Abstract
Purpose
Vigorous physical activity after diagnosis of localized prostate cancer may reduce risk of disease progression and prostate cancer-specific mortality. The molecular mechanisms by which physical activity may exert protective effects in the prostate remain unknown.
Methods
We examined the associations between self-reported physical activity and gene expression patterns in morphologically normal prostate tissue of 71 men with low risk prostate cancer on active surveillance. Differential gene expression, gene set, and pathway analyses were performed comparing dichotomous groups defined by type, intensity, and amount of physical activity reported.
Results
Cell cycling and DNA repair pathways were up-regulated in men who participated in ≥3 h/wk vigorous activity compared to men who did not. In addition, canonical pathways involved in cell signaling and metabolism, the cellular effects of sildenafil (Viagra), and the Nrf2-mediated oxidative stress response were modulated in men who reported ≥3 h/wk of vigorous activity. Differential expression analysis at the individual gene level revealed modest differences between men who performed vigorous activity for ≥3 h/wk and those who did not. There were no differences in prostate gene expression in comparisons of exercise groupings that did not consider both duration and intensity of activity.
Conclusions
Prostate gene expression and pathway analyses revealed sets of transcripts that may be modulated in normal prostate tissue by participating in ≥3 h/wk of vigorous activity after diagnosis of low risk prostate cancer. These findings suggest potential biologic mechanisms by which vigorous activity may reduce risk of prostate cancer progression, and warrant further study and validation.
Keywords: physical activity, exercise, gene expression, prostate cancer
INTRODUCTION
Approximately 240,000 men in the United States (US) are estimated to be diagnosed with prostate cancer in 2013 adding to the nearly 2.5 million US men who currently live with prostate cancer. The vast majority (93%) of newly diagnosed cases have non-metastatic prostate cancer at diagnosis and a 5-year disease-specific survival that approaches 100% (1). A large subset of these patients are eligible for active surveillance, a management strategy that offers close monitoring of localized cancers and avoids the comorbidity of invasive treatments such as surgery or radiation (2). The active surveillance protocol of repeat biopsies provides a unique opportunity to investigate the effects of lifestyle interventions, such as diet and physical activity, after diagnosis on prostate cancer biology (3–5).
A growing body of evidence suggests vigorous physical activity [i.e. activities that require six or more times the resting metabolic rate (≥6 metabolic equivalent tasks (METs)), such as jogging and cycling] or brisk walking after diagnosis may deter or delay prostate cancer progression. Our group reported for the first time that engaging in three or more hours per week of vigorous physical activity was associated with a 61% lower risk of prostate cancer-specific mortality among 2,705 men initially diagnosed with clinically localized prostate cancer (6). There was no benefit observed among men who engaged in vigorous activity for less than three hours per week. However, in a distinct cohort of 1455 men with clinically localized prostate cancer, men who walked at a brisk pace [≥3 miles per hour (MPH)] after diagnosis had a 48% lower risk of disease progression (primarily biochemical recurrence) compared to men who walked at an easy pace (<2 MPH)(7).
While these observational results are intriguing, the biological mechanisms underlying the potential relation between physical activity and prostate cancer progression remain unknown. Hypothesized mechanisms include modulation of circulating factors (e.g. insulin, growth factors, sex-steroid hormones), improved immune function, lower systemic inflammation and oxidative stress, and changes in tumor vascularization (8). Yet, no study has examined whether physical activity after diagnosis is associated with gene expression of these, or other, pathways in human prostate tissue.
Therefore, we performed an exploratory analysis to examine associations between physical activity global gene and expression patterns in the normal prostate tissue of men with low risk prostate cancer who elected active surveillance as their primary treatment strategy. Based on our prior studies, we hypothesized that men who engaged in vigorous physical activity for ≥3 h/wk would have different prostate gene expression patterns compared to men who engaged in less or no vigorous activity.
METHODS
Patient and gene expression data
Gene expression profiles (cDNA microarray dataset #GSE27140) were obtained at baseline from morphologically normal prostate tissue of 84 men with low risk prostate cancer on active surveillance who subsequently participated in a randomized controlled trial of nutritional supplements [Molecular Effects of Nutritional Supplements (MENS) Trials, NCT00402285] (3, 5). Details of the recruitment strategy, patient demographics (5), biopsy collection, and methods for cDNA microarray analysis have been previously described (3–5). The median time from diagnosis to study biopsy was three months; the median time from study biopsy to the physical activity questionnaire was two months.
Of the 84 participants in the MENS trial with gene expression data, 72 completed the baseline physical activity questions. We excluded one man who reported having bronchitis in the week prior to completing the questionnaire because the questionnaire asked men to report their activity during the last seven days, leaving 71 men eligible for analysis. Only the baseline gene expression data obtained from pre-intervention biopsy were used for this study (Table S1).
This study was approved by the University of California San Francisco Institutional Review Board. All patients gave written informed consent to participate.
Physical activity assessment
Participants were asked to respond to the following questions: “What form of aerobic exercise do you do, if any? (e.g. walking, biking, swimming, etc.)”, “How many days in the last 7 days have you done aerobic exercise?”, and “On those days you exercised, how many minutes (on average) did you spend exercising?”.
Activities that require six or more times the resting metabolic rate (e.g. have a metabolic equivalent task (MET) value of six or higher) were classified as vigorous. Vigorous activities reported by the participants included: running, jogging, swimming, squash/racquetball, cycling, cardio equipment, tennis, team sports, and rowing. If a man did not report the types of activities he engaged in, it was assumed that he did not engage in aerobic activity. Duration of vigorous activity (h/wk) was calculated by multiplying the average minutes per exercise session by the number of days per week of exercise and dividing by 60. One man reported an atypical average exercise session duration (e.g. >240 minutes/session); we assigned 240 minutes as his average exercise session duration. In addition, two men reported engaging in aerobic activities and reported the frequency of exercise (n=1) or the average exercise session duration (n=1), but not the other; we imputed the missing data using the median from all other men that engaged in aerobic activity (40 minutes/session or 3 sessions/week). Unfortunately, the questionnaire did not ask about usual walking pace, so we were unable to examine whether brisk walking was associated with differential gene expression.
In addition, we examined the United States (US) Physical Activity Guidelines for Older Adults (9) as a binary exposure. The guidelines state: “older adults should do at least 150 minutes a week of moderate-intensity, or 75 minutes a week of vigorous-intensity aerobic physical activity, or an equivalent combination of moderate- and vigorous-intensity aerobic activity. The guidelines also recommend participating in strength training activities on 2 or more days a week; however we did not collect data on resistance or balance exercises and thus our analysis only included the aerobic exercise component of the guidelines.
Associations between gene expression and dichotomous groupings
We explored associations between normal prostate gene expression and each of the dichotomous groupings (Table 1): (1) men who engaged in vigorous activities for ≥3 h/wk versus those who did not, (2) men who engaged in any vigorous activity versus those who did not, and (3) men who met the US Physical Activity Guidelines for Older Adults (aerobic component) versus those who did not. In addition to the three physical activity categorizations, we also included a comparison between overweight/obese (body mass index (BMI) >25 kg/m2) versus healthy weight men (BMI ≤25 kg/m2).
Table 1.
Clinical characteristics of participants and classification based on self-reported physical activity.
| Characteristic1 | Mean ± SD or N (%) |
|---|---|
| Age at diagnosis, y | 61 ± 8 |
| Body mass index, kg/m2 | 26 ± 5 |
| Race | |
| Caucasian | 55 (77) |
| African American | 3 (4) |
| Asian | 2 (3) |
| Not reported | 11 (15) |
| Prostate specific antigen | 4.4 ± 2.3 |
| Gleason sum | |
| 3 | 1 (4) |
| 5 | 1 (4) |
| 6 | 23 (82) |
| 7 | 3 (11) |
| Clinical T-stage | |
| T1c | 55 (77) |
| T2a | 16 (23) |
|
| |
| Physical activity classification | |
|
| |
| ≥3 h/wk vigorous activity2 | 16 (23) |
| Any vigorous activity2 | 33 (46) |
| Met US Physical Activity Guidelines for Older Adults3 | 35 (49) |
| Overweight or obese4 | 38 (57) |
Clinical characteristics and physical activity are from the baseline assessment of the MENS trial. The baseline biopsy and clinical data were collected a median of 3 months after diagnosis and the physical activity data were collected a median of 2 months after the baseline clinical visit.
A man was classified as engaging in vigorous activity if he reported engaging in any activities that require 6+ METS.
A man was classified as meeting the US Physical Activity Guidelines for Older Adults if he reported engaging in 150 min/wk of moderate-intensity, or 75 min/wk of vigorous-intensity aerobic activity, or some combination thereof.
A man was classified as overweight/obese if he had a body mass index (BMI) ≥25 kg/m2. Eight men of these men had BMI ≥30 kg/m2. BMI data was missing for 4 men.
First, we performed differential expression analysis using Significance Analysis of Microarrays (SAM) (10) to determine which individual genes in our array were significantly associated to a particular group. Next, we used an extension of SAM, called Gene Set Analysis (11, 12) to the explore associations of each the groups with a priori defined gene sets. After the gene set analysis, we utilized Ingenuity Pathway Analysis (IPA) to explore canonical pathways and networks and to identify those that were significantly associated with each group.
Differential expression analysis using SAM
Gene expression data was analyzed using the Significance Analysis of Microarrays (SAM) package. First, Genbank Accession numbers were mapped to Unigene Symbols using SOURCE (13). A two-class unpaired analysis was used to compare groups in the four dichotomous groupings described above (Table 1). Genbank accessions that were not mapped to genes were removed from the analysis. Genes were considered significant at a false discovery rate (FDR) <0.10. A heatmap was generated using SAMSTER (http://falkow.stanford.edu/whatwedo/software/programs/samster.pdf).
Gene set analysis using SAM
Gene set analysis (12, 14) was also performed using SAM to determine gene sets that were up- or down-regulated between groups. The gene sets (n=871) used for the analysis were obtained from Broad Institute’s Molecular Signature Database (MSigDB) version 3.0. Included were the curated canonical pathways from the Kyoto Encyclopedia of Genes and Genomes (KEGG; n=186), Biocarta (n=217), Reactome (n=430), the Signal Transduction Knowledge Environment (ST; n=28), and the Sigma Aldrich Cell Biology Learning Center (SA; n=10). Permutation (n=1000) was utilized to estimate the FDR for every gene set. Gene sets with FDR <0.20 were considered statistically significant.
Canonical pathway analysis using Ingenuity
We performed canonical pathway and network analysis (see next section) on the two groupings that yielded differentially expressed genes [i.e., ≥3 h/wk vigorous physical activity (yes/no) and met the US Physical Activity Guidelines for Older Adults (yes/no)]. A table containing 20,209 cDNA probes and their corresponding Genbank accession ID and expression values (fold change and FDR) were uploaded into Ingenuity Pathways Analysis (IPA) software (Ingenuity Systems, Redwood City, CA). IPA was able to map 12,169 Genbank accession IDs to genes (or molecules) present in the Ingenuity Pathways Knowledge Base. IPA then assigned gene ontology descriptions of biological function and generated networks of gene interactions on the basis of information retrieved from the software’s literature database. Up-regulated and down-regulated genes with FDR ≤0.20 were included in the pathway analysis. Ninety-four genes and 41 genes passed the cut-off of FDR ≤0.20 for inclusion into the pathway and network analysis for the following comparisons “met the US Physical Activity Guidelines for Older Adults (yes/no)” and “≥3 h/wk vigorous physical activity (yes/no)]”, respectively. Fisher’s exact tests were used to calculate a p-value to indicate association between genes in our dataset and a canonical pathway. Due to the exploratory nature of our study, in our primary analyses, we used the unadjusted p-value to determine statistically significant associations. Pathways with an unadjusted p-value <0.05 were considered statistically significant. To account for multiple comparisons, we performed a secondary analysis using the Benjamini-Hochberg method to estimate the FDR for every pathway (15). In the analysis, the cutoff for significance was an FDR <0.05. In contrast to gene set analysis by SAM, results of the IPA canonical pathway analysis do not specify whether the pathways were up- or down-regulated; therefore, we report them as “modulated”.
Network analysis using Ingenuity
We performed further bioinformatic analyses using IPA to calculate the approximate “fit” between network eligible genes (see previous section) and networks, defined as collections of genes that are highly interconnected based on Ingenuity Pathways Knowledge Base findings. Briefly, genes from the input dataset, focus genes, were overlaid onto a global molecular network developed from information contained in Ingenuity Pathways Knowledge Base. Networks of these focus genes were then algorithmically generated based on their connectivity to generate a score used to rank networks according to how relevant they are to the genes in the input dataset. IPA uses a right-tailed Fisher’s test to calculate the p-value for networks. For example, a score of 10 indicates that there is a 10−10 probability of observing the observed data if the null hypothesis of no association were true. We report networks with an IPA score ≥7.
RESULTS
Description of the study population
On average, the 71 men in our study were 61 years old at diagnosis, overweight (mean body mass index (BMI) = 26 kg/m2), and Caucasian (77%). As expected based on the clinical eligibility criteria of the original study, the men had low risk disease; the mean prostate specific antigen (PSA) was 4.4 ng/ml, 82% had Gleason sum = 6, and 77% had clinical stage = T1c (Table 1).
Sixteen men (23%) reported engaging in vigorous physical activity for ≥3 h/wk (Table 1). Thirty-three men (46%) reported engaging in any vigorous activity and 35 men (49%) met the US Physical Activity Guidelines for Older Adults (Table 1).
Individual gene analysis
Nine genes were differentially expressed among men who reported participating in ≥3 h/wk of vigorous physical activity after diagnosis compared to men who did not (Figure 1). Up-regulated genes included FNTA, CENPF, CPA4 and a SKP2 homologue and down-regulated genes included SLC25A4, SORD, SH3BGR, ALDHA13, and C2orf69 (Table S2). Two expressed sequence tags (EST) were significantly down-regulated in men who met the US Physical Activity Guidelines for Older Adults compared to those who did not. ESTs AA777930 and W94714 have significant homology to the transmembrane EMP24 protein transport domain containing 4 (TMED4) and the forkhead box C1 (FOXC1), respectively. No significant differences were observed when we compared men who participated in any vigorous physical activity to men who did none.
Figure 1. Differential expression analysis.
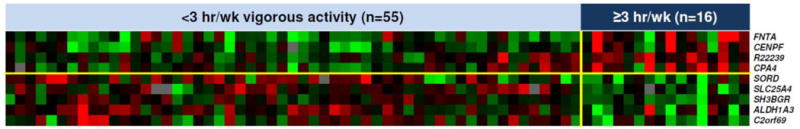
Heatmap showing up-regulated (red) genes (upper right quadrant) and down-regulated (green) genes (lower right quadrant) in normal prostate of 71 men with low-risk prostate cancer who participated in ≥3 h/wk vigorous activity compared to men who engaged in <3 h/wk vigorous activity.
Gene set analysis
We observed 25 up-regulated gene sets, including genes regulating cell cycle and DNA repair pathways, and two down-regulated gene sets involved in the semaphorin signaling pathway when comparing men who reported engaging in vigorous activity for ≥3 h/wk to men who engaged in less or no vigorous activity (Table 2, Table S3). Analysis comparing men who reported engaging in any vigorous activities v. none revealed one up-regulated gene set (deadenylation of mRNA), and men who met the US Physical Activity Guidelines for Older Adults had four positively modulated gene sets, including pathways involved in EGFR and insulin receptor signaling (Table 3, Table S4). The relation between physical activity and prostate cancer outcomes may be partially mediated through body weight; therefore we also compared the gene expression of overweight/obese v. normal weight men. Twelve gene sets were up regulated and two were down-regulated in men who were overweight/obese compared to normal weight (Table 4, Table S5). Up-regulated gene sets included pathways involved in retinol and steroid hormone metabolism. The most commonly shared genes among the perturbed gene sets are shown in Figure S1.
Table 2.
Up- and down-regulated gene sets in the normal prostate tissue of 16 men with low-risk prostate cancer who engaged in vigorous physical activity ≥3 h/wk compared to 55 men who engaged in <3 h/wk vigorous activity. Gene sets with a false discovery rate (FDR) <0.20 were considered significant.
| Gene set name | Source | p-value | FDR |
|---|---|---|---|
| Up-regulated | |||
| G1 Pathway | BIOCARTA | >0.001 | 0.0 |
| Formation of Fibrin Clot Clotting Cascade | REACTOME | >0.001 | 0.0 |
| Inactivation of APC via Direct Inhibition of the APcomplex | REACTOME | >0.001 | 0.0 |
| Cell Cycle | KEGG | 0.001 | 0.10 |
| TOB1 Pathway | BIOCARTA | 0.001 | 0.10 |
| G2 M Checkpoints | REACTOME | 0.001 | 0.10 |
| Phosphorylation of the APC | REACTOME | 0.001 | 0.10 |
| APCDC20 Mediated Degradation of Cyclin B | REACTOME | 0.002 | 0.13 |
| Chylomicron Mediated Lipid Transport | REACTOME | 0.002 | 0.13 |
| E2F Transcriptional Targets at G1 S | REACTOME | 0.002 | 0.13 |
| GA12 Pathway | STKE | 0.002 | 0.13 |
| E2F Mediated Regulation of DNA Replication | REACTOME | 0.003 | 0.15 |
| Homologous Recombination Repair | REACTOME | 0.003 | 0.15 |
| Cysteine And Methionine Metabolism | KEGG | 0.004 | 0.15 |
| Activation of ATR In Response to Replication Stress | REACTOME | 0.004 | 0.15 |
| Complement and Coagulation Cascades | KEGG | 0.005 | 0.15 |
| NKT Pathway | BIOCARTA | 0.005 | 0.15 |
| Cell Cycle Mitotic | REACTOME | 0.005 | 0.15 |
| Cyclin A1 Associated Events During G2 M Transition | REACTOME | 0.005 | 0.15 |
| Double Strand Break Repair | REACTOME | 0.005 | 0.15 |
| Fanconi Anemia Pathway | REACTOME | 0.005 | 0.15 |
| Lipoprotein Metabolism | REACTOME | 0.005 | 0.15 |
| Platelet Activation | REACTOME | 0.005 | 0.15 |
| NO2-IL12 Pathway | BIOCARTA | 0.006 | 0.17 |
| Formation of Platelet Plug | REACTOME | 0.006 | 0.17 |
| Down-regulated | |||
| SEMA4D in Semaphorin Signaling | REACTOME | >0.001 | 0.0 |
| SEMA4D Induced Cell Migration and Growth Cone Collapse | REACTOME | >0.001 | 0.0 |
Table 3.
Up- and down-regulated gene sets in the normal prostate tissue of 35 men who met the US Physical Activity Guidelines for Older Adults1 compared to 46 men who did not meet the guidelines. Gene sets with a false discovery rate (FDR) <0.20 were considered significant.
| Gene set name | Source | p-value | FDR |
|---|---|---|---|
| Up-regulated | |||
| EGFR/ErbB Signaling Pathway | KEGG | >0.001 | 0.00 |
| Insulin Receptor Pathway in Cardiac Myocytes | SIG | >0.001 | 0.00 |
| Phosphoinositide 3 Kinase Pathway | ST | >0.001 | 0.00 |
| Wnt Ca2 Cyclic GMP Pathway | ST | 0.001 | 0.18 |
The US Physical Activity Guidelines for Older Adults are a minimum of 150 min/wk of moderate-intensity, or 75 min/wk of vigorous-intensity aerobic physical activity, or an equivalent combination of moderate- and vigorous-intensity aerobic activity.
Table 4.
Up- and down-regulated gene sets in the normal prostate tissue of 38 men with low-risk prostate cancer who were considered overweight or obese1 compared to 29 men with normal BMI. Gene sets with a false discovery rate (FDR) <0.20 were considered significant.
| Gene set name | Source | p-value | FDR |
|---|---|---|---|
| Positively modulated | |||
| Retinol Metabolism | KEGG | >0.001 | 0.00 |
| IL3 Pathway | BIOCARTA | >0.001 | 0.00 |
| GRB2:SOS Provides Linkage to MAPK Signaling for Intergrins | REACTOME | >0.001 | 0.00 |
| Integrin AlphaIIbbeta3 Signaling | REACTOME | >0.001 | 0.00 |
| Xenobiotics | REACTOME | >0.001 | 0.00 |
| Amine Compound SLC Transporters | REACTOME | >0.001 | 0.00 |
| Steroid Hormone Biosynthesis | KEGG | 0.001 | 0.09 |
| Platelet Aggregation Plug Formation | REACTOME | 0.001 | 0.09 |
| TPO Pathway | BIOCARTA | 0.002 | 0.12 |
| P130Cas Linkage to MAPK Signaling for Integrins | REACTOME | 0.002 | 0.12 |
| Steroid Hormones | REACTOME | 0.002 | 0.12 |
| Shcmediated Cascade | REACTOME | 0.002 | 0.12 |
| Negatively modulated | |||
| mCalpain Pathway | BIOCARTA | >0.001 | 0.00 |
| Th1/Th2 Pathway | BIOCARTA | >0.001 | 0.00 |
Men were classified as overweight or obese if they had a body mass index (BMI) ≥ 25 kg/m2.
Canonical pathway and network analyses
Canonical pathway analysis revealed 43 pathways that were differentially modulated in men who engaged in ≥3 h/wk of vigorous activity (Table 5), including cell signaling (e.g. integrin and RhoA signaling) and metabolism (e.g. tryptophan and putrescine degradation). After correction for multiple testing, 28 pathways had a FDR <0.05 (Table 5). Of note, a pathway involved in the cellular effects of sildenafil (Viagra) (unadjusted p-value=0.0003, FDR=0.0056) and the Nrf2-mediated oxidative stress response pathway (unadjusted p-value=0.0105, FDR=0.0355) were significantly modulated in men engaging in high levels of vigorous activity. Lastly, top-scoring networks among men engaging in ≥3 h/wk of vigorous activity involved functions concerning cancer, cellular development, and cell growth and proliferation (Table S2).
Table 5.
Canonical pathways modulated in normal prostate tissue in 16 men with low-risk prostate cancer who performed vigorous physical activity for ≥3 h/wk. Canonical pathways with unadjusted p-value <0.05 were considered statistically significant. Genes present in our dataset belonging to significant canonical pathways are listed. Adjustment for multiple comparisons was also performed in IPA using the Benjamini-Hochberg method to assess false discovery rate (FDR).
| Canonical Pathways | Genes | Unadjusted p-value |
FDR |
|---|---|---|---|
| Regulation of Actin-based Motility by Rho | MYL9,ACTA2,PPP1R12B,ACTG2 | 0.0001 | 0.0044 |
| Integrin Signaling | MYL9,PARVA,ACTA2,PPP1R12B,ACTG2 | 0.0001 | 0.0051 |
| RhoA Signaling | MYL9,ACTA2,PPP1R12B,ACTG2 | 0.0002 | 0.0055 |
| Cellular Effects of Sildenafil (Viagra) | MYL9,ACTA2,PPP1R12B,ACTG2 | 0.0003 | 0.0056 |
| Tryptophan Degradation X (Mammalian, via Tryptamine) | MAOB,ALDH1A3 | 0.0008 | 0.0100 |
| Putrescine Degradation III | MAOB,ALDH1A3 | 0.0008 | 0.0100 |
| RhoGDI Signaling | MYL9,ACTA2,PPP1R12B,ACTG2 | 0.0010 | 0.0107 |
| ILK Signaling | MYL9,PARVA,ACTA2,ACTG2 | 0.0011 | 0.0112 |
| Dopamine Degradation | MAOB,ALDH1A3 | 0.0013 | 0.0115 |
| Actin Cytoskeleton Signaling | MYL9,ACTA2,PPP1R12B,ACTG2 | 0.0020 | 0.0135 |
| LPS/IL-1 Mediated Inhibition of RXR Function | MAOB,ALDH1A3,FABP1,ABCC4 | 0.0020 | 0.0135 |
| Paxillin Signaling | PARVA,ACTA2,ACTG2 | 0.0020 | 0.0135 |
| Sorbitol Degradation I | SORD | 0.0026 | 0.0151 |
| Signaling by Rho Family GTPases | MYL9,ACTA2,PPP1R12B,ACTG2 | 0.0027 | 0.0151 |
| Noradrenaline and Adrenaline Degradation | MAOB,ALDH1A3 | 0.0030 | 0.0158 |
| Mechanisms of Viral Exit from Host Cells | ACTA2,ACTG2 | 0.0047 | 0.0234 |
| Epithelial Adherens Junction Signaling | MYL9,ACTA2,ACTG2 | 0.0059 | 0.0263 |
| MSP-RON Signaling Pathway | ACTA2,ACTG2 | 0.0059 | 0.0263 |
| Tight Junction Signaling | MYL9,ACTA2,ACTG2 | 0.0069 | 0.0288 |
| Serotonin Degradation | MAOB,ALDH1A3 | 0.0072 | 0.0288 |
| Melatonin Degradation II | MAOB | 0.0102 | 0.0355 |
| NAD Biosynthesis III | NMNAT2 | 0.0102 | 0.0355 |
| NRF2-mediated Oxidative Stress Response | ACTA2,ACTG2,ABCC4 | 0.0105 | 0.0355 |
| Remodeling of Epithelial Adherens Junctions | ACTA2,ACTG2 | 0.0123 | 0.0389 |
| Agrin Interactions at Neuromuscular Junction | ACTA2,ACTG2 | 0.0129 | 0.0389 |
| NAD Salvage Pathway III | NMNAT2 | 0.0129 | 0.0389 |
| Caveolar-mediated Endocytosis Signaling | ACTA2,ACTG2 | 0.0148 | 0.0427 |
| NAD Biosynthesis from 2-amino-3-carboxymuconate Semialdehyde | NMNAT2 | 0.0155 | 0.0437 |
| TR/RXR Activation | NRGN,NCOA3 | 0.0200 | 0.0525 |
| FAK Signaling | ACTA2,ACTG2 | 0.0204 | 0.0525 |
| Crosstalk between Dendritic Cells and Natural Killer Cells | ACTA2,ACTG2 | 0.0214 | 0.0525 |
| Virus Entry via Endocytic Pathways | ACTA2,ACTG2 | 0.0219 | 0.0525 |
| VEGF Signaling | ACTA2,ACTG2 | 0.0219 | 0.0525 |
| Fcγ Receptor-mediated Phagocytosis in Macrophages and Monocytes | ACTA2,ACTG2 | 0.0240 | 0.0550 |
| Histamine Degradation | ALDH1A3 | 0.0302 | 0.0692 |
| NAD biosynthesis II (from tryptophan) | NMNAT2 | 0.0331 | 0.0708 |
| Fatty Acid α-oxidation | ALDH1A3 | 0.0331 | 0.0708 |
| Phenylalanine Degradation IV (Mammalian, via Side Chain) | MAOB | 0.0355 | 0.0741 |
| Oxidative Ethanol Degradation III | ALDH1A3 | 0.0380 | 0.0759 |
| Ethanol Degradation IV | ALDH1A3 | 0.0427 | 0.0832 |
| Cdc42 Signaling | MYL9,PPP1R12B | 0.0437 | 0.0832 |
| Aryl Hydrocarbon Receptor Signaling | ALDH1A3,NCOA3 | 0.0457 | 0.0851 |
| Hepatic Fibrosis / Hepatic Stellate Cell Activation | MYL9,ACTA2 | 0.0479 | 0.0891 |
Eight canonical pathways were modulated among men with low-risk prostate cancer who met the US Physical Activity Guidelines for Older Adults (Table 6). Modulated pathways included those involved in lipid metabolism (e.g. diphthamide and L-carnitine biosynthesis), cardiovascular signaling (e.g. cardiac β-adrenergic signaling), and cytokine signaling (e.g. role of IL-17F in allergic inflammatory airway diseases and T helper cell differentiation). Note, however, that when correction for multiple comparisons was applied, none of these pathways had a FDR <0.05 (Table 6). Three networks were highly represented in genes modulated in the normal prostate tissue of men who met the US Physical Activity Guidelines for Older Adults (Table S6). The top-scoring networks encompassed functions involved in inflammatory disease, organismal injury and abnormalities, and respiratory disease. Overlapping genes associated to the different modulated canonical pathways are shown in Figure S2.
Table 6.
Canonical pathways modulated in normal prostate tissue in 35 men with low-risk prostate cancer who met the minimum recommended amount of physical activity by the US Physical Activity Guidelines for Older Adults1. Canonical pathways with unadjusted p-value <0.05 were considered statistically significant. Genes present in our dataset belonging to significant canonical pathways are listed. Adjustment for multiple comparisons was also performed in IPA using the Benjamini-Hochberg method to assess false discovery rate (FDR).
| Canonical Pathways | Genes | Unadjusted p-value |
FDR |
|---|---|---|---|
| Diphthamide Biosynthesis | EEF2 | 0.0105 | 0.5848 |
| L-carnitine Biosynthesis | TMLHE | 0.0158 | 0.5848 |
| Role of IL-17F in Allergic Inflammatory Airway Diseases | CXCL1,CREB5 | 0.0200 | 0.5848 |
| Cardiac β-adrenergic Signaling | PPP1R10,GNG2,ATP2A2 | 0.0324 | 0.5848 |
| Inositol Pyrophosphates Biosynthesis | IP6K1 | 0.0363 | 0.5848 |
| Calcium-induced T Lymphocyte Apoptosis | HLA-DQB1,ATP2A2 | 0.0380 | 0.5848 |
| Calcium Transport I | ATP2A2 | 0.0468 | 0.5848 |
| T Helper Cell Differentiation | IL10RB,HLA-DQB1 | 0.0490 | 0.5848 |
The Physical Activity Guidelines for Older Adults recommend at least 150 min/wk of moderate-intensity, or 75 min/wk of vigorous-intensity aerobic activity, or an equivalent combination of moderate and vigorous intensity aerobic activity.
DISCUSSION
In this exploratory analysis, we observed associations between engaging in ≥3 h/wk of vigorous activity after diagnosis and gene expression patterns in the normal prostate tissue of men diagnosed with low risk prostate cancer. Specifically, pathways involved in cell cycling, DNA repair, cell signaling and metabolism, the cellular effects of sildenafil (Viagra®), and the Nrf2-mediated oxidative stress response pathway were modulated in men engaging in ≥3 h/wk of vigorous activity. There were few differences observed when comparing men who performed moderate intensity activity (meeting the guidelines) to men who did little or no activity, nor when comparing men who did any vigorous activity to men who did none. These observations support the findings of our prior reports examining physical activity in relation to clinical outcomes among men with prostate cancer (6, 7), and suggest that high levels of vigorous activity (≥3 h/wk) may be needed to affect prostate cancer prognosis.
Gene set analysis revealed the modulation of several gene sets including those involved in cell cycle control and DNA repair pathways (e.g. mitotic checkpoints and homologous recombination repair). Protein and gene expression studies have also linked increased DNA repair activity (16, 17), as well as modulation of pathways involved in immune function, metabolism (e.g. growth factors), sex hormones, and oxidative stress (e.g. endogenous antioxidant enzyme system) as potential mechanisms through which physical activity might delay or deter prostate cancer progression (18). The elucidation of these mechanisms may aid the development of strategies to prevent or delay prostate cancer progression, guide the discovery of new biomarkers for monitoring effects of lifestyle interventions, and provide targets for pharmaceutical interventions designed to enhance the potential beneficial biological effects of physical activity.
Two intriguing results were observed in our canonical pathway analysis. First, we observed the modulation of the Nrf2-mediated oxidative stress response pathway by vigorous physical activity. In our previous study (3), we also observed the modulation of this pathway following lycopene and fish oil supplementation in low-risk prostate cancer patients. Second, we observed modulation of a pathway involving the cellular effects of sildenafil (Viagra®), a drug commonly used to treat erectile dysfunction. Although these may be chance findings and we cannot rule out potential confounding factors (e.g. sildenafil use), this observation is intriguing and consistent with prior observational and clinical data indicating that lack of physical activity increases the risk of erectile dysfunction and engaging in exercise improves erectile dysfunction (19, 20). Impotence or erectile dysfunction is a common condition among men in this age range (e.g. 20–40% in men age 60–69, 50–100% in men over age 70 (20)), and these data underscore the multiple potential benefits of engaging in exercise.
Studies comparing gene set/pathway analysis methods have shown commonalities as well as considerable discrepancies in the results gleaned from the different statistical tools tested (11, 21, 22). In our study, we did not observe a substantial overlap in the results of the SAM-based gene set analysis versus IPA-based canonical pathway analysis for each of the dichotomous groupings analyzed.
Exploratory prostate gene expression analysis in overweight/obese men did not yield individual genes that were differentially expressed compared to normal weight men. However, gene set analysis revealed the modulation of gene sets involved in retinol metabolism and steroid hormone biosynthesis. Interestingly, there were no shared gene sets that were up-regulated/down-regulated between the body mass index and physical activity comparisons, suggesting that the increased risk of prostate cancer progression associated with overweight/obesity and inactivity act through different biological pathways. This is supported by the observational studies of physical activity after diagnosis and prostate cancer progression in which adjustment for body mass index did not attenuate the inverse association between physical activity and prostate cancer recurrence or mortality (6, 7). Unfortunately, we were not able to contrast obese and non-obese subjects due to our small sample size and few obese participants (only eight men).
Limitations of this exploratory study include our small sample size, less detailed physical activity assessment than prior reports, and inability to consider potential confounding factors such as smoking, diet, or medication use (e.g. sildenafil). Also, our study was only able to focus on aerobic exercise and could not address potential molecular effects of resistance or balance training on the normal prostate microenvironment (23). Additionally, due to the lack of availability of RNA samples, we were unable to validate the results using an independent method such as quantitative-polymerase chain reaction (QPCR) analysis. Thus, our novel findings require confirmation if the biologic underpinnings for promoting lifestyle changes in cancer treatment are to be understood and more widely accepted.
CONCLUSIONS
Our data implicate several pathways, including DNA repair and oxidative stress pathways, as possible mechanisms by which vigorous physical activity after diagnosis may lower risk of prostate cancer progression. These data suggest biologic plausibility for an inverse relation between vigorous physical activity and risk of prostate cancer progression, and support the development of clinical trials to examine whether increasing physical activity after diagnosis affects prostate cancer biology and disease prognosis.
Supplementary Material
Acknowledgments
We thank the patients who participated in this study; Ritu Roy for providing assistance with preprocessing the raw gene expression data; Adam Olshen for the critical review of the manuscript; the Prostate Cancer Foundation, and the National Cancer Institute/National Institutes of Health for funding.
Funding: NIH/NCI R01CA101042 and R25CA112355; Prostate Cancer Foundation.
References
- 1.Prostate. SEER Stat Fact Sheets. November 2011 SEER data submission. National Cancer Institute; 2012. [Google Scholar]
- 2.Dall’Era MA, Cooperberg MR, Chan JM, et al. Active surveillance for early-stage prostate cancer: review of the current literature. Cancer. 2008;112:1650–9. doi: 10.1002/cncr.23373. [DOI] [PubMed] [Google Scholar]
- 3.Magbanua MJ, Roy R, Sosa EV, et al. Gene expression and biological pathways in tissue of men with prostate cancer in a randomized clinical trial of lycopene and fish oil supplementation. PLoS One. 2011;6:e24004. doi: 10.1371/journal.pone.0024004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ornish D, Magbanua MJ, Weidner G, et al. Changes in prostate gene expression in men undergoing an intensive nutrition and lifestyle intervention. Proc Natl Acad Sci U S A. 2008;105:8369–74. doi: 10.1073/pnas.0803080105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chan JM, Weinberg V, Magbanua MJ, et al. Nutritional supplements, COX-2 and IGF-1 expression in men on active surveillance for prostate cancer. Cancer Causes Control. 2011;22:141–50. doi: 10.1007/s10552-010-9684-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kenfield SA, Stampfer MJ, Giovannucci E, Chan JM. Physical activity and survival after prostate cancer diagnosis in the health professionals follow-up study. J Clin Oncol. 2011;29:726–32. doi: 10.1200/JCO.2010.31.5226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Richman EL, Kenfield SA, Stampfer MJ, Paciorek A, Carroll PR, Chan JM. Physical activity after diagnosis and risk of prostate cancer progression: data from the cancer of the prostate strategic urologic research endeavor. Cancer Res. 2011;71:3889–95. doi: 10.1158/0008-5472.CAN-10-3932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jones LW, Antonelli J, Masko EM, et al. Exercise modulation of the host-tumor interaction in an orthotopic model of murine prostate cancer. Journal of applied physiology. 2012;113:263–72. doi: 10.1152/japplphysiol.01575.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Phyical Activity Guidelines Advisory Committee. Physical Activity Advisory Committee Report, 2008. Washington DC: Department of Health and Human Services; 2008. [Google Scholar]
- 10.Tusher VG, Tibshirani R, Chu G. Significance analysis of microarrays applied to the ionizing radiation response. Proc Natl Acad Sci U S A. 2001;98:5116–21. doi: 10.1073/pnas.091062498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dinu I, Potter JD, Mueller T, et al. Improving gene set analysis of microarray data by SAM-GS. BMC bioinformatics. 2007;8:242. doi: 10.1186/1471-2105-8-242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Efron B, Tibshirani R. On Testing the Significance of Sets of Genes. Ann Appl Stat. 2007;1:107–29. [Google Scholar]
- 13.Diehn M, Sherlock G, Binkley G, et al. SOURCE: a unified genomic resource of functional annotations, ontologies, and gene expression data. Nucleic Acids Res. 2003;31:219–23. doi: 10.1093/nar/gkg014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Subramanian A, Tamayo P, Mootha VK, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102:15545–50. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Benjamini Y, Hochberg Y. Controlling the False Discovery Rate - a Practical and Powerful Approach to Multiple Testing. J Roy Stat Soc B Met. 1995;57:289–300. [Google Scholar]
- 16.Radak Z, Naito H, Kaneko T, et al. Exercise training decreases DNA damage and increases DNA repair and resistance against oxidative stress of proteins in aged rat skeletal muscle. Pflugers Archiv: European journal of physiology. 2002;445:273–8. doi: 10.1007/s00424-002-0918-6. [DOI] [PubMed] [Google Scholar]
- 17.Siu PM, Pei XM, Teng BT, Benzie IF, Ying M, Wong SH. Habitual exercise increases resistance of lymphocytes to oxidant-induced DNA damage by upregulating expression of antioxidant and DNA repairing enzymes. Experimental physiology. 2011;96:889–906. doi: 10.1113/expphysiol.2011.058396. [DOI] [PubMed] [Google Scholar]
- 18.Rundle A. Molecular epidemiology of physical activity and cancer. Cancer epidemiology, biomarkers & prevention: a publication of the American Association for Cancer Research, cosponsored by the American Society of Preventive Oncology. 2005;14:227–36. [PubMed] [Google Scholar]
- 19.La Vignera S, Condorelli R, Vicari E, D’Agata R, Calogero AE. Physical activity and erectile dysfunction in middle-aged men. Journal of andrology. 2012;33:154–61. doi: 10.2164/jandrol.111.013649. [DOI] [PubMed] [Google Scholar]
- 20.Shamloul R, Ghanem H. Erectile dysfunction. Lancet. 2013;381:153–65. doi: 10.1016/S0140-6736(12)60520-0. [DOI] [PubMed] [Google Scholar]
- 21.Goeman JJ, Buhlmann P. Analyzing gene expression data in terms of gene sets: methodological issues. Bioinformatics. 2007;23:980–7. doi: 10.1093/bioinformatics/btm051. [DOI] [PubMed] [Google Scholar]
- 22.Tian L, Greenberg SA, Kong SW, Altschuler J, Kohane IS, Park PJ. Discovering statistically significant pathways in expression profiling studies. Proc Natl Acad Sci U S A. 2005;102:13544–9. doi: 10.1073/pnas.0506577102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Strasser B, Steindorf K, Wiskemann J, Ulrich CM. Impact of Resistance Training in Cancer Survivors: a Meta-analysis. Medicine and science in sports and exercise. 2013 doi: 10.1249/MSS.0b013e31829a3b63. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


