Abstract
Background:
Patients with UICC/AJCC stage II colon cancer have a high 5-year overall survival rate after surgery. Nevertheless, a significant subgroup of patients develops tumour recurrence. Currently, there are no clinically established biomarkers available to identify this patient group. We applied reverse-phase protein arrays (RPPA) for phosphatidylinositide-3-kinase pathway activation mapping to stratify patients according to their risk of tumour recurrence after surgery.
Methods:
Full-length proteins were extracted from formalin-fixed, paraffin-embedded tissue samples of 118 patients who underwent curative resection. RPPA technology was used to analyse expression and/or phosphorylation levels of six major factors of the phosphatidylinositide-3-kinase pathway. Oncogenic mutations of KRAS and BRAF, and DNA microsatellite status, currently discussed as prognostic markers, were analysed in parallel.
Results:
Expression of phospho-AKT (HR=3.52; P=0.032), S6RP (HR=6.3; P=0.044), and phospho-4E-BP1 (HR=4.12; P=0.011) were prognostic factors for disease-free survival. None of the molecular genetic alterations were significantly associated with prognosis.
Conclusions:
Our data indicate that activation of the PI3K/AKT pathway evidenced on the protein level might be a valuable prognostic marker to stratify patients for their risk of tumour recurrence. Beside adjuvant chemotherapy targeting of upregulated PI3K/AKT signalling may be an attractive strategy for treatment of high-risk patients.
Keywords: colon cancer, biomarker, reverse-phase protein array, PI3K, AKT, prognosis, survival
Colon cancer is one of the most common malignancies in the western world with more than one million new cases each year and a disease-specific mortality rate of about 33% (Ferlay et al, 2010). Although death rates are decreasing in the most developed countries owing to screening programs and therapy, colon cancer still holds the third place in cancer death statistics for women and the second for men (Jemal et al, 2009; Siegel et al, 2013). About 40% of colon carcinomas are diagnosed in TNM stage II (T3-4N0M0), according to the global UICC/AJCC standard (International Union Against Cancer /American Joint Committee on Cancer). This group of patients has a high 5-year overall survival rate of 80% and is mainly treated solely by surgery without further adjuvant treatment, whereas patients with UICC stage III or IV receive adjuvant chemotherapy (Catenacci et al, 2011). Nevertheless, a subgroup of patients with stage II carcinomas (about 20%) develops tumour recurrence and has dismal prognosis. For those patients the currently used staging fails to predict the prognosis of the patients (Mutch, 2007). Moreover, the latest edition of the TNM staging system has shown to be of limited clinical use (Nitsche et al, 2011).
Adjuvant treatment of stage II patients is currently recommended on the basis of clinical risk parameters, as described in ASCO15 or ESMO16 guidelines (e.g., poor differentiation, T4 stage, tumour perforation, inadequate lymphadenectomy) (Benson et al, 2004; Van Cutsem and Oliveira, 2009).
Therefore, evidence-based biomarkers are eagerly awaited for risk stratification in colorectal cancer. Knowledge about the molecular biology and pathogenesis of colon cancer has increased over the last years and important developments were made towards the discovery of new prognostic and predictive markers. We have shown previously that integrative analysis of molecular genetic markers, or analysis of an expression signature (ColoPrint), can improve risk stratification for stage II colon cancer patients, outperforming clinical parameters (Nitsche et al, 2011; Maak et al, 2013).
The phosphatidylinositide-3-kinase (PI3K)/proteinkinase B (Akt) pathway (Supplementary Figure 1) is known to have an important role in the development and progression of many solid cancers (Dozmorov et al, 2010; Song et al, 2011; Jiao and Nan, 2012; Vredeveld et al, 2012; Wang et al, 2012; Christgen et al, 2013). As this pathway is important for proliferation and survival of cells, it might well be involved in a more aggressive behaviour and higher metastatic capacity of colon carcinomas. Thus, analysis of activation of this pathway might provide potential biomarkers associated with dismal prognosis. In addition, recent studies of PI3K/mammalian target of rapamycin (mTOR) inhibitors for cancer treatment show promising results in pancreatic cancer, melanomas and B-cell malignancies (Britten, 2013; Roschewski et al, 2013; Wolin, 2013). These findings give rise to the hypothesis that this pathway, besides providing potential prognostic factors, could also reveal additional novel targets for colon cancer therapy. This is of special interest as the benefit of classical chemotherapy for stage II colon cancer patients has not been proven univocally so far.
The PI3K/Akt pathway is one of the major signalling cascades in colon cancers downstream of EGFR (Efferth, 2012). Its activation can has been reported in 60%–70% of colorectal cancers and inhibitors of pathway components are suggested as therapeutic agents by a number of studies. A recent review by Pandurangan (2013) summarises the contemporary developments in this field. Whereas most currently discussed prognostic and predictive markers for stage II colon cancer are based on analysis of nucleic acids, for example, by expression analysis or mutation detection, protein activity-based assays offer the advantage of gaining deeper insights in the actual cellular activities within a cancer. To address protein expression, as well as activation by post-translational modification we utilised reverse-phase protein arrays (RPPA). This technology enables the analysis of numerous proteins in a multitude of samples under standardised experimental conditions (Paweletz et al, 2001; Grubb et al, 2003). With this approach, samples can be analysed in serial dilutions, thus enabling reproducible quantitative detection of protein expression in clinical samples. The generation of technical replicates additionally contributes to assay reliability. RPPA is feasible for the analysis of cryopreserved clinical samples (Espina et al, 2003; Liotta et al, 2003; Wulfkuhle et al, 2006). Moreover, it can be applied for formalin-fixed and paraffin-embedded (FFPE) patient samples, a prerequisite for clinical routine application (Berg et al, 2011; Wolff et al, 2011; Guo et al, 2012). Thus, RPPA is an adequate tool to identify the expression of potential biomarkers within patient samples.
The aim of this study was (1) to analyse the expression of major components of the PI3K/AKT pathway, notably PI3K, Akt, glycogen synthase kinase 3 (GSK3-beta), mTOR, S6 ribosomal protein (S6RP), 4E-BP1, and their respective phosphorylated forms to map activation of the PI3K/Akt pathway in stage II colon carcinomas, and (2) to correlate expression and activation of functional PI3K/Akt pathway components with clinical and molecular parameters, such as tumour size, anatomical location, microsatellite instability (MSI) status, mutational status (BRAF, KRAS) of the tumour and post-operative survival of the patients.
Materials and methods
Tissue samples
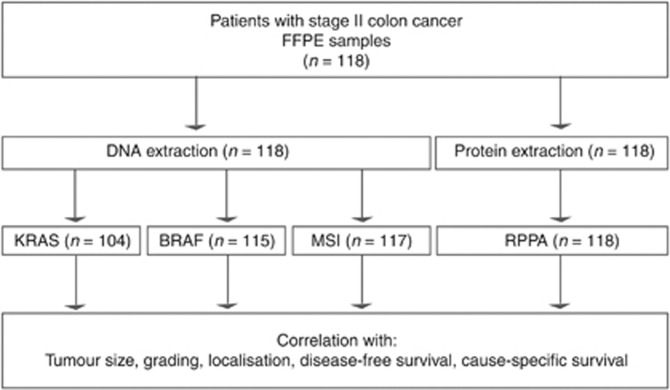
Tissue samples derived from 118 patients with stage II primary colon carcinomas, who underwent curative surgery (R0) at the Department of Surgery, Klinikum rechts der Isar, Munich from 1990 to 2010 (Figure 1), were analysed. For 71 patients adjacent normal tissue was also investigated for protein analysis. None of the patients in this study received adjuvant or neoadjuvant therapy. Formalin-fixed paraffin-embedded samples were used. All samples showed a tumour cellularity of >70%. Patient tumour characteristics are summarised in Table 1. Disease recurrence was observed in 11 patients (9.3%) after a median follow-up of 8.3 years. For 16 patients no data on disease-free survival was available. Cause-specific death occurred in 10 cases (8.5%) after a median follow-up of 8.1 years. The study was approved by the ethics committee of the Klinikum rechts der Isar (study no. 1926/2007). Informed written consent of the patients had been obtained before conducting the study.
Figure 1.
Analyses of the patient samples.
Table 1. Clinicopathological characteristics of the patients.
| Category | Subcategory | Total (%) |
|---|---|---|
| Total |
|
118 (100%) |
| Sex | Male | 67 (57) |
| |
Female |
51 (43) |
| Age |
|
65 (median) 31–90 (range) |
| Localisationa | Right sided | 56 (48) |
| |
Left sided |
61 (52) |
| Histological subtype | Classical adenocarcinoma | 111 (94) |
| |
Mucinous |
7 (6) |
| pT | T3 | 99 (84) |
| |
T4 |
19 (16) |
| pNb |
N0 |
118 (100) |
| cM |
M0 |
118 (100) |
| Grading | G1, G2 | 84 (71) |
| |
G3 |
34 (29) |
| Lymphatic invasion | Not found (L0) | 105 (89) |
| |
Present (L1) |
13 (11) |
| Venous invasion | Not found (V0) | 115 (97) |
| |
Present (V1) |
3 (3) |
| Recurrence | None | 107 (91) |
| Distant | 11 (9) | |
| |
Local |
0 (0) |
| Survival information | Alive | 92 (79) |
| Tumour-related death | 11 (9) | |
| Non-tumour-related death | 15 (12) |
Correlations between the clinicopathological factors were analysed and no confounding factors were identified.
One patient had tumours in both sides of the colon.
A median of 21 lymph nodes per patient was analysed (range 10–72).
DNA isolation
DNA was isolated from manually dissected tumour areas from 5 μm sections of FFPE tissues with a tumour cell content of at least 70%. The QIAamp DNAMini Kit (Qiagen, Hilden, Germany) was used according to the manufacturer's protocol. DNA concentration and quality was checked with an ND-1000 NanoDrop Spectrophotometer (Thermo Fisher, Schwerte, Germany).
Microsatellite instability determination
Microsatellite instability was tested with the MSI Analysis System, Version 1.2 (Promega, Mannheim, Germany) for 83 patients. This assay co-amplifies the five mononucleotide repeat markers BAT-25, BAT-26, NR-21, NR-24, and MONO-27 to determine the MSI status. Thirty-five cases were analysed with the Bethesda panel (BAT25, BAT26, D2S123, D5S346, D17S250) using the Type-it Microsatellite PCR kit (Qiagen). MSI was defined when at least two of the five markers tested showed instability. The results of these assays have been previously compared and proven highly sensitive for MSI determination (Murphy et al, 2006).
Overall, the MSI status was determined for 117 patients. For one patient the assay result was not conclusive.
BRAF
For 83 patients mutational status of the oncogene BRAF (V600E, GTG>GAG substitution in exon 15) was assessed by high-resolution melting analysis of genomic DNA on a Lightcycler 480 II platform (Roche, Mannheim, Germany; SYBR Green I/HRM Dye Protocol), in a modification of published protocols (Pichler et al, 2009). Briefly, 20 ng of genomic DNA (10 ng μl–1) were amplified in total volume of 20 μl with 10 μl high-resolution master mix, 2.4 mM MgCl2, and 0.25 mM each of oligonucleotide primers, 2 μl template DNA and 5.2 μl dH2O. Primer sequences were BRAF Exon 15 For: 5′-GGTGATTTTGGTCTAGCTACAG-3′, BRAF Exon 15 Rev: 5′-AGTAACTCAGCAGCATCTCAGG-3′. After pre-incubation (95 °C, 10 min), amplification of a 147-bp product was carried out in 42 cycles (95 °C for 15 s/61 °C for 15 s/72 °C for 15 s), followed by melting point analysis with an initial phase: 95 °C, 5 s, and 72 °C, 90 s, followed by a melting profile ranging from 72 °C to 95 °C in 19.2 min. As a positive control, genomic DNA from the BRAF-mutated colon cancer cell line HT29 was used.
Mutational status of 35 tumours was determined using high-resolution melting curve analysis and direct sequencing of aberrant cases as described by (Bettstetter et al, 2013). As a negative control, DNA isolated from a colon carcinoma, which had been demonstrated to be wild type by direct sequencing, was used.
KRAS
For 83 patients, the mutational status of the genes KRAS (codon 12 and 13 in exon 2) was analysed by high-resolution melting analysis of genomic DNA on a LightCycler 480 II platform (Roche; SYBR Green I/HRM Dye Protocol) as described previously in detail (Nitsche et al, 2012). Analysis of genomic DNA from colon cancer cell lines with and without KRAS and BRAF mutations was performed in each run as control.
KRAS mutation analysis of exon 2 was performed for 35 cases by direct sequencing of PCR products using cycle sequencing with fluorescent-labelled dye terminators and separation with an automated sequencing system (Genetic Analyzer 2100, Applied Biosystems, Darmstadt, Germany) as previously described (Langer et al, 2011).
For both methods a high sensitivity and a high concordance for the detection of somatic mutations in tumours has been reported (Gonzalez-Bosquet et al, 2011).
Protein extraction
Protein extraction was performed as previously described (Wolff et al, 2011). Briefly, 10 μm FFPE tissue sections were deparaffinised and proteins were extracted using EXB Plus (Qiagen). Tissue areas of ∼0.25 cm2 from three 10-μm-thick sections were processed in 100 μl of extraction buffer. The Bradford protein assay (Bio-Rad, Hercules, FL, USA) was used according to the manufacturer's instructions to determine protein concentrations. A western blot probing for beta-actin was performed from randomly selected lysates (n=12) to demonstrate successful protein extraction and suitability for RPPA analysis. All protein lysates tested produced a clear beta-actin band on the western blot.
Analysis of protein expression by reverse-phase protein arrays (RPPA)
Expression of 36 proteins including 15 phosphorylated proteins was determined by RPPA. Antibodies and experimental conditions are summarised in Supplementary Table 1. RPPAs were generated using the SpotBot Extreme microarrayer according to the manufacturer's instructions (Arrayit, Sunnyvale, CA, USA). For every lysate and every dilution (undiluted, 1 : 2, 1 : 4, 1 : 8, 1 : 16 buffer), two replicates were applied onto a nitrocellulose-coated glass slide (Grace Bio-Labs, Bend, OR, USA), which produced 12 data points per sample.
Immunodetection was performed similar to western blot analysis and as previously described (Wolff et al, 2011). For estimation of total protein amounts, arrays were stained in parallel with Sypro Ruby Protein Blot Stain (Molecular Probes, Eugene, OR, USA) according to the manufacturer's instructions. All specific signals were normalised to the total protein level detected by Sypro Rubystaining. Further details of the RPPA methodology and its validation have been previously described by Wolff et al (2011). All antibodies used in this study were validated for specificity by western blot analysis. The reproducibility of the method was assessed previously. There was a high reproducibility of protein expression from independent extraction procedures (n=10 samples, 3 replicates per sample) with a coefficient of variation ⩽14% for the four exemplary proteins HER2, pHER2, uPA and PAI-1. All pair-wise correlations showed a Spearman's ρ⩾0.98 (Malinowsky et al, 2012). All antibodies and conditions of use are specified in Supplementary Table 1.
Statistical analysis
IBM SPSS 20.0 Statistics statistical software (SPSS Inc., Chicago, IL, USA) and R Software version 2.11.1 (R Foundation for Statistical Computing, Vienna, Austria) were used for statistical analysis. Associations between protein expression and pathological features were given in crosstabs and were evaluated using the χ2-test and the Mann–Whitney U-test.
In order to derive optimal cutoff values of the analysed phosphorylated and non-phosphorylated proteins for recurrence risk stratification, maximally selected log-rank statistics were used. To consider multiple test issue within these analyses, the R-function ‘maxstat.test' was employed (Hothorn and Zeileis, 2008). Survival analysis was performed using Kaplan–Meier estimates, log-rank tests and Cox's proportional hazards regression analysis. All tests were two-sided, and the significance level was set at 5%.
Positive predictive values (PPV) for the factors analysed were calculated as number of patient samples correctly classified as progressive tumours divided by all patient samples classified as progressive (correctly or incorrectly).
The study was performed following the REMARK criteria (McShane et al, 2005). As the study was completely retrospective no data on treatments or patient flow is included.
Results
We determined levels of total cellular proteins and phosphorylated forms of Akt, GSK3-beta, mTOR, PI3K, S6RP, 4E-BP1 and in a cohort of 118 stage II colon cancer patients using RPPA, for 71 patients adjacent normal tissue was also analysed for protein expression. Relative expression units were calculated for each protein using MicroVigene software (VigeneTech, Carlisle, MA, USA). Supplementary Figure 2 depicts an exemplary stained slide and the dilution curves for two exemplary tumour samples, one expressing the protein at low levels and the other with high expression of the protein.
Analysis of protein expression and phosphorylation in tumours and adjacent normal tissues
The phosphorylated forms of all markers correlated significantly with each other indicating a possible coregulation of the pathway. Only the correlation between phospho-PI3K and phospho-S6RP was weak with a correlation coefficient of 0.298. Correlation coefficients are summarised in Supplementary Table 2.
Mean expression levels built over all samples did not differ much between tumours and normal tissue in the 71 cases comprising both sample types. In single patients, however, significant changes between normal and tumour samples were found. Expression differences between tumours and adjacent normal tissue larger than 1.5-fold were considered significant. Interestingly, high inter-patient variability was found for expression levels in tumours as well as in normal samples indicating high biological diversity. Changes between normal tissues and tumours are also patient specific. Although some patients show similar expression levels in tumours and adjacent normal tissue, expression levels vary largely in others. Results of the comparison between tumours and normal tissue are summarised in Supplementary Table 3.
Cutoff derivation of functional PI3K/Akt pathway components for distant recurrence
The best discriminating cutoff values were determined at 204 relative expression units (REU) for Akt, 104 REU for phospho-Akt, 151 REU for GSK3-beta, 157 REU for mTOR, 114 REU for phospho-mTOR, 114 REU for PI3K, 152 REU for phospho-PI3K, 329 REU for S6RP, 107 REU for pS6RP, 291 REU for 4E-BP1 and 121 REU for phospho-4E-BP1. Patients were classified into high- and low-expression groups for each factor according to the respective cutoff value. All cutoff values and the respective patient deviations are depicted in Table 2. Sixty-three per cent of all patients were classified Akt positive and 35% phospho-Akt positive. Eighty-three per cent fell in the high-expression group for GSK3-beta. For mTOR and phospho-mTOR 32% and 76% of all patients were classified as positive, respectively. Eighty-one per cent and 53% of tumours expressed high levels of PI3K and phospho-PI3K. S6RP and phospho-S6RP levels were elevated in 62% and 39% of all patients and expression of 4E-BP1 and phospho-4E-BP1 were above the cutoff values in 18% and 27% of all patients analysed.
Table 2. Discriminative cutoff values in relative expression units (REU) for the functional pathway components.
| |
|
Patient deviation (%) |
|
|---|---|---|---|
| Protein | Cutoff value | Negative | Positive |
| Akt | 204 | 44 (37) | 74 (63) |
| Phospho-Akt | 104 | 77 (65) | 41 (35) |
| GSK-3beta | 151 | 20 (17) | 98 (83) |
| mTOR | 157 | 80 (68) | 38 (32) |
| Phospho-mTOR | 114 | 28 (24) | 90 (76) |
| PI3K | 114 | 23 (19) | 95 (81) |
| Phospho-PI3K | 152 | 56 (47) | 62 (53) |
| S6RP | 329 | 45 (38) | 73 (62) |
| Phospho-S6RP | 107 | 72 (61) | 46 (39) |
| 4E-BP1 | 291 | 97 (82) | 21 (18) |
| Phospho-4E-BP1 | 121 | 86 (73) | 32 (27) |
Abbreviations: mTOR=mammalian target of rapamycin; S6RP=S6 ribosomal protein.
Correlation of functional factors of the PI3K/Akt-signalling pathway with disease-free and cause-specific survival
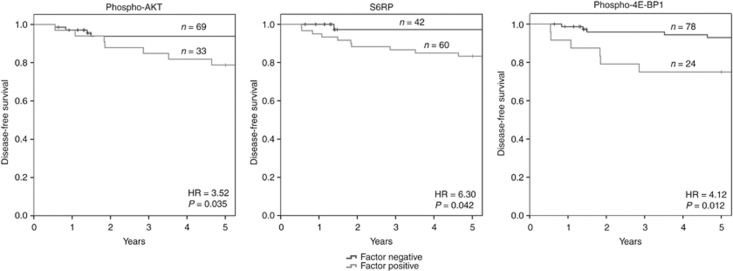
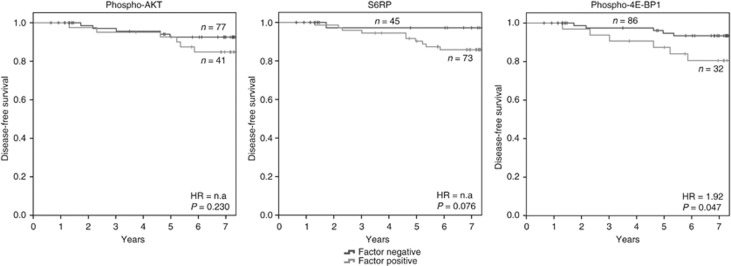
Patients with low phospho-Akt, S6RP or phospho-4E-BP1 had significantly longer disease-free survival times than patients with high expression of one of these factors (P=0.032, 0.046 and 0.011, respectively) (Figure 2). For cause-specific survival, only phospho-4E-BP1 expression showed a significant negative correlation with survival times (P=0.045). None of the other proteins (AKT, GSK3-beta, mTOR, PI3K, 4E-BP1) or phospho-proteins (phospho-mTOR, phospho-PI3K, pS6PR) reached statistical significance in this setting (Figure 3).
Figure 2.
Correlation of phospho-Akt, S6RP and phospho-4E-BP1 with disease-free survival. For phospho-Akt, 33 patients showed expression levels above the optimal cutoff of 104 and 69 patients had expression levels below this value. Survival rates were significantly distinct for both groups with P=0.032. For S6RP, 60 patients showed high expression levels >329 and 42 patients had low expression levels. Survival differed significantly in both groups (P=0.044). Twenty-four patients showed expression of phospho-4E-BP1 above the optimal cutoff value of 121 and 78 patients yielded lower protein levels. Correlation of phospho-4E-BP1 expression with disease-free survival was highly significant (P=0.012). For 16 patients no information on disease-free survival was available.
Figure 3.
Correlation of phospho-Akt, S6RP and phospho-4E-BP1 with cause-specific survival. For phospho-Akt, 77 patients showed expression levels above the optimal cutoff of 104 and 41 patients had expression of the factor below this value. For S6RP, 73 patients showed high expression levels >329 and 45 patients had low expression levels. Neither of the two factors correlated significantly with cause-specific survival (P=0.230 and 0.076, respectively). Thirty-two patients showed expression of phospho-4E-BP1 above the optimal cutoff value of 121 and 86 patients yielded lower protein levels. Correlation of phospho-4E-BP1 expression with cause-related survival was significant (P=0.047).
The independence of prognostic ability of phospho-Akt, S6RP and phospho-4E-BP1 recurrence risk stratification was further evaluated by pair-wise analysis of the proteins against each other and the highly relevant clinicopathologic factor histopathological grading (Van Cutsem and Oliveira, 2009). Here, we found phospho-4E-BP1 and histopathological grading to be independent predictors of disease-free survival. Phospho-Akt was independent of histological grade, and S6RP showed a trend towards independency of the histological grade. Results of the pair-wise analysis are summarised in Table 3.
Table 3. Pair-wise analysis of phospho-Akt, S6RP, phosphor-4E-BP1 and grading by Cox-regression (disease-free survival).
| |
|
|
95% CI for Exp (B) |
|
|
|---|---|---|---|---|---|
| Factor | Against | Exp | Min | Max | Significance |
| Phospho-Akt | S6RP | 3.459 | 1.012 | 11.820 | 0.048 |
| Phospho-4E-BP1 | 2.702 | 0.630 | 11.589 | 0.181 | |
| |
Histograding |
3.439 |
1.006 |
11.757 |
0.049 |
| S6RP | Phospho-Akt | 4.440 | 0:512 | 38.546 | 0.176 |
| Phospho-4E-BP1 | 4.235 | 0.490 | 36.589 | 0.190 | |
| |
Histograding |
6.270 |
0.802 |
49.025 |
0.080 |
| Phospho-4E-BP1 | Phospho-Akt | 4.067 | 1.241 | 13.330 | 0.021 |
| S6RP | 4.067 | 1.241 | 13.330 | 0.021 | |
| |
Histograding |
4.067 |
1.241 |
13.330 |
0.021 |
| Histograding | Phospho-Akt | 3.688 | 1.124 | 12.095 | 0.031 |
| S6RP | 3.688 | 1.124 | 12.095 | 0.031 | |
| Phospho-4E-BP1 | 3.490 | 1.062 | 11.467 | 0.040 | |
Abbreviations: CI=confidence interval; mTOR=mammalian target of rapamycin; S6RP=S6 ribosomal protein.
Parameter phospho-Akt expression lower than 104 RIU versus higher than 104 RIU; S6RP expression lower than 329 RIU versus higher than 329 RIU; phospho-4E-BP1 expression lower than 121 RIU versus higher than 121 RIU; Grading G1 and G2 versus G3. Statistically significant values are depicted in bold italic letters.
In addition, potential influence of KRAS and BRAF mutational status was addressed, as mutational activation of these oncogenes occurs frequently in colon cancer, and is known to exert cross-talk with the PI3K/AKT-signalling cascade. KRAS mutational status was available for 104 and BRAF status for 115 patients. Thirty-eight (37%) patients were mutated for KRAS, 66 (63%) patients were KRAS wild type. For BRAF, 12 (10%) were mutated and 103 (90%) patients showed the wild type. Kaplan–Meier analysis showed no significant correlation of KRAS mutation (P=0.064) or BRAF mutation (P=0.355) with disease-free survival (Supplementary Figure 3a). KRAS and BRAF mutational status did not correlate significantly with cause-specific survival (Supplementary Figure 3b). As MSI was shown to be associated with beneficial prognosis in colon cancer (Merok et al, 2012), we analysed all patients of the collective also for MSI. Twenty-six patients (22%) were microsatellite instable and 91 patients (78%) were microsatellite stable. In our cohort MSI was not significantly correlated with disease-free or cause-specific survival, most likely owing to the relatively small sample size. Thus, MSI was excluded from further analysis. Neither the analysis of functional components of the PI3K/Akt-signalling pathway by Mann–Whitney U-testing nor a χ2-test of optimal cutoff values revealed any correlation with mutational status or microsatellite instability.
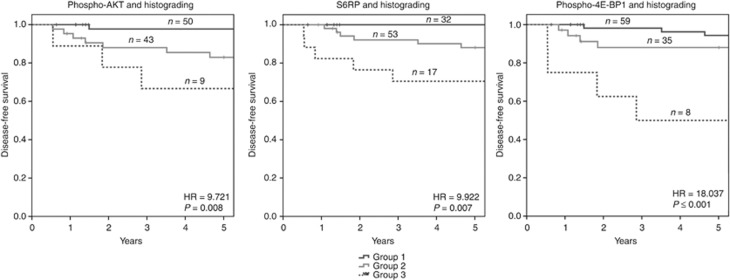
Although phospho-AKT, S6RP and phospho-4E-BP1 correlated significantly with disease-free survival, PPV of the factors are low (PPV=0.212, 0.167 and 0.250, respectively). Thus, we analysed combinations of these factors with histopathological grading for improvement of the predictive values. For all factors in combination with grading, three distinct patient groups could be identified: (1) very good disease-free survival, negative for the respective protein and grade 1 or 2, (2) intermediate survival, either positive for the respective protein and grade 1 or 2 or negative for the protein and grade 3, and (3) with poor disease-free survival positive for the protein and grade 3 (Figure 4; Supplementary Table 4).
Figure 4.
Correlation of combined protein expression and histological grading with disease-free survival. Group 1: both factors low; group 2: one factor high; Group 3: both factors high. Phospho-Akt expression was defined as low with RIU⩽104 and as high with RIU >104. S6RP expression was defined as low with RIU⩽329 and as high with RIU >329. Phospho-4E-BP1 was defined as low with RIU ⩽121 and as high with RIU >121. Histograding was defined as low for G1 and G2 and as high for G3. In the combined analysis of phospho-Akt and histograding, 50 patients belonged to group a, 43 to group b and 9 to group c. In the combined analysis of S6RP and histograding, 32 patients belonged to group a, 53 to group b and 17 to group c. In the combined analysis of phospho-4E-BP1 and histograding, 59 belonged to group 1, 35 to group 2 and 8 to group 3. For 16 patients no data on disease-free survival was available. All factor combinations correlated significantly with disease-free survival (P⩽0.008).
Correlation of functional factors of the PI3K/Akt-signalling pathway with clinical parameters
The comparison of functional components of the PI3K/Akt-signalling pathway with the clinical parameters tumour size, localisation and histological grading by Mann–Whitney U-testing did not reveal any statistically significant correlations. Also χ2-test of optimal cutoff values revealed no correlations with tumour size, localisation and histological grading. Results of the analysis are summarised in Table 4; Supplementary Table 5.
Table 4. Significance values for correlation of Akt, phospho-Akt, GSK3-beta, mTOR, phospho-mTOR, PI3K, phospho-PI3K, S6RP, phospho-S6RP, 4E-BP1 and phospho-4E-BP1 with tumour size, location and grading.
| |
P-value for correlation |
||
|---|---|---|---|
| Protein | pT category | Tumour localisation | Grading |
| Akt | 0.965 | 0.431 | 0.161 |
| phospho-Akt | 0.834 | 0.593 | 0.818 |
| GSK-3beta | 0.032 | 0.483 | 0.384 |
| mTOR | 0.256 | 0.607 | 0.298 |
| Phospho-mTOR | 0.374 | 0.862 | 0.172 |
| PI3K | 0.087 | 0.062 | 0.208 |
| Phospho-PI3K | 0.622 | 0.504 | 0.337 |
| S6RP | 0.697 | 0.838 | 0.805 |
| Phospho-S6RP | 0.761 | 0.578 | 0.369 |
| 4E-BP1 | 0.366 | 0.980 | 0.094 |
| Phospho-4E-BP1 | 0.516 | 0.185 | 0.344 |
Abbreviations: mTOR=mammalian target of rapamycin; S6RP=S6 ribosomal protein.
No significant correlations could be found.
Discussion
This study demonstrates the impact of functional components of the PI3K/Akt pathway as prognostic markers on stage II colon cancers in a retrospective single-center study. The patient cohort used here showed high similarity with cohorts used in earlier biomarker studies regarding its clinicopathological parameters indicating that patient recruitment was not biased. Our findings are essentially in line with previous studies reporting the PI3K/Akt-signalling pathway to have important roles in the development of numerous solid cancers (Dozmorov et al, 2010; Song et al, 2011; Jiao and Nan, 2012; Vredeveld et al, 2012; Wang et al, 2012; Christgen et al, 2013). Activation of the PI3K/Akt pathway was recently associated with increased cell growth, migration and invasion in colon cancer cell lines, processes underlying enhanced metastasis (Wang et al, 2013).
Although a number of clinical staging systems is available to identify patients with high risk of recurrence (Astler and Coller, 1954; Gunderson et al, 2004; O'Connell et al, 2004; Kozak and Moody, 2008), none of the systems suggested so far can identify patients with a high risk of recurrence in the group of stage II cancer patients with sufficient accuracy (O'Connell et al, 2004; Van Cutsem and Oliveira, 2009). Thus, a number of approaches were recently proposed to identify biomarkers for risk stratification of these patients. Numerous genomic markers like KRAS and TP53 mutations, activation of the canonical WNT-pathway, loss of heterozygosity of chromosome 18 and genomic instability (Fearon and Vogelstein, 1990; Locker et al, 2006; Tejpar et al, 2010) were described as typical features of colorectal cancer. However, none of the suggested markers have clearly proven its prognostic value in stage II colon cancer. Interestingly, in our study neither KRAS mutation nor BRAF mutation correlate with survival. In addition to the analysis of single genetic markers, also ‘-omics' approaches were undertaken to identify expression signatures correlated with patient outcome (Arango et al, 2005; Barrier et al, 2007; Lin et al, 2007; Webber et al, 2010; Salazar et al, 2011). However, inter-patient and even intratumoural heterogeneity, as well as cost factors and a lack of validation have precluded wide-scale clinical application (Shibayama et al, 2011).
Moreover, approaches of integrated molecular profiling, taking into account a limited number of genetic alterations have been proven useful for identification of high-risk patients with stage II colon cancer (Nitsche et al, 2012; Maak et al, 2013). As focusing on so-called ‘cancer hallmarks' present in virtually all cancer cells is a promising approach to circumvent heterogeneity, we analysed the enzymatic activities and protein expression of cyclin-dependent kinases (CDKs), the main drivers of cell-cycle progression (Zeestraten et al, 2012) in a previous study. Although the findings of this study were very promising and specific enzyme activity of CDK1 could be proven to be a robust marker for recurrence in stage II patients and for cause-specific survival of the patients, the assay format and the exclusive use of cryopreserved tissue may be difficult to integrate in the routine clinical settings. As post-translational phosphorylation of proteins is a good surrogate for their activation status, we choose an approach measuring the expression and phosphorylation of proteins belonging to the cancer relevant PI3K/Akt-signalling pathway. With a protocol adapted and validated in our laboratory, it is possible to quantify protein yields in formalin-fixed paraffin-embedded tissues, thus using standard material provided in clinical settings (Malinowsky et al, 2010).
Here, we analysed expression of molecules belonging to the PI3K/Akt-signalling pathway in 118 stage II colon cancers and 71 adjacent normal tissue samples. Although differences between the mean expression of the proteins between tumour and normal samples were not as pronounced as one might expect, we found significant changes of expression between tumour and normal tissues in individual samples (Supplementary Table 2). These data indicate that changes at the single-patient level might be more relevant to outcome than changes of mean expression levels in a group of individuals. In addition, we found high variance of protein expression when comparing individual tumours indicating different regulation of the PI3K/Akt pathway that might coincide with different tumour biology and thus different outcome. Indeed, we could identify phospho-Akt, S6RP and phospho-4E-BP1 as markers of disease recurrence in patients with stage II colon cancers. Robust cutoff values for these factors could be obtained utilising maximum log-rank statistics for threshold definition (Hothorn and Zeileis, 2008). Nervetheless, as only 11 patients show recurrence of the disease in this study the statistical power of the tests preformed is of course limited and the found cutoff values must be validated in further independent studies.
Neither the currently accepted clinical risk factors tumour size, histological grading and tumour localisation nor the mutational status of BRAF and KRAS could be identified as confounding factors by pair-wise analysis. Interestingly, the prognostic usefulness of KRAS mutations in colon cancer is discussed controversially, whereas BRAF mutations have been associated with bad prognosis in sporadic colon cancer (Deschoolmeester et al, 2010). Thus, we conclude that phospho-Akt, S6RP and phospho-4E-BP1 are potential biomarkers for the risk stratification of patients with stage II colon cancer.
Although phospho-Akt, S6RP and phospho-4E-BP1 were significantly associated with patient survival the PPV of the markers was relatively low. That would result in overtreatment of a large group of patients stratified to the high-risk group. We thought of potential strategies to overcome this problem. One possibility to enhance the PPV is the combination of markers. We analysed combinations of the proteins with histological grading, which came up as an additional independent predictor of disease-free survival in the pair-wise analysis. The results provide evidence that by using the classical histological grading system in combination with protein biomarkers, patients with stage II colon cancer can be subdivided into three groups with different risks of cancer recurrence. Although the PPV remain relatively low even after combination of factors (Supplementary Table 4), at least the patients with the highest risks of recurrence could clearly benefit from additional therapy. Whether classical chemotherapy or combination with targeted therapies against components of the activated PI3K pathway would be the best option here has to be addressed by further trials. At the moment different targeted therapies against Akt, PI3K and mTOR are under investigation in clinical trials (Britten, 2013; Roschewski et al, 2013; Wolin, 2013). Such therapeutics might enhance patient survival in the subgroup of stage II colon cancer patients that show enhanced activation of this specific pathway in our study. With help of the method introduced here it may be possible to stratify patients with activated PI3K/Akt-signalling pathway for targeted therapy.
Acknowledgments
This study received funding from the German Federal Ministry of Education and Research (BMBF) Leading-Edge Cluster m4 Personalised Medicine and Targeted Therapies, grant no. 01EX1020D. We would like to thank Christina Schott, Christian Beese Ruth Wichnalek and Susanne Plaschke for excellent technical support. Anna Munzig contributed to the analysis of MSI and Johanna Donhauser contributed to the analysis of BRAF and KRAS mutational status.
Author contributions
K-FB and KM conceived the study, carried out experiments and analysed and interpreted data. UN and KM did the statistical analysis. UN, K-PJ and CS collected the clinical and follow-up data. ED and JS-H did pathological evaluation of the samples. K-PJ, FGB, JS-H, GK and HH participated in study design and data interpretation. All authors were involved in writing the paper and approved the final version.
Footnotes
Supplementary Information accompanies this paper on British Journal of Cancer website (http://www.nature.com/bjc)
This work is published under the standard license to publish agreement. After 12 months the work will become freely available and the license terms will switch to a Creative Commons Attribution-NonCommercial-Share Alike 3.0 Unported License.
Supplementary Material
References
- Arango D, Laiho P, Kokko A, Alhopuro P, Sammalkorpi H, Salovaara R, Nicorici D, Hautaniemi S, Alazzouzi H, Mecklin JP, Järvinen H, Hemminki A, Astola J, Schwartz S, Jr, Aaltonen LA. Gene-expression profiling predicts recurrence in Dukes' C colorectal cancer. Gastroenterology. 2005;129 (3:874–884. doi: 10.1053/j.gastro.2005.06.066. [DOI] [PubMed] [Google Scholar]
- Astler VB, Coller FA. The prognostic significance of direct extension of carcinoma of the colon and rectum. Ann Surg. 1954;139 (6:846–852. doi: 10.1097/00000658-195406000-00015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrier A, Roser F, Boëlle PY, Franc B, Tse C, Brault D, Lacaine F, Houry S, Callard P, Penna C, Debuire B, Flahault A, Dudoit S, Lemoine A. Prognosis of stage II colon cancer by non-neoplastic mucosa gene expression profiling. Oncogene. 2007;26 (18:2642–2648. doi: 10.1038/sj.onc.1210060. [DOI] [PubMed] [Google Scholar]
- Benson AB, 3rd, Schrag D, Somerfield MR, Cohen AM, Figueredo AT, Flynn PJ, Krzyzanowska MK, Maroun J, McAllister P, Van Cutsem E, Brouwers M, Charette M, Haller DG. American Society of Clinical Oncology recommendations on adjuvant chemotherapy for stage II colon cancer. J Clin Oncol. 2004;22 (16:3408–3419. doi: 10.1200/JCO.2004.05.063. [DOI] [PubMed] [Google Scholar]
- Berg D, Malinowsky K, Reischauer B, Wolff C, Becker KF. Use of formalin-fixed and paraffin-embedded tissues for diagnosis and therapy in routine clinical settings. Methods Mol Biol. 2011;785:109–122. doi: 10.1007/978-1-61779-286-1_8. [DOI] [PubMed] [Google Scholar]
- Bettstetter M, Berezowska S, Keller G, Walch A, Feuchtinger A, Slotta-Huspenina J, Feith M, Drecoll E, Höfler H, Langer R. Epidermal growth factor receptor, phosphatidylinositol-3-kinase catalytic subunit/PTEN, and KRAS/NRAS/BRAF in primary resected esophageal adenocarcinomas: loss of PTEN is associated with worse clinical outcome. Hum Pathol. 2013;44 (5:829–836. doi: 10.1016/j.humpath.2012.08.005. [DOI] [PubMed] [Google Scholar]
- Britten CD. PI3K and MEK inhibitor combinations: examining the evidence in selected tumor types. Cancer Chemother Pharmacol. 2013;71 (6:1395–1409. doi: 10.1007/s00280-013-2121-1. [DOI] [PubMed] [Google Scholar]
- Catenacci DV, Kozloff M, Kindler HL, Polite B. Personalized colon cancer care in 2010. Semin Oncol. 2011;38 (2:284–308. doi: 10.1053/j.seminoncol.2011.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christgen M, Noskowicz M, Schipper E, Christgen H, Heil C, Krech T, Länger F, Kreipe H, Lehmann U. Oncogenic PIK3CA mutations in lobular breast cancer progression. Genes Chromosomes Cancer. 2013;52 (1:69–80. doi: 10.1002/gcc.22007. [DOI] [PubMed] [Google Scholar]
- Deschoolmeester V, Baay M, Specenier P, Lardon F, Vermorken JB. A review of the most promising biomarkers in colorectal cancer: one step closer to targeted therapy. Oncologist. 2010;15 (7:699–731. doi: 10.1634/theoncologist.2010-0025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dozmorov MG, Azzarello JT, Wren JD, Fung KM, Yang Q, Davis JS, Hurst RE, Culkin DJ, Penning TM, Lin HK. Elevated AKR1C3 expression promotes prostate cancer cell survival and prostate cell-mediated endothelial cell tube formation: implications for prostate cancer progression. BMC Cancer. 2010;10:672. doi: 10.1186/1471-2407-10-672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Efferth T. Signal transduction pathways of the epidermal growth factor receptor in colorectal cancer and their inhibition by small molecules. Curr Med Chem. 2012;19 (33:5735–5744. doi: 10.2174/092986712803988884. [DOI] [PubMed] [Google Scholar]
- Espina V, Mehta AI, Winters ME, Calvert V, Wulfkuhle J, Petricoin EF, 3rd, Liotta LA. Protein microarrays: molecular profiling technologies for clinical specimens. Proteomics. 2003;3 (11:2091–2100. doi: 10.1002/pmic.200300592. [DOI] [PubMed] [Google Scholar]
- Fearon ER, Vogelstein B. A genetic model for colorectal tumorigenesis. Cell. 1990;61 (5:759–767. doi: 10.1016/0092-8674(90)90186-i. [DOI] [PubMed] [Google Scholar]
- Ferlay J, Shin HR, Bray F, Forman D, Mathers C, Parkin DM. Estimates of worldwide burden of cancer in 2008: GLOBOCAN 2008. Int J Cancer. 2010;127 (12:2893–2917. doi: 10.1002/ijc.25516. [DOI] [PubMed] [Google Scholar]
- Gonzalez-Bosquet J, Calcei J, Wei JS, Garcia-Closas M, Sherman ME, Hewitt S, Vockley J, Lissowska J, Yang HP, Khan J, Chanock S. Detection of somatic mutations by high-resolution DNA melting (HRM) analysis in multiple cancers. PLoS One. 2011;6 (1:e14522. doi: 10.1371/journal.pone.0014522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grubb RL, Calvert VS, Wulkuhle JD, Paweletz CP, Linehan WM, Phillips JL, Chuaqui R, Valasco A, Gillespie J, Emmert-Buck M, Liotta LA, Petricoin EF. Signal pathway profiling of prostate cancer using reverse phase protein arrays. Proteomics. 2003;3 (11:2142–2146. doi: 10.1002/pmic.200300598. [DOI] [PubMed] [Google Scholar]
- Gunderson LL, Sargent DJ, Tepper JE, Wolmark N, O'Connell MJ, Begovic M, Allmer C, Colangelo L, Smalley SR, Haller DG, Martenson JA, Mayer RJ, Rich TA, Ajani JA, MacDonald JS, Willett CG, Goldberg RM. Impact of T and N stage and treatment on survival and relapse in adjuvant rectal cancer: a pooled analysis. J Clin Oncol. 2004;22 (10:1785–1796. doi: 10.1200/JCO.2004.08.173. [DOI] [PubMed] [Google Scholar]
- Guo H, Liu W, Ju Z, Tamboli P, Jonasch E, Mills GB, Lu Y, Hennessy BT, Tsavachidou D. An efficient procedure for protein extraction from formalin-fixed, paraffin-embedded tissues for reverse phase protein arrays. Proteome Sci. 2012;10 (1:56. doi: 10.1186/1477-5956-10-56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hothorn T, Zeileis A. Generalized maximally selected statistics. Biometrics. 2008;64 (4:1263–1269. doi: 10.1111/j.1541-0420.2008.00995.x. [DOI] [PubMed] [Google Scholar]
- Jemal A, Siegel R, Ward E, Hao Y, Xu J, Thun MJ. Cancer statistics, 2009. CA Cancer J Clin. 2009;59 (4:225–249. doi: 10.3322/caac.20006. [DOI] [PubMed] [Google Scholar]
- Jiao M, Nan KJ. Activation of PI3 kinase/Akt/HIF-1alpha pathway contributes to hypoxia-induced epithelial-mesenchymal transition and chemoresistance in hepatocellular carcinoma. Int J Oncol. 2012;40 (2:461–468. doi: 10.3892/ijo.2011.1197. [DOI] [PubMed] [Google Scholar]
- Kozak KR, Moody JS. The impact of T and N stage on long-term survival of rectal cancer patients in the community. J Surg Oncol. 2008;98 (3:161–166. doi: 10.1002/jso.21107. [DOI] [PubMed] [Google Scholar]
- Langer R, Becker K, Feith M, Friess H, Höfler H, Keller G. Genetic aberrations in primary esophageal melanomas: molecular analysis of c-KIT, PDGFR, KRAS, NRAS and BRAF in a series of 10 cases. Mod Pathol. 2011;24 (4:495–501. doi: 10.1038/modpathol.2010.220. [DOI] [PubMed] [Google Scholar]
- Lin YH, Friederichs J, Black MA, Mages J, Rosenberg R, Guilford PJ, Phillips V, Thompson-Fawcett M, Kasabov N, Toro T, Merrie AE, van Rij A, Yoon HS, McCall JL, Siewert JR, Holzmann B, Reeve AE. Multiple gene expression classifiers from different array platforms predict poor prognosis of colorectal cancer. Clin Cancer Res. 2007;13 (2 Pt 1:498–507. doi: 10.1158/1078-0432.CCR-05-2734. [DOI] [PubMed] [Google Scholar]
- Liotta LA, Espina V, Mehta AI, Calvert V, Rosenblatt K, Geho D, Munson PJ, Young L, Wulfkuhle J, Petricoin EF., 3rd Protein microarrays: meeting analytical challenges for clinical applications. Cancer Cell. 2003;3 (4:317–325. doi: 10.1016/s1535-6108(03)00086-2. [DOI] [PubMed] [Google Scholar]
- Locker GY, Hamilton S, Harris J, Jessup JM, Kemeny N, Macdonald JS, Somerfield MR, Hayes DF, Bast RC, Jr, ASCO ASCO 2006 update of recommendations for the use of tumor markers in gastrointestinal cancer. J Clin Oncol. 2006;24 (33:5313–5327. doi: 10.1200/JCO.2006.08.2644. [DOI] [PubMed] [Google Scholar]
- Maak M, Simon I, Nitsche U, Roepman P, Snel M, Glas AM, Schuster T, Keller G, Zeestraten E, Goossens I, Janssen KP, Friess H, Rosenberg R. Independent validation of a prognostic genomic signature (ColoPrint) for patients with stage II colon cancer. Ann Surg. 2013;257 (6:1053–1058. doi: 10.1097/SLA.0b013e31827c1180. [DOI] [PubMed] [Google Scholar]
- Malinowsky K, Raychaudhuri M, Buchner T, Thulke S, Wolff C, Höfler H, Becker KF, Avril S. Common protein biomarkers assessed by reverse phase protein arrays show considerable intratumoral heterogeneity in breast cancer tissues. PLoS One. 2012;7 (7:e40285. doi: 10.1371/journal.pone.0040285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malinowsky K, Wolff C, Gündisch S, Berg D, Becker K. Targeted therapies in cancer—challenges and chances offered by newly developed techniques for protein analysis in clinical tissues. J Cancer. 2010;2:26–35. doi: 10.7150/jca.2.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McShane LM, Altman DG, Sauerbrei W, Taube SE, Gion M, Clark GM, Statistics Subcommittee of the NCI-EORTC Working Group on Cancer Diagnostics REporting recommendations for tumour MARKer prognostic studies (REMARK) Br J Cancer. 2005;93 (4:387–391. doi: 10.1038/sj.bjc.6602678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merok MA, Ahlquist T, Røyrvik EC, Tufteland KF, Hektoen M, Sjo OH, Mala T, Svindland A, Lothe RA, Nesbakken A. Microsatellite instability has a positive prognostic impact on stage II colorectal cancer after complete resection: results from a large, consecutive Norwegian series. Ann Oncol. 2012;24 (5:1274–1282. doi: 10.1093/annonc/mds614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy KM, Zhang S, Geiger T, Hafez MJ, Bacher J, Berg KD, Eshleman JR. Comparison of the microsatellite instability analysis system and the Bethesda panel for the determination of microsatellite instability in colorectal cancers. J Mol Diagn. 2006;8 (3:305–311. doi: 10.2353/jmoldx.2006.050092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mutch MG. Molecular profiling and risk stratification of adenocarcinoma of the colon. J Surg Oncol. 2007;96 (8:693–703. doi: 10.1002/jso.20915. [DOI] [PubMed] [Google Scholar]
- Nitsche U, Maak M, Schuster T, Künzli B, Langer R, Slotta-Huspenina J, Janssen KP, Friess H, Rosenberg R.2011Prediction of prognosis is not improved by the seventh and latest edition of the TNM classification for colorectal cancer in a single-center collective Ann Surg 254(5793–800.discussion 800-791. [DOI] [PubMed] [Google Scholar]
- Nitsche U, Rosenberg R, Balmert A, Schuster T, Slotta-Huspenina J, Herrmann P, Bader FG, Friess H, Schlag PM, Stein U, Janssen KP.2012Integrative marker analysis allows risk assessment for metastasis in stage II colon cancer Ann Surg 256(5763–771.discussion 771. [DOI] [PubMed] [Google Scholar]
- O'Connell JB, Maggard MA, Ko CY. Colon cancer survival rates with the new American Joint Committee on Cancer sixth edition staging. J Natl Cancer Inst. 2004;96 (19:1420–1425. doi: 10.1093/jnci/djh275. [DOI] [PubMed] [Google Scholar]
- Pandurangan AK. Potential targets for prevention of colorectal cancer: a focus on PI3K/Akt/mTOR and Wnt pathways. Asian Pac J Cancer Prev. 2013;14 (4:2201–2205. doi: 10.7314/apjcp.2013.14.4.2201. [DOI] [PubMed] [Google Scholar]
- Paweletz CP, Charboneau L, Bichsel VE, Simone NL, Chen T, Gillespie JW, Emmert-Buck MR, Roth MJ, Petricoin EF, III, Liotta LA. Reverse phase protein microarrays which capture disease progression show activation of pro-survival pathways at the cancer invasion front. Oncogene. 2001;20 (16:1981–1989. doi: 10.1038/sj.onc.1204265. [DOI] [PubMed] [Google Scholar]
- Pichler M, Balic M, Stadelmeyer E, Ausch C, Wild M, Guelly C, Bauernhofer T, Samonigg H, Hoefler G, Dandachi N. Evaluation of high-resolution melting analysis as a diagnostic tool to detect the BRAF V600E mutation in colorectal tumors. J Mol Diagn. 2009;11 (2:140–147. doi: 10.2353/jmoldx.2009.080100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roschewski M, Farooqui M, Aue G, Wilhelm F, Wiestner A. Phase I study of ON 01910.Na (Rigosertib), a multikinase PI3K inhibitor in relapsed/refractory B-cell malignancies. Leukemia. 2013;27 (9:1920–1923. doi: 10.1038/leu.2013.79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salazar R, Roepman P, Capella G, Moreno V, Simon I, Dreezen C, Lopez-Doriga A, Santos C, Marijnen C, Westerga J, Bruin S, Kerr D, Kuppen P, van de Velde C, Morreau H, Van Velthuysen L, Glas AM, Van't Veer LJ, Tollenaar R. Gene expression signature to improve prognosis prediction of stage II and III colorectal cancer. J Clin Oncol. 2011;29 (1:17–24. doi: 10.1200/JCO.2010.30.1077. [DOI] [PubMed] [Google Scholar]
- Shibayama M, Maak M, Nitsche U, Gotoh K, Rosenberg R, Janssen KP. Prediction of metastasis and recurrence in colorectal cancer based on gene expression analysis: ready for the clinic. Cancers. 2011;3:2858–2869. doi: 10.3390/cancers3032858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegel R, Naishadham D, Jemal A. Cancer statistics, 2013. CA Cancer J Clin. 2013;63 (1:11–30. doi: 10.3322/caac.21166. [DOI] [PubMed] [Google Scholar]
- Song L, Xiong H, Li J, Liao W, Wang L, Wu J, Li M. Sphingosine kinase-1 enhances resistance to apoptosis through activation of PI3K/Akt/NF-kappaB pathway in human non-small cell lung cancer. Clin Cancer Res. 2011;17 (7:1839–1849. doi: 10.1158/1078-0432.CCR-10-0720. [DOI] [PubMed] [Google Scholar]
- Tejpar S, Bertagnolli M, Bosman F, Lenz HJ, Garraway L, Waldman F, Warren R, Bild A, Collins-Brennan D, Hahn H, Harkin DP, Kennedy R, Ilyas M, Morreau H, Proutski V, Swanton C, Tomlinson I, Delorenzi M, Fiocca R, Van Cutsem E, Roth A. Prognostic and predictive biomarkers in resected colon cancer: current status and future perspectives for integrating genomics into biomarker discovery. Oncologist. 2010;15 (4:390–404. doi: 10.1634/theoncologist.2009-0233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Cutsem E, Oliveira J. Primary colon cancer: ESMO clinical recommendations for diagnosis, adjuvant treatment and follow-up. Ann Oncol. 2009;20 (Suppl 4:49–50. doi: 10.1093/annonc/mdp126. [DOI] [PubMed] [Google Scholar]
- Vredeveld LC, Possik PA, Smit MA, Meissl K, Michaloglou C, Horlings HM, Ajouaou A, Kortman PC, Dankort D, McMahon M, Mooi WJ, Peeper DS. Abrogation of BRAFV600E-induced senescence by PI3K pathway activation contributes to melanomagenesis. Genes Dev. 2012;26 (10:1055–1069. doi: 10.1101/gad.187252.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D, Chen J, Chen H, Duan Z, Xu Q, Wei M, Wang L, Zhong M. Leptin regulates proliferation and apoptosis of colorectal carcinoma through PI3K/Akt/mTOR signalling pathway. J Biosci. 2012;37 (1:91–101. doi: 10.1007/s12038-011-9172-4. [DOI] [PubMed] [Google Scholar]
- Wang G, Wang F, Ding W, Wang J, Jing R, Li H, Wang X, Wang Y, Ju S, Wang H. APRIL induces tumorigenesis and metastasis of colorectal cancer cells via activation of the PI3K/Akt pathway. PLoS One. 2013;8 (1:e55298. doi: 10.1371/journal.pone.0055298. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Webber EM, Lin JS, Evelyn P Whitlock.2010Oncotype DX tumor gene expression profiling in stage II colon cancer. Application: prognostic, risk prediction PLoS Curr 2piiRRN1177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolff C, Malinowsky K, Berg D, Schragner K, Schuster T, Walch A, Bronger H, Höfler H, Becker KF. Signalling networks associated with urokinase-type plasminogen activator (uPA) and its inhibitor PAI-1 in breast cancer tissues: new insights from protein microarray analysis. J Pathol. 2011;223 (1:54–63. doi: 10.1002/path.2791. [DOI] [PubMed] [Google Scholar]
- Wolff C, Schott C, Malinowsky K, Berg D, Becker KF. Producing reverse phase protein microarrays from formalin-fixed tissues. Methods Mol Biol. 2011;785:123–140. doi: 10.1007/978-1-61779-286-1_9. [DOI] [PubMed] [Google Scholar]
- Wolin EM. PI3K/Akt/mTOR pathway inhibitors in the therapy of pancreatic neuroendocrine tumors. Cancer Lett. 2013;335 (1:1–8. doi: 10.1016/j.canlet.2013.02.016. [DOI] [PubMed] [Google Scholar]
- Wulfkuhle JD, Edmiston KH, Liotta LA, Petricoin EF., 3rd Technology insight: pharmacoproteomics for cancer–promises of patient-tailored medicine using protein microarrays. Nat Clin Pract Oncol. 2006;3 (5:256–268. doi: 10.1038/ncponc0485. [DOI] [PubMed] [Google Scholar]
- Zeestraten EC, Maak M, Shibayama M, Schuster T, Nitsche U, Matsushima T, Nakayama S, Gohda K, Friess H, van de Velde CJ, Ishihara H, Rosenberg R, Kuppen PJ, Janssen KP. Specific activity of cyclin-dependent kinase I is a new potential predictor of tumour recurrence in stage II colon cancer. Br J Cancer. 2012;106 (1:133–140. doi: 10.1038/bjc.2011.504. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.