Abstract
There is surprising confusion surrounding the concept of biological totipotency, both within the scientific community and in society at large. Increasingly, ethical objections to scientific research have both practical and political implications. Ethical controversy surrounding an area of research can have a chilling effect on investors and industry, which in turn slows the development of novel medical therapies. In this context, clarifying precisely what is meant by “totipotency” and how it is experimentally determined will both avoid unnecessary controversy and potentially reduce inappropriate barriers to research. Here, the concept of totipotency is discussed, and the confusions surrounding this term in the scientific and nonscientific literature are considered. A new term, “plenipotent,” is proposed to resolve this confusion. The requirement for specific, oocyte-derived cytoplasm as a component of totipotency is outlined. Finally, the implications of twinning for our understanding of totipotency are discussed.
Highlights
• Inaccurate use of the term “totipotent” by scientists creates unnecessary ethical controversy.
• Public concern over producing embryos by reprogramming reflects confusion over totipotency.
• Twinning by blastocyst splitting does not provide scientific evidence for totipotency.
What Is Totipotency?
The medical dictionary administered by the National Institutes of Health gives two contrasting definitions for the term totipotent: “capable of developing into a complete organism” or “differentiating into any of its cells or tissues” (www.merriam-webster.com/medlineplus/totipotent; accessed 6/23/2013). Much of the confusion surrounding the term totipotency centers on the important differences between these two definitions. A one-cell embryo (zygote) is “totipotent” in both senses; yet, some authors characterize tumors [1,2] and stem cells [3,4] as “totipotent,” based only on the second definition (ie, the ability of these cells to produce a wide range of cell types).
The difference between these two definitions is not trivial. Producing a mature organism requires the ability to both generate all the cells of the body and to organize them in a specific temporal and spatial sequence, that is, to undergo a coordinated process of development. Totipotency in this strict sense is demonstrated by the ability of an isolated cell to produce a fertile, adult individual. Consequently, a cell that is totipotent is also a one-cell embryo; that is, a cell that is capable of generating a globally coordinated developmental sequence.
While stem cells, tumors, and embryos have many molecular features in common, embryos are clearly organisms [5–8]. Embryos develop in a predictable manner toward a species-specific adult form (human embryos do not mature into mice, monkeys, or tumors). Embryos repair injury. They adapt to changing environmental conditions. Most importantly, they show coordinated interactions between parts (molecules, cells, tissues, structures, and organs) that promote the survival, health, and continued development of the organism as a whole; that is, interactions that are characteristic of “an individual constituted to carry on the activities of life by means of organs separate in function but mutually dependent: a living being,” (www.merriam-webster.com/medlineplus/organism; accessed 6/23/2013). In contrast, stem cells and tumors do not produce cells or structures in the functionally integrated progression that is characteristic of an organism. They are not capable of development.
The ability to produce an orchestrated developmental sequence should not be misconstrued as some kind of mystical element that is merely attributed to an embryo. The fact that the embryo undergoes a self-directed process of maturation is entirely a matter of empirical observation; embryos construct themselves. Scientists tend to view this developmental capability as a series of cellular/molecular events. Others may view human development in more spiritual or poetic terms. But neither of these views alters the scientific facts; embryos manifest a unique molecular composition and pattern of behavior that is characteristic of an organism (ie, “a living being”) that has not been observed in tumors or other human cells.
Regardless how individuals or societies ultimately weigh the value of the embryo relative to the value of scientific research, it is important to appreciate that in all cases, the ethical consideration given to human embryos does not reflect the status they will achieve at some point in the future (ie, what they will mature into). If this was the case, then there could be no possible objection to embryo-destructive research since, by definition, adult status is never attained in such situations. Rather, ethical consideration is given to human embryos based on the status they already possess; that is, their unique and fully operative ability to function as a human organism. Therefore, ethical controversy regarding totipotent human cells only concerns cells that are totipotent in the strict, organismal sense; that is, a cell that is a human embryo.
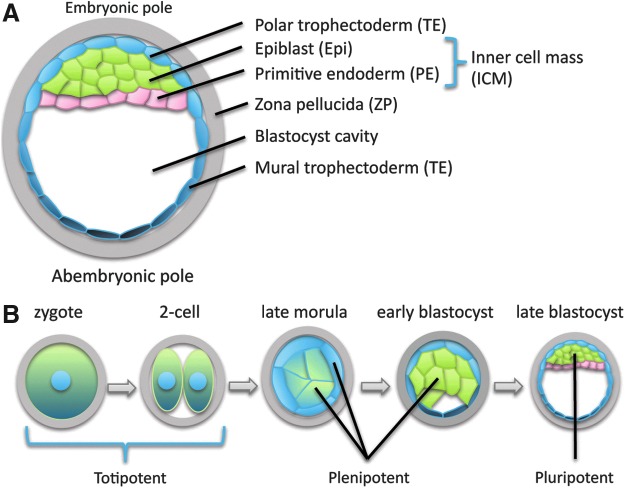
The term totipotent describes the properties of an individual cell (not a group of cells) with the two meanings of this term roughly corresponding to the progressive restriction in potential cells exhibit during normal development. At the expanded blastocyst stage (Fig. 1A), cells in specific regions of the embryo are restricted to produce only a subset of the cells observed at more-mature stages (Table 1). Restrictions in potency occur gradually as development proceeds, with the ability to independently initiate a developmental sequence (ie, totipotency in the first sense) being lost relatively early during development, and individual cells of the late morula and early blastocyst producing a wider range of derivatives than cells at the expanded blastocyst stage (Fig. 1B).
FIG. 1.
Cell types and potency at different developmental stages. (A) Anatomy and cell types of the expanded blastocyst embryo (human development, ∼5–6 days; shown as a mid-sagittal section). The position of the ICM (green and pink) defines the embryonic-abembryonic axis, which has a consistent relationship to the animal-vegetal axis of the oocyte, and may be determined by both the shape of the zygote [153,189] and the early cleavage patterns of the embryo [156,164]. The entire embryo is surrounded by an acellular protein layer known as the zona pellucida (gray). TE associated with the embryonic pole (light blue) induces the formation of PE (pink), that is initially interspersed with the presumptive epiblast (green), and segregates by cell sorting [77]. Together, epiblast and PE constitute the ICM. The blastocyst cavity is a fluid-filled space. Polar (light blue) and mural (dark blue) TE have distinct molecular properties [61,190,191] and distinct developmental fates [162]. (B) During human development, only the zygote and early cleavage-stage blastomeres (possibly up to the four-cell stage) remain totipotent, that is, capable of independently initiating a developmental sequence (Table 2). Cells of the late morula/early blastocyst are plenipotent; that is, they are able to produce all or most of the cells of the body, but not organize them into a coherent body plan (Table 2). Cells of the epiblast at the expanded blastocyst stage (the cells from which many embryonic stem cell lines are derived) are pluripotent; that is, they are able to produce cell types found in the mature body, but are not derivatives of the TE and PE (Table 2). PE, primitive endoderm; TE, trophectoderm.
Table 1.
Derivatives of Specific Blastocyst Cell Types in Human Development
| Blastocyst stage cell type | Embryonic derivatives | Fetal derivatives |
|---|---|---|
| ICM | ||
| Epi | Extraembryonic Ectoderm | Ectoderm of the amniotic membrane |
| Extraembryonic Mesoderm | Mesoderm of the amniotic membrane, chorionic membrane and yolk sac and fetal vessels of the placenta | |
| Ectoderm, Mesoderm and Endoderm | All tissues of the postnatal body | |
| PE | Extraembryonic Endoderm | Endoderm of the yolk sac |
| Polar TE | Syncytiotrophoblast | Placental tissue |
| Cytotrophoblast | ||
| Mural TE | Cytotrophoblast | Placental tissue |
In humans, PE is also referred to as hypoblast.
Epi, epiblast; ICM, inner cell mass; PE, primitive endoderm; TE, trophectoderm.
The confusion between the two senses of totipotency has led some to propose new terminology, with one author suggesting the term “totipotent” be reserved for organisms, while stem cells and tumors that produce all cell types but do not organize them into a coherent body plan would be referred to as “omnipotent” [9]. Yet the strong connotation of this word outside the field of science compromises its utility. “Plenipotent” (from the Latin plenus, or “full”) would be preferable for individual cells that are totipotent in the cellular or weak sense, reserving “pluripotent” (from the Latin pluris, or “more”) for cells that make only a subset of the normal derivatives of the embryo (Fig. 1B and Table 2). These terms would better reflect the progressive restrictions in potency that occur during normal development (Fig. 1B), and avoid the unfortunate application of the term “totipotent” to cells that are not embryos.
Table 2.
Terms, Definitions, Experimental Tests, and Examples of Cells with Different Degrees of Potency
| Term | Definition | Experimental test | Examples |
|---|---|---|---|
| Totipotent | An isolated cell that is able to produce a fertile adult individual. | An isolated cell is transferred to a uterus (after inserting into an empty zona pellucida or after developing to the blastocyst stage in culture) and it gives rise to a fertile adult. | Zygotes Some early cleavage-stage blastomeres |
| Plenipotent | An isolated cell that is able to produce all or most of the derivatives of the ICM and some or all of the TE- and PE-derived cells, but is unable to organize these cells into an integrated body plan. | The cell produces all the structures of the mature body (including germ line) with substantial contributions to TE- and/or PE-derived structures in a complementation assay. The cell makes substantial contributions to all three germ layers and to TE- and/or PE-derived structures in a chimeric embryo/animal. The cell is able to form type I yolk-sac-containing teratomas after injection into an immune-compromised mouse, including tissues with gene expression patterns and histology characteristic of the derivatives of all three germ layers plus TE- and/or PE-derived cells. The cell is able to differentiate in culture into cells with gene expression patterns and physiology characteristic of the derivatives of all three germ layers as well as TE- and/or PE-derived structures. |
ICM cells in normal embryos, prior to segregation into epiblast and PE Late morula-stage blastomeres in normal embryos prior to commitment to TE or ICM Mouse and human embryonal carcinoma cellsa Most hESCs and human induced pluripotent stem cellsa Primordial germ cells and some germ cell linesb |
| Pluripotent | An isolated cell that is able to produce all or most of the derivatives of the epiblast, but is unable to organize these cells into an integrated body plan or to produce derivatives of TE and/or PE. | The cell produces all of the structures of the mature body (including germ line), with minimal contribution to the TE- and/or PE-derived structures in a complementation assay. The cell makes substantial contributions to all three germ layers with minimal contribution to TE- and/or PE-derived structures in a chimeric embryo/animal. The cell is able to form type I non-yolk-sac-containing teratomas, including derivatives of all three germ layers, with minimal representation of TE- and/or PE-derived cells. The cell is able to differentiate in culture into cells with gene expression patterns and physiology characteristic of the derivatives of all three germ layers. |
Epiblast cells in normal embryos mESCs and mouse induced pluripotent cells under standard culture conditionsc |
Cell types that have met some or all of the experimental tests listed are given as examples, with cases in which there is some debate over the evidence being given in italics. The tests are listed in rough order of stringency, with the first test being the only definitive proof for the specified level of potency. Teratoma classification follows the designation of Cunningham et al. [193]. Note: These terms apply only to individual cells, not to groups of cells.
Testing the developmental potency of human cells is limited by ethical constraints. hESCs, iPSCs, and embryonal carcinomas are plenipotent based primarily on the last two tests and on limited data from chimeric mice.
Primordial germ cells are plenipotent based primarily on yolk-sac tumor/teratoma formation.
Some mESCs and iPSCs are able to form PE-derived cells under some culture conditions and can make limited contributes to both TE- and PE-derived structures in chimeric mice, suggesting that they may be plenipotent.
hESCs, human embryonic stem cells; iPSCs, induced pluripotent stem cells; mESCs, mouse embryonic stem cells.
In many cases, there are significant limitations in the data available to classify the developmental capabilities of cell types, particularly for human cells. Moreover, not all studies agree on the potency of the same cell type. Consequently, as is the case for all classifications, those suggested in Table 2 can only be taken as provisional, and must be interpreted relative to a specific body of work. Yet individual studies can be evaluated based on the evidence they present. For example, a recent study of embryonic stem cells (ESCs) [10] suggests neither human nor mouse cells produce trophectoderm (TE) derivatives, a finding that would classify both cell types as pluripotent. In contrast, other studies indicate that human ESCs (hESCs) are able to produce both trophoblast and syncytiotrophoblast, a finding that would classify hESCs as plenipotent [11,12]. Similarly, several recent studies indicate that subpopulations of mouse ESCs (mESCs) are able to differentiate as TE derivatives [4,13,14], which would classify this subset as plenipotent. In contrast, other studies of mESCs either indicate that TE derivatives can only be induced by artificial manipulation of specific genes [15] or, conversely, that the great majority of mESCs are able to produce TE derivatives under specific culture conditions [16,17], further illustrating that the potency of specific types of stem cells has not been conclusively resolved in the literature. Nonetheless, relating the degrees of potency observed in cell lines to the progressive restrictions in potency observed in development (Fig. 1B and Table 1) provides a more accurate classification and avoids the confusing misapplication of the term “totipotent.”
Currently, most scientists refer to ESCs from both mice and humans as “pluripotent,” without attempting to distinguish between the progressive restrictions in potency that are seen in embryos (Fig. 1B). Consequently, in the current analysis, the term totipotent (without quotes) will be used in the organismal or strict sense, and pluripotent will be used as it is currently used in the literature, that is, to refer to cells that produce, but do not organize, all or most of the cell types found in a mature individual (ie, to refer to cells that are both “plenipotent” and “pluripotent,” as defined in Table 2).
Confounding Usage of the Term “Totipotent” in the Scientific Literature
Although many scientific articles use “totipotent” in a consistent manner [18–21], a surprising number use the term less rigorously, conflating the two meanings and/or accepting inadequate evidence as proof of strict totipotency. These misapplications fall into four general classes: (1) equating participation in development with the ability to independently generate a developmental sequence, (2) equating the ability of groups of cells to collectively generate a developmental sequence with totipotency of individual cells, (3) equating the expression of early embryonic markers with totipotency, and (4) taking a partial or superficial resemblance to an embryo as evidence for totipotency.
Participation in development is not totipotency
The most common misapplication of the term totipotent in both the scientific and nonscientific literatures is to equate the ability of a cell to participate in an embryonic process with the ability to generate all of the structures of the body independently (Fig. 2). For example, the observation that stem cells will generate all or most of the tissues of the postnatal body after injection into an early stage embryo [22,23] or in a tetraploid complementation assay [24] is often misinterpreted as evidence that stem cells are totipotent [25–28]. Indeed, the generation of live mice from tetraploid complementation using reprogrammed pluripotent cells [29] has been cited as evidence that “somatic cells alone have the potential to become an adult human being” [30]. Some go so far as to conclude, “a reprogrammed human cell is not fundamentally different from a nuclear-transfer or natural fertilization zygote in its ability to become a fetus” [31].
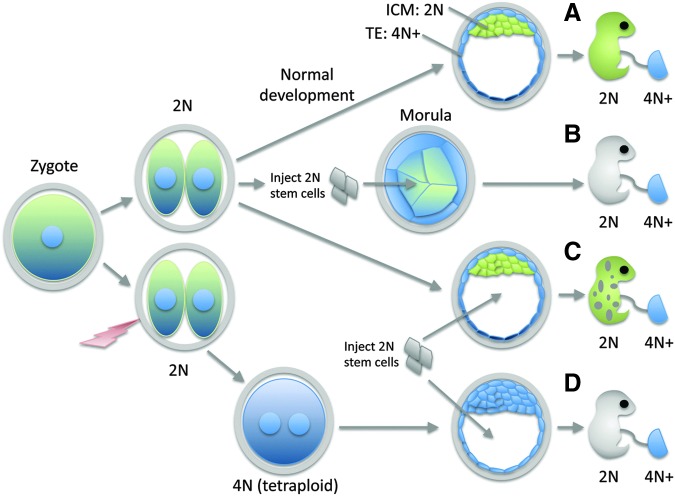
FIG. 2.
Contribution of diploid and tetraploid cells to development. (A) In normal development, cells of the TE (blue) naturally become tetraploid (or higher orders of ploidy) and contribute predominantly to the placenta. ICM cells (green) remain diploid and contribute primarily to the structures of the postnatal body. (B) Injection of diploid pluripotent stem cells with ICM-like properties into a morula-stage embryo prior to commitment of ICM cells results in stem cells contributing primarily to the ICM (and therefore the structures of the postnatal body), with the cells of the embryo largely producing TE-derived structures. (C) Injection of stem cells into a normal blastocyst-stage embryo in which the ICM has already formed results in a chimeric animal, with cells of the postnatal body derived both from the ICM and from the stem cells. (D) If embryos are electrically shocked at the two-cell stage to induce cell fusion, then an abnormally tetraploid embryo results. In most cases, the tetraploid embryo produces only TE derivatives, although live-born tetraploid mice [192] and humans [33,34] have been reported. Injection of diploid pluripotent stem cells at the blastocyst stage complements this defect, with stem cells producing most of the postnatal structures. ICM, inner cell mass.
Yet, this assertion ignores the fact that in such experimental procedures, stem cells are merely participating in an embryonic process, not originating it. Clearly an unmanipulated embryo will produce a fully formed individual all on its own, without any assistance from stem cells (Fig. 2A). The incorporation of injected cells into a preexisting embryo merely reflects the epigenetic state of pluripotent cells that biases them to contribute to the inner cell mass (ICM) lineage at both morula (Fig. 2B) and blastocyst stages (Fig. 2C).
Similarly, tetraploid embryos are still embryos, although with a significant defect that in most cases proves fatal. However, fully tetraploid mice frequently complete embryogenesis, and occasionally survive to live birth (reviewed in Eakin and Behringer [32]). Similarly, there are reports of live-born, tetraploid humans [33,34], although mosaicism has not been strictly ruled out in most cases. These results demonstrate that the capacity to generate a full developmental program (ie, to progress through the complete sequence of developmental events, despite abnormalities) persists in tetraploid embryos, and that injected diploid stem cells are merely “complementing” the developmental defect arising from the tetraploid state. In these assays (Fig. 2D), the injected stem cells produce “most” (but not all) [32] of the cells in the postnatal body. Importantly, in these experiments, stem cells respond to developmental signals and participate in developmental processes but they do not originate these processes on their own (ie, they are not totipotent), or they would also be capable of generating a full developmental sequence independently, without the “assistance” of either a morula-stage or tetraploid embryo.
Some of the confusion regarding the usage of the term totipotent in this context reflects the arbitrary distinction between structures viewed as “embryonic” (ie, those derived from the ICM and persisting in the postnatal body) and those typically referred to as “extraembryonic” (ie, tissues and organs that have primarily gestational functions; see Table 1). This distinction has a long history in human medical embryology, with the term “extraembryonic” being in common use by the early 1890s [35]. Medically, it is reasonable to distinguish between transient structures that function only during gestation and structures that have functions after birth, since only the latter are the subject of medical intervention (although with the rapidly expanding use of exutero intrapartum therapy, or EXIT, to treat fetal anomalies [36], this distinction may eventually become less medially relevant.).
Yet the term “extraembryonic” does not mean these structures are “nonembryonic.” Structures such as the amnion and placenta are clearly part of the embryo, not part of the mother or of some other entity that coexists with the embryo. Like all embryonic structures, placenta and membranes are (1) generated by the embryo, (2) physically continuous with the embryo, (3) genetically identical to the embryo, and (4) critically required for the function and survival of the embryo as a whole (ie, vital organs of the embryo itself). Consequently, “extraembryonic” structures are best seen as transient structures of the embryo and fetus; that is, organs or tissues that function only during prenatal life.
Many structures have similarly transient functions, including the lungs (functioning only during postnatal life), the thymus (functioning in a greatly diminished capacity at adult stages, compared with its function in the fetus), and the uterus (functioning as a reproductive organ only between puberty and menopause). Due to the purely gestational functions of extraembryonic structures, they are physically discarded at birth, but this is not fundamentally different from the atrophy of the ductus arteriosis (a transient vascular structure that has essential functions only prenatally) in the neonate, or the loss of primary teeth in childhood.
While injected stem cells make minor contributions to placenta and membranes in tetraploid complementation assays (indicating that they are capable of differentiating into these tissues to a limited extent), the failure of stem cells to produce the embryo entirely on their own (including all of the “extraembryonic” organs) indicates that stem cells are not totipotent in either sense, but rather are merely pluripotent, producing only part of the whole embryo in the context of an embryonic process that they do not independently initiate.
Collectively generating a developmental sequence is not totipotency
Scientific authors occasionally equate the ability of groups of cells to collectively generate a full developmental sequence with totipotency of individual cells within the group (Fig. 3). For example, based on the failure to generate live-born mice from aggregates of cells derived from 32-cell-stage embryos [37], the authors assert that blastomere “totipotency” is lost after the fifth round of cell division. Similar results were obtained and similar conclusions were drawn from the observation that “outer” cells of human blastocysts form blastocyst-like structures that initiate expression of ICM-associated genes following reaggregation [38]. Finally, several authors [27,39–42] have asserted that blastomeres isolated from morula-stage embryos are “totipotent,” because when they are reaggregated with tetraploid cells from a freshly dissociated embryo, either live-born animals or what appear to be normal blastocysts can be produced. Yet, there is no evidence in any of these studies that cells in the aggregate are “totipotent” in either sense of the term; that is, either able to produce all cells of the embryo or to organize these cells into a mature body.
FIG. 3.
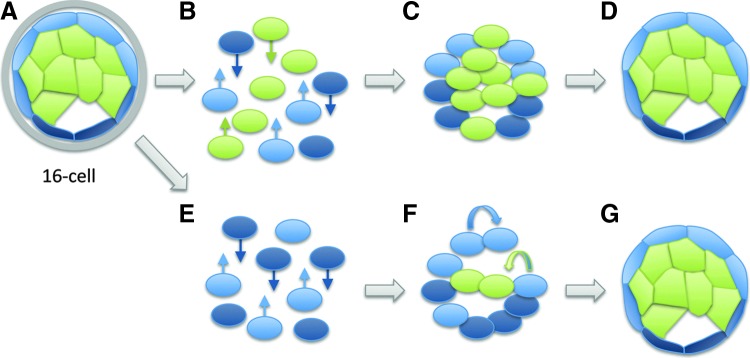
Dissociation and reaggregation of morula-stage blastomeres. (A) There is clear evidence for molecular differences between cells (ie, specification) as early as the four- to eight-cell stage [61–71,157,158]; yet, cell fates remain plastic. (B, C) Following dissociation, it is likely that a combination of cell sorting (arrows) and respecification restores the original status of the embryo. (D) Once cell–cell contacts have been reformed, development proceeds from the blastocyst stage. (E) If only “inner” or “outer” (shown) cells are reaggregated, cell sorting (arrows) is still likely to occur. (F) Outer cells remain competent to produce both ICM cells and additional TE cells (curved arrows), as they do in normal development. (G) A combination of cell sorting, regeneration of ICM cells from outer cells, and (potentially) respecification of outer cells to an ICM lineage restores the original status of the embryo.
Primary cells isolated from a freshly dissociated embryos are likely to be a mixed population with different developmental capacities (Fig. 3A) and are clearly not equivalent to stem cells [43–47]. Zygotic transcription commences during the first cell cycle in both mouse [48–51] and human [52–59] embryos. While some studies have seen no difference in gene expression between early blastomeres [60], there is evidence from several groups suggesting that as early as the 4–8-cell stage, blastomeres have unique molecular properties [61–71] that translate into distinct developmental capabilities after dissociation and reaggregation [72,73]. Substantial evidence stretching back over 60 years indicates that when cells with distinct developmental states are aggregated, cell sorting plays a significant role in re-establishing the initial pattern [74,75]. In mice, cell sorting plays a significant role in the segregation of cells within normal 16-cell [76] and 32-cell embryos [77]. Therefore, the cell sorting that has been observed in some studies [37] is likely to reorganize the spatial relationships of blastomeres with limited developmental competency to reconstitute a full embryo (Fig. 3B, E).
Yet even with the most generous interpretation of cell aggregation studies (that cell sorting does not occur and blastomeres at the 16-cell stage contribute randomly to the reconstituted embryo), the ability to respecify as part of a group (ie, the ability to make “any” cell of an embryo) is distinct from totipotency (ie, the ability to autonomously make all cells of an embryo and to organize them into a coherent whole). The significant role of “community effects” in development, first described by Sir John Gurdon in 1988 [78], clearly illustrates that the behavior of cells in groups is distinct from the behavior of the individual cells comprising the group. If the concept of totipotency is not restricted to individual cells and is instead applied to groups of cells that are collectively capable of undergoing development, then all of the cells of any organism at any stage of maturation must be considered “totipotent.” Applying the term “totipotent” in this manner reduces it to a synonym for “organism” and significantly compromises its scientific utility.
An important insight from cell aggregation experiments is that there is more than one way to make an embryo. Embryos produced by either fertilization or by cloning begin as a single-cell zygote that is (by definition) totipotent. Yet, when embryos are reconstituted from collections of morula-stage blastomeres (either diploid or a mixture of diploid and tetraploid), the embryo as a whole must be considered “totipotent” in some sense (ie, it must be considered an organism), but the individual cells within the embryo are not totipotent.
To date, there is no evidence that groups of cells derived from any source other than a freshly dissociated morula-stage embryo can collectively generate a full developmental sequence on their own. Even when freshly isolated ICM [79] or TE [80,81] from blastocyst-stage embryos are transferred to the uterus or other supportive locations, they do not develop as embryos.
Groups of stem cells are also not totipotent. mES lines were first isolated in 1981 [82,83]. Yet, despite thousands of articles that describe the production of stem cell aggregates over the last three decades, there have been no reports of spontaneous generation of embryos from ESCs. Similarly, there have been no reports of generating embryos from a combination of mES and trophoblast stem cells [84]. The failure of aggregated stem cell lines derived thus far to spontaneously form embryos is likely to reflect the fact that stem cell lines have a range of properties [85–87] that are not identical either to zygotes [88,89] or to cells that have been freshly isolated from embryos [43–47].
While these negative findings do not strictly rule out the possibility that later-stage blastomeres or pluripotent stem cells “could” combine in some way to generate an embryo (it is logically impossible to prove something from a negative finding), they strongly suggest that collective reconstitution of an embryo from cells of a reaggregated morula-stage embryo (Fig. 3) reflects a unique balance of developmental properties within those freshly dissociated cells—a balance that is not present in cultured stem cell lines or in cells derived from later-stage embryos.
Expression of molecular markers found in early embryos is not totipotency
Scientific authors occasionally conflate expression of molecular markers characteristic of specific embryonic stages with the corresponding developmental capacities of cells at those stages. For example, several studies have determined that most or all mESCs cycle in and out of a unique transcriptional state that shares some elements in common with blastomeres at the two-cell (2C) stage [14,90–92]. Cells in this 2C-like state lack pluripotency markers normally associated with the ICM, as do human zygotes and embryos at the two-cell stage [88]. The “2C-like” cells also show expanded developmental potential when injected into embryos. The authors present no evidence that isolated 2C-like cells are capable of generating embryos on their own and are careful not to pronounce their 2C-like cells totipotent in the organismal sense, but the repeated observation that the 2C stage correlates with a period in which “blastomeres are totipotent” [14] is strongly suggestive of this unwarranted conclusion and illustrates the confusing usage of the term “totipotent” in the scientific literature. Similarly, the ability of stem cells [3,4] or carcinomas [1,2] to express a molecular marker of “extraembryonic” cell lineages in addition to markers associated with ICM has been taken as evidence of totipotency, despite there being no indication that these cells can initiate a developmental sequence on their own.
Behaving like part of an embryo or merely looking like an embryo is not totipotency
Finally, some scientific authors suggest that the ability of stem cells to replicate limited aspects of normal embryonic development is evidence for totipotency, or something very close to it. For example, a number of studies have shown that ESC aggregates treated with specific signaling molecules exhibit some of the molecular cascades and cell behaviors observed during normal gastrulation [93–97]. While these studies do not equate stem cells with embryos, several authors conclude that stem cell aggregates are surprisingly “embryo like.” For example, one article states that aggregates “resemble normal embryonic development much closer than previously thought,” exhibiting “an unexpected degree of self-organization” [97]. Yet stem cell aggregates do not resemble organisms so much as they resemble tumors, which can also show a surprising degree of self-organization, producing well-formed teeth [98] and, even in one case, a remarkably normal eye [99].
Similarly, ESC aggregates [100,101] and tumors [102] occasionally generate cystic structures that have some visual similarity to a blastocyst-stage embryo, which has led some authors to ask whether ESCs and tumors might also be embryos [9,103]. Yet many cell types with clearly restricted potency will form such cystic structures, including liver, heart, and cartilage [104]; neurons [105]; fibroblasts [106]; kidney [107]; and umbilical cells [108], indicating that the mere formation of such structures is not sufficient evidence for totipotency.
Confusions Regarding Totipotency in the Nonscientific Literature: Reprogramming “Too Far”
Since the advent of cellular reprogramming [109–111], the concern that embryos might be inadvertently generated during reprogramming [112] has frequently been raised, with authors asking, for example, “how to ensure that dedifferentiation goes only so far and no further?” [113]. The possibility of producing embryos by reprogramming has been raised by some scientific authors as well [31,103].
The belief that reprogramming could “accidentally” bring a cell into a totipotent state seems to stem from the stubbornly compelling notion that unipotent, pluripotent, and totipotent represent points along a continuum [114] and that reprogramming progressively rewinds development backward toward the beginning. In this view, if reprogramming goes one step beyond pluripotency, then a totipotent zygote will accidentally be produced. Some authors have speculated that this could be done intentionally, generating an “induced totipotent stem” cell, [31] despite the obvious illogic of this assertion; totipotent zygotes are manifestly not capable of self-renewal, and are therefore not “stem cells.”
Reprogramming does not merely “rewind” development
As plausible as this concern may seem, it is not possible for reprogramming to accidently produce a totipotent cell for at least two important reasons. First, reprogramming does not simply “reverse” development, like rewinding an old audiocassette tape reverses the recording; that is, reprogramming does not move a cell backward along the developmental pathway that initially produced its mature state. Instead, reprogramming is more of a single leap from one cellular state to another [115,116]. For example, reprogramming a skin cell into a heart cell [117,118] undoubtedly involves intermediate steps between these two states, but it does not rewind development in any rational sense, since cardiac cells are not part of the developmental history of skin cells.
Pluripotency is as much of a specific cellular state as any other. To reprogram an adult cell to pluripotency, factors are introduced that result in a highly specific cascade of gene expression that is not seen in a totipotent zygote [88]. For example, in human embryos, the transcription factors Nanog and Pou5f1 (also called Oct3/4) are expressed in the ICM and are not found in the zygotes [119–121]. Consequently, a cell that expresses such factors cannot be identical to a zygote [122].
The state of the cytoplasm
The second important reason a totipotent zygote cannot be accidently produced during reprogramming involves the nature of totipotency—specifically the reasons pluripotent and totipotent are radically different cellular states, even though they occur close together in developmental time. Totipotency is not merely a state of the cell's nucleus; it also requires a very specific type of cellular cytoplasm that is a critical component of totipotency.
At this time, the only known totipotent cytoplasm is produced by an oocyte and contributed to the embryo at fertilization. The fact that oocytes produce the cytoplasmic factors that are required for an embryo to be totipotent is the reason oocytes are used for cloning. During cloning, oocyte components can, over the first several days of development, induce an adult nucleus to assume a state similar to that of a normal embryo (ie, a state that is capable of driving a complete developmental sequence), mimicking the processes that naturally occur after sperm-egg fusion [123]. If a competent nuclear state is achieved, then the oocyte-specific components in the cytoplasm work with the reprogrammed nucleus to produce an embryonic pattern of development, despite the significant differences observed between cloned embryos and embryos derived from fertilization [124–127].
Oocytes are highly structured cells that are uniquely produced by the complex process of oogenesis, which involves a characteristic sequence of gene activation [128] that is distinct from the pattern observed in the maternal pronucleus after fertilization or following zygotic gene activation [58,59]. Oogenesis also requires information from other cells in the ovary [129–131]. Moreover, recent work clearly documents multiple oocyte-derived components that are essential for mammalian embryonic development (reviewed in Li et al. [132] and Matzuk and Burns [133]). For example, oocyte-expressed Ooep [134], PADi6 [135], Nlrp5 [136], Ecat1 [137], and Tle6 [138] are required for the normal function of a critical subcortical cytoplasmic complex, with loss of any of these genes resulting in embryo lethality at the two-cell stage. Maternal expression of Kdm1B [139], Dmap1 [140], Dppa3 [141], and several others is required for correct DNA methylation and maintenance of genomic imprinting, with maternal gene deletions resulting in death at early embryonic stages. Finally, maternally supplied Brg1 [142] and Brwd1 [143] are required for zygotic gene activation, with loss of these genes resulting in arrest at the two-cell stage.
Simple reprogramming of a somatic nucleus does not establish the cytoplasmic components required for totipotency and these factors are not produced during normal embryonic development; that is, the transcriptomes of oocytes and early embryos are clearly distinct [58,59]. Without the required cytoplasmic components contributed by oocyte, there can be no zygote—regardless of the state of the nucleus [19].
In other words, even if the nucleus of a somatic cell is fully reprogrammed to be identical in every respect to that of a zygote (a state that is distinct from a pluripotent stem cell and one that would require different reprogramming factors), it would still not be totipotent, because it lacks the nongenetic factors (proteins, RNA, miRNA, and specific macromolecular complexes) that are critical components of totipotency. Conversely, if a nucleus from a bona fide zygote produced by fertilization was transferred to a differentiated cell (eg, a muscle cell), it would not continue to be a zygotic nucleus in this new location. It would be (or would rapidly become) a muscle cell nucleus that functions in the manner specified by the muscle cell cytoplasm [144].
The requirement for totipotent cytoplasm in order for a cell to actually be a zygote is precisely the reason that totipotency persists for such a short time in development. In most mammals, only the first two cells of the embryo remain totipotent, that is, able to generate a complete embryo on their own when separated [145–147], perhaps in part because cells no longer have sufficient mass to continue development [148,149]. In rare cases, totipotency is preserved until the four-cell stage [150], with there being one reported case of a child born after transfer of a four-cell embryo in which only one of the blastomeres was judged to be alive [151]. There has also been one report in pigs of totipotency persisting until the eight-cell stage [152]. But there is no evidence for totipotent cells persisting beyond this stage. As soon as a cell has only a subset of the specialized cytoplasmic components contributed by the oocyte, it is no longer totipotent.
The cell fate restrictions observed in mice and other species from the two-cell stage onward are likely to reflect inheritance of required oocyte-derived components as well as cell–cell interactions and stochastic events [72,153–156]. There is clear evidence from multiple laboratories that by the four- to eight-cell stage these two processes have resulted in blastomeres that have distinct molecular, functional, and developmental properties [61–71,157,158], indicating that from the four-cell stage onward, mammalian development is mosaic to some extent.
The requirement for totipotent cytoplasm in no way denies the clearly regulative aspects of mammalian development that depend on cell–cell signaling, cell interaction, and random events. Neither does it imply that inheritance of maternal factors is the only mechanism underlying cell differentiation in mammals, or that such factors must be differentially inherited by specific blastomeres, although differential inheritance is possible mechanism underlying early lineage restrictions. Rather it suggests that the distinction between regulative and mosaic development is a false dichotomy [159]. Cell fate decisions are produced by a continuum of overlapping and redundant mechanisms [76], with oocyte-derived factors clearly playing a critical role in the developmental competency of early blastomeres [160–164].
Is it possible to make a cloned embryo from an adult cell by reprogramming?
Reprogramming can generate pluripotent stem cells because it initiates a chain of events that brings an adult cell nucleus into a pluripotent state and this “converted” nucleus subsequently produces the non-nuclear factors that are required to actually be a pluripotent stem cell. Yet totipotent zygotes are unusual, because unlike stem cells or any other cells in the body, zygotes do not produce the highly specialized cytoplasm they require on their own. Rather they inherit this specialized cytoplasm from a very different kind of cell with a very different nuclear state, that is, from the oocyte. Once a cell nucleus has the configuration of a totipotent zygote, it does not produce the same factors produced by the oocyte.
This does not mean that it would be impossible to make an embryo by reprogramming, but it does mean that it cannot happen “accidently.” And converting an adult cell into an embryo using reprogramming (making an “induced totipotent cell”) would be difficult to accomplish, even intentionally (Fig. 4).
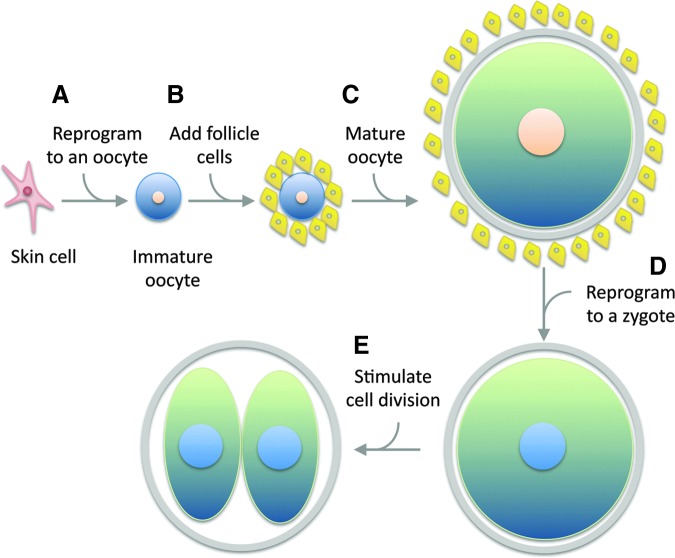
FIG. 4.
The multiple steps to reprogram a mature cell to a zygote cannot happen unintentionally. (A) A somatic cell must first be reprogrammed to an immature oocyte. (B) The oocyte must be provided with the necessary cell–cell interactions to fully mature. (C) Meiosis and genomic reprogramming must be intentionally suppressed during the maturation process. (D) Once mature, the oocyte nucleus must be reprogrammed to a zygotic state. (E) The newly formed zygote stimulated to divide. Cells are not drawn to scale; oocytes have roughly 1000× greater volume than skin cells (illustration is ∼200×greater).
To convert an adult cell into a zygote, it would first have to be reprogrammed to become a cell that is capable of providing the factors that are normally generated during the process of oogenesis. The simplest way of accomplishing this would be to reprogram the adult cell into an immature oocyte (a distinct state from a pluripotent stem cell that would require different reprogramming factors). The immature oocyte would then have to be provided with all of the cell interactions and ovarian factors required for it to become a mature oocyte. During this process, the normal epigenetic reprogramming and meiotic divisions that occur as part of oogenesis [123,165] would have to be suppressed in order to preserve the nucleus in a state that is capable of driving human development (this may not be technically or even logically possible.). Once this unnaturally suppressed oocyte had been made, the nucleus would again have to be reprogrammed to a zygotic state, a significant remodeling that normally reflects factors derived from both sperm and egg [21,123,166]. If all of this could be achieved, then the “secondarily reprogrammed” totipotent cell would have to be activated to begin the process of development. Then, and only then, would a cloned embryo be produced from an adult cell by reprogramming. And this could hardly happen “by accident.”
Twinning
It is sometimes asserted that because twinning can occur by splitting of a blastocyst-stage embryo (Fig. 5), this “proves” that the blastocyst contains a mixture of totipotent and pluripotent cells [167–169]. This view raises the concern that because pluripotent stem cells are similar to cells of the ICM, stem cell cultures could also contain a mixture of pluripotent and totipotent cells.
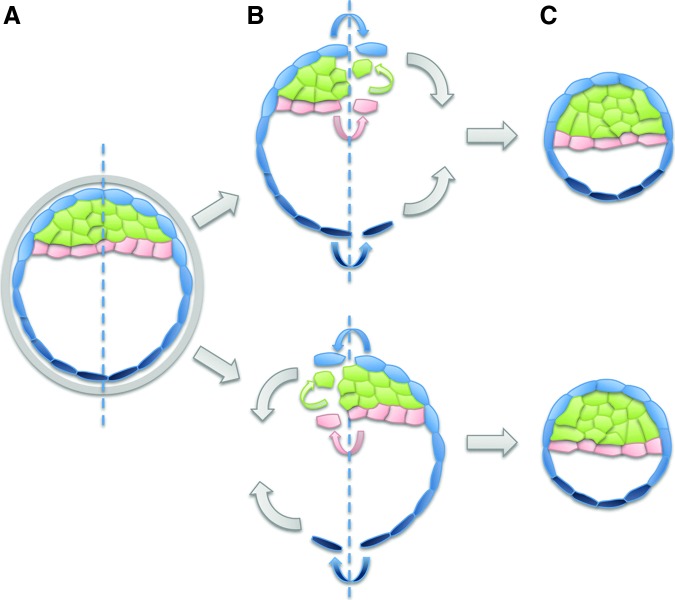
FIG. 5.
Twinning at the blastocyst stage does not require totipotent cells to be present. (A) Cells of the blastocyst have distinct molecular properties and restricted developmental capabilities. Subsequent panels show the result of splitting at the dotted line. (B) After splitting, a closed sphere rapidly reforms (curved gray arrows), and within the sphere, cells of each of the embryonic lineages replace cells within their own tissues (colored arrows). Cells within specific lineages (TE, epiblast, or PE) are likely to assume a new positional identity that reflects their new location. There is no evidence for respecification across lineages contributing to regeneration of the blastocyst. (C) The smaller, “demi-embryos” resulting from splitting have approximately half the number of cells as the original blastocyst, and proceed from the blastocyst stage in synchrony with unsplit sibling controls [184].
Twinning can potentially occur by a number of different mechanisms (a topic that has recently been re-examined in an excellent critical review; [170]). However, there is no scientific evidence to date that twinning at the blastocyst stage involves totipotent cells, and multiple studies have concluded exactly the opposite; that is, that blastocyst cells are quite unlike totipotent zygotes on multiple parameters [43–47,85–87] and that blastocyst cells are completely incapable of producing a whole embryo when isolated from each other [145–147], instead producing only stem cell lines. These observations argue strongly that twinning does not involve totipotent cells, but rather relies on some other developmental mechanism.
The simplest way twinning could occur at the blastocyst stage is by the process of “regulation,” first described by Driesch over 100 years ago [171]. Regulation does not mean that when an embryo is split, cells revert to totipotency and start the process of development all over again. It simply means that a split embryo remains an embryo. Just as an individual who loses a limb continues to be a human being, although a damaged one, a blastocyst-stage embryo that has lost half of its cells is still an organism. And one of the characteristics of organisms is that they repair injuries. Yet unlike adult human organisms, who have limited ability to regenerate lost tissue, human organisms at the embryonic stage are very good at regenerating missing parts (Fig. 5).
When an embryo is split, each half can (in some cases) become a “demi-embryo” that proceeds with development [172–175]. Direct observation indicates that in such demi-embryos, the ratio of cells in the ICM and TE is either maintained or restored by cell proliferation [176–178]. Lineage analysis indicates that cells of the TE or ICM largely contribute cells to their own lineage (Fig. 5B), just as they do in normal development [179–181]. There is no evidence for totipotent cells producing most or all of the structures of the twin after embryo splitting.
Similar results are seen for even more catastrophic injuries. When the cells of the ICM are isolated from blastocyst-stage embryos, the outer cell layer [either TE or primitive endoderm (PE)] can be partially regenerated in some cases. Detailed analysis of these imperfectly restored embryos shows that the regenerated tissues are not produced by “totipotent” cells within the ICM, but rather by committed TE or PE cells that were isolated along with the ICM [182,183].
After blastocyst splitting, embryonic development is not reinitiated (Fig. 5C), but rather proceeds in synchrony with normal (un-split), sibling embryos [184]. There is no recapitulation of the events of early development. The embryo repairs itself by the same kind of process an adult human uses to repair injury; cells in each of the embryo's specialized tissues replace the specialized cells that have been lost, and once the damage has been repaired, development proceeds from the blastocyst stage forward without returning to a zygote-like state. Twinning appears to be another example of a community effect, whereby a group of cells has properties that exceed the potency of any of the individual cells comprising the group. Importantly, twinning at the blastocyst-stage provides absolutely no evidence for the presence of totipotent cells within the blastocyst. Therefore, the ability of embryos to generate twins raises no credible concerns about the potency of stem cells derived either from reprogramming or from blastocyst-stage embryos.
Conclusions
Totipotent zygotes are distinct from pluripotent stem cells or tumors because they can originate development. The ability to both produce all cell types and to organize them into a coherent body plan is the defining feature of an organism [5,6] and also the strict definition of totipotency. Misapplication of this term in the scientific literature creates artificial controversy over areas of research that are ethically unproblematic. In the interest of both scientific accuracy and public education, scientists should confine the use of the term totipotent to embryos, and apply a rigorous scientific standard to defining totipotency; the ability of an isolated cell to mature into a fully formed individual when placed in a supportive environment (for mammals, a uterus). To describe cells that produce all the derivatives of the zygote, yet are not capable of undergoing development (eg, hESCs and embryonal carcinomas), either new terminology, such as “plenipotent,” or a qualified use of “pluripotent” would be preferable to a misleading application of the term “totipotent” to cells that are not organisms.
Clearly, there is more than one way to make an embryo. Naturally conceived human embryos are produced by fusion of sperm and egg in a woman's reproductive tract, a process that can be replicated, although imperfectly [185–187], by in vitro fertilization. Recent work has shown that human embryos capable of surviving to the blastocyst stage can be produced by cloning [188]. All three of these approaches require oocyte cytoplasm to generate a totipotent one-cell embryo.
Importantly, all other ways of “reconstituting” an embryo start with an embryo produced by one of these three methods (natural fertilization, in vitro fertilization, or cloning). Injecting pluripotent cells into an existing morula, blastocyst, or tetraploid embryo generates a chimeric animal, with injected cells contributing in varying degrees to the postnatal body (Fig. 2). Under some conditions, freshly dissociated early embryos (up to the 16-cell stage) can be reaggregated and produce live-born animals (Fig. 3). Splitting an existing embryo can result in the regeneration of two demi-embryos (Fig. 5). Yet none of these methods produce an embryo—they merely reconfigure an existing embryo or contribute additional cells to an ongoing embryonic process.
Ultimately, we may discover additional ways of constituting a full organism from cells that are not themselves totipotent or identify new ways of producing a totipotent cell. Yet in all cases (both actual and theoretical), the unique capacity of an organism to undergo development clearly distinguishes an embryo from a cell that is not totipotent. And it is the capacity of an organism to generate a complete developmental program that warrants serious ethical concern. We do not owe any particular ethical consideration to human gametes, skin cells, or stem cells. Yet, if human cells are manipulated so that they participate in the development of an existing embryo (eg, by tetraploid complementation) or are used to generate a human embryo de novo (eg, by fertilization or cloning), then the embryo merits substantial ethical consideration due to the fact that it is a human organism, that is, a human being.
Acknowledgments
The author thanks Drs. H.J. Yost and D. Prentice for suggestions on the article and figures.
Author Disclosure Statement
No competing financial interests exist.
References
- 1.Feigin ME. and Malbon CC. (2008). OSTM1 regulates beta-catenin/Lef1 interaction and is required for Wnt/beta-catenin signaling. Cell Signal 20:949–957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Honecker F, Stoop H, Mayer F, Bokemeyer C, Castrillon DH, Lau YF, Looijenga LH. and Oosterhuis JW. (2006). Germ cell lineage differentiation in non-seminomatous germ cell tumours. J Pathol 208:395–400 [DOI] [PubMed] [Google Scholar]
- 3.Abad M, Mosteiro L, Pantoja C, Canamero M, Rayon T, Ors I, Grana O, Megias D, Dominguez O, et al. (2013). Reprogramming in vivo produces teratomas and iPS cells with totipotency features. Nature 502: 340–345 [DOI] [PubMed] [Google Scholar]
- 4.Morgani SM, Canham MA, Nichols J, Sharov AA, Migueles RP, Ko MS. and Brickman JM. (2013). Totipotent embryonic stem cells arise in ground-state culture conditions. Cell Rep 3:1945–1957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Condic M. (2011). Pre-implantation stages of human development: the biological and moral status of early embryos. In: Is This Cell a Human Being?: Exploring the Status of Embryos, Stem Cells and Human-Animal Hybrids. Suarez A, Huarte J, eds. Springer, New York, p 209 [Google Scholar]
- 6.Condic ML. (2008). When does human life begin? A scientific perspective. In: Westchester Institute White Paper Series. The Westchester Institute for Ethics and the Human Person, Thornwood, NY, pp 1–18 [Google Scholar]
- 7.Ingebrigtsen R. (1912). Studies upon the characteristics of different culture media and their influence upon the growth of tissue outside of the organism. J Exp Med 16:421–431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Murray HA. (1926). Physiological ontogeny: A. Chicken embryos. Viii. Accelerations of integration and differentiation during the embryonic period. J Gen Physiol 9:603–619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Denker HW. (2004). Early human development: new data raise important embryological and ethical questions relevant for stem cell research. Naturwissenschaften 91:1–21 [DOI] [PubMed] [Google Scholar]
- 10.Bernardo AS, Faial T, Gardner L, Niakan KK, Ortmann D, Senner CE, Callery EM, Trotter MW, Hemberger M, et al. (2011). BRACHYURY and CDX2 mediate BMP-induced differentiation of human and mouse pluripotent stem cells into embryonic and extraembryonic lineages. Cell Stem Cell 9:144–155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Amita M, Adachi K, Alexenko AP, Sinha S, Schust DJ, Schulz LC, Roberts RM. and Ezashi T. (2013). Complete and unidirectional conversion of human embryonic stem cells to trophoblast by BMP4. Proc Natl Acad Sci U S A 110:E1212–E1221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Das P, Ezashi T, Schulz LC, Westfall SD, Livingston KA. and Roberts RM. (2007). Effects of fgf2 and oxygen in the bmp4-driven differentiation of trophoblast from human embryonic stem cells. Stem Cell Res 1:61–74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Canham MA, Sharov AA, Ko MS. and Brickman JM. (2010). Functional heterogeneity of embryonic stem cells revealed through translational amplification of an early endodermal transcript. PLoS Biol 8:e1000379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Macfarlan TS, Gifford WD, Driscoll S, Lettieri K, Rowe HM, Bonanomi D, Firth A, Singer O, Trono D. and Pfaff SL. (2012). Embryonic stem cell potency fluctuates with endogenous retrovirus activity. Nature 487:57–63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lu R, Yang A. and Jin Y. (2011). Dual functions of T-box 3 (Tbx3) in the control of self-renewal and extraembryonic endoderm differentiation in mouse embryonic stem cells. J Biol Chem 286:8425–8436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hayashi Y, Furue MK, Tanaka S, Hirose M, Wakisaka N, Danno H, Ohnuma K, Oeda S, Aihara Y, et al. (2010). BMP4 induction of trophoblast from mouse embryonic stem cells in defined culture conditions on laminin. In Vitro Cell Dev Biol Anim 46:416–430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mfopou JK, Geeraerts M, Dejene R, Van Langenhoven S, Aberkane A, Van Grunsven LA. and Bouwens L. (2013). Efficient definitive endoderm induction from mouse embryonic stem cell adherent cultures: a rapid screening model for differentiation studies. Stem Cell Res 12:166–177 [DOI] [PubMed] [Google Scholar]
- 18.Do JT, Han DW. and Scholer HR. (2006). Reprogramming somatic gene activity by fusion with pluripotent cells. Stem Cell Rev 2:257–264 [DOI] [PubMed] [Google Scholar]
- 19.Chan MM, Smith ZD, Egli D, Regev A. and Meissner A. (2012). Mouse ooplasm confers context-specific reprogramming capacity. Nat Genet 44:978–980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Messerschmidt DM. (2012). Should I stay or should I go: protection and maintenance of DNA methylation at imprinted genes. Epigenetics 7:969–975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wasson JA, Ruppersburg CC. and Katz DJ. (2013). Restoring totipotency through epigenetic reprogramming. Brief Funct Genomics 12:118–128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Huang J, Deng K, Wu H, Liu Z, Chen Z, Cao S, Zhou L, Ye X, Keefe DL. and Liu L. (2008). Efficient production of mice from embryonic stem cells injected into four- or eight-cell embryos by piezo micromanipulation. Stem Cells 26:1883–1890 [DOI] [PubMed] [Google Scholar]
- 23.Poueymirou WT, Auerbach W, Frendewey D, Hickey JF, Escaravage JM, Esau L, Dore AT, Stevens S, Adams NC, et al. (2007). F0 generation mice fully derived from gene-targeted embryonic stem cells allowing immediate phenotypic analyses. Nat Biotechnol 25:91–99 [DOI] [PubMed] [Google Scholar]
- 24.Nagy A, Rossant J, Nagy R, Abramow-Newerly W. and Roder JC. (1993). Derivation of completely cell culture-derived mice from early-passage embryonic stem cells. Proc Natl Acad Sci U S A 90:8424–8428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hiriart MI, Bevacqua RJ, Canel NG, Fernandez-Martin R. and Salamone DF. (2013). Production of chimeric embryos by aggregation of bovine egfp eight-cell stage blastomeres with two-cell fused and asynchronic embryos. Theriogenology 80:357–364 [DOI] [PubMed] [Google Scholar]
- 26.Sagan A. and Singer P. (2007). The moral status of stem cells. Metaphilosophy 38:264–284 [Google Scholar]
- 27.Tarkowski AK, Ozdzenski W. and Czolowska R. (2001). Mouse singletons and twins developed from isolated diploid blastomeres supported with tetraploid blastomeres. Int J Dev Biol 45:591–596 [PubMed] [Google Scholar]
- 28.Wang ZQ, Kiefer F, Urbanek P. and Wagner EF. (1997). Generation of completely embryonic stem cell-derived mutant mice using tetraploid blastocyst injection. Mech Dev 62:137–145 [DOI] [PubMed] [Google Scholar]
- 29.Zhao XY, Li W, Lv Z, Liu L, Tong M, Hai T, Hao J, Guo CL, Ma QW, et al. (2009). iPS cells produce viable mice through tetraploid complementation. Nature 461:86–90 [DOI] [PubMed] [Google Scholar]
- 30.Sagan A. and Singer P. (2009). Embryos, stem cells and moral status: a response to George and Lee. EMBO Rep 10:1283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Magill G. and Neaves WB. (2009). Ontological and ethical implications of direct nuclear reprogramming. Kennedy Inst Ethics J 19:23–32 [DOI] [PubMed] [Google Scholar]
- 32.Eakin GS. and Behringer RR. (2003). Tetraploid development in the mouse. Dev Dyn 228:751–766 [DOI] [PubMed] [Google Scholar]
- 33.Guc-Scekic M, Milasin J, Stevanovic M, Stojanov LJ. and Djordjevic M. (2002). Tetraploidy in a 26-month-old girl (cytogenetic and molecular studies). Clin Genet 61:62–65 [DOI] [PubMed] [Google Scholar]
- 34.Nakamura Y, Takaira M, Sato E, Kawano K, Miyoshi O. and Niikawa N. (2003). A tetraploid liveborn neonate: cytogenetic and autopsy findings. Arch Pathol Lab Med 127:1612–1614 [DOI] [PubMed] [Google Scholar]
- 35.Marshall A. (1893). Vertebrate Embryology: A Text-Book for Students and Practitioners. G.P. Putnam's sons, New York [Google Scholar]
- 36.Moldenhauer JS. (2013). Ex utero intrapartum therapy. Semin Pediatr Surg 22:44–49 [DOI] [PubMed] [Google Scholar]
- 37.Suwinska A, Czolowska R, Ozdzenski W. and Tarkowski AK. (2008). Blastomeres of the mouse embryo lose totipotency after the fifth cleavage division: expression of Cdx2 and Oct4 and developmental potential of inner and outer blastomeres of 16- and 32-cell embryos. Dev Biol 322:133–144 [DOI] [PubMed] [Google Scholar]
- 38.De Paepe C, Cauffman G, Verloes A, Sterckx J, Devroey P, Tournaye H, Liebaers I. and Van. de Velde H. (2013). Human trophectoderm cells are not yet committed. Hum Reprod 28:740–749 [DOI] [PubMed] [Google Scholar]
- 39.Schramm RD. and Paprocki AM. (2004). In vitro development and cell allocation following aggregation of split embryos with tetraploid or developmentally asynchronous blastomeres in rhesus monkeys. Cloning Stem Cells 6:302–314 [DOI] [PubMed] [Google Scholar]
- 40.Shinozawa T, Sugawara A, Matsumoto A, Han YJ, Tomioka I, Inai K, Sasada H, Kobayashi E, Matsumoto H. and Sato E. (2006). Development of rat tetraploid and chimeric embryos aggregated with diploid cells. Zygote 14:287–297 [DOI] [PubMed] [Google Scholar]
- 41.Tarkowski AK, Ozdzenski W. and Czolowska R. (2005). Identical triplets and twins developed from isolated blastomeres of 8- and 16-cell mouse embryos supported with tetraploid blastomeres. Int J Dev Biol 49:825–832 [DOI] [PubMed] [Google Scholar]
- 42.Tarkowski AK, Suwinska A, Czolowska R. and Ozdzenski W. (2010). Individual blastomeres of 16- and 32-cell mouse embryos are able to develop into foetuses and mice. Dev Biol 348:190–198 [DOI] [PubMed] [Google Scholar]
- 43.Brink TC, Sudheer S, Janke D, Jagodzinska J, Jung M. and Adjaye J. (2008). The origins of human embryonic stem cells: a biological conundrum. Cells Tissues Organs 188:9–22 [DOI] [PubMed] [Google Scholar]
- 44.O'Leary T, Heindryckx B, Lierman S, van Bruggen D, Goeman JJ, Vandewoestyne M, Deforce D, de Sousa Lopes SM. and De Sutter P. (2012). Tracking the progression of the human inner cell mass during embryonic stem cell derivation. Nat Biotechnol 30:278–282 [DOI] [PubMed] [Google Scholar]
- 45.Reijo Pera RA, DeJonge C, Bossert N, Yao M, Hwa Yang JY, Asadi NB, Wong W, Wong C. and Firpo MT. (2009). Gene expression profiles of human inner cell mass cells and embryonic stem cells. Differentiation 78:18–23 [DOI] [PubMed] [Google Scholar]
- 46.Tang F, Barbacioru C, Bao S, Lee C, Nordman E, Wang X, Lao K. and Surani MA. (2010). Tracing the derivation of embryonic stem cells from the inner cell mass by single-cell RNA-Seq analysis. Cell Stem Cell 6:468–478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Varela E, Schneider RP, Ortega S. and Blasco MA. (2011). Different telomere-length dynamics at the inner cell mass versus established embryonic stem (ES) cells. Proc Natl Acad Sci U S A 108:15207–15212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Martin-McCaffrey L, Willard FS, Oliveira-dos-Santos AJ, Natale DR, Snow BE, Kimple RJ, Pajak A, Watson AJ, Dagnino L, et al. (2004). RGS14 is a mitotic spindle protein essential from the first division of the mammalian zygote. Dev Cell 7:763–769 [DOI] [PubMed] [Google Scholar]
- 49.Bouniol C, Nguyen E. and Debey P. (1995). Endogenous transcription occurs at the 1-cell stage in the mouse embryo. Exp Cell Res 218:57–62 [DOI] [PubMed] [Google Scholar]
- 50.Aoki F, Worrad DM. and Schultz RM. (1997). Regulation of transcriptional activity during the first and second cell cycles in the preimplantation mouse embryo. Dev Biol 181:296–307 [DOI] [PubMed] [Google Scholar]
- 51.Zeng F. and Schultz RM. (2005). RNA transcript profiling during zygotic gene activation in the preimplantation mouse embryo. Dev Biol 283:40–57 [DOI] [PubMed] [Google Scholar]
- 52.Ao A, Erickson RP, Winston RM. and Handyside AH. (1994). Transcription of paternal Y-linked genes in the human zygote as early as the pronucleate stage. Zygote 2:281–287 [DOI] [PubMed] [Google Scholar]
- 53.Daniels R, Zuccotti M, Kinis T, Serhal P. and Monk M. (1997). XIST expression in human oocytes and preimplantation embryos. Am J Hum Genet 61:33–39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Dobson AT, Raja R, Abeyta MJ, Taylor T, Shen S, Haqq C. and Pera RA. (2004). The unique transcriptome through day 3 of human preimplantation development. Hum Mol Genet 13:1461–1470 [DOI] [PubMed] [Google Scholar]
- 55.Daniels R, Kinis T, Serhal P. and Monk M. (1995). Expression of the myotonin protein kinase gene in preimplantation human embryos. Hum Mol Genet 4:389–393 [DOI] [PubMed] [Google Scholar]
- 56.Fiddler M, Abdel-Rahman B, Rappolee DA. and Pergament E. (1995). Expression of SRY transcripts in preimplantation human embryos. Am J Med Genet 55:80–84 [DOI] [PubMed] [Google Scholar]
- 57.Daniels R, Lowell S, Bolton V. and Monk M. (1997). Transcription of tissue-specific genes in human preimplantation embryos. Hum Reprod 12:2251–2256 [DOI] [PubMed] [Google Scholar]
- 58.Xue Z, Huang K, Cai C, Cai L, Jiang CY, Feng Y, Liu Z, Zeng Q, Cheng L, et al. (2013). Genetic programs in human and mouse early embryos revealed by single-cell RNA sequencing. Nature 500:593–597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Yan L, Yang M, Guo H, Yang L, Wu J, Li R, Liu P, Lian Y, Zheng X, et al. (2013). Single-cell RNA-Seq profiling of human preimplantation embryos and embryonic stem cells. Nat Struct Mol Biol 20:1131–1139 [DOI] [PubMed] [Google Scholar]
- 60.VerMilyea MD, Maneck M, Yoshida N, Blochberger I, Suzuki E, Suzuki T, Spang R, Klein CA. and Perry AC. (2011). Transcriptome asymmetry within mouse zygotes but not between early embryonic sister blastomeres. EMBO J 30:1841–1851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Antczak M. and Van Blerkom J. (1997). Oocyte influences on early development: the regulatory proteins leptin and STAT3 are polarized in mouse and human oocytes and differentially distributed within the cells of the preimplantation stage embryo. Mol Hum Reprod 3:1067–1086 [DOI] [PubMed] [Google Scholar]
- 62.Hansis C, Grifo JA. and Krey LC. (2004). Candidate lineage marker genes in human preimplantation embryos. Reprod Biomed Online 8:577–583 [DOI] [PubMed] [Google Scholar]
- 63.Jedrusik A, Parfitt DE, Guo G, Skamagki M, Grabarek JB, Johnson MH, Robson P. and Zernicka-Goetz M. (2008). Role of Cdx2 and cell polarity in cell allocation and specification of trophectoderm and inner cell mass in the mouse embryo. Genes Dev 22:2692–2706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Torres-Padilla ME, Parfitt DE, Kouzarides T. and Zernicka-Goetz M. (2007). Histone arginine methylation regulates pluripotency in the early mouse embryo. Nature 445:214–218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Sun JH, Zhang Y, Yin BY, Li JX, Liu GS, Xu W. and Tang S. (2012). Differential expression of Axin1, Cdc25c and Cdkn2d mRNA in 2-cell stage mouse blastomeres. Zygote 20:305–310 [DOI] [PubMed] [Google Scholar]
- 66.Plachta N, Bollenbach T, Pease S, Fraser SE. and Pantazis P. (2011). Oct4 kinetics predict cell lineage patterning in the early mammalian embryo. Nat Cell Biol 13:117–123 [DOI] [PubMed] [Google Scholar]
- 67.Galan A, Montaner D, Poo ME, Valbuena D, Ruiz V, Aguilar C, Dopazo J. and Simon C. (2010). Functional genomics of 5- to 8-cell stage human embryos by blastomere single-cell cDNA analysis. PLoS One 5:e13615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Hartshorn C, Eckert JJ, Hartung O. and Wangh LJ. (2007). Single-cell duplex RT-LATE-PCR reveals Oct4 and Xist RNA gradients in 8-cell embryos. BMC Biotechnol 7:87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.May A, Kirchner R, Muller H, Hartmann P, El Hajj N, Tresch A, Zechner U, Mann W. and Haaf T. (2009). Multiplex rt-PCR expression analysis of developmentally important genes in individual mouse preimplantation embryos and blastomeres. Biol Reprod 80:194–202 [DOI] [PubMed] [Google Scholar]
- 70.Wang CW, Yao DS, Horng SG, Chiu HC, Chen CK, Lee CL, Huang HY, Wang HS, Soong YK. and Pao CC. (2004). Feasibility of human telomerase reverse transcriptase mRNA expression in individual blastomeres as an indicator of early embryo development. J Assist Reprod Genet 21:163–168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Roberts RM, Katayama M, Magnuson SR, Falduto MT. and Torres KE. (2011). Transcript profiling of individual twin blastomeres derived by splitting two-cell stage murine embryos. Biol Reprod 84:487–494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Piotrowska-Nitsche K, Perea-Gomez A, Haraguchi S. and Zernicka-Goetz M. (2005). Four-cell stage mouse blastomeres have different developmental properties. Development 132:479–490 [DOI] [PubMed] [Google Scholar]
- 73.Lorthongpanich C, Yang SH, Piotrowska-Nitsche K, Parnpai R. and Chan AW. (2008). Development of single mouse blastomeres into blastocysts, outgrowths and the establishment of embryonic stem cells. Reproduction 135:805–813 [DOI] [PubMed] [Google Scholar]
- 74.Townes P. and Holtfreter J. (1955). Directed movements and selective adhesion of embryonic amphibian cells. J Exp Zool 128:53–120 [DOI] [PubMed] [Google Scholar]
- 75.Amack JD. and Manning ML. (2012). Knowing the boundaries: extending the differential adhesion hypothesis in embryonic cell sorting. Science 338:212–215 [DOI] [PubMed] [Google Scholar]
- 76.McDole K, Xiong Y, Iglesias PA. and Zheng Y. (2011). Lineage mapping the pre-implantation mouse embryo by two-photon microscopy, new insights into the segregation of cell fates. Dev Biol 355:239–249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Meilhac SM, Adams RJ, Morris SA, Danckaert A, Le Garrec JF. and Zernicka-Goetz M. (2009). Active cell movements coupled to positional induction are involved in lineage segregation in the mouse blastocyst. Dev Biol 331:210–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Gurdon JB. (1988). A community effect in animal development. Nature 336:772–774 [DOI] [PubMed] [Google Scholar]
- 79.Rossant J. (1975). Investigation of the determinative state of the mouse inner cell mass. II. The fate of isolated inner cell masses transferred to the oviduct. J Embryol Exp Morphol 33:991–1001 [PubMed] [Google Scholar]
- 80.Carr DH. (1977). An experimental study of trophoblast growth in the lung. Obstet Gynecol 50:473–478 [PubMed] [Google Scholar]
- Tutton DA. and Carr DH. (1984). The fate of trophoblast retained within the oviduct in the mouse. Gynecol Obstet Invest 17:18–24 [DOI] [PubMed] [Google Scholar]
- 82.Evans MJ. and Kaufman MH. (1981). Establishment in culture of pluripotential cells from mouse embryos. Nature 292:154–156 [DOI] [PubMed] [Google Scholar]
- 83.Martin GR. (1981). Isolation of a pluripotent cell line from early mouse embryos cultured in medium conditioned by teratocarcinoma stem cells. Proc Natl Acad Sci U S A 78:7634–7638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Tanaka S, Kunath T, Hadjantonakis AK, Nagy A. and Rossant J. (1998). Promotion of trophoblast stem cell proliferation by FGF4. Science 282:2072–2075 [DOI] [PubMed] [Google Scholar]
- 85.Kim C, Park J, Amano T, Xu RH, Lin G, Carter MG. and Tian XC. (2012). Established preblastocyst- and blastocyst-derived ES cell lines have highly similar gene expression profiles, despite their differing requirements for derivation culture conditions. Cell Reprogram 14:1–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Tesar PJ. (2005). Derivation of germ-line-competent embryonic stem cell lines from preblastocyst mouse embryos. Proc Natl Acad Sci U S A 102:8239–8244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Tesar PJ, Chenoweth JG, Brook FA, Davies TJ, Evans EP, Mack DL, Gardner RL. and McKay RD. (2007). New cell lines from mouse epiblast share defining features with human embryonic stem cells. Nature 448:196–199 [DOI] [PubMed] [Google Scholar]
- 88.Cauffman G, De Rycke M, Sermon K, Liebaers I. and Van de Velde H. (2009). Markers that define stemness in ESC are unable to identify the totipotent cells in human preimplantation embryos. Hum Reprod 24:63–70 [DOI] [PubMed] [Google Scholar]
- 89.Galan A, Diaz-Gimeno P, Poo ME, Valbuena D, Sanchez E, Ruiz V, Dopazo J, Montaner D, Conesa A. and Simon C. (2013). Defining the genomic signature of totipotency and pluripotency during early human development. PLoS One 8:e62135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Falco G, Lee SL, Stanghellini I, Bassey UC, Hamatani T. and Ko MS. (2007). Zscan4: a novel gene expressed exclusively in late 2-cell embryos and embryonic stem cells. Dev Biol 307:539–550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Hung SS, Wong RC, Sharov AA, Nakatake Y, Yu H. and Ko MS. (2013). Repression of global protein synthesis by Eif1a-like genes that are expressed specifically in the two-cell embryos and the transient Zscan4-positive state of embryonic stem cells. DNA Res 20:391–402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Zalzman M, Falco G, Sharova LV, Nishiyama A, Thomas M, Lee SL, Stagg CA, Hoang HG, Yang HT, et al. (2010). Zscan4 regulates telomere elongation and genomic stability in ES cells. Nature 464:858–863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Arima N, Uchida Y, Yu R, Nakayama K. and Nishina H. (2013). Acetylcholine receptors regulate gene expression that is essential for primitive streak formation in murine embryoid bodies. Biochem Biophys Res Commun 435:447–453 [DOI] [PubMed] [Google Scholar]
- 94.Ding H, Keller KC, Martinez IK, Geransar RM, zur Nieden KO, Nishikawa SG, Rancourt DE. and zur Nieden NI. (2012). NO-beta-catenin crosstalk modulates primitive streak formation prior to embryonic stem cell osteogenic differentiation. J Cell Sci 125:5564–5577 [DOI] [PubMed] [Google Scholar]
- 95.Gadue P, Huber TL, Paddison PJ. and Keller GM. (2006). Wnt and TGF-beta signaling are required for the induction of an in vitro model of primitive streak formation using embryonic stem cells. Proc Natl Acad Sci U S A 103:16806–16811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Nakanishi M, Kurisaki A, Hayashi Y, Warashina M, Ishiura S, Kusuda-Furue M. and Asashima M. (2009). Directed induction of anterior and posterior primitive streak by Wnt from embryonic stem cells cultured in a chemically defined serum-free medium. FASEB J 23:114–122 [DOI] [PubMed] [Google Scholar]
- 97.ten Berge D, Koole W, Fuerer C, Fish M, Eroglu E. and Nusse R. (2008). Wnt signaling mediates self-organization and axis formation in embryoid bodies. Cell Stem Cell 3:508–518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Devoize L, Collangettes D, Le Bouedec G, Mishellany F, Orliaguet T, Dallel R. and Baudet-Pommel M. (2008). Giant mature ovarian cystic teratoma including more than 300 teeth. Oral Surg Oral Med Oral Pathol Oral Radiol Endod 105:e76–e79 [DOI] [PubMed] [Google Scholar]
- 99.Sergi C, Ehemann V, Beedgen B, Linderkamp O. and Otto HF. (1999). Huge fetal sacrococcygeal teratoma with a completely formed eye and intratumoral DNA ploidy heterogeneity. Pediatr Dev Pathol 2:50–57 [DOI] [PubMed] [Google Scholar]
- 100.Joza N, Susin SA, Daugas E, Stanford WL, Cho SK, Li CY, Sasaki T, Elia AJ, Cheng HY, et al. (2001). Essential role of the mitochondrial apoptosis-inducing factor in programmed cell death. Nature 410:549–554 [DOI] [PubMed] [Google Scholar]
- 101.Thomson JA, Kalishman J, Golos TG, Durning M, Harris CP. and Hearn JP. (1996). Pluripotent cell lines derived from common marmoset (Callithrix jacchus) blastocysts. Biol Reprod 55:254–259 [DOI] [PubMed] [Google Scholar]
- 102.Andrews PW. (2002). From teratocarcinomas to embryonic stem cells. Philos Trans R Soc Lond B Biol Sci 357:405–417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Denker HW. (2008). Human embryonic stem cells: the real challenge for research as well as for bioethics is still ahead of us. Cells Tissues Organs 187:250–256 [DOI] [PubMed] [Google Scholar]
- 104.Kelm JM. and Fussenegger M. (2004). Microscale tissue engineering using gravity-enforced cell assembly. Trends Biotechnol 22:195–202 [DOI] [PubMed] [Google Scholar]
- 105.Suslov ON, Kukekov VG, Ignatova TN. and Steindler DA. (2002). Neural stem cell heterogeneity demonstrated by molecular phenotyping of clonal neurospheres. Proc Natl Acad Sci U S A 99:14506–14511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Armstrong MT. and Armstrong PB. (2003). Growth factor modulation of the extracellular matrix. Exp Cell Res 288:235–245 [DOI] [PubMed] [Google Scholar]
- 107.Guo Q, Xia B, Moshiach S, Xu C, Jiang Y, Chen Y, Sun Y, Lahti JM. and Zhang XA. (2008). The microenvironmental determinants for kidney epithelial cyst morphogenesis. Eur J Cell Biol 87:251–266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Zaibak F, Bello P, Kozlovski J, Crombie D, Ang H, Dottori M. and Williamson R. (2009). Unrestricted somatic stem cells from human umbilical cord blood grow in serum-free medium as spheres. BMC Biotechnol 9:101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Takahashi K. and Yamanaka S. (2006). Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell 126:663–676 [DOI] [PubMed] [Google Scholar]
- 110.Takahashi K, Tanabe K, Ohnuki M, Narita M, Ichisaka T, Tomoda K. and Yamanaka S. (2007). Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell 131:861–872 [DOI] [PubMed] [Google Scholar]
- 111.Yu J, Vodyanik MA, Smuga-Otto K, Antosiewicz-Bourget J, Frane JL, Tian S, Nie J, Jonsdottir GA, Ruotti V, et al. (2007). Induced pluripotent stem cell lines derived from human somatic cells. Science 318:1917–1920 [DOI] [PubMed] [Google Scholar]
- 112.Power C. and Rasko JE. (2011). Will cell reprogramming resolve the embryonic stem cell controversy? A narrative review. Ann Intern Med 155:114–121 [DOI] [PubMed] [Google Scholar]
- 113.Neri D. (2011). The race toward ‘ethically universally acceptable’ human pluripotent (embryonic-like) stem cells: only a problem of sources? Bioethics 25:260–266 [DOI] [PubMed] [Google Scholar]
- 114.Baertschi B. and Mauron A. (2010). Moral status revisited: the challenge of reversed potency. Bioethics 24:96–103 [DOI] [PubMed] [Google Scholar]
- 115.Takahashi K. (2012). Cellular reprogramming—lowering gravity on Waddington's epigenetic landscape. J Cell Sci 125:2553–2560 [DOI] [PubMed] [Google Scholar]
- 116.Worsdorfer P, Thier M, Kadari A. and Edenhofer F. (2013). Roadmap to cellular reprogramming—manipulating transcriptional networks with DNA, RNA, proteins and small molecules. Curr Mol Med 13:868–878 [DOI] [PubMed] [Google Scholar]
- 117.Ieda M, Fu JD, Delgado-Olguin P, Vedantham V, Hayashi Y, Bruneau BG. and Srivastava D. (2010). Direct reprogramming of fibroblasts into functional cardiomyocytes by defined factors. Cell 142:375–386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Song K, Nam YJ, Luo X, Qi X, Tan W, Huang GN, Acharya A, Smith CL, Tallquist MD, et al. (2012). Heart repair by reprogramming non-myocytes with cardiac transcription factors. Nature 485:599–604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Guo G, Huss M, Tong GQ, Wang C, Li Sun L, Clarke ND. and Robson P. (2010). Resolution of cell fate decisions revealed by single-cell gene expression analysis from zygote to blastocyst. Dev Cell 18:675–685 [DOI] [PubMed] [Google Scholar]
- 120.Cauffman G, Van de Velde H, Liebaers I. and Van Steirteghem A. (2005). Oct-4 mRNA and protein expression during human preimplantation development. Mol Hum Reprod 11:173–181 [DOI] [PubMed] [Google Scholar]
- 121.Niakan KK. and Eggan K. (2013). Analysis of human embryos from zygote to blastocyst reveals distinct gene expression patterns relative to the mouse. Dev Biol 375:54–64 [DOI] [PubMed] [Google Scholar]
- 122.Mitalipov S. and Wolf D. (2009). Totipotency, pluripotency and nuclear reprogramming. Adv Biochem Eng Biotechnol 114:185–199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Morgan HD, Santos F, Green K, Dean W. and Reik W. (2005). Epigenetic reprogramming in mammals. Hum Mol Genet 14 Spec No 1:R47–R58 [DOI] [PubMed] [Google Scholar]
- 124.Fukuda A, Cao F, Morita S, Yamada K, Jincho Y, Tane S, Sotomaru Y. and Kono T. (2010). Identification of inappropriately reprogrammed genes by large-scale transcriptome analysis of individual cloned mouse blastocysts. PLoS One 5:e11274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Jincho Y, Sotomaru Y, Kawahara M, Ono Y, Ogawa H, Obata Y. and Kono T. (2008). Identification of genes aberrantly expressed in mouse embryonic stem cell-cloned blastocysts. Biol Reprod 78:568–576 [DOI] [PubMed] [Google Scholar]
- 126.Somers J, Smith C, Donnison M, Wells DN, Henderson H, McLeay L. and Pfeffer PL. (2006). Gene expression profiling of individual bovine nuclear transfer blastocysts. Reproduction 131:1073–1084 [DOI] [PubMed] [Google Scholar]
- 127.Tong GQ, Heng BC, Tan LG. and Ng SC. (2006). Aberrant profile of gene expression in cloned mouse embryos derived from donor cumulus nuclei. Cell Tissue Res 325:231–243 [DOI] [PubMed] [Google Scholar]
- 128.Grondahl ML, Borup R, Vikesa J, Ernst E, Andersen CY. and Lykke-Hartmann K. (2013). The dormant and the fully competent oocyte: comparing the transcriptome of human oocytes from primordial follicles and in metaphase II. Mol Hum Reprod 19:600–617 [DOI] [PubMed] [Google Scholar]
- 129.Li R. and Albertini DF. (2013). The road to maturation: somatic cell interaction and self-organization of the mammalian oocyte. Nat Rev Mol Cell Biol 14:141–152 [DOI] [PubMed] [Google Scholar]
- 130.Sanchez F. and Smitz J. (2012). Molecular control of oogenesis. Biochim Biophys Acta 1822:1896–1912 [DOI] [PubMed] [Google Scholar]
- 131.Mtango NR, Potireddy S. and Latham KE. (2008). Oocyte quality and maternal control of development. Int Rev Cell Mol Biol 268:223–290 [DOI] [PubMed] [Google Scholar]
- 132.Li L, Zheng P. and Dean J. (2010). Maternal control of early mouse development. Development 137:859–870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Matzuk MM. and Burns KH. (2012). Genetics of mammalian reproduction: modeling the end of the germline. Annu Rev Physiol 74:503–528 [DOI] [PubMed] [Google Scholar]
- 134.Tashiro F, Kanai-Azuma M, Miyazaki S, Kato M, Tanaka T, Toyoda S, Yamato E, Kawakami H, Miyazaki T. and Miyazaki J. (2010). Maternal-effect gene Ces5/Ooep/Moep19/Floped is essential for oocyte cytoplasmic lattice formation and embryonic development at the maternal-zygotic stage transition. Genes Cells 15:813–828 [DOI] [PubMed] [Google Scholar]
- 135.Yurttas P, Vitale AM, Fitzhenry RJ, Cohen-Gould L, Wu W, Gossen JA. and Coonrod SA. (2008). Role for PADI6 and the cytoplasmic lattices in ribosomal storage in oocytes and translational control in the early mouse embryo. Development 135:2627–2636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Fernandes R, Tsuda C, Perumalsamy AL, Naranian T, Chong J, Acton BM, Tong ZB, Nelson LM. and Jurisicova A. (2012). NLRP5 mediates mitochondrial function in mouse oocytes and embryos. Biol Reprod 86:138, 1–10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Zheng P. and Dean J. (2009). Role of Filia, a maternal effect gene, in maintaining euploidy during cleavage-stage mouse embryogenesis. Proc Natl Acad Sci U S A 106:7473–7478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Li L, Baibakov B. and Dean J. (2008). A subcortical maternal complex essential for preimplantation mouse embryogenesis. Dev Cell 15:416–425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Ciccone DN, Su H, Hevi S, Gay F, Lei H, Bajko J, Xu G, Li E. and Chen T. (2009). KDM1B is a histone H3K4 demethylase required to establish maternal genomic imprints. Nature 461:415–418 [DOI] [PubMed] [Google Scholar]
- 140.Mohan KN, Ding F. and Chaillet JR. (2011). Distinct roles of DMAP1 in mouse development. Mol Cell Biol 31:1861–1869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Payer B, Saitou M, Barton SC, Thresher R, Dixon JP, Zahn D, Colledge WH, Carlton MB, Nakano T. and Surani MA. (2003). Stella is a maternal effect gene required for normal early development in mice. Curr Biol 13:2110–2117 [DOI] [PubMed] [Google Scholar]
- 142.Bultman SJ, Gebuhr TC, Pan H, Svoboda P, Schultz RM. and Magnuson T. (2006). Maternal BRG1 regulates zygotic genome activation in the mouse. Genes Dev 20:1744–1754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Philipps DL, Wigglesworth K, Hartford SA, Sun F, Pattabiraman S, Schimenti K, Handel M, Eppig JJ. and Schimenti JC. (2008). The dual bromodomain and WD repeat-containing mouse protein BRWD1 is required for normal spermiogenesis and the oocyte-embryo transition. Dev Biol 317:72–82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Blau HM, Pavlath GK, Hardeman EC, Chiu CP, Silberstein L, Webster SG, Miller SC. and Webster C. (1985). Plasticity of the differentiated state. Science 230:758–766 [DOI] [PubMed] [Google Scholar]
- 145.Katayama M, Ellersieck MR. and Roberts RM. (2010). Development of monozygotic twin mouse embryos from the time of blastomere separation at the two-cell stage to blastocyst. Biol Reprod 82:1237–1247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Rossant J. (1976). Postimplantation development of blastomeres isolated from 4- and 8-cell mouse eggs. J Embryol Exp Morphol 36:283–290 [PubMed] [Google Scholar]
- 147.Tarkowski AK. and Wroblewska J. (1967). Development of blastomeres of mouse eggs isolated at the 4- and 8-cell stage. J Embryol Exp Morphol 18:155–180 [PubMed] [Google Scholar]
- 148.Tarkowski AK, Ozdzenski W. and Czolowska R. (2001). How many blastomeres of the 4-cell embryo contribute cells to the mouse body? Int J Dev Biol 45:811–816 [PubMed] [Google Scholar]
- 149.Van de Velde H, Cauffman G, Tournaye H, Devroey P. and Liebaers I. (2008). The four blastomeres of a 4-cell stage human embryo are able to develop individually into blastocysts with inner cell mass and trophectoderm. Hum Reprod 23:1742–1747 [DOI] [PubMed] [Google Scholar]
- 150.Johnson WH, Loskutoff NM, Plante Y. and Betteridge KJ. (1995). Production of four identical calves by the separation of blastomeres from an in vitro derived four-cell embryo. Vet Rec 137:15–16 [DOI] [PubMed] [Google Scholar]
- 151.Veiga A, Calderon G, Barri PN. and Coroleu B. (1987). Pregnancy after the replacement of a frozen-thawed embryo with less than 50% intact blastomeres. Hum Reprod 2:321–323 [DOI] [PubMed] [Google Scholar]
- 152.Saito S. and Niemann H. (1991). Effects of extracellular matrices and growth factors on the development of isolated porcine blastomeres. Biol Reprod 44:927–936 [DOI] [PubMed] [Google Scholar]
- 153.Gardner RL. and Davies TJ. (2006). An investigation of the origin and significance of bilateral symmetry of the pronuclear zygote in the mouse. Hum Reprod 21:492–502 [DOI] [PubMed] [Google Scholar]
- 154.Held E, Salilew-Wondim D, Linke M, Zechner U, Rings F, Tesfaye D, Schellander K. and Hoelker M. (2012). Transcriptome fingerprint of bovine 2-cell stage blastomeres is directly correlated with the individual developmental competence of the corresponding sister blastomere. Biol Reprod 87:154. [DOI] [PubMed] [Google Scholar]
- 155.Plusa B, Hadjantonakis AK, Gray D, Piotrowska-Nitsche K, Jedrusik A, Papaioannou VE, Glover DM. and Zernicka-Goetz M. (2005). The first cleavage of the mouse zygote predicts the blastocyst axis. Nature 434:391–395 [DOI] [PubMed] [Google Scholar]
- 156.Bischoff M, Parfitt DE. and Zernicka-Goetz M. (2008). Formation of the embryonic-abembryonic axis of the mouse blastocyst: relationships between orientation of early cleavage divisions and pattern of symmetric/asymmetric divisions. Development 135:953–962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Skamagki M, Wicher KB, Jedrusik A, Ganguly S. and Zernicka-Goetz M. (2013). Asymmetric localization of Cdx2 mRNA during the first cell-fate decision in early mouse development. Cell Rep 3:442–457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Niwa H, Toyooka Y, Shimosato D, Strumpf D, Takahashi K, Yagi R. and Rossant J. (2005). Interaction between Oct3/4 and Cdx2 determines trophectoderm differentiation. Cell 123:917–929 [DOI] [PubMed] [Google Scholar]
- 159.Zernicka-Goetz M. and Huang S. (2010). Stochasticity versus determinism in development: a false dichotomy? Nat Rev Genet 11:743–744 [DOI] [PubMed] [Google Scholar]
- 160.Lawrence PA. and Levine M. (2006). Mosaic and regulative development: two faces of one coin. Curr Biol 16:R236–R239 [DOI] [PubMed] [Google Scholar]
- 161.Tabansky I, Lenarcic A, Draft RW, Loulier K, Keskin DB, Rosains J, Rivera-Feliciano J, Lichtman JW, Livet J, et al. (2013). Developmental bias in cleavage-stage mouse blastomeres. Curr Biol 23:21–31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Takaoka K. and Hamada H. (2012). Cell fate decisions and axis determination in the early mouse embryo. Development 139:3–14 [DOI] [PubMed] [Google Scholar]
- 163.Johnson MH. (2009). From mouse egg to mouse embryo: polarities, axes, and tissues. Annu Rev Cell Dev Biol 25:483–512 [DOI] [PubMed] [Google Scholar]
- 164.Zernicka-Goetz M. (2005). Cleavage pattern and emerging asymmetry of the mouse embryo. Nat Rev Mol Cell Biol 6:919–928 [DOI] [PubMed] [Google Scholar]
- 165.Stringer JM, Barrand S. and Western P. (2013). Fine-tuning evolution: germ-line epigenetics and inheritance. Reproduction 146:R37–R48 [DOI] [PubMed] [Google Scholar]
- 166.Potok ME, Nix DA, Parnell TJ. and Cairns BR. (2013). Reprogramming the maternal zebrafish genome after fertilization to match the paternal methylation pattern. Cell 153:759–772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Hsu YC. and Gonda MA. (1980). Monozygotic twin formation in mouse embryos in vitro. Science 209:605–606 [DOI] [PubMed] [Google Scholar]
- 168.Koch-Hershenov R. (2006). Totipotency, twinning, and ensoulment at fertilization. J Med Philos 31:139–164 [DOI] [PubMed] [Google Scholar]
- 169.Velez JR. (2005). Immediate animation: thomistic principles applied to Norman Ford's objections. Ethics Med 21:11–28 [PubMed] [Google Scholar]
- 170.Herranz G. (2013). The timing of monozygotic twinning: a criticism of the common model. Zygote [Epub ahead of print]; DOI: 10.1017/S0967199413000257 [DOI] [PubMed] [Google Scholar]
- 171.Driesch H. (1892). The potency of the first two cleavage cells in echinoderm development: experimental production of partial and double formations. In: Foundations of Experimental Embryology, 1974. Willier B, Oppenheimer J, eds. Hafner, New York [Google Scholar]
- 172.Gatica R, Boland MP, Crosby TF. and Gordon I. (1984). Micromanipulation of sheep morulae to produce monozygotic twins. Theriogenology 21:555–560 [DOI] [PubMed] [Google Scholar]
- 173.Mitalipov SM, Yeoman RR, Kuo HC. and Wolf DP. (2002). Monozygotic twinning in rhesus monkeys by manipulation of in vitro-derived embryos. Biol Reprod 66:1449–1455 [DOI] [PubMed] [Google Scholar]
- 174.Reichelt B. and Niemann H. (1994). Generation of identical twin piglets following bisection of embryos at the morula and blastocyst stage. J Reprod Fertil 100:163–172 [DOI] [PubMed] [Google Scholar]
- 175.Wang ZJ, Trounson A. and Dziadek M. (1990). Developmental capacity of mechanically bisected mouse morulae and blastocysts. Reprod Fertil Dev 2:683–691 [DOI] [PubMed] [Google Scholar]
- 176.Hardy K. and Handyside AH. (1993). Cell allocation in twin half mouse embryos bisected at the 8-cell stage: implications for preimplantation diagnosis. Mol Reprod Dev 36:16–22 [DOI] [PubMed] [Google Scholar]
- 177.Rho GJ, Johnson WH. and Betteridge KJ. (1998). Cellular composition and viability of demi- and quarter-embryos made from bisected bovine morulae and blastocysts produced in vitro. Theriogenology 50:885–895 [DOI] [PubMed] [Google Scholar]
- 178.Tao T, Reichelt B. and Niemann H. (1995). Ratio of inner cell mass and trophoblastic cells in demi- and intact pig embryos. J Reprod Fertil 104:251–258 [DOI] [PubMed] [Google Scholar]
- 179.Papaioannou VE. (1982). Lineage analysis of inner cell mass and trophectoderm using microsurgically reconstituted mouse blastocysts. J Embryol Exp Morphol 68:199–209 [PubMed] [Google Scholar]
- 180.Rossant J, Croy BA, Clark DA. and Chapman VM. (1983). Interspecific hybrids and chimeras in mice. J Exp Zool 228:223–233 [DOI] [PubMed] [Google Scholar]
- 181.Zheng YL, Jiang MX, OuYang YC, Sun QY. and Chen DY. (2005). Production of mouse by inter-strain inner cell mass replacement. Zygote 13:73–77 [DOI] [PubMed] [Google Scholar]
- 182.Gardner RL. (1985). Regeneration of endoderm from primitive ectoderm in the mouse embryo: fact or artifact? J Embryol Exp Morphol 88:303–326 [PubMed] [Google Scholar]
- 183.Szczepanska K, Stanczuk L. and Maleszewski M. (2011). Isolated mouse inner cell mass is unable to reconstruct trophectoderm. Differentiation 82:1–8 [DOI] [PubMed] [Google Scholar]
- 184.Morris SA, Guo Y. and Zernicka-Goetz M. (2012). Developmental plasticity is bound by pluripotency and the Fgf and Wnt signaling pathways. Cell Rep 2:756–765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Farhi A, Reichman B, Boyko V, Mashiach S, Hourvitz A, Margalioth EJ, Levran D, Calderon I, Orvieto R, et al. (2013). Congenital malformations in infants conceived following assisted reproductive technology in comparison with spontaneously conceived infants. J Matern Fetal Neonatal Med 26:1171–1179 [DOI] [PubMed] [Google Scholar]
- 186.Hansen M, Kurinczuk JJ, Milne E, de Klerk N. and Bower C. (2013). Assisted reproductive technology and birth defects: a systematic review and meta-analysis. Hum Reprod Update 19:330–353 [DOI] [PubMed] [Google Scholar]
- 187.Lu YH, Wang N. and Jin F. (2013). Long-term follow-up of children conceived through assisted reproductive technology. J Zhejiang Univ Sci B 14:359–371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Tachibana M, Amato P, Sparman M, Gutierrez NM, Tippner-Hedges R, Ma H, Kang E, Fulati A, Lee HS, et al. (2013). Human embryonic stem cells derived by somatic cell nuclear transfer. Cell 153:1228–1238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Gray D, Plusa B, Piotrowska K, Na J, Tom B, Glover DM. and Zernicka-Goetz M. (2004). First cleavage of the mouse embryo responds to change in egg shape at fertilization. Curr Biol 14:397–405 [DOI] [PubMed] [Google Scholar]
- 190.Hierholzer A. and Kemler R. (2009). Cdx1::Cre allele for gene analysis in the extraembryonic ectoderm and the three germ layers of mice at mid-gastrulation. Genesis 47:204–209 [DOI] [PubMed] [Google Scholar]
- 191.Matsumoto H, Daikoku T, Wang H, Sato E. and Dey SK. (2004). Differential expression of ezrin/radixin/moesin (ERM) and ERM-associated adhesion molecules in the blastocyst and uterus suggests their functions during implantation. Biol Reprod 70:729–736 [DOI] [PubMed] [Google Scholar]
- 192.Snow MH. (1975). Embryonic development of tetraploid mice during the second half of gestation. J Embryol Exp Morphol 34:707–721 [PubMed] [Google Scholar]
- 193.Cunningham JJ, Ulbright TM, Pera MF. and Looijenga LH. (2012). Lessons from human teratomas to guide development of safe stem cell therapies. Nat Biotechnol 30:849–857 [DOI] [PubMed] [Google Scholar]