Abstract
Approximately twelve percent of all human cancers are caused by oncoviruses. Human viral oncogenesis is complex and only a small percentage of the infected individuals develop cancer and often many years to decades after initial infection. This reflects the multistep nature of viral oncogenesis, host genetic variability and the fact that viruses contribute to only a portion of the oncogenic events. In this review, the Hallmarks of Cancer framework of Hanahan & Weinberg (2000 and 2011) is used to dissect the viral, host and environmental co-factors that contribute to the biology of multistep oncogenesis mediated by established human oncoviruses. The viruses discussed include Epstein Barr Virus (EBV), high-risk Human Papillomaviruses (HPV16/18), Hepatitis B and C viruses (HBV, HCV respectively), Human T-cell lymphotropic virus-1 (HTLV-1) and Kaposi’s sarcoma herpesvirus (KSHV).
Keywords: viral carcinogenesis, oncovirus, multi-step carcinogenesis, molecular mechanisms of oncogenesis, EBV, KSHV, HCV, HBV, HTLV-1, HPV
Introduction
Approximately 12% of human cancers worldwide are caused by oncovirus infection with more than 80% of cases occurring in the developing world (Bouvard et al., 2009; Boyle et al., 2008; de Martel et al., 2012). Despite their prevalence, public health importance, and suitability for immunoprophylaxis and targeted therapies, understanding and managing virus-induced cancers still faces formidable challenges. This is due to limited animal models of disease, the disparate nature of viral induced cancers, the very distinct types of viruses that cause them, and the complex nature of the virus-host cell interactions leading to cancer development (zur Hausen, 2009).
Human viral oncogenesis has common traits (Bouvard et al., 2009; zur Hausen, 2009). 1) Oncoviruses are necessary but not sufficient for cancer development, so cancer incidence is much lower than virus prevalence in human populations 2) Viral cancers appear in the context of persistent infections and occur many years to decades after acute infection 3) The immune system can play a deleterious or a protective role, with some human virus-associated cancers increasing with immunosuppression and others appearing in the context of chronic inflammation. The Hallmarks of Cancer framework developed by Weinberg & Hanahan allows the dissection of the malignant phenotype into specific cellular capabilities that are acquired during the carcinogenic process (Hanahan and Weinberg, 2000, 2011). Each cancer “hallmark” represents a biological consequence of oncogenic alteration/s that underlies the tumor’s phenotypic characteristics (Figure 1). For instance mutations that constitutively activate the Ras oncogene, a potent activator of the MAPK and PI3K-AKT-mTOR cascades, convey proliferation, survival, angiogenesis and metabolic related hallmarks to a tumor cell. Mutations that inactivate the tumor suppressor p53, a DNA-damage induced activator of growth-inhibition and apoptosis, enables uncontrolled growth and genetic instability hallmarks. The Hallmarks of Cancer framework also helps to explain the multistep nature of human carcinogenesis (Vogelstein and Kinzler, 1993), which outlines the time dependence for the development of a cancer that requires the progressive acquisition of all necessary cellular hallmarks that constitute a malignant phenotype. This results from the accumulation of somatic oncogenic alterations, sometimes referred to as “oncogenic hits”, caused by spontaneous mutations or mutations as a consequence of exposure to environmental carcinogenic factors in the context of the genetic background of the host and the selective pressures imposed by the tissue microenvironment (Hanahan and Weinberg, 2000, 2011). The cancer hallmark model, therefore, is also a powerful tool to organize and understand the process of human virus associated carcinogenesis. Early studies with acutely transforming viruses suggested that viruses are sufficient to cause cancer because they encode powerful oncogenes. Human oncoviruses however appear to be necessary but not sufficient to cause cancer and are rarely fully oncogenic per se. This indicates that within the context of multistep carcinogenesis, viral infection provides only a subset of the required oncogenic hits. Additional co-factors; such as immunosuppression, chronic inflammation or environmental mutagens are generally necessary for malignant transformation (Bouvard et al., 2009; zur Hausen, 2009). The Cancer Hallmarks framework helps to discern the contribution to the oncogenic process of viral genes, of the host response to the infection, and of acquired somatic mutations. In this review, we will analyze oncogenesis mechanisms and identify host and environmental cofactors contributing to full development of malignancy for each of the major human viruses defined in the last IARC report as Group 1 Biological carcinogenic agents for which there is “sufficient evidence of carcinogenicity in humans” (Bouvard et al., 2009). These include hepatitis B virus, (HBV) hepatitis C virus (HCV), Epstein Barr Virus (EBV), high-risk Human Papillomaviruses (HPVs), Human T lymphotropic Virus-1 (HTLV-1), HIV and Kaposi’s sarcoma herpesvirus (KSHV). Since HIV acts as a co-factor of AIDS-defining cancers that are associated with EBV and KSHV, it will be discussed within the context of these other viruses.
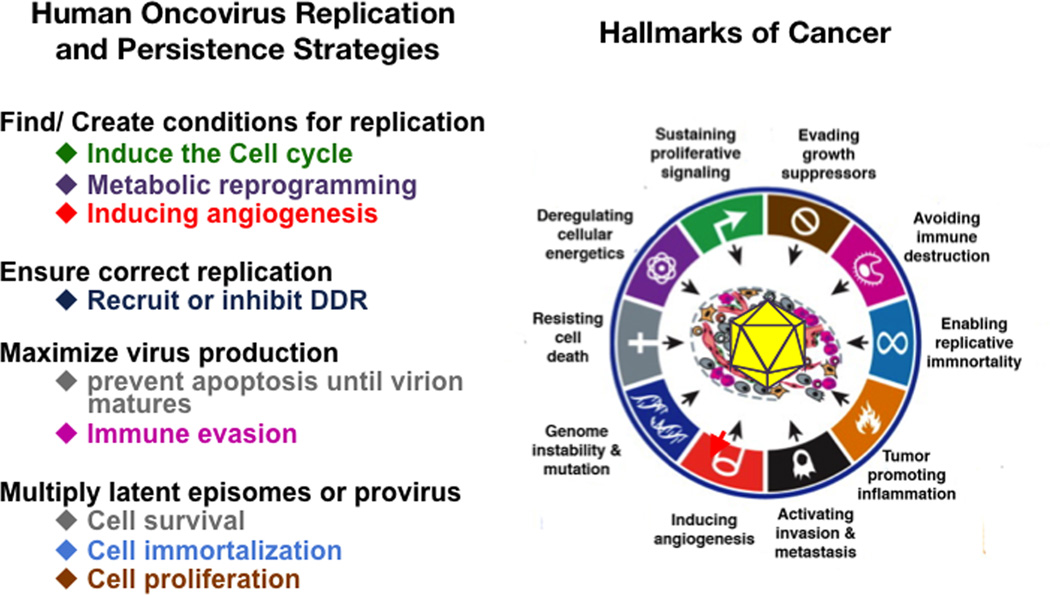
Figure 1. Oncogenic risks of viral replication and persistence strategies.
Many of the molecular mechanisms deployed by human oncoviruses to maximize replication and persistence imply hijacking the host cell’s signaling machinery leading to acquisition of cancer hallmarks. The main strategies for viral replication and persistence are listed (on the left). Below each of them are the cellular responses that the virus induces to undertake each of these strategies. Virally induced cell responses are color coded according to the Hallmarks of Cancer to which they correspond (on the right). The picture on the right is reproduced from the review from Hanahan and Weinberg (2011).
Oncovirus replication and persistence strategies involve activation of cancer-causing pathways
Co-evolution of oncoviruses and their hosts is a fight for survival. Hosts evolved immune defenses against viral infections, while viruses have co-evolved to evade host immune response and other host restrictions. Human oncogenic viruses rely on persistence to disseminate and thus deploy powerful immune evasion programs to establish long-term infections. As part of their replication and immune evasion strategies, human oncoviruses have evolved powerful anti-apoptotic and proliferative programs that can directly induce cancer hallmarks in the infected cell (Figure 1) (Moore and Chang, 2010; zur Hausen, 2009). When these viruses overcome the ability of the host to maintain homeostasis, they trigger cellular changes ultimately leading to cancer. The underlying mechanisms include 1) Signaling mimicry: viruses encode proteins that subvert in a dominant manner, host signaling mechanisms that regulate cell growth and survival (Figure 1). These are generally the same signaling pathways that, when deregulated, provide anti apoptotic and proliferative hallmark capabilities in non-viral cancers (Figure 1, Table 1). 2) Effects on the DNA damage response (DDR): Recognition of viral genomes or replicative intermediates by the host leads to induction of DDR, which many oncoviruses need for their replication. As a consequence, however, host cells acquire genetic instability, which increases their mutation rate, and accelerates acquisition of oncogenic host chromosomal alterations (McFadden and Luftig, 2013) (also see review in this issue by Weitzman and Weitzman) 3) Chronic inflammatory responses to persistent viral infection: inflammation drives reactive oxygen species (ROS) generation that promotes the acquisition of mutations. This is observed in chronic HBV and HCV infections, where virus triggered inflammatory responses lead to hepatitis, fibrosis, cirrhosis and eventually hepatocellular carcinoma. (Arzumanyan et al., 2013).
Table 1. Cancer Hallmarks activation by human viral oncogenes.
The table shows established viral oncogenes, the main cellular pathways they regulate and the cancer hallmarks they can potentially induce. Hallmarks are color coded as in Figure 1. Data from tumor models were given preference. HBV and HCV are potentially able to activate all hallmarks depending on HCC stage as described in the text and Figure 4.
| VIRUS | CANCER | V-ONC | PATHWAYS | CANCER HALLMARK |
|---|---|---|---|---|
| EBV | BL | EBNA | 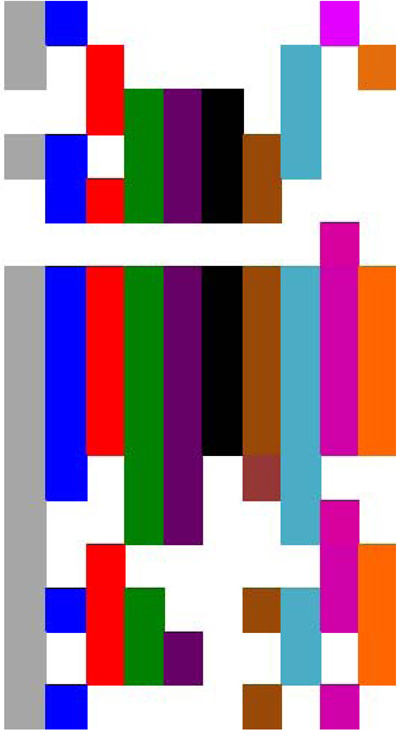 |
|
| NHL, NPC | LMP-1 | NFkB | ||
| LMP-2A | PI3K, ERK1/2 | |||
| HPV | CxCa, HNCC | E6 | p53, mTOR, hTERT | |
| E7 | Rb | |||
| E5 | ||||
| HBV | HCC | HBx | p53. Rb, Wnt, src, DNMTs, ras, PI3K JNK, NF-κB, ERK1/2. TGFβ, HDACs | |
| HCV | HCC | Core, NS3, Ns5A | p53, PARP, hTERT, TGFβ, HDACs, | |
| HTLV-1 | ATLL | Tax | NFkB, CREB, PI3K, DDR | |
| HZB | c-jun, E2F | |||
| KSHV | KS | vFLIP | NFkB | |
| LANA | p53, Rb, HIF, Notcti, Wnt | |||
| vGPCR | PI3K-AKT-mTOR, ERK, p38, JNK, NFkB, | |||
| vIRF-1 | αIFN, p53, ATM, Bim |
Epstein Barr Virus (EBV): posing oncogenic dangers by adopting a B-cell lifestyle
EBV is an oncogenic gamma-1 herpes virus implicated in several lymphoid malignancies including several B-cell, T, NK lymphomas and epithelial carcinomas (Cesarman and Mesri, 2007; Kutok and Wang, 2006). EBV mimics B-cell proliferative and survival signaling allowing it to replicate its genome while remaining latent and immune-silent in the host B-cells, thus establishing lifelong persistence which favors its transmission (Cesarman and Mesri, 2007; Young and Rickinson, 2004). In certain scenarios the B-cell growth-promoting abilities of EBV infection have oncogenic consequences. Different transcription programs are established to maintain a lifelong EBV infection. Since the same transcriptional programs are found in EBV lymphomas and cancers, the patterns of latency determining which EBV genes are expressed, are key for understanding the role of EBV infection in inducing cancer hallmarks (Cesarman and Mesri, 2007; Kutok and Wang, 2006). EBV first infects a naïve B cell where it deploys the growth program or latency III (Lat III) pattern, whereby EBV expresses EBNA 1–6, as well as LMP1, LMP2A and LMP2B. This pattern of expression forces infected cells to become proliferating B cells, allowing replication of EBV episomes. Latency III is; however, very immunogenic, and cells displaying this pattern are eliminated by cytotoxic T lymphocytes (CTLs). This drives positive selection of infected B-cells that are able to switch to a “default program” or latency II (Lat II) pattern, expressing only EBNA1, LMP1 and LMP2A of subdominant immunogenicity. In these cells, the ability of LMP1 and LMP2A to mimic CD40 and IgG receptor signaling drive B cells to differentiate into resting memory B cells (Thorley-Lawson, 2001; Young and Rickinson, 2004). These infected cells in turn can switch to either of two immunologically silent latency programs. A Latency 0 (Lat 0) pattern, where only EBV-encoded RNAs (EBER 1 and 2) are expressed, can be observed in resting memory B cells where no viral proteins need to be expressed (Thorley-Lawson, 2001). EBV-infected dividing memory B cells need to switch to the EBNA1 only program/ Latency I (Lat I) pattern, whereby EBNA1 allows the EBV episome to be segregated and retained in dividing B-cells.
Molecular mechanisms of EBV oncogenesis: Cancer hallmarks activation at every stage
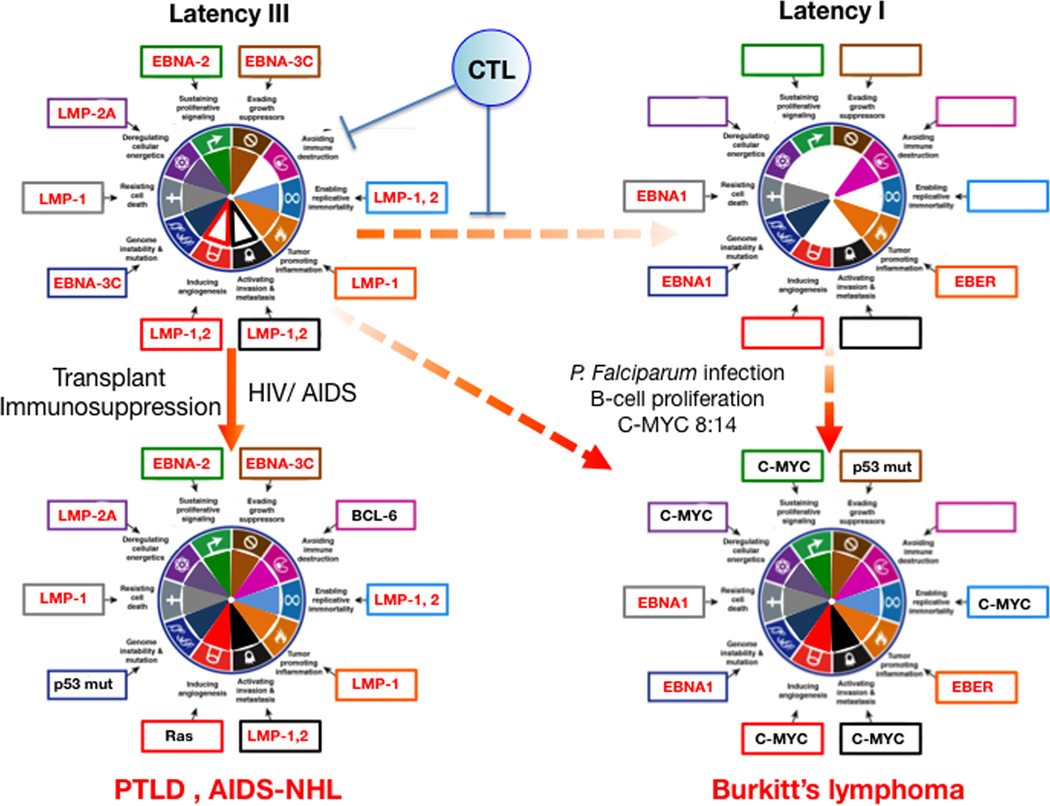
The latency patterns of EBV are associated with specific lymphoma subtypes. EBV latent gene expression can be linked to specific molecular and oncogenic cancer hallmarks of the lymphoid malignancies depicted in Figure 2.
Figure 2. Hallmarks of Cancer analysis of EBV mediated lymphomagenesis.
When EBV infects B-cells it establishes a “growth program” or Latency III pattern, which expresses EBV proteins with sufficient oncogenic potential to cause immortalization in vitro, and B-cell growth and differentiation in vivo. However this program is too immunogenic due to EBNA-2,3 protein expression. The resulting immune pressure from cytotoxic T lymphocytes (CTL) promotes virus first switching to the default program (see text) and then the latency I programs that is not immunogenic but it is also not oncogenic since it only expresses EBNA1 and EBER. In immunosuppressed and AIDS patients Lat III cells can thrive and lead to PTLD or AIDS-NHL where host somatic mutations such as Bcl-6 overexpression, p53 mutation or Ras mutations further contributes to all the hallmarks for lymphomagenesis. In malaria affected regions P. falciparum induced B cell proliferation favors the occurrence of 8:14 translocations that lead to c-myc overexpression. c-myc expression with further host somatic mutations such as p53 inactivation may complement the oncogenicity of Latency I pattern leading to Burkitt’s lymphomagenesis. Viral genes are depicted in red and host genes in black. The figure is not a strict representation of experimental data but rather a compilation of published information analyzed in the context of the Hallmarks of Cancer. Genes chosen for hallmarks activation represent one of the available examples and are based on published data with preference for tumor relevant systems where available. Filled portions within the hallmark pie are used to represent stronger or well-documented hallmark activation. Empty portions represent weaker effects or lack of activation/evidence. See text for explanations and references.
Latency I
Burkitt's lymphoma (BL). EBNA1 function in latency is to leash EBV episomes to the host chromosome thus allowing its retention and segregation during cell division. In addition EBNA1 is essential for lymphoma survival by preventing cell death (Kirchmaier and Sugden, 1997). It may also increase genomic instability by regulating Rag-1 and Rag-2 (Tsimbouri et al., 2002) and increasing ROS. EBERs are noncoding RNAs with abundant dsRNA pairing that trigger Toll-like receptor TLR-3 leading to upregulation of IFN and proinflammatory cytokines (Iwakiri et al., 2009).
Latency II
Hodgkin’s disease (HD) and nasopharyngeal carcinoma (NPC). In addition to the Lat I genes, this pattern includes expression of LMP-1 and LMP-2A, two EBV oncogenes that mimic key survival and proliferative signals in B-cells. LMP1 mimics a constitutively active CD40 receptor (Mosialos et al., 1995). LMP1 constitutively aggregates and recruits TRAFs and TRADD leading to activation of NF-kB, a key transcription factor in viral and non-viral lymphomagenesis (Mosialos et al., 1995). NFkB promotes cell survival by upregulation of the anti-apoptotic molecules A20 and Bcl-2, and a variety of cellular genes related to B-cell proliferation and malignancy. They include the inflammatory related genes IL-6, ICAM-1, LFA-3, CD40, EBI3, and the matrix metalloproteinase-9, which can contribute to invasion and metastasis. LMP-1 may promote genetic instability through telomerase activation via JNK, and inhibition of the DDR leading to micronuclei formation and chromosomal aberrations. LMP-2A is a 12 transmembrane domain protein able to mimic signaling via activated B-cell Ig receptors (Merchant et al., 2000). Its tail contains phosphotyrosines that recruit cytoplasmic tyrosine kinases such as Lyn via the SH2 domain, as well as ITAM motifs homologous to the gamma-chain Ig receptor that can recruit Syk kinase. Mimicking a crosslinked Ig Receptor, LMP-2A signaling promotes mature B-cell differentiation, survival and cell growth in absence of Ig Receptor expression, by activating the PI3K-AKT pathway and pathways mediating cell mobility and invasion (Merchant et al., 2000; Portis and Longnecker, 2004; Young and Rickinson, 2004).
Latency III
Most AIDS-associated non-Hodgkin lymphomas (NHLs) and post-transplant lymphoproliferative disorder (PTLD). In addition to Latency II’s EBNA-1, LMP-1 and LMP-2A, this pattern expresses a full oncogenic component of nuclear proteins EBNA-2, 3A, 3B, 3C, LP and the EBV microRNAs. EBNA-2 is a nuclear phosphoprotein that mimics cleaved Notch and can associates with RBP-Jκ to activate the expression of Notch target genes (Hofelmayr et al., 2001). Deregulated Notch signaling is known to drive non-viral lymphoid malignancies. EBNA-2 is able to deregulate expression of c-myc further increasing cell proliferation by up-regulating cyclin Ds and E, and by down-regulating CDK2 inhibitors such as p21CIP1 and p27KIP1 (Kaiser et al., 1999). EBNA-3C is also able target cell cycle checkpoints by engaging the SCF (skp, Cullin, F-Box) ubiquitin ligase complex that may target the retinoblastoma tumor suppressor for proteosomal degradation (Knight et al., 2005).
Lytic cycle genes
Increasing evidence suggest that expression of lytic cycle genes (Kutok and Wang, 2006), particularly early lytic genes particularly those encoding homologues of Bcl-2 (BALF1), IL-10 (BCRF1) and c-fms receptor (BARF1) may have oncogenic consequences (Thompson and Kurzrock, 2004). In a recent study of 700 EBV-immortalized lymphoblastoid cell lines that were characterized for host and viral transcriptome and epigenomes, viral lytic genes were co-expressed with cellular cancer-associated pathways, strongly suggesting that EBV lytic genes play a role in oncogenesis (Arvey et al., 2012).
EBV encoded miRNA
EBV encodes two clusters of miRNA, one adjacent to the BHRF1 gene and the other with the BART transcript. They are differentially expressed in epithelial and B-cell lineages and can play roles in oncogenesis by targeting host genes regulating pathways implicated in cell growth and apoptosis (Vereide et al., 2013; Zhu et al., 2013).
EBV mediated Lymphomagenesis
The Lat II and Lat III patterns are oncogenic in B and epithelial cells. However these patterns of EBV gene expression elicit an immune response because EBNA proteins, except EBNA1, are immunogenic (Cesarman and Mesri, 2007). In immunocompetent individuals Lat III B-cells are eliminated by the immune system (Thorley-Lawson, 2001; Young and Rickinson, 2004) (Figure 2). Thus, lymphomas with EBV Lat III will only develop in immunodeficient individuals, while EBV-lymphomas and cancers in immunocompetent hosts will display Lat I or II patterns (Cesarman and Mesri, 2007; Kutok and Wang, 2006). Nevertheless, development of all EBV induced cancers requires co-factors. For latency I cancers, co-factors facilitate the acquisition of further oncogenic hits, either by host somatic mutation or by another virus infection. In BL, Lat I (with sporadic LMP-2A expression) is found in the 90% of the cases with a t(8;14) chromosomal translocation that places the c-myc gene under the control of immunoglobulin heavy chain genes enhancer (Cesarman and Mesri, 2007; Kutok and Wang, 2006). The endemicity of BL is coincident with the Malaria belt in Africa and Plasmodium falciparum infection is a co-factor for development of BL (Rochford et al., 2005). Plasmodium infection prompts polyclonal B-cell proliferation, which in EBV infected B-cells, may increase the likelihood of chromosomal translocations that cause aberrant c-myc expression. Such translocations may allow a latency III/II infected B-cell to switch to Lat I without losing the ability to proliferate. The lymphomagenic gene, c-myc can complement Lat I oncogenic deficiencies, since it is drives cell proliferation, metabolic reprogramming and oxidative stress (Figure 2). Primary Effusion Lymphoma (PEL) is another example. In this case the oncogenic complement of EBV Lat I is latent KSHV infection with a variable percentage of lytic replication. KSHV latent genes are potent regulators of cell proliferation and survival that could complement the limited oncogenicity of EBV Lat I, reviewed in (Cesarman and Mesri, 2007) (see also section on KSHV), while lytic replication gene expression may account for some key phenotypic characteristics of PEL such as angiogenicity and permeability promoted by VEGF.
The most important co-factor for development of malignancies displaying Lat III expression patterns is immunosuppression, which is generally found in PTLD and in NHLs associated with HIV/AIDS (Figure 2). In PTLD, transplant related immunosuppression allows immunogenic Lat III infected cells to proliferate and, in time, acquire host somatic oncogenic alterations such as Bcl-6 overexpression and mutations, A20 overexpression and p53 inactivation (Cesarman and Mesri, 2006; Kutok and Wang, 2006). AIDS-NHL is a complex group of cancers that prior to the implementation of anti-retroviral therapy (ART) affected between 4 and 10% of HIV/AIDS patients. The incidence of at least some subsets appears to be decreasing with ART. In addition to immunosuppression and similar host oncogenic alterations to PTLD, other co-factors such as HIV proteins, chronic antigenic stimulation, and cytokine dysregulation may play a role (Casper, 2011; Kutok and Wang, 2006). Finally Latency II pattern is expressed in two EBV associated malignancies: Hodgkin’s Disease (HD) and NPC. HD is associated with EBV infection in approximately 40%-50% of cases depending on the geographical location. Hodgkin's-Reed-Sternberg (HRS) cells are a minority of transformed B-cells that are supporting normal lymphoid proliferation in a paracrine/ cytokine-driven manner. They are derived from germinal B-cells destined to undergo apoptosis that are protected by EBV Lat II genes LMP-1 and LMP-2 (Kuppers, 2009). NPC are epithelial cancers that also expresses Lat II patterns, where LMP-1 and LMP-2A play several well established roles in epithelial carcinogenesis (Raab-Traub, 2002). The endemicity of NPC is strongly suggestive of environmental co-factors. In Southern China one of these co-factors may be the consumption of salted fish.
Human Papillomaviruses (HPVs): small viruses that pack a big punch
Approximately 5% of all human cancers worldwide are caused by HPVs. Papillomaviridae are a large family of epitheliotropic non-enveloped viruses with double stranded circular ~8kB DNA genomes. Of the ~150 HPV genotypes that have been characterized, a small group of “high-risk” alpha HPVs that specifically infect mucosal epithelial cells cause almost all cases of cervical carcinoma, a leading cause of cancer death in women, as well as other anogenital cancers including vulvar, vaginal, penile and anal carcinomas and a fraction of oral carcinomas. In contrast, low-risk alpha HPVs cause benign genital warts. HPV16 and HPV18 are the most abundant high-risk, and HPV6 and HPV11 are the most frequent low-risk HPVs.
HPVs infect basal epithelial cells where viral genomes are persistently maintained as low-copy number episomes. Viral genome amplification, late gene expression and viral progeny synthesis are confined to terminally differentiated layers of the infected epithelium. Since terminally differentiated epithelial cells are growth arrested and do not express DNA replication enzymes needed for viral genome replication, HPVs have evolved to retain differentiated keratinocytes into a DNA-synthesis competent state. The strategies that high-risk alpha HPVs have developed to accomplish this have the potential to trigger cancer development (McLaughlin-Drubin et al., 2012; Moody and Laimins, 2010).
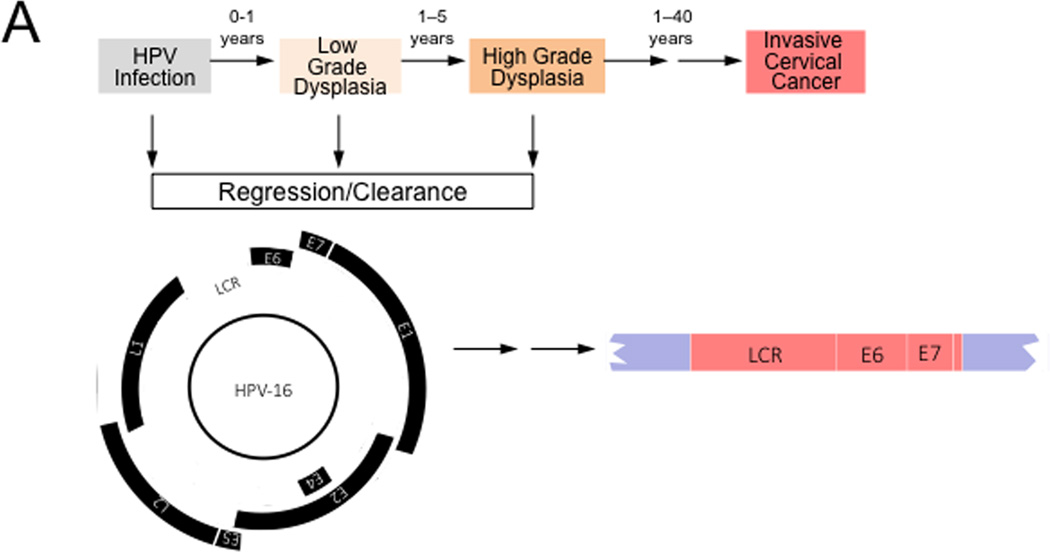
While the oncogenicity of high-risk HPVs represent consequences of their replication strategies, HPV-associated cancers generally represent “non-productive infections”, where some viral proteins are expressed but no infectious viral progeny is produced. Many high-risk HPV-positive cervical carcinomas contain integrated HPV sequences. Integration frequently occurs at common fragile sites of the human genome and in some cases can lead to increased expression of cellular protoncogenes such as MYC (Durst et al., 1987). Integration retains the integrity of the long control region (LCR), as well as the E6 and E7 coding region, while other viral proteins, including the E2 transcriptional repressor that negatively regulates E6/E7 expression, are no longer expressed, causing dysregulated E6/E7 expression (Figure 3A). In cases where HPV episomes are maintained in cancers, E6/E7 expression may be dysregulated due to epigenetic alterations of the viral genome.
Figure 3. Hallmarks of Cancer analysis of HPV associated cervical carcinogenesis.
High-risk HPV infections can give rise to low-grade dysplasia (also referred to as cervical intraepithelial neoplasia 1 (CIN1)), which can progress to high-grade dysplasia (also referred to as CIN2/3). Many of these lesions spontaneously regress, presumably because of immune clearance by the host. These lesions contain episomal HPV genomes and expression of the viral genes is tightly controlled by the interplay of cellular and viral factors. Malignant progression to invasive cervical cancer is often a very slow process and cervical cancers can arise years or decades after the initial infection. Cervical carcinomas frequently contain integrated HPV sequences. Expression of the viral E2 transcriptional repressor is generally lost upon viral genome integration resulting in dysregulated viral gene expression from the viral Long Control Region (LCR). HPV E6 and E7 are consistently expressed even after genome integration and expression of these proteins is necessary for the maintenance of the transformed phenotype. See text for detail.
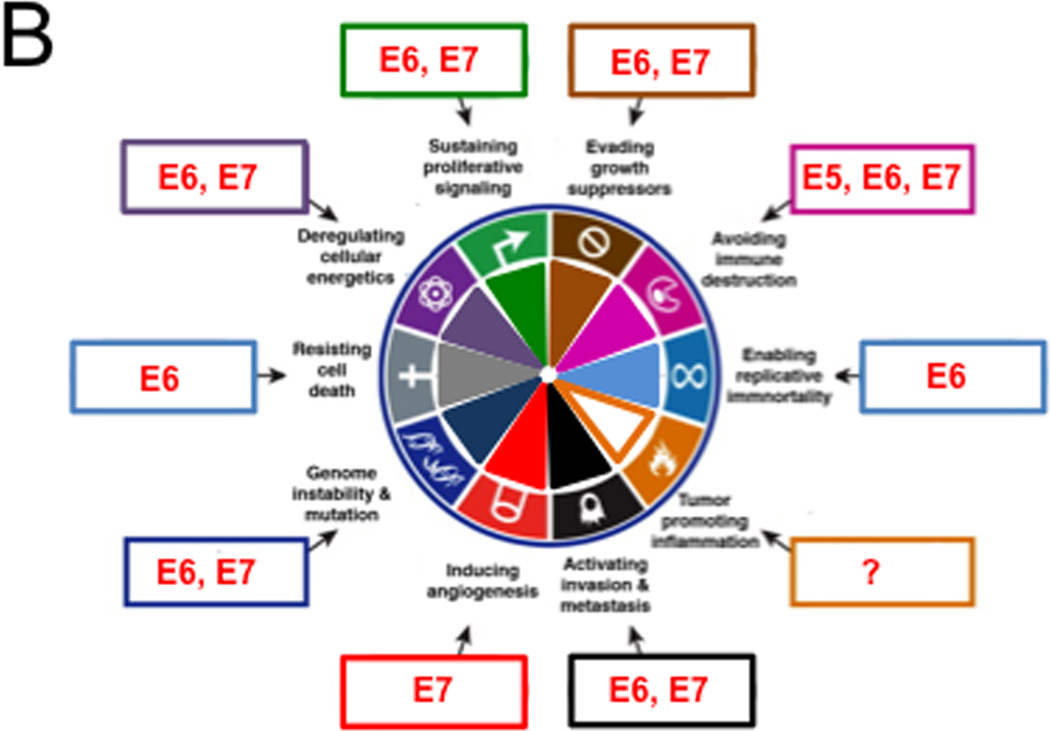
(B) High-risk HPV proteins target almost the entire spectrum of the Cancer Hallmarks. HPV genes that affect each hallmark are indicated within boxes around the wheel. See text for details
Molecular mechanisms of HPV carcinogenesis: HPV oncogenes activate multiple cancer hallmarks
High-risk HPV E6 and E7 genes encode potent oncoproteins and E5 is also oncogenic. HPV positive cervical carcinoma lines are “addicted” to E6/E7 expression and cellular regulatory pathways that are targeted by E6 and E7 are rendered “dormant” but remain intrinsically functional (Goodwin et al., 2000). The following discussion summarizes some of the biological activities of HPV E6, E7 and E5 proteins in the context of the “hallmarks of cancer” framework (Figure 3B). The readers are referred to other review articles for more comprehensive summaries of the biological and biochemical activities of HPV proteins (DiMaio and Petti, 2013; Roman and Munger, 2013; Vande Pol and Klingelhutz, 2013).
Retaining differentiating keratinocytes in a DNA synthesis competent state is key to the HPV life cycle. High-risk alpha HPV E7 proteins target the retinoblastoma tumor suppressor pRB for proteasomal degradation. This causes aberrant, persistent S-phase entry, thereby sustaining proliferative signaling. Degradation of pRB also blunts the oncogene-induced senescence response that is triggered by E7 expression and is signaled by the CDK4/CDK6 inhibitor p16INK4A. Hence pRB degradation also serves to evade growth suppressors. Degradation of pRB and uncontrolled S-phase entry leads to p53 tumor suppressor activation, which triggers apoptosis. To resist cell death, high-risk HPV E6 proteins target p53 for degradation through the associated cellular ubiquitin ligase UBE3A (E6AP). In addition, the HPV E6 protein stimulates telomerase expression and activity, thereby enabling replicative immortality (McLaughlin-Drubin et al., 2012; Moody and Laimins, 2010).
High-risk HPVs can establish long-term persistent infection of epithelial cells, therefore successfully avoiding immune destruction. E6 and E7 can each hinder innate immunity by inhibiting interferon signaling (Roman and Munger, 2013; Vande Pol and Klingelhutz, 2013) and E5 proteins can downregulate MHC class I expression. Inflammation has been linked to HPV associated cancers but it is unknown whether and/or how HPVs may trigger inflammation and/or whether inflammation may be tumor promoting or anti-tumorigenic in the context of an HPV infection. E6 and E7 each contribute to epithelial to mesenchymal transition (EMT), a key step in activating invasion and metastasis (Duffy et al., 2003; Hellner et al., 2009). Moreover, E7 has been reported to induce angiogenesis (Chen et al., 2007) and E6 and E7 also deregulate cellular energetics. E7 causes the Warburg effect, a shift of the standard oxidative phosphorylation based metabolism to aerobic fermentation (Zwerschke et al., 1999) and E6 proteins activate mTORC1 signaling by prolonging receptor protein tyrosine kinase signaling, thereby keeping cells “blissfully ignorant” regarding their energetic states (Spangle and Munger, 2013). This is thought to ensure that cellular enzymes and metabolites necessary for viral genome synthesis are available in differentiated, presumably growth factor depleted epithelial cells.
HPV16 E6/E7 can immortalize primary human epithelial cells. The resulting cell lines are non-tumorigenic but oncogenic clones emerge after prolonged passaging, and they show evidence of chromosomal abnormalities. The high-risk HPV E6 and E7 proteins actively contribute to this process by inducing genome instability and mutation. For E6 this has been linked to p53 inactivation and for E7 induction of genomic instability is a consequence of dysregulation of multiple cellular pathways. Significantly, high-risk HPV E7 proteins cause aberrant centrosome synthesis giving rise to multipolar mitoses, which are a known hallmark of high-risk HPV associated lesions and cancers (Munger et al., 2006). How induction of genomic instability may be linked to the viral life cycle is unknown, although recent studies have shown that cellular DNA repair mechanisms play a central role in papillomavirus genome synthesis (Moody and Laimins, 2009)
HPV-multistep carcinogenesis
Although the replication strategy of high-risk human papillomaviruses requires modulation of cellular pathways that impinge on multiple cancer hallmarks (Table I), expression of the viral transforming proteins is tightly controlled during normal productive HPV infections and infected cells only rarely undergo malignant transformation. Dysregulated HPV E6 and E7 expression as a consequence of integration of viral sequences into the host genome or due to epigenetic alterations of the viral genome, however, puts cells at a higher risk for undergoing oncogenic transformation (Figure 3A). The ability of the E6 and E7 proteins to induce genomic instability, thus accelerating the establishment and expansion of cells with tumor promoting host cell mutations, is at the very core of the oncogenic activities of the high-risk HPVs.
Hepatitis B virus (HBV) and Hepatitis C virus (HCV): direct and inflammation-mediated hepatocellular carcinogenesis
HBV and HCV are major etiological agents of hepatocellular carcinoma (HCC). HCC is the fifth most prevalent tumor type worldwide and the third leading cause of cancer deaths (de Martel et al., 2012). Both viruses establish chronic infections, and when accompanied by inflammation of the liver (hepatitis), hepatocellular destruction triggers regeneration and scarring (fibrosis), which can evolve into cirrhosis and HCC.
Molecular mechanisms of viral hepatocarcinogenesis
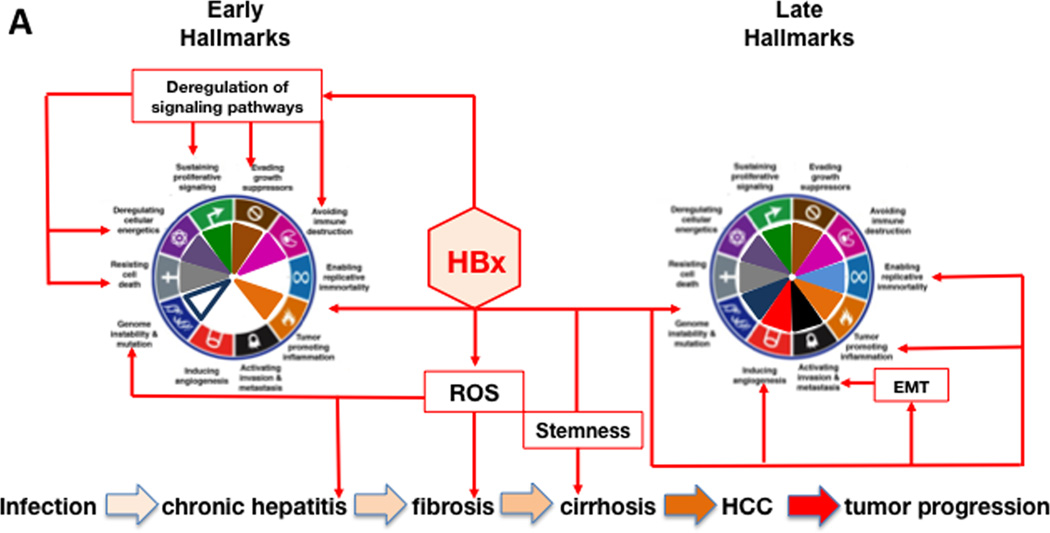
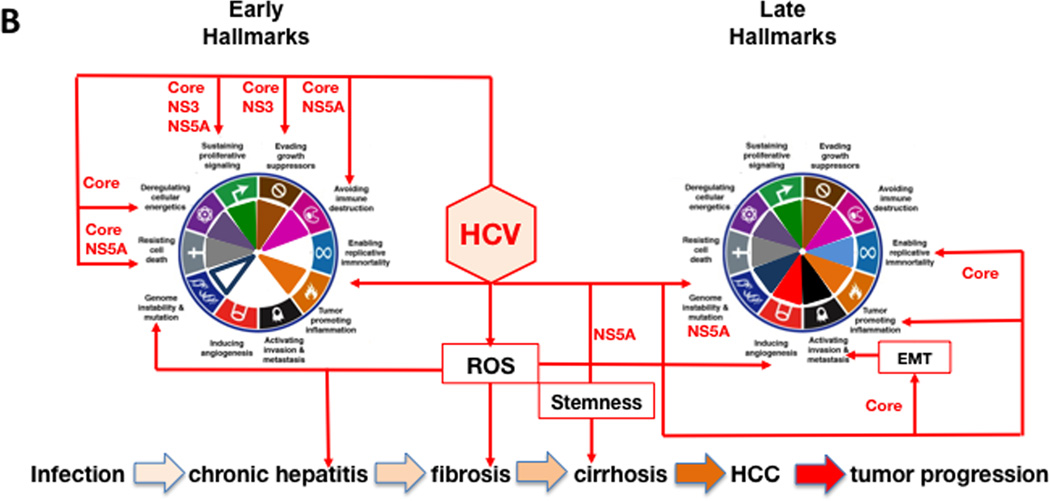
The pathogenesis of HCC is a combination of direct and indirect mechanisms, which results from chronic oxidative damage that promotes the development of mutations. HBV encoded X antigen (HBx) and HCV encoded core, nonstructural protein 5A (NS5A) and NS3 directly promote HCC by altering host gene expression, while immune mediated inflammation contributes indirectly to tumorigenesis. Although HBV and HCV contribute to cancer via different mechanisms (Brechot, 2004), they result in changes recognized as hallmark characteristics of cancer (Figure 4A and 4B).
Figure 4. Hallmarks of Cancer activation during the progression of HBV and HCV infection to HCC.
The diagram shows hallmarks activation during HBV infection, primarily by the viral HBx protein (panel A), and by HCV infection, mainly by the viral core, NS5A and NS3 proteins (panel B). The host mediators that contribute to each of the hallmarks are indicated in black text boxes. Hallmarks achieved early (left wheel) and late (right wheel) over the course of HBV- and HCV-associated HCC pathogenesis is shown for each panel. The major steps in the progression of HBV and HCV infection to HCC are shown at the bottom of each panel. The progression of these steps is indicated by increasingly darker tan-colored arrows. Please see the text for additional details.
Sustained proliferative signaling indirectly promotes virus replication through successive bouts of hepatitis, resulting in liver damage without virus clearance. For HBV, HBx stimulates cell cycle entry by activating selected cyclins and cyclin dependent kinase pathways (Zemel et al., 2011) as well as pathways such as Wnt, ras, PI3K, JAK/STAT, NF-κB and Hedgehog that promote survival and growth (Martin-Vilchez et al., 2011). Nuclear HBx regulates transcription by activating CREB, ATF-2, ATF-3, NFAT, C/EBPβ, and SMAD4 complexes, and facilitates epigenetic changes (Tian et al., 2013) that affect host cell gene expression. HBx mediated changes on miRNAs levels modulate oncogene and tumor suppressor gene expression (Yip et al., 2011). HCV encodes core, NS3 and NS5A that promote liver cell proliferation via the β-catenin pathway (Jeong et al., 2012). HCV core promotes cyclin E and cdk2 expression. Both core and NS3 also activate multiple signal transduction pathways that promote cell growth.
Both HBV and HCV evade growth suppression and avoid immune destruction by blocking apoptosis. Apoptosis is triggered by virus generated oxidative stress (intrinsic apoptosis), and from immune mediated apoptosis (extrinsic apoptosis). This permits virus persistence during chronic liver disease (CLD). HBx blocks the activation of mitochondrial antiviral signaling protein (MAVS), a key mediator of innate antiviral signaling. Further, HBx prevents extrinsic apoptosis triggered by TNFα, TGFβ, and Fas by blocking caspases 8 and 3 and activating NF-κB, the latter of which is hepatoprotective. HBx overrides TGFβ negative growth regulation, converting it to a tumor promoter (Jeong et al., 2012; Zemel et al., 2011). Avoiding immune destruction provides a survival and growth advantage for HBx-expressing hepatocytes (Wang et al., 1991). The finding that HBx up-regulates proinflammatory cytokines (Martin-Vilchez et al., 2011), also favors regeneration of HBx expressing hepatocytes, which is consistent with the observation that HBx expression correlates with CLD (Jin et al., 2001). HCV infection triggers innate immunity, but viral proteins block the signaling that triggers interferon beta (IFNβ) (Kumthip et al., 2012) as well as IFN alpha signaling by targeting JAK/STAT (Rehermann, 2009). In addition to avoiding immune destruction, HBV and HCV survive by inhibiting apoptosis. For HBV, promotion or inhibition of apoptosis is context dependent (Assrir et al., 2010). HBx suppresses p53 by direct binding and transcriptional down-regulation, promotes phosphorylation (inactivation) of Rb, and down-regulates several cdk inhibitors (Zemel et al., 2011). Mitochrondria associated HBx increases ROS. The latter, combined with cell cycle progression, increases the risk for the appearance and propagation of mutations (Zemel et al., 2011). In cases where HBx appears to promote apoptosis, this may reflect a cellular response to inappropriate growth signaling triggered by HBx (Feitelson et al., 2005). For HCV, core and NS3 inactivate multiple tumor suppressors (Brechot, 2004; Zemel et al., 2011). HCV core blocks immune mediated apoptosis by inhibiting caspase 8. NS5A binding to cellular signaling molecules suppresses immune responses, tumor suppressors, and apoptosis. HCV transgenic mice are resistant to Fas mediated apoptosis by blocking multiple caspases (Zemel et al., 2011). Both HBV and HCV up-regulate miR-181, which promotes the appearance of “stemness” markers in HCC (Jeong et al., 2012).
Replicative immortality is another cancer hallmark influenced by HBV and HCV. The HBx gene has been found integrated near the human telomerase reverse transcriptase (hTERT) gene, resulting in over-expression of hTERT, which is also a transcriptional target for HBx (Feitelson and Lee, 2007). For HCV, stable transfection of human hepatocytes with HCV core promoted dedifferentiation, continuous growth, and upregulated expression of telomerase, which can promote immortalization (Ray et al., 2000).
HBV and HCV are known to promote pathways that induce angiogenesis. In HBV carriers, cirrhotic nodules are often hypoxic and most are strongly positive for HBx. HBx may promote survival and growth by transcriptionally activating and stabilizing hypoxia inducible factor 1α (HIF1α) (Yoo et al., 2003). HIF1α transcriptionally activates angiopoietin-2 (Ang-2) and vascular endothelial growth factor (VEGF) (Martin-Vilchez et al., 2011). HBx up-regulates matrix metalloproteinases (MMPs) that digest the fibrous capsules of early tumors, resulting in increased angiogenesis and metastasis. Thus, HBx promotes the cellular response to hypoxia and stimulates angiogenesis. HCV promotes angiogenesis by generating ROS, which activates multiple signaling pathways that stabilize HIF1α. In both HBV and HCV infections, fibrogenesis stimulated angiogenesis, because CLD is characterized by elevated levels of the angiogenic cytokines, including VEGF (Vrancken et al., 2012), which may be an important therapeutic target that could delay the onset and/or progression of HCC.
Invasion and metastases is important in malignant progression of HCC and elevated hepatocyte growth factor (HGF) receptor, c-met, can lead to scattering, angiogenesis, proliferation, enhanced cell motility, invasion, and metastasis. HBx may promote c-met expression (Xie et al., 2010), but it is not clear whether this occurs with HCV. HBx also promotes invasion and metastasis by stimulating expression of MMPs, which degrade fibrous capsules around tumor nodules (Zemel et al., 2011). HCV core abrogates TGFβ–dependent tumor suppression via activation of JNK/pSmad3L signaling, resulting in EMT, and tumor invasion.
Tumor promoting inflammation is critical to HCC development. ROS generated in this context promotes a microenvironment conducive to mutagenesis, tumor development and survival. CLD results in tissue damage, regeneration, fibrosis and eventually hypoxia. In hypoxia, stem-like and progenitor cells survive and expand (Keith and Simon, 2007), and may form carcinoma in situ. In HCV, NS5A promotes expression the stem cell factor Nanog (Machida et al., 2012). For HBV, HBx also induces the expression of Nanog and other “stemness” promoting factors (Arzumanyan et al., 2011). Genome instability and mutation contribute to HBV and HCV driven tumor initiation and progression. During CLD, hepatocellular regeneration is accompanied by integration of HBV DNA fragments most often spanning the X gene. Although integration is common and appears random, there is clustering near or within fragile sites and other cancer associated regions that are prone to genetic instability (Feitelson and Lee, 2007). Cis-acting mechanisms in HCC have been reported, although the only host gene with integration events in tumors from different patients is hTERT (Feitelson and Lee, 2007). Many integration events encode functional HBx (Kew, 2011). HBx inactivates p53, thereby compromising genome integrity (Matsuda and Ichida, 2009). HBx also inhibits damage DNA binding protein 1 (DDB1) in nucleotide excision repair. HBx promotes the appearance of multinucleated cells, chromosomal rearrangement and micronuclei formation (Martin-Vilchez et al., 2011). HBx interacts with proteins that regulate centrosomal integrity, such as chromosome maintenance region 1 (Crm1) and HBXIP (Martin-Vilchez et al., 2011). Crm1 transports HBx into the cytoplasm, but when HBx inactivates Crm1, HBx accumulates in the nucleus where it alters host gene expression that promotes cell growth. HBXIP binding to HBx results in excessive centrosome replication, tripolar and multipolar spindles and subsequent aberrant chromosome segregation. Thus, HBx is centrally involved in genome instability.
DNA repair and apoptosis is also regulated by poly(ADP-ribose) polymerase 1 (PARP-1) (Langelier and Pascal, 2013). HBx stabilizes PARP, which protects infected cells from apoptosis. HCV NS5A stabilizes PARP-1 by blocking caspase 3 mediated-cleavage (Zemel et al., 2011). PARP-1 cleavage is also accelerated by HCV (Zhao et al., 2012). The latter may permit the propagation of mutations and promote genetic instability in HCV infected cells.
Deregulating cellular energetics permits dysplastic cells and cells in in situ tumors to survive and grow. In aerobic glycolysis, where pyruvate is converted into lactate, the latter is used to accelerate biosynthesis for growth in the absence of oxidative phosphorylation. The underlying switch (Warburg effect) involves cross-talk between metabolic pathways in the mitochondria and epigenetic mechanisms in the nucleus (Martinez-Pastor et al., 2013). Protein acetylation derives from the intracellular pool of acetyl-CoA, most of which derives from citrate (in the Krebs cycle) in mitochondria. HBx and HCV core in mitochondria may affect the intracellular citrate levels by triggering chronic oxidative stress that disrupts membrane potential (mtΔΨ) and organelle function (Brault et al., 2013). This may reduce the levels of citrate that are available for conversion to acetyl-CoA, and thus for epigenetic modification of chromatin through protein acetylation. HBx and HCV activate histone deacetylase 1 (HDAC1) and up-regulated metastasis associated protein 1 (MTA-1) (Yoo et al., 2003), both of which contribute to HCC. HCV core promoted steatosis and HBx induced β-catenin, in part, by attenuating expression of SIRT1, an NAD dependent histone deacetylase (Srisuttee et al., 2012). When glucose is plentiful, NAD is converted to NADH, and SIRT1 is inhibited, so these viruses may mimic conditions that promote tumor cell growth via epigenetic modification of host gene expression. HBV and HCV also up-regulate DNA methyl-transferases (DNMTs), which may block the expression of tumor suppressor genes that are methylated in HCC (Tian et al., 2013).
Human T-cell Leukemia Virus-1 (HTLV-1): causal agent of Adult T-cell Leukemia /lymphoma (ATL)
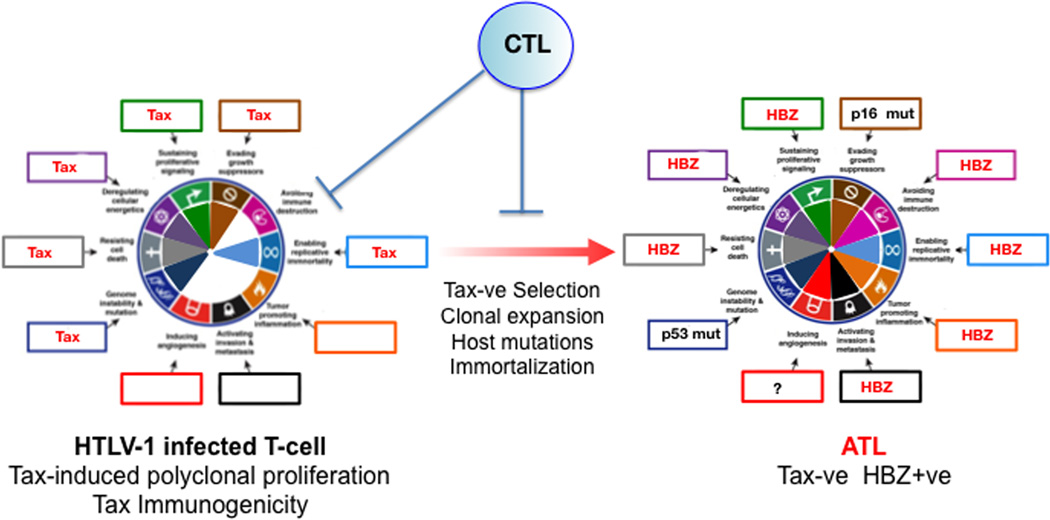
HTLV-1, the first described human lymphotropic retrovirus is the etiologic agent for Adult T-cell lymphoma (ATL), an incurable malignancy of mature CD4+ T-cells, which is endemic to Japan and certain areas of Africa, Central and South America (Cesarman and Mesri, 2007; Matsuoka and Jeang, 2007; Yoshida, 2001). HTLV-1 driven oncogenesis is a two-step process. A first step in which the viral Tax transactivator induces polyclonal T-cell proliferation that then switches to a Tax-repressed or deleted second step which is supported by the HTLV-1 bZip (HBZ) protein and RNA, together with host oncogenic alterations (Matsuoka and Jeang, 2007) (Figure 5)
Figure 5. Hallmarks of Cancer analysis for HTLV-1 induced Adult T-cell Lymphoma.
Tax activation of survival and proliferation hallmarks in HTLV-1 infected T-cells leads to polyclonal expansion of infected cells leading to immortalization. However since Tax is immunogenic, host CTLs select for infected cells with downregulated or deleted Tax. After a prolonged asymptomatic period of 20–40 years, that involves acquisition of host mutations, immortalization and clonal expansion, adult T-cell leukemia (ATL) cells emerge in approximately 5% of infected individuals. Tax functions are compensated by HZB-mediated hallmark activation and by host mutations such as p16INK4A and p53. Most HTLV-1-infected individuals remain asymptomatic carriers.
Activation of cancer hallmarks by HTLV-1
Tax
Tax is a pleiotropic viral oncogene that induces T-cell expansion by activating cell proliferation and survival pathways such as NF-kB, PI3K-AKT, and CREB (Cesarman and Mesri, 2007; Matsuoka and Jeang, 2007; Yoshida, 2001). Tax activates the IKK complex by directly interacting with NEMO through a mechanism that involves Tax ubiquitination via Ubc13 (Shembade et al., 2007), and by blocking PP2A inactivation of IKK signaling. In T-cells NF-kB activates genes related to T-cell proliferation, growth and survival such as c-rel, c-myc, and the ligand and the receptor for the T-cell growth factor IL-2, leading to an IL-2 autocrine loop (Leung and Nabel, 1988; Maruyama et al., 1987). This, together with hTERT activation, which enables replicative immortality is thought to mediate Tax -induced immortalization (Grassmann et al., 2005). Tax also activates CREB-mediated transcription (Kwok et al., 1996) via a PKA independent pathway and regulates CBP/p300. Tax activates the PI3K-AKT prosurvival and growth pathway (Matsuoka and Jeang, 2007). Tax oncogenicity is evidenced by its ability to promote cell cycle entry by CDK activation, its ability to inhibit p53 via the phosphatase Wip1-mediated modulation of p300/CBP (Zane et al., 2012), and by the disc large (DLG) tumor suppressor (Grassmann et al., 2005; Matsuoka and Jeang, 2007). Furthermore Tax inhibits the DDR and interacts with the mitotic apparatus (Ching et al., 2006) causing chromosomal aberrations and genetic instability, thus increasing the likelihood of acquiring somatic oncogenic alterations (Grassmann et al., 2005; Matsuoka and Jeang, 2007).
HBZ
Although many HTLV-1 genomes are deleted in their 5’ end, all of them are intact in the 3’ LTR from where they are able to stably transcribe HBZ mRNA throughout the process of infection and transformation (Matsuoka and Jeang, 2007; Satou et al., 2006). HBZ is also able to activate cancer hallmarks, and thus to partially compensate for Tax downregulation. HBZ protein and RNA appear to be critical for supporting the survival and immune evasion of ATL cells (Matsuoka and Jeang, 2007; Satou et al., 2006). In addition to activating transcription of JunD, c-jun and ATF pro-survival genes, the HBZ RNA may be able to directly activate cell proliferation (Satou et al., 2006) by promoting transcription of the cell cycle promoter E2F. HBZ also activates hTERT expression, supporting replicative immortality (Matsuoka and Jeang, 2007).
HTLV-1 oncogenesis
Tax is a target of host CTLs (Matsuoka and Jeang, 2007). Thus cells that loose Tax expression by silencing or deletion will evade immune elimination and be favorably selected (Fig. 5). Survival of Tax-negative cells is supported by HTLV-1 HBZ protein and RNA, and by host somatic mutations (Matsuoka and Jeang, 2007), all of which lead to immortalization. This preneoplastic state sets the stage for further acquisition of somatic host alterations including deletion or mutations of the CDKN2/p16INK4A tumor and p53 suppressor genes (Sato et al., 2010; Uchida et al., 1998) and chromosomal aberrations, generally correlating with tumor progression, and eventually resulting in T-cell malignant transformation (Matsuoka and Jeang, 2007). Consistent with Tax expressing HTLV-1 infected cells being controlled by T-cell responses, known cofactors for ATL development in HTLV-1 infected individuals are related to the individual’s ability to control HTLV-1 infection including immune status and co-infection with certain parasites (Matsuoka and Jeang, 2007).
Kaposi’s sarcoma herpesvirus (KSHV): Tumorigenesis by classic and paracrine oncogenesis
KSHV or human herpesvirus-8 (HHV-8) is a γ2-herpesvirus that is the causal agent of Kaposi’s sarcoma (KS) (Chang et al., 1994), reviewed in (Ganem, 2010; Mesri et al., 2010). KS is characterized by the proliferation of infected spindle cells of vascular and lymphatic endothelial origin, accompanied by intense angiogenesis with erythrocyte extravasation and inflammatory infiltration (Ganem, 2010; Mesri et al., 2010). KSHV can infect a variety of cells including endothelial lineage, monocytes and B cells. KSHV encodes many oncogenic viral homologues of host proteins with the potential to drive cell survival, proliferation, immune evasion and angiogenesis (Coscoy, 2007; Ganem, 2010; Mesri et al., 2010). Similar to other herpesviruses, such as EBV discussed above, KSHV can undergo latent or lytic stages of replication. Latency is a state whereby KSHV replicates along with the host by expressing KSHV latency-associated nuclear antigen (LANA), which tethers the KSHV episome to the host chromosome thus assuring its maintenance and segregation during host cell division. In the KSHV lytic cycle the virus expresses the full viral program necessary for replication and assembly of infectious virus with cytopathic cell lysis (Ganem, 2010; Mesri et al., 2010). Both KSHV lytic and latent gene expression programs are thought to be necessary for KS development, and KS lesions contain mostly latently-infected cells with a variable percentage of lytically-infected cells (Ganem, 2010; Mesri et al., 2010). The potential contributions of individual latent and lytic KSHV gene products to KS and to specific cancer hallmarks are discussed below and depicted in Figure 6.
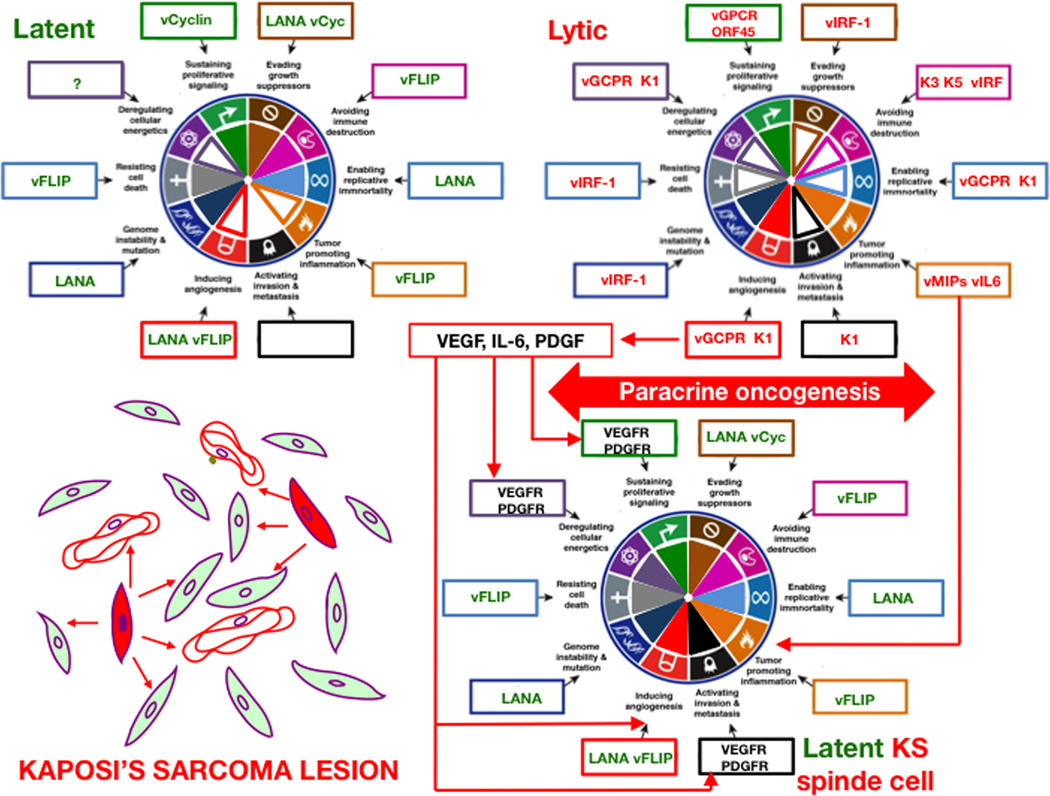
Figure 6. Hallmarks of Cancer Analysis for Kaposi’s sarcoma pathogenesis based on the paracrine oncogenesis hypothesis.
KSHV infections could be either latent or lytic. Latently infected cells express the LANA transcript and the KSHV miRNA. Although these genes can activate KS cancer hallmarks they are insufficient to transform the cell and to induce the characteristic KS angiogenic phenotype. Lytic infected cells on the other hand express viral genes that make the highly angiogenic and inflammatory but they are doomed for cytopathic death and under immune control. In immunosuppressed and AIDS individuals, lytic infection is more permissible and it can combine with latently infected cells to form tumors via the mechanism of paracrine oncogenesis (see text for details). Bottom left is a schematic representation of a KS tumor showing latently infected cells (filled in green), lytically infected cells (filled in red), blood vessels (unfilled in red) and paracrine effects (depicted as red arrows). Within the boxes around the wheel, latent viral genes are in green, lytic viral genes in red and host genes are in black. The figure is not a strict representation of experimental data but a compilation of published information analyzed in the context of the Hallmarks of Cancer and the paracrine oncogenesis hypothesis. Genes chosen for hallmarks activation represent one of the available examples and are based on published data with preference for tumor relevant systems where available. Filled portions of hallmark pie are used to represent stronger or well documented hallmark activation. Empty portions represent weaker effects or lack of activation/ evidence. See text for explanations and references.
Cancer Hallmarks activation during latent KSHV infection
KSHV latent transcripts include genes and miRNAs that favor viral persistence and replication while promoting host-cell proliferation and survival. LANA and v-cyclin affect cell cycle checkpoint mediators. LANA inhibits both the p53 (Friborg et al., 1999) and the RB tumor suppressor pathways (Radkov et al., 2000), thus allowing the infected cell to become insensitive to anti-growth signals, avoiding cell cycle arrest and promoting genetic instability (Si and Robertson, 2006). KSHV promotes cell proliferation via its cyclin-D homolog, v-cyclin, a constitutive activator of CDK4/6 (Swanton et al., 1997), that is able to directly downregulate the p27KIP1 cdk inhibitor (Ellis et al., 1999; Sarek et al., 2006). LANA antagonizes growth inhibitory WNT signaling by stabilizing β-catenin through sequestration of its negative regulator GSK-3β, upregulating via this and other pathways the expression of c-myc (Fujimuro et al., 2003). Furthermore, LANA targets the ubiquitin ligase Sel10 to stabilize the Notch protooncogene, leading to cell proliferation (Lan et al., 2007). Thus, through the concerted activity of LANA and v-cyclin, latently infected cells become self-sufficient in growth signals and proliferate. Latent KSHV evades apoptosis by promoting NFκB signaling through different mechanisms. The FLICE/caspase 8-inhibitory protein (vFLIP) is a potent constitutive activator of NF-kB anti apoptotic signaling that is critical for survival of latently infected cells (Guasparri et al., 2004). In addition, KSHV encoded miRNAs promote survival and tumorigenesis by targeting regulatory members of the NF-kB pathway (Zhu et al., 2013).
LANA supports a pro-angiogenic program by stabilizing the VEGF transcription factor hypoxia-inducible factor 1 (HIF-1) via targeting its ubiquitin ligase, the von Hippel-Lindau (VHL) tumor suppressor, for degradation (Cai et al., 2006), In addition, vFLIP-activation of NF-kB leads to the induction and secretion of pro-angiogenic cytokines such as IL-6 (Grossmann et al., 2006),. The viral miRNA miRK1–5 has been reported to dowregulate the anti-angiogenic protein thrombospondin-1 (TSP-1) (Samols et al., 2007) and this may also help promote angiogenesis. LANA can increase the lifespan of human umbilical vein endothelial cells and is an activator of hTERT expression, which can confer cells with limitless replicative potential (Verma et al., 2004).
Metabolic profiling of KSHV infected cells indicates that the virus reprograms the metabolism of host cell to resemble that of cancer cells (Delgado et al., 2012). Specifically, KSHV infection shifts endothelial cells towards aerobic glycolysis and lactic acid production while decreasing oxygen consumption, thus inducing the Warburg effect, a metabolic characteristic of tumor cells (Delgado et al., 2010).
vFLIP activated NF-kB signaling drives inflammation via overexpression of many cytokines and chemokines implicated in KS pathogenesis (Grossmann et al., 2006), while viral Kaposin B can upregulate cytokine mRNA levels by activating a kinase that inhibits the mRNA decay pathway (McCormick and Ganem, 2005). Interestingly, KSHV upregulation of inflammatory mediators such as COX-2 and its metabolites were found to play a role in maintenance of viral latency through LANA upregulation (George Paul et al., 2010). In addition to being less immunogenic, latently infected cells can evade the immune response by resisting Fas induced-apoptosis through vFLIP-induced activation of NF-kB, which leads to upregulation of anti-apoptotic members of the Bcl-2 family (Guasparri et al., 2004).
Cancer Hallmarks activation during lytic KSHV infection
During lytic infection, KSHV expresses the full replication program to produce new virions. Several of these gene products including vGPCR, vIRF1, and vBCL2 are viral homologs of host signaling and growth regulators. The KSHV lytic program induce cancer hallmarks through the expression of genes that favor viral replication by affecting the DNA damage response, reprogramming metabolism, promoting survival, and mediating immune evasion. KSHV lytic genes also induce host responses for recruiting cells of the endothelial and monocytic lineages and thus promote angiogenesis and inflammation, which are characteristic hallmarks of KS (Figure 6).
Insensitivity to anti-growth signals
A viral homolog of the interferon inducible factor, vIRF-1, inhibits the tumor suppressor p53 via interaction with its co-activator p300/CBP promoting its monoubiquitination and delocalization to the cytoplasm (Nakamura et al., 2001).
Self-sufficiency in growth signals
KSHV encodes a constitutively active G-protein coupled receptor homolog (vGPCR) of the angiogenic chemokines IL-8 and Gro-a receptors. vGPCR, the ITAM-like sequence bearing K1 protein, and ORF45 all constitutively induce ERK and PI3K/AKT/mTOR cascades(Bais et al., 2003; Chang and Ganem, 2013; Wang et al., 2006). vGPCR activates secretion of VEGF, IL-6 and PDGF (Jensen et al., 2005), that together with the vIL-6 viral chemokine (or virokine) (Nicholas et al., 1997) are autocrine growth factors that can also drive latent cell proliferation by paracrine mechanisms (Montaner et al., 2006).
Evading apoptosis
vGPCR activates PI3K/AKT and NF-kB anti-apoptotic pathways that promote endothelial cell survival (Bais et al., 2003). In addition to reducing p53-driven cell death, vIRF-1 can prevent apoptosis during viral replication by direct inhibition of pro-apoptotic BH3 domain-containing proteins such as BIM (Choi and Nicholas, 2010).
Genetic instability
vIRF-1 inhibits ATM activation of p53 to suppress DNA damage-induced apoptosis, a response that can lead to accumulation of mutations and genetic instability (Shin et al., 2006). vGPCR activates ROS production via Rac1 activation of NADPH-oxidase (Ma et al., 2013). ROS can cause oxidative DNA damage leading to increased mutation rate and accumulation of chromosomal alterations.
Sustained angiogenesis
Prominent angiogenesis is a typical characteristic of KS. Several lytic genes have the potential to cause an angiogenic switch in infected cells. vGPCR activates VEGF expression and secretion (Bais et al., 1998) via MAPK phosphorylation of HIF-1α (Sodhi et al., 2000) and through activation of the PI3K-AKT-mTORC1 pathway (Sodhi et al., 2006). vGCPR is also able of upregulating numerous proangiogenic factors such as PDGF, Angpt-2 and IL-6 (Jensen et al., 2005). vGPCR is a potent angiogenic oncogene able to induce angioproliferative KS-like lesions in several transgenic mouse models (Jensen et al., 2005; Montaner et al., 2003), and it was shown to be necessary for KSHV tumorigenesis and angiogenesis (Mutlu et al., 2007). Other angiogenic lytic genes are K1, which activates secretion of VEGF and MMP-9 (Wang et al., 2004), which together with vGPCR mediates endothelial to mesenchymal transition (EnMT) that may favor cell invasion and metastasis (Cheng et al., 2011). Virokines such as vMIP-I, II and III(Stine et al., 2000) (also refered to as vCCLs) and vIL-6 (Aoki et al., 1999) have also been shown to have pro-angiogenic properties in several experimental systems.
Limitless replicative potential/Immortalization
vGPCR-mediated autocrine activation of VEGF-R2/KDR can immortalize human EC via alternative lengthening of telomeres (ALT) (Bais et al., 2003). Ectopic expression of K1 in endothelial cells has also been reported to cause cellular immortalization (Wang et al., 2004).
Metabolic reprogramming
Several lytic proteins including vGPCR, K1 and ORF45 activate the master metabolic regulator mTORC1 (Chang and Ganem, 2013; Sodhi et al., 2006; Wang et al., 2006). mTORC1 activation drives protein and nucleotide synthesis needed for viral replication by switching glucose metabolism towards aerobic glycolysis; an enabling characteristic of prolferative cancer cells. ORF45 activates mTORC1 via ERK2/RSK(Chang and Ganem, 2013) while vGPCR activates mTORC1 by directly triggering the PI3K-AKT pathway and through paracrine interactions by growth factors such as VEGF and PDGF that act on mTORC1 activating receptors (Sodhi et al., 2006). The viral K3 ubiquitin ligase may induce glucose metabolic reprogramming in lytically infected cells by affecting stability and signaling via receptor tyrosine kinases (Karki et al., 2011).
Inflammation
vGPCR induces inflammatory signaling and cytokine secretion (Bais et al., 1998; Sodhi et al., 2006). KSHV encoded virokines such as vMIP-III can chemo-attract inflammatory cells (Stine et al., 2000). K15, a predicted transmembrane viral protein, can promote inflammatory signalling via JNK and NF-kB signaling leading to secretion of various chemokines and cytokines (Brinkmann et al., 2007).
Immune evasion
Lytic KSHV expresses many immune-regulatory genes including vIRF-1-4, which display variable specificities for inhibition of the Type I interferon response (Offermann, 2007). ORF63 encodes for a viral homolog of human NLRP1, which is able to inhibit inflammasome activation (Gregory et al., 2011). Similar to other herpesviruses, KSHV has evolved unique abilities to downregulate immune-recognition receptors. They include the K3 and K5 ubiquitin ligases that are able to downregulate MHC-I, ICAM and NK receptors (Cadwell and Coscoy, 2005; Coscoy, 2007)
Mechanisms of KSHV induced oncogenesis
As outlined above, KSHV’s comprehensive oncogenic arsenal conveys all cancer hallmarks. Yet the ability of KSHV to cause cancer is limited to four clinically defined populations: HIV/AIDS patients (Epidemic or AIDS-associated KS), immunosuppressed individuals (transplant KS), people living in geographical areas in sub-Saharan Africa (Endemic KS) and elderly individuals of Mediterranean or Azhkenazi origin (classic KS) (Ganem, 2010; Mesri et al., 2010). To explain the high incidence of KS in HIV/AIDS patients, HIV-1 and immune-related mechanisms have been proposed. Changes in cytokine profiles which are characteristic of HIV/AIDS-associated immune activation as well as the presence of HIV-1 Tat can favor KS development via increased KSHV lytic reactivation and also by direct angiogenesis mechanisms(Aoki and Tosato, 2007). During AIDS, decrease in CD4+ T-helper cells hampers immune-control of KSHV infected cells. Lytic patterns of expression include more oncogenic genes (Fig. 6), but they are also more immunogenic (Ganem, 2010; Mesri et al., 2010). Both CTLs and a type of suppressor T-cells are able to either eliminate or suppress lytic infection (Myoung and Ganem, 2011). Thus, in immunocompetent hosts, lytic KSHV infected cells are suppressed by the immune system. In immunosuppressed patients, however,, KSHV infected cells are not subject to immune clearance, thus allowing KSHV to express its full oncogenic repertoire including genes able to induce characteristic KS hallmarks such as angiogenesis and inflammation. Latently infected cells that also express the oncogenic early lytic genes but would not complete the lytic cycle, referred to as abortive lytic replication(Bais et al., 2003), may become progressively transformed and acquire somatic oncogenic alterations that would allow the infected cells to switch back to less immunogenic latent forms, similar to what has been discussed for HTLV-1 and for EBV Lat I cancers. A second scenario would have to take into account the paradoxical fact that while KSHV latently infected cells, which are a majority of the spindle cells in KS lesions, are not fully transformed, lytic infection where KSHV angiogenic oncogenes are expressed is cytopathic and thus lytically infected cells cannot be a stable part of the transformed state. A “paracrine oncogenesis” hypothesis has been developed that is based on the occurrence in KS lesions of either lytically infected cells or latently infected cells expressing lytic genes (Bais et al., 2003; Chang and Ganem, 2013; Montaner et al., 2003) (Figure 6). These cells express KSHV genes such as vGPCR, K1 and ORF 45 that stimulate the production of host cytokines and angiogenesis growth factors which, in concert with virokines vIL-6 and vMIPs/vCCLs will stimulate angiogenesis and support latently infected cell proliferation (Figure 6) (Mesri et al., 2010). Hence, the full KSHV-induced oncogenic complement enabling the acquisition of all cancer hallmarks for KS tumor formation may be achieved by a paracrine connection between latent and lytically infected cells (Figure 6). This is supported by laboratory findings that cells expressing KSHV lytic genes enable tumorigenicity of latent KSHV genes via paracrine mechanisms (Montaner et al., 2006). This model also accounts for the higher incidence of KS in immunosuppressed and HIV/AIDS patients. Clinical findings consistent with the necessity of lytically infected cells for AIDS-KS tumor formation and the role of the immune system in their control include the steep decline in AIDS-KS incidence since ART implementation, the anti-AIDS-KS response to ART upon immune reconstitution, and the observation that some anti-viral drugs targeting KSHV lytic replication were able to prevent AIDS-KS, reviewed in (Casper, 2011; Mesri et al., 2010).
Preventing and targeting viral oncogenesis in the clinical setting
Virus-induced cancers are amenable to immunoprophylaxis by vaccines (Schiller and Lowy, 2010). Further, viral oncogenes constitute attractive and sometimes druggable non-host targets. This is exemplified by the major success in vaccinating against HBV, the promise of prophylactic HPV vaccines (Schiller and Lowy, 2010) as well as the new generation nucleoside analogs to treat HCV. Current clinical efforts in preventing or treating virus-induced cancers are a combination of anti-viral agents, agents that induce virus-related cytotoxicity events, agents that target virus-induced cancer hallmarks and agents that are used to successfully treat cancers with similar histological classification but of non-viral etiology. Table 2 summarizes some of the most promising and successful approaches that are either FDA approved or have already been clinically tested. Despite these major successes, the treatment of virus-induced cancers remains challenging. Major obstacles are the limited availability of validated preclinical animal models for some virally induced cancers, and the complexity of simultaneously targeting the viral and the host oncogenome, which on one hand may increase the number of potential therapeutic targets but also provides two sources of potential resistance to therapy.
Table 2.
Therapeutic and preventive approaches successfully used to prevent and target oncoviral infections or their cancer associated hallmarks.
| Virus | Cancer | Intervention or Drug | Target | Hallmark or Process |
|---|---|---|---|---|
| Pharmacological approaches | ||||
| HBV/HCV | HCC | Sorafenib | VEGFR, PDGFR, Raf | Angiogenesis Proliferation |
| KSHV | Transp KS, AIDS-KS | Rapamycin | mTORCl | Angiogenesis |
| AIDS-KS | Imatinib | PDGFR, c-kit | Proliferation | |
| HTLV-1 | ATLL | AZT + αIFN | NFkB | Survival |
| EBV | NHL | Butvrate + GCV | HDAQ viral TK DNA polymerase | Survival |
| HBV | Viral CLD leading to HCC | Nucleoside analogs, IFN | Viral polymerase Protein translation | Chronic infection |
| HCV | Viral CLD leading to HCC | ribavirin; IFN; protease inhibitors | virus replication; protein translation | Chronic infection |
| Immunoprophylaxis and Immunotherapy | ||||
| EBV | PTLD | EBV Specific CTL | Lat III infected cells | Survival Immune evasion |
| HPV | CxCC NHC | L1 VLP vaccine | Infection | Infection |
| HBV | HCC | HBsAe VLP vaccine | HBsAe | Infection |
Conclusion
Human viral cancers are pathobiological consequences of infection with viruses that evolved powerful mechanisms to persist and replicate through deregulation of host oncogenic pathways, conveying cancer hallmarks to the infected cell. The six major human oncoviruses are able to target many of these pathways. Natural barriers to viral oncogenesis are the immune response and the innate safeguard for human cancer constituted by the multi-hit nature of the carcinogenesis process, for which viral infection contributes only partially. Environmental and host co-factors such as immunosuppression, genetic predisposition or mutagens can each affect this delicate equilibrium and could accelerate the development of these cancers. The infectious nature and long incubation periods of human viral cancers provide unique windows of opportunity for prevention and clinical intervention.
Acknowledgements
We thank Drs. Douglas Hanahan and Robert Weinberg for developing this insightful concept of cancer development. It constitutes the inspiration and conceptual framework of this review. We also thank our colleagues for their contributions to our understanding of virally induced cancers. We regret that for space reasons we were not able to comprehensively reference all their contributions. EAM is supported by PHS NIH NCI grants CA 136387, by OHAM/ NCI supplements to the Miami CFAR, by the PHS NIAID MIAMI CFAR 5P30AI073961 and by the Bankhead Coley Florida Biomedical Foundation grant 3BB05. MAF is supported by PHS grants CA104025 and AI076535 and by Temple University. KM is supported by PHS grants CA081135, CA066980, and CA141583.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Contributor Information
Enrique A. Mesri, Email: emesri@med.miami.edu.
Mark Feitelson, Email: feitelso@temple.edu.
Karl Munger, Email: kmunger@rics.bwh.harvard.edu.
REFERENCES
- Aoki Y, Jaffe ES, Chang Y, Jones K, Teruya-Feldstein J, Moore PS, Tosato G. Angiogenesis and hematopoiesis induced by Kaposi's sarcoma-associated herpesvirus-encoded interleukin-6. Blood. 1999;93:4034–4043. [PubMed] [Google Scholar]
- Aoki Y, Tosato G. Interactions between HIV-1 Tat and KSHV. Current topics in microbiology and immunology. 2007;312:309–326. doi: 10.1007/978-3-540-34344-8_12. [DOI] [PubMed] [Google Scholar]
- Arvey A, Tempera I, Tsai K, Chen HS, Tikhmyanova N, Klichinsky M, Leslie C, Lieberman PM. An atlas of the Epstein-Barr virus transcriptome and epigenome reveals host-virus regulatory interactions. Cell host & microbe. 2012;12:233–245. doi: 10.1016/j.chom.2012.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arzumanyan A, Friedman T, Ng IO, Clayton MM, Lian Z, Feitelson MA. Does the hepatitis B antigen HBx promote the appearance of liver cancer stem cells? Cancer research. 2011;71:3701–3708. doi: 10.1158/0008-5472.CAN-10-3951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arzumanyan A, Reis HM, Feitelson MA. Pathogenic mechanisms in HBV- and HCV-associated hepatocellular carcinoma. Nature reviews Cancer. 2013;13:123–135. doi: 10.1038/nrc3449. [DOI] [PubMed] [Google Scholar]
- Assrir N, Soussan P, Kremsdorf D, Rossignol JM. Role of the hepatitis B virus proteins in pro- and anti-apoptotic processes. Front Biosci (Landmark Ed) 2010;15:12–24. doi: 10.2741/3602. [DOI] [PubMed] [Google Scholar]
- Bais C, Santomasso B, Coso O, Arvanitakis L, Raaka EG, Gutkind JS, Asch AS, Cesarman E, Gershengorn MC, Mesri EA. G-protein-coupled receptor of Kaposi's sarcoma-associated herpesvirus is a viral oncogene and angiogenesis activator. Nature. 1998;391:86–89. doi: 10.1038/34193. [DOI] [PubMed] [Google Scholar]
- Bais C, Van Geelen A, Eroles P, Mutlu A, Chiozzini C, Dias S, Silverstein RL, Rafii S, Mesri EA. Kaposi's sarcoma associated herpesvirus G protein-coupled receptor immortalizes human endothelial cells by activation of the VEGF receptor-2/ KDR. Cancer cell. 2003;3:131–143. doi: 10.1016/s1535-6108(03)00024-2. [DOI] [PubMed] [Google Scholar]
- Bouvard V, Baan R, Straif K, Grosse Y, Secretan B, El Ghissassi F, Benbrahim- Tallaa L, Guha N, Freeman C, Galichet L, et al. A review of human carcinogens--Part B: biological agents. The lancet oncology. 2009;10:321–322. doi: 10.1016/s1470-2045(09)70096-8. [DOI] [PubMed] [Google Scholar]
- Boyle P, Levin B International Agency for Research on Cancer; World Health Organization. World cancer report 2008. Lyon Geneva: International Agency for Research on Cancer ; Distributed by WHO Press; 2008. [Google Scholar]
- Brault C, Levy PL, Bartosch B. Hepatitis C virus-induced mitochondrial dysfunctions. Viruses. 2013;5:954–980. doi: 10.3390/v5030954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brechot C. Pathogenesis of hepatitis B virus-related hepatocellular carcinoma: old and new paradigms. Gastroenterology. 2004;127:S56–S61. doi: 10.1053/j.gastro.2004.09.016. [DOI] [PubMed] [Google Scholar]
- Brinkmann MM, Pietrek M, Dittrich-Breiholz O, Kracht M, Schulz TF. Modulation of host gene expression by the K15 protein of Kaposi's sarcoma-associated herpesvirus. Journal of virology. 2007;81:42–58. doi: 10.1128/JVI.00648-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cadwell K, Coscoy L. Ubiquitination on nonlysine residues by a viral E3 ubiquitin ligase. Science. 2005;309:127–130. doi: 10.1126/science.1110340. [DOI] [PubMed] [Google Scholar]
- Cai QL, Knight JS, Verma SC, Zald P, Robertson ES. EC5S ubiquitin complex is recruited by KSHV latent antigen LANA for degradation of the VHL and p53 tumor suppressors. PLoS pathogens. 2006;2:e116. doi: 10.1371/journal.ppat.0020116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casper C. The increasing burden of HIV-associated malignancies in resourcelimited regions. Annual review of medicine. 2011;62:157–170. doi: 10.1146/annurev-med-050409-103711. [DOI] [PubMed] [Google Scholar]
- Cesarman E, Mesri EA. Pathogenesis of viral lymphomas. Cancer treatment and research. 2006;131:49–88. doi: 10.1007/978-0-387-29346-2_2. [DOI] [PubMed] [Google Scholar]
- Cesarman E, Mesri EA. Kaposi sarcoma-associated herpesvirus and other viruses in human lymphomagenesis. Current topics in microbiology and immunology. 2007;312:263–287. doi: 10.1007/978-3-540-34344-8_10. [DOI] [PubMed] [Google Scholar]
- Chang HH, Ganem D. A unique herpesviral transcriptional program in KSHV-infected lymphatic endothelial cells leads to mTORC1 activation and rapamycin sensitivity. Cell host & microbe. 2013;13:429–440. doi: 10.1016/j.chom.2013.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang Y, Cesarman E, Pessin MS, Lee F, Culpepper J, Knowles DM, Moore PS. Identification of herpesvirus-like DNA sequences in AIDSassociated Kaposi's sarcoma. Science. 1994;266:1865–1869. doi: 10.1126/science.7997879. [DOI] [PubMed] [Google Scholar]
- Chen W, Li F, Mead L, White H, Walker J, Ingram DA, Roman A. Human papillomavirus causes an angiogenic switch in keratinocytes which is sufficient to alter endothelial cell behavior. Virology. 2007;367:168–174. doi: 10.1016/j.virol.2007.05.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng F, Pekkonen P, Laurinavicius S, Sugiyama N, Henderson S, Gunther T, Rantanen V, Kaivanto E, Aavikko M, Sarek G, et al. KSHV-initiated notch activation leads to membrane-type-1 matrix metalloproteinase-dependent lymphatic endothelial-to-mesenchymal transition. Cell host & microbe. 2011;10:577–590. doi: 10.1016/j.chom.2011.10.011. [DOI] [PubMed] [Google Scholar]
- Ching YP, Chan SF, Jeang KT, Jin DY. The retroviral oncoprotein Tax targets the coiled-coil centrosomal protein TAX1BP2 to induce centrosome overduplication. Nat Cell Biol. 2006;8:717–724. doi: 10.1038/ncb1432. [DOI] [PubMed] [Google Scholar]
- Choi YB, Nicholas J. Bim nuclear translocation and inactivation by viral interferon regulatory factor. PLoS pathogens. 2010;6:e1001031. doi: 10.1371/journal.ppat.1001031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coscoy L. Immune evasion by Kaposi's sarcoma-associated herpesvirus. Nature reviews Immunology. 2007;7:391–401. doi: 10.1038/nri2076. [DOI] [PubMed] [Google Scholar]
- de Martel C, Ferlay J, Franceschi S, Vignat J, Bray F, Forman D, Plummer M. Global burden of cancers attributable to infections in 2008: a review and synthetic analysis. The lancet oncology. 2012;13:607–615. doi: 10.1016/S1470-2045(12)70137-7. [DOI] [PubMed] [Google Scholar]
- Delgado T, Carroll PA, Punjabi AS, Margineantu D, Hockenbery DM, Lagunoff M. Induction of the Warburg effect by Kaposi's sarcoma herpesvirus is required for the maintenance of latently infected endothelial cells. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:10696–10701. doi: 10.1073/pnas.1004882107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delgado T, Sanchez EL, Camarda R, Lagunoff M. Global metabolic profiling of infection by an oncogenic virus: KSHV induces and requires lipogenesis for survival of latent infection. PLoS pathogens. 2012;8:e1002866. doi: 10.1371/journal.ppat.1002866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DiMaio D, Petti LM. The E5 proteins. Virology. 2013;445:99–114. doi: 10.1016/j.virol.2013.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duffy CL, Phillips SL, Klingelhutz AJ. Microarray analysis identifies differentiation-associated genes regulated by human papillomavirus type 16 E6. Virology. 2003;314:196–205. doi: 10.1016/s0042-6822(03)00390-8. [DOI] [PubMed] [Google Scholar]
- Durst M, Croce CM, Gissmann L, Schwarz E, Huebner K. Papillomavirus sequences integrate near cellular oncogenes in some cervical carcinomas. Proceedings of the National Academy of Sciences of the United States of America. 1987;84:1070–1074. doi: 10.1073/pnas.84.4.1070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellis M, Chew YP, Fallis L, Freddersdorf S, Boshoff C, Weiss RA, Lu X, Mittnacht S. Degradation of p27(Kip) cdk inhibitor triggered by Kaposi's sarcoma virus cyclin-cdk6 complex. Embo J. 1999;18:644–653. doi: 10.1093/emboj/18.3.644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feitelson MA, Lee J. Hepatitis B virus integration, fragile sites, and hepatocarcinogenesis. Cancer letters. 2007;252:157–170. doi: 10.1016/j.canlet.2006.11.010. [DOI] [PubMed] [Google Scholar]
- Feitelson MA, Reis HM, Liu J, Lian Z, Pan J. Hepatitis B virus X antigen (HBxAg) and cell cycle control in chronic infection and hepatocarcinogenesis. Frontiers in bioscience : a journal and virtual library. 2005;10:1558–1572. doi: 10.2741/1640. [DOI] [PubMed] [Google Scholar]
- Friborg J, Jr, Kong W, Hottiger MO, Nabel GJ. p53 inhibition by the LANA protein of KSHV protects against cell death. Nature. 1999;402:889–894. doi: 10.1038/47266. [DOI] [PubMed] [Google Scholar]
- Fujimuro M, Wu FY, ApRhys C, Kajumbula H, Young DB, Hayward GS, Hayward SD. A novel viral mechanism for dysregulation of beta-catenin in Kaposi's sarcoma-associated herpesvirus latency. Nat Med. 2003;9:300–306. doi: 10.1038/nm829. [DOI] [PubMed] [Google Scholar]
- Ganem D. KSHV and the pathogenesis of Kaposi sarcoma: listening to human biology and medicine. The Journal of clinical investigation. 2010;120:939–949. doi: 10.1172/JCI40567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- George Paul A, Sharma-Walia N, Kerur N, White C, Chandran B. Piracy of prostaglandin E2/EP receptor-mediated signaling by Kaposi's sarcoma-associated herpes virus (HHV-8) for latency gene expression: strategy of a successful pathogen. Cancer research. 2010;70:3697–3708. doi: 10.1158/0008-5472.CAN-09-3934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodwin EC, Yang E, Lee CJ, Lee HW, DiMaio D, Hwang ES. Rapid induction of senescence in human cervical carcinoma cells. Proceedings of the National Academy of Sciences of the United States of America. 2000;97:10978–10983. doi: 10.1073/pnas.97.20.10978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grassmann R, Aboud M, Jeang KT. Molecular mechanisms of cellular transformation by HTLV-1 Tax. Oncogene. 2005;24:5976–5985. doi: 10.1038/sj.onc.1208978. [DOI] [PubMed] [Google Scholar]
- Gregory SM, Davis BK, West JA, Taxman DJ, Matsuzawa S, Reed JC, Ting JP, Damania B. Discovery of a viral NLR homolog that inhibits the inflammasome. Science. 2011;331:330–334. doi: 10.1126/science.1199478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grossmann C, Podgrabinska S, Skobe M, Ganem D. Activation of NF-kappaB by the latent vFLIP gene of Kaposi's sarcoma-associated herpesvirus is required for the spindle shape of virus-infected endothelial cells and contributes to their proinflammatory phenotype. Journal of virology. 2006;80:7179–7185. doi: 10.1128/JVI.01603-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guasparri I, Keller SA, Cesarman E. KSHV vFLIP is essential for the survival of infected lymphoma cells. The Journal of experimental medicine. 2004;199:993–1003. doi: 10.1084/jem.20031467. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Hanahan D, Weinberg RA. The hallmarks of cancer. Cell. 2000;100:57–70. doi: 10.1016/s0092-8674(00)81683-9. [DOI] [PubMed] [Google Scholar]
- Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- Hellner K, Mar J, Fang F, Quackenbush J, Munger K. HPV16 E7 oncogene expression in normal human epithelial cells causes molecular changes indicative of an epithelial to mesenchymal transition. Virology. 2009;391:57–63. doi: 10.1016/j.virol.2009.05.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofelmayr H, Strobl LJ, Marschall G, Bornkamm GW, Zimber-Strobl U. Activated Notch1 can transiently substitute for EBNA2 in the maintenance of proliferation of LMP1-expressing immortalized B cells. Journal of virology. 2001;75:2033–2040. doi: 10.1128/JVI.75.5.2033-2040.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwakiri D, Zhou L, Samanta M, Matsumoto M, Ebihara T, Seya T, Imai S, Fujieda M, Kawa K, Takada K. Epstein-Barr virus (EBV)-encoded small RNA is released from EBV-infected cells and activates signaling from Toll-like receptor 3. The Journal of experimental medicine. 2009;206:2091–2099. doi: 10.1084/jem.20081761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen KK, Manfra DJ, Grisotto MG, Martin AP, Vassileva G, Kelley K, Schwartz TW, Lira SA. The human herpes virus 8-encoded chemokine receptor is required for angioproliferation in a murine model of Kaposi's sarcoma. J Immunol. 2005;174:3686–3694. doi: 10.4049/jimmunol.174.6.3686. [DOI] [PubMed] [Google Scholar]
- Jeong SW, Jang JY, Chung RT. Hepatitis C virus and hepatocarcinogenesis. Clinical and molecular hepatology. 2012;18:347–356. doi: 10.3350/cmh.2012.18.4.347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin YM, Yun C, Park C, Wang HJ, Cho H. Expression of hepatitis B virus X protein is closely correlated with the high periportal inflammatory activity of liver diseases. Journal of viral hepatitis. 2001;8:322–330. doi: 10.1046/j.1365-2893.2001.00308.x. [DOI] [PubMed] [Google Scholar]
- Kaiser C, Laux G, Eick D, Jochner N, Bornkamm GW, Kempkes B. The proto-oncogene c-myc is a direct target gene of Epstein-Barr virus nuclear antigen 2. Journal of virology. 1999;73:4481–4484. doi: 10.1128/jvi.73.5.4481-4484.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karki R, Lang SM, Means RE. The MARCH family E3 ubiquitin ligase K5 alters monocyte metabolism and proliferation through receptor tyrosine kinase modulation. PLoS pathogens. 2011;7:e1001331. doi: 10.1371/journal.ppat.1001331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keith B, Simon MC. Hypoxia-inducible factors, stem cells, and cancer. Cell. 2007;129:465–472. doi: 10.1016/j.cell.2007.04.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kew MC. Hepatitis B virus x protein in the pathogenesis of hepatitis B virus-induced hepatocellular carcinoma. Journal of gastroenterology and hepatology 26 Suppl. 2011;1:144–152. doi: 10.1111/j.1440-1746.2010.06546.x. [DOI] [PubMed] [Google Scholar]
- Kirchmaier AL, Sugden B. Dominant-negative inhibitors of EBNA-1 of Epstein-Barr virus. Journal of virology. 1997;71:1766–1775. doi: 10.1128/jvi.71.3.1766-1775.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knight JS, Sharma N, Robertson ES. Epstein-Barr virus latent antigen 3C can mediate the degradation of the retinoblastoma protein through an SCF cellular ubiquitin ligase. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:18562–18566. doi: 10.1073/pnas.0503886102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumthip K, Chusri P, Jilg N, Zhao L, Fusco DN, Zhao H, Goto K, Cheng D, Schaefer EA, Zhang L, et al. Hepatitis C virus NS5A disrupts STAT1 phosphorylation and suppresses type I interferon signaling. Journal of virology. 2012;86:8581–8591. doi: 10.1128/JVI.00533-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuppers R. The biology of Hodgkin's lymphoma. Nature reviews Cancer. 2009;9:15–27. doi: 10.1038/nrc2542. [DOI] [PubMed] [Google Scholar]
- Kutok JL, Wang F. Spectrum of Epstein-Barr virus-associated diseases. Annual review of pathology. 2006;1:375–404. doi: 10.1146/annurev.pathol.1.110304.100209. [DOI] [PubMed] [Google Scholar]
- Kwok RP, Laurance ME, Lundblad JR, Goldman PS, Shih H, Connor LM, Marriott SJ, Goodman RH. Control of cAMP-regulated enhancers by the viral transactivator Tax through CREB and the co-activator CBP. Nature. 1996;380:642–646. doi: 10.1038/380642a0. [DOI] [PubMed] [Google Scholar]
- Lan K, Verma SC, Murakami M, Bajaj B, Kaul R, Robertson ES. Kaposi's sarcoma herpesvirus-encoded latency-associated nuclear antigen stabilizes intracellular activated Notch by targeting the Sel10 protein. Proceedings of the National Academy of Sciences of the United States of America. 2007;104:16287–16292. doi: 10.1073/pnas.0703508104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langelier MF, Pascal JM. PARP-1 mechanism for coupling DNA damage detection to poly(ADP-ribose) synthesis. Current opinion in structural biology. 2013;23:134–143. doi: 10.1016/j.sbi.2013.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leung K, Nabel GJ. HTLV-1 transactivator induces interleukin-2 receptor expression through an NF-kappa B-like factor. Nature. 1988;333:776–778. doi: 10.1038/333776a0. [DOI] [PubMed] [Google Scholar]
- Ma Q, Cavallin LE, Leung HJ, Chiozzini C, Goldschmidt-Clermont PJ, Mesri EA. A role for virally induced reactive oxygen species in Kaposi's sarcoma herpesvirus tumorigenesis. Antioxidants & redox signaling. 2013;18:80–90. doi: 10.1089/ars.2012.4584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machida K, Chen CL, Liu JC, Kashiwabara C, Feldman D, French SW, Sher L, Hyeongnam JJ, Tsukamoto H. Cancer stem cells generated by alcohol, diabetes, and hepatitis C virus. Journal of gastroenterology and hepatology. 2012;27(Suppl 2):19–22. doi: 10.1111/j.1440-1746.2011.07010.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin-Vilchez S, Lara-Pezzi E, Trapero-Marugan M, Moreno-Otero R, Sanz- Cameno P. The molecular and pathophysiological implications of hepatitis B X antigen in chronic hepatitis B virus infection. Reviews in medical virology. 2011 doi: 10.1002/rmv.699. [DOI] [PubMed] [Google Scholar]
- Martinez-Pastor B, Cosentino C, Mostoslavsky R. A tale of metabolites: the cross-talk between chromatin and energy metabolism. Cancer discovery. 2013;3:497–501. doi: 10.1158/2159-8290.CD-13-0059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maruyama M, Shibuya H, Harada H, Hatakeyama M, Seiki M, Fujita T, Inoue J, Yoshida M, Taniguchi T. Evidence for aberrant activation of the interleukin-2 autocrine loop by HTLV-1-encoded p40x and T3/Ti complex triggering. Cell. 1987;48:343–350. doi: 10.1016/0092-8674(87)90437-5. [DOI] [PubMed] [Google Scholar]
- Matsuda Y, Ichida T. Impact of hepatitis B virus X protein on the DNA damage response during hepatocarcinogenesis. Medical molecular morphology. 2009;42:138–142. doi: 10.1007/s00795-009-0457-8. [DOI] [PubMed] [Google Scholar]
- Matsuoka M, Jeang KT. Human T-cell leukaemia virus type 1 (HTLV-1) infectivity and cellular transformation. Nature reviews Cancer. 2007;7:270–280. doi: 10.1038/nrc2111. [DOI] [PubMed] [Google Scholar]
- McCormick C, Ganem D. The kaposin B protein of KSHV activates the p38/MK2 pathway and stabilizes cytokine mRNAs. Science. 2005;307:739–741. doi: 10.1126/science.1105779. [DOI] [PubMed] [Google Scholar]
- McFadden K, Luftig MA. Interplay between DNA tumor viruses and the host DNA damage response. Current topics in microbiology and immunology. 2013;371:229–257. doi: 10.1007/978-3-642-37765-5_9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLaughlin-Drubin ME, Meyers J, Munger K. Cancer associated human papillomaviruses. Current opinion in virology. 2012;2:459–466. doi: 10.1016/j.coviro.2012.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merchant M, Caldwell RG, Longnecker R. The LMP2A ITAM is essential for providing B cells with development and survival signals in vivo. Journal of virology. 2000;74:9115–9124. doi: 10.1128/jvi.74.19.9115-9124.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mesri EA, Cesarman E, Boshoff C. Kaposi's sarcoma and its associated herpesvirus. Nature reviews Cancer. 2010;10:707–719. doi: 10.1038/nrc2888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montaner S, Sodhi A, Molinolo A, Bugge TH, Sawai ET, He Y, Li Y, Ray PE, Gutkind JS. Endothelial infection with KSHV genes in vivo reveals that vGPCR initiates Kaposi's sarcomagenesis and can promote the tumorigenic potential of viral latent genes. Cancer cell. 2003;3:23–36. doi: 10.1016/s1535-6108(02)00237-4. [DOI] [PubMed] [Google Scholar]
- Montaner S, Sodhi A, Ramsdell AK, Martin D, Hu J, Sawai ET, Gutkind JS. The Kaposi's sarcoma-associated herpesvirus G protein-coupled receptor as a therapeutic target for the treatment of Kaposi's sarcoma. Cancer research. 2006;66:168–174. doi: 10.1158/0008-5472.CAN-05-1026. [DOI] [PubMed] [Google Scholar]
- Moody CA, Laimins LA. Human papillomaviruses activate the ATM DNA damage pathway for viral genome amplification upon differentiation. PLoS pathogens. 2009;5:e1000605. doi: 10.1371/journal.ppat.1000605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moody CA, Laimins LA. Human papillomavirus oncoproteins: pathways to transformation. Nature reviews Cancer. 2010;10:550–560. doi: 10.1038/nrc2886. [DOI] [PubMed] [Google Scholar]
- Moore PS, Chang Y. Why do viruses cause cancer? Highlights of the first century of human tumour virology. Nature reviews Cancer. 2010;10:878–889. doi: 10.1038/nrc2961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mosialos G, Birkenbach M, Yalamanchili R, VanArsdale T, Ware C, Kieff E. The Epstein-Barr virus transforming protein LMP1 engages signaling proteins for the tumor necrosis factor receptor family. Cell. 1995;80:389–399. doi: 10.1016/0092-8674(95)90489-1. [DOI] [PubMed] [Google Scholar]
- Munger K, Hayakawa H, Nguyen CL, Melquiot NV, Duensing A, Duensing S. Viral carcinogenesis and genomic instability. Exs. 2006:179–199. doi: 10.1007/3-7643-7378-4_8. [DOI] [PubMed] [Google Scholar]
- Mutlu AD, Cavallin LE, Vincent L, Chiozzini C, Eroles P, Duran EM, Asgari Z, Hooper AT, La Perle KM, Hilsher C, et al. In vivo-restricted and reversible malignancy induced by human herpesvirus-8 KSHV: a cell and animal model of virally induced Kaposi's sarcoma. Cancer cell. 2007;11:245–258. doi: 10.1016/j.ccr.2007.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myoung J, Ganem D. Active lytic infection of human primary tonsillar B cells by KSHV and its noncytolytic control by activated CD4+ T cells. The Journal of clinical investigation. 2011;121:1130–1140. doi: 10.1172/JCI43755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura H, Li M, Zarycki J, Jung JU. Inhibition of p53 tumor suppressor by viral interferon regulatory factor. Journal of virology. 2001;75:7572–7582. doi: 10.1128/JVI.75.16.7572-7582.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicholas J, Ruvolo VR, Burns WH, Sandford G, Wan X, Ciufo D, Hendrickson SB, Guo HG, Hayward GS, Reitz MS. Kaposi's sarcoma-associated human herpesvirus-8 encodes homologues of macrophage inflammatory protein-1 and interleukin-6. Nat Med. 1997;3:287–292. doi: 10.1038/nm0397-287. [DOI] [PubMed] [Google Scholar]
- Offermann MK. Kaposi sarcoma herpesvirus-encoded interferon regulator factors. Current topics in microbiology and immunology. 2007;312:185–209. doi: 10.1007/978-3-540-34344-8_7. [DOI] [PubMed] [Google Scholar]
- Portis T, Longnecker R. Epstein-Barr virus (EBV) LMP2A mediates B-lymphocyte survival through constitutive activation of the Ras/PI3K/Akt pathway. Oncogene. 2004;23:8619–8628. doi: 10.1038/sj.onc.1207905. [DOI] [PubMed] [Google Scholar]
- Raab-Traub N. Epstein-Barr virus in the pathogenesis of NPC. Seminars in cancer biology. 2002;12:431–441. doi: 10.1016/s1044579x0200086x. [DOI] [PubMed] [Google Scholar]
- Radkov SA, Kellam P, Boshoff C. The latent nuclear antigen of Kaposi sarcoma-associated herpesvirus targets the retinoblastoma-E2F pathway and with the oncogene Hras transforms primary rat cells. Nature Medicine. 2000;6:1121–1127. doi: 10.1038/80459. [DOI] [PubMed] [Google Scholar]
- Ray RB, Meyer K, Ray R. Hepatitis C virus core protein promotes immortalization of primary human hepatocytes. Virology. 2000;271:197–204. doi: 10.1006/viro.2000.0295. [DOI] [PubMed] [Google Scholar]
- Rehermann B. Hepatitis C virus versus innate and adaptive immune responses: a tale of coevolution and coexistence. The Journal of clinical investigation. 2009;119:1745–1754. doi: 10.1172/JCI39133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rochford R, Cannon MJ, Moormann AM. Endemic Burkitt's lymphoma: a polymicrobial disease? Nature reviews Microbiology. 2005;3:182–187. doi: 10.1038/nrmicro1089. [DOI] [PubMed] [Google Scholar]
- Roman A, Munger K. The papillomavirus E7 proteins. Virology. 2013;445:138–168. doi: 10.1016/j.virol.2013.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samols MA, Skalsky RL, Maldonado AM, Riva A, Lopez MC, Baker HV, Renne R. Identification of cellular genes targeted by KSHV-encoded microRNAs. PLoS pathogens. 2007;3:e65. doi: 10.1371/journal.ppat.0030065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarek G, Jarviluoma A, Ojala PM. KSHV viral cyclin inactivates p27KIP1 through Ser10 and Thr187 phosphorylation in proliferating primary effusion lymphomas. Blood. 2006;107:725–732. doi: 10.1182/blood-2005-06-2534. [DOI] [PubMed] [Google Scholar]
- Sato H, Oka T, Shinnou Y, Kondo T, Washio K, Takano M, Takata K, Morito T, Huang X, Tamura M, et al. Multi-step aberrant CpG island hyper-methylation is associated with the progression of adult T-cell leukemia/lymphoma. The American journal of pathology. 2010;176:402–415. doi: 10.2353/ajpath.2010.090236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satou Y, Yasunaga J, Yoshida M, Matsuoka M. HTLV-I basic leucine zipper factor gene mRNA supports proliferation of adult T cell leukemia cells. Proceedings of the National Academy of Sciences of the United States of America. 2006;103:720–725. doi: 10.1073/pnas.0507631103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schiller JT, Lowy DR. Vaccines to prevent infections by oncoviruses. Annual review of microbiology. 2010;64:23–41. doi: 10.1146/annurev.micro.112408.134019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shembade N, Harhaj NS, Yamamoto M, Akira S, Harhaj EW. The human T-cell leukemia virus type 1 Tax oncoprotein requires the ubiquitin-conjugating enzyme Ubc13 for NF-kappaB activation. Journal of virology. 2007;81:13735–13742. doi: 10.1128/JVI.01790-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin YC, Nakamura H, Liang X, Feng P, Chang H, Kowalik TF, Jung JU. Inhibition of the ATM/p53 signal transduction pathway by Kaposi's sarcomaassociated herpesvirus interferon regulatory factor 1. Journal of virology. 2006;80:2257–2266. doi: 10.1128/JVI.80.5.2257-2266.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Si H, Robertson ES. Kaposi's sarcoma-associated herpesvirus-encoded latency-associated nuclear antigen induces chromosomal instability through inhibition of p53 function. Journal of virology. 2006;80:697–709. doi: 10.1128/JVI.80.2.697-709.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sodhi A, Chaisuparat R, Hu J, Ramsdell AK, Manning BD, Sausville EA, Sawai ET, Molinolo A, Gutkind JS, Montaner S. The TSC2/mTOR pathway drives endothelial cell transformation induced by the Kaposi's sarcoma-associated herpesvirus G protein-coupled receptor. Cancer cell. 2006;10:133–143. doi: 10.1016/j.ccr.2006.05.026. [DOI] [PubMed] [Google Scholar]
- Sodhi A, Montaner S, Patel V, Zohar M, Bais C, Mesri EA, Gutkind JS. The Kaposi's sarcoma-associated herpes virus G protein-coupled receptor upregulates vascular endothelial growth factor expression and secretion through mitogen-activated protein kinase and p38 pathways acting on hypoxia-inducible factor 1alpha. Cancer research. 2000;60:4873–4880. [PubMed] [Google Scholar]
- Spangle JM, Munger K. The HPV16 E6 oncoprotein causes prolonged receptor protein tyrosine kinase signaling and enhances internalization of phosphorylated receptor species. PLoS pathogens. 2013;9:e1003237. doi: 10.1371/journal.ppat.1003237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srisuttee R, Koh SS, Kim SJ, Malilas W, Boonying W, Cho IR, Jhun BH, Ito M, Horio Y, Seto E, et al. Hepatitis B virus X (HBX) protein upregulates beta-catenin in a human hepatic cell line by sequestering SIRT1 deacetylase. Oncology reports. 2012;28:276–282. doi: 10.3892/or.2012.1798. [DOI] [PubMed] [Google Scholar]
- Stine JT, Wood C, Hill M, Epp A, Raport CJ, Schweickart VL, Endo Y, Sasaki T, Simmons G, Boshoff C, et al. KSHV-encoded CC chemokine vMIP-III is a CCR4 agonist, stimulates angiogenesis, and selectively chemoattracts TH2 cells. Blood. 2000;95:1151–1157. [PubMed] [Google Scholar]
- Swanton C, Mann DJ, Fleckenstein B, Neipel F, Peters G, Jones N. Herpes viral cyclin/Cdk6 complexes evade inhibition by CDK inhibitor proteins. Nature. 1997;390:184–187. doi: 10.1038/36606. [DOI] [PubMed] [Google Scholar]
- Thompson MP, Kurzrock R. Epstein-Barr virus and cancer. Clinical cancer research : an official journal of the American Association for Cancer Research. 2004;10:803–821. doi: 10.1158/1078-0432.ccr-0670-3. [DOI] [PubMed] [Google Scholar]
- Thorley-Lawson DA. Epstein-Barr virus: exploiting the immune system. Nature reviews Immunology. 2001;1:75–82. doi: 10.1038/35095584. [DOI] [PubMed] [Google Scholar]
- Tian Y, Yang W, Song J, Wu Y, Ni B. Hepatitis B virus X protein-induced aberrant epigenetic modifications contributing to human hepatocellular carcinoma pathogenesis. Molecular and cellular biology. 2013;33:2810–2816. doi: 10.1128/MCB.00205-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsimbouri P, Drotar ME, Coy JL, Wilson JB. bcl-xL and RAG genes are induced and the response to IL-2 enhanced in EmuEBNA-1 transgenic mouse lymphocytes. Oncogene. 2002;21:5182–5187. doi: 10.1038/sj.onc.1205490. [DOI] [PubMed] [Google Scholar]
- Uchida T, Kinoshita T, Murate T, Saito H, Hotta T. CDKN2 (MTS1/P16(INK4A)) GENE ALTERATIONS IN ADULT T-CELL LEUKEMIA/LYMPHOMA [Review] Leukemia & lymphoma. 1998;29:27–35. doi: 10.3109/10428199809058379. [DOI] [PubMed] [Google Scholar]
- Vande Pol SB, Klingelhutz AJ. Papillomavirus E6 oncoproteins. Virology. 2013;445:115–137. doi: 10.1016/j.virol.2013.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vereide DT, Seto E, Chiu YF, Hayes M, Tagawa T, Grundhoff A, Hammerschmidt W, Sugden B. Epstein-Barr virus maintains lymphomas via its miRNAs. Oncogene. 2013 doi: 10.1038/onc.2013.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verma SC, Borah S, Robertson ES. Latency-associated nuclear antigen of Kaposi's sarcoma-associated herpesvirus up-regulates transcription of human telomerase reverse transcriptase promoter through interaction with transcription factor Sp1. Journal of virology. 2004;78:10348–10359. doi: 10.1128/JVI.78.19.10348-10359.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogelstein B, Kinzler KW. The multistep nature of cancer. Trends in genetics : TIG. 1993;9:138–141. doi: 10.1016/0168-9525(93)90209-z. [DOI] [PubMed] [Google Scholar]
- Vrancken K, Paeshuyse J, Liekens S. Angiogenic activity of hepatitis B and C viruses. Antiviral chemistry & chemotherapy. 2012;22:159–170. doi: 10.3851/IMP1987. [DOI] [PubMed] [Google Scholar]
- Wang L, Dittmer DP, Tomlinson CC, Fakhari FD, Damania B. Immortalization of primary endothelial cells by the K1 protein of Kaposi's sarcomaassociated herpesvirus. Cancer research. 2006;66:3658–3666. doi: 10.1158/0008-5472.CAN-05-3680. [DOI] [PubMed] [Google Scholar]
- Wang L, Wakisaka N, Tomlinson CC, DeWire SM, Krall S, Pagano JS, Damania B. The Kaposi's sarcoma-associated herpesvirus (KSHV/HHV-8) K1 protein induces expression of angiogenic and invasion factors. Cancer research. 2004;64:2774–2781. doi: 10.1158/0008-5472.can-03-3653. [DOI] [PubMed] [Google Scholar]
- Wang WL, London WT, Lega L, Feitelson MA. HBxAg in the liver from carrier patients with chronic hepatitis and cirrhosis. Hepatology. 1991;14:29–37. doi: 10.1002/hep.1840140106. [DOI] [PubMed] [Google Scholar]
- Xie B, Xing R, Chen P, Gou Y, Li S, Xiao J, Dong J. Downregulation of c-Met expression inhibits human HCC cells growth and invasion by RNA interference. The Journal of surgical research. 2010;162:231–238. doi: 10.1016/j.jss.2009.04.030. [DOI] [PubMed] [Google Scholar]
- Yip WK, Cheng AS, Zhu R, Lung RW, Tsang DP, Lau SS, Chen Y, Sung JG, Lai PB, Ng EK, et al. Carboxyl-terminal truncated HBx regulates a distinct microRNA transcription program in hepatocellular carcinoma development. PloS one. 2011;6:e22888. doi: 10.1371/journal.pone.0022888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo YG, Oh SH, Park ES, Cho H, Lee N, Park H, Kim DK, Yu DY, Seong JK, Lee MO. Hepatitis B virus X protein enhances transcriptional activity of hypoxia-inducible factor-1alpha through activation of mitogen-activated protein kinase pathway. The Journal of biological chemistry. 2003;278:39076–39084. doi: 10.1074/jbc.M305101200. [DOI] [PubMed] [Google Scholar]
- Yoshida M. Multiple viral strategies of HTLV-1 for dysregulation of cell growth control. Annual review of immunology. 2001;19:475–496. doi: 10.1146/annurev.immunol.19.1.475. [DOI] [PubMed] [Google Scholar]
- Young LS, Rickinson AB. Epstein-Barr virus: 40 years on. Nature reviews Cancer. 2004;4:757–768. doi: 10.1038/nrc1452. [DOI] [PubMed] [Google Scholar]
- Zane L, Yasunaga J, Mitagami Y, Yedavalli V, Tang SW, Chen CY, Ratner L, Lu X, Jeang KT. Wip1 and p53 contribute to HTLV-1 Tax-induced tumorigenesis. Retrovirology. 2012;9:114. doi: 10.1186/1742-4690-9-114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zemel R, Issachar A, Tur-Kaspa R. The role of oncogenic viruses in the pathogenesis of hepatocellular carcinoma. Clinics in liver disease. 2011;15:261–279. doi: 10.1016/j.cld.2011.03.001. vii-x. [DOI] [PubMed] [Google Scholar]
- Zhao P, Han T, Guo JJ, Zhu SL, Wang J, Ao F, Jing MZ, She YL, Wu ZH, Ye LB. HCV NS4B induces apoptosis through the mitochondrial death pathway. Virus research. 2012;169:1–7. doi: 10.1016/j.virusres.2012.04.006. [DOI] [PubMed] [Google Scholar]
- Zhu Y, Haecker I, Yang Y, Gao SJ, Renne R. gamma-Herpesvirus-encoded miRNAs and their roles in viral biology and pathogenesis. Current opinion in virology. 2013;3:266–275. doi: 10.1016/j.coviro.2013.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- zur Hausen H. The search for infectious causes of human cancers: where and why (Nobel lecture) Angew Chem Int Ed Engl. 2009;48:5798–5808. doi: 10.1002/anie.200901917. [DOI] [PubMed] [Google Scholar]
- Zwerschke W, Mazurek S, Massimi P, Banks L, Eigenbrodt E, Jansen-Durr P. Modulation of type M2 pyruvate kinase activity by the human papillomavirus type 16 E7 oncoprotein. Proceedings of the National Academy of Sciences of the United States of America. 1999;96:1291–1296. doi: 10.1073/pnas.96.4.1291. [DOI] [PMC free article] [PubMed] [Google Scholar]