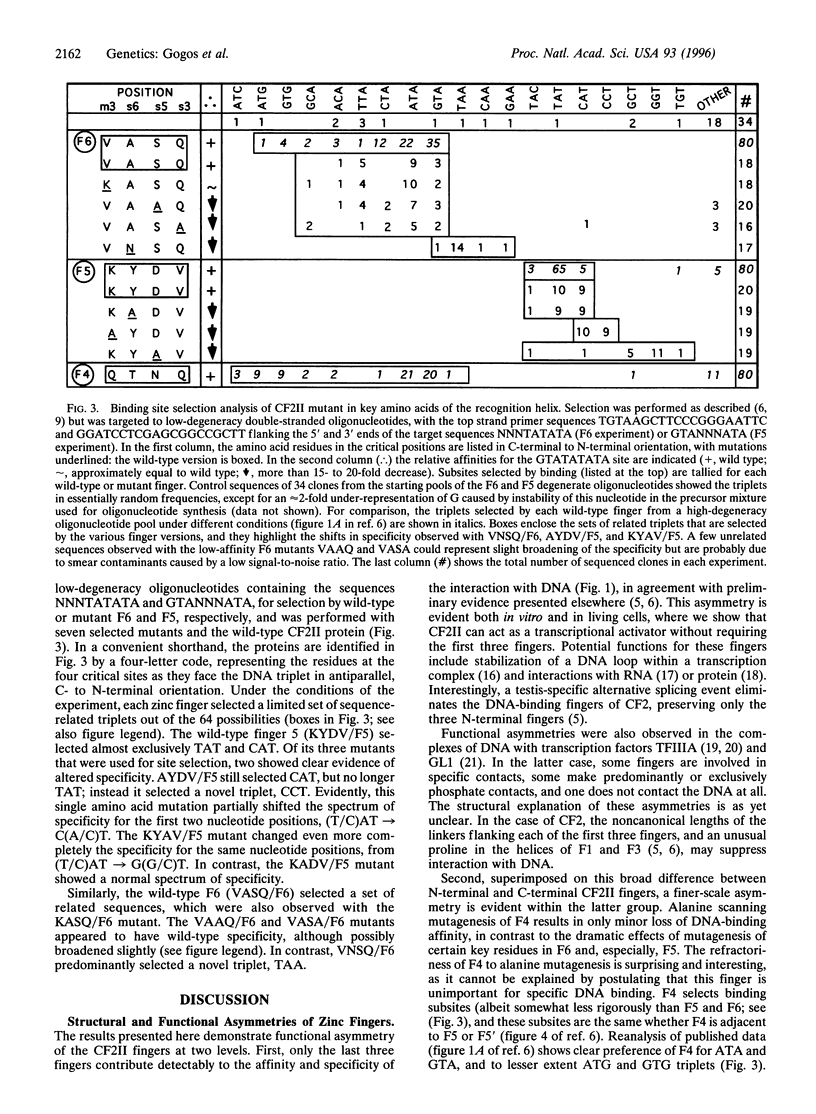
Abstract
The Drosophila CF2II protein, which contains zinc fingers of the Cys2His2 type and recognizes an A+T-rich sequence, behaves in cell culture as an activator of a reporter chloramphenicol acetyltransferase gene. This activity depends on C-terminal but not N-terminal zinc fingers, as does in vitro DNA binding. By site-specific mutagenesis and binding site selection, we define the critical amino acid-base interactions. Mutations of single amino acid residues at the leading edge of the recognition helix are rarely neutral: many result in a slight change in affinity for the ideal DNA target site; some cause major loss of affinity; and others change specificity for as many as two bases in the target site. Compared to zinc fingers that recognize G+C-rich DNA, CF2II fingers appear to bind to A+T-rich DNA in a generally similar manner, but with additional flexibility and amino acid-base interactions. The results illustrate how zinc fingers may be evolving to recognize an unusually diverse set of DNA sequences.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Abel T., Bhatt R., Maniatis T. A Drosophila CREB/ATF transcriptional activator binds to both fat body- and liver-specific regulatory elements. Genes Dev. 1992 Mar;6(3):466–480. doi: 10.1101/gad.6.3.466. [DOI] [PubMed] [Google Scholar]
- Brown N. H., Kafatos F. C. Functional cDNA libraries from Drosophila embryos. J Mol Biol. 1988 Sep 20;203(2):425–437. doi: 10.1016/0022-2836(88)90010-1. [DOI] [PubMed] [Google Scholar]
- Choo Y., Klug A. Selection of DNA binding sites for zinc fingers using rationally randomized DNA reveals coded interactions. Proc Natl Acad Sci U S A. 1994 Nov 8;91(23):11168–11172. doi: 10.1073/pnas.91.23.11168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choo Y., Klug A. Toward a code for the interactions of zinc fingers with DNA: selection of randomized fingers displayed on phage. Proc Natl Acad Sci U S A. 1994 Nov 8;91(23):11163–11167. doi: 10.1073/pnas.91.23.11163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Churchill M. E., Tullius T. D., Klug A. Mode of interaction of the zinc finger protein TFIIIA with a 5S RNA gene of Xenopus. Proc Natl Acad Sci U S A. 1990 Jul;87(14):5528–5532. doi: 10.1073/pnas.87.14.5528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Del Rio S., Setzer D. R. The role of zinc fingers in transcriptional activation by transcription factor IIIA. Proc Natl Acad Sci U S A. 1993 Jan 1;90(1):168–172. doi: 10.1073/pnas.90.1.168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desjarlais J. R., Berg J. M. Redesigning the DNA-binding specificity of a zinc finger protein: a data base-guided approach. Proteins. 1992 Feb;12(2):101–104. doi: 10.1002/prot.340120202. [DOI] [PubMed] [Google Scholar]
- Desjarlais J. R., Berg J. M. Toward rules relating zinc finger protein sequences and DNA binding site preferences. Proc Natl Acad Sci U S A. 1992 Aug 15;89(16):7345–7349. doi: 10.1073/pnas.89.16.7345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fairall L., Schwabe J. W., Chapman L., Finch J. T., Rhodes D. The crystal structure of a two zinc-finger peptide reveals an extension to the rules for zinc-finger/DNA recognition. Nature. 1993 Dec 2;366(6454):483–487. doi: 10.1038/366483a0. [DOI] [PubMed] [Google Scholar]
- Gogos J. A., Hsu T., Bolton J., Kafatos F. C. Sequence discrimination by alternatively spliced isoforms of a DNA binding zinc finger domain. Science. 1992 Sep 25;257(5078):1951–1955. doi: 10.1126/science.1290524. [DOI] [PubMed] [Google Scholar]
- Gogos J. A., Kafatos F. C. Determination of sequence preferences of DNA binding proteins using pooled solid-phase sequencing of low degeneracy oligonucleotide mixtures. Methods Mol Biol. 1994;30:295–312. doi: 10.1385/0-89603-256-6:295. [DOI] [PubMed] [Google Scholar]
- Harrison S. C. A structural taxonomy of DNA-binding domains. Nature. 1991 Oct 24;353(6346):715–719. doi: 10.1038/353715a0. [DOI] [PubMed] [Google Scholar]
- Hsu T., Gogos J. A., Kirsh S. A., Kafatos F. C. Multiple zinc finger forms resulting from developmentally regulated alternative splicing of a transcription factor gene. Science. 1992 Sep 25;257(5078):1946–1950. doi: 10.1126/science.1411512. [DOI] [PubMed] [Google Scholar]
- Jacobs G. H. Determination of the base recognition positions of zinc fingers from sequence analysis. EMBO J. 1992 Dec;11(12):4507–4517. doi: 10.1002/j.1460-2075.1992.tb05552.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jamieson A. C., Kim S. H., Wells J. A. In vitro selection of zinc fingers with altered DNA-binding specificity. Biochemistry. 1994 May 17;33(19):5689–5695. doi: 10.1021/bi00185a004. [DOI] [PubMed] [Google Scholar]
- Joho K. E., Darby M. K., Crawford E. T., Brown D. D. A finger protein structurally similar to TFIIIA that binds exclusively to 5S RNA in Xenopus. Cell. 1990 Apr 20;61(2):293–300. doi: 10.1016/0092-8674(90)90809-s. [DOI] [PubMed] [Google Scholar]
- Kunkel T. A. Rapid and efficient site-specific mutagenesis without phenotypic selection. Proc Natl Acad Sci U S A. 1985 Jan;82(2):488–492. doi: 10.1073/pnas.82.2.488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee M. S., Gippert G. P., Soman K. V., Case D. A., Wright P. E. Three-dimensional solution structure of a single zinc finger DNA-binding domain. Science. 1989 Aug 11;245(4918):635–637. doi: 10.1126/science.2503871. [DOI] [PubMed] [Google Scholar]
- Mastrangelo I. A., Courey A. J., Wall J. S., Jackson S. P., Hough P. V. DNA looping and Sp1 multimer links: a mechanism for transcriptional synergism and enhancement. Proc Natl Acad Sci U S A. 1991 Jul 1;88(13):5670–5674. doi: 10.1073/pnas.88.13.5670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakaseko Y., Neuhaus D., Klug A., Rhodes D. Adjacent zinc-finger motifs in multiple zinc-finger peptides from SWI5 form structurally independent, flexibly linked domains. J Mol Biol. 1992 Nov 20;228(2):619–636. doi: 10.1016/0022-2836(92)90845-b. [DOI] [PubMed] [Google Scholar]
- Nardelli J., Gibson T. J., Vesque C., Charnay P. Base sequence discrimination by zinc-finger DNA-binding domains. Nature. 1991 Jan 10;349(6305):175–178. doi: 10.1038/349175a0. [DOI] [PubMed] [Google Scholar]
- Nardelli J., Gibson T., Charnay P. Zinc finger-DNA recognition: analysis of base specificity by site-directed mutagenesis. Nucleic Acids Res. 1992 Aug 25;20(16):4137–4144. doi: 10.1093/nar/20.16.4137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavletich N. P., Pabo C. O. Crystal structure of a five-finger GLI-DNA complex: new perspectives on zinc fingers. Science. 1993 Sep 24;261(5129):1701–1707. doi: 10.1126/science.8378770. [DOI] [PubMed] [Google Scholar]
- Pavletich N. P., Pabo C. O. Zinc finger-DNA recognition: crystal structure of a Zif268-DNA complex at 2.1 A. Science. 1991 May 10;252(5007):809–817. doi: 10.1126/science.2028256. [DOI] [PubMed] [Google Scholar]
- Rebar E. J., Pabo C. O. Zinc finger phage: affinity selection of fingers with new DNA-binding specificities. Science. 1994 Feb 4;263(5147):671–673. doi: 10.1126/science.8303274. [DOI] [PubMed] [Google Scholar]
- Richardson J. S., Richardson D. C. Amino acid preferences for specific locations at the ends of alpha helices. Science. 1988 Jun 17;240(4859):1648–1652. doi: 10.1126/science.3381086. [DOI] [PubMed] [Google Scholar]
- Shea M. J., King D. L., Conboy M. J., Mariani B. D., Kafatos F. C. Proteins that bind to Drosophila chorion cis-regulatory elements: a new C2H2 zinc finger protein and a C2C2 steroid receptor-like component. Genes Dev. 1990 Jul;4(7):1128–1140. doi: 10.1101/gad.4.7.1128. [DOI] [PubMed] [Google Scholar]
- Thukral S. K., Morrison M. L., Young E. T. Mutations in the zinc fingers of ADR1 that change the specificity of DNA binding and transactivation. Mol Cell Biol. 1992 Jun;12(6):2784–2792. doi: 10.1128/mcb.12.6.2784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vrana K. E., Churchill M. E., Tullius T. D., Brown D. D. Mapping functional regions of transcription factor TFIIIA. Mol Cell Biol. 1988 Apr;8(4):1684–1696. doi: 10.1128/mcb.8.4.1684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H., Yang W. P., Barbas C. F., 3rd Building zinc fingers by selection: toward a therapeutic application. Proc Natl Acad Sci U S A. 1995 Jan 17;92(2):344–348. doi: 10.1073/pnas.92.2.344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon C., Privé G. G., Goodsell D. S., Dickerson R. E. Structure of an alternating-B DNA helix and its relationship to A-tract DNA. Proc Natl Acad Sci U S A. 1988 Sep;85(17):6332–6336. doi: 10.1073/pnas.85.17.6332. [DOI] [PMC free article] [PubMed] [Google Scholar]




