Abstract
Termites digest wood and other lignocellulosic substrates with the help of their intestinal microbiota. While the functions of the symbionts in the digestive process are slowly emerging, the origin of the bacteria colonizing the hindgut bioreactor is entirely unknown. Recently, our group discovered numerous representatives of bacterial lineages specific to termite guts in a closely related omnivorous cockroach, but it remains unclear whether they derive from the microbiota of a common ancestor or were independently selected by the gut environment. Here, we studied the bacterial gut microbiota in 34 species of termites and cockroaches using pyrotag analysis of the 16S rRNA genes. Although the community structures differed greatly between the major host groups, with dramatic changes in the relative abundances of particular bacterial taxa, we found that the majority of sequence reads belonged to bacterial lineages that were shared among most host species. When mapped onto the host tree, the changes in community structure coincided with major events in termite evolution, such as acquisition and loss of cellulolytic protists and the ensuing dietary diversification. UniFrac analysis of the core microbiota of termites and cockroaches and construction of phylogenetic tree of individual genus level lineages revealed a general host signal, whereas the branching order often did not match the detailed phylogeny of the host. It remains unclear whether the lineages in question have been associated with the ancestral cockroach since the early Cretaceous (cospeciation) or are diet-specific lineages that were independently acquired from the environment (host selection).
INTRODUCTION
Termites digest wood and other lignocellulosic substrates with the help of their intestinal microbiota—a symbiosis that has fascinated biologists for more than a century (1). The ability to mineralize lignocellulose and humus lends termites an important place in carbon and nitrogen cycling in tropical soils (2) and makes them promising models for the industrial conversion of lignocellulose into microbial products and the production of biofuels (3).
The ancestors of termites were presumably detritivorous subsocial cockroaches (4, 5). About 130 million years ago, they gained the ability to digest wood through acquisition of cellulolytic flagellates (6, 7). These eukaryotic protists, which fill up the bulk of the hindgut volume, are the major habitat of the prokaryotic community present in the digestive tracts of all phylogenetically lower termites (1, 8). The complete loss of all gut flagellates in the youngest termite family, the Termitidae—another hallmark in the evolutionary history of termites—led to dietary diversification and enormous ecological success (6, 9). While the Macrotermitinae established a unique symbiosis with a lignocellulolytic fungus (10), other lineages of higher termites started to exploit diets of increasing humification, a development accompanied by further differentiation of the hindgut (9) and its entirely prokaryotic microbiota (8, 11).
While the role of the cellulolytic flagellates in lower termites is well defined, the functions of the mostly uncultivated bacterial symbionts in the digestive process, particularly in the flagellate-free higher termites, are just emerging (1, 8). Most importantly, the origin of the bacteria colonizing the hindgut bioreactor is entirely unknown (11).
Although the gut microbiota differs substantially between termite species, it comprises many phylogenetic clusters that are unique to termites 12–14). The origin of these lineages remains unclear, but their detection also in the guts of several cockroaches (15–17), the closest relatives of termites, together with occasional evidence of cocladogenesis with the termite hosts (18, 19) has given rise to the hypothesis that the bacterial microbiota of extant termites and cockroaches is derived from their common dictyopteran ancestors.
In this study, we used a cultivation-independent high-throughput approach to characterize the diversity and structure of the intestinal microbiota in a broad selection of termites and cockroaches. 16S rRNA gene fragments (the V3-V4 region) were amplified with universal bar-coded primers and classified using a comprehensive reference database of all homologs previously obtained from insect guts, which had been optimized to resolve termite- and cockroach-specific groups (20). Comparative analysis of the data sets was employed to detect the presence and distribution of common bacterial lineages across the major host groups.
MATERIALS AND METHODS
Insect samples.
Termites were taken from colonies maintained in the laboratory or were collected in the field. Cockroaches and other insects were purchased from commercial breeders, and the hindguts were dissected immediately upon arrival (17, 20). In some cases, field-collected termites had to be preserved in ethanol for transport. Since the entire guts of ethanol-preserved specimens were processed within less than 1 week, detrimental effects of this treatment on community structure can be excluded (21). Moreover, our data set includes several closely related species that differ with respect to this pretreatment but yielded highly similar profiles, which further precludes a potential bias introduced by the inclusion of ethanol-stored samples. Details on the nature and origin of each sample are shown in Table 1. Field-collected specimens were routinely identified by sequencing their mitochondrial cytochrome oxidase II (COII) genes (26).
TABLE 1.
Characteristics of 16S rRNA gene amplicon libraries of bacterial hindgut microbiota of each host species
| Sample no. | Host species | Origina | Total no. of reads | No. of genus level taxa | No. of OTUs (3% dissimilarity) | Coverage (%)b | Diversity indexc |
NCBI accession no.d | ||
|---|---|---|---|---|---|---|---|---|---|---|
| Richness | Diversity | Evenness | ||||||||
| Cockroaches | ||||||||||
| 1 | Ergaula capucina | B1 | 6,020 | 232 | 891 | 70.5 | 1,266 | 5.52 | 0.84 | 068 |
| 2 | Symploce macroptera | B1 | 5,045 | 135 | 499 | 80.9 | 431 | 4.70 | 0.82 | 069 |
| 3 | Rhyparobia maderae | B1 | 12,164 | 268 | 1,346 | 70.8 | 2,540 | 5.42 | 0.78 | 070 |
| 4 | Elliptorhina chopardi | B1 | 6,794 | 200 | 663 | 79.6 | 798 | 5.25 | 0.83 | 071 |
| 5 | Panchlora sp. | B1 | 11,889 | 212 | 2,042 | 66.6 | 1,064 | 4.41 | 0.72 | 072 |
| 6 | Diploptera punctata | B1 | 5,708 | 161 | 543 | 80.8 | 627 | 4.93 | 0.82 | 073 |
| 7 | Opisthoplatia orientalis | B1 | 11,707 | 291 | 1,515 | 70.5 | 3,153 | 5.72 | 0.80 | 074 |
| 8 | Panesthia angustipennis | B1 | 5,394 | 202 | 1,141 | 72.5 | 1,710 | 6.01 | 0.88 | 075 |
| 9 | Salganea esakii | B1 | 17,412 | 296 | 1,916 | 80.8 | 2,955 | 6.27 | 0.84 | 076 |
| 10 | Eublaberus posticus | B1 | 103,530 | 416 | 5,743 | 79.9 | 12,034 | 5.34 | 0.64 | 077 |
| 11 | Schultesia lampyridiformis | B1 | 5,085 | 217 | 857 | 70.3 | 1,482 | 5.42 | 0.83 | 078 |
| 12 | Eurycotis floridana | B1 | 41,336 | 354 | 3,410 | 77.2 | 6,855 | 5.80 | 0.75 | 079 |
| 13 | Shelfordella lateralis | B1 | 6,226 | 186 | 714 | 82.4 | 674 | 5.30 | 0.86 | 080 |
| 14 | Blatta orientalis | B1 | 8,024 | 246 | 1,069 | 68.6 | 2,045 | 5.14 | 0.76 | 081 |
| 15 | Cryptocercus punctulatus | F1 | 6,715 | 180 | 715 | 75.5 | 884 | 4.90 | 0.78 | 082 |
| Lower termites | ||||||||||
| 16 | Mastotermes darwiniensis | L1 | 7,596 | 137 | 398 | 86.3 | 583 | 3.94 | 0.68 | 083 |
| 17 | Zootermopsis nevadensis | L2 | 6,129 | 278 | 1,617 | 72.6 | 3,451 | 5.18 | 0.72 | 084 |
| 18 | Hodotermopsis sjoestedti | L1 | 7,600 | 272 | 1,584 | 73.9 | 3,569 | 5.25 | 0.73 | 085 |
| 19 | Hodotermes mossambicus | F2 | 16,520 | 204 | 978 | 74.3 | 1,840 | 5.33 | 0.79 | 086 |
| 20 | Incisitermes marginipennis | L1 | 16,491 | 299 | 2,807 | 79.0 | 6,354 | 4.27 | 0.56 | 087 |
| 21 | Neotermes jouteli | F3 | 6,256 | 276 | 2,354 | 78.4 | 4,547 | 4.70 | 0.63 | 088 |
| 22 | Reticulitermes santonensis | L2 | 48,066 | 112 | 427 | 85.1 | 602 | 3.92 | 0.67 | 089 |
| 23 | Coptotermes niger | L1 | 53,003 | 91 | 166 | 87.2 | 202 | 2.26 | 0.45 | 090 |
| Higher termites | ||||||||||
| 24 | Odontotermes sp.e | F4 | 12,898 | 307 | 1,005 | 63.1 | 1,391 | 5.77 | 0.86 | 091 |
| 25 | Macrotermes sp. | L1 | 12,073 | 260 | 1,358 | 69.5 | 2,790 | 5.34 | 0.76 | 092 |
| 26 | Macrotermes subhyalinuse | F5 | 27,297 | 211 | 4,805 | 68.4 | 1,182 | 5.23 | 0.84 | 093 |
| 27 | Alyscotermes trestuse | F5 | 24,582 | 550 | 3,203 | 78.6 | 5,940 | 6.57 | 0.82 | 094 |
| 28 | Cubitermes ugandensis | F6 | 22,832 | 211 | 5,413 | 49.7 | 2,020 | 6.49 | 0.97 | 095 |
| 29 | Ophiotermes sp.e | F7 | 8,418 | 328 | 1,336 | 76.0 | 2,026 | 6.13 | 0.85 | 096 |
| 30 | Amitermes meridionalise | F8 | 23,840 | 354 | 1,556 | 85.5 | 2,246 | 5.04 | 0.70 | 097 |
| 31 | Microcerotermes sp.e | F5 | 34,626 | 291 | 2,358 | 79.1 | 4,407 | 4.55 | 0.61 | 098 |
| 32 | Nasutitermes corniger | L3 | 10,363 | 175 | 1,998 | 65.5 | 1,208 | 4.15 | 0.67 | 099 |
| 33 | Nasutitermes takasagoensis | F9 | 16,619 | 198 | 1,602 | 77.4 | 3,607 | 4.04 | 0.56 | 100 |
| 34 | Trinervitermes sp. | F5 | 25,173 | 232 | 1,103 | 84.2 | 1,943 | 4.68 | 0.67 | 101 |
| Others | ||||||||||
| 35 | Pachnoda ephippiata | B2 | 10,033 | 339 | 1,325 | 80.2 | 1,335 | 5.77 | 0.85 | 102 |
| 36 | Acheta domesticus | B2 | 5,326 | 104 | 241 | 84.2 | 276 | 4.14 | 0.80 | 103 |
| 37 | Gryllus assimilis | B1 | 26,800 | 190 | 669 | 90.3 | 712 | 4.13 | 0.70 | 104 |
Origins of samples: B, commercial breeders (B1, Jörg Bernhardt, Helbigsdorf, Germany [http://www.schaben-spinnen.de]; B2, b.t.b.e. Insektenzucht, Schnürpflingen, Germany); F, field collections (F1, Heywood County, NC, USA [by C. Nalepa]; F2, near Pretoria, South Africa [by J. Rohland]; F3, Fort Lauderdale, FL, USA [by R. H. Scheffrahn]; F4, near Kajiado, Kenya; F5, near Nairobi, Kenya [by J. O. Nonoh]; F6, Lhiranda Hill, Kakamega, Kenya; F7, Kalunja Glade, Kakamega, Kenya [by D. K. Ngugi]; F8, Lakefield NP, Cape York, Australia [by A. Brune]; F9, near Nishihara, Japan [by G. Tokuda]); L, laboratory colonies (L1, R. Plarre, Federal Institute for Materials Research and Testing, Berlin, Germany; L2, MPI Marburg; L3, R. H. Scheffrahn, University of Florida, Fort Lauderdale, FL, USA).
Good's coverage estimator (22).
Based on OTUs. Richness, Chao1 estimator (23); diversity, nonparametric Shannon index (24); evenness index (25).
All data sets were submitted to the Sequence Read Archive of NCBI (http://www.ncbi.nlm.nih.gov; BioProject PRJNA217467). The individual accession numbers are in the format SAMN02228nnn, with the last three digits indicated in the table.
Ethanol-preserved specimen; the entire gut was used for DNA extraction.
Pyrotag sequencing.
DNA was extracted from the pooled gut homogenates of 3 to 10 individuals of each species (depending on gut volume) using a bead-beating protocol with phenol-chloroform purification (27). PCR amplification of the V3-V4 region of the bacterial 16S rRNA genes with a bar-coded primer set (343Fmod-784Rmod) modified to optimize coverage of the taxa known to prevail in termite and cockroach guts was performed as previously described (20). Amplicons were mixed in equimolar amounts and commercially sequenced (454 GS FLX Titanium technology; GATC Biotech, Constance, Germany). The pyrotag sequences were preprocessed and aligned using the Mothur software suite (28) (version 1.27.0) under stringent conditions (29) (reads of >200 bp, no ambiguous bases, and a maximum number of homopolymers of ≤8). The sequences in each sample where denoised with the Acacia program (30) using default parameters, except that the standard deviation from the mean read length was set to 5 to avoid the loss of entire taxa from individual data sets due to sequence length heterogeneity between phylotypes. Denoising reduced the number of operational taxonomic units (OTUs) (3% sequence dissimilarity) in the samples by 0 to 5%.
Classification.
Sequence reads were classified with the Naive Bayesian Classifier implemented in Mothur, using a bootstrap value of 60% as the cutoff. Since classification success with public reference databases was limited due to lack of taxonomic resolution, particularly in the groups represented in termites and cockroaches (20) (Table 2), we used a customized reference database to improve resolution (DictDb v. 2.3). The reference database was built on the basis of the Silva database (Silva SSU Ref NR release 114; http://www.arb-silva.de), to which additional sequences from bacterial microbiota of dictyopteran insects were added, including both sequences from published studies and unpublished data from our laboratory. The taxonomy of relevant lineages was refined by incorporating genus level taxa that have been identified either in published phylogenies of relevant groups (13, 31) or additional hitherto unresolved monophyletic groups. The reference database is available upon request; publication of the latest version, documenting the detailed classification of termite- and cockroach-specific clusters, is in preparation.
TABLE 2.
Improvement of classification successa
| Host species | Classification successb (%) |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Phylum |
Class |
Order |
Family |
Genus |
||||||
| RDP | This study | RDP | This study | RDP | This study | RDP | This study | RDP | This study | |
| Cockroaches | ||||||||||
| Ergaula capucina | 94.9 | 99.3 | 79.3 | 96.1 | 73.4 | 93.1 | 63.3 | 88.0 | 33.2 | 61.6 |
| Panesthia angustipennis | 95.9 | 99.3 | 79.3 | 96.1 | 77.2 | 94.2 | 59.0 | 86.0 | 24.8 | 61.0 |
| Salganea esakii | 93.8 | 99.4 | 79.7 | 95.3 | 78.8 | 93.5 | 71.1 | 90.1 | 40.6 | 74.6 |
| Blatta orientalis | 96.2 | 99.3 | 79.8 | 97.7 | 77.9 | 96.3 | 70.5 | 93.4 | 48.0 | 66.6 |
| Cryptocercus punctulatus | 93.8 | 99.0 | 74.8 | 93.4 | 72.5 | 90.2 | 59.0 | 82.8 | 33.1 | 67.7 |
| Termites | ||||||||||
| Reticulitermes santonensis | 85.5 | 99.2 | 80.9 | 96.2 | 79.2 | 95.6 | 76.1 | 94.5 | 68.0 | 93.3 |
| Cubitermes ugandensis | 92.0 | 98.7 | 84.4 | 97.4 | 78.4 | 94.9 | 64.7 | 83.3 | 24.3 | 69.4 |
| Nasutitermes corniger | 87.8 | 98.9 | 83.0 | 97.4 | 81.6 | 96.5 | 77.9 | 95.3 | 40.9 | 91.0 |
Improvement of classification success using our curated reference database, which included all homologs previously obtained from insect guts and was optimized to resolve all termite- and cockroach-specific groups (20), over that using the Ribosomal Database Project (RDP) database (release 10, update 30).
The proportions of classified sequences in representative samples are reported for different taxonomic levels. Classification success for all samples is shown in Table S2 in the supplemental material.
Statistical analyses.
All samples were subsampled to the smallest number of reads per sample in the data set (5,045 reads). Classification-dependent ordinations (genus level) were based on the Bray-Curtis dissimilarity coefficient (32). To reduce the dimensions of the data set, the results were displayed using nonmetric multidimensional scaling (NMDS). Classification-independent ordinations were carried out using the same strategy, with reads grouped into OTUs, or a phylogeny-based analysis of the reads with UniFrac (33) displayed using principal-coordinate analysis (PCoA). For all analyses, the significance of clusters was tested by analysis of variance using distance matrices (ADONIS). The significance of clusters in OTU- and taxon-based analyses was tested independently with the multiresponse permutation procedure (MRPP). To determine the contributions of the genera to the ordination patterns, we carried out principal-component analysis (PCA) of the entire data set (frequency of reads at the genus level) and calculated the contribution of each genus to all dimensions relative to all other genera (34). Multivariate statistics were carried out using the R software (version 2.15.1) with the vcd and vegan packages (35–37). Phylogeny-based analysis of community similarity (unweighted UniFrac) of the core microbiota of cockroaches and termites was conducted with the genus level taxa that were present in >70% of the species in each of the major host groups; to account for differences in read numbers, each taxon was randomly subsampled to 10 sequences per sample. A cladogram was constructed based on the resulting dissimilarity matrix using a neighbor-joining algorithm.
Phylogenetic analysis of the pyrotag reads.
After random subsampling to 5,045 reads per sample, all sequences were classified and sorted into genus level bins. All samples in the same bin were grouped into OTUs (3% dissimilarity), and one representative sequence per OTU was selected for each sample using the Mothur command get.oturep, which also returns the number of reads in each OTU. Maximum-likelihood trees were calculated for each genus level lineage with FastTree 2 (38), transformed into ultrametric trees using PATHd8 (39), and visualized using the R package APE (40).
Accession numbers.
The COII gene sequences of all species that were not represented in public databases have been submitted to NCBI GenBank (accession numbers KF372028 to KF372033). The pyrotag data sets were submitted to the NCBI Sequence Read Archive (BioProject PRJNA217467; the individual accession numbers are listed in Table 1).
RESULTS AND DISCUSSION
The bacterial 16S rRNA genes in hindgut DNA of 34 termites and cockroaches and a few other insects were amplified with universal, bar-coded primers for the V3-V4 region. For each host species, we obtained an average of 10,000 high-quality sequence reads (Table 1). This is the first such data for most of these species, and classification against the RDP database (41) yielded large fractions of unclassified sequences, particularly in lower taxonomic ranks (Table 2). Our curated reference database (20) significantly improved classification, increasing the fraction of classified sequences in the different samples at the genus level from 24 to 68% (RDP) to 61 to 93% (our database) (Table 2).
Classification yielded 200 to 300 genus level taxa for the majority of samples (between 90 and 550 in extreme cases) (Table 1). The detailed classification results for all taxonomic ranks can be found in interactive Table S1 in the supplemental material. The number of OTUs obtained by similarity-based clustering of the sequences (3% sequence dissimilarity) was 2- to 10-fold higher, indicating additional diversity at the species level (Table 1). Predictions of species richness and coverage (Good's coverage and Chao1 estimators) that are based on the abundance of singletons in a data set underline the fact that high-throughput sequencing also fails to cover the entire bacterial diversity in a gut community (Table 1); even the samples with the largest numbers of reads (>100,000) still contain a large fraction of populations present only in low abundance (see Table S1 in the supplemental material).
The bacterial communities of each host group already differed greatly at the phylum level (Fig. 1). Spirochaetes were rare in cockroaches but abundant in lower termites and wood-feeding higher termites, often representing the majority of the reads, which is in agreement with the general notion that spirochetes are the most characteristic element of the termite gut microbiota (42). Firmicutes and Bacteroidetes were generally more abundant in cockroaches than in termites, except for a large proportion of Firmicutes in all soil-feeding higher termites and Bacteroidetes in the lower termite Coptotermes niger, again confirming results previously obtained for selected species (17, 43–45). Members of the Elusimicrobia were highly represented only in lower termites.
FIG 1.
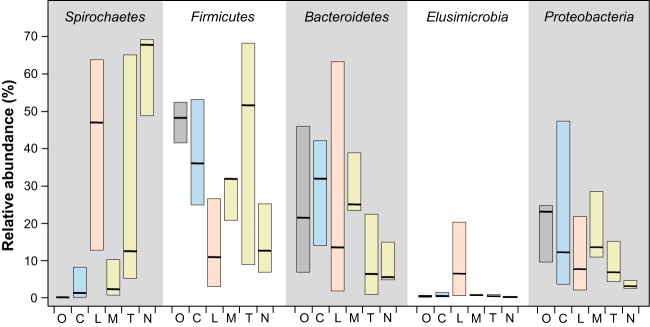
Relative abundances of the most prevalent bacterial phyla in the gut microbiota of different host groups. O, other insects; C, cockroaches; L, lower termites; M, T, and N, the higher termites Macrotermitinae, Termitinae, and Nasutitermitinae, respectively. Cryptocercidae and Apicotermitinae were not included because each group was represented by only a single species. The bars and horizontal lines show the ranges and median numbers of sequence reads assigned to the respective phyla. Detailed results for all bacterial phyla and individual host species are shown in Fig. S1 in the supplemental material.
Ordination analysis revealed high similarities among the bacterial microbiota of the different host groups. The robust clustering of samples based on genus level classification (Fig. 2) was also found with classification-independent (OTU-based) and phylogeny-based (UniFrac) approaches (see Fig. S2 in the supplemental material). In all cases, cockroaches were clearly separated from termites and lower termites from higher termites. Also, the different subfamilies of the Termitidae formed discrete clusters, with the fungus-cultivating Macrotermitinae showing strong affinity for the cockroaches. A notable exception was the wood-feeding cockroach Cryptocercus punctulatus, the closest relative of termites (4, 6). It did not cluster among the other cockroaches but was always more similar to lower termites, with which it shares the presence of cellulolytic flagellates. The (unrelated) wood-feeding cockroaches Panesthia angustipennis and Salganea esakii (family Blaberidae), whose gut microbiota lacks such flagellates, clustered with the omnivorous cockroaches.
FIG 2.
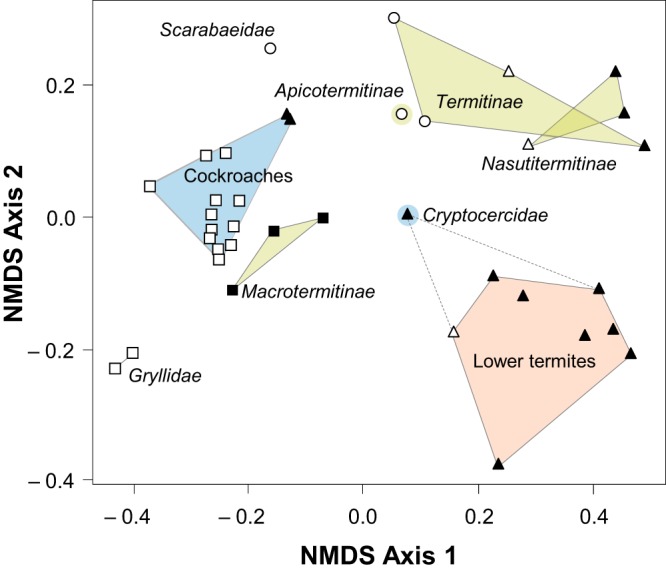
Phylogenetic patterns in the community structures of the bacterial gut microbiota of the different host species. Community similarities (Bray-Curtis) were calculated based on the distributions of genus level taxa (see Table S1 in the supplemental material) and visualized by NMDS (stress, 11.3%). The symbols indicate feeding habits: generalists (□), wood feeding (▲), grass feeding (△), soil/humus feeding (○), and fungus cultivating (■). Two species of crickets (Gryllidae) and a beetle larva (Scarabaeidae) were included as outgroups. The species identifier for each data point is included in Fig. S2a in the supplemental material. The clusters were supported by both ADONIS and MRPP analyses (P < 0.001).
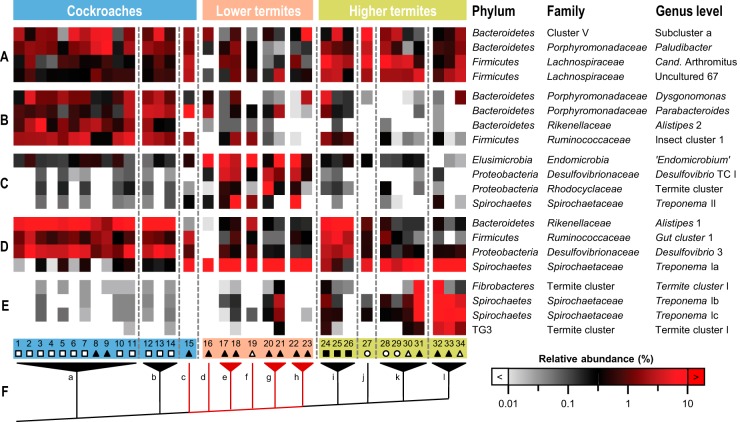
When we ranked all 884 genus level taxa in the data set according to their contributions to the ordination results, it became apparent that the top 100 genera alone were responsible for almost 70% of the pattern and represented 90% of the sequences in the data set (see Table S3 in the supplemental material). Many genus-level taxa occurred in all major host lineages, extending the previously postulated presence of termite-specific bacterial lineages to all cockroaches (17, 19), but with distinct differences in their relative distributions (Fig. 3, groups A and D). An obvious break in the pattern between cockroaches and termites (Fig. 3, groups B, C, and D) indicated that the transition from an omnivorous to a wood-feeding lifestyle had a strong impact on bacterial community structure. Bacterial lineages abundant in cockroaches decreased in frequency in lower termites, and rare lineages dramatically increased. The latter was most obvious in the spirochetal cluster Treponema Ia and matches the dominance of Spirochaetes in the guts of wood-feeding termites (11, 42). Since this cluster comprises the homoacetogenic Treponema primitia (46), its upshift is also consistent with changes in the distributions of several functional marker genes (formyltetrahydrofolate synthetase, CO dehydrogenase, and hydrogenase) (47–49), which indicated that the bacteria responsible for reductive acetogenesis in omnivorous cockroaches are not the same as those in wood-feeding termites and C. punctulatus.
FIG 3.
Relative abundances of selected bacterial lineages in the gut microbiota that contribute strongly to the separation of cockroaches, lower termites, and higher termites in the ordination analyses. The lineages were selected from the top 50 taxa (see Table S3 in the supplemental material) and subjectively sorted according to patterns (A to E) as explained in the text. The color scale is logarithmic to emphasize rare taxa. The numbers indicate host species (see Table 1). The symbols indicate feeding habits (see the legend to Fig. 2). The tree (F) illustrates a simplified phylogeny of major host lineages (a, other cockroaches; b, Blattidae; c, Cryptocercidae; d, Mastotermitidae; e, Termopsidae; f, Hodotermitidae; g, Kalotermitidae; h, Rhinotermitidae; i, Macrotermitinae; j, Apicotermitinae; k, Termitinae; l, Nasutitermitinae). The branches connecting species that harbor gut flagellates are in red.
Several of the genus level taxa that predominated only in lower termites (Fig. 3, group C) represent lineages that harbor specific symbionts of termite gut flagellates. Taxa comprising ectosymbiotic spirochetes (Treponema II) (50) and endosymbiotic “Candidatus Endomicrobium” (15, 51) and Desulfovibrio spp. (TC I) (52, 53) were abundant only in those termites that harbor the respective host flagellates. However, low numbers of endomicrobia were also consistently present in cockroaches and higher termites, corroborating the presence of putatively free-living relatives (54) that were recruited as endosymbionts, presumably long after the flagellates had established their symbiosis with lower termites (55). Also the dynamic patterns of cluster V Bacteroidetes among the lower termites (Fig. 3, groups A and C), which harbor several lineages of symbionts that have strictly cospeciated with their respective flagellate hosts (19, 56), is in agreement with their recruitment from free-living relatives that are present but in low abundance in termites lacking these flagellates (19).
The second obvious break in the community patterns was between lower and higher termites, marking a decrease in abundance of the flagellate-associated bacterial lineages and a strong increase in several other taxa (Fig. 3, groups C and E). The dominance of termite-specific clusters of Fibrobacteres, the TG3 phylum, and certain Treponema lineages (Ib and Ic) in wood- and grass-feeding termites is consistent with previous reports on the distribution of these groups (13). There is strong evidence from enzymatic (57) and metagenomic (58, 59) studies of Nasutitermes and Amitermes spp. that bacterial members of the gut microbiota—particularly Fibrobacteres (possibly including the related TG3 phylum) and Spirochaetes—took over the function of the flagellates in fiber digestion. Our results indicate that these putative cellulose-digesting bacteria are apparently represented among lower termites but cannot form large populations because the protists sequester all wood particles in their food vacuoles, restricting the bacteria to soluble substrates. Thus, the dramatic changes in the bacterial community between lower and higher termites are probably due to both the gain of new substrates and the loss of the flagellate niche.
Also, the resurgence in the Macrotermitinae of taxa that prevail in cockroaches may be related to the dietary diversification of higher termites following the loss of flagellates. The high similarity between the gut microbiota of omnivorous cockroaches and Macrotermitinae, first discovered in a study of the gut microbiota of the cockroach Shelfordella lateralis (17), is rooted in the shared presence of lineages that are only at low abundance in wood- or soil-feeding termites (Fig. 3, group D). Predominance of Bacteroidetes and Firmicutes seems to be a common feature of omnivorous animals (60, 61); their abundance in the Macrotermitinae may be caused by the protein-rich fungal biomass included in the diet of the fungus-cultivating species (62).
Most genus-level taxa were unevenly distributed across cockroaches and lower and higher termites, but many of them were consistently represented among the members of the major host groups (Fig. 4). Although the variable taxa were at least five times more numerous than these core taxa, they represented less than a quarter of the reads in the respective data sets (Table 3). The 30 taxa common to the three groups included members of the Spirochaetes (see Table S3 in the supplemental material), underlining the fact that small populations of the most typical element of the termite gut microbiota are also present in cockroaches. Although these core taxa represented only a small fraction of the genus level diversity, they made up almost half of the reads in the entire data set (Fig. 5). Taxa common to termites but not regularly present in cockroaches (see Table S3 in the supplemental material) represented only 8.7% of the reads. It is important to note that the core taxa are not always restricted to dictyopteran hosts. Almost half of the core taxa (14 of 30) were also present in the three other insect species included in this study, representing a large proportion of the reads in these samples (Pachnoda ephippiata, 22%; Acheta domesticus, 45%; Gryllus assimilis, 26%). The most abundant lineages in these insects that were shared with most dictyopteran samples were gut cluster 2 (Lachnospiraceae) in the scarab beetle larva (P. ephippiata) and Alistipes 2 (Rikenellaceae) and Dysgonomonas (Porphyromonadacae) in the crickets ( A. domesticus and G. assimilis).
FIG 4.
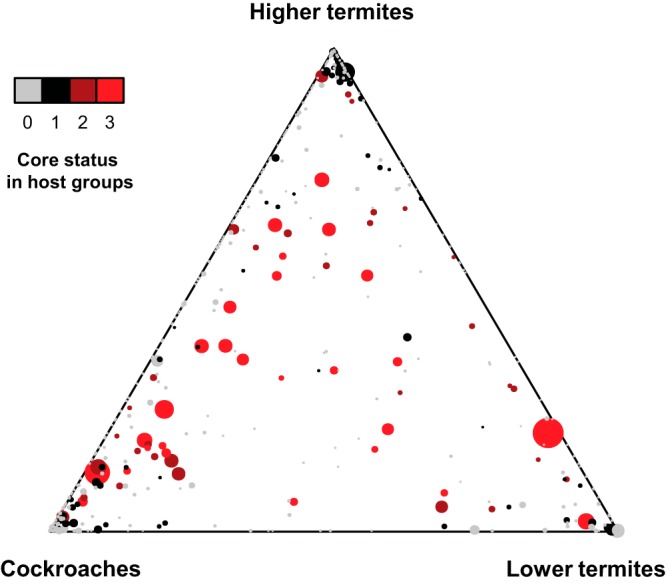
Ternary plot of the distribution of genus level taxa across the major host groups. The area of each circle represents the relative abundance of the reads in the entire data set, the position specifies their average abundance in the respective host groups, and the colors indicate the number of host groups in which core status is attained (presence in >70% of the hosts) (the data are from Table S3 in the supplemental material). An interactive version that allows identification of the genus behind each data point of the figure is included as a supplemental file (see Fig. S3 at http://www.termites.de/brune/publ/suppl/AEM04206-13_Figure_S3.html).
TABLE 3.
Numbers of genus level taxa considered variable and core taxa and their average contributions to the gut communities in the major host groups
| Parameter | Valuea |
|||||
|---|---|---|---|---|---|---|
| Cockroaches |
Lower termites |
Higher termites |
||||
| Variable | Core | Variable | Core | Variable | Core | |
| No. of taxa | 363 | 67 | 270 | 50 | 746 | 87 |
| Percentage of reads | 21.2 | 78.8 | 22.1 | 77.9 | 20.9 | 79.1 |
Core taxa were defined as those present in >70% of the species of the respective host groups (see Table S3 in the supplemental material).
FIG 5.
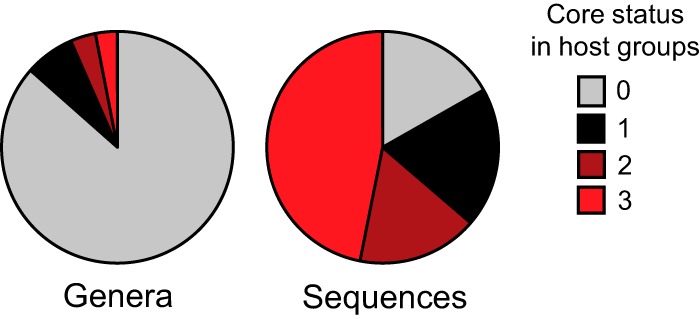
Representation of core taxa in the data set based on the number of genera or sequence reads in the entire data set. Core status was assigned if a taxon was represented by >70% of the host species in a major host group (cockroaches, lower termites, and higher termites).
Despite the abundant presence of lineages that are not restricted to dictyopteran hosts, a UniFrac analysis of the core taxa retained a clear host signal in the phylogeny of its components (Fig. 6). Cockroaches formed a sister group of the Cryptocercus/termite clade, and higher termites were apical to all lineages with gut flagellates. However, the internal topology of the cladogram often did not match the branching order of the host tree (Fig. 3F), particularly in the cockroaches and lower termites, indicating that the dictyopteran core microbiota is not caused by cospeciation. Rather, the lack of clustering among the gut microbiota of blattid cockroaches and the proximity of wood-feeding blaberid cockroaches (Panesthia angustipennis and Salganea esakii) to the Cryptocercus/termite clade suggest that factors other than host phylogeny must shape the bacterial community structure.
FIG 6.
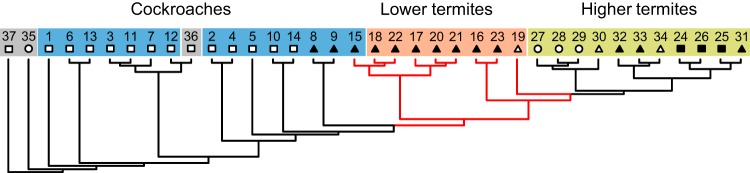
Cladogram of community similarities based on the core taxa common to all major host groups (unweighted UniFrac analysis of the sequences belonging to the core genera). For the sample numbers, see Table 1. The symbols indicate lifestyle: generalists (□), wood feeding (▲), grass feeding (△), soil/humus feeding (○), and fungus cultivating (■). The branches connecting species that harbor gut flagellates are in red.
Closer inspection of the genus level taxa that contribute most to the separation of the major host groups revealed that phylogenetic clustering is often restricted to sequences from particular host groups (see Fig. S4O and R in the supplemental material). Although the results of phylogenetic analyses based on short sequences must be interpreted with caution, it is noteworthy that the basal position of sequences from cockroach guts relative to those from termites and the apical position of sequences from higher termites are frequent themes. However, it remains unclear whether the lineages in question were already associated with the ancestral cockroaches (cospeciation) or are diet-specific lineages that were independently acquired from the environment (host selection). The latter would explain the frequently observed quasirandom occurrence of the same genus level lineages among different host groups. A prominent example is Alistipes 2, which is also highly abundant in the cricket (Achaeta domesticus) and contributes to the similarity of its core microbiota to that of several cockroaches (see Fig. S4C in the supplemental material). The unexpected presence of cockroach clusters in higher termites (see Fig. S4H in the supplemental material) suggests that the horizontal transfer of microbiota between different host lineages also has to be considered. A puzzling phenomenon is the presence of a small number of phylotypes with identical sequences within sequence clusters of entirely different hosts (see Fig. S4N in the supplemental material). Here, horizontal transfer and environmental uptake are unlikely explanations, and the possibility of methodological artifacts (e.g., mistagging of templates during the emulsion PCR [63]) has to be considered.
It remains to be investigated whether traces of host phylogeny can also be found in the archaeal microbiota in the guts of termites and cockroaches. Although archaea are much less abundant than bacteria (0.1 to 3% of prokaryotes in termite guts [64]), methanogens seem to be present in all dictyopteran lineages (65). The diversity of the archaeal community is much smaller than that of bacteria and comprises both termite-specific clusters and lineages with representatives from many environments (27, 66).
Conclusions.
This study provides a new view of the complex bacterial communities in the gut microbiota of termites. Clearly, phylogeny is not the only driver of community structure in the dictyopteran microbiota. Changes in the quality of the diet (lignin and fiber content or humification state) or the provision of new niches for nitrogen-fixing or -upgrading symbionts promoted bacteria from different functional guilds that were either already present in the microbial seed bank of the gut (19) or newly acquired from the environment and caused their decline when such services were no longer required.
The results of our study provide a foundation for future studies targeting the specific roles of important bacterial populations by metagenomic and metatranscriptomic analysis or single-cell approaches. Of particular interest will be mechanisms of bacterial cellulose degradation and humus digestion in higher termites (59), microbial interactions in hydrogen metabolism and methanogenesis (67), and the emerging role of the flagellate symbionts in the nitrogen economy of digestive symbiosis (8). Also, in keeping with Dobzhansky's famous dictum (68), the complex patterns in the gut microbiota of this ancient group of insects make sense only in the light of evolution.
Supplementary Material
ACKNOWLEDGMENTS
We thank Kiyoto Maekawa (University of Toyama), Christine Nalepa (North Carolina State University), Rudy Plarre (Federal Institute for Materials Research and Testing, Berlin, Germany), Rudolf Scheffrahn (University of Florida), Gaku Tokuda (University of the Ryukyus), and our coworkers David Ngugi and James Nonoh (MPI Marburg) for providing insect samples and Katja Meuser for technical assistance.
Footnotes
Published ahead of print 31 January 2014
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.04206-13.
REFERENCES
- 1.Brune A, Ohkuma M. 2011. Role of the termite gut microbiota in symbiotic digestion, p 439–475 In Bignell DE, Roisin Y, Lo N. (ed), Biology of termites: a modern synthesis. Springer, Dordrecht, The Netherlands [Google Scholar]
- 2.Jouquet P, Traoré S, Choosai C, Hartmann C, Bignell D. 2011. Influence of termites on ecosystem functioning. Ecosystem services provided by termites. Eur. J. Soil Biol. 47:215–222. 10.1016/j.ejsobi.2011.05.005 [DOI] [Google Scholar]
- 3.Ni J, Tokuda G. 2013. Lignocellulose-degrading enzymes from termites and their symbiotic microbiota. Biotechnol. Adv. 31:838–850. 10.1016/j.biotechadv.2013.04.005 [DOI] [PubMed] [Google Scholar]
- 4.Inward D, Beccaloni G, Eggleton P. 2007. Death of an order: a comprehensive molecular phylogenetic study confirms that termites are eusocial cockroaches. Biol. Lett. 3:331–335. 10.1098/rsbl.2007.0102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nalepa CA, Bignell DE, Bandi C. 2001. Detritivory, coprophagy, and the evolution of digestive mutualisms in Dictyoptera. Insect. Soc. 48:194–201. 10.1007/PL00001767 [DOI] [Google Scholar]
- 6.Engel MS, Grimaldi DA, Krishna K. 2009. Termites (Isoptera): their phylogeny, classification, and rise to ecological dominance. Am. Mus. Novit. 3650:1–27. 10.1206/651.1 [DOI] [Google Scholar]
- 7.Ohkuma M, Noda S, Hongoh Y, Nalepa CA, Inoue T. 2009. Inheritance and diversification of symbiotic trichonymphid flagellates from a common ancestor of termites and the cockroach Cryptocercus. Proc. R. Soc. B 276:239–245. 10.1098/rspb.2008.1094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hongoh Y. 2011. Toward the functional analysis of uncultivable, symbiotic microorganisms in the termite gut. Cell. Mol. Life Sci. 68:1311–1325. 10.1007/s00018-011-0648-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bignell DE, Eggleton P. 2000. Termites in ecosystems, p 363–387 In Abe T, Bignell DE, Higashi M. (ed), Termites: evolution, sociality, symbiosis, ecology. Kluwer Academic Publishers, Dordrecht, The Netherlands [Google Scholar]
- 10.Nobre T, Rouland-Lefèvre C, Aanen DK. 2011. Comparative biology of fungus cultivation in termites and ants, p 193–210 In Bignell DE, Roisin Y, Lo N. (ed), Biology of termites: a modern synthesis. Springer, Dordrecht, The Netherlands [Google Scholar]
- 11.Ohkuma M, Brune A. 2011. Diversity, structure, and evolution of the termite gut microbial community, p 413–438 In Bignell DE, Roisin Y, Lo N. (ed), Biology of termites: a modern synthesis. Springer, Dordrecht, The Netherlands [Google Scholar]
- 12.Lilburn TG, Schmidt TM, Breznak JA. 1999. Phylogenetic diversity of termite gut spirochaetes. Environ. Microbiol. 1:331–345. 10.1046/j.1462-2920.1999.00043.x [DOI] [PubMed] [Google Scholar]
- 13.Hongoh Y, Deevong P, Hattori S, Inoue T, Noda S, Noparatnaraporn N, Kudo T, Ohkuma M. 2006. Phylogenetic diversity, localization and cell morphologies of the candidate phylum TG3 and a subphylum in the phylum Fibrobacteres, recently found bacterial groups dominant in termite guts. Appl. Environ. Microbiol. 72:6780–6788. 10.1128/AEM.00891-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hongoh Y, Ekpornprasit L, Inoue T, Moriya S, Trakulnaleamsai S, Ohkuma M, Noparatnaraporn N, Kudo T. 2006. Intracolony variation of bacterial gut microbiota among castes and ages in the fungus-growing termite Macrotermes gilvus. Mol. Ecol. 15:505–516. 10.1111/j.1365-294X.2005.02795.x [DOI] [PubMed] [Google Scholar]
- 15.Ohkuma M, Sato T, Noda S, Ui S, Kudo T, Hongoh Y. 2007. The candidate phylum ‘Termite Group 1' of bacteria: phylogenetic diversity, distribution, and endosymbiont members of various gut flagellated protists. FEMS Microbiol. Ecol. 60:467–476. 10.1111/j.1574-6941.2007.00311.x [DOI] [PubMed] [Google Scholar]
- 16.Geissinger O, Herlemann DPR, Mörschel E, Maier UG, Brune A. 2009. The ultramicrobacterium “Elusimicrobium minutum” gen. nov., sp. nov., the first cultivated representative of the termite group 1 phylum. Appl. Environ. Microbiol. 75:2831–2840. 10.1128/AEM.02697-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schauer C, Thompson CL, Brune A. 2012. The bacterial community in the gut of the cockroach Shelfordella lateralis reflects the close evolutionary relatedness of cockroaches and termites. Appl. Environ. Microbiol. 78:2758–2767. 10.1128/AEM.07788-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hongoh Y, Deevong P, Inoue T, Moriya S, Trakulnaleamsai S, Ohkuma M, Vongkaluang C, Noparatnaraporn N, Kudo T. 2005. Intra- and interspecific comparisons of bacterial diversity and community structure support coevolution of gut microbiota and termite host. Appl. Environ. Microbiol. 71:6590–6599. 10.1128/AEM.71.11.6590-6599.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Noda S, Hongoh Y, Sato T, Ohkuma M. 2009. Complex coevolutionary history of symbiotic Bacteroidales bacteria of various protists in the gut of termites. BMC Evol. Biol. 9:158. 10.1186/1471-2148-9-158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Köhler T, Dietrich C, Scheffrahn RH, Brune A. 2012. High-resolution analysis of gut environment and bacterial microbiota reveals functional compartmentation of the gut in wood-feeding higher termites (Nasutitermes spp.). Appl. Environ. Microbiol. 78:4691–4701. 10.1128/AEM.00683-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Deevong P, Hongoh Y, Inoue T, Trakulnaleamsai S, Kudo T, Noparatnaraporn N, Ohkuma M. 2006. Effect of temporal sample preservation on the molecular study of a complex microbial community in the gut of the termite Microcerotermes sp. Microbes Environ. 21:78–85. 10.1264/jsme2.21.78 [DOI] [Google Scholar]
- 22.Good IJ. 1953. The population frequencies of species and the estimation of population parameters. Biometrika 40:237–264 [Google Scholar]
- 23.Chao A. 1984. Nonparametric estimation of the number of classes in a population. Scand. J. Stat. 11:265–270 [Google Scholar]
- 24.Chao A, Shen TJ. 2003. Nonparametric estimation of Shannon's index of diversity when there are unseen species in sample. Environ. Ecol. Stat. 10:429–443. 10.1023/A:1026096204727 [DOI] [Google Scholar]
- 25.Legendre P, Legendre L. 1998. Numerical ecology, 2nd ed. Elsevier, Montreal, Canada [Google Scholar]
- 26.Pester M, Brune A. 2006. Expression profiles of fhs (FTHFS) genes support the hypothesis that spirochaetes dominate reductive acetogenesis in the hindgut of lower termites. Environ. Microbiol. 8:1261–1270. 10.1111/j.1462-2920.2006.01020.x [DOI] [PubMed] [Google Scholar]
- 27.Paul K, Nonoh JO, Mikulski L, Brune A. 2012. “Methanoplasmatales,” Thermoplasmatales-related archaea in termite guts and other environments, are the seventh order of methanogens. Appl. Environ. Microbiol. 78:8245–8253. 10.1128/AEM.02193-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schloss PD, Westcott SL, Ryabin T, Hall JR, Hartmann M, Hollister EB, Lesniewski RA, Oakley BB, Parks DH, Robinson CJ, Sahl JW, Stres B, Thallinger GG, Van Horn DJ, Weber CF. 2009. Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl. Environ. Microbiol. 75:7537–7541. 10.1128/AEM.01541-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schloss PD, Gevers D, Westcott SL. 2011. Reducing the effects of PCR amplification and sequencing artifacts on 16S rRNA-based studies. PLoS One 6:e27310. 10.1371/journal.pone.0027310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bragg L, Stone G, Imelfort M, Hugenholtz P, Tyson GW. 2012. Fast, accurate error-correction of amplicon pyrosequences using Acacia. Nat. Methods 9:425–426. 10.1038/nmeth.1990 [DOI] [PubMed] [Google Scholar]
- 31.Thompson CL, Vier R, Mikaelyan A, Wienemann T, Brune A. 2012. ‘Candidatus Arthromitus' revised: segmented filamentous bacteria in arthropod guts are members of Lachnospiraceae. Environ. Microbiol. 14:1454–1465. 10.1111/j.1462-2920.2012.02731.x [DOI] [PubMed] [Google Scholar]
- 32.Bray JR, Curtis JT. 1957. An ordination of the upland forest communities of southern Wisconsin. Ecol. Monogr. 27:325–349. 10.2307/1942268 [DOI] [Google Scholar]
- 33.Lozupone C, Knight R. 2005. UniFrac: a new phylogenetic method for comparing microbial communities. Appl. Environ. Microbiol. 71:8228–8235. 10.1128/AEM.71.12.8228-8235.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Abdi H, Williams LJ. 2010. Principal component analysis. Wiley Interdiscip. Rev. Comput. Stat. 2:433–459. 10.1002/wics.101 [DOI] [Google Scholar]
- 35.R Development Core Team. 2011. R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria: http://www.r-project.org [Google Scholar]
- 36.Meyer D, Zeileis A, Hornik K. 2013. vcd: visualizing categorical data. R package version 1.2–13. http://cran.r-project.org/package=vcd [Google Scholar]
- 37.Oksanen J, Blanchet JG, Kindt R, Legendre P, Minchin PR, O'Hara RB, Simpson GL, Solymos P, Stevens MHH, Wagner H. 2013. vegan: community ecology package. http://CRAN.R-project.org/package=vegan [Google Scholar]
- 38.Price MN, Dehal PS, Arkin AP. 2010. FastTree 2: approximately maximum-likelihood trees for large alignments. PLoS One 5:e9490. 10.1371/journal.pone.0009490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Britton T, Anderson C, Jacquet D, Lundqvist S, Bremer K. 2007. Estimating divergence times in large phylogenetic trees. Syst. Biol. 56:741–752. 10.1080/10635150701613783 [DOI] [PubMed] [Google Scholar]
- 40.Paradis E, Claude J, Strimmer K. 2004. APE: analyses of phylogenetics and evolution in R language. Bioinformatics 20:289–290. 10.1093/bioinformatics/btg412 [DOI] [PubMed] [Google Scholar]
- 41.Cole JR, Wang Q, Cardenas E, Fish J, Chai B, Farris RJ, Kulam-Syed-Mohideen AS, McGarrell DM, Marsh T, Garrity GM, Tiedje JM. 2009. The Ribosomal Database Project: improved alignments and new tools for rRNA analysis. Nucleic Acids Res. 37:D141–D145. 10.1093/nar/gkn879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Breznak JA. 2000. Ecology of prokaryotic microbes in the guts of wood- and litter-feeding termites, p 209–231 In Abe T, Bignell DE, Higashi M. (ed), Termites: evolution, sociality, symbiosis, ecology. Kluwer Academic Publishers, Dordrecht, The Netherlands [Google Scholar]
- 43.Schmitt-Wagner D, Friedrich MW, Wagner B, Brune A. 2003. Phylogenetic diversity, abundance, and axial distribution of bacteria in the intestinal tract of two soil-feeding termites (Cubitermes spp.). Appl. Environ. Microbiol. 69:6007–6017. 10.1128/AEM.69.10.6007-6017.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Noda S, Iida T, Kitade O, Nakajima H, Kudo T, Ohkuma M. 2005. Endosymbiotic Bacteroidales bacteria of the flagellated protist Pseudotrichonympha grassii in the gut of the termite Coptotermes formosanus. Appl. Environ. Microbiol. 71:8811–8817. 10.1128/AEM.71.12.8811-8817.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Thongaram T, Hongoh Y, Kosono S, Ohkuma M, Trakulnaleamsai S, Noparatnaraporn N, Kudo T. 2005. Comparison of bacterial communities in the alkaline gut segment among various species of higher termites. Extremophiles 9:229–238. 10.1007/s00792-005-0440-9 [DOI] [PubMed] [Google Scholar]
- 46.Graber JR, Leadbetter JR, Breznak JA. 2004. Description of Treponema azotonutricium sp. nov. and Treponema primitia sp. nov., the first spirochetes isolated from termite guts. Appl. Environ. Microbiol. 70:1315–1320. 10.1128/AEM.70.3.1315-1320.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Matson EG, Gora KG, Leadbetter JR. 2011. Anaerobic carbon monoxide dehydrogenase diversity in the homoacetogenic hindgut microbial communities of lower termites and the wood roach. PLoS One 6:e19316. 10.1371/journal.pone.0019316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ottesen EA, Leadbetter JR. 2011. Formyltetrahydrofolate synthetase gene diversity in the guts of higher termites with different diets and lifestyles. Appl. Environ. Microbiol. 77:3461–3467. 10.1128/AEM.02657-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ballor NR, Leadbetter JR. 2012. Analysis of extensive [FeFe] hydrogenase gene diversity within the gut microbiota of insects representing five families of Dictyoptera. Microb. Ecol. 63:586–595. 10.1007/s00248-011-9941-5 [DOI] [PubMed] [Google Scholar]
- 50.Iida T, Ohkuma M, Ohtoko K, Kudo T. 2000. Symbiotic spirochetes in the termite hindgut: phylogenetic identification of ectosymbiotic spirochetes of oxymonad protists. FEMS Microbiol. Ecol. 34:17–26. 10.1111/j.1574-6941.2000.tb00750.x [DOI] [PubMed] [Google Scholar]
- 51.Stingl U, Radek R, Yang H, Brune A. 2005. ‘Endomicrobia’: cytoplasmic symbionts of termite gut protozoa form a separate phylum of prokaryotes. Appl. Environ. Microbiol. 71:1473–1479. 10.1128/AEM.71.3.1473-1479.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sato T, Hongoh Y, Noda S, Hattori S, Ui S, Ohkuma M. 2009. Candidatus Desulfovibrio trichonymphae, a novel intracellular symbiont of the flagellate Trichonympha agilis in termite gut. Environ. Microbiol. 11:1007–1015. 10.1111/j.1462-2920.2008.01827.x [DOI] [PubMed] [Google Scholar]
- 53.Strassert JFH, Köhler T, Wienemann THG, Ikeda-Ohtsubo W, Faivre N, Franckenberg S, Plarre R, Radek R, Brune A. 2012. ‘Candidatus Ancillula trichonymphae', a novel lineage of endosymbiotic Actinobacteria in termite gut flagellates of the genus Trichonympha. Environ. Microbiol. 14:3259–3270. 10.1111/1462-2920.12012 [DOI] [PubMed] [Google Scholar]
- 54.Ikeda-Ohtsubo W, Faivre N, Brune A. 2010. Putatively free-living ‘Endomicrobia’—ancestors of the intracellular symbionts of termite gut flagellates? Environ. Microbiol. Rep. 2:554–559. 10.1111/j.1758-2229.2009.00124.x [DOI] [PubMed] [Google Scholar]
- 55.Ikeda-Ohtsubo W, Brune A. 2009. Cospeciation of termite gut flagellates and their bacterial endosymbionts: Trichonympha species and ‘Candidatus Endomicrobium trichonymphae'. Mol. Ecol. 18:332–342. 10.1111/j.1365-294X.2008.04029.x [DOI] [PubMed] [Google Scholar]
- 56.Desai M, Strassert JFH, Meuser K, Hertel H, Ikeda-Ohtsubo W, Radek R, Brune A. 2010. Strict cospeciation of devescovinid flagellates and Bacteroidales ectosymbionts in the gut of dry-wood termites (Kalotermitidae). Environ. Microbiol. 12:2120–2132. 10.1111/j.1462-2920.2009.02080.x [DOI] [PubMed] [Google Scholar]
- 57.Tokuda G, Watanabe H. 2007. Hidden cellulases in termites: revision of an old hypothesis. Biol. Lett. 3:336–339. 10.1098/rsbl.2007.0073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Warnecke F, Luginbühl P, Ivanova N, Ghassemian M, Richardson TH, Stege JT, Cayouette M, McHardy AC, Djordjevic G, Aboushadi N, Sorek R, Tringe SG, Podar M, Martin HG, Kunin V, Dalevi D, Madejska J, Kirton E, Platt D, Szeto E, Salamov A, Barry K, Mikhailova N, Kyrpides NC, Matson EG, Ottesen EA, Zhang X, Hernández M, Murillo C, Acosta LG, Rigoutsos I, Tamayo G, Green BD, Chang C, Rubin EM, Mathur EJ, Robertson DE, Hugenholtz P, Leadbetter JR. 2007. Metagenomic and functional analysis of hindgut microbiota of a wood-feeding higher termite. Nature 450:560–565. 10.1038/nature06269 [DOI] [PubMed] [Google Scholar]
- 59.He S, Ivanova N, Kirton E, Allgaier M, Bergin C, Scheffrahn RH, Kyrpides NC, Warnecke F, Tringe SG, Hugenholtz P. 2013. Comparative metagenomic and metatranscriptomic analysis of hindgut paunch microbiota in wood- and dung-feeding higher termites. PLoS One 8:e61126. 10.1371/journal.pone.0061126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Eckburg PB, Bik EM, Bernstein CN, Purdom E, Dethlefsen L, Sargent M, Gill SR, Nelson KE, Relman DA. 2005. Diversity of the human intestinal microbial flora. Science 308:1635–1638. 10.1126/science.1110591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Leser TD, Amenuvor JZ, Jensen TK, Lindecrona RH, Boye M, Møller K. 2002. Culture-independent analysis of gut bacteria: the pig gastrointestinal tract microbiota revisited. Appl. Environ. Microbiol. 68:673–690. 10.1128/AEM.68.2.673-690.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Hyodo F, Tayasu I, Inoue T, Azuma JI, Kudo T, Abe T. 2003. Differential role of symbiotic fungi in lignin degradation and food provision for fungus-growing termites (Macrotermitinae: Isoptera). Funct. Ecol. 17:186–193. 10.1046/j.1365-2435.2003.00718.x [DOI] [Google Scholar]
- 63.Carlsen T, Aas AB, Lindner D, Vrålstad T, Schumacher T, Kauserud H. 2012. Don't make a mista(g)ke: is tag switching an overlooked source of error in amplicon pyrosequencing studies? Fung. Ecol. 5:747–749. 10.1016/j.funeco.2012.06.003 [DOI] [Google Scholar]
- 64.Brauman A, Dore J, Eggleton P, Bignell D, Breznak JA, Kane MD. 2001. Molecular phylogenetic profiling of prokaryotic communities in guts of termites with different feeding habits. FEMS Microbiol. Ecol. 35:27–36. 10.1111/j.1574-6941.2001.tb00785.x [DOI] [PubMed] [Google Scholar]
- 65.Hackstein JHP, van Alen TA, Rosenberg J. 2006. Methane production by terrestrial arthropods, p 155–180 In König H, Varma A. (ed), Intestinal microorganisms of termites and other invertebrates. Springer, Berlin, Germany [Google Scholar]
- 66.Brune A. 2010. Methanogens in the digestive tract of termites, p 81–100 In Hackstein JHP. (ed), (Endo)symbiotic methanogenic archaea. Springer, Heidelberg, Germany [Google Scholar]
- 67.Pester M, Brune A. 2007. Hydrogen is the central free intermediate during lignocellulose degradation by termite gut symbionts. ISME J. 1:551–565. 10.1038/ismej.2007.62 [DOI] [PubMed] [Google Scholar]
- 68.Dobzhansky T. 1973. Nothing in biology makes sense except in the light of evolution. Am. Biol. Teacher 35:125–129. 10.2307/4444260 [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.