Abstract
We report the first electrochemical SNP detection system that can accurately discriminate homozygous and heterozygous genotypes using microfluidics technology. To achieve this, our system performs real-time melt-curve analysis of surface immobilized hybridization probe. As an example, we used our sensor to genotype two SNPs in the apolipoprotein E (ApoE) gene, where homozygous and heterozygous mutations greatly affect the risk of late-onset Alzheimer’s disease. Using probes specific for each SNP, we simultaneously acquired melt curves for probe-target duplexes at two different loci and thereby accurately distinguish all six possible ApoE allele combinations. Since the design of our device and probes can be readily adapted for targeting other loci, we believe that our method offers a modular platform for SNP-based disease diagnosis and personalized medicine.
Keywords: DNA, electrochemistry, microfluidics, molecular diagnostics
Single nucleotide polymorphisms (SNP) are important diagnostic indicators for inheritable diseases[1, 2] and predicting drug responses.[3] Genetically-informed treatment and dosing decisions will require effective platforms for SNP detection that are rapid, portable, and cost effective.[4] Such platforms must also support multiplexed detection and accurately differentiate between homozygous and heterozygous mismatches – a critical factor for many diagnoses.[5, 6]
Electrochemical DNA sensors offer robustness, portability, and compatibility with microfluidics and microelectronics,[7, 8] and many varieties of electrochemical DNA sensors (as reviewed by Palecek and Bartosik)[9] have been developed for SNP detection, including various enzymatic[10–12] and hybridization-based assays.[13–15] Surface-immobilized, redox-reporter labeled probes offer many advantages in this arena as they require no exogenous reagents, exhibit low background, and can operate directly in complex mixtures.[16] Innovative probe designs based on triple-stem probes,[17] polarity-switching probes,[18] and base-pair stacking probes[19] have all proven successful at SNP detection. Unfortunately, none of these sensors have shown the capacity to accurately discriminate heterozygous mismatches, as such samples contain a mixture of matched and mismatched DNA, and these probes cannot accurately resolve the minute differences in hybridization energy. Fluorescence-based, solution-phase methods such as dynamic allele-specific hybridization (DASH) can, in contrast, distinguish homozygous and heterozygous SNPs via melting curve analysis in solution phase.[20] This has spurred interest in integrating melt-curve analysis with electrochemical detection, but these efforts have been confounded by difficulties in obtaining accurate electrochemical and temperature measurements with sufficient speed and resolution.[21–23]
We present a novel microfluidic device that performs electrochemical melt-curve measurements with unprecedented temporal resolution at speeds that enable “real-time” analysis, and can accurately discriminate between homozygous and heterozygous SNP genotypes. Our ‘microE-DASH’ chip employs surface-bound, redox-tagged DNA probes[24] complementary to a SNP-containing target sequence. As we heat the chip, we can obtain a melt-curve by continuously measuring changes in the electrochemical signal. Furthermore, microE-DASH can achieve multiplexed detection of homozygous and heterozygous genotypes at multiple loci in a single step by integrating multiple probes and electrodes within the chip. As a model, we used microE-DASH to genotype two distinct SNP loci associated with the ApoE gene, where specific alleles strongly affect individual risk of late-onset Alzheimer’s disease.[25]
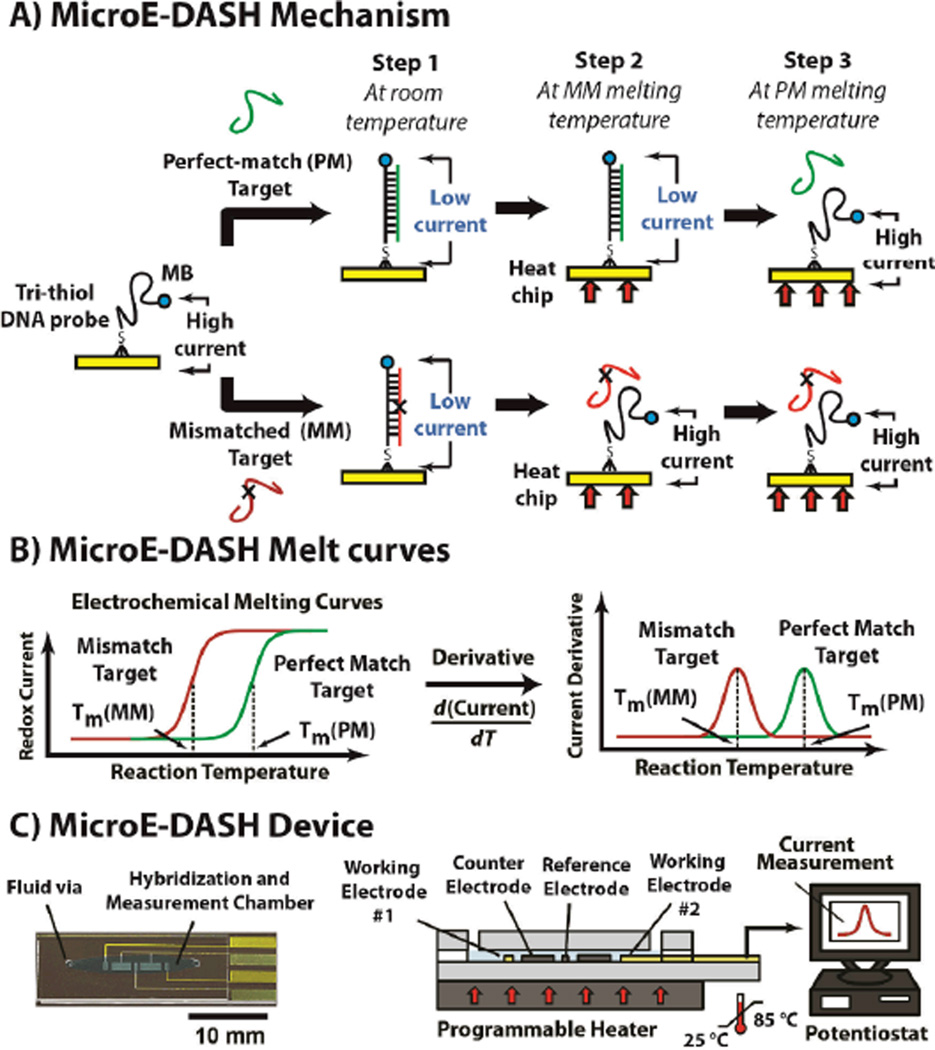
MicroE-DASH achieves accurate SNP discrimination by monitoring thermal melting of duplexes formed by surface-bound probes and their DNA targets in real time (Figure 1A). The probes are linear, single-stranded DNA, modified with a redox reporter (methylene blue; MB) at their 3' end and self-assembled onto gold working electrodes via a tri-thiol modification at their 5' end. In the absence of target, these probes are unstructured and flexible, allowing the MB reporter to readily approach the gold electrode, generating a redox current that can be measured using alternating-current voltammetry (ACV). However, probe-target hybridization decreases the current because the stiffer double-stranded duplex greatly reduces reporter access to the electrode.[26]
Figure 1.
Micro-Electrochemical Dynamic Allele-Specific Hybridization (microE-DASH). A) MicroE-DASH obtains real-time electrochemical melt curves from linear, single-stranded DNA probes complementary to the SNP target. The probes are linked to electrodes via a 5’ tri-thiol anchor with a methylene blue (MB) redox reporter at the 3’ terminus. The flexible unbound probe allows MB to readily approach the working electrode, generating high current. Rigid target-probe duplexes restrict MB-electrode interaction, resulting in a current decrease. Temperature-dependent changes in redox current reveal differences in melting temperature (Tm) between perfectly-matched (PM) and mismatched (MM) targets. B) The Tm corresponds to the temperature at which the rate of current change is greatest. We can thus determine the Tm by the peak of the first derivative of the redox current as a function of temperature (dI/dT). C) The microE-DASH chip consists of two glass pieces separated by a PDMS gasket, mounted on a programmable Peltier heater. The lower glass piece features two working electrodes, coupled with counter and reference electrodes, which form an electrochemical cell.
To distinguish perfectly-matched (PM) targets from those containing single-base mismatches (MM), we ramp the temperature until the duplex completely melts. Due to its higher hybridization energy, the PM target (Figure 1A, top) melts at a higher temperature than the MM target (Figure 1A, bottom), and MicroE-DASH can readily distinguish the two by continuously measuring redox current as a function of temperature (Figure 1B, left). Furthermore, we can accurately determine the melting temperature of the duplex (Tm) by taking the derivative of the current as a function of temperature (dI/dT) as described below (Figure 1B, right). Heaton et al.[27] and Mahajan et al.[28] have shown that strong DC electric fields can influence DNA duplex formation and dyhybridization. However, given the lower magnitude and shorter duration of our AC measurements, we suspect that electric fields have negligible effect on the measured melt temperature of our duplexes.
The MicroE-DASH chip contains a single reaction chamber that incorporates platinum reference and counter electrodes and two probe-conjugated gold working electrodes to form a multiplexed electrochemical measurement cell (Figure 1C; see Supporting Information for fabrication details). The chip is mounted on a Peltier heater, which controls the temperature in a pre-programmed sequence while a potentiostat continuously records the peak redox current from the two working electrodes. Importantly, the chip is designed for minimal thermal resistance, so that the temperature of the electrodes and reaction chamber can equilibrate in seconds (Figure S1). This enables near-real-time acquisition of melt-curves, such that the entire assay can be completed within 30 minutes without the need to return to room temperature for measurement.[22, 29] To ensure probe stability, we immobilized them onto the electrodes via 5-tri-thiol modification.[30] This dramatically improves their thermal stability; mono-thiol probes detach from the electrodes at temperatures as low as 75 °C, whereas tri-thiol probes are stable at temperatures up to 85 °C, maintaining >99% of the maximum signal (Figure S2).
As proof of concept, we designed probes for SNPs within the ApoE gene that serve as important clinical diagnostic indicators for Alzheimer’s disease.[25] In addition to the normal isoform (ε3), this gene has two variants (ε4 and ε2) arising from SNPs rs429358 (T:C) and rs7412 (C:T), referred to as T1 and T2 respectively in this work. Carriers of the ε4 allele – and ε4-ε4 homozygotes in particular – exhibit greater risk of developing Alzheimer’s.[31] Conversely, the ε2 allele is associated with a reduced likelihood for Alzheimer’s.[32] Accurate identification of the six possible allele combinations (Table 1) is thus important for identifying high-risk individuals.
Table 1.
ApoE genotypes and their associated MicroE-DASH readout
| Genotype | Alzheimer’s Risk | T1 Melt Curve | T2 Melt Curve |
|---|---|---|---|
| ε3-ε3 | Normal | PM | PM |
| ε2-ε2 | Decreased | PM | MM |
| ε4-ε4 | Increased | MM | PM |
| ε3-ε2 | Decreased | PM | Heterozygous |
| ε3–ε4 | Increased | Heterozygous | PM |
| ε2–ε4 | Normal | Heterozygous | Heterozygous |
We designed probes complementary to the T1 and T2 sequences associated with the ε3 isoform. This allowed us to readily identify homozygotes for ε3, ε2, ε4, or any heterozygous combination of alleles, based on whether T1 and T2 form PM or MM duplexes with their respective probes (Table 1). We considered three key criteria in designing the probes. First, we ensured that Tm occurs between 45–65 °C, which is sufficiently high for distinguishing secondary melt transitions but within the thermal stability range of the tri-thiolated probes. Second, we designed probes with minimal secondary structure because self- hybridization can result in complex melting behavior and compete with target hybridization. Finally, we designed the two probes with minimal sequence overlap to minimize potential cross-reactivity. We performed modeling with mfold[33] software to ensure our probe design satisfied the first two criteria (see Supporting Information for sequences), and verified their target specificity by immobilizing the probes onto gold electrodes and incubating them with non-matching targets (i.e., T1 target with T2 probe, and vice versa), resulting in minimal signal change compared to matched targets (Figure S3).
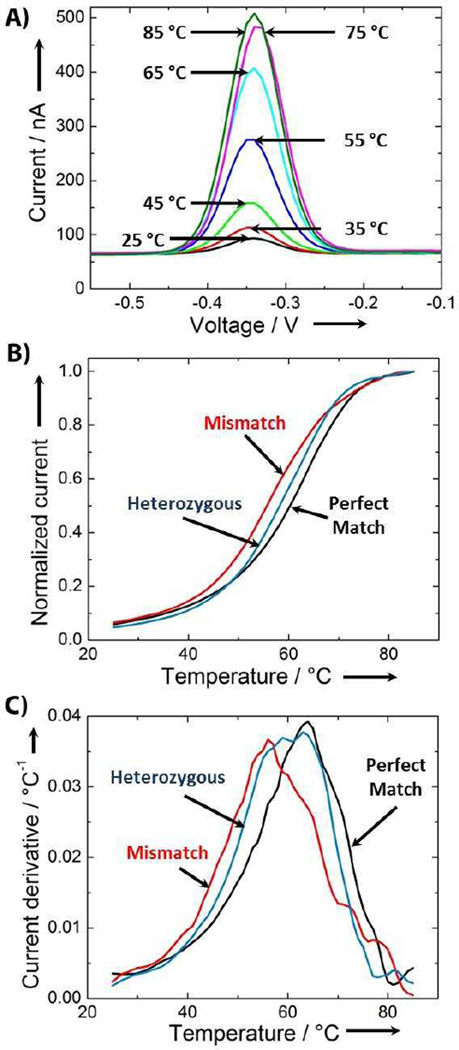
ACV measurements taken as a function of temperature can precisely track the melting characteristics of DNA targets. To demonstrate this, we hybridized the PM target to the T2 probe and measured the electrochemical current from 25 to 85 °C in 1 °C intervals. We observed a dramatic increase in redox current between 45 and 65 °C (Figure 2A), consistent with the sudden transition expected during melting of the T2-PM duplex. In the absence of target, instead of the sudden current increase expected from a duplex melting transition, we saw a current increase resulting from increased thermal motion of probe molecules at higher temperatures,[23] which in turn results in more frequent interaction between redox reporter molecules and the electrode (Figure S4).
Figure 2.
DNA sample genotyping using microE-DASH. A) Peak current traces taken in 10°C increments during temperature ramp of T2 probe incubated with a PM target reveal melting-dependent increases in current. B) Plotting the peak currents measured for T2 probe incubated with PM, MM and a heterozygous 1:1 mixture of PM and MM at 1°C increments reveals distinct melt-curves for each genotype. Based on the half-maximal current, the PM (black) target forms the most stable duplex with the highest Tm. The MM (red) target forms a less stable duplex with a lower Tm, while the heterozygous sample (blue) exhibits an intermediate Tm. C) By plotting the derivative of the current as a function of temperature (dI/dT) versus temperature, we can determine Tm with a precision of approximately ±1 °C. The target trace from each genotype has a unique peak.
Differences in melting behavior can be directly observed in real-time by plotting the peak redox current as a function of temperature. We demonstrated this after challenging T2 probes with PM, MM or a heterozygous (HET) mixture of PM and MM targets (1:1 ratio) (Figure 2B). We normalized peak-current readings to a 0 to 1 scale using the initial and maximum current values in each run, with five-point averaging to smooth the melt curve (see Methods). As a general trend, we observed clear differences among the three melt curves. The PM target melted at a higher temperature (Tm = 61 °C) than the MM target (Tm = 56 °C), while the curve for the HET mixture fell in between, with a Tm of 58 °C.
To determine accurate Tm we calculated the derivative of the current with respect to temperature (dI/dT) and defined Tm as the point at which this derivative is largest, analogus to established fluorescence melting approaches.[34] PM and MM targets produced single, distinct peaks with Tm of 63 °C and 56 °C, respectively (Figure 2C, red and black). This large difference enabled accurate and reproducible detection of SNPs in homozygous samples. Analysis of HET samples is challenging because the resulting dI/dT plot consists of two peaks from the PM and MM targets that can potentially overlap (Figure 2C, blue). To de-convolve the contribution of each probe-target duplex, we adopted an analytical strategy used for extracting peaks from multiple melt-curves (see Methods). We applied a curve-fitting algorithm to our single-peak PM and MM melt-curves, and determined that a Lorentzian function yields the best fit to our experimental data (Figure S5).[35, 36] When we applied this fit to the T2 heterozygous melt-curve (Figure 2C), we identified two curves with peaks at 65.4 °C and 56.6 °C. These correspond well to the measured Tm of PM and MM, respectively, confirming the validity of this approach.
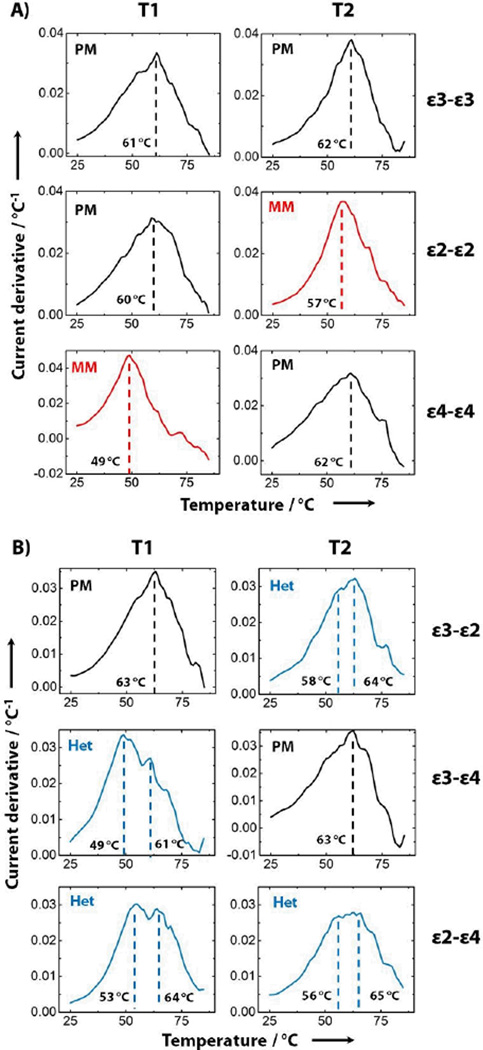
MicroE-DASH’s capacity to simultaneously differentiate multiple homozygous and heterozygous SNP samples enabled us to accurately genotype all six ApoE SNP combinations in a single-step assay. We assembled a duplex microE-DASH chip with a different probe immobilized on each of the two sensors, allowing us to simultaneously determine melt temperatures for both T1 and T2. We first obtained melt curves for each of the three homozygous ApoE genotypes (Figure 3A). As expected, these samples generated single peaks for both T1 and T2, since SNP mismatches occur in both alleles for any given homozygous genotype. The single-peak Tm measured in these duplex devices agree with repeated measurements from single-target experiments of T1 (PM: 61 ± 1.5 °C, MM: 50 ± 1 °C) and T2 (PM: 62 ± 0.6 °C, MM: 56 ± 0.6 °C). Furthermore, the shifts in the melt curve observed for the ε2-ε2 and ε4-ε4 samples corresponded with our predictions based on the target-probe mismatches identified in Table 1.
Figure 3.
ApoE genotyping via multiplexed real-time melt-curve analysis. Plots of independent melt-curve derivatives from samples containing PCR-length ApoE targets simulating different A) homozygous (PM and MM) and B) heterozygous (Het) genotypes, with the corresponding Tm reported in black. Each row represents a different allele combination, associated with a different response in the T1 or T2 melting curves. Black plots indicate perfect match between probe and target, whereas red plots indicate SNP mismatch. Blue plots represent profiles with dual peaks arising from heterozygous samples.
We subsequently used microE-DASH to accurately identify all three possible heterozygous genotypes. We obtained T1 and T2 melt curves as described above, and determined that each heterozygous combination yielded the predicted melt-curve shifts (Figure 3B). The ε3-ε2 and ε3-ε4 genotypes resulted in heterozygous melt curves for T2 and T1, respectively, while the ε2–ε4 genotype yielded heterozygous melt curves for both probes. We used deconvolution analysis on the heterozygous melt curves to extract individual Tm for each target-probe combination, which were in agreement with the PM and MM Tm obtained from homozygous samples. For example, the T2 melt curve for the ε3-ε2 target yielded peaks of 58°C and 64°C, which correspond well with our previous MM and PM Tm measurements of 57°C and 63°C respectively. The ΔTm for these heterozygous samples matches well with the individually measured single-peak Tm. This confirms that microE-DASH can accurately report both homozygous and heterozygous genotypes independent of the specific sample composition.
The microE-DASH microfluidic electrochemical SNP biosensor is thus capable of discriminating homozygous and heterozygous samples within ~30 minutes. Demonstrating the potential diagnostic utility of this, we have accurately resolved the six different SNP genotypes commonly associated with ApoE, an important diagnostic biomarker for Alzheimer’s disease. microE-DASH can be readily expanded to incorporate microfluidic PCR and single-strand generation in an integrated device.[37] Since the design of our probes is relatively straightforward, we believe that our microE-DASH may offer a modular platform for SNP-based disease diagnostics and personalized medicine.
Supplementary Material
Footnotes
We are grateful for financial support from the Otis Williams Fellowship, National Institutes of Health, and the Institute of Collaborative Biotechnologies through the Army Research Office. Microfabrication was carried out in the Nanofabrication Facility at UCSB.
Supporting information for this article is available on the WWW under http://dx.doi.org/10.1002/anie.201xxxxxx.
Contributor Information
Allen H. J. Yang, Department of Mechanical Engineering, University of California Santa Barbara (USA).
Kuangwen Hsieh, Department of Mechanical Engineering, University of California Santa Barbara (USA).
Adriana S. Patterson, Department of Chemistry and Biochemistry and Biomolecular Science and Engineering Program, University of California, Santa Barbara (USA)
B. Scott Ferguson, Department of Mechanical Engineering, University of California Santa Barbara (USA).
Michael Eisenstein, Department of Mechanical Engineering, University of California Santa Barbara (USA).
Kevin W. Plaxco, Department of Chemistry and Biochemistry and Biomolecular Science and Engineering Program, University of California, Santa Barbara (USA)
H. Tom Soh, Email: tsoh@engr.ucsb.edu, Materials Department and Department of Mechanical Engineering University of California, Santa Barbara, Santa Barbara, CA 93106 (USA); Department of Mechanical Engineering, University of California Santa Barbara (USA).
References
- 1.Suh Y, Vijg J. Mutation Research-Fundamental and Molecular Mechanisms of Mutagenesis. 2005;573:41–53. [Google Scholar]
- 2.Naylor SL. Frontiers in Bioscience. 2007;12:4111–4131. doi: 10.2741/2375. [DOI] [PubMed] [Google Scholar]
- 3.Johnson JA, Evans WE. Trends in Molecular Medicine. 2002;8:300–305. doi: 10.1016/s1471-4914(02)02354-7. [DOI] [PubMed] [Google Scholar]
- 4.Ginsburg GS, McCarthy JJ. Trends in Biotechnology. 2001;19:491–496. doi: 10.1016/s0167-7799(01)01814-5. [DOI] [PubMed] [Google Scholar]
- 5.Abou-Sleiman PM, Muqit MMK, McDonald NQ, Yang YX, Gandhi S, Healy DG, Harvey K, Harvey RJ, Deas E, Hatia K, Quinn N, Lees A, Latchman DS, Wood NW. Ann Neurol. 2006;60:414–419. doi: 10.1002/ana.20960. [DOI] [PubMed] [Google Scholar]
- 6.van Rijsingen IAW, Hermans-van Ast JF, Arens YHJM, Schalla SM, de Die-Smulders CEM, van den Wijngaard A, Pinto YM. Neth Heart J. 2009;17:458–463. doi: 10.1007/BF03086304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Drummond TG, Hill MG, Barton JK. Nature Biotechnology. 2003;21:1192–1199. doi: 10.1038/nbt873. [DOI] [PubMed] [Google Scholar]
- 8.Mir M, Homs A, Samitier J. Electrophoresis. 2009;30:3386–3397. doi: 10.1002/elps.200900319. [DOI] [PubMed] [Google Scholar]
- 9.Palecek E, Bartosik M. Chem Rev. 2012;112:3427–3481. doi: 10.1021/cr200303p. [DOI] [PubMed] [Google Scholar]
- 10.Feng KJ, Zhao JJ, Wu ZS, Jiang JH, Shen GL, Yu RQ. Biosensors & Bioelectronics. 2011;26:3187–3191. doi: 10.1016/j.bios.2010.12.024. [DOI] [PubMed] [Google Scholar]
- 11.Shin S, Won BY, Jung C, Shin SC, Cho DY, Leec SS, Park HG. Chem Commun. 2011;47:6611–6613. doi: 10.1039/c1cc11476j. [DOI] [PubMed] [Google Scholar]
- 12.Huang Y, Zhu J, Li GY, Chen ZC, Jiang JH, Shen GL, Yu RQ. Biosensors & Bioelectronics. 2013;42:526–531. doi: 10.1016/j.bios.2012.10.033. [DOI] [PubMed] [Google Scholar]
- 13.Ikeda R, Kobayashi S, Chiba J, Inouye M. Chem-Eur J. 2009;15:4822–4828. doi: 10.1002/chem.200802729. [DOI] [PubMed] [Google Scholar]
- 14.Wan Y, Lao RJ, Liu G, Song SP, Wang LH, Li D, Fan CH. J Phys Chem B. 2010;114:6703–6706. doi: 10.1021/jp100871u. [DOI] [PubMed] [Google Scholar]
- 15.Wu Y, Lai RY. Chem Commun. 2013;49:3422–3424. doi: 10.1039/c3cc41281d. [DOI] [PubMed] [Google Scholar]
- 16.Lubin AA, Plaxco KW. Accounts Chem Res. 2010;43:496–505. doi: 10.1021/ar900165x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Xiao Y, Lou XH, Uzawa T, Plakos KJI, Plaxco KW, Soh HT. J Am Chem Soc. 2009;131:15311–15316. doi: 10.1021/ja905068s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hsieh K, White RJ, Ferguson BS, Plaxco KW, Xiao Y, Soh HT. Angew Chem Int Edit. 2011;50:11176–11180. doi: 10.1002/anie.201103482. [DOI] [PubMed] [Google Scholar]
- 19.Boon EM, Barton JK. Bioconjugate Chem. 2003;14:1140–1147. doi: 10.1021/bc034139l. [DOI] [PubMed] [Google Scholar]
- 20.Howell WM, Jobs M, Gyllensten U, Brookes AJ. Nature Biotechnology. 1999;17:87–88. doi: 10.1038/5270. [DOI] [PubMed] [Google Scholar]
- 21.Surkus AE, Flechsig GU. Electroanalysis. 2009;21:1119–1123. [Google Scholar]
- 22.Nasef H, Beni V, O'Sullivan CK. Anal Methods-Uk. 2010;2:1461–1466. [Google Scholar]
- 23.Wohlgamuth CH, McWilliams MA, Slinker JD. Anal Chem. 2013;85:1462–1467. doi: 10.1021/ac302508f. [DOI] [PubMed] [Google Scholar]
- 24.Fan CH, Plaxco KW, Heeger AJ. Proceedings of the National Academy of Sciences of the United States of America. 2003;100:9134–9137. doi: 10.1073/pnas.1633515100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Strittmatter WJ, Roses AD. Annu Rev Neurosci. 1996;19:53–77. doi: 10.1146/annurev.ne.19.030196.000413. [DOI] [PubMed] [Google Scholar]
- 26.Uzawa T, Cheng RR, White RJ, Makarov DE, Plaxco KW. J Am Chem Soc. 2010;132:16120–16126. doi: 10.1021/ja106345d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Heaton RJ, Peterson AW, Georgiadis RM. Proceedings of the National Academy of Sciences of the United States of America. 2001;98:3701–3704. doi: 10.1073/pnas.071623998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mahajan S, Richardson J, Brown T, Bartlett PN. J Am Chem Soc. 2008;130:15589–15601. doi: 10.1021/ja805517q. [DOI] [PubMed] [Google Scholar]
- 29.Umek RM, Lin SW, Vielmetter J, Terbrueggen RH, Irvine B, Yu CJ, Kayyem JF, Yowanto H, Blackburn GF, Farkas DH, Chen YP. J Mol Diagn. 2001;3:74–84. doi: 10.1016/S1525-1578(10)60655-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Phares N, White RJ, Plaxeo KW. Anal Chem. 2009;81:1095–1100. doi: 10.1021/ac8021983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Verghese PB, Castellano JM, Holtzman DM. Lancet Neurol. 2011;10:241–252. doi: 10.1016/S1474-4422(10)70325-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Qiu C, Kivipelto M, Aguero-Torres H, Winblad B, Fratiglioni L. J Neurol Neurosur Ps. 2004;75:828–833. doi: 10.1136/jnnp.2003.021493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Vercoutere W, Winters-Hilt S, Olsen H, Deamer D, Haussler D, Akeson M. Nature Biotechnology. 2001;19:248–252. doi: 10.1038/85696. [DOI] [PubMed] [Google Scholar]
- 34.Reed GH, Kent JO, Wittwer CT. Pharmacogenomics. 2007;8:597–608. doi: 10.2217/14622416.8.6.597. [DOI] [PubMed] [Google Scholar]
- 35.Belozerova I, Levicky R. J Am Chem Soc. 2012;134:18667–18676. doi: 10.1021/ja3066368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wienken CJ, Baaske P, Duhr S, Braun D. Nucleic Acids Res. 2011;39 doi: 10.1093/nar/gkr035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ferguson BS, Buchsbaum SF, Swensen JS, Hsieh K, Lou X, Soh HT. Anal Chem. 2009;81:6503–6508. doi: 10.1021/ac900923e. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.