Abstract
Protective antibodies play an essential role in immunity to infection by neutralizing microbes or their toxins and recruiting microbicidal effector functions. Identification of the protective B-cell epitopes, those parts of microbial antigens that contact the variable regions of the protective antibodies, can lead to development of antibody therapeutics, guide vaccine design, enable assessment of protective antibody responses in infected or vaccinated individuals, and uncover or localize pathogenic microbial functions that could be targeted by novel antimicrobials. Monoclonal antibodies are required to link in vivo or in vitro protective effects to specific epitopes and may be obtained from experimental animals or from humans, and their binding can be localized to specific regions of antigens by immunochemical assays. The epitopes are then identified with mapping methods such as X-ray crystallography of antigen–antibody complexes, antibody inhibition of hydrogen–deuterium exchange in the antigen, antibody-induced alteration of the nuclear magnetic resonance spectrum of the antigen, and experimentally validated computational docking of antigen–antibody complexes. The diversity in shape, size and structure of protective B-cell epitopes, and the increasing importance of protective B-cell epitope discovery to development of vaccines and antibody therapeutics are illustrated through examples from different microbe categories, with emphasis on epitopes targeted by broadly neutralizing antibodies to pathogens of high antigenic variation. Examples include the V-shaped Ab52 glycan epitope in the O-antigen of Francisella tularensis, the concave CR6261 peptidic epitope in the haemagglutinin stem of influenza virus H1N1, and the convex/concave PG16 glycopeptidic epitope in the gp120 V1/V2 loop of HIV type 1.
Keywords: antibodies, antigens/peptides/epitopes, human, structural biology/crystallography
Introduction
Antibodies participate in the immune response to microbes by interacting with surface or secreted microbial antigens. Each antibody binds to an epitope, defined as the three-dimensional structure of the amino acids, sugars or other residues in an antigen that can be contacted by the variable (V) regions of an antibody.1,2 The most protective antibodies against viruses and toxins target those epitopes on microbial antigens that interact with host receptors to invade host cells, and block invasion. They are therefore neutralizing antibodies.3–7 The most protective antibodies against extracellular microbes target carbohydrate epitopes on capsular or other cell surface polysaccharides,7 enabling microbe-killing through Fc-mediated effector mechanisms: complement-dependent killing, phagocytosis, and antibody-dependent cellular cytotoxicity.8 These antibody functions are major determinants for the success of vaccination at preventing infectious disease.
Currently licensed vaccines are directed against: viruses, including measles, mumps, rubella, rabies, poliovirus, varicella virus, papillomavirus; extracellular bacteria, such as Streptococcus, Meningococcus and Haemophilus influenzae type b; and bacterial toxins, such as diphtheria and tetanus toxins (http://www.cdc.gov/vaccines/schedules/).7 These vaccines work by inducing microbe-specific or toxin-specific protective antibodies,3–7 although T cells are required for somatic hypermutation to produce high-affinity IgG and IgA antibodies, and are important for the generation of B-cell memory.9
The essential role of antibodies against infectious diseases is further evidenced by the effectiveness of passively administered intravenous immunoglobulin for long-term treatment of immune deficiencies like X-linked agammaglobulinaemia and hyper-IgM syndrome.10 Specific immunoglobulins, such as hepatitis B immunoglobulin (http://www.cdc.gov/mmwr/preview/mmwrhtml/00022736.htm), tetanus immunoglobulin11 and rabies immunoglobulin12 are successfully used as post-exposure prophylaxis. In recent years, monoclonal antibodies (mAbs) have been used for prophylaxis against infections with respiratory syncytial virus (RSV)13 and rabies,12 and for treatment of inhalational anthrax.14
Despite the clinical success of licensed vaccines and passively administered antibody preparations, the development of effective vaccines and therapeutic antibodies against viruses and extracellular bacteria that exhibit high antigenic variation, against non-viral intracellular pathogens like fungi and intracellular bacteria, and against microbes with multi-stage life-cycles like protozoan and metazoan parasites has proven to be challenging.6,7 Furthermore, vaccines, immunotherapeutics and other antimicrobials are needed for prophylaxis and treatment of diseases caused by emerging and re-emerging infectious agents and potential agents of bioterrorism, including naturally evolving or intentionally engineered drug-resistant variants (http://www.niaid.nih.gov/topics/emerging/pages/list.aspx).
Development of vaccines and antibody therapeutics is greatly aided by identification of microbial epitopes targeted by protective antibodies – protective B-cell epitopes. This knowledge can lead directly to development of therapeutic antibodies, as it has for infections with RSV,13 rabies12 and anthrax.14 It could also guide the design of subunit vaccines to include protective epitopes and exclude any identified pathogenic epitopes that might induce cross-reactive autoimmune15 or infection-enhancing antibodies.16–18 Furthermore, known protective B-cell epitopes could be used to monitor the quality of antibody responses in infected or vaccinated individuals.19–21 Lastly, identification of protective B-cell epitopes may uncover or localize pathogenic microbial functions which, as has been suggested22,23 and demonstrated,24 may lead to the development of novel antimicrobials. We review here recent approaches to discovery of protective microbial B-cell epitopes, based largely on examples tabulated at the end of the article.
Strategies to identify and characterize anti-microbial protective mAbs
Identification of protective B-cell epitopes requires protective mAbs which, through their interaction with antigen, prevent or contribute to prevention of microbial pathogenesis. How are protective mAbs obtained? In some cases one or more protective antigens in a given microbe are known and mAbs to a target antigen, or fragments thereof, are generated and tested for efficacy against the microbe in vivo and/or in vitro. In other cases no knowledge of protective microbial antigens is available, or such knowledge is ignored, and a collection of mAbs to microbial surface components and/or secreted products is first generated and divided into groups in which all group members bind to the same antigen in immunoassays. The target antigens are identified and representative mAbs against each antigen are then tested for efficacy against the microbe.25
The mAbs are derived from lymphocyte-containing samples obtained from immunized experimental animals, most often mice, usually by hybridoma production,25–32 or from naturally infected or vaccinated humans by one of several methods. These include: cloning of V region genes from bone marrow or peripheral blood B cells or plasma cells into phage-display22,33,34 or yeast-display35 vectors, in vitro activation and expansion of memory B cells,21 cloning of the V region genes from single memory B cells21,22,36–40 or plasma cells41,42 into IgG expression vectors, or Epstein–Barr virus transformation of B cells40,43 optionally followed by fusion with myeloma cells.44 The target antigen of each mAb is identified by immunochemical assays including ELISA and Western blot analysis on purified candidate microbial antigens.25,45,46 Whether or not purified (native or recombinant) antigen is available, the protein and/or carbohydrate nature of the target epitope can be determined by pre-treatment of the antigen or antigen mixture with proteases25 or glycosidases37,47,48 in ELISA or Western blot. Protein antigens can be identified by proteome microarray analysis, in which reactivity of each mAb to the recombinantly expressed microbial proteins is assessed;25 or by mass spectrometric analysis of an SDS–polyacrylamide gel band immuneprecipitated from a microbial extract by the mAb49 or of a mAb-reactive spot on a two-dimensional gel.50 Alternatively, mAbs are screened directly in a functional assay such as neutralization,21,36–38 or memory B cells are selected by FACS for binding to the antigen but not to an antigen variant in which the epitope region of interest has been deleted.39
Monoclonal antibodies specific for the same microbial antigen can be subgrouped by the germline genes that encode their VH and VL regions22,29,38,42,44,51 and by their ability to block each other's binding to the antigen in competition immunoassays.29,37–40,45,46 Hence, sharing of the same VH, D, JH, VL and JL genes, or even just the same VH and VL genes is reflected in high amino acid sequence homology (http://www.imgt.org/) and indicates specificity for the same epitope. For antigens with sequentially repeating epitopes, as often found in capsular and outer-membrane microbial polysaccharides, mAbs that block each other's binding to the antigen target the same or an overlapping epitope.29,32,52 Although this is also true for protein antigens, some anti-protein mAbs that block each other's binding to the antigen may do so not by targeting an overlapping epitope but by interfering with each other's binding sterically or allosterically.38 However, in a group of mAbs specific for the same antigen, those mAbs that do not block each other's antigen-binding, or that show different cross-blocking profiles with third-party mAbs, define different epitopes.
In vivo testing of mAbs for anti-microbial efficacy is done in animal models of infection, including mice,22,28,29,33,53–55 rats,36 guinea pigs,56 non-human primates,57,58 and humanized mice,59 in which protective mAbs are identified by their ability to confer or prolong survival or reduce microbial burden. In addition, or alternatively, mAbs are functionally evaluated in vitro for their ability to cause killing of target microbes or a reduction in the microbes' replication,27,30 block host-cell invasion by microbes or their products (neutralize),21,22,33,36–39,42,44,46,56,58,60–63 block the binding of microbial factors to host components,30,33 or interfere with assembly of microbial toxins.64 Use of human immune components in these in vitro assays, such as human cell lines,33,36–38,46,55 can validate results obtained in vivo in animal models. Although efficacy indicates that the targeted epitope is protective, lack of efficacy does not necessarily mean that the epitope is non-protective because in addition to epitope specificity the protective efficacy of mAbs depends on their avidity and isotype.29,52,54 A flow diagram of general strategies for generation and identification of protective mAbs is shown in Fig. 1.
Figure 1.
Flow diagram of general strategies for identification of protective monoclonal antibodies (mAbs).
Types, definitions and documentation of microbial B-cell epitopes
Microbial B-cell epitopes are located mainly on the exposed parts of microbial antigens,65 generally protein and carbohydrate components, and may consist entirely of amino acids in peptidic epitopes, sugar residues in glycan epitopes, or combinations thereof in glycopeptidic epitopes. Many glycan epitopes and a small minority of peptidic epitopes consist of contiguous residues and are, therefore, referred to as continuous (or linear). However, the vast majority of peptidic epitopes are discontinuous, consisting of one to several amino acids that are separated by a few or many non-contact amino acids in the primary sequence but are brought together by the folding of the protein.2 Therefore, the existence of discontinuous peptidic epitopes depends on a supporting scaffold of non-contact residues. The minimal contiguous amino acid sequence containing all residues of a discontinuous epitope, which is required for proper conformation of the contact residues, has been termed the ‘contact residue span’ and may range from 20 to 400 amino acids (most frequently 50–79) in native proteins.66 Hence, a B-cell epitope is defined both by the identities of the contact residues and by their conformation, which is determined by the three-dimensional fold of the contact residue span.
Information on published and investigator-submitted microbial B-cell epitopes can be found at the Immune Epitope Database and Analysis Resource (IEDB) (http://www.iedb.org/), which houses B-cell and T-cell epitope data and makes them accessible and searchable.1,67 This includes the contact residues of the epitopes, antibody-binding and protection assays, and tools for mapping of discontinuous epitopes onto three-dimensional antigen structures and for epitope prediction.67 Among the tens of thousands of documented microbial B-cell epitopes, the vast majority were defined based on antigen-derived synthetic peptides used to induce antibodies in experimental animals. Some of the antibodies to these peptides cross-react with the native and/or recombinant microbial antigen (http://www.iedb.org/), but their affinities for the native microbial antigen are generally several orders of magnitude lower than those of antibodies induced by immunization with the native antigen.2 Among the discontinuous peptidic and the non-peptidic epitopes only some have been shown to be protective, and no or few B-cell epitopes have been studied for several of the microbes considered to be emerging or re-emerging infectious agents or potential agents of bioterrorism (http://www.iedb.org/).
Methods of B-cell epitope mapping
Epitope mapping refers to identification of the antigen residues that are contacted by a complementary antibody during antigen–antibody binding. As a first step in epitope mapping, the binding of protective mAbs (often referred to as just antibodies) can be localized to specific regions of antigens by testing the reactivity of the mAbs to antigen subunits or fragments.26,30,68,69 For microbial carbohydrate chains with repeating units, antibodies that bind to unique epitopes at the non-reducing end can be distinguished from those that bind to repeating internal epitopes by their Western blot patterns, where the binding intensity of the latter but not the former increases with increasing chain size. This is exemplified in Fig. 2 for O-antigen chains that comprise a capsular polysaccharide and part of the lipopolysaccharide of Francisella tularensis, a potential bioterrorism agent.29,32
Figure 2.
Western blot distinction between unique terminal and repeating internal epitopes of microbial carbohydrate chains with repeating units. The N62 monoclonal antibody (mAb), specific for a terminal epitope in the O-antigen (O-Ag) of the Francisella tularensis lipopolysaccharide (which consists of variable numbers of a tetrasaccharide repeat, represented as ABCD) binds with equal intensity to short and long lipopolysaccharide chains (relative to the abundance of each chain). But the binding intensity of the Ab52 mAb, specific for a repeating internal epitope, increases with increasing chain length, as more mAb molecules are bound. The basis for the differential binding pattern is illustrated schematically for the (bracketed) lower part of the Western blot, with N62 represented by grey ovals and Ab52 by cyan ovals. Western blot lanes from Lu et al.29
Finer mapping methods for peptidic, glycan and glycopeptidic epitopes include: X-ray crystallography of antigen–antibody (Ag–Ab) complexes, antibody inhibition of hydrogen-deuterium exchange in the antigen, antibody-induced alteration of the nuclear magnetic resonance (NMR) spectrum of the antigen, glycan microarray probing of the antibody, oligosaccharide (glycan) competition with antigen for antibody-binding, selection or screening of antigen-derived proteolytic fragments or peptides for antibody-binding, testing the Ag–Ab reactivity of site-directed (antigen or antibody) or random (antigen) mutants, electron and cryoelectron microscopy of Ag–Ab complexes, and experimentally validated computational docking of Ag–Ab complexes.
X-ray crystallographic analysis of Ag–Ab complexes reveals the identities of the contact residues and conformations of both the epitope and the complementary binding-site of the antibody.21,30,33,42,48,52,62,70–75 It can be applied to any type of antigen, including protein and carbohydrate antigens. In the first step of this method, favourable conditions for nucleation and growth of high-quality crystals of the Ag–Ab complex are identified by testing a wide variety of crystallization solutions. The crystal of choice is exposed in multiple orientations to a beam of monochromatic X-rays, which are scattered by the electrons of its atoms, and the resulting diffraction patterns of spots (or reflections) are recorded. The information in the diffraction data is then used to calculate a three-dimensional electron-density map of the molecules that make up the crystal, in which the known (non-hydrogen) atoms of the antigen and antibody (from primary sequence analysis) are positioned to create a model of the complex. The contact residues that comprise the epitope can be defined by identifying all antigen residues within 4–5 Å of the antibody, a distance used by the Molecular Modeling Database to define contacts.76,77 X-ray crystallography requires large amounts of highly pure antigen and antibody, usually Fab antibody fragment, and is limited by inability to obtain Ag–Ab co-crystals with some antigens, especially membrane proteins.
Antibody inhibition of hydrogen–deuterium exchange in the antigen, which involves the deuterium exchange – mass spectrometry (DXMS) technology,78,79 reveals small segments containing contact residues in protein antigens.30,44 DXMS exploits the continuous reversible exchange of peptide-amide hydrogens in proteins with water hydrogens, the exchange rate of each hydrogen correlating directly with the extent to which it is exposed (accessible) to solvent. The exchange rates are determined by incubating the protein in buffer with deuterated water (D2O) for graded time periods followed by proteolysis into overlapping peptides, which are separated chromatographically. The deuterium content of each peptide is then analysed by mass spectrometry to obtain a ‘heat’ map of the exchange rate for the entire protein. Subtraction of the DXMS heat map of the antibody-bound antigen from the DXMS heat map of the free antigen reveals the sequences(s) of the antigen where deuterium exchange was inhibited by antibody binding. Although the spatial resolution of DXMS is not at the single residue level, the antigen segments comprising the epitope are localized to within a few amino acids.79 DXMS can be performed with lower-purity preparations of antigen and antibody than X-ray crystallography, does not require crystals, and intact antibody molecules, rather than Fab fragments, can be routinely used.
Antibody-induced alteration of the NMR spectrum of the antigen reveals contact residues of both protein and carbohydrate antigens.48,80–83 NMR spectroscopy is based on the ability of the nuclei of some isotopes, like 1H, 2H, 13C and 15N, to absorb and re-emit electromagnetic radiation at a specific resonance frequency when placed in a magnetic field (http://www.cis.rit.edu/htbooks/nmr/). The resonance frequency of different nuclei of the same isotope is altered by the electrons of neighbouring atoms. This results in different signals (chemical shifts) relative to a standard reference molecule, generating a spectrum that yields information about the chemical environment of each nucleus, from which its identity can be determined. The complexity of NMR spectra makes it difficult to determine the structure of large Ag–Ab complexes. However, the antigen residues within 5 Å of the antibody (the contact residues) can be determined if the antigen spectrum does not overlap with the antibody spectrum, as will be the case for carbohydrate antigens or for recombinant protein antigens that have been labelled, for example, with 2H, 13C and 15N.81,84 Subtraction of the NMR spectrum of the antibody-bound antigen from the NMR spectrum of the free antigen is then used to identify contact residues by the changes in chemical shifts that occur when residues exposed to solvent in the free antigen are buried in the Ag–Ab complex. This technique, called saturation transfer difference (STD) NMR, is similar in principle to DXMS except that both peptidic and non-peptidic epitopes can be mapped and the actual contact residues are identified in STD NMR. However, only relatively small antigen fragments can be used for epitope mapping by STD NMR, and labelled recombinant protein fragments are needed.
Oligosaccharides, synthesized or purified from microbial sources, can be used to probe the binding of antibody in glycan microarrays37 or in competition immunoassays.29,37,54,80,85 For glycan microarray probing, large numbers of different glycans are immobilized (printed) on glass slides in microspots, and antibody-binding is detected with fluorescent reagents, which are quantified in a fluorescence scanner.86 For oligosaccharide competition, short antigen-derived oligosaccharides of different length and structure are used as competitors in immunoassays that measure binding of the antibody to the antigen. Per cent binding-inhibition versus competitor concentration is plotted for each competitor and the most potent competitor, which requires the lowest concentration for half-maximal binding-inhibition, is deduced to comprise the glycan epitope. These methods are limited by the difficulty of synthesizing some oligosaccharides or purifying sufficient quantities from microbial sources. Furthermore, the spatial resolution will not be at the single residue level if oligosaccharides that differ by single sugar residues are not available.29,54
Antigen-derived overlapping or non-overlapping proteolytic fragments and peptides can be displayed on the surface of phage particles30 or yeast cells26,68 for selection by the antibody or can be used in Western blot or ELISA or microarrays to screen for antibody binding (peptide scanning).21,30,40,75 This approach is limited by the dependence of many epitopes on the three-dimensional structure of the larger antigen75 and the likelihood of identifying only partial epitopes.2,30
Site-directed mutations (point mutations, insertions, deletions) can be introduced into recombinant versions of either the antigen21,38,40,43,44,62,87 or the antibody21,48,52,74 and the mutants can be tested for loss of Ag–Ab binding or other functions. Mutants are often obtained by changing entire segments of the antigen or specific residues to alanine (alanine shaving)88 and verifying proper folding of the mutant proteins by their binding to mAbs that target other epitopes and/or to polyclonal antibodies.39,46 Alternatively, systematic random mutations to alanine can be introduced along an entire protein antigen (alanine scanning) and mutants probed for antibody binding, to obtain partial epitope information.21,26,38,46,48,87
Electron microscopy (EM) allows visualization of Ag–Ab complexes.44,52 The Ag–Ab mixtures are spread on a metal grid and introduced into a high-vacuum column in the electron microscope. There the sample is exposed to and diffracts an electron beam, which is then focused by electrostatic and electromagnetic lenses to yield electron-density maps that can be converted into an image. Stains and fixatives are used to protect the sample from radiation damage, but these alter the fine structure of the macromolecules. In a modification called cryoEM, Ag–Ab complexes can be observed in physiological buffers, without stains and fixatives, by preserving the complexes in a frozen hydrated state by rapid freezing at near liquid nitrogen temperatures, which protects the sample from radiation damage. The resolution of most EM and cryoEM methods is too low to reveal contact residues and, therefore, Ag–Ab models obtained by other methods are used to interpret the EM and cryoEM maps.
Computational docking of Ag–Ab complexes can be used to predict the contact residues using Ag–Ab models from either homology modelling or, preferably, X-ray crystallography of both or at least one of the partners. In the docking protocol, the antigen and antibody structures are tested in a large number of orientations and then each orientation is scored by energy, seeking the global minimum orientation. The scoring function can be greatly enhanced in accuracy if experimental data from the methods discussed above are used to guide (constrain) the selection, referred to as experimentally validated computational docking.20,40,43,44,54,80,82,83
Because epitope information from different mapping methods differs in extent and resolution (summarized in Table 1), multiple mapping methods are often used to obtain complementary or supporting data. As exemplified in Fig. 3 for the mapping of a protective peptidic B-cell epitope in Neisseria meningitides factor H binding protein,30 X-ray crystallography usually provides the most complete highest-resolution map. But other methods such as mutational analysis of both antigen and antibody contact residues and testing the binding of intact antibody to antigen under physiological conditions may be needed to reveal the critical contacts, without which Ag–Ab binding is abolished or greatly reduced, or those that make one antibody more potent or more cross-reactive than another at a particular function like neutralization.19,27,33,35,38,56,60,61,71 Furthermore, the epitope core responsible for a pathogenic function may be shared by antibodies that target overlapping epitopes with slightly different registers of contact residues.81 Therefore, for initial identification of protective B-cell epitope cores, the lower resolution maps obtained using DXMS for peptidic epitopes, and oligosaccharide competition and/or glycan microarray probing for glycan epitopes, may suffice.
Table 1.
Information obtained from different epitope mapping methods
| Method | Epitope type(s) mapped | Extent and resolution of map |
|---|---|---|
| X-ray crystallography of Ag–Ab complex | any1 | entire epitope, contact residues, epitope conformation |
| DXMS (Ab-inhibition of hydrogen-deuterium exchange in the Ag) | peptidic | entire epitope, small segments containing the contact residues |
| STD NMR (Ab-induced alteration of the NMR spectrum of the Ag) | any | contact residues |
| Glycan array probing or oligosaccharide competition for Ab-binding | glycan and glycopeptidic | entire epitope (glycan) or partial epitope (glycopeptidic), contact residues if oligosaccharides differing by one sugar are available; otherwise – small segments containing the contact residues |
| Peptide scanning (of Ag peptides) or testing of Ag fragments for Ab binding | peptidic | usually partial epitope, small segments containing some of the contact residues |
| Mutagenesis (alanine shaving, alanine scanning, point or deletion mutations) and testing for Ag–Ab binding | peptidic and glycopeptidic | partial epitope, critical contacts |
| EM or cryoEM of Ag–Ab complex | any | epitope region |
| Experimentally validated computational docking of Ag–Ab complex (constrained by data from other methods) | any | entire epitope, contact residues, epitope conformation |
Ab, antibody; Ag, antigen; cryoEM, cryoelectron microscopy; DXMS, deuterium exchange/mass spectrometry; EM, electron microscopy; STD NMR, saturation transfer difference nuclear magnetic resonance.
Including peptidic, glycan, lipid, nucleic acid, or combinations thereof.
Figure 3.
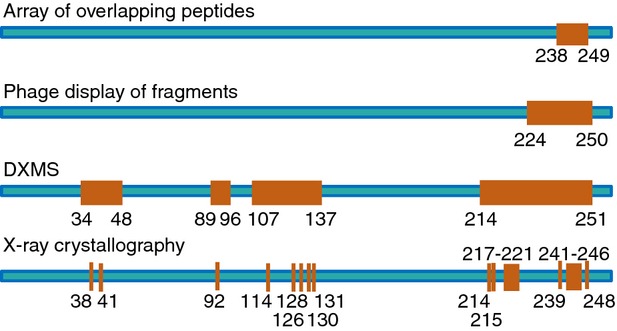
Linear representation of mapping results for a B-cell epitope in Neisseria meningitides factor H binding protein, obtained using different methods. Figure adapted with modifications from Malito et al.30 Epitope segments and contact residues are indicated by amino acid numbers and represented in orange.
Relation of shape, size and structure of microbial B-cell epitopes to protective antibody function
B-cell epitopes come in a variety of shapes and sizes, reflecting the complementary shapes and sizes of the binding-sites of antibodies, as illustrated in Fig. 4 for five protective epitopes and outlined in Table 2 for one or more protective epitope examples from each microbe category including bacteria, fungi, parasites and viruses. Epitopes can be convex, concave, relatively flat (with minor protrusions and depressions), or mixtures thereof, vary in surface area from < 200 to > 1500 Å2, and consist of from fewer than two to more than 34 residues (Fig. 4 and Table 2). They can be glycan, peptidic or glycopeptidic, include secondary structure segments of α-helices, β-strands and loops, and encompass or depend on the conformation of multiple antigen subunits (quaternary epitopes72,73) (Fig. 4 and Table 2), or even span two antigens (hybrid epitopes43).
Figure 4.
Examples of protective B-cell epitope structures. Antigens are represented as solid molecular surfaces (left panels and top right) and ribbon diagrams (right panels) rotated away from the reader approximately 90° about the horizontal axis, coloured with cyan carbons except for contact residues which are coloured orange. Antibody V regions are shown as wire-mesh molecular surfaces coloured grey (VH) and purple (VL). Images are clipped front and back to more clearly show interactions of antibodies and antigens. Selected contact residues are indicated in each panel for reference. The four sugars of the tetrasaccharide repeat in Francisella tularensis O-Ag108 are represented as ABCD for one and A'B'C'D' for the other of the two repeats shown. S, serine; E, glutamic acid; Q, glutamine; K, lysine; N, asparagine; D, aspartic acid; Man, mannose; Sia, sialic acid. Images were generated using Maestro (version 9 3 5, Schrödinger, Inc., New York, NY).
Table 2.
Examples of protective B-cell epitope discovery
 |
A terminal and a repeating-internal epitope in F. tularensis O-antigen exemplify convex glycan epitopes, the former a 185-Å2 two-sugar epitope that fits in a cavity-type antibody-binding site29 and the latter a 308-Å2 six-sugar V-shaped epitope that fits in a groove-type binding site that has a small central pocket that anchors the vertex of the epitope89 (Fig. 4a and Table 2). The protective efficacy of the two targeting antibodies is probably due mainly to mediation of effector functions. A 680-Å2 20-amino acid hydrophobic pocket formed by parts of a highly conserved α-helix and loop in the stem of influenza virus haemagglutinin is a concave peptidic epitope, which interacts with a convex binding-site formed by an exceptionally long CDR3 in the VH region of a broadly neutralizing Ab (bNAb).71 This bNAb can inactivate six of the 16 influenza virus haemagglutinin subtypes by blocking the low pH-induced conformational change required for fusion of the virus with the host cell endocytic membrane during infection71 (Fig. 4b and Table 2). A 1013-Å2 18-amino acid epitope with convex and concave parts, consisting of both helical and loop segments of the trimeric RSV fusion (F) protein in its pre-fusion conformation, interacts with a complementary antibody that locks the F protein in the pre-fusion conformation, preventing the rearrangement required for exposure of the fusion peptide and fusion of the viral and host cell membranes73 (Table 2 and Fig. 4c; the post-fusion conformation of the F protein is shown for comparison). A 1535-Å2 glycopeptidic epitope with both convex and concave parts, and consisting of β-strand amino acids and both high mannose-type and complex-type N-linked glycans in gp120 of the HIV-1 envelope spike, interacts with a bNAb38,48 (Table 2 and Fig. 4d). The bNAb can inactivate 73% of 162 strains representing major HIV-1 clades,48 possibly by preventing envelope conformations that allow binding to the host cell receptor CD4.
Clinical applications or implications of protective microbial B-cell epitopes
Translation of protective B-cell epitope mapping results into clinical applications is occurring and will continue to occur. As outlined for the examples given in Table 2, the influenza A bNAb CR6261, specific for a conserved epitope in the haemagglutinin stem, is in Phase I clinical trials as a therapeutic, and sera from patients with tularaemia,20 malaria19 and HIV infection21 have been tested for the presence of antibodies to mapped protective epitopes by competition ELISA for antigen-binding with the protective mAbs. Two antigens for which protective epitopes have been mapped are in Phase I–II clinical trials as malaria vaccines (Table 2), and one is a major component of a meningococcal serogroup B vaccine that has been recently licensed for use by the European Medicines Agency90 (see Table 2). The sera of vaccinees could be tested for the presence of antibodies to mapped protective epitopes as a correlate of vaccine protection.
Other protective antibodies could also be developed as therapeutics, especially for infections with drug-resistant microbes like methicillin-resistant S. aureus33 or Oseltamivir-resistant H1N1 influenza viruses,42 or for long-term vectored immunoprophylaxis against HIV infection by injection of adeno-associated virus transduced with an expression vector encoding several non-overlapping bNAbs.59 In addition, or alternatively, the mapped epitopes of protective antibodies could be engineered into new or improved vaccine designs. For example, an influenza virus haemagglutinin comprised of the stem only, without the highly variable head domain, is efficacious in mouse models and has been proposed as a candidate for a universal flu vaccine.91,92 Development of engineered improved versions of bNAbs could be considered for therapeutic or prophylactic treatment of dengue virus infection, if combined with modifications to the Fc region to reduce interaction with Fcγ receptors and hence the possibility of antibody-dependent enhancement of infection, which occurs at sub-neutralizing antibody concentrations.46,55 And, in view of the high number of somatic mutations found in the V regions of HIV-1 bNAbs21,93,94 and the discovery that the germline V region progenitors may not bind the same antigen,39 the development of HIV vaccines that would guide the immune system, stepwise, from immunoglobulin gene rearrangement, through affinity maturation and production of bNAbs, has been proposed6,74,95 (see Table 2).
Concluding remarks
Mapping the protective B-cell epitomes of microbes, the totality of protective epitope cores on microbial antigens, will continue to provide the best mAb candidates to be used for antimicrobial therapy and prophylaxis. Furthermore, such epitome knowledge will enable the design of subunit vaccines enriched in protective epitopes. Analysis of the molecular interactions between bNAbs and microbial antigens will inform the engineering of vaccine subunits to guide the immune system towards production of similar antibodies. Such vaccine strategies have a chance for success against pathogens of high antigenic variation that can replicate inside Fc receptor-bearing host cells if multiple protective epitopes are used68,96 and, most importantly, if combined with strategies to elicit both cytotoxic and helper T cells that can kill or stimulate the microbicidal activity of microbe-infected host cells.
Acknowledgments
B-cell epitope discovery studies from the authors' laboratories, included in this review, were supported with Federal funds from the US National Institute of Allergy and Infectious Diseases, National Institutes of Health, Department of Health and Human Services, under Contract No. HHSN272200900054C.
Glossary
- bNAb
broadly neutralizing antibody
- cryoEM
cryoelectron microscopy
- DXMS
deuterium exchange/mass spectrometry
- HIV
human immunodeficiency virus
- IC50
half-maximal inhibitory concentration
- LPS
lipopolysaccharide
- NMR
nuclear magnetic resonance
- RSV
respiratory syncitial virus
- STD
saturation transfer difference
- scFv
single-chain variable fragment
Disclosures
The authors have no potential conflicts of interest.
References
- 1.Greenbaum JA, Andersen PH, Blythe M, et al. Towards a consensus on datasets and evaluation metrics for developing B-cell epitope prediction tools. J Mol Recognit. 2007;20:75–82. doi: 10.1002/jmr.815. [DOI] [PubMed] [Google Scholar]
- 2.Regenmortel Van MH. What is a B-cell epitope? Methods Mol Biol. 2009;524:3–20. doi: 10.1007/978-1-59745-450-6_1. [DOI] [PubMed] [Google Scholar]
- 3.Corti D, Lanzavecchia A. Broadly neutralizing antiviral antibodies. Annu Rev Immunol. 2013;31:705–42. doi: 10.1146/annurev-immunol-032712-095916. [DOI] [PubMed] [Google Scholar]
- 4.Amanna IJ, Messaoudi I, Slifka MK. Protective immunity following vaccination: how is it defined? Hum Vaccin. 2008;4:316–9. doi: 10.4161/hv.4.4.5751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Plotkin SA. Correlates of protection induced by vaccination. Clin Vaccine Immunol. 2010;17:1055–65. doi: 10.1128/CVI.00131-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Burton DR, Poignard P, Stanfield RL, Wilson IA. Broadly neutralizing antibodies present new prospects to counter highly antigenically diverse viruses. Science. 2012;337:183–6. doi: 10.1126/science.1225416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nabel GJ. Designing tomorrow's vaccines. N Engl J Med. 2013;368:551–60. doi: 10.1056/NEJMra1204186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Desjarlais JR, Lazar GA. Modulation of antibody effector function. Exp Cell Res. 2011;317:1278–85. doi: 10.1016/j.yexcr.2011.03.018. [DOI] [PubMed] [Google Scholar]
- 9.Tarlinton D, Good-Jacobson K. Diversity among memory B cells: origin, consequences, and utility. Science. 2013;341:1205–11. doi: 10.1126/science.1241146. [DOI] [PubMed] [Google Scholar]
- 10.Jolles S, Sewell WA, Misbah SA. Clinical uses of intravenous immunoglobulin. Clin Exp Immunol. 2005;142:1–11. doi: 10.1111/j.1365-2249.2005.02834.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cooke MW. Are current UK tetanus prophylaxis procedures for wound management optimal? Emerg Med J. 2009;26:845–8. doi: 10.1136/emj.2008.070268. [DOI] [PubMed] [Google Scholar]
- 12.Both L, Banyard AC, Dolleweerd van C, Horton DL, Ma JK, Fooks AR. Passive immunity in the prevention of rabies. Lancet Infect Dis. 2012;12:397–407. doi: 10.1016/S1473-3099(11)70340-1. [DOI] [PubMed] [Google Scholar]
- 13.Group TI-RS. Palivizumab, a humanized respiratory syncytial virus monoclonal antibody, reduces hospitalization from respiratory syncytial virus infection in high-risk infants. The IMpact-RSV Study Group. Pediatrics. 1998;102:531–7. [PubMed] [Google Scholar]
- 14.Fox JL. Anthrax drug first antibacterial mAb to win approval. Nat Biotechnol. 2013;31:8. doi: 10.1038/nbt0113-8. [DOI] [PubMed] [Google Scholar]
- 15.Haynes BF, Fleming J, St Clair EW, et al. Cardiolipin polyspecific autoreactivity in two broadly neutralizing HIV-1 antibodies. Science. 2005;308:1906–8. doi: 10.1126/science.1111781. [DOI] [PubMed] [Google Scholar]
- 16.Kapikian AZ, Mitchell RH, Chanock RM, Shvedoff RA, Stewart CE. An epidemiologic study of altered clinical reactivity to respiratory syncytial (RS) virus infection in children previously vaccinated with an inactivated RS virus vaccine. Am J Epidemiol. 1969;89:405–21. doi: 10.1093/oxfordjournals.aje.a120954. [DOI] [PubMed] [Google Scholar]
- 17.Kim HW, Canchola JG, Brandt CD, Pyles G, Chanock RM, Jensen K, Parrott RH. Respiratory syncytial virus disease in infants despite prior administration of antigenic inactivated vaccine. Am J Epidemiol. 1969;89:422–34. doi: 10.1093/oxfordjournals.aje.a120955. [DOI] [PubMed] [Google Scholar]
- 18.Halstead SB. Neutralization and antibody-dependent enhancement of dengue viruses. Adv Virus Res. 2003;60:421–67. doi: 10.1016/s0065-3527(03)60011-4. [DOI] [PubMed] [Google Scholar]
- 19.Mugyenyi CK, Elliott SR, McCallum FJ, Anders RF, Marsh K, Beeson JG. Antibodies to polymorphic invasion-inhibitory and non-inhibitory epitopes of Plasmodium falciparum apical membrane antigen 1 in human malaria. PLoS ONE. 2013;8:e68304. doi: 10.1371/journal.pone.0068304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lu Z, Perkins HM, Sharon J. Antibodies to both terminal and internal B-cell epitopes of Francisella tularensis O-polysaccharide produced by patients with tularemia. Clin Vaccine Immunol. doi: 10.1128/CVI.00626-13. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Huang J, Ofek G, Laub L, et al. Broad and potent neutralization of HIV-1 by a gp41-specific human antibody. Nature. 2012;491:406–12. doi: 10.1038/nature11544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Throsby M, van den Brink E, Jongeneelen M, et al. Heterosubtypic neutralizing monoclonal antibodies cross-protective against H5N1 and H1N1 recovered from human IgM+ memory B cells. PLoS ONE. 2008;3:e3942. doi: 10.1371/journal.pone.0003942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Xu R, Krause JC, McBride R, Paulson JC, Crowe JE, Jr, Wilson IA. A recurring motif for antibody recognition of the receptor-binding site of influenza hemagglutinin. Nat Struct Mol Biol. 2013;20:363–70. doi: 10.1038/nsmb.2500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fleishman SJ, Whitehead TA, Ekiert DC, Dreyfus C, Corn JE, Strauch EM, Wilson IA, Baker D. Computational design of proteins targeting the conserved stem region of influenza hemagglutinin. Science. 2011;332:816–21. doi: 10.1126/science.1202617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lu Z, Roche MI, Hui JH, Unal B, Felgner PL, Gulati S, Madico G, Sharon J. Generation and characterization of hybridoma antibodies for immunotherapy of tularemia. Immunol Lett. 2007;112:92–103. doi: 10.1016/j.imlet.2007.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sukupolvi-Petty S, Brien JD, Austin SK, et al. Functional analysis of antibodies against dengue virus type 4 reveals strain-dependent epitope exposure that impacts neutralization and protection. J Virol. 2013;87:8826–42. doi: 10.1128/JVI.01314-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Giuliani MM, Santini L, Brunelli B, et al. The region comprising amino acids 100 to 255 of Neisseria meningitidis lipoprotein GNA 1870 elicits bactericidal antibodies. Infect Immun. 2005;73:1151–60. doi: 10.1128/IAI.73.2.1151-1160.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Han Y, Kozel TR, Zhang MX, MacGill RS, Carroll MC, Cutler JE. Complement is essential for protection by an IgM and an IgG3 monoclonal antibody against experimental, hematogenously disseminated candidiasis. J Immunol. 2001;167:1550–7. doi: 10.4049/jimmunol.167.3.1550. [DOI] [PubMed] [Google Scholar]
- 29.Lu Z, Rynkiewicz MJ, Yang CY, et al. The binding sites of monoclonal antibodies to the nonreducing end of Francisella tularensis O-antigen accommodate mainly the terminal saccharide. Immunology. 2013;140:374–89. doi: 10.1111/imm.12150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Malito E, Faleri A, Lo Surdo P, et al. Defining a protective epitope on factor H binding protein, a key meningococcal virulence factor and vaccine antigen. Proc Natl Acad Sci U S A. 2013;110:3304–9. doi: 10.1073/pnas.1222845110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Morens DM, Halstead SB. Measurement of antibody-dependent infection enhancement of four dengue virus serotypes by monoclonal and polyclonal antibodies. J Gen Virol. 1990;71(Pt 12):2909–14. doi: 10.1099/0022-1317-71-12-2909. [DOI] [PubMed] [Google Scholar]
- 32.Roche MI, Lu Z, Hui JH, Sharon J. Characterization of monoclonal antibodies to terminal and internal O-antigen epitopes of Francisella tularensis lipopolysaccharide. Hybridoma (Larchmt) 2011;30:19–28. doi: 10.1089/hyb.2010.0083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Foletti D, Strop P, Shaughnessy L, et al. Mechanism of action and in vivo efficacy of a human-derived antibody against Staphylococcus aureus α-hemolysin. J Mol Biol. 2013;425:1641–54. doi: 10.1016/j.jmb.2013.02.008. [DOI] [PubMed] [Google Scholar]
- 34.Maruyama T, Rodriguez LL, Jahrling PB, et al. Ebola virus can be effectively neutralized by antibody produced in natural human infection. J Virol. 1999;73:6024–30. doi: 10.1128/jvi.73.7.6024-6030.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Garcia-Rodriguez C, Geren IN, Lou J, et al. Neutralizing human monoclonal antibodies binding multiple serotypes of botulinum neurotoxin. Protein Eng Des Sel. 2011;24:321–31. doi: 10.1093/protein/gzq111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kwakkenbos MJ, Diehl SA, Yasuda E, et al. Generation of stable monoclonal antibody-producing B cell receptor-positive human memory B cells by genetic programming. Nat Med. 2010;16:123–8. doi: 10.1038/nm.2071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Walker LM, Huber M, Doores KJ, et al. Broad neutralization coverage of HIV by multiple highly potent antibodies. Nature. 2011;477:466–70. doi: 10.1038/nature10373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Walker LM, Phogat SK, Chan-Hui PY, et al. Broad and potent neutralizing antibodies from an African donor reveal a new HIV-1 vaccine target. Science. 2009;326:285–9. doi: 10.1126/science.1178746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wu X, Yang ZY, Li Y, et al. Rational design of envelope identifies broadly neutralizing human monoclonal antibodies to HIV-1. Science. 2010;329:856–61. doi: 10.1126/science.1187659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kong R, Li H, Georgiev I, et al. Epitope mapping of broadly neutralizing HIV-2 human monoclonal antibodies. J Virol. 2012;86:12115–28. doi: 10.1128/JVI.01632-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lantto J, Haahr Hansen M, Rasmussen SK, et al. Capturing the natural diversity of the human antibody response against vaccinia virus. J Virol. 2011;85:1820–33. doi: 10.1128/JVI.02127-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Whittle JR, Zhang R, Khurana S, et al. Broadly neutralizing human antibody that recognizes the receptor-binding pocket of influenza virus hemagglutinin. Proc Natl Acad Sci U S A. 2011;108:14216–21. doi: 10.1073/pnas.1111497108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lewis GK, Fouts TR, Ibrahim S, et al. Identification and characterization of an immunogenic hybrid epitope formed by both HIV gp120 and human CD4 proteins. J Virol. 2011;85:13097–104. doi: 10.1128/JVI.05072-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Thornburg NJ, Nannemann DP, Blum DL, et al. Human antibodies that neutralize respiratory droplet transmissible H5N1 influenza viruses. J Clin Invest. 2013;123:4405–9. doi: 10.1172/JCI69377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cole KS, Alvarez M, Elliott DH, et al. Characterization of neutralization epitopes of simian immunodeficiency virus (SIV) recognized by rhesus monoclonal antibodies derived from monkeys infected with an attenuated SIV strain. Virology. 2001;290:59–73. doi: 10.1006/viro.2001.1144. [DOI] [PubMed] [Google Scholar]
- 46.Costin JM, Zaitseva E, Kahle KM, et al. Mechanistic study of broadly neutralizing human monoclonal antibodies against dengue virus that target the fusion loop. J Virol. 2013;87:52–66. doi: 10.1128/JVI.02273-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cevallos AM, Bhat N, Verdon R, et al. Mediation of Cryptosporidium parvum infection in vitro by mucin-like glycoproteins defined by a neutralizing monoclonal antibody. Infect Immun. 2000;68:5167–75. doi: 10.1128/iai.68.9.5167-5175.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pancera M, Shahzad-Ul-Hussan S, Doria-Rose NA, et al. Structural basis for diverse N-glycan recognition by HIV-1-neutralizing V1-V2-directed antibody PG16. Nat Struct Mol Biol. 2013;20:804–13. doi: 10.1038/nsmb.2600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Savitt AG, Mena-Taboada P, Monsalve G, Benach JL. Francisella tularensis infection-derived monoclonal antibodies provide detection, protection, and therapy. Clin Vaccine Immunol. 2009;16:414–22. doi: 10.1128/CVI.00362-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hsu CA, Lin WR, Li JC, Liu YL, Tseng YT, Chang CM, Lee YS, Yang CY. Immunoproteomic identification of the hypothetical protein NMB1468 as a novel lipoprotein ubiquitous in Neisseria meningitidis with vaccine potential. Proteomics. 2008;8:2115–25. doi: 10.1002/pmic.200700574. [DOI] [PubMed] [Google Scholar]
- 51.Wu X, Zhou T, Zhu J, et al. Focused evolution of HIV-1 neutralizing antibodies revealed by structures and deep sequencing. Science. 2011;333:1593–602. doi: 10.1126/science.1207532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Pejchal R, Doores KJ, Walker LM, et al. A potent and broad neutralizing antibody recognizes and penetrates the HIV glycan shield. Science. 2011;334:1097–103. doi: 10.1126/science.1213256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Han Y, Riesselman MH, Cutler JE. Protection against candidiasis by an immunoglobulin G3 (IgG3) monoclonal antibody specific for the same mannotriose as an IgM protective antibody. Infect Immun. 2000;68:1649–54. doi: 10.1128/iai.68.3.1649-1654.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lu Z, Madico G, Roche MI, et al. Protective B-cell epitopes of Francisella tularensis O-polysaccharide in a mouse model of respiratory tularaemia. Immunology. 2012;136:352–60. doi: 10.1111/j.1365-2567.2012.03589.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tharakaraman K, Robinson LN, Hatas A, et al. Redesign of a cross-reactive antibody to dengue virus with broad-spectrum activity and increased in vivo potency. Proc Natl Acad Sci U S A. 2013;110:E1555–64. doi: 10.1073/pnas.1303645110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Parren PW, Geisbert TW, Maruyama T, Jahrling PB, Burton DR. Pre-and postexposure prophylaxis of Ebola virus infection in an animal model by passive transfer of a neutralizing human antibody. J Virol. 2002;76:6408–12. doi: 10.1128/JVI.76.12.6408-6412.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hessell AJ, Rakasz EG, Poignard P, et al. Broadly neutralizing human anti-HIV antibody 2G12 is effective in protection against mucosal SHIV challenge even at low serum neutralizing titers. PLoS Pathog. 2009;5:e1000433. doi: 10.1371/journal.ppat.1000433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Oswald WB, Geisbert TW, Davis KJ, Geisbert JB, Sullivan NJ, Jahrling PB, Parren PW, Burton DR. Neutralizing antibody fails to impact the course of Ebola virus infection in monkeys. PLoS Pathog. 2007;3:e9. doi: 10.1371/journal.ppat.0030009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Balazs AB, Chen J, Hong CM, Rao DS, Yang L, Baltimore D. Antibody-based protection against HIV infection by vectored immunoprophylaxis. Nature. 2012;481:81–4. doi: 10.1038/nature10660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Coley AM, Parisi K, Masciantonio R, et al. The most polymorphic residue on Plasmodium falciparum apical membrane antigen 1 determines binding of an invasion-inhibitory antibody. Infect Immun. 2006;74:2628–36. doi: 10.1128/IAI.74.5.2628-2636.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lazarou M, Guevara Patino JA, Jennings RM, et al. Inhibition of erythrocyte invasion and Plasmodium falciparum merozoite surface protein 1 processing by human immunoglobulin G1 (IgG1) and IgG3 antibodies. Infect Immun. 2009;77:5659–67. doi: 10.1128/IAI.00167-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Modis Y, Ogata S, Clements D, Harrison SC. Structure of the dengue virus envelope protein after membrane fusion. Nature. 2004;427:313–9. doi: 10.1038/nature02165. [DOI] [PubMed] [Google Scholar]
- 63.Thullier P, Lafaye P, Megret F, Deubel V, Jouan A, Mazie JC. A recombinant Fab neutralizes dengue virus in vitro. J Biotechnol. 1999;69:183–90. doi: 10.1016/S0168-1656(99)00037-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Pelat T, Hust M, Laffly E, et al. High-affinity, human antibody-like antibody fragment (single-chain variable fragment) neutralizing the lethal factor (LF) of Bacillus anthracis by inhibiting protective antigen–LF complex formation. Antimicrob Agents Chemother. 2007;51:2758–64. doi: 10.1128/AAC.01528-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ramaraj T, Angel T, Dratz EA, Jesaitis AJ, Mumey B. Antigen–antibody interface properties: composition, residue interactions, and features of 53 non-redundant structures. Biochim Biophys Acta. 2012;1824:520–32. doi: 10.1016/j.bbapap.2011.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Stave JW, Lindpaintner K. Antibody and antigen contact residues define epitope and paratope size and structure. J Immunol. 2013;191:1428–35. doi: 10.4049/jimmunol.1203198. [DOI] [PubMed] [Google Scholar]
- 67.Kim Y, Ponomarenko J, Zhu Z, et al. Immune epitope database analysis resource. Nucleic Acids Res. 2012;40:W525–30. doi: 10.1093/nar/gks438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Levy R, Forsyth CM, LaPorte SL, Geren IN, Smith LA, Marks JD. Fine and domain-level epitope mapping of botulinum neurotoxin type A neutralizing antibodies by yeast surface display. J Mol Biol. 2007;365:196–210. doi: 10.1016/j.jmb.2006.09.084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Mazzoni MR, Porchia F, Hamm HE. Proteolytic fragmentation for epitope mapping. Methods Mol Biol. 2009;524:77–86. doi: 10.1007/978-1-59745-450-6_6. [DOI] [PubMed] [Google Scholar]
- 70.Coley AM, Gupta A, Murphy VJ, Bai T, Kim H, Foley M, Anders RF, Batchelor AH, et al. Structure of the malaria antigen AMA1 in complex with a growth-inhibitory antibody. PLoS Pathog. 2007;3:1308–19. doi: 10.1371/journal.ppat.0030138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Ekiert DC, Bhabha G, Elsliger MA, Friesen RH, Jongeneelen M, Throsby M, Goudsmit J, Wilson IA. Antibody recognition of a highly conserved influenza virus epitope. Science. 2009;324:246–51. doi: 10.1126/science.1171491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Lee JE, Fusco ML, Hessell AJ, Oswald WB, Burton DR, Saphire EO. Structure of the Ebola virus glycoprotein bound to an antibody from a human survivor. Nature. 2008;454:177–82. doi: 10.1038/nature07082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.McLellan JS, Chen M, Leung S, et al. Structure of RSV fusion glycoprotein trimer bound to a prefusion-specific neutralizing antibody. Science. 2013;340:1113–7. doi: 10.1126/science.1234914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Zhou T, Georgiev I, Wu X, et al. Structural basis for broad and potent neutralization of HIV-1 by antibody VRC01. Science. 2010;329:811–7. doi: 10.1126/science.1192819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Matho MH, Maybeno M, Benhnia MR, et al. Structural and biochemical characterization of the vaccinia virus envelope protein D8 and its recognition by the antibody LA5. J Virol. 2012;86:8050–8. doi: 10.1128/JVI.00836-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Haste AndersenP, Nielsen M, Lund O. Prediction of residues in discontinuous B-cell epitopes using protein 3D structures. Protein Sci. 2006;15:2558–67. doi: 10.1110/ps.062405906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Madej T, Addess KJ, Fong JH, et al. MMDB: 3D structures and macromolecular interactions. Nucleic Acids Res. 2012;40:D461–4. doi: 10.1093/nar/gkr1162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Woods VLJr, Hamuro Y. High resolution, high-throughput amide deuterium exchange-mass spectrometry (DXMS) determination of protein binding site structure and dynamics: utility in pharmaceutical design. J Cell Biochem Suppl. 2001;37:89–98. doi: 10.1002/jcb.10069. [DOI] [PubMed] [Google Scholar]
- 79.Zhang Z, Smith DL. Determination of amide hydrogen exchange by mass spectrometry: a new tool for protein structure elucidation. Protein Sci. 1993;2:522–31. doi: 10.1002/pro.5560020404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Johnson MA, Cartmell J, Weisser NE, Woods RJ, Bundle DR. Molecular recognition of Candida albicans (1→2)-β-mannan oligosaccharides by a protective monoclonal antibody reveals the immunodominance of internal saccharide residues. J Biol Chem. 2012;287:18078–90. doi: 10.1074/jbc.M112.355578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Morgan WD, Frenkiel TA, Lock MJ, Grainger M, Holder AA. Precise epitope mapping of malaria parasite inhibitory antibodies by TROSY NMR cross-saturation. Biochemistry. 2005;44:518–23. doi: 10.1021/bi0482957. [DOI] [PubMed] [Google Scholar]
- 82.Scarselli M, Cantini F, Santini L, et al. Epitope mapping of a bactericidal monoclonal antibody against the factor H binding protein of Neisseria meningitidis. J Mol Biol. 2009;386:97–108. doi: 10.1016/j.jmb.2008.12.005. [DOI] [PubMed] [Google Scholar]
- 83.Simonelli L, Pedotti M, Beltramello M, Livoti E, Calzolai L, Sallusto F, Lanzavecchia A, Varani L. Rational engineering of a human anti-dengue antibody through experimentally validated computational docking. PLoS ONE. 2013;8:e55561. doi: 10.1371/journal.pone.0055561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Rosen O, Anglister J. Epitope mapping of antibody–antigen complexes by nuclear magnetic resonance spectroscopy. Methods Mol Biol. 2009;524:37–57. doi: 10.1007/978-1-59745-450-6_3. [DOI] [PubMed] [Google Scholar]
- 85.Nitz M, Ling CC, Otter A, Cutler JE, Bundle DR. The unique solution structure and immunochemistry of the Candida albicans β-1,2-mannopyranan cell wall antigens. J Biol Chem. 2002;277:3440–6. doi: 10.1074/jbc.M109274200. [DOI] [PubMed] [Google Scholar]
- 86.Heimburg-Molinaro J, Song X, Smith DF, Cummings RD. Preparation and analysis of glycan microarrays. Curr Protoc Protein Sci. 2011;64:12.10.1–29. doi: 10.1002/0471140864.ps1210s64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Tang H, Robinson JE, Gnanakaran S, et al. Epitopes immediately below the base of the V3 loop of gp120 as targets for the initial autologous neutralizing antibody response in two HIV-1 subtype B-infected individuals. J Virol. 2011;85:9286–99. doi: 10.1128/JVI.02286-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Thullier P, Avril A, Mathieu J, Behrens CK, Pellequer JL, Pelat T. Mapping the epitopes of a neutralizing antibody fragment directed against the lethal factor of Bacillus anthracis and cross-reacting with the homologous edema factor. PLoS ONE. 2013;8:e65855. doi: 10.1371/journal.pone.0065855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Rynkiewicz MJ, Lu Z, Hui JH, Sharon J, Seaton BA. Structural analysis of a protective epitope of the Francisella tularensis O-polysaccharide. Biochemistry. 2012;51:5684–94. doi: 10.1021/bi201711m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Frosi G, Biolchi A, Sapio ML, et al. Bactericidal antibody against a representative epidemiological meningococcal serogroup B panel confirms that MATS underestimates 4CMenB vaccine strain coverage. Vaccine. 2013;31:4968–74. doi: 10.1016/j.vaccine.2013.08.006. [DOI] [PubMed] [Google Scholar]
- 91.Bommakanti G, Citron MP, Hepler RW, et al. Design of an HA2-based Escherichia coli expressed influenza immunogen that protects mice from pathogenic challenge. Proc Natl Acad Sci U S A. 2010;107:13701–6. doi: 10.1073/pnas.1007465107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Steel J, Lowen AC, Wang TT, Yondola M, Gao Q, Haye K, Garcia-Sastre A, Palese P. Influenza virus vaccine based on the conserved hemagglutinin stalk domain. MBio. 2010;1:e00018–10. doi: 10.1128/mBio.00018-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Klein F, Diskin R, Scheid JF, et al. Somatic mutations of the immunoglobulin framework are generally required for broad and potent HIV-1 neutralization. Cell. 2013;153:126–38. doi: 10.1016/j.cell.2013.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Bonsignori M, Alam SM, Liao HX, Verkoczy L, Tomaras GD, Haynes BF, Moody MA. HIV-1 antibodies from infection and vaccination: insights for guiding vaccine design. Trends Microbiol. 2012;20:532–9. doi: 10.1016/j.tim.2012.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Klein F, Mouquet H, Dosenovic P, Scheid JF, Scharf L, Nussenzweig MC. Antibodies in HIV-1 vaccine development and therapy. Science. 2013;341:1199–204. doi: 10.1126/science.1241144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Pal P, Dowd KA, Brien JD, et al. Development of a highly protective combination monoclonal antibody therapy against Chikungunya virus. PLoS Pathog. 2013;9:e1003312. doi: 10.1371/journal.ppat.1003312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Schneider MC, Exley RM, Chan H, Feavers I, Kang YH, Sim RB, Tang CM. Functional significance of factor H binding to Neisseria meningitidis. J Immunol. 2006;176:7566–75. doi: 10.4049/jimmunol.176.12.7566. [DOI] [PubMed] [Google Scholar]
- 98.Madico G, Welsch JA, Lewis LA, et al. The meningococcal vaccine candidate GNA1870 binds the complement regulatory protein factor H and enhances serum resistance. J Immunol. 2006;177:501–10. doi: 10.4049/jimmunol.177.1.501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Cantini F, Savino S, Scarselli M, et al. Solution structure of the immunodominant domain of protective antigen GNA1870 of Neisseria meningitidis. J Biol Chem. 2006;281:7220–7. doi: 10.1074/jbc.M508595200. [DOI] [PubMed] [Google Scholar]
- 100.Inoshima I, Inoshima N, Wilke GA, Powers ME, Frank KM, Wang Y, Bubeck Wardenburg J. A Staphylococcus aureus pore-forming toxin subverts the activity of ADAM10 to cause lethal infection in mice. Nat Med. 2011;17:1310–4. doi: 10.1038/nm.2451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Bhakdi S, Tranum-Jensen J. α-toxin of Staphylococcus aureus. Microbiol Rev. 1991;55:733–51. doi: 10.1128/mr.55.4.733-751.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Song L, Hobaugh MR, Shustak C, Cheley S, Bayley H, Gouaux JE. Structure of staphylococcal α-hemolysin, a heptameric transmembrane pore. Science. 1996;274:1859–66. doi: 10.1126/science.274.5294.1859. [DOI] [PubMed] [Google Scholar]
- 103.Walker B, Bayley H. Key residues for membrane binding, oligomerization, and pore forming activity of staphylococcal α-hemolysin identified by cysteine scanning mutagenesis and targeted chemical modification. J Biol Chem. 1995;270:23065–71. doi: 10.1074/jbc.270.39.23065. [DOI] [PubMed] [Google Scholar]
- 104.Vinogradov EV, Shashkov AS, Knirel YA, Kochetkov NK, Tochtamysheva NV, Averin SF, Goncharova OV, Khlebnikov VS. Structure of the O-antigen of Francisella tularensis strain 15. Carbohydr Res. 1991;214:289–97. doi: 10.1016/0008-6215(91)80036-m. [DOI] [PubMed] [Google Scholar]
- 105.Wang Q, Shi X, Leymarie N, Madico G, Sharon J, Costello CE, Zaia J. A typical preparation of Francisella tularensis O-antigen yields a mixture of three types of saccharides. Biochemistry. 2011;50:10941–50. doi: 10.1021/bi201450v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Apicella MA, Post DM, Fowler AC, et al. Identification, characterization and immunogenicity of an O-antigen capsular polysaccharide of Francisella tularensis. PLoS ONE. 2010;5:e11060. doi: 10.1371/journal.pone.0011060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Okan NA, Kasper DL. The atypical lipopolysaccharide of Francisella. Carbohydr Res. 2013;378:79–83. doi: 10.1016/j.carres.2013.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Gunn JS, Ernst RK. The structure and function of Francisella lipopolysaccharide. Ann N Y Acad Sci. 2007;1105:202–18. doi: 10.1196/annals.1409.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Nelson RD, Shibata N, Podzorski RP, Herron MJ. Candida mannan: chemistry, suppression of cell-mediated immunity, and possible mechanisms of action. Clin Microbiol Rev. 1991;4:1–19. doi: 10.1128/cmr.4.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Cummings RD, Doering TL. Fungi. In: Varki A, Cummings RD, Esko JD, Freeze HH, Stanley P, Bertozzi CR, Hart GW, Etzler ME, editors. Essentials of Glycobiology. 2nd edn. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 2009. pp. 309–21. [PubMed] [Google Scholar]
- 111.Riglar DT, Richard D, Wilson DW, et al. Super-resolution dissection of coordinated events during malaria parasite invasion of the human erythrocyte. Cell Host Microbe. 2011;9:9–20. doi: 10.1016/j.chom.2010.12.003. [DOI] [PubMed] [Google Scholar]
- 112.Bai T, Becker M, Gupta A, Strike P, Murphy VJ, Anders RF, Batchelor AH. Structure of AMA1 from Plasmodium falciparum reveals a clustering of polymorphisms that surround a conserved hydrophobic pocket. Proc Natl Acad Sci U S A. 2005;102:12736–41. doi: 10.1073/pnas.0501808102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Pizarro JC, Vulliez-Le Normand B, Chesne-Seck ML, et al. Crystal structure of the malaria vaccine candidate apical membrane antigen 1. Science. 2005;308:408–11. doi: 10.1126/science.1107449. [DOI] [PubMed] [Google Scholar]
- 114.Coley AM, Campanale NV, Casey JL, Hodder AN, Crewther PE, Anders RF, Tilley LM, Foley M. Rapid and precise epitope mapping of monoclonal antibodies against Plasmodium falciparum AMA1 by combined phage display of fragments and random peptides. Protein Eng. 2001;14:691–8. doi: 10.1093/protein/14.9.691. [DOI] [PubMed] [Google Scholar]
- 115.Votiakov VI, Mishaeva NP. The validation of the use of acarid antigens in developing vaccinal-serum preparations for the prevention of tick-borne encephalitis. Zh Mikrobiol Epidemiol Immunobiol. 1990;6:103–9. [PubMed] [Google Scholar]
- 116.Morgan WD, Birdsall B, Frenkiel TA, et al. Solution structure of an EGF module pair from the Plasmodium falciparum merozoite surface protein 1. J Mol Biol. 1999;289:113–22. doi: 10.1006/jmbi.1999.2753. [DOI] [PubMed] [Google Scholar]
- 117.Gamblin SJ, Skehel JJ. Influenza hemagglutinin and neuraminidase membrane glycoproteins. J Biol Chem. 2010;285:28403–9. doi: 10.1074/jbc.R110.129809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Bullough PA, Hughson FM, Skehel JJ, Wiley DC. Structure of influenza haemagglutinin at the pH of membrane fusion. Nature. 1994;371:37–43. doi: 10.1038/371037a0. [DOI] [PubMed] [Google Scholar]
- 119.Walsh EE, Hruska J. Monoclonal antibodies to respiratory syncytial virus proteins: identification of the fusion protein. J Virol. 1983;47:171–7. doi: 10.1128/jvi.47.1.171-177.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Sugrue RJ, Brown C, Brown G, Aitken J, Mc LRHW. Furin cleavage of the respiratory syncytial virus fusion protein is not a requirement for its transport to the surface of virus-infected cells. J Gen Virol. 2001;82:1375–86. doi: 10.1099/0022-1317-82-6-1375. [DOI] [PubMed] [Google Scholar]
- 121.Guirakhoo F, Hunt AR, Lewis JG, Roehrig JT. Selection and partial characterization of dengue 2 virus mutants that induce fusion at elevated pH. Virology. 1993;194:219–23. doi: 10.1006/viro.1993.1252. [DOI] [PubMed] [Google Scholar]
- 122.Modis Y, Ogata S, Clements D, Harrison SC. A ligand-binding pocket in the dengue virus envelope glycoprotein. Proc Natl Acad Sci U S A. 2003;100:6986–91. doi: 10.1073/pnas.0832193100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Crill WD, Roehrig JT. Monoclonal antibodies that bind to domain III of dengue virus E glycoprotein are the most efficient blockers of virus adsorption to Vero cells. J Virol. 2001;75:7769–73. doi: 10.1128/JVI.75.16.7769-7773.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Cockburn JJ, Navarro Sanchez ME, Fretes N, et al. Mechanism of dengue virus broad cross-neutralization by a monoclonal antibody. Structure. 2012;20:303–14. doi: 10.1016/j.str.2012.01.001. [DOI] [PubMed] [Google Scholar]
- 125.Thullier P, Demangel C, Bedouelle H, Megret F, Jouan A, Deubel V, Mazie JC, Lafaye P. Mapping of a dengue virus neutralizing epitope critical for the infectivity of all serotypes: insight into the neutralization mechanism. J Gen Virol. 2001;82:1885–92. doi: 10.1099/0022-1317-82-8-1885. [DOI] [PubMed] [Google Scholar]
- 126.Takada A, Robison C, Goto H, Sanchez A, Murti KG, Whitt MA, Kawaoka Y. A system for functional analysis of Ebola virus glycoprotein. Proc Natl Acad Sci U S A. 1997;94:14764–9. doi: 10.1073/pnas.94.26.14764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Wool-Lewis RJ, Bates P. Characterization of Ebola virus entry by using pseudotyped viruses: identification of receptor-deficient cell lines. J Virol. 1998;72:3155–60. doi: 10.1128/jvi.72.4.3155-3160.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Sanchez A, Yang ZY, Xu L, Nabel GJ, Crews T, Peters CJ. Biochemical analysis of the secreted and virion glycoproteins of Ebola virus. J Virol. 1998;72:6442–7. doi: 10.1128/jvi.72.8.6442-6447.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Volchkov VE, Feldmann H, Volchkova VA, Klenk HD. Processing of the Ebola virus glycoprotein by the proprotein convertase furin. Proc Natl Acad Sci U S A. 1998;95:5762–7. doi: 10.1073/pnas.95.10.5762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Chandran K, Sullivan NJ, Felbor U, Whelan SP, Cunningham JM. Endosomal proteolysis of the Ebola virus glycoprotein is necessary for infection. Science. 2005;308:1643–5. doi: 10.1126/science.1110656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Schornberg K, Matsuyama S, Kabsch K, Delos S, Bouton A, White J. Role of endosomal cathepsins in entry mediated by the Ebola virus glycoprotein. J Virol. 2006;80:4174–8. doi: 10.1128/JVI.80.8.4174-4178.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Pantophlet R, Burton DR. GP120: target for neutralizing HIV-1 antibodies. Annu Rev Immunol. 2006;24:739–69. doi: 10.1146/annurev.immunol.24.021605.090557. [DOI] [PubMed] [Google Scholar]
- 133.Wyatt R, Sodroski J. The HIV-1 envelope glycoproteins: fusogens, antigens, and immunogens. Science. 1998;280:1884–8. doi: 10.1126/science.280.5371.1884. [DOI] [PubMed] [Google Scholar]
- 134.Kwong PD, Wyatt R, Robinson J, Sweet RW, Sodroski J, Hendrickson WA. Structure of an HIV gp120 envelope glycoprotein in complex with the CD4 receptor and a neutralizing human antibody. Nature. 1998;393:648–59. doi: 10.1038/31405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Guex N, Peitsch MC. SWISS-MODEL and the Swiss-PdbViewer: an environment for comparative protein modeling. Electrophoresis. 1997;18:2714–23. doi: 10.1002/elps.1150181505. [DOI] [PubMed] [Google Scholar]
- 136.Krissinel E, Henrick K. Inference of macromolecular assemblies from crystalline state. J Mol Biol. 2007;372:774–97. doi: 10.1016/j.jmb.2007.05.022. [DOI] [PubMed] [Google Scholar]