Abstract
The microbial composition of artisan and industrial animal rennet pastes was studied by using both culture-dependent and -independent approaches. Pyrosequencing targeting the 16S rRNA gene allowed to identify 361 operational taxonomic units (OTUs) to the genus/species level. Among lactic acid bacteria (LAB), Streptococcus thermophilus and some lactobacilli, mainly Lactobacillus crispatus and Lactobacillus reuteri, were the most abundant species, with differences among the samples. Twelve groups of microorganisms were targeted by viable plate counts revealing a dominance of mesophilic cocci. All rennets were able to acidify ultrahigh-temperature-processed (UHT) milk as shown by pH and total titratable acidity (TTA). Presumptive LAB isolated at the highest dilutions of acidified milks were phenotypically characterized, grouped, differentiated at the strain level by randomly amplified polymorphic DNA (RAPD)-PCR analysis, and subjected to 16S rRNA gene sequencing. Only 18 strains were clearly identified at the species level, as Enterococcus casseliflavus, Enterococcus faecium, Enterococcus faecalis, Enterococcus lactis, Lactobacillus delbrueckii, and Streptococcus thermophilus, while the other strains, all belonging to the genus Enterococcus, could not be allotted into any previously described species. The phylogenetic analysis showed that these strains might represent different unknown species. All strains were evaluated for their dairy technological performances. All isolates produced diacetyl, and 10 of them produced a rapid pH drop in milk, but only 3 isolates were also autolytic. This work showed that animal rennet pastes can be sources of LAB, mainly enterococci, that might contribute to the microbial diversity associated with dairy productions.
INTRODUCTION
Several traditional Italian cheeses are manufactured in small farms with raw milk from animals of indigenous breeds. Most of these productions are carried out without the addition of commercial or natural starter cultures. However, the presence of certain species of lactic acid bacteria (LAB) is of paramount importance to transform milk into cheese (1). Unlike the majority of lactic acid fermentations, which are generally characterized by a given dominant group of LAB driving almost the entire process of transformation of the raw materials into final products, the production of cheese relies on two distinct groups of LAB species: starter LAB (SLAB), which participate in the fermentation process, and nonstarter LAB (NSLAB), which are responsible for maturation of the cheese (2). The successful fermentation of raw milk cheeses made without the addition of starter cultures strongly depends on the presence of SLAB in the raw materials.
In this context, the main sources of microbial contamination that might provide desirable LAB for the fermentation are generally considered to be the milk, the equipment used during processing, and the dairy environment (3, 4). Other sources of microbial contamination during cheese making that might affect the microbial community of milk at different extents are represented by the different ingredients added, especially the spices (5, 6). However, a relevant contribution to the microbial complexity of cheeses might be made by the rennet used for milk curdling. Animal rennets are coagulant enzyme preparations extracted from the abomasum of ruminants, mainly veal, kid, and lamb (7). They have been used as coagulant agents in the production of the majority of cheese varieties for centuries. In some cases, the microbial loads of animal rennets represent the major part of the natural microflora responsible for the fermentation process (8). The microbiological investigations of animal rennets are focused mainly on their stability and their role in the generation of defects (taste and flavor, putrefaction, disintegration, and blowing) in cheese (9, 10), as well as in their hygienic quality (11). In contrast, some studies were performed to evaluate the possible role of rennet to act as a vector for the transfer of microencapsulated bacteria with probiotic properties into cheese (12, 13).
Studies on the microbial composition of animal rennets, using mainly culture-dependent methods, highlighted the presence of aerobic mesophilic bacteria, coliforms, anaerobic spore-forming bacteria, LAB, staphylococci, yeasts, and molds (8, 10, 14–16). In this research, following the scheme reported in Fig. 1, we studied the microbial ecology of different artisanal and industrial animal rennets commonly used for the production of traditional cheeses in Sicily (southern Italy). The microbiota of rennets was assessed by culture-independent 16S rRNA gene sequencing, and the LAB diversity was evaluated by culture-dependent methods; the acidifying capacity of rennet pastes was evaluated in milk to select dairy LAB for cheese making; the most promising isolates were identified at the species and strain levels and characterized for their technological traits.
FIG 1.
Scheme of the analysis performed on rennet samples and LAB.
MATERIALS AND METHODS
Sample collection.
The animal rennet pastes (Table 1) were collected in several dairy factories located in western Sicily (Italy) producing two different pasta-filata cheese types, Caciocavallo Palermitano and PDO Vastedda della valle del Belìce, obtained with raw cows' and raw ewes' milk, respectively. Sample collection occurred twice from each dairy at an interval of 15 days. The rennets were sampled before their addition into bulk milks, transferred into sterile plastic bags, and transported under refrigeration with a portable refrigerator to the laboratory, where they were immediately subjected to microbiological analysis. Rennets of each dairy were pooled before DNA extraction.
TABLE 1.
Characteristics of the animal rennet pastes
| Rennet sample ID | Animal | Production level | Amt (g/100 liters) added for cheese making | Cheese or use | City (province)a |
|---|---|---|---|---|---|
| 1 | Lamb | Artisanal | 45 | Caciocavallo Palermitano | Godrano (PA) |
| 2 | Lamb | Artisanal | 50 | Caciocavallo Palermitano | Godrano (PA) |
| 3 | Lamb | Industrial | 40 | Vastedda della valle del Belìce | Santa Margherita del Belìce (AG) |
| 4 | Lamb | Artisanal | 40 | Vastedda della valle del Belìce | Menfi (AG) |
| 5 | Lamb | Artisanal | 30 | Caciocavallo Palermitano | Terrasini (PA) |
| 6 | Lamb | Artisanal | 50 | Caciocavallo Palermitano | Cinisi (PA) |
| 7 | Lamb | Artisanal | 50 | Caciocavallo Palermitano | Godrano (PA) |
| 8 | Lamb | Artisanal | 20 | Caciocavallo Palermitano | Terrasini (PA) |
| 9 | Lamb | Artisanal | 30 | Vastedda della valle del Belìce | Salemi (TP) |
| 10 | Lamb | Artisanal | 30 | Vastedda della valle del Belìce | Partanna (TP) |
| 11 | Kid | Industrial | 50 | Caciocavallo Palermitano | Godrano (PA) |
| Naturen | Calf | Industrial | Used for milk clotting activity comparison |
PA, Palermo; AG, Agrigento; TP, Trapani.
Microbiological analysis.
Serial decimal dilutions of rennet samples (10 g) were prepared in Ringer's solution (Oxoid, Milan, Italy). The first dilutions were homogenized in a stomacher (BagMixer 400; Interscience, Saint Nom, France) for 2 min at the highest speed. The cell suspensions were plated and incubated as follows: on plate count agar (PCA) with 1 g/liter added skimmed milk (SkM), incubated aerobically at 30°C for 72 h, for total mesophilic counts (TMC); on PCA-SkM, incubated aerobically at 7°C for 7 days, for total psychrotrophic counts (TPC); on violet red bile glucose agar (VRBGA), incubated anaerobically for 24 h at 37°C, for members of the Enterobacteriaceae family; on kanamycin esculin azide (KAA) agar, incubated aerobically at 37°C for 24 h, for enterococci; on Pseudomonas agar base (PAB) supplemented with 10 mg/ml cetrimide fucidin, incubated aerobically at 20°C for 48 h, for pseudomonads; on Baird-Parker agar (BP) with added rabbit plasma fibrinogen (RPF) supplement, incubated aerobically at 37°C for 48 h, for coagulase-negative streptococci (CNS) and, when a clear halo surrounded the colonies, for coagulase-positive streptococci (CPS); on de Man-Rogosa-Sharpe (MRS) agar, acidified at pH 5.4 with lactic acid (5 M), incubated anaerobically for 48 h at 30 and 44°C, for mesophilic and thermophilic rod LAB, respectively; on M17 agar, incubated anaerobically for 48 h at 30 and 44°C, for mesophilic and thermophilic coccus LAB, respectively; and on dichloran rose Bengal chloramphenicol (DRBC) agar, incubated aerobically at 25°C for 48 h, for yeasts. Clostridial content was estimated by the most-probable-number (MPN) technique using a 3 × 3 scheme following the methodology reported by Franciosi et al. (17): undiluted samples and decimal dilutions were pasteurized at 85°C for 15 min and inoculated into reinforced clostridium medium (RCM) supplemented with 1.4% (vol/vol) Na-lactate (Merck, Darmstadt, Germany); after that, test tubes were sealed with paraffin-vaseline (1:6) and incubated for 7 days at 37°C. All media and supplements were purchased from Oxoid. Microbiological counts were carried out in triplicate for all samples at each collection time.
DNA extraction, amplicon library preparation, and pyrosequencing.
Total DNA extraction from the 11 rennet pools was carried out using the NucleoSpin Food kit (Macherey-Nagel GmbH & Co. KG, Düren, Germany) according to the manufacturer's instructions. The microbial diversity was studied by pyrosequencing of the amplified V1-to-V3 region of the 16S rRNA gene by using primers Gray28f (5′-TTTGATCNTGGCTCAG) and Gray519r (5′-GTNTTACNGCGGCKGCTG) amplifying a fragment of 520 bp (18). Roche 454 sequencing adaptors were included in the forward primer followed by a 10-bp sample-specific multiplex identifier (MID). Each PCR mixture (final volume, 50 μl) contained 50 ng of template DNA, 0.4 μM each primer, 0.50 mM each deoxynucleoside triphosphate, 2.5 mM MgCl2, 5 μl of 10× PCR buffer, and 2.5 U of native Taq polymerase (Invitrogen, Milan, Italy). The following PCR conditions were used: 94°C for 2 min, 35 cycles of 95°C for 20 s, 56°C for 45 s, and 72°C for 5 min, and a final extension at 72°C for 7 min. Duplicate amplicons were produced and pooled before library preparation. After agarose gel electrophoresis, PCR products were purified twice with the Agencourt AMPure kit (Beckman Coulter, Milan, Italy) and quantified using QuantiFluor (Promega, Milan, Italy), and an equimolar pool was obtained prior to further processing. The amplicon pool was used for pyrosequencing on a GS Junior platform (454 Life Sciences, Roche Diagnostics, Italy) according to the manufacturer's instructions by using titanium chemistry.
Bioinformatics and data analysis.
Raw reads were first filtered according to the 454 processing pipeline. Sequences were then analyzed and further filtered by using QIIME 1.6.0 software (19). In order to guarantee a higher level of accuracy in terms of operational taxonomic unit (OTU) detection, after the split library script performed by QIIME, the reads were excluded from the analysis if they had an average quality score lower than 25, if they were shorter than 300 bp, and if there were ambiguous base calls. Sequences that passed the quality filter were denoised (20), and singletons were excluded. OTUs defined by a similarity of 97% were picked using the uclust method (21), and the representative sequences were submitted to the RDPII classifier (22) to obtain the taxonomy assignment and the relative abundance of each OTU using the Greengenes 16S rRNA gene database (23).
Alpha diversity was evaluated through QIIME to generate rarefaction curves and Good's coverage, Chao1 richness (24), and Shannon diversity (25) indices. The OTU taxonomy table generated by QIIME was used to produce a heatmap by using the software TMeV v 4.8 (26).
Milk clotting activity.
Clotting time (r), curd firming time (k20), and curd firmness measured 30 min after the addition of rennet (a30) were measured by the Formagraph system (Maspress apparatus; Foss Italia, Padua, Italy) according to the manufacturer's instructions. The rennet samples were diluted 1:10 (wt/vol) in distilled H2O (dH2O) and kept under agitation with a magnetic stirrer for 5 min, and 200 μl of dilution was added to 10 ml of pasteurized cows' milk heated at 35°C. Each rennet sample was analyzed five times with two distinct Formagraph appliances, for a total of 10 repetitions. For comparison, the commercial liquid calf rennet Naturen (Chr. Hansen's Laboratory, Parma, Italy), containing chymosin (≤30%, wt/vol) and having a strength of 130 international milk clotting units (IMCU)/ml, was diluted 1:10 (vol/vol) in dH2O, and 200 μl was added to 10 ml of pasteurized cows' milk heated at 35°C.
Acidifying capacity of rennet pastes.
The contribution of the rennet pastes to the acidification of curds was evaluated in full-fat ultrahigh-temperature-processed (UHT) milk. The 11 rennet samples (0.15 g) were inoculated individually in 300 ml of milk and incubated at 30°C for 6 days. Values of pH (determined electrometrically using the pHmeter BASIC 20+ [Crison Instrument S.A., Barcelona, Spain]) and total titratable acidity (TTA; determined by titration with 0.1 N NaOH and expressed in terms of ml of NaOH) were measured on 10 ml of each sample (aseptically collected) immediately after inoculation and at 24-h intervals for the 6 days. Microbiological analyses (TMC, TPC, and mesophilic and thermophilic rod and coccus LAB) were carried out on 10 ml of each sample immediately after inoculation and after 6 days. A control test was obtained with the addition of 0.15 ml of filter-sterilized (0.20-μm-pore-size filter; Sartorius, AubagneCedex, France) liquid calf rennet (Clerici Sacco International, Cadorago, Italy) in milk. All trials were carried out in duplicate in two consecutive weeks.
Isolation and phenotypical screening of rennet LAB.
The presumptive LAB grown on MRS and M17, after 48 h of incubation at 30 and 44°C, were collected from the plates containing the colonies well separated from one another and resulting from the analysis of all 11 milks acidified for 6 days with the rennet pastes. The colonies were selected on the basis of their appearance, and at least 4 colonies per morphology were isolated. The isolates were tested for the Gram reaction (Gregersen KOH method) and catalase activity (assayed with H2O2, 5% [wt/vol]) and purified after consecutive subculturing on the same media and under the same growth conditions as those used for plate counts.
Phenotypic characterization was carried out in order to obtain an initial grouping of the isolates. The cell morphology of LAB isolates was determined by an optical microscope. Subsequently, LAB were subjected to further phenotypic assays. Rods and cocci were grouped on the basis of cell disposition, growth at 15 and 45°C, hydrolysis of arginine and esculin, acid production from arabinose, ribose, xylose, fructose, galactose, lactose, sucrose, and glycerol, and CO2 production from glucose tested with Durham's tubes. Cocci were also grouped for their growth at pH 9.2 and in the presence of 6.5 g/liter NaCl to separate enterococci from other dairy cocci.
Strain differentiation and identification.
The isolates were genetically processed to be differentiated at the strain level. The DNA from overnight cultures grown in the optimal conditions was extracted by the Instagene Matrix kit (Bio-Rad, Hercules, CA) as described by the manufacturer. Crude cell extracts were used as the template DNAs for PCR. Strain differentiation was performed by random amplification of polymorphic DNA-PCR (RAPD-PCR) following the scheme reported by Settanni et al. (27) using the primers M13, AB111, and AB106. Amplifications were performed by means of the T1 Thermocycler (Biometra, Göttingen, Germany), and the PCR products were separated by electrophoresis on 1.5% (wt/vol) agarose gel (Gibco BRL, Cergy Pontoise, France) and visualized by UV transillumination after staining with the SYBR safe DNA gel stain (Molecular Probes, Eugene, OR). GeneRuler 100 bp Plus DNA ladder (M-Medical Srl, Milan, Italy) was used as a molecular size marker. RAPD-PCR profiles were analyzed with the pattern analysis software package Gelcompare II Version 6.5 (Applied Maths, Sin-Martens-Latem, Belgium). Calculation of similarities of band profiles was based on the Pearson product moment correlation coefficient. Dendrograms were obtained by means of the unweighted pair group method using an arithmetic average clustering algorithm.
The genotypic identification of the different strains of LAB was carried out by 16S rRNA gene sequencing. PCRs were performed as described by Weisburg et al. (28). DNA fragments were visualized, and amplicons of about 1,600 bp were purified by the QIAquick purification kit (Qiagen S.p.a., Milan, Italy) and sequenced using the same primers as those employed for PCR amplification. DNA sequencing reactions were performed by PRIMM (Milan, Italy). The sequences were compared with those available in the GenBank/EMBL/DDBJ (http://www.ncbi.nlm.nih.gov) (29) and EzTaxon-e (http://eztaxon-e.ezbiocloud.net/) (30) databases. The last database compares a given sequence to those of type strains only. The isolates were considered to represent the species in question if 97% or higher similarity was detected. The multiplex PCR assay based on the sodA gene reported by Jackson et al. (31) was applied to the enterococci not identified to the species level.
Technological screening.
All LAB were characterized for their capacity to acidify milk, to undergo autolysis, and to generate diacetyl. Overnight cultures were centrifuged at 5,000 × g for 5 min, washed twice, and resuspended in Ringer's solution to an optical density of ca. 1.0 at 600 nm (OD600), which approximately corresponds to a concentration of 109 CFU/ml (32), to standardize bacterial inocula. The acidifying capacity was tested in 100 ml UHT milk inoculated with 1% (vol/vol) of cell suspension and incubated at 30 or 44°C for the mesophilic and thermophilic isolates, respectively. Values of pH were measured out at 2-h intervals for the first 8 h and then 24, 48, and 72 h after inoculation on aliquots of 4 ml, aseptically collected from each test flask.
The autolysis of whole cells was determined in KH2PO4 (50 mM, pH 6.5), applying the method described by Mora et al. (33). OD600 was measured at 2-h intervals for the first 8 h and then 24, 48, and 72 h after inoculation.
Diacetyl production was determined as described by King (34). Briefly, LAB suspensions prepared in Ringer's solution as reported above were inoculated in UHT milk for 24 h at 30°C, and to aliquots of 1 ml was added 0.5 ml of α-naphthol (1%, wt/vol) and KOH (16%, wt/vol), followed by incubation at 30°C for 10 min. Diacetyl generation was indicated by the formation of a red ring at the top of the tubes.
Acidification, autolysis, and diacetyl generation tests were performed in duplicate with two independent LAB inocula.
Improvement of acidifying properties of rennets by LAB addition.
To evaluate the aptitudes of rennet LAB to turn rennets into good acidifiers, the fastest acidifier LAB were added to the rennet samples showing the slowest kinetics of acidification. LAB were selected on the basis of their milk acidification capacity, prepared in Ringer's solution as reported above, and added to the rennet pastes at a final concentration of approximately 104 CFU/g. The acidifying capacity of rennet pastes was determined as reported above, and the pH values were registered after 8 and 24 h from inoculation. The tests were carried out in duplicate.
Statistical analyses.
Microbial loads, milk clotting activities, changes of pH and TTA, and microbial evolutions during acidification were statistically analyzed using the generalized linear model (GLM) procedure, including the effects of sample, with the program SAS 2008, version 9.2 (Statistical Analysis System Institute Inc., Cary, NC, USA). The Student t test was used for mean comparisons. The post hoc Tukey method was applied for pairwise comparison between each rennet paste and control. Significance level was set at P values of <0.05.
Nucleotide sequence accession numbers.
The sequences determined in this study have been deposited in GenBank under accession numbers KF286609 to KF286618, KF826008 to KF826010, KF826012 to KF826022, KF826025 to KF826027, and KF856609 to KF856613. The pyrosequencing results are available at NCBI's Sequence Read Archive (accession number SRP026104).
RESULTS
Microbiological analysis of the rennet pastes.
The viable counts of the 12 microbial groups investigated in the rennet samples are reported in Table 2. CPS were undetectable in all samples, while CNS were found in samples 3 and 12. Enterobacteriaceae, enterococci, yeasts, and mesophilic and thermophilic LAB were cultivable only in a few samples. Pseudomonads were estimated at between 3.0 and 3.4 log CFU/g in six samples. All samples resulted positive for the presence of mesophilic cocci, whose counts were in the range of 3.5 to 5.1 log CFU/g. The levels of mesophilic cocci were almost superimposable to those of TMC. On average, TPC were at 1 to 1.5 log CFU/g lower than TMC. Clostridia, although at very low levels in some cases, were found in six rennets. The highest levels of viable populations were registered for rennet 12.
TABLE 2.
Microbial loads of rennet samples
| Microbial group | Microbial loada (log CFU/g) in rennet sample: |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | |
| TPC | 2.6 ± 0.1 Aa | 3.2 ± 0.8 A Bb | 3.0 ± 0.4 Ab | 3.6 ± 0.3 Bc | 3.7 ± 0.4 Bc | 3.0 ± 0.1 Ab | 3.0 ± 0.2 Ab | 3.0 ± 0.1 Ab | 3.3 ± 0.1 Bc | 2.9 ± 0.2 Ab | 3.5 ± 0.4 Bc |
| TMC | 4.1 ± 3.7 Aa | 4.1 ± 0.5 Aa | 3.9 ± 0.1 Aa | 4.6 ± 0.7 Ab | 4.5 ± 0.1 Ab | 4.6 ± 0.5 Ab | 3.9 ± 0.1 Aa | 4.2 ± 0.2 Aa | 4.6 ± 0.6 Ab | 4.1 ± 0.3 Aa | 5.3 ± 0.6 Bc |
| Enterobacteriaceae | <1 Aa | <1 Aa | <1 Aa | <1 Aa | <1 Aa | <1 Aa | <1 Aa | 3.9 ± 0.3 Bb | <1 Aa | 3.4 ± 0.2 Bb | <1 Aa |
| Enterococci | <2 Aa | <2 Aa | 3.6 ± 0.2 Bb | 3.4 ± 0.6 Bb | <2 Aa | <2 Aa | <2 Aa | <2 Aa | <2 Aa | <2 Aa | <2 Aa |
| Pseudomonads | <2 Aa | 3.4 ± 0.1 Bb | <2 Aa | 3.4 ± 0.5 Bb | <2 Aa | 3.4 ± 0.7 Bb | <2 Aa | <2 Aa | 3.0 ± 0.4 Bb | 3.1 ± 0.3 Bb | 3.4 ± 0.8 Bb |
| CNS | <2 Aa | <2 Aa | 3.1 ± 0.2Cc | <2 Aa | <2 Aa | <2 Aa | <2 Aa | <2 Aa | <2 Aa | <2 Aa | 2.2 ± 0.3 Bb |
| CPS | <2 Aa | <2 Aa | <2 Aa | <2 Aa | <2 Aa | <2 Aa | <2 Aa | <2 Aa | <2 Aa | <2 Aa | <2 Aa |
| Mesophilic rod LAB | <1 Aa | <1 Aa | <1 Aa | 2.7 ± 0.7 Bb | <1 Aa | <1 Aa | <1 Aa | <1 Aa | <1 Aa | <1 Aa | 4.1 ± 0.1Cc |
| Thermophilic rod LAB | <1 Aa | <1 Aa | <1 Aa | <1 Aa | <1 Aa | <1 Aa | <1 Aa | <1 Aa | <1 Aa | <1 Aa | 3.0 ± 0.1 Bb |
| Mesophilic coccus LAB | 3.6 ± 0.1 Aa | 3.5 ± 0.3 Aa | 3.8 ± 0.2 Aa | 4.0 ± 0.3 Aa | 4.2 ± 0.2 Ab | 4.3 ± 0.1 Ab | 3.7 ± 0.8 Aa | 3.7 ± 0.2 Aa | 4.0 ± 0.1 Aa | 4.1 ± 0.1 Ab | 5.1 ± 0.6 Ac |
| Thermophilic coccus LAB | <1 Aa | <1 Aa | 3.1 ± 0.1 Bb | <1 Aa | 4.4 ± 0.3 Bc | <1 Aa | <1 Aa | <1 Aa | <1 Aa | <1 Aa | 3.7 ± 0.4 Bb |
| Yeasts | <2 Aa | <2 Aa | <2 Aa | <2 Aa | <2 Aa | <2 Aa | <2 Aa | <2 Aa | <2 Aa | <2 Aa | 4.0 ± 0.1 Bb |
| Clostridiab | 1.4 | 0.3 | 2.6 | 0 | 2.6 | 0.4 | 0 | 0 | 0 | 0 | 0.1 |
The results represent mean values of six plate counts (carried out in triplicate for two independent sample collections within each location). Lowercase (a, b) and uppercase (A, B) letters indicate different statistical significances for pairwise comparison with control at P values of <0.05 and <0.01, respectively.
As estimated by most probable number (MPN).
Microbial diversity.
A total of 46,266 raw reads were obtained for the rennet samples and analyzed; 40,597 reads passed the quality filters applied through the QIIME split_library.py script, with an average value of 2,963 reads/sample and an average length of 519 bp. The number of reads analyzed, the number of OTUs, the Good's estimated sample coverage (ESC), and the Chao1 and Shannon indices obtained for the samples are reported in Table 3. The rarefaction analysis and the ESC indicated that more than 90% of the microbial diversity was covered in most of the samples.
TABLE 3.
Number of reads, observed diversity, and estimated sample coverage for 16S rRNA amplicons analyzed in this studya
| Rennet sample no. | No. of reads | No. of OTUs | Chao1 index | Shannon index | ESC (%) |
|---|---|---|---|---|---|
| 1 | 3,308 | 155 | 464.79 | 2.79 | 97 |
| 2 | 3,530 | 249 | 586.53 | 3.27 | 96 |
| 3 | 3,119 | 312 | 803.05 | 3.73 | 93 |
| 4 | 3,150 | 496 | 1491.51 | 5.19 | 89 |
| 5 | 4,366 | 411 | 979.47 | 3.89 | 94 |
| 6 | 2,272 | 588 | 1767.95 | 6.21 | 81 |
| 7 | 3,391 | 481 | 1702.50 | 4.80 | 90 |
| 8 | 3,298 | 457 | 1465.06 | 4.20 | 90 |
| 9 | 1,118 | 259 | 630.34 | 4.70 | 84 |
| 10 | 3,056 | 376 | 1110.27 | 4.02 | 91 |
| 11 | 1,989 | 121 | 382.07 | 2.63 | 96 |
Abbreviations: OTU, operational taxonomic unit; ESC, estimated sample coverage. Chao1, Shannon, and ESC were calculated with QIIME at the 3% distance level. More details on bioinformatics analysis are given in Materials and Methods.
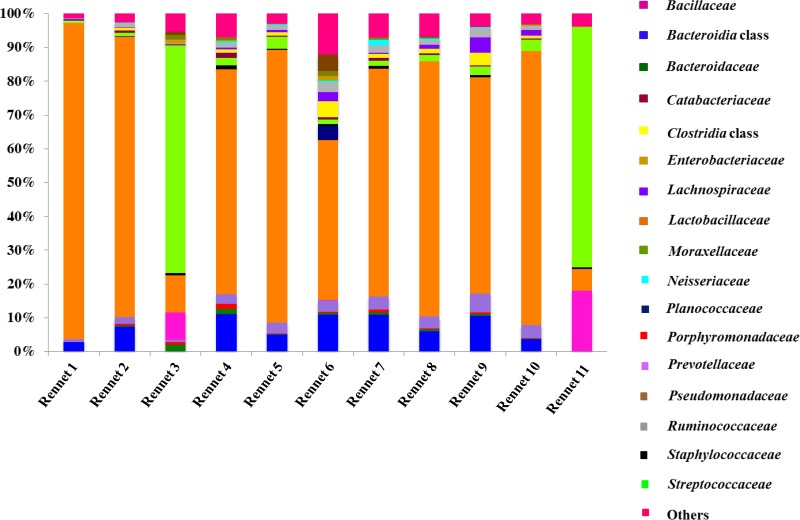
Pyrosequencing allowed to identify a total of 121 OTUs at the family level. The relative abundances of bacterial families identified in animal rennet pastes are reported in Fig. 2, where only groups with an incidence of 1% in at least one sample were considered. Seventeen bacterial families were detected in the rennet samples considered. The major families were Lactobacillaceae and Streptococcaceae. Lactobacillaceae represented the highest proportion (ranging from 47.4 to 93.5%) of bacteria in almost all samples, with the exception of rennets 3 and 12, where Streptococcaceae represented 67.5 and 71.1% of the bacterial microbiota, respectively. The other families or classes (when the consensus on the particular nomenclature was lacking) detected at a certain percentage of abundance (>2% in several rennet samples) were Bacteroidia, Clostridia, Prevotellaceae, Lachnospiraceae, and Ruminococcaceae (Fig. 2).
FIG 2.
Relative abundances (%) of bacterial families identified by pyrosequencing in animal rennet pastes. Only families occurring at >1% abundance in at least one sample were included.
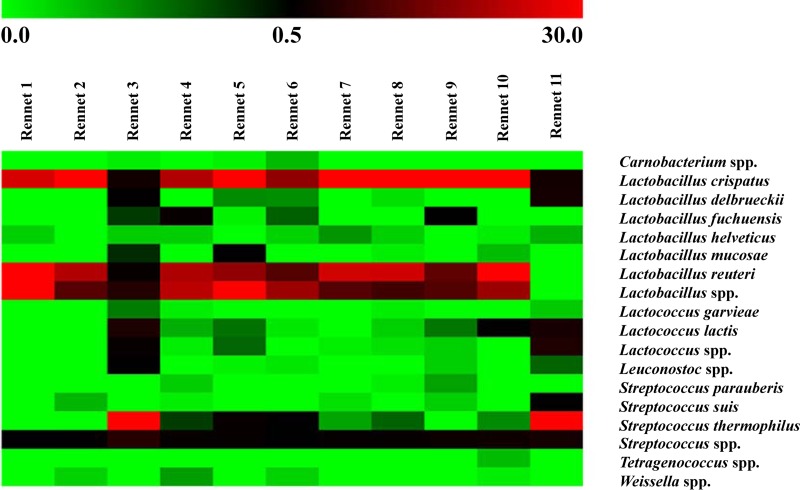
Analyzing the microbial diversity to deeper taxonomic assignment, genera and species identified by pyrosequencing can be observed in the heatmap shown in Fig. 3. Only OTUs belonging to the LAB group and occurring at >0.1% abundance in at least one sample were included. Eighteen OTUs were identified. Among LAB, Streptococcus thermophilus and some lactobacilli were the most abundant species, with differences among the samples. S. thermophilus was the most abundant species in rennets 3 and 12 only (56.4 and 59.8%, respectively). In contrast, all other samples were characterized by the prevalence of Lactobacillus crispatus and Lactobacillus reuteri, with abundances ranging within 20 to 30% of the total OTUs. Other LAB detected were Lactobacillus delbrueckii (3.2%) in rennet 12 and Lactococcus lactis (4.0%) in rennet 3. In addition, further Lactobacillus spp. were identified in most of the samples, but a species level assignment was not obtained (Fig. 3).
FIG 3.
Distribution of bacterial genera and species as identified by pyrosequencing in animal rennet pastes. Only OTUs belonging to the lactic acid bacteria group and occurring at >0.1% abundance in at least one sample were included. The color scale indicates the relative abundance of each OTU within the samples.
Rennet coagulation.
The animal rennet pastes were evaluated for their coagulation parameters (Table 4) in comparison with the liquid rennet Naturen, commonly used as an internal control for the Formagraph instrument (35). The rennet samples showed different r, k20, and a30 values. However, some artisanal rennets showed Formagraph parameters comparable to those characterizing the Naturen preparation.
TABLE 4.
Milk clotting activities of animal rennet pastes
| Rennet sample ID | Valuea (mean ± SEM) for Formagraph parameter: |
||
|---|---|---|---|
| r (min) | k20 (min) | a30 (mm) | |
| 1 | 6.13 ± 0.53 Bb | 3.15 ± 0.16 Aa | 49.11 ± 2.30 Aa |
| 2 | 9.15 ± 0.74 Bb | 3.59 ± 0.37 Bb | 46.55 ± 1.66 Aa |
| 3 | 4.83 ± 0.26 Bb | 3.22 ± 0.30 Aa | 48.97 ± 2.38 Aa |
| 4 | 6.36 ± 0.18 Bb | 3.26 ± 0.12 Aa | 47.91 ± 2.44 Aa |
| 5 | 3.33 ± 0.17 Bb | 2.50 ± 0.28 Bb | 50.13 ± 2.55 Ba |
| 6 | 4.03 ± 0.34 Aa | 2.57 ± 0.33 Bb | 50.03 ± 2.21 Ba |
| 7 | 12.60 ± 0.65 Bb | 3.96 ± 0.38 Bb | 41.27 ± 2.28 Bb |
| 8 | 3.08 ± 0.29 Bb | 2.30 ± 0.12 Bb | 51.46 ± 2.04 Bb |
| 9 | 8.33 ± 0.53 Bb | 3.35 ± 0.17 Bb | 47.99 ± 2.06 Aa |
| 10 | 4.25 ± 0.31 Aa | 2.57 ± 0.37 Bb | 50.96 ± 2.43 Bb |
| 11 | 5.48 ± 0.14 Bb | 3.05 ± 0.25 Aa | 49.59 ± 1.72 Bb |
| Naturen | 4.15 ± 0.57 Aa | 3.01 ± 0.57 Aa | 47.02 ± 2.75 Aa |
Lowercase (a, b) and uppercase (A, B) letters indicate different statistical significances for pairwise comparison with Naturen at P values of <0.05 and <0.01, respectively.
Acidifying capacity of rennet pastes.
The capacity of the rennet pastes to acidify UHT milk was evaluated by determining the changes of pH and TTA (see Table S1 in the supplemental material). During the 6 days of observation, the pH of the control trial, containing the filter-sterilized liquid rennet, decreased slower (from 6.70 at time zero [T0] to 6.29 after 6 days) than that of the trials containing added rennet pastes. Consistently, increasing TTA was observed in all trials. The pH of the trials T1 to T12 decreased more rapidly than that of the control trial; after 2 days from inoculation, all trials except trial T11 showed a pH of milk below 5.0. However, only trials T3 and T12 showed pH values below 4.0 and the highest TTA levels (19.3 and 21.0, respectively) at the sixth day of fermentation. In several trials, the pH registered on the second day did not significantly vary until the end of the observation. At any time of pH and TTA evaluation, all trials had statistically different results from one another. Furthermore, all trials differed significantly from the control.
The microbiological analysis of milk (see Table S2 in the supplemental material) showed the increase of all microbial groups monitored (TMC, TPC, and mesophilic and thermophilic rod and coccus LAB) at the sixth day of fermentation, even though most of them were undetectable at T0. The highest levels of microbial loads were registered on M17 incubated both at 30°C and at 44°C. Despite the high values observed for TPC and TMC, trial T11 displayed a low final concentration of LAB (5.1 log CFU/ml). Trials T7 and T8 reached a final LAB load of approximately 107 CFU/ml, while 108 CFU/ml of LAB were found for all other trials. With the exception of thermophilic coccus LAB for trial T8 and psychrotrophic bacteria for trials T1 to T3, T7 to T9, and T12, all differences from the control trial were found to be statistically significant.
Isolation and phenotypic grouping of rennet LAB.
Eighty-nine presumptive LAB colonies were isolated from the milks acidified with the animal rennet pastes, which were subjected to the high-throughput sequencing before addition to milk. Only 29 cocci and 3 rods were Gram positive and catalase negative and considered presumptive LAB. The combination of the phenotypic characters considered (growth temperature, hydrolysis of arginine and esculin, acid production from different carbon sources, and fermentation metabolism) allowed the separation of the 32 cultures into seven groups (see Table S3 in the supplemental material). The most numerous group was group IV, which included 14 isolates. No isolate produced showed a heterofermentative metabolism. All three rod-shaped isolates grouped together (group I) and were obligately homofermentative since the growth was scored negative in the presence of pentose sugars. The isolates forming long chains of cocci formed group II.
Strain typing and identification of LAB.
All presumptive LAB were analyzed by RAPD-PCR. The combination of the three RAPD patterns for each isolate indicated the presence of 32 different strains in the resulting dendrogram (see Fig. S1 in the supplemental material). At the similarity level of 50%, four main clusters were observed. The clusters A and C, with eight and five strains, respectively, included LAB characterized by the phenotypic profiles IV to VII, and cluster D contained only the strains included in the phenotypic group IV, while cluster E contained those included in the phenotypic groups III and IV.
The 32 LAB were subsequently subjected to 16S rRNA gene sequencing. The sequences were compared with those available in two distinct databases, and 18 strains were clearly identified to the species level, i.e., Enterococcus casseliflavus, Enterococcus faecium, Enterococcus faecalis, Enterococcus lactis, Lactobacillus delbrueckii, and Streptococcus thermophilus, since sequence similarity was higher than 97% in both databases (Table 5). Thirteen strains were allotted into the genus Enterococcus, but none of them shared a 16S rRNA gene sequence identity of at least 97% with any type strain of the known species within this genus. The multiplex PCR based on the sodA gene of these strains (results not shown) did not recognize any Enterococcus species. The type strain of the closest relative for all the unidentified strains was E. lactis DSM 23655T, with which they shared different percentages (92.12 to 96.94%) of identity on the 16S rRNA gene (GenBank accession no. GU983697). A phylogram (see Fig. S2 in the supplemental material) was constructed with these sequences and all those corresponding to the E. lactis identified in this work, in order to evaluate their phylogenetic distances. All 13 unidentified Enterococcus strains formed distinct branches of the phylogenetic tree. In particular, the strains CGLBL221 and CGLBL225 were phylogenetically far distant from the other 11 unidentified enterococci. The distances evidenced by phylogenetic analysis reflected the differences showed by the RAPD dendrogram.
TABLE 5.
Identification of the fastest acidifier rennet LAB
| Strain | Species | Rennet sample no. | Phenotypic group | RAPD cluster | % similarity (accession no. of closest relative) by: |
Accession no. | Sequence length (bp) | |
|---|---|---|---|---|---|---|---|---|
| BLAST | EzTaxon | |||||||
| CGLBL3 | Streptococcus thermophilus | 3 | II | K | 99 (KC545904) | 98.16 (AY188354) | KF856613 | 1,490 |
| CGLBL58 | Enterococcus casseliflavus | 11 | IV | F | 99 (KF060255) | 98.45 (AJAM01000006) | KF826012 | 1,492 |
| CGLBL73 | Lactobacillus delbrueckii | 11 | I | L | 98 (KF060256) | 97.77 (AEXU01000148) | KF856612 | 1,486 |
| CGLBL85 | Enterococcus faecalis | 3 | VII | G | 99 (HF558530) | 99.26 (AE016830) | KF826013 | 1,497 |
| CGLBL100 | Enterococcus sp. | 1 | III | E | 97 (AY675247) | 96.79 (GU983697) | KF286610 | 1,494 |
| CGLBL106 | Enterococcus sp. | 2 | VII | A | 96 (KC478514) | 96.38(GU983697) | KF826009 | 1,498 |
| CGLBL109 | Enterococcus sp. | 2 | VII | A | 96 (HM058854) | 95.82 (DQ411813) | KF826010 | 1,436 |
| CGLBL115 | E. faecalis | 3 | IV | H | 99 (KC692178) | 98.79 (AE016830) | KF826008 | 1,503 |
| CGLBL118 | Enterococcus sp. | 4 | VI | A | 96 (KC478513) | 96.73 (DQ411813) | KF856609 | 1,444 |
| CGLBL137 | Enterococcus sp. | 8 | VI | A | 96 (AY675247) | 95.69 (GU983697) | KF826026 | 1,502 |
| CGLBL139 | Enterococcus faecium | 9 | IV | A | 98 (KC478514) | 98.13 (DQ411813) | KF826025 | 1,416 |
| CGLBL140 | Enterococcus lactis | 9 | V | A | 97 (AY683836) | 97.27 (GU983697) | KF826027 | 1,485 |
| CGLBL145 | E. lactis | 11 | V | B | 99 (AY683836) | 99.51 (GU983697) | KF826014 | 1,498 |
| CGLBL146 | E. faecalis | 11 | IV | H | 99 (KC692178) | 98.99 (AE016830) | KF826015 | 1,500 |
| CGLBL153 | E. lactis | 1 | IV | D | 97 (AY683836) | 97.49 (GU983697) | KF826016 | 1,494 |
| CGLBL155 | Enterococcus sp. | 2 | IV | D | 96 (KC478514) | 96.10 (GU983697) | KF826017 | 1,505 |
| CGLBL159 | E. lactis | 3 | IV | D | 97 (AY683836) | 97.56 (GU983697) | KF826018 | 1,503 |
| CGLBL160 | E. lactis | 3 | IV | A | 98 (AY683836) | 97.43 (GU983697) | KF826019 | 1,494 |
| CGLBL186 | Enterococcus sp. | 2 | IV | C | 97 (AY675247) | 96.66 (GU983697) | KF286611 | 1,511 |
| CGLBL188 | E. faecalis | 11 | IV | E | 99 (HQ721272) | 98.72 (AB012212) | KF286612 | 1,494 |
| CGLBL189 | S. thermophilus | 11 | II | K | 99 (KC545895) | 98.11 (AY188354) | KF856611 | 1,487 |
| CGLBL193 | L. delbrueckii subsp. delbrueckii | 3 | I | I | 99(KF060256) | 99.81 (AY050172) | KF286613 | 1,060 |
| CGLBL198 | L. delbrueckii | 6 | I | B | 99 (KC545930) | 97.77 (AY050172) | KF826020 | 1,517 |
| CGLBL203a | Enterococcus sp. | 5 | IV | C | 99 (AB795648) | 98.65 (GU983697) | KF286614 | 1,053 |
| CGLBL204 | Enterococcus sp. | 7 | V | C | 92 (EU337116) | 92.12 (GU983697) | KF286615 | 1,497 |
| CGLBL208 | S. thermophilus | 11 | II | M | 99 (FR875178) | 99.05 (AY188354) | KF286609 | 1,482 |
| CGLBL213 | Enterococcus sp. | 4 | VI | C | 97 (AY675247) | 96.73 (GU983697) | KF286616 | 1,499 |
| CGLBL221 | Enterococcus sp. | 6 | VII | C | 95 (KC478514) | 95.20 (GU983697) | KF286617 | 1,495 |
| CGLBL223 | Enterococcus sp. | 9 | IV | A | 97 (KC478514) | 96.94 (GU983697) | KF826021 | 1,496 |
| CGLBL225 | Enterococcus sp. | 9 | V | J | 95 (AY675247) | 94.57 (GU983697) | KF286618 | 1,496 |
| CGLBL253 | E. lactis | 10 | IV | G | 99 (AY683836) | 99.58 (GU983697) | KF826022 | 1,500 |
| CGLBL274 | E. faecium | 10 | IV | E | 99 (FJ378689) | 99.46 (DQ411813) | KF856610 | 947 |
The strain CGLBL203 was identified as Enterococcus sp. due to the discordance found between BLAST and EzTaxon.
Technological characterization of LAB.
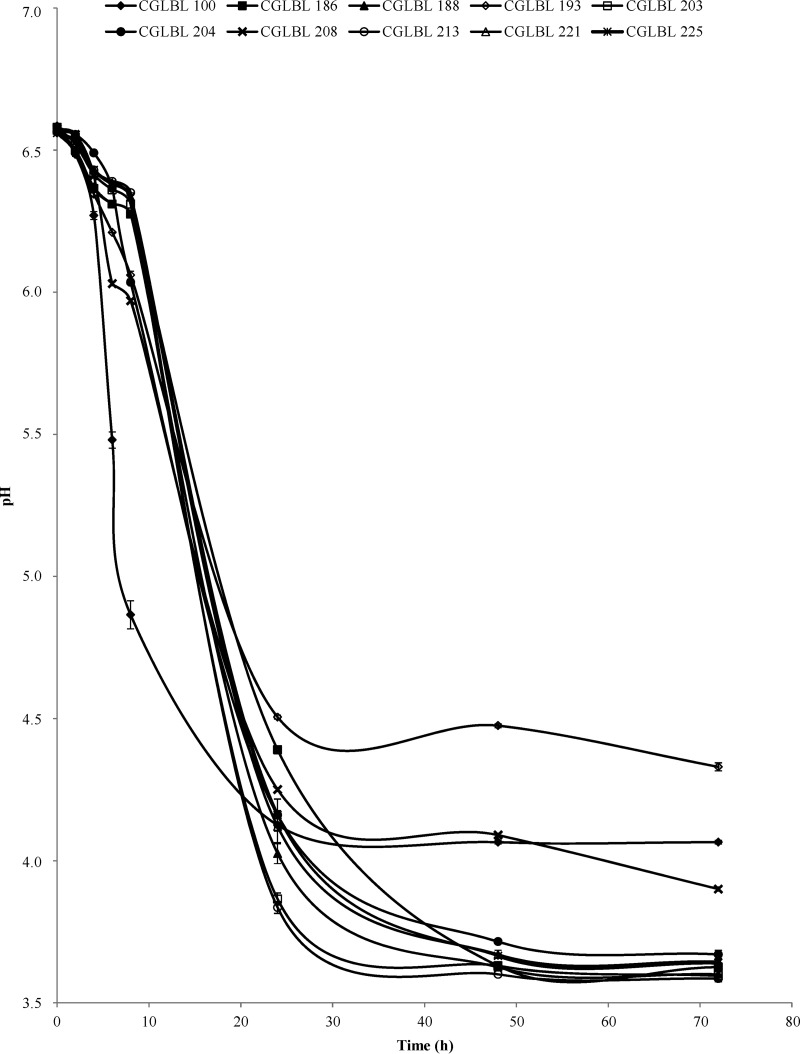
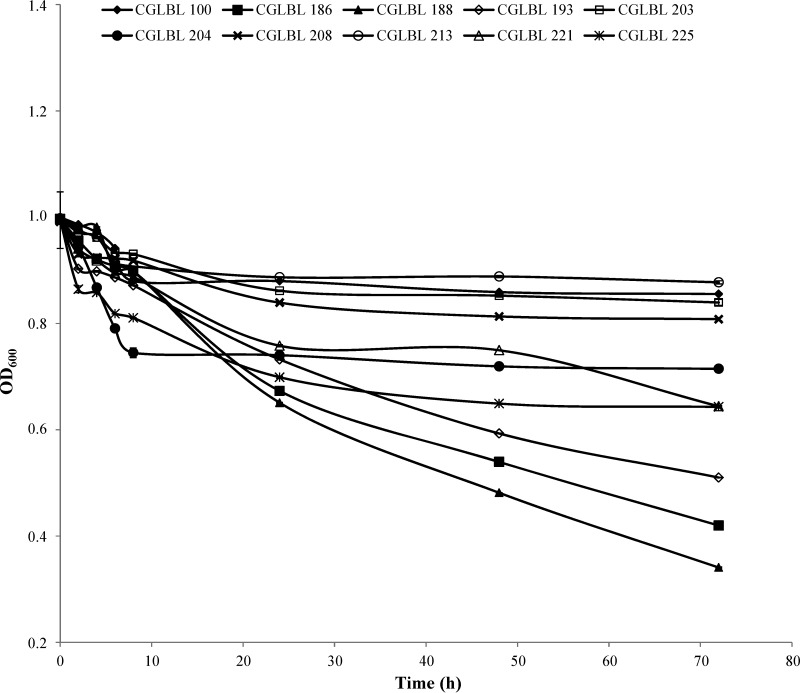
All LAB were subjected to technological screening. The results of the acidification evaluated in UTH milk at the optimal growth temperature of each LAB (results not shown) indicated that 10 strains effected a rapid pH drop (Fig. 4). In particular, Enterococcus sp. strain CGLBL100 determined a significantly (P < 0.05) more rapid drop of pH than other LAB within 8 h, even though after 24 h the lowest pHs were registered for Enterococcus species strains CGLBL203 and CGLBL213. Despite the high percentage of Lactobacillaceae detected by pyrosequencing (Fig. 2), no strain from the rennet samples 8 and 11 was selected as acidifier. In fact, rennets 8 and 11 determined the highest values of pH registered after 6 days of milk acidification (see Table S1 in the supplemental material). Even though Enterococcus sp. strain CGLBL100 was characterized by the fastest acidification kinetics within 8 h from inoculation in milk, it was isolated from a 6-day fermented milk that did not show a good acidification (see Table S1 in the supplemental material). On the contrary, the strains CGLBL203 and CGLBL213, which showed the lowest endpoint pH, were originating from the milk fermented with rennets 5 and 4, respectively, which were among the most acidifying rennet paste samples (see Table S1 in the supplemental material).
FIG 4.
Evolution of milk pH determined by the fastest acidifier rennet LAB. Bars represent standard deviations of the means. Tests were performed in duplicate. Vertical bars not visible are smaller than the symbol.
Autolysis of LAB showing acidifying aptitudes is reported in Fig. 5. Only Enterococcus sp. strain CGLBL186, E. faecalis CGLBL188, and L. delbrueckii subsp. delbrueckii CGLBL193 were characterized by a consistent decrease of OD within the 72 h of observation. All isolates were able to produce diacetyl (results not shown).
FIG 5.
Autolysis of the fastest acidifier rennet LAB. Bars represent standard deviations of the means. Tests were performed in duplicate. Vertical bars not visible are smaller than the symbol.
Improvement of acidifying properties of rennets by LAB addition.
Enterococcus species strains CGLBL100, CGLBL203, and CGLBL213 were chosen to evaluate the capacity of rennet LAB to turn rennet paste 11, characterized by the slowest kinetics of acidification, into a good acidifying rennet. The level of LAB inocula was at about 104 CFU/g in order to contaminate the rennet paste at the same initial concentration of mesophilic coccus LAB detected (Table 2). After 8 h from inoculation, the strain CGLBL100 showed a drop of milk pH to 5.75 ± 0.05, that of strain CGLBL203 to 5.74 ± 0.03, and that of strain CGLBL213 to 5.69 ± 0.03. A similar trend for the three strains was observed at 24 h from rennet addition, since milk inoculated with rennet 11 inoculated with strain CGLBL100 reached a final pH of 4.03 ± 0.04; a final pH of 4.02 ± 0.07 was observed with strain CGLBL203 and one of 3.98 ± 0.11 with strain CGLBL213. These results, compared to those reported in Fig. 4, demonstrated that LAB strains CGLBL100, CGLBL203, and CGLBL213 turned a slow-acidifying rennet into a good acidifier but showed that strain CGLBL100, after mixing with the indigenous LAB of rennet 11, had a different acidifying capacity from that registered when inoculated in milk singly.
DISCUSSION
The aim of this work was to evaluate the microbial communities of animal rennet pastes as a source of interesting LAB populations. To pursue this goal, we applied culture-dependent and -independent techniques. Although there is no universal strategy to investigate the microbial biodiversity of complex matrices, the combination of more methodologies might provide a global overview of the microbial composition. This approach is particularly important for the food ingredients that have not been deeply investigated yet, because several microorganisms with beneficial properties can be lost with the classical methods of detection, based on the cultivation and isolation of the viable cells, that are currently available.
The viable counts showed that all samples hosted mesophilic cocci and their concentrations were approximately at the same levels of TMC, while mesophilic and thermophilic rods and thermophilic cocci were cultivable in only a few samples. The viable counts of LAB estimated in this work are in the same order of magnitude of those reported by Voidarou et al. (8) but are consistently lower than those estimated by Flórez et al. (15). However, the last authors analyzed samples of rennet extracts prepared “from dried kid stomachs cut into strips and left to steep in acidified cheese whey for at least 24 h” (15), and this can explain the high levels of LAB found (about 108 CFU/ml), since cheese whey is a source of viable LAB (36).
CPS were undetectable in all samples, but CNS were found in the two industrial rennets and were in the range 2.2 to 3.1 log CFU/g. The presence of CNS at detectable levels only in the industrial rennets indicates that the microbial composition of the rennets might strictly depend on the method used for production. Similar levels of concentration were found for some rennets analyzed by Flórez et al. (15). The liquid rennet studied by Temelli et al. (16) hosted very low levels of staphylococci, and the rennet paste “pytia” contained barely 0.3 log CFU/g of Staphylococcus aureus (8).
Not all samples were positive for the presence of Enterobacteriaceae, enterococci, yeasts, pseudomonads, and clostridia, confirming previous studies carried out on liquid rennets that reported undetectable levels of this microbial groups in several samples (10, 14–16). Low concentrations of yeasts were registered also by Voidarou et al. (8).
The analysis of the uncultivable component of the microbiota of a given raw material assumes a paramount importance when the microorganisms relevant during the transformation are present in a dormant/viable but not cultivable (VBNC) state, not detectable by culture-dependent methods. The VBNC community may include both the microorganisms technologically useful for the transformation process and those undesired (pathogenic/spoilage) for the stability and safety of the final food products. In this study, all rennet pastes were analyzed by culture-independent pyrosequencing as an up-to-date, sensitive approach for the evaluation of microbial diversity (37). Pyrosequencing shows a higher sensitivity and efficiency for the evaluation of microbial biodiversity than other culture-independent PCR-based approaches such as electrophoretic methods (37, 38). The microbiota of the rennets analyzed in this study consisted of 361 OTUs. Seventeen main families represented 92.7 to 98.9% of the bacterial communities of the samples. Within the group of LAB, Lactobacillaceae represented the highest proportion of bacteria in all artisanal samples, followed by Streptococcaceae, which dominated the two industrial samples. Bacteroidia, Clostridia, Prevotellaceae, Lachnospiraceae, and Ruminococcaceae were also found to contaminate the samples. Prevotellaceae, Lachnospiraceae, and Ruminococcaceae are reported to be present at high abundances within the bovine abomasal microbiota (39). Although none of the rennet samples analyzed in this work had a bovine origin, the presence of the three above families at high abundance levels in lamb and kid rennets is due to their high adaptation to the ruminant abomasum environment.
In general, high-throughput sequencing evidenced a different microbial composition of industrial and artisanal animal rennets. This finding might be dependent on the methods applied to produce the rennet pastes, which need to be investigated in order to retrieve the origin of the different microbial groups.
Seventeen LAB OTUs were identified to the species level, with S. thermophilus being the main LAB species for the industrial rennets, while Lactobacillus crispatus and Lactobacillus reuteri were found at consistent levels in the artisanal samples. These data suggest a possible competition between streptococci and lactobacilli. Several LAB species, mainly Lactococcus lactis and lactobacilli, were also previously identified in rennet by a culture-independent PCR-denaturing gradient gel electrophoresis (DGGE) approach (40).
Voidarou et al. (8) stated that artisan traditional rennets are safe to use and technologically beneficial to dairy processes and may provide characteristic flavor in the case of Cabrales cheese. In order to test the suitability of rennet as source of starter LAB, we evaluated the acidifying capacity of the rennet pastes. Changes of milk pH and TTA during 6 days of observation showed that the industrial rennets determined a faster and stronger acidification than the artisanal rennets, which is not in agreement with the results previously obtained by Voidarou et al. (8). The microbiological analysis of the acidified milks showed final LAB loads of approximately 107 to 108 CFU/ml of LAB for almost all trials, showing that all rennets hosted LAB able to acidify milk. However, after 24 h, only three artisan rennets showed an acidifying ability similar to that expressed by the industrial samples.
In order to isolate the most promising strains to act as starter cultures, various presumptive LAB colonies were isolated from the highest dilutions of the milks acidified with the animal rennet pastes. The 89 presumptive LAB were phenotypically separated into seven groups. The 89 isolates were confirmed to be different strains by using genotyping, but only 32 strains belonged to the group of LAB and were identified as E. casseliflavus, E. faecium, E. faecalis, E. lactis, L. delbrueckii, and S. thermophilus, or they were identified as Enterococcus species that remained unidentified. The phylogenetic analysis revealed that these enterococci had E. lactis as the closest related species and could represent more unknown species.
Overall, the culture-independent analysis indicated high levels of S. thermophilus in the industrial rennets, while L. crispatus and L. reuteri were the major species in the artisanal rennets. These species of lactobacilli are typically not recognized as SLAB species (41). The species dominating in culture-independent analyses were not always isolated after milk acidification by the different rennets. Indeed, S. thermophilus was isolated at high levels from acidified milk only when the milk was acidified with rennet 12. This inconsistency may be due to either the VBNC state of the microbiota identified in the rennets by pyrosequencing or by the fact that other bacteria such as enterococci or L. delbrueckii may outcompete other lactobacilli or streptococci during the growth in milk. Furthermore, L. crispatus and L. reuteri belong to the normal bacterial microbiota of gastrointestinal and genital tracts of humans and animals (42) and might not be adapted to the rennet environment. Even though both Lactobacillus species have been reported to be associated with several fermented foods in past, their detection was often due to a phenotypic misidentification. Hence, after 6 days of milk fermentation, the absence of L. crispatus and L. reuteri from the dominating group of LAB could be imputable to competition with other, faster-acidifying species that are better adapted to milk.
Most of the dominant species present after milk fermentation were, surprisingly, of the Enterococcus genus and in some rennets were not detected by plate count and/or were revealed to be present at very low percentages by pyrosequencing. However, once in milk, these strains were able to develop rapidly and overcame the other species that were found to be more abundant in rennets.
The isolates were characterized for their technological features. Besides the acidifying capability, other general traits relevant for SLAB are the rapid cell autolysis and the capacity to generate diacetyl (43). Ten isolates determined a rapid drop of milk pH and produced diacetyl, but only three of them underwent a rapid autolysis. No isolate from the rennet samples 8 and 11, due to their low acidifying power observed in milk, was selected as technologically interesting. Other studies found that only a small percentage of acidifier LAB showed autolytic properties (32). The addition of fast-acidifying strains turned slow-acidifying rennets into good acidifiers.
From an ecological point of view, this study provided the microbial composition of several animal rennet pastes produced at different levels and investigated the technological role of their LAB.
In conclusion, although artisan and industrial animal rennet pastes showed a considerable microbial diversity, this work proved that animal rennet pastes are sources of LAB strains with technological traits useful in cheese making. Furthermore, animal rennets also represented sources of isolation of new species of Enterococcus, which need further studies to be better classified. However, the high abundances of LAB families found in rennets in this work, which have not been detected at high proportions in the ruminant abomasal microbiota (39), and the different microbial communities detected for industrial and artisanal rennets suggest the need for better investigating the process of rennet paste production.
Supplementary Material
ACKNOWLEDGMENT
This research was financially supported by the Italian Ministry of Health, project IZS SI 06/11 RC, “Prodotti lattiero caseari siciliani: tecniche di produzione e rischio microbiologico.”
Footnotes
Published ahead of print 17 January 2014
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.03837-13.
REFERENCES
- 1.Parente E, Cogan TM. 2004. Starter cultures: general aspects, p 123–148 In Fox PF, McSweeney PLH, Cogan TM, Guinee TP. (ed), Cheese: chemistry, physics and microbiology. Elsevier, London, England [Google Scholar]
- 2.Settanni L, Moschetti G. 2010. Non-starter lactic acid bacteria used to improve cheese quality and provide health benefits. Food Microbiol. 27:691–697. 10.1016/j.fm.2010.05.023 [DOI] [PubMed] [Google Scholar]
- 3.Beresford TP, Fitzsimons NA, Brennan NL, Cogan TM. 2001. Recent advances in cheese microbiology. Int. Dairy J. 11:259–274. 10.1016/S0958-6946(01)00056-5 [DOI] [Google Scholar]
- 4.Franciosi E, Settanni L, Carlin S, Cavazza A, Poznanski E. 2008. A factory-scale application of secondary adjunct cultures selected from lactic acid bacteria during “Puzzone di Moena” cheese ripening. J. Dairy Sci. 91:2981–2991. 10.3168/jds.2007-0764 [DOI] [PubMed] [Google Scholar]
- 5.Bhowmick BK, Saha ML, Khan MR. 2006. Microbial study of some milk with special reference to coli form bacteria. Int. J. Dairy Sci. 1:57–62. 10.3923/ijds.2006.57.62 [DOI] [Google Scholar]
- 6.Krumov K, Ivanov G, Slavchev A, Nenov N. 2010. Improving the processed cheese quality by the addition of natural spice extracts. Adv. J. Food Sci. Technol. 2:335–339 http://maxwellsci.com/print/ajfst/v2-335-339.pdf [Google Scholar]
- 7.Salvadori del Prato O. 1998. Trattato di tecnologia casearia. Edagricole, Bologna, Italy [Google Scholar]
- 8.Voidarou C, Tzora A, Malamou O, Akrida-Demertzi K, Demertzis PG, Vassos D, Rozos G, Alexopoulos A, Plessas S, Stavropoulou E, Skoufou M, Bezirtzoglou E, Riganakos G. 2011. Chemical and microbiological characterization of artisan inoculants used for the fermentation of traditional dairy products in Epirus area (Greece). Anaerobe 17:354–357. 10.1016/j.anaerobe.2011.07.004 [DOI] [PubMed] [Google Scholar]
- 9.Upadhyay VK, McSweeney PLH, Magboul AAA, Fox PF. 2004. Proteolysis in cheese during ripening, p 391–434 In Fox PF, McSweeney PLH, Cogan TM, Guinee TP. (ed), Cheese: chemistry, physics and microbiology. Elsevier, London, England [Google Scholar]
- 10.Hofi M, Azza Farahat M, Zayan AF. 2011. Quality assessment of locally produced Egyptian liquid rennet. Internet J. Food Saf. 13:182–187 http://www.internetjfs.org/articles/Quality%20Assessment%20of%20Locally%20Produced%20Egyptian%20Liquid%20Rennet.pdf [Google Scholar]
- 11.European Commission. 2002. The safety of animal rennet in regard to risks from animal TSE and BSE in particular: adopted by the Scientific Steering Committee at its meeting of 16 May 2002. http://ec.europa.eu/food/fs/sc/ssc/out265_en.pdf [Google Scholar]
- 12.Heidebach T, Först P, Kulozik U. 2009. Microencapsulation of probiotic cells by means of rennet-gelation of milk proteins. Food Hydrocoll. 23:1670–1677. 10.1016/j.foodhyd.2009.01.006 [DOI] [Google Scholar]
- 13.Santillo A, Albenzio M, Bevilacqua A, Corbo MR, Sevi A. 2012. Encapsulation of probiotic bacteria in lamb rennet paste: effects on the quality of Pecorino cheese. J. Dairy Sci. 95:3489–3500. 10.3168/jds.2011-4814 [DOI] [PubMed] [Google Scholar]
- 14.Cakmaki S, Boroúlu E. 2004. Some quality characteristics of commercial liquid rennet samples. Turk. J. Vet. Anim. Sci. 28:501–505 http://journals.tubitak.gov.tr/veterinary/issues/vet-04-28-3/vet-28-3-7-0210-21.pdf [Google Scholar]
- 15.Flórez AB, Hernández-Barranco AM, Marcos I, Mayo B. 2006. Biochemical and microbiological characterization of artisan kid rennet extracts used for Cabrales cheese manufacture. LWT Food Sci. Technol. 39:605–612. 10.1016/j.lwt.2005.03.019 [DOI] [Google Scholar]
- 16.Temelli S, Anar S, Sen C, Akyuva P. 2006. Determination of microbiological contamination sources during Turkish white cheese production. Food Control 17:856–861. 10.1016/j.foodcont.2005.05.012 [DOI] [Google Scholar]
- 17.Franciosi E, Settanni L, Cologna N, Cavazza A, Poznanski E. 2011. Microbial analysis of raw cows' milk used for cheese-making: influence of storage treatments on microbial composition and other technological traits. World J. Microbiol. Biotechnol. 27:171–180. 10.1007/s11274-010-0443-2 [DOI] [Google Scholar]
- 18.Ercolini D, De Filippis F, La Storia A, Iacono M. 2012. “Remake” by high-throughput sequencing of the microbiota involved in the production of water buffalo mozzarella cheese. Appl. Environ. Microbiol. 78:8142–8145. 10.1128/AEM.02218-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, Fierer N, Peña AG, Goodrich JK, Gordon JI, Huttley GA, Kelley ST, Knights D, Koenig JE, Ley RE, Lozupone CA, McDonald D, Muegge BD, Pirrung M, Reeder J, Sevinsky JR, Turnbaugh PJ, Walters WA, Widmann J, Yatsunenko T, Zaneveld J, Knight R. 2010. QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 7:335–336. 10.1038/nmeth.f.303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Reeder J, Knight R. 2010. Rapidly denoising pyrosequencing amplicon reads by exploiting rank-abundance distributions. Nat. Methods 7:668–669. 10.1038/nmeth0910-668b [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Edgar RC. 2010. Search and clustering orders of magnitude faster than BLAST. Bioinformatics 26:2460–2461. 10.1093/bioinformatics/btq461 [DOI] [PubMed] [Google Scholar]
- 22.Wang Q, Garrity GM, Tiedje JM, Cole JR. 2007. Naïve Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl. Environ. Microbiol. 73:5261–5267. 10.1128/AEM.00062-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.McDonald D, Price MN, Goodrich J, Nawrocki EP, De Santis TZ, Probst A, Andersen GL, Knight R, Hugenholtz P. 2012. An improved Greengenes taxonomy with explicit ranks for ecological and evolutionary analyses of bacteria and archaea. ISME J. 6:610–618. 10.1038/ismej.2011.139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chao A, Bunge J. 2002. Estimating the number of species in a stochastic abundance model. Biometrics 58:531–539. 10.1111/j.0006-341X.2002.00531.x [DOI] [PubMed] [Google Scholar]
- 25.Shannon CE, Weaver W. 1949. The mathematical theory of communication. University of Illinois Press, Urbana, IL [Google Scholar]
- 26.Saeed AI, Sharov V, White J, Li J, Liang W, Bhagabati N, Braisted J, Klapa M, Currier T, Thiagarajan M, Sturn A, Snuffin M, Rezantsev A, Popov D, Ryltsov A, Kostukovich E, Borisovsky I, Liu Z, Vinsavich A, Trush V, Quackenbush J. 2003. TM4: a free, open-source system for microarray data management and analysis. Biotechniques 34:374–378 ftp://occams.dfci.harvard.edu/pub/bio/TM4_Biotechniques_2003.pdf [DOI] [PubMed] [Google Scholar]
- 27.Settanni L, Di Grigoli A, Tornambé G, Bellina V, Francesca N, Moschetti G, Bonanno A. 2012. Persistence of wild Streptococcus thermophilus strains on wooden vat and during the manufacture of a Caciocavallo type cheese. Int. J. Food Microbiol. 155:73–81. 10.1016/j.ijfoodmicro.2012.01.022 [DOI] [PubMed] [Google Scholar]
- 28.Weisburg WG, Barns SM, Pelletier DA, Lane DJ. 1991. 16S ribosomal DNA amplification for phylogenetic study. J. Bacteriol. 173:697–703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W. 1997. Grapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389–3402. 10.1093/nar/25.17.3389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chun J, Lee J-H, Jung Y, Kim M, Kim S, Kim BK, Lim YW. 2007. EzTaxon: a web-based tool for the identification of prokaryotes based on 16S ribosomal RNA gene sequences. Int. J. Syst. Evol. Microbiol. 57:2259–2261. 10.1099/ijs.0.64915-0 [DOI] [PubMed] [Google Scholar]
- 31.Jackson CR, Fedorka-Cray PJ, Barrett JB. 2004. Use of a genus- and species-specific multiplex PCR for identification of enterococci. J. Clin. Microbiol. 42:3558–3565. 10.1128/JCM.42.8.3558-3565.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Settanni L, Gaglio R, Guarcello R, Francesca N, Carpino S, Sannino C, Todaro M. 2013. Selected lactic acid bacteria as a hurdle to the microbial spoilage of cheese: application on a traditional raw ewes' milk cheese. Int. Dairy J. 32:126–132. 10.1016/j.idairyj.2013.04.010 [DOI] [Google Scholar]
- 33.Mora D, Musacchio F, Fortina MG, Senini L, Manachini PL. 2003. Autolytic activity and pediocin-induced lysis in Pediococcus acidilactici and Pediococcus pentosaceus strains. J. Appl. Microbiol. 94:561–570. 10.1046/j.1365-2672.2003.01868.x [DOI] [PubMed] [Google Scholar]
- 34.King N. 1948. Modification of Voges-Proskauer test for rapid colorimetric determination of acetyl methyl carbimol plus diacetyl in butter. Dairy Ind. 13:860–866 [Google Scholar]
- 35.Klandar AH, Lagaude A, Chevalier-Lucia D. 2007. Assessment of the rennet coagulation of skim milk: a comparison of methods. Int. Dairy J. 17:1151–1160. 10.1016/j.idairyj.2007.03.005 [DOI] [Google Scholar]
- 36.Rossetti L, Fornasari ME, Gatti M, Lazzi C, Neviani E, Giraffa G. 2008. Grana Padano cheese whey starters: microbial composition and strain distribution. Int. J. Food Microbiol. 127:168–171. 10.1016/j.ijfoodmicro.2008.06.005 [DOI] [PubMed] [Google Scholar]
- 37.Ercolini D. 2013. High-throughput sequencing and metagenomics: moving forward in the culture-independent analysis of food microbial ecology. Appl. Environ. Microbiol. 79:3148–3155. 10.1128/AEM.00256-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ercolini D, Ferrocino I, Nasi A, Ndagijimana M, Vernocchi P, La Storia A, Laghi L, Mauriello G, Guerzoni ME, Villani F. 2011. Monitoring of microbial metabolites and bacterial diversity in beef stored in different packaging conditions. Appl. Environ. Microbiol. 77:7372–7381. 10.1128/AEM.05521-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Li RW, Wu S, Li W, Huang Y, Gasbarre LC. 2011. Metagenome plasticity of the bovine abomasal microbiota in immune animals in response to Ostertagia ostertagi infection. PLoS One 6:e24417. 10.1371/journal.pone.0024417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Flórez AB, Mayo B. 2006. PCR–DGGE as a tool for characterizing dominant microbial populations in the Spanish blue-veined Cabrales cheese. Int. Dairy J. 16:1205–1210. 10.1016/j.idairyj.2005.11.008 [DOI] [Google Scholar]
- 41.Fox PF, McSweeney PLH, Cogan TM, Guinee TP. 2004. Cheese: chemistry, physics and microbiology. Elsevier, London, England [Google Scholar]
- 42.Beasley S. 2004. Isolation, identification and exploitation of lactic acid bacteria from human and animal microbiota. Ph.D. thesis University of Helsinki, Helsinki, Finland [Google Scholar]
- 43.Franciosi E, Settanni L, Cavazza A, Poznanski E. 2009. Biodiversity and technological potential of wild lactic acid bacteria from raw cows' milk. Int. Dairy J. 19:3–11. 10.1016/j.idairyj.2008.07.008 [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.