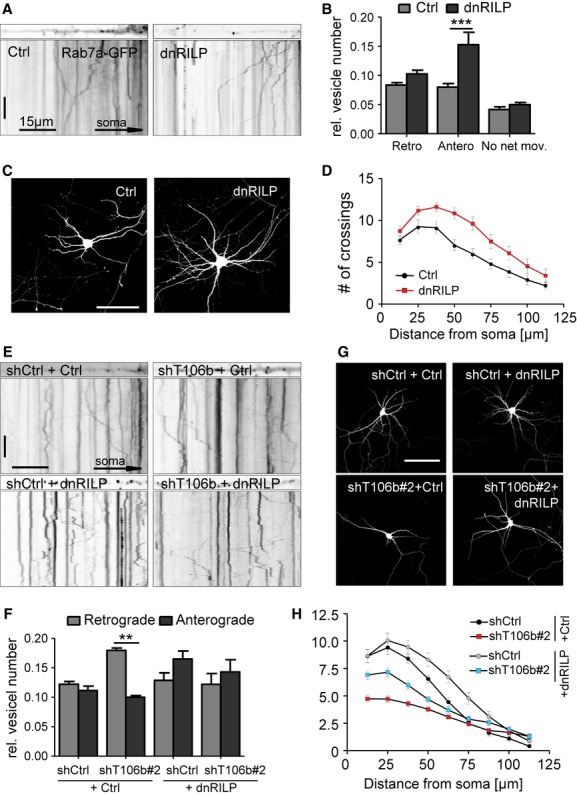
Overexpression of dominant-negative RILP increases anterograde movement of lysosomes and restores dendritic branching in TMEM106B knockdown neurons.
A Primary hippocampal neurons (DIV6+3) were transfected with Rab7a-GFP and either empty vector (Ctrl) or dnRILP and live-imaged every second for 5 min to visualize late-endosomal/lysosomal trafficking in dendrites. Dendrite segments and kymographs of dendritic movement of Rab7a-GFP labeled vesicles. Scale bar represents 60 s (vertical) and 15 μm (horizontal).
B Quantitative analysis of vesicle movement from 5 min kymographs. Vesicles were manually classified according to their motility. At least seven neurons per condition were analyzed per experiment in at least three independent experiments. Mean ± s.e.m., unpaired t-test: *** denotes P < 0.001.
C, D Primary rat hippocampal neurons (DIV7+5) were cotransfected with an empty vector (Ctrl) or dnRILP together with GFP. Sholl analysis to quantify dendritic complexity. (n = 25 per condition; three independent experiments, mean ± s.e.m., two-way ANOVA: 50 μm P < 0.001, 62.5 μm P < 0.01, 75 μm P < 0.05). Scale bar represents 100 μm.
E Primary hippocampal neurons (DIV6+3) were transfected with combinations of control shRNA (shCtrl) or TMEM106B shRNA#2 (shT106b) and dnRILP or an empty vector (Ctrl) and Rab7a-GFP to visualize late endosomal/lysosomal trafficking. Dendrite segments and kymographs showing dendritic movement of Rab7a-GFP-labeled vesicles. Scale bars represent 60 s and 15 μm.
F Quantitative analysis of vesicle movement from 5 min kymographs. Vesicles were manually classified according to their motility. At least seven neurons per condition were analyzed per experiment in at least three independent experiments, mean ± s.e.m., unpaired t-test: ** denotes P < 0.01.
G, H Primary rat hippocampal neurons (DIV7+5) were cotransfected with combinations of control shRNA (shCtrl) or TMEM106B shRNA#2 (shT106b) and dnRILP or an empty vector (Ctrl) together with GFP. Sholl analysis as above. Coexpression of dnRILP ameliorates the branching phenotype of TMEM106B knockdown (n > 25 per condition; 3 independent experiments, mean ± s.e.m., two-way ANOVA: shCtrl+Ctrl versus shT106b#2 + Ctrl: from 12.5 to 50 μm radius P < 0.001. shT106b#2 + Ctrl versus shT106b#2 + dnRILP: from 12.5 μm P < 0.001, 25 μm P < 0.01. shCtrl+Ctrl versus shCtrl+dnRILP: from 62.5 to 75 μm P < 0.01). Scale bar represents 100 μm.
We therefore asked whether promoting anterograde lysosomal transport may rescue the impaired branching in TMEM106B knockdown neurons. Expression of dnRILP restored the balance of anterograde and retrograde lysosomal transport in neurons transfected with TMEM106B shRNA (Fig 8E and F). Importantly, dnRILP expression also enhanced dendritic complexity in TMEM106B knockdown neurons (Fig 8G and H). The functional rescue of TMEM106B knockdown by promoting anterograde lysosomal transport indicates that the misbalanced lysosomal trafficking directly reduces dendrite complexity.
Discussion
Together, our data implicate TMEM106B in dendrite morphogenesis and maintenance by controlling lysosomal trafficking through its novel interacting partner MAP6. Vesicular trafficking, microtubule dynamics, and neurite development and maintenance are tightly connected and all have been linked to neurodegeneration (Garcia & Cleveland, 2001; Hoogenraad & Bradke, 2009). Thus, TMEM106B risk variants may contribute to FTLD pathogenesis through lysosomal misrouting particularly in GRN mutation carriers.
Neuronal phenotype of TMEM106B knockdown
Our data confirm the predominant localization of TMEM106B to late-endosomes/lysosomes in primary neurons (Brady et al, 2012; Chen-Plotkin et al, 2012; Lang et al, 2012). TMEM106B overexpression is reported to inhibit lysosomal function (Brady et al, 2012). We find that TMEM106B knockdown has no effect on the maturation of the lysosomal protease Cathepsin B indicating that lysosomal acidification is normal in the absence of TMEM106B. Thus, impaired lysosomal function upon TMEM106B overexpression may be attributed to the pronounced TMEM106B aggregation seen upon strong overexpression (Brady et al, 2012). To avoid unspecific effects, we carefully titrated the TMEM106B levels back to endogenous levels for rescue experiments in HeLa cells and neurons.
TMEM106B knockdown dramatically alters the neuronal architecture without affecting viability. While axon length in immature neurons is increased by 40%, dendritic arborization is severely blunted in mature neurons. The dominant-negative Rab7a T22N inhibits lysosomal biogenesis and leads to dendrite loss similar to TMEM106B knockdown showing that functional lysosomes are required for dendritic arborization. In contrast, enhancing anterograde transport of lysosomes in dendrites by expression of dnRILP promotes dendritic growth. Moreover, dendritic spines appear less mature and the levels of synaptic marker proteins are reduced in TMEM106B knockdown neurons, which has also been observed in the brains of FTLD patients (Clare et al, 2010). These changes imply weakened synaptic strength and impaired synaptic plasticity, which are common attributes of neurodegenerative diseases and awaits further study in FTLD/ALS (Tackenberg et al, 2009; Petkau et al, 2012). These findings argue for an important role of TMEM106B in regulating neuronal morphology through subtle alteration in lysosomal function, most likely through altered transport.
TMEM106B/MAP6 interaction
We identified MAP6 as TMEM106B interacting partner by mass spectrometry and confirmed the interaction using bidirectional coimmunoprecipitation, immunoblotting and live imaging of lysosomal trafficking. The C-terminal repeat region of the neuron-enriched splice variant of MAP6 binds to the cytoplasmic N-terminus of TMEM106B preferentially. MAP6 is a microtubule-associated protein that has been implicated in the cold-stability of microtubules, cellular morphology, cognition and mood (Bosc et al, 1996; Andrieux et al, 2002; Arama et al, 2012; Fournet et al, 2012). Posttranslational modifications, such as phosphorylation and palmitoylation, as well as overall protein levels have been shown to regulate MAP6 localization and target the protein also to cellular compartments other than microtubules i.e. Golgi apparatus, dendritic branch points and spines (Baratier et al, 2006; Gory-Faure et al, 2006). Moreover, MAP6 coaggregates with neurofilaments in spheroid axonal aggregates in ALS (Letournel et al, 2003). Given the dramatic changes upon TMEM106B knockdown on dendritic arborization, linking TMEM106B to microtubules is intriguing. We show that overexpressing MAP6 phenocopies the effect of TMEM106B knockdown on dendrite morphology and that knockdown of MAP6 fully restores dendritic arborization upon TMEM106B knockdown, suggesting these morphological changes depend on the interaction of TMEM106B with MAP6.
Lysosome trafficking in dendrites
TMEM106B knockdown and MAP6 overexpression increase the number and speed of retrogradely moving lysosomes in dendrites suggesting this may be the common cause for the impaired dendritic branching. In HeLa cells, altered trafficking even leads to dramatic perinuclear clustering of lysosomes at the microtubule-organizing center, where most of the microtubules nucleate in these cells (Akhmanova & Steinmetz, 2008; Tamura & Draviam, 2012). Restoring the balance of dendritic trafficking of lysosomes through MAP6 knockdown and nocodazole treatment rescue dendritic branching. Interestingly, comparable low concentrations of nocodazole also enhance transport of adenovirus particles (Giannakakou et al, 2002). Moreover, promoting anterograde lysosomal transport in dendrites independent of TMEM106B and MAP6 also rescues dendritic branching in TMEM106B knockdown cells, indicating that imbalanced lysosomal transport causes this phenotype.
Ample evidence suggests that an imbalance between anterograde and retrograde transport of lysosomes may impair protein and membrane turnover in the distal dendrites and thus impair dendrite and spine maintenance. The secretory pathway in general and endosomal/lysosomal trafficking in particular are important for neurite outgrowth (Horton et al, 2005; Sann et al, 2009; Jan & Jan, 2010). Interestingly, ALS-causing mutants in the ER-targeted protein VAPB impair membrane delivery to dendrites through its interacting protein YIF1A and thereby inhibit dendritic branching (Nishimura et al, 2004; Kuijpers et al, 2013). These findings further support overlapping pathomechanisms in the FTLD/ALS disease spectrum. Moreover, fusion of lysosomes with the plasma membrane (and secretion of lysosomal content) supplies membrane components during wound sealing and cell growth (Reddy et al, 2001; Chakrabarti et al, 2003; Huynh et al, 2004). Promoting anterograde lysosomal transport using dnRILP strongly enhances dendritic complexity. Thus, a shift towards retrograde transport of lysosomes may contribute to dendrite withering through loss of lipid membranes in distal dendrites in TMEM106B knockdown neurons.
Additionally, disturbed lysosomal trafficking may impair growth factor signaling through altered transport of receptors, such as the signaling of GRN via sortilin or tumor necrosis factor receptor (Hu et al, 2010; Tang et al, 2011). Intriguingly, MAP6 was recently also reported to interact with Intersectin1 (Morderer et al, 2012), a protein important for endocytosis and signal transduction, suggesting MAP6 may play a broader role in endosomal and lysosomal trafficking. Altered signal transduction may also contribute to increased axonal length upon TMEM106B knockdown, since axons are less dependent on membrane supply through the secretory pathway than dendrites but strongly respond to guidance cues (Ye et al, 2007).
Model
We propose the following model for TMEM106B action centered on its role on lysosomal trafficking (Fig 9). Under normal conditions, binding of lysosomal TMEM106B to microtubule-attached MAP6 inhibits active retrograde transport of lysosomes along dendrites (Fig 9A). Upon TMEM106B knockdown, retrogradely moving vesicles no longer bind to MAP6 and are transported with increased speed and fewer stops along the dendrites. This imbalance in late-endosomal/lysosomal trafficking with a shift towards the somatic compartment may impair dendrite and synapse stability through loss of membranes (Fig 9B). Overexpression of MAP6 saturates the binding sites on microtubules and excess MAP6 binds to TMEM106B on lysosomes without anchoring them to microtubules which causes a dominant-negative effect mimicking the knockdown of TMEM106B and leads to enhanced retrograde transport (Fig 9B). This model is supported by live imaging experiments showing MAP6-GFP moving together with LAMP1-RFP labeled lysosomes (Fig S9). Knockdown of MAP6 alone or double knockdown of TMEM106B and MAP6 slightly accelerates lysosome trafficking, but affects retrograde and anterograde trafficking to the same extent, thus preventing membrane loss in the dendrite (Fig 9C). Therefore, additional mechanisms (such as the interaction of Rab7a and RILP) likely control bidirectional transport of lysosomes in dendrites. Enhanced microtubule dynamics upon MAP6 knockdown (Bosc et al, 1996) may add to this effect because increasing microtubule dynamics with nocodazole also enhances overall motility of lysosomes and restores the balance of retrograde and anterograde trafficking in TMEM106B knockdown neurons. Thus, low concentrations of the anti-cancer drug Vincristine, which also enhances microtubule dynamics, may be beneficial for patients with TMEM106B variants or GRN mutations, where lysosomes accumulate in the soma and proximal dendrites (Chen-Plotkin et al, 2012).
A similar transport mechanism has already been shown for the docking of mitochondria to the axonal cytoskeleton. Interaction of the mitochondrial protein Syntaphilin with dynein light chain LC8 (independently of its motor function) stalls mitochondrial transport along axonal microtubules (Kang et al, 2008; Chen et al, 2009). Our data suggest that TMEM106B together with its binding partner MAP6 act as a molecular brake for retrogradely moving late endosomes and lysosomes in a similar manner without affecting mitochondrial transport. Since TMEM106B is predominantly localized in the late-endosomal/lysosomal compartment, the trafficking function of MAP6/TMEM106B is likely specific to this compartment.
Disease implication
We and others find no effect of TMEM106B knockdown on the expression of FTLD-associated proteins GRN, TDP-43, FUS and Tau or neuronal viability suggesting that TMEM106B risk variants may sensitize neurons to GRN or TDP-43-dependent pathomechanisms, rather than causing neurodegeneration directly (Lang et al, 2012). Emerging pathological and genetic evidence supports lysosomal impairment in FTLD, particularly in patients with GRN mutations in whom TMEM106B variants confer the strongest risk (Van Deerlin et al, 2010). Homozygous loss of GRN in mice and patients leads to lipofuscin accumulation, a common feature of lysosomal dysfunction (Ahmed et al, 2010; Smith et al, 2012). Moreover, CLN3 mutations that cause juvenile onset neuronal ceroid lipofuscinosis induce perinuclear clustering of lysosomes similar to the phenotype of TMEM106B knockdown in HeLa cells (Tuxworth et al, 2009; Uusi-Rauva et al, 2012). Interestingly, TMEM106B and presumably lysosomes accumulate in the soma and proximal dendrites in FTLD patients, most prominently in those with GRN mutations (Chen-Plotkin et al, 2012). Thus, lysosomal misrouting may be a common pathomechanism of FTLD that is further boosted by TMEM106B risk variants.
We identified a molecular interaction partner and a very specific function in lysosomal transport for TMEM106B, a protein of previously unknown function, opening new therapeutic strategies for FTLD treatment.
Materials and Methods
Antibodies and reagents
Rabbits were immunized with an MBP-TMEM106B-NT fusion protein (rat amino acids 1–91) or with MBP-MAP6-CT (amino acids 793-952 according to NP_058900). The serum was purified with corresponding GST fusion proteins cross-linked to glutathione sepharose with Bis(sulfosuccinimidyl)suberate (BS3; Pierce, Rockford, IL, USA), eluted by pH shift (0.1 M glycine 0.5 M NaCl, pH 2.5) and immediately neutralized (1/10 volume 1 M Tris, pH 9.5). The monoclonal TMEM106B antibody (6F2) was described previously (Lang et al, 2012). Antibodies for LAMP1 (Enzo Life Sciences, Lörrach, Germany), LAMP2 (Developmental Studies Hybridoma Bank), β-catenin (Sigma-Aldrich, St. Louis, MO, USA), Cathepsin B (Santa-Cruz Biotechnology, Dallas, TX, USA), myc (9E10, Santa-Cruz Biotechnology), HA (3F10; Roche Applied Science, Mannheim, Germany), GFP (Neuromab, Davis, CA, USA), Rab7 (Cell Signaling Technologies, Danver, MA, USA), MAP6 (Cell Signaling Technologies and Abcam), βIII-tubulin (Sigma-Aldrich), β-actin (Sigma-Aldrich), FUS (Bethyl), TDP-43 (Cosmo Bio, Tokyo, Japan), total-Tau (Dako, Hamburg, Germany), Tau-1 (Millipore, Billerica, MA, USA), Synaptophysin (Millipore), PSD-95 (Neuromab), Na+/K+-ATPase (Developmental Studies Hybridoma Bank), Tom-20 (Santa-Cruz Biotechnology), calnexin (Enzo Life Sciences), γ-adaptin (BD Transduction Laboratories, San Jose, CA, USA), TfR (Invitrogen, Grand Island, NY, USA), SV2 (Developmental Studies Hybridoma Bank) are commercially available. Nocodazole was from Sigma-Aldrich.
DNA constructs and lentivirus production
Rat TMEM106B cDNA was expressed from a lentiviral vector driven by human synapsin promoter. shRNAs were cloned into pSUPER (target sequences: TMEM106B #1 GCAGATTGATTATACGGTA, TMEM106B #2 GTGGAAGGAACACGACTTA, MAP6 GGTGCAGATCAGCGTGACA and luciferase control CGTACGCGGAATACTTCGA). For lentiviral knockdown the H1 promoter shRNA cassette was subcloned in a modified pLentilox 3.7 with a GFP-cassette driven by human synapsin promoter (Edbauer et al, 2010). The TMEM106B N-terminal fragment (AA 1-93) was cloned into the pEGFP-C1 vector. Rat MAP6 (NP_058900.1), human Map6 (isoform 1: NP_149052.1, isoform 2: NP_997460.1) and dnRILP (rat AA 201-401) were cloned into GW1-myc expression vector driven by the CMV promoter (British Biotechnology, Oxford, UK). All constructs were verified by sequencing. siRNA against human TMEM106B was purchased from Thermo Scientific (Waltham, MA, USA) (target sequence GATCAGAGATTAAGGCCAA).
Lentivirus was produced in HEK293FT cells cotransfected with psPAX2, pVSVg and the respective overexpression or knockdown constructs (Orozco et al, 2012). After harvest, the supernatant was concentrated by ultracentrifugation and the virus particles resuspended in Neurobasal medium.
Cell culture and transfection
Hippocampal and cortical neurons were prepared from embryonic day 18 Sprague–Dawley rats and transfected with Lipofectamine 2000 or transduced with lentivirus as described before (Orozco et al, 2012). For the analysis of axonal morphology and lysosomal trafficking in axons, hippocampal neurons were held on astrocyte feeder cultures using N2 medium and transfected before plating with an Amaxa 4D-Nucleofector (Lonza, Basel, Switzerland) with primary culture kit P3 (Orozco et al, 2012). Immunofluorescence and immunoblotting was performed as described previously (Orozco et al, 2012). Images were taken on Zeiss LSM 510 or 710 laser scanning microscopes (Zeiss, Jena, Germany) using 40× and 63× objectives with 1 Airy unit pinhole. HeLa cells were transfected with X-tremeGENE 9 DNA Transfection Reagent (Roche Applied Science) for 72 h and either processed for immunoblotting or immunocytochemistry as described before (Lang et al, 2012). For coimmunoprecipitation experiments, HEK293FT cells were transfected with Lipofectamine 2000 according to the manufacturer's instructions.
Cell viability assay
Neurons were plated in 96-well plates and transduced with the indicated lentiviruses for 5 days. XTT assay (Roche Applied Science) was performed according to the manufacturer's instructions.
Quantitative analysis of colocalization
The amount of colocalization of two fluorescent signals in a confocal image was analyzed using Pearson′s coefficient estimated with the JaCoP plugin (Bolte & Cordelieres, 2006) of ImageJ.
Live cell imaging and quantification of organelle movement
Time lapse images were taken on a Zeiss Cell observer SD spinning-disc microscope with an air-cooled Evolve 512 EMCCD camera at 1 Hz for 5 min. Neurons were kept in a climate chamber (37°C, 5% CO2) during image acquisition. Kymographs of vesicular movement from the axon or 3–5 dendrite segments per cell were generated and manually analyzed using ImageJ software (Multiple Kymograph plugin by J. Rietdorf and A. Seitz). For the analysis of average run length and velocity only vesicles with more than 2.5 μm run length were included.
Morphological analysis
For the automatic analysis of lysosomes, projected images were segmented with Definiens Developer XD (version 2.02; Definiens AG, Munich, Germany). Nuclei regions were identified in the DAPI channel by automatic thresholding. Touching objects were separated by using the shape split function and subsequent erosion and dilation of the nucleus objects. Cell regions were identified based on β-catenin staining by a watershed algorithm using nucleus objects as seeds. Cell borders were smoothed by subsequent erosion and dilation. Lysosome region was identified in each cell object by automatic thresholding of the LAMP2 channel. The mean distance of lysosomes from nucleus was calculated by averaging the distance of each pixel belonging to the lysosome region to nucleus border. Aggregation of lysosomes was quantified with the Clark aggregation index (CAI). For each object, D(x) denotes the spatial distance to the next neighbor of pixels belonging to the lysosome region, N(x) denotes the number of pixels belonging to the lysosome region and A denotes the number of pixels of the cell object. The CAI is defined as [sum(D(x))/N(x))*sqrt(N(x)/A)].
Dendritic complexity was quantified using manual Sholl analysis using MetaMorph software (Molecular Devices, Sunnyvale, CA, USA). Concentric circles were laid around the cell soma from 12.5 to 112.5 μm (in 12.5 μm intervals) from the soma. The number of dendrites crossing each circle was counted. For morphological analysis of axons, the length of the tau-1 positive neurite was measured using the AxioVision software. All image acquisition and quantification for morphological analyses were done blind to the experimental conditions.
Immunoprecipitation from rat brain
P15 Sprague-Dawley-rat brains were homogenized in 0.32 M sucrose, 4 mM Hepes, 2 mM EDTA (pH 7.4). After centrifugation (75,600 g, 30 min) the pellet was lysed with 1% Triton X-100 in PBS for 20 min at 4°C. Subsequently, the soluble fraction (100,000 g, 20 min) was subjected to immunoprecipitation with TMEM106B, MAP6 or an unspecific control antibody (anti-GST) coupled (BS3; Thermo Scientific) to Dynabeads (Life-Technologies, Carlsbad, CA, USA) for 1.5 h at room temperature. After several washing steps with lysis buffer protein was eluted from the beads with 50 mM glycine, pH 2.8 for Coomassie staining (NOVEX colloidal blue staining kit; Invitrogen) and subsequent LC-MS/MS analysis, or directly boiled in sample buffer (4% SDS, 20% glycerol, 5% β-mercaptoethanol, 200 mM sodium phosphate pH 7.4) for immunoblotting. Protease and phosphatase inhibitor were present in all steps of the immunoprecipitation.
Coimmunoprecipitation from HEK293FT cells
HEK293FT (Invitrogen) cells were transfected with the respective constructs for 48 h using Lipofectamine 2000 (Invitrogen). Cells were washed with PBS, lysed with 1% Triton X-100 in PBS and the lysate centrifuged (40 min, 17 ,000 g). The supernatant was diluted with PBS to 0.5% Triton X-100, precleared for 30 min with Protein A sepharose beads and afterwards subjected to immunoprecipitation with 30 μl myc-agarose beads (Sigma-Aldrich). Samples were washed four times with 0.5% Triton X-100 in PBS before loading. In all steps, protease and phosphatase inhibitors were present.
Mass spectrometry
Tryptic in-gel digestion of the proteins was performed as described (Shevchenko et al, 2006). Peptides were analyzed by an LC-MS/MS set-up coupling a Proxeon Easy nLCII (Thermo Fisher Scientific) with in-house packed 15 cm columns (2.4 μm C18 beads, Dr. Maisch GmbH) to an LTQ Velos Orbitrap mass spectrometer (Thermo Scientific). A bilinear gradient of 60 to 85 min was applied for peptide separation; collision induced dissociation (CID) was applied for fragmentation of TOP14 peptides. Data analysis was performed using the Proteome Discoverer 1.2 (Thermo Scientific) with the embedded SEQUEST algorithm for peptide identification. The International Protein Index database for rat (version 3.87) was used for the database search with carbamidomethylation of cysteine as a static and oxidation of methionine as a dynamic modification. Only full tryptic peptides with a maximum of 2 missed cleavages and an FDR below 5% were allowed.
Subcellular fractionation
An adult rat brain was homogenized in 20 ml buffer (10 mM Hepes, 1 mM EDTA, 0.32 M sucrose, pH 7.4). Postnuclear supernatant (1500 g, 10 min) was centrifuged (100 000 g, 30 min) and the membrane fraction was resuspended in homogenization buffer (10 mM Hepes, 1 mM EDTA, 0.25 M sucrose, pH 7.4). 1/10 volume was loaded on a step gradient (2.5, 5, 7.5, 10, 12.5, 15, 17.5, 20 and 30% iodixanol in homogenization buffer). After centrifugation (40,000 rpm, 2.5 h in a TH-641 rotor) 1 ml fractions were collected by needle puncture at the bottom.
Acknowledgments
We thank J. Banzhaf-Strathmann, Eva Bentmann, S. Hassan, B. Schmid and C. Wahl-Schott for critical comments. We thank Andrea Wenninger-Weinzierl, and Nagore Astola for technical assistance. Automated image analysis was performed by C. Moehl at the DZNE light microscopy facility. CML was supported by a fellowship of the Hans and Ilse Breuer Foundation. DE was supported by the Helmholtz Young Investigator program HZ-NG-607. We thank the Competence Network for Neurodegenerative Diseases (KNDD) of the Bundesministerium für Bildung und Forschung (BMBF) for support to CH and SFL. The research leading to these results has received funding from the European Research Council under the European Union's Seventh Framework Programme (FP7/2007-2013) / ERC grant agreement no. 321366-Amyloid to CH. SFL was supported by the JPND-RiMOD program.
Author contributions
BMS and DE conceived the experiments. BMS performed and analyzed most experiments in neurons and rat brain. CML performed the analysis in heterologous cells. SH and SFL performed mass spectrometry analyses. DO and ST analyzed the axonal phenotype. KR and DE generated reagents. CCH, AC, CH and DE supervised research. BMS and DE wrote the manuscript with input from all coauthors.
Conflict of interest
The authors declare that they have no conflict of interest.
Supplementary information
for this article is available online: http://emboj.embopress.org
References
- Ahmed Z, Sheng H, Xu YF, Lin WL, Innes AE, Gass J, Yu X, Wuertzer CA, Hou H, Chiba S, Yamanouchi K, Leissring M, Petrucelli L, Nishihara M, Hutton ML, McGowan E, Dickson DW, Lewis J. Accelerated lipofuscinosis and ubiquitination in granulin knockout mice suggest a role for progranulin in successful aging. Am J Pathol. 2010;177:311–324. doi: 10.2353/ajpath.2010.090915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akhmanova A, Steinmetz MO. Tracking the ends: a dynamic protein network controls the fate of microtubule tips. Nat Rev Mol Cell Biol. 2008;9:309–322. doi: 10.1038/nrm2369. [DOI] [PubMed] [Google Scholar]
- Andrieux A, Salin PA, Vernet M, Kujala P, Baratier J, Gory-Faure S, Bosc C, Pointu H, Proietto D, Schweitzer A, Denarier E, Klumperman J, Job D. The suppression of brain cold-stable microtubules in mice induces synaptic defects associated with neuroleptic-sensitive behavioral disorders. Genes Dev. 2002;16:2350–2364. doi: 10.1101/gad.223302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arama J, Boulay AC, Bosc C, Delphin C, Loew D, Rostaing P, Amigou E, Ezan P, Wingertsmann L, Guillaud L, Andrieux A, Giaume C, Cohen-Salmon M. Bmcc1s, a novel brain-isoform of Bmcc1, affects cell morphology by regulating MAP6/STOP functions. PLoS ONE. 2012;7:e35488. doi: 10.1371/journal.pone.0035488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baratier J, Peris L, Brocard J, Gory-Faure S, Dufour F, Bosc C, Fourest-Lieuvin A, Blanchoin L, Salin P, Job D, Andrieux A. Phosphorylation of microtubule-associated protein STOP by calmodulin kinase II. J Biol Chem. 2006;281:19561–19569. doi: 10.1074/jbc.M509602200. [DOI] [PubMed] [Google Scholar]
- Bolte S, Cordelieres FP. A guided tour into subcellular colocalization analysis in light microscopy. J Microsc. 2006;224:213–232. doi: 10.1111/j.1365-2818.2006.01706.x. [DOI] [PubMed] [Google Scholar]
- Bosc C, Cronk JD, Pirollet F, Watterson DM, Haiech J, Job D, Margolis RL. Cloning, expression, and properties of the microtubule-stabilizing protein STOP. Proc Natl Acad Sci USA. 1996;93:2125–2130. doi: 10.1073/pnas.93.5.2125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brady OA, Zheng Y, Murphy K, Huang M, Hu F. The frontotemporal lobar degeneration risk factor, TMEM106B, regulates lysosomal morphology and function. Hum Mol Genet. 2012;22:685–695. doi: 10.1093/hmg/dds475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bucci C, Thomsen P, Nicoziani P, McCarthy J, van Deurs B. Rab7: a key to lysosome biogenesis. Mol Biol Cell. 2000;11:467–480. doi: 10.1091/mbc.11.2.467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Busch J, Martinez-Lage M, Ashbridge E, Grossman M, Van Deerlin V, Hu F, Lee V, Trojanowski J, Chen-Plotkin A. Expression of TMEM106B, the frontotemporal lobar degeneration-associated protein, in normal and diseased human brain. Acta Neuropathol Commun. 2013;1:36. doi: 10.1186/2051-5960-1-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cantalupo G, Alifano P, Roberti V, Bruni CB, Bucci C. Rab-interacting lysosomal protein (RILP): the Rab7 effector required for transport to lysosomes. EMBO J. 2001;20:683–693. doi: 10.1093/emboj/20.4.683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakrabarti S, Kobayashi KS, Flavell RA, Marks CB, Miyake K, Liston DR, Fowler KT, Gorelick FS, Andrews NW. Impaired membrane resealing and autoimmune myositis in synaptotagmin VII-deficient mice. J Cell Biol. 2003;162:543–549. doi: 10.1083/jcb.200305131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen YM, Gerwin C, Sheng ZH. Dynein light chain LC8 regulates syntaphilin-mediated mitochondrial docking in axons. J Neurosci. 2009;29:9429–9438. doi: 10.1523/JNEUROSCI.1472-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen-Plotkin AS, Unger TL, Gallagher MD, Bill E, Kwong LK, Volpicelli-Daley L, Busch JI, Akle S, Grossman M, Van Deerlin V, Trojanowski JQ, Lee VM. TMEM106B, the risk gene for frontotemporal dementia, is regulated by the microRNA-132/212 cluster and affects progranulin pathways. J Neurosci. 2012;32:11213–11227. doi: 10.1523/JNEUROSCI.0521-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clare R, King VG, Wirenfeldt M, Vinters HV. Synapse loss in dementias. J Neurosci Res. 2010;88:2083–2090. doi: 10.1002/jnr.22392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark PJ, Evans FC. Distance to nearest neighbor as a measure of spatial relationships in populations. Ecology. 1954;35:445–453. [Google Scholar]
- Cruchaga C, Graff C, Chiang HH, Wang J, Hinrichs AL, Spiegel N, Bertelsen S, Mayo K, Norton JB, Morris JC, Goate A. Association of TMEM106B gene polymorphism with age at onset in granulin mutation carriers and plasma granulin protein levels. Arch Neurol. 2011;68:581–586. doi: 10.1001/archneurol.2010.350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cruts M, Gijselinck I, van der Zee J, Engelborghs S, Wils H, Pirici D, Rademakers R, Vandenberghe R, Dermaut B, Martin JJ, van Duijn C, Peeters K, Sciot R, Santens P, De Pooter T, Mattheijssens M, Van den Broeck M, Cuijt I, Vennekens K, De Deyn PP, et al. Null mutations in progranulin cause ubiquitin-positive frontotemporal dementia linked to chromosome 17q21. Nature. 2006;442:920–924. doi: 10.1038/nature05017. [DOI] [PubMed] [Google Scholar]
- Dehmelt L, Halpain S. The MAP2/Tau family of microtubule-associated proteins. Genome Biol. 2005;6:204. doi: 10.1186/gb-2004-6-1-204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeJesus-Hernandez M, Mackenzie IR, Boeve BF, Boxer AL, Baker M, Rutherford NJ, Nicholson AM, Finch NA, Flynn H, Adamson J, Kouri N, Wojtas A, Sengdy P, Hsiung GY, Karydas A, Seeley WW, Josephs KA, Coppola G, Geschwind DH, Wszolek ZK, et al. Expanded GGGGCC hexanucleotide repeat in noncoding region of C9ORF72 causes chromosome 9p-linked FTD and ALS. Neuron. 2011;72:245–256. doi: 10.1016/j.neuron.2011.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edbauer D, Neilson JR, Foster KA, Wang CF, Seeburg DP, Batterton MN, Tada T, Dolan BM, Sharp PA, Sheng M. Regulation of synaptic structure and function by FMRP-associated microRNAs miR-125b and miR-132. Neuron. 2010;65:373–384. doi: 10.1016/j.neuron.2010.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filimonenko M, Stuffers S, Raiborg C, Yamamoto A, Malerod L, Fisher EM, Isaacs A, Brech A, Stenmark H, Simonsen A. Functional multivesicular bodies are required for autophagic clearance of protein aggregates associated with neurodegenerative disease. J Cell Biol. 2007;179:485–500. doi: 10.1083/jcb.200702115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finch N, Carrasquillo MM, Baker M, Rutherford NJ, Coppola G, Dejesus-Hernandez M, Crook R, Hunter T, Ghidoni R, Benussi L, Crook J, Finger E, Hantanpaa KJ, Karydas AM, Sengdy P, Gonzalez J, Seeley WW, Johnson N, Beach TG, Mesulam M, et al. TMEM106B regulates progranulin levels and the penetrance of FTLD in GRN mutation carriers. Neurology. 2011;76:467–474. doi: 10.1212/WNL.0b013e31820a0e3b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fournet V, de Lavilleon G, Schweitzer A, Giros B, Andrieux A, Martres MP. Both chronic treatments by epothilone D and fluoxetine increase the short-term memory and differentially alter the mood status of STOP/MAP6 KO mice. J Neurochem. 2012;123:982–996. doi: 10.1111/jnc.12027. [DOI] [PubMed] [Google Scholar]
- Garcia ML, Cleveland DW. Going new places using an old MAP: tau, microtubules and human neurodegenerative disease. Curr Opin Cell Biol. 2001;13:41–48. doi: 10.1016/s0955-0674(00)00172-1. [DOI] [PubMed] [Google Scholar]
- Giannakakou P, Nakano M, Nicolaou KC, O'Brate A, Yu J, Blagosklonny MV, Greber UF, Fojo T. Enhanced microtubule-dependent trafficking and p53 nuclear accumulation by suppression of microtubule dynamics. Proc Natl Acad Sci USA. 2002;99:10855–10860. doi: 10.1073/pnas.132275599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gory-Faure S, Windscheid V, Bosc C, Peris L, Proietto D, Franck R, Denarier E, Job D, Andrieux A. STOP-like protein 21 is a novel member of the STOP family, revealing a Golgi localization of STOP proteins. J Biol Chem. 2006;281:28387–28396. doi: 10.1074/jbc.M603380200. [DOI] [PubMed] [Google Scholar]
- Hoogenraad CC, Bradke F. Control of neuronal polarity and plasticity–a renaissance for microtubules? Trends Cell Biol. 2009;19:669–676. doi: 10.1016/j.tcb.2009.08.006. [DOI] [PubMed] [Google Scholar]
- Horton AC, Racz B, Monson EE, Lin AL, Weinberg RJ, Ehlers MD. Polarized secretory trafficking directs cargo for asymmetric dendrite growth and morphogenesis. Neuron. 2005;48:757–771. doi: 10.1016/j.neuron.2005.11.005. [DOI] [PubMed] [Google Scholar]
- Hu F, Padukkavidana T, Vaegter CB, Brady OA, Zheng Y, Mackenzie IR, Feldman HH, Nykjaer A, Strittmatter SM. Sortilin-mediated endocytosis determines levels of the frontotemporal dementia protein, progranulin. Neuron. 2010;68:654–667. doi: 10.1016/j.neuron.2010.09.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huynh C, Roth D, Ward DM, Kaplan J, Andrews NW. Defective lysosomal exocytosis and plasma membrane repair in Chediak-Higashi/beige cells. Proc Natl Acad Sci USA. 2004;101:16795–16800. doi: 10.1073/pnas.0405905101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jan YN, Jan LY. Branching out: mechanisms of dendritic arborization. Nat Rev Neurosci. 2010;11:316–328. doi: 10.1038/nrn2836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jordens I, Fernandez-Borja M, Marsman M, Dusseljee S, Janssen L, Calafat J, Janssen H, Wubbolts R, Neefjes J. The Rab7 effector protein RILP controls lysosomal transport by inducing the recruitment of dynein-dynactin motors. Curr Biol. 2001;11:1680–1685. doi: 10.1016/s0960-9822(01)00531-0. [DOI] [PubMed] [Google Scholar]
- Ju JS, Fuentealba RA, Miller SE, Jackson E, Piwnica-Worms D, Baloh RH, Weihl CC. Valosin-containing protein (VCP) is required for autophagy and is disrupted in VCP disease. J Cell Biol. 2009;187:875–888. doi: 10.1083/jcb.200908115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang JS, Tian JH, Pan PY, Zald P, Li C, Deng C, Sheng ZH. Docking of axonal mitochondria by syntaphilin controls their mobility and affects short-term facilitation. Cell. 2008;132:137–148. doi: 10.1016/j.cell.2007.11.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapitein LC, Hoogenraad CC. Which way to go? Cytoskeletal organization and polarized transport in neurons. Mol Cell Neurosci. 2011;46:9–20. doi: 10.1016/j.mcn.2010.08.015. [DOI] [PubMed] [Google Scholar]
- Kuijpers M, Yu KL, Teuling E, Akhmanova A, Jaarsma D, Hoogenraad CC. The ALS8 protein VAPB interacts with the ER-Golgi recycling protein YIF1A and regulates membrane delivery into dendrites. EMBO J. 2013;32:2056–2072. doi: 10.1038/emboj.2013.131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lang CM, Fellerer K, Schwenk BM, Kuhn PH, Kremmer E, Edbauer D, Capell A, Haass C. Membrane orientation and subcellular localization of transmembrane protein 106B (TMEM106B), a major risk factor for frontotemporal lobar degeneration. J Biol Chem. 2012;287:19355–19365. doi: 10.1074/jbc.M112.365098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Letournel F, Bocquet A, Dubas F, Barthelaix A, Eyer J. Stable tubule only polypeptides (STOP) proteins co-aggregate with spheroid neurofilaments in amyotrophic lateral sclerosis. J Neuropathol Exp Neurol. 2003;62:1211–1219. doi: 10.1093/jnen/62.12.1211. [DOI] [PubMed] [Google Scholar]
- Morderer D, Nikolaienko O, Skrypkina I, Cherkas V, Tsyba L, Belan P, Rynditch A. Endocytic adaptor protein intersectin 1 forms a complex with microtubule stabilizer STOP in neurons. Gene. 2012;505:360–364. doi: 10.1016/j.gene.2012.06.061. [DOI] [PubMed] [Google Scholar]
- Mukhopadhyay A, Funato K, Stahl PD. Rab7 regulates transport from early to late endocytic compartments in Xenopus oocytes. J Biol Chem. 1997;272:13055–13059. doi: 10.1074/jbc.272.20.13055. [DOI] [PubMed] [Google Scholar]
- Neumann M, Sampathu DM, Kwong LK, Truax AC, Micsenyi MC, Chou TT, Bruce J, Schuck T, Grossman M, Clark CM, McCluskey LF, Miller BL, Masliah E, Mackenzie IR, Feldman H, Feiden W, Kretzschmar HA, Trojanowski JQ, Lee VM. Ubiquitinated TDP-43 in frontotemporal lobar degeneration and amyotrophic lateral sclerosis. Science. 2006;314:130–133. doi: 10.1126/science.1134108. [DOI] [PubMed] [Google Scholar]
- Nicholson AM, Finch NA, Wojtas A, Baker MC, Perkerson RB, III, Castanedes-Casey M, Rousseau L, Benussi L, Binetti G, Ghidoni R, Hsiung GY, Mackenzie IR, Finger E, Boeve BF, Ertekin-Taner N, Graff-Radford NR, Dickson DW, Rademakers R. TMEM106B p.T185S regulates TMEM106B protein levels: implications for frontotemporal dementia. J Neurochem. 2013;126:781–791. doi: 10.1111/jnc.12329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimura AL, Mitne-Neto M, Silva HC, Richieri-Costa A, Middleton S, Cascio D, Kok F, Oliveira JR, Gillingwater T, Webb J, Skehel P, Zatz M. A mutation in the vesicle-trafficking protein VAPB causes late-onset spinal muscular atrophy and amyotrophic lateral sclerosis. Am J Hum Genet. 2004;75:822–831. doi: 10.1086/425287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orozco D, Tahirovic S, Rentzsch K, Schwenk BM, Haass C, Edbauer D. Loss of fused in sarcoma (FUS) promotes pathological Tau splicing. EMBO Rep. 2012;13:759–764. doi: 10.1038/embor.2012.90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petkau TL, Neal SJ, Milnerwood A, Mew A, Hill AM, Orban P, Gregg J, Lu G, Feldman HH, Mackenzie IR, Raymond LA, Leavitt BR. Synaptic dysfunction in progranulin-deficient mice. Neurobiol Dis. 2012;45:711–722. doi: 10.1016/j.nbd.2011.10.016. [DOI] [PubMed] [Google Scholar]
- Press B, Feng Y, Hoflack B, Wandinger-Ness A. Mutant Rab7 causes the accumulation of cathepsin D and cation-independent mannose 6-phosphate receptor in an early endocytic compartment. J Cell Biol. 1998;140:1075–1089. doi: 10.1083/jcb.140.5.1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rademakers R, Neumann M, Mackenzie IR. Advances in understanding the molecular basis of frontotemporal dementia. Nat Rev Neurol. 2012;8:423–434. doi: 10.1038/nrneurol.2012.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy A, Caler EV, Andrews NW. Plasma membrane repair is mediated by Ca(2 + )-regulated exocytosis of lysosomes. Cell. 2001;106:157–169. doi: 10.1016/s0092-8674(01)00421-4. [DOI] [PubMed] [Google Scholar]
- Renton AE, Majounie E, Waite A, Simon-Sanchez J, Rollinson S, Gibbs JR, Schymick JC, Laaksovirta H, van Swieten JC, Myllykangas L, Kalimo H, Paetau A, Abramzon Y, Remes AM, Kaganovich A, Scholz SW, Duckworth J, Ding J, Harmer DW, Hernandez DG, et al. A hexanucleotide repeat expansion in C9ORF72 is the cause of chromosome 9p21-linked ALS-FTD. Neuron. 2011;72:257–268. doi: 10.1016/j.neuron.2011.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritz D, Vuk M, Kirchner P, Bug M, Schutz S, Hayer A, Bremer S, Lusk C, Baloh RH, Lee H, Glatter T, Gstaiger M, Aebersold R, Weihl CC, Meyer H. Endolysosomal sorting of ubiquitylated caveolin-1 is regulated by VCP and UBXD1 and impaired by VCP disease mutations. Nat Cell Biol. 2011;13:1116–1123. doi: 10.1038/ncb2301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rutherford NJ, Carrasquillo MM, Li M, Bisceglio G, Menke J, Josephs KA, Parisi JE, Petersen RC, Graff-Radford NR, Younkin SG, Dickson DW, Rademakers R. TMEM106B risk variant is implicated in the pathologic presentation of Alzheimer disease. Neurology. 2012;79:717–718. doi: 10.1212/WNL.0b013e318264e3ac. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sann S, Wang Z, Brown H, Jin Y. Roles of endosomal trafficking in neurite outgrowth and guidance. Trends Cell Biol. 2009;19:317–324. doi: 10.1016/j.tcb.2009.05.001. [DOI] [PubMed] [Google Scholar]
- Shevchenko A, Tomas H, Havlis J, Olsen JV, Mann M. In-gel digestion for mass spectrometric characterization of proteins and proteomes. Nat Protoc. 2006;1:2856–2860. doi: 10.1038/nprot.2006.468. [DOI] [PubMed] [Google Scholar]
- Sholl DA. Dendritic organization in the neurons of the visual and motor cortices of the cat. J Anat. 1953;87:387–406. [PMC free article] [PubMed] [Google Scholar]
- Smith KR, Damiano J, Franceschetti S, Carpenter S, Canafoglia L, Morbin M, Rossi G, Pareyson D, Mole SE, Staropoli JF, Sims KB, Lewis J, Lin WL, Dickson DW, Dahl HH, Bahlo M, Berkovic SF. Strikingly different clinicopathological phenotypes determined by progranulin-mutation dosage. Am J Hum Genet. 2012;90:1102–1107. doi: 10.1016/j.ajhg.2012.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sreedharan J, Blair IP, Tripathi VB, Hu X, Vance C, Rogelj B, Ackerley S, Durnall JC, Williams KL, Buratti E, Baralle F, de Belleroche J, Mitchell JD, Leigh PN, Al-Chalabi A, Miller CC, Nicholson G, Shaw CE. TDP-43 mutations in familial and sporadic amyotrophic lateral sclerosis. Science. 2008;319:1668–1672. doi: 10.1126/science.1154584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tackenberg C, Ghori A, Brandt R. Thin, stubby or mushroom: spine pathology in Alzheimer's disease. Curr Alzheimer Res. 2009;6:261–268. doi: 10.2174/156720509788486554. [DOI] [PubMed] [Google Scholar]
- Tamura N, Draviam VM. Microtubule plus-ends within a mitotic cell are ‘moving platforms’ with anchoring, signalling and force-coupling roles. Open Biol. 2012;2:120132. doi: 10.1098/rsob.120132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang W, Lu Y, Tian QY, Zhang Y, Guo FJ, Liu GY, Syed NM, Lai Y, Lin EA, Kong L, Su J, Yin F, Ding AH, Zanin-Zhorov A, Dustin ML, Tao J, Craft J, Yin Z, Feng JQ, Abramson SB, et al. The growth factor progranulin binds to TNF receptors and is therapeutic against inflammatory arthritis in mice. Science. 2011;332:478–484. doi: 10.1126/science.1199214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuxworth RI, Vivancos V, O'Hare MB, Tear G. Interactions between the juvenile Batten disease gene, CLN3, and the Notch and JNK signalling pathways. Hum Mol Genet. 2009;18:667–678. doi: 10.1093/hmg/ddn396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urwin H, Authier A, Nielsen JE, Metcalf D, Powell C, Froud K, Malcolm DS, Holm I, Johannsen P, Brown J, Fisher EM, van der Zee J, Bruyland M, Van Broeckhoven C, Collinge J, Brandner S, Futter C, Isaacs AM. Disruption of endocytic trafficking in frontotemporal dementia with CHMP2B mutations. Hum Mol Genet. 2010;19:2228–2238. doi: 10.1093/hmg/ddq100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uusi-Rauva K, Kyttala A, van der Kant R, Vesa J, Tanhuanpaa K, Neefjes J, Olkkonen VM, Jalanko A. Neuronal ceroid lipofuscinosis protein CLN3 interacts with motor proteins and modifies location of late endosomal compartments. Cell Mol Life Sci. 2012;69:2075–2089. doi: 10.1007/s00018-011-0913-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Deerlin VM, Sleiman PM, Martinez-Lage M, Chen-Plotkin A, Wang LS, Graff-Radford NR, Dickson DW, Rademakers R, Boeve BF, Grossman M, Arnold SE, Mann DM, Pickering-Brown SM, Seelaar H, Heutink P, van Swieten JC, Murrell JR, Ghetti B, Spina S, Grafman J, et al. Common variants at 7p21 are associated with frontotemporal lobar degeneration with TDP-43 inclusions. Nat Genet. 2010;42:234–239. doi: 10.1038/ng.536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van der Zee J, Van Langenhove T, Kleinberger G, Sleegers K, Engelborghs S, Vandenberghe R, Santens P, Van den Broeck M, Joris G, Brys J, Mattheijssens M, Peeters K, Cras P, De Deyn PP, Cruts M, Van Broeckhoven C. TMEM106B is associated with frontotemporal lobar degeneration in a clinically diagnosed patient cohort. Brain. 2011;134:808–815. doi: 10.1093/brain/awr007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vass R, Ashbridge E, Geser F, Hu WT, Grossman M, Clay-Falcone D, Elman L, McCluskey L, Lee VM, Van Deerlin VM, Trojanowski JQ, Chen-Plotkin AS. Risk genotypes at TMEM106B are associated with cognitive impairment in amyotrophic lateral sclerosis. Acta Neuropathol. 2011;121:373–380. doi: 10.1007/s00401-010-0782-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wils H, Kleinberger G, Pereson S, Janssens J, Capell A, Van Dam D, Cuijt I, Joris G, De Deyn PP, Haass C, Van Broeckhoven C, Kumar-Singh S. Cellular ageing, increased mortality and FTLD-TDP-associated neuropathology in progranulin knockout mice. J Pathol. 2012;228:67–76. doi: 10.1002/path.4043. [DOI] [PubMed] [Google Scholar]
- Ye B, Zhang Y, Song W, Younger SH, Jan LY, Jan YN. Growing dendrites and axons differ in their reliance on the secretory pathway. Cell. 2007;130:717–729. doi: 10.1016/j.cell.2007.06.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


