Abstract
The formation of body segments (somites) in vertebrate embryos is accompanied by molecular oscillations (segmentation clock). Interaction of this oscillator with a wave traveling along the body axis (the clock-and-wavefront model) is generally believed to control somite number, size, and axial identity. Here we show that a clock-and-wavefront mechanism is unnecessary for somite formation. Non-somite mesoderm treated with Noggin generates many somites that form simultaneously, without cyclic expression of Notch-pathway genes, yet have normal size, shape, and fate. These somites have axial identity: The Hox code is fixed independently of somite fate. However, these somites are not subdivided into rostral and caudal halves, which is necessary for neural segmentation. We propose that somites are self-organizing structures whose size and shape is controlled by local cell-cell interactions.
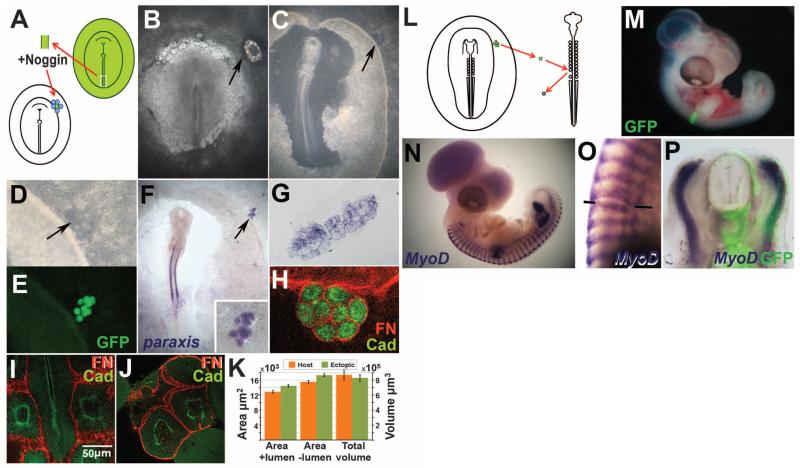
The mesoderm of the embryo, from which the cardiovascular and musculoskeletal systems arise, derives from the primitive streak (PS) during gastrulation. A high level of bone morphogenetic protein (BMP) at the posterior PS generates ventral mesoderm (blood vessels, lateral and extraembryonic mesoderm), whereas lower levels near the anterior tip generate paraxial mesoderm, from which somites (future striated muscle and axial skeleton) develop (1). Somites are epithelial spheres that form sequentially from head to tail on either side of the spinal cord. The combination of a molecular clock (cell-autonomous Notch and Wnt oscillations) and a wave traveling the length of the paraxial mesoderm (2, 3) is thought to regulate the number, size, timing of formation, and axial identity (4-6) of somites. Because the BMP antagonist Noggin is sufficient to transform ventral cells to a dorsal (somite) fate (7, 8), we applied Noggin as evenly as possible (9) to dorsalize posterior PS explants from quail or green fluorescent protein (GFP)–transgenic chick embryos and thus to test whether somites could be generated independently of a segmentation clock (10, 11). Explants from stage-5 (12) embryos were incubated in Noggin for 3 hours, then grafted into a remote (extraembryonic) region of a host chick embryo surrounded by Noggin-soaked beads (Fig. 1, A and B). A few hours later (total 9 to 12 hours), 6 to 14 somite-like structures had formed, arranged as a “bunch of grapes” (Fig. 1, C to E) rather than in linear sequence. Like normal somites, these structures express paraxis (8) (Fig. 1, F and G) and consist of epithelial cells around a lumen (Fig. 1, G to J), with apical N-cadherin and a Fibronectin-positive basal lamina (Fig. 1, H to J). The size of each somite-like structure is normal: Fig. 1K compares ectopic and normal somite volumes calculated from living embryos and multiphoton cross-sectional areas with and without the lumen (t tests P = 0.496, 0.401, and 0.493, respectively).
Fig. 1. BMP inhibition generates normal somites.
(A to E) Experimental design. The PS of a donor quail or GFP-transgenic embryo is excised; exposed to Noggin; and grafted, surrounded by Noggin-beads, to the periphery of a host chick embryo [(A and B), arrows]. After overnight incubation, a group of somite-like structures—arranged as a bunch of grapes—appears [(C and D), arrows]. These structures fluoresce if the donor is a GFP-transgenic embryo (E). (F to P) The ectopic structures are real somites: They express paraxis (F and G) and N-cadherin [green in (H) to (J)] and are surrounded by a Fibronectin matrix [red in (H) to (J)]. Multiphoton confocal sections through normal (I) and ectopic (J) somites were used to estimate somite sizes (K). When an ectopic somite is grafted instead of a somite in an older embryo (L), the graft incorporates well (M). After 2 to 3 days, the grafted somite appropriately expresses MyoD (N to P).
To test whether the ectopic somites can give rise to normal somite derivatives, we replaced individual recently formed somites in 10 to 14 somite secondary hosts with ectopic GFP-transgenic somites (Fig. 1L). After 2 to 3 days (stages 19 to 25), the grafted somite was well integrated (Fig. 1M) and expressed the sclerotome/vertebral marker Pax1 (fig. S1) (n = 6 experiments) and the dermomyotome/muscle marker MyoD (Fig. 1, N to P) (n = 4) in the correct positions. Some blood vessels were also generated (fig. S1), which may be normal somite derivatives (13, 14) or cells retaining their original lateral fate. Thus, the structures in the “bunch of grapes” are indeed somites.
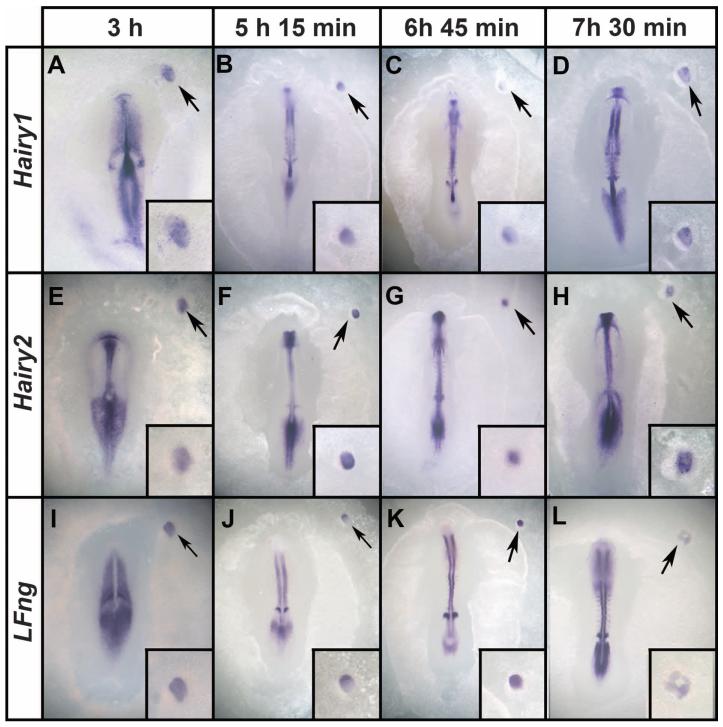
To test whether somites form sequentially or simultaneously, we used time-lapse microscopy to film ectopic GFP-transgenic somite formation. About 6 to 14 somites form in just 2 hours (9 to 11 hours after grafting) (fig. S2 and movies S1 and S2). The finding that so many somites can form almost synchronously suggests that the ectopic somites form independently of a clock. To assess the molecular clock, we examined embryos at different time points before ectopic somite formation for expression of clock genes Hairy1 (Fig. 2, A to D), Hairy2 (Fig. 2, E to H), and LFng (Fig. 2, I to L) at 45-min intervals between 3 and 7.5 hours after exposure of PS explants to Noggin. Although host embryos displayed typical (10) strong variations in the pattern of expression, the explants showed only subtle variations, not like a prepattern of the somites that would later form. Moreover, when examining many embryos for each marker at a particular time point (fig. S3), oscillatory expression was evident in the host embryo, but the explants (insets) show comparatively uniform expression. Examination of Dapper1 and -2 expression suggests that Noggin-treated mesoderm can generate somites without passing through a presomitic-like state (fig. S4). These results strongly suggest that the ectopic somites form simultaneously and without cyclic expression of clock genes.
Fig. 2. Ectopic somites form without cyclic expression of segmentation clock genes.
Embryos were fixed at 45-min intervals (examples shown at 3, 5.15, 6.45, and 7.5 hours after grafting to a host embryo) and stained for expression of Hairy1 (A to D), Hairy2 (E to H), and LFng (I to L). The in situ embryos were developed to reveal the segmentation clock in the presomitic cells of the host. Although patterns of expression in the presomitic mesoderm of the host are dynamic, no major differences in expression are seen in the graft (insets). Arrows mark the graft region, which is shown magnified in the insets.
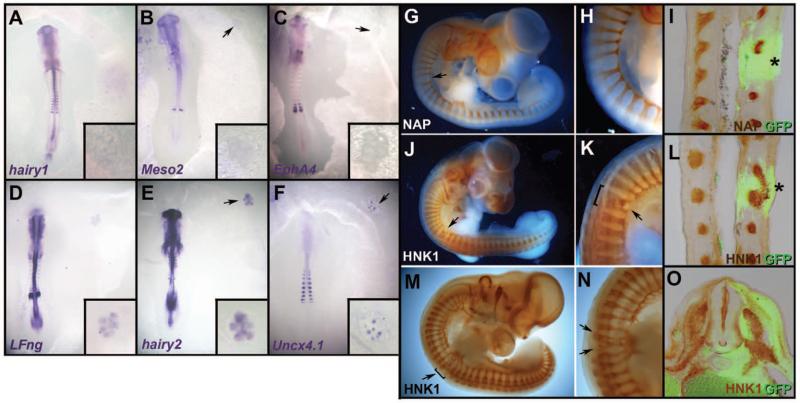
Each somite is normally subdivided into two halves, rostral and caudal, a property subsequently required for segmentation of the peripheral nervous system (15). To test whether the ectopic somites are subdivided, we examined expression of caudal (Hairy1, Hairy2, LFng, Uncx4.1, and Meso2) and rostral (EphA4) markers. None of them revealed subdivision of the ectopic somites. Hairy1 [0 of 22 embryos (0/22)], Meso2 (0/22), and EphA4 (0/19) were not expressed (Fig. 3, A to C); LFng (22/24) (Fig. 3D) and Hairy2 (8/8) (Fig. 3E) were expressed weakly and uniformly throughout the somites; and Uncx4.1 (13/19) (Fig. 3F) was patchy (Fig. 3F). Therefore, ectopic somites seem to lack coherent rostrocaudal identity, because the patterns of different genes are inconsistent with each other. As neural crest cells and motor axons normally only migrate through the rostral half of the sclerotome (15), we used this as an additional test of somite patterning. An ectopic GFP-somite was grafted instead of a normal somite in a secondary host (Fig. 1L). At stage 22 to 25, the patterns of motor axon growth (Fig. 3, G to I) and neural crest migration (Fig. 3, J to O) were disrupted. Abnormalities included an enlarged gap between motor roots (Fig. 3, G to I), fusion of adjacent ventral roots and dorsal root ganglia (Fig. 3, J to L), or several small ganglia formed within a grafted somite (Fig. 3, M to O), as if the somite contained random islands of permissive (non-caudal) cells exploited by axons and crest cells. These results suggest that the ectopic somites are not subdivided into rostral and caudal halves, consistent with the proposal (16) that the clock is required for this feature of segmentation.
Fig. 3. Ectopic somites are not subdivided into rostral and caudal halves.
(A to F) Ectopic somites were analyzed for expression of caudal (Hairy1, Meso2, LFng, Hairy2, and Uncx4.1) and rostral (EphA4) markers. Hairy1 (A), Meso2 (B), and EphA4 (C) are not expressed; LFng and Hairy2 (D and E) are weak and uniform; and Uncx4.1 is expressed as random patches (F). Insets show a magnified view of the graft. (G to O) As a further test of rostrocaudal patterning, embryos grafted as in Fig. 1L were stained for motor axons [neurofilament-associated protein NAP, (G to I), brown] or neural crest [HNK1, (J to O), brown] and anti-GFP [green in (I), (L), and (O)]. A large gap (G to I), fused roots (J to L), or multiple small ganglia (M to O) form in the ectopic somite (arrows, asterisks). Sections (I) and (L) are coronal, (O) is transverse at the level of the graft.
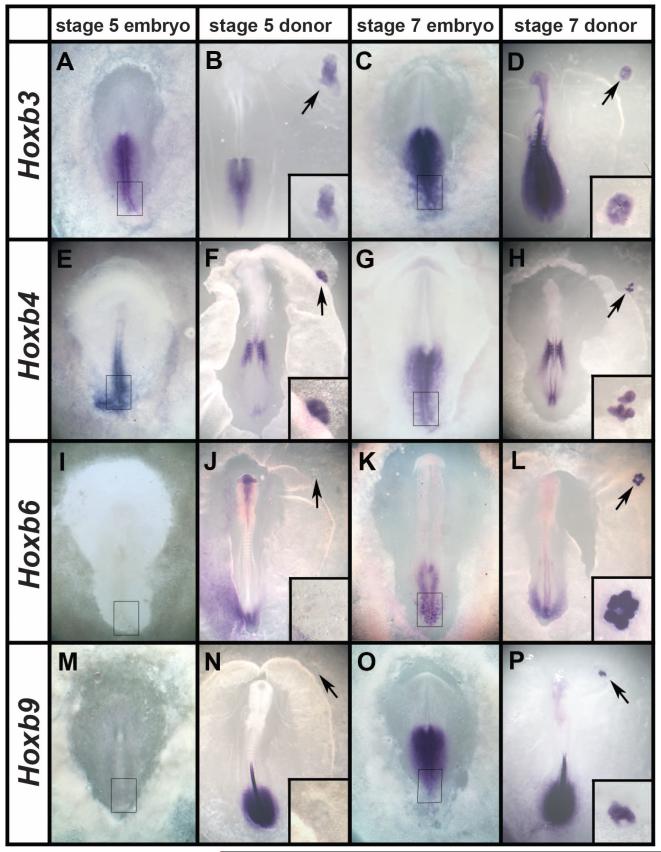
During normal development, the occipital somites (the most cranial four or five somites) form almost simultaneously rather than in sequence (movie S3) and lack expression of some rostral and/or caudal markers (17-19). Could the ectopic somites be occipital? We examined expression of Hox genes (20, 21) (Fig. 4, A to P): Hoxb3 (Fig. 4, A and C) and Hoxb4 (Fig. 4, E and G) were both expressed (Fig. 4, B, D, F, and H), suggesting that they are not occipital. Hoxb6 and Hoxb9 were not expressed (Fig. 4, F and J), suggesting that they are cervical (somite eight or nine). The posterior PS of stage-5 donor embryos expresses similar genes: Hoxb3 and b4, but not b6 or b9 (Fig. 4, A, E, I, and M); the latter start to be expressed later (stage 7 or 8) (Fig. 4, C, G, K, and O). We therefore tested whether somites made from PSs from older embryos (stage 8) express these markers. Indeed, they do (Fig. 4, L and P). This confirms that the Hox code imparting axial identity to cells is already present in the PS (22), independently of the segmentation clock (6), and suggests that the axial identity of the ectopic somites is specified according to which Hox genes are expressed in the posterior PS at the time of explantation, even though this region does not normally contribute to somites. Therefore, either exit of cells from the PS or, more likely, inhibition of BMP by Noggin arrests the molecular clock controlling expression of Hox genes that impart axial identity (23). In vivo, this may happen as presomitic cells leave the BMP-expressing PS and lie next to the notochord, the endogenous source of Noggin.
Fig. 4. Ectopic somites have trunk identity, fixed according to the Hox genes expressed in the donor PS.
(A to P) At stage 5, the posterior PS expresses Hoxb3 (A) and b4 (E), but not b6 (I) or b9 (M). Ectopic somites made from posterior streak explants from these stages show a similar pattern of expression (B, F, J, and N). At stages 7 and 8, the posterior streak expresses all four genes (C, G, K, and O), as do the ectopic somites formed from it (D, H, L, and P). Arrows point to the graft region, shown magnified in the insets.
The clock-and-wavefront model requires both an oscillator and a wave. In zebrafish, changing the period of molecular oscillations affects somite number and size (5, 6). We show that somites can form without oscillations of segmentation clock genes; all of their properties are normal, except for their rostrocaudal subdivision. Moreover, waves and gradients are also unnecessary, because the spatial organization and simultaneous formation of the ectopic somites does not seem compatible with this. We therefore propose that the main function of the clock is to subdivide somites into rostral and caudal halves and to couple this to somite formation.
If clock-and-wavefront mechanisms are not required to control somite formation, what does? Our observations implicate local cell-cell interactions. Embryological experiments (24) suggest that somites are self-organizing structures, regulated by intrinsic properties of the cells and packing constraints for cells undergoing mesenchymal-to-epithelial conversion as they form spheres. We tested this in computer simulations using CompuCell-3D (25, 26), with the following assumptions: (i) A cell mass is exposed to Noggin evenly and simultaneously; (ii) in response, cells polarize and elongate; (iii) polarized cells secrete extracellular matrix; (iv) polarized cells have to be exposed to extracellular space at both their apical and basal surfaces; (v) tight junctions form at the apical ends; and (vi) misplaced cells rearrange their polarity and attach to their appropriate ends (27). This causes cells to become arranged in spherical masses around a lumen (movie S4) (9). After a transition period of intense cell rearrangement, the somites stabilize. The number of cells they contain is relatively invariant, and their structure is similar to that seen in vivo. There is no tendency to merge into a giant structure, nor do very small stable somites form. We propose that somite size and shape can be controlled entirely by local cell interactions, such as adhesion and packing constraints of cells transitioning between the mesenchyme and a polarized epithelium (28). Inhibition of BMP by Noggin may be a trigger for this conversion, consistent with the abnormal somite formation in Noggin-null mice (29), and may also “freeze” molecular determinants of axial identity (Hox code). In normal embryos, the segmentation clock and associated wave are likely to play a role in regulating the timing of somite formation and coupling this to the subdivision of each somite into rostral and caudal subcompartments.
Supplementary Material
Acknowledgments
This work was funded by Medical Research Council (grant G0700095) and the NIH (National Institute of General Medical Sciences grants R01GM076692 and R01GM077138). N. De-Oliveira helped with cloning, C. Thrasivoulou with confocal microscopy, and A. Norris with vibratome sectioning. S. Evans, J. Sharpe, A. Streit, and S. Thorsteinsdóttir provided information and constructive comments.
Footnotes
References and Notes
- 1.Muñoz-Sanjuán I, Brivanlou AH. In: Gastrulation: From Cells to Embryo. Stern CD, editor. Cold Spring Harbor Press; New York: 2004. pp. 475–489. [Google Scholar]
- 2.Dubrulle J, McGrew MJ, Pourquié O. Cell. 2001;106:219–232. doi: 10.1016/s0092-8674(01)00437-8. [DOI] [PubMed] [Google Scholar]
- 3.Dubrulle J, Pourquié O. Nature. 2004;427:419–422. doi: 10.1038/nature02216. [DOI] [PubMed] [Google Scholar]
- 4.Cooke J, Zeeman EC. J. Theor. Biol. 1976;58:455–476. doi: 10.1016/s0022-5193(76)80131-2. [DOI] [PubMed] [Google Scholar]
- 5.Harima Y, Takashima Y, Ueda Y, Ohtsuka T, Kageyama R. Cell Rep. 2013;3:1–7. doi: 10.1016/j.celrep.2012.11.012. [DOI] [PubMed] [Google Scholar]
- 6.Schröter C, Oates AC. Curr. Biol. 2010;20:1254–1258. doi: 10.1016/j.cub.2010.05.071. [DOI] [PubMed] [Google Scholar]
- 7.Tonegawa A, Funayama N, Ueno N, Takahashi Y. Development. 1997;124:1975–1984. doi: 10.1242/dev.124.10.1975. [DOI] [PubMed] [Google Scholar]
- 8.Streit A, Stern CD. Mech. Dev. 1999;85:85–96. doi: 10.1016/s0925-4773(99)00085-4. [DOI] [PubMed] [Google Scholar]
- 9.See materials and methods and other supplementary materials on Science Online.
- 10.Palmeirim I, Henrique D, Ish-Horowicz D, Pourquié O. Cell. 1997;91:639–648. doi: 10.1016/s0092-8674(00)80451-1. [DOI] [PubMed] [Google Scholar]
- 11.Oates AC, Morelli LG, Ares S. Development. 2012;139:625–639. doi: 10.1242/dev.063735. [DOI] [PubMed] [Google Scholar]
- 12.Hamburger V, Hamilton HL. J. Morphol. 1951;88:49–92. [PubMed] [Google Scholar]
- 13.Beddington RS, Martin P. Mol. Biol. Med. 1989;6:263–274. [PubMed] [Google Scholar]
- 14.Stern CD, Fraser SE, Keynes RJ, Primmett DR. Development. 1988;104(suppl.):231–244. doi: 10.1242/dev.104.Supplement.231. [DOI] [PubMed] [Google Scholar]
- 15.Keynes RJ, Stern CD. Nature. 1984;310:786–789. doi: 10.1038/310786a0. [DOI] [PubMed] [Google Scholar]
- 16.Takahashi Y, Inoue T, Gossler A, Saga Y. Development. 2003;130:4259–4268. doi: 10.1242/dev.00629. [DOI] [PubMed] [Google Scholar]
- 17.Jouve C, Iimura T, Pourquie O. Development. 2002;129:1107–1117. doi: 10.1242/dev.129.5.1107. [DOI] [PubMed] [Google Scholar]
- 18.Lim TM, Lunn ER, Keynes RJ, Stern CD. Development. 1987;100:525–533. doi: 10.1242/dev.100.3.525. [DOI] [PubMed] [Google Scholar]
- 19.Rodrigues S, Santos J, Palmeirim I. Gene Expr. Patterns. 2006;6:673–677. doi: 10.1016/j.modgep.2006.01.004. [DOI] [PubMed] [Google Scholar]
- 20.Burke AC, Nelson CE, Morgan BA, Tabin C. Development. 1995;121:333–346. doi: 10.1242/dev.121.2.333. [DOI] [PubMed] [Google Scholar]
- 21.Gaunt SJ. Int. J. Dev. Biol. 1994;38:549–552. [PubMed] [Google Scholar]
- 22.Iimura T, Pourquié O. Nature. 2006;442:568–571. doi: 10.1038/nature04838. [DOI] [PubMed] [Google Scholar]
- 23.Wacker SA, McNulty CL, Durston AJ. Dev. Biol. 2004;266:123–137. doi: 10.1016/j.ydbio.2003.10.011. [DOI] [PubMed] [Google Scholar]
- 24.Stern CD, Bellairs R. J. Embryol. Exp. Morphol. 1984;81:75–92. [PubMed] [Google Scholar]
- 25.Hester SD, Belmonte JM, Gens JS, Clendenon SG, Glazier JA. PLOS Comput. Biol. 2011;7:e1002155. doi: 10.1371/journal.pcbi.1002155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Swat MH, et al. Methods Cell Biol. 2012;110:325–366. doi: 10.1016/B978-0-12-388403-9.00013-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Martins GG, et al. PLOS ONE. 2009;4:e7429. doi: 10.1371/journal.pone.0007429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nakaya Y, Kuroda S, Katagiri YT, Kaibuchi K, Takahashi Y. Dev. Cell. 2004;7:425–438. doi: 10.1016/j.devcel.2004.08.003. [DOI] [PubMed] [Google Scholar]
- 29.McMahon JA, et al. Genes Dev. 1998;12:1438–1452. doi: 10.1101/gad.12.10.1438. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.