Abstract
Rhizobia have a versatile catabolism that allows them to compete successfully with other microorganisms for nutrients in the soil and in the rhizosphere of their respective host plants. In this study, Bradyrhizobium japonicum USDA 110 was found to be able to utilize oxalate as the sole carbon source. A proteome analysis of cells grown in minimal medium containing arabinose suggested that oxalate oxidation extends the arabinose degradation branch via glycolaldehyde. A mutant of the key pathway genes oxc (for oxalyl-coenzyme A decarboxylase) and frc (for formyl-coenzyme A transferase) was constructed and shown to be (i) impaired in growth on arabinose and (ii) unable to grow on oxalate. Oxalate was detected in roots and, at elevated levels, in root nodules of four different B. japonicum host plants. Mixed-inoculation experiments with wild-type and oxc-frc mutant cells revealed that oxalotrophy might be a beneficial trait of B. japonicum at some stage during legume root nodule colonization.
INTRODUCTION
Rhizobia have a broad metabolic capacity and can use a multitude of carbon and nitrogen sources which allow them to be successful and competitive in soil and in the rhizosphere of host plants (1). Within root nodules, differentiated rhizobia (bacteroids) reduce N2 to ammonium, which is secreted to the plant in return for C4-dicarboxylic acids as carbon and energy sources (2). The C4-dicarboxylic acids malate, succinate, and fumarate have been shown to be the primary carbon sources of bacteroids and can actively cross the peribacteroid membrane (3–5). In fact, transport of C4-dicarboxylic acids is required for nitrogen fixation (6–8). C4-dicarboxylic acids are directly fed into the tricarboxylic acid (TCA) cycle to supply the bacteroid with enough energy to perform nitrogen fixation (3).
However, despite the fact that dicarboxylic acids were shown to be the major carbon sources for N2-fixing bacteroids, large quantities of hexose and pentose sugars are also found in nodules (9), suggesting an important role in nodule metabolism and N2 fixation. Enzymes for hexose and pentose transport and metabolism have been reported to be present in bacteroids (10–12). The pentose l-arabinose is a well-known substrate of B. japonicum (13, 14), and this sugar is routinely used for in vitro growth studies. Previous work has shown that l-arabinose is degraded by a pathway that conceptually resembles the Entner-Doudoroff pathway (14). By analogy, l-arabinose is first oxidized to l-2-keto-3-deoxyarabonate (l-KDA) (15–17), which is then converted into α-ketoglurate in the case of fast-growing rhizobia (18–20) or to glycolaldehyde and pyruvate in the case of slow-growing species like B. japonicum (14). While pyruvate most likely is oxidized in the TCA cycle, the fate of glycolaldehyde has not been resolved yet. In this study, through proteomic analysis of B. japonicum cells grown either in minimal medium with l-arabinose as the sole carbon and energy source or in complete medium containing l-arabinose, we identified highly expressed products of candidate genes involved in the degradation of arabinose (17). Based on protein expression data, we suggest that glycolaldehyde is oxidized to glyoxylate and then reduced to glycerate through a glyoxylate carboligase and a tartronate semialdehyde reductase for assimilation and/or is converted to oxalate and then oxidized to formate and CO2 through the formyl-coenzyme A (CoA) transferase (Frc), oxalyl-CoA decarboxylase (Oxc), and formate dehydrogenase reactions for energy generation (Fig. 1). Results from previous transcriptome and proteome studies in B. japonicum nodules (12, 21) suggested that glyoxylate is preferentially degraded via oxalate oxidation during symbiosis. In fact, in contrast to the glyoxylate carboligase and tartronate semialdehyde reductase, the Frc and Oxc enzymes were detected in nodules where oxalate presence also could be measured. In agreement with these results, the Δfrc-oxc mutant strain constructed in the present work is not able to grow on oxalate and is partially defective in free-living growth in minimal medium containing arabinose. We also show that the mutant has a disadvantage when competing for nodule occupancy against the wild-type strain.
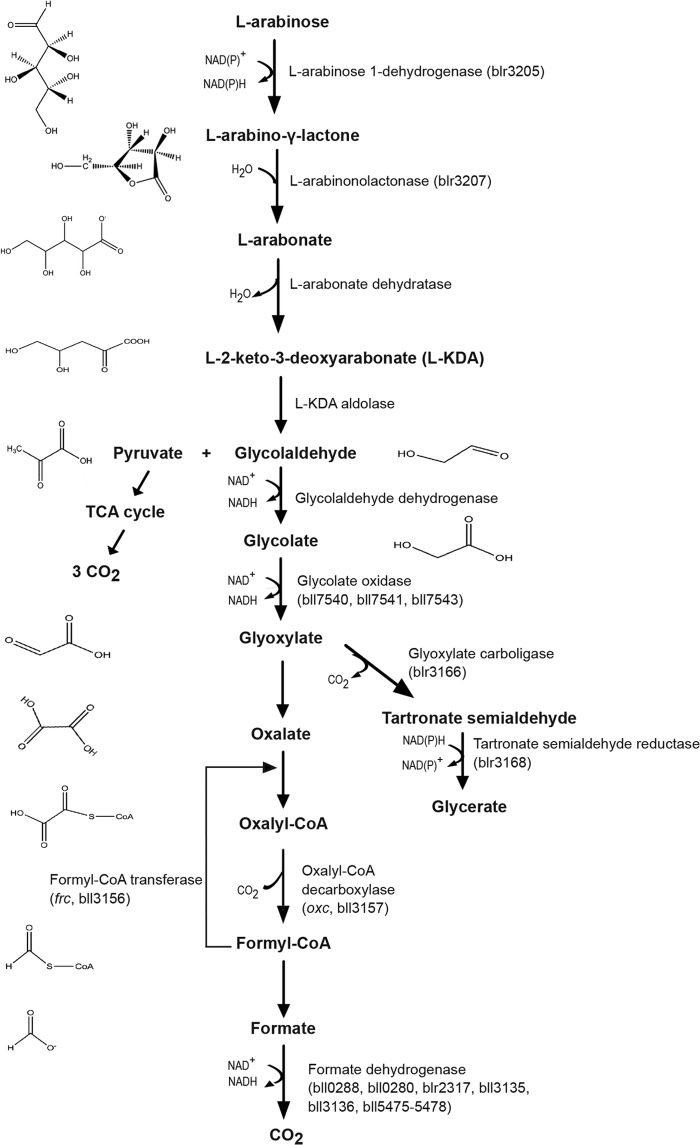
FIG 1.
Proposed pathway for the catabolism of l-arabinose and its link to the oxalate degradation pathway (based on reference 17). Gene names and enzyme assignments were made according to amino acid sequence similarity to previously identified enzymes. The chemical structures have been created with ChemDraw.
MATERIALS AND METHODS
Bacterial strains, media, and growth conditions.
The bacterial strains and plasmids used in this work are listed in Table S1 in the supplemental material. Escherichia coli cells were cultivated in Luria-Bertani medium (22) at 37°C using the following concentrations of antibiotics (in μg/ml): ampicillin (200), kanamycin (30), chloramphenicol (20), and tetracycline (10). Bradyrhizobium japonicum cells were routinely cultivated at 30°C on peptone-salts-yeast extract (PSY) medium (23) supplemented with 0.1% arabinose or in defined buffered Vincent's minimal medium (BVM) (24, 25), referred to as minimal medium. Carbon sources used in defined media were filter sterilized and used at a final concentration of 20 mM arabinose or 20 mM succinate. The appropriate antibiotic concentrations (in μg/ml) were added: spectinomycin (100) and kanamycin (100). Aerobic cultures for phenotypic growth analysis in PSY medium or BVM were grown in 500-ml Erlenmeyer flasks containing 25 ml medium supplemented with spectinomycin (100 μg ml−1) and the respective C source on a shaker (160 rpm) at 30°C. For each strain or condition, the growth of three independent cultures was analyzed.
Sample preparation and liquid chromatography-tandem MS (LC-MS/MS) analysis for proteomics.
Sample preparation and mass spectrometric (MS) analysis were performed as described in detail elsewhere (12). In brief, proteins extracted from three replicates of B. japonicum cells grown in complex and minimal medium with l-arabinose until mid-exponential phase were separated on a Tris-HCl polyacrylamide gel. After reduction and carbamidomethylation the proteins were digested with trypsin (Promega, Madison, WI, USA), and the resulting peptides were separated by reverse-phase high-performance liquid chromatography (RP-HPLC) and analyzed by a hybrid LTQ-Orbitrap XL mass spectrometer (Thermo Fisher Scientific, Waltham, MA, USA) interfaced with a nanoelectrospray source (12). Mass spectra were further processed with an in-house processing pipeline (26). In brief, fragment ion mass spectra were extracted from Thermo RAW files using msconvert (ProteoWizard, version 3.0.3831), merged in one MGF file per gel and searched against a composite B. japonicum USDA 110 protein database (RefSeq NC_004463.1, 22 July 2013) containing 256 common contaminants (e.g., human keratin and trypsin). The search was performed with the powerful search engine MS-GF+ (MS-GFDB v7747) for a match to fully tryptic and semitryptic peptides with up to two missed cleavage sites and a mass tolerance of 25 ppm. Oxidation (M), deamidation (NQ), and methylation (DE) were used as variable modifications and carbamidomethylation (C) as a fixed modification. Using the decoy option of MS-GF+, we filtered the list of peptide spectrum matches (PSMs) to an estimated overall false discovery rate (FDR) of 0.2%, classified the PSMs with PeptideClassifier (27), and further considered only peptides (tryptic or semitryptic) that unambiguously imply one bacterial protein sequence. The FDR at the peptide level amounted to about 1.5%, and that at the protein level amounted to about 2.5% when requiring two peptides or three spectra for protein identification. A total of 3,390 and 3,149 proteins were identified in complex and minimal medium, respectively.
Protein abundance calculation.
Relative protein abundance (in ppm) was estimated based on spectral counts as described by Schrimpf and colleagues (28) and extended recently (29).
qPCR analysis.
RNA was extracted from B. japonicum 110spc4 cultures grown in minimal medium, with 20 mM arabinose or 20 mM succinate as the C source, to mid-exponential phase. Cell harvest, RNA extraction, cDNA synthesis, and quantitative PCR (qPCR) analysis were done as described previously (30, 31).
Metabolite extraction and HPLC-MS analysis.
Several intermediates of the l-arabinose degradation pathway (l-2-keto-3-deoxyarabonate, glycolaldehyde, and glyoxylate) as well as oxalate could be identified by a metabolomics approach (using nanoflow ion-pair RP-HPLC coupled with nanospray high-resolution MS with a split-free nano-LC Ultra system connected to an LTQ-Orbitrap mass spectrometer). Sampling, quenching, and central metabolite extraction were carried out as described by Schneider and colleagues (32). The data were normalized to the biomass produced in the cultures.
Oxalotrophic growth.
The B. japonicum wild type and mutant strain Δfrc-oxc were grown on double-layered Schlegel mineral medium plates supplemented with either Ca-oxalate or Na-oxalate (33). The first layer contains the following ingredients: Na2HPO4·12H2O, 9.0 g/liter; KH2PO4, 1.5 g/liter; NH4Cl, 1.0 g/liter; MgSO4·7H2O, 0.2 g/liter; ammonium ferric citrate, 0.005 g/liter; CaCl2, 0.01 g/liter; ZnSO4·7H2O, 50 μg/liter; MnCl2·4H2O, 15 μg/liter; H3BO3, 150 μg/liter; CoCl2·6H2O, 100 μg/liter; CuCl2·2H2O, 50 μg/liter; NiCl2·6H2O, 10 μg/liter; NaMoO4·2H2O, 15 μg/liter; and agar, 15 g/liter. The second layer is composed of Schlegel mineral medium to which Ca-oxalate or Na-oxalate (7 g/liter or 2.7 g/liter; Sigma-Aldrich, Steinheim, Germany) was added (33). For this test, wild-type and mutant cultures were pregrown in minimal medium. Cells were washed and set to an optical density at 600 nm (OD600) of 4. A cotton swab was used to streak the cells on the plate. Plates were incubated at 30°C for 7 days. The assays were repeated three times.
Isothermal calorimetry.
Isothermal calorimetry analyses were performed according to Bravo et al. (34). Briefly, solid Angle's medium (35) was supplemented with different carbon sources at a final concentration of 35 mM Ca-oxalate. The media were poured into sterile microcalorimetric ampoules to obtain slants. These slants were inoculated with B. japonicum wild-type and Δfrc-oxc strains using a loop with an inoculum size sufficient to grow bacteria as a lawn. The ampoules were then sealed and introduced in the microcalorimeter (TAM48; Waters/TA, DE). After the thermal equilibration procedure, measurements were taken for at least 7 days. The data were recorded continuously by the microcalorimeter and resampled to obtain an effective sampling rate of 1 data point every 5 min. Samples were removed from the microcalorimeter and visually inspected in order to check that bacteria did form a lawn.
Construction of mutant strains.
DNA was isolated from the B. japonicum wild-type strain 110spc4 as previously described (36). Plasmid DNA from E. coli strains was obtained by using the NucleoSpin plasmid kit (Macherey-Nagel, Düren, Germany). Mutagenesis of genes was done by marker replacement. To construct a deletion mutant in the bll3156-3157 genes, PCR fragments of the 5′ and 3′ flanking regions of bll3156-3157 were amplified using the following primer pairs: bll3156-1_rev (TACGGCTGGCGTCCGGCGAAC) and bll3156-2_for (GTCTCTGGTCGAACCGCTTACTCGGCG) for the 3′ region of bll3156 and bll3156-3_for (TGTCTCCCTGGTCTCAATACTG) and bll3156-4_rev (GTCGAGCATCACCGGCTCG) for the 5′ region of bll3157. PCR products were cloned in the pGEM-T Easy vector (Promega, Madison, WI, USA), and the correct sequence was verified by sequence analysis. Up- and downstream regions were subcloned into the pSUP202pol4 vector (37), and a kanamycin resistance cassette (aphII) derived from pBSL15 (38) was inserted between both regions. The resulting plasmid, pRJ6243, was used for conjugation with B. japonicum strain 110spc4. The correct genomic integration was verified by PCR. The resulting Δfrc-oxc deletion mutant was named 6243 (Fig. 2).
FIG 2.
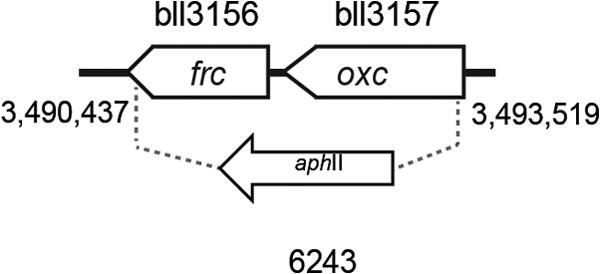
Physical map of the B. japonicum genomic region that harbors genes of the oxalate metabolism. Gene bll3156 (frc) codes for a formyl-CoA transferase and bll3157 (oxc) for an oxalyl-CoA decarboxylase. The precise structure of the B. japonicum deletion mutant is indicated with an arrow along with the strain number. Numbers below the vertical lines represent genome coordinates (the map is drawn to scale).
Plant material, inoculation, and cultivation.
Sterilization of soybean (Glycine max [L.] Merr. cv. Williams), mungbean (Vigna radiata), cowpea (Vigna unguiculata), and siratro (Macroptilium atropurpureum) seeds was done as described earlier (21). Plants were inoculated with cultures of B. japonicum that had been grown for 5 days in full medium and diluted to approximately 100 bacteria per plant. Nitrogenase activity was determined by using the acetylene reduction assay 21 days postinfection (dpi) for soybean, mungbean, and cowpea and 31 dpi for siratro. Bacteria were isolated from randomly selected nodules to confirm the presence of appropriate genetic markers.
Competitiveness in symbiosis.
Soybean, mungbean, cowpea, and siratro plants were infected with a mixture of the wild type and the Δfrc-oxc mutant strain containing a total of 100 bacteria per plant. Cultures of the wild type and mutant were grown and diluted to the same CFU per milliliter. A 90:10 ratio (Δfrc-oxc to wild type) was chosen to assess symbiotic competition. Serial dilutions of the mixed inoculum were plated on selective agar to control the number of inoculated cells. All nodules from one plant were harvested at the peak of nitrogenase activity 21 (soybean, mungbean, and cowpea) and 31 (siratro) dpi. Nodules were surface sterilized (100% ethanol for 5 min) and rinsed in sterile distilled water. Nodules were then crushed in 1 ml PSY medium using a mortar, and this suspension was serially diluted and spotted on plates containing the appropriate selection for strain differentiation. The plates were incubated for 4 days at 30°C, and the ratio of the mutant to wild-type strain in nodule extracts was determined and compared to the initial inoculum ratio. As a control, plants infected with either the wild type or mutant strain only were processed. At least three independent plants were processed per host. The nodule extracts were spotted in duplicates. Data were evaluated for statistical significance using Student's t test and SPSS 17.0 software.
Determination of the oxalate content of roots and nodules of legumes inoculated with B. japonicum.
Root nodules of soybean, mungbean, cowpea, and sirato infected with B. japonicum as well as root-only material were collected, immediately flash-frozen in liquid nitrogen, and stored at −80°C until use. To assess the soluble oxalate content in nodules, 50 mg of nodule material was homogenized in 200 μl distilled water (dH2O) using a TissueLyzer (Qiagen, Valencia, CA, USA) (1.3 min at 30 Hz) with a tungsten carbide bead (3 mm; Qiagen). One hundred mg of root material was ground in liquid nitrogen using a mortar and pestle. The resulting powder was collected and resuspended in 200 μl of dH2O. To ensure optimal destruction, the material was further homogenized in a TissueLyzer (1.3 min at 30 Hz). The supernatant of nodule and root samples was collected by a low-speed centrifugation step. The oxalate concentration in each sample was determined using a urinalysis diagnostic kit (TrinityBiotech, St. Louis, MO) according to the manufacturer's instructions. Measurements were done in triplicate on three independent plants.
Proteomics data accession number.
Proteomics data associated with the manuscript can be downloaded from ProteomeXchange under accession number PXD000487.
RESULTS AND DISCUSSION
Proteome analysis of B. japonicum grown in minimal medium containing arabinose.
The proteome of cells grown in minimal medium containing 20 mM l-arabinose as the carbon and energy source revealed the presence of several enzymes involved in l-arabinose degradation (Fig. 1 and Table 1). Apart from detecting l-arabinose 1-dehydrogenase (Blr3205), l-arabinolactonase (Blr3207), and glycolate oxidase (Bll7540-41, Bll7543), the enzymes glyoxylate carboligase (Blr3166) as well as the tartronate semialdehyde reductase (Blr3168) (Fig. 1) were expressed abundantly in minimal medium containing l-arabinose. Remarkably, large amounts of two enzymes involved in oxalate degradation were also detected: a formyl-CoA transferase (Frc) (bll3156) and an oxalyl-CoA decarboxylase (Oxc) (bll3157) that share 71% and 78% amino acid sequence identity with the previously studied Frc and Oxc of Oxalobacter formigenes (39–41). This led us to speculate that glycolaldehyde is oxidized to glyoxylate, which can be fed into at least two pathways: (i) reduction to glycerate through the activity of glyoxylate carboligase (blr3166) (42) and tartronate semialdehyde reductase (blr3168), or (ii) oxidation to oxalate followed by the stepwise, complete oxidation to formate and CO2 through the activities of Oxc and Frc (43, 44) and formate dehydrogenase. We previously showed that only the enzymes Oxc and Frc involved in the second pathway were detected in bacteroids during symbiosis with all different host plants (12, 21) (Table 1). Interestingly, using qPCR analysis, an elevated expression of bll3157 and bll3156 was not observed in B. japonicum cultures grown in minimal medium with succinate as the carbon source. The induction factors in arabinose-grown cells were measured as 45 for bll3157 and 116 for bll3156 compared to levels for succinate-grown cells. Thus, it appeared as if the oxalate oxidation pathway was switched on when l-arabinose was offered as the carbon source.
TABLE 1.
Normalized protein abundance data for enzymes involved in l-arabinose degradation and oxalate catabolism in B. japonicumd
| Enzymec | Designationa | Normalized protein abundanceb (ppm) |
||||
|---|---|---|---|---|---|---|
| Symbiotic |
Free living |
|||||
| Soybean | Cowpea | Siratro | BVM plus arabinose | PSY plus arabinose | ||
| l-Arabinose 1-DH | Blr3205 | 23 | 38 | 42 | 522 | 502 |
| l-Arabinonolactonase | Blr3207 | 31 | 55 | 13 | 555 | 458 |
| Glycolate oxidase | Bll7540 | 114 | 81 | 91 | 98 | 68 |
| Bll7541 | 9 | 47 | 34 | 73 | 80 | |
| Bll7543 | 5 | 11 | 46 | 45 | ||
| Oxalyl-CoA decarboxylase (Oxc) | Bll3157 | 342 | 237 | 205 | 1,389 | 519 |
| Formyl-CoA transferase (Frc) | Bll3156 | 364 | 394 | 211 | 800 | 604 |
| Formate DH | Bll0288 | |||||
| Bll0280 | ||||||
| Blr2317 | 63 | 56 | ||||
| Bll3135 | ||||||
| Bll3136 | 2 | 2 | ||||
| Bll5475 | ||||||
| Bll5476 | 18 | 66 | 207 | |||
| Bll5477 | 19 | 108 | 2 | |||
| Bll5478 | 27 | 49 | 145 | 4 | ||
| Glyoxylate carboligase | Blr3166 | 526 | 13 | |||
| Tartronate semialdehyde reductase | Blr3168 | 774 | 28 | |||
Construction and growth analysis of a Δfrc-oxc deletion mutant.
To further analyze the hypothesis that oxalate degradation is linked to arabinose utilization, a Δfrc-oxc mutant strain was constructed (strain 6243) (Fig. 2). Growth of B. japonicum wild-type and mutant strains was analyzed in minimal and complex medium. The minimal medium was supplemented with either 20 mM l-arabinose, 20 mM succinate, or 20 mM Na-oxalate.
First, we compared the ability of both the wild type and the Δfrc-oxc mutant to catabolize oxalate using plate assays and microcalorimetry (see Materials and Methods). We initially aimed at monitoring oxalotrophic growth in liquid cultures using Ca- or Na-oxalate as the sole source of carbon. Even though various concentrations (5, 10, and 20 mM) of oxalate were used, these cultivation attempts failed. Therefore, we examined growth on Schlegels minimal medium agar plates in the presence of 20 mM Na-oxalate or 35 mM Ca-oxalate as the sole carbon source (data not shown). For this test, strains were first grown in minimal medium and then streaked out on agar plates. This test revealed that the B. japonicum wild type was able to grow on plates supplemented with 20 mM Na-oxalate, whereas the Δfrc-oxc mutant did not grow. Likewise, microcalorimetric analyses on Angle's medium supplemented with 35 mM Ca-oxalate revealed that the B. japonicum wild type was able to grow on Ca-oxalate, whereas no growth was observed for the Δfrc-oxc mutant (Fig. 3C). The metabolic heat production resulting from the oxidation of oxalate according to the reaction 2C2H2O4 + O2 → 4CO2 + 2H2O with a reaction enthalpy (ΔH°rxn) of −499 KJ/mol is directly proportional to the oxalate consumption rate. At this point, it must be noted that biomass production can easily be neglected (34), because biomass yield is extremely low with oxalate. Thus, each curve represents the overall metabolic activity related to the growth of B. japonicum. Such measurements can be compared using common metabolic assays, such as the 2,3,5-triphenyltetrazolium chloride assay, for example (45). The metabolic activity pattern for the B. japonicum wild-type strain was similar to what could be observed in other studies (34).
FIG 3.
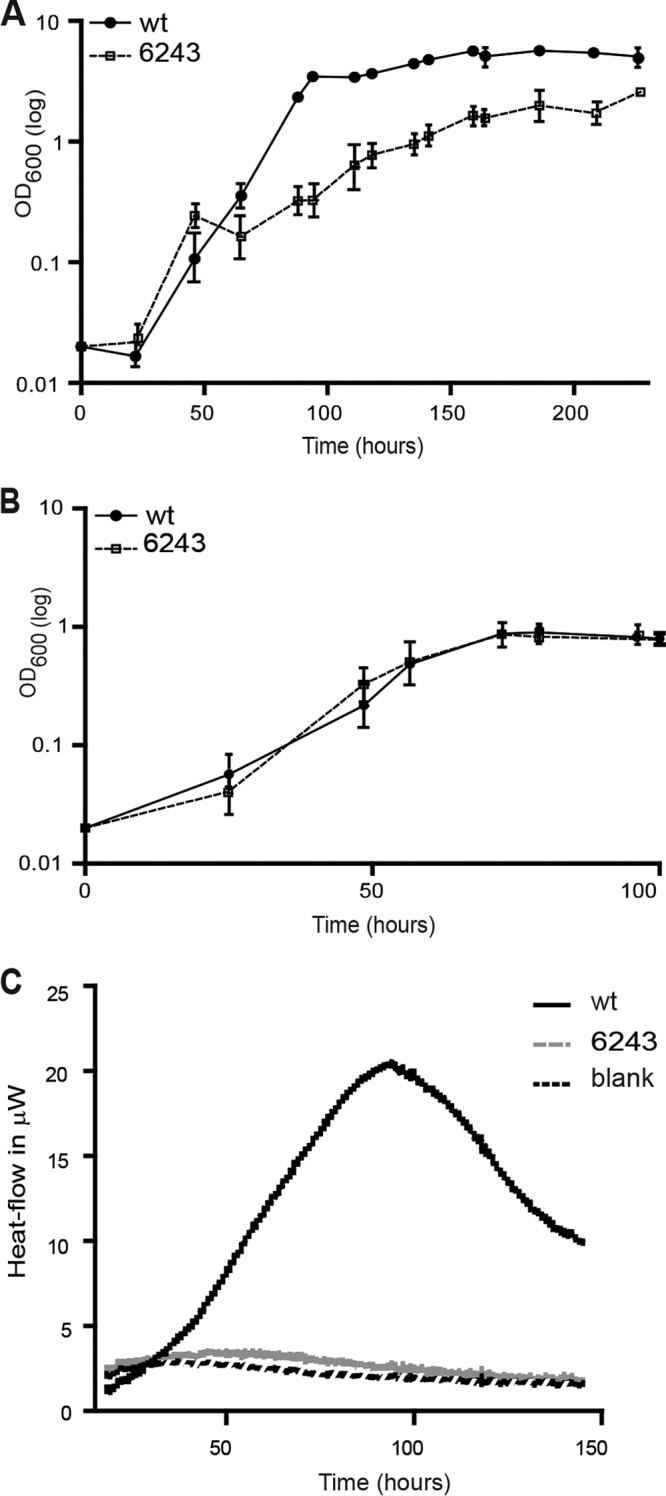
Growth analysis of the B. japonicum wild type and the Δfrc-oxc mutant in minimal medium supplemented with two different carbon sources. (A) Twenty millimolar arabinose; (B) 20 mM succinate. Shown are mean values for all taken time points ± standard deviations. (C) The heat production related to the metabolic activity during oxalotrophic growth of B. japonicum wild-type and mutant cells was monitored using isothermal microcalorimetry.
When grown on minimal medium with l-arabinose as the carbon source, the Δfrc-oxc mutant displayed a diminished growth rate compared to that of the wild type (Fig. 3A). The mean generation time of the parental strain was 23.4 h, compared to 37.3 h for the Δfrc-oxc mutant. Moreover, wild-type cells reached a final optical density of 5.6, while the mutant strain did not exceed an OD of 2. In contrast, the Δfrc-oxc strain exhibited growth behavior similar to that of the wild-type strain when cultivated either in minimal medium with the dicarboxylic acid succinate (Fig. 3B) or in complex medium (data not shown). Thus, the results have shown that a deletion of two specific oxalate degradation genes caused a partial defect in growth with arabinose. This can be explained in at least two different ways: (i) in the mutant, l-arabinose may not be consumed as efficiently as in the wild type for lack of oxalate degradation and partial energy conservation through the formate dehydrogenase reaction, and (ii) the mutant may accumulate inhibitory concentrations of oxalate and its precursors. We tested the latter scenario by measuring the cellular contents of oxalate, glyoxylate, and glycolaldehyde and found these compounds to be elevated by a factor of up to 2 in the mutant compared to the wild type (data not shown). Whether or not this slight increase is inhibitory to growth has not been further investigated. Regardless of which of the two explanations is true, the degradation of oxalate appears to be a necessary requirement for the complete utilization of arabinose for growth and biomass formation.
Oxalate content in roots and root nodules induced by B. japonicum.
Since both Frc and Oxc had been detected in soybean, cowpea, and siratro bacteroids using a global proteomics approach (12, 21) (Table 1), we became interested in exploring whether oxalate is present in nodules as a potential C source for the bacteroids. Notably, it has been hypothesized previously that oxalate is a potential energy-yielding substrate sufficient to sustain nitrogen fixation in Vicia faba (46).
The oxalate content of roots and root nodules of four B. japonicum host plants (soybean, siratro, cowpea, and mungbean) was analyzed using an oxalate oxidase assay (Table 2). In general, three times higher oxalate concentrations were detected in root nodules than in the roots of uninfected plants (Table 2). This showed that oxalate is indeed present in nodules and might be available as a C source together with other compounds, such as succinate and malate.
TABLE 2.
Oxalate content of root material and root nodules of cowpea, mungbean, siratro, and soybean plants infected with the B. japonicum wild type
| Host plant | Oxalate level (mg g−1 wet wt) ina: |
|
|---|---|---|
| Root | Root nodules | |
| Cowpea | 0.10 ± 0.01 | 0.37 ± 0.08 |
| Mungbean | 0.09 ± 0.01 | 0.31 ± 0.05 |
| Siratro | 0.09 ± 0.01 | 0.22 ± 0.01 |
| Soybean | 0.03 ± 0.01 | 0.18 ± 0.02 |
Results are presented as means ± standard deviations where n ≥ 2.
Plant symbiosis and competition for nodule occupancy.
The fact that B. japonicum host plants displayed different concentrations of soluble oxalate prompted us to investigate the role of the oxalate degradation genes in symbiosis. The symbiotic efficiency of the wild type and the Δfrc-oxc mutant was tested on all plant hosts. Plants infected with Δfrc-oxc grew as healthy as the wild type, displaying green leaves, normal nodule development, and wild-type-like nitrogenase activity 21 dpi for soybean, mungbean, and cowpea and 31 dpi for siratro (see Table S2 in the supplemental material). Moreover, the same number of nodules was observed in plants inoculated with the Δfrc-oxc mutant and with the parental strain. Reisolation of bacteroids from plants used in this test revealed comparable viable cell counts for wild-type and mutant strains (data not shown).
It was previously shown that mutations in genes responsible for the catabolism of carbon sources, such as rhamnose and myo-inositol, and other used nutrients, such as mimosine, are correlated with a decreased ability to compete for nodule occupancy (47–50). Therefore, we investigated if the presence of the oxalate catabolic pathway could represent a competitive advantage for nodulation occupancy on all host plants. In order to test nodule colonization, 50 CFU/ml of B. japonicum Δfrc-oxc and wild-type cells were mixed at a 90:10 ratio and used for inoculation of soybean, siratro, cowpea, and mungbean seedlings. After 21 dpi for soybean, mungbean, and cowpea and 31 dpi for siratro, all nodules from one plant were collected. In total, three independent experiments including at least three plants were performed per strain and host. To isolate bacteroids, root nodules were homogenized and the wild type-to-mutant ratio was investigated by comparing the recovered viable cell counts of Δfrc-oxc and wild-type strains using genetic markers. In all host plants the Δfrc-oxc mutant was affected in nodule occupancy when competing with the parental strain (Fig. 4), despite the fact that the mutant was present in 10-fold excess over the wild type in the inoculant mixture. A possible impact of the antibiotic resistance cassette (aphII in strain Δfrc-oxc) on the nodulation competition phenotype had been excluded by results presented recently (51), where an ACC deaminase mutant containing the identical aphII antibiotic resistance cassette could compete as well as the wild type for nodule occupancy.
FIG 4.
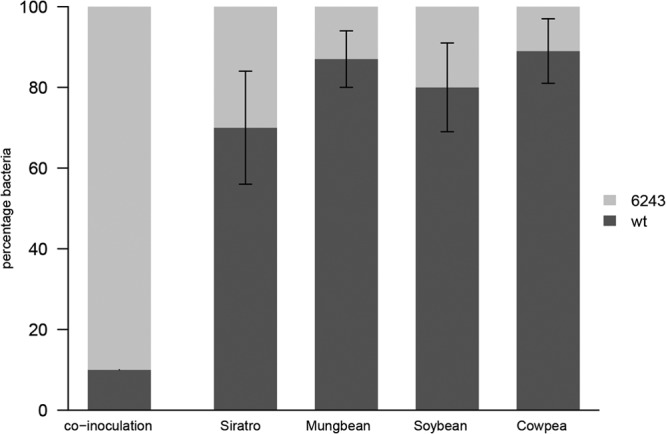
Symbiosis and competition for root-nodule colonization. Soybean, cowpea, siratro, and mungbean seedlings were coinoculated into 3 plants at a ratio of 10:90 B. japonicum wild-type (wt) to Δfrc-oxc cells. Percentages of wild-type and mutant strains recovered from plant nodules at different inoculum ratios were compared. The tested host plants are shown along the x axis. The y axis represents mean values ± standard deviations of percentages of bacteroids recovered from crushed nodules. Experimental data were assessed for statistical significance by means of the Student t test. Statistical analyses were performed using SPSS 17.0 software. Recovered percentages were significantly different between the wild type and mutant (P < 0.05).
Concluding remarks.
In this study, we demonstrated that B. japonicum is capable of entertaining an oxalotrophic lifestyle. Sequence analyses indicated that the oxc and frc genes involved in oxalate degradation are present and conserved in other Bradyrhizobium strains (52) but not in fast-growing rhizobia such as Sinorhizobium meliloti, Rhizobium leguminosarum, and R. etli. In this study, based on proteomics and growth analysis on different carbon sources, we showed that the degradation of l-arabinose creates intermediates that most likely are fed into the oxalate degradation pathway (Fig. 1). Disrupting the oxalate degradation branch by mutation leads to compromised growth on arabinose, because either the additive benefit from reductant formation in the formate dehydrogenase reaction is lacking or inhibitory amounts of oxalate and its precursors build up in the mutant. While the presence of functional formyl-CoA transferase (frc) and oxalyl-CoA decarboxylase (oxc) genes in B. japonicum is dispensable for the establishment of an effective symbiosis, these enzymes nevertheless seem to provide an advantage in the process of root-nodule colonization by B. japonicum. It can be speculated that at some point during rhizobial infection and nodule occupation the ability to degrade oxalate, which is present in roots and root nodules, represents a beneficial trait for B. japonicum.
Supplementary Material
ACKNOWLEDGMENTS
We thank Pilar Junier for advice on oxalotrophy analyses, Andrea Lindemann for initial work on l-arabinose degradation, Martina Lardi for help with statistical analysis, and Hans-Martin Fischer for constructive discussions.
This work was supported by ETH Zurich and by a grant from the Swiss National Foundation for Scientific Research.
We are grateful to Dulce-Nombre Rodríguez-Navarro and Franscisco Temprano (Las Torres-Tomejil, Seville, Spain) for providing soybean seeds and to William Broughton (University of Geneva, Switzerland) for cowpea and siratro seeds.
Footnotes
Published ahead of print 24 January 2014
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.03314-13.
REFERENCES
- 1.Udvardi M, Poole PS. 2013. Transport and metabolism in legume-rhizobia symbioses. Annu. Rev. Plant Biol. 64:781–805. 10.1146/annurev-arplant-050312-120235 [DOI] [PubMed] [Google Scholar]
- 2.Lodwig E, Poole P. 2003. Metabolism of Rhizobium bacteroids. Crit. Rev. Plant Sci. 22:37–78. 10.1080/713610850 [DOI] [Google Scholar]
- 3.Prell J, Poole P. 2006. Metabolic changes of rhizobia in legume nodules. Trends Microbiol. 14:161–168. 10.1016/j.tim.2006.02.005 [DOI] [PubMed] [Google Scholar]
- 4.Udvardi MK, Price GD, Gresshoff PM, Day DA. 1988. A dicarboxylate transporter on the peribacteroid membrane of soybean nodules. FEBS Lett. 231:36–40. 10.1016/0014-5793(88)80697-5 [DOI] [Google Scholar]
- 5.Udvardi MK, Day DA. 1997. Metabolite transport across symbiotic membranes of legume nodules. Annu. Rev. Plant Physiol. Plant Mol. Biol. 48:493–523. 10.1146/annurev.arplant.48.1.493 [DOI] [PubMed] [Google Scholar]
- 6.Ronson CW, Lyttleton P, Robertson JG. 1981. C4-dicarboxylate transport mutants of Rhizobium trifolii form ineffective nodules on Trifolium repens. Proc. Natl. Acad. Sci. U. S. A. 78:4284–4288. 10.1073/pnas.78.7.4284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ronson CW, Astwood PM, Downie JA. 1984. Molecular cloning and genetic organization of C4-dicarboxylate transport genes from Rhizobium leguminosarum. J. Bacteriol. 160:903–909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yurgel SN, Kahn ML. 2004. Dicarboxylate transport by rhizobia. FEMS Microbiol. Rev. 28:489–501. 10.1016/j.femsre.2004.04.002 [DOI] [PubMed] [Google Scholar]
- 9.Streeter JG. 1980. Carbohydrates in soybean nodules. II. Distribution of compounds in seedlings during the onset of nitrogen fixation. Plant Physiol. 66:471–476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Glenn AR, Mckay IA, Arwas R, Dilworth MJ. 1984. Sugar metabolism and the symbiotic properties of carbohydrate mutants of Rhizobium leguminosarum. J. Gen. Microbiol. 130:239–245 [Google Scholar]
- 11.Salminen SO, Streeter JG. 1987. Uptake and metabolism of carbohydrates by Bradyrhizobium japonicum bacteroids. Plant Physiol. 83:535–540. 10.1104/pp.83.3.535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Delmotte N, Ahrens CH, Knief C, Qeli E, Koch M, Fischer HM, Vorholt JA, Hennecke H, Pessi G. 2010. An integrated proteomics and transcriptomics reference data set provides new insights into the Bradyrhizobium japonicum bacteroid metabolism in soybean root nodules. Proteomics 10:1391–1400. 10.1002/pmic.200900710 [DOI] [PubMed] [Google Scholar]
- 13.Stowers MD. 1985. Carbon metabolism in Rhizobium species. Annu. Rev. Microbiol. 39:89–108. 10.1146/annurev.mi.39.100185.000513 [DOI] [PubMed] [Google Scholar]
- 14.Pedrosa FO, Zancan GT. 1974. L-Arabinose metabolism in Rhizobium japonicum. J. Bacteriol. 119:336–338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Novick NJ, Tyler ME. 1982. L-Arabinose metabolism in Azospirillum brasiliense. J. Bacteriol. 149:364–367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Watanabe S, Kodaki T, Makino K. 2006. Cloning, expression, and characterization of bacterial L-arabinose 1-dehydrogenase involved in an alternative pathway of L-arabinose metabolism. J. Biol. Chem. 281:2612–2623. 10.1074/jbc.M506477200 [DOI] [PubMed] [Google Scholar]
- 17.Watanabe S, Shimada N, Tajima K, Kodaki T, Makino K. 2006. Identification and characterization of L-arabonate dehydratase, L-2-keto-3-deoxyarabonate dehydratase, and L-arabinolactonase involved in an alternative pathway of L-arabinose metabolism. Novel evolutionary insight into sugar metabolism. J. Biol. Chem. 281:33521–33536. 10.1074/jbc.M606727200 [DOI] [PubMed] [Google Scholar]
- 18.Duncan MJ, Fraenkel DG. 1979. α-Ketoglutarate dehydrogenase mutant of Rhizobium meliloti. J. Bacteriol. 137:415–419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Watanabe S, Kodaki T, Makino K. 2006. A novel α-ketoglutaric semialdehyde dehydrogenase: evolutionary insight into an alternative pathway of bacterial L-arabinose metabolism. J. Biol. Chem. 281:28876–28888. 10.1074/jbc.M602585200 [DOI] [PubMed] [Google Scholar]
- 20.Poysti NJ, Loewen ED, Wang Z, Oresnik IJ. 2007. Sinorhizobium meliloti pSymB carries genes necessary for arabinose transport and catabolism. Microbiology 153:727–736. 10.1099/mic.0.29148-0 [DOI] [PubMed] [Google Scholar]
- 21.Koch M, Delmotte N, Rehrauer H, Vorholt JA, Pessi G, Hennecke H. 2010. Rhizobial adaptation to hosts, a new facet in the legume root-nodule symbiosis. Mol. Plant Microbe Interact. 23:784–790. 10.1094/MPMI-23-6-0784 [DOI] [PubMed] [Google Scholar]
- 22.Miller J. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 23.Regensburger B, Hennecke H. 1983. RNA polymerase from Rhizobium japonicum. Arch. Microbiol. 135:103–109. 10.1007/BF00408017 [DOI] [PubMed] [Google Scholar]
- 24.Serventi F, Youard ZA, Murset V, Huwiler S, Bühler D, Richter M, Luchsinger R, Fischer HM, Brogioli R, Niederer M, Hennecke H. 2012. Copper starvation-inducible protein for cytochrome oxidase biogenesis in Bradyrhizobium japonicum. J. Biol. Chem. 287:38812–38823. 10.1074/jbc.M112.406173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Vincent JM. 1970. A manual for the practical study of root nodule bacteria. Blackwell Scientific Publications, Oxford, United Kingdom [Google Scholar]
- 26.Omasits U, Quebatte M, Stekhoven DJ, Fortes C, Roschitzki B, Robinson MD, Dehio C, Ahrens CH. 2013. Directed shotgun proteomics guided by saturated RNA-seq identifies a complete expressed prokaryotic proteome. Genome Res. 23:1916–1927. 10.1101/gr.151035.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Qeli E, Ahrens CH. 2010. PeptideClassifier for protein inference and targeted quantitative proteomics. Nat. Biotechnol. 28:647–650. 10.1038/nbt0710-647 [DOI] [PubMed] [Google Scholar]
- 28.Schrimpf SP, Weiss M, Reiter L, Ahrens CH, Jovanovic M, Malmstrom J, Brunner E, Mohanty S, Lercher MJ, Hunziker PE, Aebersold R, von Mering C, Hengartner MO. 2009. Comparative functional analysis of the Caenorhabditis elegans and Drosophila melanogaster proteomes. PLoS Biol. 7:e48. 10.1371/journal.pbio.1000048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Carlier AL, Omasits U, Ahrens CH, Eberl L. 2013. Proteomics analysis of Psychotria leaf nodule symbiosis: improved genome annotation and metabolic predictions. Mol. Plant Microbe Interact. 26:1325–1333. 10.1094/MPMI-05-13-0152-R [DOI] [PubMed] [Google Scholar]
- 30.Pessi G, Ahrens CH, Rehrauer H, Lindemann A, Hauser F, Fischer HM, Hennecke H. 2007. Genome-wide transcript analysis of Bradyrhizobium japonicum bacteroids in soybean root nodules. Mol. Plant Microbe Interact. 20:1353–1363. 10.1094/MPMI-20-11-1353 [DOI] [PubMed] [Google Scholar]
- 31.Lindemann A, Moser A, Pessi G, Hauser F, Friberg M, Hennecke H, Fischer HM. 2007. New target genes controlled by the Bradyrhizobium japonicum two-component regulatory system RegSR. J. Bacteriol. 189:8928–8943. 10.1128/JB.01088-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schneider K, Skovran E, Vorholt JA. 2012. Oxalyl-coenzyme A reduction to glyoxylate is the preferred route of oxalate assimilation in Methylobacterium extorquens AM1. J. Bacteriol. 194:3144–3155. 10.1128/JB.00288-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Aragno MS, Schlegel HG. 1991. The mesophilic hydrogen-oxidizing (Knallgas) bacteria. Springer Verlag, Berlin, Heidelberg, New York [Google Scholar]
- 34.Bravo D, Braissant O, Solokhina A, Clerc M, Daniels AU, Verrecchia E, Junier P. 2011. Use of an isothermal microcalorimetry assay to characterize microbial oxalotrophic activity. FEMS Microbiol. Ecol. 78:266–274. 10.1111/j.1574-6941.2011.01158.x [DOI] [PubMed] [Google Scholar]
- 35.Angle JS, McGrath SP, Chaney RL. 1991. New culture medium containing ionic concentrations of nutrients similar to concentrations found in the soil solution. Appl. Environ. Microbiol. 57:3674–3676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hahn M, Hennecke H. 1984. Localized mutagenesis in Rhizobium japonicum. Mol. Gen. Genet. 193:46–52. 10.1007/BF00327412 [DOI] [Google Scholar]
- 37.Fischer HM, Babst M, Kaspar T, Acuña G, Arigoni F, Hennecke H. 1993. One member of a groESL-like chaperonin multigene family in Bradyrhizobium japonicum is co-regulated with symbiotic nitrogen fixation genes. EMBO J. 12:2901–2912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Alexeyev MF. 1995. Three kanamycin resistance gene cassettes with different polylinkers. Biotechniques 18:52–56 [PubMed] [Google Scholar]
- 39.Lung HY, Baetz AL, Peck AB. 1994. Molecular cloning, DNA sequence, and gene expression of the oxalyl-coenzyme A decarboxylase gene, oxc, from the bacterium Oxalobacter formigenes. J. Bacteriol. 176:2468–2472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sidhu H, Ogden SD, Lung HY, Luttge BG, Baetz AL, Peck AB. 1997. DNA sequencing and expression of the formyl coenzyme A transferase gene, frc, from Oxalobacter formigenes. J. Bacteriol. 179:3378–3381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sahin N. 2003. Oxalotrophic bacteria. Res. Microbiol. 154:399–407. 10.1016/S0923-2508(03)00112-8 [DOI] [PubMed] [Google Scholar]
- 42.Chang YY, Wang AY, Cronan JE., Jr 1993. Molecular cloning, DNA sequencing, and biochemical analyses of Escherichia coli glyoxylate carboligase, an enzyme of the acetohydroxy acid synthase-pyruvate oxidase family. J. Biol. Chem. 268:3911–3919 [PubMed] [Google Scholar]
- 43.Stewart CS, Duncan SH, Cave DR. 2004. Oxalobacter formigenes and its role in oxalate metabolism in the human gut. FEMS Microbiol. Lett. 230:1–7. 10.1016/S0378-1097(03)00864-4 [DOI] [PubMed] [Google Scholar]
- 44.Svedruzic D, Jonsson S, Toyota CG, Reinhardt LA, Ricagno S, Linqvist Y, Richards NGJ. 2005. The enzymes of oxalate metabolism: unexpected structures and mechanisms. Arch. Biochem. Biophys. 433:176–192. 10.1016/j.abb.2004.08.032 [DOI] [PubMed] [Google Scholar]
- 45.Braissant O, Wirz D, Gopfert B, Daniels AU. 2010. Use of isothermal microcalorimetry to monitor microbial activities. FEMS Microbiol. Lett. 303:1–8. 10.1111/j.1574-6968.2009.01819.x [DOI] [PubMed] [Google Scholar]
- 46.Trinchant JC, Guerin V, Rigaud J. 1994. Acetylene reduction by symbiosomes and free bacteroids from broad bean (Vicia faba L.) nodules. Plant Physiol. 105:555–561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Fry J, Wood M, Poole PS. 2001. Investigation of myo-inositol catabolism in Rhizobium leguminosarum bv. viciae and its effect on nodulation competitiveness. Mol. Plant Microbe Interact. 14:1016–1025. 10.1094/MPMI.2001.14.8.1016 [DOI] [PubMed] [Google Scholar]
- 48.Jimenez-Zurdo JI, van Dillewijn P, Soto MJ, de Felipe MR, Olivares J, Toro N. 1995. Characterization of a Rhizobium meliloti proline dehydrogenase mutant altered in nodulation efficiency and competitiveness on alfalfa roots. Mol. Plant Microbe Interact. 8:492–498. 10.1094/MPMI-8-0492 [DOI] [PubMed] [Google Scholar]
- 49.Oresnik IJ, Pacarynuk LA, O'Brien SAP, Yost CK, Hynes MF. 1998. Plasmid-encoded catabolic genes in Rhizobium leguminosarum bv. trifolii: evidence for a plant-inducible rhamnose locus involved in competition for nodulation. Mol. Plant Microbe Interact. 11:1175–1185. 10.1094/MPMI.1998.11.12.1175 [DOI] [Google Scholar]
- 50.Soedarjo M, Borthakur D. 1998. Mimosine, a toxin produced by the tree-legume Leucaena provides a nodulation competition advantage to mimosine-degrading Rhizobium strains. Soil Biol. Biochem. 30:1605–1613. 10.1016/S0038-0717(97)00180-6 [DOI] [Google Scholar]
- 51.Murset V, Hennecke H, Pessi G. 2012. Disparate role of rhizobial ACC deaminase in root-nodule symbioses. Symbiosis 57:43–50. 10.1007/s13199-012-0177-z [DOI] [Google Scholar]
- 52.Bravo D, Martin G, David MM, Cailleau G, Verrecchia E, Junier P. 2013. Identification of active oxalotrophic bacteria by bromodeoxyuridine DNA-labeling in a microcosm soil experiments. FEMS Microbiol. Lett. 348:103–111. 10.1111/1574-6968.12244 [DOI] [PubMed] [Google Scholar]
- 53.Kaneko T, Nakamura Y, Sato S, Minamisawa K, Uchiumi T, Sasamoto S, Watanabe A, Idesawa K, Iriguchi M, Kawashima K, Kohara M, Matsumoto M, Shimpo S, Tsuruoka H, Wada T, Yamada M, Tabata S. 2002. Complete genomic sequence of nitrogen-fixing symbiotic bacterium Bradyrhizobium japonicum USDA110. DNA Res. 9:189–197. 10.1093/dnares/9.6.189 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.