Abstract
Rare cells are low-abundance cells in a much larger population of background cells. Conventional benchtop techniques have limited capabilities to isolate and analyze rare cells because of their generally low selectivity and significant sample loss. Recent rapid advances in microfluidics have been providing robust solutions to the challenges in the isolation and analysis of rare cells. In addition to the apparent performance enhancements resulting in higher efficiencies and sensitivity levels, microfluidics provides other advanced features such as simpler handling of small sample volumes and multiplexing capabilities for high-throughput processing. All of these advantages make microfluidics an excellent platform to deal with the transport, isolation, and analysis of rare cells. Various cellular biomarkers, including physical properties, dielectric properties, as well as immunoaffinities, have been explored for isolating rare cells. In this Focus article, we discuss the design considerations of representative microfluidic devices for rare cell isolation and analysis. Examples from recently published works are discussed to highlight the advantages and limitations of the different techniques. Various applications of these techniques are then introduced. Finally, a perspective on the development trends and promising research directions in this field are proposed.
Introduction
Cell types with an abundance of less than 1000 in one milliliter sample are considered as rare.1 Rare cells are highly important for various applications such as the diagnosis and prognosis of many cancers, prenatal diagnosis, and the diagnosis of viral infections. Typical rare cells in blood samples are circulating tumor cells (CTCs), circulating fetal cells, stem cells, and cells infected by virus or parasites. Rare cells in water samples include various pathogenic bacteria and those infected by viruses. Since preparation techniques for rare cell isolation depend on the source from where the cell samples are obtained, this article mainly focuses on isolation of rare cells from biological samples such as blood and other bodily fluids.
Microfluidics is a technology that enables transport and manipulation of fluids and particles such as cells in the microscale. A typical microfluidic device consists of a microchannel network integrated with various sensors and actuators. Common microchannels have dimensions on the order of hundreds of microns, while the size of cells ranges from several to tens of micrometers. Therefore, strong interactions between cells, the fluid flow and the microchannels are expected because of the similar length scales. The small size of microfluidic devices also allows for the implementation of new protocols such as single-cell analysis or on-chip cell culture, which is previously not possible with macroscale devices.
Microfluidics has been used for the isolation, enrichment and analysis of rare cells. These cells are isolated from a large population of other cell types based on one or several unique properties. A number of reviews on the isolation of rare cells, especially CTCs, have been published recently. Pratt et al. categorized rare cell isolation concepts as non-electrokinetic and electrokinetic methods.2 Several papers have been published to review the various label-free isolation methods.3-8 Distinguishing physical properties such as cell size, deformability, compressibility, shape, density, size, surface properties, electrical polarizeability, magnetic susceptibility and refractive index have been considered as biomarkers. A combination of these physical properties can form a unique profile for a given cell type. Multiple separation techniques can be used to match a specific profile. Karimi et al. recently reviewed hydrodynamic methods for cell isolation.9 The authors categorized the isolation methods according to hydrodynamic phenomena such as inertial effects, viscoelastic focusing, Dean flows, cavitation and hydrodynamic vorticity. Dharmasiri et al. reviewed different microdevices for rare cell isolation and categorized them as either macroscale or microscale techniques.1 The sorting mechanisms are further subcategorized based on immunoaffinity, physical separation, dielectrophoresis, or magnetic and fluorescence activated sorting. Yu et al. also reviewed different approaches to the isolation of CTCs.10 The authors categorized the isolation methods as either nucleic acid-based approach, or physical-properties-based approach. Danova et al. discussed both the technical aspects and clinical implications of CTC isolation.11 This review focused on existing macroscale commercial systems, rather than microfluidic systems. Smith et al. discusses the impact of flow in a microfluidic device on rare cell isolation based on immunoaffinities.12 Design considerations from the engineering perspective were discussed in their paper. Hyun and Jung reviewed the different microfluidic devices for rare cell isolation with the focus on affinity-based methods, along with the use of dielectrophoresis and hydrophoresis as label-free methods.13 Chen et al. reviewed microfluidic approaches for cancer cells detection, characterization and separation.14 The authors mainly reviewed methods based on surface affinity and dielectrophoresis. Alix-Panabieres briefly discusses various existing techniques for enrichment and detection of CTCs based on physical and biological properties.15 Muluneh and Issadore reviewed immunomagnetic detection of CTCs.16 Hong and Zu discussed current challenges for detecting CTCs including the requirements for point-of-care testing and the use of microdevices.17
The present Focus article looks at isolation, enrichment and analysis of rare cells from an engineering perspective. Design considerations based on the various properties are discussed and compared. Current challenges that limit efficient isolation of rare cells are first discussed. Properties of rare cells that can be used to distinguish them from the background cells are analyzed. Specific parameters are then defined in order to evaluate and compare various isolation approaches. These different isolation and analysis techniques are illustrated using recently published works. The paper concludes with a perspective on the development trends and promising research directions in the field of microfluidic-based rare cell isolation and analysis.
Design considerations
The rarity of low-abundance cells represents a challenge in the design of microfluidic devices. First, the frequency of a cell type in a given population depends on other parameters such as the sampling location, the age of the sample, and the stage of the disease. Second, rarity means that a large sample volume is needed for a high statistical confidence. Thus, the required sample volume may vary for even the same cell type. The large sample volume and the small microchannel dimensions inevitably lead to a long processing time or a low throughput.1 Increasing the flow rate to increase throughput brings associated problems such as higher shear stress on the cells, a high pressure drop across the fluidic system, and in the case of isolation based on biochemical affinity, possible cell detachment. To preserve the structural integrity of the cell, the fluidic shear stress should be only on the order of few millipascals.12 A logical solution for processing a large volume with a high throughput is to increase the number of microfluidic devices to perform sorting in parallel. Besides the problem of low throughput, the rarity also means that minimizing sample losses is an important design consideration. If isolated cells need to be transported to different platforms to perform various subsequent steps of the assay, microfluidics can provide an integrated solution that reduces this loss.
In addition to the problem of the small population to be isolated, rare cells often exist with many subtypes that may require multiple isolation steps to sort them.18 For example, there are a number of healthy and benign types of circulating epithelial cells that could be isolated when CTCs are actually targeted. Some rare cells originate from a specific source such as a tumor and circulate with the blood. Hence, the site where the blood is sampled may affect the outcome of the rare cell count. Due to their larger sizes, rare cells may be filtered and blocked by small capillaries before reaching the site of sampling. Rare cells may also be masked by the presence of platelets or coagulation factors, prohibiting isolation concepts based on affinity. Depending on the subsequent analysis steps, the isolation concept should be gentle enough to preserve the integrity of the cell membrane and the genome in the nucleus. Ideally, the cells should be viable for subsequent culturing or single-cell molecular analysis. Isolation based on affinity relies on biochemical markers that may be variably expressed on a cell membrane, leading to missing rare cells or their subtypes. Clogging and damage caused by sample transport in the microchannel present additional challenges to the design of microfluidic devices for isolation and analysis of rare cells.
Because of the above challenges, a comparison between different isolation methods and the use of isolated rare cells as biomarkers require a standardized protocol for sampling and pre-isolation processing. Several parameters can be used to evaluate the performance of a microfluidic device for rare cell isolation, namely: efficiency, recovery rate, yield, purity, and viability. The efficiency (which is also termed as the recovery rate or yield) is the ratio between the number of isolated cells at the exit of the device over the number of known targeted cells introduced at the inlet. Thus, the efficiency can only be determined in experiments with spiked cell samples. Another common measure of efficiency is enrichment, which is the ratio of the volumetric concentration of the isolated cells in the device with respect to the original supplied sample. The purity is the ratio of isolated target cells with respect to the total number of captured cells. The viability is the percentage of cells surviving the isolation process with respect to the total number of target cells. The performance of cell isolation may depend on the signature used in a given method. Following, common signatures that can be used for isolating rare cells are discussed.
Hydrodynamic signatures: size, density, deformability (Young's modulus), morphology
Rare cells can be distinguished by several physical properties including their size, density, deformability and morphology. Most epithelial cells such as CTCs have sizes on the order of 15 to 25 microns, and are larger than red blood cells (RBCs) (6-9 microns) and white blood cells (WBCs) (8-14 microns). However, in contrast to blood cells with a narrow size distribution, rare cells may have a wide size range. To further complicate isolation based on size, the size of rare cells is often dynamic since the same cell type may change in size over time depending on environmental conditions. Mechanical filtration is an effective method to separate cells based on size. A number of microfilter designs are available. Microfilters are also well suited for the separation of cells with different deformabilities. A mismatch in density also allows for isolation based on buoyancy or inertial forces. The availability of high-speed, digital microscopy allows for real-time monitoring of cells in a sample. Differences in shape or morphology can be automatically captured using real-time image processing.
Dielectrophoretic signature: specific membrane capacitance
Cells become polarized in a non-uniform electric field and can be manipulated by the induced dielectropheresis (DEP) force. DEP signatures not only vary among different cell types, but also among different activation states of the same cell type. The DEP force is tuned by varying the applied electric field strength and the alternating current (AC) frequency. Varying these two parameters would allow for the selective sorting of a given cell type.
Immunochemical signature: specific antibodies
The adhesion force between a cell and a surface labeled with antibodies is proportional to the capturing surface area and the contact area with the cell. The proportionality factor is the bond strength between a single antigen-antibody complex.19 The adhesion force needs to balance the drag force of the flow. Thus, an upper limit for the flow rate and the throughput can be estimated for isolation based on this affinity. The shear stress needed to overcome this cell adhesion is on the order of few millipascals. Isolation methods based on the immunochemical signature is further subcategorized as either positive or negative isolation. In positive isolation, the target cells are captured and remain inside the microfluidic device. In negative isolation, all the other cell types are captured or filtered inside the device allowing only the intact target cells to proceed to the exit.
Magnetophoretic signature: magnetic susceptibility, or immunospecific bonding to magnetic nanoparticles
The magnetic force is a body force. However, in the length scale of microfluidic devices, similar to the inertial force, magnetic forces are useful for cell isolation.20 Magnetophoresis is the motion of a magnetic particle in a diamagnetic medium, or conversely, of a diamagnetic particle in a magnetic medium. In order to make the target cells magnetic, magnetic particles coated with specific antibodies can be used to selectively bond to a particular cell type. The magnetic susceptibility of a cell type can also work as a magnetophoretic signature for isolation. Since most cells are diamagnetic, the sample should be mixed with a magnetic fluid to provide a mismatch in the magnetic susceptibility and the corresponding magnetophoretic force.
Rare cell isolation methods and devices
The design process of a microfluidic device for rare cell isolation may start with determining the key operating parameters such as sample size, throughput, separation time and rarity. These parameters will guide to a decision on channel dimensions and possible parallelization. With the given geometry and fabrication constrains, a suitable isolation concept and device design based on one of the above signatures can then be selected. The following section discusses various design examples of recently published works according to the respective signatures. The highlighted advantages and disadvantages could be a good decision support for researchers tasked with designing, fabricating and testing a microfluidic device for rare cell isolation.
Isolation based on hydrodynamic signatures
As mentioned above, isolation of rare cells can take advantage of the difference in their physical properties (size, deformability, density, etc.), which are independent of their biochemical markers. Cell separation techniques based on physical properties share some common advantages. First of all, the approach is label free. These separation methods only rely on the physical properties of the cells, requiring no immunostaining or antibody labeling. Expensive chemical reagents are not needed, decreasing the sample preparation time as well as the risk of cell damage. Second, a high sorting throughput is possible for cell separation based on physical characteristics. With a proper design, this separation method is able to handle a large amount of cellular samples in a short time. For example, an advanced filtration technique can process 7 ml of whole blood within minutes.21 Third, these separation techniques are highly reproducible and convenient. The flow rate, sample concentration, and device geometry are usually the key factors for the successful separation of a given cell type. Once the working conditions are optimized for a cellular sample, the protocol could be validated for many clinical applications. With these advantages, the sorting approach based on physical properties has been applied to a variety of cells such as isolation of CTCs from blood cells, parasites from blood cells, as well as WBCs and platelets from whole blood samples.22-25 On the other hand, there are limitations in selectivity due to the overlap in physical properties of the target and non-target cells.26,27 Furthermore, external factors such as the medium viscosity (plasma in separation of blood cells) and cell-cell interactions in high concentrations may undermine the separation performance. As a result, more efforts are still needed to overcome these challenges. In the past few years, many microfluidic devices have been developed for separating rare cells by their size and deformability. This section discusses particularly three approaches: filtration, deterministic lateral displacement (DLD), and inertial-flow-based methods. All three methods share the advantages mentioned above, but have their own distinct features and technical challenges, which will be addressed in detail.
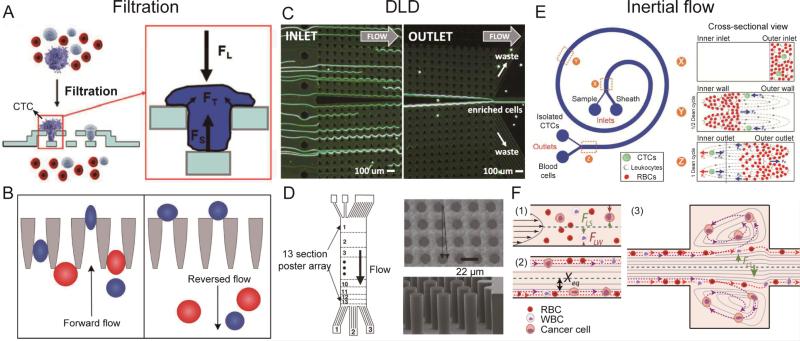
The filtration technique separates cells based on variations in both size and deformability.28 The cellular sample flows through filters made as micropillars, porous membranes, or microweirs.29-31 Cells larger than a critical size or with lower deformability are trapped on the inlet side of the filters or retained in the filter arrays. Smaller cells or those with a higher deformability can pass through the filter and are collected at the outlet.32 Two advantages are often highlighted for the use of filtration techniques: the ability to scale up for a high throughput by numbering up filtration units, and the simple design geometry for separating different cell types. Recently, interesting works have been reported on filtration technique with improved performance.115 However, this technique still needs to overcome some challenges. The major issues of filtration are the integrity of cells that squeeze through the filtration pores. The filtering process often results in changes in the cytoskeleton and cell damage. Zheng et al. developed a 3D filtration system with two porous PDMS layers (Fig. 1A).21 When tumor cells were trapped in the filtration pores of the top layer, the bottom layer provides support in the opposite direction and thus effectively reduces the concentrated tension stress on the cells. This design allows the collection with high percentage of intact of tumor cells (>85%) at a high driving pressure (0.5 psi) as well as flow rate (3-5 min for 9 ml of blood sample). Clogging and saturation of the filter is another problem which may result in irregular flows and a loss of the filtration efficiency. To solve this problem, McFaul et al. employed the ratchet structure and an oscillatory flow to prevent clogging (Fig. 1B).33 With the help of a reverse flow, no clogging and degradation of the device functionality was observed during continuous operation of more than 4 hours.
Fig. 1.
(A) Filtration using a 3D microfilter device and (inset) the applied forces on a trapped cell. FL: force caused by the fluidic flow pressure. FS: supporting force from the bottom membrane. FT: tensional stress force on the plasma membrane. (B) Microfluidic ratchet cell-sorting mechanism. Smaller and more deformable cells can squeeze through the funnel constrictions during forward flow. However, they are unable to pass back through the funnels when the flow direction is reversed periodically to unclog the filter. (C) A DLD device with one inlet and two outlets (collection and waste). Using a symmetrical design, large cells dispersed in the inlet are focused against the central channel wall, where they can be collected at the collection outlet, while smaller cells enter the waste outlet. (D) A DLD device designed to separate WBCs from RBCs and platelets. 13 sections of post arrays with different critical diameters and spacings were used to separate cells with a range of diameters. (E) Schematic illustration of the separation principle for high-throughput CTC isolation using Dean Flow Fractionation. Under the influence of Dean drag forces (blue arrows), the smaller hematologic cells migrate along the Dean vortices towards the inner wall, then back towards the outer wall again. The larger CTCs will experience additional strong inertial lift forces (red arrows) and focus along the microchannel inner wall, thus achieving separation. (F) The principle of a vortex chip based on inertial forces. At the channel inlet, cells are randomly distributed and experience two opposing lift forces, the wall effect FLW and the shear-gradient lift force FLS. As a result, cells migrate to dynamic lateral equilibrium positions, Xeq. Upon entrance into the reservoir, the wall effect is reduced. Larger cells still experience a large FLS and are pushed away from the channel centerline into the vortices. Smaller cells do not experience enough FLS and remain in the main flow. Images reproduced from Refs. 21, 33, 35, 25, 38, and 39.
In the DLD technique, the fluid flows past an array of microposts. Each row of the posts has a lateral offset from the previous row.34 Particles with a size below a critical hydrodynamic diameter follow the periodic streamline patterns through the gaps, and are able to navigate past the posts. Particles with a size above the critical hydrodynamic diameter cannot follow a streamline but bump against the posts, and are displaced laterally, opposite to the small particles. Hence, particles with different diameters can be continuously separated and collected at different outlets. DLD has demonstrated its capabilities in high-throughput CTC separation from a blood sample. Loutherback et al. showed that an optimized DLD array (Fig. 1C) can isolate spiked cancer cells from blood cells with efficiency greater than 85% at a flow rate up to 10 mL/min.35 The DLD approach can effectively separate cells with different sizes, as well as cells with different shapes and deformability. This technique provides a morphology-based method for cell classification and identification.36 Furthermore, DLD has shown one unique property where a simple array design can be used to separate multiple types of cells simultaneously. To separate cells with a range of sizes, a number of different post arrays were designed one immediately after another. Each array has a slightly different critical diameter. Using the method shown in Fig. 1D, Davis et al. successfully separated white blood cells, red blood cells, and platelets from plasma.25 The discriminating resolution or the difference in size of DLD separation can be as small as 10 nm.34
Rare cells can be separated using the inertial lift force, which is related to the particle velocity in a confined microfluidic channel. Since the particle size determines the magnitude of the lift force, microfluidic devices with special designs are used to induce inertial forces on cells with different sizes and deformability.37 Inertial-based separation is either performed by continuously separating multiple types of cells, or by enriching one specific cell type. In the first category, Hou et al. demonstrated Dean Flow Fractionation (DFF) where a spiral microchannel (Fig. 1E) was used to generate centrifugal forces for continuous, size-based, one-step separation of CTCs from blood.38 Using cancer cell lines, the device performance was optimized to achieve a recovery rate of more than 85%, and a throughput of 3 mL/h. The device was also validated by positive CTC enumeration in samples from patients with metastatic lung cancer. In the second category, Sollier et al. developed multiple expansion-contraction reservoirs placed in series and parallel to a straight main channel (Fig. 1F).39 When the sample flow had a high Reynolds number (Re), cells larger than a critical diameter were isolated in the reservoirs, while any cells smaller than the critical diameter were washed down the channel. This design successfully extracted and enumerated CTCs from the blood of patients with breast and lung cancer, which were enriched to a high purity with limited leukocyte contamination (purity ranged from 57 to 94%). A short processing time (20 min for 7.5 mL of whole blood), and a high final concentration (final volume down to 300 μL) were achieved. Inertial microfluidics has also been used to separate cells with different deformabilities. Hur et al. designed an inertial microfluidic channel that took advantage of the balance between a deformability-induced force and an inertial lift force.40 In contrast to benign breast cancer cells (MCF7), those with increased metastatic potential (i.e., modMCF7 cells) tend to migrate towards the flow centerline despite the similarity of their diameters. Compared with the other two separation methods, inertia-based sorting is a contactless approach. During separation, cells suspend in the fluid and do not contact any solid surface, minimizing cell damage.41 On the other hand, the magnitude and direction of the lift force in inertia-based cell separation are influenced by multiple parameters including the channel dimensions, the cross-sectional aspect ratio, the particle diameter, flow rate, and particle-particle interactions.37 Thus, the design and operation of inertia-based fluidic microdevices for rare cell separation requires a detailed theoretical background and practical experience.
Isolation based on dielectrohporestic signature
Another label-free isolation approach is dielectrophoresis (DEP), which has been widely studied in microfluidics. DEP has been demonstrated to be capable of separating cells based on the variations in the dielectric properties of different cell types.42-46 Recently, DEP-based separation of rare cells, including oral cancer cells, colorectal cancer cells, prostate tumor-initiating cells, and melanomas, has been reported.47-51 By using a DEP technique, a high recovery rate and purity can be achieved with an optimized flow rate, where the DEP force acting on a target cell is larger than the fluidic drag force. The dominant DEP force allows the cell sample to move towards or away from the electrodes, rather than following the sample flow.
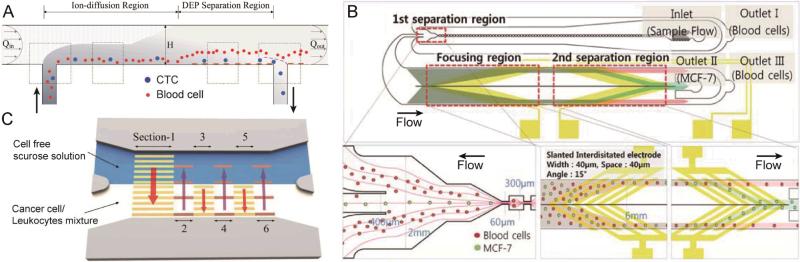
In order to obtain a significant number of rare cells for analysis, the minimum volume of a sample that needs to be processed is usually about 5-10 mL. This leads to issues associated with processing throughput. In order to improve the throughput, DEP-based sorting techniques combined with hydrodynamic approaches have been proposed. Shim et al.52 recently reported a continuous flow-based dielectrophoretic field-flow-fractionation (DEP-FFF) technique53-56 as shown in Fig. 2A. This technique is capable of processing 10 mL of sample in less than 1 hour. As cells passed over the electrodes, the target cells were attracted towards the bottom of the channel because of a positive DEP force, while non-target cells were forced away from the bottom because of the negative DEP force. Thus, the two cell types were brought to different heights in the channel, and eventually were separated due to the balance between sedimentation, hydrodynamic lift, and DEP forces. The target cells, after being restored to an equilibrium height, were finally collected at one outlet. This technique was tested with various tumor cell types spiked into peripheral blood mononuclear cells (PBMNs) with a resulting average recovery rate of ~75%. In their design, cells were separated in the vertical direction with little restriction on the width of the microchannel. Therefore, increasing the channel width can potentially further increase the throughput, which is usually difficult to realize with conventional lateral DEP separation methods.57-59
Fig. 2.
(A) A continuous-flow chamber based on dielectrophoretic field-flow-fractionation (DEP-FFF) to isolate tumor cells from peripheral blood mononuclear cells (PBMNs). (B) Schematic diagram of a microfluidic device for cancer cell separation using multi-orifice flow fractionation (MOFF) and DEP. In the first separation region, the relatively larger MCF-7 cells and a few blood cells pass into the center channel and enter the DEP channel, while most blood cells exit at Outlet I. In the focusing region, all cells experience a positive DEP force and then align along both sides of the channel. Finally, the second separation region selectively isolates MCF-7 cells via DEP. (C) An illustration of a microfluidic device using an optically induced-dielectrophoretic (ODEP) force for cancer cell isolation. Six sections of animated (moving in the direction of the red arrows) light-bar screens were digitally projected onto the CTC isolation zone. Images reproduced from Refs. 52, 62, and 66.
Another strategy to improve the performance of DEP techniques is to employ a pre-separation step prior to DEP isolation. By integrating DEP with multi-orifice flow fractionation (MOFF),60,61 Moon et al.62 were able to separate human breast cancer cells from diluted blood as shown in Fig. 2B. In this work, rare cells were serially separated in two steps. First, a majority of the blood cells were removed by MOFF, while most of the CTCs with a small number of blood cells were delivered into a subsequent separation region. In this region, DEP forces were used to further purify and enrich the target sample by isolating the CTCs into a smaller outlet channel. This separation scheme showed that after two serial sorting processes, CTCs were separated from blood cells with a recovery rate of 76%, and enriched by ~160 fold. In addition, the use of a sheath flow was not needed because an array of DEP electrodes was implemented for cell alignment before the second separation step. However, when adopting a multi-step separation method, the accumulated loss of the target cells inherent in each step may become significant and should be taken into account.
In addition to physical electrodes, an optically-induced-dielectrophoresis (ODEP) technique has also been proposed,63-65 allowing for tunable and dynamic electrode patterning in real time. Huang et al.66 utilized virtual dynamic electrodes to separate CTCs from a leukocyte background with a novel design, as shown in Fig. 3C. Six moving light-bar screens defined the separation region, which were divided into two groups (identified as sections 1, 3, and 5; and sections 2, 4, and 6 in Fig. 3C). These two groups of light-bars screened in opposite lateral directions. As the sample passed through the separation region, the cancer cells were gradually pulled out into the buffer flow, while the leukocytes were confined in the sample stream, due to the differences in their sizes and dielectric properties. The novelty of this design is the generation of a series of independent and localized ODEP electrodes. This allows for the introduction of alternating, opposing forces in the separation region to improve the selection sensitivity, which was rarely reported in previous studies. Although the throughput still needs to be improved before it can be applied to handle larger sample volumes, this concept is worth further investigation.
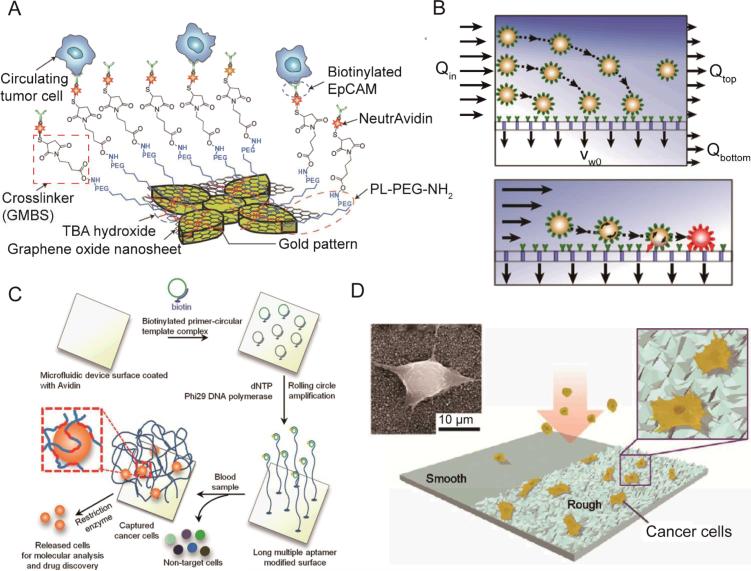
Fig. 3.
(A) Schematic showing the chemical conjugation between the functionalized graphene oxide nanosheets and the EpCAM antibodies. Graphene oxide nanosheets are adsorbed onto the gold pattern. The N-γ-maleimidobutyryloxy succinimide ester (GMBS) crosslinker binds to PL–PEG–NH2 on the graphene oxide nanosheets. The NeutrAvidin is connected to the GMBS and biotinylated EpCAM. (B) Enhanced cell transport to a fluid-permeable capture surface is achieved by diverging streamlines. Gentle cell rolling and arrest on the capture surface occurs due to reduced shear and increased cell-surface interactions. (C) Isolation and detection of cancer cells in whole blood using a long, multivalent DNA aptamer-based microfluidic device. The magnified box illustrates a captured cell bound by several long DNA molecules via multiple aptamer domains (red colored sections). (D) Schematic of a nanotopography generated by RIE on a glass surface. The inserts show a magnified illustration (right) and SEM (left) images of cancer cells captured on the nanoroughened glass surfaces. Images reproduced from Refs. 90, 91, 92, and 99.
Isolation based on immunochemical signature through surface interaction
The interaction between a solid surface and cells has been explored as another successful strategy for rare cell isolation. The adoption of this strategy has been accelerated by the recent development of microfluidic technologies. The large surface to volume ratio of microfluidic channels significantly increases the possibility of cell-to-surface interactions and leads to a better isolation performance. Compared to other approaches using immunochemical signature, surface-based cell separation requires fewer or no sample preparation steps.67-74 Labeling and modification occurs on the surface of the device instead of on the surface of the cells. This approach shortens the overall assay time and simplifies the overall isolation protocol. Fewer sample preparation steps are crucial for practical applications as the loss of the precious rare cells can be minimized. Generally, surface-based cell separations rely on the differential adhesion potential between target cells and non-target cells. The differential adhesion between cells can be created either through immobilizing immunoaffinity-binding molecules or implementing specific surface patterns. In positive selection mode, target cells should preferably bind to the surface, whereas non-target cells have minimum specific interactions with the surface. After applying an optimum shear stress (typically by adjusting the flow rate), the separation of target cells from non-target cells can be achieved. The “optimum shear stress” is the balance between the separation efficiency and the purity. In practice, it is almost impossible to find a shear stress level for both best separation efficiency and highest purity. Thus, it is always important to understand which parameter is the more critical for the specific cell separation application. A higher shear stress tends to give a better purity, as it is able to remove non-specifically bound cells more efficiently. Lower shear stress allows for better interaction between cells and the surface and favors higher separation efficiency. For rare cell isolation, finding all the rare cells is always the highest priority, so separation efficiency is more important than purity. However, in practice, the shear stress or flow rate cannot be too low and takes too much time, especially for isolation of rare cells that requires screening a large sample volume. Therefore, the key for using surface-based separation in rare cell applications is to increase the throughput as much as possible while maintaining a high efficiency.
Due to the dominant laminar flow regime, cells tend to follow the streamline in microchannels. However, laminar flow also prevents effective interaction between cells and the surface. Thus, many previous studies focused on modifying the surface of microchannels to enhance cell-surface interactions. Nagrath et al. fabricated 78,000 silicon microposts in a microchannel and reported the successful capture of CTCs using the anti-EpCAM antibody from clinical samples.75 The large amount of microposts significantly increases the surface area for cell capture compared to flat straight channels. Moreover, the possibility of cell-surface interaction is further enhanced if laminar flow streams are disrupted by the staggered array of microposts. Gleghorn et al. used a micropost array to create size dependent enhancement of cell-surface interactions.76 The array of microposts favored the capture of larger cancer cells over the smaller blood cells, resulting in better capture purity. Sheng et al. also reported the use of micropost structures for capturing CTCs using an aptamer coated surface.77 In addition to the micropost strategy, Soper and coworkers reported a CTC capture device with high aspect ratio (35 μm width and 150 μm height) and sinusoidal-shaped channels.78-81 These optimized microchannels lead to the successful capture of CTCs from 1 mL blood sample in 37 min. Stott et al. implemented herringbone structures in a microfluidic channel to enhance cell-surface interaction.82 The herringbone structures generate chaotic advection enabling rapid fluid mixing. The resulting chaotic flow also helps to increase the chance of cell-surface interaction. Stott et al. showed that a capture efficiency of 92% was achieved with a similar flow rate to the micropost-based CTC-chip. The herringbone chip not only improves the performance of the previous device but also simplifies the fabrication process. This design has been adopted by other surface-based rare cell capture devices.83,84 Recently, Sheng et al. further enhanced the capture efficiency and purity of the herringbone chip by increasing the groove widths, and applied the optimized device to monitor patients’ response to therapy.85
Since specific binding only occurs on the surface, modifying the surface is another important direction to improve the performance of isolation devices based on surface interactions. Improved efficiency has been reported from using surfaces patterned with nanostructures. Nanostructures on the surface facilitate cell binding by increasing the surface area and disturbing the flow. Wang et al. patterned the surface with silicon nanopillars and showed improved cell capture as compared to a flat surface.86 Combining herringbone structures in a microfluidic channel, a capture efficiency of 95% for MCF-7 cells were achieved with a flow rate of 1 mL/h.84 Halloysite nanotubes, multivalent DNA nanospheres, and TiO2 nanoparticles have been studied and demonstrated their abilities to enhance cell capture on the surface.83,87-89 Recently, Yoon et al. reported the isolation of CTCs on functionalized graphene oxide nanosheets.90 Flower-shaped gold patterns (58,957 flowers over 100×100 μm) were first deposited on a flat silicon substrate (Fig. 3A). Graphene oxide was then adsorbed onto the gold surface. After coated with anti-EpCAM, the device isolated MCF-7 cells with a capture efficiency of 82% and a throughput of 3 mL/h. Mittal et al. fabricated a porous membrane for cell capture in a sandwiched microfluidic flow channel (Fig. 3B).91 The pore size of the membrane is 200 nm, allowing the fluid to pass while retaining cells on the surface. Since the fluid is able to pass along the bottom surface, an additional flow towards the surface exists and forces cells to interact with it. As a result, this device allows for a much higher flow rate while maintaining the same level of capture efficiency. The authors reported a 20-fold increase in flow rate as compared to a solid surface device. Inspired by creatures of nature such as jelly fish, which use long tentacles to capture flowing particles, Zhao et al. immobilized long chain DNA molecules (10−100 micrometers) on the channel surface (Fig. 3C).92 The DNA molecules contain repeated specific aptamer sequences to recognize target cells. The long-chain DNA molecules extend into the flow and increase the accessibility of aptamer to cells, avoiding the detrimental boundary effects at the surface. The authors compared the aptamer-coated device with a CTC-chip and a silicon nanopillar chip, and demonstrated a higher capture efficiency at a flow rate of 2 mL/h.
One major limitation for surface-based isolation of rare cells is the difficulty in releasing the viable cells after capture. Dead cells after separation may affect subsequent analysis. Releasing with elevated shear stress has a low efficiency and causes excessive dilution of the sample. More importantly, a high shear stress very likely damages the cells. Another common option for releasing isolated cells is enzyme digestion. However, enzyme digestion may require a prolonged digestion time and damages cells during the digestion process. Shen et al. replaced the capture antibody with an aptamer for capturing lung cancer CTCs.93 Because an aptamer is an oligonucleotide, it can be cleaved using a nuclease solution without disturbing the cells. Several groups employed thermal responsive materials to achieve the highly efficient and viable release of cells.94-97 Poly(Nisopropyacrylamide) (PNIPAAm) is a widely used thermal responsive material for releasing captured cells. PNIPAAm is hydrophobic at a temperature above its lower critical solution temperature (LCST) of 32 °C. When the temperature is below 32 °C, the material becomes hydrophilic due to the hydration of its molecules. A hydrophilic surface is not preferable for protein adsorption, so cells can be removed with a mildly elevated shear stress. After release, a viability of 94% has been reported. Plouffe et al. utilized an aqueous alginate solution and achieved the release of viable cells.98 At room temperature, aqueous alginate can form a hydrogel after adding calcium ions. This process is reversible. A calcium ion chelator, such as EDTA, can transform the hydrogel back to its liquid state and allow for the release of the cells. No significant change in cell viability was reported from using this process.
Fu et al. reported the successful specific capture of cancer cells on glass surfaces with a random nano-roughness (Fig. 3D).99 The separation of cancer cells from blood cells was achieved through their differential adhesion preference. Cancer cells are more likely to attach on the rough glass surface generated by reactive ion etching (RIE). Compared to other surface-based cell capture methods, this method is unique because it does not require specific recognition molecules. The method has the potential to become an important complementary approach to the existing antibody-based surface capture methods. However, this strategy still needs more validation using clinical samples before it can be adopted widely.
Isolation based on magnetophoretic signature
Isolation method based on magnetophoresis has been developed for years as a relatively mature and reliable technique for microfluidic-based rare cell isolation.100-104 This concept uses a magnetic field gradient to selectively isolate magnetic-particle-labeled cells from the sample flow, and thus is also called immunomagnetic technique. Compared to the passive hydrodynamic approaches, such as inertial microfluidics and DLD, immunomagnetic-based separation actively applies a strong magnetic force to extract the magnetic-particle-labeled cells, which is less dependent on the flow conditions and particle-particle interactions. Therefore, this technique is able to achieve a higher specificity, and is possible to process whole blood in a high-throughput manner.105-107 Furthermore, the immunomagnetic technique is a biocompatible approach with low damage to the cells. Although a high-gradient magnetic field is used, the forces are not directly applied on cells. It has been reported that almost 100% viability can be achieved with this method.108 The most obvious limitation of this technique is the requirement for a label, and hence additional sample preparation steps are required.
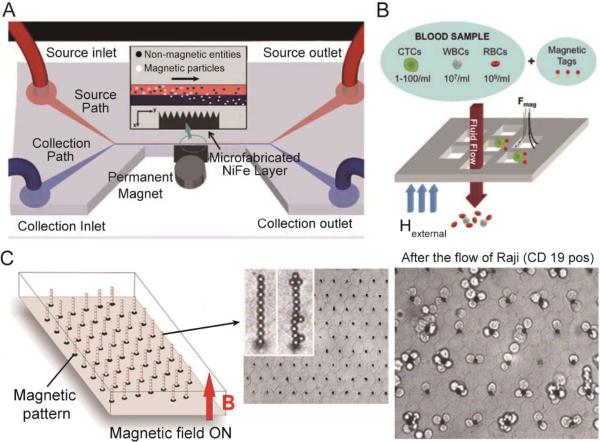
Among the immunomagentic approaches, lateral magnetophoresis is a common method for cell separation. This method utilizes a horizontal magnetic field to drag labeled target cells from the sample flow into the buffer flow (Fig. 4A).109 Lateral magnetophoresis has been demonstrated as a robust technique for rare cell separation with a high throughput and a high recovery rate. A typical example is the magnetophoretic microseparator reported recently by Kim et al. to isolate CTCs from human peripheral blood.108 After specifically binding to immunomagnetic beads, CTC isolation was performed via lateral magnetophoresis. Diluted blood samples spiked with CTCs were tested with this device, achieving a high recovery rate of ~90% and a purity of 97% at a flow rate up to 5 mL/h. However, this design requires a buffer flow and may dilute the collected target sample. Zhang et al. replaced the lateral magnetic field gradient by a vertical version, and isolate rare cells from a continuous blood sample flow. This method does not require any buffer flow and allows for simultaneous sample concentration with a high capture efficiency.106, 110
Fig. 4.
(A) Schematic of a magnetophoretic microdevice with two inlets and two outlets. Inset shows how magnetic beads flowing from the upper source path are pulled across the laminar streamline boundary into the lower collection path when subjected to a magnetic field gradient. (B) Capture principle of a magnetic sifter. A whole blood sample is labeled with magnetic tags and pumped through the pores by an applied external magnetic field. Magnetically labeled target cells are captured at the pore edges where high magnetic field gradients exist. Unlabeled cells pass through the pores. (C) Operating principle and practical implementation of the Ephesia system. A hexagonal array of magnetic ink is patterned on the bottom of a microfluidic channel. The application of an external vertically-aligned magnetic field induces the formation of a regular array of magnetic bead columns localized on top of the ink dots. After the passage of 400 Raji cells, numerous cells are captured on the columns. Images reproduced from Refs. 109, 107, and 111.
Unlike conventional magnetophoresis,108 which images and analyzes the isolated rare cells on a separate cover slip, Earhart et al. recently developed a magnetic sifter for integrated on-chip separation, imaging, and analysis of rare cells as shown in Fig. 4B.107 A magnetic porous structure enabled the passage of cellular samples and generated sufficient magnetic forces to capture the labeled cells. The isolation of rare cells was simply performed by passing the sample through the magnetic sifter. Upon the application of an external magnetic field, CTCs labeled with magnetic beads were trapped at the pore edges, while non-labeled blood cells were removed by the flow. CTC isolation from human whole blood was realized with this magnetic sifter at a flow rate of 10 mL/h, achieving a high recovery rate of ~90%. The direct contact between the magnetic material and the sample flow is crucial for improving the isolation efficiency. Moreover, by removing the external magnetic field, isolated rare cells were released and collected at the outlet after on-chip imaging and analysis.
As mentioned at the beginning of this section, the requirement for a label in this technique requires an additional sample pretreatment step. To address this issue, Viovy et al. invented an unconventional immunomagnetic separation method shown in Fig. 4C.111 Instead of labeling the target cells, antibody-conjugated magnetic beads were dynamically assembled into an array of columns in the microfluidic channel with the help of the pre-patterned magnetic traps on the substrate. As a result, when the sample flows through the channel, cells with specific biomarkers are captured by the assembled magnetic beads. Recently, this device has been applied to CTC isolation by Svobodova et al.112,113 Though the sample preparation step was avoided by this method, labeled magnetic beads are still needed. The mechanism of this method is similar to the immunocapture-based cell isolation. However, the superiority of this method lies in the dynamically assembled microstructures (magnetic microbeads), allowing for controllable release and reconstruction using an external magnetic field. To achieve high-throughput rare cell isolation, the magnetic forces for assembly need to be strong enough to overcome the high flow velocity.
Summary on rare cell isolation methods and devices
Table 1 summarizes the performance of each rare cell isolation technique introduced in this section in terms of volumetric flow rate, efficiency, purity, and cell viability. Due to the differences in cell types and sample concentrations in each study, we are unable to draw a definitive conclusion about which technique is superior based on the data reported. Even using the same mechanism, devices with different designs varied in their results. However, some general patterns are observed and can be summarized into design rules. The cell samples used in these studies reveals that many research works have focused on CTC isolation from whole human blood, because of their great clinical significance in cancer diagnostics and therapy. Nevertheless, these techniques have also been applied to isolate stem cells, parasites, and bacteria, demonstrating their general applicability. From an engineering point of view, cell isolation based on physical properties can obtain a higher volumetric flow rate than other isolation techniques. Filtration, DLD, and inertial fluidic techniques have been reported with an operating flow rate of more than 1 mL/min. This high flow rate is rarely found in other techniques. However, the flow rate does not reflect an actual throughput, which is defined as the total amount of cells processed per unit time. As a result, the sample concentration should also be considered. In this respect, the immunomagnetic and immunoaffinity-based methods show a higher ability to process human whole blood without any dilution. Other techniques including physical-property-based isolation, as well as DEP approaches, usually require sample dilution, and some of them need sheath flows or buffer flows as well. Most of the studies have characterized the efficiency but only some of them reported the purity, because the capture efficiency is more important for rare cell isolation. Generally, a higher efficiency can be achieved by decreasing the flow rate. Therefore, a compromise should be found between the isolation efficiency and throughput. The purities in different studies were varied from 10% to 90%, while an efficiency higher than 80% is usually preferred. The average efficiency of all the studies listed in Table 1 is calculated to be ~82%. Moreover, most of the reported cell viabilities are higher than 90%.
Table 1.
Summary of various rare cell isolation techniques
| Techniques | Sample type | Flow rate | Efficiency | Purity | Viability | Ref. |
|---|---|---|---|---|---|---|
| Filtration | MCF-7 btrast cancer cells | 3 mL/min | 87% | 85% | 21 | |
| Breast, gastric, and colon tumor cells | 1 mL/min | >80% | 98% | 22 | ||
| Breast and colon cancer cells | 12 μl/min | >80% | >80% | 32 | ||
| Hematopoietic stem cells | 17.2 μL/min | >98% | >90% | 114 | ||
| MCF-7 breast cancer cells | 25-100 μl/min | 90%-50% | 115 | |||
| DLD | MDAMB231, PC3, MCF10A cancer cells | 10 mL/min | >85% | >95% | 35 | |
| T. brucei parasites in blood | 60 nL/min | 99% | 23 | |||
| Inertial fluidics | HeLa and MCF7 cancer cells | 23% | 85% | 116 | ||
| MCF-7, MDA-MB-231, HeLa cells | 50 μL/min | >85% | ~10% | 98% | 38 | |
| MCF-7, T24 and MDA-MB-231 cells | ~1 mL/min | >80% | >90% | 117 | ||
| Nucleated cells in blood | >30 μL/min | >90% | 118 | |||
| MDA-MB-231 breast cancer cells | 565 μL/min | 85% | 119 | |||
| DEP | SKOV3 and MDA-MB-231 cancer cells | 18-25 μL/min | 68.3% | 97.1% | 54 | |
| rare bacteria in blood | 300 μL/h | 95% | 42 | |||
| MDA-MB cancer cells | 301 μL/h | 70%–80% | 52 | |||
| MDA-MB cancer cells | >90% | 10% | ~90% | 56 | ||
| prostate tumor initiating cells | 4 μL/min | 61%-70% | 50 | |||
| Immunocapture | leukemia cells | 36 μL/min | ~95% | ~81% | ~93% | 77 |
| Circulating tumor cells | 200 μL/min | 92% | 14% | 95% | 82 | |
| Lymphoblast CCRF-CEM cells | 60 μL/h | ~80% | ~38% | 66% | 92 | |
| Circulating tumor cells | 34 μL/min | 65% | 50% | 98% | 75 | |
| Prostate cancer circulating tumor cells | 17 μL/min | >80% | 62% | 76 | ||
| Immunomagnetic | M6C breast cancer cells | 200 μL/min | 90% | > 90% | 103 | |
| MCF7 breast cancer cells | 60% | 50% | 100 | |||
| C. albicans fungi in blood | 330 μL/min | 80% | 105 | |||
| H1650 cancer cells | 170 μL/min | 96% | 107 | |||
| COLO205 and SKBR3 cancer cells | 170 μL/min | 86%-90% | 106 | |||
Obviously, every technique has its own limitations. For example, filtration for cell separation is a label-free, high-throughput approach, but its specificity is relatively low due to the possible overlap in cell sizes, making it not applicable to all sample types. On the other hand, cell isolation based on immunocapture has a high specificity, but the throughput is limited by effective cell-surface interactions. One strategy to improve the performance of rare cell separation is the combination of two or more separation mechanisms. Several works have explored this direction, including the use of DEP with immunocapture,120,121 DEP with DLD,122 DLD with immunocapture,123 as well as immunoaffinity with hydrodynamic force.124 Another strategy is to explore novel separation techniques and physical phenomena to overcome the existing challenges in rare cell isolation. For example, in the past few years, acoustofluidics has paved a new way for particle/cell separation.125-127 Recently, Adams et al. demonstrated an ultra high-throughput (1 L/h) acoustophorestic microdevice, which was able to remove RBCs from human whole blood with an efficiency up to 95%.128 Acoustic waves have also been utilized to separate prostate cancer cells from white blood cells (WBCs),129 and sorting of viable MCF-7 breast tumor cells from nonviable cells.130 Although acoustic-wave-based rare cell separation is still at an early research stage, further development and optimization may lead to a useful technique for rare cell separation because of its advantages of high throughput, label-free nature, and noninvasiveness.131-135
Rare cell analysis methods and devices
The subsequent step after isolation of rare cells is their analysis. Cell analysis is important for fundamental cellular studies, drug discovery, diagnostics, and prognostics. The analysis is usually conducted at the molecular level (DNA, RNA, protein, secreted molecules, etc.) or at the cellular level (cell metabolism, cell morphology, cell-cell interactions, etc.). For the analysis of rare cells, a high throughput and a high sensitivity are often required for the chosen analytical methods and devices, because of their extremely low abundance and the associated high level of noise. In this respect, microfluidics provides an ideal platform for rare cell studies, due to the simplified handling of small sample volumes, multiplexing capabilities, and large surface-to-volume ratios.136,137 Rare cell studies using microfluidics are carried out either in stationary fluids or in continuous flows. Both methods have their own scope of corresponding applications. Stationary fluids are usually preferred for cell studies at the molecular level, while both methods have been adopted in the studies at the cellular level. Over the past decade, a variety of microfluidic techniques have been developed based on these two modes, which will be introduced and illustrated below by representative examples. A small number of these techniques have been applied to study specific biological questions, while most of them still remain on the proof-of-concept stage.
Rare cell analysis in stationary microfluidics
In stationary microfluidics, the cell sample is fixed in the microfluidic device for long-term cell culture, biochemical reactions, and signal detection.138 This analysis approach allows for the detection of low-intensity signals by enhancing the exposure time, and also allows for monitoring the dynamic cellular responses with time-lapsed imaging. Here, the advantages of miniaturization techniques over conventional approaches (e.g., cells in a Petri dish culture) manifest in terms of a high-throughput capability, usually by isolating cells in a large multiplex microarray for parallel processing, even at single-cell resolution.139,140 These high-density arrays are often microstructures (microwells, microchamber, etc.) fabricated by photolithography, or microdroplets generated in-situ to encapsulate multiple cells or single cells.136 One example developed by the Quake group using the microstructure-based approach is the fabrication of dynamic arrays on a chip for genetic analysis.141 In their design, thousands of PDMS-based microchambers were created with controllable microvalves for performing single-cell real-time PCR.142-144 This technique has recently been applied to single-cell profiling of CTCs, and provided an early insight into CTC heterogeneity.145
One of the challenges for rare cell analysis at the molecular level is the ability to detect low-intensity signals. The small volumes (from pL to nL) in microdevices restricts signal dilution in the fluidic environment. Some new microfluidic-based bioengineered approaches for the enhancement of detection sensitivities have also been reported in the past few years.146-148 The on-chip rolling-circle-enhanced enzyme activity detection (REEAD) technique presented by Juul et al. is a typical example (Fig. 5A).149 In their studies, the REEAD technique was used to quantify single enzymatic events (hTopl or Flp recombinase activities) in rare cells with a background of wild-type cells. The single cells were encapsulated into an array of picoliter droplets for lysis and enzymatic reactions (top image in Fig. 5A). Since the recombinase reaction products (circular DNA) were too small to be detected using microscopes, an isothermal rolling circle amplification (RCA) was then carried out to convert the circular DNA into a fluorescently-labeled, long-chain tandem repeat products (left bottom image in Fig. 5A). This critical signal-amplification step enabled the visualization of fluorescence signals from the tandem repeat products, and hence the direct quantification of single enzyme activities (right bottom image in Fig. 5A). Although the REEAD assay has been demonstrated in an off-chip manner before, the spreading of signals in a large reaction volume (~9 mm2) makes it less sensitive.150 The on-chip REEAD assay in this work was able to achieve single-cell resolution and the limit of detection (LOD) was increased by >100 fold compared with the traditional “large-volume” assay.
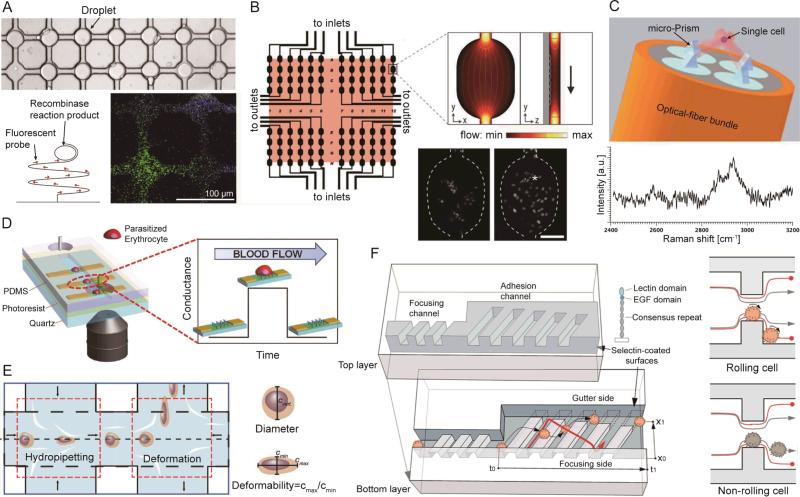
Fig. 5.
(A) A microdevice for rolling-circle-enhanced enzyme activity detection (REEAD) assay for the specific detection of single enzymatic events. The droplets are confined in a drop-trap on a primer-coated glass slide on which isothermal rolling circle amplification (RCA) takes place. (B) Schematic of the microarray layout with 96 wells with the 6 × 5 mm2 scanning area shaded. Inset shows a finite element model of fluid flow through a microwell. The fluorescence images show the H2B-EYFP-labeled PtK2 cells in a single microwell after seeding (left); 12 h later (right), they attached, spread, and divided (*). Scale bar: 100 μm. (C) Schematic of optical trapping of a single cell for Raman spectroscopy. (D) (Left) A microdevice for graphene-based detection of single Plasmodium falciparum-infected erythrocyte. (Right) Specific binding between ligands located on positively charged membrane knobs of infected erythrocyte and CD36 receptors on the graphene channel produces a distinct conductance change. The conductance returns to a baseline value when the parasite-infected erythrocyte exits the graphene channel. (E) A microdevice for hydrodynamic deformability cytometry. The first opposing pair of microchannel branched flows impact the cell suspension perpendicularly to perform hydropipetting. The second pair of channels flows towards the cell suspension, forming an extensional flow to perform deformability cytometry. (F) (Left) Schematic of the microfluidic cell rolling cytometer, in which cells are forced into contact with adhesion molecule-coated ridges. (Right) Cross-section views of the cell rolling cytometer. Specific adhesion interactions retard the cell and change its trajectory. Without specific interactions, a cell quickly travels through the channel, following the focusing trajectory. Images reproduced from Refs. 149, 152, 154, 165, 168, and 170.
Rare cell studies at the cellular level are usually applied for cell identification, monitoring of cellular kinetics, as well as the measurement of cell properties, such as mechanical, optical, and immunochemical properties.137,151 In stationary microfluidics, the microenvironment can better mimic in vivo conditions. Moreover, the miniaturized microdevices allow for fully-integrated parallel processing with improved efficiency and throughput, which are especially important for rare cell events. In a traditional macro-scale setup, cellular kinetic studies are only optimized for either high spatio-temporal resolution or high experimental throughput. To find a balance between imaging resolution and experimental throughput, Albrecht et al. developed a microfludic-based time-lapsed imaging platform for rare cell event detection, as illustrated in Fig. 5B.152 This microdevice contains a 96-microwell array (6×5 mm2 scanning area), divided into four separate fluidic circuits. Each circuit had a common inlet and outlet for sample loading and medium exchange using a pipette. The platform can be used to study the dynamic responses of cells, such as mitotic kinetics and spindle orientation dynamics. At a high imaging resolution, this microdevice was able to achieve a much higher throughput as compared to other traditional approaches. For example, a 5-day experiment conducted on this microfluidic platform achieved comparable results that would have taken more than a month to finish processing using the single-well microscopy. In addition, the day-to-day variability was also suppressed with this platform owing to the high-resolution parallel analysis.
In order to measure the optical properties of cells, it is sometimes necessary to immobilize cells using microfluidics. Raman scattering is a label-free method for cell identification and characterization.153 However, this method requires a longer time to measure the signal compared with fluorescence detection. Recently, Liberale et al. demonstrated an integrated optical tweezers for three-dimensional (3D) immobilization of single cells as shown in Fig. 5C.154 In this setup, a four optical fiber bundle was incorporated into the substrate of the microdevice to generate four independent laser beams. Four micro prisms located at the top of the fibers then refracted the laser lines towards a focal point for 3D trapping of the cells (top image of Fig. 5C). This miniaturized optical tweezers allowed for the long-term interrogation of single cells by immobilizing them at a stable position in a non-contact manner, with more control in the vertical direction compared with conventional 2D optical tweezers. Using this tool, the distinct Raman signature of a colon cancer cell was successfully measured (bottom image of Fig. 5C). However, the throughput was relatively low. To improve the throughput for rare cell scanning, a large array of micro-optical tweezes for simultaneous trapping and measurement is preferred.
Rare cell analysis in continuous-flow microfluidics
Microfluidic platforms have become a powerful tool to investigate rare cells in continuous flow or microfluidic cytometry. Because of its single-cell resolution, the rare cell sample can directly go through the system without prior separation or purification. Fluorescence intensity, wavelength, and light scatter are the most commonly used parameters in conventional flow cytometry.155 Microfluidic cytometry shows a greater diversity in the biomarkers, which enables the analysis of cells from different perspectives. In addition to the conventional optical parameters,156 microfluidic cytometry is also capable of measuring other parameters, such as electrical properties (conductance, impedance),157-159 mechanical properties (deformability, stiffness),155,156 magnetic properties (magnetic resistance, Hall effect),162,163 adhesion behaviors,164 and other optical properties (brightness with two-photon excitation, fluorescence lifetime).155
Electrical signals have been widely used as a characteristic signature in microfluidic cytometry for cell identification, cell counting, and differentiation of cell states, due to its high sensitivity and compatibility to be integrated with microfluidic systems.157-159 Ang et al. developed a graphene-based flow sensor to detect malaria-infected red blood cells in a “flow-catch-release” method as shown in Fig. 5D.165 As the blood sample flows through the microchannel which was integrated with a graphene-transistor array, malaria-infected red blood cells were temporarily captured by the surface-functionalized graphene, resulting in changes in the electrical conductance. By characterizing the conductance dwell times, cells at different stages of infection could be identified. The electrical signal-based flow cytometers can lead to a highly compact and cost-effective system for label-free rare cell sensing by eliminating the need of a high-power laser and an expensive optics system. Moreover, an optics-free approach is much easier to develop a parallel processing model by implementing large-scale integrated electronic circuits.
In the past few years, the mechanical properties of cells has drawn more attention as a biomarker arising from its high correlation to certain diseases.166 A variety of works have been reported to explore the mechanical properties of tumor cells, parasite-infected red blood cells, and stem cells.160,161,167 Recently, Dudani et al. developed a high-throughput (65,000 cells/s) microfluidic cytometer for label-free, cellular-mechanical phenotyping based on pinched-flow hydrodynamic stretching as shown in Fig. 5E.168 The inertially-focused cells were deformed by hydrodynamic forces when they were delivered to the center of an extensional flow. The cell deformability was then characterized with the assistance of high-speed microscopic imaging and an automated image analysis program. The throughput of this work was higher by over an order of magnitude than that reported previously by the same group. This improvement was achieved by the integration of a “hydropipetting” unit, which squeezed cells with perpendicular high-speed pinch flows and thus reducing their residence time at the stagnation point of the extensional flow.
The dynamic adhesion behavior of cells has also been applied as a specific biomarker for phenotyping of rare cells. However, the conventional parallel-plate chambers have poor tethering or contact efficiency, which might reduce their sensitivities and performance.169 To overcome this limitation, Choi et al. recently reported a novel microfluidic cell-rolling cytometer to quantify surface adhesion dynamics of mesenchymal stem cells (MSC) as shown in Fig. 5F.170 This cytometer was constructed by creating an array of 3D ridges, coated with adhesion molecules (receptors) in a microchannel. The cells first flowed into a narrow focusing channel, in which the high shear stress limited cell rolling on the surface. As cells went into the wider adhesion channel, the decreased sheer stress resulted in the interaction between cells and the adhesion ridges. The cells with specific ligands were able to roll on the channel surface due to the combined action of the ligand-receptor interaction and the fluidic drag force. These cells thereby followed the orientation of the ridges into the gutter side. Conversely, the non-interacting cells just followed the flow direction and stayed in the focusing side. Using microscope observations, the transit times and lateral positions of the rolling cells were characterized with single-cell resolution. This rolling cell cytometer was sensitive enough to quantify different levels of cellular adhesion, revealing the differentiation states of MSCs, which is beyond the ability of a conventional flow cytometer.
Applications of rare cell isolation and analysis
The previous sections discussed various microfluidic-based methods and devices for rare cell isolation and analysis. These techniques will need to be further optimized before being applied to different cell types. Besides CTCs, other types of rare cells also have significant value in biomedical research, diagnostics, and therapeutics. In this section, we will briefly introduce the background and properties of several categories of rare cells in order to provide some direction for future applications and optimization of these techniques.
Circulating tumor cells (CTCs)
Carcinoma cells shed from primary tumor sites which circulate in the blood stream are circulating tumor cells. They are considered to be closely related to cancer metastasis which is the leading cause of cancer mortality. Preliminary studies have indicated the role of CTCs in the prognosis and treatment of metastatic cancers.171-173 The first challenge in isolating CTCs is the low abundance of CTCs in a blood sample (normally 1-100 cells/mL), which calls for methods with high throughput, recovery rate, and sensitivity. Second, isolation is more complicated due to the heterogeneity of CTCs. For example, many immunoaffnity methods rely on the expression of epithelial markers (i.e., EpCAM) by CTCs. However, tumor cells that are able to enter the blood stream may undergo an epithelial-mesenchymal transition, resulting in the down-regulation or negative expression of EpCAM.174 Even within the same patient sample, there may be different subpopulations of cancer cells, e.g., cancer stem cells (CSCs).18 To avoid the bias from biochemical detection methods, CTCs can also be isolated based on physical parameters, such as size and deformability. The sizes of most CTCs are estimated in the range of 12-25 μm, which are larger than most of the blood cells.30 But it should also be noted that different types of cancers have different size distributions for CTCs, some of which may overlap with blood cells.175,176 The deformability is another possible parameter to differentiate CTCs and blood cells, but researchers should be aware that the same population of CTCs may have different deformabilities depending on the state of the cells. Zhang et al. reported that cancer cells with a higher metastatic potential showed a high deformability.160
Circulating endothelial cells (CECs) and circulating endothelial progenitor cells (EPCs)
The disturbance of endothelial function and integrity is closely related with many pathological processes, including the presence of an infection, cancer, or cardiovascular disease. One potential non-invasive approach to evaluate the integrity of the endothelium is by detecting circulating endothelial cells in the blood. It has been reported that the number of circulated endothelial cells increases with the level of vascular damage due to a disease.177 Circulating endothelial progenitor cells are considered to originate from bone marrow and play a role in the repair of damaged blood vessels.178 Depending on the detection methods and the specific disease, the concentration of CECs and EPCs in a human blood sample varies significantly with a typical range between 1-10,000 cells/mL.179 There is still no single surface biomarker to identify CECs and EPCs.180 Generally, CECs should have negative expression of a pan-leukocyte marker (CD45) and a primitive haemopoietic cell marker (CD133) and positive expression of endothelial markers (e.g., von Willebrand factor, VE-cadherin, CD 105, and CD146).178,181 CD146 is the most widely used marker for immunomagnetic isolation of CECs.182 For EPCs, they should also be positive for immaturity markers (CD34 and CD133) in addition to the expression of endothelial markers.180 The sizes of CECs range from 20 to 50 μm in diameter, whereas EPCs are normally less than 20 μm since they are immature cells.178
Fetal cells in maternal blood
Prenatal diagnosis of fetal cytogenetic characteristics is critical to the identification of fetal autosomal abnormalities. Currently, this is achieved through amniocentesis or chorionic villus sampling (CVS).183 However, these approaches are invasive and thus increase the risk of fetal loss or maternal injury. Therefore, developing non-invasive methods for prenatal diagnosis is of great importance. Fetal cells circulating in the maternal blood are the ideal candidates for non-invasive examination of the genetic information of a fetus. There are several types of fetal cells in maternal blood that can be used for diagnosis, including trophoblasts, leukocytes, and erythroblasts (nucleated red blood cells (NRBCs)). Among them, NRBCs are a more favored target as they are relative more abundant and have a shorter life-span, which avoids contamination between pregnancies.184 These fetal cells are extremely rare. Current estimates range from 1-2 cells per 1 mL to 1-3 cells per 30 mL.185 Furthermore, the isolation of fetal cells is complicated by the lack of surface makers. For NRBCs, immunomagnetic separation relies on the depletion of CD45+ cells, or the positive enrichment of CD71+ cells.186 NRBCs have diameters ranging from 9 to 12 μm and are reported to be more deformable than white blood cells.29
Stem cells
Stem cells are undifferentiated cells with the potential to develop into many different cell types. Stem cells can be broadly classified into two types: embryonic stem cells and adult stem cells. Adult stem cells are mostly isolated from bone marrow, adipose tissue, and blood, from which there are several different subpopulations. Among the different types of stem cells, mesenchymal stem cells (MSCs) and hemopoietic stem cells (HSCs) are highly attractive and have been intensively studied due to their promise in biomedical applications. Stem cells have a great value in medical therapies because they provide potential solutions for many human diseases. For example, stem cells have been applied to the treatment of leukemia via bone marrow transplantation. High cell purity is often required in these clinical applications. However, high-purity isolation of stem cells is challenging because of their rarity and various subpopulations. Since HSCs are larger than other cells in the bone marrow, size-based techniques are possible for HSC isolation.114 Besides, CD34 has been demonstrated as a useful biomarker for HSCs for immunoaffinity-based or immunomagnetic isolation.187
Infected cells
Cells infected by viruses, bacteria, or parasites may cause cell dysfunction and eventually cell death, resulting in serious health complications. HIV-infected T cells (AIDS) and Plasmodium falciparum-infected RBCs (malaria) are examples of infected blood cells. Early diagnosis of these infections can lead to effective treatment for the diseases. However, the low number of infected cells at the early stage of infection requires techniques with a high sensitivity and throughput. These infections usually induce changes in cell properties, such as mechanical or biochemical properties, which may provide useful biomarkers to isolate infected cells from the healthy ones. Malaria, with a relatively high mortality rate worldwide, has been widely studied using microfludic techniques. The Plasmodium falciparum-infected RBCs have significant changes in cell deformability, as well as changes in magnetic and electrical properties. As a result, different mechanisms have been reported to isolate or analyze infected RBCs.161,165,188,189 Notably, because malaria-infected RBCs become paramagnetic due to a malaria byproduct (hemozoin), a magnetic field gradient can be used to directly isolate infected cells without the need for magnetic beads as labels.188
Bacteria
Bacteria are single-cell microorganisms. Most bacteria have a diameter between 0.5 and 5 μm, which is smaller than most human cells (> 5 μm). Their morphologies vary, including spheres, rods, and spirals. Rapid detection of pathogenic bacteria, such as nontyphoidal Salmonella, E. coli, Vibrio cholera, Shigella, and Campylobacter, is highly important in food safety. Moreover, bacteria circulating in the blood may cause bacteremia and blood poisoning, posing a threat to human health. The current standard method for diagnosis of these diseases is based on a blood culture, which is time-consuming and can easily be contaminated. Some studies have also been reported to enrich and identify bacteria via microfluidic devices, which can be divided into two strategies. One is to enrich bacteria from blood cells based on their differences, such as differences in physical or dielectrical properties.59,190 Another method is conducted at the molecular level by directly amplifying target nucleic acids with microfluidic polymerase chain reaction (μPCR) techniques, followed by an identification process.191
Sperm cells
Sperm cells are produced in the male gonads by meiotic division during the process of spermatogenesis. As male gametes, the morphology of sperm cells is quite different from the somatic cells. Sperm cells have a tadpole-like shape with a head (~5 μm in diameter), a connecting piece, and a tail (~55 μm in length). They contain only one set of chromosomes; therefore, they cannot divide and have a limited life span. As a rare cell, sperm cells are important in forensic investigations, especially in cases involving sexual assault. DNA extracted from sperm cells can be critical evidence used to identify the perpetrator. However, the sperm cells collected via vaginal swabs are usually contaminated with a much larger background of the victims’ epithelial cells. Before DNA profiling, pretreatments of the sample, such as purification and concentration of the sperm cells are necessary.192,193 Differential extraction is the conventional method for sperm cell selection. Sperm cells can also be isolated from epithelial cells based on their significant difference in size.
Conclusions and perspectives
The present paper discusses the various techniques for isolation and analysis of rare cells as well as their applications. Table 1 indicates that it is difficult to compare the different isolation methods quantitatively. A relatively high efficiency above 80% has been reported for most of the reviewed isolation methods. However, a benchmarking standard is lacking to effectively compare the different methods based on efficiency. With a standardized benchmarking method, the results from different isolation methods could be collected into a design bank for effective comparisons of isolation devices. Most of the reviewed works report only proof-of-concept devices. In order to be translated for use in a clinical environment, these devices need to have a high efficiency, a high purity, and a high throughput. Improving cell integrity is another unmet need for future microfluidic devices focused on rare cell isolation. For extremely rare cells, there is also a need to multiply the number of cells via an on-chip culture. Post-isolation cell cultures would increase the number of viable samples and generate enough material for subsequent analyses. Another problem of proof-of-concept prototypes is that the devices were tested with spiked immortalized cell lines, which does not truly represent a clinical sample. Furthermore, the isolation and enumeration of rare cells currently does not take advantage of all the benefits provided by microfluidics. More quantitative approaches can be applied to the target cells on the same device. Since captured cells may represent a variety of subtypes or the same cell type at different stages of growth, subsequent molecular analysis to characterize the genotype and the associated phenotype would improve profiling of the isolated cells. These subsequent analysis steps would be highly dependent on the viability of the captured cells. A gentle and efficient isolation method is therefore needed in the near future. The possibility of isolating different growth stages of the same cell type promises a direct and reliable comparison of the gene expression, which is highly valuable for applications such as personalized cancer treatment.
Although microfluidic devices provide a number of advantages for isolating and analyzing rare cells, their throughput is still an issue when processing a large sample volume. For instance, most of the existing microfluidic devices would need several hours to completely process a standard 7.5 mL blood sample. In general, microfiltration does not require a very small flow channel and may offer a higher throughput than other methods. However, the high flow rate and large gradients in shear stress may damage the isolated cells. Thus, microfiltration is more suitable for enumeration of rare cells in situations without subsequent analysis steps. When optimizing for a high throughput, more gentle capturing methods such as dielectrophoresis, magnetophoresis, and acoustophoresis are better alternatives. However, there have not been many reported works using clinical samples, based on these concepts. Hence, developing microfluidic devices for rare cell isolation based on dielectrophoretic, magnetophoretic and acoustophoretic signatures could be an interesting area for future research. Isolation based on inertial microfluidics represents an approach which is both label-free and capable of high throughput. However, this technique is currently not fully exploited. In combination with one of the other reviewed techniques, isolation of rare cells based on inertial microfluidics may lead to unique label-free and high-throughput solutions. Realizing a gentle and high-throughput isolation device may also open the possibility for continuous sorting and isolation. In this case, a blood sample could be removed from the body for rare cell isolation and then re-introduced back to the body. Continuous isolation could allow for the identification of extremely rare and healthy stem cells, which was previously impossible to detect due to the upper limit of the volume of a human blood sample than can be safely drawn at one time. A high throughput is also important for biophysical analysis due to the limited time that the captured target cells are viable. Furthermore, a high-throughput analysis concept that is compatible with isolation devices based on dielectrophoresis, magnetophoresis, and acoustophoresis is a highly desirable future development area. Electrical impedance spectroscopy, ultrasonic inspection, magnetic resonance imaging, and other magnetic detection methods are good examples of these high-throughput analysis techniques.
Another approach to increase throughput is to optimize a sequence of multiple isolation methods on a single chip. This would also lead to improved efficiency and purity. Most reported works in the past were focused on a single isolation method. Although the design and fabrication of a microfluidic device with multiple isolation steps could be complex, the resulting benefits in efficiency and purity may be attractive enough for future research and commercial development in this direction. As mentioned previously, a cascaded isolation system may allow for the separation of different subtypes or different stages of the same cell lines. The analysis of these isolated cells would be highly valuable, for instance, for cancer research. Finally, an important related niche area for rare cell isolation devices is drug screening. Captured target cells can be transferred to an on-chip cell culture platform and exposed to drug candidates. These types of devices would accelerate the process of drug screening and development. More importantly, drug screening on a chip would allow for the implementation of truly personalized treatment options for cancer patients.
Acknowledgement
This research was supported by the National Institutes of Health (NIH) Director's New Innovator Award (1DP2OD007209-01).
References
- 1.Dharmasiri U, Witek MA, Adams AA, Soper SA. Annu. Rev. Anal. Chem. 2010;3:409–431. doi: 10.1146/annurev.anchem.111808.073610. [DOI] [PubMed] [Google Scholar]
- 2.Pratt ED, Huang C, Hawkins BG, Gleghorn JP, Kirby BJ. Chem. Eng. Sci. 2011;66:1508–1522. doi: 10.1016/j.ces.2010.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gossett DR, Weaver WM, Mach AJ, Hur SC, Tse HTK, Lee W, Amini H, Di Carlo D. Anal. Bioanal. Chem. 2010;397:3249–3267. doi: 10.1007/s00216-010-3721-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Harouaka RA, Nisic M, Zheng SY. J. Lab. Autom. 2013 doi: 10.1177/2211068213494391. DOI: 10.1177/2211068213494391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jin C, McFaul SM, Duffy SP, Deng X, Tavassoli P, Black PC, Ma H. Lab Chip. 2013;14:32–44. doi: 10.1039/c3lc50625h. [DOI] [PubMed] [Google Scholar]
- 6.Yu L, Ng SR, Xu Y, Dong H, Wang YJ, Li CM. Lab Chip. 2013;13:3163–3182. doi: 10.1039/c3lc00052d. [DOI] [PubMed] [Google Scholar]
- 7.Arya SK, Lim B, Rahman ARA. Lab Chip. 2013;13:1995–2027. doi: 10.1039/c3lc00009e. [DOI] [PubMed] [Google Scholar]
- 8.Li P, Stratton ZS, Dao M, Ritz J, Huang TJ. Lab Chip. 2013;13:602–609. doi: 10.1039/c2lc90148j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Karimi A, Yazdi S, Ardekani AM. Biomicrofluid. 2013;7:021501. doi: 10.1063/1.4799787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yu M, Stott S, Toner M, Maheswaran S, Haber DA. J. Cell Bio. 2011;192:373–382. doi: 10.1083/jcb.201010021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Danova M, Torchio M, Mazzini G. Expert Rev. Mol. Diagn. 2011;11:473–485. doi: 10.1586/erm.11.33. [DOI] [PubMed] [Google Scholar]
- 12.Smith JP, Barbati AC, Santana SM, Gleghorn JP, Kirby BJ. Electrophoresis. 2012;33:3133–3142. doi: 10.1002/elps.201200263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hyun KA, Jung HI. Electrophoresis. 2013;34:1028–1041. doi: 10.1002/elps.201200417. [DOI] [PubMed] [Google Scholar]
- 14.Chen J, Li J, Sun Y. Lab Chip. 2012;12:1753–1767. doi: 10.1039/c2lc21273k. [DOI] [PubMed] [Google Scholar]
- 15.Alix-Panabieres C, Schwarzenbach H, Pantel K. Ann. Rev. Med. 2012;63:199–215. doi: 10.1146/annurev-med-062310-094219. [DOI] [PubMed] [Google Scholar]
- 16.Muluneh M, Issadore D. Adv. Drug Delivery Rev. 2013 doi: 10.1016/j.addr.2013.09.013. DOI: 10.1016/j.addr.2013.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hong B, Zu YL. Theranostics. 2013;3:377–394. doi: 10.7150/thno.5195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Plaks V, Koopman CD, Werb Z. Science. 2013;341:1186–1188. doi: 10.1126/science.1235226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schwesinger F, Ros R, Strunz T, Anselmetti D, Güntherodt HJ, Honegger A, Jermutus L, Tiefenauer L, Pluckthun A. Proc. Natl. Acad. Sci. U.S.A. 2000;97:9972–9977. doi: 10.1073/pnas.97.18.9972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nguyen NT. Microfluid. Nanofluid. 2012;12:1–16. [Google Scholar]
- 21.Zheng S, Lin HK, Lu B, Williams A, Datar R, Cote RJ, Tai YC. Biomed. Microdevices. 2011;13:203–213. doi: 10.1007/s10544-010-9485-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hosokawa M, Hayata T, Fukuda Y, Arakaki A, Yoshino T, Tanaka T, Matsunaga T. Anal. Chem. 2010;82:6629–6635. doi: 10.1021/ac101222x. [DOI] [PubMed] [Google Scholar]
- 23.Holm SH, Beech JP, Barrett MP, Tegenfeldt JO. Lab Chip. 2011;11:1326–1332. doi: 10.1039/c0lc00560f. [DOI] [PubMed] [Google Scholar]
- 24.Wu Z, Willing B, Bjerketorp J, Janssonb JK, Hjort K. Lab Chip. 2009;9:1193–1199. doi: 10.1039/b817611f. [DOI] [PubMed] [Google Scholar]
- 25.Davis JA, Inglis DW, Morton KJ, Lawrence DA, Huang LR, Chou SY, Sturm JC, Austin RH. Proc. Natl. Acad. Sci. U.S.A. 2006;103:14779–14784. doi: 10.1073/pnas.0605967103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kim MS, Sim TS, Kim YJ, Kim SS, Jeong H, Park J-M, Moon H-S, Kim SI, Gurel O, Lee SS, Lee J-G, Park JC. Lab Chip. 2012;12:2874–2880. doi: 10.1039/c2lc40065k. [DOI] [PubMed] [Google Scholar]
- 27.Park J-M, Lee J-Y, Lee J-G, Jeong H, Oh J-M, Kim YJ, Park D, Kim MS, Lee HJ, Oh JH, Lee SS, Lee W-Y, Huh N. Anal. Chem. 2012;84:7400–7407. doi: 10.1021/ac3011704. [DOI] [PubMed] [Google Scholar]
- 28.Mohamed H, Murray M, Turner JN, Caggana M. J. Chromatogr. A. 2009;1216:8289–8295. doi: 10.1016/j.chroma.2009.05.036. [DOI] [PubMed] [Google Scholar]
- 29.Mohamed H, Turner JN, Caggana M. J. Chromatogr. A. 2007;1162:187–192. doi: 10.1016/j.chroma.2007.06.025. [DOI] [PubMed] [Google Scholar]
- 30.Zheng S, Lin H, Liu JQ, Balic M, Datar R, Cote RJ, Tai YC. J. Chromatogr. A. 2007;1162:154–161. doi: 10.1016/j.chroma.2007.05.064. [DOI] [PubMed] [Google Scholar]
- 31.Chen X, Cui DF, Liu CC, Li H. Sens. Actuators. B: Chem. 2008;130:216–221. [Google Scholar]
- 32.Tan SJ, Yobas L, Lee GYH, Ong CN, Lim CT. Biomed. Microdevices. 2009;11:883–892. doi: 10.1007/s10544-009-9305-9. [DOI] [PubMed] [Google Scholar]
- 33.McFaul SM, Lin BK, Ma H. Lab Chip. 2012;12:2369–2376. doi: 10.1039/c2lc21045b. [DOI] [PubMed] [Google Scholar]
- 34.Huang LR, Cox EC, Austin RH, Sturm JC. Science. 2004;304:987–989. doi: 10.1126/science.1094567. [DOI] [PubMed] [Google Scholar]
- 35.Loutherback K, D'Silva J, Liu L, Wu A, Austin RH. AIP Adv. 2012;2:042107. doi: 10.1063/1.4758131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Beech JP, Holm SH, Adolfsson K, Tegenfeldt JO. Lab Chip. 2012;12:1048–1051. doi: 10.1039/c2lc21083e. [DOI] [PubMed] [Google Scholar]
- 37.Di Carlo D. Lab Chip. 2009;9:3038–3046. doi: 10.1039/b912547g. [DOI] [PubMed] [Google Scholar]
- 38.Hou HW, Warkiani ME, Khoo BL, Li ZR, Soo RA, Tan DSW, Lim WT, Han J, Bhagat AAS, Lim CT. Sci. Rep. 2013;3:1259. doi: 10.1038/srep01259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sollier E, Go DE, Che J, Gossett DR, O'Byrne S, Weaver WM, Kummer N, Rettig M, Goldman J, Nickols N, McCloskey S, Kulkarni RP, Di Carlo D. Lab Chip. 2014;14:63–77. doi: 10.1039/c3lc50689d. [DOI] [PubMed] [Google Scholar]
- 40.Hur SC, Henderson-MacLennan NK, McCabe ERB, Di Carlo D. Lab Chip. 2011;11:912–920. doi: 10.1039/c0lc00595a. [DOI] [PubMed] [Google Scholar]
- 41.Amini H, Sollier E, Weaver WM, Di Carlo D. Proc. Natl. Acad. Sci. U.S.A. 2012;109:11593–11598. doi: 10.1073/pnas.1207550109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hu X, Bessette PH, Qian J, Meinhart CD, Daugherty PS, Soh HT. Proc. Natl. Acad. Sci. U.S.A. 2005;102:15757–15761. doi: 10.1073/pnas.0507719102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gascoyne PRC, Nohsari J, Anderson TJ, Becker FF. Electrophoresis. 2009;30:1388–1398. doi: 10.1002/elps.200800373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Han KH, Han SI, Frazier AB. Lab Chip. 2009;9:2958–2964. doi: 10.1039/b909753h. [DOI] [PubMed] [Google Scholar]
- 45.Moon HS, Kwon K, Kim SI, Han H, Sohn J, Lee S, Jung HI. Lab Chip. 2011;11:1118–1125. doi: 10.1039/c0lc00345j. [DOI] [PubMed] [Google Scholar]
- 46.Ho CT, Lin RZ, Chang WY, Chang HY, Liu CH. Lab Chip. 2006;6:724–734. doi: 10.1039/b602036d. [DOI] [PubMed] [Google Scholar]
- 47.Broche LM, Bhadal N, Lewis MP, Porter S, Hughes MP, Labeed FH. Oral Oncol. 2007;43:199–203. doi: 10.1016/j.oraloncology.2006.02.012. [DOI] [PubMed] [Google Scholar]
- 48.Mulhall HJ, Labeed FH, Kazmi B, Costea DE, Hughes MP, Lewis MP. Anal. Bioanal. Chem. 2011;401:2455–2463. doi: 10.1007/s00216-011-5337-0. [DOI] [PubMed] [Google Scholar]
- 49.Yang F, Yang XM, Jiang H, Bulkhaults P, Wood P, Hrushesky W, Wang GR. Biomicrofluidics. 2010;4:013204. doi: 10.1063/1.3279786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Salmanzadeh A, Romero L, Shafiee H, Gallo-Villanueva RC, Stremler MA, Cramer SD, Davalos RV. Lab Chip. 2012;12:182–189. doi: 10.1039/c1lc20701f. [DOI] [PubMed] [Google Scholar]
- 51.Sabuncu AC, Liu JA, Beebe SJ, Beskok A. Biomicrofluidics. 2010;4:021101. doi: 10.1063/1.3447702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Shim S, Stemke-Hal K, Tsimberidou AM, Noshari J, Anderson TE. Biomicrofludics. 2013;7:011807. doi: 10.1063/1.4774304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Vykoukal J, Vykoukal DM, Freyberg S, Alt EU, Gascoyne PRC. Lab Chip. 2008;8:1386–1393. doi: 10.1039/b717043b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Gupta V, Jafferji I, Garza M, Melnikova VO, Hasegawa DK, Pethig R, Davis DW. Biomirofluidics. 2012;6:024133. doi: 10.1063/1.4731647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wang XB, Vykoukal J, Becker FF, Gascoyne PR. Biophys. J. 1998;74:2689–2701. doi: 10.1016/S0006-3495(98)77975-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Gascoyne PRC, Noshari J, Anderson TJ, Becker FF. Electrophoresis. 2009;30:1388–1398. doi: 10.1002/elps.200800373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Park S, Zhang Y, Wang T-H, Yang S. Lab Chip. 2011;11:2893–2900. doi: 10.1039/c1lc20307j. [DOI] [PubMed] [Google Scholar]
- 58.Cheng I-F, Chang H-C, Hou D, Chang H-C. Biomicrofluidics. 2007;1:021503. doi: 10.1063/1.2723669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kuczenski RS, Chang H-C, Revzin A. Biomicrofluidics. 2011;5:032005. doi: 10.1063/1.3608135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Park J, Jung H. Anal. Chem. 2009;81:8280–8288. doi: 10.1021/ac9005765. [DOI] [PubMed] [Google Scholar]
- 61.Park J, Song S, Jung H. Lab Chip. 2009;9:939–948. doi: 10.1039/b813952k. [DOI] [PubMed] [Google Scholar]
- 62.Moon HS, Kwon K, Kim SI, Han H, Sohn J, Lee S, Jung H. Lab Chip. 2011;11:1118–1125. doi: 10.1039/c0lc00345j. [DOI] [PubMed] [Google Scholar]
- 63.Ohta AT, Garcia M, Valley JK, Banie L, Hsu HY, Jamshidi A, Neale SL, Lue T, Wu MC. Lab Chip. 2010;10:3213–3217. doi: 10.1039/c0lc00072h. [DOI] [PubMed] [Google Scholar]
- 64.Lin YH, Yang YW, Chen YD, Wang SS, Chang YH, Wu MH. Lab Chip. 2012;12:1164–1173. doi: 10.1039/c2lc21097e. [DOI] [PubMed] [Google Scholar]
- 65.Valley JK, Swinton P, Boscardin WJ, Lue TF, Rinaudo PF, Wu MC, Garcia MM. PLoS One. 2010;5:e10160. doi: 10.1371/journal.pone.0010160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Huang SB, Wu MH, Lin YH, Hsieh CH, Yang CL, Lin HC, Tseng CP, Lee GB. Lab Chip. 2013;13:1371–1383. doi: 10.1039/c3lc41256c. [DOI] [PubMed] [Google Scholar]
- 67.Cheng XH, Irimia D, Dixon M, Sekine K, Demirci U, Zamir L, Tompkins RG, Rodriguez W, Toner M. Lab Chip. 2007;7:170–178. doi: 10.1039/b612966h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Li P, Gao Y, Pappas D. Anal. Chem. 2012;84:8140–8148. doi: 10.1021/ac302002a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Didar TF, Tabrizian M. Lab Chip. 2010;10:3043–3053. doi: 10.1039/c0lc00130a. [DOI] [PubMed] [Google Scholar]
- 70.Li P, Gao Y, Pappas D. Anal. Chem. 2011;83:7863–7869. doi: 10.1021/ac201752s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Li P, Tian Y, Pappas D. Anal. Chem. 2011;83:774–781. doi: 10.1021/ac102975g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Hyun K-A, Lee TY, Jung H-I. Anal. Chem. 2013;85:4439–4445. doi: 10.1021/ac3037766. [DOI] [PubMed] [Google Scholar]
- 73.Wang K, Marshall MK, Garza G, Pappas D. Anal. Chem. 2008;80:2118–2124. doi: 10.1021/ac702553w. [DOI] [PubMed] [Google Scholar]
- 74.Hatch A, Pesko DM, Murthy SK. Anal. Chem. 2012;84:4618–4621. doi: 10.1021/ac300496q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Nagrath S, Sequist LV, Maheswaran S, Bell DW, Irimia D, Ulkus L, Smith MR, Kwak EL, Digumarthy S, Muzikansky A, Ryan P, Balis UJ, Tompkins RG, Haber DA, Toner M. Nature. 2007;450:1235–1239. doi: 10.1038/nature06385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Gleghorn JP, Pratt ED, Denning D, Liu H, Bander NH, Tagawa ST, Nanus DM, Giannakakou PA, Kirby BJ. Lab Chip. 2010;10:27–29. doi: 10.1039/b917959c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Sheng W, Chen T, Kamath R, Xiong X, Tan W, Fan ZH. Anal. Chem. 2012;84:4199–4206. doi: 10.1021/ac3005633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Dharmasiri U, Njoroge SK, Witek MA, Adebiyi MG, Kamande JW, Hupert ML, Barany F, Soper SA. Anal. Chem. 2011;83:2301–2309. doi: 10.1021/ac103172y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Kamande JW, Hupert ML, Witek MA, Wang H, Torphy RJ, Dharmasiri U, Njoroge SK, Jackson JM, Aufforth RD, Snavely A, Yeh JJ, Soper SA. Anal. Chem. 2013;85:9092–9100. doi: 10.1021/ac401720k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Dharmasiri U, Balamurugan S, Adams AA, Okagbare PI, Obubuafo A, Soper SA. Electrophoresis. 2009;30:3289–3300. doi: 10.1002/elps.200900141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Adams AA, Okagbare PI, Feng J, Hupert ML, Patterson D, Göttert J, McCarley RL, Nikitopoulos D, Murphy MC, Soper SA. J. Am. Chem. Soc. 2008;130:8633–8641. doi: 10.1021/ja8015022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Stott SL, Hsu C-H, Tsukrov DI, Yu M, Miyamoto DT, Waltman BA, Rothenberg SM, Shah AM, Smas ME, Korir GK, et al. Proc. Natl. Acad. Sci. U.S.A. 2010;107:18392–18397. doi: 10.1073/pnas.1012539107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Sheng W, Chen T, Tan W, Fan ZH. ACS Nano. 2013;7:7067–7076. doi: 10.1021/nn4023747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Wang S, Liu K, Liu J, Yu ZTF, Xu X, Zhao L, Lee T, Lee EK, Reiss J, Lee Y-K, et al. Angew. Chem. Int. Ed. 2011;50:3084–3088. doi: 10.1002/anie.201005853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Sheng W, Ogunwobi OO, Chen T, Zhang J, George TJ, Liu C, Fan ZH. Lab Chip. 2014;14:89–98. doi: 10.1039/c3lc51017d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Wang S, Wang H, Jiao J, Chen K-J, Owens GE, Kamei K.-i., Sun J, Sherman DJ, Behrenbruch CP, Wu H, Tseng H-R. Angew. Chem. Int. Ed. 2009;48:8970–8973. doi: 10.1002/anie.200901668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Hughes AD, King MR. Langmuir. 2010;26:12155–12164. doi: 10.1021/la101179y. [DOI] [PubMed] [Google Scholar]
- 88.Han W, Allio BA, Foster DG, King MR. ACS Nano. 2009;4:174–180. doi: 10.1021/nn900442c. [DOI] [PubMed] [Google Scholar]
- 89.He R, Zhao L, Liu Y, Zhang N, Cheng B, He Z, Cai B, Li S, Liu W, Guo S, Chen Y, Xiong B, Zhao X-Z. Biomed. Microdevices. 2013;15:617–626. doi: 10.1007/s10544-013-9781-9. [DOI] [PubMed] [Google Scholar]
- 90.Yoon HJ, Kim TH, Zhang Z, Azizi E, Pham TM, Paoletti C, Lin J, Ramnath N, Wicha MS, Hayes DF, Simeone DM, Nagrath S. Nat. Nano. 2013;8:735–741. doi: 10.1038/nnano.2013.194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Mittal S, Wong IY, Deen WM, Toner M. Biophys. J. 2012;102:721–730. doi: 10.1016/j.bpj.2011.12.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Zhao W, Cui CH, Bose S, Guo D, Shen C, Wong WP, Halvorsen K, Farokhzad OC, Teo GSL, Phillips JA, Dorfman DM, Karnik R, Karp JM. Proc. Natl. Acad. Sci. U.S.A. 2012;109:19626–19631. doi: 10.1073/pnas.1211234109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Shen Q, Xu L, Zhao L, Wu D, Fan Y, Zhou Y, OuYang W-H, Xu X, Zhang Z, Song M, Lee T, Garcia MA, Xiong B, Hou S, Tseng H-R, Fang X. Adv. Mater. 2013;25:2368–2373. doi: 10.1002/adma.201300082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Gurkan UA, Anand T, Tas H, Elkan D, Akay A, Keles HO, Demirci U. Lab Chip. 2011;11:3979–3989. doi: 10.1039/c1lc20487d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Gurkan UA, Tasoglu S, Akkaynak D, Avci O, Unluisler S, Canikyan S, MacCallum N, Demirci U. Adv. Healthc. Mater. 2012;1:661–668. doi: 10.1002/adhm.201200009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Liu H, Liu X, Meng J, Zhang P, Yang G, Su B, Sun K, Chen L, Han D, Wang S, Jiang L. Adv. Mater. 2013;25:922–927. doi: 10.1002/adma.201203826. [DOI] [PubMed] [Google Scholar]
- 97.Tang Z, Akiyama Y, Itoga K, Kobayashi J, Yamato M, Okano T. Biomaterials. 2012;33:7405–7411. doi: 10.1016/j.biomaterials.2012.06.077. [DOI] [PubMed] [Google Scholar]
- 98.Plouffe BD, Brown MA, Iyer RK, Radisic M, Murthy SK. Lab Chip. 2009;9:1507–1510. doi: 10.1039/b823523f. [DOI] [PubMed] [Google Scholar]
- 99.Chen W, Weng S, Zhang F, Allen S, Li X, Bao L, Lam RHW, Macoska JA, Merajver SD, Fu J. ACS Nano. 2012;7:566–575. doi: 10.1021/nn304719q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Chen C-L, Chen K-C, Pan Y-C, Lee T-P, Hsiung L-C, Lin C-M, Chen C-Y, Lin C-H, Chiang B-L, Wo AM. Lab Chip. 2011;11:474–483. doi: 10.1039/c0lc00332h. [DOI] [PubMed] [Google Scholar]
- 101.Furdui VI, Harrison DJ. Lab Chip. 2004;4:614–618. doi: 10.1039/b409366f. [DOI] [PubMed] [Google Scholar]
- 102.Adams JD, Soh HT. JALA Charlottesv Va. 2009;14:331–340. doi: 10.1016/j.jala.2009.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Kang JH, Krause S, Tobin H, Mammoto A, Kanapathipillai M, Ingber DE. Lab Chip. 2012;12:2175–2181. doi: 10.1039/c2lc40072c. [DOI] [PubMed] [Google Scholar]
- 104.Plouffe BD, Mahalanabis M, Lewis LH, Klapperich CM, Murthy SK. Anal. Chem. 2012;84:1336–1344. doi: 10.1021/ac2022844. [DOI] [PubMed] [Google Scholar]
- 105.Yung CW, Fiering J, Mueller AJ, Ingber DE. Lab Chip. 2009;9:1171–1177. doi: 10.1039/b816986a. [DOI] [PubMed] [Google Scholar]
- 106.Hoshino K, Huang Y-Y, Lane N, Huebschman M, Uhr JW, Frenkel EP, Zhang X. Lab Chip. 2011;11:3449–3457. doi: 10.1039/c1lc20270g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Earhart CM, Hughes CE, Gaster RS, Ooi CC, Wilson RJ, Zhou LY, Humke EW, Xu L, Wong DJ, Willingham SB, et al. Lab Chip. 2013;14:78–88. doi: 10.1039/c3lc50580d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Kim S, Han S-I, Park M-J, Jeon C-W, Joo Y-D, Choi I-H, Han K-H. Anal. Chem. 2013;85:2779–2786. doi: 10.1021/ac303284u. [DOI] [PubMed] [Google Scholar]
- 109.Xia N, Hunt TP, Mayers BT, Alsberg E, Whitesides GM, Westervelt RM, Ingber DE. Biomed Microdevices. 2006;8:299–308. doi: 10.1007/s10544-006-0033-0. [DOI] [PubMed] [Google Scholar]
- 110.Hoshino K, Chen P, Huang Y-Y, Zhang X. Anal. Chem. 2012;84:4292–4299. doi: 10.1021/ac2032386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Saliba A-E, Saias L, Psychari E, Minc N, Simon D, Bidard F-C, Mathiot C, Piergac J-Y, Fraisier V, Salamero J, et al. Proc. Natl. Acad. Sci. U.S.A. 2010;107:14524–14529. doi: 10.1073/pnas.1001515107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Svobodova Z, Kucerova J, Autebert J, Horak D, Bruckova L, Viovy J-L, Bilkova Z. Electrophoresis. 2013 doi: 10.1002/elps.201300196. DOI: 10.1002/elps.201300196. [DOI] [PubMed] [Google Scholar]
- 113.Horák D, Svobodová Z, Autebert J, Coudert B, Plichta Z, Královec K, Bílková Z, Viovy J-L, Biomed J. Mater. Res. A. 2013;101:23–32. doi: 10.1002/jbm.a.34297. [DOI] [PubMed] [Google Scholar]
- 114.Schirhagl R, Fuereder I, Hall EW, Medeiros BC, Zare RN. Lab Chip. 2011;11:3130–3135. doi: 10.1039/c1lc20353c. [DOI] [PubMed] [Google Scholar]
- 115.Kuo JS, Zhao Y, Schiro PG, Ng L, Lim DSW, Shelby JP, Chiu DT. Lab Chip. 2010;10:837–842. doi: 10.1039/b922301k. [DOI] [PubMed] [Google Scholar]
- 116.Hur SC, Mach AJ, Di Carlo D. Biomicrofluidics. 2011;5:022206. doi: 10.1063/1.3576780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Warkiani ME, Guan G, Luan KB, Lee WC, Bhagat AAS, Chaudhuri PK, Tan DS-W, Lim WT, Lee SC, Chen PCY, Lim CT, Han J. Lab Chip. 2013;14:128–137. doi: 10.1039/c3lc50617g. [DOI] [PubMed] [Google Scholar]
- 118.Parichehreh V, Medepallai K, Babbarwal K, Sethu P. Lab Chip. 2013;13:892–900. doi: 10.1039/c2lc40663b. [DOI] [PubMed] [Google Scholar]
- 119.Tanaka T, Ishikawa T, Numayama-Tsuruta K, Imai Y, Ueno H, Matsuki N, Yamaguchi T. Lab Chip. 2012;12:4336–4343. doi: 10.1039/c2lc40354d. [DOI] [PubMed] [Google Scholar]
- 120.Huang C, Liu H, Bander NH, Kirby BJ. Biomed. Microdevices. 2013;15:941–948. doi: 10.1007/s10544-013-9784-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Yang L, Banada PP, Chatni MR, Lim KS, Bhunia AK, Ladischde M, Bashir R. Lab Chip. 2006;6:896–905. doi: 10.1039/b607061m. [DOI] [PubMed] [Google Scholar]
- 122.Beech JP, Jönsson P, Tegenfeldt JO. Lab Chip. 2009;9:2698–2706. doi: 10.1039/b823275j. [DOI] [PubMed] [Google Scholar]
- 123.Liu Z, Zhang W, Huang F, Feng H, Shu W, Xu X, Chen Y. Biosens. Bioelectron. 2013;47:113–119. doi: 10.1016/j.bios.2013.03.017. [DOI] [PubMed] [Google Scholar]
- 124.Choi S, Karp JM, Karnik R. Lab Chip. 2012;12:1427–1430. doi: 10.1039/c2lc21225k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Petersson F, Aberg L, Swärd-Nilsson A-M, Laurell T. Anal. Chem. 2007;79:5117–5123. doi: 10.1021/ac070444e. [DOI] [PubMed] [Google Scholar]
- 126.Shi J, Huang H, Stratton Z, Lawit A, Huang Y, Huang TJ. Lab Chip. 2009;9:3354–3359. doi: 10.1039/b915113c. [DOI] [PubMed] [Google Scholar]
- 127.Ding X, Li P, Lin S-CS, Stratton ZS, Nama N, Guo F, Slotcavage D, Mao X, Shi J, Costanzo F, Huang TJ. Lab Chip. 2013;13:3626–3649. doi: 10.1039/c3lc50361e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Adams JD, Ebbesen CL, Barnkob R, Yang AHJ, Soh HT, Bruus H. J. Micromech. Microeng. 2012;22:075017. [Google Scholar]
- 129.Augustsson P, Magnusson C, Nordin M, Lilja H, Laurell T. Anal. Chem. 2012;84:7954–7962. doi: 10.1021/ac301723s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Yang AHJ, Soh HT. Anal. Chem. 2012;84:10756–10762. doi: 10.1021/ac3026674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Ding X, Lin S-CS, Kiraly B, Yue H, Li S, Chiang IK, Shi J, Benkovic SJ, Huang TJ. Proc. Natl. Acad. Sci. U. S. A. 2012;109:11105–11109. doi: 10.1073/pnas.1209288109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Shi J, Ahmed D, Mao X, Lin S-CS, Lawit A, Huang TJ. Lab Chip. 2009;9:2890–2895. doi: 10.1039/b910595f. [DOI] [PubMed] [Google Scholar]
- 133.Li S, Ding X, Guo F, Chen Y, Lapsley M, Lin S-CS, Wang L, McCoy JP, Cameron C, Huang TJ. Anal. Chem. 2013;85:5468–5474. doi: 10.1021/ac400548d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Ding X, Lin S-CS, Lapsley MI, Li S, Guo X, Chan CY, Chiang I-K, McCoy JP, Huang TJ. Lab Chip. 2012;12:4228–4231. doi: 10.1039/c2lc40751e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Chen Y, Ding X, Lin S-CS, Yang S, Huang P-H, Nama N, Zhao Y, Nawaz AA, Guo F, Wang W, et al. ACS Nano. 2013;7:3306–3314. doi: 10.1021/nn4000034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Vincent ME, Liu W, Haney EB, Ismagilov RF. Chem. Soc. Rev. 2010;39:974–984. doi: 10.1039/b917851a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Chen J, Li J, Sun Y. Lab Chip. 2012;12:1753–1767. doi: 10.1039/c2lc21273k. [DOI] [PubMed] [Google Scholar]
- 138.Lecault V, White AK, Singhal A, Hansen CL. Curr. Opin. Chem. Biol. 2012;16:381–390. doi: 10.1016/j.cbpa.2012.03.022. [DOI] [PubMed] [Google Scholar]
- 139.Wheeler AR, Throndset WR, Whelan RJ, Leach AM, Zare RN, Liao YH, Farrell K, Manger ID, Daridon A. Anal. Chem. 2003;75:3581–3586. doi: 10.1021/ac0340758. [DOI] [PubMed] [Google Scholar]
- 140.Yin H, Marshall D. Curr. Opin. Chem. Biol. 2012;23:110–119. doi: 10.1016/j.copbio.2011.11.002. [DOI] [PubMed] [Google Scholar]
- 141.Sanchez-Freire V, Ebert AD, Kalisky T, Quake SR, Wu JC. Nat. Protoc. 2012;7:829–838. doi: 10.1038/nprot.2012.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Unger MA, Chou H-P, Thorsen T, Scherer A, Quake SR. Science. 2000;288:113–116. doi: 10.1126/science.288.5463.113. [DOI] [PubMed] [Google Scholar]
- 143.Thorsen T, Maerkl SJ, Quake SR. Science. 2002;298:580–584. doi: 10.1126/science.1076996. [DOI] [PubMed] [Google Scholar]
- 144.Ottesen EA, Hong JW, Quake SR, Leadbetter JR. Science. 2006;314:1464–1467. doi: 10.1126/science.1131370. [DOI] [PubMed] [Google Scholar]
- 145.Powell1 AA, Talasaz AH, Zhang H, Coram MA, Reddy A, Deng G, Telli ML, Advani RH, Carlson RW, Mollick JA, et al. PLoS One. 2012;7:e33788. doi: 10.1371/journal.pone.0033788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Tinnefeld P. Nat. Nanotechnol. 2013;8:480–482. doi: 10.1038/nnano.2013.122. [DOI] [PubMed] [Google Scholar]
- 147.Loveland AB, Habuchi S, Walter JC, van Oijen AM. Nat. Methods. 2012;9:987–992. doi: 10.1038/nmeth.2174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Walt DR. Anal. Chem. 2013;85:1258–1263. doi: 10.1021/ac3027178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Juul S, Ho Y-P, Koch J, Andersen FF, Stougaard M, Leong KW, Knudsen BR. ACS Nano. 2011;5:8305–8310. doi: 10.1021/nn203012q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Stougaard M, Lohmann JS, Mancino A, Celik S, Andersen FF, Koch J, Knudsen BR. ACS Nano. 2009;3:223–233. doi: 10.1021/nn800509b. [DOI] [PubMed] [Google Scholar]
- 151.Primiceri E, Chiriaco MS, Rinaldi R, Maruccio G. Lab Chip. 2013;13:3789–3802. doi: 10.1039/c3lc50550b. [DOI] [PubMed] [Google Scholar]
- 152.Albrecht DR, Underhill GH, Resnikoff J, Mendelson A, Bhatia SN, Shah JV. Integr. Biol. 2010;2:278–287. doi: 10.1039/b923699f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Li M, Xu J, Romero-Gonzalez M, Banwart SA, Huang WE. Curr. Opin. Chem. Biol. 2012;23:56–63. doi: 10.1016/j.copbio.2011.11.019. [DOI] [PubMed] [Google Scholar]
- 154.Liberale C, Cojoc G, Bragheri F, Minzioni P, Perozziello G, Rocca RL, Ferrara L, Rajamanickam V, Fabrizio ED, Cristiani I. Sci. Rep. 2013;3:1258. doi: 10.1038/srep01258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Davis LM, Lubbeck JL, Dean K,M, Palmer AE, Jimenez R. Lab Chip. 2013;13:2320–2327. doi: 10.1039/c3lc50191d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Mao X, Nawaz AA, Lin S-CS, Lapsley MI, Zhao Y, McCoy JP, El-Deiry WS, Huang TJ. Biomicrofluidics. 2012;6:024113. doi: 10.1063/1.3701566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Song H, Wang Y, Rosano JM, Prabhakarpandian B, Garson C, Pant K, Lai E. Lab Chip. 2013;13:2300–2310. doi: 10.1039/c3lc41321g. [DOI] [PubMed] [Google Scholar]
- 158.Watkins NN, Sridhar S, Cheng X, Chen GD, Toner M, Rodriguez W, Bashir R. Lab Chip. 2011;11:1437–1447. doi: 10.1039/c0lc00556h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Choi H, Kim KB, Jeon CS, Hwang I, Lee S, Kim HK, Kim HC, Chung TD. Lab Chip. 2013;13:970–977. doi: 10.1039/c2lc41376k. [DOI] [PubMed] [Google Scholar]
- 160.Zhang W, Kai K, Choi DS, Iwamoto T, Nguyen YH, Wong H, Landis MD, Ueno NT, Chang J, Qin L. Proc. Natl. Acad. Sci. U S A. 2012;109:18707–18712. doi: 10.1073/pnas.1209893109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Bow H, Pivkin IV, Diez-Silva M, Goldfless SJ, Dao M, Niles JC, Suresh S, Han J. Lab Chip. 2011;11:1065–1073. doi: 10.1039/c0lc00472c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Helou M, Reisbeck M, Tedde SF, Richter L, Bar L, Bosch JJ, Stauber RH, Quandtd E, Hayden O. Lab Chip. 2013;13:1035–1038. doi: 10.1039/c3lc41310a. [DOI] [PubMed] [Google Scholar]
- 163.Issadore D, Chung J, Shao H, Liong M, Ghazani AA, Castro CM, Weissleder R, Lee H. Sci. Transl. Med. 2012;4:141ra92. doi: 10.1126/scitranslmed.3003747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Overby DR, Matthews BD, Alsberg E, Ingber DE. Acta Biomater. 2005;1:295–303. doi: 10.1016/j.actbio.2005.02.003. [DOI] [PubMed] [Google Scholar]
- 165.Ang PK, Li A, Jaiswal M, Wang Y, Hou HW, Thong JTL, Lim CT, Loh KP. Nano Lett. 2011;11:5240–5246. doi: 10.1021/nl202579k. [DOI] [PubMed] [Google Scholar]
- 166.Mao X, Huang TJ. Lab Chip. 2012;12:4006–4009. doi: 10.1039/c2lc90100e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.González-Cruz RD, Fonseca VC, Darling EM. Proc. Natl. Acad. Sci. 2012;109:E1523–E1529. doi: 10.1073/pnas.1120349109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Dudani JS, Gossett DR, Tse HTK, Di Carlo D. Lab Chip. 2013;13:3728–3734. doi: 10.1039/c3lc50649e. [DOI] [PubMed] [Google Scholar]
- 169.Sackstein R, Merzaban JS, Cain DW, Dagia NM, Spencer JA, Lin CP, Wohlgemuth R. Nat. Med. 2008;14:181–187. doi: 10.1038/nm1703. [DOI] [PubMed] [Google Scholar]
- 170.Choi S, Levy O, Coelho MB, Cabral JMS, Karp JM, Karnik R. Lab Chip. 2013 doi: 10.1039/c3lc50923k. DOI: 10.1039/C3LC50923K. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Cristofanilli M, Budd GT, Ellis MJ, Stopeck A, Matera J, Miller MC, Reuben JM, Doyle GV, Allard WJ, Terstappen LW, Hayes DF. N. Engl. J. Med. 2004;351:781–791. doi: 10.1056/NEJMoa040766. [DOI] [PubMed] [Google Scholar]
- 172.de Bono JS, Scher HI, Montgomery RB, Parker C, Miller MC, Tissing H, Doyle GV, Terstappen LW, Pienta KJ, Raghavan D. Clin. Cancer Res. 2008;14:6302–6309. doi: 10.1158/1078-0432.CCR-08-0872. [DOI] [PubMed] [Google Scholar]
- 173.Alix-Panabières C, Pantel K. Clin. Chem. 2013;59:110–118. doi: 10.1373/clinchem.2012.194258. [DOI] [PubMed] [Google Scholar]
- 174.Zhang L, Ridgway LD, Wetzel MD, Ngo J, Yin W, Kumar D, Goodman JC, Groves MD, Marchetti D. Sci. Transl. Med. 2013;5:180ra48. doi: 10.1126/scitranslmed.3005109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Vona G, Sabile A, Louha M, Sitruk V, Romana S, Schütze K, Capron F, Franco D, Pazzagli M, Vekemans M, Lacour B, Bréchot C, Paterlini-Bréchot P. Am. J. Pathol. 2000;156:57–63. doi: 10.1016/S0002-9440(10)64706-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Maheswaran S, Haber DA. Curr. Opin. Genet. Dev. 2010;20:96–99. doi: 10.1016/j.gde.2009.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Goon PK, Lip GY, Boos CJ, Stonelake PS, Blann AD. Neoplasia. 2006;8:79–88. doi: 10.1593/neo.05592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Blann AD, Woywodt A, Bertolini F, Bull TM, Buyon JP, Clancy RM, Haubitz M, Hebbel RP, Lip GY, Mancuso P, Sampol J, Solovey A, Dignat-George F. J. Thromb. Haemost. 2005;93:228–235. doi: 10.1160/TH04-09-0578. [DOI] [PubMed] [Google Scholar]
- 179.Khan SS, Solomon MA, McCoy JP. Cytometry B Clin. Cytom. 2005;64B:1–8. doi: 10.1002/cyto.b.20040. [DOI] [PubMed] [Google Scholar]
- 180.Duda DG, Cohen KS, Scadden DT, Jain RK. Nat. Protoc. 2007;2:805–810. doi: 10.1038/nprot.2007.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Damani S, Bacconi A, Libiger O, Chourasia AH, Serry R, Gollapudi R, Goldberg R, Rapeport K, Haaser S, Topol S, et al. Sci. Transl. Med. 2012;4:126ra33. doi: 10.1126/scitranslmed.3003451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Woywodt A, Blann AD, Kirsch T, Erdbruegger U, Banzet N, Haubitz M, Dignat-George F. J. Thromb. Haemost. 2006;4:671–677. doi: 10.1111/j.1538-7836.2006.01794.x. [DOI] [PubMed] [Google Scholar]
- 183.Bianchi DW. Br. J. Haematol. 1999;105:574–583. doi: 10.1046/j.1365-2141.1999.01383.x. [DOI] [PubMed] [Google Scholar]
- 184.Pongsritasana T, Wongratanacheewin S, Prasertcharoensuk V, Sermswan RW. Asian Pac. J. Allergy Immunol. 2006;24:65–71. [PubMed] [Google Scholar]
- 185.Bischoff FZ, Marquéz-Do DA, Martinez DI, Dang D, Horne C, Lewis D, Simpson JL. Clin. Genet. 2003;63:483–489. doi: 10.1034/j.1399-0004.2003.00087.x. [DOI] [PubMed] [Google Scholar]
- 186.Christensen B, Philip J, Kølvraa S, Lykke-Hansen L, Hromadnikova L, Gohel D, Lorch T, Plesch A, Bang J, Smidt-Jensen S, Hertz J, Djursing H. Fetal Diagn. Ther. 2005;20:106–112. doi: 10.1159/000082432. [DOI] [PubMed] [Google Scholar]
- 187.Liang X, Xu K, Xu J, Chen W, Shen H, Liu J. J. Magn. Magn. Mater. 2009;321:1885–1888. [Google Scholar]
- 188.Nam J, Huang H, Lim H, Lim C, Shin S. Anal. Chem. 2013;85:7316–7323. doi: 10.1021/ac4012057. [DOI] [PubMed] [Google Scholar]
- 189.Gascoyne P, Mahidol C, Ruchirawat M, Satayavivad J, Watcharasit P, Beckera FF. Lab Chip. 2002;2:70–75. doi: 10.1039/b110990c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Podszun S, Vulto P, Heinz H, Hakenberg S, Hermann C, Hankemeier T, Urban GA. Lab Chip. 2012;12:451–457. doi: 10.1039/c1lc20575g. [DOI] [PubMed] [Google Scholar]
- 191.Patterson AS, Heithoff DM, Ferguson BS, Soh HT, Mahan MJ, Plaxco KW. Appl. Environ. Microbiol. 2013;79:2302–2311. doi: 10.1128/AEM.03882-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Horsman KM, Barker SLR, Ferrance JP, Forrest KA, Koen KA, Landers JP. Anal. Chem. 2005;77:742–749. doi: 10.1021/ac0486239. [DOI] [PubMed] [Google Scholar]
- 193.Liu P, Li X, Greenspoon SA, Scherer JR, Mathies RA. Lab Chip. 2011;11:1041–1048. doi: 10.1039/c0lc00533a. [DOI] [PubMed] [Google Scholar]