Abstract
Although Plasmodium vivax is a leading cause of malaria around the world, only a handful of vivax antigens are being studied for vaccine development. Here, we investigated genetic signatures of selection and geospatial genetic diversity of two leading vivax vaccine antigens – Plasmodium vivax merozoite surface protein 1 (pvmsp-1) and Plasmodium vivax circumsporozoite protein (pvcsp). Using scalable next-generation sequencing, we deep-sequenced amplicons of the 42 kDa region of pvmsp-1 (n = 44) and the complete gene of pvcsp (n = 47) from Cambodian isolates. These sequences were then compared with global parasite populations obtained from GenBank. Using a combination of statistical and phylogenetic methods to assess for selection and population structure, we found strong evidence of balancing selection in the 42 kDa region of pvmsp-1, which varied significantly over the length of the gene, consistent with immune-mediated selection. In pvcsp, the highly variable central repeat region also showed patterns consistent with immune selection, which were lacking outside the repeat. The patterns of selection seen in both genes differed from their P. falciparum orthologs. In addition, we found that, similar to merozoite antigens from P. falciparum malaria, genetic diversity of pvmsp-1 sequences showed no geographic clustering, while the non-merozoite antigen, pvcsp, showed strong geographic clustering. These findings suggest that while immune selection may act on both vivax vaccine candidate antigens, the geographic distribution of genetic variability differs greatly between these two genes. The selective forces driving this diversification could lead to antigen escape and vaccine failure. Better understanding the geographic distribution of genetic variability in vaccine candidate antigens will be key to designing and implementing efficacious vaccines.
Author Summary
Plasmodium vivax causes tens of millions of malaria cases each year. Although some vaccines against P. vivax are being developed, little is known about the geospatial genetic diversity and selective constraints of the parasite surface antigens that these vaccines target. In order to create vaccines that are both efficacious and useful in diverse regions of the world, the strain diversity of these potential vaccine targets must be well understood. Specifically, we must understand whether and how the human immune system develops immunity against these antigens as well as understanding whether these antigens are similar in geographically diverse parasite populations. Here, using next-generation sequencing and population-genetic analyses, we found evidence of likely immune selection in specific regions of two leading vivax vaccine candidate antigens, PvMSP-1 and PvCSP. At the pvmsp-1 locus, we also found more genetic variability within populations than between populations, with some DNA sequences from geographically diverse populations being highly similar. In contrast, pvcsp sequences from geographically diverse populations are very distinct from one another, with specific sequence patterns occurring in certain geographic regions. Our findings provide new insights into the geographic genetic diversity of these two antigens and can help inform the development of effective P. vivax vaccines.
Introduction
Plasmodium vivax causes 80 to 300 million infections per year and over 2.5 billion people remain at risk of infection despite malaria elimination efforts [1]. Now, concern over P. vivax is growing due to reports of increasingly severe disease [2], emerging chloroquine resistance [3], and multi-drug resistance [4]. Ultimately, an effective vaccine will be important for controlling P. vivax malaria [5]. The fact that humans naturally develop partial immunity to P. vivax and P. falciparum lends hope for effective vaccines against these parasites; however, because the majority of global malaria research funding targets P. falciparum [6], [7], only a handful of P. vivax antigens are currently being considered for vaccine development [8]. Among these are P. vivax merozoite surface protein 1 (pvmsp-1) and circumsporozoite protein (pvcsp).
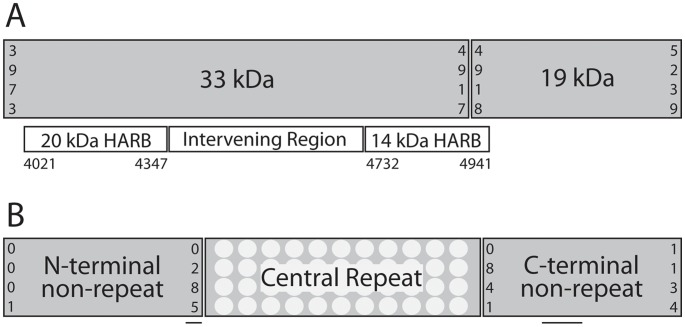
PvMSP-1, an erythrocytic vaccine candidate, plays an important role in reticulocyte invasion [9]. Its C-terminus contains a 42 kDa region, which is processed into 33 and 19 kDa fragments ( Figure 1A ). The 33 kDa fragment contains two high-affinity reticulocyte binding clusters (HARBs) (20 kDa and 14 kDa), and antibodies against the HARBs confer protection in monkeys [10]. In humans, antibodies to the 42 kDa region have also been associated with clinical protection, making this region an attractive vaccine candidate [11]–[14]. Another vivax protein, PvCSP, is a pre-erythrocytic vaccine candidate and is critical in sporozoite motility and hepatocyte invasion [15]. P. vivax circumsporozoite protein has an immunogenic central repeat, consisting of two major types of nonapeptide repeats (VK210 and VK247 – there is also a rarer repeat type termed vivax-like) flanked by highly conserved 5′ and 3′ regions ( Figure 1B ). The P. falciparum ortholog of pvcsp, as formulated in RTS,S, is the most advanced P. falciparum vaccine candidate to date, showing modest efficacy at one year interim analysis in a Phase III trial [16].
Figure 1. Protein domains and immunologically-relevant regions of pvmsp-1 42 kDa region and pvcsp.
For both genes, numbers indicate coordinates according to the Sal1 reference genes. Sequences for pvmsp-1 (PVX_099980) and pvcsp (PVX_119355) were accessed August 14, 2012 from PlasmoDB.org. (A) The pvmsp-1 42 kDa region is composed of two primary subunits – a 33 kDa and a 19 kDa subunit. Other sub-regions, including the 20 kDa and 14 kDa HARBs have been previously defined and studied. Here, we define the region between the HARBs as the “intervening region.” (B) The pvcsp gene is composed of three regions – an N-terminal non-repeat region, a central repeat region, and a C-terminal non-repeat region. The central repeat region consists of two major nonapeptide repeat types, termed VK210 and VK247. Approximate locations of pvcsp regions I and II are noted with horizontal lines in the N- and C-terminal non-repeat regions, respectively.
Despite this knowledge of PvMSP-1 and PvCSP, little is known about the geospatial genetic diversity of these antigens. Variation in these antigens may become a mechanism of vaccine resistance if strain-specific immunity is important in protection, as has been seen in some P. falciparum vaccine candidates [17]. Vaccine trials of P. falciparum AMA1 and MSP2 as well as genetic crosses using P. chabaudi underscore the importance of strain-specific immunity as a determinant of outcome [18]–[21]. Additionally, despite initial evidence that strain-specific immunity may not impact RTS,S efficacy [22]–[25], the incomplete protection afforded by the RTS,S vaccine in Phase II and III trials [16], [26], [27] has prompted a careful examination of strain-specific responses to this vaccine. Thus, as momentum grows for field trials of P. vivax vaccine antigens, carefully designed population genetic studies of P. vivax vaccine candidates will be key to assess the need for multivalent vaccine formulations.
To better understand the selective forces on, and geospatial genetic diversity associated with pvmsp-1 and pvcsp, we used the Illumina sequencing platform to determine haplotypes for 42 kDa region of pvmsp-1 (n = 44) and we used the PacBio and Illumina platforms to sequence the complete pvcsp gene (n = 47) from Cambodian isolates [28]. To dissect the immune selection acting on these regions, we studied these sequences using population genetic tests of selection and models of tandem repeat evolution. To evaluate the global genetic diversity of pvmsp-1 and pvcsp, we extracted worldwide pvmsp-1 and pvcsp sequence data available in GenBank (n = 238 for pvmsp-1 and n = 412 for pvcsp) (Figure S1), and studied our sequence data alongside the sequences from GenBank msp-1. Finally, we compare the performance of Illumina and PacBio sequencing to traditional Sanger sequencing, and discuss the potential and challenges of next-generation sequencing for population genetic studies of malaria parasite antigens.
Methods
Parasite isolates
Clinical samples from a previous study were used for this study [29]. Written informed consent was acquired from each individual and the study was approved by the IRB at University of North Carolina, the IRB of the Naval Medical Research Unit #2, Jakarta, Indonesia, and the Cambodian National Ethical Committee for Health Research. Briefly, blood spots were collected from 109 patients with uncomplicated vivax malaria, presenting to a clinic in Chumkiri, Cambodia during 2006–07. We selected 48 subjects with a multiplicity of infection (MOI) of one (n = 20) or two (n = 28) for sequencing. MOI was determined by heteroduplex tracking assay (HTA) [28], [30]. Briefly, in an HTA, radiolabeled DNA probes are annealed to genomic DNA and drawn through a non-denaturing gel matrix. The number of bands observed represents the number of conformation differences present among heteroduplexes, and is a proxy for the number of infection clones (MOI). Details of the method have been published elsewhere [31].
Amplification of pvmsp-1 and pvcsp
The pvmsp-1 42 kDa region (nucleotides 3973–5239 of Sal1 PVX_099980, www.PlasmoDB.org) was amplified using primers F: 5′-CAG GAC TAC GCC GAG GAC TA-3′ and R: 5′-GGA GGA AAA GCA ACA TGA GC-3′ and an Eppendorf Mastercycler (Eppendorf, Hauppauge, NY) in 50 µL reactions containing 5 µL 10× Qiagen Hotstar Master Mix (Qiagen, Valencia, CA), 0.25 µL Qiagen Hotstar Taq, 300 nM forward primer, 300 nM reverse primer, 1 µL 10 mM dNTPs, and 5 µL 5–10 mM template. Cycling conditions were: 95°C×15 m; 35 cycles of 95°C×45 s, 55°C×45 s, 72°C×3 m; and 72°C×10 m. The pvcsp gene (PVX_119355) was performed by nested PCR. The outer step used primers F: 5′-GGC AAA CTC ACA AAC ATC CA-3′ and R: 5′-TGC GTA AGC GCA TAA TGT GT-3′. Reactions were as above except for 600 nM forward primer, 600 nM reverse primer, 1 µL 10 mM dNTPs, 5 µL 5–10 mM template, 6 µL of 25 mM MgCl2, and 28.75 µL H2O. Cycling conditions were: 95°C×15 m; 25 cycles of 95°C×45 s, 45°C×45 s, 72°C×3 m; and 72°C×10 m. The inner step used 600 nM of each of the primers F: 5′-AAA CAG CCA AAG GCC TAC AA-3′ and R: 5′-GAC GCC GAA AAT ATT GGA TG-3′ using 5–10 µL of the initial amplification. The cycling conditions were: 95°C×15 m; 25 cycles of 95°C×45 s, 54°C×45 s, 72°C×3 m; and 72°C×10 m.
Amplicon sequencing and sequence determination
pvmsp-1 and pvcsp amplicons were fragmented by acoustic shearing (Covaris, Woburn, MA) using the following settings: 10% duty cycle, 5.0 intensity, 200 cycles per burst, and frequency sweeping mode. Forty-eight barcoded libraries were prepared using the NEXTflex multiplex library kit (Bioo Scientific, Austin, Texas), each containing the pooled pvmsp-1 and pvcsp amplicons from one patient. Libraries were sequenced on the Illumina HiSeq2000, using the paired-end 100 base pair chemistry (Illumina, San Diego, CA).
We used Lasergene SeqMan NGen v.3.1.1 (DNASTAR, Madison, WI) to assemble pvmsp-1 short reads de novo and to determine SNP frequency within each assembly. For purposes of comparison and confirmation, we re-sequenced 9 pvmsp-1 amplicons with differing MAFs: 3 samples with all MAFs>90%; 3 samples with all MAFs between 60% and 90%; 3 samples with MAF<60% for at least one SNP. Sanger-sequence haplotypes were compared to predicted Illumina haplotypes. Based on these comparisons, only predicted pvmsp-1 haplotypes with MAF>60% at all polymorphic sites were used in our analysis.
In addition to Illumina sequencing, pvcsp amplicons were sequenced using PacBio Circular Consensus Sequencing (Pacific Biosciences, Menlo Park, CA). One PacBio SMRT cell produced a total of 12103 reads with a minimum of 3× circular consensus coverage, which were used for this study. These were further filtered, removing truncated reads or reads with errors in the barcode. This left 8430 reads (3979 forward and 4451 reverse). Clustering attempted to minimize false positive haplotypes due to erroneous base calls and PCR slippage within the tandem repeat region. For each sample, haplotypes were created by clustering reads, allowing reads differing only by indels of 1 and 2 bases and low quality mismatches to collapse. Low quality was defined as either a mismatching base Q<30 or any Q<25 within an 11 basepair region centered on the mismatch, as has been applied previously to rigorous SNP discovery from shotgun data [32]. To overcome artifacts of PCR infidelity due to slippage events leading to shortened repeats and false haplotypes, we set a high threshold requiring that co-occurring haplotypes of the same repeat type be at high frequency in order to exclude the low frequency variation/stuttering in the repeat region. Haplotype repeat type was then determined by translation and the most frequent haplotype of each major repeat type (VK210 and VK247) present was kept >0.5%. Additional haplotypes of major repeat types were kept if they were common (>20%) and thus unlikely to be due simply to low frequency slippage events. In total across all samples 4081 of the 8430 reads clustered contributed to utilized haplotypes.
The long-read haplotypes determined through consensus clustering were used as templates for short-read alignment using Bowtie2 v 2.1.0 [33], with very-sensitive alignment parameters and stringent filtering for Mapping Quality Score and Alignment Score. Final sequence predictions were used for the analyses in this paper and were deposited in GenBank under accession numbers JX461243-JX461285 and KJ173797- KJ173802 for pvcsp, and JX461286-JX461333 for pvmsp-1.
Rarefaction curves of haplotypes were calculated using EstimateS v9.0. Individual-based curves using sampling without replacement were estimated [34] and extrapolated to 2× the actual sample number [35]. Rarefaction plots were visualized in the R base package (http://cran.us.r-project.org/).
Acquisition of published sequences for inter-population comparisons
GenBank was queried for population sets published prior to August 1, 2013, which included sequence data for the 42 kDa region of pvmsp-1 and the whole-gene of pvcsp. Sequences from a recent publication [36] were excluded because the isolates were collected over the course of a 12 year period. The authors provide evidence that the haplotype distribution of this population changed substantially over time, making this population inappropriate for our analysis of selection.
Assessing selection on pvmsp-1 and pvcsp
Population datasets with >25 sequences that were collected over a span of ≤4 years were included for analysis of selection. We used DnaSP v5.1 to perform tests of selection [37]. We calculated polymorphism and Tajima's D across pvmsp-1 and the pvcsp constant regions using a 50 bp sliding window with a 25 bp step size. We also performed 1000 coalescent simulations with recombination to determine a 95% confidence interval and centile for each Tajima's D estimate [38]. To test for long-term selection, we used the McDonald-Kreitman (MK) test [39]. Skew was calculated using Fisher's exact test (two tailed). For the pvmsp-1 42 kDa region amplicons reported here and by others, 15 Plasmodium knowlesi pkmsp-1 isolates from Thailand [40] (Accession Nos. JF837339-JF837353) were used as the interspecies outgroup. Three insertions and deletions occurred in the 42 kDa region of pvmsp-1 relative to pkmsp-1, and were not considered. We could not obtain MK estimates for pvcsp sequences due to numerous insertions and deletions relative to pkcsp.
For analysis of pvcsp repeats, we performed pairwise comparisons of untranslated repeat units within individual pvcsp sequences [41]. We calculated skewness and mean nucleotide differences between repeat units, as previously reported [42]. Similar to the methods of Dias et al., 2013, we also calculated dN/dS on the first 1–459 bases of all 32 VK210 repeat regions and the first 1–540 bases of all 15 VK247 repeat regions. This analysis was performed in MEGA5, using the Nei-Gojobori method [43].
Phylogenetics and statistics to determine population structure
Interpopulation heterogeneity was first assessed using Wright's fixation index (F ST). Pairwise fixation values between pvmsp-1 populations were calculated in DnaSP. Site-specific fixation values for pairwise comparisons among Cambodia, NW Thailand, S Thailand, India, and Turkey were generated using the analysis of molecular variance (AMOVA) function within Arlequin v3.11 [44].
Neighbor-joining trees for pvmsp-1, pvcsp VK210, and pvcsp VK247 were drawn using the APE package for R [45]. To generate trees based off pvmsp-1, distance calculations between haplotypes were performed in MEGA5 using the maximum composite likelihood method to construct a neighbor-joining tree file. For the pvcsp CR, we used MS_Align (v.2.0) [46], [47] to create genetic distance matrices separately comparing both the VK210 and VK247 repeat arrays. MS_Align generates an event-based genetic distance using a model of tandem repeat evolution (expansion, deletion, substitution). Cost parameters for MS_Align were set to 0.1 for amplification or contraction and 5 for repeat insertion or deletion. A pairwise cost table of repeat-to-repeat mutations was created in MEGA5 using the maximum composite likelihood method and used as input for MS_Align [41], [48]. MS_Align output matrices were used by FastME [49], [50] to construct neighbor-joining trees with balanced branch-length estimation.
To cluster geographic groups, we calculated Hudson's nearest-neighbor statistic (SNN) [51]. Input was in the form of a pairwise distance matrix between all haplotypes for each phylogeny. For this statistic, highly distant populations have values approaching 1 while panmictic populations have values near 0.5. To test the reproducibility of the geographic clustering predicted by SNN, 1000 jackknife samplings were constructed for both pvmsp-1 and pvcsp VK210 and VK247 populations using Fast UniFrac [52]. For each jackknife replicate, 5 individuals, based on the size of the smallest population, were randomly selected from each population and used to redraw trees. Observed splits between geographic populations were quantified and used to assign confidence to predicted geographic clusters. To evaluate potential mutational paths connecting all pvmsp-1 haplotypes, we constructed a median-joining network using NETWORK v4.6 (Fluxus Engineering, Suffolk, England) [53]. This method expresses multiple plausible evolutionary paths in the form of cycles. A similar analysis was not completed for pvcsp due to the variable length of CR haplotypes.
Results
pvmsp-1 sequences
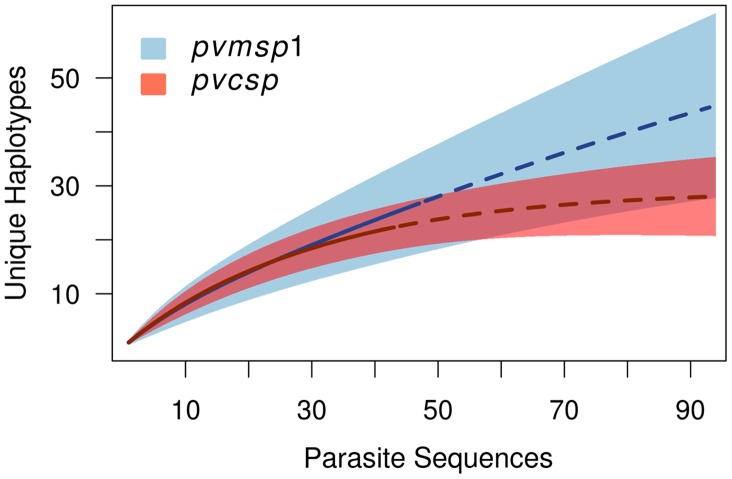
We Illumina sequenced pvmsp-1 42 kDa-fragments ( Figure 1A ) from 48 patients, and compared these to Sanger sequencing data for selected samples. Illumina haplotypes with a major allele frequency of >60% agreed with Sanger haplotypes in every case tested (n = 6). Illumina haplotypes with a major allele frequency of <60% did not consistently agree with Sanger haplotypes (n = 3). Thus, we were able to build 44 complete pvmsp-1 42 kDa haplotypes (26 unique haplotypes) with a major allele frequency of >60% at all polymorphic sites ( Table 1 ). The average coverage depth for all isolates was >800 reads per base, with all bases having ≥100 reads of coverage. Haplotype accumulation (rarefaction) curves were estimated, and then further extrapolated to show that our sample captured fewer than half the total pvmsp-1 haplotypes in this region of Cambodia ( Figure 2 ). In addition to these isolates, we identified 238 submissions in GenBank [54]–[58] (Table S1) containing either the whole-gene or 42 kDa-region sequence information.
Table 1. Summary population genetic data for Plasmodium vivax antigens.
| Country of Origin | n 1 | S 2 | K 3 | π 4 | H 5 | Hd 6 | Tajima's D |
| pvmsp-1 : 42 kDa region | |||||||
| Cambodia | 44 | 62 | 24.8 | 0.020 | 26 | 0.950 | 2.08* |
| India | 28 | 64 | 24.9 | 0.021 | 27 | 0.997 | 1.32* |
| NW Thailand | 65 | 62 | 24.9 | 0.020 | 34 | 0.968 | 2.42* |
| S Thailand | 67 | 42 | 6.46 | 0.005 | 5 | 0.336 | −0.986 |
| Turkey | 30 | 33 | 8.33 | 0.007 | 3 | 0.536 | −0.001 |
| pvcsp : N- and C-terminal non-repeat regions | |||||||
| Cambodia | 47 | - | - | - | 24 | - | - |
| N-terminal non-repeat | 3 | 0.971 | 0.003 | 3 | 0.500 | 0.901 | |
| C-terminal non-repeat | 2 | 0.318 | 0.001 | 2 | 0.159 | −0.538 | |
| Columbia | 27 | - | - | - | 27 | - | - |
| N-terminal non-repeat | 2 | 0.285 | 0.001 | 2 | 0.143 | −0.954 | |
| C-terminal non-repeat | 0 | - | - | 1 | 0.000 | - | |
This table includes all population sequence sets which contained sufficient numbers to perform allele-based tests of neutrality. Population sets which included sequence data only for pvcsp repeat regions alone are not summarized here.
*p<0.05;
number of haplotypes;
within-population variant sites;
average number of nucleotide differences;
nucleotide diversity;
number of haplotypes;
haplotype diversity.
Figure 2. Haplotype rarefaction curves for the Cambodian cohort.
Calculated rarefaction curves are depicted by solid blue (pvmsp-1) and red (pvcsp) lines. Dotted lines represent rarefaction values extrapolated according to the methods of Cowell, et al. The 95% CIs of rarefaction estimates for pvmsp-1 and pvcsp are demarked by light blue and light red shaded areas, respectively.
Detecting signatures of selection within pvmsp-1
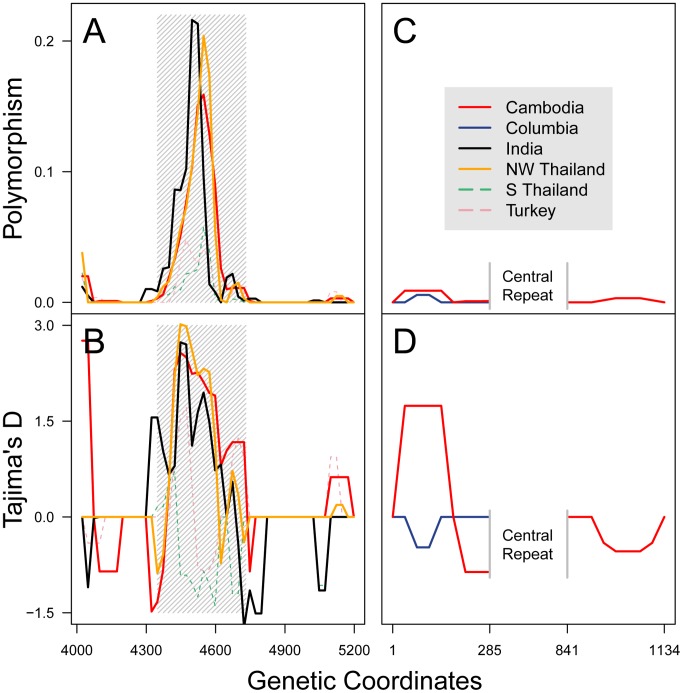
The interaction between human host and the parasite has had a profound impact on the parasite genome, leaving behind characteristic “signatures” of natural selection [59], which are detectable using population genetics approaches to examine sequence diversity. We first assessed nucleotide diversity ( Figure 3A ), and observed a spike of polymorphism in the region between the two HARBs (positions 4348–4731 in the Sal1 reference). We termed this the “intervening region”. To test whether the diversity in the intervening region is due to long-term selection, we used the McDonald-Kreitman (MK) test [39] to compare the ratio of non-synonymous to synonymous nucleotide polymorphisms between the Cambodian P. vivax population and a Thai P. knowlesi population [40]. We observed a highly elevated MK ratio (p = 0.00427) in the intervening region but not in the HARBs (data not shown) or the entire 42 kDa region (p = 0.681), suggesting that the intervening region is under long-term selective pressure ( Table 2 ).
Figure 3. Nucleotide diversity and Tajima's D across the pvmsp-1 42 kDa region and the whole pvcsp gene.
Polymorphism (nucleotide diversity, π) (A) and Tajima's D (B) were calculated across the pvmsp-1 amplicon for five diverse populations. A sliding window (50 bp window and 25 bp step size) was used to achieve a high resolution analysis. Grey hatches demark the intervening region (nucleotides 4348–4731). For pvcsp, N-terminal and C-terminal non-repeat regions were analyzed for nucleotide polymorphism (C) and evidence of balancing selection (D) using a sliding window. Putatively panmictic populations are marked with a solid line, while populations known to be subject to strong selective forces are marked with dotted lines. All coordinates are based on Sal1 pvmsp-1 and pvcsp reference sequences.
Table 2. McDonald-Kreitman test for selection in pvmsp-1.
| McDonald-Kreitman Comparisons | ||||
| 42 kDa region | 42 kDa intervening region | |||
| Synonymous | Non-synonymous | Synonymous | Non-synonymous | |
| Fixed | 96 | 93 | 32 | 28 |
| Polymorphic | 34 | 39 | 10 | 31 |
| p = 0.681 | p = 0.00427 | |||
Evidence for long-term selective pressure on the pvmsp-1 42 kDa region and the 42 kDa intervening region was assessed with the McDonald-Kreitman test, using P. knowlesi msp1 as the outgroup comparator. A Fisher's exact test (two tailed) was used to determine significance.
To determine whether the long-term selective pressure shaping the intervening region is potentially due to human immunity, we assessed balancing selection in this region, as balancing selection within a malaria antigen suggests that the antigen is a target of the human immune system [59]. We applied Tajima's D test of neutrality [60] to five geographically distinct P. vivax populations (all populations with n>25, accounting for 190 of 238 available sequences) ( Table 1 , Figure 3B ). In panmictic populations with an uncomplicated demographic history [59], the Tajima's D statistic can indicate whether a nucleotide sequence is under directional (D<0) or balancing selection (D>0). Populations not subjected to recent bottlenecks (i.e. Cambodia, India, and NW Thailand, [54], [58]) demonstrated a significant signature of balancing selection in the pvmsp-1 42 kDa region ( Table 1 ). This signature occurred specifically in the intervening region ( Figure 3B ), and is consistent with the conclusion that human immunity targets the intervening region.
The three regions of the pvmsp-1 fragment that are considered vaccine candidates were each assessed for diversity in the Cambodian population [9], [61]. In contrast to the intervening region, the 20 kDa HARB (Sal1 positions 4021–4347) and 14 kDa HARB (Sal1 positions 4732–4941) showed no coding polymorphisms and no evidence of balancing selection, similar to recent reports [61]. The 19 kDa fragment (Sal1 nucleotide positions 4918–5239) also showed limited diversity, with only a K1709E substitution, and no evidence of balancing selection.
Geospatial genetic diversity at the pvmsp-1 42 kDa region
Although the pvmsp-1 42 kDa region contains potential vaccine candidates [9], [61], the 42 kD region's global genetic diversity has not been carefully evaluated. To study pvmsp-1 42 kDa diversity, we calculated Wright's Fixation index (F ST) [62] for each pairwise comparison between five diverse populations ( Table 3 ). F ST values between naturally evolving parasite populations (Cambodia, NW Thailand, and India) approached zero, showing a high degree of genetic similarity, while comparisons with populations that have undergone a recent bottleneck (S Thailand and Turkey) showed a high degree of genetic distance due to their limited number of haplotypes. Similarly, F ST values calculated for each variable site demonstrate a high degree of homogeneity in pairwise comparisons between the Cambodia, NW Thailand, and India populations across all sites, and substantial heterogeneity between S Thailand and Turkey across all sites (Figure S2). This is evidence that balancing selection maintains a similar range of alleles in the pvmsp-1 42 kDa region of multiple geographically diverse naturally evolving P. vivax populations.
Table 3. Interpopulation F-statistics for pvmsp-1.
| pvmsp-1 Global F ST 0.340 | ||||
| pvmsp-1 Pairwise | Cambodia | India | NW Thailand | S Thailand |
| India | 0.031 | |||
| NW Thailand | 0.000 | 0.043 | ||
| S. Thailand | 0.449 | 0.433 | 0.366 | |
| Turkey | 0.361 | 0.329 | 0.403 | 0.796 |
F ST values compare the relatedness of a gene among different populations of the same species. Reported values compare the relatedness of pvmsp-1 42 kDa alleles for pairwise comparisons between Cambodia, India, NW Thailand, S Thailand, and Turkey. F ST values approaching 0 indicate greater relatedness, while values approaching 1 indicate substantial inter-population variability. Global F ST statistic calculated between all pvmsp-1 populations with n>25 indicates that relatively little genetic distance exists between the sampled populations. However, pairwise comparisons demonstrate that some populations exhibit a high degree of genetic similarity (Cambodia and India, for example) while other populations are more dissimilar (S Thailand and Turkey, for example).
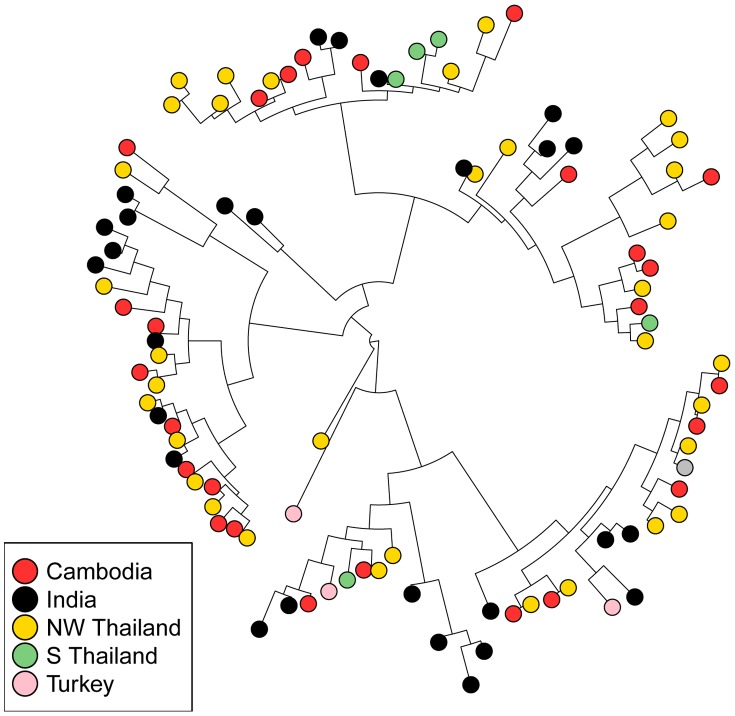
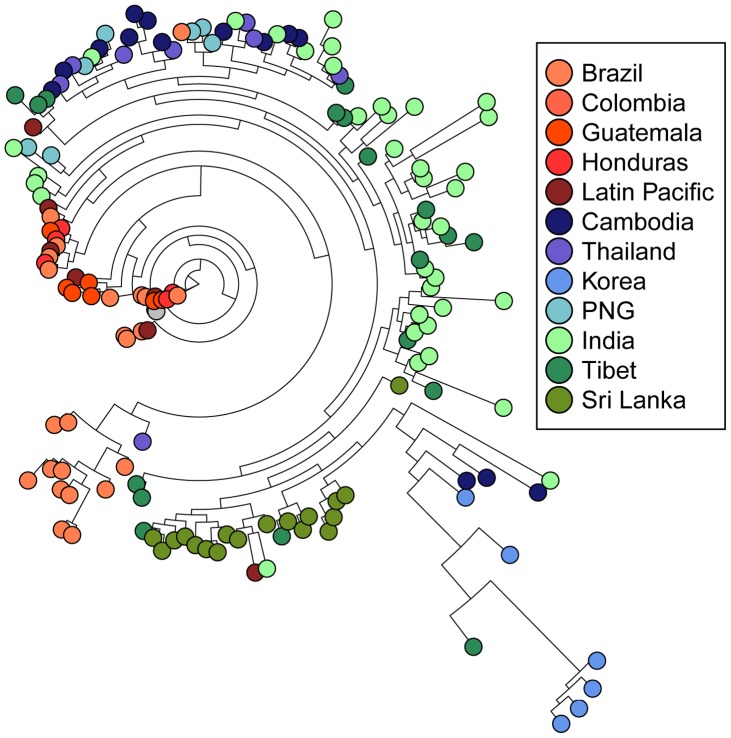
To visualize whether 42 kDa sequences cluster according to geography, we compared all unique haplotypes in a single neighbor-joining tree, which revealed little clustering according to geographic origin ( Figure 4 ). We quantified the extent of this clustering using Hudson's nearest-neighbor statistic (SNN), which assesses how frequently a variant's nearest neighbor is from the same population [51]. In both global and pairwise comparisons, pvmsp-1 42 kDa sequences from naturally evolving populations in Cambodia, India, and NW Thailand showed no evidence of strong geographic clustering ( Table 4 ). To further confirm this finding, a neighbor-joining consensus tree was created and underwent 1000 jackknifed replicates ( Figure 5A ). Results showed that the predicted splits between most populations occurred only less than 50% of the time, providing strong evidence that there is minimal geographic clustering of pvmsp-1 42 kDa sequences.
Figure 4. Neighbor-joining tree of 42 kDa regions from pvmsp-1 isolates.
All unique 42pvmsp-1 population set with n>25 were plotted on a single unrooted, neighbor-joining phylogenetic tree. The Sal1 reference sequence is marked in grey.
Table 4. Snn statistics for the pvmsp-1 42 kDa region and the pvcsp central repeat region.
| pvmsp-1 Global Snn 0.410* | Cambodia | NW Thailand | S Thailand | India | |||||
| Cambodia | |||||||||
| NW Thailand | 0.318 | ||||||||
| S Thailand | 0.780 | 0.865 | |||||||
| India | 0.673 | 0.779* | 0.865 | ||||||
| Turkey | 0.897 | 0.946 | 0.750 | 0.917 |
Snn values approaching 1 indicate genetic isolation while values near 0.5 indicate that two geographically disparate populations may approximate panmixia. Global and pairwise Snn values show stronger geographic clustering among pvcsp VK210 and VK247 repeats than among pvmsp-1 42 kDa regions.
* indicates significance to (p≤0.05) after Bonferroni correction for multiple comparisons.
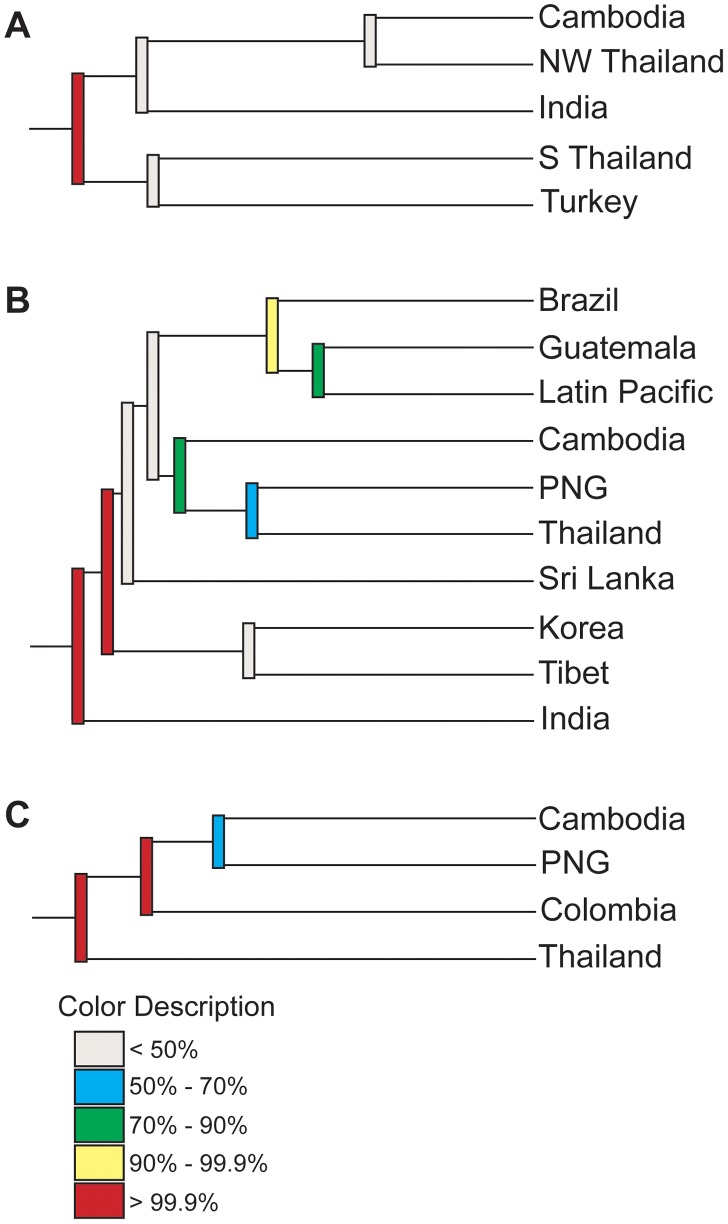
Figure 5. Jackknifed consensus trees demonstrate reproducible geographic clustering in pvcsp VK210 and VK247 isolates, but not pvmsp-1.
The reproducibility of population clustering was assessed using 1000 jackknifed phylogenies. Individual populations clustered together or apart in each of the 1000 jackknifed phylogenies, and the frequency of a split between any two populations was quantified. Populations with grey bars (<50% splits) were genetically similar, while populations with red bars (>99.9% splits) were highly genetically distinct. Phylogenies were built from the pvmsp-1 42 kDa region (A), the pvcsp VK210 central repeat (B), and pvcsp VK247 central repeat (C).
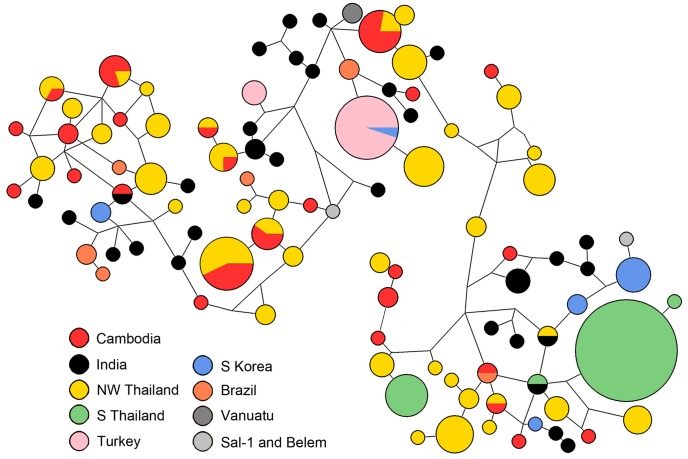
To better understand the evolutionary relationships between pvmsp-1 haplotypes from around the world, we employed a median-joining network to describe the set of potential mutational paths between all available global pvmsp-1 42 kDa sequences [53]. The network shows extensive admixture of parasite populations from diverse locales, with numerous mutational paths connecting haplotypes ( Figure 6 ). With the exception of populations from S Thailand and Turkey, which have undergone recent bottlenecks, these data provide further evidence that there is no clustering by geography.
Figure 6. Median-joining network of diverse pvmsp-1 populations proposes multiple mutational paths between geographically diverse populations.
286 pvmsp-1 42 kDa sequences from diverse geographical regions were used as input to create an unrooted median-joining network. This network is a visual representation of the mutational paths that may explain the observed sequence diversity. Each node represents an allele, node size represents the frequency of that allele (range n = 1 to n = 54), and node color corresponds to country of origin. Cycles within the diagram represent alternative evolutionary pathways. Corners represent obligate intermediate sequences that were not observed among the sampled alleles. Line length is not proportional to genetic distance.
pvcsp sequences
We sequenced the complete pvcsp gene from 43 isolates using the PacBio and Illumina platforms. de novo assembly of the Illumina paired-end short reads was not possible, due to over-collapse in the central repeat (CR) region, resulting in inappropriately short CRs. In contrast, PacBio long reads allowed the gene to be sequenced in its entirety and, after clustering, predicted 47 pvcsp haplotypes within the 43 samples. Reported error rates for PacBio sequencing have been high, especially for indels [63]; however, the use of Circular Consensus Sequencing allows single DNA fragments to be read multiple times, decreasing the error rate of the final predicted sequence. To check the accuracy of PacBio pvcsp haplotypes, individual haplotypes were used as a template for alignment of Illumina reads from the same clinical isolate. The addition of Illumina reads corrected only a single 1-bp deletion in a single haplotype. Therefore, after clustering, PacBio-predicted haplotypes have an error rate of 1/(∼1200 basepairs/sequence ×47 sequences), or approximately 0.002%.
Considering the entire gene, there were 24 unique haplotypes at the nucleotide level, and most genetic diversity was within the CR ( Figure 1 ). Both nonapeptide repeat array types – VK210 (total n = 32, range 17–21 repeat units) and VK247 (total n = 15, range 20–21 repeat units) – were represented in our Cambodian population, with no VK210–VK247 hybrids (reviewed in [64]). The average Illumina short-read depth for each isolate was >1000, with all bases having ≥5 reads of coverage. In addition to our isolates, we identified one cohort of nearly complete pvcsp sequences (n = 27), and 12 cohorts of CR sequences (n = 385) [65]–[70] ( Table 1 ). An extrapolated rarefaction curve showed that we sampled more than two thirds of the pvcsp CR haplotypes in this part of Cambodia, and that there are significantly fewer pvcsp CR variants in this region of Cambodia than pvmsp-1 42 kDa variants ( Figure 2 ).
Detecting signatures of selection within pvcsp
In contrast to pvmsp-1, the 5′ and 3′ non-repeat regions of pvcsp had no significant signatures of selection either by the MK test (data not shown) or Tajima's D test ( Table 1 ). The 5′ non-repeat region in the Cambodian cohort showed a non-significant signature of balancing selection ( Table 1 and Figure 3D ), which was due to a G38N amino acid polymorphism. This polymorphism also was observed in 6/16 parasites from the Latin Pacific region (JQ511263-JQ511276, JQ511279, JQ511286) and 2/27 parasites from Colombia (GU339072 and GU339085). The 3′ non-repeat region had little evidence of balancing selection, with Tajima's D values ∼0 ( Table 1 and Figure 3D ). Within pvcsp, an 18 amino-acid C-terminal motif known as Region II (amino acid residues 311–328 in Sal1) is important for parasite invasion of hepatocytes [71] and purportedly contains both B and T-cell epitopes [72], [73]. Among all Cambodia and Colombia parasite isolates, this motif is completely conserved at the nucleotide and protein level, with an amino-acid sequence of EWTPCSVTCGVGVRVRRR, similar to previous reports [61].
To better understand the selective forces acting upon the pvcsp CR, we assessed the dN/dS ratio for Cambodian VK210 and VK247 [66]. Strikingly, synonymous substitutions were strongly favored in both VK210 (dN/dS = 0.267; Z test p<0.001) and VK247 (dN/dS = 0.166; Z test p<0.001) repeats. This is consistent with the finding that VK210 and VK247 isolates from around the world consistently demonstrate a depressed dN/dS ratio, suggesting that the VK210 and VK247 repeat regions are both under strong purifying selection [66].
The CR of P. falciparum csp is thought to evolve by slipped-strand mispairing [42]. To understand if a similar mechanism works in the pvcsp repeats, we studied the mismatch distribution of pairwise genetic distances between untranslated repeat units within each VK210 and VK247 repeat array type in Cambodia. Consistent with another study [68], we observed a strong right skew in the proportion of genetic differences between pairwise VK210 repeat comparisons, and between pairwise VK247 repeat comparisons, evidence that pvcsp repeats have a high proportion of identical or nearly identical repeats (data not shown). This finding is consistent with a continuous and rapid expansion and contraction of repeats by slipped-strand mispairing, which may be a mechanism to evade host immunity [42].
Geospatial genetic diversity at the pvcsp central repeat
A recent study assessed global genetic diversity in the pvcsp CR, but did not define the correlates of differentiation between populations [66]. Moreover, this report investigated CR diversity by using a subset of the repeat region that was invariant in length. This approach may not reflect true population structure as it only assesses repeats early in the CR. Indeed, we have found that certain repeat types do cluster in locations within the repeat arrays (data not shown).
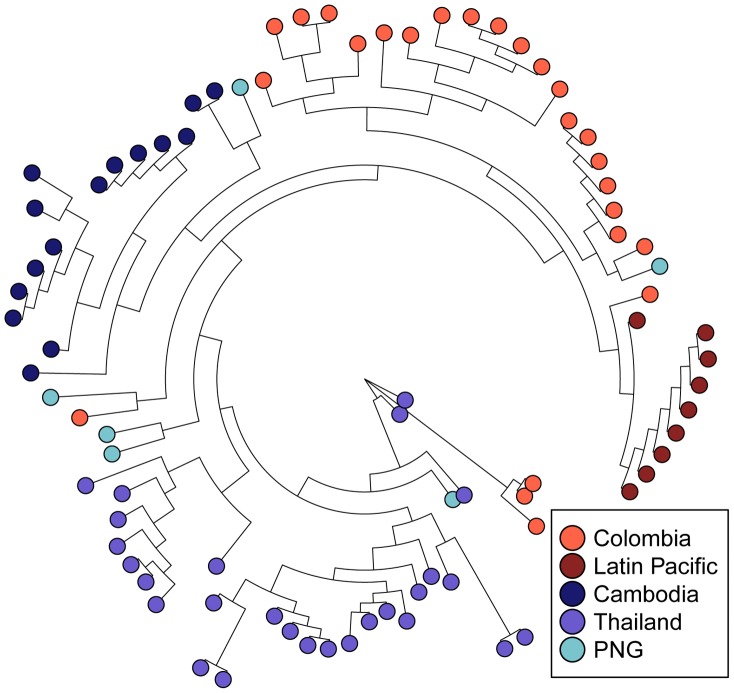
To more rigorously study the global diversity of the pvcsp CR, we modeled CR repeat expansion, contraction, and substitution using MS_Align, which calculates an event-based genetic distance between CR haplotypes [46]. From these data, we constructed neighbor-joining trees for global VK210 and VK247 repeat arrays isolates ( Figures 7 – 8 ). In contrast to pvmsp-1, the VK210 and VK247 trees revealed striking geographic clustering by country and continent. We quantified clustering using Hudson's SNN, and observed strong genetic differentiation between most geographically diverse parasite populations, in contrast to pvmsp-1 ( Table 4 ). To confirm this finding, neighbor-joining consensus trees for both VK210 and VK247 were subjected to 1000 jackknife replicates and the reproducibility of predicted splits between populations was tested demonstrating a strong correlation between genetic distance and geography ( Figure 5B–C ).
Figure 7. Neighbor-joining tree of pvcsp VK210 repeat arrays.
All unique repeat array haplotypes from each pvcsp VK210 population set were plotted on a single unrooted, neighbor-joining phylogenetic tree. Visual inspection reveals strong geographic clustering by region and country. Latin American sequences are in shades of red, South East Asian sequences are in shades of blue, and South and Central Asian sequences are in shades of green.
Figure 8. Neighbor-joining tree of pvcsp VK247 repeat arrays.
All repeat array haplotypes from each pvcsp VK247 population set were plotted on a single unrooted, neighbor-joining phylogenetic tree. Latin American sequences are in shades of red, South East Asian sequences are in shades of blue.
We were able to define the peptide sequence basis of the clustering observed among pvcsp CR repeats. For VK210 repeats, almost all (81/84) Latin American repeat arrays contained either a 5′ (GDRADGQPA)4 or an internal (GDRADGQPA)3–4, while very few (11/278) of the Asian sequences contained one or both of these features. Similarly, for VK247 repeat arrays, all (34/34) Latin American sequences began with a single EDGAGDQPG repeat, while only one (1/44) Asian sequence began with this repeat. These sequence features may represent a reliable method to assign sequences to a geographic region.
Discussion
This study (1) presents the first population set of pvmsp-1 and pvcsp sequences from Cambodia, (2) identifies a signature of putative immune-mediated, frequency-dependent selection in the pvmsp-1 42 kDa region and the pvcsp CR, and (3) provides the most comprehensive evaluation to date of geospatial genetic diversity for these genes. We also demonstrate the feasibility of using a next-generation sequencing approach to study the genetic diversity of malaria antigens.
A distinguishing feature of this study is the use of next-generation sequencing methods to generate P. vivax amplicon sequence data from clinical isolates. This work represents a first step into this largely unexplored territory. As a relatively new technology, next-generation sequencing methods must be validated before use in molecular epidemiological studies. We provide evidence that the dominant Illumina-predicted pvmsp-1 haplotypes are consistent with Sanger sequencing, and are fit for comparison with population sets generated by traditional sequencing methods. Methods for predicting multiple haplotypes from short-read sequencing are under development and will need further validation. We also demonstrate the ability of combined PacBio-Illumina haplotypes to predict pvcsp VK210 and VK247 haplotypes out of individual mixed infections. As next-generation sequencing methods are utilized more frequently for population genetic studies of infectious diseases, the methods introduced here will be further improved and will help to provide greater insight into Plasmodia population genetics.
Evidence of selection in both pvmsp-1 and pvcsp
We found compelling genetic evidence that the pvmsp-1 42 kDa intervening region is under strong immune pressure in multiple panmictic populations. Results from the MK test suggested that this region is under sustained selective pressure ( Table 2 ); however, because a positive MK test can signify balancing selection or weak negative selection [74], [75], we tested the hypothesis that this region is under balancing selection using Tajima's D test of neutrality. Since multiple populations showed strong evidence of balancing selection by Tajima's D ( Table 1 , Figure 3B ), we conclude that the intervening region is undergoing continual diversifying, balancing selection. An alternative hypothesis is that the positive Tajima's D values are an artifact of recent population contractions. Because (1) a positive Tajima's D was observed in multiple populations, and (2) other regions of pvmsp-1 contained negative Tajima's D values, we conclude that the 42 kDa intervening region of pvmsp-1 undergoes frequency-dependent (and likely immune-mediated) balancing selection.
Because PvMSP-1 is a merozoite surface antigen, it is highly accessible to antibodies and complement. The predicted structure of the 42 kDa region shows that the 33 kDa fragment covers the 19 kDa fragment [11], [76], limiting its exposure to the human immune system relative to the 33 kDa fragment. This observation could explain the extensive balancing selection present in the 33 kDa fragment (specifically, the intervening region) but not in the 19 kDa fragment. Additionally, this finding suggests that the sliding window approach for evaluating polymorphism and balancing selection may help generate hypotheses about functionally important (19 kDa fragment, for example) or immunologically dominant (the intervening region, for example) regions of P. vivax proteins.
For pvmsp-1, Tajima's D and F ST were inversely correlated. Populations with strong evidence of high Tajima's D in the pvmsp-1 intervening region showed a low genetic differentiation by F ST. This suggests that in naturally evolving populations, diversification of this region is extensive and maintains a similar range of genetic diversity despite geographic distance. Populations that have undergone a recent bottleneck show a low Tajima's D with relatively few variants and strong genetic differentiation from more diverse populations. This suggests that if strain-specific immune responses are important in vaccine efficacy, vaccines may work more effectively if other interventions can be used to bottleneck the population, thus decreasing its genetic diversity [54].
The central repeat region (CR) is a primary immunodominant region of PvCSP. Though alignment-based methods to assess for selection (Tajima's D, for example) cannot be employed in a tandem repeat region, there is wide-ranging evidence that selective pressures shape the genetic composition of the pvcsp CR [77]–[81], including new evidence hinting that hosts develop strain-specific immunity to P. falciparum NANP repeats of varying lengths [82]. Indeed, the presence of two distinct repeat types (VK210 and VK247) may itself be evidence of selection as suggested in a study of the P. cynomolgi csp CR [80].
Our analysis of the two CR array types, VK210 and VK247, also suggests that selection is occurring in this region. In pairwise comparisons of nucleotide and amino acid differences we observed a positive skew showing decreased differences among repeat units. This finding is consistent with Patil et al.'s study of pvcsp isolates from Brazil [68], and provides further evidence that both VK210 and VK247 repeat arrays may continuously evolve via slipped-strand mispairing [42]. Furthermore, consistent with a recent study of selection in worldwide pvcsp isolates [66], we found that Cambodian pvcsp VK210 and VK247 isolates have a strong bias toward synonymous substitutions. This signature of purifying selection is consistent with reports from pfcsp [83]–[85] and suggests that there are a limited number of amino acid polymorphisms allowable within this repeat region. Taken together, these findings suggest that expansion, contraction, and rearrangement of repeat units, rather than generation of novel repeat units through mutation, maintain genetic diversity at the pvcsp locus in both VK210 and VK247 variants. This phenomenon may be responsible for immune evasion [68], [86].
Although these two vivax genes are orthologs of well-characterized vaccine candidate antigens from P. falciparum malaria, substantial differences are seen in the effects of immune selection between these genes and their orthologs. Previous reports have shown that the functionally similar pfmsp-1 42 kDa fragment has relatively low nucleotide diversity and lacks evidence of balancing selection by Tajima's D [87]. pfcsp, on the other hand, shows a high level of nucleotide diversity [88]–[90] and modest Tajima's D elevations in the C-terminal T cell epitopes [88], [91]. These patterns are in stark contrast to our observations in P. vivax, and this highlights the need for P. vivax-specific studies to determine appropriate candidate vaccine antigens.
Finally, our analysis of the pvmsp-1 42 kDa region underscores the importance of selecting an appropriate parasite population for population-genetic studies. We did not observe signatures of balancing selection in pvmsp-1 populations from S Thailand or Turkey. This is likely due to bottlenecks secondary to robust malaria control measures employed in S Thailand [54] and limited human migration in Turkey [58]. Thus, appropriate selection of panmictic populations for these studies is critical.
Differing patterns of geospatial genetic diversity at pvmsp-1 and pvcsp
Using both tree-based and statistical methods [92], we found that pvcsp, but not pvmsp-1, showed strong clustering by geography ( Tables 3 – 4 and Figures 4 – 8 ). For pvmsp-1, we observed little geographic clustering among naturally evolving parasite populations, suggesting that immune selection maintains similar pvmsp-1 alleles around the globe. Notably similar findings have been described in Duffy Binding Protein and Thrombospondin-related anonymous protein in vivax malaria [61], while a recent global survey of diversity in the Apical Membrane Antigen 1 found evidence of geographically restricted haplotypes [93]. In contrast to pvmsp-1, we found that pvcsp variants demonstrate strong evidence of geographic clustering. This juxtaposition between pvmsp-1 and pvcsp sequences is similar to what has previously been described for merozoite and sporozoite antigens in P. falciparum [94]. The population sets included in this survey were collected in different years. While it is known that novel P. vivax surface antigen types can appear in the course of a decade [57], it is difficult to assess the magnitude of this effect on our analyses. As more pvmsp-1 and pvcsp population sets are collected, this will become clearer.
It is interesting that the CR of pvcsp shows evidence of multiple forms of selection: (1) the depressed number of non-synonymous mutations suggests purifying selection, (2) the differences in CR genotypes between geographic locations suggests directional selection, and (3) the genetic composition of the repeats suggests rapid expansion and contraction, possibly due to immune selection. It is unclear what drives the first two signatures of selection. We hypothesize a model in which purifying selection within a population limits the amino acid composition of repeats due to functional constraints of the protein, while directional selection between populations is driven by environmental factors.
One environmental factor that may explain both the purifying and directional selection of parasite pvcsp CR sequences is the mosquito vector. The circumsporozoite protein is expressed in the mosquito during oocyst development [95] and in the salivary glands [96], [97]. It is also critical in sporozoite motility [15]. We found no overlap in the distribution of Anopheline species between the countries from Asia and Latin America included in this study (data not shown) [98]–[100]. Furthermore, there is substantial evidence that different Anopheline species and strains show differential ability to be infected by malaria [101]–[104].
Regardless of the cause of the differing patterns of geospatial genetic diversity we observed in pvmsp-1 and pvcsp, the observation itself has significance for vaccine design. The malaria vaccine field is just beginning to unravel how antigenic diversity within a single parasite population can reduce vaccine efficacy [105]. Our findings highlight an additional level of complexity that will hinder the implementation of a vivax vaccine – antigenic variability. While the effects of immune cross-reactivity against different antigenic variants aren't fully known, the extensive intrapopulation variability seen in pvmsp-1 may necessitate a highly multivalent pvmsp-1 vaccine, while the dramatic interpopulation variability seen in pvcsp suggests that a PvCSP-based vaccine that is effective in one part of the globe may not be effective in other regions. Thus, a thorough understanding of the geospatial genetic diversity of candidate vaccine antigens must inform antigen selection for vaccine design.
GenBank accession numbers
pvcsp sequences: JX461243-JX461285 and KJ173797- KJ173802
pvmsp-1 sequences: JX461286-JX461333
Supporting Information
Geographic distribution of P. vivax populations contributing to this study. In total, we identified 13 populations with pvmsp-1 42 kDa fragment sequences and 13 populations with pvcsp central repeat or whole-gene sequences. These populations were collected from 14 countries, pictured above. For countries with n≥10 isolates, the total number of pvmsp-1 and pvcsp isolates is marked.
(TIF)
F ST values at polymorphic sites within the pvmsp-1 42 kDa intervening region. Available parasite populations with n>25 individuals (Cambodia, India, NW Thailand, S Thailand, and Turkey) share 42 variable sites within the 42 kDa intervening region of pvmsp-1. F ST values for each variable site were calculated in a pairwise manner between all five populations. F ST values approaching 0 indicate limited inter-population variability at that site, while values approaching 1 indicate substantial inter-population variability. Coordinates are reported for every third polymorphic site.
(TIF)
pvmsp-1 and pvcsp sequences included in this study. The PlasmoDB gene identifier is PVX_099980 for pvmsp-1 and PVX_119355 for pvcsp. *Indicates the year the sequences were made available in GenBank. †Indicates study unpublished but sequences available in GenBank.
(DOCX)
Funding Statement
This work was supported by the US Department of Defense Global Emerging Infections Surveillance and Response System (DoD-GEIS) Program (for funding of the clinical trial), the University of North Carolina Research Council (UL1TR000083) and from the National Institutes of Health (AI089819 to JJJ). CMP was supported by the UNC MD/PhD Program (T32 GM008719) and Genetics Curriculum (T32 GM007092) and a grant from the Infectious Disease Society of America Medical Scholars Program. The views expressed in this paper are those of the authors and do not represent the official position of the U.S. Department of Defense, NIH, or UNC Chapel Hill. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Mueller I, Galinski MR, Baird JK, Carlton JM, Kochar DK, et al. (2009) Key gaps in the knowledge of Plasmodium vivax, a neglected human malaria parasite. Lancet Infect Dis 9: 555–566 doi:10.1016/S1473-3099(09)70177-X [DOI] [PubMed] [Google Scholar]
- 2. Tjitra E, Anstey NM, Sugiarto P, Warikar N, Kenangalem E, et al. (2008) Multidrug-resistant Plasmodium vivax associated with severe and fatal malaria: a prospective study in Papua, Indonesia. PLoS Med 5: e128 doi:10.1371/journal.pmed.0050128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Price RN, Douglas NM, Anstey NM (2009) New developments in Plasmodium vivax malaria: severe disease and the rise of chloroquine resistance. Curr Opin Infect Dis 22: 430–435 doi:10.1097/QCO.0b013e32832f14c1 [DOI] [PubMed] [Google Scholar]
- 4. Marfurt J, de Monbrison F, Brega S, Barbollat L, Müller I, et al. (2008) Molecular markers of in vivo Plasmodium vivax resistance to amodiaquine plus sulfadoxine-pyrimethamine: mutations in pvdhfr and pvmdr1. J Infect Dis 198: 409–417 doi:10.1086/589882 [DOI] [PubMed] [Google Scholar]
- 5. Greenwood B, Targett G (2009) Do we still need a malaria vaccine? Parasite Immunol 31: 582–586 doi:10.1111/j.1365-3024.2009.01140.x [DOI] [PubMed] [Google Scholar]
- 6. Baird JK (2013) Evidence and Implications of Mortality Associated with Acute Plasmodium vivax Malaria. Clin Microbiol Rev 26: 36–57 doi:10.1128/CMR.00074-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. PATH Malaria Vaccine Initiative (2011) Staying the course? Malaria research and development in a time of economic uncertainty [Google Scholar]
- 8. Arévalo-Herrera M, Chitnis C, Herrera S (2010) Current status of Plasmodium vivax vaccine. Hum Vaccin 6: 124–132. [DOI] [PubMed] [Google Scholar]
- 9. Espinosa AM, Sierra AY, Barrero CA, Cepeda LA, Cantor EM, et al. (2003) Expression, polymorphism analysis, reticulocyte binding and serological reactivity of two Plasmodium vivax MSP-1 protein recombinant fragments. Vaccine 21: 1033–1043. [DOI] [PubMed] [Google Scholar]
- 10. Collins WE, Kaslow DC, Sullivan JS, Morris CL, Galland GG, et al. (1999) Testing the efficacy of a recombinant merozoite surface protein (MSP-1(19) of Plasmodium vivax in Saimiri boliviensis monkeys. Am J Trop Med Hyg 60: 350–356. [DOI] [PubMed] [Google Scholar]
- 11. Blackman MJ, Heidrich HG, Donachie S, McBride JS, Holder AA (1990) A single fragment of a malaria merozoite surface protein remains on the parasite during red cell invasion and is the target of invasion-inhibiting antibodies. J Exp Med 172: 379–382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Guevara Patiño JA, Holder AA, McBride JS, Blackman MJ (1997) Antibodies that inhibit malaria merozoite surface protein-1 processing and erythrocyte invasion are blocked by naturally acquired human antibodies. J Exp Med 186: 1689–1699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Udhayakumar V, Anyona D, Kariuki S, Shi YP, Bloland PB, et al. (1995) Identification of T and B Cell Epitopes Recognized by Humans in the C- Terminal 42-kDa Domain of the Plasmodium Falciparum Merozoite Surface Protein (MSP)-1. J Immunol 154: 6022–6030. [PubMed] [Google Scholar]
- 14. Nwuba RI, Sodeinde O, Anumudu CI, Omosun YO, Odaibo AB, et al. (2002) The Human Immune Response to Plasmodium falciparum Includes Both Antibodies That Inhibit Merozoite Surface Protein 1 Secondary Processing and Blocking Antibodies. Infect Immun 70: 5328–5331 doi:10.1128/IAI.70.9.5328-5331.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Sultan AA (1999) Molecular mechanisms of malaria sporozoite motility and invasion of host cells. Int Microbiol Off J Span Soc Microbiol 2: 155–160. [PubMed] [Google Scholar]
- 16. Agnandji ST, Lell B, Soulanoudjingar SS, Fernandes JF, Abossolo BP, et al. (2011) First results of phase 3 trial of RTS,S/AS01 malaria vaccine in African children. N Engl J Med 365: 1863–1875 doi:10.1056/NEJMoa1102287 [DOI] [PubMed] [Google Scholar]
- 17. Takala SL, Plowe CV (2009) Genetic diversity and malaria vaccine design, testing and efficacy: preventing and overcoming “vaccine resistant malaria.”. Parasite Immunol 31: 560–573 doi:10.1111/j.1365-3024.2009.01138.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Thera MA, Doumbo OK, Coulibaly D, Laurens MB, Ouattara A, et al. (2011) A field trial to assess a blood-stage malaria vaccine. N Engl J Med 365: 1004–1013 doi:10.1056/NEJMoa1008115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Genton B, Betuela I, Felger I, Al-Yaman F, Anders RF, et al. (2002) A Recombinant Blood-Stage Malaria Vaccine Reduces Plasmodium falciparum Density and Exerts Selective Pressure on Parasite Populations in a Phase 1-2b Trial in Papua New Guinea. J Infect Dis 185: 820–827 doi:10.1086/339342 [DOI] [PubMed] [Google Scholar]
- 20. Pattaradilokrat S, Cheesman SJ, Carter R (2007) Linkage Group Selection: Towards Identifying Genes Controlling Strain Specific Protective Immunity in Malaria. PLoS ONE 2: e857 doi:10.1371/journal.pone.0000857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Martinelli A, Cheesman S, Hunt P, Culleton R, Raza A, et al. (2005) A genetic approach to the de novo identification of targets of strain-specific immunity in malaria parasites. Proc Natl Acad Sci U S A 102: 814–819 doi:10.1073/pnas.0405097102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Enosse S, Dobaño C, Quelhas D, Aponte JJ, Lievens M, et al. (2006) RTS,S/AS02A malaria vaccine does not induce parasite CSP T cell epitope selection and reduces multiplicity of infection. PLoS Clin Trials 1: e5 doi:10.1371/journal.pctr.0010005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Kumkhaek C, Phra-ek K, Rénia L, Singhasivanon P, Looareesuwan S, et al. (2005) Are Extensive T Cell Epitope Polymorphisms in the Plasmodium falciparum Circumsporozoite Antigen, a Leading Sporozoite Vaccine Candidate, Selected by Immune Pressure? J Immunol 175: 3935–3939. [DOI] [PubMed] [Google Scholar]
- 24. Alloueche A, Milligan P, Conway DJ, Pinder M, Bojang K, et al. (2003) Protective Efficacy of the Rts,s/as02 Plasmodium Falciparum Malaria Vaccine Is Not Strain Specific. Am J Trop Med Hyg 68: 97–101. [PubMed] [Google Scholar]
- 25. Kumkhaek C, Phra-Ek K, Rénia L, Singhasivanon P, Looareesuwan S, et al. (2005) Are extensive T cell epitope polymorphisms in the Plasmodium falciparum circumsporozoite antigen, a leading sporozoite vaccine candidate, selected by immune pressure? J Immunol Baltim Md 1950 175: 3935–3939. [DOI] [PubMed] [Google Scholar]
- 26. Abdulla S, Salim N, Machera F, Kamata R, Juma O, et al. (2013) Randomized, controlled trial of the long term safety, immunogenicity and efficacy of RTS,S/AS02(D) malaria vaccine in infants living in a malaria-endemic region. Malar J 12: 11 doi:10.1186/1475-2875-12-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Olotu A, Fegan G, Wambua J, Nyangweso G, Awuondo KO, et al. (2013) Four-Year Efficacy of RTS,S/AS01E and Its Interaction with Malaria Exposure. N Engl J Med 368: 1111–1120 doi:10.1056/NEJMoa1207564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Lin JT, Juliano JJ, Kharabora O, Sem R, Lin F-C, et al. (2012) Individual Plasmodium vivax msp1 Variants within Polyclonal P. vivax Infections Display Different Propensities for Relapse. J Clin Microbiol 50: 1449–1451 doi:10.1128/JCM.06212-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Rogers WO, Sem R, Tero T, Chim P, Lim P, et al. (2009) Failure of artesunate-mefloquine combination therapy for uncomplicated Plasmodium falciparum malaria in southern Cambodia. Malar J 8: 10 doi:10.1186/1475-2875-8-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Givens MB, Lin JT, Lon C, Gosi P, Char MC, et al. (2014) Development of a Capillary Electrophoresis-Based Heteroduplex Tracking Assay To Measure In-Host Genetic Diversity of Initial and Recurrent Plasmodium vivax Infections in Cambodia. J Clin Microbiol 52: 298–301 doi:10.1128/JCM.02274-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Ngrenngarmlert W, Kwiek JJ, Kamwendo DD, Ritola K, Swanstrom R, et al. (2005) Measuring Allelic Heterogeneity in Plasmodium Falciparum by a Heteroduplex Tracking Assay. Am J Trop Med Hyg 72: 694–701. [PubMed] [Google Scholar]
- 32. Altshuler D, Pollara VJ, Cowles CR, Van Etten WJ, Baldwin J, et al. (2000) An SNP map of the human genome generated by reduced representation shotgun sequencing. Nature 407: 513–516 doi:10.1038/35035083 [DOI] [PubMed] [Google Scholar]
- 33. Langmead B, Salzberg SL (2012) Fast gapped-read alignment with Bowtie 2. Nat Methods 9: 357–359 doi:10.1038/nmeth.1923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Colwell RK, Mao CX, Chang J (2004) INTERPOLATING, EXTRAPOLATING, AND COMPARING INCIDENCE-BASED SPECIES ACCUMULATION CURVES. Ecology 85: 2717–2727 doi:10.1890/03-0557 [Google Scholar]
- 35. Colwell RK, Chao A, Gotelli NJ, Lin S-Y, Mao CX, et al. (2012) Models and estimators linking individual-based and sample-based rarefaction, extrapolation and comparison of assemblages. J Plant Ecol 5: 3–21 doi:10.1093/jpe/rtr044 [Google Scholar]
- 36. Kang J-M, Ju H-L, Kang Y-M, Lee D-H, Moon S-U, et al. (2012) Genetic polymorphism and natural selection in the C-terminal 42 kDa region of merozoite surface protein-1 among Plasmodium vivax Korean isolates. Malar J 11: 206 doi:10.1186/1475-2875-11-206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Librado P, Rozas J (2009) DnaSP v5: a software for comprehensive analysis of DNA polymorphism data. Bioinforma Oxf Engl 25: 1451–1452 doi:10.1093/bioinformatics/btp187 [DOI] [PubMed] [Google Scholar]
- 38. Hudson RR (1987) Estimating the recombination parameter of a finite population model without selection. Genet Res 50: 245–250. [DOI] [PubMed] [Google Scholar]
- 39. McDonald JH, Kreitman M (1991) Adaptive protein evolution at the Adh locus in Drosophila. Nature 351: 652–654 doi:10.1038/351652a0 [DOI] [PubMed] [Google Scholar]
- 40. Jongwutiwes S, Buppan P, Kosuvin R, Seethamchai S, Pattanawong U, et al. (2011) Plasmodium knowlesi Malaria in humans and macaques, Thailand. Emerg Infect Dis 17: 1799–1806 doi:10.3201/eid1710.110349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Tamura K, Peterson D, Peterson N, Stecher G, Nei M, et al. (2011) MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 28: 2731–2739 doi:10.1093/molbev/msr121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Hughes AL (2004) The evolution of amino acid repeat arrays in Plasmodium and other organisms. J Mol Evol 59: 528–535 doi:10.1007/s00239-004-2645-4 [DOI] [PubMed] [Google Scholar]
- 43. Nei M, Gojobori T (1986) Simple methods for estimating the numbers of synonymous and nonsynonymous nucleotide substitutions. Mol Biol Evol 3: 418–426. [DOI] [PubMed] [Google Scholar]
- 44. Excoffier L, Laval G, Schneider S (2005) Arlequin (version 3.0): an integrated software package for population genetics data analysis. Evol Bioinforma Online 1: 47–50. [PMC free article] [PubMed] [Google Scholar]
- 45. Paradis E, Claude J, Strimmer K (2004) APE: Analyses of Phylogenetics and Evolution in R language. Bioinformatics 20: 289–290 doi:10.1093/bioinformatics/btg412 [DOI] [PubMed] [Google Scholar]
- 46. Bérard S, Nicolas F, Buard J, Gascuel O, Rivals E (2006) A fast and specific alignment method for minisatellite maps. Evol Bioinforma Online 2: 303–320. [PMC free article] [PubMed] [Google Scholar]
- 47. Bérard S, Rivals E (2003) Comparison of minisatellites. J Comput Biol J Comput Mol Cell Biol 10: 357–372 doi:10.1089/10665270360688066 [DOI] [PubMed] [Google Scholar]
- 48. Tamura K, Nei M, Kumar S (2004) Prospects for inferring very large phylogenies by using the neighbor-joining method. Proc Natl Acad Sci U S A 101: 11030–11035 doi:10.1073/pnas.0404206101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Desper R, Gascuel O (2002) Fast and accurate phylogeny reconstruction algorithms based on the minimum-evolution principle. J Comput Biol J Comput Mol Cell Biol 9: 687–705 doi:10.1089/106652702761034136 [DOI] [PubMed] [Google Scholar]
- 50. Saitou N, Nei M (1987) The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 4: 406–425. [DOI] [PubMed] [Google Scholar]
- 51. Hudson RR (2000) A new statistic for detecting genetic differentiation. Genetics 155: 2011–2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Hamady M, Lozupone C, Knight R (2010) Fast UniFrac: facilitating high-throughput phylogenetic analyses of microbial communities including analysis of pyrosequencing and PhyloChip data. ISME J 4: 17–27 doi:10.1038/ismej.2009.97 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Bandelt HJ, Forster P, Röhl A (1999) Median-joining networks for inferring intraspecific phylogenies. Mol Biol Evol 16: 37–48. [DOI] [PubMed] [Google Scholar]
- 54. Jongwutiwes S, Putaporntip C, Hughes AL (2010) Bottleneck effects on vaccine-candidate antigen diversity of malaria parasites in Thailand. Vaccine 28: 3112–3117 doi:10.1016/j.vaccine.2010.02.062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Putaporntip C, Jongwutiwes S, Sakihama N, Ferreira MU, Kho W-G, et al. (2002) Mosaic organization and heterogeneity in frequency of allelic recombination of the Plasmodium vivax merozoite surface protein-1 locus. Proc Natl Acad Sci U S A 99: 16348–16353 doi:10.1073/pnas.252348999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Thakur A, Alam MT, Sharma YD (2008) Genetic diversity in the C-terminal 42 kDa region of merozoite surface protein-1 of Plasmodium vivax (PvMSP-1(42)) among Indian isolates. Acta Trop 108: 58–63 doi:10.1016/j.actatropica.2008.08.011 [DOI] [PubMed] [Google Scholar]
- 57. Han E-T, Wang Y, Lim CS, Cho JH, Chai J-Y (2011) Genetic diversity of the malaria vaccine candidate merozoite surface protein 1 gene of Plasmodium vivax field isolates in Republic of Korea. Parasitol Res 109: 1571–1576 doi:10.1007/s00436-011-2413-5 [DOI] [PubMed] [Google Scholar]
- 58. Zeyrek FY, Tachibana S-I, Yuksel F, Doni N, Palacpac N, et al. (2010) Limited polymorphism of the Plasmodium vivax merozoite surface protein 1 gene in isolates from Turkey. Am J Trop Med Hyg 83: 1230–1237 doi:10.4269/ajtmh.2010.10-0353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Weedall GD, Conway DJ (2010) Detecting signatures of balancing selection to identify targets of anti-parasite immunity. Trends Parasitol 26: 363–369 doi:10.1016/j.pt.2010.04.002 [DOI] [PubMed] [Google Scholar]
- 60. Tajima F (1989) Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics 123: 585–595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Chenet SM, Tapia LL, Escalante AA, Durand S, Lucas C, et al. (2012) Genetic diversity and population structure of genes encoding vaccine candidate antigens of Plasmodium vivax. Malar J 11: 68 doi:10.1186/1475-2875-11-68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Hudson RR, Slatkin M, Maddison WP (1992) Estimation of levels of gene flow from DNA sequence data. Genetics 132: 583–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Carneiro MO, Russ C, Ross MG, Gabriel SB, Nusbaum C, et al. (2012) Pacific biosciences sequencing technology for genotyping and variation discovery in human data. BMC Genomics 13: 375 doi:10.1186/1471-2164-13-375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Lim CS, Tazi L, Ayala FJ (2005) Plasmodium vivax: recent world expansion and genetic identity to Plasmodium simium. Proc Natl Acad Sci U S A 102: 15523–15528 doi:10.1073/pnas.0507413102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Henry-Halldin CN, Sepe D, Susapu M, McNamara DT, Bockarie M, et al. (2011) High-throughput molecular diagnosis of circumsporozoite variants VK210 and VK247 detects complex Plasmodium vivax infections in malaria endemic populations in Papua New Guinea. Infect Genet Evol J Mol Epidemiol Evol Genet Infect Dis 11: 391–398 doi:10.1016/j.meegid.2010.11.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Dias S, Wickramarachchi T, Sahabandu I, Escalante AA, Udagama PV (2013) Population genetic structure of the Plasmodium vivax circumsporozoite protein (Pvcsp) in Sri Lanka. Gene 518: 381–387 doi:10.1016/j.gene.2013.01.003 [DOI] [PubMed] [Google Scholar]
- 67. Hernández-Martínez MÁ, Escalante AA, Arévalo-Herrera M, Herrera S (2011) Antigenic diversity of the Plasmodium vivax circumsporozoite protein in parasite isolates of Western Colombia. Am J Trop Med Hyg 84: 51–57 doi:10.4269/ajtmh.2011.09-0785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Patil A, Orjuela-Sánchez P, da Silva-Nunes M, Ferreira MU (2010) Evolutionary dynamics of the immunodominant repeats of the Plasmodium vivax malaria-vaccine candidate circumsporozoite protein (CSP). Infect Genet Evol J Mol Epidemiol Evol Genet Infect Dis 10: 298–303 doi:10.1016/j.meegid.2010.01.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Santos-Ciminera PD, Alecrim M das GC, Roberts DR, Quinnan GV Jr (2007) Molecular epidemiology of Plasmodium vivax in the State of Amazonas, Brazil. Acta Trop 102: 38–46 doi:10.1016/j.actatropica.2007.02.013 [DOI] [PubMed] [Google Scholar]
- 70. Lopez AC, Ortiz A, Coello J, Sosa-Ochoa W, Torres REM, et al. (2012) Genetic diversity of Plasmodium vivax and Plasmodium falciparum in Honduras. Malar J 11: 391 doi:10.1186/1475-2875-11-391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Cerami C, Kwakye-Berko F, Nussenzweig V (1992) Binding of malarial circumsporozoite protein to sulfatides [Gal(3-SO4)beta 1-Cer] and cholesterol-3-sulfate and its dependence on disulfide bond formation between cysteines in region II. Mol Biochem Parasitol 54: 1–12. [DOI] [PubMed] [Google Scholar]
- 72. Sinnis P, Clavijo P, Fenyö D, Chait BT, Cerami C, et al. (1994) Structural and functional properties of region II-plus of the malaria circumsporozoite protein. J Exp Med 180: 297–306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Seth RK, Bhat AA, Rao DN, Biswas S (2010) Acquired immune response to defined Plasmodium vivax antigens in individuals residing in northern India. Microbes Infect 12: 199–206 doi:10.1016/j.micinf.2009.12.006 [DOI] [PubMed] [Google Scholar]
- 74. Tetteh KKA, Stewart LB, Ochola LI, Amambua-Ngwa A, Thomas AW, et al. (2009) Prospective Identification of Malaria Parasite Genes under Balancing Selection. PLoS ONE 4: e5568 doi:10.1371/journal.pone.0005568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Charlesworth J, Eyre-Walker A (2008) The McDonald–Kreitman Test and Slightly Deleterious Mutations. Mol Biol Evol 25: 1007–1015 doi:10.1093/molbev/msn005 [DOI] [PubMed] [Google Scholar]
- 76. Blackman MJ, Whittle H, Holder AA (1991) Processing of thePlasmodium falciparum major merozoite surface protein-1: identification of a 33-kilodalton secondary processing product which is shed prior to erythrocyte invasion. Mol Biochem Parasitol 49: 35–44 doi:10.1016/0166-6851(91)90128-S [DOI] [PubMed] [Google Scholar]
- 77. Arnot DE, Barnwell JW, Tam JP, Nussenzweig V, Nussenzweig RS, et al. (1985) Circumsporozoite protein of Plasmodium vivax: gene cloning and characterization of the immunodominant epitope. Science 230: 815–818 doi:10.1126/science.2414847 [DOI] [PubMed] [Google Scholar]
- 78. Arevalo-Herrera M, Roggero MA, Gonzalez JM, Vergara J, Corradin G, et al. (1998) Mapping and comparison of the B cell epitopes recognized on the Plasmodium vivax circumsporozoite protein by immune Colombians and immunized Aotus monkeys. Ann Trop Med Parasitol 92: 539–551 doi:10.1080/00034989859230 [PubMed] [Google Scholar]
- 79. Herrera S, Escobar P, Plata Cde, Avila GI, Corradin G, et al. (1992) Human recognition of T cell epitopes on the Plasmodium vivax circumsporozoite protein. J Immunol 148: 3986–3990. [PubMed] [Google Scholar]
- 80. Hughes AL (1991) Circumsporozoite protein genes of malaria parasites (Plasmodium spp.): evidence for positive selection on immunogenic regions. Genetics 127: 345–353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Nardin E, Clavijo P, Mons B, Belkum Avan, Ponnudurai T, et al. (1991) T cell epitopes of the circumsporozoite protein of Plasmodium vivax. Recognition by lymphocytes of a sporozoite-immunized chimpanzee. J Immunol 146: 1674–1678. [PubMed] [Google Scholar]
- 82. Bowman NM, Congdon S, Mvalo T, Patel JC, Escamilla V, et al. (2013) Comparative population structure of Plasmodium falciparum circumsporozoite protein NANP repeat lengths in Lilongwe, Malawi. Sci Rep 3: 1990 doi:10.1038/srep01990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Escalante AA, Lal AA, Ayala FJ (1998) Genetic polymorphism and natural selection in the malaria parasite Plasmodium falciparum. Genetics 149: 189–202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Rich SM, Hudson RR, Ayala FJ (1997) Plasmodium falciparum antigenic diversity: Evidence of clonal population structure. Proc Natl Acad Sci U S A 94: 13040–13045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Hartl DL (2004) The origin of malaria: mixed messages from genetic diversity. Nat Rev Microbiol 2: 15–22 doi:10.1038/nrmicro795 [DOI] [PubMed] [Google Scholar]
- 86. Ferreira MU, Hartl DL (2007) Plasmodium falciparum: Worldwide sequence diversity and evolution of the malaria vaccine candidate merozoite surface protein-2 (MSP-2). Exp Parasitol 115: 32–40 doi:10.1016/j.exppara.2006.05.003 [DOI] [PubMed] [Google Scholar]
- 87. Pacheco MA, Poe AC, Collins WE, Lal AA, Tanabe K, et al. (2007) A comparative study of the genetic diversity of the 42 kDa fragment of the merozoite surface protein 1 in Plasmodium falciparum and P. vivax. Infect Genet Evol 7: 180–187 doi:10.1016/j.meegid.2006.08.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Bailey JA, Mvalo T, Aragam N, Weiser M, Congdon S, et al. (2012) Use of Massively Parallel Pyrosequencing to Evaluate the Diversity of and Selection on Plasmodium falciparum csp T-Cell Epitopes in Lilongwe, Malawi. J Infect Dis 206: 580–587 doi:10.1093/infdis/jis329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Jalloh A, Jalloh M, Matsuoka H (2009) T-cell epitope polymorphisms of the Plasmodium falciparum circumsporozoite protein among field isolates from Sierra Leone: age-dependent haplotype distribution? Malar J 8: 120 doi:10.1186/1475-2875-8-120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Gandhi K, Thera MA, Coulibaly D, Traoré K, Guindo AB, et al. (2012) Next generation sequencing to detect variation in the Plasmodium falciparum circumsporozoite protein. Am J Trop Med Hyg 86: 775–781 doi:10.4269/ajtmh.2012.11-0478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Weedall GD, Preston BMJ, Thomas AW, Sutherland CJ, Conway DJ (2007) Differential evidence of natural selection on two leading sporozoite stage malaria vaccine candidate antigens. Int J Parasitol 37: 77–85 doi:10.1016/j.ijpara.2006.09.001 [DOI] [PubMed] [Google Scholar]
- 92. Zárate S, Pond SLK, Shapshak P, Frost SDW (2007) Comparative Study of Methods for Detecting Sequence Compartmentalization in Human Immunodeficiency Virus Type 1. J Virol 81: 6643–6651 doi:10.1128/JVI.02268-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Arnott A, Mueller I, Ramsland PA, Siba PM, Reeder JC, et al. (2013) Global Population Structure of the Genes Encoding the Malaria Vaccine Candidate, Plasmodium vivax Apical Membrane Antigen 1 (PvAMA1). PLoS Negl Trop Dis 7: e2506 doi:10.1371/journal.pntd.0002506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Barry AE, Schultz L, Buckee CO, Reeder JC (2009) Contrasting population structures of the genes encoding ten leading vaccine-candidate antigens of the human malaria parasite, Plasmodium falciparum. PloS One 4: e8497 doi:10.1371/journal.pone.0008497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Boulanger N, Charoenvit Y, Krettli A, Betschart B (1995) Developmental changes in the circumsporozoite proteins of Plasmodium berghei and P. gallinaceum in their mosquito vectors. Parasitol Res 81: 58–65. [DOI] [PubMed] [Google Scholar]
- 96. Posthuma G, Meis JF, Verhave JP, Hollingdale MR, Ponnudurai T, et al. (1988) Immunogold localization of circumsporozoite protein of the malaria parasite Plasmodium falciparum during sporogony in Anopheles stephensi midguts. Eur J Cell Biol 46: 18–24. [PubMed] [Google Scholar]
- 97. Golenda CF, Starkweather WH, Wirtz RA (1990) The distribution of circumsporozoite protein (CS) in Anopheles stephensi mosquitoes infected with Plasmodium falciparum malaria. J Histochem Cytochem Off J Histochem Soc 38: 475–481. [DOI] [PubMed] [Google Scholar]
- 98.WHO World Malaria Report (2011). Available: http://www.who.int/malaria/publications/atoz/9789241564403/en/index.html.
- 99. Foley DH, Klein TA, Lee I-Y, Kim M-S, Wilkerson RC, et al. (2011) Mosquito species composition and Plasmodium vivax infection rates on Baengnyeong-do (island), Republic of Korea. Korean J Parasitol 49: 313–316 doi:10.3347/kjp.2011.49.3.313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Yoo D-H, Shin E-H, Park M-Y, Kim HC, Lee D-K, et al. (2013) Mosquito species composition and Plasmodium vivax infection rates for Korean army bases near the demilitarized zone in the Republic of Korea, 2011. Am J Trop Med Hyg 88: 24–28 doi:10.4269/ajtmh.2012.11-0755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Adak T, Singh OP, Das MK, Wattal S, Nanda N (2005) Comparative susceptibility of three important malaria vectors Anopheles stephensi, Anopheles fluviatilis, and Anopheles sundaicus to Plasmodium vivax. J Parasitol 91: 79–82 doi:10.1645/GE-3514 [DOI] [PubMed] [Google Scholar]
- 102. Marrelli MT, Honório NA, Flores-Mendoza C, Lourenço-de-Oliveira R, Marinotti O, et al. (1999) Comparative susceptibility of two members of the Anopheles oswaldoi complex, An. oswaldoi and An. konderi, to infection by Plasmodium vivax. Trans R Soc Trop Med Hyg 93: 381–384. [DOI] [PubMed] [Google Scholar]
- 103. Joshi D, Choochote W, Park M-H, Kim J-Y, Kim T-S, et al. (2009) The susceptibility of Anopheles lesteri to infection with Korean strain of Plasmodium vivax. Malar J 8: 42 doi:10.1186/1475-2875-8-42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Da Silva ANM, Santos CCB, Lacerda RN, Machado RLD, Póvoa MM (2006) Susceptibility of Anopheles aquasalis and an. darlingi to Plasmodium vivax VK210 and VK247. Mem Inst Oswaldo Cruz 101: 547–550. [DOI] [PubMed] [Google Scholar]
- 105. Takala SL, Coulibaly D, Thera MA, Batchelor AH, Cummings MP, et al. (2009) Extreme Polymorphism in a Vaccine Antigen and Risk of Clinical Malaria: Implications for Vaccine Development. Sci Transl Med 1: 2ra5 doi:10.1126/scitranslmed.3000257 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Geographic distribution of P. vivax populations contributing to this study. In total, we identified 13 populations with pvmsp-1 42 kDa fragment sequences and 13 populations with pvcsp central repeat or whole-gene sequences. These populations were collected from 14 countries, pictured above. For countries with n≥10 isolates, the total number of pvmsp-1 and pvcsp isolates is marked.
(TIF)
F ST values at polymorphic sites within the pvmsp-1 42 kDa intervening region. Available parasite populations with n>25 individuals (Cambodia, India, NW Thailand, S Thailand, and Turkey) share 42 variable sites within the 42 kDa intervening region of pvmsp-1. F ST values for each variable site were calculated in a pairwise manner between all five populations. F ST values approaching 0 indicate limited inter-population variability at that site, while values approaching 1 indicate substantial inter-population variability. Coordinates are reported for every third polymorphic site.
(TIF)
pvmsp-1 and pvcsp sequences included in this study. The PlasmoDB gene identifier is PVX_099980 for pvmsp-1 and PVX_119355 for pvcsp. *Indicates the year the sequences were made available in GenBank. †Indicates study unpublished but sequences available in GenBank.
(DOCX)