Abstract
Usher syndrome 1B (USH1B) is a severe, autosomal recessive, deaf–blind disorder caused by mutations in myosin 7A (MYO7A). Patients are born profoundly deaf and exhibit progressive loss of vision starting in their first decade. MYO7A is expressed in human photoreceptors and retinal pigment epithelium, but disease pathology begins in photoreceptors, highlighting the need to develop a gene replacement strategy that effectively targets this cell type. For its safety and efficacy in clinical trials and ability to transduce postmitotic photoreceptors, we have focused on developing a clinically applicable adeno-associated virus (AAV) platform for delivering full-length MYO7A cDNA (∼6.7 kb). Packaging of full-length MYO7A cDNA in AAV produces vectors with heterogeneous, fragmented genomes (“fAAV”) capable of reconstituting full-length cDNA postinfection. We previously showed that fAAV vectors effectively delivered full-length MYO7A in vitro and in vivo. However, fAAV vectors are relatively inefficient and their heterogeneous genomes preclude definitive characterization, a drawback for clinical translatability. The aim of this study was to overcome these limitations by creating dual-AAV-vector platforms for USH1B with defined genomes. Human MYO7A was cloned in AAV vector pairs, each containing genomes <5 kb and intact inverted terminal repeats. One vector contained a promoter and 5′ portion of the cDNA and the partner vector contained a 3′ portion and polyadenylation signal. “Simple overlap” vectors share a central part of the MYO7A cDNA sequence. “Trans-splicing” and “hybrid” vectors utilize splice donor and acceptor sites with and without an additional central recombinogenic sequence, respectively. Vector pairs expressed full-length MYO7A in vitro and in vivo with equal or higher efficiency than fAAV, with a hybrid platform being most efficient. Importantly, analysis of MYO7A mRNA derived from each dual-vector platform revealed 100% fidelity to the predicted sequence. Our results suggest that dual AAV vectors with defined genetic payloads are a potential treatment option for USH1B.
Introduction
Usher syndrome (USH) is a clinically and genetically heterogeneous group of autosomal recessive disorders that make up the most frequent cause of combined deafness–blindness (Smith et al., 1994). Of the three clinical subtypes, Usher syndrome type 1 (USH1) is the most severe (Keats and Corey, 1999). Patients are born profoundly deaf, have vestibular dysfunction, and exhibit prepubertal onset of retinitis pigmentosa. The most common form of USH1 accounting for at least half of all cases is USH1B, caused by mutations in the myosin 7A (MYO7A) gene (Weil et al., 1995; Astuto et al., 2000; Bharadwaj et al., 2000; Ouyang et al., 2005). This gene encodes MYO7A, an unconventional myosin that is expressed both in cochlear hair cells of the inner ear and in photoreceptors/retinal pigment epithelial (RPE) cells of the retina (Hasson et al., 1995; Weil et al., 1995; Wolfrum et al., 1998; Gibbs et al., 2003). In the RPE it has been reported that MYO7A participates in melanosome localization (Liu et al., 1998), contributes to the removal of phagosomes, and aids in the light-dependent translocation of the visual cycle enzyme RPE65 (Lopes et al., 2011). Within photoreceptors, MYO7A is thought to play a role in the movement of proteins from the inner to outer segments (Liu et al., 1999). Its subcellular localization in mouse photoreceptors is controversial; however, most reports indicate that it is associated with connecting cilia (Williams, 2008). A recent study of primate photoreceptors indicates that MYO7A and other Usher proteins are expressed specifically at membrane interfaces between the inner and outer segments of rods, and between the microvillus-like calycal processes and basolateral outer segments of rods and cones (Sahly et al., 2012). The finding that MYO7A localizes to photoreceptor calycal processes and that these structures are absent in mouse retina may explain the lack of a retinal degeneration phenotype in Myo7a mutant, shaker1 mice as well as other mouse models of USH (Williams, 2008; Saihan et al., 2009). In the absence of a reliable animal model, analysis of USH1B disease pathology is restricted to patients. Optical imaging and psychophysical testing of young USH1B patients revealed that photoreceptors are the first detectable site of disease (Jacobson et al., 2008), highlighting the need to develop a gene replacement strategy that effectively targets both RPE and photoreceptors. Because the two allelic variants of human MYO7A are both relatively large, with coding sequences of 6534 and 6648 bp, respectively (Weil et al., 1996), treatment of USH1B will require a vector platform capable of delivering a relatively large cDNA.
Adeno-associated virus (AAV) has emerged as the gold standard in retinal gene therapy because of its ability to efficiently transduce postmitotic cells, high safety profile, and demonstrated efficacy in proof-of-concept studies and clinical trials (Bainbridge et al., 2008; Hauswirth et al., 2008; Cideciyan et al., 2009; Maguire et al., 2009; Simonelli et al., 2010; Flotte et al., 2011; Nathwani et al., 2011; Bowles et al., 2012). AAV payloads persist as episomal DNA that are stable within postmitotic cells like photoreceptors and RPE over extended periods of time. One limitation of conventional AAV is its relatively small packaging capacity (∼5 kb). Recently, it was reported that AAV is capable of delivering cDNAs over 6 kb in length. Although the initial report indicated that the entire oversized genome was encapsidated (Allocca et al., 2008), this has proven to be unreproducible. It was later shown independently by several groups that large genomes are randomly truncated during packaging (Dong et al., 2010; Lai et al., 2010; Wu et al., 2010; Kapranov et al., 2012). Upon coinfection, these fragmented AAV genomes (fAAV) most likely undergo homologous recombination leading ultimately to expression of full-length protein. This process is inefficient since the packaging of oversized genomes results in low vector titers and poor transduction efficiency compared with standard-sized constructs (Dong et al., 2010; Lai et al., 2010). Most importantly, the heterogeneous nature of fAAV genomes precludes the ability to discretely characterize genetic payload in any vector preparation. This potentially complicates regulatory approval for therapeutic application. Defined dual AAV vectors provide an alternative approach. It was shown for minidystrophin and other large genes (Halbert et al., 2002; Lai et al., 2005, 2008; Lostal et al., 2010; Zhang and Duan, 2012) that expression of full-length protein can be achieved by splitting the cDNA between two individual vectors. Vector 1 contains the 5′ portion of the gene under the control of a promoter and vector 2 contains the 3′ portion of the gene and polyadenylation signal. After coinfection, intermolecular concatemerization followed by homologous recombination is mediated through overlapping regions of the cDNA, hence the term “simple overlap” system. However, inverted terminal repeats (ITRs), at the ends of vector genomes, are naturally recombinogenic so that many products of two vector recombination contain a central ITR. In the trans-splicing system, splice donor and acceptor sites are incorporated into respective vectors. Thus, the intervening RNA arising from ITR sequence is spliced out from the pre-mRNA. Simple overlap and trans-splicing dual-vector systems have been shown to lead to expression of full-length protein (Yan et al., 2000, 2002; Duan et al., 2001, 2003). Several improvements over the original dual-vector platforms have been devised. Hirsch et al. (2009) designed oligonucleotides to function as a tether to direct concatemerization of the 5′ and 3′ portions of the cDNA in respective vectors. Also, a “hybrid” vector system, so called because it utilizes both a highly recombinogenic sequence shared by the 5′ and the 3′ vector to direct homologous recombination and the aforementioned splice donor and acceptor sites for splicing, has also been developed. The hybrid system has showed increased expression relative to trans-splicing-only vectors (Lai et al., 2006; Ghosh et al., 2008, 2011). In this study, we evaluate multiple dual-AAV-vector systems with defined genetic payloads and show that they are capable of expressing MYO7A in vitro and in vivo with equal or higher efficiency than fAAV. The dual-vector systems compared were “simple overlap,” in which vector pairs share a central 1365 bp overlapping region of the MYO7A cDNA that mediates recombination; “trans-splicing,” in which vectors contain nonoverlapping 5′ and 3′ portions of MYO7A combined with either splice donor or splice acceptor sites derived from alkaline phosphatase (AP), respectively; “hybrid,” in which vectors contain the splice donor and splice acceptor component of the trans-splicing platform and an overlapping region of either AP sequence or intronic MYO7A sequence, either of which is intended to promote recombination; and the “native intron” hybrid vector, in which the MY07A intronic sequence contains native splice signals.
In this study, we sought to directly compare dual-AAV-vector platforms for their ability to drive MYO7A expression in vitro and in vivo. First, we assessed the ability of each platform to express full-length gene product in HEK293 cells and quantified relative efficiencies. Second, we characterized the fidelity of cDNA generated from recovered mRNA arising from each dual-vector platform by completely sequencing the region of overlap and/or splicing. Finally, for the two best performing dual-vector systems in vitro, we evaluated localization of MYO7A in retina relative to that mediated by the fAAV system.
Materials and Methods
AAV vector plasmid design and cloning
The full-length coding sequence of MYO7A (human isoform2; NM_001127180) was cloned into a vector plasmid containing the strong, ubiquitous CMV/chicken β-actin (smCBA) promoter (Haire et al., 2006), a polyadenylation signal, and the AAV2 ITRs. Packaging of this plasmid generated the fAAV vector (Fig. 1A). In all systems the 5′ vectors share the smCBA promoter and a 5′ portion of MYO7A, whereas the 3′ vectors contain a 3′ portion of MYO7A and a bovine growth hormone (bGH) polyadenylation signal. Oligonucleotides used for vector construction are listed in Table 1. The simple overlap contains nucleotides 1 through 3644 of MYO7A cDNA from the ATG in the 5′ vector and nucleotides 2279 through 6534 in the 3′ vector. The fragments were amplified with P1 and P3 by polymerase chain reaction (PCR) and cloned into the 5′ vector via NotI and NheI and the 3′ vector with P3 (AflII) and P4 (KpnI), respectively. The resulting two vector plasmids share 1365 bp of overlapping MYO7A sequence (Fig. 1B). The trans-splicing and hybrid vectors utilize splice junctions composed of either ideal splice donor and acceptor sites derived from AP coding sequence or native MYO7A splice junctions from exons 23 and 24 (Yan et al., 2002). To create the 5′ trans-splicing vector, the splice acceptor site was amplified by use of P5 and P6 (NheI), and the amplicon was then used in a second reaction with P7 (NsiI) to add a part of the MYO7A coding sequence for cloning. The corresponding 3′ vector was similarly created by amplifying the splice acceptor site with P8 (AflII) and P9 in a first PCR and adding part of the 3′ MYO7A coding sequence with P10 (AgeI) in a second PCR (Fig. 1C). The AP hybrid vectors were created by adding 270 bp of AP overlap sequence to the respective trans-splicing vectors (Ghosh et al., 2011). The sequence was amplified by PCR and, in so doing, appropriate restriction endonuclease sites were added. For the 5′ vector P11 (NheI) and P12 (SalI) were used and for the 3′ vector P13 (NotI) and P14 (AflII) were used (Fig. 1D). A fourth vector pair, “native intron hybrid” vector, was created to utilize the natural sequence in and around intron 23 of MYO7A as a recombination locus and subsequent splicing signal. The 5′ portion was created by amplifying intron 23 with P15 and P16 (NheI) first and using the resulting amplicon in a second reaction with P7 (NsiI) for cloning. The corresponding 3′ vector was constructed by amplifying the intron 23 with P17 and P18 (AflII) and the resulting amplicon with P10 (AgeI) in a second reaction (Fig. 1E).
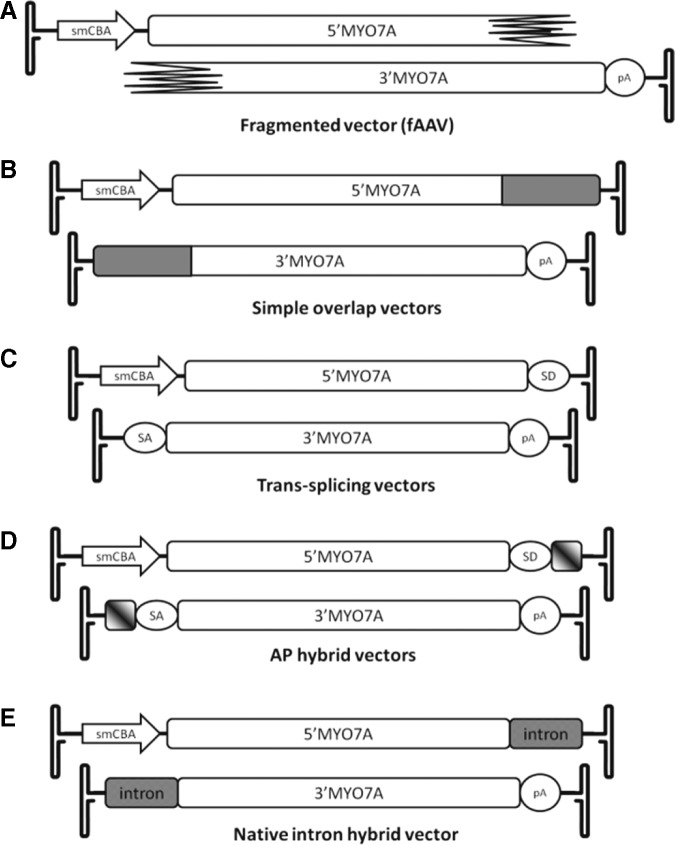
FIG. 1.
Schematic representation of the dual-AAV-vector pairs created for this study. (A) Fragmented vector (fAAV). (B) Simple overlap: the 1365 bp shared between the two vectors is shaded gray. (C) Trans-splicing vector. (D) AP hybrid vector: the 270 bp element shared between the two vectors is marked with diagonal gradient shading (1/3 head as described by Ghosh et al., 2011). (E) The native intron hybrid vectors utilizing the natural intron 23 of MYO7A sharing 250 bp overlapping sequence. 3′MYO7A, 3′ portion of MYO7A; 5′MYO7A, 5′ portion of MYO7A; AAV, adeno-associated virus; AP, alkaline phosphatase; intron, intron 23 of MYO7A; pA, polyadenylation signal; SA, splice acceptor site; SD, splice donor site; smCBA, cytomegalovirus immediate early/chicken β-actin chimeric promoter.
Table 1.
Oligonucleotides Used in This Study
| Oligo | 5′–3′ sequence (restriction sites underlined) | Restriction site |
|---|---|---|
| P1 | GCGGCGGCCGCCACCATGGTGATTCTTCAGCAGGGGGAC | NotI |
| P2 | GCGGCTAGCGAAGTTCCGCAGGTACTTGAC | NheI |
| P3 | GCGCTTAAGCAGGTCTAACTTTCTGAAGCTG | AflII |
| P4 | GCGGGTACCTCACTTGCCGCTCCTGGAGCC | KpnI |
| P5 | GGCACCTAGTGGCTTTGAGGTAAGTATCAAGGTTACAAGAC | |
| P6 | GCGGCTAGCTCAGAAACGCAAGAGTCTTC | NheI |
| P7 | CTTCTTTGTGCGATGCATCAAG | NsiI |
| P8 | GCGCTTAAGCGACGCATGCTCGCGATAG | AflII |
| P9 | CGCCCTCGCTCCAGGTCCTGTGGAGAGAAAGGCAAAG | |
| P10 | GAACCCGAACCGGTCCTTG | AgeI |
| P11 | GCGGCTAGCCCCCGGGTGCGCGGCG | NheI |
| P12 | GCGGTCGACGAAACGGTCCAGGCTATGTG | SalI |
| P13 | GCGGCGGCCGCCCCCGGGTGCGCGGCG | NotI |
| P14 | GCGCTTAAGGAAACGGTCCAGGCTATGTG | AflII |
| P15 | CAGGCACCTAGTGGCTTTGAGGTACCAGGCTAGGGACAGG | |
| P16 | GCGGCTAGCCGCCTGAGCCCAGAAGTTC | NheI |
| P17 | CGCCCTCGCTCCAGGTCCTGAAGGAGACAAGAGGTATG | |
| P18 | GCGCTTAAGCACCGCTTGTGTTGATCCTC | AflII |
| P19 | GCCAGGGAAGGATCCCATG | BamHI |
| P20 | GCGGGTACCTCATGCGTAATCCGGTACATCGTAAGGGTACTTGCCGCTCCTGGAGCC | KpnI |
| P21 | AGCTTCGTAGAGTTTGTGGAGCGG | |
| P22 | GAGGGGCAAACAACAGATG |
Oligonucleotides were used to make 5′ and 3′ vectors of the dual-vector platforms (P1–P20). Oligonucleotides were used to characterize the fidelity of the overlap in simple overlap, trans-splicing and AP hybrid vector platforms (P21–P22). Restriction sites used for cloning are underlined and the introduced HA tag is noted in italics (P19).
For in vivo studies, a human influenza hemagglutinin (HA) tag was added to the 3′ termini of the full-length, simple overlap, trans-splicing, and hybrid 3′ vectors by utilizing a unique BamHI site (P19) and replacing the nontagged 3′ end with an HA-tagged (P20) version. All constructs were sequence verified by Sanger sequencing (University of Florida ICBR Sequencing Core, Gainesville, FL).
Reverse transcription and characterization of overlap region
HEK293 cells were infected with dual vectors and total RNA was extracted with the RNeasy kit (Qiagen, Hilden, Germany) according to manufacturer's protocol. Two micrograms of RNA was then subjected to DNaseI (NEB) digest for 30 min at 37°C followed by heat inactivation at 75°C for 10 min. Reverse transcription to cDNA was achieved with the SuperscriptIII kit (Life Technologies, Grand Island, NY) according to protocol utilizing the oligo dT primer. Two microliters of cDNA was used as template in a PCR (95°C 3 min initial denature, 35 cycles of 95°C 45 sec, 55°C 45 sec, 72°C 12 min, and final 72°C 15 min) using primers P21 and P22 (see Table 1). Annealing sites for these primers are located 5′ and 3′, respectively, of the area of cDNA overlap (in other words, outside the region of overlap) in the simple overlap and hybrid vector pairs. The 3′ primer anneals to sequence complimentary to the bGH polyA. Resulting products were then digested with either PpuMI or BglII and separated on a 1.5% agarose gel and subsequently analyzed on a UV screen. Separately, products were digested with KpnI and AgeI and subsequently cloned into a pUC vector for sequencing of the entire overlap region. M13 forward and reverse primers specific for the vector were used to obtain sense and antisense reads resulting in ∼140 bp for which sense and antisense reads overlapped. To confirm that our methods were capable of detecting aberrant sequence (quality control), we generated MYO7A sequence with either an artificial insertion (HindIII fill in at position 2635) or a point mutation (T-C) at position 2381 and repeated the analyses.
Vector production, transfections, and infections
All vectors intended for in vitro analyses were separately packaged in AAV2 or AAV2(tripleY-F) capsid containing three phenylalanine-to-tyrosine substitutions (Y444F, Y500F, Y730F) (Petrs-Silva et al., 2011). AAV2-based vectors were chosen for in vitro experiments based on their increased transduction efficiency relative to other serotypes (Ryals et al., 2011). Vectors intended for in vivo analyses were packaged in AAV8(Y733F) capsid (Petrs-Silva et al., 2009). This vector was chosen for in vivo experiments based on its increased transduction of photoreceptors/RPE (the two sites of documented MYO7A expression) relative to other serotypes. All vectors were packaged, purified, and titered by standard methods as previously described (Zolotukhin et al., 2002; Jacobson et al., 2006). Human embryonic kidney (HEK293) cells were transfected by calcium phosphate method with vector plasmid carrying the full-length MYO7A coding sequence of variant 2 (plasmid used to package fAAV). These transfected cells were used as a positive control throughout immunoblot analyses to indicate the appropriate size of full-length MYO7A protein. Vector infections were carried out in HEK293 cells with titer-matched AAV vectors. In brief, cells were grown to 60–70% confluency. All vectors were diluted in balanced salt solution to achieve the desired multiplicity of infection (MOI). If not specifically mentioned, cells were infected at 10,000 genome-containing particles/cell of each vector, resulting in an MOI of 20,000 total for each vector pair. Cells were incubated in 10% serum containing medium for 3 days postinfection at 37°C under 7% CO2 and were then analyzed via immunoblot.
Animals
Vectors were subretinally injected in 1-month-old C57BL/6 mice (Jackson Laboratory, Bar Harbor, ME). All mice were maintained in the University of Florida Health Science Center's animal care facilities and were handled in accordance with the statement of Association for Research in Vision and Ophthalmology for Use of Animals in Ophthalmic and Vision Research and the guidelines of the Institutional Animal Care and Use Committee of the University of Florida.
Subretinal injections
One microliter of the following AAV8(Y733F)-based vectors was injected subretinally in one eye of C57BL/6 mice: single fAAV (1×1013 vg/ml), front and back half “hybrid” vectors combined equally (each vector=1×1013 vg/ml), or front and back half “simple overlap” vectors combined equally (each vector=1×1013 vg/ml). Subretinal injections were performed as previously described (Timmers et al., 2001). Further analysis was carried out only on animals that received comparable, successful injections (>60% retinal detachment with minimal surgical complications).
Protein extraction and immunoblotting
Transfected and infected HEK293 cells were harvested and washed twice in PBS and processed as previously reported with minor modifications (Boye et al., 2012). The cells were lysed by 3×30 sec pulses of sonication in 200 μl of sucrose buffer (0.23 M sucrose, 2 mM EDTA, 5 mM Tris-HCl, pH 7.5) containing protease inhibitors (Roche, Mannheim, Germany). Unlysed cells and cell debris were removed by centrifugation at 14,000 rpm for 10 min. The protein concentration of the supernatant was measured with BCA (Thermo Fisher Scientific, Rockland, IL). Equal amounts of protein were then loaded on 7.5% sodium dodecyl sulfate polyacrylamide gel electrophoresis gels (BioRad, Hercules, CA) and transferred in CAPS buffer (pH 11) onto PVDF membranes (Millipore, Billerica, MA). Blots were then labeled with antibodies against MYO7A (monoclonal antibody raised against amino acids 11–70 of human MYO7A; Santa Cruz, Dallas, TX; 1:1000) or HA (MMS-101P; Covance, Gaithersburg, MD; 1:500) and beta-actin (ab 34731; Abcam, Cambridge, MA; 1:5000). For visualization with the Odyssey system (Li-Cor, Lincoln, NE), an antimouse and an antirabbit secondary antibody conjugated with CW800 and IR680 dyes (Li-Cor), respectively, were used. Semiquatitative densitometric measurements were performed with Odyssey acquisition and analysis software (Li-Cor). The dual-color images were separated in their respective channels and converted to gray scale for presentation purposes. Size markers present in one channel of each blot were added to both channels for visualization of protein sizes.
Tissue processing and immunohistochemistry
Six weeks postinjection, C57BL/6 mice were enucleated and their eyes processed and immunostained as previously described (Boye et al., 2011) with minor modifications. Retinas were immunostained with an antibody specific for HA (monoclonal Ab clone 12CA5; Roche), counterstained with DAPI, and imaged with a spinning disk confocal microscope (Nikon Eclipse TE2000 microscope equipped with Perkin Elmer Ultraview Modular Laser System and Hamamatsu O-RCA-R2 camera). Images were obtained sequentially using a 20×(air) objective lens. All settings (exposure, gain, laser power) were identical across images. All image analysis was performed using Volocity 5.5 software (Perkin Elmer, Waltham, MA).
Results
Expression of MYO7A with simple overlap vectors
AAV2-based simple overlap vectors were evaluated in vitro at a variety of MOIs to evaluate how the concentration of vector pairs related to MYO7A expression. We also evaluated how levels of MYO7A changed over time in infected cells. HEK293 cells were infected with simple overlap vector pairs packaged in AAV2(tripleY-F) vector (Fig. 2). A preliminary coinfection with AAV2(tripleY-F) simple overlap vectors (MOI of 10,000 for each vector) indicated that MYO7A is expressed and that migration of the protein on gel is identical to a full-length transfection control (Fig. 2A). Coinfection at MOIs of 400, 2000, and 10,000 of each vector shows that the efficiency of the simple overlap system is proportional to the amount of 5′ and 3′ vectors used (Fig. 2B). MYO7A expression increased as a function of incubation time up to 5 days postinjection in HEK293 cells (Fig. 2C). The visible expression decline was because of a reduction of viable cells in the culture vessel at the later times (data not shown).
FIG. 2.
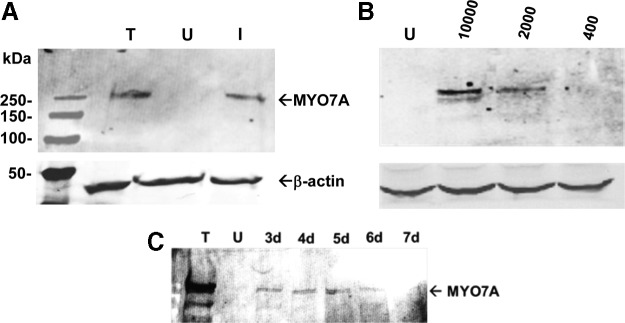
Human embryonic kidney (HEK293) cells express human MYO7A after infection with simple overlap vectors (MOI of 10,000 for both vectors) packaged in AAV2(tripleY-F). Equal amounts of protein were separated on 7.5% sodium dodecyl sulfate polyacrylamide gel electrophoresis and stained for MYO7A (A). HEK293 cells infected with AAV2(tripleY-F) at MOIs of 10,000, 2000, and 400 (B). Time course of MYO7A expressed in HEK293 cells. Cells were harvested 3–7 days after infection (C). I, HEK293 cells infected with AAV2 dual vectors; MOI, multiplicity of infection; T, HEK293 cells transfected with full-length MYO7A plasmid; U, untreated HEK293 cells.
Comparison of fAAV-MYO7A to dual-AAV-MYO7A expression and evaluation of AAV serotype efficiency
Previously, we and others have shown that fAAV-MYO7A is able to ameliorate the retinal phenotype of the shaker1 mouse (Colella et al., 2013; Lopes et al., 2013; Trapani et al., 2013). To provide a basis for comparison we evaluated dual-AAV-vector expression relative to fAAV in vitro. After infection in HEK293 cells, all dual-vector systems expressed MYO7A more efficiently than fAAV (Fig. 3). The AP hybrid platform showed the strongest expression, followed by the simple overlap system.
FIG. 3.
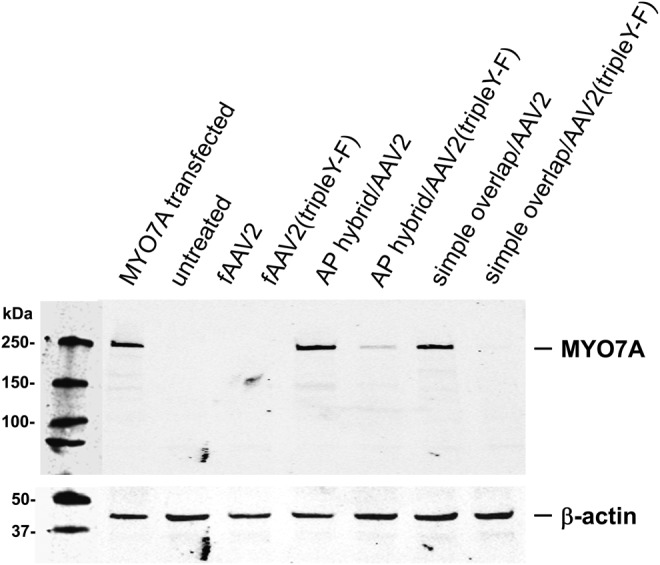
Comparison of AAV2 and AAV2(tripleY-F)-based vectors in HEK293 cells. Cells were infected with single fAAV, AP hybrid, and simple overlap MYO7A dual-vector platforms packaged in AAV2 or AAV2(tripleY-F) at an MOI of 10,000. HEK293 cells transfected with MYO7A plasmid were used as positive control.
Other studies have shown, in the context of a conventionally sized DNA payload, that the transduction efficiency and kinetics of AAV2(tripleY-F) vectors are increased relative to standard AAV2 both in vitro and in vivo (Li et al., 2010; Markusic et al., 2010; Ryals et al., 2011). We directly compared efficiency of AAV2 versus AAV2(tripleY-F) dual vectors in HEK293 cells. To our surprise, standard AAV2-mediated MYO7A expression was higher than that seen with titer-matched AAV2(tripleY-F) (Fig. 3). Identical results were obtained when comparing different AAV2 and AAV2(tripleY-F) dual-vector preparation packaged with identical vector plasmid (data not shown). The basis for this difference in efficiencies in vitro remains to be elucidated.
Comparison of relative efficiencies and specificity of full-length MYO7A expression
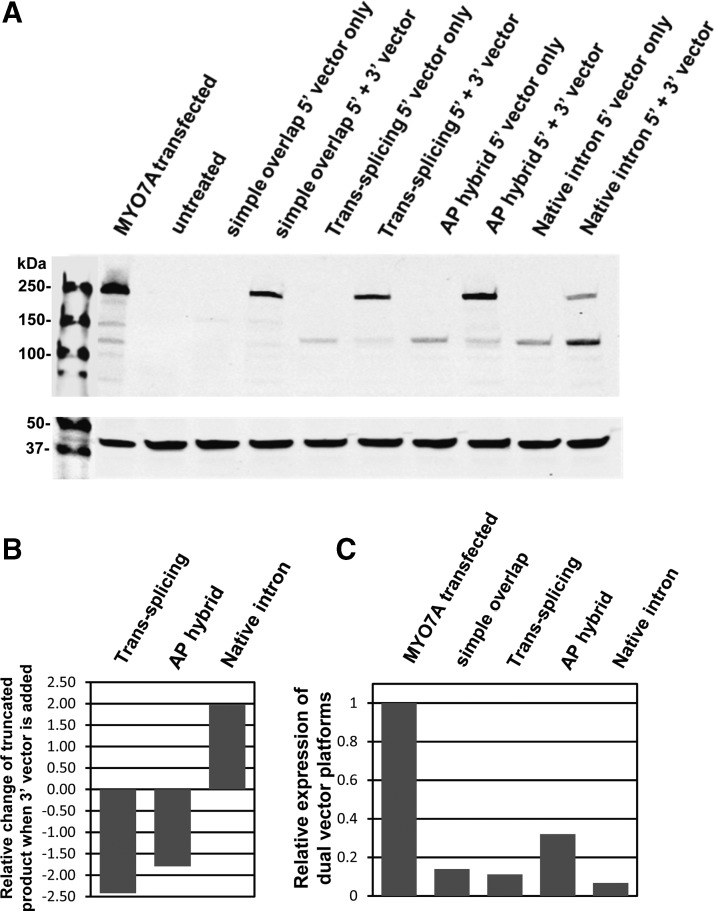
To quantitatively evaluate the relative expression efficiencies of the dual-vector platforms and to assess specificity of full-length protein, we infected HEK293 cells with either the 5′ and 3′ AAV2-based vector pairs combined or the corresponding 5′ vector alone. We included an additional hybrid vector pair that incorporated native MYO7A intronic sequence (intron 23) that served as overlapping sequence and provided appropriate splicing signals. All 5′ vectors produced low amounts of a defined, less than full-length peptide detectable on Western blot with the exception of the simple overlap vector (Fig. 4A). However, the trans-splicing and the AP hybrid platforms revealed a distinct decrease of this undesired product when the 3′ vector was added to the sample (Fig. 4A). The native intron hybrid platform also showed this secondary band on Western blots, again suggestive of a truncated protein originating from the 5′ vector alone. In contrast to all other platforms tested, this band intensity increased with the addition of the 3′ vector. Each platform's relative ability to promote reconstitution was compared by quantifying the amount of 5′ vector-mediated truncated protein product in the presence or absence of the respective 3′ vector (Fig. 4B). Full-length MYO7A expression on Western blot was then quantified relative to transfection control (Fig. 4C). AP hybrid-mediated MYO7A was the strongest followed by simple overlap, trans-splicing, and native intron hybrid (Fig. 4C).
FIG. 4.
Human MYO7A expressed in HEK293 cells. Cells were infected with AAV2-based vector platforms. For each of the dual-vector systems, the corresponding 5′ and 3′ vectors or only the 5′ vector were used for infection. HEK293 cells transfected with MYO7A plasmid were used as positive control. Cells were infected with the MYO7A dual-vector pairs at an MOI of 10,000 for each vector. Protein samples were analyzed on Western blot with an antibody against MYO7A (A). Each platform's relative ability to promote reconstitution was compared by quantifying the amount of 5′ vector-mediated truncated protein product in the presence or absence of the respective 3′ vector (B). Full-length MYO7A expression mediated by dual vectors was quantified relative to transfection control (C).
Characterization of the overlap/splice region of the expressed MYO7A
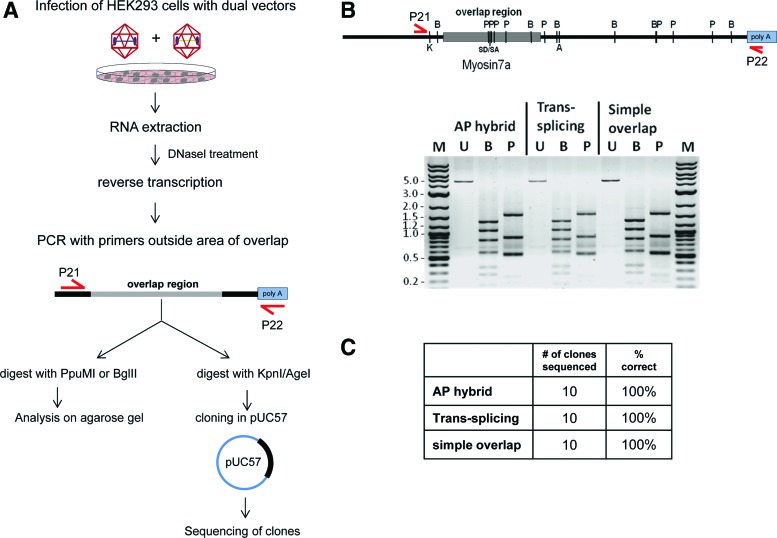
To characterize the fidelity of the mRNA arising from dual vectors, HEK293 cells were infected with dual vectors and RNA extracted, reverse transcribed, and subjected to PCR utilizing primers binding upstream of the overlap region and in the bGH polyA signal region producing a 4.5 kb PCR fragment (Fig. 5A). An identically treated sample not containing reverse transcriptase was used as control for chromosomal DNA contamination. Plasmid containing the full-length MYO7A coding sequence was used as positive control for PCR. A preliminary screen of AAV-mediated MYO7A mRNA was performed by analyzing the pattern of fragment migration on agarose gel following restriction endonuclease digests with PpuMI and BglII (Fig. 5A). Identical banding patterns, consistent with the predicted pattern (PpuMI: 1591, 876, 556, 548, 541, 238, 168, 42, and 36 bp; BglII: 1335, 1074, 827, 583, 360, 272, and 146 bp), were observed following digests of amplicons from each dual-vector platform tested, indicating that no gross alterations (deletions/insertions) occurred as a consequence of either homologous recombination of vector pairs and/or RNA splicing (Fig. 5B). To further characterize the fidelity of the overlap region, we restricted a fragment containing the complete overlap area (1829 bp) and cloned them into pUC57 (Fig. 5A). Sequencing results of 10 clones picked at random per vector platform revealed that the overlap region was 100% identical to the consensus/predicted MYO7A sequence (Fig. 5C). This indicates that, in the context of the simple overlap platform, homologous recombination was accurate. Additionally, in the context of trans-splicing vectors, accurate splicing occurred. Finally, for the AP hybrid vectors, a combination of accurate homologous recombination and/or splicing took place. To determine whether this protocol was capable of detecting aberrant sequence in reconstituted MYO7A, we generated a sequence that contained either an insertion of a HindIII recognition site (TAGC) at position 2635 or a point mutation (T-C) at position 2381. Using the same methods, we were able to reliably detect an aberrant sequence within the overlap region. Representative chromatograms of the MYO7A sequence following reconstitution of AP hybrid vectors with or without the aforementioned mutations are shown in Supplementary Fig. S1 (Supplementary Data are available online at www.liebertpub.com/hgtb).
FIG. 5.
Characterization of MYO7A dual vectors' restoration of coding sequence. Experimental plan is shown in (A). HEK293 cells were infected with AAV2-based dual-vector platforms and RNA was extracted and PCR-amplified with gene-specific primers. Control digests with BglII (B) and PpuMI (P) revealed the predicted banding pattern shown in (B). Undigested (U) PCR product is shown as control and a DNA size marker for reference (M). Separately, products were digested with KpnI and AgeI and cloned into pUC57 for sequencing of the entire overlap region (C). Ten clones per vector platform were analyzed. M13 forward and reverse primers specific for the subclone vector were used to obtain sense and antisense reads (each ∼1000 bp) resulting in ∼140 bp for which sense and antisense reads overlapped. PCR, polymerase chain reaction. Color images available online at www.liebertpub.com/hgtb
MYO7A expression mediated by dual vectors in mouse retina
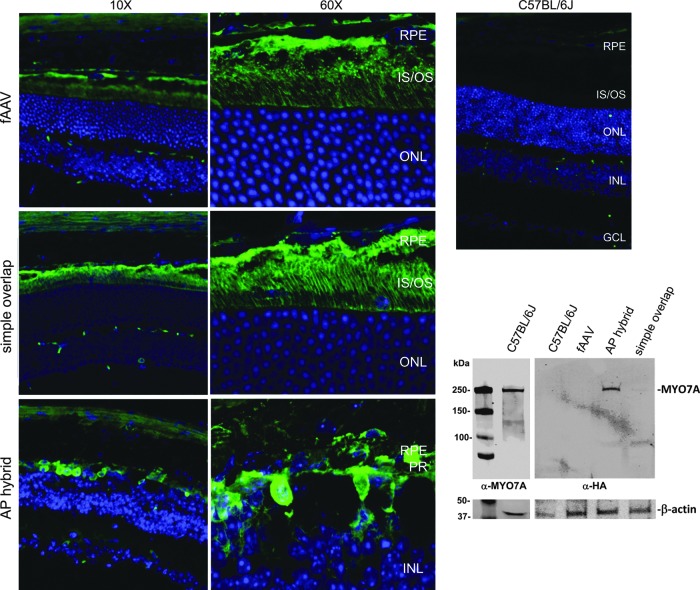
To investigate the expression of MYO7A from the two best performing dual-vector platforms in vivo, C57BL/6J mice were subretinally injected with 1×1010 vector genomes per eye of simple overlap and AP hybrid systems packaged in AAV8(733) and analyzed 4 weeks later by Western blot and immunohistochemistry (Fig. 6). AAV8(733)-fAAV-MYO7A vector was also injected to provide a basis for comparison. To distinguish between endogenous MYO7A and exogenous expression mediated by vectors, sequence coding for an HA tag was added to the C′ terminus of the MYO7A cDNA in all constructs. Resulting retinas were immunostained for HA to reveal that fAAV vector along with both dual-vector platforms mediated expression of MYO7A in photoreceptors and RPE. A recent report concluded that simple overlap vectors were more efficient for gene transfer to the RPE than photoreceptors (Trapani et al., 2013). We observed simple overlap-mediated MYO7A expression in both RPE and photoreceptors (Fig. 6). In contrast to our previous results showing “spotty” MYO7A expression mediated by AAV2-based simple overlap vectors (Lopes et al., 2013), we found, when packaged in AAV8(733), that simple overlap vectors mediated MYO7A expression in the majority of RPE and photoreceptor cells (Fig. 6). Photoreceptor degeneration/outer nuclear layer thinning was apparent in eyes injected with the AP hybrid vector system (Fig. 6). Despite the observed degeneration, AP hybrid-mediated MYO7A was clearly detected in residual PR cell bodies and RPE and was sufficient to be detected by immunoblot (Fig. 6). By Western blot analysis using HA antibody, simple overlap-mediated MYO7A was present in just detectable amounts. In contrast, fAAV-mediated protein levels were insufficient to be detected in this assay (Fig. 6). Using an antibody against MYO7A, immunoblot of WT mouse retina revealed that both endogenous MYO7A and dual-vector-mediated, HA-tagged MYO7A migrated similarly (Fig. 6).
FIG. 6.
Dual-vector-mediated MYO7A (HA) expression in vivo. C57BL/6J mice were injected subretinally with AAV2-based dual vectors containing a C′ terminal HA tag. Retinal protein expression was analyzed 4 weeks later by immunohistochemistry and Western blot. Ten-micrometer frozen retinal cross sections were imaged at 10×and 60×. Equal amounts of protein were separated on a 4–15% polyacrylamide gel and stained with an HA antibody. For comparison, endogenous MYO7A from C57BL/6J retina was probed with an antibody against MYO7A to confirm that HA-tagged MYO7A migrated at the appropriate size. GCL, ganglion cell layer; INL, inner nuclear layer; IS, inner segments; ONL, outer nuclear layer; OS, outer segments; PR, photoreceptors; RPE, retinal pigment epithelium.
Discussion
AAV is an efficient vehicle for gene transfer to the retina in animal models and humans (Boye et al., 2013). One limitation, however, is its relatively small packaging capacity. It is possible to package cDNAs exceeding conventional size limitations (i.e., >5 kb), but resulting vector is composed of randomly truncated fragments differing greatly in size. Enrichment for packaged genomes of maximum size is feasible and can increase the efficiency of the system (Hirsch et al., 2013). In general, however, the heterogeneous nature of the fAAV platform may complicate regulatory approval before clinical application.
Here we show that dual AAV vectors with defined genetic payloads can be used to deliver a large transgene in vitro and in vivo. Our initial experiments using the simplest of all dual-vector platforms revealed that efficiency of AAV2-based simple overlap vectors is proportional to the amount of 5′ and 3′ vectors used and that MYO7A expression mediated by this system increased as a function of incubation time in HEK293 cells. Next, we evaluated three distinct dual-vector platforms and compared them to single, fragmented fAAV vector in vitro. All dual vectors analyzed drove higher levels of MYO7A expression than fAAV. Of all platforms tested, a hybrid vector system containing overlapping, recombinogenic sequence and splice donor/acceptor sites from the AP gene (AP hybrid) was the most efficient.
Regarding the specificity with which the dual-vector platforms express the correct-sized gene product, we noted in vitro that trans-splicing and hybrid dual-vector platforms generated an additional band of lower molecular weight as detected by immunoblot (monoclonal antibody used was raised against the amino terminus MYO7A). The expression of this truncated product was much more pronounced for infections with 5′ vectors alone. What might account for this additional band? After entry into the host cell, the virus capsid is removed and the single-stranded DNA payload is released. The ITRs carried by the single strand serve as primer for DNA polymerases to produce a double strand. The resulting circular intermediates consist mainly of monomers that, over time, convert into multimeric concatemers through intermolecular recombination (Duan et al., 1998; Yang et al., 1999). The dual-vector systems created in this study utilize this strategy to achieve full-length protein expression. A limiting factor lies in the fact that the highly recombinogenic ITRs flanking the expression cassettes are identical in nature leading to a random recombination and consequently a random orientation of the vector parts relative to each other. This random recombination inevitably results in reduced efficiency because only concatemers that have the two vector parts in 5′–3′ orientation are able to express the full-length protein. This concatemerization over time is consistent with our observation that the amount of single-vector product is reduced in favor of the full-length protein when both 5′ and 3′ vectors are combined. Interestingly, the simple overlap system does not generate truncated product, even when only the 5′ vector is used for infections. In contrast to the trans-splicing and hybrid vectors, there is virtually no intervening sequence between the end of the MYO7A coding sequence and the right-hand ITR. It may be that splice donor sequences enhance the likelihood of truncated product through some as-yet-to-be-determined mechanism.
Moving forward, how can we overcome the issue of random concatemerization and thereby increase specificity as well as efficiency of these dual-vector platforms? A number of strategies have been devised to circumvent this problem. First, the addition of a highly recombinogenic sequences such as that used in the AP hybrid vector here has resulted in significantly increased protein expression compared with the trans-splicing system. Ghosh et al. provide a detailed analysis of the 270 bp AP sequence used in this study as well as other sequences derived from AP that direct recombination and lead to significant improvement over trans-splicing vectors (Ghosh et al., 2011). Our finding that AP hybrid vectors are more efficient than trans-splicing vectors supports that the AP sequence directs at least some of the concatemerization events toward the proper orientation with recombination then occurring via this sequence or via the ITRs. Regardless, with more concatemers properly aligned, the AP hybrid system mediates more efficient MYO7A expression. Another approach for directing concatemerization is the use of single-strand oligonucleotides that are capable of tethering the back end of the 5′ vector and the front end of the 3′ vector together (Hirsch et al., 2009). However, this strategy requires efficient delivery of the oligonucleotide to the nucleus of the target cells timed with the dual vectors. Finally, dual vectors utilizing mismatched ITRs can be used to direct concatemerization in a head-to-tail orientation (Yan et al., 2005). This may require optimization of the AAV packaging machinery, but is worth exploring.
Before clinical application of dual AAV vectors, the integrity of reconstituted full-length transcript will need to be carefully evaluated. Notably, in our study we found that the sequence in the overlap region of all dual vectors tested in vitro was 100% identical to the consensus/predicted MYO7A sequence. This indicates that homologous recombination and/or splicing was accurate in each dual-vector platform. Similar experiments to evaluate the region of overlap are warranted for each large transgene evaluated. Ideally, this characterization should also be performed in vivo.
Similar to our in vitro results, we found the highest levels of MYO7A expression in retinas of mice subretinally injected with AAV8-based AP hybrid vectors (as assessed by probing for HA on Western blot). Notably, no truncated proteins were evident in retinas expressing either simple overlap or AP-hybrid mediated MYO7A. The reason for this observed difference remains to be elucidated but may involve differences in the DNA repair machinery that mediate recombination in actively dividing cells versus postmitotic photoreceptors/RPE (Hirsch et al., 2013). Dual-vector-mediated MYO7A-HA expression was observed in the photoreceptors and RPE of WT mice, locations where MYO7A is thought to have a functional role (Williams and Lopes, 2011). In eyes injected with AP hybrid vectors, we observed marked thinning of the outer nuclear layer. It has previously been shown that vector-mediated overexpression of MYO7A leads to retinal toxicity (Hashimoto et al. 2007). Taken together with the high efficiency of transduction observed in vitro for the AP hybrid platform (current study), the most likely explanation for the observed pathology is excessive production of MYO7A. However, this remains to be definitively determined. Despite the marked degeneration, significant amounts of AP hybrid-mediated, full-length MYO7A-HA were detected on Western blot. As high concentrations of vectors were used in these experiments, a simple solution to circumvent toxicity could be to reduce vector genomes injected or replace the strong, ubiquitous smCBA promoter with an endogenous or homologous promoter, and/or a promoter with attenuated strength. Future studies will encompass these experiments. An alternative explanation for toxicity of the strong AP hybrid platform is expression of undesired products, like the observed protein expressed from the 5′ vectors alone in vitro. However, we note that only full-size MYO7A-HA was apparent on Western blot of the AP hybrid-treated retina.
An obvious question raised is whether AP hybrid vectors such as those used here will be the most efficient platform for delivering large genes in general. It is clear that efficiency of simple overlap and hybrid dual-AAV-vector systems is dependent on the overlapping sequence used to mediate recombination. In contrast to simple overlap-mediated MYO7A expression, we have observed that expression of another large protein (ATP-binding cassette, subfamily A, member 4—“ABCA4”) via the simple overlap system was highly inefficient relative to other dual-vector platforms in vitro (unpublished results). This suggests that the performance of dual vectors containing overlap is intrinsically linked to the recombinogenic potential of the portions of the gene used as overlap. Therefore, caution should be taken in extending the predictive value of comparisons made for dual-AAV-vector platforms using “generic” overlap elements. For example, one may find that a simple overlap system containing overlapping sequence from a reporter gene is far less efficient than a hybrid system incorporating the same overlapping sequence. This does not necessarily mean that a simple overlap system containing overlapping regions of therapeutic genes like MYO7A, ABCA4, or CEP290 will be less efficient than a hybrid system containing generic overlapping sequence like AP. Other important considerations are the level of efficiency required of each transgene (i.e., how much protein is required to affect therapy?) and the specificity of each platform for each transgene (i.e., will simple overlap vectors containing genes other than MYO7A lead to production of truncated proteins?). For all reasons stated, comparisons of dual-vector platforms unique to each large transgene are warranted.
With the goal of developing an AAV-based treatment for USH1B, we look to what has been accomplished in animal models of this disease. Similar to our previous observation that fAAV-MYO7A and simple overlap dual vectors are capable of restoring melanosome migration and opsin localization in the shaker1 mouse (Lopes et al., 2013), a recent study indicates that hybrid and trans-splicing vectors similar to those used here were also capable of restoring the ultrastructural retinal phenotypes in this model. Notably, shaker1 mice lack retinal degeneration and the severe functional abnormalities seen in USH1B patients (Liu et al., 1997). This fact renders in vivo analysis of therapeutic outcomes in the shaker1 retina problematic. Alternative animal models for evaluating a treatment for this devastating disease warrant consideration.
Our results demonstrate that MYO7A can be efficiently expressed using dual-AAV-vector systems. We show that the platforms containing overlapping elements, namely, the simple overlap system and the AP hybrid system, were both highly efficient. AP hybrid vectors showed the strongest expression of all systems tested, with little observable truncated protein in vitro and none observed in vivo. Simple overlap vectors showed good expression and were the most specific (no truncated protein products were observed) even when the 5′-only vector was used to infect cells. AAV has emerged as the preferred clinical vector and it efficiently transduces both photoreceptors and RPE. Because we have demonstrated that MYO7A sequence fidelity is preserved following recombination and/or splicing of dual-AAV-vector platforms and because only full-length MYO7A was detectable in mouse retinas injected with dual vectors, we propose that a dual-AAV-vector strategy may be a valid option for the treatment of retinal disorders associated with mutations in large genes such as USH1B. In general, USH1B patients exhibit a progressive decline in visual acuity, retinal function, and photoreceptor structure with age (Jacobson et al., 2011). However, like other retinal degenerations, disease severity varies between patients depending on the MYO7A mutation(s) they carry (Jacobson et al., 2011). Many USH1B patients retain central retinal structure and function as well as islands of peripheral/temporal field until late stages of the disease (Jacobson et al., 2011). Detailed analyses of patient genotype and phenotype will therefore be required to understand the safest/most effective temporal and spatial parameters for clinical application of this technology.
Supplementary Material
Acknowledgments
The authors thank Dr. Dongsheng Duan for generously sharing the sequence of “AP head” and Dr. Seok-Hong Min, Dr. James Peterson, Santhi Mani, and Jingfen Sun for their excellent technical assistance. They also acknowledge grants from the Foundation Fighting Blindness, Research to Prevent Blindness, Inc., and National Eye Institute (P30EY021721) for partial support of this work.
Author Disclosure Statement
W.W.H. and the University of Florida have a financial interest in the use of AAV therapies and own equity in a company (AGTC Inc.) that might, in the future, commercialize some aspects of this work.
References
- Allocca M., Doria M., Petrillo M., et al. (2008). Serotype-dependent packaging of large genes in adeno-associated viral vectors results in effective gene delivery in mice. J. Clin. Invest. 118, 1955–1964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Astuto L.M., Weston M.D., Carney C.A., et al. (2000). Genetic heterogeneity of Usher syndrome: analysis of 151 families with Usher type I. Am. J. Hum. Genet. 67, 1569–1574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bainbridge J.W., Smith A.J., Barker S.S., et al. (2008). Effect of gene therapy on visual function in Leber's congenital amaurosis. N. Engl. J. Med. 358, 2231–2239 [DOI] [PubMed] [Google Scholar]
- Bharadwaj A.K., Kasztejna J.P., Huq S., et al. (2000). Evaluation of the myosin VIIA gene and visual function in patients with Usher syndrome type I. Exp. Eye Res. 71, 173–181 [DOI] [PubMed] [Google Scholar]
- Bowles D.E., McPhee S.W., Li C., et al. (2012). Phase 1 gene therapy for Duchenne muscular dystrophy using a translational optimized AAV vector. Mol. Ther. 20, 443–455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boye S.L., Conlon T., Erger K., et al. (2011). Long-term preservation of cone photoreceptors and restoration of cone function by gene therapy in the guanylate cyclase-1 knockout (GC1KO) mouse. Invest. Ophthalmol.Vis. Sci. 52, 7098–7108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boye S.L., Peshenko I.V., Huang W.C., et al. (2012). AAV-mediated gene therapy in the guanylate cyclase (RetGC1/RetGC2) double knockout mouse model of Leber congenital amaurosis. Hum. Gene Ther. 24, 189–202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boye S.E., Boye S.L., Lewin A.S., and Hauswirth W.W. (2013). A comprehensive review of retinal gene therapy. Mol. Ther. 21, 509–519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cideciyan A.V., Hauswirth W.W., Aleman T.S., et al. (2009). Human RPE65 gene therapy for Leber congenital amaurosis: persistence of early visual improvements and safety at 1 year. Hum. Gene Ther. 20, 999–1004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colella P., Sommella A., Marrocco E., et al. (2013). Myosin7a deficiency results in reduced retinal activity which is improved by gene therapy. PLoS One 8, e72027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong B., Nakai H., and Xiao W. (2010). Characterization of genome integrity for oversized recombinant AAV vector. Mol. Ther. 18, 87–92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan D., Sharma P., Yang J., et al. (1998). Circular intermediates of recombinant adeno-associated virus have defined structural characteristics responsible for long-term episomal persistence in muscle tissue. J. Virol. 72, 8568–8577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan D., Yue Y., and Engelhardt J.F. (2001). Expanding AAV packaging capacity with trans-splicing or overlapping vectors: a quantitative comparison. Mol. Ther. 4, 383–391 [DOI] [PubMed] [Google Scholar]
- Duan D., Yue Y., Yan Z., and Engelhardt J.F. (2003). Trans-splicing vectors expand the packaging limits of adeno-associated virus for gene therapy applications. Methods Mol. Med. 76, 287–307 [DOI] [PubMed] [Google Scholar]
- Flotte T.R., Trapnell B.C., Humphries M., et al. (2011). Phase 2 clinical trial of a recombinant adeno-associated viral vector expressing alpha1-antitrypsin: interim results. Hum. Gene Ther. 22, 1239–1247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh A., Yue Y., Lai Y., and Duan D. (2008). A hybrid vector system expands adeno-associated viral vector packaging capacity in a transgene-independent manner. Mol. Ther. 16, 124–130 [DOI] [PubMed] [Google Scholar]
- Ghosh A., Yue Y., and Duan D. (2011). Efficient transgene reconstitution with hybrid dual AAV vectors carrying the minimized bridging sequences. Hum. Gene Ther. 22, 77–83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibbs D., Kitamoto J., and Williams D.S. (2003). Abnormal phagocytosis by retinal pigmented epithelium that lacks myosin VIIa, the Usher syndrome 1B protein. Proc. Natl. Acad. Sci. USA 100, 6481–6486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haire S.E., Pang J., Boye S.L., et al. (2006). Light-driven cone arrestin translocation in cones of postnatal guanylate cyclase-1 knockout mouse retina treated with AAV-GC1. Invest. Ophthalmol. Vis. Sci. 47, 3745–3753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halbert C.L., Allen J.M., and Miller A.D. (2002). Efficient mouse airway transduction following recombination between AAV vectors carrying parts of a larger gene. Nat. Biotechnol. 20, 697–701 [DOI] [PubMed] [Google Scholar]
- Hashimoto T., Gibbs D., Lillo C., Azarian S.M., Legacki E., Zhang X.M., et al. (2007). Lentiviral gene replacement therapy of retinas in a mouse model for Usher syndrome type 1B. Gene Ther. 14, 584–594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasson T., Heintzelman M.B., Santos-Sacchi J., et al. (1995). Expression in cochlea and retina of myosin VIIa, the gene product defective in Usher syndrome type 1B. Proc. Natl. Acad. Sci. USA 92, 9815–9819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hauswirth W.W., Aleman T.S., Kaushal S., et al. (2008). Treatment of leber congenital amaurosis due to RPE65 mutations by ocular subretinal injection of adeno-associated virus gene vector: short-term results of a phase I trial. Hum. Gene Ther. 19, 979–990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirsch M.L., Storici F., Li C., et al. (2009). AAV recombineering with single strand oligonucleotides. PLoS One 4, e7705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirsch M.L., Li C., Bellon I., et al. (2013). Oversized AAV transduction is mediated via a DNA-PKcs-independent, Rad51C-dependent repair pathway. Mol. Ther. 21, 2205–2216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobson S.G., Acland G.M., Aguirre G.D., et al. (2006). Safety of recombinant adeno-associated virus type 2-RPE65 vector delivered by ocular subretinal injection. Mol. Ther. 13, 1074–1084 [DOI] [PubMed] [Google Scholar]
- Jacobson S.G., Cideciyan A.V., Aleman T.S., et al. (2008). Usher syndromes due to MYO7A, PCDH15, USH2A or GPR98 mutations share retinal disease mechanism. Hum. Mol. Genet. 17, 2405–2415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobson S.G., Cideciyan A.V., Gibbs D., et al. (2011). Retinal disease course in Usher syndrome 1B due to MYO7A mutations. Invest. Ophthalmol. Vis. Sci. 52, 7924–7936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapranov P., Chen L., Dederich D., et al. (2012). Native molecular state of adeno-associated viral vectors revealed by single-molecule sequencing. Hum. Gene Ther. 23, 46–55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keats B.J., and Corey D.P. (1999). The usher syndromes. Am. J. Med. Genet. 89, 158–166 [PubMed] [Google Scholar]
- Lai Y., Yue Y., Liu M., et al. (2005). Efficient in vivo gene expression by trans-splicing adeno-associated viral vectors. Nat. Biotechnol. 23, 1435–1439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai Y., Yue Y., Liu M., and Duan D. (2006). Synthetic intron improves transduction efficiency of trans-splicing adeno-associated viral vectors. Hum. Gene Ther. 17, 1036–1042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai Y., Li D., Yue Y., and Duan D. (2008). Design of trans-splicing adeno-associated viral vectors for Duchenne muscular dystrophy gene therapy. Methods Mol. Biol. 433, 259–275 [DOI] [PubMed] [Google Scholar]
- Lai Y., Yue Y., and Duan D. (2010). Evidence for the failure of adeno-associated virus serotype 5 to package a viral genome > or=8.2 kb. Mol. Ther. 18, 75–79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M., Jayandharan G.R., Li B., et al. (2010). High-efficiency transduction of fibroblasts and mesenchymal stem cells by tyrosine-mutant AAV2 vectors for their potential use in cellular therapy. Hum. Gene Ther. 21, 1527–1543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X.Z., Walsh J., Mburu P., et al. (1997). Mutations in the myosin VIIA gene cause non-syndromic recessive deafness. Nat. Genet. 16, 188–190 [DOI] [PubMed] [Google Scholar]
- Liu X., Ondek B., and Williams D.S. (1998). Mutant myosin VIIa causes defective melanosome distribution in the RPE of shaker-1 mice. Nat. Genet. 19, 117–118 [DOI] [PubMed] [Google Scholar]
- Liu X., Udovichenko I.P., Brown S.D., et al. (1999). Myosin VIIa participates in opsin transport through the photoreceptor cilium. J. Neurosci. 19, 6267–6274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopes V.S., Gibbs D., Libby R.T., et al. (2011). The Usher 1B protein, MYO7A, is required for normal localization and function of the visual retinoid cycle enzyme, RPE65. Hum. Mol. Genet. 20, 2560–2570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopes V.S., Boye S.E., Louie C.M., et al. (2013). Retinal gene therapy with a large MYO7A cDNA using adeno-associated virus. Gene Ther. 20, 824–833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lostal W., Bartoli M., Bourg N., et al. (2010). Efficient recovery of dysferlin deficiency by dual adeno-associated vector-mediated gene transfer. Hum. Mol. Genet. 19, 1897–1907 [DOI] [PubMed] [Google Scholar]
- Maguire A.M., High K.A., Auricchio A., et al. (2009). Age-dependent effects of RPE65 gene therapy for Leber's congenital amaurosis: a phase 1 dose-escalation trial. Lancet 374, 1597–1605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markusic D.M., Herzog R.W., Aslanidi G.V., et al. (2010). High-efficiency transduction and correction of murine hemophilia B using AAV2 vectors devoid of multiple surface-exposed tyrosines. Mol. Ther. 18, 2048–2056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nathwani A.C., Tuddenham E.G., Rangarajan S., et al. (2011). Adenovirus-associated virus vector-mediated gene transfer in hemophilia B. N. Engl. J. Med. 365, 2357–2365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ouyang X.M., Yan D., Du L.L., et al. (2005). Characterization of Usher syndrome type I gene mutations in an Usher syndrome patient population. Hum. Genet. 116, 292–299 [DOI] [PubMed] [Google Scholar]
- Petrs-Silva H., Dinculescu A., Li Q., et al. (2009). High-efficiency transduction of the mouse retina by tyrosine-mutant AAV serotype vectors. Mol. Ther. 17, 463–471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrs-Silva H., Dinculescu A., Li Q., et al. (2011). Novel properties of tyrosine-mutant AAV2 vectors in the mouse retina. Mol. Ther. 19, 293–301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryals R.C., Boye S.L., Dinculescu A., et al. (2011). Quantifying transduction efficiencies of unmodified and tyrosine capsid mutant AAV vectors in vitro using two ocular cell lines. Mol. Vis. 17, 1090–1102 [PMC free article] [PubMed] [Google Scholar]
- Sahly I., Dufour E., Schietroma C., et al. (2012). Localization of Usher 1 proteins to the photoreceptor calyceal processes, which are absent from mice. J. Cell Biol. 199, 381–399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saihan Z., Webster A.R., Luxon L., and Bitner-Glindzicz M. (2009). Update on Usher syndrome. Curr. Opin. Neurol. 22, 19–27 [DOI] [PubMed] [Google Scholar]
- Simonelli F., Maguire A.M., Testa F., et al. (2010). Gene therapy for Leber's congenital amaurosis is safe and effective through 1.5 years after vector administration. Mol. Ther. 18, 643–650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith R.J., Berlin C.I., Hejtmancik J.F., et al. (1994). Clinical diagnosis of the Usher syndromes. Usher Syndrome Consortium. Am. J. Med. Genet. 50, 32–38 [DOI] [PubMed] [Google Scholar]
- Timmers A.M., Zhang H., Squitieri A., and Gonzalez-Pola C. (2001). Subretinal injections in rodent eyes: effects on electrophysiology and histology of rat retina. Mol. Vis. 7, 131–137 [PubMed] [Google Scholar]
- Trapani I., Colella P., Sommella A., et al. (2013). Effective delivery of large genes to the retina by dual AAV vectors. EMBO Mol. Med. 6, 194–211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weil D., Blanchard S., Kaplan J., et al. (1995). Defective myosin VIIA gene responsible for Usher syndrome type 1B. Nature 374, 60–61 [DOI] [PubMed] [Google Scholar]
- Weil D., Levy G., Sahly I., et al. (1996). Human myosin VIIA responsible for the Usher 1B syndrome: a predicted membrane-associated motor protein expressed in developing sensory epithelia. Proc. Natl. Acad. Sci. USA 93, 3232–3237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams D.S. (2008). Usher syndrome: animal models, retinal function of Usher proteins, and prospects for gene therapy. Vis. Res. 48, 433–441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams D.S., and Lopes V.S. (2011). The many different cellular functions of MYO7A in the retina. Biochem. Soc. Trans. 39, 1207–1210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfrum U., Liu X., Schmitt A., et al. (1998). Myosin VIIa as a common component of cilia and microvilli. Cell Motil. Cytoskeleton 40, 261–271 [DOI] [PubMed] [Google Scholar]
- Wu Z., Yang H., and Colosi P. (2010). Effect of genome size on AAV vector packaging. Mol. Ther. 18, 80–86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan Z., Zhang Y., Duan D., and Engelhardt J.F. (2000). Trans-splicing vectors expand the utility of adeno-associated virus for gene therapy. Proc. Natl. Acad. Sci. USA 97, 6716–6721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan Z., Ritchie T.C., Duan D., and Engelhardt J.F. (2002). Recombinant AAV-mediated gene delivery using dual vector heterodimerization. Methods Enzymol. 346, 334–357 [DOI] [PubMed] [Google Scholar]
- Yan Z., Zak R., Zhang Y., and Engelhardt J.F. (2005). Inverted terminal repeat sequences are important for intermolecular recombination and circularization of adeno-associated virus genomes. J. Virol. 79, 364–379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J., Zhou W., Zhang Y., et al. (1999). Concatamerization of adeno-associated virus circular genomes occurs through intermolecular recombination. J. Virol. 73, 9468–9477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., and Duan D. (2012). Novel mini-dystrophin gene dual adeno-associated virus vectors restore neuronal nitric oxide synthase expression at the sarcolemma. Hum. Gene Ther. 23, 98–103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zolotukhin S., Potter M., Zolotukhin I., et al. (2002). Production and purification of serotype 1, 2, and 5 recombinant adeno-associated viral vectors. Methods 28, 158–167 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.