Abstract
The RING-in-between-RING (RBR) E3s are a curious family of ubiquitin E3-ligases, whose mechanism of action is unusual in several ways. Their activities are auto-inhibited, causing a requirement for activation by protein-protein interactions or posttranslational modifications. They catalyse ubiquitin conjugation by a concerted RING/HECT-like mechanism in which the RING1 domain facilitates E2-discharge to directly form a thioester intermediate with a cysteine in RING2. This short-lived, HECT-like intermediate then modifies the target. Uniquely, the RBR ligase HOIP makes use of this mechanism to target the ubiquitin amino-terminus, by presenting the target ubiquitin for modification using its distinctive LDD region.
Keywords: RBR, E3 ligase, TRIAD, autoinhibition, ubiquitination, mechanism
Introduction
The role of E3-ligases in ubiquitin conjugation is to mediate the transfer of ubiquitin from an E2 ubiquitin-conjugating enzyme to the target. This step is uncoupled from the first steps in ubiquitination—which involve the E1- and ATP-dependent activation of ubiquitin and its transfer to E2 enzymes—because E1 and E3 use overlapping surfaces of the E2 enzymes. The E3s are therefore involved solely in the chemical transfer of the C-terminus of ubiquitin from a thioester on a cysteine to the isopeptide linkage in the target.
Traditionally, the E3s have been separated into two classes, RING/U-box and HECT type ligases, both of which activate the E2 reaction and recognize the target molecule. The HECT E3-ligases form a thioester-intermediate on an active-site cysteine before transferring it onto its target, whereas RING E3-ligases facilitate the direct transfer of the ubiquitin from the E2 cysteine (Fig 1). More recently, a third class of E3-ligases was identified, the RING-in-between-RING (RBR) E3-ligases, which contain a highly conserved catalytic unit consisting of a RING1, an in-between RING (IBR), and a RING2 domain (Figs 1 and 2) 1.
Figure 1.
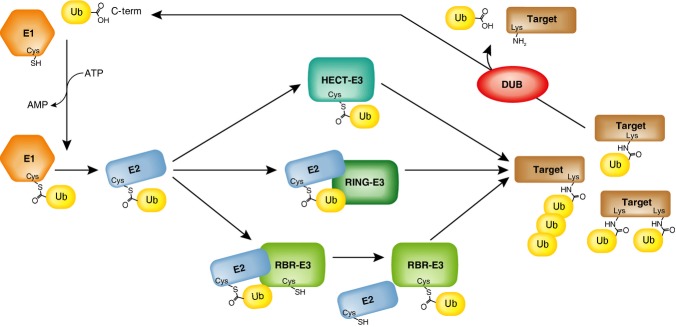
RBR E3-ligases have a unique mode of transferring ubiquitin to a target.
The ubiquitin (Ub) C-terminus is activated in an ATP-dependent manner by an E1 activating enzyme, and is subsequently transferred to form a thioester intermediate on an E2 conjugase. The final transfer of ubiquitin onto its target is mediated by E3-ligases that either form a thioester intermediate with the ubiquitin (HECT E3-ligases), mediate a direct transfer of the ubiquitin from the E2 onto its target (RING E3-ligases), or function as RING/HECT-type hybrids (RBR E3-ligases). Through this cascade of E1, E2 and E3 enzymes, the ubiquitination machinery mediates the formation of mono-ubiquitination, multi-mono-ubiquitination, or ubiquitin chain formation on its targets. The ubiquitin signal can be removed by de-ubiquitination enzymes (DUBs).
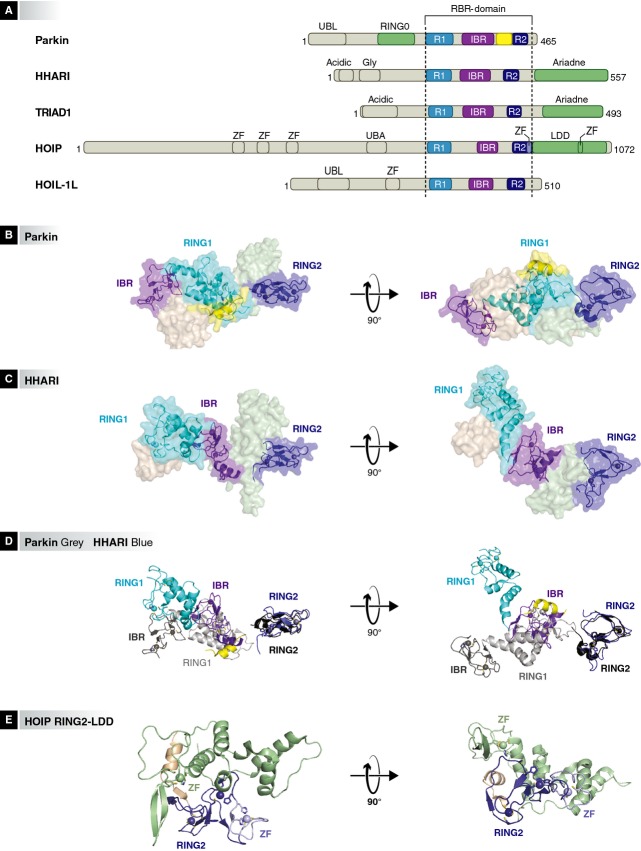
Figure 2.
Domain arrangements in RBR E3-ligases.
The domain borders are drawn to scale according to Uniprot definitions (http://www.uniprot.org). Ubiquitin like domain (UBL), acidic region (Acidic), glycine-rich region (Gly), zinc finger (ZF), ubiquitin-associated domain (UBA). The RBRs are represented in RING1:cyan (R1), purple (IBR) and blue (R2). The Parkin RING0, C-terminal Ariadne domain and linear ubiquitin determining domain (LDD) are represented in pale green and the Parkin linker/tether helix (also called PUB or REP) is yellow. B) Crystal structure of full-length Parkin (surface representation) (PDB 4k95). The RBR (cartoon) is autoinhibited by the N-terminal regions of Parkin. The colors correspond to the colors in the schematic representation in Figure 2A. C) Surface representation of the crystal structure of full length HHARI (PDB 4kbl). The RBR is shown as cartoon. D) A cartoon of superposed RBR domains of Parkin and HHARI (SSM (WinCoot 0.7.1) superposition of RING2 domain of HHARI on the RING2 domain of Parkin). Parkin RING1:light grey80, IBR:dark grey40, PUB domain: yellow, RING2:black. HHARI RING1:cyan, IBR:purple, RING2:blue. E) Crystal structure of HOIP RING2-LDD (PDB 4ljo).
The RBR class of proteins was first described in 1999 by two separate groups that identified the conservation of a triple-RING/zinc finger motif in eukaryotic species, including animals, plants, fungi and protists 2, 3. Further analysis of the pattern of cysteines and histidines in the RING/zinc fingers indicated that the RBR domain has arisen only once in evolution 4. There are 14 separate human RBR proteins that have been assigned to 8 distinct subfamilies of the RBR family 5 (Table 1).
Table 1.
Human RBR E3-ligases
| Subfamily | Name | Alternative names | Uniprot |
|---|---|---|---|
| Ariadne | ARIH1 | ARI1, HHARI | Q9Y4X5 |
| Ariadne | ARIH2 | ARI2, TRIAD1 | O95376 |
| Ariadne | CUL9 | PARC, H7-AP1, KIAA0708 | Q8IWT3 |
| Ariadne | ANKIB1 | KIAA1386 | Q9P2G1 |
| Parkin | PARK2 | PRKN, PARKIN | O60260 |
| RNF144 | RNF144A | KIAA0161, hUIP4, UBCE7IP4 | P50876 |
| RNF144 | RNF144B | P53RFP, IBRDC2 | Q7Z419 |
| XAP3 | RBCK1 | HOIL-1L, RNF54, XAP3 | Q9BYM8 |
| Dorfin | RNF19A | DORFIN | Q9NV58 |
| Dorfin | RNF19B | NKLAM, IBRDC3, DJ174N9.1 | Q6ZMZ0 |
| Paul | RNF31 | HOIP, PAUL, ZIBRA | Q96EP0 |
| TRIAD3 | RNF216 | ZIN, TRIAD3, UBCE7IP1 | Q9NWF9 |
| ARA54 | RNF14 | ARA54 | Q9UBS8 |
| Not assigned | RNF217 | C6orf172, IBRDC1, FLJ16403 | Q8TC41 |
The RBR E3-ligases were shown to use both RING and HECT-like mechanisms 1. The ubiquitin transfer is initiated by the interaction of an E2∼ubiquitin with the RBR 1, 6, 7, similar to the interaction between E2s and classical RING E3-ligases 5, 6, 7, but in RBRs this interaction is used to facilitate the formation of a HECT-like thioester intermediate between the C-terminus of the ubiquitin and an active-site cysteine on RING2 before it is coupled to its substrate. Furthermore, most, if not all, of these ligases are distinguished by the tight regulation of their enzymatic activity by auto-inhibition 6, 7, 8, 9, 10, 11. In this review, we discuss how RBR E3-ligases transfer ubiquitin to their targets.
Cellular functions of RBRs
The few RBRs that have been analyzed in detail—Parkin, HHARI, TRIAD1, HOIP and HOIL-1L—are involved in important cellular events: transcription and RNA metabolism, translation, subcellular tethering, regulation of posttranslational modifications and protein stability, cellular and stress signalling, cell-cycle control, and response to microbial infection 12. Consequently, the misregulation of the activity of RBR proteins is important in disease 5, 13, 14, 15, which makes them interesting potential drug targets. Here, we briefly discuss their function before a more in-depth analysis of the molecular details of their enzymatic mechanisms.
Parkin
Parkin (PARK2) mutations cause familial autosomal-recessive juvenile Parkinson disease and are a frequent cause of sporadic early- and late-onset Parkinson disease (PD) 16, 17, 18. Parkin is found in many tissues, but is primarily expressed in the brain, including the substantia nigra, indicating a possible involvement in the loss of dopaminergic neurons that form the hallmark of PD 17. In addition, it is a putative tumour suppressor 19, 20.
Parkin recruitment to the outer mitochondrial membrane is dependent on Pink1 21 and its own ligase activity 22. The Parkin E3-ligase activity is crucial for its function, and possible targets include α-synuclein 23, CDCrel-1 24, Pael-R 25, and misfolded DJ1/PARK7 26, which accumulate in patients with PD. Therefore, the Parkin E3-ligase activity is thought to be crucial for the prevention of PD pathogenic Lewy body formation, which mainly contain ubiquitin and α-synuclein 27. Furthermore, Parkin plays important roles in mitophagy upon stimulation by the mitochondrial uncoupler CCCP and possibly in other forms of autophagy in the cell, such as the susceptibility to intracellular bacterial pathogens 21, 28, 29. A growing number of targets are being identified, including mitofusin-1, mitofusin-2 and HKI, which are involved in maintaining mitochondrial integrity 30, 31, 32; the p38 subunit of aminoacyl-tRNA synthase 33; RanBP2, which is part of the nucleocytoplasmic transport machinery 34; transcription factor SIM2 35; anti-apoptotic and autophagy inhibitory protein Bcl-2 36; and Parkin itself 1, 24. An in-depth proteomics analysis of the Parkin dependent proteome will be important to validate and identify more direct Parkin targets 37. Although the understanding of most Parkin interactions is still superficial, a major role for the Parkin E3-ligase function seems to be the protection of cells against the accumulation of misfolded and unfolded proteins.
HHARI
Parkin and HHARI share substantial sequence identity 38. In PD with mutated Parkin, the E3-ligase activity of Parkin is abolished in all cells. Nevertheless, only the dopaminergic neurons in the brain seem to be sensitive to the loss of Parkin function. Thus, it is likely that other cells express a redundant E3-ligase to compensate for the loss of Parkin. HHARI is a likely candidate, as it binds many of the same protein partners, such as CDCrel-1, synphilin-1, and CASK 39, and may target synphilin-1 and SIM2 for degradation in cells 35. Interestingly, HHARI is also found in Lewy bodies in dopaminergic neurons in PD, indicating that it cannot compensate the loss of Parkin activity in these cells 39. Nevertheless, HHARI is not mutated in patients with PD.
In addition to the description of the possible overlapping interactions of HHARI and Parkin, two studies have analyzed the cellular function of HHARI. First, HHARI is suggested to play a role in the regulation of protein translation by targeting the eukaryotic mRNA cap-binding protein 4EHP for proteasomal degradation 40. Second, HHARI has been shown to positively regulate cellular proliferation, which functionally correlates with its over-expression in head-and-neck squamous cell carcinoma biopsies 41. Neddylated Cullin-RING ligase complexes have been recently shown to bind and activate HHARI, which has important implications for its cellular function 42.
TRIAD1
Loss of TRIAD1 in mice is associated with embryonic death and causes the degradation of nuclear IκBβ, leading to excessive NF-κB signalling in dendritic cells 43. Furthermore, the depletion of TRIAD1 causes a defect in membrane trafficking that leads to the accumulation of the growth hormone receptor and the epidermal growth factor receptor in intracellular vesicles and at the plasma membrane 44. Finally, TRIAD1 plays a role in the regulation of myeloid progenitor cell proliferation, by modulating HoxA10 activity 45 and stabilizing proteins such as Gfi1B and p53 46, 47. TRIAD1 cellular concentrations seem to be tightly regulated. TRIAD1 levels are up-regulated during granulocytic and monocytic differentiation 48, 49. In contrast, it is negatively controlled by proteasomal degradation, for which it is ubiquitinated by, for example, Mdm2 46, 48.
Direct targets of the TRIAD1 E3-ligase activity have not been identified in vivo, but it seems to regulate the stability of various proteins indirectly, in the sense that it does not directly ubiquitinate them for degradation. For example, TRIAD1 inhibits the proteasome-dependent degradation of Gfi1B in myeloid progenitor cells through an interaction between its RING2 domain and Gfi1B, thereby inhibiting myeloid progenitor cell proliferation 47, 50. This interaction possibly competes with binding of other E3-ligases that target Gfi1B, or it might recruit DUBs that deubiquitinate Gfi1B. In addition, TRIAD1 has been suggested to compete with Mdm2 for p53 binding, thus preventing the ubiquitin-dependent degradation of p53 46. TRIAD1 has also been linked to the regulation of the activity of HoxA10. High expression levels of HoxA10 correlate with a poor prognosis in acute myeloid leukemia (AML) 51, 52, and induce TRIAD1 levels in myeloid progenitor cells, which increases total protein ubiquitination levels 45, 53. Interestingly, TRIAD1 antagonizes HoxA10-induced cellular proliferation, but whether HoxA10 is a direct target for TRIAD1 remains to be determined 51, 52, 54. Nevertheless, the inhibitory effect of TRIAD1 on the proliferation of myeloid progenitor cells critically relies on its RING domains and is inhibited by proteasome inhibitors 50. Therefore, the TRIAD1 E3-ligase activity is likely to target HoxA10 or regulators of HoxA10 for proteasomal degradation. Finally, it has recently been shown that TRIAD1 is recruited to and activated by CUL5-RBX2 complexes, which might be critical for identifying additional cellular functions 42.
HOIP and HOIL-1L
Two RBR E3-ligases, HOIP and HOIL-1L, are part of the linear ubiquitin assembly complex (LUBAC), which is essential for the activation of the NF-κB pathway. LUBAC comprises SHARPIN, HOIP and HOIL-1L, of which HOIP forms a critical catalytic centre 55, 56, 57, 58, 59. LUBAC has the unique capability to mediate the formation of linear ubiquitin chains 57. Interestingly, HOIP contains the linear ubiquitin chain specificity of the complex 57, 60, but either HOIL-1L or SHARPIN are needed to release its autoinhibited state and direct the activity of the complex towards its targets. Weak linear chain-forming activity was also reported for HOIL-1L 7, based on the fact that His-Ub could not be modified. However, as HOIL-1L requires an intact N-terminus also on the donor ubiquitin 61, further experiments are required to elucidate its independent activity. The LUBAC E3-ligase targets its activity towards NEMO, RIP1, RIP2 and K63-linked ubiquitin chains in the NF-κB pathway 55, 58, 62, 63, 64. Modification of NEMO with linear chains under stress conditions is further regulated by Parkin, which interacts with the LUBAC complex 65. Mechanistically, besides the RBR of HOIP, the catalytic cysteine in the RBR domain of HOIL-1L is required for the attachment of the first ubiquitin of the linear ubiquitin chain to NEMO 61.
In the NF-κB pathway, conjugated linear ubiquitin chains are selectively recognized by the UBAN domain of NEMO and the NZF domains of HOIL-1L and SHARPIN 55, 66, 67, 68, 69, which are believed to stabilize the TNF-R1/NEMO/LUBAC signalling complex, co-localize LUBAC with the TAK1-complex, recruit additional NEMO molecules, and facilitate NF-κB-dependent gene expression. However, the DUBs A20 and OTULIN/Gumby are also recruited to the linear ubiquitin chains to negatively regulate LUBAC-induced NF-κB activation by preventing the interaction between NEMO and LUBAC 64, 70, 71, 72. The role of LUBAC in the immune response is reviewed in this EMBO reports ubiquitin series 73.
The RBR mechanism
RBR E3-ligases follow a two-step mechanism whereby the interaction of a ubiquitin-charged E2-enzyme with RING1 promotes the transfer of ubiquitin to a cysteine on RING2, to form a thioester intermediate, prior to the transfer of ubiquitin to its target protein. Since the thioester intermediate is reminiscent of the HECT E3-enzymes, the mechanism has been named HECT-like, although it should be noted that the structures of these domains do not resemble HECT domains (see below). This combined RING/HECT-like molecular mechanism underlying the RBR-mediated ubiquitination of proteins was uncovered in HHARI based on a study on Parkin and HHARI in 2011 1, and confirmed for HOIP in 2012 6, 7 and Parkin in 2013 22. As several RBR E3-ligases have been shown to function via this two-step mechanism, it is likely that all RBR E3-ligases function in this manner.
The RBRs share a number of features that distinguish them from other RING E3s. First, the requirement for an E2 in vitro is less strict than in most other E3-ligases. RBRs are able to use the E1 as the donating thioester directly, in a step that is also independent of the RING1 domain of the RBR 6, 7, 74. Second, the thioester bond between ubiquitin and the cysteine on the RING2 domain is very transient and difficult to detect, indicating that this step is not rate-limiting in the reaction. Finally—unlike classical RING E3-ligases in which the interaction between the RING domain and the E2 activates the E2∼ubiquitin thioester 75, 76—the RBR RING1 is not sufficient to allow the discharge of ubiquitin from the E2, but additionally requires the presence of the RING2 catalytic cysteine 1, 6, 77. Altogether, this suggests that the RING1 domain may not facilitate the allosteric activation of the E2∼ubiquitin as occurs in classical RINGs 78, 79, 80, but possibly catalyzes the transfer of the ubiquitin onto RING2 by a different mechanism.
RBR structures
Recent structural information of the RBR regions of Parkin, HHARI and HOIP provides insight into the molecular details of the architecture of the RBR proteins (Fig 2) 9, 10, 11, 77, 81. The crystal structures of Parkin and HHARI show that the relative orientation of the RING1, IBR and RING2 domains of the RBR unit is highly variable. The RBR of Parkin was crystallized in its auto-inhibited form as a compact structure with extensive inter-domain interactions 9, 10, 11, while the RBR of HHARI adopts an extended conformation in its autoinhibited state 77. A HOIP construct lacking RING1 and the IBR reveals the tight interaction between RING2 and the C-terminal linear ubiquitin chain determining domain (LDD) 82.
In the crystal structures of Parkin, the RING2 domain of the RBR is positioned at the opposite side of the protein from the IBR, placing the two domains 49 Å apart by a linker. In this conformation, the E2∼ubiquitin bound to RING1 is positioned > 50 Å away from the active- site C431 in RING2, too far for trans-thiolation of the ubiquitin from the E2 onto the E3 9, 10, 11. In HHARI, the C-terminal Ariadne domain blocks the active-site cysteine in RING2, preventing the transfer of the ubiquitin from an E2 onto the RBR RING2 77. Thus, the RBRs require conformational changes for their activation. Additional studies are needed to reveal the precise orientation of the RING1, IBR and RING2 domains in catalytically active forms of the RBR proteins, and it remains to be seen how long-lived such states will be.
If one analyzes the individual domains in these structures, the RING1 domain in HHARI and Parkin has the typical C3HC4 topology of classical RING domains, coordinating two zinc-ions in a cross-brace structure that contains all the necessary features for the interaction with E2s. Neither the IBR domain nor the RING2 domain has a RING fold, but they are zinc fingers that coordinate two zinc ions in a similar manner. They share a common IBR-fold that is also found in the APC/C inhibitors Emi1 (FBXO5) and Emi2 (FBXO43) 83. Interestingly, the RING2 of HOIP differs from other RBR proteins, since it has an additional zinc finger incorporated near the end of RING2 (Fig 2E), which is structurally important for the positioning of the target ubiquitin in linear ubiquitin chain formation 82.
The crystal structures of Parkin and HHARI show that the IBR forms multiple interactions with RING1, as well as with regions N- and C-terminal of the RBR unit. Furthermore, models of the E2-bound RBR unit suggest that parts of the IBR contribute to the RING1 binding surface of the E3 on which the E2∼ubiquitin docks 11. Therefore, the IBR forms an important structural part of the RBR that is probably involved in the regulation of the activity and accessibility of the RBR unit.
The RING2 domain is the catalytic unit of the RBR. It contains a conserved cysteine residue, which forms the active site with which the ubiquitin C-terminus can form a thioester bond during the transfer of the ubiquitin from the E2 onto the substrate. The final transfer of the ubiquitin from the E2 cysteine onto the E3 RING2 domain is suggested by the crystal structure of Parkin and HHARI to be facilitated by a catalytic triad that consists of C431 (catalytic cysteine), H433 and E444 (numbering according to the Parkin sequence) 9, 10, 11, 77. These residues are not conserved throughout the RBR family and the details of the reaction of the subsequent step, the transfer of a donor ubiquitin from the RING2 cysteine onto a target ubiquitin, seem to be subtly different between Parkin (or HHARI) and HOIP. In the latter, the arrangement of the catalytic site is different, due to the addition of a second zinc finger at this position, and only the corresponding RING2 C885 and H887 of the proposed catalytic triad are essential, whereas Q896 (which aligns with Parkin E444) is not involved in the chain-forming reaction 6, 11. In addition, the residues around the N-terminus on the target ubiquitin, to which the C-terminus of the donor ubiquitin is attached, may contribute to the chemistry of the reaction 61, 82. Despite these differences in the catalytic site, all three RBRs can react with ubiquitin that is modified with a reactive group on its C-terminus, such as VME or propargyl 11, indicating that they share features that differ from other thioester forming molecules such as E2s and HECT E3s.
Autoinhibition and regulation of the RBR domain
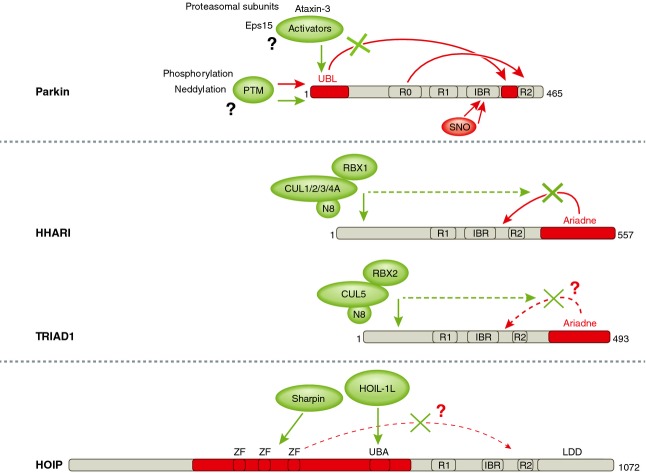
The activity of the different RBR E3-ligases is highly regulated (Fig 3). The proteins can be regulated at the transcriptional level, but are also activated and inactivated by posttranslational modifications and protein-protein interactions.
Figure 3.
Regulation of the E3-ligase activity of Parkin, HHARI, TRIAD1 and HOIP.
Schematic representation of the RBR E3-ligases. The domains involved in autoinhibitory interactions are shown in red, whereas activators are shown in green. Autoinhibitory interactions between domains are depicted by the red arrows, and green arrows indicate interactions and domains that are influenced by the activators. Dotted lines are used for interactions that have not been fully characterized yet. See text for details. IBR = in between ring; LDD = linear-ubiquitin-chain determining domain; N8 = Nedd8; PTM = post-translational modification; SNO = s-nitrosylation; R = RING; UBA = ubiquitin associated domain; ZF = zinc finger.
All of the characterized RBR E3-ligases are autoinhibited by domains surrounding the RING1, IBR and RING2 domains. The E3-ligase activity of Parkin is embedded within the RBR domain in its C-terminus. Its catalytic activity was reportedly auto-inhibited by its N-terminal ubiquitin-like domain (UBL) 8, which binds to a linker/tether region between the IBR and RING2 of the RBR domain. This interaction is important for Parkin function, as multiple pathogenic mutations are found in the UBL 8. At least three targets of Parkin contain domains that interact with the auto-inhibitory UBL domain, suggesting a target-induced Parkin activation 84, 85. Structural studies reveal that the linker/tether region obstructs E2 enzyme access to the RING1 domain even in the absence of the UBL, indicating that this linker region is critical to relieve the autoinhibition 9, 10, 11, 77 (Fig 3). In addition, RING0 was found to obstruct access to the catalytic cysteine and ligase activation requires its rearrangement 9, 10, 11.
Alternative activation may occur through the posttranslational modification of Parkin with Nedd8, which was reported to induce its E3-ligase activity 86. Also phosphorylation of Ser65 by PINK1 activates Parkin 87, 88, 89, in a mechanism where the ubiquitin-ligase Fbxo7 is important 28. In contrast, tyrosine-phosphorylation by c-Abl seems to inhibit Parkin 90. Furthermore, Parkin activity is regulated by the modification of multiple cysteines within its IBR with nitric oxide (s-nitrosylation) 91, 92, which is possibly induced by the increased levels of nitrosative stress in PD 93. However, the precise effects of the s-nitrosylation on Parkin activity remain to be resolved.
In HHARI, the Ariadne domain that is C-terminal to its RBR provides an auto-inhibitory function. This autoinhibition is released upon interaction of the N-terminus of HHARI with a NEDD8-CUL1-RBX1 Cullin-RING complex 42 (Fig 3). The Ariadne RBR E3-ligase TRIAD1 is also strongly activated upon an interaction with a Neddylated Cullin-RING complex consisting of NEDD8-CUL5-RBX2 (Fig 3) 42. The interaction with Neddylated Cullin-RING complexes might be a common feature of Ariadne RBR E3-ligases, which would link their function to that of Cullin-RING complexes. Interestingly, the details of the autoinhibition of HHARI and TRIAD1 must be different from Parkin autoinhibition, as E2 interaction is not impeded 42.
The RBR of HOIP is inhibited by domains that are located N-terminally to the RBR 6, 7. HOIP activation takes place through interactions between its UBA-domain and/or NZF2 domains with the UBL-domain of either HOIL-1L or Sharpin, its partners in the LUBAC complex (Fig 3) 7, 56, 57, 59. In the absence of its N-terminal domains, the RBR unit is constitutively active and HOIL-1L no longer provides additional activation, indicating that the N-terminal domains mediate the auto-inhibition. However, NEMO ubiquitination still requires the presence of additional LUBAC components 61. The precise mechanism by which the HOIP N-terminus blocks the RBR activity is uncertain, as the RBR is not inhibited when the N-terminus is added to in vitro reactions in trans and there are no crystal structures of inhibited HOIP available. Consequently, the exact mechanism by which the catalytic domain is kept in an inactive state in full length HOIP remains to be resolved.
Interestingly, in both Parkin and HOIP, the domains N-terminal and/or within the RBR autoinhibit the catalytic activity 6, 7, 8, 9, 10, 11, whereas in HHARI the C-terminal domain constrains the active conformation of the RBR unit by blocking the RING2 domain 77. Thus, regulation of RBR activity by domains outside the RBR is a general feature of this class of E3 ligase, but the specific mechanism by which the catalytic activity is inhibited varies for each individual protein. Nevertheless, the interactions of Parkin, HOIP, HHARI and TRIAD1 N-termini with other proteins may release the auto-inhibitory state of the RBRs 42, 84, 85.
Chain formation specificity by RBRs
The formation of the HECT-like ubiquitin∼E3 intermediate by RBR proteins would suggest that the target specificity of the RBRs resembles that of the HECT E3-ligases, in which the E2s do not play a role in the final transfer of the ubiquitin onto its target. Indeed, this is the case for HOIP, for which the linear-chain-forming ability overrules the E2 chain-type specificity 57.
HOIP ligase activity, either in the context of LUBAC or as an N-terminal deletion construct that is constitutively active (HOIPRBR-LDD), is induced by a variety of E2-enzymes. All E2s induce the selective formation of linear ubiquitin chains, indicating that the E2s do not contribute to the chain formation specificity of HOIP (Table 2). Instead, the transfer of the ubiquitin from the E3 onto the N-terminal amine-group of the target ubiquitin is mediated by the specific positioning of the target ubiquitin by the HOIP C-terminal LDD 6, 7 (Fig 4). The structure of the HOIP RING2-LDD region in complex with ubiquitin was recently solved 82, and interestingly it reveals that the LDD embraces the RING2 domain. The RING2 and LDD together create the binding site for the target ubiquitin and position it to present the amino-terminus to the RING2∼donor ubiquitin thioester for the formation of a peptide bond between the two ubiquitins (Fig 4). This LDD extension to the RBR domain is not found in other RBR E3-ligases, suggesting that this mechanism of chain formation is unique to HOIP.
Table 2.
E2 interactions with human RBR E3-ligases
| Name | E3 Activitya | Y2Hb | Pull down/interactionsc | Functionald | Ubiquitin Chain typese | References |
|---|---|---|---|---|---|---|
| HHARI | Yes | L3, L6 | L3 (R1+20AA), L6 (R1,IBR) | D3, L3 | 1, 38, 40, 103, 106 | |
| TRIAD1 | Yes | D1, D2, D3, D4, E1, E2, E3, L3, L6, R2, T, V1 | L3 (R1), E1 (R1), N (R2) | D3, E1, E2, L3, N/V2 | K48 (L3), K63 (N/V2), K* (D3) | 42, 47, 48, 50, 107, 108 |
| PARC | Yes | L3 | 109 | |||
| ANKIB1 | nd | |||||
| PARKIN | Yes | L3(R2), L6(R2) | A, D2, D3, L3, L6 | K48, K63 (N/2V) | 1, 24, 25, 26, 36, 94, 95, 99, 100, 110 | |
| RNF144A | nd | H, L3, L6, V1, V2 | L3(R1), L6(R1) | 108, 111 | ||
| RNF144B | Yes | I, L3, L6, T, U, Z | L3 (RBR), L6(RBR) | 108, 112, 113, 114 | ||
| HOIL-1L | Yes | D4, G1, L3, L6, N, S, U | D3 | 108, 115 | ||
| RNF19A | Yes | L3(RBR), L6(RBR) | 116, 117, 118 | |||
| RNF19B | Yes | L3(FL), L6(FL) | 119, 120 | |||
| HOIP | Yes | L6 (FL) | D3, L3 | Linear (all tested E2s) | 6, 121 | |
| RNF216 | Yes | L3, L6 | K48 (E2 not identified) | 122 | ||
| RNF14 | Yes | D1, D2, D3, D4, E1, E2, E3, U, V1,W | E1(R1), E2(R1), E3(R1) | E1, E2, E3 | 108, 114, 123, 124 | |
| RNF217 | nd | |||||
| LUBAC | Yes | B, D1, D2, D3, K, L3 | Linear (all tested E2s) | 55, 57, 58, 62 |
Experimental evidence for E3-ligase activity yes/not done.
E2 interaction-partners identified by yeast-two-hybrid, Ube2 names are shown.
E2 interaction-partners identified by pull down assays or by other methods, the column contains the Ube2 names and the RBR interaction-sites between brackets.
Ube2 names of the E2s that have been shown to be functionally active with the RBR E3.
Ubiquitin chain types formed in cooperation with specific E2s of which the Ube2 name is shown between brackets.
various different ubiquitin chain types formed.
Figure 4.
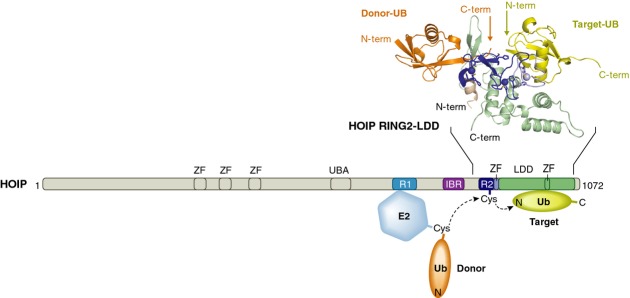
Linear ubiquitin chain formation by HOIP.
The positioning of the target ubiquitin (yellow) by the HOIP LDD (green) is critical for the transfer of the donor-ubiquitin (orange) from the E2 (blue), via a cysteine on RING2 onto the N-terminus of the target ubiquitin. The crystal structure of HOIP RING2-LDD in complex with a donor and a target ubiquitin (PDB 4ljo) shows a snapshot of the orientation of the proteins just before the two ubiquitins are linked together by HOIP. IBR = in between ring; LDD = linear-ubiquitin-chain determining domain; R = RING; UBA = ubiquitin associated domain; ZF = zinc finger.
The targeting of a lysine in NEMO by the HOIP/HOIL-1L complex requires NEMO recognition sites in LUBAC 58 as well as the presence of both an active HOIP RBR and an active HOIL-1L RING2 catalytic site 61. Thus, also in this case the E3 determines which lysines are modified. Understanding the details of this two RBR mechanism will require further studies but, intriguingly, the process also depends on an intact amino terminus of the donor ubiquitin 61, as modifications at the N-terminus or mutation of the adjacent E16 affect the process. If other RBRs have such a requirement, studies using tagged ubiquitin may not reveal their true activity. Lysine targeting by LUBAC could be rare in cells, as most of the linear chains on NEMO are attached to K63 chains, conjugated to the ubiquitin amino-termini in these chains 63.
In contrast to the E3-dependent targeting of LUBAC, the current literature suggests that ubiquitin chain formation by the RBRs ARIH2, Parkin, and RNF216 relies on the ubiquitin chain formation specificity of the E2 that is used in the reaction (Table 2).
The Parkin RBR domain is functionally active with the E2s Ube2A 94, Ube2D2 95, Ube2D3 1, 24, 96, Ube2L3 86, 97, 98, 99, and Ube2L6 24, with which it mediates the formation of various ubiquitin signals (Table 2). Parkin mediates the formation of K63-linked ubiquitin chains in cooperation with the K63-specific E2 Ube2N/Ube2V2 100, 101, to target misfolded DJ1 for dynein-mediated transport to aggresomes 26. It also catalyses the formation of K48-linked ubiquitin chains 100, 101, and works with Ube2L3 to target RanBP2 for degradation 86. Finally, Parkin has been suggested to mediate the mono-ubiquitination of targets in an IBR-RING2 dependent manner in cooperation with Ube2L3 and Ube2N/Ube2V2 98, targeting p38 97, Hsp70/Hsc70 102, and Bcl-2 36. A comparison of the Parkin-mediated ubiquitin chain formation activities in the various studies suggests that the E2s play a major role in the specificity of ubiquitin chain formation. Consequently, the E2 enzyme that is used by Parkin strongly determines the cellular outcome of the proteins that are targeted.
The E3-ligase HHARI has been shown to interact through its RING1/IBR domains to the E2s Ube2L3 and Ube2L6 (Table 2) 38, 40, 103. However, only the functionality of the E2/HHARI interaction with Ube2L3 has been validated 1. Interestingly, also Ube2D3 has been shown to be functionally active with HHARI, even though is has not been identified as a binding partner in yeast two-hybrid studies 1, 38, 40. Unfortunately, there are no data available about the ubiquitin chain formation specificity and target selection by this E3, leaving the precise mechanism by which HHARI ubiquitinates its targets to be resolved.
Sidebar: In need of answers
The past 2 years have seen a rapid increase in insight into the structure and function of RBR E3-ligases, revealing how these ligases perform the ubiquitination reaction. The field now appreciates the importance of their two-step RING/HECT-like mechanism, their autoinhibition and its release by protein partners, and the first insights into the transfer to different target amino groups have emerged.
Nevertheless, important mechanistic and functional questions remain: how do RBRs achieve E2 activation, given that the process is different from other RING domains? What do the RBRs look like in the activated state? Does the chain-specificity in the activated state follow E2 or E3 preferences? As the amino-group specificity of HOIP is regulated by its unique LDD, it is unlikely to be mirrored in other RBR ligases. When studying the chain type and lysine specificity of RBRs, it may be valuable to keep in mind the surprising importance of an intact donor-ubiquitin N-terminus for lysine targeting by HOIL-1L.
Ultimately, insights into mechanism and activation may help to address the most important question of all: what are the genuine targets of these RBR ligases in cells. The identification of these targets is a prerequisite for full appreciation of their function.
The E3-ligase activity of TRIAD1 in cells can potentially be mediated by a large variety of E2s that have been found as TRIAD1 interaction partners (Table 2). Of these E2s, Ube2L3 and Ube2E1 have been shown to interact with the TRIAD1 RING1 domain, and Ube2N binds to TRIAD1 RING2 50. The functional relevance of the interaction has been validated for several E2s in auto-ubiquitination and free ubiquitin chain formation assays, showing that TRIAD1 mediates the formation of K48-ubiquitin chains with Ube2L3, K63-ubiquitin chains in cooperation with Ube2N/Ube2V2, and various different chains in cooperation with Ube2D3 42, 47, 48. Thus, the E3-ligase activity of TRIAD1 follows the ubiquitin chain formation specificity of the E2 that is used in the reaction. Nevertheless, it is not clear if TRIAD1 follows the E2 chain-type choice on its cellular targets.
“Ubiquitylation: mechanism and functions” Review series
Previous issues of EMBO reports include:
Building and remodeling Cullin-RING E3 ubiquitin ligases, by Wade Harper et al
Ubiquitin in the immune system, by Henning Walczak et al
Other reviews in this series, which will be published in consecutive issues of EMBO reports, will cover:
Dynamic survey of mitochondria by ubiquitin, by Mafalda Escobar-Henriques and Thomas Langer
Ubiquitylation in stem cells, by Iannis Aifantis et al
Understanding ubiquitylation one structure at a time, by Ronald Hay et al
So far, many of these assays rely on autoubiquitination as a read-out for activity. Moreover, most have been performed without the full activation of the E3 ligase, as details of the activation mechanisms of RBRs have only recently been revealed. It will be interesting to reconstitute reactions with the different E2 enzymes in the activated states of the RBR enzymes and follow the behaviour on genuine targets. Thus far, it appears as if the final transfer of the ubiquitin by these RBR E3-ligases is mediated by the E2 determining the chain type, despite the dependence on a HECT-like transfer to the target. This suggests that the E2s stay in the complex to facilitate chain formation, which puts an interesting mechanistic constraint on the reaction. Possibly, the E2s cooperate directly with RING2 to position the target ubiquitin close to the active site cysteine of the RBR E3-ligase. Alternatively, the role for the E2 in the ubiquitin chain formation specificity might be explained by a mechanism in which the E2 and E3 collaborate to mediate the specific formation of ubiquitin chains on the E2 active site cysteine 104, before the RBR RING2 mediates the en-bloc transfer of the ubiquitin chain from the E2 onto the target protein. These options require further studies. In addition, the precise role of the RING domains in the RBR needs further analysis, as interaction studies identified that some RBRs interact with E2s through their RING2 domains (Table 2), or bind possible targets via the RBR 24, 35, 105. Consequently, the combined RING/HECT type mechanism might be modulated in these reactions, and further studies are needed to elucidate their functional significance.
Acknowledgments
We thank group members and colleagues for discussion, an unknown reviewer for the suggestion of en-bloc transfer in RBR mechanisms and Francesca Mattiroli and Thangavelu Kaliyappan for critical reading of the manuscript. This work was supported by an ERC advanced grant ‘Ubiquitin balance’, grant number 24997.
Glossary
Glossary
- 4EHP
Translation initiation factor 4E homologous protein
- DUBs
De-ubiquitination enzymes
- E1
Ubiquitin activating enzyme
- E2
Ubiquitin conjugase
- E3
Ubiquitin ligase
- Gfi1
Growth factor independence 1
- HECT
Homologous to the E6-AP carboxy terminus
- HHARI
Human homologue of Drosophila Ariadne
- HOIL-1L
Heme-oxidized IRP2 ubiquitin ligase-1
- IBR
In-between RING
- LDD
Linear ubiquitin chain determining domain
- LUBAC
Linear ubiquitin chain assembly complex
- NEMO
NF-κB essential modulator
- NF-κB
Nuclear factor kappa-light-chain-enhancer of activated B cells
- NZF
Npl4 zinc-finger
- RBR
RING-IBR-RING ligase
- RING
Really interesting new gene
- UBAN
Ubiquitin-binding domain in ABIN and NEMO
Conflict of interest
The authors declare that they have no conflict of interest.
References
- Wenzel DM, Lissounov A, Brzovic PS, Klevit RE. UBCH7 reactivity profile reveals parkin and HHARI to be RING/HECT hybrids. Nature. 2011;474:105–108. doi: 10.1038/nature09966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morett E, Bork P. A novel transactivation domain in parkin. Trends Biochem Sci. 1999;24:229–231. doi: 10.1016/s0968-0004(99)01381-x. [DOI] [PubMed] [Google Scholar]
- van der Reijden BA, Erpelinck-Verschueren CA, Lowenberg B, Jansen JH. TRIADs: a new class of proteins with a novel cysteine-rich signature. Protein Sci. 1999;8:1557–1561. doi: 10.1110/ps.8.7.1557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marin I, Ferrus A. Comparative genomics of the RBR family, including the Parkinson's disease-related gene parkin and the genes of the ariadne subfamily. Mol Biol Evol. 2002;19:2039–2050. doi: 10.1093/oxfordjournals.molbev.a004029. [DOI] [PubMed] [Google Scholar]
- Marin I, Lucas JI, Gradilla AC, Ferrus A. Parkin and relatives: the RBR family of ubiquitin ligases. Physiol Genomics. 2004;17:253–263. doi: 10.1152/physiolgenomics.00226.2003. [DOI] [PubMed] [Google Scholar]
- Smit JJ, Monteferrario D, Noordermeer SM, van Dijk WJ, van der Reijden BA, Sixma TK. The E3 ligase HOIP specifies linear ubiquitin chain assembly through its RING-IBR-RING domain and the unique LDD extension. EMBO J. 2012;31:3833–3844. doi: 10.1038/emboj.2012.217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stieglitz B, Morris-Davies AC, Koliopoulos MG, Christodoulou E, Rittinger K. LUBAC synthesizes linear ubiquitin chains via a thioester intermediate. EMBO Rep. 2012;13:840–846. doi: 10.1038/embor.2012.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaugule VK, Burchell L, Barber KR, Sidhu A, Leslie SJ, Shaw GS, Walden H. Autoregulation of Parkin activity through its ubiquitin-like domain. EMBO J. 2011;30:2853–2867. doi: 10.1038/emboj.2011.204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riley BE, Lougheed JC, Callaway K, Velasquez M, Brecht E, Nguyen L, Shaler T, Walker D, Yang Y, Regnstrom K, Diep L, Zhang Z, Chiou S, Bova M, Artis DR, Yao N, Baker J, Yednock T, Johnston JA. Structure and function of Parkin E3 ubiquitin ligase reveals aspects of RING and HECT ligases. Nat Commun. 2013;4:1982. doi: 10.1038/ncomms2982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trempe JF, Sauve V, Grenier K, Seirafi M, Tang MY, Menade M, Al-Abdul-Wahid S, Krett J, Wong K, Kozlov G, Nagar B, Fon EA, Gehring K. Structure of parkin reveals mechanisms for ubiquitin ligase activation. Science. 2013;340:1451–1455. doi: 10.1126/science.1237908. [DOI] [PubMed] [Google Scholar]
- Wauer T, Komander D. Structure of the human Parkin ligase domain in an autoinhibited state. EMBO J. 2013;32:2099–2112. doi: 10.1038/emboj.2013.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisenhaber B, Chumak N, Eisenhaber F, Hauser MT. The ring between ring fingers (RBR) protein family. Genome Biol. 2007;8:209. doi: 10.1186/gb-2007-8-3-209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belgnaoui SM, Paz S, Samuel S, Goulet ML, Sun Q, Kikkert M, Iwai K, Dikic I, Hiscott J, Lin R. Linear ubiquitination of NEMO negatively regulates the interferon antiviral response through disruption of the MAVS-TRAF3 complex. Cell Host Microbe. 2012;12:211–222. doi: 10.1016/j.chom.2012.06.009. [DOI] [PubMed] [Google Scholar]
- Seymour RE, Hasham MG, Cox GA, Shultz LD, Hogenesch H, Roopenian DC, Sundberg JP. Spontaneous mutations in the mouse Sharpin gene result in multiorgan inflammation, immune system dysregulation and dermatitis. Genes Immun. 2007;8:416–421. doi: 10.1038/sj.gene.6364403. [DOI] [PubMed] [Google Scholar]
- Boisson B, Laplantine E, Prando C, Giliani S, Israelsson E, Xu Z, Abhyankar A, Israël L, Trevejo-Nunez G, Bogunovic D, Cepika AM, MacDuff D, Chrabieh M, Hubeau M, Bajolle F, Debré M, Mazzolari E, Vairo D, Agou F, Virgin HW, et al. Immunodeficiency, autoinflammation and amylopectinosis in humans with inherited HOIL-1 and LUBAC deficiency. Nat Immunol. 2012;13:1178–1186. doi: 10.1038/ni.2457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darios F, Corti O, Lucking CB, Hampe C, Muriel MP, Abbas N, Gu WJ, Hirsch EC, Rooney T, Ruberg M, Brice A. Parkin prevents mitochondrial swelling and cytochrome c release in mitochondria-dependent cell death. Hum Mol Genet. 2003;12:517–526. doi: 10.1093/hmg/ddg044. [DOI] [PubMed] [Google Scholar]
- Kitada T, Asakawa S, Hattori N, Matsumine H, Yamamura Y, Minoshima S, Yokochi M, Mizuno Y, Shimizu N. Mutations in the parkin gene cause autosomal recessive juvenile parkinsonism. Nature. 1998;392:605–608. doi: 10.1038/33416. [DOI] [PubMed] [Google Scholar]
- Matsumine H, Saito M, Shimoda-Matsubayashi S, Tanaka H, Ishikawa A, Nakagawa-Hattori Y, Yokochi M, Kobayashi T, Igarashi S, Takano H, Sanpei K, Koike R, Mori H, Kondo T, Mizutani Y, Schäffer AA, Yamamura Y, Nakamura S, Kuzuhara S, Tsuji S, et al. Localization of a gene for an autosomal recessive form of juvenile Parkinsonism to chromosome 6q25.2-27. Am J Hum Genet. 1997;60:588–596. [PMC free article] [PubMed] [Google Scholar]
- Poulogiannis G, McIntyre RE, Dimitriadi M, Apps JR, Wilson CH, Ichimura K, Luo F, Cantley LC, Wyllie AH, Adams DJ, Arends MJ. PARK2 deletions occur frequently in sporadic colorectal cancer and accelerate adenoma development in Apc mutant mice. Proc Natl Acad Sci USA. 2010;107:15145–15150. doi: 10.1073/pnas.1009941107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veeriah S, Morris L, Solit D, Chan TA. The familial Parkinson disease gene PARK2 is a multisite tumor suppressor on chromosome 6q25.2-27 that regulates cyclin E. Cell Cycle. 2010;9:1451–1452. doi: 10.4161/cc.9.8.11583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narendra D, Tanaka A, Suen DF, Youle RJ. Parkin is recruited selectively to impaired mitochondria and promotes their autophagy. J Cell Biol. 2008;183:795–803. doi: 10.1083/jcb.200809125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazarou M, Narendra DP, Jin SM, Tekle E, Banerjee S, Youle RJ. PINK1 drives Parkin self-association and HECT-like E3 activity upstream of mitochondrial binding. J Cell Biol. 2013;200:163–172. doi: 10.1083/jcb.201210111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimura H, Schlossmacher MG, Hattori N, Frosch MP, Trockenbacher A, Schneider R, Mizuno Y, Kosik KS, Selkoe DJ. Ubiquitination of a new form of alpha-synuclein by parkin from human brain: implications for Parkinson's disease. Science. 2001;293:263–269. doi: 10.1126/science.1060627. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Gao J, Chung KK, Huang H, Dawson VL, Dawson TM. Parkin functions as an E2-dependent ubiquitin- protein ligase and promotes the degradation of the synaptic vesicle-associated protein, CDCrel-1. Proc Natl Acad Sci USA. 2000;97:13354–13359. doi: 10.1073/pnas.240347797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imai Y, Soda M, Inoue H, Hattori N, Mizuno Y, Takahashi R. An unfolded putative transmembrane polypeptide, which can lead to endoplasmic reticulum stress, is a substrate of Parkin. Cell. 2001;105:891–902. doi: 10.1016/s0092-8674(01)00407-x. [DOI] [PubMed] [Google Scholar]
- Olzmann JA, Li L, Chudaev MV, Chen J, Perez FA, Palmiter RD, Chin LS. Parkin-mediated K63-linked polyubiquitination targets misfolded DJ-1 to aggresomes via binding to HDAC6. J Cell Biol. 2007;178:1025–1038. doi: 10.1083/jcb.200611128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenner P, Olanow CW. Understanding cell death in Parkinson's disease. Ann Neurol. 1998;44:S72–S84. doi: 10.1002/ana.410440712. [DOI] [PubMed] [Google Scholar]
- Burchell VS, Nelson DE, Sanchez-Martinez A, Delgado-Camprubi M, Ivatt RM, Pogson JH, Randle SJ, Wray S, Lewis PA, Houlden H, Abramov AY, Hardy J, Wood NW, Whitworth AJ, Laman H, Plun-Favreau H. The Parkinson's disease-linked proteins Fbxo7 and Parkin interact to mediate mitophagy. Nat Neurosci. 2013;16:1257–1265. doi: 10.1038/nn.3489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manzanillo PS, Ayres JS, Watson RO, Collins AC, Souza G, Rae CS, Schneider DS, Nakamura K, Shiloh MU, Cox JS. The ubiquitin ligase parkin mediates resistance to intracellular pathogens. Nature. 2013;501:512–516. doi: 10.1038/nature12566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gegg ME, Cooper JM, Chau KY, Rojo M, Schapira AH, Taanman JW. Mitofusin 1 and mitofusin 2 are ubiquitinated in a PINK1/parkin-dependent manner upon induction of mitophagy. Hum Mol Genet. 2010;19:4861–4870. doi: 10.1093/hmg/ddq419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okatsu K, Iemura S, Koyano F, Go E, Kimura M, Natsume T, Tanaka K, Matsuda N. Mitochondrial hexokinase HKI is a novel substrate of the Parkin ubiquitin ligase. Biochem Biophys Res Commun. 2012;428:197–202. doi: 10.1016/j.bbrc.2012.10.041. [DOI] [PubMed] [Google Scholar]
- Ziviani E, Tao RN, Whitworth AJ. Drosophila parkin requires PINK1 for mitochondrial translocation and ubiquitinates mitofusin. Proc Natl Acad Sci USA. 2010;107:5018–5023. doi: 10.1073/pnas.0913485107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corti O, Hampe C, Koutnikova H, Darios F, Jacquier S, Prigent A, Robinson JC, Pradier L, Ruberg M, Mirande M, Hirsch E, Rooney T, Fournier A, Brice A. The p38 subunit of the aminoacyl-tRNA synthetase complex is a Parkin substrate: linking protein biosynthesis and neurodegeneration. Hum Mol Genet. 2003;12:1427–1437. doi: 10.1093/hmg/ddg159. [DOI] [PubMed] [Google Scholar]
- Um JW, Chung KC. Functional modulation of parkin through physical interaction with SUMO-1. J Neurosci Res. 2006;84:1543–1554. doi: 10.1002/jnr.21041. [DOI] [PubMed] [Google Scholar]
- Okui M, Yamaki A, Takayanagi A, Kudoh J, Shimizu N, Shimizu Y. Transcription factor single-minded 2 (SIM2) is ubiquitinated by the RING-IBR-RING-type E3 ubiquitin ligases. Exp Cell Res. 2005;309:220–228. doi: 10.1016/j.yexcr.2005.05.018. [DOI] [PubMed] [Google Scholar]
- Chen D, Gao F, Li B, Wang H, Xu Y, Zhu C, Wang G. Parkin mono-ubiquitinates Bcl-2 and regulates autophagy. J Biol Chem. 2010;285:38214–38223. doi: 10.1074/jbc.M110.101469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarraf SA, Raman M, Guarani-Pereira V, Sowa ME, Huttlin EL, Gygi SP, Harper JW. Landscape of the PARKIN-dependent ubiquitylome in response to mitochondrial depolarization. Nature. 2013;496:372–376. doi: 10.1038/nature12043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ardley HC, Tan NG, Rose SA, Markham AF, Robinson PA. Features of the parkin/ariadne-like ubiquitin ligase, HHARI, that regulate its interaction with the ubiquitin-conjugating enzyme, Ubch7. J Biol Chem. 2001;276:19640–19647. doi: 10.1074/jbc.M011028200. [DOI] [PubMed] [Google Scholar]
- Parelkar SS, Cadena JG, Kim C, Wang Z, Sugal R, Bentley B, Moral L, Ardley HC, Schwartz LM. The Parkin-like human homolog of Drosophila ariadne-1 (HHARI) can induce aggresome formation in mammalian cells and is immunologically detectable in Lewy bodies. J Mol Neurosci. 2012;46:109–121. doi: 10.1007/s12031-011-9535-1. [DOI] [PubMed] [Google Scholar]
- Tan NG, Ardley HC, Scott GB, Rose SA, Markham AF, Robinson PA. Human homologue of ariadne promotes the ubiquitylation of translation initiation factor 4E homologous protein, 4EHP. FEBS Lett. 2003;554:501–504. doi: 10.1016/s0014-5793(03)01235-3. [DOI] [PubMed] [Google Scholar]
- Elmehdawi F, Wheway G, Szymanska K, Adams M, High AS, Johnson CA, Robinson PA. Human Homolog of Drosophila Ariadne (HHARI) is a marker of cellular proliferation associated with nuclear bodies. Exp Cell Res. 2013;319:161–172. doi: 10.1016/j.yexcr.2012.10.002. [DOI] [PubMed] [Google Scholar]
- Kelsall IR, Duda DM, Olszewski JL, Hofmann K, Knebel A, Langevin F, Wood N, Wightman M, Schulman BA, Alpi AF. TRIAD1 and HHARI bind to and are activated by distinct neddylated Cullin-RING ligase complexes. EMBO J. 2013;32:2848–2860. doi: 10.1038/emboj.2013.209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin AE, Ebert G, Ow Y, Preston SP, Toe JG, Cooney JP, Scott HW, Sasaki M, Saibil SD, Dissanayake D, Kim RH, Wakeham A, You-Ten A, Shahinian A, Duncan G, Silvester J, Ohashi PS, Mak TW, Pellegrini M. ARIH2 is essential for embryogenesis, and its hematopoietic deficiency causes lethal activation of the immune system. Nat Immunol. 2013;14:27–33. doi: 10.1038/ni.2478. [DOI] [PubMed] [Google Scholar]
- Hassink G, Slotman J, Oorschot V, Van Der Reijden BA, Monteferrario D, Noordermeer SM, Van Kerkhof P, Klumperman J, Strous GJ. Identification of the ubiquitin ligase Triad1 as a regulator of endosomal transport. Biol Open. 2012;1:607–614. doi: 10.1242/bio.2012778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Bei L, Shah CA, Horvath E, Eklund EA. HoxA10 influences protein ubiquitination by activating transcription of ARIH2, the gene encoding Triad1. J Biol Chem. 2011;286:16832–16845. doi: 10.1074/jbc.M110.213975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bae S, Jung JH, Kim K, An IS, Kim SY, Lee JH, Park IC, Jin YW, Lee SJ, An S. TRIAD1 inhibits MDM2-mediated p53 ubiquitination and degradation. FEBS Lett. 2012;586:3057–3063. doi: 10.1016/j.febslet.2012.07.022. [DOI] [PubMed] [Google Scholar]
- Marteijn JA, van der Meer LT, van Emst L, van Reijmersdal S, Wissink W, de Witte T, Jansen JH, Van der Reijden BA. Gfi1 ubiquitination and proteasomal degradation is inhibited by the ubiquitin ligase Triad1. Blood. 2007;110:3128–3135. doi: 10.1182/blood-2006-11-058602. [DOI] [PubMed] [Google Scholar]
- Marteijn JA, van Emst L, Erpelinck-Verschueren CA, Nikoloski G, Menke A, de Witte T, Lowenberg B, Jansen JH, van der Reijden BA. The E3 ubiquitin-protein ligase Triad1 inhibits clonogenic growth of primary myeloid progenitor cells. Blood. 2005;106:4114–4123. doi: 10.1182/blood-2005-04-1450. [DOI] [PubMed] [Google Scholar]
- Wang W, Yoder JH. Hox-mediated regulation of doublesex sculpts sex-specific abdomen morphology in Drosophila. Dev Dyn. 2012;241:1076–1090. doi: 10.1002/dvdy.23791. [DOI] [PubMed] [Google Scholar]
- Marteijn JA, van der Meer LT, Smit JJ, Noordermeer SM, Wissink W, Jansen P, Swarts HG, Hibbert RG, de Witte T, Sixma TK, Jansen JH, van der Reijden BA. The ubiquitin ligase Triad1 inhibits myelopoiesis through UbcH7 and Ubc13 interacting domains. Leukemia. 2009;23:1480–1489. doi: 10.1038/leu.2009.57. [DOI] [PubMed] [Google Scholar]
- Kawagoe H, Humphries RK, Blair A, Sutherland HJ, Hogge DE. Expression of HOX genes, HOX cofactors, and MLL in phenotypically and functionally defined subpopulations of leukemic and normal human hematopoietic cells. Leukemia. 1999;13:687–698. doi: 10.1038/sj.leu.2401410. [DOI] [PubMed] [Google Scholar]
- Camos M, Esteve J, Jares P, Colomer D, Rozman M, Villamor N, Costa D, Carrio A, Nomdedeu J, Montserrat E, Campo E. Gene expression profiling of acute myeloid leukemia with translocation t(8;16)(p11;p13) and MYST3-CREBBP rearrangement reveals a distinctive signature with a specific pattern of HOX gene expression. Cancer Res. 2006;66:6947–6954. doi: 10.1158/0008-5472.CAN-05-4601. [DOI] [PubMed] [Google Scholar]
- Sauvageau G, Lansdorp PM, Eaves CJ, Hogge DE, Dragowska WH, Reid DS, Largman C, Lawrence HJ, Humphries RK. Differential expression of homeobox genes in functionally distinct CD34+ subpopulations of human bone marrow cells. Proc Natl Acad Sci USA. 1994;91:12223–12227. doi: 10.1073/pnas.91.25.12223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armstrong SA, Staunton JE, Silverman LB, Pieters R, den Boer ML, Minden MD, Sallan SE, Lander ES, Golub TR, Korsmeyer SJ. MLL translocations specify a distinct gene expression profile that distinguishes a unique leukemia. Nat Genet. 2002;30:41–47. doi: 10.1038/ng765. [DOI] [PubMed] [Google Scholar]
- Gerlach B, Cordier SM, Schmukle AC, Emmerich CH, Rieser E, Haas TL, Webb AI, Rickard JA, Anderton H, Wong WW, Nachbur U, Gangoda L, Warnken U, Purcell AW, Silke J, Walczak H. Linear ubiquitination prevents inflammation and regulates immune signalling. Nature. 2011;471:591–596. doi: 10.1038/nature09816. [DOI] [PubMed] [Google Scholar]
- Ikeda F, Deribe YL, Skånland SS, Stieglitz B, Grabbe C, Franz-Wachtel M, van Wijk SJ, Goswami P, Nagy V, Terzic J, Tokunaga F, Androulidaki A, Nakagawa T, Pasparakis M, Iwai K, Sundberg JP, Schaefer L, Rittinger K, Macek B, Dikic I. SHARPIN forms a linear ubiquitin ligase complex regulating NF-kappaB activity and apoptosis. Nature. 2011;471:637–641. doi: 10.1038/nature09814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirisako T, Kamei K, Murata S, Kato M, Fukumoto H, Kanie M, Sano S, Tokunaga F, Tanaka K, Iwai K. A ubiquitin ligase complex assembles linear polyubiquitin chains. EMBO J. 2006;25:4877–4887. doi: 10.1038/sj.emboj.7601360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tokunaga F, Sakata S, Saeki Y, Satomi Y, Kirisako T, Kamei K, Nakagawa T, Kato M, Murata S, Yamaoka S, Yamamoto M, Akira S, Takao T, Tanaka K, Iwai K. Involvement of linear polyubiquitylation of NEMO in NF-kappaB activation. Nat Cell Biol. 2009;11:123–132. doi: 10.1038/ncb1821. [DOI] [PubMed] [Google Scholar]
- Tokunaga F, Nakagawa T, Nakahara M, Saeki Y, Taniguchi M, Sakata S, Tanaka K, Nakano H, Iwai K. SHARPIN is a component of the NF-kappaB-activating linear ubiquitin chain assembly complex. Nature. 2011;471:633–636. doi: 10.1038/nature09815. [DOI] [PubMed] [Google Scholar]
- Hostager BS, Fox DK, Whitten D, Wilkerson CG, Eipper BA, Francone VP, Rothman PB, Colgan JD. HOIL-1L interacting protein (HOIP) as an NF-kappaB regulating component of the CD40 signaling complex. PLoS ONE. 2010;5:e11380. doi: 10.1371/journal.pone.0011380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smit JJ, van Dijk WJ, El Atmioui D, Merkx R, Ovaa H, Sixma TK. Target Specificity of the E3 Ligase LUBAC for Ubiquitin and NEMO Relies on Different Minimal Requirements. J Biol Chem. 2013;288:31728–31737. doi: 10.1074/jbc.M113.495846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haas TL, Emmerich CH, Gerlach B, Schmukle AC, Cordier SM, Rieser E, Feltham R, Vince J, Warnken U, Wenger T, Koschny R, Komander D, Silke J, Walczak H. Recruitment of the linear ubiquitin chain assembly complex stabilizes the TNF-R1 signaling complex and is required for TNF-mediated gene induction. Mol Cell. 2009;36:831–844. doi: 10.1016/j.molcel.2009.10.013. [DOI] [PubMed] [Google Scholar]
- Emmerich CH, Ordureau A, Strickson S, Arthur JS, Pedrioli PG, Komander D, Cohen P. Activation of the canonical IKK complex by K63/M1-linked hybrid ubiquitin chains. Proc Natl Acad Sci USA. 2013;110:15247–15252. doi: 10.1073/pnas.1314715110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiil BK, Damgaard RB, Wagner SA, Keusekotten K, Fritsch M, Bekker-Jensen S, Mailand N, Choudhary C, Komander D, Gyrd-Hansen M. OTULIN restricts Met1-linked ubiquitination to control innate immune signaling. Mol Cell. 2013;50:818–830. doi: 10.1016/j.molcel.2013.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller-Rischart AK, et al. The E3 ligase parkin maintains mitochondrial integrity by increasing linear ubiquitination of NEMO. Mol Cell. 2013;49:908–921. doi: 10.1016/j.molcel.2013.01.036. [DOI] [PubMed] [Google Scholar]
- Müller-Rischart AK, Pilsl A, Beaudette P, Patra M, Hadian K, Funke M, Peis R, Deinlein A, Schweimer C, Kuhn PH, Lichtenthaler SF, Motori E, Hrelia S, Wurst W, Trümbach D, Langer T, Krappmann D, Dittmar G, Tatzelt J, Winklhofer KF. Specific recognition of linear ubiquitin chains by NEMO is important for NF-kappaB activation. Cell. 2009;136:1098–1109. doi: 10.1016/j.cell.2009.03.007. [DOI] [PubMed] [Google Scholar]
- Sato Y, Fujita H, Yoshikawa A, Yamashita M, Yamagata A, Kaiser SE, Iwai K, Fukai S. Specific recognition of linear ubiquitin chains by the Npl4 zinc finger (NZF) domain of the HOIL-1L subunit of the linear ubiquitin chain assembly complex. Proc Natl Acad Sci USA. 2011;108:20520–20525. doi: 10.1073/pnas.1109088108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner S, Carpentier I, Rogov V, Kreike M, Ikeda F, Lohr F, Wu CJ, Ashwell JD, Dotsch V, Dikic I, Beyaert R. Ubiquitin binding mediates the NF-kappaB inhibitory potential of ABIN proteins. Oncogene. 2008;27:3739–3745. doi: 10.1038/sj.onc.1211042. [DOI] [PubMed] [Google Scholar]
- Kensche T, Tokunaga F, Ikeda F, Goto E, Iwai K, Dikic I. Analysis of nuclear factor-kappaB (NF-kappaB) essential modulator (NEMO) binding to linear and lysine-linked ubiquitin chains and its role in the activation of NF-kappaB. J Biol Chem. 2012;287:23626–23634. doi: 10.1074/jbc.M112.347195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verhelst K, Carpentier I, Kreike M, Meloni L, Verstrepen L, Kensche T, Dikic I, Beyaert R. A20 inhibits LUBAC-mediated NF-kappaB activation by binding linear polyubiquitin chains via its zinc finger 7. EMBO J. 2012;31:3845–3855. doi: 10.1038/emboj.2012.240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keusekotten K, Elliott PR, Glockner L, Fiil BK, Damgaard RB, Kulathu Y, Wauer T, Hospenthal MK, Gyrd-Hansen M, Krappmann D, Hofmann K, Komander D. OTULIN antagonizes LUBAC signaling by specifically hydrolyzing Met1-linked polyubiquitin. Cell. 2013;153:1312–1326. doi: 10.1016/j.cell.2013.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivkin E, Almeida SM, Ceccarelli DF, Juang YC, MacLean TA, Srikumar T, Huang H, Dunham WH, Fukumura R, Xie G, Gondo Y, Raught B, Gingras AC, Sicheri F, Cordes SP. The linear ubiquitin-specific deubiquitinase gumby regulates angiogenesis. Nature. 2013;498:318–324. doi: 10.1038/nature12296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zinngrebe J, Montinaro A, Peltzer N, Walczak H. Ubiquitin in the immune system. EMBO Rep. 2014 doi: 10.1002/embr.201338025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chew KC, Matsuda N, Saisho K, Lim GG, Chai C, Tan HM, Tanaka K, Lim KL. Parkin mediates apparent E2-independent monoubiquitination in vitro and contains an intrinsic activity that catalyzes polyubiquitination. PLoS ONE. 2011;6:e19720. doi: 10.1371/journal.pone.0019720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petroski MD, Deshaies RJ. Mechanism of lysine 48-linked ubiquitin-chain synthesis by the cullin-RING ubiquitin-ligase complex SCF-Cdc34. Cell. 2005;123:1107–1120. doi: 10.1016/j.cell.2005.09.033. [DOI] [PubMed] [Google Scholar]
- Seol JH, Feldman RM, Zachariae W, Shevchenko A, Correll CC, Lyapina S, Chi Y, Galova M, Claypool J, Sandmeyer S, Nasmyth K, Deshaies RJ. Cdc53/cullin and the essential Hrt1 RING-H2 subunit of SCF define a ubiquitin ligase module that activates the E2 enzyme Cdc34. Genes Dev. 1999;13:1614–1626. doi: 10.1101/gad.13.12.1614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duda DM, Olszewski JL, Schuermann JP, Kurinov I, Miller DJ, Nourse A, Alpi AF, Schulman BA. Structure of HHARI, a RING-IBR-RING ubiquitin ligase: autoinhibition of an Ariadne-family E3 and insights into ligation mechanism. Structure. 2013;21:1030–1041. doi: 10.1016/j.str.2013.04.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plechanovova A, Jaffray EG, Tatham MH, Naismith JH, Hay RT. Structure of a RING E3 ligase and ubiquitin-loaded E2 primed for catalysis. Nature. 2012;489:115–120. doi: 10.1038/nature11376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pruneda JN, Littlefield PJ, Soss SE, Nordquist KA, Chazin WJ, Brzovic PS, Klevit RE. Structure of an E3:E2∼Ub complex reveals an allosteric mechanism shared among RING/U-box ligases. Mol Cell. 2012;47:933–942. doi: 10.1016/j.molcel.2012.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dou H, Buetow L, Hock A, Sibbet GJ, Vousden KH, Huang DT. Structural basis for autoinhibition and phosphorylation-dependent activation of c-Cbl. Nat Struct Mol Biol. 2012;19:184–192. doi: 10.1038/nsmb.2231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spratt DE, Martinez-Torres RJ, Noh YJ, Mercier P, Manczyk N, Barber KR, Aguirre JD, Burchell L, Purkiss A, Walden H, Shaw GS. A molecular explanation for the recessive nature of parkin-linked Parkinson's disease. Nat Commun. 2013;4:1983. doi: 10.1038/ncomms2983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stieglitz B, Rana RR, Koliopoulos MG, Morris-Davies AC, Schaeffer V, Christodoulou E, Howell S, Brown NR, Dikic I, Rittinger K. Structural basis for ligase-specific conjugation of linear ubiquitin chains by HOIP. Nature. 2013;503:422–426. doi: 10.1038/nature12638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frye JJ, Brown NG, Petzold G, Watson ER, Grace CR, Nourse A, Jarvis MA, Kriwacki RW, Peters JM, Stark H, Schulman BA. Electron microscopy structure of human APC/C(CDH1)-EMI1 reveals multimodal mechanism of E3 ligase shutdown. Nat Struct Mol Biol. 2013;20:827–835. doi: 10.1038/nsmb.2593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fallon L, Belanger CM, Corera AT, Kontogiannea M, Regan-Klapisz E, Moreau F, Voortman J, Haber M, Rouleau G, Thorarinsdottir T, Brice A, van Bergen En Henegouwen PM, Fon EA. A regulated interaction with the UIM protein Eps15 implicates parkin in EGF receptor trafficking and PI(3)K-Akt signalling. Nat Cell Biol. 2006;8:834–842. doi: 10.1038/ncb1441. [DOI] [PubMed] [Google Scholar]
- Tsai YC, Fishman PS, Thakor NV, Oyler GA. Parkin facilitates the elimination of expanded polyglutamine proteins and leads to preservation of proteasome function. J Biol Chem. 2003;278:22044–22055. doi: 10.1074/jbc.M212235200. [DOI] [PubMed] [Google Scholar]
- Um JW, Han KA, Im E, Oh Y, Lee K, Chung KC. Neddylation positively regulates the ubiquitin E3 ligase activity of parkin. J Neurosci Res. 2012;90:1030–1042. doi: 10.1002/jnr.22828. [DOI] [PubMed] [Google Scholar]
- Kondapalli C, Kazlauskaite A, Zhang N, Woodroof HI, Campbell DG, Gourlay R, Burchell L, Walden H, Macartney TJ, Deak M, Knebel A, Alessi DR, Muqit MM. PINK1 is activated by mitochondrial membrane potential depolarization and stimulates Parkin E3 ligase activity by phosphorylating Serine 65. Open Biol. 2012;2:120080. doi: 10.1098/rsob.120080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiba-Fukushima K, Imai Y, Yoshida S, Ishihama Y, Kanao T, Sato S, Hattori N. PINK1-mediated phosphorylation of the Parkin ubiquitin-like domain primes mitochondrial translocation of Parkin and regulates mitophagy. Sci Rep. 2012;2:1002. doi: 10.1038/srep01002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sha D, Chin LS, Li L. Phosphorylation of parkin by Parkinson disease-linked kinase PINK1 activates parkin E3 ligase function and NF-kappaB signaling. Hum Mol Genet. 2010;19:352–363. doi: 10.1093/hmg/ddp501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imam SZ, Zhou Q, Yamamoto A, Valente AJ, Ali SF, Bains M, Roberts JL, Kahle PJ, Clark RA, Li S. Novel regulation of parkin function through c-Abl-mediated tyrosine phosphorylation: implications for Parkinson's disease. J Neurosci. 2011;31:157–163. doi: 10.1523/JNEUROSCI.1833-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung KK, Thomas B, Li X, Pletnikova O, Troncoso JC, Marsh L, Dawson VL, Dawson TM. S-nitrosylation of parkin regulates ubiquitination and compromises parkin's protective function. Science. 2004;304:1328–1331. doi: 10.1126/science.1093891. [DOI] [PubMed] [Google Scholar]
- Ozawa K, Komatsubara AT, Nishimura Y, Sawada T, Kawafune H, Tsumoto H, Tsuji Y, Zhao J, Kyotani Y, Tanaka T, Takahashi R, Yoshizumi M. S-nitrosylation regulates mitochondrial quality control via activation of parkin. Sci Rep. 2013;3:2202. doi: 10.1038/srep02202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ischiropoulos H. Oxidative modifications of alpha-synuclein. Ann N Y Acad Sci. 2003;991:93–100. doi: 10.1111/j.1749-6632.2003.tb07466.x. [DOI] [PubMed] [Google Scholar]
- Haddad DM, Vilain S, Vos M, Esposito G, Matta S, Kalscheuer VM, Craessaerts K, Leyssen M, Nascimento RM, Vianna-Morgante AM, De Strooper B, Van Esch H, Morais VA, Verstreken P. Mutations in the intellectual disability gene Ube2a cause neuronal dysfunction and impair parkin-dependent mitophagy. Mol Cell. 2013;50:831–843. doi: 10.1016/j.molcel.2013.04.012. [DOI] [PubMed] [Google Scholar]
- Rankin CA, Joazeiro CA, Floor E, Hunter T. E3 ubiquitin-protein ligase activity of Parkin is dependent on cooperative interaction of RING finger (TRIAD) elements. J Biomed Sci. 2001;8:421–429. doi: 10.1007/BF02255952. [DOI] [PubMed] [Google Scholar]
- Capili AD, Edghill EL, Wu K, Borden KL. Structure of the C-terminal RING finger from a RING-IBR-RING/TRIAD motif reveals a novel zinc-binding domain distinct from a RING. J Mol Biol. 2004;340:1117–1129. doi: 10.1016/j.jmb.2004.05.035. [DOI] [PubMed] [Google Scholar]
- Hampe C, Ardila-Osorio H, Fournier M, Brice A, Corti O. Biochemical analysis of Parkinson's disease-causing variants of Parkin, an E3 ubiquitin-protein ligase with monoubiquitylation capacity. Hum Mol Genet. 2006;15:2059–2075. doi: 10.1093/hmg/ddl131. [DOI] [PubMed] [Google Scholar]
- Matsuda N, Kitami T, Suzuki T, Mizuno Y, Hattori N, Tanaka K. Diverse effects of pathogenic mutations of Parkin that catalyze multiple monoubiquitylation in vitro. J Biol Chem. 2006;281:3204–3209. doi: 10.1074/jbc.M510393200. [DOI] [PubMed] [Google Scholar]
- Staropoli JF, McDermott C, Martinat C, Schulman B, Demireva E, Abeliovich A. Parkin is a component of an SCF-like ubiquitin ligase complex and protects postmitotic neurons from kainate excitotoxicity. Neuron. 2003;37:735–749. doi: 10.1016/s0896-6273(03)00084-9. [DOI] [PubMed] [Google Scholar]
- Doss-Pepe EW, Chen L, Madura K. Alpha-synuclein and parkin contribute to the assembly of ubiquitin lysine 63-linked multiubiquitin chains. J Biol Chem. 2005;280:16619–16624. doi: 10.1074/jbc.M413591200. [DOI] [PubMed] [Google Scholar]
- Lim KL, Chew KC, Tan JM, Wang C, Chung KK, Zhang Y, Tanaka Y, Smith W, Engelender S, Ross CA, Dawson VL, Dawson TM. Parkin mediates nonclassical, proteasomal-independent ubiquitination of synphilin-1: implications for Lewy body formation. J Neurosci. 2005;25:2002–2009. doi: 10.1523/JNEUROSCI.4474-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore DJ, West AB, Dikeman DA, Dawson VL, Dawson TM. Parkin mediates the degradation-independent ubiquitination of Hsp70. J Neurochem. 2008;105:1806–1819. doi: 10.1111/j.1471-4159.2008.05261.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moynihan TP, Ardley HC, Nuber U, Rose SA, Jones PF, Markham AF, Scheffner M, Robinson PA. The ubiquitin-conjugating enzymes UbcH7 and UbcH8 interact with RING finger/IBR motif-containing domains of HHARI and H7-AP1. J Biol Chem. 1999;274:30963–30968. doi: 10.1074/jbc.274.43.30963. [DOI] [PubMed] [Google Scholar]
- Li W, Tu D, Li L, Wollert T, Ghirlando R, Brunger AT, Ye Y. Mechanistic insights into active site-associated polyubiquitination by the ubiquitin-conjugating enzyme Ube2g2. Proc Natl Acad Sci USA. 2009;106:3722–3727. doi: 10.1073/pnas.0808564106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inn KS, Gack MU, Tokunaga F, Shi M, Wong LY, Iwai K, Jung JU. Linear ubiquitin assembly complex negatively regulates RIG-I- and TRIM25-mediated type I interferon induction. Mol Cell. 2011;41:354–365. doi: 10.1016/j.molcel.2010.12.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aguilera M, Oliveros M, Martinez-Padron M, Barbas JA, Ferrus A. Ariadne-1: a vital Drosophila gene is required in development and defines a new conserved family of ring-finger proteins. Genetics. 2000;155:1231–1244. doi: 10.1093/genetics/155.3.1231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stelzl U, Worm U, Lalowski M, Haenig C, Brembeck FH, Goehler H, Stroedicke M, Zenkner M, Schoenherr A, Koeppen S, Timm J, Mintzlaff S, Abraham C, Bock N, Kietzmann S, Goedde A, Toksöz E, Droege A, Krobitsch S, Korn B. A human protein-protein interaction network: a resource for annotating the proteome. Cell. 2005;122:957–968. doi: 10.1016/j.cell.2005.08.029. [DOI] [PubMed] [Google Scholar]
- Markson G, Kiel C, Hyde R, Brown S, Charalabous P, Bremm A, Semple J, Woodsmith J, Duley S, Salehi-Ashtiani K, Vidal M, Komander D, Serrano L, Lehner P, Sanderson CM. Analysis of the human E2 ubiquitin conjugating enzyme protein interaction network. Genome Res. 2009;19:1905–1911. doi: 10.1101/gr.093963.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikolaev AY, Li M, Puskas N, Qin J, Gu W. Parc: a cytoplasmic anchor for p53. Cell. 2003;112:29–40. doi: 10.1016/s0092-8674(02)01255-2. [DOI] [PubMed] [Google Scholar]
- Shimura H, Hattori N, Kubo S, Mizuno Y, Asakawa S, Minoshima S, Shimizu N, Iwai K, Chiba T, Tanaka K, Suzuki T. Familial Parkinson disease gene product, parkin, is a ubiquitin-protein ligase. Nat Genet. 2000;25:302–305. doi: 10.1038/77060. [DOI] [PubMed] [Google Scholar]
- Martinez-Noel G, Muller U, Harbers K. Identification of molecular determinants required for interaction of ubiquitin-conjugating enzymes and RING finger proteins. Eur J Biochem. 2001;268:5912–5919. doi: 10.1046/j.0014-2956.2001.02541.x. [DOI] [PubMed] [Google Scholar]
- Ng CC, Arakawa H, Fukuda S, Kondoh H, Nakamura Y. p53RFP, a p53-inducible RING-finger protein, regulates the stability of p21WAF1. Oncogene. 2003;22:4449–4458. doi: 10.1038/sj.onc.1206586. [DOI] [PubMed] [Google Scholar]
- Huang J, Xu LG, Liu T, Zhai Z, Shu HB. The p53-inducible E3 ubiquitin ligase p53RFP induces p53-dependent apoptosis. FEBS Lett. 2006;580:940–947. doi: 10.1016/j.febslet.2005.09.105. [DOI] [PubMed] [Google Scholar]
- van Wijk SJ, de Vries SJ, Kemmeren P, Huang A, Boelens R, Bonvin AM, Timmers HT. A comprehensive framework of E2-RING E3 interactions of the human ubiquitin-proteasome system. Mol Syst Biol. 2009;5:295. doi: 10.1038/msb.2009.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tatematsu K, Yoshimoto N, Okajima T, Tanizawa K, Kuroda S. Identification of ubiquitin ligase activity of RBCK1 and its inhibition by splice variant RBCK2 and protein kinase Cbeta. J Biol Chem. 2008;283:11575–11585. doi: 10.1074/jbc.M706961200. [DOI] [PubMed] [Google Scholar]
- Niwa J, Ishigaki S, Doyu M, Suzuki T, Tanaka K, Sobue G. A novel centrosomal ring-finger protein, dorfin, mediates ubiquitin ligase activity. Biochem Biophys Res Commun. 2001;281:706–713. doi: 10.1006/bbrc.2001.4414. [DOI] [PubMed] [Google Scholar]
- Ito T, Niwa J, Hishikawa N, Ishigaki S, Doyu M, Sobue G. Dorfin localizes to Lewy bodies and ubiquitylates synphilin-1. J Biol Chem. 2003;278:29106–29114. doi: 10.1074/jbc.M302763200. [DOI] [PubMed] [Google Scholar]
- Niwa J, Ishigaki S, Hishikawa N, Yamamoto M, Doyu M, Murata S, Tanaka K, Taniguchi N, Sobue G. Dorfin ubiquitylates mutant SOD1 and prevents mutant SOD1-mediated neurotoxicity. J Biol Chem. 2002;277:36793–36798. doi: 10.1074/jbc.M206559200. [DOI] [PubMed] [Google Scholar]
- Fortier JM, Kornbluth J. NK lytic-associated molecule, involved in NK cytotoxic function, is an E3 ligase. J Immunol. 2006;176:6454–6463. doi: 10.4049/jimmunol.176.11.6454. [DOI] [PubMed] [Google Scholar]
- Ambrose EC, Kornbluth J. Downregulation of uridine-cytidine kinase like-1 decreases proliferation and enhances tumor susceptibility to lysis by apoptotic agents and natural killer cells. Apoptosis. 2009;14:1227–1236. doi: 10.1007/s10495-009-0385-z. [DOI] [PubMed] [Google Scholar]
- Ehrlund A, Anthonisen EH, Gustafsson N, Venteclef N, Robertson Remen K, Damdimopoulos AE, Galeeva A, Pelto-Huikko M, Lalli E, Steffensen KR, Gustafsson JA, Treuter E. E3 ubiquitin ligase RNF31 cooperates with DAX-1 in transcriptional repression of steroidogenesis. Mol Cell Biol. 2009;29:2230–2242. doi: 10.1128/MCB.00743-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuang TH, Ulevitch RJ. Triad3A, an E3 ubiquitin-protein ligase regulating Toll-like receptors. Nat Immunol. 2004;5:495–502. doi: 10.1038/ni1066. [DOI] [PubMed] [Google Scholar]
- Kang HY, Yeh S, Fujimoto N, Chang C. Cloning and characterization of human prostate coactivator ARA54, a novel protein that associates with the androgen receptor. J Biol Chem. 1999;274:8570–8576. doi: 10.1074/jbc.274.13.8570. [DOI] [PubMed] [Google Scholar]
- Ito K, Adachi S, Iwakami R, Yasuda H, Muto Y, Seki N, Okano Y. N-Terminally extended human ubiquitin-conjugating enzymes (E2s) mediate the ubiquitination of RING-finger proteins, ARA54 and RNF8. Eur J Biochem. 2001;268:2725–2732. doi: 10.1046/j.1432-1327.2001.02169.x. [DOI] [PubMed] [Google Scholar]