Abstract
Chemical exposures in fish have been linked to loss of olfaction leading to an inability to detect predators and prey and decreased survival. However, the mechanisms underlying olfactory neurotoxicity are not well characterized, especially in environmental exposures which involve chemical mixtures. We used zebrafish to characterize olfactory transcriptional responses by two model olfactory inhibitors, the pesticide chlorpyrifos (CPF) and mixtures of CPF with the neurotoxic metal copper (Cu). Microarray analysis was performed on RNA from olfactory tissues of zebrafish exposed to CPF alone or to a mixture of CPF and Cu. Gene expression profiles were analyzed using Principal Component Analysis and hierarchical clustering, whereas gene set analysis was used to identify biological themes in the microarray data. Microarray results were confirmed by real-time PCR on genes serving as potential biomarkers of olfactory injury. In addition, we mined our previously published Cu-induced zebrafish olfactory transcriptional response database (Tilton et al., 2008) for the purposes of discriminating pathways of olfaction impacted by either the individual agents or the CPF-Cu mixture transcriptional signatures. CPF exposure altered the expression of gene pathways associated with cellular morphogenesis and odorant binding, but not olfactory signal transduction, a known olfactory pathway for Cu. The mixture profiles shared genes from the Cu and CPF datasets, whereas some genes were altered only by the mixtures. The transcriptional signature of the mixtures was more similar to that in zebrafish exposed to Cu alone then for CPF. In conclusion, exposure to a mixture containing a common environmental metal and pesticide causes a unique transcriptional signature that is heavily influenced by the metal, even when organophosphate predominates. Our findings support using zebrafish microarray analysis to elucidate mechanisms of olfactory loss and to identify the components of mixtures which most strongly contribute to olfactory injury.
Keywords: copper, chlorpyrifos, organophosphate pesticide, zebrafish, olfaction, toxicity, mixtures
Introduction
Exposure to several classes of chemicals has been linked to impaired chemosensory function and loss of olfaction in vertebrates (Deamer and Genter, 1995; Tierney et al., 2007b; Tierney et al., 2008). The inhalation of certain metals such as copper (Cu) (Trombley and Shepherd, 1996; Horning and Trombley, 2001), cadmium (Rydzewski et al., 1998) and zinc (van Denderen et al., 2001; Slotnick et al., 2007), as well as solvents and other occupational chemicals (Yu et al., 2004) can result in olfactory epithelial injury and loss of sense of smell. Chemical neurobehavioral injury in fish resulting from exposures to pesticides and metals can be problematic due to the loss of olfactory-driven behaviors such as homing, predator avoidance, prey selection and reproduction which are critical for survival (Sandahl et al., 2004; Sandahl et al., 2005; Tierney et al., 2010). The mechanisms by which pollutants may cause these chemosensory injuries are complex and are poorly understood, especially in the context of mixture exposures, a common scenario in aquatic environments.
The multi-tissue olfactory system of fish extends from the olfactory rosettes that are in contact with the outside environment, to the olfactory bulb (OB) and neuronal networks within the brain. Odorant binding to olfactory receptors in the olfactory epithelium triggers a conserved G-protein signaling cascade. This process of olfactory signal transduction (OST) produces electrical impulses in peripheral sensory neurons that are propagated to the OB for processing and integration prior to the relay of sensory information to more central networks (Laberge and Hara, 2001). Despite the complexities and data gaps underlying the olfactory system of fish, the neuroanatomy and physiology of the olfactory system in zebrafish is comparatively well described, and includes recent insights into the understanding of calcium signaling in the olfactory bulb (OB), neuronal connections with the brain, and olfactory cell regeneration (Byrd and Brunjes, 2001; Tabor et al., 2004; Sato et al., 2007). Accordingly, zebrafish provide a powerful experimental tool to advance our understanding of chemically induced olfactory injury in mammals as well as in ecological studies.
Although a number of toxicants can disrupt olfactory processes in vertebrates, metals and pesticides are among the most strongly linked to olfactory injuries in aquatic species (Lurling and Scheffer, 2007). The common trace metal Cu potently disrupts olfaction in salmonids (Baldwin et al., 2003; Sandahl et al., 2004; McIntyre et al., 2008), and we have shown in a microarray study using zebrafish that Cu-olfactory impairment may be largely attributed to a transcriptional repression of conserved signal transduction genes in the zebrafish olfactory system (Tilton et al., 2008). The pesticide chlorpyrifos (CPF) is an organophosphate pesticide that alters neuronal development, synaptic stability and growth in rat and human neuronal cells (Garcia et al., 2002; Howard et al., 2005; Chen et al., 2008; Roegge et al., 2008) and is highly toxic to the fish olfactory system (Sandahl et al., 2004; Tierney et al., 2007a). In the current study, we investigated gene expression patterns underlying olfactory injury caused by exposure to CPF alone, or as a binary mixture in combination with Cu. In addition, we analyzed zebrafish olfactory transcriptional signatures associated with CPF with those from our previous dataset obtained on exposure to Cu (Tilton et . al, 2008) to better understand mechanisms of olfactory injury, and to characterize the contribution of the two agents toward olfactory injury in the mixture. Specifically, we exposed zebrafish to a series of CPF concentrations to establish a molar equivalent transcriptional response in the olfactory system in order to compare to a mixture response using the more widely studied trace metal Cu. By mining our previously published Cu-induced zebrafish olfactory transcriptional response database (Tilton et al., 2008), we were able to identify and discriminate biochemical pathways of olfaction impacted by either the individual agents or the CPF-Cu mixture transcriptional signatures generated from this study.
Materials and Methods
Animal care and maintenance
One year old adult AB strain zebrafish were a generous gift from Dr. Robert Tanguay at Oregon State University. The animals were maintained in 10 gallon aquaria with re-circulating filtration at a density of 4 animals/L. Fish were fed flake food in the morning and artemia in the afternoon. Ammonia, nitrite, pH, and temperature were recorded at least once daily. The water source was city municipal water that was passed through a treatment system containing 0.2 μm filtration, activated carbon, and ionic- and mixed-bed filters. The resulting water pH was 5.5 and devoid of conductivity and chlorine. Water for fish was reconstituted freshwater using Instant Ocean® salts at 1000±100 μS, and the pH was adjusted to 7.0 using Na2HCO3 and heated to 27°C in a holding reservoir. Tanks received a 20% daily water change or more as necessary.
Exposures
Zebrafish were exposed for 24 hr in three replicate groups of five animals for each experimental concentration and control group. Exposures were conducted in 3L of water held in a sealed 3.5L glass jar maintained at 28°C using a water bath. A CPF stock solution (ChemService Inc., West Chester, PA) was made up in dimethylsulfoxide (DMSO). The final concentration of DMSO for each exposure (i.e. CPF alone, Cu alone, CPF + Cu) as well as for controls was 0.001%. Zebrafish were exposed to nominal CPF concentrations of 0.1, 0.25 or 0.6 μM (35, 88 or 220 ug/L). Although the CPF concentrations used in this study were higher than typically encountered in surface waters (0.1-10 ppb) (Gilliom, 2007), the doses were selected to create molar ratio exposures between Cu and CPF in this study in which Cu predominated the mixture, CPF dominated the mixture, or the molar ratios of the two compounds were equivalent (described below). The exposure jars were pretreated with the respective CPF concentrations for 24 hr prior to use and reused for each of the treatments of CPF to minimize loss to glass surface adherence. Water samples were collected and analyzed for the concentrations of CPF and CPF-oxon by the Pacific Agricultural Laboratory (Portland, OR). Our previous study confirmed a close association among the nominal and actual Cu exposure concentrations (Tilton et al., 2008).
For the binomial mixtures, the CuCl2 (Alpha Aeser, Ward Hill, MA) stock solution was prepared in distilled water and exposure concentrations were held constant at 0.25 μM (16 ug/L) in combination with each of the three CPF concentrations. As a result, this experimental design allowed us to expose zebrafish to three mixture scenarios in which, 1) Cu was in molar excess (Mix A, 2.5:1 for Cu:CPF), 2) Cu was equimolar (Mix B, 1:1), and 3) CPF was in excess (Mix C, 1:2.5). All Cu exposures were also spiked with 0.001% DMSO to control for possible carrier solvent effects. Exposures were conducted in sealed glass jars, maintained at 26°C for 24 hr, and were staggered by 1 hr to allow for tissue collection. Each replicate pool had 2 males and 3 females (or vice versa) that were alternated so that the ratios across replicates approximated 1:1 (males: females). After 24 hr of exposure, water quality parameters (pH, dissolved oxygen, and temperature) were recorded. Water quality parameters did not appreciably change over the course of the experiment.
Olfactory tissue collection and total RNA isolation
Individual fish were euthanized by cervical dislocation. The olfactory rosettes, OB, and telencephalon were removed by dissection, immediately placed in Trizol (Invitrogen, Carlsbad, CA), and stored at -80°C until processing for total RNA isolation. Olfactory RNA was isolated from tissue pooled from 5 animals in each group. A total of 3 replicate pools of RNA were generated for each experimental condition resulting in a total of 21 pooled samples, each of which was arrayed separately. Total olfactory RNA was isolated, purified and quantitated as described previously (Tilton et al., 2008). RNA quality was determined with an Agilent 2100 Bioanalyzer (Santa Clara, CA) prior to further processing. All olfactory RNA samples were devoid of contamination or RNA degradation.
Microarray analysis
5 μg of pooled total RNA was processed and hybridized to Affymetrix GeneChip Zebrafish Genome Arrays (Affymetrix, Inc., Santa Clara, CA). Zebrafish Genome coverage for this array was selected from the following public data sources: RefSeq (July 2003), GenBank® (release 136.0, June 2003), dbEST (July 2003), and UniGene (Build 54, June 2003). The array was designed in collaboration with representative members of the Zebrafish community and the National Institutes of Health. Sample and microarray processing, scanning and feature extraction were conducted as described previously (Tilton et al., 2008). All samples arrayed for the current and our previously published study (Tilton et al., 2008) were processed simultaneously to minimize the number of control animals, arrays, and technical variability between studies. This approach allowed for a direct comparison of the microarray analyses from this and our previous investigation into the olfactory toxicity of Cu alone. Microarray data were processed with several Bioconductor software packages (http://www.bioconductor.org/) (Gentleman et al., 2004). Stringent microarray data quality control was carried out and included visual inspection of the GCOS DAT chip images as well as the chip pseudo-images generated with the Bioconductor affyPLM package, assessment of percent present calls and average background signals with the Bioconductor Simpleaffy package, inspection of histograms of raw signal intensities, and comparison of the relative log expression and normalized unscaled standard errors using the Bioconductor affyPLM package. The data were normalized with the Bioconductor gcrma package Ananth. The Bioconductor limma package (Gentleman et al., 2004) was used to calculate fold-changes and p-values. P-values were adjusted for multiplicity with the Bioconductor package q value (Tusher et al., 2001), which allows for selecting statistically significant genes while limiting the estimated false discovery rate. Principal component analysis (PCA) was carried out with JMP Genomics software (SAS Institute Inc., Cary, NC). Hierarchical clustering was performed with the Bioconductor stats package. This type of unsupervised clustering visualizes how the samples cluster together based on Cu and CPF content, as shown in Figure 1b. Pearson correlations were calculated with GraphPad Prism (GraphPad Software, La Jolla, CA). Functional annotations were based on the published zebrafish literature, ZFIN (www.zfin.org) zebrafish expression database, and putative homology from Gene Ontology (GO; www.geneontology.org) and/or OMIM™ (www.ncbi.nlm.nih.gov/omim/) databases.
Figure 1. Principal component analysis (PCA), hierarchical clustering and Venn diagrams of chlorpyrifos (CPF), copper (Cu), and binomial mixture (Mix) microarray data.
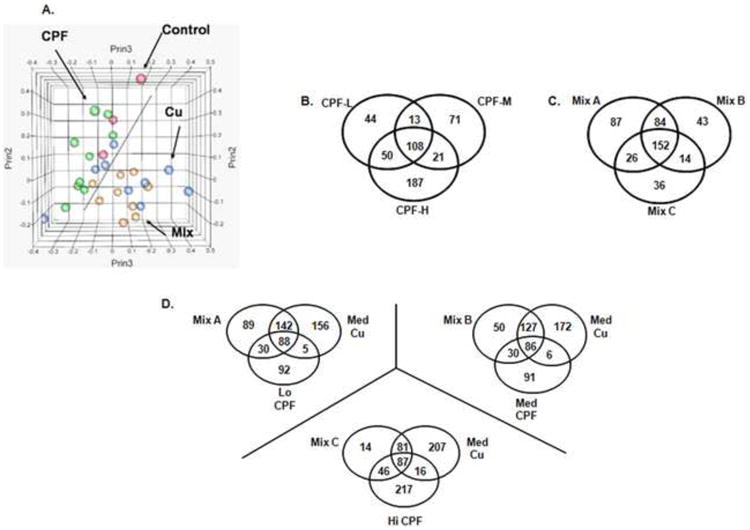
A) PCA reduces a complex data set to a lower dimension that can reveal hidden, simplified structures in the data. Data from all genes on the array were included in the PCA and the three top principal components were used to generate the three dimensional figure shown. Red circles represent controls, green circles CPF, blue circles Cu, and yellow circles mixtures. PCA clearly separates the Cu exposed microarray data sets from those originating from the CPF exposures. This separation is partially obscured by the two dimensional representation of the three dimensional Figure 1A and the following description is intended to remedy this limitation. The data points representing Cu treatments (blue) move forward and to the lower right of the figure with increasingly higher doses, whereas the CPF data points (green) extend to the back and to the upper left of the figure. By contrast, the controls and the samples treated with the low Cu or low CPF concentrations congregate along the center dotted line. The mixture data points (gold dots) associated closer with the Cu treatments. B-D) Venn diagrams showing the number of shared genes between treatments significantly affected (p<0.05) in at least one treatment at >1.5 fold greater than controls. B) Between 213-366 total transcripts were perturbed by CPF at any one concentration, and 108 differentially expressed transcripts were shared between all three CPF concentrations. C) Although the mixtures altered the expression of a roughly similar number of transcripts (228-349 transcripts), the number of altered genes decreased as the concentration of CPF in the mixtures increased. The majority of altered transcripts were shared (152) among all three mixtures or between any two combinations. Each mixture exhibited a unique response which decreased as the concentration of CPF in the mixtures increased. D) Venn diagrams of mixture treatments compared to the signatures of individual constituents. Overall Mix A (upper left) and Mix B (upper right) have similar relationships between treatments while Mix C (bottom) differs.
Gene ontology (GO) analysis
The hypergeometric distribution method was used to determine enhanced GO categories (Camon et al., 2003) using the Bioconductor package topGO (Alexa et al., 2006). Using the classic, elim, and weighted method approaches, we determined the number of GO terms that were significantly different from controls (p<0.05) for each treatment.
Gene set analysis (GSA)
We used the freely available GSA R-code to identify categories of genes showing coordinated changes in individual gene expression over the experimental conditions (Subramanian et al., 2005). In contrast to the cumulative hypergeometric distribution method described above for GO analysis, GSA considers all genes in the experiment and identifies gene sets/pathways with genes that show concordant changes in expression, even if such changes are modest. We used three databases for GSA: GO Biological Process, GO Molecular Function (Camon et al., 2003), and C2 from the Molecular Signatures Database. We also conducted GSA analysis of custom designed olfactory signal transduction pathway gene sets. The Kyoto Encyclopedia of Genes and Genomes (KEGG; http://www.genome.jp/kegg) specifies 16 genes that are involved in the olfactory signal transduction cascade. We identified 110 Affymetrix probe set IDs that share sequence similarity with these 16 genes (OST-110, supplemental table 1). The 110 probe set IDs formed the basis for custom designing six additional gene sets covering various aspects of the olfactory signal transduction pathway by removing genes least likely to contribute to OST within the olfactory neurons of the olfactory rosettes, or by applying increasing stringency of sequence similarity (see (Tilton et al., 2008) for a more detailed explanation). One of these included all possible genes found on the array with similarity to the 5 genes making up the fundamental signal transduction pathway (OST-root) (i.e. olfactory receptors, adenylate cyclase, g-proteins, cyclic-nucleotide and chloride channels) and another gene set contained all possible genes which act as inhibitors within the OST pathway (OST-Inhib). The aforementioned custom designed olfactory gene sets were included in the BP gene ontology database and for GSA.
SybrGreen quantitative real-time PCR confirmation of microarray data
Primers for microarray validation were designed using the Affymetrix probe set sequence and Oligo Primer Analysis Software, v. 6.71 (Cascade, CO) (Supplemental table 2). Genes found to be significantly differentially expressed by microarray analysis, or genes with utility as potential molecular biomarkers of olfactory impairment were selected for quantitative PCR analysis. Each primer pair produced a single PCR product as evidenced by melt curve analysis and gel electrophoresis. Expression of β-actin did not differ among treatments based on either microarray analysis or quantitative real time–PCR (qPCR) and was used for qPCR data normalization.
Results
Waterborne CPF and Cu concentrations
CPF concentrations in water were determined at 0 and 24 hours of exposure. At 24 hours, CPF concentrations in exposure jars containing fish had decreased by more than 80% for each concentration, whereas they were essentially unchanged in control jars which did not contain fish (Supplemental Table 3). These findings indicate that CPF was taken up effectively by the zebrafish. In addition, only a small amount of CPF-oxon was detected at the different CPF concentrations (0.6-2% of nominal CPF concentrations measured at 0 hours; data not shown). The measured Cu concentrations generally agreed closely with the nominal Cu concentrations (Tilton et al., 2008). None of the exposures were lethal.
Principal component analysis and hierarchal clustering of CPF, Cu and CPF/Cu mixture transcriptional profiles
We previously reported the effects of Cu exposures on olfactory transcription in zebrafish (Tilton et al., 2008). The experimental conditions and methods used in the previous and current studies were identical and thus allowed for a direct comparison of the CPF, Cu and Cu/CPF mixture transcriptional profiles. In the current study, we used principal component analysis (PCA) to investigate how the CPF, Cu and Cu/CPF mixture transcriptional profiles inter-related. PCA reduces a complex data set to a lower dimension that can reveal hidden, simplified structures in the data. Data from all the genes on the array were included in the current analysis, without any prior filtering. Figure 1A. shows the principal components of the CPF, Cu and CPF/Cu-mixtures in 3 dimensions. PCA clearly separates the Cu exposed microarray data sets from those originating from the CPF exposures. PCA also indicates that the Cu/CPF mixture data points associated closer with the Cu treatments than the CPF treatments.
Data for genes exhibiting at least 1.8-fold differential expression at p<0.05 in any of the treatment groups relative to controls were used in hierarchical cluster analysis. This analysis included our previous Cu dataset (Tilton et al., 2008). As observed in Figure 2, there was a distinct clustering among CPF and Cu transcriptional profiles, reflecting unique transcriptional signatures and suggesting distinct biochemical mechanisms of olfactory perturbation. Consistent with the PCA (Figure 1A), the CPF/Cu mixture samples clustered with the medium dose Cu (Cu-M) samples, but not with any of the CPF samples.
Figure 2.
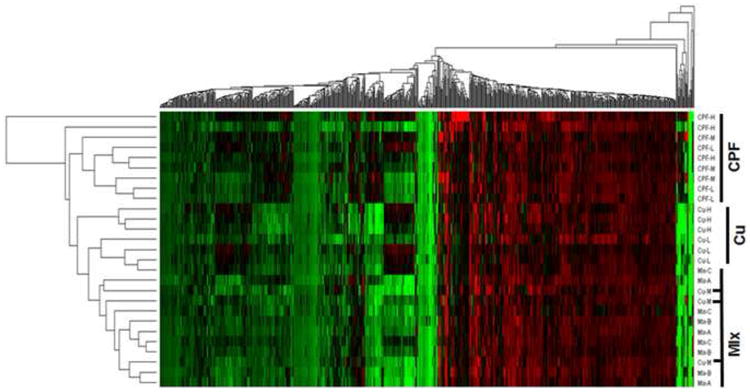
Hierarchical clustering of transcripts significantly changed (p<0.05 and ≥1.8 fold) vs. controls. The cluster labeled “Mix” contains the CPF and Cu mixture exposures and the three Cu-M (medium dose copper) samples are indicated with horizontal black lines.
Single gene analysis
The low (0.1 uM; CPF-L), medium (0.25 uM; CPF-M) and high (0.6 uM; CPF-H) pesticide doses significantly (p<0.05) modulated the expression of 215, 213, and 366 transcripts, respectively, using a 1.5-fold cutoff. The choice of a 1.5-fold cut-off was based on our previous experience with the Affymetrix array platform; i.e. we could confirm the majority of gene expression changes of 1.5-fold or greater with q-PCR, but not changes that were smaller than 1.5-fold. Of these aforementioned genes, 108 transcripts were affected by all three CPF doses (Figure 1B). The majority of the responses associated with the low and medium doses were shared with at least one other CPF dose (i.e. 80% of 215 differentially expressed genes in the CPF-L exposure group, and 67% of 213 differentially expressed genes in the CPF-M exposure group). The exception was the high CPF dose with 51% unique transcriptional changes (187 of 366 differentially-expressed genes). In addition, we observed a dose-dependent increase in the number of unique transcripts with 44, 71, and 187 of each at the low, medium, and high doses respectively (Figure 1B).
The number of transcripts uniquely associated with each of the three mixtures (Mix-A, 0.1 uM CPF+0.25 uM Cu; Mix-B 0.25 uM CPF+0.25 uM Cu; Mix-C, 0.6 uM CPF+0.25 uM Cu) decreased as the CPF concentration increased (Figure 1C). At the same time, expression of 152 common transcripts was changed by each of the three mixtures. Many transcripts were only affected by the combined CPF/Cu treatments and not by exposure to either constituent alone (Figure 1D). Transcript changes associated with each mixture treatment were shared proportionally more with Cu relative to CPF treatments. Specifically, mixtures A, B and C shared 1.95-fold, 1.84-fold and 1.26-fold more transcripts, respectively, with the corresponding Cu exposures than for CPF. Complete lists of all differentially regulated genes (≥1.5-fold, p < 0.05) for all CPF and mixture treatment groups are available as supplemental data [Supplemental Tables 4 and 5].
To evaluate the mixture relationships, Pearson correlation coefficients were determined between the mixture treatments and their respective constituents (Figures 3A-C). All the mixtures showed strong correlations to Cu, with Pearson coefficients of R = 0.92, 0.90, and 0.85 for Mix A, Mix B and Mix C, and two-tailed linear regression values of r2 = 0.84, 0.81, and 0.72, respectively. The correlation to the respective CPF treatment in each mixture was not nearly as strong (R = 0.81, 0.71, 0.75 and r2 = 0.65, 0.50, 0.56, respectively). Data presented in Figures 3D-G represent an evaluation of whether the mixture responses were reflective of a theoretical additive response of Cu and CPF constituents. Many of the mixture transcriptional responses were less than predicted for the theoretical additive value of the constituents suggesting they largely resulted from exposure to one of the two constituent compounds. As observed, virtually no transcripts showed greater than additive responses (Figures 3E-G).
Figure 3. Correlations of gene profiles from each binomial mixture relative to individual components and to a theoretical expectation of response-addition.
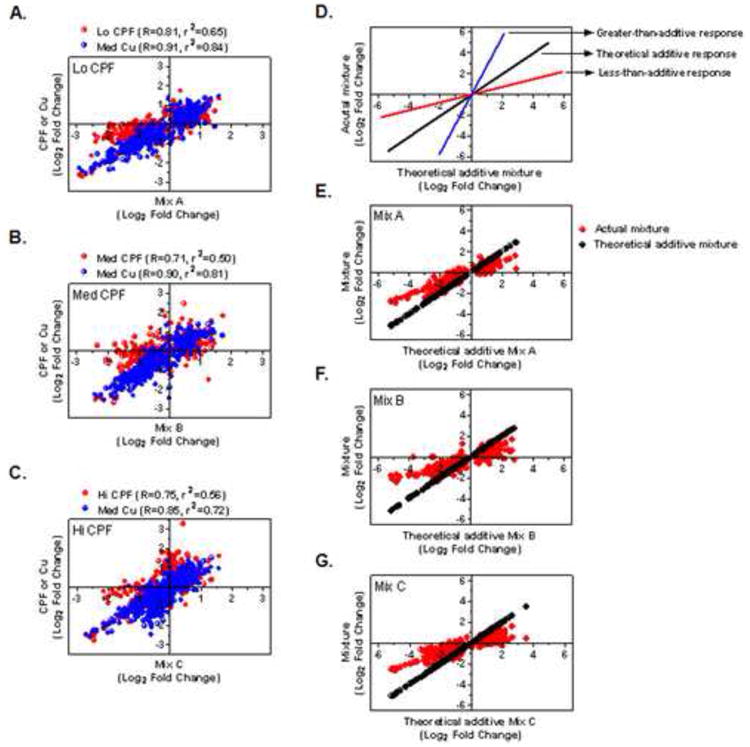
Values are expressed as fold change (Log2) compared to vehicle-treated controls and were plotted to generate Pearson correlation coefficients (R) among treatments and least-squares linear regression (r2 values, p <0.0001). A) Mix A versus CPF-L or Med Cu, B) Mix B versus Med CPF or Med Cu, C) Mix C versus Hi CPF or Med Cu, D) theoretical additive, greater-than-additive, or less-than-additive response of the actual binomial mixtures relative to theoretical mixture values calculated from addition of each constituent, E) Mix A versus theoretical additive Mix A, F) Mix B versus theoretical additive Mix B, and G) Mix C versus theoretical additive Mix C. All binomial mixtures show a less-than-additive response.
Identification of gene pathways altered by mixtures and their components
Unlike single gene analysis, category analysis allows for determination of statistical significance of gene pathways and identification of biological themes. We used two complementary category analysis methods, including Gene Set Analysis (GSA) and the cumulative hypergeometric distribution method topGO. In contrast to the cumulative hypergeometric method, GSA does not require filtering (p-value and/or fold-change cutoffs) to define differentially expressed genes that can be further analyzed while evaluating all genes in the experiment (Subramanian et al., 2005).
GSA was employed to identify Biological Process (BP) and Molecular Function (MF) Gene Ontology gene sets that were statistically significant in the CPF and CPF/Cu mixture data sets relative to the controls (Figure 4 and Supplemental Tables 6, 7, 8, 9). Morphogenesis processes were disproportionately represented in the CPF treatments (18% of 118 gene sets, p ≤ 0.01) (Figure 4A), and the majority of these gene sets had a negative score, indicating a coordinated downregulation of genes in these pathways. Furthermore, exposure to CPF largely downregulated transcription of gene sets related to metabolic processes, cell growth or replication, and olfactory sensory cell function (Figure 4). The number of altered gene sets related to morphogenesis, cell growth, and cell replication increased as CPF concentrations increased (Figure 4E). In contrast, the number of metabolic and catabolic process gene sets decreased as CPF concentration increased (Figure 4E). The addition of Cu strongly impacted the transcriptional signature of CPF in the mixtures, resulting in gene sets with primarily positive scores and up-regulation of transcription (Figure 4B). Specifically, ‘metabolic processes’ gene sets showed positive relationships between CPF and mixtures (99 total gene sets, p ≤ 0.01). In contrast to the CPF exposures, GSA did not identify impact on morphogenesis gene sets in mixtures exposures. In general, the transcriptional effects of the mixtures are more consistent with those previously observed with Cu alone.
Figure 4. Gene Set Analysis (GSA) identification of Gene Ontology categories significantly over represented in the CPF or Cu/CPF mixture treatments relative to controls.
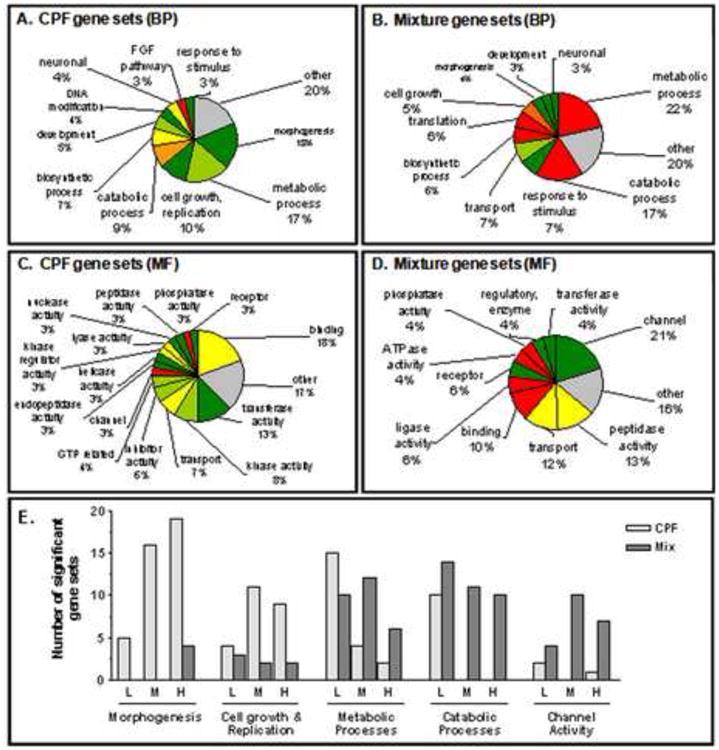
Pie graphs show the percentage of gene sets altered by category. Color indicates the shift of the treated gene sets based on the GSA score and the intensity reflects the overall shift of all the gene sets within the category. Dark green, all gene sets in group were significantly down regulated. Lime green, a majority of down regulated gene sets in the categorization. Yellow, no dominant pattern either up or down in the categorization. Red, all gene sets in the categorization were significantly up-regulated. Orange, a majority of gene sets in the categorization were up-regulated. Other unrelated gene sets totaling <3% each were included in the Other category (grey). A) Percentage of BP gene sets altered by category out of a total of 118 unique gene sets, p <0.01, with CPF treatment. B) Percentage of biological process (BP) gene sets altered by category out of a total of 99 unique gene sets, p <0.01, in the mixture treatments. C) Percentage of molecular function (MF) gene sets altered by category out of a total of 73 unique gene sets, p<0.05, with CPF treatment. D) Percentage of MF gene sets altered by category out of a total of 70 unique gene sets, p<0.01, in any mixture treatment. E) Comparison of number of gene sets related to morphogenesis, cell growth and cell replication, ion channel activities, and cellular metabolic and catabolic processes affected by CPF or the CPF/Cu mixture (L, M and H refer to the low, medium and high doses of CPF respectively).
Analysis of gene regulation in CPF-exposed zebrafish using the MF Gene Ontology database revealed gene sets associated with DNA-, RNA-, metal- and protein- binding that were significantly altered by CPF alone (73 total gene sets, p ≤ 0.05; Figure 4C) or by CPF in combination with Cu (71 total gene sets, p ≤ 0.01; Figure 4D). Furthermore, gene sets related to olfactory signal transduction (OST), including receptor and channel groupings, were also significant in both treatment groups. No gene set in the MF database showed a clear dose response to CPF alone. However, channel gene sets (e.g. related to ion channel activity, calcium channel activity, voltage-gated ion channel activity; for detailed list see supplemental tables 6, 7, 8, 9) were more affected with increasing CPF concentrations in the mixture treatments (Figure 4E).
TopGO analysis identified between 8 and 22 BP and MF gene ontology categories that were significantly (p <0.05) enriched within the three CPF treatment groups (Figure 4, Supplemental tables 10, 11). Interesting examples of over-enriched GO terms include neuron migration, morphogenesis, and ion homeostasis related gene sets. However, a consistent pattern in the gene sets was not readily apparent. Among the mixture treatments, GO terms which were significantly over-enriched (24 to 37 GO terms), included calcium and other ion binding or homeostatic processes. Also present were OST related gene sets. Interestingly, the presence of gene sets related to neuron migration, positive regulation of axon extension, neural plate morphogenesis and integrin-mediated signaling pathway suggest impairment to neuronal growth and possibly to neuronal regeneration pathways in the mixture group.
GSA of custom designed olfactory signal transduction pathway gene sets
We used GSA to investigate seven custom-designed gene sets targeting different numbers of genes found on the array with strong similarity to the 16 genes generally considered the core of olfactory G-protein signaling (i.e. the OST pathway (Supplemental Table 1). Using these custom gene sets no discernable coordinated shift by CPF was observed (Table 1) Upon closer inspection several genes within the pathway were impaired by CPF relative to controls and in some cases differently compared to the CPF/Cu mixture or Cu alone (e.g. CLC, OR2 G-proteins and arrestins (Supplemental table 12). These data indicate that the OST pathway showed greater impairment with increasing CPF only in the presence of Cu. In all Cu/CPF mixture groups, gene sets were generally down-regulated significantly in a manner similar to that previously observed with exposure to Cu alone (Tilton et al., 2008).
Table 1.
Gene Set Analysis results showing the relative shift from controls (score) and significance of custom pathways targeting olfactory signal transduction (OST) at each chlorpyrifos or mixture treatment. Significant pathways (p<0.05) are colored green or red. Negative shifts in the treated datasets relative to controls are indicated in green. Positive shifts are in red.
| Gene set number | Gene set name | Gene set length | Score | p-value | Score | p-value |
|---|---|---|---|---|---|---|
|
| ||||||
| CPF L | CPF L | Mix A | Mix A | |||
|
| ||||||
| 1792 | OST110 Pathway | 110 | -0.23 | 0.14 | -0.51 | 0.05 |
| 1791 | OST60 Pathway | 60 | -0.20 | 0.19 | -0.23 | 0.16 |
| 1790 | OST40 Pathway | 40 | -0.12 | 0.29 | -0.45 | 0.10 |
| 1789 | OST21 Pathway | 21 | 0.29 | 0.10 | -0.42 | 0.03 |
| 1788 | OST16 Pathway | 16 | 0.08 | 0.41 | 0.25 | 0.21 |
| 1787 | OST12 Pathway | 12 | 0.14 | 0.31 | 0.33 | <0.001 |
| 1794 | OSTroot Pathway | 18 | -0.37 | 0.04 | -0.86 | <0.001 |
| 1793 | OSTinhib Pathway | 31 | -0.10 | 0.24 | -0.14 | 0.31 |
|
| ||||||
| CPF M | CPF M | Mix B | Mix B | |||
|
| ||||||
| 1792 | OST110 Pathway | 110 | -0.12 | 0.20 | -0.63 | <0.001 |
| 1791 | OST60 Pathway | 60 | -0.09 | 0.34 | -0.37 | 0.05 |
| 1790 | OST40 Pathway | 40 | -0.01 | 0.44 | -0.67 | <0.001 |
| 1789 | OST21 Pathway | 21 | 0.02 | 0.47 | -0.47 | <0.001 |
| 1788 | OST16 Pathway | 16 | -0.05 | 0.41 | 0.03 | 0.45 |
| 1787 | OST12 Pathway | 12 | -0.13 | 0.32 | -0.16 | 0.29 |
| 1794 | OSTroot Pathway | 18 | -0.10 | 0.20 | -1.05 | <0.001 |
| 1793 | OSTinhib Pathway | 31 | 0.01 | 0.45 | 0.17 | 0.40 |
|
| ||||||
| CPF H | CPF H | Mix C | Mix C | |||
|
| ||||||
| 1792 | OST110 Pathway | 110 | -0.14 | 0.19 | -0.70 | <0.001 |
| 1791 | OST60 Pathway | 60 | -0.04 | 0.35 | -0.51 | <0.001 |
| 1790 | OST40 Pathway | 40 | -0.05 | 0.40 | -0.87 | <0.001 |
| 1789 | OST21 Pathway | 21 | -0.46 | <0.001 | -0.68 | <0.001 |
| 1788 | OST16 Pathway | 16 | -0.25 | 0.26 | -0.03 | 0.44 |
| 1787 | OST12 Pathway | 12 | -0.33 | 0.05 | -0.08 | 0.40 |
| 1794 | OSTroot Pathway | 18 | -0.14 | 0.36 | -1.55 | <0.001 |
| 1793 | OSTinhib Pathway | 31 | -0.06 | 0.44 | 0.15 | 0.31 |
Quantitative PCR studies and analysis of olfactory biomarkers
Several genes identified as potential markers of olfactory cellular injury and functional impairment were investigated by qPCR. Overall, the more sensitive qPCR technique confirmed the microarray response detecting even greater transcriptional changes than found by array, which has been reported (Morey et al., 2006). The expression of gefiltin 3 (gef), a neuronal intermediate filament protein important in neuronal regeneration in olfactory epithelial tissue (Niloff et al., 1998), was significantly repressed by CPF (the opposite response from the Cu-induced transcriptional induction reported previously [22]). As observed in figure 5A, exposure to CPF appeared to drive the transcriptional response of gef in the mixtures. Another marker under investigation, Neuropeptide FF-amide peptide precursor-like mRNA (npffl), encodes a protein belonging to a group of neuromodulatory peptides (Panula et al., 2006). Exposure to the lowest CPF concentration or to the mixtures induced npffl, consistent with the response to Cu alone (Tilton et al., 2008) (Figure 5A). Another gene purported to be highly expressed in the OB, ferritin (Connor et al., 1990) showed little response to CPF, but was induced on exposure to Cu alone, and also by the Cu/CPF mixtures (Figure 4B). The pvalb8 protein is important in maintaining calcium ion balance in neuronal cells (Mukhopadhyay et al., 2009). As observed in figure 5B, expression of the pvalb8 transcript was impacted by CPF, but was much more repressed in the presence of Cu. Interestingly, the transcriptional response to Cu alone was nearly identical to the response of the mixtures for both ferritin and pvalb8 (Figure 5B). Furthermore, the modest depression (<2-fold decrease) of olfactory receptor 2.1 on exposure to CPF as evidenced by qPCR was substantially different than the significant repression (> 10-fold) of this gene observed by the mixture exposures (Figure 5C). Transcripts of calbindin 2-like(calb2), a calcium binding protein in the olfactory epithelium, were not affected by CPF exposure. In contrast, the mixtures (or Cu alone) decreased calb2l mRNA levels significantly (> 5-fold) relative to controls (Figure 5C). Two other markers, olfactory marker protein(omp) and s100 encode proteins important in the cellular function of sensory cells (Bastianelli et al., 1995). Exposure to CPF decreased the expression of these genes, and this inhibitory transcriptional effect was increased in the mixture exposures (Figure 5D). Collectively, our qPCR analyses yielded results that were consistent with those from the microarrays and reinforced a role for Cu as the primary driver for the transcriptional changes in the zebrafish olfactory system.
Figure 5. Quantitative real-time PCR confirmation of the microarray data.
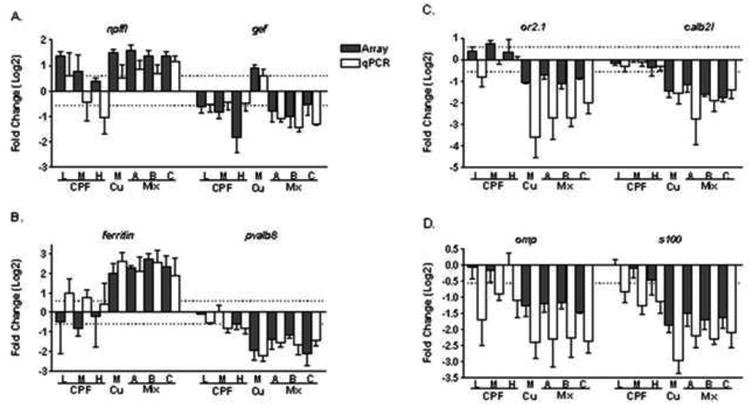
Values are expressed as average fold change (Log2) with standard error compared to controls. The array response (shaded bars) for each treatment is grouped with the qPCR results (white bars) and the two methods compare favorably in both the direction and magnitude of the response. A) neuropeptide FF-amide peptide precursor-like (npffl) and neuronal protein gefiltin (gef); B) parvalbumin 8 (pvalb8) and a putative ferritin heavy chain (ferritin); C) calbindin 2, like (calb2l) and olfactory receptor 2.1 (or2.1); D) S100 protein (s100) and olfactory marker protein (omp). Dashed line across all data illustrates 1.5-fold change from controls.
Discussion
In the present study, we have shown that exposure to the pesticide olfactory toxicant CPF alters the expression of a diverse number of transcripts in the olfactory system of zebrafish, and have identified important gene pathways preferentially targeted by CPF exposure. We previously reported that exposure to Cu, another common environmental pollutant and olfactory toxicant, depresses the transcription of key gene elements comprising the zebrafish olfactory signal transduction pathway consistent with inhibitory effects of Cu on the responsiveness of olfactory receptor neurons to natural odorants. The fact that exposure to binary mixtures of the two agents resulted in olfactory transcriptional signatures that were strongly influenced by Cu, regardless of its molar ratio with CPF, has implications for exposures of chemical classes comprising complex mixtures and identifying those chemicals responsible for functional chemosensory deficits. Specifically, the fact that the presence of Cu caused the CPF response to shift towards the transcriptional profile produced by exposure to Cu alone suggests that it may be the major driving force in olfactory transcriptional responses in the zebrafish. We would be interested if similar results are observed with mixtures of other metal and organophosphate neurotoxicants.
The use of two complementary bioinformatics approaches (i.e. TopGO analysis and GSA) revealed molecular interactions among biochemical pathways impacted by exposure to our model neurotoxicants. The observed downregulation of gene sets related to morphogenesis pathways, cell growth and replication, and neuronal processes with increasing CPF concentrations by GSA suggests a CPF-induced loss in the regenerative capacity of the olfactory system. This may include, for example, the replacement of receptor neurons in the peripheral sensory epithelium. In this regard, the olfactory bulb and telencephalon also contain a host of neuronal cell types that undergo regeneration. This process occurs as a result of toxicant exposure, but also under homeostatic physiological conditions as part of normal maintenance and development (Byrd and Brunjes, 2001; Grandel et al., 2006). The fact that exposure to CPF alone reduces the expression of genes encoding specific neuronal proteins such as gefiltin, which is linked to neuronal regeneration, neurogenesis, and axonal outgrowth, is consistent with a previous study showing that CPF inhibits axonal outgrowth in rat sympathetic neurons and sensory neurons (Yang et al., 2008). Conversely, the fact that exposure to Cu alone induced gefiltin mRNA expression is illustrative of how the two toxicants may have distinct and even opposite effects on regeneration in the olfactory system, and indicate that multiple processes, both shared and unique, are impaired by these two very different olfactory toxicants. Our results are also supportive of a role of monitoring gefiltin mRNA expression as a potential biomarker of nerve regeneration following injury. Furthermore, while the doses of the two agents used in our study were likely not cytotoxic, the high dose of Cu employed approximated concentrations shown to elicit cell death in the olfactory epithelium of fish (Julliard et al., 1993). Accordingly, the observed Cu-induced positive modulation of gefiltin mRNA expression may have been associated with an active regeneration of olfactory sensory neurons resulting from a toxic effect under higher Cu concentrations.
A more detailed analysis of gene pathways underlying the functional properties of sensory neurons was accomplished by monitoring a custom collection of conserved gene sets belonging to the OST cascade. We specifically focused on the expression of genes that code for proteins essential for odorant binding or the electrochemical cascades that underlie olfactory signal transduction (Tilton et al., 2008). These pathways are strongly impacted on exposure to Cu and appear to be an important mechanism by which Cu interferes with fish behaviors that depend on a normally functioning olfactory system (Tilton et al., 2008). Although we did not detect significant net impacts or shifts in gene expression following exposure to CPF, it is important to note that we did observe a differential expression of some genes within the OST pathway. This suggests that olfactory impairment by the organophosphate may involve effects on signal transduction, which would be consistent with the previous finding that CPF inhibits olfactory responses to chemical cues (e.g., bile salts and amino acids (Sandahl et al., 2004)) that activate non-overlapping olfactory receptor proteins in fish. The fact that OST genes impacted by CPF are in some cases different than those impacted by Cu, and in many cases are positively regulated (as opposed to downregulated), is supportive of different mechanisms of olfactory injury by these two classes of chemicals.
We used qPCR to assess the effect of CPF on the expression of several genes that code for cellular markers in the OB and olfactory epithelium, including known components of the olfactory signal transduction cascade. Overall, CPF exposure elicited relatively few significant changes in expression of OB genes elicited by exposure to Cu, including putative ferritin heavy chain or pvalb8 protein. However, there was a negative trend similar to that observed by the mixture exposures. Olfactory receptors and the calcium binding protein calbin2-like were unresponsive to CPF despite being strongly repressed in the mixtures. Interestingly, other receptors found on the array such as OR 5.1, showed decreases with CPF exposure (Supplemental table 12). The genes encoding olfactory marker protein, a marker of mature olfactory neurons important in modulation of cAMP levels in OST signaling (Bastianelli et al., 1995; Reisert et al., 2007), as well as s100 protein (Yamagishi et al., 1994), a marker for mature olfactory crypt cells, showed little change in the array experiment. Although the fact that relatively high environmental concentrations of CPF caused little modulation of zebrafish olfactory signal transduction gene pathways, it will be important to determine the functional significance of CPF on zebrafish olfactory signal transduction.
Our microarray results for the CPF/Cu mixtures provide some insight into how toxic mechanisms can shift when contaminants co-occur in mixtures. The effects of CPF/Cu mixtures on gene expression were closely correlated with the effects of Cu alone and, to a lesser extent, to CPF, indicating a dominating effect by Cu. Furthermore, there were many interesting gene targets shared between both Cu and CPF, including those in gene sets related to ion transport, ion binding, and channel activities. These functional groups share many of the same gene sets, as well as a similar direction of the relative shift in transcription from controls. Metals such as zinc and Cu are known to interact with voltage- and ligand-gated ion channels, including calcium channels (Horning and Trombley, 2001). Altered gene sets related to the binding of different cations including calcium in the mixture treatments point to alterations in ion balance and membrane potentials as potential targets for both of these classes of contaminants. There is recent evidence to suggest that cytotoxicity from organophosphates observed in primary neuronal cultures involves disturbances in intracellular homeostasis of calcium (Giordano et al., 2007). Collectively, the evidence presented herein is supportive of the hypothesis that metals and organophosphates likely target similar processes such as calcium ion balance and specifically ion binding and channels. Additional studies addressing the impacts of Cu and CPF on neuronal ion balance in sensory neurons of the olfactory system could more closely define and differentiate molecular impacts from these two chemical classes.
In conclusion, CPF exposure to zebrafish impairs cellular pathways and gene sets related to morphogenesis, growth and other neuronal processes in the olfactory system. Therefore, this organophosphate insecticide may modulate multiple biochemical pathways that underlie the normal electrochemical transduction of olfactory sensory information as well as genes required for neuronal maintenance and regeneration. Although exposure to Cu and CPF both affect olfactory function and depress olfactory mediated behaviors, these compounds appear to act via different mechanisms. Moreover, relative to CPF, the effects of Cu on gene transcription in olfactory networks are much more pronounced. Exposure to mixtures of the two toxicants produced a unique transcriptional signature that is heavily influenced by Cu. Collectively, the results of the study provide a basis for understanding mixture effects on olfactory toxicity and also mechanisms of action of neurotoxic metals and pesticides. Future studies will investigate the effects of Cu, CPF and a mixture on olfactory mediated behavior and olfactory injury.
Supplementary Material
Acknowledgments
Funding information. This work was supported in part by NIEHS Superfund Research program (P42-ES04696), and also by the University of Washington NIEHS Center for Ecogenetics and Environmental Health (P30-ES07033). SCT was the recipient of an NIEHS postdoctoral fellowship in Environmental Pathology and Toxicology (T32-ES07032).
Footnotes
Supporting Information Available. Supplemental Tables EST.xl. contains 12 Supplemental Tables, including; Table S1 showing GSA olfactory signal transduction (OST) pathways, Table S2 with sequences of PCR primers, Table S3 indicating chemical sampling measurements, Table S4 showing differentially expressed genes in at least one CPF treatment, Table S5 showing differentially expressed genes in at least one CPF/Cu mixture treatment, Tables S6 and S7 with GSA results of the CPF treatments for the Biological Process and Molecular Function Gene Ontology categories, Table S8 and S9 showing GSA results of the chlorpyrifos/Cu treatments for the Biological Process and Molecular Function Gene Ontology categories, Tables S10 and S11 showing TopGO analysis results for CPF and mixture exposures, and Table S12 showing average -fold change gene lists for genes in the OST pathways.
References
- Alexa A, Rahnenfuhrer J, Lengauer T. Improved scoring of functional groups from gene expression data by decorrelating GO graph structure. Bioinformatics. 2006;22:1600–1607. doi: 10.1093/bioinformatics/btl140. [DOI] [PubMed] [Google Scholar]
- Baldwin DH, Sandahl JF, Labenia JS, Scholz NL. Sublethal effects of copper on coho salmon: impacts on nonoverlapping receptor pathways in the peripheral olfactory nervous system. Environ Toxicol Chem. 2003;22:2266–2274. doi: 10.1897/02-428. [DOI] [PubMed] [Google Scholar]
- Bastianelli E, Polans AS, Hidaka H, Pochet R. Differential distribution of six calcium-binding proteins in the rat olfactory epithelium during postnatal development and adulthood. J Comp Neurol. 1995;354:395–409. doi: 10.1002/cne.903540308. [DOI] [PubMed] [Google Scholar]
- Byrd CA, Brunjes PC. Neurogenesis in the olfactory bulb of adult zebrafish. Neuroscience. 2001;105:793–801. doi: 10.1016/s0306-4522(01)00215-9. [DOI] [PubMed] [Google Scholar]
- Blechinger SR, Kusch RC, Haugo K, Matz C, Chivers DP, Krone PH. Brief embryonic cadmium exposure induces a stress response and cell death in the developing olfactory system followed by long-term olfactory deficits in juvenile zebrafish. Toxicol Appl Pharmacol. 2007;224:72–80. doi: 10.1016/j.taap.2007.06.025. [DOI] [PubMed] [Google Scholar]
- Camon E, Barrell D, Brooksbank C, Magrane M, Apweiler R. The Gene Ontology Annotation (GOA) Project-Application of GO in SWISS-PROT, TrEMBL and InterPro. Comp Funct Genomics. 2003;4:71–74. doi: 10.1002/cfg.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen YH, Huang FL, Cheng YC, Wu CJ, Yang CN, Tsay HJ. Knockdown of zebrafish Nav1.6 sodium channel impairs embryonic locomotor activities. J Biomed Sci. 2008;15:69–78. doi: 10.1007/s11373-007-9200-4. [DOI] [PubMed] [Google Scholar]
- Connor JR, Menzies SL, St Martin SM, Mufson EJ. Cellular distribution of transferrin, ferritin, and iron in normal and aged human brains. J Neurosci Res. 1990;27:595–611. doi: 10.1002/jnr.490270421. [DOI] [PubMed] [Google Scholar]
- Deamer NJ, Genter MB. Olfactory toxicity of diethyldithiocarbamate (DDTC) and disulfiram and the protective effect of DDTC against the olfactory toxicity of dichlobenil. Chem Biol Interact. 1995;95:215–226. doi: 10.1016/0009-2797(94)03561-l. [DOI] [PubMed] [Google Scholar]
- Deamer NJ, O'Callaghan JP, Genter MB. Olfactory toxicity resulting from dermal application of 2,6-dichlorobenzonitrile (dichlobenil) in the C57Bl mouse. Neurotoxicology. 1994;15:287–293. [PubMed] [Google Scholar]
- Eddins D, Cerutti D, Williams P, Linney E, Levin ED. Zebrafish provide a sensitive model of persisting neurobehavioral effects of developmental chlorpyrifos exposure: c deficits. Neurotoxicol Teratol. 2010;32:99–108. doi: 10.1016/j.ntt.2009.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia SJ, Seidler FJ, Qiao D, Slotkin TA. Chlorpyrifos targets developing glia: effects on glial fibrillary acidic protein. Brain Res Dev Brain Res. 2002;133:151–161. doi: 10.1016/s0165-3806(02)00283-3. [DOI] [PubMed] [Google Scholar]
- Gentleman RC, Carey VJ, Bates DM, Bolstad B, Dettling M, Dudoit S, Ellis B, Gautier L, Ge Y, Gentry J, Hornik K, Hothorn T, Huber W, Iacus S, Irizarry R, Leisch F, Li C, Maechler M, Rossini AJ, Sawitzki G, Smith C, Smyth G, Tierney L, Yang JY, Zhang J. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol. 2004;5:R80. doi: 10.1186/gb-2004-5-10-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilliom RJ. Pesticides in U.S. streams and groundwater. Environ Sci Technol. 2007;41:3408–3414. doi: 10.1021/es072531u. [DOI] [PubMed] [Google Scholar]
- Giordano G, Afsharinejad Z, Guizzetti M, Vitalone A, Kavanagh TJ, Costa LG. Organophosphorus insecticides chlorpyrifos and diazinon and oxidative stress in neuronal cells in a genetic model of glutathione deficiency. Toxicol Appl Pharmacol. 2007;219:181–189. doi: 10.1016/j.taap.2006.09.016. [DOI] [PubMed] [Google Scholar]
- Grandel H, Kaslin J, Ganz J, Wenzel I, Brand M. Neural stem cells and neurogenesis in the adult zebrafish brain: origin, proliferation dynamics, migration and cell fate. Dev Biol. 2006;295:263–277. doi: 10.1016/j.ydbio.2006.03.040. [DOI] [PubMed] [Google Scholar]
- Horning MS, Trombley PQ. Zinc and copper influence excitability of rat olfactory bulb neurons by multiple mechanisms. J Neurophysiol. 2001;86:1652–1660. doi: 10.1152/jn.2001.86.4.1652. [DOI] [PubMed] [Google Scholar]
- Howard AS, Bucelli R, Jett DA, Bruun D, Yang D, Lein PJ. Chlorpyrifos exerts opposing effects on axonal and dendritic growth in primary neuronal cultures. Toxicol Appl Pharmacol. 2005;207:112–124. doi: 10.1016/j.taap.2004.12.008. [DOI] [PubMed] [Google Scholar]
- Julliard AK, Saucier D, Astic L. Effects of chronic low-level copper exposure on ultrastructure of the olfactory system in rainbow trout (Oncorhynchus mykiss) Histol Histopathol. 1993;8:655–672. [PubMed] [Google Scholar]
- Laberge F, Hara TJ. Neurobiology of fish olfaction: a review. Brain Res Brain Res Rev. 2001;36:46–59. doi: 10.1016/s0165-0173(01)00064-9. [DOI] [PubMed] [Google Scholar]
- Lurling M, Scheffer M. Info-disruption: pollution and the transfer of chemical information between organisms. Trends Ecol Evol. 2007;22:374–379. doi: 10.1016/j.tree.2007.04.002. [DOI] [PubMed] [Google Scholar]
- McIntyre JK, Baldwin DH, Meador JP, Scholz NL. Chemosensory Deprivation in Juvenile Coho Salmon Exposed to Dissolved Copper under Varying Water Chemistry Conditions. Environ Sci Technol. 2008;42:1352–1358. doi: 10.1021/es071603e. [DOI] [PubMed] [Google Scholar]
- Morey JS, Ryan JC, Van Dolah FM. Microarray validation: factors influencing correlation between oligonucleotide microarrays and real-time PCR. Biol Proced Online. 2006;8:175–193. doi: 10.1251/bpo126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukhopadhyay A, McGuire T, Peng CY, Kessler JA. Differential effects of BMP signaling on parvalbumin and somatostatin interneuron differentiation. Development. 2009;136:2633–2642. doi: 10.1242/dev.034439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niloff MS, Dunn RJ, Levine RL. The levels of retinal mRNA for gefiltin, a neuronal intermediate filament protein, are regulated by the tectum during optic fiber regeneration in the goldfish. Brain Res Mol Brain Res. 1998;61:78–89. doi: 10.1016/s0169-328x(98)00204-6. [DOI] [PubMed] [Google Scholar]
- Panula P, Sallinen V, Sundvik M, Kolehmainen J, Torkko V, Tiittula A, Moshnyakov M, Podlasz P. Modulatory neurotransmitter systems and behavior: towards zebrafish models of neurodegenerative diseases. Zebrafish. 2006;3:235–247. doi: 10.1089/zeb.2006.3.235. [DOI] [PubMed] [Google Scholar]
- Reisert J, Yau KW, Margolis FL. Olfactory marker protein modulates the cAMP kinetics of the odour-induced response in cilia of mouse olfactory receptor neurons. J Physiol. 2007;585:731–740. doi: 10.1113/jphysiol.2007.142471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roegge CS, Timofeeva OA, Seidler FJ, Slotkin TA, Levin ED. Developmental diazinon neurotoxicity in rats: later effects on emotional response. Brain Res Bull. 2008;75:166–172. doi: 10.1016/j.brainresbull.2007.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rydzewski B, Sulkowski W, Miarzynska M. Olfactory disorders induced by cadmium exposure: a clinical study. Int J Occup Med Environ Health. 1998;11:235–245. [PubMed] [Google Scholar]
- Sandahl JF, Baldwin DH, Jenkins JJ, Scholz NL. Odor-evoked field potentials as indicators of sublethal neurotoxicity in juvenile coho salmon exposed to copper, chlorpyrifos, or esfenvalerate. Canadian Journal of Fisheries and Aquatic Science. 2004;61:404–413. [Google Scholar]
- Sandahl JF, Baldwin DH, Jenkins JJ, Scholz NL. Comparative thresholds for acetylcholinesterase inhibition and behavioral impairment in coho salmon exposed to chlorpyrifos. Environ Toxicol Chem. 2005;24:136–145. doi: 10.1897/04-195r.1. [DOI] [PubMed] [Google Scholar]
- Sato Y, Miyasaka N, Yoshihara Y. Hierarchical regulation of odorant receptor gene choice and subsequent axonal projection of olfactory sensory neurons in zebrafish. J Neurosci. 2007;27:1606–1615. doi: 10.1523/JNEUROSCI.4218-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selderslaghs IW, Hooyberghs J, De Coen W, Witters HE. Locomotor activity in zebrafish embryos: a new method to assess developmental neurotoxicity. Neurotoxicol Teratol. 2010;32:460–471. doi: 10.1016/j.ntt.2010.03.002. [DOI] [PubMed] [Google Scholar]
- Slotnick B, Sanguino A, Husband S, Marquino G, Silberberg A. Olfaction and olfactory epithelium in mice treated with zinc gluconate. Laryngoscope. 2007;117:743–749. doi: 10.1097/MLG.0b013e318033006b. [DOI] [PubMed] [Google Scholar]
- Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, Mesirov JP. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabor R, Yaksi E, Weislogel JM, Friedrich RW. Processing of odor mixtures in the zebrafish olfactory bulb. J Neurosci. 2004;24:6611–6620. doi: 10.1523/JNEUROSCI.1834-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tierney K, Casselman M, Takeda S, Farrell T, Kennedy C. The relationship between cholinesterase inhibition and two types of swimming performance in chlorpyrifos-exposed coho salmon (Oncorhynchus kisutch) Environ Toxicol Chem. 2007a;26:998–1004. doi: 10.1897/06-459r.1. [DOI] [PubMed] [Google Scholar]
- Tierney KB, Baldwin DH, Hara TJ, Ross PS, Scholz NL, Kennedy CJ. Olfactory toxicity in fishes. Aquatic toxicology. 2010 doi: 10.1016/j.aquatox.2009.09.019. In press. [DOI] [PubMed] [Google Scholar]
- Tierney KB, Ross PS, Kennedy CJ. Linuron and carbaryl differentially impair baseline amino acid and bile salt olfactory responses in three salmonids. Toxicology. 2007b;231:175–187. doi: 10.1016/j.tox.2006.12.001. [DOI] [PubMed] [Google Scholar]
- Tierney KB, Sampson JL, Ross PS, Sekela MA, Kennedy CJ. Salmon olfaction is impaired by an environmentally realistic pesticide mixture. Environ Sci Technol. 2008;42:4996–5001. doi: 10.1021/es800240u. [DOI] [PubMed] [Google Scholar]
- Tilton F, Tilton S, Bammler T, Beyer R, Farin F, Stapleton PL, Gallagher EP. Transcriptional biomarkers and mechanisms of copper-induced olfactory injury in zebrafish. Environmental Science and Technology. 2008;42:9404–9411. doi: 10.1021/es801636v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trombley PQ, Shepherd GM. Differential modulation by zinc and copper of amino acid receptors from rat olfactory bulb neurons. J Neurophysiol. 1996;76:2536–2546. doi: 10.1152/jn.1996.76.4.2536. [DOI] [PubMed] [Google Scholar]
- Tusher VG, Tibshirani R, Chu G. Significance analysis of microarrays applied to the ionizing radiation response. Proc Natl Acad Sci U S A. 2001;98:5116–5121. doi: 10.1073/pnas.091062498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Denderen JC, van Wieringen GW, Hillen B, Bleys RL. Zinc sulphate-induced anosmia decreases the nerve fibre density in the anterior cerebral artery of the rat. Auton Neurosci. 2001;94:102–108. doi: 10.1016/S1566-0702(01)00354-X. [DOI] [PubMed] [Google Scholar]
- Yamagishi M, Fujiwara M, Nakamura H. Olfactory mucosal findings and clinical course in patients with olfactory disorders following upper respiratory viral infection. Rhinology. 1994;32:113–118. [PubMed] [Google Scholar]
- Yang D, Howard A, Bruun D, Ajua-Alemanj M, Pickart C, Lein PJ. Chlorpyrifos and chlorpyrifos-oxon inhibit axonal growth by interfering with the morphogenic activity of acetylcholinesterase. Toxicol Appl Pharmacol. 2008;228:32–41. doi: 10.1016/j.taap.2007.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu IT, Lee NL, Zhang XH, Chen WQ, Lam YT, Wong TW. Occupational exposure to mixtures of organic solvents increases the risk of neurological symptoms among printing workers in Hong Kong. J Occup Environ Med. 2004;46:323–330. doi: 10.1097/01.jom.0000121367.69269.07. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


