Abstract
Bacterial microcompartments (BMCs) are organelles that encapsulate functionally linked enzymes within a proteinaceous shell. The prototypical example is the carboxysome, which functions in carbon fixation in cyanobacteria and some chemoautotrophs. It is increasingly apparent that diverse heterotrophic bacteria contain BMCs that are involved in catabolic reactions, and many of the BMCs are predicted to have novel functions. However, most of these putative organelles have not been experimentally characterized. In this study, we sought to discover the function of a conserved BMC gene cluster encoded in the majority of the sequenced planctomycete genomes. This BMC is especially notable for its relatively simple genetic composition, its remote phylogenetic position relative to characterized BMCs, and its apparent exclusivity to the enigmatic Verrucomicrobia and Planctomycetes. Members of the phylum Planctomycetes are known for their morphological dissimilarity to the rest of the bacterial domain: internal membranes, reproduction by budding, and lack of peptidoglycan. As a result, they are ripe for many discoveries, but currently the tools for genetic studies are very limited. We expanded the genetic toolbox for the planctomycetes and generated directed gene knockouts of BMC-related genes in Planctomyces limnophilus. A metabolic activity screen revealed that BMC gene products are involved in the degradation of a number of plant and algal cell wall sugars. Among these sugars, we confirmed that BMCs are formed and required for growth on l-fucose and l-rhamnose. Our results shed light on the functional diversity of BMCs as well as their ecological role in the planctomycetes, which are commonly associated with algae.
INTRODUCTION
Bacterial microcompartments (BMCs) are organelles bounded by a proteinaceous shell. They were first observed in electron micrographs of cyanobacteria (1) and later in some chemoautotrophs (2). Subsequent characterization showed they contain ribulose bisphosphate carboxylase/oxygenase (RuBisCO) and were required for carbon fixation at atmospheric concentrations of CO2, and they were dubbed carboxysomes (3). Apparently icosahedral, carboxysome shells are formed by three types of proteins. BMC-H proteins contain a single copy of the Pfam00936 domain and form hexamers (4), while BMC-T proteins are a fusion of two Pfam00936 domains and form trimers (pseudohexamers) (5, 6). BMC-H and BMC-T proteins are thought to compose the facets of the icosahedron, while BMC-P proteins, composed of a single Pfam03319 domain, form pentamers that cap the vertices (7, 8). Several years after the identification of the proteins that make up the carboxysome shell, operons involved in 1,2-propanediol (PDU) and ethanolamine (EUT) degradation in Salmonella enterica were sequenced, and it was discovered that these loci contain homologs to genes encoding carboxysomal BMC-H, BMC-T, and BMC-P proteins (9–12). The formation of polyhedral bodies in S. enterica was subsequently confirmed by electron microscopy and shown to be regulated by the availability of 1,2-propanediol or ethanolamine (12, 13). These catabolic BMCs are referred to as metabolosomes, in contrast to the carbon-fixing carboxysomes.
Several lines of evidence suggest that at least one common function of carboxysomes and metabolosomes is to concentrate a volatile or toxic metabolite (14–17). The carboxysome shell acts as a CO2 diffusion barrier, increasing the local concentration of RuBisCO substrate and thus improving carbon fixation (17). The PDU metabolosome houses enzymes that form and degrade propionaldehyde, a toxic metabolite (15). Similarly, the EUT metabolosome encapsulates enzymes that generate and degrade acetaldehyde, a volatile and reactive metabolite (14). It has been proposed that substrate channeling due to the close packing of these functionally sequential enzymes prevents the buildup of toxic and/or volatile metabolites (13).
The PDU and EUT metabolosomes likewise appear to share common core biochemistry. In both, an aldehyde is generated that is then oxidized by an aldehyde dehydrogenase to form an acyl coenzyme A (CoA) species, reducing NAD+ to NADH in the process (18, 19). Because this reaction is isolated from the cytosolic substrate pool, the NAD+ and CoA need to be regenerated within the metabolosome. In order to regenerate NAD+, an alcohol dehydrogenase reduces a second aldehyde to an alcohol, which exits the BMC (20). To regenerate CoA, a phosphotransacylase replaces the CoA moiety with a phosphate (21). The acyl phosphate then presumably participates in substrate level phosphorylation via an acyl kinase also encoded within the BMC locus (22).
Bioinformatic surveys of sequenced bacterial genomes have revealed that roughly 20% contain gene clusters with the potential to form BMCs, for which only speculative functions have been assigned, based on gene annotations (23–25). One of these loci has recently been investigated further in Clostridium phytofermentans and has been shown to utilize a glycyl radical enzyme (GRE) to degrade 1,2-propanediol (26); this is in contrast to the PDU metabolosome of S. enterica, which uses a vitamin B12-dependent enzyme. The recent C. phytofermentans study marked the first reverse-genetics approach to confirm the presence of a bioinformatically predicted BMC, and surprisingly, it revealed an alternative mechanism with which to degrade 1,2-propanediol. However, the majority of bioinformatically identified BMC loci remain with tentative or no functional prediction whatsoever.
We identified one such locus conserved in the majority of sequenced planctomycete genomes and in one clade of the phylum Verrucomicrobia. Depending on the species, the locus contains between 11 and 13 genes, which we refer to as Planctomycetes and Verrucomicrobia metabolosome genes A through O (pvmA through pvmO) (see Table S1 in the supplemental material). The phyla Planctomycetes and Verrucomicrobia are closely related; they are united by their morphological similarity, in particular intracellular membrane organization (27, 28), and were among the first bacteria to challenge the view that bacteria are simple organisms lacking organizational complexity. The planctomycetes form three distinct clades: autotrophic species that contain the anammoxosome (reviewed by van Niftrik and Jetten [29]); the class Phycisphaerae, which are facultative aerobes and divide by binary fission (30); and the class Planctomycetia, members of which are predominantly aerobic or facultatively aerobic, lack anammoxosomes, and divide by budding. Nine members from the last clade representing seven genera isolated from very different habitats have sequenced genomes; eight contain the BMC gene cluster. This locus is apparently restricted to the Planctomycetes and Verrucomicrobia, and its function provides clues to the ecological role of the ubiquitous planctomycetes, as well as the diversity of BMCs.
MATERIALS AND METHODS
Strains, media, and culture growth conditions.
The strains used in this study are described in Table 1. P. limnophilus Mü 290T (31) was grown in YVT, a modified PYGV medium (DSMZ medium 621 [http://www.dsmz.de]): 0.1% yeast extract, 10 mM Tris-HCl (pH 7.5), vitamin solution, and salt solution. YVT medium was supplemented with a 10 mM concentration of an additional carbon source as needed. Agar (1.5%) was added to make solid medium. Liquid cultures for all experiments were grown aerobically in a volume of medium that was one-fifth the volume of the flask: generally 50 ml of medium in a 250-ml Erlenmeyer flask. Flasks were grown in the dark at 30°C with shaking at 150 rpm. l-Fucose and l-rhamnose were purchased from Sigma-Aldrich, and Laminaria fucoidan was purchased from Glycomix.
TABLE 1.
Strains used in this study
| Strain name | Genotype | Term used in text | Reference or source |
|---|---|---|---|
| Mü 290T | Wild type | Wild type | 31 |
| OE3 | Δplim_1104::npt | Δplim_1104, control strain | This work |
| OE4 | Δplim_1751::IN(npt) | ΔpvmJ::IN(npt) | This work |
| OE22 | Δlim_1751::npt | ΔpvmJ | This work |
| OE23 | Δ(plim_1756-plim_1755)::npt | ΔpvmDE, shell-less mutant | This work |
| OE24 | Δplim_1747::npt | ΔpvmN | This work |
Genetic transformation of P. limnophilus.
The kanamycin resistance gene, including 137 bp upstream of the ATG start codon, was amplified from plasmid pUH-37 via PCR using primers OE53 (TATAGAATTCAGATCTCGGAATTGCCAGCT) and OE54 (TATAGGATCCTCAGAAGAACTCGTC). EcoRI and BglII restriction sites were added to the 5′ end, a BamHI restriction site was added to the 3′ end, and the gene was cloned into the p2741 cloning vector. This vector was chosen for its small size, paucity of restriction enzyme sites, and high copy number; these features make it amenable to cloning of multipart constructs.
For homologous recombination, genomic DNA sequences 500 to 1,000 bp upstream and downstream of the gene(s) to be replaced with the kanamycin resistance cassette were amplified via PCR with EcoRI and BglII restriction sites on the 5′ end and a BamHI restriction site on the 3′ end (primer information is in Table S2 in the supplemental material). Knockout cassettes were constructed in the p2741 vector using the BglBricks cloning strategy (32). Circular plasmids were used for electroporation. Linearized plasmids and PCR fragments were also tested and could also transform P. limnophilus. Aliquots (100 μl) of P. limnophilus competent cells were prepared and electroporated as described previously (33). Electroporated cells were recovered in 1 ml of liquid YVT supplemented with d-glucose for 1.5 h with shaking at 30°C, and all cells were plated onto YVT plates supplemented with d-glucose and 30 μg/ml of kanamycin for the selection. Plates were incubated at 30°C in the dark until colonies formed, after 5 to 7 days. Colonies were struck onto fresh selection plates, grown for an additional 3 to 5 days, and then genotyped by colony PCR or PCR using purified genomic DNA as the template.
Phylogenetic analysis.
For the 16S rRNA species tree, full-length 16S rRNA sequences were obtained from IMG (http://img.jgi.doe.gov) and NCBI (http://www.ncbi.nlm.nih.gov), aligned using MUSCLE (34, 35), and curated with GBLOCKs (36), and phylogeny was constructed using PhyML 3.0 (37) with 500 bootstraps using the Phylogeny.fr web interface (38, 39). For the gene concatenation tree, orthologs of pduL (see gene tree in Fig. S1A in the supplemental material) in S. enterica were concatenated to orthologs of pduP (gene tree in Fig. S1B) present in a list of 67 genomes that contain a BMC gene cluster (see Table S3 in the supplemental material), sequences were aligned using MUSCLE as described above, and gapped positions were manually trimmed using Jalview (40). The phylogeny was constructed using PhyML as described above, with 100 bootstraps. The gene clusters were selected with the goal of maximizing the number of phyla represented. Metabolosome type was assigned based on the identity of the defining enzyme of the gene cluster: propanediol dehydratase (PDU) or ethanolamine ammonia lyase (EUT). Both the PDU2 and glycyl radical enzyme (GRE) clades contain a GRE homolog; the gene concatenations that branched close to the PDU clade were defined as PDU2, and those that branched close to the EUT clade were designated GRE. The presence of an aldolase and the absence of any of the aforementioned signature enzymes are the defining features of the Planctomycetes and Verrucomicrobia (PV) and PV-like BMC gene clusters. The PV-like gene clusters have the same features (gene annotations) as the PV type, but the gene order is different.
Gene comparisons.
Pairwise BLAST comparisons were performed using the IMG genome BLASTP utility with a minimum E value of 1e−2 and minimum identity of 30% as criteria for a positive hit, and orthologs were defined as those genes that returned bidirectional BLAST hits by the IMG server (see above).
Biolog assay.
Cells were grown for 5 days on solid YVT medium supplemented with glucose, then scraped off using a sterile cotton swab, and resuspended in IF0a medium, obtained from Biolog, to an optical density at 600 nm (OD600) of 0.24 as measured in by a NanoDrop 2000c in the cuvette holder. Although the cells were not washed in fresh medium to control for nutrient transfer when resuspended, the A1 well of each Biolog plate contains no carbon source and was used as a blank when measuring dye reduction, to serve as a control. Cell suspension (1.76 ml) was added to 22.24 ml of inoculating fluid (YVT with 0.005% yeast extract instead of 0.1% and 240 μl of dye G, obtained from Biolog). Phenotypic microarray plates 1 and 2 were inoculated with 100 μl of final cell suspension, wrapped in aluminum foil to retard evaporation, and incubated at 30°C for 5 days. The absorbance at 590 nm was recorded on a BioTek EON microplate spectrophotometer.
Growth curves.
P. limnophilus strains were grown on plates for 5 days and then resuspended into YVT medium using a sterile cotton swab to scrape cells from the plate. Cultures were resuspended in liquid YVT to an OD600 of about 0.5, as measured by a BioTek EON microplate spectrophotometer, and inoculated into liquid medium for growth curves to a starting OD600 of 0.01 ± 0.02. Cultures were grown as described above.
Electron microscopy.
Cultures were grown as described above (YVT medium supplemented with a 10 mM concentration of an additional carbon source) for 9 days prior to sample preparation for electron microscopy. Cells were concentrated by centrifugation and loaded into 50-μm-deep specimen carriers (catalog no. 390; Wohlwend Engineering, Sennwald, Switzerland) and ultrarapidly frozen in a Bal-Tec HPM 010 high-pressure freezer (Bal-Tec Ag, Liechtenstein). Frozen samples were freeze substituted over a period of 2 h using the superquick-freeze substitution (SQFS) method of McDonald and Webb (41). Cells were rinsed in pure acetone, infiltrated with Epon resin (25, 50, 75, and 100% [the last value for three times]) for 15 min at each concentration, and then polymerized for 2 days in a 60°C oven. Sections 60 nm thick were cut and poststained for 4 min in 2% aqueous uranyl acetate and 2 min in lead citrate (42). Images were taken with a Gatan Ultrascan 1000 camera (Gatan, Inc., Pleasanton, CA) bottom mounted on a Tecnai Spirit transmission electron microscope with an accelerating voltage of 120 kV (FEI, Hillsboro, OR). All original electron micrographs were adjusted in the same manner using Adobe Photoshop CS6 as follows: brightness was decreased 20%, and contrast was increased 90%.
RESULTS
A conserved BMC gene cluster is present in members of the phyla Planctomycetes and Verrucomicrobia.
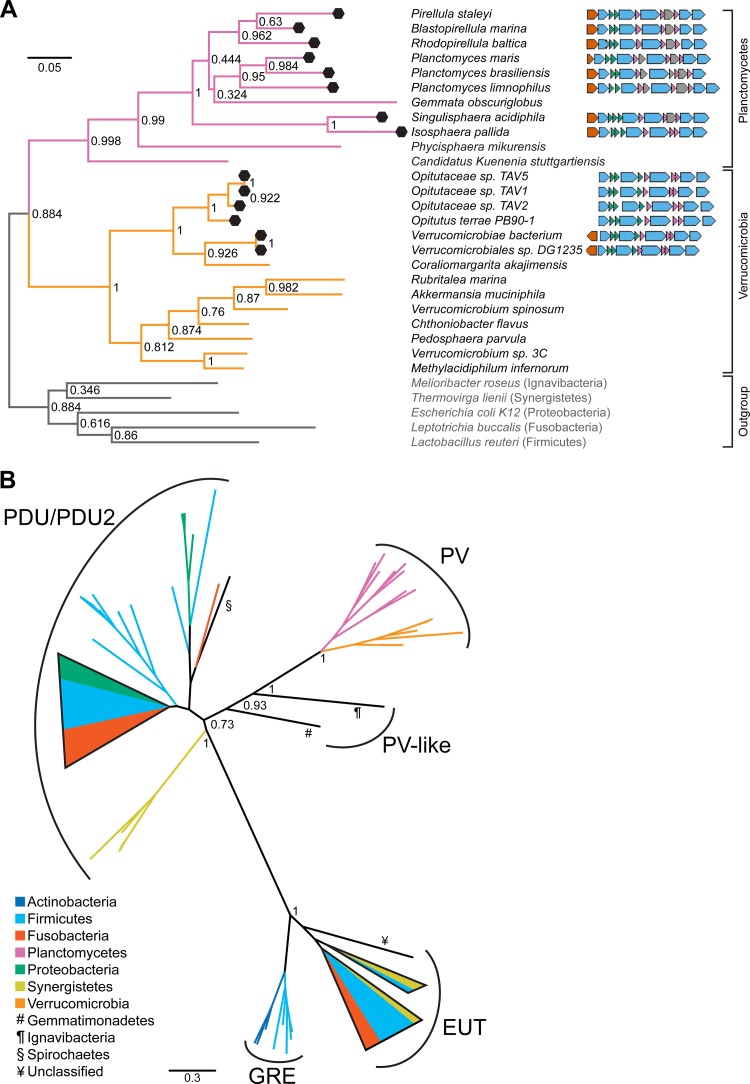
The Planctomycetes and Verrucomicrobia-type (PV) BMC gene cluster is defined by the presence of two (or three in Singulisphaera acidiphila and Isosphaera pallida) genes encoding BMC-H proteins and three genes encoding BMC-P proteins (Fig. 1A). In addition to the shell proteins, five genes code for putative enzymes, and the cluster is preceded by a transcriptional regulator, suggestive of operon-like regulation (Fig. 1A). Interestingly, the gene clusters present in the Verrucomicrobia either do not have this transcriptional regulator or have it in the opposite orientation relative to the rest of the gene cluster; this may reflect a different type of regulation between the two phyla.
FIG 1.
Phylogenetic trees. (A) 16S rRNA tree of representative species of the phyla Planctomycetes and Verrucomicrobia with sequenced genomes (as of June 2013) and five outgroup species. Bootstrap values are shown at nodes, and black hexagons indicate species with a BMC gene cluster. Each species' BMC gene cluster is shown schematically to the right of the species name, color-coded by gene annotation as follows: orange, transcriptional regulator; blue, enzyme; green, BMC-H shell protein; pink, BMC-P shell protein; gray, hypothetical protein. (B) Gene tree of the concatenation of aldehyde dehydrogenase (pduP) and phosphotransferase (pduL) homologs (Table 2; see also Table S3 in the supplemental material). Branches with less than 50% bootstrap support were collapsed and drawn in cartoon form. Branch color or symbol corresponds to phylum. Collapsed branches were similarly colored to represent phyla present within. Scale bar represents number of substitutions per site. Clades are annotated by the key enzymes present in the gene clusters (see Materials and Methods). PDU, vitamin B12-dependent propanediol utilization; PDU2, vitamin B12-independent (glycyl radical enzyme-utilizing) propanediol utilization; PV, Planctomycetes and Verrucomicrobia type; EUT, vitamin B12-dependent ethanolamine utilization; GRE, locus of unknown function containing a glycyl radical enzyme. Bootstrap values of branches separating major clades are shown.
Based on the species tree, it appears as though the PV gene cluster was present in the last common ancestor of the classes Phycisphaerae and Planctomycetia, with Gemmata obscuriglobus having lost the gene cluster (Fig. 1A). A similar situation is apparent in the Verrucomicrobia, where Coraliomargarita akajimensis apparently lost the gene cluster. The bootstrap values for the nodes at which G. obscuriglobus bifurcates within the class Planctomycetia are very low, and this could be because the “true” position of G. obscuriglobus is most likely basally located within the class, as has been shown in other, more phylogenetically focused studies (43–45). As a result, we cannot definitively determine whether the G. obscuriglobus genome lost the BMC locus or never contained it. However, in either case the species tree is consistent with vertical inheritance of the gene cluster within the respective phyla, as opposed to horizontal gene transfer, highlighting the retention of the loci over evolutionary timescales. Conservation of genes in a specific order across species typically suggests that they function together in a given biological phenomenon (46); accordingly, we hypothesized that the PV gene cluster encodes a bona fide BMC.
Bioinformatic comparison of PV BMC genes to other metabolosome genes.
We focused on the gene complement in P. limnophilus as the representative BMC-associated gene cluster because this species is rapidly becoming a model organism with which to study planctomycete biology (33, 47). We compared its composition to that of the EUT and vitamin B12-dependent PDU BMC loci found in S. enterica and to the glycyl radical enzyme utilizing the PDU (hereafter referred to as PDU2) BMC locus found in C. phytofermentans (Table 2). While the PDU, PDU2, and EUT loci encode at least one BMC-T protein, P. limnophilus does not have any, nor does any planctomycete or verrucomicrobium. Furthermore, the PV locus contains three genes encoding BMC-P proteins, while the EUT and PDU BMC loci each have only one. In addition to the differences in shell protein composition, it also appears that all four BMCs use distinct enzymes to generate an aldehyde (Table 2). Downstream of aldehyde generation, all four types of BMC loci are similar, encoding enzymes required to participate in the core BMC biochemistry (Table 2). Furthermore, all of the aldehyde-generating enzymes as well as the aldehyde dehydrogenases encoded in all of the loci contain a BMC encapsulation peptide (23) not present in non-BMC-associated homologs. Thus, it appears that an aldehyde can be generated and partially catabolized in the putative PV BMC, as in the PDU, EUT, and PDU2 BMCs.
TABLE 2.
Protein compositions of functionally distinct metabolosomes
| BMC type | No. and name of shell protein gene(s) |
Gene(s) for aldehyde-generating enzymes |
Gene for core metabolosome enzymes |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| BMC-H | BMC-T | BMC-P | 1,2-Propanediol oxidoreductase | Aldolase | Propionaldehyde generating | Acetaldehyde generating | Aldehyde dehydrogenase | Phosphotransferase | Acyl kinase | Alcohol dehydrogenase | |
| PDU (S. enterica) | 3; pduA, -J, -K, -U | 2; pduB, -T | 1; pduN | None | None | pduCDEa (B12-dependent diol dehydratase) | None | pduPa | pduLa | pduWa | pduQa |
| EUT (S. enterica) | 3; eutK, -M, -S | 1; eutL | 1; eutN | None | None | None | eutBCa (ethanolamine ammonia lyase) | eutEa | eutDa | acka | eutGa |
| PDU2 (C. phytofermentans) | 4; Cphy_1176, Cphy_1180, Cphy_1181, Cphy_1182 | 1; Cphy_1186 | 1; Cphy_1184 | Cphy_1185 | Cphy_1177 | Cphy_1174 (B12-independent diol dehydratase) | None | Cphy_1178 | Cphy_1183 | Cphy_1327 | Cphy_1179 |
| PV (P. limnophilus) | 2; pvmD, -E | 0 | 3; pvmH, -K, -M | None | pvmN | None | None | pvmJ | pvmB | pvmG | pvmO |
Experimentally verified function. Accession numbers for genes are provided in Tables S1 and S4 in the supplemental material.
The aldolase encoded in the PDU2 locus has been reported to generate lactaldehyde from l-fucose and l-rhamnose (26) and is homologous to the aldolase encoded in the PV locus, suggesting a similar function in these BMCs. While pairwise sequence comparisons (see Fig. S2 in the supplemental material) supported a possible common function in the BMCs, a few key differences are apparent. First, the propanol dehydrogenase encoded in the PDU2 BMC gene cluster (Cphy_1179) does not have any bidirectional hits to genes in the P. limnophilus genome, indicating that no ortholog is present (48) (see Fig. S2). Second, the gene order within the PV BMC gene cluster is markedly different from that in other BMC gene clusters; such dissimilarity has been proposed to be indicative of operon evolution associated with diverging function (49). Finally and most critically, both the PV BMC gene cluster and the P. limnophilus genome lack any homologs to the propanediol dehydratase (forming propionaldehyde from 1,2-propanediol) genes found in the PDU and PDU2 loci (see Fig. S2), so it is unlikely that propanediol is processed by this BMC. Given these observations, we posited that the PV BMC is involved in the degradation of l-fucose, l-rhamnose, and/or related sugars via a pathway different from that of C. phytofermentans.
Phylogenetic analysis of PV-type BMC genes.
Only two protein domain families are conserved across all BMC types found in microbial genomes: Pfam00936 (in BMC-H and BMC-T) and Pfam03319 (BMC-P). Because these domains are small (approximately 100 amino acids) and are present in different copy numbers in different BMC gene clusters, sequence alignments result in low phylogenetic signal. However, for catabolic BMCs, the three enzymes involved in the core biochemistry are common to all (Table 2). Accordingly, we searched for homologs of these enzymes in sequenced microbial genomes and constructed a phylogeny using a concatenation of the aldehyde dehydrogenase and phosphotransacylase genes from 67 diverse genomes (see Table S3 in the supplemental material). Because some gene clusters encoded multiple putative alcohol dehydrogenases, that core enzyme was not included in the concatenation.
In the resulting gene tree, a number of distinct clades were apparent (Fig. 1B). BMCs containing glycyl radical enzymes (GREs) either grouped near those involved in metabolism of propanediol (PDU2) or formed a distinct clade close to representative sequences of the EUT metabolosome (Fig. 1B, GRE). In addition to the PDU/PDU2, GRE, and EUT groups, PV enzymes form a well-defined clade (Fig. 1B). The branch length separating the PV-type clade from the PDU/PDU2 clade is comparable to that separating the EUT and PDU/PDU2 clades, suggesting that the enzymes within the PV-type BMC are functionally divergent from propanediol metabolism. Within the PV clade, the tree topology is essentially congruent with the species tree (data not shown), supporting our hypothesis that the BMC-associated genes have descended vertically through the Planctomycetes and Verrucomicrobia. In contrast, the other major clades are paraphyletic, indicating horizontal gene transfer (Fig. 1B). Due to the clear distinction of the PV lineage, we inferred that the PV BMC has a novel function distinct from any other characterized to date.
The PV BMC appears to be involved in several saccharide degradation pathways.
In order to experimentally identify the substrate of the PV BMC and the related biochemical reactions that take place, we conducted an aerobic metabolic screen using Biolog phenotype microarrays (Biolog, Hayward, CA). In this assay, 96-well plates have prospective substrates dried in each well and are inoculated with a cell suspension containing a redox-sensitive dye. Reducing equivalents are generated when a sample has metabolic activity in the well; these reduce the dye, turning it purple. Given that all BMCs characterized to date are involved in carbon metabolism, we selected phenotype microarrays containing a library of carbon sources. One of the presumed core PV BMC enzymes, encoded by pvmJ, is a NAD+-dependent dehydrogenase, which should produce NADH when the BMC substrate is degraded. We predicted that a mutant of P. limnophilus deficient in this gene would display little to no metabolism-associated signal, and comparison with wild-type growth would be an effective method to rapidly generate a list of candidate functions for the BMC. pvmJ was deleted using homologous recombination by replacing the gene with the npt kanamycin resistance gene under the control of its own promoter, in an orientation opposite to that of the rest of the gene cluster (Fig. 2A). This marks the first instance a directed gene knockout has been made in the phylum Planctomycetes; all other studies have employed random transposon insertion (33, 47). We confirmed that the selectable marker replaced pvmJ by PCR genotyping (Fig. 2B). All tested clones were double recombinants, not requiring a secondary selection step, highlighting the potential of reverse genetics to study planctomycetal biology. In order to control for any adverse metabolic effects that expressing the npt gene may have, we also generated a control strain, with the npt gene replacing plim_1104, a pseudogene in a relatively barren region of the chromosome (see Fig. S3A and B in the supplemental material). Thus, we have also identified a neutral site in the P. limnophilus genome in which to introduce genetic constructs for heterologous expression for other types of genetic studies.
FIG 2.
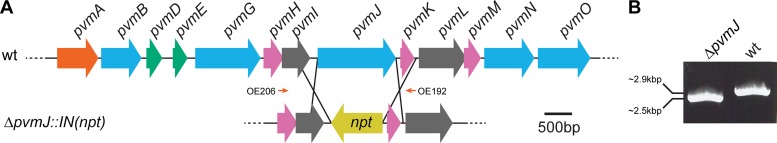
Homologous recombination to generate knockout mutants in P. limnophilus. (A) Schematic of the BMC gene cluster in P. limnophilus and the homologous recombination used to create the ΔpvmJ::IN(npt) mutant. Orange, transcriptional regulator; blue, enzyme; green, BMC-H shell protein; pink, BMC-P shell protein; gray, hypothetical gene; yellow, kanamycin resistance gene (npt). Orange arrows represent primers OE206 and OE192 (see Table S2 in the supplemental material) used for PCR genotyping. OE206 is outside the homology region, showing that recombination happened at the desired locus. (B) PCR amplification using primers OE206 and OE192 showing that amplicons are different sizes for the wild-type (wt) and knockout strains.
Surprisingly, the aldehyde dehydrogenase ΔpvmJ mutant displayed a metabolic deficiency under several conditions, as opposed to one or two (see Fig. S4 in the supplemental material). There were also some compounds with which the mutant appeared to produce a stronger signal than the control strain, but we did not expect these compounds to readily inform about the function of the BMC and so they were not further investigated. The substrates that displayed the strongest metabolic deficiency were pectin, mannose-containing compounds (d-mannose, mannan, α-methyl-d-mannoside), turanose-containing compounds (turanose and melezitose), d-raffinose, and l-fucose (see Fig. S4). For all of these sugars, the mutant still displayed a basal level of metabolic signal except l-fucose, which was completely devoid of metabolic signal (see Fig. S5 in the supplemental material). Furthermore, we observed only minimal metabolic signal in the well containing propanediol (PM1 well D5) (see Fig. S5), as well as no significant difference in signal between the mutant and control strain with this substrate. Thus, the PV BMC aldehyde dehydrogenase appears to be involved in the catabolism of a number of carbon sources and is absolutely necessary for the degradation of l-fucose and l-rhamnose, which appears to proceed via a pathway independent of propanediol.
PV-type BMC genes are required to degrade l-fucose and l-rhamnose.
In order to focus on the physiological roles of the putative aldehyde dehydrogenase (encoded by pvmJ) and the putative aldolase (encoded by pvmN), we constructed deletion mutants designed specifically to avoid any polarity effects (see Fig. S3C to F in the supplemental material). We confirmed that these mutations produced no transcript of the deleted gene and did not significantly disrupt the expression of downstream genes when cells were grown on l-fucose (data not shown).
We investigated the growth of the ΔpvmJ and ΔpvmN strains in the presence or absence of l-fucose and the structurally similar deoxy sugar l-rhamnose. Due to the slow doubling time of P. limnophilus (about 20 h on d-glucose; data not shown), it was not feasible to conduct the growth experiments with either l-fucose or l-rhamnose as the sole carbon source. Instead, we continued to supplement the medium with 0.1% yeast extract to reach suitable growth rates. The growth profiles of the wild type and the Δplim_1104 control strain were comparable, allowing us to control for any metabolic stress associated with the expression of the npt gene product (see Fig. S6 in the supplemental material).
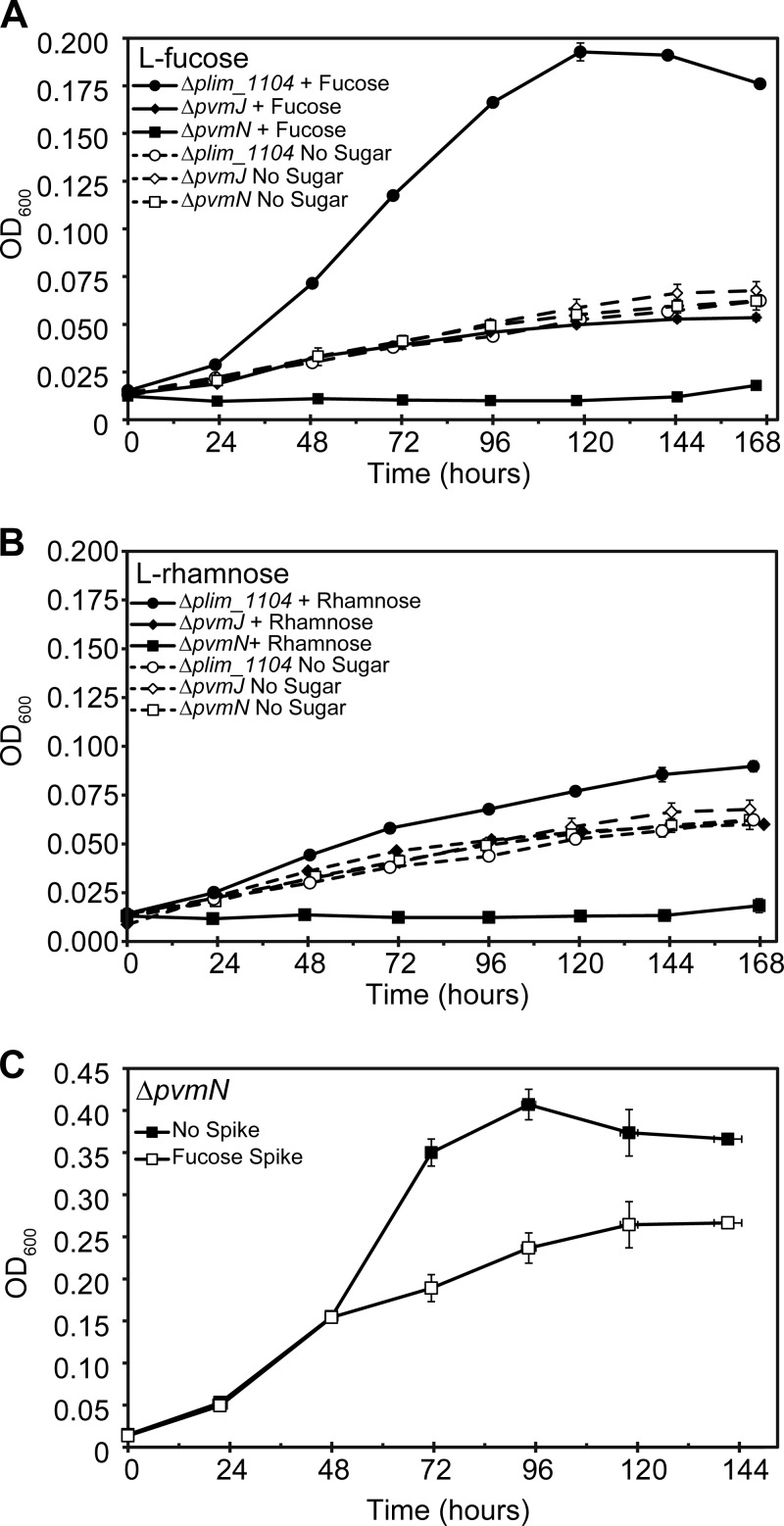
The ΔpvmJ mutant had a detectable growth defect in the presence of l-fucose and a slight defect for l-rhamnose, but it had a growth rate similar to that of the control strain on yeast extract alone, indicating that this aldehyde dehydrogenase mutant could not utilize those sugars for energy but could grow on nutrients supplied by the yeast extract (Fig. 3A and B). Interestingly, the ΔpvmN (aldolase mutant) strain did not grow at all in the presence of either l-fucose or l-rhamnose (Fig. 3A and B). This is suggestive of a toxic effect, since nutrition from the yeast extract is present. We verified this by spiking d-glucose-grown ΔpvmN cultures with l-fucose when they reached early exponential growth. The cultures that were challenged with l-fucose displayed a dramatically reduced growth rate, and the cell density after 6 days of growth was substantially lower than that of the control (Fig. 3C). These observations are consistent with an accumulation of an inhibitory factor when the ΔpvmN strain is grown on l-fucose or l-rhamnose. We further suggest that the pvmN gene product serves to detoxify this inhibitory factor. In addition, it appears that this gene cluster is not catabolically repressed by the presence of d-glucose. Collectively, the data indicate that the putative aldolase and aldehyde dehydrogenase of the PV BMC gene cluster are involved in the catabolic pathway of l-fucose and l-rhamnose and that this pathway produces toxic intermediates.
FIG 3.
Growth curves of Δplim_1104, ΔpvmJ, and ΔpvmN strains grown on l-fucose (A) and l-rhamnose (B). “No sugar” data in panel B are reproduced from panel A. (C) Spiking experiment in which the ΔpvmN strain was grown on 5 mM d-glucose for 2 days, at which point 5 mM l-fucose was added to half of the cultures. Solid squares represent nonspiked cultures, and open squares represent cultures with l-fucose added. In all graphs, each data point is an average of three independently grown cultures (n = 3); each error bar represents 1 standard deviation.
Planctomycetes assemble a metabolosome to degrade l-fucose and l-rhamnose.
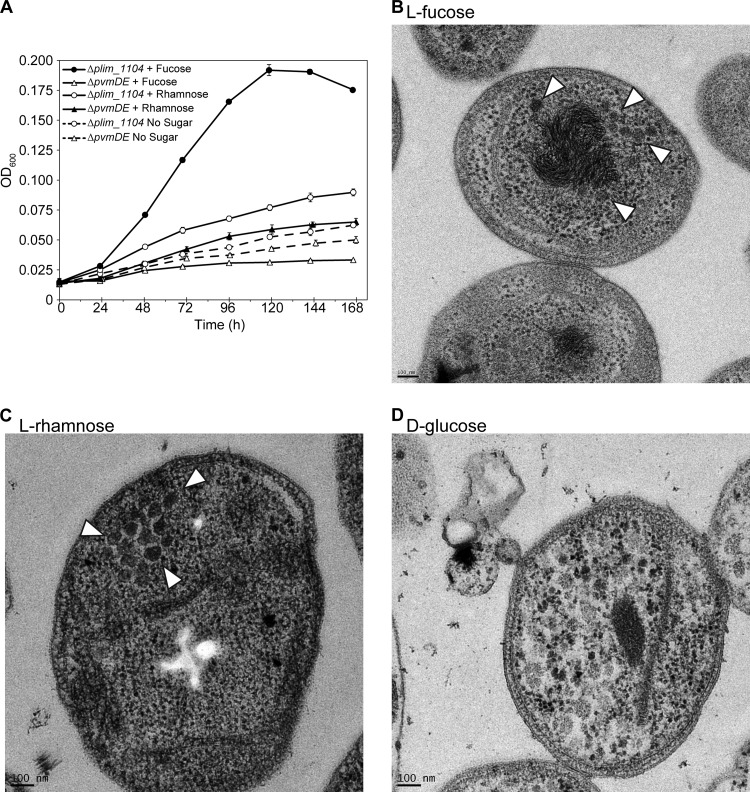
Since metabolosomes have been proposed to sequester reactions that produce toxic intermediates (15), we examined whether the formation of a BMC (composed of a shell and encapsulated enzymes) was involved in the metabolism of l-fucose and l-rhamnose. In order to confirm a role for the BMC shell proteins, we deleted pvmD and pvmE, the only two BMC-H shell protein genes present in the gene cluster (Table 2; see also Fig. S3G and H in the supplemental material); this deletion was expected to abolish metabolosome shell formation or integrity. In the presence of l-rhamnose, the ΔpvmDE mutant grew slower than the control, mimicking cells grown on yeast extract alone. However, when grown in the presence of l-fucose, the mutant exhibited growth inhibition similar to that of the ΔpvmN mutant (the putative aldolase mutant) and grew roughly 1 order of magnitude slower than the control strain (Fig. 4A). This is in contrast to previous studies on the EUT and PDU metabolosome in which individual shell proteins were deleted and relatively minor growth defects were observed (14, 50). These data suggest that the shell proteins are required for the optimal metabolism of both l-fucose and l-rhamnose.
FIG 4.
BMCs are expressed during growth on fucose and rhamnose. (A) Growth curves using l-fucose or l-rhamnose of the ΔpvmDE and Δplim_1104 strains grown with or without sugars. Data for the Δplim_1104 strain are reproduced from Fig. 3A and B. Each data point is an average of three independently grown cultures (n = 3); each error bar represents 1 standard deviation. (B to D) Electron microscopy of wild-type P. limnophilus showing BMCs (indicated by white arrowheads) when grown for 9 days on l-fucose (B) and l-rhamnose (C) but not d-glucose (D).
To confirm that a BMC is formed, we visualized wild-type P. limnophilus cells grown on l-fucose, l-rhamnose, or d-glucose by electron microscopy. It has been reported that BMCs are most apparent in stationary-phase cultures of S. enterica (12, 13) and C. phytofermentans (26), so we grew P. limnophilus to stationary phase before preparing thin sections. BMC-like structures were present in l-fucose- and l-rhamnose-grown cell populations but not in d-glucose-grown cultures (Fig. 4B, C, and D). These structures are 55 ± 9 nm (n = 47) in diameter and resemble in size and morphology the BMCs observed in S. enterica (51) and C. phytofermentans (26). Although amorphous bodies are present in d-glucose-grown cells, they do not have the distinct edges seen for the BMCs and do not otherwise resemble the structures of metabolosomes (Fig. 4D). Collectively, these data strongly suggest that a bona fide BMC is synthesized in P. limnophilus in order to degrade fucose and rhamnose.
BMC genes are required for growth on fucoidan, a sulfated polysaccharide from macroalgae.
l-Fucose and l-rhamnose rarely occur in nature as monosaccharides; they are usually components of polysaccharides. We next considered possible sources of these saccharides that may be encountered by planctomycetes. Planctomycete genomes are known to encode more sulfatase genes than those of most organisms; moreover, some planctomycetes have been shown to grow on sulfated polysaccharides (52). The sulfated polysaccharide fucoidan is composed of 95% l-fucose (53), and we hypothesized that P. limnophilus could utilize this compound as a source for l-fucose. We conducted a growth assay using the Δplim_1104 control strain, the ΔpvmDE shell mutant, and the ΔpvmN aldolase mutant on YVT medium supplemented with fucoidan extract from the Laminaria genus of brown algae. While the Δplim_1104 strain exhibited appreciable growth, the shell protein ΔpvmDE mutant appeared to grow about the same as on yeast extract alone, indicating that the fucoidan could not be metabolized and that no inhibitory factor was accumulating (Fig. 5). The ΔpvmN aldolase mutant exhibited the same growth suppression seen on l-fucose and l-rhamnose (Fig. 5), suggesting that fucoidan is desulfated and broken down into fucose monomers and then further metabolized to an inhibitory metabolite.
FIG 5.
Growth curves of Δplim_1104, ΔpvmDE, and ΔpvmN strains grown on fucoidan from Laminaria spp. “No sugar” data are reproduced from Fig. 3A and 4A. Each data point is an average of three independently grown cultures (n = 3); each error bar represents 1 standard deviation.
DISCUSSION
We have shown that bacterial microcompartments are formed in P. limnophilus and that they are required for aerobic growth on l-fucose, l-rhamnose, and the sulfated polysaccharide fucoidan. In order to study the BMC, we generated the first reported series of directed gene knockouts in a planctomycete, advancing the genetic toolset available for the phylum. Analysis of these mutants suggests that the organelle serves to detoxify products generated in the aerobic degradation pathway(s) of those sugars. In addition, we have observed a BMC mutant to be deficient in the degradation of several other carbon sources present in a metabolic screen.
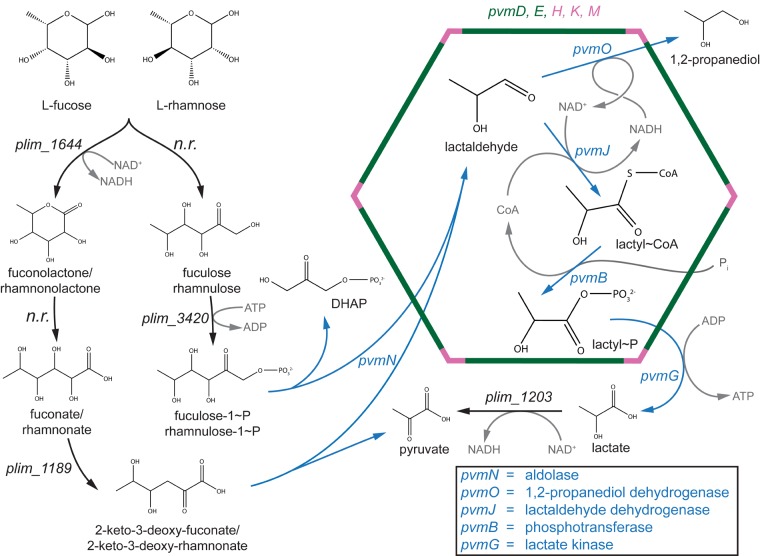
Based on our bioinformatic and experimental data, and previous genetic characterization of homologs in the EUT, PDU, and clostridial PDU2 metabolosomes, we propose a biochemical model for the function of the PV BMC, with two alternative upstream pathways to generate lactaldehyde from either l-rhamnose or l-fucose: phosphorylative (54) and nonphosphorylative (55–57) (Fig. 6). It is not clear which of these strategies P. limnophilus uses, as we could not identify homologs for the full enzyme complement for either pathway in its genome (Fig. 6; see also Fig. S2 in the supplemental material); these activities may be provided via nonorthologous gene displacement (58). The BMC-encoded aldolase is capable of acting on intermediates of both pathways to produce lactaldehyde as well as a compound that can readily enter central metabolism: either dihydroxyacetone phosphate or pyruvate (55, 59–61). Downstream of the aldolase reaction, the remaining four enzymes in the PV BMC gene cluster participate in reactions that degrade lactaldehyde to lactate while allowing for cofactor recycling within the compartment; this is required because the BMC shell prevents the diffusion of NAD+/H and coenzyme A to the cytosol (20, 21) (Fig. 6). The lactate can then enter glycolysis via pyruvate and be respired to generate several reducing equivalents for cellular energy. The propanediol is produced as a result of cofactor recycling and may be excreted; there is no obvious evidence of P. limnophilus being able to metabolize this compound (see Fig. S5 in the supplemental material, PM1 well D4), an observation corroborated by other investigators (62). Due to the PV BMC metabolism being aerobic, the end products of the BMC metabolism (DHAP and/or pyruvate) likely enter the tricarboxylic acid (TCA) cycle and generate ATP through oxidative phosphorylation. We posit that the competitive advantage provided by the PV BMC outweighs the potential loss of energy in the form of excreted propanediol. Furthermore, although we grew P. limnophilus aerobically only, our model also supports fermentative growth on l-fucose and l-rhamnose, as the pathway would net one NADH or ATP, depending on if the phosphorylative or nonphosphorylative pathway was being utilized, respectively.
FIG 6.
Proposed model of l-fucose metabolism in P. limnophilus. Two potential pathways to reach lactaldehyde are shown. Enzymes encoded by the pvm gene cluster are shown in blue, and shell proteins are shown in green (BMC-H) or pink (BMC-P). Cofactors are depicted in gray. Enzymes that potentially catalyze reactions not directly involved in the BMC chemistry are shown in black. “n.r.” stands for no result, meaning that there was no significant BLAST hit using characterized enzymes as a query. DHAP, dihydroxyacetone phosphate.
Both pvmN, which generates an aldehyde, and pvmJ, which degrades the aldehyde, are predicted to encode a BMC encapsulation peptide (23). Similar peptides are found in a subset of enzymes associated with PDU (63, 64), PDU2, and EUT BMCs that are shown or predicted to generate or degrade an aldehyde intermediate (23). These peptides interact with shell proteins (23, 65), although it is not clear whether they associate with the inner or the outer surface of the shell; for example, it has recently been proposed that ethanolamine ammonia lyase associates with the outside the shell and “injects” acetaldehyde into the lumen (21). It is possible that pvmN is similarly associated with the outside of the shell, as it presumably would be difficult for its six-carbon substrate to efficiently diffuse across a shell designed to sequester metabolites (25). Although some crystal structures of shell proteins contain large gated pores that could allow larger metabolites across (5, 6, 66), these are thus far confined to BMC-T proteins, which PV BMC loci lack (Table 2). As ADP and ATP are slightly larger than l-fucose, we predict that the final product of the core BMC chemistry, lactyl phosphate, diffuses out of the compartment and is converted to lactate by the cytosolic product of pvmG. It must be noted, however, that the PV BMC shell gene complement is unique and could allow for hetero-oligomeric shell units that permit the larger substrates and cofactors to diffuse across the shell, functionally replacing BMC-T shell units (discussed below).
Phylogeny and evolution of metabolosomes.
An accurate phylogeny of the various BMC types has not been previously reported. In this study, we have used a core metabolic gene concatenation strategy to begin to explore the evolutionary history of functionally distinct BMCs (Fig. 1B). Although useful for our purposes, our analysis has limitations, mainly that the sample size is small (n = 67; see Table S3 in the supplemental material) and that carboxysome and EUT operons were excluded due to the lack of pduL homologs in those loci. Nevertheless, we observed that metabolosomes distinctly group by function, potentially allowing predicted catabolic BMCs to be preliminarily assigned a function based on the phylogenetic position of their core enzymes. Supporting these assignments, two classes of BMCs that use glycyl radical enzymes (GREs) emerged, and they are consistent with two different functional assignments: one presumably involved in 1,2-propanediol utilization (26, 67) (PDU2, grouping with PDU loci) and another clade, GRE, closely related to the EUT BMC, proposed to metabolize choline, a compound structurally similar to ethanolamine (68).
Notably, the PV BMC appears to be restricted to the phyla Planctomycetes and Verrucomicrobia (Fig. 1). This is a curiosity, as BMC gene clusters appear to be prone to horizontal gene transfer, explaining their widespread presence in bacterial genomes (25, 69, 70). However, there are multiple biochemical strategies used by bacteria to degrade l-fucose and l-rhamnose (55, 57, 61, 71), and the PV BMC would not be expected to provide any further fitness advantage to organisms that could already degrade those compounds; this perhaps explains their restricted distribution. A corollary to these assumptions is that ancient planctomycetes could not fully metabolize l-fucose and l-rhamnose until acquiring a BMC locus via horizontal gene transfer.
Environmental sources of PV BMC substrates.
Optimal l-fucose and l-rhamnose metabolism could be very important for planctomycete niche specialization. These sugars are commonly found in the extrapolysaccharides of bacteria and fungi (72), lipids (73), and plant and animal saccharides (53, 74, 75). Planctomycetes have been shown to attach to detritus (marine snow), which is composed of decaying plant and animal materials that most likely contain l-fucose and l-rhamnose, and are thought to be involved in carbon cycling in oceans (76). Furthermore, a recent study has shown that two planctomycetes, one marine and one freshwater, are capable of degrading several other plant-derived sugars (62). Seaweeds and other marine algae contain an abundance of sulfated polysaccharides (77), two of which are fucoidan and ulvan, which are composed of 95% l-fucose (53) and 55% l-rhamnose (75), respectively. Members of the phylum Planctomycetes dominate seaweed-associated biofilms (78), frequently co-occur with algal blooms (79), and contain an abnormal abundance of sulfatase genes that are used to degrade sulfated polysaccharides (52); all of these observations are consistent with the prediction that many Planctomycetes species use a BMC in degrading algal polysaccharides.
Sulfated polysaccharides have been associated with a plant response to salinity (80), and almost all identified sulfated polysaccharides are derived from marine algae, so it is logical that marine planctomycetes would encode the PV BMC for use in polysaccharide degradation. In addition to planctomycetes, a marine verrucomicrobium of relatively close relation to Opitutus terrae, a species that contains a pvm locus, was shown to degrade fucoidan from several algal sources (81). Intriguingly, we have shown that P. limnophilus, a freshwater planctomycete, can grow on fucoidan and that BMC components are required for its degradation. This strongly suggests that the organism is exposed to sulfated polysaccharides in its typical niche. Furthermore, several other freshwater planctomycetes also contain the pvm locus. It could be that the abundance of sulfated polysaccharides in freshwater systems has been underestimated; at least one freshwater phytoplankton species has been observed to possess a sulfated fucan (82).
Recently, a comprehensive metabolic screen of P. limnophilus suggested that the organism is specialized to degrade plant-derived sugars and polysaccharides (62). Our results confirmed these observations and further implicated the involvement of the BMC in degrading some of these compounds. Notably, the ΔpvmJ::IN(npt) strain displayed reduced metabolism on pectin, mannan, and raffinose. These sugars do not yield lactaldehyde or propionaldehyde in known degradation pathways, but they do produce some compounds, such as aldols and glyceraldehyde, that could be substrates of pvm gene products. Thus, assuming that shell formation could be regulated, enzymes within the gene cluster would still be beneficial to planctomycetes as soluble proteins, even when l-fucose- and l-rhamnose-rich compounds are not present. Whether or not the organism uses just a subset of enzymes from the gene cluster or an entire BMC is synthesized to degrade those sugars remains to be investigated.
Diversity of bacterial microcompartments.
Compared to other BMC loci, the pvm locus appears to be relatively simple, encoding only a transcriptional regulator, enzymes, and BMC-H and BMC-P shell proteins. Other BMC loci encode BMC-T shell proteins and several accessory proteins that are required to form functional organelles (23, 50, 83–86). Although some pvm loci contain hypothetical genes that could encode accessory proteins (Fig. 1A), their presence is not conserved across the various pvm loci. It will be interesting to determine if these hypothetical genes are required for BMC function and, if so, why some planctomycetes need them while others do not. Also perplexing is why the pvm locus contains three genes for BMC-P. BMC-P proteins are minor components of BMC shells: only 60 BMC-P polypeptide chains are required per icosahedral organelle to form the vertices (7, 87), and PDU, PDU2, and EUT operons each contain only one BMC-P gene. It may be the case that the BMC-P proteins encoded in the pvm locus assemble into heteropentamers to cap the vertices or incorporate into the facets of the shell with the BMC-H subunits.
The PV BMC marks the sixth experimentally characterized function for a BMC, joining the carboxysome, PDU, EUT, PDU2, and ethanol-utilizing ETU (88, 89) BMCs. Metabolosome specialization can be correlated with the environmental niche of the organism. For example, S. enterica and E. coli are enteric bacteria that are generally exposed to anaerobic conditions in the gut. When they are provided with l-fucose as a carbon source, the sugar is anaerobically degraded to 1,2-propanediol, which is then excreted (71). When the pdu operon was acquired by an ancestral strain, it most likely provided a substantial fitness advantage; the organism could now utilize the propanediol previously wasted (69). Similarly, C. phytofermentans is an anaerobe associated with plant cell wall degradation and would also be expected to ferment l-fucose and l-rhamnose, which are common components of plant cell wall polysaccharides, such as the pectin rhamnogalacturonan I. On the other hand, Planctomycetes and Verrucomicrobia that contain pvm loci are usually associated with aerobic habitats, and some are even strict aerobes and would not need to ferment l-fucose and l-rhamnose but instead degrade them (and potentially other polysaccharides) aerobically through the PV BMC.
With continued sequencing of microbial genomes, the phylogenetic and functional diversity of metabolosomes is increasingly apparent, while the core biochemistry is strikingly conserved. Based on our comparison of the PV metabolosome to other characterized examples, the key difference among metabolosome types is the identity of the enzyme that generates the aldehyde; the biochemical transformations downstream of aldehyde generation are remarkably similar. Coupled with the observation that BMCs tend to be organized in readily transmissible units (i.e., tightly clustered loci), it is possible to envision that new functional types of BMCs could have evolved simply by horizontally transferring a section of a metabolosome locus that encodes core enzymes and shell proteins. All the core enzymes would need to be transferred, as cofactor recycling is necessary for the macromolecular biochemical machine to proceed. Assuming a certain degree of enzymatic promiscuity, the transplanted BMC would degrade short-chain aldehydes (similar to propionaldehyde, lactaldehyde, and acetaldehyde), provide a new metabolic function for the host microbe, and over time be specialized by selective pressures into a new functional type of BMC. This could account for both the apparent biochemical diversity and the pervasiveness of metabolosomes among the bacteria and could have implications for bacterial speciation. Carboxysomes, in contrast, are essentially anabolic BMCs; they do not share the core biochemistry of the catabolic metabolosomes. Likewise, understanding their position in the evolutionary history of BMCs is required to develop a comprehensive theory of BMC evolution and phylogenesis.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by the NSF (EF1105892).
The p2741 vector was a generous gift from Christopher Anderson. We thank the Kerfeld laboratory for critical and collegial discussions and Jan Zarzycki for a critical reading of the manuscript. We thank Rick I. Webb for helping to interpret electron micrographs.
Footnotes
Published ahead of print 31 January 2014
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.03887-13.
REFERENCES
- 1.Drews G, Niklowitz W. 1956. Beiträge zur Cytologie der Blaualgen II. Mitteilung Zentroplasma und granuläre Einschlüsse von Phormidium uneinatum. Arch. Mikrobiol. 24:147–162 [PubMed] [Google Scholar]
- 2.Shively JM, Decker GL, Greenawalt JW. 1970. Comparative ultrastructure of the thiobacilli. J. Bacteriol. 101:618–627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Shively JM, Ball F, Brown DH, Saunders RE. 1973. Functional organelles in prokaryotes: polyhedral inclusions (carboxysomes) of Thiobacillus neapolitanus. Science 182:584–586. 10.1126/science.182.4112.584 [DOI] [PubMed] [Google Scholar]
- 4.Kerfeld CA, Sawaya MR, Tanaka S, Nguyen CV, Phillips M, Beeby M, Yeates TO. 2005. Protein structures forming the shell of primitive bacterial organelles. Science 309:936–938. 10.1126/science.1113397 [DOI] [PubMed] [Google Scholar]
- 5.Klein MG, Zwart P, Bagby SC, Cai F, Chisholm SW, Heinhorst S, Cannon GC, Kerfeld CA. 2009. Identification and structural analysis of a novel carboxysome shell protein with implications for metabolite transport. J. Mol. Biol. 392:319–333. 10.1016/j.jmb.2009.03.056 [DOI] [PubMed] [Google Scholar]
- 6.Cai F, Sutter M, Cameron JC, Stanley DN, Kinney JN, Kerfeld CA. 2013. The structure of CcmP, a tandem bacterial microcompartment domain protein from the β-carboxysome, forms a subcompartment within a microcompartment. J. Biol. Chem. 288:16055–16063. 10.1074/jbc.M113.456897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tanaka S, Kerfeld CA, Sawaya MR, Cai F, Heinhorst S, Cannon GC, Yeates TO. 2008. Atomic-level models of the bacterial carboxysome shell. Science 319:1083–1086. 10.1126/science.1151458 [DOI] [PubMed] [Google Scholar]
- 8.Sutter M, Wilson SC, Deutsch S, Kerfeld CA. 2013. Two new high-resolution crystal structures of carboxysome pentamer proteins reveal high structural conservation of CcmL orthologs among distantly related cyanobacterial species. Photosynth. Res. 10.1007/s11120-013-9909-z [DOI] [PubMed] [Google Scholar]
- 9.Kofoid E, Rappleye C, Stojiljkovic I, Roth J. 1999. The 17-gene ethanolamine (eut) operon of Salmonella typhimurium encodes five homologues of carboxysome shell proteins. J. Bacteriol. 181:5317–5329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Stojiljkovic I, Bäumler AJ, Heffron F. 1995. Ethanolamine utilization in Salmonella typhimurium: nucleotide sequence, protein expression, and mutational analysis of the cchA cchB eutE eutJ eutG eutH gene cluster. J. Bacteriol. 177:1357–1366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen P, Andersson DI, Roth JR. 1994. The control region of the pdu/cob regulon in Salmonella typhimurium. J. Bacteriol. 176:5474–5482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bobik TA, Havemann GD, Busch RJ, Williams DS, Aldrich HC. 1999. The propanediol utilization (pdu) operon of Salmonella enterica serovar Typhimurium LT2 includes genes necessary for formation of polyhedral organelles involved in coenzyme B12-dependent 1,2-propanediol degradation. J. Bacteriol. 181:5967–5975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Brinsmade SR, Paldon T, Jorge C, Escalante-Semerena JC. 2005. Minimal functions and physiological conditions required for growth of Salmonella enterica on ethanolamine in the absence of the metabolosome. J. Bacteriol. 187:8039–8046. 10.1128/JB.187.23.8039-8046.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Penrod JT, Roth JR. 2006. Conserving a volatile metabolite: a role for carboxysome-like organelles in Salmonella enterica. J. Bacteriol. 188:2865–2874. 10.1128/JB.188.8.2865-2874.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sampson EM, Bobik TA. 2008. Microcompartments for B12-dependent 1,2-propanediol degradation provide protection from DNA and cellular damage by a reactive metabolic intermediate. J. Bacteriol. 190:2966–2971. 10.1128/JB.01925-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Price GD, Badger MR. 1989. Expression of human carbonic anhydrase in the cyanobacterium Synechococcus PCC7942 creates a high CO2-requiring phenotype; evidence for a central role for carboxysomes in the CO2 concentrating mechanism. Plant Physiol. 91:505–513. 10.1104/pp.91.2.505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dou Z, Heinhorst S, Williams EB, Murin CD, Shively JM, Cannon GC. 2008. CO2 fixation kinetics of Halothiobacillus neapolitanus mutant carboxysomes lacking carbonic anhydrase suggest the shell acts as a diffusional barrier for CO2. J. Biol. Chem. 283:10377–10384. 10.1074/jbc.M709285200 [DOI] [PubMed] [Google Scholar]
- 18.Roof DM, Roth JR. 1988. Ethanolamine utilization in Salmonella typhimurium. J. Bacteriol. 170:3855–3863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Leal NA, Havemann GD, Bobik TA. 2003. PduP is a coenzyme-A-acylating propionaldehyde dehydrogenase associated with the polyhedral bodies involved in B12-dependent 1,2-propanediol degradation by Salmonella enterica serovar Typhimurium LT2. Arch. Microbiol. 180:353–361. 10.1007/s00203-003-0601-0 [DOI] [PubMed] [Google Scholar]
- 20.Cheng S, Fan C, Sinha S, Bobik TA. 2012. The PduQ enzyme is an alcohol dehydrogenase used to recycle NAD+ internally within the Pdu microcompartment of Salmonella enterica. PLoS One 7:e47144. 10.1371/journal.pone.0047144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Huseby DL, Roth JR. 2013. Evidence that a metabolic microcompartment contains and recycles private cofactor pools. J. Bacteriol. 195:2864–2879. 10.1128/JB.02179-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Palacios S, Starai VJ, Escalante-Semerena JC. 2003. Propionyl coenzyme A is a common intermediate in the 1,2-propanediol and propionate catabolic pathways needed for expression of the prpBCDE operon during growth of Salmonella enterica on 1,2-propanediol. J. Bacteriol. 185:2802–2810. 10.1128/JB.185.9.2802-2810.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kinney JN, Salmeen A, Cai F, Kerfeld CA. 2012. Elucidating the essential role of the conserved carboxysomal protein CcmN reveals a common feature of bacterial microcompartment assembly. J. Biol. Chem. 287:17729–17736. 10.1074/jbc.M112.355305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jorda J, Lopez D, Wheatley NM, Yeates TO. 2013. Using comparative genomics to uncover new kinds of protein-based metabolic organelles in bacteria. Protein Sci. 22:179–195. 10.1002/pro.2196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kerfeld CA, Heinhorst S, Cannon GC. 2010. Bacterial microcompartments. Annu. Rev. Microbiol. 64:391–408. 10.1146/annurev.micro.112408.134211 [DOI] [PubMed] [Google Scholar]
- 26.Petit E, Latouf WG, Coppi MV, Warnick TA, Currie D, Romashko I, Deshpande S, Haas K, Alvelo-Maurosa JG, Wardman C, Schnell DJ, Leschine SB, Blanchard JL. 2013. Involvement of a bacterial microcompartment in the metabolism of fucose and rhamnose by Clostridium phytofermentans. PLoS One 8:e54337. 10.1371/journal.pone.0054337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lee K-C, Webb RI, Janssen PH, Sangwan P, Romeo T, Staley JT, Fuerst JA. 2009. Phylum Verrucomicrobia representatives share a compartmentalized cell plan with members of bacterial phylum Planctomycetes. BMC Microbiol. 9:5. 10.1186/1471-2180-9-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wagner M, Horn M. 2006. The Planctomycetes, Verrucomicrobia, Chlamydiae and sister phyla comprise a superphylum with biotechnological and medical relevance. Curr. Opin. Biotechnol. 17:241–249. 10.1016/j.copbio.2006.05.005 [DOI] [PubMed] [Google Scholar]
- 29.van Niftrik L, Jetten MSM. 2012. Anaerobic ammonium-oxidizing bacteria: unique microorganisms with exceptional properties. Microbiol. Mol. Biol. Rev. 76:585–596. 10.1128/MMBR.05025-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fukunaga Y, Kurahashi M, Sakiyama Y, Ohuchi M, Yokota A, Harayama S. 2009. Phycisphaera mikurensis gen. nov., sp. nov., isolated from a marine alga, and proposal of Phycisphaeraceae fam. nov., Phycisphaerales ord. nov. and Phycisphaerae classis nov. in the phylum Planctomycetes. J. Gen. Appl. Microbiol. 55:267–275. 10.2323/jgam.55.267 [DOI] [PubMed] [Google Scholar]
- 31.Hirsch P, Muller M. 1985. Planctomyces limnophilus sp. nov., a stalked and budding bacterium from freshwater. Syst. Appl. Microbiol. 6:276–280. 10.1016/S0723-2020(85)80031-X [DOI] [Google Scholar]
- 32.Anderson JC, Dueber JE, Leguia M, Wu GC, Goler JA, Arkin AP, Keasling JD. 2010. BglBricks: a flexible standard for biological part assembly. J. Biol. Eng. 4:1–12. 10.1186/1754-1611-4-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jogler C, Glöckner FO, Kolter R. 2011. Characterization of Planctomyces limnophilus and development of genetic tools for its manipulation establish it as a model species for the phylum Planctomycetes. Appl. Environ. Microbiol. 77:5826–5829. 10.1128/AEM.05132-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Edgar RC. 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32:1792–1797. 10.1093/nar/gkh340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Edgar RC. 2004. MUSCLE: a multiple sequence alignment method with reduced time and space complexity. BMC Bioinformatics 5:113. 10.1186/1471-2105-5-113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Castresana J. 2000. Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol. Biol. Evol. 17:540–552. 10.1093/oxfordjournals.molbev.a026334 [DOI] [PubMed] [Google Scholar]
- 37.Guindon S, Dufayard J-F, Lefort V, Anisimova M, Hordijk W, Gascuel O. 2010. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst. Biol. 59:307–321. 10.1093/sysbio/syq010 [DOI] [PubMed] [Google Scholar]
- 38.Dereeper A, Guignon V, Blanc G, Audic S, Buffet S, Chevenet F, Dufayard J-F, Guindon S, Lefort V, Lescot M, Claverie J-M, Gascuel O. 2008. Phylogeny.fr: robust phylogenetic analysis for the non-specialist. Nucleic Acids Res. 36:W465–W469. 10.1093/nar/gkn180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dereeper A, Audic S, Claverie J-M, Blanc G. 2010. BLAST-EXPLORER helps you building datasets for phylogenetic analysis. BMC Evol. Biol. 10:8. 10.1186/1471-2148-10-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Waterhouse AM, Procter JB, Martin DMA, Clamp M, Barton GJ. 2009. Jalview version 2—a multiple sequence alignment editor and analysis workbench. Bioinformatics 25:1189–1191. 10.1093/bioinformatics/btp033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.McDonald KL, Webb RI. 2011. Freeze substitution in 3 hours or less. J. Microsc. 243:227–233. 10.1111/j.1365-2818.2011.03526.x [DOI] [PubMed] [Google Scholar]
- 42.Reynolds ES. 1963. The use of lead citrate at high pH as an electron-opaque stain in electron microscopy. J. Cell Biol. 17:208–212. 10.1083/jcb.17.1.208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kamneva OK, Knight SJ, Liberles DA, Ward NL. 2012. Analysis of genome content evolution in PVC bacterial super-phylum: assessment of candidate genes associated with cellular organization and life-style. Genome Biol. Evol. 4:1–50. 10.1093/gbe/evr123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ward NL, Rainey FA, Hedlund BP, Staley JT, Ludwig W, Stackebrandt E. 2000. Comparative phylogenetic analyses of members of the order Planctomycetales and the division Verrucomicrobia: 23S rRNA gene sequence analysis supports the 16S rRNA gene sequence-derived phylogeny. Int. J. Syst. Evol. Microbiol. 50:1965–1972. 10.1099/00207713-50-6-1965 [DOI] [PubMed] [Google Scholar]
- 45.Gupta RS, Bhandari V, Naushad HS. 2012. Molecular signatures for the PVC clade (Planctomycetes, Verrucomicrobia, Chlamydiae, and Lentisphaerae) of bacteria provide insights into their evolutionary relationships. Front. Microbiol. 3:327. 10.3389/fmicb.2012.00327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yin Y, Zhang H, Olman V, Xu Y. 2010. Genomic arrangement of bacterial operons is constrained by biological pathways encoded in the genome. Proc. Natl. Acad. Sci. U. S. A. 107:6310–6315. 10.1073/pnas.0911237107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Schreier HJ, Dejtisakdi W, Escalante JO, Brailo M. 2012. Transposon mutagenesis of Planctomyces limnophilus and analysis of a pckA mutant. Appl. Environ. Microbiol. 78:7120–7123. 10.1128/AEM.01794-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wolf YI, Koonin EV. 2012. A tight link between orthologs and bidirectional best hits in bacterial and archaeal genomes. Genome Biol. Evol. 4:1286–1294. 10.1093/gbe/evs100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Fondi M, Emiliani G, Fani R. 2009. Origin and evolution of operons and metabolic pathways. Res. Microbiol. 160:502–512. 10.1016/j.resmic.2009.05.001 [DOI] [PubMed] [Google Scholar]
- 50.Cheng S, Sinha S, Fan C, Liu Y, Bobik TA. 2011. Genetic analysis of the protein shell of the microcompartments involved in coenzyme B12-dependent 1,2-propanediol degradation by Salmonella. J. Bacteriol. 193:1385–1392. 10.1128/JB.01473-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Cheng S, Liu Y, Crowley CS, Yeates TO, Bobik TA. 2008. Bacterial microcompartments: their properties and paradoxes. Bioessays 30:1084–1095. 10.1002/bies.20830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wegner C-E, Richter-Heitmann T, Klindworth A, Klockow C, Richter M, Achstetter T, Glöckner FO, Harder J. 2012. Expression of sulfatases in Rhodopirellula baltica and the diversity of sulfatases in the genus Rhodopirellula. Mar. Genomics 9:51–61. 10.1016/j.margen.2012.12.001 [DOI] [PubMed] [Google Scholar]
- 53.Morya VK, Kim J, Kim E-K. 2012. Algal fucoidan: structural and size-dependent bioactivities and their perspectives. Appl. Microbiol. Biotechnol. 93:71–82. 10.1007/s00253-011-3666-8 [DOI] [PubMed] [Google Scholar]
- 54.Boronat A, Aguilar J. 1981. Metabolism of l-fucose and l-rhamnose in Escherichia coli: differences in induction of propanediol oxidoreductase. J. Bacteriol. 147:181–185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yew WS, Fedorov AA, Fedorov EV, Rakus JF, Pierce RW, Almo SC, Gerlt JA. 2006. Evolution of enzymatic activities in the enolase superfamily: l-fuconate dehydratase from Xanthomonas campestris. Biochemistry 45:14582–14597. 10.1021/bi061687o [DOI] [PubMed] [Google Scholar]
- 56.Watanabe S, Piyanart S, Makino K. 2008. Metabolic fate of l-lactaldehyde derived from an alternative l-rhamnose pathway. FEBS J. 275:5139–5149. 10.1111/j.1742-4658.2008.06645.x [DOI] [PubMed] [Google Scholar]
- 57.Watanabe S, Makino K. 2009. Novel modified version of nonphosphorylated sugar metabolism—an alternative l-rhamnose pathway of Sphingomonas sp. FEBS J. 276:1554–1567. 10.1111/j.1742-4658.2009.06885.x [DOI] [PubMed] [Google Scholar]
- 58.Koonin EV, Mushegian AR, Bork P. 1996. Non-orthologous gene displacement. Trends Genet. 12:334–336 [PubMed] [Google Scholar]
- 59.Chiu T, Feingoldt DS. 1969. l-Rhamnulose 1-phosphate aldolase from Escherichia coli. Crystallization and properties. Biochemistry 8:98–108 [DOI] [PubMed] [Google Scholar]
- 60.Ghalambor MA, Heath EC. 1962. The metabolism of l-fucose. II. The enzymatic cleavage of l-fuculose 1-phosphate. J. Biol. Chem. 237:2427–2433 [PubMed] [Google Scholar]
- 61.Watanabe S, Saimura M, Makino K. 2008. Eukaryotic and bacterial gene clusters related to an alternative pathway of nonphosphorylated l-rhamnose metabolism. J. Biol. Chem. 283:20372–20382. 10.1074/jbc.M801065200 [DOI] [PubMed] [Google Scholar]
- 62.Jeske O, Jogler M, Petersen J, Sikorski J, Jogler C. 2013. From genome mining to phenotypic microarrays: Planctomycetes as source for novel bioactive molecules. Antonie Van Leeuwenhoek 104:551–567. 10.1007/s10482-013-0007-1 [DOI] [PubMed] [Google Scholar]
- 63.Fan C, Cheng S, Liu Y, Escobar CM, Crowley CS, Jefferson RE, Yeates TO, Bobik TA. 2010. Short N-terminal sequences package proteins into bacterial microcompartments. Proc. Natl. Acad. Sci. U. S. A. 107:7509–7514. 10.1073/pnas.0913199107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Fan C, Bobik TA. 2011. The N-terminal region of the medium subunit (PduD) packages adenosylcobalamin-dependent diol dehydratase (PduCDE) into the Pdu microcompartment. J. Bacteriol. 193:5623–5628. 10.1128/JB.05661-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Fan C, Cheng S, Sinha S, Bobik TA. 2012. Interactions between the termini of lumen enzymes and shell proteins mediate enzyme encapsulation into bacterial microcompartments. Proc. Natl. Acad. Sci. U. S. A. 109:14995–15000. 10.1073/pnas.1207516109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Tanaka S, Sawaya MR, Yeates TO. 2010. Structure and mechanisms of a protein-based organelle in Escherichia coli. Science 327:81–84. 10.1126/science.1179513 [DOI] [PubMed] [Google Scholar]
- 67.Scott KP, Martin JC, Campbell G, Mayer C-D, Flint HJ. 2006. Whole-genome transcription profiling reveals genes up-regulated by growth on fucose in the human gut bacterium “Roseburia inulinivorans.” J. Bacteriol. 188:4340–4349. 10.1128/JB.00137-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Craciun S, Balskus EP. 2012. Microbial conversion of choline to trimethylamine requires a glycyl radical enzyme. Proc. Natl. Acad. Sci. U. S. A. 109:21307–21312. 10.1073/pnas.1215689109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Lawrence JG, Roth JR. 1996. Selfish operons: horizontal transfer may drive the evolution of gene clusters. Genetics 143:1843–1860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Hess WR, Rocap G, Ting CS, Larimer F, Stilwagen S, Lamerdin J, Chisholm SW. 2001. The photosynthetic apparatus of Prochlorococcus: insights through comparative genomics. Photosynth. Res. 70:53–71. 10.1023/A:1013835924610 [DOI] [PubMed] [Google Scholar]
- 71.Cocks GT, Aguilar J, Lin ECC. 1974. Evolution of l-1,2-propanediol catabolism in Escherichia coli by recruitment of enzymes for l-fucose and l-lactate metabolism. J. Bacteriol. 118:83–88 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Vanhooren PT, Vandamme EJ. 1999. l-Fucose: occurrence, physiological role, chemical, enzymatic and microbial synthesis. J. Chem. Technol. Biotechnol. 74:479–497. [DOI] [Google Scholar]
- 73.Linhardt RJ, Bakhit R, Daniels L, Mayerl F, Pickenhagen W. 1989. Microbially produced rhamnolipid as a source of rhamnose. Biotechnol. Bioeng. 33:365–368. 10.1002/bit.260330316 [DOI] [PubMed] [Google Scholar]
- 74.Staudacher E, Altmann F, Wilson IB, März L. 1999. Fucose in N-glycans: from plant to man. Biochim. Biophys. Acta 1473:216–236. 10.1016/S0304-4165(99)00181-6 [DOI] [PubMed] [Google Scholar]
- 75.Jaulneau V, Lafitte C, Jacquet C, Fournier S, Salamagne S, Briand X, Esquerré-Tugayé M-T, Dumas B. 2010. Ulvan, a sulfated polysaccharide from green algae, activates plant immunity through the jasmonic acid signaling pathway. J. Biomed. Biotechnol. 2010:525291. 10.1155/2010/525291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Fuerst JA, Sagulenko E. 2011. Beyond the bacterium: planctomycetes challenge our concepts of microbial structure and function. Nat. Rev. Microbiol. 9:403–413. 10.1038/nrmicro2578 [DOI] [PubMed] [Google Scholar]
- 77.Jiao G, Yu G, Zhang J, Ewart HS. 2011. Chemical structures and bioactivities of sulfated polysaccharides from marine algae. Mar. Drugs 9:196–223. 10.3390/md9020196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Lage OM, Bondoso J. 2011. Planctomycetes diversity associated with macroalgae. FEMS Microbiol. Ecol. 78:366–375. 10.1111/j.1574-6941.2011.01168.x [DOI] [PubMed] [Google Scholar]
- 79.Morris RM, Longnecker K, Giovannoni SJ. 2006. Pirellula and OM43 are among the dominant lineages identified in an Oregon coast diatom bloom. Environ. Microbiol. 8:1361–1370. 10.1111/j.1462-2920.2006.01029.x [DOI] [PubMed] [Google Scholar]
- 80.Aquino RS, Grativol C, Mourão PAS. 2011. Rising from the sea: correlations between sulfated polysaccharides and salinity in plants. PLoS One 6:e18862. 10.1371/journal.pone.0018862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Sakai T, Ishizuka K, Kato I. 2003. Isolation and characterization of a fucoidan-degrading marine bacterium. Mar. Biotechnol. 5:409–416. 10.1007/s10126-002-0118-6 [DOI] [PubMed] [Google Scholar]
- 82.Giroldo D, Vieira AAH. 2002. An extracellular sulfated fucose-rich polysaccharide produced by a tropical strain of Cryptomonas obovata (Cryptophyceae). J. Appl. Phycol. 14:185–191. 10.1023/A:1019972109619 [DOI] [Google Scholar]
- 83.Rae BD, Long BM, Badger MR, Price GD. 2012. Structural determinants of the outer shell of β-carboxysomes in Synechococcus elongatus PCC 7942: roles for CcmK2, K3-K4, CcmO, and CcmL. PLoS One 7:e43871. 10.1371/journal.pone.0043871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Long BM, Badger MR, Whitney SM, Price GD. 2007. Analysis of carboxysomes from Synechococcus PCC7942 reveals multiple Rubisco complexes with carboxysomal proteins CcmM and CcaA. J. Biol. Chem. 282:29323–29335. 10.1074/jbc.M703896200 [DOI] [PubMed] [Google Scholar]
- 85.Sinha S, Cheng S, Fan C, Bobik TA. 2012. The PduM protein is a structural component of the microcompartments involved in coenzyme B12-dependent 1,2-propanediol degradation by Salmonella enterica. J. Bacteriol. 194:1912–1918. 10.1128/JB.06529-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Cameron JC, Wilson SC, Bernstein SL, Kerfeld CA. 2013. Biogenesis of a bacterial organelle: the carboxysome assembly pathway. Cell 155:1131–1140. 10.1016/j.cell.2013.10.044 [DOI] [PubMed] [Google Scholar]
- 87.Cai F, Menon BB, Cannon GC, Curry KJ, Shively JM, Heinhorst S. 2009. The pentameric vertex proteins are necessary for the icosahedral carboxysome shell to function as a CO2 leakage barrier. PLoS One 4:e7521. 10.1371/journal.pone.0007521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Lurz R, Mayer F, Gottschalk G. 1979. Electron microscopic study on the quaternary structure of the isolated particulate alcohol-acetaldehyde dehydrogenase complex and on its identity with the polygonal bodies of Clostridium kluyveri. Arch. Microbiol. 120:255–262. 10.1007/BF00423073 [DOI] [Google Scholar]
- 89.Heldt D, Frank S, Seyedarabi A, Ladikis D, Parsons JB, Warren MJ, Pickersgill RW. 2009. Structure of a trimeric bacterial microcompartment shell protein, EtuB, associated with ethanol utilization in Clostridium kluyveri. Biochem. J. 423:199–207. 10.1042/BJ20090780 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.