Abstract
Background information
Commitment to splicing occurs co-transcriptionally, but a major unanswered question is the extent to which various modifications of chromatin, the template for transcription in vivo, contribute to the regulation of splicing.
Results
Here we perform genome-wide analyses showing that inhibition of specific marks – H2B ubiquitylation, H3K4 methylation, and H3K36 methylation – perturbs splicing in budding yeast, with each modification exerting gene-specific effects. Furthermore, semi-quantitative mass spectrometry on purified nuclear mRNPs and chromatin immunoprecipitation analysis on intron-containing genes indicated that H2B ubiquitylation, but not Set1-, Set2- or Dot1-dependent H3 methylation, stimulates recruitment of the early splicing factors, namely U1 and U2 snRNPs, onto nascent RNAs.
Conclusions
These results suggest that histone modifications impact splicing of distinct subsets of genes using distinct pathways.
INTRODUCTION
Transcriptional control of gene expression has long been thought to require the coordinated modification of histones (Suganuma and Workman 2011) but recent evidence suggests an additional role for these modifications in controlling the eventual fate of an mRNA after it is transcribed (Carrillo Oesterreich et al., 2011; Hnilicova and Stanek 2011; Luco et al., 2011; Nino et al., 2013). A number of correlative studies have shown that nucleosomes at DNA encoding exons and introns bear distinct covalent modifications in metazoa (reviewed in (Carrillo Oesterreichet al., 2011), and yeast (Shieh et al., 2011) These observations raise the possibility that epigenetic marks may have a widespread role in providing direct regulatory input into co-transcriptional splicing decisions.
In Saccharomyces cerevisiae, splicing occurs co-transcriptionally for the vast majority of intron-containing genes (Carrillo Oesterreich et al., 2010), and consistently, splicing factors are recruited to nascent transcripts (Kotovic et al., 2003; Gornemann et al., 2005; Lacadie and Rosbash 2005; Moore et al., 2006; Tardiff et al., 2006; Aitken et al., 2011). Interestingly, the histone acetyltransferase catalytic subunit of the SAGA complex, Gcn5, has been shown to control the co-transcriptional recruitment of the U2 snRNP (Gunderson and Johnson 2009). However, the extent to which additional histone modifications of the chromatin landscape regulate the co-transcriptional recruitment of the spliceosome is still unclear.
Transcription-associated histone H2B mono-ubiquitylation (Ub-H2B) and the downstream histone H3 methylation events have established roles in transcription activation and nucleosome dynamics (Robzyk et al., 2000; Sun and Allis 2002; Wood et al., 2003; Vitaliano-Prunier et al., 2008). In addition, we recently showed that Ub-H2B influences export of mRNPs by promoting the recruitment of the nuclear export machinery to nascent transcripts (Babour et al., 2012; Vitaliano-Prunier et al., 2012). Furthermore, we recently reported genetic and functional interactions between the Ub-H2B machinery and the SR-like spliceosome-associated factor Npl3 (Moehle et al., 2012), suggesting that the role of Ub-H2B in gene expression is not limited to directing transcription itself. This result prompted us to determine the contribution of transcription-dependent chromatin marks, and in particular Ub-H2B, on spliceosome assembly and function on nascent transcripts.
RESULTS
Defects in Ub-H2B, H3K4me, H3K36me cause introns to accumulate for distinct subsets of transcripts
Transcripts in S. cerevisiae do not generally undergo alternative splicing, but the constitutive splicing reaction is sensitive to a number of environmental perturbations (Pleiss et al., 2007; Munding et al., 2010; Bergkessel et al., 2011). While relatively few genes are spliced, intron-containing genes account for nearly one third of total cellular transcription (Ares et al., 1999), so it is critical for yeast to appropriately control the efficiency of this step in gene expression. We recently reported that in a genetic background sensitized by loss of Npl3, a protein known to promote splicing of a subset of genes, a short 37°C temperature shift revealed a modest dependence of pre-mRNA splicing on Ub-H2B (Moehle et al., 2012).
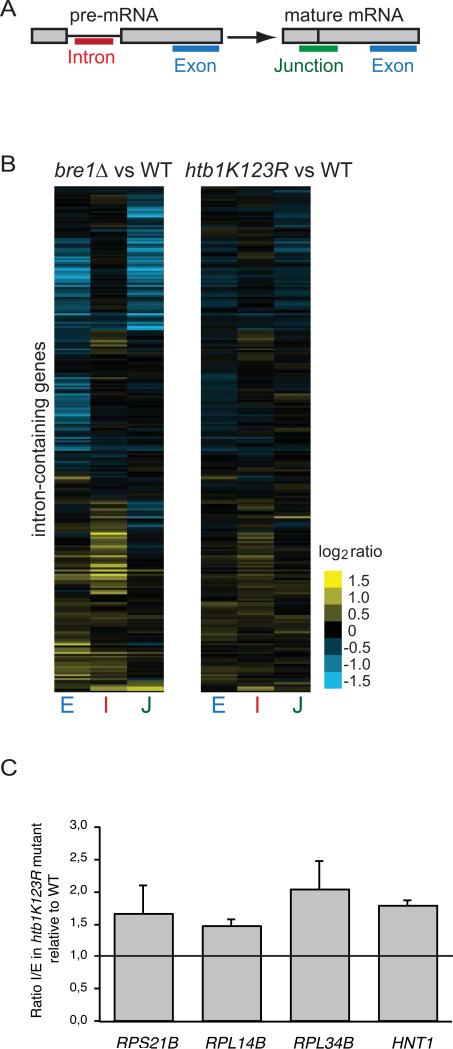
Here, we further explored the potential connection between chromatin modification and splicing by capitalizing on the observation that nuclear export factor assembly onto nascent mRNPs is very tightly regulated by Ub-H2B during a 3-hour shift to 39°C, an experimental condition that challenges mRNA biogenesis without affecting genome-wide expression (Babour et al., 2012; Vitaliano-Prunier et al., 2012). Indeed, using splicing-sensitive microarrays (figure 1A), we see that at 39°C, abrogating Ub-H2B by deleting the H2B E3 ligase, BRE1, or mutating the targeted residue in H2B (htb1K123R) led to increases in the levels of intron for many genes, consistent with a defect in the splicing of those transcripts (figure 1B and 2C, Table S3). To more easily compare these datasets, we calculated intron/exon ratios, an established approach to normalize for differences in transcription (Clark et al., 2002). We observed that, importantly, genes affected by H2B mutation extensively overlapped with genes affected by BRE1 deletion (figure 1D). While the ribosomal protein genes (RPGs) are a category of spliced genes often regulated together (Pleiss et al., 2007; Bergkessel et al., 2011), Ub-H2B-dependent effects on splicing were not enriched for RPG transcripts. Validation of the microarray data by using RT-qPCR to measure relative intron and exon abundance of several transcripts confirmed Ub-H2B-mediated changes with respect to a wild-type strain (figure 1C). Taken together, our data show that loss of Ub-H2B has clear gene-specific effects on intron accumulation and, thus, prompt the conclusion that Bre1-dependent Ub-H2B is important for splicing at 39°C.
Figure 1. Defects in Ub-H2B promote splicing defects.
(A) Schematic of probes contained on the microarray for each intron-containing gene. (B) Heat maps of log2 ratios of each gene feature in bre1Δ or htb1K123R compared to isogenic WT strains after a 3-hour shift to 39°C. Gene order is different for each sub-panel. (C) RT-qPCR measurements of unspliced mRNAs using single-locus RT-qPCR. Percent unspliced RNA is represented as fold change compared to wild-type.
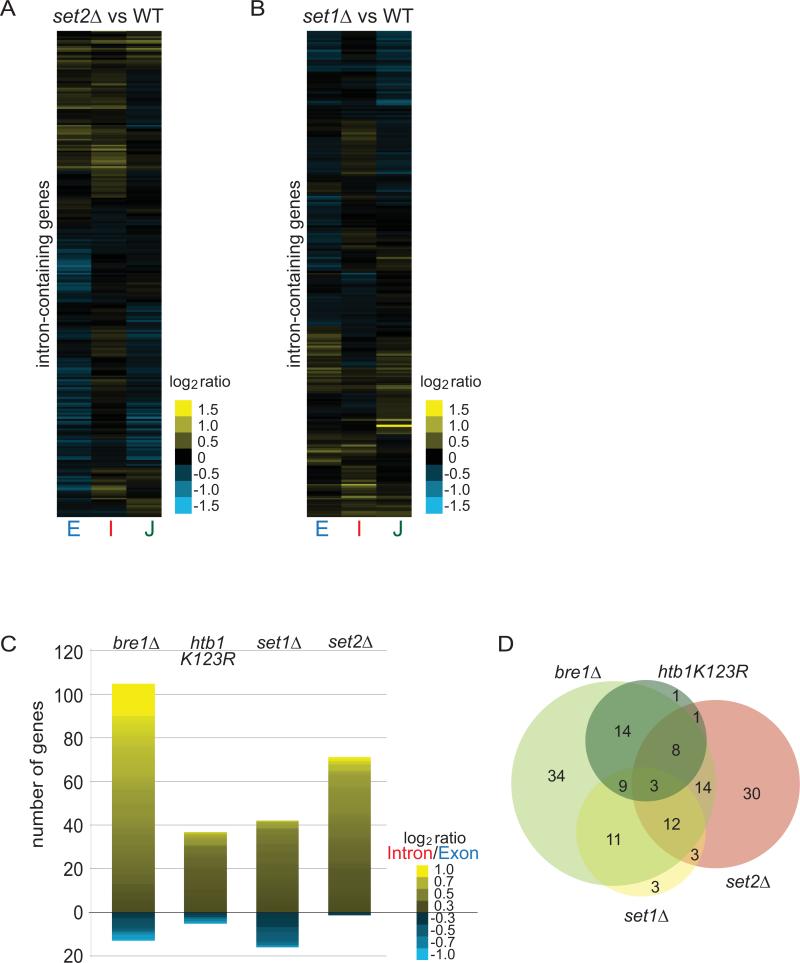
Figure 2. Defects in Ub-H2B, H3K4me, H3K36me cause introns to accumulate for distinct subsets of transcripts.
(A, B) Heat maps of log2 ratios of each gene feature in set2Δ or set1Δ strains compared to isogenic WT strains after a 3-hour shift to 39°C. Gene order is different for each sub-panel. (C) Histogram of number of genes exhibiting log2 (Intron/Exon) ratio greater than 0.3 or less than -0.3. Heat map within bar shows degree of splicing change of those genes. (D) Venn diagram of genes from histograms for comparison of each genotype directly. Note: set1Δ and htb1K123R have 1 gene overlap which cannot be shown. The circle sizes are representative of the numbers of genes with log2I/E>0.3 but the overlaps are not to scale.
Ub-H2B is strictly required for other histone marks such as the trimethylation of histone H3 on both lysine 4 by the Set1-containing COMPASS complex (Sun and Allis 2002), and lysine 79 by Dot1 (Briggs et al., 2002; Ng et al., 2002), and facilitates the Set2-mediated methylation of H3K36 on some intron-containing genes (Shieh et al., 2011) (not shown). Surprisingly, we found that deletion of SET2 also causes accumulation of intron and an increase in the intron/exon ratio for many transcripts (figure 2A and 2C), but the observation that 83% of Set2-dependent genes are not also dependent on Ub-H2B (figure 2D) suggests Set2 is working separately from the Ub-H2B pathway. Microarray results from a strain lacking SET1 were consistent with a mild splicing defect as gauged both by intron accumulation and intron/exon ratios (figure 2B and 2C). However only a small number of genes (13) overlap with those affected in htb1K123R (figure 2D), indicating that the effects of Ub-H2B on intron/exon ratios were not strictly mediated by H3K4 methylation. We observe a comparatively larger effect from BRE1 deletion than mutation of the H2B target residue, which is consistent with an additional Bre1 target or function that also promotes splicing. Bre1 targets many of the same genes as COMPASS component Set1, but whether ubiquitylation of the Swd2 component of the COMPASS complex by Bre1 might be involved in this process remains to be determined (Vitaliano-Prunier et al., 2008).
Importantly, no global changes of gene expression were observed upon inhibition of Ub-H2B or downstream H3 methylations either at 30°C (Lenstra et al., 2011; Margaritis et al., 2012) or 39°C (Vitaliano-Prunier et al., 2012). Only a minority of genes exhibited altered expression in the different mutant strains, none of which encoded components of the splicing machinery. This argues against direct transcriptional control of the splicing machinery expression by Ub-H2B.
Together, these results suggest that splicing efficiencies – inferred by changes in pre-mRNA and total mRNA levels – are dependent on contributions from multiple transcription-coupled histone marks, with the relative contribution being different from one intron-containing gene to another. We reasoned that the decrease in splicing efficiency we observed was unlikely to be caused by a wholesale block in spliceosome function, but rather could relate to a delay in the onset of the splicing reaction. We therefore sought to determine whether Ub-H2B might influence the ability of splicing factors to associate with transcripts.
Preventing ubiquitylation of H2B alters the recruitment of early splicing factors to Cap-binding complex-associated mRNPs
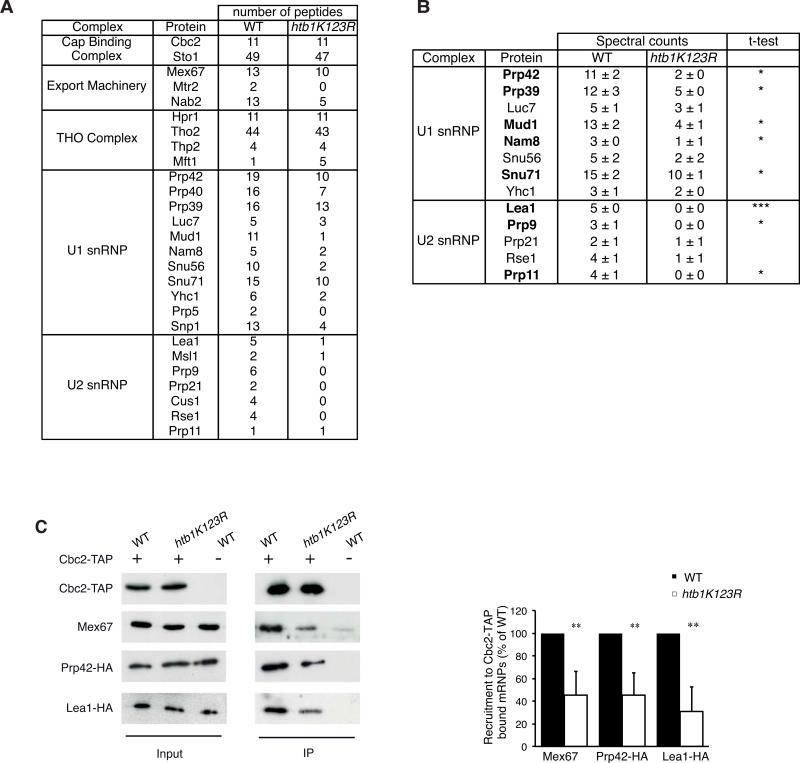
Since Ub-H2B-mediated splicing is likely mechanistically separated from that driven by downstream histone methylations, what step in splicing is impacted by loss of Ub-H2B? To address this question, nucle mRNPs were purified from temperature-shifted wild-type (WT) and htb1K123R cells using a genomically TAP-tagged Cbc2 of the nuclear cap-binding complex (CBC), and their proteomes were analyzed by tandem mass spectrometry (Oeffinger et al., 2007). Because the CBC is associated with nuclear mRNPs from early synthesis to nuclear exit, the composition of purified mRNPs reflects the sum of all biogenesis events, including transcription, mRNA processing and mRNA packaging that lead to their formation (Oeffinger et al., 2007). The proteome of CBC-associated mRNPs was enriched in splicing factors (snRNPs), 3' end processing machinery, mRNA export factors and other factors that are recruited during transcription elongation such as the THO complex (table S4; (Oeffinger et al., 2007)). A recent transcriptome-wide analysis of of CBC-associated mRNAs reveals a clear enrichment of unspliced versus spliced mRNA, consistent with an interaction occuring at an early step of transcription (Tuck and Tollervey 2013). In addition, multiple subunits of RNA Polymerase II (RNAPII) were detected in CBC-interacting mRNPs, confirming that some nascent transcripts were associated with TAP-tagged Cbc2 (table S4).
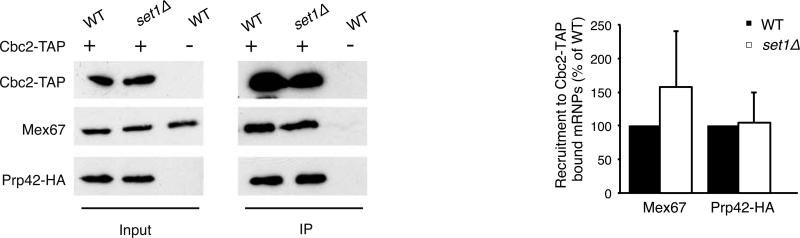
This global approach showed that preventing Ub-H2B impaired the association of the nuclear export machinery as previously described (Vitaliano-Prunier et al., 2012), but maintained WT levels of other factors known to bind mRNAs, including the transcription elongation THO complex (figure 3A, Table S4). Early spliceosome assembly onto premRNAs entails binding of the U1 and U2 snRNPs: consistent with the observed splicing defect, the number of peptides corresponding to U1 and U2 was significantly lower in the htb1K123R strain compared to WT, while the overall protein level of these factors was similar in both strains (figure 3A and 3C). This defect was also seen using a semi-quantitative analysis based on a spectral counting approach (Heintz et al., 2009) (figure 3B, Tables S5 and S6). To confirm this decrease in CBC-associated U1 and U2, we directly assayed the co-immunoprecipitation of an endogenously HA-tagged version of either Prp42 (U1 snRNP) or Lea1 (U2 snRNP) with TAP-tagged Cbc2 and observed a reproducible decrease in both Prp42-HA and Lea1-HA (figure 3C). While we cannot rule out that some factors pulled down by TAP-tagged Cbc2 might be directly bound to the CBC, as has been observed for the tri-snRNP in mammals (Pabis et al., 2013), the decreased association of U1 and U2 proteins in the pull-down reported here is consistent with the splicing defect seen in strains lacking Ub-H2B. In contrast, this defect was not phenocopied by deletion of SET1 (figure 4), further arguing that the control of U1 and U2 recruitment by Ub-H2B is not mediated by downstream methylation by Set1.
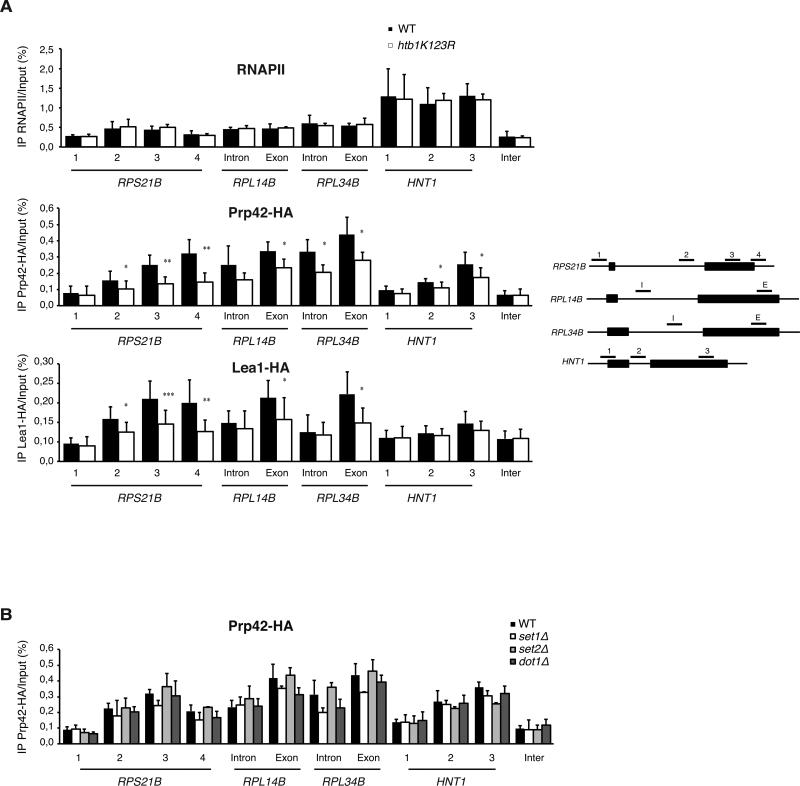
Figure 3. Preventing ubiquitylation of H2B alters the recruitment of early splicing factors to CBC-associated mRNPs.
(A) Nuclear mRNPs were purified from Cbc2-TAP-tagged WT or htb1K123R cells after a 3-hour shift to 39°C and associated proteins were analyzed by MS-MS after SDS-PAGE. In order to highlight relative differences in the protein composition, the number of unique peptides for each indicated protein has been considered as index of abundance (Merz et al., 2007). Complete identification of the CBC proteomes is shown in Table S4. (B) Components of the U1 and U2 snRNPs associated with nuclear mRNPs were semi-quantified using a spectral counting approach. The mean ± SD of spectral counts corresponding to three injections are indicated. Significance of the differences observed between both strains was evaluated using Student t-test (*P 0.01–0.05; ***P <0.001). Significant differences are indicated in bold. (C) Nuclear mRNPs were purified from WT or htb1K123R cells expressing Cbc2-TAP and Prp42-HA or Lea1-HA. Co-purifying proteins were detected by Western Blot with anti-HA or anti-Mex67 antibodies (left panel). The ratio of mRNP-associated proteins relative to the WT cells and to the immuno-purified Cbc2-TAP was determined from at least 3 independent experiments (mean ± SD) (right panel). Significance of the differences observed between both strains was evaluated using Student t-test (**P 0.001–0.01).
Figure 4. Loss of Set1-dependent methylation does not affect the recruitment of U1 snRNP to CBC-associated mRNPs.
Nuclear mRNPs were purified from WT or set1Δ cells expressing Cbc2-TAP and Prp42-HA. Co-purifying proteins were detected by Western Blot with anti-HA or anti-Mex67 antibodies (left panel). The ratio of mRNP-associated proteins relative to the WT cells and to the immuno-purified Cbc2-TAP was determined from at least 3 independent experiments (mean ± SD) (right panel). Significance of the differences observed between both strains was evaluated using Student t-test (**P 0.001–0.01).
Loss of H2B ubiquitylation impairs recruitment of early splicing factors to transcribing genes
It has been suggested that the splicing activity can be facilitated by the cotranscriptional recruitment of the splicing machinery (Tardiff et al., 2006; Aitken et al., 2011); thus, we reasoned that if the defect seen in recruitment of U1 and U2 to mRNPs occurs co-transcriptionally, this may further account for the intron accumulation phenotype of a strain lacking Ub-H2B. To ask this, we used chromatin immunoprecipitation (ChIP) to study early splicing factor association with on four intron-containing genes whose splicing is sensitive to loss of Ub-H2B: 3 ribosomal protein genes RPS21B, RPL14B and RPL34B and the non-ribosome protein gene HNT1 (Table S3). Of note, HNT1 splicing was also sensitive to deletion of SET1 or SET2 (Table S3). We found a marked decrease in Prp42 association at these genes in the htb1K123R strain. Importantly, this decrease in U1 association occurred at genes where RNAPII levels and resulting mRNA expression remained unchanged (figure 5A and data not shown), and does not occur in cells lacking the SET1, SET2 and DOT1 methyltransferases (figure 5B). Furthermore, in agreement with data from the CBC pulldown (figure 3), Lea1 exhibited weaker association with these genes in the htb1K123R strain, which was revealed primarily at the 3’ ends (figure 5A). Our data reveal that loss of Ub-H2B adversely affects recruitment of the early splicing machinery to nascent transcripts.
Figure 5. Loss of H2B ubiquitylation, but not Set1-, Set2- or Dot1- dependent methylation of H3, impairs recruitment of early splicing factors to transcribing genes.
ChIP experiments were performed on extracts prepared from the indicated HA-tagged strains after a 3-hour shift to 39°C, using anti-RNAPII or -HA antibodies. Four intron-containing genes were considered, 3 ribosome protein genes RPS21B, RPL14B and RPL34B and the non-ribosome protein gene HNT1 (Supplementary Table S1). Histograms depict the mean and standard deviations of at least three independent experiments. The significance of the differences of recruitment observed between WT and mutant cells was evaluated using Student t-test (*P 0.01–0.05; **P 0.001–0.01).
Loss of H2B ubiquitylation marginally affects recruitment of Npl3
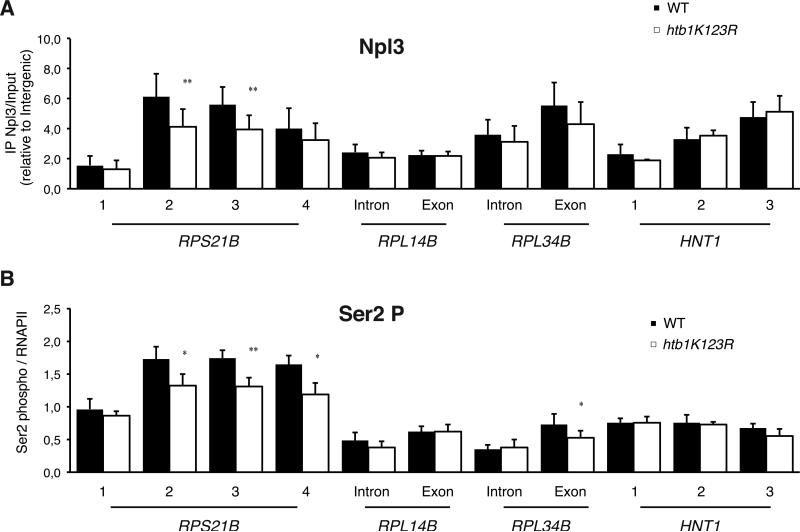
We previously showed that the SR protein Npl3 promotes U1 and U2 association with nascent transcripts and physically interacts with the Ub-H2B machinery (Kress et al., 2008; Moehle et al., 2012). While we do see a modest decrease in Npl3 ChIP (figure 6A) at genes whose splicing is promoted by Npl3 (Kress et al., 2008), this cannot account for the broad consequences of losing Ub-H2B on pre-mRNA splicing shown here, in particular on non-RPGs.
Figure 6. H2B ubiquitylation promotes to RNAPII Ser2 phosphorylation and Npl3 recruitment on a subset of intron-containing genes.
(A) Phosphorylation of RNAPII Ser2 on indicated intron-containing genes was analyzed by ChIP assay in WT and htb1K123R cells using antibodies to RNAPII and phosphoSer2 specific antibodies. The RNAPII Ser2P/RNAPII ratio is shown. (B) Recruitment of Npl3 on indicated intron-containing genes was analyzed by ChIP assay in WT and htb1K123R cells using anti Npl3 antibodies and normalized to the intergenic region. Significance of the differences observed between both strains was evaluated using Student t-test (*P 0.01– 0.05; **P 0.001–0.01).
Similar to SR proteins in metazoa, Npl3 is known to associate with the C-terminal domain (CTD) of RNAPII upon Serine 2 phosphorylation (Dermody et al., 2008) but we only observed a weak decrease in Serine 2 phosphorylation on the RNAPII CTD at RPS21B and RPL34B in the htb1K123R strain (figure 6B).
DISCUSSION
Early splicing factors associate with nascent pre-mRNAs (Kotovic et al., 2003; Gornemann et al., 2005; Lacadie and Rosbash 2005; Moore et al., 2006; Tardiff et al., 2006), and, therefore, it is critical to understand how the chromatin landscape contributes to cotranscriptional spliceosome assembly and function. Our results indicate that in budding yeast, an organism with relatively simple intron/exon architecture and limited splice site variation, multiple histone modifications (Ub-H2B, H3K4me and H3K36me) are required for optimal spliceosome function at distinct subsets of genes. In agreement with this, we also show that Ub-H2B facilitates the association of the early splicing machinery to mRNPs while the RNA is still being transcribed, in a molecular pathway that is distinct from Set1- or Set2-mediated splicing. Thus, our data support the idea that the chromatin landscape of a locus, as defined by these modifications, can impact the fate of a transcript, reflecting the capacity of the spliceosome to interpret and integrate multiple inputs from the chromatin landscape. This idea is further supported by evidence that histone acetylation by Gcn5 and deacetylation by Hos2/3 (Gunderson and Johnson 2009; Gunderson et al., 2011), as well as incorporation of the variant histone H2A.Z (Albulescu et al., 2012) can also influence spliceosome assembly onto nascent transcripts and splicing efficiency of those transcripts.
In metazoa, where the recognition of and discrimination between alternative, degenerate splice sites must be tightly regulated, histone modifications, including Ub-H2B, have been shown to reflect intron/exon structure (Kolasinska-Zwierz et al., 2009; Schwartz et al., 2009; Spies et al., 2009; Dhami et al., 2010; Huff et al., 2010; Jung et al., 2012). In fact, modulation of the levels of the Ub-H2B machinery in human cells has revealed that this mark promotes recognition of splice sites, and importantly, as we show here in budding yeast, does so in context-dependent ways (Figures 1-3; (Jung et al., 2012; Zhang et al., 2013). While Ub-H2B also reflects intron/exon structure on budding yeast genes (Shieh et al., 2011), it is remarkable that the one to two differentially marked nucleosomes associated with yeast introns, typically 100-400bp in budding yeast (Spingola et al., 1999), can impact splicing. The conservation of the connection between Ub-H2B and splicing from yeast to humans suggests that this is a universal strategy for spliceosome regulation.
How does Ub-H2B promote splicing factor association? It is possible that Ub-H2B-mediated spliceosome association is, in part, influenced by a minor impairment in the recruitment of the CBC to some genes (not shown), as the CBC and early splicing factors physically interact in yeast (Colot et al., 1996). In addition, it has been recently shown that CBC depletion in mammalian cells led to defects in co-transcriptional spliceosome assembly via both RNA-dependent interaction with U1 and U2 snRNPs and RNA-independent interactions with U4/U6 and U5 snRNPs (Pabis et al., 2013). We could barely detect components of the U4/U6 and U5 snRNPs in our yeast CBC proteome, suggesting that this interaction may not occur in yeast. However, Pabis et al. hypothesized that CBC, U1 and U2 bind cooperatively onto RNA to promote splicing efficiency (Pabis et al., 2013), a process that could be influenced by Ub-H2B. Conversely, Ub-H2B and H3K36 methylation levels are highly dependent on an intact CBC (Hossain et al., 2009; Hossain et al., 2013), highlighting two examples of the high degree of coupling between mRNA processing and chromatin structure.
In metazoans, it has been suggested that histone modifications may directly recruit splicing factors via specific histone mark-specific adaptors (Sims et al., 2007; Luco et al., 2010); this has been previously proposed to occur in yeast as well (Gunderson et al., 2011). Thus it is possible that ubiquitylated H2B also directly or indirectly recruits a splicing factor in budding yeast. Alternatively, as histone modifications can influence RNAPII dynamics, it is possible that changes in elongation speed may explain the splicing defects seen in the chromatin mutants tested here (Howe et al., 2003; Braberg et al., 2013; Dujardin et al., 2013). Therefore, it is interesting that multiple reports have suggested that the ubiquitin modification stabilizes nucleosomes during elongation (Chandrasekharan et al., 2009); given the splicing defects that can occur when RNA polymerase elongates too quickly (Braberg et al., 2013), perhaps uncontrolled RNAPII elongation in the htb1K123R strain accounts for the decrease in splicing efficiency reported here.
An intriguing aspect of chromatin-based splicing regulation is the complexity of the chromatin template; as our microarrays revealed, Ub-H2B, H3K4me and H3K36me influence splicing at somewhat overlapping subsets of genes. As the field moves forward, a challenge will be to understand how the spliceosome integrates the signals from individual histone modifications to achieve the appropriate splicing outcomes for the needs of the cell. The many histone post-translational modifications, and their combined effects on transcription, RNAPII CTD phosphorylation state, and mRNA processing perhaps provide each emerging transcript with a specific mRNA “identity” that allows dynamic modulation of splicing, mRNA export and quality control.
MATERIALS AND METHODS
Yeast strains and culture
Strains used in this study are listed in Supplementary Table 1. The derivative strains were obtained using PCR based homologous recombination as described in (Longtine et al., 1998). Yeast cells were grown overnight at 30°C in YPD medium (Yeast extract, peptone, dextrose). To perform the 3-hours shift at 39°C, when the 30°C overnight culture reached OD600=1, one volume of medium preheated to 48°C was added and cultures were incubated at 39°C for 3 hours.
Microarray analysis
Strains were grown for microarray analysis as described above with a 3-hour shift to 39°C, at which point they were collected by centrifugation. Microarrays were performed as in (Moehle et al., 2012). For each microarray shown, results from two biological replicates were averaged. In addition, each biological replicate contains 6 technical replicates per probe, as well as dye-flipped replicates. The heat maps in Figure 1 were created using Java Treeview (Saldanha 2004) and show the log2-based fold change in the indicated strain as compared to an isogenic WT. Intron/Exon ratios were calculated for each intron-containing gene (log2 (Intron/Exon) = log2 (Intronmutant/IntronWT) – log2 (Exonmutant/ExonWT); these values were converted into a histogram for any gene with Intron/Exon value <-0.3 or >0.3.
RNA Isolation and RT-qPCR
Total RNA isolation was performed by the hot acid phenol method (Sigma Aldrich). cDNA from total RNAs were obtained by retro-transcription with random oligonucleotides (Roche) using the SuperScriptTM II reverse Transcriptase (Invitrogen). Real time qPCR was then performed using the SYBR Green mix (Roche) and the Light Cycler 480 system (Roche) with gene specific primers described in Supplementary Table 2.
Antibodies
Commercial antibodies used in this study were anti-RNA polymerase II (RNAPII ChIP 12 μg, 8WG16, MMS126R, Covance), anti-phosphorylated Ser 2 of RNAPII CTD (ChIP 12 μg, clone 3E10, 04-1571, Millipore), anti-HA (ChIP 8 μg, WB 1:2000, clone HA-11, MMS-101R, Covance), anti-H3 (ChIP 4 μg, ab1791, abcam), anti-H3K36me3 (ChIP 4 μg, ab9050, abcam). Polyclonal anti-Mex67 (WB 1:20,000) and anti-Npl3 (ChIP 1,5 μL, WB 1:10,000) antibodies were previously described (Siebel and Guthrie 1996; Gwizdek et al., 2005). TAP-tagged proteins were immunoprecipitated using IgG sepharose beads (50 μL for ChIP analysis). Western blot analyses were performed using appropriate HRP-coupled secondary antibodies and chemi-luminescence protein immunoblotting reagents (Pierce).
Immunoaffinity Purification of nuclear mRNPs from frozen cell grindate
Cells shifted at 39°C for 3hrs in YPD were rapidly frozen in liquid nitrogen before cryolysis (Ossareh-Nazari et al., 2010). Immunoaffinity purification of mRNPs was performed as described in (Oeffinger et al., 2007) with minor modification. Frozen cell grindates were rapidly thawed into nine volumes of RNP buffer (20mM Hepes, pH7.4, 110mM KOAc, 0.5%Triton, 0.1% Tween 20, 1:100 Solution P, 1:5000 RNasin (Promega), 1:5000 Antifoam B (Sigma); 1:1000 DTT). The resulting extracts were centrifuged 5 min at low speed to eliminate the large cellular debris before clarification by filtration. Immunoprecipitation was performed using magnetic beads (Dynal) conjugated with rabbit IgG (Sigma) (Alber et al., 2007). Resulting eluates were lyophilized, resuspended in SDS-PAGE sample buffer, separated by SDS-PAGE on a 4–12% NuPAGE Novex Bis-Tris precast gel (Invitrogen) according to the manufacturer's specifications and visualized by Coomassie blue staining.
Alternatively, nuclear mRNPs were purified from glass-beads lyzed cells as previously described (Iglesias et al., 2010; Vitaliano-Prunier et al., 2012).
SDS-PAGE and in-gel digestion
After electrophoresis, gels were stained by colloidal Coomassie Blue (BioSafe coomassie stain; Bio-Rad) and whole lanes were cut into 14 5x4 mm slices using a disposable grid-cutter (the gel-company, Tübingen, Germany). Slices were divided into three and in-gel digestion using trypsin (Promega, Madison, WI) was performed overnight at 37 °C, after in-gel reduction and alkylation using the MassPrep Station (Waters, Milford, MA, USA). Tryptic peptides were extracted using 60 % ACN in 0.1 % formic acid for 1h at room temperature. The volume was reduced in a vacuum centrifuge and adjusted to 10 μl using 0.1 % formic acid in water before nanoLC-MS/MS (nanoliquid chromatography coupled to tandem mass spectrometry) analysis (see Supplementary information).
Liquid chromatography/Mass spectrometry
NanoLC-MS/MS was performed using a nanoACQUITY ultra performance liquid chromatography (UPLC®) system (Waters, Milford, MA, USA) coupled to a maXis 4G QTOF mass spectrometer (BrukerDaltonics, Bremen, Germany). Detailed procedure is presented in Supplementary information.
Peptide counts
The peptide count of unique peptides was performed using Scaffold 3 software (version 3.6.5; Proteome Software Inc., Portland, OR, USA) after having filtered the results at 1% < FDR.
Spectral counts
The label-free semi-quantitation was performed using Scaffold 3 software (version 3.6.5; Proteome Software Inc., Portland, OR, USA) exporting the un-weighed spectral count into an excel file after having filtered the result at 1% < FDR as described in the Supplementary data. The three replicates of each biological sample shown in Table S6 (except the negative control) were horizontally normalized to the highest spectral count value obtained for nuclear cap-binding protein complex subunit (WT injection 1: 429 un-weighed spectra counts). The following procedure as described in (Gokce et al., 2011; Miguet et al., 2013) has been applied for the six independent injections: the number of spectra obtained for the nuclear cap-binding complex has been divided by 429 and the ratio obtained was multiplied independently by the number of spectral counts for each protein, in order to reduce the variance observed between samples and replicates.
Chromatin Immunoprecipitation (ChIP) and qPCR
ChIPs were performed as described previously (Gwizdek et al., 2006). 8 OD units of cell lysate were immunoprecipitated with the amount of antibodies mentioned above. Immunoprecipitated DNA was analyzed by quantitative PCR using primers referenced in Table S2. Non-specific signals were assessed by analyzing immunoprecipitated DNA with primers against an intergenic region. The specificity of the signal was also evaluated using untagged strains. The resulting amplifications were similar to those observed for the intergenic region (not shown). Results correspond to the mean of at least 3 independent experiments.
Supplementary Material
ACKNOWLEDGEMENTS
We are grateful to Thanasis Margaritis and Frank Holstege for helpful advices. This study was supported by the National Research Agency (grant 2010 BLAN1227-01 to CD), the Who am I? laboratory of excellence [ANR-11-LABX-0071 to CD] funded by the “Investments for the Future” program [ANR-11-IDEX-0005-01], the NIH grant NIHGM21119 to CG, the Proteomic French Infrastructure [ProFI to AVD and CSR] and the Fondation pour la Recherche Médicale [to AVD and CSR]. CG is an American Cancer Society Research Professor of Molecular Genetics. LH is supported by the University Paris Descartes.
ABBREVIATIONS
- CBC
cap-binding complex
- ChIP
Chromatin Immunoprecipitation
- CTD
C-terminal domain
- H3K4me
Lysine 4 methylation of Histone H3
- mRNP
messenger ribonucleoprotein
- RNAPII
RNA Polymerase II
- RPG
ribosomal protein gene
- snRNP
small nuclear ribonucleoprotein
- Ub-H2B
histone H2B mono-ubiquitylation
- WT
wild-type
Footnotes
Author Contribution
LH, EAM, DB performed experiments; LH, EAM, DB, AVD, CS, CG, CD designed the experiments and analyzed the data; CD, EAM, and CG wrote the manuscript.
SUPPLEMENTAL MATERIAL
Supplemental material is available for this article
The authors declare that they have no conflict of interest.
REFERENCES
- Aitken S, Alexander RD, Beggs JD. Modelling reveals kinetic advantages of co-transcriptional splicing. PLoS Comput Biol. 2011;7:e1002215. doi: 10.1371/journal.pcbi.1002215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alber F, Dokudovskaya S, Veenhoff LM, Zhang W, Kipper J, Devos D, Suprapto A, Karni-Schmidt O, Williams R, Chait BT, Sali A, Rout MP. The molecular architecture of the nuclear pore complex. Nature. 2007;450:695–701. doi: 10.1038/nature06405. [DOI] [PubMed] [Google Scholar]
- Albulescu LO, Sabet N, Gudipati M, Stepankiw N, Bergman ZJ, Huffaker TC, Pleiss JA. A quantitative, high-throughput reverse genetic screen reveals novel connections between Pre-mRNA splicing and 5′ and 3′ end transcript determinants. PLoS Genet. 2012;8:e1002530. doi: 10.1371/journal.pgen.1002530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ares M, Jr., Grate L, Pauling MH. A handful of intron-containing genes produces the lion's share of yeast mRNA. RNA. 1999;5:1138–1139. doi: 10.1017/s1355838299991379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babour A, Dargemont C, Stutz F. Ubiquitin and assembly of export competent mRNP. Biochim Biophys Acta. 2012;1819:521–530. doi: 10.1016/j.bbagrm.2011.12.006. [DOI] [PubMed] [Google Scholar]
- Bergkessel M, Whitworth GB, Guthrie C. Diverse environmental stresses elicit distinct responses at the level of pre-mRNA processing in yeast. RNA. 2011;17:1461–1478. doi: 10.1261/rna.2754011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braberg H, Jin H, Moehle EA, Chan YA, Wang S, Shales M, Benschop JJ, Morris JH, Qiu C, Hu F, Tang LK, Fraser JS, Holstege FC, Hieter P, Guthrie C, Kaplan CD, Krogan NJ. From Structure to Systems: High-Resolution, Quantitative Genetic Analysis of RNA Polymerase II. Cell. 2013;154:775–788. doi: 10.1016/j.cell.2013.07.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Briggs SD, Xiao T, Sun ZW, Caldwell JA, Shabanowitz J, Hunt DF, Allis CD, Strahl BD. Gene silencing: trans-histone regulatory pathway in chromatin. Nature. 2002;418:498. doi: 10.1038/nature00970. [DOI] [PubMed] [Google Scholar]
- Carrillo Oesterreich F, Bieberstein N, Neugebauer KM. Pause locally, splice globally. Trends Cell Biol. 2011;21:328–335. doi: 10.1016/j.tcb.2011.03.002. [DOI] [PubMed] [Google Scholar]
- Carrillo Oesterreich F, Preibisch S, Neugebauer KM. Global analysis of nascent RNA reveals transcriptional pausing in terminal exons. Mol Cell. 2010;40:571–581. doi: 10.1016/j.molcel.2010.11.004. [DOI] [PubMed] [Google Scholar]
- Chandrasekharan MB, Huang F, Sun ZW. Ubiquitination of histone H2B regulates chromatin dynamics by enhancing nucleosome stability. Proc Natl Acad Sci U S A. 2009;106:16686–16691. doi: 10.1073/pnas.0907862106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark TA, Sugnet CW, Ares M., Jr. Genomewide analysis of mRNA processing in yeast using splicing-specific microarrays. Science. 2002;296:907–910. doi: 10.1126/science.1069415. [DOI] [PubMed] [Google Scholar]
- Colot HV, Stutz F, Rosbash M. The yeast splicing factor Mud13p is a commitment complex component and corresponds to CBP20, the small subunit of the nuclear cap-binding complex. Genes Dev. 1996;10:1699–1708. doi: 10.1101/gad.10.13.1699. [DOI] [PubMed] [Google Scholar]
- Dermody JL, Dreyfuss JM, Villen J, Ogundipe B, Gygi SP, Park PJ, Ponticelli AS, Moore CL, Buratowski S, Bucheli ME. Unphosphorylated SR-like protein Npl3 stimulates RNA polymerase II elongation. PLoS One. 2008;3:e3273. doi: 10.1371/journal.pone.0003273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhami P, Saffrey P, Bruce AW, Dillon SC, Chiang K, Bonhoure N, Koch CM, Bye J, James K, Foad NS, Ellis P, Watkins NA, Ouwehand WH, Langford C, Andrews RM, Dunham I, Vetrie D. Complex exon-intron marking by histone modifications is not determined solely by nucleosome distribution. PLoS One. 2010;5:e12339. doi: 10.1371/journal.pone.0012339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dujardin G, Lafaille C, Petrillo E, Buggiano V, Gomez Acuna LI, Fiszbein A, Godoy Herz MA, Nieto Moreno N, Munoz MJ, Allo M, Schor IE, Kornblihtt AR. Transcriptional elongation and alternative splicing. Biochim Biophys Acta. 2013;1829:134–140. doi: 10.1016/j.bbagrm.2012.08.005. [DOI] [PubMed] [Google Scholar]
- Gokce E, Shuford CM, Franck WL, Dean RA, Muddiman DC. Evaluation of normalization methods on GeLC-MS/MS label-free spectral counting data to correct for variation during proteomic workflows. J Am Soc Mass Spectrom. 2011;22:2199–2208. doi: 10.1007/s13361-011-0237-2. [DOI] [PubMed] [Google Scholar]
- Gornemann J, Kotovic KM, Hujer K, Neugebauer KM. Cotranscriptional spliceosome assembly occurs in a stepwise fashion and requires the cap binding complex. Mol Cell. 2005;19:53–63. doi: 10.1016/j.molcel.2005.05.007. [DOI] [PubMed] [Google Scholar]
- Gunderson FQ, Johnson TL. Acetylation by the transcriptional coactivator Gcn5 plays a novel role in co-transcriptional spliceosome assembly. PLoS Genet. 2009;5:e1000682. doi: 10.1371/journal.pgen.1000682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunderson FQ, Merkhofer EC, Johnson TL. Dynamic histone acetylation is critical for cotranscriptional spliceosome assembly and spliceosomal rearrangements. Proc Natl Acad Sci U S A. 2011;108:2004–2009. doi: 10.1073/pnas.1011982108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gwizdek C, Hobeika M, Kus B, Ossareh-Nazari B, Dargemont C, Rodriguez MS. The mRNA nuclear export factor Hpr1 is regulated by Rsp5-mediated ubiquitylation. J Biol Chem. 2005;280:13401–13405. doi: 10.1074/jbc.C500040200. [DOI] [PubMed] [Google Scholar]
- Gwizdek C, Iglesias N, Rodriguez MS, Ossareh-Nazari B, Hobeika M, Divita G, Stutz F, Dargemont C. Ubiquitin-associated domain of Mex67 synchronizes recruitment of the mRNA export machinery with transcription. Proc Natl Acad Sci U S A. 2006;103:16376–16381. doi: 10.1073/pnas.0607941103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heintz D, Gallien S, Wischgoll S, Ullmann AK, Schaeffer C, Kretzschmar AK, van Dorsselaer A, Boll M. Differential membrane proteome analysis reveals novel proteins involved in the degradation of aromatic compounds in Geobacter metallireducens. Mol Cell Proteomics. 2009;8:2159–2169. doi: 10.1074/mcp.M900061-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hnilicova J, Stanek D. Where splicing joins chromatin. Nucleus. 2011;2:182–188. doi: 10.4161/nucl.2.3.15876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hossain MA, Chung C, Pradhan SK, Johnson TL. The yeast cap binding complex modulates transcription factor recruitment and establishes proper histone H3K36 trimethylation during active transcription. Mol Cell Biol. 2013;33:785–799. doi: 10.1128/MCB.00947-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hossain MA, Claggett JM, Nguyen T, Johnson TL. The cap binding complex influences H2B ubiquitination by facilitating splicing of the SUS1 pre-mRNA. RNA. 2009;15:1515–1527. doi: 10.1261/rna.1540409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howe KJ, Kane CM, Ares M., Jr. Perturbation of transcription elongation influences the fidelity of internal exon inclusion in Saccharomyces cerevisiae. RNA. 2003;9:993–1006. doi: 10.1261/rna.5390803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huff JT, Plocik AM, Guthrie C, Yamamoto KR. Reciprocal intronic and exonic histone modification regions in humans. Nat Struct Mol Biol. 2010;17:1495–1499. doi: 10.1038/nsmb.1924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iglesias N, Tutucci E, Gwizdek C, Vinciguerra P, Von Dach E, Corbett AH, Dargemont C, Stutz F. Ubiquitin-mediated mRNP dynamics and surveillance prior to budding yeast mRNA export. Genes Dev. 2010;24:1927–1938. doi: 10.1101/gad.583310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung I, Kim SK, Kim M, Han YM, Kim YS, Kim D, Lee D. H2B monoubiquitylation is a 5′-enriched active transcription mark and correlates with exon-intron structure in human cells. Genome Res. 2012;22:1026–1035. doi: 10.1101/gr.120634.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolasinska-Zwierz P, Down T, Latorre I, Liu T, Liu XS, Ahringer J. Differential chromatin marking of introns and expressed exons by H3K36me3. Nat Genet. 2009;41:376–381. doi: 10.1038/ng.322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotovic KM, Lockshon D, Boric L, Neugebauer KM. Cotranscriptional recruitment of the U1 snRNP to intron-containing genes in yeast. Mol Cell Biol. 2003;23:5768–5779. doi: 10.1128/MCB.23.16.5768-5779.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kress TL, Krogan NJ, Guthrie C. A single SR-like protein, Npl3, promotes pre-mRNA splicing in budding yeast. Mol Cell. 2008;32:727–734. doi: 10.1016/j.molcel.2008.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lacadie SA, Rosbash M. Cotranscriptional spliceosome assembly dynamics and the role of U1 snRNA:5′ss base pairing in yeast. Mol Cell. 2005;19:65–75. doi: 10.1016/j.molcel.2005.05.006. [DOI] [PubMed] [Google Scholar]
- Lenstra TL, Benschop JJ, Kim T, Schulze JM, Brabers NA, Margaritis T, van de Pasch LA, van Heesch SA, Brok MO, Groot Koerkamp MJ, Ko CW, van Leenen D, Sameith K, van Hooff SR, Lijnzaad P, Kemmeren P, Hentrich T, Kobor MS, Buratowski S, Holstege FC. The specificity and topology of chromatin interaction pathways in yeast. Mol Cell. 2011;42:536–549. doi: 10.1016/j.molcel.2011.03.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longtine MS, McKenzie A, 3rd, Demarini DJ, Shah NG, Wach A, Brachat A, Philippsen P, Pringle JR. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast. 1998;14:953–961. doi: 10.1002/(SICI)1097-0061(199807)14:10<953::AID-YEA293>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- Luco RF, Allo M, Schor IE, Kornblihtt AR, Misteli T. Epigenetics in alternative pre-mRNA splicing. Cell. 2011;144:16–26. doi: 10.1016/j.cell.2010.11.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luco RF, Pan Q, Tominaga K, Blencowe BJ, Pereira-Smith OM, Misteli T. Regulation of alternative splicing by histone modifications. Science. 2010;327:996–1000. doi: 10.1126/science.1184208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margaritis T, Oreal V, Brabers N, Maestroni L, Vitaliano-Prunier A, Benschop JJ, van Hooff S, van Leenen D, Dargemont C, Geli V, Holstege FC. Two distinct repressive mechanisms for histone 3 lysine 4 methylation through promoting 3′-end antisense transcription. PLoS Genet. 2012;8:e1002952. doi: 10.1371/journal.pgen.1002952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merz C, Urlaub H, Will CL, Luhrmann R. Protein composition of human mRNPs spliced in vitro and differential requirements for mRNP protein recruitment. RNA. 2007;13:116–128. doi: 10.1261/rna.336807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miguet L, Lennon S, Baseggio L, Traverse-Glehen A, Berger F, Perrusson N, Chenard MP, Galoisy AC, Eischen A, Mayeur-Rousse C, Maar A, Fornecker L, Herbrecht R, Felman P, Van Dorsselaer A, Carapito C, Cianferani S, Mauvieux L. Cell-surface expression of the TLR homolog CD180 in circulating cells from splenic and nodal marginal zone lymphomas. Leukemia. 2013;27:1748–1750. doi: 10.1038/leu.2013.3. [DOI] [PubMed] [Google Scholar]
- Moehle EA, Ryan CJ, Krogan NJ, Kress TL, Guthrie C. The Yeast SR-Like Protein Npl3 Links Chromatin Modification to mRNA Processing. PLoS Genet. 2012;8:e1003101. doi: 10.1371/journal.pgen.1003101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore MJ, Schwartzfarb EM, Silver PA, Yu MC. Differential recruitment of the splicing machinery during transcription predicts genome-wide patterns of mRNA splicing. Mol Cell. 2006;24:903–915. doi: 10.1016/j.molcel.2006.12.006. [DOI] [PubMed] [Google Scholar]
- Munding EM, Igel AH, Shiue L, Dorighi KM, Trevino LR, Ares M., Jr. Integration of a splicing regulatory network within the meiotic gene expression program of Saccharomyces cerevisiae. Genes Dev. 2010;24:2693–2704. doi: 10.1101/gad.1977410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng HH, Xu RM, Zhang Y, Struhl K. Ubiquitination of histone H2B by Rad6 is required for efficient Dot1-mediated methylation of histone H3 lysine 79. J Biol Chem. 2002;277:34655–34657. doi: 10.1074/jbc.C200433200. [DOI] [PubMed] [Google Scholar]
- Nino CA, Herissant L, Babour A, Dargemont C. mRNA Nuclear Export in Yeast. Chem Rev. 2013 doi: 10.1021/cr400002g. [DOI] [PubMed] [Google Scholar]
- Oeffinger M, Wei KE, Rogers R, DeGrasse JA, Chait BT, Aitchison JD, Rout MP. Comprehensive analysis of diverse ribonucleoprotein complexes. Nat Methods. 2007;4:951–956. doi: 10.1038/nmeth1101. [DOI] [PubMed] [Google Scholar]
- Ossareh-Nazari B, Bonizec M, Cohen M, Dokudovskaya S, Delalande F, Schaeffer C, Van Dorsselaer A, Dargemont C. Cdc48 and Ufd3, new partners of the ubiquitin protease Ubp3, are required for ribophagy. EMBO Rep. 2010;11:548–554. doi: 10.1038/embor.2010.74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pabis M, Neufeld N, Steiner MC, Bojic T, Shav-Tal Y, Neugebauer KM. The nuclear cap-binding complex interacts with the U4/U6.U5 tri-snRNP and promotes spliceosome assembly in mammalian cells. RNA. 2013;19:1054–1063. doi: 10.1261/rna.037069.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pleiss JA, Whitworth GB, Bergkessel M, Guthrie C. Rapid, transcript-specific changes in splicing in response to environmental stress. Mol Cell. 2007;27:928–937. doi: 10.1016/j.molcel.2007.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robzyk K, Recht J, Osley MA. Rad6-dependent ubiquitination of histone H2B in yeast. Science. 2000;287:501–504. doi: 10.1126/science.287.5452.501. [DOI] [PubMed] [Google Scholar]
- Saldanha AJ. Java Treeview--extensible visualization of microarray data. Bioinformatics. 2004;20:3246–3248. doi: 10.1093/bioinformatics/bth349. [DOI] [PubMed] [Google Scholar]
- Schwartz S, Meshorer E, Ast G. Chromatin organization marks exonintron structure. Nat Struct Mol Biol. 2009;16:990–995. doi: 10.1038/nsmb.1659. [DOI] [PubMed] [Google Scholar]
- Shieh GS, Pan CH, Wu JH, Sun YJ, Wang CC, Hsiao WC, Lin CY, Tung L, Chang TH, Fleming AB, Hillyer C, Lo YC, Berger SL, Osley MA, Kao CF. H2B ubiquitylation is part of chromatin architecture that marks exon-intron structure in budding yeast. BMC Genomics. 2011;12:627. doi: 10.1186/1471-2164-12-627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siebel CW, Guthrie C. The essential yeast RNA binding protein Np13p is methylated. Proc Natl Acad Sci U S A. 1996;93:13641–13646. doi: 10.1073/pnas.93.24.13641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sims RJ, 3rd, Millhouse S, Chen CF, Lewis BA, Erdjument-Bromage H, Tempst P, Manley JL, Reinberg D. Recognition of trimethylated histone H3 lysine 4 facilitates the recruitment of transcription postinitiation factors and pre-mRNA splicing. Mol Cell. 2007;28:665–676. doi: 10.1016/j.molcel.2007.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spies N, Nielsen CB, Padgett RA, Burge CB. Biased chromatin signatures around polyadenylation sites and exons. Mol Cell. 2009;36:245–254. doi: 10.1016/j.molcel.2009.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spingola M, Grate L, Haussler D, Ares M., Jr. Genome-wide bioinformatic and molecular analysis of introns in Saccharomyces cerevisiae. RNA. 1999;5:221–234. doi: 10.1017/s1355838299981682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suganuma T, Workman JL. Signals and combinatorial functions of histone modifications. Annu Rev Biochem. 2011;80:473–499. doi: 10.1146/annurev-biochem-061809-175347. [DOI] [PubMed] [Google Scholar]
- Sun ZW, Allis CD. Ubiquitination of histone H2B regulates H3 methylation and gene silencing in yeast. Nature. 2002;418:104–108. doi: 10.1038/nature00883. [DOI] [PubMed] [Google Scholar]
- Tardiff DF, Lacadie SA, Rosbash M. A genome-wide analysis indicates that yeast pre-mRNA splicing is predominantly posttranscriptional. Mol Cell. 2006;24:917–929. doi: 10.1016/j.molcel.2006.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuck AC, Tollervey D. A Transcriptome-wide Atlas of RNP Composition Reveals Diverse Classes of mRNAs and lncRNAs. Cell. 2013;154:996–1009. doi: 10.1016/j.cell.2013.07.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vitaliano-Prunier A, Babour A, Herissant L, Apponi L, Margaritis T, Holstege FC, Corbett AH, Gwizdek C, Dargemont C. H2B ubiquitylation controls the formation of export-competent mRNP. Mol Cell. 2012;45:132–139. doi: 10.1016/j.molcel.2011.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vitaliano-Prunier A, Menant A, Hobeika M, Geli V, Gwizdek C, Dargemont C. Ubiquitylation of the COMPASS component Swd2 links H2B ubiquitylation to H3K4 trimethylation. Nat Cell Biol. 2008;10:1365–1371. doi: 10.1038/ncb1796. [DOI] [PubMed] [Google Scholar]
- Wood A, Krogan NJ, Dover J, Schneider J, Heidt J, Boateng MA, Dean K, Golshani A, Zhang Y, Greenblatt JF, Johnston M, Shilatifard A. Bre1, an E3 ubiquitin ligase required for recruitment and substrate selection of Rad6 at a promoter. Mol Cell. 2003;11:267–274. doi: 10.1016/s1097-2765(02)00802-x. [DOI] [PubMed] [Google Scholar]
- Zhang Z, Jones A, Joo HY, Zhou D, Cao Y, Chen S, Erdjument-Bromage H, Renfrow M, He H, Tempst P, Townes TM, Giles KE, Ma L, Wang H. USP49 deubiquitinates histone H2B and regulates cotranscriptional pre-mRNA splicing. Genes Dev. 2013;27:1581–1595. doi: 10.1101/gad.211037.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.