Abstract
Recognition of microbial challenges leads to enhanced immunity at both the local and systemic levels. In Arabidopsis, EFR and PEPR1/PEPR2 act as the receptor for the bacterial elongation factor EF-Tu (elf18 epitope) and for the endogenous PROPEP-derived Pep epitopes, respectively. The PEPR pathway has been described to mediate defence signalling following microbial recognition. Here we show that PROPEP2/PROPEP3 induction upon pathogen challenges is robust against jasmonate, salicylate, or ethylene dysfunction. Comparative transcriptome profiling between Pep2-and elf18-treated plants points to co-activation of otherwise antagonistic jasmonate-and salicylate-mediated immune branches as a key output of PEPR signalling. Accordingly, as well as basal defences against hemibiotrophic pathogens, systemic immunity is reduced in pepr1 pepr2 plants. Remarkably, PROPEP2/PROPEP3 induction is essentially restricted to the pathogen challenge sites during pathogen-induced systemic immunity. Localized Pep application activates genetically separable jasmonate and salicylate branches in systemic leaves without significant PROPEP2/PROPEP3 induction. Our results suggest that local PEPR activation provides a critical step in connecting local to systemic immunity by reinforcing separate defence signalling pathways.
Keywords: DAMP, MAMP, pattern recognition receptor, PEPR, systemic immunity
Introduction
In multicellular organisms, recognition of non-self or altered self molecules by pattern recognition receptors (PRRs) leads to a first line of inducible defences that restrict microbial propagation (Boller & Felix, 2009; Kawai & Akira, 2011; Segonzac & Zipfel, 2011). The PRR ligands include microbial signatures typically conserved within a class of microbes, termed microbe-associated molecular patterns (MAMPs), and endogenous elicitors generated upon perturbations of host cellular processes, termed danger-associated molecular patterns (DAMPs). In plants, PRRs described to date are limited to membrane-localized receptors. The Arabidopsis Leu-rich repeat (LRR)-receptor kinases (RKs) FLS2 and EFR recognize the bacterial MAMPs flagellin (epitope flg22) and elongation factor EF-Tu (epitope elf18), respectively. Likewise, the LRR-RKs PEPR1 and PEPR2 recognize the elicitor-active Pep epitopes conserved in the endogenous PROPEP polypeptides (Yamaguchi & Huffaker, 2011).
The significance of MAMP-triggered immunity (MTI) has been well documented in plant immunity. Loss of FLS2 or EFR significantly reduces basal immunity to the infection of adapted and non-adapted bacterial pathogens (Zipfel et al, 2004, 2006; Nekrasov et al, 2009; Saijo et al, 2009). MTI is also functionally connected to another layer of plant immunity triggered upon the recognition of an avirulent pathogen effector, designated effector-triggered immunity (ETI), and to systemic acquired resistance (SAR) (Mishina & Zeier, 2007; Shen & Schulze-Lefert, 2007). However, the molecular links between local MAMP perception and effective activation of local and systemic immunity remain poorly understood.
During MTI, PRRs trigger a stereotypic set of defence-related outputs. Changes of ion fluxes across the plasma membranes, reactive oxygen species (ROS) bursts, and MAPK activation are typically detectable within minutes. They are followed, within several hours to days, by ethylene (ET) production, extensive transcriptional reprogramming, cell wall remodelling, and metabolic changes including biosynthesis of anti-microbial compounds (Boller & Felix, 2009; Segonzac & Zipfel, 2011). Genetic studies on Arabidopsis have revealed evolutionarily conserved components in a protein quality control pathway in the endoplasmic reticulum (ER) that defines the biogenesis route for EFR (Saijo, 2010). In an ER-resident glucosidase IIa allele, designated rsw3, EFR-triggered immunity and sustained activation of defence-related genes are impaired, although the receptor accumulation and the other tested MTI-associated outputs remain unaffected (Lu et al, 2009). Such phase separation of MTI signalling, of which the late phase is closely associated with immune activation, has been also described for the FLS2 pathway (Tsuda et al, 2009; Serrano et al, 2012). These findings point to the importance of sustained transcriptional reprogramming as a critical step for coupling initial MAMP perception with effective MTI activation. However, the target genes, their functions, and the molecular basis for sustained transcriptional reprogramming in MTI remain to be elucidated. In this respect, DAMP sensing and signalling has been postulated as an amplification system for MAMP-triggered signalling (Fontana & Vance, 2011). This model has received much attention in recent studies on both plants and animals (Kawai & Akira, 2011; Yamaguchi & Huffaker, 2011).
The Arabidopsis PROPEP family (PROPEP1–PROPEP6) has a conserved elicitor-active epitope (designated Pep1-Pep6, respectively) that is thought to act as a DAMP (Huffaker & Ryan, 2007). Of note, despite the lack in PROPEPs for an N-terminal signal peptide for entering the canonical secretory pathway, Pep epitope recognition occurs through the cell surface receptors PEPR1/PEPR2 (Yamaguchi et al, 2006, 2010). This implies a model in which PROPEPs (and/or their elicitor-active derivatives) accumulate in the cytoplasm, but are released to the extracellular spaces upon the disruption of cell membrane integrity, thereby eliciting PEPR-mediated signalling (Huffaker & Ryan, 2007). Given massive up-regulation of PROPEP2 and PROPEP3 upon MAMPs or pathogen challenges (Huffaker et al, 2006; Logemann et al, 2013), this model further postulates that the PEPR pathway serves to intensify and/or propagate defence signalling following MAMP perception (Ryan et al, 2007).
Consistent with this model, Pep perception leads to enhanced plant immunity: Exogenous Pep peptide application confers resistance to the bacterial phytopathogen Pseudomonas syringae pv tomato (Pst) and the fungal phytopathogen Botrytis cinerea in a PEPR-dependent manner (Yamaguchi et al, 2010; Liu et al, 2013); Arabidopsis plants overexpressing PROPEP1 or PROPEP2 better retain root growth in the presence of the oomycete phytopathogen Pythium irregular (Huffaker et al, 2006; Yamaguchi et al, 2010). Functional interactions have been also documented between MAMP and PEPR signalling pathways: PEPRs are required for maximal activation of FLS2-and EFR-triggered signalling and immunity to bacterial infection (Ma et al, 2012; Tintor et al, 2013); Pre-exposure to bacterial and fungal MAMPs enhances a ROS burst upon subsequent Pep application (Flury et al, 2013; Klauser et al, 2013); PEPR interacts with and phosphorylates BIK1, a central regulator for both MAMP and ET signalling (Liu et al, 2013). However, much remains to be learned about the role for the PEPR pathway in the control of overall host immunity.
In both local and systemic immunity, defence-related hormone pathways play a vital role for defence execution and fine-tuning (Robert-Seilaniantz et al, 2011; Pieterse et al, 2012). In general, salicylate (SA)-dependent defences are effective against biotrophic and hemi-biotrophic pathogens, whilst defences based on jasmonates (JA) and ET are effective against necrotrophic pathogens and insect herbivores. An antagonistic relationship has been well documented between SA and JA pathways. When SA and JA are supplied at nearly saturated high concentrations, SA signalling activation typically overrides JA signalling in Arabidopsis. However, the outcome of the SA-JA interactions differs according to the timing of elicitation and relative signal flux levels between the two hormone pathways, and is also influenced by other hormones. For example, timely application of ET renders the JA response resistant to the negative effect of SA (Leon-Reyes et al, 2010). Such a complex network of defence hormone signalling allows plants to coordinate between different defence pathways and optimize overall host immunity according to the type of the pathogens encountered and environmental conditions. Extensive genetic studies on separate or simultaneous disruptions of SA, JA, and ET pathways in Arabidopsis have revealed their synergistic interactions in promoting MTI (Tsuda et al, 2009). However, the mechanisms that utilize such cooperative connectivity of these hormone pathways remain to be identified.
In this study we further pursue the role for the PEPR pathway in plant immunity following recent publications for its role in MTI. Our results indicate that sustained activation of PROPEP2 and PROPEP3 upon pathogen challenges is robust against hormone imbalances. Genome-wide transcriptome profiling on Pep2-and elf18-treated plants reveals an inventory of PEPR-regulated genes, and points to co-activation of JA-and SA-mediated branches as a distinctive output of the PEPR pathway. In good accordance, genetic evidence indicates a contribution of PEPRs to basal defences against hemi-biotrophic pathogens, systemic immunity, and systemic propagation of MAMP-triggered signalling. Remarkably, active PEPR signalling seems to be essentially restricted to the sites of direct pathogen challenges, implying that the PEPR pathway primarily acts locally and thereby promotes systemic signalling. Together, our findings point to the functional significance for the PEPR pathway in coupling local and systemic immunity.
Results
Robust PROPEP2/PROPEP3 induction during local immune responses
To better understand the mechanisms that link MAMP recognition to PEPR signalling, we tested possible alterations in PROPEP2 (At5g64890) and PROPEP3 (At5g64905) expression in rsw3 plants. In the wild-type (WT) seedlings, both transcript levels dramatically increased upon elf18 application, which persisted for 24 h (Fig 1A). However, elf18-induced activation of the two genes was impaired in rsw3 seedlings (Fig 1A). On the other hand, rsw3 plants were indistinguishable from WT plants in Pep2-induced activation for PROPEP3 and pathogenesis related1 (PR1) and PR2, encoding defence-related proteins (van Loon et al, 2006) (supplementary Fig S1A). Given the requirement for PEPRs in elf18-as well as flg22-induced activation of defence-related genes (Ma et al, 2012; Tintor et al, 2013), these results imply that the previously described defects of rsw3 plants in elf18-induced transcriptional reprogramming (Lu et al, 2009) might be in part attributed to the impaired PROPEP2/PROPEP3 induction. Together, these findings support the notion that PROPEP2/PROPEP3 induction provides a critical step for the engagement of the PEPR pathway during MTI.
Figure 1.
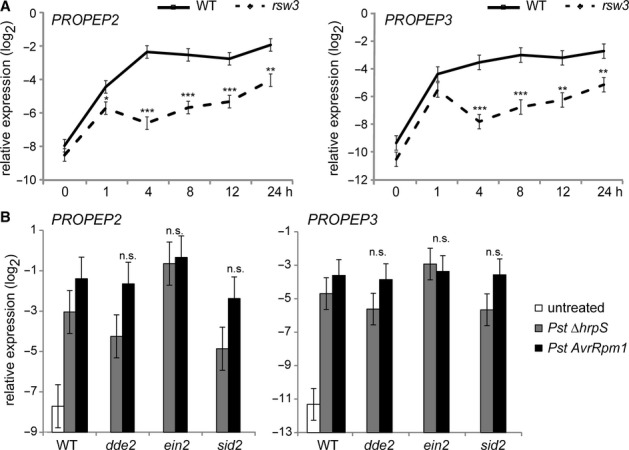
- Quantitative reverse-transcription-PCR (qRT-PCR) analysis for PROPEP2 and PROPEP3 in 10-day-old seedlings in response to 1 μM elf18. On the vertical axis, the log2 expression levels relative to that of At4g26410 are shown. Lines represent means and SE of three biological replicates calculated by the mixed linear model. Asterisks indicate significant differences from the WT plants at the corresponding time points (*q < 0.05, **q < 0.01, ***q < 0.001, two-tailed tests).
- qRT-PCR analysis for PROPEP2 and PROPEP3 in the leaves of 4-week-old plants challenged with 108 cfu/ml Pst DC3000 ΔhrpS or 107 cfu/ml Pst DC3000 AvrRpm1 for 24 h. Bars represent means and SE of two biological replicates calculated by the mixed linear model. Statistical analysis (two-tailed t-tests) indicates no significant differences (n.s.) in the transcript levels for PROPEP2 or PROPEP3 between the tested mutant plants and WT plants upon the challenges with the corresponding Pst strains. On the vertical axis, the log2 expression levels relative to that of At4g26410 are shown.
We also verified that PROPEP2 and PROPEP3 are induced upon the challenges with the Pst DC3000 ΔhrpS strain that is deficient in the type III effector secretion system and hence considered a trigger for MTI (Yuan & He, 1996) or with Pst DC3000 AvrRpm1 that rapidly triggers potent ETI via the resistance protein RPM1 (Grant et al, 1995) (Fig 1B). It has been described that PROPEP2 is induced upon exogenous application of methyl-JA (MeJA) or MeSA and that PROPEP3 is induced upon MeSA (Huffaker & Ryan, 2007). Our earlier work revealed ET-dependent PROPEP2 induction and ET-independent PROPEP3 induction in response to elf18 (Tintor et al, 2013). Following these findings, we further assessed the requirements for JA, ET, or SA in pathogen-induced activation of PROPEP2 and PROPEP3 in dde2,ein2, and sid2 plants. DDE2, EIN2, and SID2 provide a critical step for JA biosynthesis (Park et al, 2002), for the vast majority of ET-mediated responses (Alonso et al, 1999), and for SA biosynthesis upon pathogen challenges (Wildermuth et al, 2001), respectively. When challenged with Pst DC3000 ΔhrpS, both PROPEP2 and PROPEP3 were induced in all these mutants without a significant decrease, pointing to the robustness of their induction during MTI against these hormone imbalances (Fig 1B). This also implies the engagement of another MAMP receptor than FLS2 and EFR that mediates PROPEP2 induction in ein2 plants, given the impairment of both FLS2 and EFR functions in the mutant (Boutrot et al, 2010; Mersmann et al, 2010; Tintor et al, 2013). Upon Pst DC3000 AvrRpm1 inoculation, PROPEP2/PROPEP3 induction was also essentially retained in dde2,ein2, and sid2 plants (Fig 1B), as expected from the compensatory interactions between these hormone pathways in ETI (Tsuda et al, 2009). The observed robustness of PROPEP2/PROPEP3 induction against perturbations of these hormone pathways, which are often associated with pathogen challenges, might imply the engagement and effectiveness of the PEPR pathway in plant immunity to a wide range of pathogens.
Consistent with this idea, enhanced growth of Pst DC3000 has been described in pepr1 pepr2 plants (Ma et al, 2012). We also showed that pepr1 pepr2 plants were more susceptible than WT plants to a less virulent path-29 strain of the hemi-biotrophic pathogen Colletotrichum higginsianum (Ch) (Huser et al, 2009) (supplementary Fig S1B). These studies demonstrate that PEPRs are required for basal immunity to these hemibiotrophic pathogens.
Genome-wide transcriptome analysis for the PEPR and EFR pathways
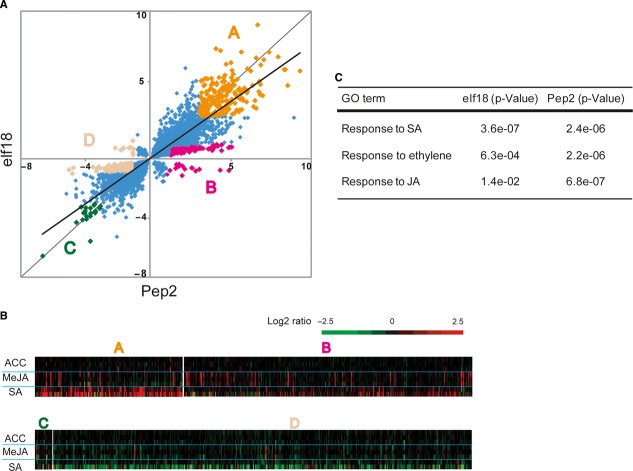
To gain insight into the molecular mechanisms by which PEPR signalling activation leads to enhanced immunity, we performed global transcriptome studies on Pep2-treated plants in comparison with elf18-treated plants. We exposed 10-day-old seedlings to 1 μM of Pep2 or elf18 for 2 and 10 h. In a whole genome microarray analysis (ATH1, Affymetrix, High Wycombe, UK), we detected the expression of a total of 15858 genes above the background levels. We plotted these genes based on their relative expression fold in response to Pep2 versus in response to elf18 (Fig 2A and supplementary Fig S2). This comparison revealed an overall high overlap between the two profiles (Fig 2A and supplementary Fig S2), consistent with the shared characteristics between Pep-and MAMP-induced responses (Yamaguchi & Huffaker, 2011).
Figure 2.
- The x-axis shows the log2 ratios of transcript levels in Pep2 (1 μM)-treated versus untreated seedlings upon for 2 h (q ≤ 0.05), and the y-axis shows the log2 ratios of transcript levels in elf18 (1 μM)-treated versus untreated seedlings (q ≤ 0.05). The regression of this scatter plot is indicated by the bold line. Genes commonly up-regulated upon both elicitors (>3 [log2], q ≤ 0.05) and those selectively up-regulated upon Pep2 (> 1 [log2] for Pep2 and < 1 [log2] for elf18, q < 0.05) are highlighted in orange (group A) or pink (group B), respectively. Genes commonly down-regulated upon both elicitors (<–3 [log2], q < 0.05) and those selectively down-regulated upon Pep2 (< –1 [log2] for Pep2 and >–1 [log2] for elf18, q < 0.05) are highlighted in green (group C) or beige (group D), respectively.
- The genes of the four groups defined in A are cross-referenced to public database for their expression responses to ACC (ET), MeJA or SA (Genevestigator v3). Each row represents the values of biologically independent different datasets. The relative expression (in log2 ratios) is colored red for induction and green for repression as illustrated in the fold change color bar.
- Gene Ontology (GO) analysis of the genes up-regulated upon elf18 or Pep2 (>1 [log2], q < 0.05) for their responses to the selected hormones (http://bioinfo.cau.edu.cn/agriGO/). The P-values were calculated by comparing the gene number ratio of [the hormone-responsive genes]/[the elicitor-responsive genes] and that of [the hormone-responsive genes]/[the genes of detectable expression], respectively.
We scored the genes exhibiting ≥2 fold changes in response to Pep2 or elf18 in WT plants as compared to untreated plants (2 h) or to the cognate receptor mutant plants (10 h). Pep2 application up-or down-regulated 1401 and 1286 genes at 2 h, whilst 234 and 164 genes at 10 h, respectively (q < 0.05) (supplementary Table S1). Likewise, elf18 application up-or down-regulated 1144 and 895 genes at 2 h, and 474 genes and 665 genes at 10 h, respectively (supplementary Table S1).
We define four different classes in the genes differentially regulated by either elicitor at 2 h (Fig 2A, supplementary Table S2): Classes A and C respectively represent the genes up-or down-regulated in response to both Pep2 and elf18 by ≥8 fold; and Classes B and D respectively represent the genes up-or down-regulated upon Pep2 (by ≥ 2fold) but not influenced upon elf18 (by <2 fold). Thus, the former and latter classes respectively represent the target genes common to both PRR pathways or specific to the PEPR pathway.
We then separately cross-referenced the genes of these classes to public databases for their expression responses to the defence-related hormones ET (1-aminocyclopropane-1-carboxylic acid [ACC]), MeJA, and SA (AtGenExpress, Genevestigator). We notice that SA-inducible genes are over-represented in Class A (Fig 2B; supplementary Table S3), suggesting that a major common output of the PEPR and EFR pathways involves to activate SA-inducible genes. This notion was also supported by the dataset obtained 10 h after elicitor application (supplementary Fig S2). The results well account for the earlier described Pst DC3000 resistance that is induced upon both elicitors (Zipfel et al, 2006; Yamaguchi et al, 2010). On the other hand, Classes B and D respectively exhibit over-representation of JA-inducible genes and SA-repressible genes (Fig 2B; supplementary Table S3). This implies that PEPR signalling distinctively co-activates subsets of JA-inducible genes (Class B) together with SA-inducible genes, and that it also more effectively represses subsets of SA-repressible genes (Class D) than EFR signalling.
A gene ontology analysis on the Pep2-or elf18-induced genes (≥ 2-fold) at 2 h indicates that over-representation for ET-and JA-responsive genes is more prominent in Pep2-induced genes compared to elf18-induced genes, whilst overrepresentation for SA-responsive genes is seen to a similar degree between Pep2-and elf18-induced genes (Fig 2C, supplementary Table S4).
These results also support the notion that the PEPR pathway facilitates co-activation of SA and JA/ET-mediated immune branches.
The PEPR pathway co-activates SA-and JA/ET-dependent immune branches
We validated this notion by quantitative reverse-transcription-PCR (qRT-PCR) analysis for an SA marker, PR1, and for JA/ET markers, PDF1.2a and PDF1.3, encoding small anti-microbial peptides termed defensins (Thomma et al, 2002). Pep2 application significantly activates all these 3 genes in WT seedlings (Fig 3A), as described earlier in detached leaves for PR1 and PDF1.2a induction (Huffaker & Ryan, 2007). Moreover, Pep2 triggers greater PDF1.2a induction as compared to elf18 or flg22 (supplementary Fig S3A), corroborating the aforementioned strength of the PEPR pathway in transcriptional activation of this JA/ET branch output. We then tested the requirements for SA or JA in Pep-triggered activation of the corresponding marker genes. Pep2-triggered activation of PR1 is impaired in sid2 plants, whilst that of both PDF1.2a and PDF1.3 is reduced in dde2 plants (Fig 3A). These results strongly suggest that PEPR signalling activation facilitates co-activation of SA-and JA-mediated immune branches. This is in good accordance with the aforementioned decrease in basal immunity to the hemi-biotrophic pathogens in pepr1 pepr2 plants (supplementary Fig S1B).
Figure 3.
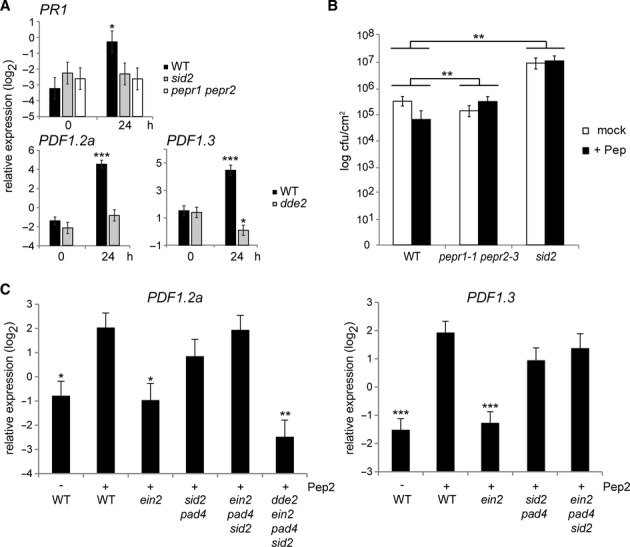
- qRT-PCR analysis for the indicated SA and JA marker genes in 10-day-old seedlings treated with 1 μM Pep2 for the indicated times.
- Leaves of 4-week-old plants pretreated with a mixture of Pep2 and Pep3 (1 μM each, +Pep) for 1 day were syringe-inoculated with Pst DC3000 (1 × 105 cfu/ml). The bacterial titer ± SD at 3 dpi is shown. Student's t test and Benjamini–Hochberg method was carried out to determine the significance of the difference in the induced resistance between the mutant and WT plants.
- qRT-PCR analysis for the JA markers in 10-day-old seedlings with or without Pep2 (1 μM) for 24 h.
Data information: For qRT-PCR analysis (A and C), bars represent means and SE of at least three biological replicates calculated by the mixed linear model. On the vertical axis, shown are the log2 expression levels relative to that of At4g26410. Asterisks indicate significant differences from the value of the corresponding genotype at 0 h (A) or those from the value in Pep-treated WT plants (C) (*q < 0.05, **q < 0.01, ***q < 0.001, two-tailed t-tests).
We then tested SA dependence in PEPR-mediated immunity to bacterial infection. Consistent with the earlier studies (Yamaguchi et al, 2010), the pretreatment of mature leaves with a mixture of Pep2 and Pep3 significantly lowered bacterial growth in a PEPR-dependent manner (Fig 3B). However, Pep-induced suppression of bacterial growth was abolished in sid2 plants, demonstrating the requirement for SA in Pep-induced resistance against Pst DC3000 (Fig 3B).
PEPR-mediated JA branch activation is enhanced by ET but antagonized by SA
Given the ET-mediated alleviation of SA-JA antagonism (Leon-Reyes et al, 2010), we further examined Pep-induced expression of PDF1.2a and PDF1.3 in the absence of DDE2, EIN2, SID2, PAD4, or combinations thereof. PAD4 has been described to contribute to SA-based immunity (Jirage et al, 1999). We found that Pep2-induced activation of the two defensin genes was reduced in ein2 plants, pointing to a positive role for ET in their induction (Fig 3C). By contrast, genetic removal of the SA branch conferred by SID2 and PAD4 substantially restored Pep2-induction of the two genes in ein2 pad4 sid2 plants, despite the ET signalling dysfunction (Fig 3C). However, the restored induction of PDF1.2a was abolished by the simultaneous disruption of the JA branch in dde2 ein2 pad4 sid2 plants, confirming the essential role for JA in this output (Fig 3C). Thus, our results suggest that ET positively influences this JA-dependent output against antagonistic SA effects in the PEPR pathway. By contrast, we were unable to detect significant PDF1.2a activation in response to elf18 even in the absence of both SID2 and PAD4 (supplementary Fig S3B). In contrast to PEPR signalling, this implies an intrinsic weakness of EFR signalling for this JA branch activation.
The PEPR pathway promotes systemic immunity
Together with earlier findings that MTI activation is sufficient to establish SAR (Mishina & Zeier, 2007) and that sequential engagement of JA-and SA-dependent processes precede SAR (Truman et al, 2007), our findings described above prompted us to investigate a possible contribution of the PEPR pathway to systemic immunity. We thus assessed whether systemic immune response is altered in pepr1 pepr2 plants. To this end, we pre-inoculated local leaves (expanded leaves in the lower layer of the plant) with Pst DC3000 AvrRpm1, and then traced the expression of PR1 and PR2 in systemic non-challenged leaves (expanded leaves in the upper layer of the plant). The transcript levels for both SAR markers were elevated in both local and systemic leaves of WT plants during SAR (Fig 4A). The induction of these genes remained largely unaffected in local leaves of pepr1 pepr2 plants upon the bacterial challenges (Fig 4A), in accordance with essentially intact RPM1-mediated cell death response in pepr1 pepr2 plants (supplementary Fig S4). However, remarkably, the induction of the two PR genes is significantly reduced in systemic leaves of pepr1 pepr2 plants (Fig 4A). Together, our results indicate that PEPRs are required for transcriptional reprogramming in systemic leaves following local pathogen challenges or ETI activation.
Figure 4.
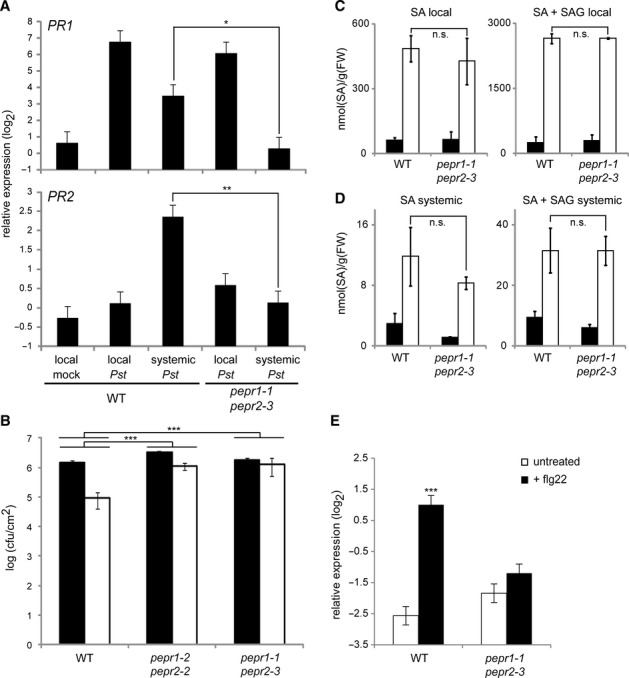
Systemic immunity is impaired in pepr1 pepr2 mutants despite WT like local PR gene activation and SA accumulation.
A qRT-PCR analysis for PR1 and PR2 in local and systemic leaves at 24 or 48 hpi, respectively, with Pst DC3000 AvrRpm1 (107 cfu) in local leaves.
B Growth of Psm ES4326 in systemic leaves (3 dpi) of Pst DC3000 AvrRpm1-preinoculated (white) or mock-treated (black) plants. The bacterial titers ± SD are shown.
C–D SA levels in local (24 hpi) and systemic (48 hpi) leaves of 4-week-old plants inoculated with Pst AvrRpm1 (white) or mock-treated (black). The means ± SD of three biological replicates (n = 4) are shown.
E qRT-PCR analysis for PR1 in systemic leaves 48 h after infiltration of 1 μM flg22 in local leaves.
Data information: For qRT-PCR analysis (A and E), bars represent means and SE of at least two biological replicates calculated by the mixed linear model. On the vertical axis, the log2 expression levels relative to that of At4g26410 are shown. Asterisks indicate significant differences from Pst-treated systemic WT plants (A) or untreated samples (E) (*q < 0.05, **q < 0.01, ***q < 0.001, two-tailed t-tests). For the pathogen inoculation assays and SA measurements (B–D), student's t-test and Benjamini-Hochberg method was carried out to determine the significance of the differences in the induced resistance (B) and in the induced SA levels between the mutant and WT plants. Double Asterisks (**) indicate significant differences with q < 0.01 and triple asterisk (***) with q < 0.001.
We then tested whether SAR is compromised in pepr1 pepr2 plants against a pathogen. To this end, 48 h after the first inoculation of local leaves with Pst DC3000 AvrRpm1, we inoculated systemic leaves with the bacterial pathogen P. s. pv maculicola (Psm) ES4326. Compared to the mock control, initial local Pst AvrRpm1 inoculation led to a great decrease of the growth of secondly challenged Psm in systemic leaves of WT plants (Fig 4B). By contrast, even after Pst AvrRpm1-preinoculation, pepr1 pepr2 plants allowed high Psm growth in systemic leaves, indicating that SAR is impaired in the absence of PEPRs (Fig 4B). This provides evidence for a critical role for the PEPR pathway in SAR. Given the aforementioned retention of local defence responses (Fig 4A and supplementary Fig S4) in pepr1 pepr2 plants, our results suggest that the PEPR pathway becomes rate-limiting in the establishment of SAR rather than in the local defence activation under these conditions.
We then assessed whether the defects in systemic activation of the tested SA markers and immunity in pepr1 pepr2 plants are associated with SA production upon pathogen challenges. Levels of total SA (free SA plus SA 2-O-β-glucoside [SAG]) and free SA were determined in local and systemic leaves 24 and 48 h, respectively, after the infiltration of Pst AvrRpm1 into local leaves. WT and pepr1 pepr2 plants were essentially indistinguishable for the basal levels or pathogen-induced levels of total and free SA in both local and systemic leaves (Fig 4C and D). This suggests that the PEPR pathway contributes to SAR independent of increasing SA levels, which might be favoured for the simultaneous activation of the SA and JA branches (Figs 2 and 3).
The PEPR pathway couples MAMP perception with systemic immune response
We next directly tested whether the PEPR pathway provides a causal link between localized MAMP perception and systemic immunity. Flg22 has been described to trigger systemic immune response (Mishina & Zeier, 2007). We thus traced the expression of PR1 in systemic leaves following local flg22 application. We confirmed that flg22 application to local leaves resulted in an elevation of PR1 transcript levels in systemic leaves of WT plants (Fig 4E). However, systemic activation of this output was significantly reduced in pepr1 pepr2 plants (Fig 4E), providing evidence that the PEPR pathway links MAMP perception to systemic immune response.
PEPR signalling predominantly operates in local pathogen-challenged sites to confer systemic immune activation
We next sought to determine whether the PEPR pathway acts in local or systemic tissues during systemic immunity. It has been described that exogenous Pep application induces the expression of all PROPEP members, thereby providing positive auto-feedback for PEPR signal amplification (Huffaker & Ryan, 2007). We thus used PROPEP2/PROPEP3 induction as a proxy for PEPR signalling activation. During SAR, we detected PROPEP2/PROPEP3 induction in local leaves that were directly challenged with Pst DC3000 AvrRpm1, but not in systemic leaves that were not directly challenged (Fig 5A). We infer from these results that the PEPR-PROPEP2/PROPEP3 auto-feedback, and thus the PEPR signalling system per se, is not significantly activated in systemic leaves.
Figure 5.
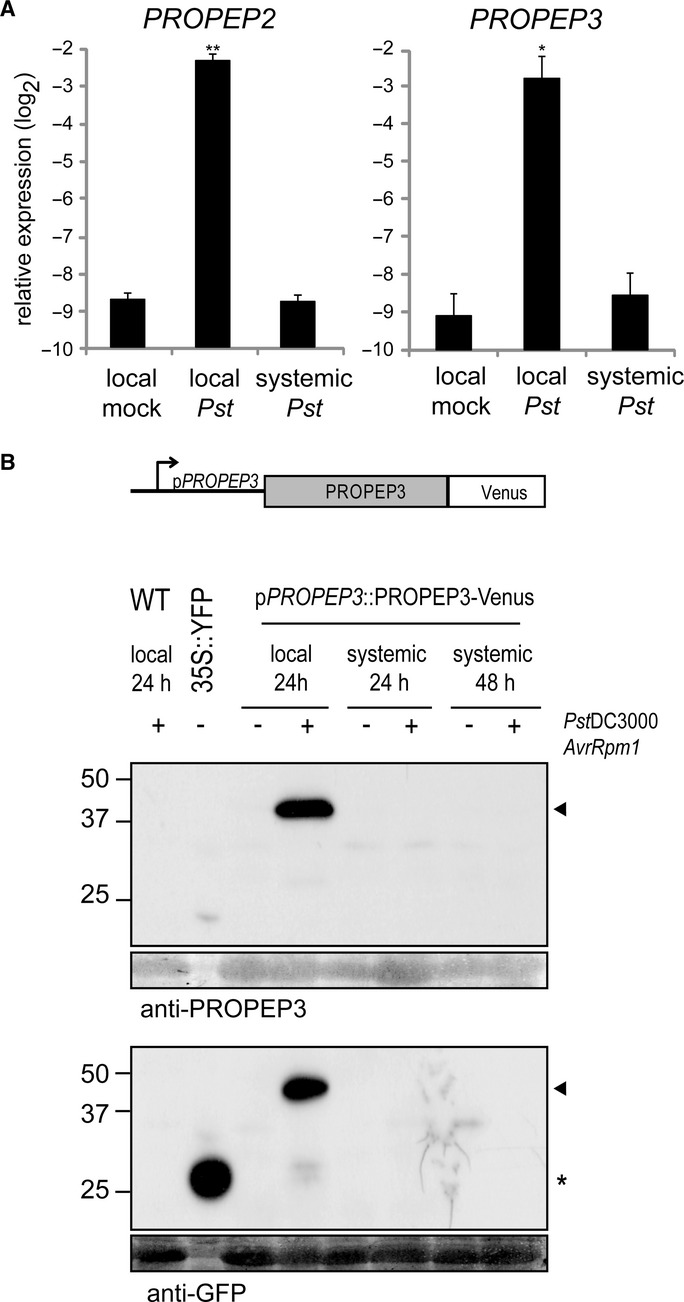
- qRT-PCR analysis for PROPEP2 and PROPEP3 in local and systemic leaves 24 hpi with Pst DC3000 (105 cfu/ml) in 4-week-old plants. Bars represent means and SE of two biological replicates calculated by the mixed linear model. On the vertical axis, the log2 expression levels relative to that of At4g26410 are shown. Asterisks indicate significant differences from mock-treated plants (*q < 0.05, **q < 0.01, two-tailed t-tests).
- The construct used for the generation of stable transgenic plants expressing PROPEP3-Venus protein under the control of the native regulatory DNA sequences. Local and systemic leaves of 4-week-old plants at 24 or 48 hpi, respectively, with Pst DC3000 AvrRpm1 (107 cfu) were subjected to immunoblot analysis with anti-PROPEP3 or anti-GFP antibodies. Non-transformed Arabidopsis plants (WT) and N. benthamiana plants transiently expressing free YFP (35S::YFP) were used as controls. The lower panels below the immunoblots show the immunoblot membranes stained with Coomassie Brilliant Blue for verifying equal loading of the Arabidopsis protein samples. The arrowheads indicate the positions of the PROPEP3-Venus protein bands, and the asterisk indicates the position of the free YFP protein.
We also traced PROPEP3 protein accumulation during SAR. For this purpose, we generated transgenic Arabidopsis lines that express a C-terminal Venus fusion of PROPEP3 under the control of its native regulatory DNA sequences. We verified that PROPEP3-Venus can provide an elicitor-active ligand for PEPRs: The preparations of the fusion protein expressed in N. benthamiana leaves inhibit Arabidopsis root growth in a PEPR-dependent manner (supplementary Fig S5B and C).
In Arabidopsis plants inoculated with Pst DC3000 AvrRpm1, our immunoblot analysis detected the PROPEP3-Venus fusion in the local challenged leaves but not in systemic non-challenged leaves at the tested time points (Fig 4B), essentially reflecting the aforementioned accumulation pattern of the endogenous PROPEP3 transcripts. The identity of the PROPEP3-Venus signal was also confirmed with anti-PROPEP3 specific antibodies that specifically recognize PROPEP3, presumably through the N-and C-terminal epitopes of the antigen (Fig 4B and supplementary Fig S5A). Under our conditions, we were not able to detect a possibly processed form of PROPEP3-Venus. We also failed to detect the endogenous PROPEP3 protein with our anti-PROPEP3 antibodies, possibly due to low stability. Nevertheless, our results imply that the PEPR ligands predominantly accumulate and elicit the receptor signalling in the pathogen challenge sites during SAR.
Local Pep application is sufficient to confer systemic immunity
We reasoned that if PEPR activation serves to generate or propagate a long-distance immune signal, local Pep application should trigger systemic immune response. Indeed, SA and JA branch markers, PR1 and PDF1.2a, respectively, were activated in systemic upper leaves upon Pep2 application in lower leaves (Fig 6A). Pep2-induced systemic induction of PDF1.2a was abolished in dde2 ein2 and pepr1 pepr2 plants, indicating that enforced PEPR signalling activation indeed confers systemic activation of this JA branch output (Fig 6B). However, we note that this occurred again without detectable activation of PROPEP2 and PROPEP3 in systemic leaves (Fig 6C), as well as during pathogen-induced SAR (Fig 5A). Given the aforementioned positive auto-feedback of PEPR signalling, the absence of significant PROPEP2/PROPEP3 induction in systemic leaves makes it unlikely that Pep2 per se travels from the primary application sites to systemic tissues at physiologically significant levels.
Figure 6.
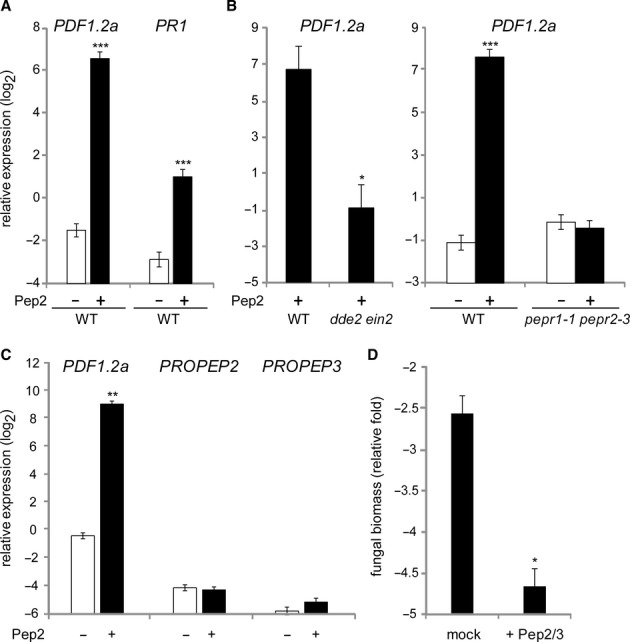
Local PEPR activation confers systemic immunity.
A–B qRT-PCR analysis for PDF1.2a and PR1 in systemic leaves of 4-week-old plants upon 1 μM Pep2 application for 24 h.
C qRT-PCR analysis for PROPEP2,PROPEP3, and PDF1.2a in systemic leaves upon local Pep2 application as performed in (A) and (B).
D Systemic, non-treated leaves of 4-week-old plants pretreated with a mixture of Pep2 and Pep3 (1 μM each) for 24 h, were spray-inoculated with Ch path-29 (5 × 105 spores/ml). Fungal biomass was determined by qRT-PCR analysis at 5 dpi.
Data information: Bars represent means and SE of at least three biological replicates calculated by the mixed linear model. On the vertical axis, the log2 expression levels relative to that of At4g26410 are shown. Asterisks indicate significant differences from mock-treated WT plants (A, B right, C, and D) or from Pep-treated WT plants (B left) (*q < 0.05, **q < 0.001, two-tailed t-tests).
We also tested whether systemic immunity is enhanced upon local Pep2 application to pathogen infection. When challenged with Psm or Pst DC3000, we did not consistently detect enhanced resistance in systemic leaves after Pep2 application in local leaves. However, we found that local Pep application significantly enhanced systemic immunity to Ch path-29 strain (Fig 6D). These results might be in part attributed to high PDF1.2 induction in systemic leaves (Fig 6A), given the previously described role for defensins in basal defences against Ch (Hiruma et al, 2011). Together, our findings support the notion that PEPR signalling predominantly occurs at the sites of the primary pathogen challenges, which is yet required to enhance systemic immunity.
Genetic separation between Pep-induced systemic JA-and SA-branch activation
To gain insight into the mechanisms by which localized activation of PEPR signalling enhances systemic immunity, we tested possible alterations in Pep-induced systemic activation of PDF1.2 and PR1 in the absence of previously defined SAR regulators. NPR1 acts as the master regulator for SA-based immunity and SAR (Durrant & Dong, 2004); ALD1 is required for pathogen-induced pipecolic acid (Pip) biosynthesis, full MTI and ETI responses, systemic SA accumulation, and SAR (Song et al, 2004; Navarova et al, 2012); Flavin-dependent monooxygenase1 (FMO1) is required for local immunity, systemic SA accumulation, and pathogen-and Pip-induced SAR (Bartsch et al, 2006; Koch et al, 2006; Mishina & Zeier, 2007; Navarova et al, 2012). We found that Pep-induced systemic PR1 activation was significantly reduced in npr1 and ald1 plants (Fig 7). However, Pep-induced PDF1.2a activation was virtually retained in ald1,fmo1, and npr1 plants (Fig 7). This verifies that Pep-induced signalling boosts these SA-and JA-related outputs in systemic leaves via separate genetic requirements. Our results thus strengthen the notion that the PEPR pathway facilitates co-activation of different immune branches at the systemic level as well.
Figure 7.
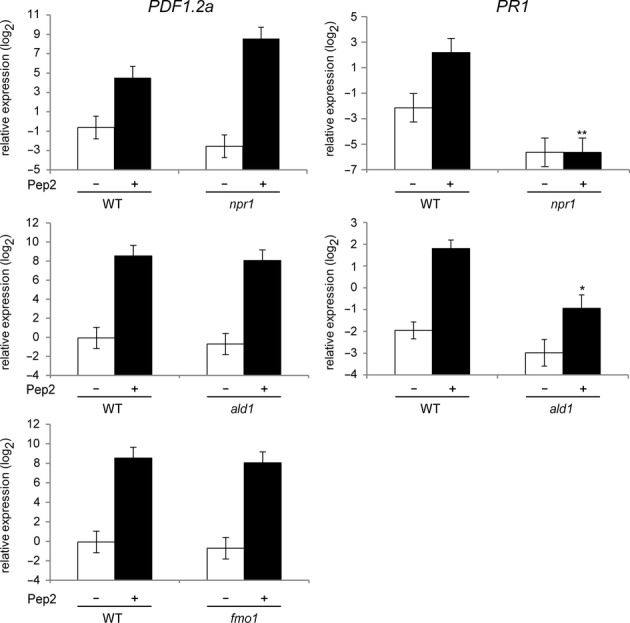
The roles for previously defined SAR regulators in Pep-induced systemic immune responses.
qRT-PCR analysis for PDF1.2a and PR1 in systemic leaves of 4-week-old plants upon 1 μM Pep2 application for 24 h. Bars represent means and SE of at least three biological replicates calculated by the mixed linear model. On the vertical axis, the log2 expression levels relative to that of At4g26410 are shown. Asterisks indicate significant differences from Pep-treated WT plants (*q < 0.05, **q < 0.001, two-tailed t-tests).
Discussion
With the focus on the PEPR pathway, we gain insight into the mechanisms that link MAMP recognition to immune activation at both the local and systemic levels. In MTI, the transition from initial to sustained signalling phases is closely associated with effective defence activation (Lu et al, 2009; Tsuda et al, 2009; Serrano et al, 2012). Given the incurred fitness costs, it is conceivable that the phase transition is facilitated by, or constrained to, the conditions in which the host is gravely threatened. Prolonged exposure to high-dose MAMPs might reflect massive and persistent growth of potentially pathogenic microbes in the plant. Following our separate study (Tintor et al, 2013), the present study further points to the importance for sustained activation of PROPEP2 and PROPEP3, a preparatory step for PEPR signalling activation, as a critical step in MTI.
However, if the PEPR pathway mediates DAMP sensing and signalling, its activation is expected to be stimulated upon pathogen challenges. In Arabidopsis seedlings, elf18-induced PR1 activation is greatly reduced in pepr1 pepr2 plants (Tintor et al, 2013), indicating that PEPR signalling contributes to this output under sterile conditions. By contrast, elf18 application alone is insufficient to induce PDF1.2a (supplementary Fig S3A) despite massive induction of PROPEP2 and PROPEP3, whereas application of the elicitor-active Pep2 epitope substantially activates this JA branch marker. Together, these results suggest that another additional cue than the MAMP is required for full activation of PEPR signalling, in terms of the repertoire and/or amplitude of outputs. This cue seems to be dispensable but to boost the receptor signalling at the level of the ligand, receptor, post-recognition signalling, or combinations thereof, which might be associated with pathogen challenges. Future studies will be required to clarify these possibilities and to elucidate the nature of the elicitor-active ligands generated from PROPEP2 and PROPEP3 in vivo.
Our genetic evidence obtained in the present and earlier studies (Tintor et al, 2013) points to the robustness of the PEPR pathway against perturbations of defence-related hormone responses: MAMP-and pathogen-induced PROPEP2 and PROPEP3 activation substantially occurs against genetic defects for SA, JA, and ET responses; PEPR1 expression and PEPR function are largely retained in ein2 plants where FLS2 and EFR functions are greatly reduced (Tintor et al, 2013). These features seem to provide an advantage for the engagement of the proposed DAMP signalling system in defence activation against pathogen-mediated interference with these hormone pathways. Indeed, the present and other earlier studies point to a role for the PEPR pathway in augmenting defence signalling following the perception of a wide range of MAMPs and in basal immunity to taxonomically unrelated pathogens at their inoculation sites (Ma et al, 2012; Flury et al, 2013; Liu et al, 2013; Tintor et al, 2013). Deciphering the mechanistic basis for the robustness of the PEPR pathway also represents an important challenge for future studies.
Our comparative transcriptome analysis on Pep2-and elf18-treated plants has revealed that the PEPR pathway is distinct from the EFR pathway in that the former up-regulates a JA-inducible branch under the control of the TF ERF1 (represented by PDF genes encoding defensins), in addition to SA-inducible PR genes. Pep-induced PDF1.2a activation also occurs in mature leaves (Fig 6) (Huffaker & Ryan, 2007), excluding the possibility that the observed differences between PEPR and EFR regulons (Fig 2 and supplementary Fig S3) merely reflect poor EFR function in seedling roots (Christensen et al, 2010). Thus, it appears that PEPR-mediated coordination of these SA and JA branches downstream of MAMP perception not only reinforces MAMP receptor-triggered signalling but also extends the spectrum of immune responses during MTI. The lowered local basal defences against the tested hemibiotrophic pathogens in pepr1 pepr2 plants can be attributed to the lack of this PEPR function.
We note that PEPR signalling keeps this JA branch active in an ET-dependent manner even in the presence of high SA branch activity (Fig 3C). Thus, the PEPR pathway defines a signalling component that utilizes the earlier described positive connectivity between the three hormone branches (Leon-Reyes et al, 2010). However, consistent with this ET dependence, PEPR signalling has the opposite effects between the ERF1-JA branch (enhanced by ET) and another JA branch mediated by the TF Myc2 (repressed by ET) (Tintor et al, 2013). In good accordance, concomitant induction of JA and ET production has been described in excised leaves upon Pep1 application (Huffaker et al, 2013), which might underscore the differential control of the two JA branches.
Localized interactions with compatible or incompatible pathogens lead to the establishment of SAR (Durrant & Dong, 2004; Dempsey & Klessig, 2012). Local MTI activation is sufficient to confer SAR via a process that remains unknown (Mishina & Zeier, 2007). SAR requires the proper generation of a mobile signal(s) at the primary challenge site, transmittance of the signal from local to distal sites through the vasculature, and perception of the signal in the distal sites. pepr1 pepr2 plants exhibited defects in both MAMP-and pathogen-induced systemic immune responses. The former defects are well correlated with the previously described impairments of MAMP-induced responses in the MAMP perception sites of the mutant (Ma et al, 2012; Tintor et al, 2013). However, the latter defects occurred where no significant alterations were observed in local immune responses to the avirulent Pst strain tested (Fig 4 and supplementary Fig S4). This might reflect the existence of a PEPR sub-function that is specific to systemic immunity or a pathogen-dependent mechanism that compensates for the loss of PEPRs in local immunity but not in systemic immunity. Alternatively, but not mutually exclusively, the requirement for PEPR-mediated signal amplification might be greater for the activation of systemic immunity in non-challenged sites compared to local immunity in directly challenged sites. This model is also supported by the earlier studies that genetic defects for the basal defence regulators ALD1 and FMO1 are more severe in systemic compared to local immunity (Song et al, 2004; Bartsch et al, 2006; Mishina & Zeier, 2007; Navarova et al, 2012).
Our results point to the restriction of PROPEP2/3 activation to the sites of direct pathogen/elicitor challenges during SAR (Figs 5 and 6). This suggests that the PEPR pathway becomes primarily engaged in the local sites, thereby promoting the generation and/or spread of a mobile long-distance signal(s). This is consistent with the hypothesis that the PEPR pathway acts in DAMP sensing and signalling within (the proximity to) the sites of pathogen attacks (Fig 8). The observed lack of active PEPR signalling in systemic non-challenged leaves also disfavours that an active PEPR ligand(s) per se is delivered in physiologically relevant levels into systemic tissues. In sum, our findings lead to a model in which local action of the PEPR pathway at the sites of direct pathogen challenges plays a pivotal role in the establishment of systemic immunity (Fig 8).
Figure 8.
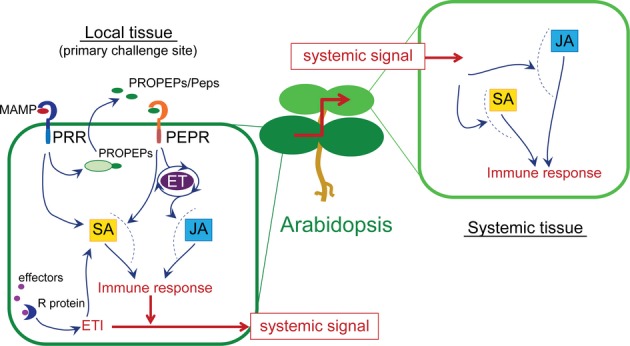
A model for the PEPR pathway in the control of local and systemic immunity.
The molecular links between PEPR-mediated signaling and the downstream SA and JA branches remain to be shown (dashed lines).
Consistent with this model, local Pep application is sufficient to enhance systemic immunity. SA-dependent SAR is mounted via a complex network of multiple signalling pathways, of which a rate-limiting step seems to differ according to the trigger or the environmental conditions (Dempsey & Klessig, 2012). JA-dependent induced systemic resistance (ISR) is mounted on root-colonization of selected, non-pathogenic Pseudomonas rhizobacteria in an NPR1-dependent manner (van Wees et al, 2000). Like Pep-induced systemic immunity as shown in this study, the simultaneous activation of SAR and ISR also allows co-activation of SA-and JA-branches in systemic tissues (van Wees et al, 2000). However, although NPR1 is required for SAR and ISR, NPR1 is dispensable for Pep-induced systemic activation of PDF1.2a, indicating the separation of the genetic requirements between these systemic immune responses. The mechanisms underlying PEPR-mediated systemic immunity will require further investigations.
Our findings on the PEPR pathway highlight several characteristics that are shared by the peptide hormone Systemin pathway in tomato (Schilmiller & Howe, 2005; Yamaguchi & Huffaker, 2011). Ligand and/or receptor activation that is critical for systemic immune activation seems to predominantly occur at the local challenged sites. Both ligands are expected to translocate across the membranes for their recognition, despite the lack of an N-terminal signal peptide in the ligand precursors. Systemin-like peptides have only been described in the Solanoideae subfamily of the Solanaceae family (Yamaguchi & Huffaker, 2011), whereas polypeptides of a Pep-related epitope are widespread in higher plants (Huffaker et al, 2006, 2011). Importantly, although Pep elicitor activity seems to be specific to the plant family, the outcome of Pep perception might be a concurrent theme in a wide range of phylogenetic lineages of higher plants (Huffaker et al, 2013). Signalling systems that scale the level of tissue damages or pathogen threats in local sites might be extensively used for fine control of systemic immune response.
Materials and Methods
Plant growth conditions
The Arabidopsis thaliana accession Col-0 was used as WT. For microarray and qRT-PCR analysis, 10-day-old seedlings (grown under 12 h light/12 h dark on 0.5 × MS agar plates containing 1% sucrose for 5 days and then transferred to a 0.5 × MS liquid medium containing 1% sucrose for another 5 days) were subjected to elicitor treatment within 1 h of the onset of the light period. For SAR analysis (qRT-PCR analysis and SA content measurement) and pathogen inoculation assays, 4-week-old plants grown on soil under 10 h light/14 h dark were used unless otherwise stated.
Plant materials
The Arabidopsis thaliana accession Col-0 was used as WT. efr-1 (Zipfel et al, 2006), fls2 (Zipfel et al, 2004), efr fls2 (Nekrasov et al, 2009), rsw3 (Lu et al, 2009), pepr1-1 pepr2-1 and pepr1-2 pepr2-2 (Yamaguchi et al, 2010), dde2-2,ein2-1,sid2-2,dde2-2 ein2-1,pad4-1 sid2-2,ein2-1 pad4-1 sid2-2,dde2-2 ein2-1 pad4-1 sid2-2 (Tsuda et al, 2009), pen2-1 (Lipka et al, 2005), npr1-1(Wang et al, 2005), fmo1 and ald1 (Navarova et al, 2012) were described earlier.
Bioassays for elicitor-induced responses
Elicitor response assays in Arabidopsis were performed as described earlier (Lu et al, 2009). For gene expression analysis, whole seedlings were treated with 1 μM elf18, flg22, Pep1, Pep2, or Pep3 for the indicated times, unless otherwise stated.
Bacterial inoculation assays
Bacterial inoculation assays were performed as described previously (Lu et al, 2009) with the following modifications. For Pep-induced resistance assays, plants were syringe-infiltrated with a mixture of 1 μM Pep2 and Pep3 or water (mock) 24 h before inoculation. Pst DC3000 suspension at 1 × 105 cfu/ml was syringe-infiltrated into 2–3 leaves of eight plants per genotype per treatment. Three days after inoculation the bacterial titer was determined as described above. These experiments have been repeated at least three times with the same conclusion.
SAR assays
To assess transcriptional changes during SAR, local leaves (expanded rosette leaves in the second top layer of the plant) of 4-week old Arabidopsis plants were infiltrated with Pst DC3000 AvrRpm1 (1 × 107 cfu/ml) or a mixture of 1 μM Pep2 and Pep3. The local leaves and systemic leaves (non-challenged expanded rosette leaves in the top layer of the plant) were harvested at the indicated times. Eight leaves of 3–4 plants were used per sample for total RNA and protein extract preparations. For bacterial growth measurement, local leaves of 4-week-old Arabidopsis plants were initially infiltrated with Pst DC3000 AvrRpm1 (1 × 107 cfu/ml) or 10 mM MgCl2 as mock control. Two days later, Pseudomonas syringae pv. maculicola (Psm) at 1 × 106 cfu/ml were infiltrated in systemic leaves, and then the bacterial titer of Psm was determined 3 days post inoculation as described above.
For Pep-induced SAR assays, local leaves were infiltrated with a mixture of 1 μM Pep2 and Pep3 or water (mock) for 24 h, and then Ch path-29 (5 × 105 spores/ml) was sprayed onto systemic leaves (n = 8). Fungal biomass was determined at 4 and 5 days post inoculation.
Quantitative RT-PCR (qRT-PCR) analysis
Total RNA was isolated from plant samples using TRI reagent following the manufacturer's instructions (Ambion, Grand Island, NY, USA). Five microgram of total RNA was reverse transcribed using an oligo(dT) primer and reverse transcriptase (Transcriptor Reverse Transcriptase; Roche, Mannheim, Germany). Ten times diluted cDNA was used as a template for quantitative PCR using a Bio-Rad iQ5 multicolor real-time PCR detection system (Bio-Rad, Munich, Germany). Real-time DNA amplification was monitored using Bio-Rad iQ5 optical system software (Bio-Rad). The expression level of genes of interest was normalized to that of an endogenous reference gene At4g26410 (Czechowski et al, 2005).
SA measurement
SA contents were measured essentially following the published procedures (Bednarek et al, 2005). Leaf material (100–200 mg fresh weight) was extracted with aqueous methanol. Leaf extracts were hydrolyzed with ß-glucosidase (EC 3.2.1.21; Sigma-Aldrich, Steinheim, Germany), and released free SA was re-extracted as described (Lee & Raskin, 1998). HPLC analyses were performed on an Agilent (Waldbronn, Germany) 1100 HPLC system.
Colletotrichum higginsianum inoculation assays
For lesion size measurements, Ch path-29 (Huser et al, 2009) was drop inoculated (5 × 105 spores/ml) on fully expanded leaves of 4-week old Arabidopsis plants 5 days before analysis (n= ˜ 30 lesions). To quantitatively assess fungal growth by qRT-PCR, 12-day-old seedlings were drop inoculated with Ch path-29 (1 × 105 spores/ml) 3 days before harvest. The Ch Actin transcript levels in relation to At4g26410 transcript levels of 12 seedlings per sample were determined to assess fungal biomass as described (Narusaka et al, 2009). The causative mutated gene in Ch path-29 remains to be determined (Huser et al, 2009).
Statistical analysis for bacterial growth assays and Ch lesion size measurements
The obtained values were compared using two-tailed t-tests and further analysed by multiple test corrections using the Benjamini–Hochberg method.
Immunoblot analysis
Protein extracts were prepared in an extraction buffer [50 mM Tris-HCl pH 7.0; 2% SDS; 2 mM DTT; 10% glycerol; 1 mM AEBSF (Sigma), 1% (v/v) P9599 protease inhibitor cocktail (Sigma)]. The supernatants were recovered after the centrifugation at 15 000 rpm for 15 min at 4°C and then subjected to immunoblot analyses with anti-PROPEP3 or anti-GFP antibodies.
Antibodies used
Anti-GFP antibodies were purchased from Invitrogen. PROPEP3 antibodies were generated in rabbits using the N- and C-terminal portion of PROPEP3 (the amino acid residues 1–12 MENLRNGEDNGS and 82–96 KTKPTPSSGKGGKHN) as antigens.
Acknowledgments
We thank Drs. Cyril Zipfel, Yube Yamaguchi, Fumiaki Katagiri, and Jürgen Zeier for published Arabidopsis mutants, Dr. Richard O'Connell for the Ch strains used, Dr. Bruno Hüttel for microarray analysis, and Dr. Kazue Kanehara for technical assistance. This work was supported in part by the Max Planck Society, grants from SFB670 (Y.S.), from the MEXT of Japan (no. 20380027; Y.T.), and PhD fellowships from the International Max Planck Research School Program (A.R. and X.L.) and from the Japan Society for the Promotion of Science (no. 10J03196; K.H.).
Author contributions
AR, KY, KH, MY-Y, XL, and YS performed experiments; AR, KY, KH, MY-Y, YT, and YS designed experiments; AR, KY, KH, MY-Y, XL, KT, and YS analysed data; AR and YS wrote the manuscript. All authors commented on the manuscript.
Conflict of interest
The authors declare that they have no conflict of interest.
Supplementary information for this article is available online: http://emboj.embopress.org
References
- Alonso JM, Hirayama T, Roman G, Nourizadeh S, Ecker JR. EIN2, a bifunctional transducer of ethylene and stress responses in Arabidopsis. Science. 1999;284:2148–2152. doi: 10.1126/science.284.5423.2148. [DOI] [PubMed] [Google Scholar]
- Bartsch M, Gobbato E, Bednarek P, Debey S, Schultze JL, Bautor J, Parker JE. Salicylic acid-independent ENHANCED DISEASE SUSCEPTIBILITY1 signaling in Arabidopsis immunity and cell death is regulated by the monooxygenase FMO1 and the Nudix hydrolase NUDT7. Plant Cell. 2006;18:1038–1051. doi: 10.1105/tpc.105.039982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bednarek P, Schneider B, Svatos A, Oldham NJ, Hahlbrock K. Structural complexity, differential response to infection, and tissue specificity of indolic and phenylpropanoid secondary metabolism in Arabidopsis roots. Plant Physiol. 2005;138:1058–1070. doi: 10.1104/pp.104.057794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boller T, Felix G. A renaissance of elicitors: perception of microbe-associated molecular patterns and danger signals by pattern-recognition receptors. Annu Rev Plant Biol. 2009;60:379–406. doi: 10.1146/annurev.arplant.57.032905.105346. [DOI] [PubMed] [Google Scholar]
- Boutrot F, Segonzac C, Chang KN, Qiao H, Ecker JR, Zipfel C, Rathjen JP. Direct transcriptional control of the Arabidopsis immune receptor FLS2 by the ethylene-dependent transcription factors EIN3 and EIL1. Proc Natl Acad Sci USA. 2010;107:14502–14507. doi: 10.1073/pnas.1003347107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christensen A, Svensson K, Thelin L, Zhang WJ, Tintor N, Prins D, Funke N, Michalak M, Schulze-Lefert P, Saijo Y, Sommarin M, Widell S, Persson S. Higher plant calreticulins have acquired specialized functions in Arabidopsis. PLoS One. 2010;5:e11342. doi: 10.1371/journal.pone.0011342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czechowski T, Stitt M, Altmann T, Udvardi MK, Scheible WR. Genome-wide identification and testing of superior reference genes for transcript normalization in Arabidopsis. Plant Physiol. 2005;139:5–17. doi: 10.1104/pp.105.063743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dempsey DA, Klessig DF. SOS – too many signals for systemic acquired resistance? Trends Plant Sci. 2012;17:538–545. doi: 10.1016/j.tplants.2012.05.011. [DOI] [PubMed] [Google Scholar]
- Durrant WE, Dong X. Systemic acquired resistance. Annu Rev Phytopathol. 2004;42:185–209. doi: 10.1146/annurev.phyto.42.040803.140421. [DOI] [PubMed] [Google Scholar]
- Flury P, Klauser D, Schulze B, Boller T, Bartels S. The anticipation of danger: microbe-associated molecular pattern perception enhances AtPep-triggered oxidative burst. Plant Physiol. 2013;161:2023–2035. doi: 10.1104/pp.113.216077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fontana MF, Vance RE. Two signal models in innate immunity. Immunol Rev. 2011;243:26–39. doi: 10.1111/j.1600-065X.2011.01037.x. [DOI] [PubMed] [Google Scholar]
- Grant MR, Godiard L, Straube E, Ashfield T, Lewald J, Sattler A, Innes RW, Dangl JL. Structure of the Arabidopsis RPM1 gene enabling dual specificity disease resistance. Science. 1995;269:843–846. doi: 10.1126/science.7638602. [DOI] [PubMed] [Google Scholar]
- Hiruma K, Nishiuchi T, Kato T, Bednarek P, Okuno T, Schulze-Lefert P, Takano Y. Arabidopsis ENHANCED DISEASE RESISTANCE 1 is required for pathogen-induced expression of plant defensins in nonhost resistance, and acts through interference of MYC2-mediated repressor function. Plant J. 2011;67:980–992. doi: 10.1111/j.1365-313X.2011.04651.x. [DOI] [PubMed] [Google Scholar]
- Huffaker A, Dafoe NJ, Schmelz EA. ZmPep1, an ortholog of Arabidopsis elicitor peptide 1, regulates maize innate immunity and enhances disease resistance. Plant Physiol. 2011;155:1325–1338. doi: 10.1104/pp.110.166710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huffaker A, Pearce G, Ryan CA. An endogenous peptide signal in Arabidopsis activates components of the innate immune response. Proc Natl Acad Sci USA. 2006;103:10098–10103. doi: 10.1073/pnas.0603727103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huffaker A, Pearce G, Veyrat N, Erb M, Turlings TC, Sartor R, Shen Z, Briggs SP, Vaughan MM, Alborn HT, Teal PE, Schmelz EA. Plant elicitor peptides are conserved signals regulating direct and indirect antiherbivore defense. Proc Natl Acad Sci USA. 2013;110:5707–5712. doi: 10.1073/pnas.1214668110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huffaker A, Ryan CA. Endogenous peptide defense signals in Arabidopsis differentially amplify signaling for the innate immune response. Proc Natl Acad Sci USA. 2007;104:10732–10736. doi: 10.1073/pnas.0703343104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huser A, Takahara H, Schmalenbach W, O'Connell R. Discovery of pathogenicity genes in the crucifer anthracnose fungus Colletotrichum higginsianum, using random insertional mutagenesis. Mol Plant Microbe Interact. 2009;22:143–156. doi: 10.1094/MPMI-22-2-0143. [DOI] [PubMed] [Google Scholar]
- Jirage D, Tootle TL, Reuber TL, Frost LN, Feys BJ, Parker JE, Ausubel FM, Glazebrook J. Arabidopsis thaliana PAD4 encodes a lipase-like gene that is important for salicylic acid signaling. Proc Natl Acad Sci USA. 1999;96:13583–13588. doi: 10.1073/pnas.96.23.13583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawai T, Akira S. Toll-like receptors and their crosstalk with other innate receptors in infection and immunity. Immunity. 2011;34:637–650. doi: 10.1016/j.immuni.2011.05.006. [DOI] [PubMed] [Google Scholar]
- Klauser D, Flury P, Boller T, Bartels S. Several MAMPs, including chitin fragments, enhance AtPep-triggered oxidative burst independently of wounding. Plant Signal Behav. 2013;8:e25346. doi: 10.4161/psb.25346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koch M, Vorwerk S, Masur C, Sharifi-Sirchi G, Olivieri N, Schlaich NL. A role for a flavin-containing mono-oxygenase in resistance against microbial pathogens in Arabidopsis. Plant J. 2006;47:629–639. doi: 10.1111/j.1365-313X.2006.02813.x. [DOI] [PubMed] [Google Scholar]
- Lee HI, Raskin I. Glucosylation of salicylic acid in Nicotiana tabacum cv. Xanthi-nc. Phytopathology. 1998;88:692–697. doi: 10.1094/PHYTO.1998.88.7.692. [DOI] [PubMed] [Google Scholar]
- Leon-Reyes A, Du Y, Koornneef A, Proietti S, Korbes AP, Memelink J, Pieterse CM, Ritsema T. Ethylene signaling renders the jasmonate response of Arabidopsis insensitive to future suppression by salicylic Acid. Mol Plant Microbe Interact. 2010;23:187–197. doi: 10.1094/MPMI-23-2-0187. [DOI] [PubMed] [Google Scholar]
- Lipka V, Dittgen J, Bednarek P, Bhat R, Wiermer M, Stein M, Landtag J, Brandt W, Rosahl S, Scheel D, Llorente F, Molina A, Parker J, Somerville S, Schulze-Lefert P. Pre-and postinvasion defenses both contribute to nonhost resistance in Arabidopsis. Science. 2005;310:1180–1183. doi: 10.1126/science.1119409. [DOI] [PubMed] [Google Scholar]
- Liu Z, Wu Y, Yang F, Zhang Y, Chen S, Xie Q, Tian X, Zhou JM. BIK1 interacts with PEPRs to mediate ethylene-induced immunity. Proc Natl Acad Sci USA. 2013;110:6205–6210. doi: 10.1073/pnas.1215543110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Logemann E, Birkenbihl RP, Rawat V, Schneeberger K, Schmelzer E, Somssich IE. Functional dissection of the PROPEP2 and PROPEP3 promoters reveals the importance of WRKY factors in mediating microbe-associated molecular pattern-induced expression. New Phytol. 2013;198:1165–1177. doi: 10.1111/nph.12233. [DOI] [PubMed] [Google Scholar]
- van Loon LC, Rep M, Pieterse CM. Significance of inducible defense-related proteins in infected plants. Annu Rev Phytopathol. 2006;44:135–162. doi: 10.1146/annurev.phyto.44.070505.143425. [DOI] [PubMed] [Google Scholar]
- Lu X, Tintor N, Mentzel T, Kombrink E, Boller T, Robatzek S, Schulze-Lefert P, Saijo Y. Uncoupling of sustained MAMP receptor signaling from early outputs in an Arabidopsis endoplasmic reticulum glucosidase II allele. Proc Natl Acad Sci USA. 2009;106:22522–22527. doi: 10.1073/pnas.0907711106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Y, Walker RK, Zhao Y, Berkowitz GA. Linking ligand perception by PEPR pattern recognition receptors to cytosolic Ca2+ elevation and downstream immune signaling in plants. Proc Natl Acad Sci USA. 2012;109:19852–19857. doi: 10.1073/pnas.1205448109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mersmann S, Bourdais G, Rietz S, Robatzek S. Ethylene signaling regulates accumulation of the FLS2 receptor and is required for the oxidative burst contributing to plant immunity. Plant Physiol. 2010;154:391–400. doi: 10.1104/pp.110.154567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishina TE, Zeier J. Pathogen-associated molecular pattern recognition rather than development of tissue necrosis contributes to bacterial induction of systemic acquired resistance in Arabidopsis. Plant J. 2007;50:500–513. doi: 10.1111/j.1365-313X.2007.03067.x. [DOI] [PubMed] [Google Scholar]
- Narusaka M, Shirasu K, Noutoshi Y, Kubo Y, Shiraishi T, Iwabuchi M, Narusaka Y. RRS1 and RPS4 provide a dual Resistance-gene system against fungal and bacterial pathogens. Plant J. 2009;60:218–226. doi: 10.1111/j.1365-313X.2009.03949.x. [DOI] [PubMed] [Google Scholar]
- Navarova H, Bernsdorff F, Doring AC, Zeier J. Pipecolic acid, an endogenous mediator of defense amplification and priming, is a critical regulator of inducible plant immunity. Plant Cell. 2012;24:5123–5141. doi: 10.1105/tpc.112.103564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nekrasov V, Li J, Batoux M, Roux M, Chu ZH, Lacombe S, Rougon A, Bittel P, Kiss-Papp M, Chinchilla D, van Esse HP, Jorda L, Schwessinger B, Nicaise V, Thomma BP, Molina A, Jones JD, Zipfel C. Control of the pattern-recognition receptor EFR by an ER protein complex in plant immunity. EMBO J. 2009;28:3428–3438. doi: 10.1038/emboj.2009.262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park JH, Halitschke R, Kim HB, Baldwin IT, Feldmann KA, Feyereisen R. A knock-out mutation in allene oxide synthase results in male sterility and defective wound signal transduction in Arabidopsis due to a block in jasmonic acid biosynthesis. Plant J. 2002;31:1–12. doi: 10.1046/j.1365-313x.2002.01328.x. [DOI] [PubMed] [Google Scholar]
- Pieterse CM, Van der Does D, Zamioudis C, Leon-Reyes A, Van Wees SC. Hormonal modulation of plant immunity. Annu Rev Cell Dev Biol. 2012;28:489–521. doi: 10.1146/annurev-cellbio-092910-154055. [DOI] [PubMed] [Google Scholar]
- Robert-Seilaniantz A, Grant M, Jones JD. Hormone crosstalk in plant disease and defense: more than just jasmonate-salicylate antagonism. Annu Rev Phytopathol. 2011;49:317–343. doi: 10.1146/annurev-phyto-073009-114447. [DOI] [PubMed] [Google Scholar]
- Ryan CA, Huffaker A, Yamaguchi Y. New insights into innate immunity in Arabidopsis. Cell Microbiol. 2007;9:1902–1908. doi: 10.1111/j.1462-5822.2007.00991.x. [DOI] [PubMed] [Google Scholar]
- Saijo Y. ER quality control of immune receptors and regulators in plants. Cell Microbiol. 2010;12:716–724. doi: 10.1111/j.1462-5822.2010.01472.x. [DOI] [PubMed] [Google Scholar]
- Saijo Y, Tintor N, Lu X, Rauf P, Pajerowska-Mukhtar K, Haweker H, Dong X, Robatzek S, Schulze-Lefert P. Receptor quality control in the endoplasmic reticulum for plant innate immunity. EMBO J. 2009;28:3439–3449. doi: 10.1038/emboj.2009.263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schilmiller AL, Howe GA. Systemic signaling in the wound response. Curr Opin Plant Biol. 2005;8:369–377. doi: 10.1016/j.pbi.2005.05.008. [DOI] [PubMed] [Google Scholar]
- Segonzac C, Zipfel C. Activation of plant pattern-recognition receptors by bacteria. Curr Opin Microbiol. 2011;14:54–61. doi: 10.1016/j.mib.2010.12.005. [DOI] [PubMed] [Google Scholar]
- Serrano M, Kanehara K, Torres M, Yamada K, Tintor N, Kombrink E, Schulze-Lefert P, Saijo Y. Repression of sucrose/ultraviolet B light-induced flavonoid accumulation in microbe-associated molecular pattern-triggered immunity in Arabidopsis. Plant Physiol. 2012;158:408–422. doi: 10.1104/pp.111.183459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen QH, Schulze-Lefert P. Rumble in the nuclear jungle: compartmentalization, trafficking, and nuclear action of plant immune receptors. EMBO J. 2007;26:4293–4301. doi: 10.1038/sj.emboj.7601854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song JT, Lu H, McDowell JM, Greenberg JT. A key role for ALD1 in activation of local and systemic defenses in Arabidopsis. Plant J. 2004;40:200–212. doi: 10.1111/j.1365-313X.2004.02200.x. [DOI] [PubMed] [Google Scholar]
- Thomma BP, Cammue BP, Thevissen K. Plant defensins. Planta. 2002;216:193–202. doi: 10.1007/s00425-002-0902-6. [DOI] [PubMed] [Google Scholar]
- Tintor N, Ross A, Kanehara K, Yamada K, Fan L, Kemmerling B, Nurnberger T, Tsuda K, Saijo Y. Layered pattern receptor signaling via ethylene and endogenous elicitor peptides during Arabidopsis immunity to bacterial infection. Proc Natl Acad Sci USA. 2013;110:6211–6216. doi: 10.1073/pnas.1216780110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Truman W, Bennett MH, Kubigsteltig I, Turnbull C, Grant M. Arabidopsis systemic immunity uses conserved defense signaling pathways and is mediated by jasmonates. Proc Natl Acad Sci USA. 2007;104:1075–1080. doi: 10.1073/pnas.0605423104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuda K, Sato M, Stoddard T, Glazebrook J, Katagiri F. Network properties of robust immunity in plants. PLoS Genet. 2009;5:e1000772. doi: 10.1371/journal.pgen.1000772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D, Weaver ND, Kesarwani M, Dong X. Induction of protein secretory pathway is required for systemic acquired resistance. Science. 2005;308:1036–1040. doi: 10.1126/science.1108791. [DOI] [PubMed] [Google Scholar]
- van Wees SC, de Swart EA, van Pelt JA, van Loon LC, Pieterse CM. Enhancement of induced disease resistance by simultaneous activation of salicylate-and jasmonate-dependent defense pathways in Arabidopsis thaliana. Proc Natl Acad Sci USA. 2000;97:8711–8716. doi: 10.1073/pnas.130425197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wildermuth MC, Dewdney J, Wu G, Ausubel FM. Isochorismate synthase is required to synthesize salicylic acid for plant defence. Nature. 2001;414:562–565. doi: 10.1038/35107108. [DOI] [PubMed] [Google Scholar]
- Yamaguchi Y, Huffaker A. Endogenous peptide elicitors in higher plants. Curr Opin Plant Biol. 2011;14:351–357. doi: 10.1016/j.pbi.2011.05.001. [DOI] [PubMed] [Google Scholar]
- Yamaguchi Y, Huffaker A, Bryan AC, Tax FE, Ryan CA. PEPR2 is a second receptor for the Pep1 and Pep2 peptides and contributes to defense responses in Arabidopsis. Plant Cell. 2010;22:508–522. doi: 10.1105/tpc.109.068874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi Y, Pearce G, Ryan CA. The cell surface leucine-rich repeat receptor for AtPep1, an endoaenous peptide elicitor in Arabidopsis, is functional in transgenic tobacco cells. Proc Natl Acad Sci USA. 2006;103:10104–10109. doi: 10.1073/pnas.0603729103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan J, He SY. The Pseudomonas syringae Hrp regulation and secretion system controls the production and secretion of multiple extracellular proteins. J Bacteriol. 1996;178:6399–6402. doi: 10.1128/jb.178.21.6399-6402.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zipfel C, Kunze G, Chinchilla D, Caniard A, Jones JD, Boller T, Felix G. Perception of the bacterial PAMP EF-Tu by the receptor EFR restricts Agrobacterium-mediated transformation. Cell. 2006;125:749–760. doi: 10.1016/j.cell.2006.03.037. [DOI] [PubMed] [Google Scholar]
- Zipfel C, Robatzek S, Navarro L, Oakeley EJ, Jones JD, Felix G, Boller T. Bacterial disease resistance in Arabidopsis through flagellin perception. Nature. 2004;428:764–767. doi: 10.1038/nature02485. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.