Abstract
The broad-complex tramtrack and bric a brac-zinc finger transcriptional regulator(BTB-ZF), promyelocytic leukemia zinc finger (PLZF), was recently shown to control the development of the characteristic innate T cell phenotype and effector functions of NK T cells. Interestingly, the ectopic expression of PLZF was shown to push conventional T cells into an activated state that seems to be proinflammatory. The factors that control the normal expression of PLZF in lymphocytes are unknown. In this study, we show that PLZF expression is not restricted to NK T cells but is also expressed by a subset of γδ T cells, functionally defining distinct subsets of this innate T cell population. A second BTB-ZF gene, ThPOK, is important for the phenotype of the PLZF-expressing γδ T cells. Most importantly, TCR signal strength and expression of inhibitor of differentiation gene 3 control the frequency of PLZF-expressing γδ T cells. This study defines the factors that control the propensity of the immune system to produce potentially disease-causing T cell subsets.
Multipotent progenitor T cells migrate from the bone marrow to the thymus and give rise to mature T cells that express an αβ or γδ TCR. Despite their emergence from a common T cell progenitor, γδ T cells are fundamentally distinct from conventional (αβ) T cells. αβ and γδ T cells transit through a multistage, differentiation program in the thymus. αβ TCR-expressing thymocytes go through a CD4+CD8+ double-positive stage prior to maturing to CD4 single-positive or CD8 single-positive T cells. In contrast, studies showed that γδ T cells emerge directly from double-negative cells, and the mature cells generally do not express CD4 or CD8 coreceptors (1, 2). αβ T cell development requires interactions between the TCR and self-peptide:MHC, primarily on the surface of stromal cells, although some hematopoietic cells can also support T cell development (3). In contrast, γδ T cells do not seem to require positive selection for full maturation, except for skin-resident dendritic epidermal T cells (4, 5).
In mice, γδ T cells appear in “waves” at defined stages throughout fetal and neonatal development. Each wave of development produces cells that migrate to specific tissues in a process that likely depends on explicit signals that shape the genetic program of these cells (6–8). It is clear that γδ T cells that express different TCRs are enriched in certain tissues and that these cells have specific roles, ranging from immunosurveillance to tissue homeostasis (8–10). Several studies showed that γδ T cells are early responders in various models of infectious disease and carry out important roles in auto-immunity and tumor surveillance, although they constitute a relatively minor population (1–5%) of total lymphocytes (9, 11–13).
Unlike naive conventional T cells, γδ T cells typically exhibit an “activated” phenotype and rapidly respond to foreign Ags (14–17). Upon activation, γδ T cells largely produce IFN-γ, which modulates innate- and adaptive-specific immune responses during infection (8, 18, 19). Recent reports showed that γδ T cells are also an important source of IL-17, another proinflammatory cytokine that is important for the recruitment of neutrophils to areas of inflammation (20–22). Notably, some γδ T cells also secrete Th2 cytokines in vitro and in vivo. Originally identified as Thy1.1dull γδ T cells, studies revealed that these IL-4–producing γδ T cells express a Vγ1.1+Vδ6.3+ TCR (16, 23). This unique subset of γδ T cells is CD44hiCD69+ CD62Llo, and nearly half express NK1.1 and CD4. Thus, based on several phenotypic and functional characteristics, Vγ1.1+Vδ6.3+ (γδ) T cells, like invariant NK T cells (NKT cells), are categorized as “innate-like” T cells and are believed to operate at the interface of the innate and adaptive immune response.
A central aim in understanding the role of γδ T cells during the immune response is identifying the molecular signals that govern their differentiation and function. Recently, we and other investigators showed that promyelocytic leukemia zinc finger (PLZF; encoded by Zbtb16) is necessary for the development of NKT cell effector functions (24, 25). PLZF belongs to the broad-complex and tramtrack bric-à-brac (BTB)-poxvirus-zinc finger (ZF) family of transcription factors, which have essential roles in various biological processes, including germ cell maintenance, specificity of neuromuscular connections, and axial limb development (26–29). A number of BTB-ZF proteins have been identified that control fundamental aspects of immune cell development. For example, LRF directs B-versus-T cell fate, whereas ThPOK is a critical factor for CD4 lineage commitment (30–34). Given the innate-like T cell features shared by γδ and NKT cells, we investigated whether PLZF plays a role in the differentiation and function of γδ T cells. We found that a specific subset of γδ T cells, defined by expression of the Vγ1.1+Vδ6.3+ TCR, expressed the innate T cell determinant PLZF and that PLZF was required for the function of these cells. Finally, recent studies indicated that strong TCR signaling is required for γδ T cell development (35, 36). Paradoxically, reduced TCR signal strength results in a dramatic expansion in PLZF-expressing Vγ1.1+ Vδ6.3+ T cells. Inhibitor of differentiation gene 3 (Id3), a molecular target of TCR signaling pathways, controls the development of PLZF-expressing Vγ1.1+Vδ6.3+ T cells. Our data support a model in which reduced TCR signaling strength alters Id3 activity and directs cells into the PLZF-expressing Vγ1.1+Vδ6.3+ T lineage.
Materials and Methods
Mice
PLZF-deficient mice have been described (24, 29). Fyn-, TCRβ-, and CD1d-deficient mice were purchased from The Jackson Laboratory (Bar Harbor, ME). ThPOKGFP/+ and ThPOKGFP/− mice (32) were housed at the Skirball Institute, New York University. Id3-deficient mice (37) were a kind gift from Dr. Robert Benezra (Memorial Sloan-Kettering Cancer Center [MSKCC]). Age-matched C57BL/6 mice (The Jackson Laboratory) and littermates from PLZF heterozygous matings were used as wild-type (WT) controls (4–8 wk). All animal work was done in compliance with MSKCC’s Internal Animal Care and Use Committee and the guidelines of the Federal Office of Laboratory Animal Welfare. Animals were housed in the Research Animal Resources Center of MSKCC. Animal housing rooms were under temperature and humidity control, the mice were not subjected to water or food restrictions, and bedding material was placed in each cage. Four full-time veterinarians and six veterinarian technicians staff the facility. The veterinary staff is located on site, and a clinical veterinarian is available at all times. Animal care staff carried out routine husbandry procedures, including changing cages, feeding, and watering.
All mice were sacrificed prior to use. Euthanasia was conducted in accordance with the American Veterinary Medical Association Guidelines on Euthanasia. Briefly, mice were sacrificed by asphyxiation with CO2 delivered into the cages at <5 pounds per square inch per second. Death of the animal was confirmed by lack of respiration and toe pinch. CO2 euthanasia stations are inspected regularly by the Internal Animal Care and Use Committee personnel. Tissues were removed for experiments after confirmation of death.
Flow cytometry and cell sorting
Single-cell suspensions were incubated with normal mouse serum, unlabeled streptavidin, and Fc receptor-blocking Ab before being stained at 4° C with specific Abs. Intracellular staining with anti-PLZF (24), cells were made permeable with an intracellular staining buffer set (eBioscience, San Diego, CA). In nonpermeabilized cell preparations, dead cells were excluded by incubating with the DNA-intercalating dye DAPI. Cell doublets were excluded by comparing side-scatter width with forward scatter area. Intestinal intraepithelial lymphocytes (iIELs) were isolated as previously described (38), with modifications. In brief, after removal of feces and Peyer’s patches, intestines were cut longitudinally, washed twice in RPMI 1640 containing 25 mM HEPES, cut into 2-mm pieces, and incubated in Ca2+- and Mg2+-free Hank’s balanced salt solution in 15 mM HEPES, 10% FCS, and 5 mM EDTA for 40 min at 37°C, with gentle manual inversion. Samples were vortexed for 15 s, and supernatants were collected through 100-μm cell strainers. iIELs were enriched using 44%:66% Percoll density gradient centrifugation. Samples were collected on an LSR II flow cytometer (BD Biosciences, San Jose, CA), and data were analyzed using FloJo software (Tree Star, Ashland, OR). Cell sorting was done on a FACSAria flow cytometer (BD Biosciences) by MSKCC’s Flow Cytometry Core Facility. mAbs anti-Vδ6.3 TCR (8F4H7B7), anti-Vγ4 (UC3-10AG), anti-Vδ4 (GL2), anti-CD3 (145-2C11), anti-CD4 (GK1.5, RM4.5), anti-CD69 (H1.2F3), anti-NK1.1 (PK136), anti-IL-4 (11B11), and streptavidin-PeCy7 were purchased from BD Biosciences; anti-γδ TCR (GL3), anti-TCRβ (H57-594), anti-CD8 (53-6.7), anti-CD122 (Tm.b1), anti-CD44 (IM7), anti-IFN-γ (XMG1.2), anti–TNF-α (MPG-XT22), and anti–IL-17A (Ebio17B7) were purchased from eBioscience; and anti-CD24 (M1/69) was purchased from (BioLegend, San Diego, CA). mAbs anti-PLZF, anti-IL4 (11B11), and anti-CD3 (500A2) were generated by MSKCC’s mAb Core Facility (24).
In vitro T cell activation
FACS-sorted γδ T cells were activated for 6 h with phorbol 12 myristate 13-acetate (100 ng/ml) and ionomycin (500 ng/ml); brefeldin A (3 μg/ml) was added for the final 5 h of the incubation. Then cells were fixed and made permeable with the intracellular staining buffer set (eBioscience), followed by intracellular staining for IFN-γ and IL-4 or TNF-α and PLZF. For cytokine-secretion assays, sorted γδ T cells were activated with plate-bound anti-CD3 (10 μg/ml) in round-bottom 96-well plates (complete media) for 72 h. Secreted cytokines were detected with cytokine bead arrays (BD Biosciences).
Statistical calculations
Statistical differences were determined with the Mann–Whitney U test, which is a parametric alternative to the Student t test that is more robust for small sample sizes for repeats of each experiment.
Results
PLZF expression is limited to certain subsets of γδ T cells
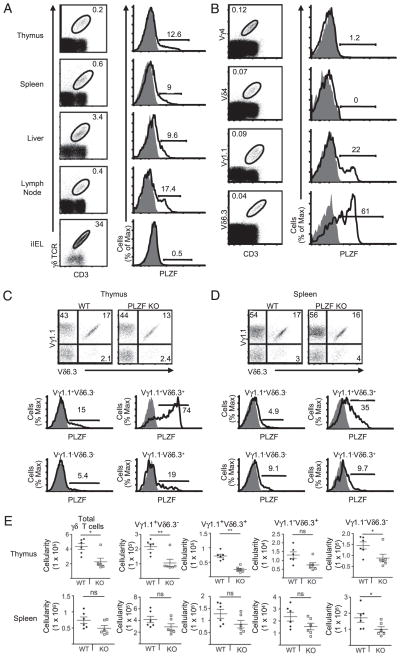
Intracellular FACS staining revealed that PLZF was expressed in 10–18% of γδ T cells from the thymus, spleen, liver, and lymph nodes. (Fig. 1A). PLZF was highly expressed by Vγ1.1 and Vδ6.3 T cells but not by Vγ4- or Vδ4-expressing T cells (Fig. 1B). Consistent with previous reports, we found that the majority of Vδ6.3 T cells were paired with Vγ1.1 (39, 40) (Fig. 1C, 1D, Supplemental Fig. 1). The bulk of PLZF expression was restricted to these Vγ1.1+Vδ6.3+ T cells (Fig. 1C), although PLZF expression was also detected in a small percentage of Vγ1.1-only and Vδ6.3-only cells and in some γδ T cells that expressed neither chain. Furthermore, PLZF was not expressed in all Vγ1.1+Vδ6.3+ T cells. In contrast to NKT cells, the percentage of Vγ1.1+Vδ6.3+ T cells was not significantly altered in the absence of PLZF (Fig. 1C). There was a decrease in the absolute numbers of γδ T cells in the thymus and spleen (Fig. 1E, Supplemental Fig. 1D). This reduction was attributable to a general decrease in the size of the thymus and spleen, as previously reported (24). PLZF expression was largely restricted to Vγ1.1+Vδ6.3+ T cells in the spleen and liver, but similar to NKT cells, the expression was reduced. (Fig. 1D, Supplemental Fig. 1A). However, Vγ1.1+Vδ6.3+ iIELs did not express PLZF (Fig. 1A, Supplemental Fig. 1B). Overall, the data suggest that there are essentially two different lineages of Vγ1.1+Vδ6.3+ γδ T cells: some that express PLZF and some that do not. This observation is consistent with the finding that Vγ1.1+Vδ6.3+ T cells developed in PLZF-deficient mice, which was in sharp contrast to the ~50-fold reduction seen for NKT cells.
FIGURE 1.
The majority of PLZF is expressed by Vγ1.1+Vδ6.3+ T cells. Intracellular staining for PLZF in γδ T cells in primary tissues, secondary tissues, and iIELs (A), in Vγ4 (FITC), Vδ4 (FITC), Vγ1.1 (Biotin), and Vδ6.3 (PE) γδ thymocytes (B), and in γδ T cell subsets bearing indicated γδ TCR heterodimers in thymus (C) and splenocytes (D) in WT (open curves) or PLZF-deficient mice (shaded curves). Numbers adjacent to outlined areas indicate the percentage of γδ T cells; bracketed lines next to graphs specify the percent total of PLZF-positive cells. γδ T cell subsets were identified with anti-CD3 PerCP-Cy5.5, anti-γδ TCR APC, anti-Vδ6.3 TCR PE, and anti-Vγ1.1 TCR biotin/streptavidin PE-Cy7. E, Absolute numbers of total γδ T cells and γδ subsets in the thymus and spleen of WT littermates (n = 6) and PLZF-deficient mice (n = 6; error bars represent the SD). Approximately 3 × 106 events were acquired for these experiments. Doublet exclusion was done as indicated in the Materials and Methods. Data are representative of more than six independent experiments. All flow cytometry plots are quantified in log10 fluorescence. Each symbol represents an individual mouse. *p = 0.05; **p < 0.05.
PLZF-negative Vγ1.1+Vδ6.3+ T cells are phenotypically and functionally distinct
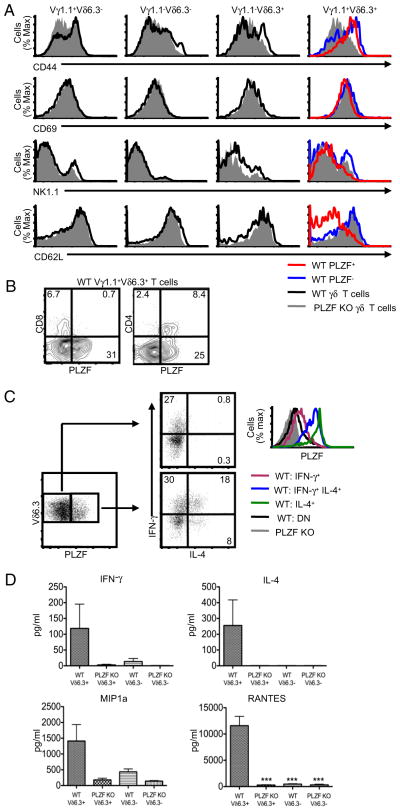
It was reported that up to half of Vδ6.3 T cells express NK1.1 (16). Consistent with this finding, ~40–50% of the Vγ1.1+Vδ6.3+ T cells in WT mice expressed NK1.1; however, we reliably found a higher proportion of NK1.1 expression among PLZF-negative Vγ1.1+Vδ6.3+ T cells compared with PLZF-positive Vγ1.1+ Vδ6.3+ T cells (Fig. 2A). PLZF-expressing Vγ1.1+Vδ6.3+ T cells had sharply reduced levels of the adhesion molecule L-selectin (CD62L) compared with WT PLZF-negative, and PLZF-deficient cells. CD44 expression was clearly reduced on PLZF-deficient Vγ1.1+Vδ6.3+ T cells, but there was only a subtle change when WT PLZF-positive and -negative Vγ1.1+Vδ6.3+ T cells were compared. The expression of NK1.1, CD69, CD122, DX5, and NKG2D was not substantially different among these cell types (Fig. 2A and data not shown). We did not observe any noteworthy change in the phenotype of the non-Vγ1.1+Vδ6.3+ γδ T cell subsets. Interestingly, PLZF-positive Vγ1.1+Vδ6.3+ thymocytes are largely CD24low compared with PLZF-negative Vγ1.1+Vδ6.3+ thymocytes (Supplemental Fig. 2). This is also consistent with NKT cells, which are nearly all CD24low in the thymus (41). Notably, nearly all CD4+ γδ T cells, but none of the CD8+ γδ T cells, expressed PLZF (Fig. 2B).
FIGURE 2.
Phenotype and function of PLZF-positive and -negative Vγ1.1+Vδ6.3+ T cells. A, Expression of CD44 (APC), CD69 (PerCP-Cy5.5), NK1.1 (PerCP-Cy5.5), and CD62L (APC-Alexa Fluor 750) by WT PLZF-positive (red), PLZF-negative (blue or black), and PLZF-deficient (gray) γδ thymocytes. B, CD4 (APC-Cy7) and CD8 (Pacific Blue) expression on WT Vγ1.1+Vδ6.3+ T cells compared with intracellular staining for PLZF. C, Intracellular staining for IFN-γ (PE-Cy7), IL-4 (APC), and PLZF (Alexa Fluor 488) in Vδ6.3 T cells following activation. Numbers indicate the percentage of cells in each quadrant. D, Cytokine analysis of supernatants of 5 × 104 FACS-sorted Vδ6.3+ and Vδ6.3− T cells from WT and PLZF-deficient splenocytes with plate-bound anti-CD3 for 3 d. Error bars represent the SD. ***p = 0.05. Data are representative of six (A and B) or five (C and D) independent experiments. γδ T cell subsets were identified with anti-CD3 PerCP-Cy5.5, anti-γδ TCR biotin, and anti-Vδ6.3 TCR PE. Doublet exclusion was done as indicated in the Materials and Methods. All flow cytometry plots are quantified in log10 fluorescence.
We found a functional disparity between PLZF-expressing and PLZF-negative Vδ6.3 T cells in WT mice (Fig. 2C). Sorted Vδ6.3 T cells from the spleens of WT mice were stimulated with PMA and ionomycin, followed by intracellular cytokine staining. More than 25% of the activated PLZF-expressing Vδ6.3 T cells produced IL-4 compared with ~1% of the PLZF-negative cells. Furthermore, similar to NKT cells (24), many PLZF-expressing γδ T cells simultaneously produced IL-4 and IFN-γ. Of particular interest, high levels of PLZF expression correlated with the ability of the γδ T cells to produce IL-4 (Fig. 2C).
Next we activated sorted Vδ6.3+ and Vδ6.3− splenocytes from WT and PLZF-deficient mice with plate-bound anti-CD3 (Fig. 2D). PLZF-deficient γδ T cells produced almost undetectable levels of IFN-γ and IL-4 compared with WT cells. Interestingly, expression of the CC chemokines MIP-1α and RANTES also was lost in the PLZF-deficient γδ T cells. These molecules are potent chemoattractants for macrophages and other immune cells in microbial and viral infections (42). Using these experimental conditions, we did not detect obvious changes in the expression of IL-1, -10, -13, and -17 or TNF-α (data not shown).
It was shown that “trans-conditioning” of γδ T cells by αβ TCR-expressing thymocytes is necessary to induce the expression of genes that regulate the differentiation and function of γδ T cells (43, 44). However, PLZF expression in Vγ1.1+Vδ6.3+ T cells is normal in TCRβ-deficient mice and, therefore, does not seem to require trans-conditioning for expression (Supplemental Fig. 3A). Finally, the expression of PLZF and the frequency of Vγ1.1+ Vδ6.3+ T cells is not significantly altered in CD1d-deficient mice, suggesting that the absence of CD1d molecules and/or NKT cells has no significant affect on the development of these cells (Supplemental Fig. 3B and data not shown). Interestingly, transgene-mediated ectopic expression of PLZF does not alter the phenotype or function of γδ T cells (Supplemental Fig. 4). It is noteworthy that endogenous levels of PLZF in WT Vγ1.1+Vδ6.3+ T cells was several fold higher compared with transgenic mice.
ThPOK expression in γδ T cells
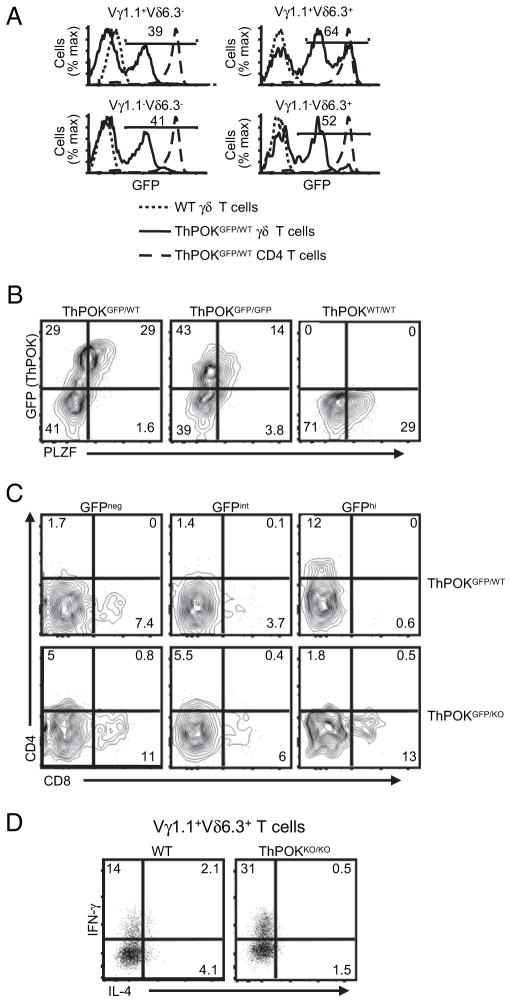
In CD4 T cells, the BTB-ZF transcription factor ThPOK is required to maintain Th effector functions (32–34). Interestingly, NKT cells, some of which express CD4, were also shown to express ThPOK (T. Egawa and E.S. Alonzo, unpublished observations). Therefore, the finding that some of the PLZF-expressing Vγ1.1+Vδ6.3+ T cells also expressed CD4 (Fig. 2B) raised the possibility that these cells also express ThPOK. Therefore, tissues from two ThPOK-reporter mice were analyzed. One line had one copy of ThPOK replaced with a cDNA encoding a GFP (ThPOKGFP/WT). The second line had the GFP allele and a null allele of ThPOK (ThPOKGFP/−); therefore, it was deficient for the expression of ThPOK (32).
GFP expression was detected in ~25% of the γδ T cells in the thymus and nearly 45% of the γδ T cells in the spleen (Supplemental Fig. 5A). Other than in Vγ1.1+Vδ6.3+ T cells, the level of ThPOK expression in γδ T cells was far lower than in αβ TCR CD4 T cells (Fig. 3A, Supplemental Fig. 5B). Staining of the ThPOK reporter mice for PLZF showed that in Vγ1.1+Vδ6.3+ T cells, all of the ThPOK high-expressing cells expressed PLZF (Fig. 3B). Interestingly, PLZF expression levels seemed to be reduced in ThPOK-deficient Vγ1.1+Vδ6.3+ T cells (Fig. 3B, right panel). The decreased PLZF levels correlate with a loss of IL-4 expression but an increase in IFN-γ expression following activation of the Vγ1.1+Vδ6.3+ T cells (Fig. 3D). However, the absence of ThPOK did not significantly alter the frequency of Vγ1.1+ Vδ6.3+ T cells (data not shown).
FIGURE 3.
Variable ThPOK expression in γδ T cell subsets. A, ThPOK-GFP reporter expression in spleen γδ T cells and CD4 T cells WT and ThPOKGFP/WT. Numbers in quadrants indicate the percentage of γδ T cells that express GFP. B, GFP (ThPOK) and PLZF expression in spleen cells from ThPOKGFP/+ and ThPOKGFP/GFP mice. C, Expression of CD4 (APC-Cy7) and CD8 (Pacific Blue) in Vγ1.1+Vδ6.3+ splenocytes from ThPOKGFP/WT and ThPOKGFP/KO reporter mice. D, INF-γ and IL-4 expression in spleen Vγ1.1+Vδ6.3+ T cells from WT or ThPOK-deficient (ThPOKko/ko) mice following activation with PMA/ionomycin. γδ T cell subsets were identified with anti-CD3 (PerCP-Cy5.5), anti-γδ TCR (APC), anti-Vδ6.3 TCR (PE), and anti-Vγ1.1 TCR (biotin/streptavidin PE-Cy7). Approximately 3 × 106 events were acquired for these experiments. Doublet exclusion was done as indicated in the Materials and Methods. B–D, Numbers indicate the percentage of cells in each quadrant. Data are representative of at least two experiments. All flow cytometry plots are quantified in log10 fluorescence.
A distinct population of CD4+ cells was found exclusively among the GFPhi Vγ1.1+Vδ6.3+ T cells from ThPOKGFP/WT mice (Fig. 3C). Interestingly, a large percentage of the GFPhi Vγ1.1+ Vδ6.3+ T cells from ThPOKGFP/KO mice failed to express CD4. In the absence of functional ThPOK, some of the GFPhi Vγ1.1+ Vδ6.3+ T cells expressed CD8 rather than CD4. Finally, the GFPint Vγ1.1+Vδ6.3+ T cells were essentially negative for the expression of CD4 or CD8 in both mouse lines.
Altered development and PLZF expression in signaling lymphocyte activation molecule-associated protein-deficient Vγ1.1-Vδ6.3 T cells
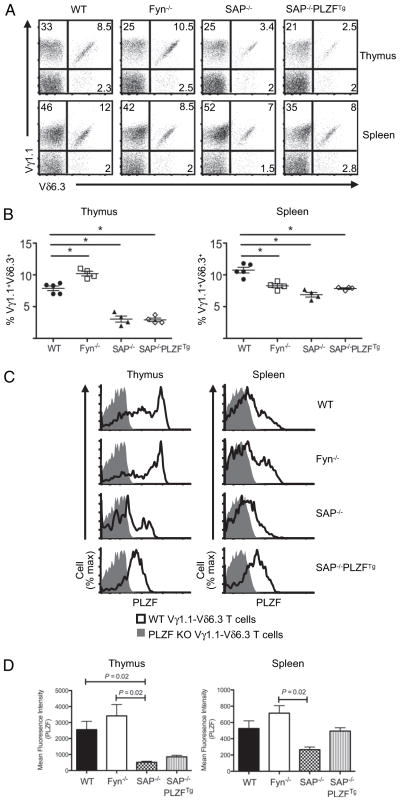
NKT cell development is severely impaired in Fyn- (45, 46) and signaling lymphocyte activation molecule (SLAM)-associated protein (SAP)-deficient (47) mice. Therefore, Fyn- and SAP-deficient mice were studied to determine whether these molecules were also necessary for the development of PLZF-expressing γδ T cells. Thymocytes and splenocytes from SAP- and Fyn-deficient mice were stained with Abs against Vγ1.1 and Vδ6.3. γδ T cell development was not overtly altered in either mouse (data not shown). The loss of Fyn correlated with an increased frequency of Vγ1.1+ Vδ6.3+ T cells (+22%) in the thymus but a decreased frequency (−23%) in the spleen (Fig. 4A, 4B). In SAP-deficient mice, there was a dramatic loss of Vγ1.1+Vδ6.3+ T cells. In the thymus and the spleen, the frequency decreased 61% and 36%, respectively, compared with age-matched WT mice (Fig. 4A, 4B). Furthermore, there was a substantial decrease in PLZF expression in the Vγ1.1+Vδ6.3+ T cells from SAP-deficient thymuses and spleens (Fig. 4C, 4D). However, PLZF expression levels in Fyn-deficient Vγ1.1+Vδ6.3+ T cells were comparable to WT levels. The transgenic expression of PLZF in SAP-deficient mice did not restore the frequency of Vγ1.1+ Vδ6.3+ T cells to WT levels (Fig. 4A, 4B). Finally, although SAP deficiency clearly impacted the development of the PLZF-expressing γδ T cells, there were no significant alterations in the expression of SLAMF1 (CD150), 2B4 (CD244), or SLAMF6 (LY108) in PLZF-negative, -positive, or -deficient Vγ1.1+Vδ6.3+ T cells (data not shown). Together, these data show that unlike for NKT cells, Fyn is largely dispensable, but SAP plays an important role in the development of Vγ1.1+Vδ6.3+ T cells.
FIGURE 4.
Vγ1.1+Vδ6.3+ T cells in Fyn- and SAP-deficient mice and SAP-deficient PLZF-transgenic mice. A, FACS analysis of indicated γδ subsets thymocytes (top) and splenocytes (bottom) in indicated mouse strains. B, Percent frequency of Vγ1.1+Vδ6.3+ T cells among thymocytes (left) and splenocytes (right) from WT (n = 5), Fyn-deficient (n = 4), SAP-deficient (n = 4), and SAP-deficient PLZF-transgenic (n = 4) mice. Each symbol represents an individual mouse. *p = 0.05. C, PLZF expression in Vγ1.1+Vδ6.3+ thymocytes (left) and splenocytes (right) in indicated mouse strains. D, Mean fluorescence intensity (W/m2) of PLZF levels in Vγ1.1+ Vδ6.3+ thymocytes: WT = 2559 ±1049; Fyn KO = 3464 ± 1407; SAP KO = 517 ± 127; SAP KO-PLZF Tg = 856 ± 171; Vγ1.1+Vδ6.3+ spleen cells: WT = 527 ±187; Fyn KO = 715 ± 186; SAP KO = 316 ± 69; SAP KO-PLZF Tg = 494 ± 72. Error bars represent the SD. γδ T cell subsets were identified with anti-CD3 (PerCP-Cy5.5), anti-γδ TCR (APC), anti-Vδ6.3 TCR (PE), and anti-Vγ1.1 TCR (biotin/streptavidin PE-Cy7). Approximately 3 × 106 events were acquired for these experiments. Doublet exclusion was done as indicated in the Materials and Methods. Data are representative of at least three experiments. All flow cytometry plots are quantified in log10 fluorescence.
Signaling requirements for the development of PLZF-expressing γδ T cells
Data suggest that developing NKT cells require strong TCR-mediated signals for proper development (48). Strength of signaling models have also been proposed for directing thymocytes into the CD8αα and regulatory T cell lineages (49). All of these T cells subsets are believed to express high-avidity, self-reactive TCRs that result in strong TCR signaling during development. It was also shown that strong TCR-mediated signals are necessary for commitment of thymocytes to the γδ T cell lineage, whereas weaker signals favor the αβ T cell lineage (35, 36). The restricted TCR profile of the PLZF-expressing γδ T cells suggested that the specificity of the TCR may play a pivotal role in directing the cells into this lineage. Therefore, the strength of the TCR signal might affect Vγ1.1+Vδ6.3+ T cell development. To test this possibility, recently characterized SLP-76 (Src homology 2-domain–containing leukocyte phosphoprotein of 76 kDa) mutant mice, which have tyrosine to phenylalanine mutations “knocked-in” at key sites of phosphorylation, were analyzed (50).
A mutation of the tyrosine at position 145 (Y145F) within the N-terminal acidic domain of SLP-76 disrupts interactions with IL-2 inducible T cell kinase (ITK) in Jurkat cells (51). Consistent with the cell line studies, SLP-76:Y145F mutant mice were found to phenocopy results from ITK-deficient mice (50). In contrast, mutations in the tyrosines at positions Y112 and Y128 (Y112:128F) within the N-terminal domain of SLP-76 disrupt functional interactions with Vav1, a ρ-family GTP exchange factor, and Nck, an adaptor protein (52–54). Thus, defects in proximal TCR signaling in Y145F and Y112:128F mutant mice differentially affect some aspects of signaling.
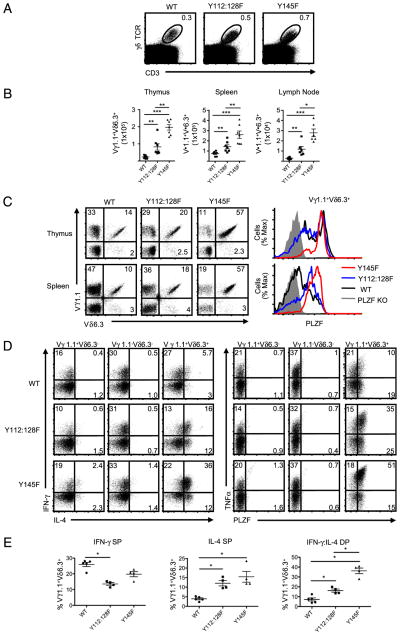
In sharp contrast to the development of NKT cells, the Y145F and Y112:128F mutations resulted in an increased percentage of γδ T cells in the thymus (Fig. 5A). The increase in absolute numbers of γδ T cells in the thymus of the mutant mice was not statistically significant; however, it was attributable to a decrease in total thymic cellularity (50). The increased percentage of γδ T cells in the thymus was due to a 2- to 5-fold increase in the frequency and absolute numbers of Vγ1.1+Vδ6.3+ T cells in Y112:128F and Y145F mice, respectively (Fig. 5B, 5C, Supplemental Fig. 6A). There also was a significant increase in the percentage and absolute numbers of the Vγ1.1+Vδ6.3+ T cell population in the spleen and lymph nodes of the two mutant mice. Furthermore, the frequency of PLZF-expressing γδ T cells was also increased, particularly in the spleen, where nearly all of the Vγ1.1+Vδ6.3+ T cells in the mutant mice expressed high levels of the transcription factor (Fig. 5C). This increase in PLZF expression was specific to Vγ1.1+Vδ6.3+ T cells, because we did not find elevated levels of PLZF expression among non-Vγ1.1+Vδ6.3+ γδ T cell subsets (Supplemental Fig. 6B).
FIGURE 5.
Reduced strength of TCR signal enhances the development of Vγ1.1+Vδ6.3+ T cells. A, Frequency of γδ T cells in the thymuses from WT, SLP-76 Y112:128F (Y112:128F), and SLP-76 Y145F mutant mice (Y145). B, The absolute numbers of indicated γδ subsets in the thymus, spleen, and lymph nodes of the indicated mouse strains. C, The frequency of γδ subsets and PLZF expression analysis in Vγ1.1+Vδ6.3+ T cells from thymocytes and splenocytes of WT, Y112:128F, and Y145F mice. D, Intracellular staining for IFN-γ (Pacific Blue) and IL-4 (Alexa Fluor 488; left panel) or TNF-α (Alexa Fluor 488) and PLZF (Pacific Blue; right panel) in indicated γδ subsets from pooled splenocytes and lymphocytes in WT (top), Y112:128F (middle), and Y145F (bottom) mice. E, The frequency of Vγ1.1+Vδ6.3+ T cells that are SPs for IFN-γ (left) and IL-4 (middle) or DPs (right) for IFN-γ and IL-4 from WT, Y112:128F, and Y145F mice. γδ T cell subsets were identified with anti-CD3 (PerCP-Cy5.5), anti-γδ TCR (APC), anti-Vδ6.3 TCR (PE), and anti-Vγ1.1 TCR (biotin/streptavidin PE-Cy7). Approximately 6 × 106 events per file were collected for these experiments. Multiple files were concatenated in FloJo software. Doublet exclusion was done as indicated in the Materials and Methods. Each symbol represents an individual mouse. *p = 0.05. Error bars represent the SD. All flow cytometry plots are quantified in log10 fluorescence.
We activated pooled Vγ1.1+Vδ6.3+ T cells from the spleen and lymph nodes of WT, Y112:128F, and Y145F mice with PMA/ionomycin, followed by intracellular cytokine staining. In both mutant mice, the Vγ1.1+Vδ6.3+ T cells, but not the other γδ T cell subsets, had significant increases in the proportion of cells that expressed IFN-γ and IL-4 simultaneously or IL-4 alone (Fig. 5D, 5E). Furthermore, there was an increase in the frequency and amount of TNF-α on a per cell basis produced by the mutant Vγ1.1+Vδ6.3+ T cells (Fig. 5D and data not shown). These findings indicate that altered TCR signaling, particularly at the SLP-76–ITK interface, dramatically affects the differentiation and function of Vγ1.1+Vδ6.3+ T cells.
Id3 controls the development of Vγ1.1+Vδ6.3+ T cells
Strong TCR-mediated signals are required to direct developing thymocytes into the γδ T cell lineage (35, 36). TCR signal strength, as measured by ERK1/2 phosphorylation, leads to increased transcription of early growth response (Egr) genes. Reduced Egr expression results in increased αβ T cell development, whereas increased Egr expression leads to enhanced γδ T cell development. Egr1 directly regulates the transcription of Id3 (55). In the absence of Id3, increased Egr expression no longer enhances the development of γδ T cells. Therefore, MAPK, ERK, Egr, and Id3 seem to be in a linear pathway that regulates the αβ versus γδ T cell lineage decision (36). Id3-deficient mice were examined to determine whether Id3 was a potential downstream gene target of altered strength of signaling in Vγ1.1+Vδ6.3+ T cells (37).
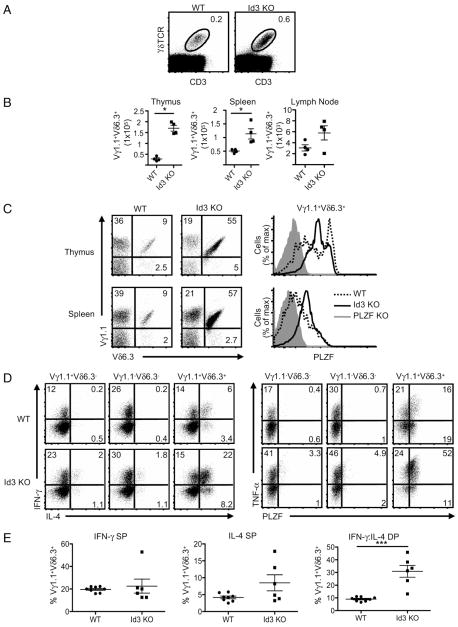
Thymus cellularity in Id3-deficient mice is not altered, suggesting that early T cell development events are not under the control of Id3, whereas the positive selection of αβ T cells is significantly reduced in the absence of Id3 (56). In sharp contrast, we found that the percentage of γδ T cells in Id3-deficient mice was markedly increased (Fig. 6A). Nearly identical to what was found in the TCR-signaling mutant mice (Fig. 5), the frequency and absolute number of Vγ1.1+Vδ6.3+ T cells were increased (Fig. 6B). Similar to the SLP-76 mutants, the percentage of Vγ1.1+Vδ6.3+ T cells expressing high levels of PLZF was also greatly increased (Fig. 6C). The phenotype of Vγ1.1+Vδ6.3+ T cells in Id3-deficient mice closely resembled what was observed in both SLP-76 mutant mice, particularly with regard to increases in the proportion of NK1.1+ and CD4+ cells, as well as upregulation of CD44 and downregulation of CD62L (Supplemental Fig. 7B, 7C).
FIGURE 6.
Id3 controls the development of PLZF-expressing Vγ1.1+Vδ6.3+ T cells. A, The frequency of γδ T cells in the thymuses from WT and Id3-deficient (Id3 KO) mice. B, The absolute numbers of Vγ1.1+Vδ6.3+ T cells in the thymus, spleen, and lymph nodes of WT and Id3 KO mice. *p = 0.0005. C, The frequency of γδ T cell subsets in WT and Id3 KO mice (left) and PLZF expression analysis of Vγ1.1+Vδ6.3+ thymocytes and splenocytes of WT and Id3 KO mice (right). D, Intracellular staining for IFN-γ (Pacific Blue) and IL-4 (Alexa Fluor 488; left panel) or TNF-α (Alexa Fluor 488) and PLZF (Pacific Blue; right panel) in indicated γδ subsets from pooled splenocytes and lymphocytes in WT (top) and Id3 KO (bottom) mice. E, The frequency of Vγ1.1+Vδ6.3+ T cells that are SPs for IFN-γ (left) and IL-4 (middle) or DPs for IFN-γ and IL-4 (right) in WT and Id3 KO mice. γδ T cell subsets were identified with anti-CD3 (PerCP-Cy5.5), anti-γδ TCR (APC), anti-Vδ6.3 TCR (PE), and anti-Vγ1.1 TCR (biotin/streptavidin PE-Cy7). Approximately 6 × 106 events per file were collected for these experiments. Multiple files were concatenated in FloJo software. Doublet exclusion was done as indicated in the Materials and Methods. Data are representative of at least four experiments. Error bars represent the SD. All flow cytometry plots are quantified in log10 fluorescence.
Similar to the SLP-76 mutants, there was a significant increase in the frequency of Vγ1.1+Vδ6.3+ T cells that simultaneously produced IFN-γ and IL4 (Fig. 6D, 6E). There was a 2-fold increase in IL-4 single producers (SPs) and a 5-fold increase in Vγ1.1+Vδ6.3+ T cells that expressed TNF-α (Fig. 6D). Overall, these data show that Id3 deficiency leads to a Vγ1.1+Vδ6.3+ T cell phenotype that is very similar to that found in mice with disrupted TCR signaling.
Discussion
In the absence of PLZF, NKT cells do not acquire innate T cell effector functions and do not express activation markers typically expressed by innate-like T cells (24, 25). In this study, we showed that in addition to NKT cells, PLZF is highly expressed by ~70% of thymic and ~40% of peripheral Vγ1.1+Vδ6.3+ T cells (Fig. 1, Supplemental Fig. 1). The presence of Vγ1.1+Vδ6.3+ T cells in PLZF-deficient mice and the significant reduction of only the PLZF-expressing Vγ1.1+Vδ6.3+ T cells in SAP-deficient mice strongly suggested that PLZF-positive and -negative Vγ1.1+ Vδ6.3+ T cells represent two distinct lineages. Interestingly, the Vγ1.1+Vδ6.3+ iIELs also did not express PLZF, although it is possible that these cells developed extrathymically (57).
PLZF-positive and -negative Vγ1.1+Vδ6.3+ T cells were phenotypically similar. Compared with the other γδ T cell subsets, both populations expressed high levels of CD44, and about half expressed NK1.1. However, the low expression of CD62L clearly distinguished PLZF-positive Vγ1.1+Vδ6.3+ T cells from PLZF-negative Vγ1.1+ Vδ6.3+ T cells. CD44 expression was also diminished in PLZF-deficient Vγ1.1+Vδ6.3+ T cells. In contrast to NKT cells, the absence of PLZF does not alter NK1.1 expression in Vγ1.1+Vδ6.3+ T cells. We did not observe any noteworthy changes in the phenotype of non-Vγ1.1+Vδ6.3+ γδ T cell subsets from PLZF-deficient mice.
Some of the activated PLZF-positive Vδ6.3 T cells simultaneously secreted IFN-γ and IL-4, whereas PLZF-negative Vδ6.3 T cells only secreted IFN-γ. Furthermore, Vδ6.3 T cells from PLZF-deficient mice produced less IFN-γ and no IL-4, reflecting functional data observed for PLZF-deficient NKT cells (24). In addition, PLZF-positive Vδ6.3 T cells secreted large amounts of MIP-1α and RANTES, chemokines important for recruiting macrophages and other cells to sites of infection (42). This finding seems to be consistent with the immunoregulatory role of Vγ1.1+Vδ6.3+ T cells during microbial infections (8, 14). Taken together, these data demonstrate that PLZF defines the functional potential of Vγ1.1+ Vδ6.3+ T cells and distinguishes these cells from other γδ T cells.
We found that a substantial percentage of γδ T cells expressed ThPOK. High ThPOK expression was almost exclusively found in the PLZF-expressing Vγ1.1+Vδ6.3+ T cells, although the majority of these cells did not express CD4. Nonetheless, ThPOK is clearly required for CD4 expression in Vγ1.1+Vδ6.3+ T cells, because in the absence of this transcription factor, the CD4-expressing cells seem to switch to CD8 expression. It was recently shown that ThPOK expression is required to maintain CD4 T cell effector functions (34). In ThPOK-deficient mice, there was an ~50% decrease in PLZF expression in Vγ1.1+Vδ6.3+ T cells. It is interesting to speculate that ThPOK is required for some aspects of Vγ1.1+ Vδ6.3+ T cell function, but its function is modified by PLZF.
PLZF-expressing Vγ1.1+Vδ6.3+ T cells and NKT cells seem to have many features in common. However, the factors that control the development of NKT cells and PLZF-expressing Vγ1.1+ Vδ6.3+ T cells seem to be different. The loss of Fyn nearly ablates NKT cell development (45, 46), but it has no impact on Vγ1.1+ Vδ6.3+ T cells. Even the loss of SAP, which is essential for NKT cells (47), only partially affects Vγ1.1+Vδ6.3+ T cell development. However, in SAP-deficient mice, there was a substantial decrease in the frequency and level of PLZF expression in Vγ1.1+Vδ6.3+ T cells.
Strong TCR-mediated signals are critical for the development of several T cell lineages, including γδ T cells and NKT cells (35, 36, 49). Therefore, we examined the development of Vγ1.1+Vδ6.3+ T cells in mice carrying mutations of key tyrosines in SLP-76 (50). The SLP-76–Y145F mutation phenocopies mice deficient for the expression of ITK (50). Remarkably, the frequency and absolute numbers of Vγ1.1+Vδ6.3+ T cells expressing high levels of PLZF were dramatically increased in these mice. The percentage of cells that produced IFN-γ and IL-4 simultaneously also increased significantly, as did the percentage of cells that produced only IL-4. The SLP-76–Y112:128F mutation also resulted in a significant increase in cells with these characteristics, but the change was not as great as with the Y145F mutation. Both mutations also resulted in a higher percentage of CD4+ and NK1.1+ PLZF-expressing cells.
Interestingly, the loss of ITK promotes the development of innate-like CD8 T cells (58–60). Therefore, reduced TCR signal strength requirements may be a common feature for some innate-like T cells (61). Indeed, ITK mice were recently shown to have increased numbers of Vδ6.3 T cells (62, 63). However, both of the SLP-76 mutant mice and the ITK-deficient mice have substantially reduced numbers of NKT cells (50, 64). The Y145F and Y112:128F mutations differentially alter TCR-induced calcium flux and MAPK/ERK phosphorylation, and both mutations also result in reduced NFAT activity (50, 65). Indeed, the reduced NFAT could be involved in the NKT cell development defect, because NFAT controls Egr2, which is necessary for NKT cell development (66).
The downstream effects of the Y112:128F mutation of SLP-76 are not entirely clear because this mutation likely disrupts the function of the guanine exchange factor Vav1, as well as the adapter protein Nck (67). Nck plays a role in actin cytoskeletal reorganization following TCR activation (68) and directly interacts with CD3 (69). Interestingly, Nck was also shown to interact with SAP (70). Therefore, it is possible that altered Nck activity in Y112:128F mutant mice might reduce SAP activity, thereby counterbalancing the effects of altered TCR signaling in these mice. This might explain why the Vγ1.1+Vδ6.3+ T cell phenotype in the SLP-76–Y112:128 mutant mice was less dramatic compared with Y145F mutant mice. In support of this, SLP-76–Y145F, SAP-deficient mice do not have increased numbers of PLZF-expressing Vγ1.1+Vδ6.3+ T cells (data not shown). Therefore, the increased frequency of Vγ1.1+Vδ6.3+ T cells that is a consequence of altered TCR signaling does not supersede the requirement for SAP-mediated signaling.
Commitment to the γδ lineage is a consequence of strong TCR signaling that results in increased MAPK/ERK activity that leads to the induction of Egr1 (36). Transcription of Id3, a molecular target of Egr1, inhibits E protein activity, which is believed to set signaling thresholds that influence αβ/γδ-lineage choice. We found that Id3-deficient mice had increased numbers of Vγ1.1+ Vδ6.3+ T cells and that nearly all of these cells express high levels of PLZF. Remarkably, the phenotype of Vγ1.1+Vδ6.3+ T cells in the Id3-deficient mice was nearly identical to that found in the SLP-76 mutant mice. Id3-deficient mice had an increased frequency of TNF-α SP and IFN-γ and IL-4 double-producer (DP) Vγ1.1+Vδ6.3+ T cells. In some ways, these data seem to contradict previously established models of TCR signaling in γδ T cell development (35, 36). For example, it was shown that the lack of Id3 impedes the development of γδ T cells between embryonic days 15 and 18 (36). However, neither Vγ1.1 nor Vδ6.3 TCRs are actively being rearranged (6, 71) during fetal development. Furthermore, several studies showed that specific γδ T cell populations rely on certain molecules for their development, but those same molecules are seemingly dispensable for other γδ T cell populations (72–74).
It remains to be addressed whether PLZF-positive and -negative Vγ1.1+Vδ6.3+ T cells have different specificities due to variability in the TCR CDR3 regions. This might lead to different strengths of signaling during selection in the thymus. For example, a recent study showed that in the absence of MHC class I-like molecules T10/T22 in β2m-KO mice, γδ T cells that normally recognize these ligands developed but had a profound change in their effector functions (22). Two studies relevant to this question were recently published (75, 76). Kreslavsky et al. (75) showed that T cells in mice carrying transgenes encoding a Vγ1.1+Vδ6.3+ TCR were strongly, but not completely, skewed toward the PLZF-expressing lineage. They also showed that PLZF is induced in γδ thymocytes after culturing on OP9-DL1 cells for 5 d in the presence of anti-γδ TCR Ab. Together with data showing that CD5 levels were higher on Vγ1.1+Vδ6.3+ thymocytes, it was proposed that strong TCR-mediated signals are involved in inducing PLZF expression. Contrary to this conclusion, our data clearly showed that, in vivo, reduced TCR signal strength results in a dramatic increase in the PLZF-expressing γδ T cell lineage. Furthermore, only ~70% of the Vγ1.1+Vδ6.3+ TCR transgenic thymocytes (75) and ~60% of WT Vγ1.1+Vδ6.3+ T cells (Fig. 1B) expressed PLZF. Therefore, despite expressing the identical TCR, PLZF was not induced in all cells. In contrast, once the TCR signal strength was reduced by the Y145F mutation to SLP-76, nearly all of the Vγ1.1+Vδ6.3+ T cells expressed high levels of PLZF (Fig. 5C). Together, these data seem to make a simple TCR avidity-based model for inducing PLZF expression tenuous.
Similar to our studies, Lauritsen et al. (76) and Ueda-Hayakawa et al. (77) recently showed that Vγ1.1 T cells are increased in the absence of Id3. Ueda-Hayakawa et al. (77) suggested that, in the absence of Id3, there is an increased window of opportunity for thymocytes to rearrange TCRγ-chains. In contrast, Lauritsen et al. (76) argued that the alteration in γδ T cells is due to the failure to negatively select thymocytes expressing the Vγ1.1 TCR. Neither of these explanations seems to fully account for our observations.
In the study by Ueda-Hayakawa et al. (77), it was proposed that double-negative cells that had failed to rearrange a functional TCRβ-chain were able to undergo further TCR rearrangement at the TCRγ-locus in the absence of Id3. Therefore, it was proposed that Id3 restricts the “window” of TCR gene rearrangement in double-negative thymocytes. However, it is not clear how the same reasoning can be used to explain the nearly identical increase of Vγ1.1 T cells that we found in the SLP-76 mutant mice (Figs. 5B, 6C), because the signaling mediated by SLP-76 at the double-negative stage is expected to be dependent on pre-TCR expression. It was shown that the complete loss of SLP-76 disrupts TCRβ allelic exclusion (78). Therefore, it remains possible that disrupted allelic exclusion allows for additional TCRγ-chain rearrangements.
Decreased negative selection due to reduced TCR signals, as proposed by Lauritsen et al. (76), at first seems to be more consistent with our data. However, a clear role for the negative selection of developing γδ T cells remains to be defined (22). Furthermore, the idea that NK1.1+ γδ T cells are self-reactive is largely based upon comparison with NKT cells. WT NKT cells have a constitutively activated phenotype, which was believed to be a consequence of their TCR specificity for a ubiquitously expressed self-Ag (79). However, this concept has fallen from favor, now that it has been shown that the activated NKT cell phenotype is a consequence of the expression of PLZF (24, 25). Furthermore, it is difficult to understand why most Vγ1.1+Vδ6.3+ TCR-expressing thymocytes are negatively selected, whereas others expressing the same TCR mature. Finally, reduced negative selection does not explain the increase in PLZF-expressing Vγ1.1+Vδ6.3+ T cells from ~60% to nearly 100% in the Id3-deficient and SLP-76 mutant mice.
We propose that the increase in Vγ1.1+Vδ6.3+ T cells in SLP-76 mutant and Id3-deficient mice is a consequence of the cells being directed into the PLZF-expressing lineage. PLZF-expressing NKT cells are known to go through an enormous proliferative burst early in development. Therefore, it is likely that the PLZF-expressing γδ T cells also go through a similar expansion, although this has not been directly tested. Furthermore, NKT cells rapidly accumulate in adult mice (80), and it is possible that Vγ1.1+Vδ6.3+ T cells also have this feature, perhaps as the result of both cell types developing during the postnatal period.
Overall, our work demonstrates the presence of two functionally distinct subsets of Vγ1.1+Vδ6.3+ T cells based on PLZF expression. Our data suggest a model in which reduced TCR signaling strength leads to decreased Id3 transcription, likely due to reduced Egr activity. The lack of Id3 then directs cells into the PLZF-expressing Vγ1.1+Vδ6.3+ T cell lineage rather than the non–PLZF-expressing γδ T cell lineage. The similarity of the phenotypes observed in the SLP-76 Y145F mice and the Id3-deficient mice is consistent with a linear relationship, analogous to what was reported in γδ T cells (36). This model is supported by recent data showing that conditional deletion of E2A in Id3-deficient T cells prevents the increase of Vγ1.1+ γδ T cells (77). Our work provides new insight into γδ T cell differentiation and function, while highlighting the role of BTB-ZF proteins in these enigmatic cells.
Supplementary Material
Acknowledgments
We thank T. Martillotti and J. Yeo for technical assistance, Drs. L. Denzin and M. Huse for critical reading of the manuscript and advice, and Drs. D. Littman and J. Allison for reagents.
This work was supported, in part, by National Institutes of Health Grant AI059739, the May and Samuel Rudin Family Foundation, and National Cancer Institute Grant P30-CA 08748, which provides partial support for Memorial Sloan-Kettering Cancer Center’s Monoclonal Antibody, Flow Cytometry, Glassware Washing core facilities, and the Research Animal Resource Center. E.S.A. is supported by the Ruth L. Kirschstein National Research Service Award F31CA130744. R.A.G. is supported by a predoctoral training grant from the Cancer Research Institute. T.E. is supported by a fellowship from the Leukemia & Lymphoma Society.
Abbreviations used in this paper
- BTB
broad-complex and tramtrack bric-à-brac
- DP
double producer
- Egr
early growth response
- Id3
inhibitor of differentiation gene 3
- iIEL
intestinal intraepithelial lymphocyte
- ITK
IL-2 inducible T cell kinase
- MSKCC
Memorial Sloan-Kettering Cancer Center
- NKT cells
NK T cells
- PLZF
promyelocytic leukemia zinc finger
- SLP-76
Src homology 2-domain–containing leukocyte phosphoprotein of 76 kDa
- SAP
signaling lymphocyte activation molecule-associated protein
- SLAM
signaling lymphocyte activation molecule
- SP
single producer
- ZF
zinc finger
Footnotes
The online version of this article contains supplemental material.
Disclosures
The authors have no financial conflicts of interest.
References
- 1.Taghon T, Yui MA, Pant R, Diamond RA, Rothenberg EV. Developmental and molecular characterization of emerging β- and gammadelta-selected pre-T cells in the adult mouse thymus. Immunity. 2006;24:53–64. doi: 10.1016/j.immuni.2005.11.012. [DOI] [PubMed] [Google Scholar]
- 2.Hayday AC. [γ][δ] cells: a right time and a right place for a conserved third way of protection. Annu Rev Immunol. 2000;18:975–1026. doi: 10.1146/annurev.immunol.18.1.975. [DOI] [PubMed] [Google Scholar]
- 3.Li W, Kim MG, Gourley TS, McCarthy BP, Sant’Angelo DB, Chang CH. An alternate pathway for CD4 T cell development: thymocyte-expressed MHC class II selects a distinct T cell population. Immunity. 2005;23:375–386. doi: 10.1016/j.immuni.2005.09.002. [DOI] [PubMed] [Google Scholar]
- 4.Lewis JM, Girardi M, Roberts SJ, Barbee SD, Hayday AC, Tigelaar RE. Selection of the cutaneous intraepithelial gammadelta+ T cell repertoire by a thymic stromal determinant. Nat Immunol. 2006;7:843–850. doi: 10.1038/ni1363. [DOI] [PubMed] [Google Scholar]
- 5.Boyden LM, Lewis JM, Barbee SD, Bas A, Girardi M, Hayday AC, Tigelaar RE, Lifton RP. Skint1, the prototype of a newly identified immunoglobulin superfamily gene cluster, positively selects epidermal gammadelta T cells. Nat Genet. 2008;40:656–662. doi: 10.1038/ng.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Havran WL, Allison JP. Developmentally ordered appearance of thymocytes expressing different T-cell antigen receptors. Nature. 1988;335:443–445. doi: 10.1038/335443a0. [DOI] [PubMed] [Google Scholar]
- 7.Allison JP, Havran WL. The immunobiology of T cells with invariant γ δ antigen receptors. Annu Rev Immunol. 1991;9:679–705. doi: 10.1146/annurev.iy.09.040191.003335. [DOI] [PubMed] [Google Scholar]
- 8.Carding SR, Egan PJ. Gammadelta T cells: functional plasticity and heterogeneity. Nat Rev Immunol. 2002;2:336–345. doi: 10.1038/nri797. [DOI] [PubMed] [Google Scholar]
- 9.Hayday A, Tigelaar R. Immunoregulation in the tissues by gammadelta T cells. Nat Rev Immunol. 2003;3:233–242. doi: 10.1038/nri1030. [DOI] [PubMed] [Google Scholar]
- 10.Jameson J, Ugarte K, Chen N, Yachi P, Fuchs E, Boismenu R, Havran WL. A role for skin gammadelta T cells in wound repair. Science. 2002;296:747–749. doi: 10.1126/science.1069639. [DOI] [PubMed] [Google Scholar]
- 11.Peng SL, Madaio MP, Hayday AC, Craft J. Propagation and regulation of systemic autoimmunity by gammadelta T cells. J Immunol. 1996;157:5689–5698. [PubMed] [Google Scholar]
- 12.Harrison LC, Dempsey-Collier M, Kramer DR, Takahashi K. Aerosol insulin induces regulatory CD8 γ δ T cells that prevent murine insulin-dependent diabetes. J Exp Med. 1996;184:2167–2174. doi: 10.1084/jem.184.6.2167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Girardi M, Oppenheim DE, Steele CR, Lewis JM, Glusac E, Filler R, Hobby P, Sutton B, Tigelaar RE, Hayday AC. Regulation of cutaneous malignancy by gammadelta T cells. Science. 2001;294:605–609. doi: 10.1126/science.1063916. [DOI] [PubMed] [Google Scholar]
- 14.Hsieh B, Schrenzel MD, Mulvania T, Lepper HD, DiMolfetto-Landon L, Ferrick DA. In vivo cytokine production in murine listeriosis. Evidence for immunoregulation by γ δ+ T cells. J Immunol. 1996;156:232–237. [PubMed] [Google Scholar]
- 15.Janis EM, Kaufmann SH, Schwartz RH, Pardoll DM. Activation of γ δ T cells in the primary immune response to. Mycobacterium tuberculosis Science. 1989;244:713–716. doi: 10.1126/science.2524098. [DOI] [PubMed] [Google Scholar]
- 16.Azuara V, Levraud JP, Lembezat MP, Pereira P. A novel subset of adult γ δ thymocytes that secretes a distinct pattern of cytokines and expresses a very restricted T cell receptor repertoire. Eur J Immunol. 1997;27:544–553. doi: 10.1002/eji.1830270228. [DOI] [PubMed] [Google Scholar]
- 17.Lees RK, Ferrero I, MacDonald HR. Tissue-specific segregation of TCRgamma δ+ NKT cells according to phenotype TCR repertoire and activation status: parallels with TCR alphabeta+NKT cells. Eur J Immunol. 2001;31:2901–2909. doi: 10.1002/1521-4141(2001010)31:10<2901::aid-immu2901>3.0.co;2-#. [DOI] [PubMed] [Google Scholar]
- 18.Belles C, Kuhl AK, Donoghue AJ, Sano Y, O’Brien RL, Born W, Bottomly K, Carding SR. Bias in the γ δ T cell response to Listeria monocytogenes. V δ 6.3+ cells are a major component of the γ δ T cell response toListeria monocytogenes. J Immunol. 1996;156:4280–4289. [PubMed] [Google Scholar]
- 19.Yin Z, Chen C, Szabo SJ, Glimcher LH, Ray A, Craft J. T-Bet expression and failure of GATA-3 cross-regulation lead to default production of IFN-γ by gammadelta T cells. J Immunol. 2002;168:1566–1571. doi: 10.4049/jimmunol.168.4.1566. [DOI] [PubMed] [Google Scholar]
- 20.Lockhart E, Green AM, Flynn JL. IL-17 production is dominated by gammadelta T cells rather than CD4 T cells during Mycobacterium tuberculosis infection. J Immunol. 2006;177:4662–4669. doi: 10.4049/jimmunol.177.7.4662. [DOI] [PubMed] [Google Scholar]
- 21.Shibata K, Yamada H, Hara H, Kishihara K, Yoshikai Y. Resident Vdelta1+ gammadelta T cells control early infiltration of neutrophils after Escherichia coli infection via IL-17 production. J Immunol. 2007;178:4466–4472. doi: 10.4049/jimmunol.178.7.4466. [DOI] [PubMed] [Google Scholar]
- 22.Jensen KD, Su X, Shin S, Li L, Youssef S, Yamasaki S, Steinman L, Saito T, Locksley RM, Davis MM, et al. Thymic selection determines gammadelta T cell effector fate: antigen-naive cells make interleukin-17 and antigen-experienced cells make interferon γ. Immunity. 2008;29:90–100. doi: 10.1016/j.immuni.2008.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gerber DJ, Azuara V, Levraud JP, Huang SY, Lembezat MP, Pereira P. IL-4-producing γ δ T cells that express a very restricted TCR repertoire are preferentially localized in liver and spleen. J Immunol. 1999;163:3076–3082. [PubMed] [Google Scholar]
- 24.Kovalovsky D, Uche OU, Eladad S, Hobbs RM, Yi W, Alonzo E, Chua K, Eidson M, Kim HJ, Im JS, et al. The BTB-zinc finger transcriptional regulator PLZF controls the development of invariant natural killer T cell effector functions. Nat Immunol. 2008;9:1055–1064. doi: 10.1038/ni.1641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Savage AK, Constantinides MG, Han J, Picard D, Martin E, Li B, Lantz O, Bendelac A. The transcription factor PLZF directs the effector program of the NKT cell lineage. Immunity. 2008;29:391–403. doi: 10.1016/j.immuni.2008.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kelly KF, Daniel JM. POZ for effect—POZ-ZF transcription factors in cancer and development. Trends Cell Biol. 2006;16:578–587. doi: 10.1016/j.tcb.2006.09.003. [DOI] [PubMed] [Google Scholar]
- 27.Costoya JA, Hobbs RM, Barna M, Cattoretti G, Manova K, Sukhwani M, Orwig KE, Wolgemuth DJ, Pandolfi PP. Essential role of Plzf in maintenance of spermatogonial stem cells. Nat Genet. 2004;36:653–659. doi: 10.1038/ng1367. [DOI] [PubMed] [Google Scholar]
- 28.Hu S, Fambrough D, Atashi JR, Goodman CS, Crews ST. The Drosophila abrupt gene encodes a BTB-zinc finger regulatory protein that controls the specificity of neuromuscular connections. Genes Dev. 1995;9:2936–2948. doi: 10.1101/gad.9.23.2936. [DOI] [PubMed] [Google Scholar]
- 29.Barna M, Hawe N, Niswander L, Pandolfi PP. Plzf regulates limb and axial skeletal patterning. Nat Genet. 2000;25:166–172. doi: 10.1038/76014. [DOI] [PubMed] [Google Scholar]
- 30.Maeda T, Merghoub T, Hobbs RM, Dong L, Maeda M, Zakrzewski J, van den Brink MR, Zelent A, Shigematsu H, Akashi K, et al. Regulation of B versus T lymphoid lineage fate decision by the proto-oncogene LRF. Science. 2007;316:860–866. doi: 10.1126/science.1140881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.He X, He X, Dave VP, Zhang Y, Hua X, Nicolas E, Xu W, Roe BA, Kappes DJ. The zinc finger transcription factor Th-POK regulates CD4 versus CD8 T-cell lineage commitment. Nature. 2005;433:826–833. doi: 10.1038/nature03338. [DOI] [PubMed] [Google Scholar]
- 32.Egawa T, Littman DR. ThPOK acts late in specification of the helper T cell lineage and suppresses Runx-mediated commitment to the cytotoxic T cell lineage. Nat Immunol. 2008;9:1131–1139. doi: 10.1038/ni.1652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Muroi S, Naoe Y, Miyamoto C, Akiyama K, Ikawa T, Masuda K, Kawamoto H, Taniuchi I. Cascading suppression of transcriptional silencers by ThPOK seals helper T cell fate. Nat Immunol. 2008;9:1113–1121. doi: 10.1038/ni.1650. [DOI] [PubMed] [Google Scholar]
- 34.Wang L, Wildt KF, Zhu J, Zhang X, Feigenbaum L, Tessarollo L, Paul WE, Fowlkes BJ, Bosselut R. Distinct functions for the transcription factors GATA-3 and ThPOK during intrathymic differentiation of CD4(+) T cells. Nat Immunol. 2008;9:1122–1130. doi: 10.1038/ni.1647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hayes SM, Li L, Love PE. TCR signal strength influences alphabeta/gammadelta lineage fate. Immunity. 2005;22:583–593. doi: 10.1016/j.immuni.2005.03.014. [DOI] [PubMed] [Google Scholar]
- 36.Haks MC, Lefebvre JM, Lauritsen JP, Carleton M, Rhodes M, Miyazaki T, Kappes DJ, Wiest DL. Attenuation of gammadeltaTCR signaling efficiently diverts thymocytes to the alphabeta lineage. Immunity. 2005;22:595–606. doi: 10.1016/j.immuni.2005.04.003. [DOI] [PubMed] [Google Scholar]
- 37.Pan L, Sato S, Frederick JP, Sun XH, Zhuang Y. Impaired immune responses and B-cell proliferation in mice lacking the Id3 gene. Mol Cell Biol. 1999;19:5969–5980. doi: 10.1128/mcb.19.9.5969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Puddington L, Olson S, Lefrançois L. Interactions between stem cell factor and c-Kit are required for intestinal immune system homeostasis. Immunity. 1994;1:733–739. doi: 10.1016/s1074-7613(94)80015-4. [DOI] [PubMed] [Google Scholar]
- 39.Kalataradi H, Eyster CL, Fry A, Vollmer MK, Fu YX, Born WK, O’Brien RL. Allelic differences in TCR γ-chains alter γ δ T cell antigen reactivity. J Immunol. 1994;153:1455–1465. [PubMed] [Google Scholar]
- 40.O’Brien RL, Fu YX, Cranfill R, Dallas A, Ellis C, Reardon C, Lang J, Carding SR, Kubo R, Born W. Heat shock protein Hsp60-reactive γ δ cells: a large, diversified T-lymphocyte subset with highly focused specificity. Proc Natl Acad Sci USA. 1992;89:4348–4352. doi: 10.1073/pnas.89.10.4348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Benlagha K, Kyin T, Beavis A, Teyton L, Bendelac A. A thymic precursor to the NK T cell lineage. Science. 2002;296:553–555. doi: 10.1126/science.1069017. [DOI] [PubMed] [Google Scholar]
- 42.Cook DN, Beck MA, Coffman TM, Kirby SL, Sheridan JF, Pragnell IB, Smithies O. Requirement of MIP-1 α for an inflammatory response to viral infection. Science. 1995;269:1583–1585. doi: 10.1126/science.7667639. [DOI] [PubMed] [Google Scholar]
- 43.Pennington DJ, Silva-Santos B, Shires J, Theodoridis E, Pollitt C, Wise EL, Tigelaar RE, Owen MJ, Hayday AC. The inter-relatedness and interdependence of mouse T cell receptor gammadelta+ and alphabeta+ cells. Nat Immunol. 2003;4:991–998. doi: 10.1038/ni979. [DOI] [PubMed] [Google Scholar]
- 44.Silva-Santos B, Pennington DJ, Hayday AC. Lymphotoxin-mediated regulation of gammadelta cell differentiation by alphabeta T cell progenitors. Science. 2005;307:925–928. doi: 10.1126/science.1103978. [DOI] [PubMed] [Google Scholar]
- 45.Eberl G, Lowin-Kropf B, MacDonald HR. Cutting edge: NKT cell development is selectively impaired in Fyn-deficient mice. J Immunol. 1999;163:4091–4094. [PubMed] [Google Scholar]
- 46.Gadue P, Morton N, Stein PL. The Src family tyrosine kinase Fyn regulates natural killer T cell development. J Exp Med. 1999;190:1189–1196. doi: 10.1084/jem.190.8.1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nichols KE, Hom J, Gong SY, Ganguly A, Ma CS, Cannons JL, Tangye SG, Schwartzberg PL, Koretzky GA, Stein PL. Regulation of NKT cell development by SAP, the protein defective in XLP. Nat Med. 2005;11:340–345. doi: 10.1038/nm1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bendelac A, Rivera MN, Park SH, Roark JH. Mouse CD1-specific NK1 T cells: development, specificity, and function. Annu Rev Immunol. 1997;15:535–562. doi: 10.1146/annurev.immunol.15.1.535. [DOI] [PubMed] [Google Scholar]
- 49.Baldwin TA, Hogquist KA, Jameson SC. The fourth way? Harnessing aggressive tendencies in the thymus. J Immunol. 2004;173:6515–6520. doi: 10.4049/jimmunol.173.11.6515. [DOI] [PubMed] [Google Scholar]
- 50.Jordan MS, Smith JE, Burns JC, Austin JE, Nichols KE, Aschenbrenner AC, Koretzky GA. Complementation in trans of altered thymocyte development in mice expressing mutant forms of the adaptor molecule SLP76. Immunity. 2008;28:359–369. doi: 10.1016/j.immuni.2008.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bunnell SC, Diehn M, Yaffe MB, Findell PR, Cantley LC, Berg LJ. Biochemical interactions integrating Itk with the T cell receptor-initiated signaling cascade. J Biol Chem. 2000;275:2219–2230. doi: 10.1074/jbc.275.3.2219. [DOI] [PubMed] [Google Scholar]
- 52.Wu J, Motto DG, Koretzky GA, Weiss A. Vav and SLP-76 interact and functionally cooperate in IL-2 gene activation. Immunity. 1996;4:593–602. doi: 10.1016/s1074-7613(00)80485-9. [DOI] [PubMed] [Google Scholar]
- 53.Bubeck Wardenburg J, Pappu R, Bu JY, Mayer B, Chernoff J, Straus D, Chan AC. Regulation of PAK activation and the T cell cytoskeleton by the linker protein SLP-76. Immunity. 1998;9:607–616. doi: 10.1016/s1074-7613(00)80658-5. [DOI] [PubMed] [Google Scholar]
- 54.Raab M, da Silva AJ, Findell PR, Rudd CE. Regulation of Vav-SLP-76 binding by ZAP-70 and its relevance to TCR ζ/CD3 induction of interleukin-2. Immunity. 1997;6:155–164. doi: 10.1016/s1074-7613(00)80422-7. [DOI] [PubMed] [Google Scholar]
- 55.Bain G, Cravatt CB, Loomans C, Alberola-Ila J, Hedrick SM, Murre C. Regulation of the helix-loop-helix proteins, E2A and Id3, by the Ras-ERK MAPK cascade. Nat Immunol. 2001;2:165–171. doi: 10.1038/84273. [DOI] [PubMed] [Google Scholar]
- 56.Rivera RR, Johns CP, Quan J, Johnson RS, Murre C. Thymocyte selection is regulated by the helix-loop-helix inhibitor protein, Id3. Immunity. 2000;12:17–26. doi: 10.1016/s1074-7613(00)80155-7. [DOI] [PubMed] [Google Scholar]
- 57.Bandeira A, Itohara S, Bonneville M, Burlen-Defranoux O, Mota-Santos T, Coutinho A, Tonegawa S. Extrathymic origin of intestinal intra-epithelial lymphocytes bearing T-cell antigen receptor γ δ. Proc Natl Acad Sci USA. 1991;88:43–47. doi: 10.1073/pnas.88.1.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hu J, Sahu N, Walsh E, August A. Memory phenotype CD8+ T cells with innate function selectively develop in the absence of active Itk. Eur J Immunol. 2007;37:2892–2899. doi: 10.1002/eji.200737311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Broussard C, Fleischacker C, Horai R, Chetana M, Venegas AM, Sharp LL, Hedrick SM, Fowlkes BJ, Schwartzberg PL. Altered development of CD8+ T cell lineages in mice deficient for the Tec kinases Itk and Rlk. Immunity. 2006;25:93–104. doi: 10.1016/j.immuni.2006.05.011. [DOI] [PubMed] [Google Scholar]
- 60.Horai R, Mueller KL, Handon RA, Cannons JL, Anderson SM, Kirby MR, Schwartzberg PL. Requirements for selection of conventional and innate T lymphocyte lineages. Immunity. 2007;27:775–785. doi: 10.1016/j.immuni.2007.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Atherly LO, Lucas JA, Felices M, Yin CC, Reiner SL, Berg LJ. The Tec family tyrosine kinases Itk and Rlk regulate the development of conventional CD8+ T cells. Immunity. 2006;25:79–91. doi: 10.1016/j.immuni.2006.05.012. [DOI] [PubMed] [Google Scholar]
- 62.Qi Q, Xia M, Hu J, Hicks E, Iyer A, Xiong N, August A. Enhanced development of CD4+ gammadelta T cells in the absence of Itk results in elevated IgE production. Blood. 2009;114:564–571. doi: 10.1182/blood-2008-12-196345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Felices M, Yin CC, Kosaka Y, Kang J, Berg LJ. Tec kinase Itk in gammadeltaT cells is pivotal for controlling IgE production in vivo. Proc Natl Acad Sci USA. 2009;106:8308–8313. doi: 10.1073/pnas.0808459106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Felices M, Berg LJ. The Tec kinases Itk and Rlk regulate NKT cell maturation, cytokine production, and survival. J Immunol. 2008;180:3007–3018. doi: 10.4049/jimmunol.180.5.3007. [DOI] [PubMed] [Google Scholar]
- 65.Jordan MS, Sadler J, Austin JE, Finkelstein LD, Singer AL, Schwartzberg PL, Koretzky GA. Functional hierarchy of the N-terminal tyrosines of SLP-76. J Immunol. 2006;176:2430–2438. doi: 10.4049/jimmunol.176.4.2430. [DOI] [PubMed] [Google Scholar]
- 66.Lazarevic V, Zullo AJ, Schweitzer MN, Staton TL, Gallo EM, Crabtree GR, Glimcher LH. The gene encoding early growth response 2, a target of the transcription factor NFAT, is required for the development and maturation of natural killer T cells. Nat Immunol. 2009;10:306–313. doi: 10.1038/ni.1696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wu JN, Koretzky GA. The SLP-76 family of adapter proteins. Semin Immunol. 2004;16:379–393. doi: 10.1016/j.smim.2004.08.018. [DOI] [PubMed] [Google Scholar]
- 68.Lettau M, Pieper J, Janssen O. Nck adapter proteins: functional versatility in T cells. Cell Commun Signal. 2009;7:1. doi: 10.1186/1478-811X-7-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Gil D, Schamel WW, Montoya M, Sánchez-Madrid F, Alarcón B. Recruitment of Nck by CD3 ε reveals a ligand-induced conformational change essential for T cell receptor signaling and synapse formation. Cell. 2002;109:901–912. doi: 10.1016/s0092-8674(02)00799-7. [DOI] [PubMed] [Google Scholar]
- 70.Li C, Schibli D, Li SS. The XLP syndrome protein SAP interacts with SH3 proteins to regulate T cell signaling and proliferation. Cell Signal. 2009;21:111–119. doi: 10.1016/j.cellsig.2008.09.014. [DOI] [PubMed] [Google Scholar]
- 71.Grigoriadou K, Boucontet L, Pereira P. Most IL-4-producing γ δ thymocytes of adult mice originate from fetal precursors. J Immunol. 2003;171:2413–2420. doi: 10.4049/jimmunol.171.5.2413. [DOI] [PubMed] [Google Scholar]
- 72.Penninger JM, V, Wallace A, Kishihara K, Mak TW. The role of p56lck and p59fyn tyrosine kinases and CD45 protein tyrosine phosphatase in T-cell development and clonal selection. Immunol Rev. 1993;135:183–214. doi: 10.1111/j.1600-065x.1993.tb00649.x. [DOI] [PubMed] [Google Scholar]
- 73.Kawai K, Kishihara K, Molina TJ, Wallace VA, Mak TW, Ohashi PS. Impaired development of V γ 3 dendritic epidermal T cells in p56lck protein tyrosine kinase-deficient and CD45 protein tyrosine phosphatase-deficient mice. J Exp Med. 1995;181:345–349. doi: 10.1084/jem.181.1.345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Kadlecek TA, van Oers NS, Lefrancois L, Olson S, Finlay D, Chu DH, Connolly K, Killeen N, Weiss A. Differential requirements for ZAP-70 in TCR signaling and T cell development. J Immunol. 1998;161:4688–4694. [PubMed] [Google Scholar]
- 75.Kreslavsky T, Savage AK, Hobbs R, Gounari F, Bronson R, Pereira P, Pandolfi PP, Bendelac A, von Boehmer H. TCR-inducible PLZF transcription factor required for innate phenotype of a subset of gammadelta T cells with restricted TCR diversity. Proc Natl Acad Sci USA. 2009;106:12453–12458. doi: 10.1073/pnas.0903895106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Lauritsen JP, Wong GW, Lee SY, Lefebvre JM, Ciofani M, Rhodes M, Kappes DJ, Zúñiga-Pflücker JC, Wiest DL. Marked induction of the helix-loop-helix protein Id3 promotes the gammadelta T cell fate and renders their functional maturation Notch independent. Immunity. 2009;31:565–575. doi: 10.1016/j.immuni.2009.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Ueda-Hayakawa I, Mahlios J, Zhuang Y. Id3 restricts the developmental potential of γ δ lineage during thymopoiesis. J Immunol. 2009;182:5306–5316. doi: 10.4049/jimmunol.0804249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Aifantis I, V, Pivniouk I, Gärtner F, Feinberg J, Swat W, Alt FW, von Boehmer H, Geha RS. Allelic exclusion of the T cell receptor β locus requires the SH2 domain-containing leukocyte protein (SLP)-76 adaptor protein. J Exp Med. 1999;190:1093–1102. doi: 10.1084/jem.190.8.1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Bendelac A. Nondeletional pathways for the development of autoreactive thymocytes. Nat Immunol. 2004;5:557–558. doi: 10.1038/ni0604-557. [DOI] [PubMed] [Google Scholar]
- 80.Dao T, Guo D, Ploss A, Stolzer A, Saylor C, Boursalian TE, Im JS, Sant’Angelo DB. Development of CD1d-restricted NKT cells in the mouse thymus. Eur J Immunol. 2004;34:3542–3552. doi: 10.1002/eji.200425546. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.