Abstract
Mitochondrial calcium uptake is a critical event in various cellular activities. Two recently identified proteins, the mitochondrial Ca2+ uniporter (MCU), which is the pore-forming subunit of a Ca2+ channel, and mitochondrial calcium uptake 1 (MICU1), which is the regulator of MCU, are essential in this event. However, the molecular mechanism by which MICU1 regulates MCU remains elusive. In this study, we report the crystal structures of Ca2+-free and Ca2+-bound human MICU1. Our studies reveal that Ca2+-free MICU1 forms a hexamer that binds and inhibits MCU. Upon Ca2+ binding, MICU1 undergoes large conformational changes, resulting in the formation of multiple oligomers to activate MCU. Furthermore, we demonstrate that the affinity of MICU1 for Ca2+ is approximately 15–20 μM. Collectively, our results provide valuable details to decipher the molecular mechanism of MICU1 regulation of mitochondrial calcium uptake.
Keywords: calcium, MCU, mechanism, MICU1, mitochondrial calcium uptake
Introduction
Mitochondria are the power plants of the cell and function as important nodes within signaling pathways to regulate various cellular activities. Calcium in the mitochondria plays a key role in several important functions, such as controlling the metabolic rate for cellular energy (i.e., ATP) production (Denton & McCormack, 1980; Balaban, 2009), the modulation of the amplitude and shape of the cytosolic Ca2+ transients (Herrington et al, 1996; Babcock et al, 1997), and the induction of apoptosis through the release of cytochrome c from the mitochondrial intermembrane space into the cytosolic space (Hajnoczky et al, 2000; Orrenius et al, 2003).
Mitochondrial Ca2+ uptake was discovered in the 1960s and was found to be mediated by the mitochondrial Ca2+ uniporter (MCU) (Deluca & Engstrom, 1961; Vasington & Murphy, 1962). MCU is a ruthenium-red-sensitive and highly selective Ca2+ ion channel that uptakes massive amounts of Ca2+ across the inner membrane (Kirichok et al, 2004). Mitochondrial Ca2+ levels are tightly linked to the regulation of cytosolic Ca2+ levels. When the inositol 1,4,5-triphosphate (IP3)-gated channels of the endoplasmic reticulum (ER) are open, the mitochondrial surface will be exposed to a higher concentration of Ca2+ than in the bulk cytosol (Rizzuto et al, 1998; Csordas et al, 2006); therefore, Ca2+ can be rapidly transferred into the mitochondria matrix by MCU, which is driven by the presence of an electrochemical proton gradient across the inner mitochondrial membrane (Reed & Bygrave, 1975; Bragadin et al, 1979). Once the cytoplasmic Ca2+ levels are again low, Ca2+ is transported out of the mitochondria by a Na+/Ca2+ exchanger (NCX) and a H+/Ca2+ exchanger (HCX) that are present in the membrane (Rizzuto et al, 2012) and potentially by a mitochondrial permeability transition pore (mPTP) (Bernardi & von Stockum, 2012).
After 50 years of research, the molecular machinery of the massive mitochondrial calcium uptake system has finally been identified (Rizzuto et al, 2012; Dedkova & Blatter, 2013). In 2010, based on clues from comparative physiology, evolutionary genomics, organelle proteomics, and genome-wide RNAi screening, the mitochondrial calcium uptake 1 (MICU1) protein was identified as an essential element of mitochondrial calcium uptake (Perocchi et al, 2010). Down-regulation of MICU1 caused significant suppression of the mitochondrial Ca2+ signal evoked by an IP3-linked agonist but did not disrupt mitochondrial respiration or the membrane potential. MICU1 is a ∼54-kDa protein with an amino-terminal mitochondrial targeting sequence, a predicted transmembrane helix (aa ∼33–52), and a cytosolic C-terminus (aa ∼53–476) containing two classical EF-hand Ca2+-binding domains (Linding et al, 2003; Aichberger et al, 2005; Perocchi et al, 2010). It localizes to the mitochondrial inner membrane and does not appear to participate in channel pore formation (Csordas et al, 2013; Sancak et al, 2013). Thus, MICU1 is likely to serve as a Ca2+ sensor that regulates MCU (Collins & Meyer, 2010; Hajnoczky & Csordas, 2010). Recent studies suggest that MICU1 acts as a threshold for MCU-mediated calcium uptake (Mallilankaraman et al, 2012; Csordas et al, 2013). The pore-forming MCU protein was identified in 2011 (Baughman et al, 2011; De Stefani et al, 2011). This protein is a 40-kDa mitochondrial inner membrane protein that is ubiquitously expressed in mammals and physically interacts with MICU1 (Baughman et al, 2011; De Stefani et al, 2011). Moreover, the MICU1 and MCU expression patterns are tightly coupled across most species (Bick et al, 2012). The uniporter complex was recently characterized to be composed of MCU, MCUb, MICU1, MICU2, and EMRE (Sancak et al, 2013).
To further understand the molecular mechanisms of MICU1 Ca2+ sensing and mitochondrial calcium uptake regulation, we determined the crystal structures of Ca2+-free and Ca2+-bound human MICU1. Our results indicated that MICU1 exists as a hexamer and possibly inhibits the function of MCU in the absence of Ca2+. Ca2+ loading of MICU1 results in the formation of multiple MICU1 oligomers, which may activate MCU.
Results
Ca2+-free and Ca2+-bound MICU1 structures
To obtain Ca2+-free MICU1 crystals, four constructs (residues 53-476, residues 62-476, residues 80-476, and residues 97-476) based on a sequence alignment of MICU1 from different species (Supplementary Fig 1) were evaluated. One of the constructs (residues 97-476, referred to as MICU1-xtal hereafter; different constructs and names are shown in Supplementary Fig 2A) successfully produced diffraction-quality crystals. The Ca2+-free MICU1-xtal structure was determined using single-wavelength anomalous dispersion (SAD) at the resolution of 3.2 Å. The Ca2+-free MICU1-xtal structure includes four regions (Fig 1A and B): the N-domain, the N-lobe, the C-lobe, and the C-helix. The N-domain (residues 103-177) consists of three α-helices and three antiparallel β-strands. The N-lobe (residues 183-318) and the C-lobe (residues 319-445) are composed of six and seven α-helices, respectively. The C-helix (residues 446-476) is one long helix that does not specifically interact with the primary Ca2+-free MICU1-xtal core.
Figure 1.
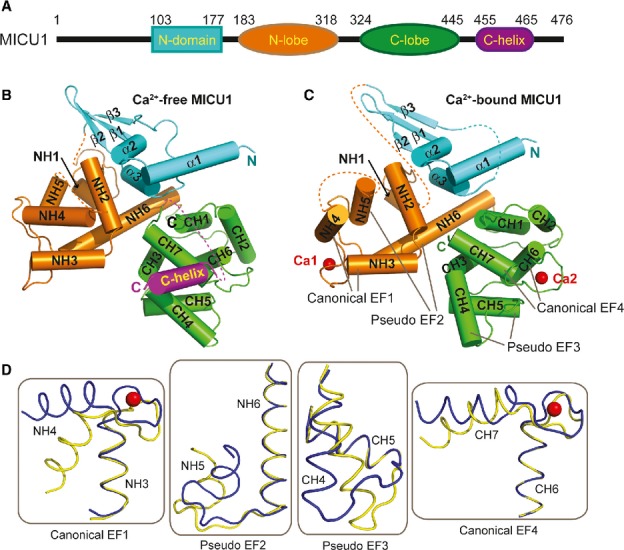
Crystal structures of Ca2+-free and Ca2+-bound human MICU1.
A A schematic drawing of human MICU1.
B, C Cartoon representation of the overall structure of MICU1 in the Ca2+-free and the Ca2+-bound state. The N-domain, N-lobe, C-lobe, C-helix, and Ca2+ are colored cyan, orange, green, purple, and red, respectively.
D Structural comparisons of four helix-loop-helix structural units before and after Ca2+ binding. The Ca2+-free form is colored yellow, and the Ca2+-bound form is colored blue. The calcium ions are shown as red spheres.
To obtain Ca2+-bound MICU1 crystals, the C-helix of MICU1 was deleted to avoid protein aggregation. This modified MICU1 protein (residues 97-444, referred to as MICU1-xtal-deltaC hereafter; Supplementary Fig 2A) was crystallized in the presence of 5 mM calcium chloride. The Ca2+-bound MICU1-xtal-deltaC structure was determined using molecular replacement at a resolution of 2.7 Å. The topology of the Ca2+-bound form is similar to the topology of the Ca2+-free form (Fig 1B and C). Two calcium ions are present in the Ca2+-bound MICU1-xtal-deltaC structure. One calcium ion is bound to the EF hand (referred to as the canonical EF1) that consists of the NH3 and NH4 helices of the N-lobe. The second calcium ion is bound to the EF hand (also known as the canonical EF4) that is composed of the CH6 and CH7 helices of the C-lobe. Both calcium ions bound to sites of the conventional EF hand (Supplementary Fig 2B), as predicted by previous studies (Perocchi et al, 2010). Furthermore, other two helix-loop-helix structural units (referred to as pseudo-EF2 and pseudo-EF3) were found in the MICU1 structure. These units are composed of the NH5 and NH6 helices for pseudo-EF2 as well as the CH4 and CH5 helices for pseudo-EF3. Apparently, these two sites were unable to bind calcium ions, although they contain helix-loop-helix structural units (Fig 1C). Therefore, each N-and C-lobe contains two helix-loop-helix structural units that were capable of binding only one calcium ion, which was also observed for the STIM1 EF-SAM domain (Stathopulos et al, 2008).
Ca2+ loading induced large conformational changes in MICU1, particularly at the EF hands (Fig 1D). In the canonical EF1, the NH4 helix rotated 45° upon Ca2+ loading. This significant movement induced the rotation of the adjacent NH5 helix to a similar degree (∼45°) in the pseudo-EF2. In sharp contrast, Ca2+ loading only caused a slight rotation of the CH7 helix in the canonical EF4. Surprisingly, the CH4 helix demonstrated a large rotation of approximately 35° in the pseudo-EF3. We assumed that this movement was caused by dimer rearrangement (see the following discussion) rather than by Ca2+ binding to the canonical EF4.
The C-helix is critical for mitochondrial calcium uptake
In the absence of Ca2+, MICU1-xtal in solution predominantly existed as a hexamer (∼237 kDa, theoretical molecular weight of 43 kDa for each molecule) (Fig 2A and Supplementary Fig 3A), whereas, in the presence of Ca2+, MICU1-xtal existed as a mixture of oligomeric forms (Fig 2B and Supplementary Fig 3B).
Figure 2.
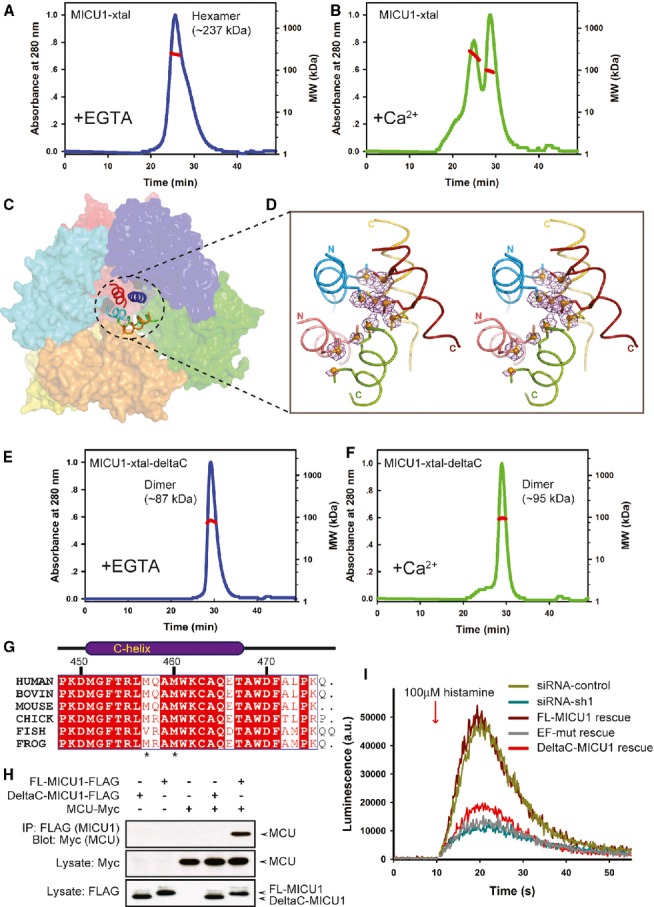
The C-helix is critical for mitochondrial calcium uptake.
A, B Multi-angle laser scattering analysis of MICU1-xtal in the absence or presence of Ca2+. MALS-determined molecular weights were calculated over prominent peaks (red line).
C The hexameric packing of Ca2+-free MICU1. The surface map of six molecules in the asymmetric unit is shown. A cartoon model of the C-helix is presented in the center of the hexamer.
D The anomalous scattering map of the selenium-substituted Ca2+-free MICU1-xtal structure. The map colored in magenta was contoured at 3.0σ. Cartoon representations of the five C-helices from different Ca2+-free MICU1 molecules are shown. One C-helix is not included in the final model due to poor electron density. The methionines are depicted as sticks, and Se atoms are depicted as spheres.
E, F Multi-angle laser scattering analysis of MICU1-xtal-deltaC in the absence or presence of Ca2+.
G Sequence alignment of the MICU1 protein from human (Q9BPX6), bovine (Q0IIL1), mouse (Q8VCX5), chick (E1BWC6), fish (A4IG32), and frog (B1H2N3). The residues that are conserved among the species are highlighted in red. The residue numbers of human MICU1 are shown in black.
H Western blot analysis of co-immunoprecipitated wild-type FL-MICU1 and mutant DeltaC-MICU1 with the MCU channel in the absence of Ca2+.
I cDNA rescue of the RNAi mitochondrial calcium uptake phenotype.
Source data are available online for this figure.
Consistent with the observations in solution, the Ca2+-free MICU1-xtal crystal structure revealed six molecules in the asymmetric unit, which are packed by a trimer of dimers (Fig 2C and Supplementary Fig 4). Unexpectedly, in the Ca2+-free MICU1-xtal structure, the C-helix is located in the center of the six molecules that are packed as a helix bundle. The C-helix was identified by selenium heavy atoms of the sequence motif (-MQAM-) in an anomalous scattering electron density map (Fig 2D). The hexamer is composed of two layers, and three molecules in each layer are loosely bound (Supplementary Fig 5). These results suggest that the C-helix is presumably an important factor in the assembly of the Ca2+-free MICU1-xtal hexamer. To verify this hypothesis, we determined the oligomerization state of MICU1-xtal-deltaC. This mutant exhibited a predominant peak corresponding to the MICU1 dimer both in the absence and in the presence of Ca2+ (Fig 2E, F and Supplementary Fig 3C), suggesting that the C-helix is necessary for the formation of the Ca2+-free MICU1-xtal hexamer and substantially contributed to the formation of multiple oligomers of Ca2+-bound MICU1-xtal.
To further analyze the physiological significance of the C-helix, we first aligned the sequence of MICU1 from different species and determined that the C-helix is highly conserved in vertebrates (Fig 2G). We then compared the binding of full-length MICU1 (FL-MICU1) and DeltaC-MICU1, which is a mutant of FL-MICU1 with the C-helix deleted, to MCU via co-immunoprecipitation (Fig 2H). In the absence of Ca2+, the deletion of the C-helix significantly weakened the binding of MICU1 to MCU.
We then further performed Ca2+ uptake measurements in mitochondria. A FL-MICU1 mutant containing EF-hand mutations (referred to as EF-mut) was constructed as a negative control (Perocchi et al, 2010). MICU1 was depleted by the transfection of HeLa cells with siRNA for 48 h and was then rescued by the co-transfection of FL-MICU1 or the mutants with a mitochondria-targeted aequorin (mt-AEQ). Luminescence from the mt-AEQ was then counted after histamine stimulation. The RNAi Ca2+ uptake phenotype can be fully rescued by FL-MICU1. In sharp contrast, DeltaC-MICU1 fails to restore mitochondrial Ca2+ uptake (Fig 2I), which was similar to the behavior of the EF-mut mutant. Collectively, our results suggested that the C-helix played an important role in assembling the Ca2+-free MICU1 hexamer and in mitochondrial calcium uptake.
Ca2+-free and Ca2+-bound MICU1 dimers
As described above, the MICU1-xtal molecule exhibited different oligomerization behaviors before and after binding Ca2+. To elucidate the underlying mechanism, we characterized the MICU1 dimer in detail. In the crystal structure of Ca2+-free MICU1-xtal, two MICU1 molecules, except for the C-helix, were packed together by 2-fold noncrystallographic symmetry. The dimer was reciprocally formed by interactions between the NH3 and NH4 helices from one monomer with the CH4 and CH5 helices of another monomer, which buried a total surface area of approximately 1,122 Å2 (Fig 3A). Ca2+ binding to MICU1 induced large conformational changes in the monomer, which led to a completely different dimer formation. The interface of the Ca2+-bound MICU1-xtal-deltaC dimer was composed of the CH4 helix from both monomers as well as the CH4-CH5 loop from one monomer with the NH3 helix from the reciprocal monomer. The buried surface area of the Ca2+-bound MICU1-xtal-deltaC dimer was approximately 434 Å2 (Fig 3B). The overall dimers of the Ca2+-free and the Ca2+-bound MICU1 structures could not be superposed. After the superposition of each monomer from the two dimers, the additional superposition of the second monomer from the two dimers required an approximately 74° rotation (Fig 3C).
Figure 3.
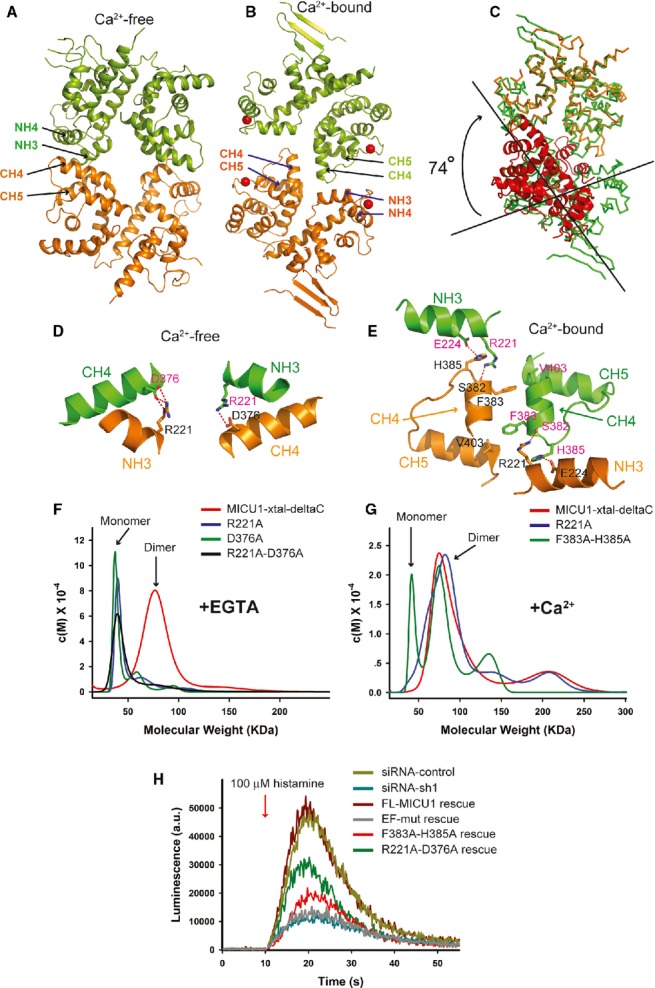
The MICU1 dimer.
A, B A cartoon representation of the MICU1 dimer in the absence or presence of Ca2+. Each molecule in one dimer is colored differently. Calcium is shown as a red sphere.
C Superposition of Ca2+-free and Ca2+-bound MICU1. The Ca2+-bound MICU1 dimer is colored green. One molecule of Ca2+-free MICU1 (orange) superposed well with Ca2+-bound MICU1. However, the second molecule of Ca2+-free MICU1 (red) required a 74° rotation for superposition onto Ca2+-bound MICU1.
D, E Detailed interactions within interfaces of Ca2+-free and Ca2+-bound MICU1 dimers. Each molecule in the dimer is colored green and orange, respectively. The oxygen and nitrogen atoms are colored red and blue, respectively. Hydrogen bonds are depicted as red dotted lines.
F, G Sedimentation velocity analysis of the MICU1-xtal-deltaC wild-type and mutants in the absence or presence of Ca2+.
H cDNA rescue of the RNAi mitochondrial calcium uptake phenotype.
Within the Ca2+-free MICU1-xtal dimer interface, the two monomers primarily interact through a salt bridge. Asp376 from one monomer interacts with Arg221 from the reciprocal monomer via a salt bridge (Fig 3D). The R221A or D376A single mutation or the R221A-D376A double mutation of MICU1-xtal-deltaC converted the dimer into the monomer in the absence of Ca2+ (Fig 3F). The Ca2+-bound MICU1-xtal-deltaC dimer interface is formed by hydrogen bonding and hydrophobic interactions (Fig 3E). Specifically, the side chain of His385 and the main chain oxygen atom of Ser382 from one monomer interact with the side chains of Glu224 and Arg221 from the reciprocal monomer via hydrogen bonds. Additionally, Phe383 from one monomer interacts with Val403 from the reciprocal monomer via hydrophobic interactions. The R221A single mutation of MICU1-xtal-deltaC was unable to disrupt the dimer formation in the presence of Ca2+. The F383A-H385A double mutation of MICU1-xtal-deltaC converted the dimer into the monomer, and the dimer mixture had an approximate ratio of 1:1 in the presence of Ca2+ (Fig 3G).
To further evaluate the physiological relevance of these two MICU1 dimers, the mitochondrial calcium uptake of the FL-MICU1 wild-type and mutants was compared (Fig 3H). We constructed the following two mutants: the F383A-H385A-FL mutant (a double mutation of F383A and H385A in FL-MICU1), which is thought to affect dimer formation in the presence of Ca2+, and the R221A-D376A-FL mutant (a double mutation of R221A and D376A in FL-MICU1), which was designed to affect dimer formation in the absence of Ca2+. In comparison with wild-type, the RNAi Ca2+ uptake phenotype was partially rescued by both mutants, indicating that the two dimers observed in our structure closely correlated with the MICU1 oligomeric states in vivo. In summary, our results suggest that Ca2+ binding to MICU1 induced the rearrangement of the dimer, which presumably converted the Ca2+-free MICU1 hexamer into a mixture of Ca2+-bound MICU1 oligomers.
MICU1 Ca2+-binding sites
In the Ca2+-bound MICU1-xtal-deltaC crystal structure, two Ca2+-binding sites are present. At the canonical EF1 site, Ca2+ is coordinated by the side chains of Asp231, Asn233, Asp235, and Glu242, a main chain oxygen atom of Glu237, and a water molecule (Fig 4A). At the canonical EF4 site, Ca2+ is coordinated by the side chains of Asp421, Asp423, Asn425, and Glu432 and a main chain oxygen atom of Glu427. Moreover, the EF1 sites are located in the periphery of the Ca2+-free MICU1 hexamer, whereas the EF4 sites lie closer to the center (Supplementary Fig 6A).
Figure 4.
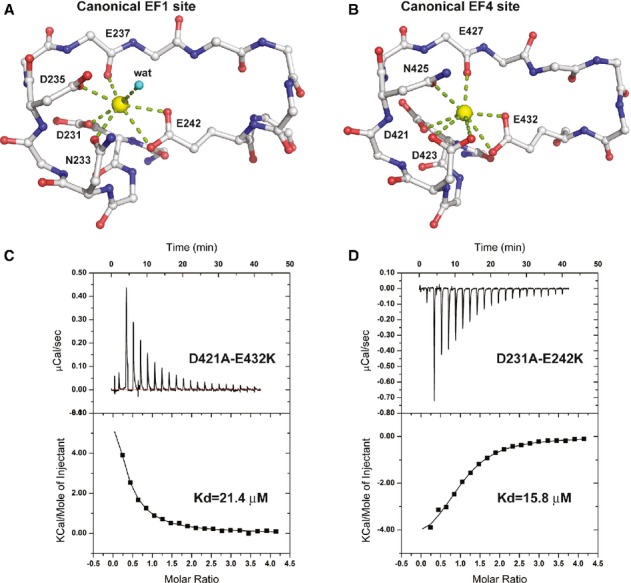
MICU1 calcium-binding sites.
A, B Detailed interactions of two calcium-binding sites of Ca2+-bound MICU1. Calcium ions are colored yellow. The oxygen and nitrogen atoms are colored red and blue, respectively. A water molecule is denoted as “wat.” The side chains of several residues are not shown for clarity.
C, D The dissociation constants of MICU1-xtal-deltaC mutants (D421A-E432K and D231A-E242K) in the presence of calcium ions were determined using ITC.
We used isothermal titration calorimetry (ITC) to further characterize the Ca2+-binding capacity of these two sites. The titration profile indicated that multiple binding sites for Ca2+ exist in the MICU1-xtal or the MICU1-xtal-deltaC proteins (Supplementary Fig 6B), and thus, the binding affinity of each site could not be resolved. We then constructed the D421A-E432K mutant to remove the Ca2+-binding site of the canonical EF4 in addition to the D231A-E242K mutant to remove another Ca2+-binding site of the canonical EF1 in MICU1-xtal-deltaC. Both mutants exhibited a titration profile that was fitted using a one-site binding model (Fig 4C and D). The Ca2+-binding affinities at the EF1 and EF4 sites were estimated to be approximately 21.4 μM and 15.8 μM, respectively, which were comparable to the affinities of the EF site of other Ca2+-binding proteins, such as the flagellar Ca2+-binding protein (Buchanan et al, 2005), nucleobindin (de Alba & Tjandra, 2004), grancalcin (Lollike et al, 2001), and others. Considering that the cytosolic Ca2+ concentration is approximately 0.1 μM in the resting cell (Babcock et al, 1997), our results indicated that MICU1 is less likely to bind Ca2+ in the resting cell.
Discussion
The present study describes the crystal structures of Ca2+-free and Ca2+-bound MICU1. Our results demonstrated that Ca2+-free MICU1 exists as a hexamer and that the C-terminal helix of MICU1 directly mediates the formation of the hexamer. These results are consistent with recent reports that MCU and MICU1 are present in a large complex (∼480 kDa) (Perocchi et al, 2010; Plovanich et al, 2013). We also observed that Ca2+-free MICU1 was capable of binding to MCU, which is similar to previous reports (Baughman et al, 2011; Mallilankaraman et al, 2012). The binding of Ca2+-free MICU1 to MCU may be mediated by the MICU1 C-helix because the deletion of this C-helix abolishes MICU1 binding to MCU. Furthermore, it has been reported that MICU1 constitutively suppressed mitochondrial calcium uptake in the cytosolic Ca2+ range of the resting cell (Mallilankaraman et al, 2012; Csordas et al, 2013). These results collectively support a model in which Ca2+-free MICU1 forms a hexamer to bind and inhibit the function of MCU.
As observed for several Ca2+-binding proteins, Ca2+ loading of MICU1 induced large conformational changes (Gifford et al, 2007). First, the EF1 site, but not the EF4 site, within the MICU1 molecule underwent large conformational changes; however, these two sites both bind calcium ions. Consequently, a large hydrophobic pocket was exposed after Ca2+ loading at the EF1 site (Supplementary Fig 7). Second, Ca2+ loading of the EF hands of MICU1 induced the rearrangement of the MICU1 dimer, which presumably led to the disassembly of the Ca2+-free MICU1 hexamer. This result is consistent with a recent report that the EF hands of MICU1 are central to the cooperative activation of MCU (Csordas et al, 2013). Third, MICU1 formed multiple oligomers after binding Ca2+. The oligomerization appears to largely depend on the MICU1 C-helix because MICU1-xtal-deltaC is predominantly a dimer in solution in the presence of Ca2+. Furthermore, in the presence of Ca2+, we observed that FL-MICU1, but not DeltaC-MICU1, associated with MCU (Supplementary Fig 8). The RNAi Ca2+ uptake phenotype can be rescued by FL-MICU1, but not DeltaC-MICU1. These results suggest that not only the EF hands but also the C-helix of MICU1 is necessary for the activation of MCU.
MICU1 faces the intermembrane space (Supplementary Fig 9) (Csordas et al, 2013) and acts as a Ca2+ sensor to regulate mitochondrial calcium uptake (Perocchi et al, 2010). We found that the affinity of MICU1 for Ca2+ is approximately 15–20 μM. These MICU1 molecular characteristics correlate with its biological function in the regulation of MCU opening. MCU is a Ca2+-selective channel with low affinity for Ca2+ (Rizzuto et al, 2012), suggesting that MCU plays a major role in the response to a high concentration of cytosolic Ca2+. Therefore, mitochondrial Ca2+ influx may be mediated by multiple channels that respond to different Ca2+ concentration ranges outside of the mitochondria to fulfill multiple tasks (O-Uchi et al, 2012). Indeed, a recent study identified multiple conductance of Ca2+ currents in isolated mitoplasts (Bondarenko et al, 2013).
Our results, together with the results of previous studies, allow us to propose a simplified scenario regarding the regulation of mitochondrial calcium uptake by MICU1. Inside the resting cell, the cytosolic Ca2+ concentration is approximately 0.1 μM. At this time, mitochondrial MICU1 is free of Ca2+ and thus binds and inhibits MCU. After a cellular response to an outside stimulus, the release of Ca2+ from within the Ca2+ stores elevates the cytosolic Ca2+ concentration. MICU1 senses and binds Ca2+ and undergoes conformational changes, which result in the activation of the MCU channel that leads to massive calcium uptake. The details concerning the activation of MCU by Ca2+-bound MICU1 await further elucidation. The decrease in the cytosolic Ca2+ concentration may result in the loss of Ca2+ from MICU1 and may inhibit the MCU channel again. Our study provides valuable details to elucidate the molecular mechanism of the machinery of mitochondrial calcium uptake.
Materials and Methods
Protein expression and Purification
The human MICU1-xtal (residues 97-476) and MICU1-xtal-deltaC (residues 97-444) constructs were cloned into the vector with a hexahistidine (6× His) and a PreScission protease cleavage site at the N-terminals. The plasmids were then transformed into Escherichia coli BL21(DE3) and over-expressed by induction with 0.2 mM isopropyl-1-thiogalactopyranoside (IPTG) at 37°C for 4 h.
The bacteria were harvested by centrifugation and were resuspended in buffer A (20 mM Tris–HCl pH 7.0, 500 mM NaCl, 20 mM imidazole, and 0.1 mM PMSF) and were then lysed by sonication. After centrifugation at 18,000 × g for 40 min, the supernatant was loaded onto Ni-NTA affinity columns and washed with buffer A. The target protein was eluted using buffer B (20 mM Tris–HCl pH 7.0, 200 mM NaCl, and 400 mM imidazole) and cleaved by PreScission protease at 4°C for 16 h (the MICU1-xtal protein was not cleaved for crystallization). The sample was desalted using a Hiprep 26/10 desalting column (GE Healthcare) with 20 mM Tris–HCl pH 7.0, 200 mM NaCl, and 5 mM DTT. The protein was then applied to a HiTrap-Q column (GE Healthcare) equilibrated with 20 mM Tris–HCl pH 7.0, 200 mM NaCl, and 5 mM DTT to remove the nonspecific binding nucleic acids. Flow through was collected and concentrated to 10 ml by ultrafiltration and was then purified by a HiLoad 26/60 Superdex 200 size-exclusion column in the presence of 20 mM MES pH 6.8, 300 mM NaCl, and 5 mM DTT with either 2 mM EGTA or 5 mM CaCl2. The MICU1-xtal protein with EGTA was concentrated to 2 mg/ml for crystallization, and MICU1-xtal-deltaC with CaCl2 was concentrated to 4 mg/ml.
The Se-Met derivative protein was produced by following the same protocol as used for the wild-type protein, with the exception that methionine auxotroph E. coli B834(DE3) cells and minimal medium were used to express the recombinant protein.
Other constructs (residues 53-476, residues 62-476, and residues 80-476) and mutants were expressed and purified by following the same protocol as used for the MICU1-xtal protein.
Crystallization and Data collection
The MICU1-xtal protein (2 mg/ml in 20 mM MES pH 6.8, 300 mM NaCl, 5 mM DTT, and 2 mM EGTA) was crystallized using the hanging drop vapor diffusion method mixed 1:1 with a reservoir solution of 8% PEG 3350 and 0.075 M ammonium citrate tribasic pH 7.0. After microseeding, the crystals grew much bigger. Crystals were grown for 1 week at 20°C and frozen in a cryoprotectant consisting of the reservoir solution supplemented with 30% ethylene glycol. The Se-Met crystals were produced in the same conditions used for the wild-type protein, with the exception that 1.5 mg/ml Se-Met protein was used for crystallization.
The MICU1-xtal-deltaC protein (4 mg/ml in 20 mM MES pH 6.8, 300 mM NaCl, 5 mM DTT, and 5 mM CaCl2) was crystallized using the sitting drop vapor diffusion method equilibrated against a 43% reservoir solution composed of (w/v) 2-methyl-2,4-pentanediol and 10 mM CaCl2. Crystals were grown for 3 weeks at 4°C, and the reservoir solution itself was used as a cryoprotectant.
Wild-type data and single anomalous data were collected at the element Se peak wavelength on the BL17U1 station of the Shanghai Synchrotron Radiation Facility (SSRF) and then were processed using the HKL2000 software (Otwinowski & Minor, 1997).
Structure determination and Refinement
For the Ca2+-free MICU1 structure, the HKL2MAP program (Pape & Schneider, 2004) was used to search the Se sites and successfully found 36 sites of a total of 90 sites. The initial phases were then calculated using the PHENIX Autosol (Adams et al, 2010). Model building was performed using the COOT program (Emsley et al, 2010). After the initial model was built, iterative refinement was performed with the PHENIX refinement program and COOT. The orientations of the amino acid side chains and bound water molecules were modeled on the basis of 2Fobs−Fcalc and Fobs−Fcalc difference Fourier maps. The final structure had an Rcrystal value of 25.4% and an Rfree value of 30.7%. The structure of the Ca2+-bound MICU1 was determined by the PHASER (McCoy et al, 2007) using the Ca2+-free MICU1 structure as the model. The structure refinement was performed with the PHENIX program, and the built model was created by COOT. The final structure had an Rcrystal value of 21.2% and an Rfree value of 28.2% with good geometry. Detailed data collection and refinement statistics are summarized in Table 1. The represented 2Fobs−Fcalc electron density map of Ca2+-free and Ca2+-bound MICU1 is shown in Supplementary Fig 10.
Table 1.
Data collection and refinement statistics for MICU1 structures
| Crystal name | Se-MICU1-xtal | MICU1-xtal-deltaC |
|---|---|---|
| Space group | P21 | I212121 |
| Unit cell | ||
| a, b, c (Å) | a = 90.88, b = 146.82, c = 115.87 | a = 99.28, b = 105.80, c = 64.80 |
| α, β, γ (°) | α = γ = 90.00, β = 111.08 | α = β = γ = 90.00 |
| Molecule/asu | 6 | 2 |
| Wavelength (Å) | 0.9791 | 0.9791 |
| Resolution range (Å) | 50–3.20 (3.31–3.20)† | 50–2.70 (2.80–2.70)† |
| No. of unique reflections | 46,641 | 22,801 |
| Redundancy | 6.2 (6.2)† | 7.4 (7.6)† |
| Rsym (%)* | 15.7 (75.4)† | 8.5 (58.7)† |
| I/σ | 17.9 (3.0)† | 32.0 (3.8)† |
| Completeness (%) | 99.9 (100.0)† | 99.1 (98.8)† |
| Figure of merit | ||
| Acentric/centric | 0.420/0.152 | |
| SAD/density modification | 0.381/0.638 | |
| Refinement | ||
| Rcrystal (%)‡ | 25.4 | 21.2 |
| Rfree (%)¶ | 30.7 | 28.2 |
| RMSDbond (Å) | 0.010 | 0.007 |
| RMSDangle(°) | 1.5 | 1.1 |
| Number of | ||
| Protein atoms | 15,350 | 4533 |
| Ligand atoms | 0 | 47 |
| Solvent atoms | 13 | 30 |
| Residues in (%) | ||
| Most favored | 80.4 | 87.5 |
| Additional allowed | 18.0 | 12.1 |
| Generously allowed | 1.7 | 0.4 |
| Disallowed | 0 | 0 |
| Average B factor (Å2) of | ||
| Chain A, B, C, D, E, F | 87.7, 74.2, 71.4, 62.5, 61.0, 62.1 | 63.3, 63.3, … |
| Ligand | – | 75.2 |
| Solvent | 22.7 | 47.5 |
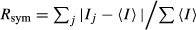 , where I_j is the intensity of the jth reflection and
, where I_j is the intensity of the jth reflection and 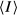 is the average intensity.
is the average intensity.
The highest resolution shell.
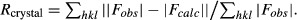
Rfree, calculated the same as Rcrystal, but from a test set containing 5% of data excluded from the refinement calculation.
Analytical ultracentrifugation
Sedimentation velocity (SV) measurements were performed in a Beckman/Coulter XL-I analytical ultracentrifuge using double-sector and sapphire windows. The SV experiments were conducted at 50,000 rpm and at 4°C using interference light detection and double-sector centerpieces loaded with approximately 10-60 μM proteins. The samples were in the buffer containing 50 mM MES pH 6.8, 250 mM NaCl, and 1 mM DTT with either 2 mM EGTA or 5 mM CaCl2. The SV data were analyzed using the SEDFIT program (Brown & Schuck, 2006) with a Continuous c(M) and c(S) Distribution model.
Isothermal titration calorimetry (ITC)
Measurements were conducted at 16°C using an ITC-200 microcalorimeter (GE Healthcare). The samples were buffered with 50 mM MES buffer pH 6.8 containing 200 mM NaCl. To determine the calcium-binding affinities of wild-type MICU1-xtal and mutants, 500 μM CaCl2 was stepwise injected into 25 μM protein samples. To determine the calcium-binding affinities of wild-type MICU1-xtal-deltaC and mutants, 1 mM CaCl2 was injected into 50 μM protein samples. The data were analyzed with the MicroCal Origin software.
Multi-angle light scattering
Multi-angle light scattering (MALS) was performed in-line with size-exclusion chromatography (SEC) using a 18° DAWN HELLOS α instrument equipped with an Optilab rEX Refractive Index Detector (Wyatt Technology). Five hundred microliters of 50–100 μM protein samples was injected into a Superdex 200 10/300 GL column (GE Healthcare) equilibrated with buffer of 20 mM Tris–HCl pH 7.2, 500 mM NaCl, 1 mM DTT and 2 mM EGTA or 5 mM CaCl2 at a flow rate of 0.5 ml/min. The protein concentrations were calculated from the absorbance at 280 nm, and the light scattering data were collected at 663 nm. Molecular weight was calculated using the ASTRA software (Wyatt Technology).
Cell culture and Transfection
HEK293T and HeLa cells (ATCC) were cultured in Dulbecco's modified Eagle's medium (DMEM, Sigma-Aldrich, Co.) supplemented with 10% v/v fetal bovine serum (FBS, HyClone Thermo Fisher Scientific, Inc.). The cells were maintained in a 95% air and 5% CO2 environment at 37°C.
Gene fragments corresponding to the full-length MICU1 were cloned into a pEGFP-N1 vector (Clontech). The EGFP gene was deleted and a FLAG tag was inserted between the Pro96 and Glu97 residues of human MICU1 using a standard PCR-based mutagenesis method. This modified plasmid was named pMICU1-96FLAG-wt. The mutant DeltaC-MICU1 was created based on pMICU1-96FLAG-wt and was confirmed by DNA sequencing. The MCU gene fragments were cloned into the pcDNA4/Myc-His A.
Co-immunoprecipitation and Western blot analysis
Transfected HEK293T cells were collected in PBS. Cells harvested from a 10-cm culture dish were lysed in 200 μl of TBS buffer (20 mM Tris–HCl, pH 7.4, 2 mM EDTA, and 150 mM NaCl) containing 1.0% n-dodecyl-β-D-maltoside (DDM) and the Roche protease inhibitor cocktail. After incubation on ice for 30 min, the lysate was diluted by a fourfold volume of TBS buffer without DDM and was then centrifuged at 13,000 × g for 30 min. A total of 40 μl of a 1:1 slurry of protein A/G agarose (Pierce) was incubated with 0.5 μg of anti-FLAG antibody for 1 h and was washed twice with cold TBS buffer containing 0.2% DDM. The cell extract supernatant (1 ml) was added to the antibody-coupled protein A/G beads and was incubated for 2 h at 4°C. The beads were washed three times for 10 min with cold TBS buffer containing 0.2% DDM followed by elution with the addition of 40 μl of SDS loading buffer. Subsequently, 10 μl of the eluent was loaded onto a 12% SDS–PAGE gel and transferred onto a PVDF membrane for Western blot analysis. Immunoblot analysis was conducted using a primary antibody to c-myc. The protein–antibody complexes were detected using enhanced chemiluminescence.
Silencing of MICU1 and cDNA rescue experiments
HeLa cells (ATCC) were cultured in DMEM with 10% FBS and were plated onto a 24-well plate (Costar). For short-term silencing, HeLa cells were transfected with the siRNA version of an identical sh1 hairpin that was used in previous reports (Perocchi et al, 2010; Csordas et al, 2013) for 48 h using Oligofectamine (Invitrogen). The FL-MICU1 wild-type and mutants were then co-transfected with a mitochondria-targeted aequorin for an additional 24 h using Fugene 6 Transfection Reagent (Promega).
Calcium luminescence assay
The transfected cells were loaded with 1 μM coelenterazine hcp in Ringer's solution containing 145 mM NaCl, 1 mM MgCl2, 4.5 mM KCl, 10 mM glucose, 20 mM HEPES, pH 7.4, and 1 mM CaCl2 for 60 min at 25°C. After loading, the cells were washed twice with Ringer's solution. The mitochondrial Ca2+ measurements were performed upon histamine treatment. Light emission was measured in a Synergy4 spectrophotometer (BioTek) at 460 nm every 0.2 s.
Proteinase K digestion
HEK293T cells were collected and resuspended in hypotonic buffer. After gentle homogenization with a Dounce homogenizer, cell lysates were subjected to differential centrifugation and gradient centrifugation. Isolated mitochondria were treated with different digitonin concentrations (0–0.8%) or 0.2% n-dodecyl-ß-maltoside along with 50 μg/ml proteinase K on ice for 30 min. Digestion was terminated with 5 mM phenylmethyl sulphonyl fluoride (Sigma) (final concentration). Mitochondrial proteins were separated by SDS–PAGE, and proteins were detected by Western blot.
Accession numbers
Atomic coordinates and structure factors for the reported crystal structures have been deposited in the Protein Data Bank with accession code 4NSC for Ca2+-free MICU1 and 4NSD for Ca2+-bound MICU1.
Acknowledgments
We are grateful to Dr. Rosario Rizzuto for the plasmid of the mitochondrial matrix targeted aequorin (mt-AEQ) construct, to the staff at the beamline BL17U1 of the Shanghai Synchrotron Radiation Facility and at the beamline BL-17A at Photon Factory (Tsukuba, Japan) for excellent technical assistance during data collection. This work was supported by the 973 Program (Grants 2012CB917200 and 2013CB910400) and the Natural Science Foundation of China (Grants 31370826, 31300628, and 31170684).
Author contributions
LW did protein purification, crystallization, and biochemical experiments; XY did structure determination, structure refinement, co-IP, and calcium imaging experiments; XY and SL did cDNA rescue experiment; ZW did AUC experiment; YL and JF did purification and crystallization; LW, XY, YZ, and YS analyzed the data. LW, XY, and YS designed the study and wrote the paper. All authors discussed the results and commented on the manuscript.
Conflict of interest
The authors declare that they have no conflict of interest.
Supplementary information
for this article is available online: http://emboj.embopress.org
References
- Adams PD, Afonine PV, Bunkoczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse-Kunstleve RW, McCoy AJ, Moriarty NW, Oeffner R, Read RJ, Richardson DC, Richardson JS, Terwilliger TC, Zwart PH. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aichberger KJ, Mittermann I, Reininger R, Seiberler S, Swoboda I, Spitzauer S, Kopp T, Stingl G, Sperr WR, Valent P, Repa A, Bohle B, Kraft D, Valenta R. Hom s 4, an IgE-reactive autoantigen belonging to a new subfamily of calcium-binding proteins, can induce Th cell type 1-mediated autoreactivity. J Immunol. 2005;175:1286–1294. doi: 10.4049/jimmunol.175.2.1286. [DOI] [PubMed] [Google Scholar]
- Alba de E, Tjandra N. Structural studies on the Ca2+-binding domain of human nucleobindin (calnuc) Biochemistry. 2004;43:10039–10049. doi: 10.1021/bi049310a. [DOI] [PubMed] [Google Scholar]
- Babcock DF, Herrington J, Goodwin PC, Park YB, Hille B. Mitochondrial participation in the intracellular Ca2+ network. J Cell Biol. 1997;136:833–844. doi: 10.1083/jcb.136.4.833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balaban RS. The role of Ca(2 + ) signaling in the coordination of mitochondrial ATP production with cardiac work. Biochim Biophys Acta. 2009;1787:1334–1341. doi: 10.1016/j.bbabio.2009.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baughman JM, Perocchi F, Girgis HS, Plovanich M, Belcher-Timme CA, Sancak Y, Bao XR, Strittmatter L, Goldberger O, Bogorad RL, Koteliansky V, Mootha VK. Integrative genomics identifies MCU as an essential component of the mitochondrial calcium uniporter. Nature. 2011;476:341–345. doi: 10.1038/nature10234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernardi P, Stockum von S. The permeability transition pore as a Ca(2 + ) release channel: new answers to an old question. Cell Calcium. 2012;52:22–27. doi: 10.1016/j.ceca.2012.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bick AG, Calvo SE, Mootha VK. Evolutionary diversity of the mitochondrial calcium uniporter. Science. 2012;336:886. doi: 10.1126/science.1214977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bondarenko AI, Jean-Quartier C, Malli R, Graier WF. Characterization of distinct single-channel properties of Ca(2 + ) inward currents in mitochondria. Pflugers Arch. 2013;465:997–1010. doi: 10.1007/s00424-013-1224-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bragadin M, Pozzan T, Azzone GF. Kinetics of Ca2+ carrier in rat liver mitochondria. Biochemistry. 1979;18:5972–5978. doi: 10.1021/bi00593a033. [DOI] [PubMed] [Google Scholar]
- Brown PH, Schuck P. Macromolecular size-and-shape distributions by sedimentation velocity analytical ultracentrifugation. Biophys J. 2006;90:4651–4661. doi: 10.1529/biophysj.106.081372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchanan KT, Ames JB, Asfaw SH, Wingard JN, Olson CL, Campana PT, Araujo AP, Engman DM. A flagellum-specific calcium sensor. J Biol Chem. 2005;280:40104–40111. doi: 10.1074/jbc.M505777200. [DOI] [PubMed] [Google Scholar]
- Collins S, Meyer T. Cell biology: A sensor for calcium uptake. Nature. 2010;467:283. doi: 10.1038/467283a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Csordas G, Golenar T, Seifert EL, Kamer KJ, Sancak Y, Perocchi F, Moffat C, Weaver D, Perez SDLF, Bogorad R, Koteliansky V, Adijanto J, Mootha VK, Hajnoczky G. MICU1 controls both the threshold and cooperative activation of the mitochondrial Ca2+ uniporter. Cell Metab. 2013;17:976–987. doi: 10.1016/j.cmet.2013.04.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Csordas G, Renken C, Varnai P, Walter L, Weaver D, Buttle KF, Balla T, Mannella CA, Hajnoczky G. Structural and functional features and significance of the physical linkage between ER and mitochondria. J Cell Biol. 2006;174:915–921. doi: 10.1083/jcb.200604016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stefani De D, Raffaello A, Teardo E, Szabo I, Rizzuto R. A forty-kilodalton protein of the inner membrane is the mitochondrial calcium uniporter. Nature. 2011;476:336–340. doi: 10.1038/nature10230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dedkova EN, Blatter LA. Calcium signaling in cardiac mitochondria. J Mol Cell Cardiol. 2013;58:125–133. doi: 10.1016/j.yjmcc.2012.12.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deluca HF, Engstrom GW. Calcium uptake by rat kidney mitochondria. Proc Natl Acad Sci USA. 1961;47:1744–1750. doi: 10.1073/pnas.47.11.1744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denton RM, McCormack JG. The role of calcium in the regulation of mitochondrial metabolism. Biochem Soc Trans. 1980;8:266–268. doi: 10.1042/bst0080266. [DOI] [PubMed] [Google Scholar]
- Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallogr D Biol Crystallogr. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gifford JL, Walsh MP, Vogel HJ. Structures and metal-ion-binding properties of the Ca2+-binding helix-loop-helix EF-hand motifs. Biochem J. 2007;405:199–221. doi: 10.1042/BJ20070255. [DOI] [PubMed] [Google Scholar]
- Hajnoczky G, Csordas G. Calcium signalling: fishing out molecules of mitochondrial calcium transport. Curr Biol. 2010;20:R888–R891. doi: 10.1016/j.cub.2010.09.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hajnoczky G, Csordas G, Madesh M, Pacher P. Control of apoptosis by IP(3) and ryanodine receptor driven calcium signals. Cell Calcium. 2000;28:349–363. doi: 10.1054/ceca.2000.0169. [DOI] [PubMed] [Google Scholar]
- Herrington J, Park YB, Babcock DF, Hille B. Dominant role of mitochondria in clearance of large Ca2+ loads from rat adrenal chromaffin cells. Neuron. 1996;16:219–228. doi: 10.1016/s0896-6273(00)80038-0. [DOI] [PubMed] [Google Scholar]
- Kirichok Y, Krapivinsky G, Clapham DE. The mitochondrial calcium uniporter is a highly selective ion channel. Nature. 2004;427:360–364. doi: 10.1038/nature02246. [DOI] [PubMed] [Google Scholar]
- Linding R, Russell RB, Neduva V, Gibson TJ. GlobPlot: Exploring protein sequences for globularity and disorder. Nucleic Acids Res. 2003;31:3701–3708. doi: 10.1093/nar/gkg519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lollike K, Johnsen AH, Durussel I, Borregaard N, Cox JA. Biochemical characterization of the penta-EF-hand protein grancalcin and identification of L-plastin as a binding partner. J Biol Chem. 2001;276:17762–17769. doi: 10.1074/jbc.M100965200. [DOI] [PubMed] [Google Scholar]
- Mallilankaraman K, Doonan P, Cardenas C, Chandramoorthy HC, Muller M, Miller R, Hoffman NE, Gandhirajan RK, Molgo J, Birnbaum MJ, Rothberg BS, Mak DO, Foskett JK, Madesh M. MICU1 is an essential gatekeeper for MCU-mediated mitochondrial Ca(2 + ) uptake that regulates cell survival. Cell. 2012;151:630–644. doi: 10.1016/j.cell.2012.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ. Phaser crystallographic software. J Appl Crystallogr. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orrenius S, Zhivotovsky B, Nicotera P. Regulation of cell death: the calcium-apoptosis link. Nat Rev Mol Cell Biol. 2003;4:552–565. doi: 10.1038/nrm1150. [DOI] [PubMed] [Google Scholar]
- Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Method Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- O-Uchi J, Pan S, Sheu SS. Perspectives on: SGP symposium on mitochondrial physiology and medicine: molecular identities of mitochondrial Ca2+ influx mechanism: updated passwords for accessing mitochondrial Ca2+-linked health and disease. J Gen Physiol. 2012;139:435–443. doi: 10.1085/jgp.201210795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pape T, Schneider TR. HKL2MAP: a graphical user interface for macromolecular phasing with SHELX programs. J Appl Crystallogr. 2004;37:843–844. [Google Scholar]
- Perocchi F, Gohil VM, Girgis HS, Bao XR, McCombs JE, Palmer AE, Mootha VK. MICU1 encodes a mitochondrial EF hand protein required for Ca(2 + ) uptake. Nature. 2010;467:291–296. doi: 10.1038/nature09358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plovanich M, Bogorad RL, Sancak Y, Kamer KJ, Strittmatter L, Li AA, Girgis HS, Kuchimanchi S, Groot De J, Speciner L, Taneja N, Oshea J, Koteliansky V, Mootha VK. MICU2, a paralog of MICU1, resides within the mitochondrial uniporter complex to regulate calcium handling. PLoS ONE. 2013;8:e55785. doi: 10.1371/journal.pone.0055785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed KC, Bygrave FL. A kinetic study of mitochondrial calcium transport. Eur J Biochem. 1975;55:497–504. doi: 10.1111/j.1432-1033.1975.tb02187.x. [DOI] [PubMed] [Google Scholar]
- Rizzuto R, Stefani De D, Raffaello A, Mammucari C. Mitochondria as sensors and regulators of calcium signalling. Nat Rev Mol Cell Biol. 2012;13:566–578. doi: 10.1038/nrm3412. [DOI] [PubMed] [Google Scholar]
- Rizzuto R, Pinton P, Carrington W, Fay FS, Fogarty KE, Lifshitz LM, Tuft RA, Pozzan T. Close contacts with the endoplasmic reticulum as determinants of mitochondrial Ca2+ responses. Science. 1998;280:1763–1766. doi: 10.1126/science.280.5370.1763. [DOI] [PubMed] [Google Scholar]
- Sancak Y, Markhard AL, Kitami T, Kovacs-Bogdan E, Kamer KJ, Udeshi ND, Carr SA, Chaudhuri D, Clapham DE, Li AA, Calvo SE, Goldberger O, Mootha VK. EMRE is an essential component of the mitochondrial calcium uniporter complex. Science. 2013;342:1379–1382. doi: 10.1126/science.1242993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stathopulos PB, Zheng L, Li GY, Plevin MJ, Ikura M. Structural and mechanistic insights into STIM1-mediated initiation of store-operated calcium entry. Cell. 2008;135:110–122. doi: 10.1016/j.cell.2008.08.006. [DOI] [PubMed] [Google Scholar]
- Vasington FD, Murphy JV. Ca ion uptake by rat kidney mitochondria and its dependence on respiration and phosphorylation. J Biol Chem. 1962;237:2670–2677. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


