Abstract
The spore-forming bacterium Paenibacillus larvae causes a severe and highly infective bee disease, American foulbrood (AFB). Despite the large economic losses induced by AFB, the virulence factors produced by P. larvae are as yet unknown. To identify such virulence factors, we experimentally infected young, susceptible larvae of the honeybee, Apis mellifera carnica, with different P. larvae isolates. Honeybee larvae were reared in vitro in 24-well plates in the laboratory after isolation from the brood comb. We identified genotype-specific differences in the etiopathology of AFB between the tested isolates of P. larvae, which were revealed by differences in the median lethal times. Furthermore, we confirmed that extracts of P. larvae cultures contain low-molecular-weight compounds, which are toxic to honeybee larvae. Our data indicate that P. larvae secretes metabolites into the medium with a potent honeybee toxic activity pointing to a novel pathogenic factor(s) of P. larvae. Genome mining of P. larvae subsp. larvae BRL-230010 led to the identification of several biosynthesis gene clusters putatively involved in natural product biosynthesis, highlighting the potential of P. larvae to produce such compounds.
INTRODUCTION
Honeybees are the most important economic and ecological pollinators of wild plants and insect-pollinated cultivated crops. About 35% of the global crop-based food production benefits from animal-mediated pollination, with bees being the primary pollinators for most of the crops requiring animal pollination (1). One factor that contributes to their high pollinator efficiency is the high population densities in bee hives. On the other hand, this high density makes bees especially vulnerable to various pathogens, such as viruses, mites, and bacteria. Among them, Paenibacillus larvae is the most virulent pathogenic bacterium. It is a spore-forming, Gram-positive, rod-shaped, and obligatory pathogenic bacterium that has to kill its host to form novel spores, which can then be transmitted further. It forms extremely tenacious spores that remain infectious for over half a century (2, 3). The bacterium has a highly pathogenic reproduction rate, producing a huge number of spores within each infected larva (4). Strains of P. larvae can be subdivided into four different genotypes that differ in their virulence (5, 6). While larvae infected with P. larvae genotypes ERIC II to ERIC IV were killed within 6 to 7 days, it took ERIC I around 12 to 14 days to kill all infected individuals. Therefore, genotype ERIC I was considered to be less virulent to bee larvae than the other three genotypes (2, 5). The different lethal times (times to 100% mortality of bees [LT100]) are relevant because infection symptoms or deaths may be detected by nurse bees more frequently. Removing infected bees earlier may be beneficial for the whole colony.
P. larvae is the only known causative pathogen of American foulbrood (AFB) (5, 7, 8). The disease has spread worldwide (9, 10) and is causing a significant decrease in honeybee populations and significant drops of honey, pollen, propolis, royal jelly, and beeswax production (11). AFB is a highly deleterious bacterial honeybee disease that only affects the larval stages of the honeybee (2, 12), with 24- to 48-h-old larvae being most susceptible to P. larvae (2). Adult worker bees become contaminated while removing dead larvae, which probably is an important infection pathway to spread the infectious spores in the colony. The infection begins when spore-contaminated food is swallowed by the larvae. After germination in the larval midgut, vegetative cells proliferate and translocate into the hemocoel, probably by penetrating the midgut epithelium, and consecutively spread within the hemolymph, causing septicemia and ultimately killing the larvae (13). After the larvae die, their tissue decays while the infected larval body changes to a brownish and viscous mass (ropy stage). It next dries to a hard scale, which tightly adheres to the lower cell wall and contains millions of spores (4, 12–15). Although it was commonly assumed that the honeybee, Apis mellifera, is the only host species, P. larvae may cause bacteremic infections in humans when the spores are unintentionally self-injected, e.g., via honey-prepared methadone by drug users with a suppressed immune system (16).
Despite the wealth of studies about AFB and P. larvae, relatively little is known about the chemicals causing larval toxicity. This is even more astonishing given that several bioactive compounds have been described from other Paenibacillus species (17–24). Other entomopathogenic bacteria (25, 26) produce and secrete many different pathogenic compounds, for example, proteins, antibiotics, or polypeptides. This has also been observed for P. larvae, which produced a broad spectrum of antibacterial compounds, probably suppressing potential competitors in the honeybee larvae cadaver (27). Moreover, it secretes proteases, which may act as virulence factors during the infection process (28).
The AB toxins Plx1 and Plx2 are among the virulence factors of P. larvae and have been identified in the genome of P. larvae ERIC I but not in the genome of ERIC II (29). They show considerable homology to MTX1 from Lysinibacillus sphaericus (30). MTX1 was described as a toxin that induces rounding up of host cells (31). This effect is needed during the P. larvae infection pathway while breaching the midgut epithelium to invade the host's hemocoel (13).
The aim of this study was to determine whether P. larvae secretes low-molecular-weight compounds which are toxic to honeybee larvae reared in vitro under controlled experimental conditions without social brood care interference. Thus, an in vitro rearing of honeybee larvae had to be established, and different strains of P. larvae were analyzed using the bee larvae as a bioassay as well as by analytical chemistry methods. Additionally, the available genomes of two strains of P. larvae were analyzed regarding gene clusters which might be involved in the biosynthesis of these virulence factors.
MATERIALS AND METHODS
Honeybees.
All experiments were carried out on larvae from Apis mellifera carnica. Larvae derived from the progeny of a single wild-mated queen were maintained in an AFB symptom-free apiary at the Institut für Bienenkunde, Oberursel, Germany. The experiments were performed between July and August 2010.
Artificial rearing.
Larvae were reared in 24-well tissue plates at 35°C and 95% rH according to published methods (6, 32). First-instar larvae (ca. 12 h; larval age was estimated by size), which were grafted from worker brood cells, were used throughout the experiments. The larvae were carefully removed from their cells using a grafting tool (Schweizer Umlarvlöffel, Carl Fritz Imkertechnik GmbH & Co. KG, Mellrichstadt, Germany). A 24-well tissue culture plate (3.4 ml/well; Orange Scientific, Braine-l'Alleud, Belgium) was selected as the larval and the postdefecation rearing container. One larva was placed into each well onto 100 μl of the larva diet. During the first 5 days, larvae were transferred every 24 h to a fresh well with a new spore-free diet. From day 6 until defecation, larvae were grafted every 48 h to reduce handling stress. The infectious larval diet was fed only during the first 24 h after grafting. During extract and supernatant tests, larvae were fed from day one until they defecated or died. Larvae were fed daily ad libitum (100 μl) with an excess of a liquid larva diet consisting of 50% (wt/wt) royal jelly (Imkerei Ullmann, Erlensee, Germany), 6% (wt/vol) glucose (Sigma-Aldrich, Deutschland), and 6% (wt/vol) fructose (Carl Roth GmbH, Karlsruhe, Deutschland) in sterile deionized water. The transfer of the growing larvae to a new well with fresh diet took place on a heating plate adjusted to about 34°C, and dead larvae were removed. For the transfer of larger larvae, a rounded sterile metal spatula was used. After defecation (approximately at day 9), the larvae were transferred to pupation wells lined with paper tissues. Prior to the transfer into pupation wells, larvae were placed and rolled briefly on absorbent paper to remove adhering larval diet and feces. The larvae were maintained in an incubator (type 6120; Heraeus Instruments) at 35°C and 96% rH. The experiments were terminated at day 15, because most bees have reached the prepupal stage by then. Clinical symptoms of AFB should have been diagnosed before this point, such that only a little additional mortality is predicted.
Vital parameter estimation.
Each day the larvae from control and infected groups were examined under a stereo microscope. The presence of larval exuvia, signs of respiration and injury, disease symptoms, color change, infestation of fungal infection, or other abnormalities were monitored and dead larvae were removed. The number of dead larvae on each plate was determined, and surviving larvae were transferred to fresh food.
Bacterial strains.
The P. larvae strains DSM 16116, DSM 16115, and DSM 17237 used in this study were obtained from the Deutsche Sammlung von Mikroorganismen und Zellkulturen GmbH (DSMZ, Braunschweig, Germany). They represent German field isolates of the year 2003 and were genotype ERIC II or AB (33, 34). NRLL B-41635 was obtained from the United States Department of Agriculture (Agricultural Research Service [ARS]). The genome of strain P. larvae subsp. larvae BRL-230010 was sequenced recently (35) and was used for the identification of biosynthesis gene clusters. NRLL B-41635 was cultivated in MYPGP media for showing negative effects on larva development as described previously (36, 37). All other strains were grown in J medium (38).
Exposure bioassay.
Cultures containing spores of all four tested P. larvae strains were diluted 1:25 in sugar solution (6% fructose, 6% glucose; described above) and subsequently mixed with 50% (vol/vol) royal jelly. This resulted in a 1:50 dilution of the P. larvae cultures in the larval diet. The spore-contaminated larval diet was prepared freshly immediately before each experiment and fed to larvae only once a day during the in vitro breeding experiment. Spore suspensions were stored at 4°C before they were used for experiments.
Due to their low water solubility, Amberlite XAD16 extracts were dissolved first in methanol (final methanol concentration in the diet, <1%). This solution was mixed with larval diet by dissolving them first in the sugar solution and mixed with 50% (vol/vol) royal jelly. To control for toxic effects of methanol on developing honeybee larvae, a 1% methanol control was run in parallel. As a further control, larvae were fed a diet containing only MYPGP or J medium.
The Amberlite XAD16 extract of the most virulent strain, DSM 16116, was fractionated further using solid-phase extraction, leading to five fractions (0.1% trifluoroacetic acid [TFA], 10% acetonitrile [ACN], 50% ACN, 99.9% ACN, and flowthrough) in addition to the full Amberlite XAD16 extract (VT1) from P. larvae strain DSM 16116.
Cultivation of P. larvae and extraction of secreted metabolites.
All tested Paenibacillus larvae strains were kept on Difco Columbia blood agar base (Becton, Dickinson and Company, Sparks, MD) containing 5% defibrinated sheep blood or J medium agar plates, and single colonies were used for the inoculation of liquid cultures. For infection of larvae with different isolates of P. larvae, bacterial strains were cultivated for a maximum of 5 days at 34°C in 75 ml of J medium (38) containing (all descriptions are in wt/vol) 0.5% tryptone, 1.5% yeast extract (both from Becton, Dickinson, and Company, Sparks, MD), 0.3% K2HPO4, and 0.5% glucose, which was separately added after sterilization (both from Carl Roth GmbH, Karlsruhe, Germany). Alternatively, strains were kept in 75 ml MYPGP medium (36, 37) containing 1.5% yeast extract, 0.3% K2PO4, 0.2% glucose, and 0.1% sodium pyruvate, as well as 1.0% Mueller-Hinton broth (CM0405; Oxoid). In the case of those cultures, which would later be used for infection of honeybee larvae, 0.01% (wt/vol) mangan sulfate was added to the cultivation media in order to maximize the sporulation activity, since infection of A. melifera larvae is caused by germinated P. larvae spores.
For metabolite extraction and successive toxicity tests, P. larvae strains were cultivated at 34°C for 48 h under permanent shaking at 120 rpm in 500-ml baffled flasks and for an additional cultivation period of 24 h under identical conditions as described above with 2% Amberlite XAD16 (Sigma-Aldrich Chemie GmbH, Steinheim, Germany) added to the culture medium. Cultures were harvested by removing the Amberlite XAD16 beads after a 72-h total cultivation time. Two culture volumes of analytical reagent-grade methanol (Thermo Scientific, Loughborough, United Kingdom) were added to the loaded Amberlite XAD16 to extract resin-bound metabolites. After 1 h under permanent shaking, the extract was decanted and centrifuged for 20 min at 4,000 rpm in a Megafuge 10R (Heraeus Sepatech, Hanau, Germany). To remove residual CFU, the supernatant was filtered through a polytetrafluoroethylene (PTFE) filter (pore size, 0.45 μm; diameter, 22 mm; Phenomenex, Aschaffenburg, Germany). The sterility of the filtrate was tested by streaking 300 μl of the filtrate on J medium agar plates containing 1.5% (wt/vol) agar. The filtered extract was concentrated by means of a rotary evaporator (Heidolph Instruments GmbH & Co. KG, Schwalbach, Germany) and dried in 4-ml glass vials using a vacuum centrifuge (concentrator 5301; Eppendorf, Hamburg, Germany). For feeding of Amberlite XAD16 extracts to larvae, the XAD16 extracts were dissolved first in methanol due to their low water solubility, such that the final methanol concentration in the larval diet was less than 1%.
To evaluate the efficiency of the metabolite extraction from liquid P. larvae cultures by means of Amberlite XAD16, the toxicity of sterile filtered P. larvae culture supernatants and identically treated but Amberlite XAD16-extracted culture supernatants was tested. For this purpose, the larval diet was prepared with the above-mentioned culture supernatants. Briefly, the sugars which were used for the sugar solution part of the larva diet were diluted in sterile culture supernatants resembling the volume of water that would have been used instead.
XAD extract from DSM 16116 was fractionated with Strata C18-E reversed-phase solid-phase extraction (RP-SPE) cartridges (20-g bed mass; Phenomenex). The extract was dissolved in 0.1% TFA and loaded onto a C18E-SPE cartridge. After washing with 200 ml 0.1% TFA, compounds were successively eluted with 200 ml of 10, 30, 50, and 99.9% ACN (0.1% TFA each). Obtained fractions were dried using a rotary evaporator and stored at −20°C.
Analyses of culture extracts.
The Amberlite XAD16 extracts of culture supernatants as well as the fractions obtained from C18E-SPE were analyzed by means of liquid chromatography electrospray ionization mass spectrometry (LC-MS) as well as matrix-assisted laser desorption-ionization mass spectrometry (MALDI-MS). Briefly, extracts were analyzed, as described previously (39), using a Dionex Ultimate 3000 system coupled to a Bruker AmaZon X mass spectrometer and an Acquity UPLC BEH C18 1.7-μm RP column (Waters) using a gradient of acetonitrile-0.1% formic acid in H2O ranging from 5 to 95% in 22 min at a flow rate of 0.6 ml min−1. For MALDI analysis, extracts were diluted in 70% ACN with 0.1% TFA. All samples were mixed 1:2 with 1 μl of 20 mM 4-chloro-α-cyanocinnamic acid (ClCCA) (40, 41) in 70% ACN, spotted onto a polished stainless steel target, and air dried. MALDI-MS and MALDI tandem mass spectrometry (MALDI-MS2) analyses were performed with a MALDI LTQ Orbitrap XL (Thermo Fisher Scientific, Inc., Waltham, MA) equipped with a nitrogen laser at 337 nm. For MALDI-MS of culture supernatants, samples were prepared as a 1:5 dilution in 70% ACN–0.1% TFA, mixed 1:1 with the above-mentioned matrix solution, and measured on a Voyager-DE STR mass spectrometer (Applied Biosystems, Darmstadt, Germany).
Genome analysis.
For identification of possible secondary metabolite biosynthesis gene clusters in the available genomes of Paenibacillus larvae subsp. larvae B-3650 and Paenibacillus larvae subsp. larvae BRL-23001, the contigs of both genome-sequencing projects were concatenated with spacers of 25 N (unspecified nucleotides) to obtain two individual linear DNA strands. The fully closed genome of P. larvae subsp. larvae DSM 25430, recently made accessible at the NCBI website, was also analyzed. An antiSMASH (42, 43) analysis of all DNA sequences was performed, and the putative biosynthesis gene clusters were consecutively analyzed manually to dismiss possible artificial clusters that resulted from concatenation or false annotation. Briefly, for B-3650 and BRL-23001, the Geneious software (version 5.6.2) was used to predict possible open reading frames (ORF) in the identified genomic loci. The start codons ATG, GTG, and CTG were considered, and to allow the identification of antibiotic and bacteriocin structural genes, 90 bp was allowed as the minimal ORF size. To identify possible functions of the encoded proteins, the protein BLAST was used to identify homologous proteins as well as conserved motifs. For the identification of nonribosomal peptide synthetase (NRPS) and polyketide synthase (PKS) domain architectures, the NRPS-PKS analysis website was used (44).
Statistics.
The mean times of death were estimated with the Kaplan-Meier test. The survival rates of control and treatment groups were compared using a log-rank test (Mantel-Cox test). All statistical tests were performed using SPSS Statistics, version 20.0 (IBM, Chicago, IL) running on a PC under Windows XP, service pack 3.
RESULTS
Artificial rearing.
In order to assess larva survival during in vitro rearing, we monitored survival daily (up to day 7) for three replicates per experimental condition. Results from the control group demonstrate successful in vitro rearing with >80% survival of larva up to day 7 (Fig. 1). Control larvae (without treatment) were reared on all plates for all experiments. The median survival time (MST) of bees was evaluated until day 15 twice in control groups with medium (for assay 1, MST = 10.79 days, n = 24 larvae and 8 plates; for assay 2, MST = 11.92 days, n = 36 larvae and 12 plates) and once with methanol (MST = 11.79 days, n = 72 larvae and 24 plates) (Table 1). In each assay, these groups did not differ from the control group without medium or methanol (degrees of freedom [df] = 1; P > 0.05 by Mantel-Cox test) (assay 1, MST = 10.17 days, n = 78 larvae and 21 plates; assay 2, MST = 11.5 days, n = 183 larvae and 43 plates; Kaplan-Meier test) (Table 1). Similarly, the median survival times between the medium and the methanol control groups in the same assay did not differ (df = 1; P > 0.05 by Mantel-Cox test) (Table 1). This indicates that 1% methanol is not toxic for the larvae and that the medium either contains no compounds harmful to the larvae or their concentrations are too low to impair development (Table 1). Therefore, for all further experiments, the control groups did not contain methanol.
FIG 1.

Honeybee larvae 9 days after eclosion. After defecation, the larvae were transferred to pupation wells lined with paper tissues (right).
TABLE 1.
Survival of artificially reared larvae fed with different treatments compared to each other and to control groups by time of death after inoculation
| Treatmentc | MSTa | SE | Significant differenceb |
|---|---|---|---|
| Infection (assay 1) | |||
| DSM 16116 (n = 45) (3 plates) | 9.47 | 0.55 | A |
| DSM 16115 (n = 63) (6 plates) | 10.11 | 0.50 | AB |
| DSM 17237 (n = 14) (2 plates) | 10.58 | 1.19 | AB |
| NRLL B-41635 (n = 63) (6 plates) | 10.11 | 0.45 | AB |
| Medium control (n = 24) (8 plates) | 10.79 | 0.92 | B |
| Control (n = 78) (21 plates) | 10.17 | 0.43 | AB |
| Supernatant (assay 2) | |||
| Amberlite XAD16-extracted supernatant (n = 21) (3 plates) | 8.10 | 0.52 | A |
| Supernatant (n = 20) (3 plates) | 5.60 | 0.42 | B |
| Control (n = 183) (43 plates) | 11.50 | 0.25 | C |
| Amberlite XAD16 extract (assay 2) | |||
| DSM 16116 (n = 34) (3 plates) | 3.82 | 0.10 | A |
| DSM 16115 (n = 36) (3 plates) | 4.11 | 0.14 | A |
| DSM 17237 (n = 36) (3 plates) | 5.61 | 0.20 | B |
| NRLL B-41635 (n = 81) (6 plates) | 12.23 | 0.39 | C |
| Methanol control (n = 72) (24 plates) | 11.79 | 0.42 | DC |
| Medium control (n = 36) (12 plates) | 11.92 | 0.59 | DC |
| Control (n = 183) (43 plates) | 11.50 | 0.25 | D |
| Fractions (3.5 mg/ml) (assay 2) | |||
| VT1 (n = 21) (3 plates) | 7.76 | 0.44 | A |
| 10% ACN (n = 25) (3 plates) | 6.28 | 0.38 | B |
| 30% ACN (n = 27) (3 plates) | 7.15 | 0.50 | AB |
| 50% ACN (n = 27) (3 plates) | 3.81 | 0.25 | C |
| 99.9% ACN (n = 27) (3 plates) | 3.07 | 0.05 | D |
| 0.1 % TFA (n = 27) (3 plates) | 4.704 | 0.139 | E |
| Flowthrough (n = 21) (3 plates) | 13.05 | 0.81 | F |
| Control (n = 183) (43 plates) | 11.50 | 0.25 | F |
Median survival time in days, estimated with the Kaplan-Meier test to day 15.
If two treatments have different letters, their MST are significantly different from each other (P < 0.05 by log-rank test and Mantel-Cox test).
n, number of honeybee larvae.
Infections with P. larvae spores.
Artificially reared honeybee larvae were infected by ingestion of spore-contaminated food. Compared to the control group, which was fed a diet without P. larvae spores, all tested strains affect larval development, because they showed the typical clinical symptoms (45). After the larvae were identified as dead, they were separated from the others and inspected daily. Their color changed to brown and got darker every day. The dead larvae finally dissolved into a viscous mass sticking to the walls of the storage vessel, as has been described for the ropy stage, and consecutively dried down to a hard scale. Although all tested strains showed the typical clinical symptoms, the different P. larvae genotypes (ERIC I and ERIC II) tend to vary in virulence even if the MST estimated with the Kaplan-Meier test did not differ significantly (df = 1; P > 0.05 by Mantel-Cox test) (Table 1; also see Fig. S1 in the supplemental material). The survival time of honeybee larvae after infection with P. larvae ERIC I strain NRLL B-41625 was similar to that of the control group. In contrast, all three tested ERIC II strains were more virulent, as described previously (6).
P. larvae toxicity induced by small molecules.
When the Amberlite XAD16 extracts were tested for toxicity, larvae fed with Amberlite XAD16 extracts from DSM 16116, DSM 16115, and DSM 17237 cultures had increased mortalities compared to the controls (MST were estimated with Kaplan-Meier test and compared to each other with the Mantel-Cox test; df = 1; P < 0.001) (Table 1 and Fig. 2). The mortality of larvae fed with Amberlite XAD16 extract from strain NRLLB-41635 (MST = 12.2 days by Kaplan-Meier test) did not differ from the control (MST = 11.5 days by Kaplan-Meier test; df = 1; P > 0.05 by Mantel-Cox test) (Table 1). Nevertheless, these larvae showed the symptoms of AFB (ropy and scale stages).
FIG 2.
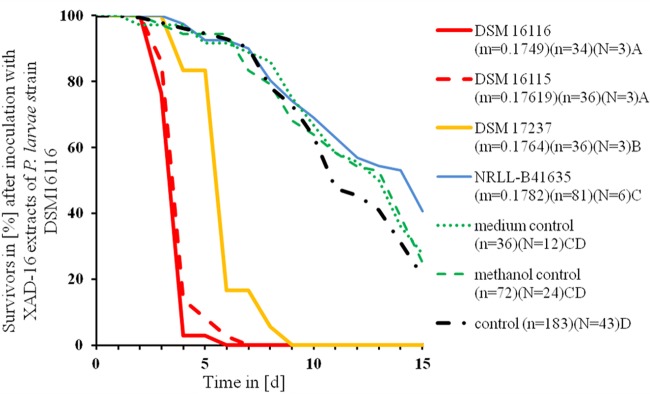
Larval survival after inoculation with Amberlite XAD16 extracts (3.5 mg/ml) from different strains of P. larvae. Strains are compared to each other and to control groups by mean time of death (MST) estimated with the Kaplan-Meier test. Statistically different MSTs are indicated by different letters; treatments sharing the same letter do not differ. P < 0.01 by log-rank test and Mantel-Cox test. m = original amount of extract (g), n = number of honeybee larvae, N = number of plates used.
In order to address whether low-molecular-weight compounds secreted by P. larvae are bound to the XAD16 resin, the supernatant of the most virulent strain, DSM 16116, was analyzed for toxicity with and without previous XAD16 extraction. For this, DSM 16116 was grown with or without the adsorber resin Amberlite XAD16 that binds typical low-molecular-weight natural products like peptides or polyketides (39, 46, 47), and the cell-free culture supernatant was consecutively used to prepare the larva diet. Adding XAD16-extracted culture supernatant or nonextracted supernatant to the diet reduced the survival time of larvae compared to that of the control larvae. Furthermore, the nonextracted supernatant showed a significantly higher toxicity toward the honeybee larvae than the extracted supernatant (df = 1; P > 0.001 by Mantel-Cox test) (Table 1 and Fig. 3). This indicates that XAD16 extraction removes virulence factors secreted by P. larvae from the supernatant.
FIG 3.
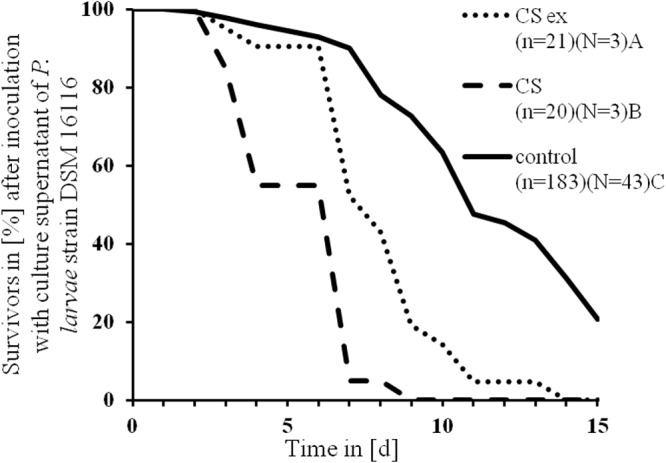
Larval survival after inoculation with spore-free culture supernatant of P. larvae DSM 16116 before (CS) and after Amberlite XAD16 extraction (CS ex). CS is compared to CS ex, where low-molecular-mass molecules are absent, and to a control group by mean time of death (MST) estimated with the Kaplan-Meier test. The different letters indicate significant differences between the MST. P < 0.001 by log-rank test and Mantel-Cox test. n = number of honeybee larvae, N = number of plates used.
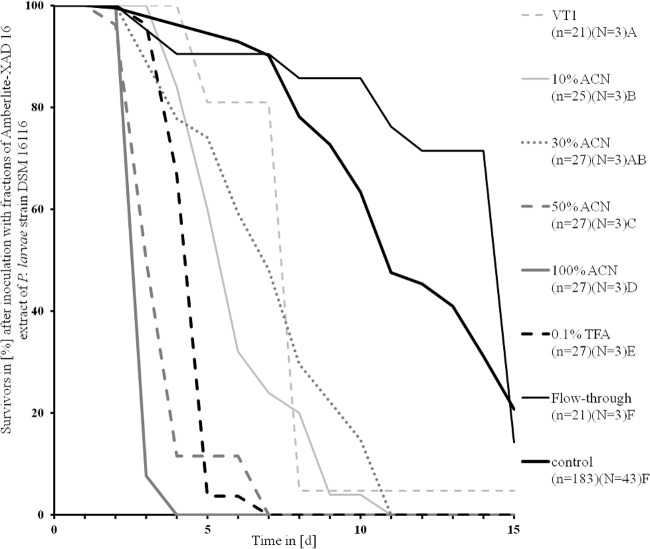
Subsequent solid-phase extraction of the Amberlite XAD16 extracts from DSM 16116 led to five fractions (0.1% trifluoroacetic acid, 10% ACN, 50% ACN, 99.9% ACN, and flowthrough) that were also analyzed for their toxicity. All fractions were fed at the identical concentration of 3.5 mg/ml, except for 99.9% ACN with 0.003 mg/ml larval diet, because only small amounts could be extracted.
All fractions were toxic except for the flowthrough (df = 1; P > 0.001 by Mantel-Cox test) (Table 1). Larvae fed with VT1, the full extract of DSM 16116, died with an MST of 7.76 days; that of the control was 11.5 days.
Larvae fed with the flowthrough fraction had an MST of 13.05 days and no negative effects on larval development. The MST of the 100% ACN fraction was only 3.07 days, the 50% ACN fraction had an MST of 6.95 days, and the 10% ACN fraction had an MST of only 5 days. It was conspicuous that larvae of the 10% ACN fraction were clearly smaller than the control larvae. The MST of the 30% ACN fraction did not differ from that of the VT1 fraction, while all other fractions differed from each other, from the control, and from the VT1 fraction (MST were estimated with Kaplan-Meier test and compared to each other with the Mantel-Cox test; df = 1; P < 0.001) (Table 1 and Fig. 4).
FIG 4.
Larval survival after inoculation with fractions (3.5 mg/ml) of Amberlite XAD16 extracts of Paenibacillus larvae DSM 16116. Fractions are compared to each other and to the control group by mean time of death (MST) estimated with the Kaplan-Meier test. The different letters indicate significant differences between the MST. P < 0.001 by log-rank test and Mantel-Cox test. n = number of honeybee larvae, N = number of plates used.
Mass spectrometry of prepurified P. larvae extracts.
Amberlite XAD16 extracts from all tested P. larvae strains (DSM 16116, DSM 16115, DSM 17237, and NRLL B-41635) were analyzed by positive-ion mode MALDI-Orbitrap mass spectrometry (see Fig. S2 in the supplemental material). Additionally, the raw culture supernatants (without Amberlite XAD16 extraction) were analyzed by MALDI-TOF mass spectrometry (see Fig. S3). In both cases, no qualitative differences were observed, which could explain the significantly different activities in the toxicity assays. MALDI-MS analysis of the fractions from DSM 16116 showed the presence of different compounds in the different fraction allowing their isolation in future experiments (see Fig. S4).
In silico analyses of the P. larvae genomes for bioactive natural product biosynthesis genes.
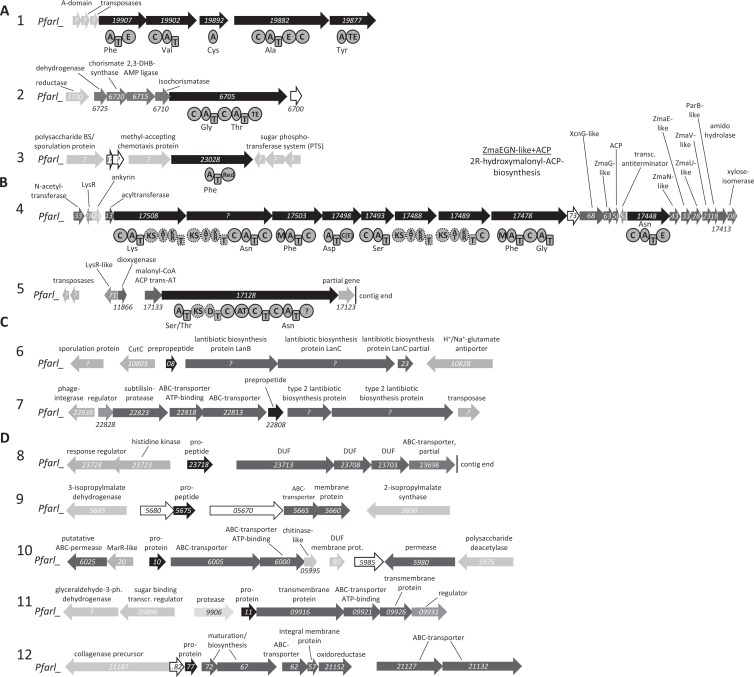
Since no genomic information of the Paenibacillus strains cultivated in this study were available, the genomes of P. larvae subsp. larvae B-3650, P. larvae subsp. larvae BRL-23001 (35), and P. larvae subsp. larvae DSM 25430 were analyzed. An antiSMASH analysis of all genomes was followed by a manual inspection of all identified putative secondary metabolite gene clusters. The genomes of B-3650 and BRL-23001 showed an identical set of natural product biosynthesis gene clusters (Fig. 5) differing from the genome of DSM 25430 (see Fig. S5 in the supplemental material). However, several similar gene clusters have been identified in these genomes (numbered identically in Fig. 5 and in Fig. S5 for simplicity). In all genomes, gene clusters responsible for the production of nonribosomally made peptides (NRP), NRP-polyketide (PK) hybrids, lantibiotics, as well as bacteriocins were identified (Fig. 5; also see Fig. S5). Gene cluster 2 resembles the paenibactin biosynthesis gene cluster responsible for the biosynthesis of the siderophore paenibactin from Paenibacillus eglii B69 (Fig. 5; also see Fig. S5) (22). Cluster 3 encodes a three-domain nonribosomal peptide synthetase (NRPS) with an adenylation (A), thiolation (T), and terminal reduction (Red) domain (Fig. 5; also see Fig. S5). Similar NRPS have been shown to be involved in the biosynthesis of piperazines or terphenylquinones from fungi (48, 49). The largest gene cluster (cluster 4) encodes an NRPS-PKS hybrid with all structural features of the prodrug activation mechanism previously identified in the biosynthesis of xenocoumacin, zwittermicin, and colibactin (39, 50–52). Additionally, a putatively incomplete NRPS-PKS hybrid gene cluster (cluster 5 in Fig. 5) responsible for the biosynthesis of bacillomycin/mycosubtilin-like lipopeptides was identified in strain B-3650. The NRPS-PKS hybrid protein Pfarl_17128 (Fig. 5B) exhibited a highly similar domain architecture and 69% sequence identity to the mycosubtilin NRPS in Bacillus atrophaeus 1942 (subunit A) as well as 68% sequence identity with BmyA from the bacillomycin biosynthesis machinery in Bacillus amyloliquefaciens FZB42 (53, 54). However, no further genes associated with the biosynthesis of a mycosubtilin- or bacillomycin-like compound could be identified on the contig. In strain DSM 25430, an additional PKS-NRPS hybrid that is encoded by cluster 5 (see Fig. S5) also shows some similarity to the mycosubtilin biosynthesis enzymes. Two and one biosynthesis gene clusters involved in the biosynthesis of lantibiotics and five and two clusters involved in the biosynthesis of bacteriocins were identified in the genomes of strains B-3650 and DSM 25430, respectively (Fig. 5; also see Fig. S5). The alignment of both lantibiotic preproproteins (gene clusters 6 and 7) (see Fig. S6) against other database-deposited lantibiotic preproprotein structures showed only weak sequence similarity to other lantibiotic preproproteins, except for the conservation of one C-terminal cysteine residue (55).
FIG 5.
Putative secondary metabolite gene clusters identified in the genome of Paenibacillus larvae subsp. larvae B-3650. (A) NRPS cluster; (B) NRPS-PKS hybrid clusters; (C) lantibiotic biosynthesis cluster; and (D) bacteriocin biosynthesis cluster. Black arrows represent genes encoding NRPS or NRPS-PKS hybrids or lantibiotic or bacteriocin propeptides. Dark gray arrows represent genes putatively involved in the biosynthesis and transport of the corresponding secondary metabolite. Gray, light gray, and white arrows represent genes encoding putative regulatory proteins, proteins probably not involved in secondary metabolite biosynthesis, and hypothetical proteins, respectively. Trans-AT, trans-acting acyltransferase; BS, biosynthesis; AT, aminotransferase; CoA, coenzyme A; ACP, acyl carrier protein; prot., protein; transcr., transcription; 3-ph., 3-phosphate.
DISCUSSION
We have shown that low-molecular-weight compounds produced by P. larvae are toxic to honeybee larvae. Whereas all strains analyzed showed a similar toxicity profile when spore suspensions were added to the food source of the larvae, extracts from different strains of P. larvae differed significantly in their toxicity, pointing to either different virulence factors or different amounts thereof. Our study indicates that DSM 16116 is the most virulent strain. Similarly virulent is DSM 16115, which was assigned to genotype ERIC II (E. Genersch, personal communication). Accordingly, the incubation times of ERIC II and ERIC IV strains are shorter than those of ERIC I strains (6). The LT100 values of ERIC II and VI were 6 to 7 days, whereas that of ERIC I was approximately 12 days. As DSM 17237, with an unknown ERIC subtype, behaves similarly to DSM 16116 and DSM 16115, it might also represent an ERIC II subtype. Interestingly, NRLL B-41635 (ERIC I) differs from the other strains (all ERIC II), as its toxic effects appear later, which might be due to its genotype difference.
The fact that Amberlite XAD16 extracts (Fig. 2) seemed to be more toxic than spores (see Fig. S1 in the supplemental material) of the same strain might be the result of the direct application of these putative virulence factors in the food source. In the case of spores, P. larvae needs to germinate in the midgut and then produces the respective toxic compounds during the infection process, which might delay the toxicity. We have also shown that not all toxic compounds bind to the XAD resin, which implies the presence of different toxic compounds. Indeed, several other protein virulence factors have previously been described from P. larvae, including metalloproteases, protein toxins, and others (29, 56–58). However, these protein virulence factors with molecular masses of up to 100 kDa would be too large to bind to the XAD resin, which is described as binding mainly low-molecular-weight compounds, such as antibiotics and other secondary metabolites, but also hydrophobic compounds of up to 40 kDa.
Unfortunately, we were not able to isolate a pure toxic compound, but we could show active fractions that can be purified in the future using established in vitro honeybee larva-rearing methods (Fig. 1). Mass spectrometry revealed the presence of several different compounds in the P. larvae crude extracts (see Fig. S2 in the supplemental material) as well as in the bioactive fractions (see Fig. S4). Interestingly, the analysis of the different P. larvae supernatants revealed the presence of highly similar or identical compounds in these strains (see Fig. S3), indicating a conserved class of compounds among the different strains.
Additionally, active compounds are not necessarily visible in the acquired mass spectra, because the acquired mass spectra do not cover the compound's mass or because the toxic substance(s) is not visible based on mass(es) overlapping those of other compounds. When P. larvae extracts were analyzed by MALDI-MS (see Fig. S2 in the supplemental material), no masses corresponding to already-known secondary metabolites from Paenibacillus were identified. Additionally, application of the recently introduced peptidogenomics approach (59) was not reasonable, since no genome sequence of the analyzed strains was available.
In order to address the potential of P. larvae to produce low-molecular-weight toxic compounds, the available genome of P. larvae BRL-230010 was analyzed for the presence of biosynthesis gene clusters of different natural product classes. Among the 12 identified gene clusters are biosynthesis gene clusters encoding NRPS, hybrids of PKS and NRPS, as well as lantibiotics and bacteriocins, all of which could be responsible for the observed toxic effect. The m/z ratios observed in the different extracts are clearly in the range one would expect for the products of the identified gene clusters. Especially interesting is the hybrid PKS-NRPS gene cluster 4, encoding a compound that is most likely activated via proteolytic cleavage, as was shown for the potent antibiotic xenocoumacin from the entomopathogenic bacterium Xenorhabdus nematophila (39) and the still-unknown compound colibactin, a potent virulence factor of pathogenic Escherichia coli strains (51).
Future work must focus on the identification of the compounds responsible for the observed toxicity and also correlate these compounds with their respective gene clusters. The latter will allow us to study the regulation of biosynthesis in the larval context and might shed more light on the molecular mechanisms leading to AFB.
Supplementary Material
ACKNOWLEDGMENTS
We are grateful to Stefan Fuchs for help with the statistical analyses and the beekeepers of the Institut für Bienenkunde Oberursel, Matthias Ullmann and Beate Springer, for expert bee rearing and grafting assistance.
H.B.B. and S.W.F. are grateful for financial support from the excellence initiative of the Hessian Ministry of Science and Art (LOEWE research focus “Insect Biotechnology”).
Footnotes
Published ahead of print 7 February 2014
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.04049-13.
REFERENCES
- 1.Klein A-M, Vaissière BE, Cane JH, Steffan-Dewenter I, Cunningham SA, Kremen C, Tscharntke T. 2007. Importance of pollinators in changing landscapes for world crops. Proc. Biol. Sci. 274:303–313. 10.1098/rspb.2006.3721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Genersch E. 2008. Paenibacillus larvae and American foulbrood—long since known and still surprising. J. Verbr. Lebensm. 3:429–434. 10.1007/s00003-008-0379-8 [DOI] [Google Scholar]
- 3.Dancer BN, Chantawannakul P. 1997. The proteases of American foulbrood scales. J. Invertebr. Pathol. 70:79–87. 10.1006/jipa.1997.4672 [DOI] [PubMed] [Google Scholar]
- 4.Gregorc A, Bowen ID. 1998. Histopathological and histochemical changes in honeybee larvae (Apis mellifera L.) after infection with Bacillus larvae, the causative agent of American foulbrood disease. Cell Biol. Int. 22:137–144. 10.1006/cbir.1998.0232 [DOI] [PubMed] [Google Scholar]
- 5.Genersch E. 2006. Reclassification of Paenibacillus larvae subsp. pulvifaciens and Paenibacillus larvae subsp. larvae as Paenibacillus larvae without subspecies differentiation. Int. J. Syst. Evol. Microbiol. 56:501–511. 10.1099/ijs.0.63928-0 [DOI] [PubMed] [Google Scholar]
- 6.Genersch E, Ashiralieva A, Fries I. 2005. Strain- and genotype-specific differences in virulence of Paenibacillus larvae subsp. larvae, a bacterial pathogen causing American foulbrood disease in honeybees. Appl. Environ. Microbiol. 71:7551–7555. 10.1128/AEM.71.11.7551-7555.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.White GF. 1906. The bacteria of the apiary, with special reference to bee diseases. U.S. Department of Agriculture, Washington, DC [Google Scholar]
- 8.Katznelson H. 1950. Bacillus pulvifaciens (n. sp.), an organism associated with powdery scale of honeybee larvae. J. Bacteriol. 59:153–155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rauch S, Ashiralieva A, Hedtke K, Genersch E. 2009. Negative correlation between individual-insect-level virulence and colony-level virulence of Paenibacillus larvae, the etiological agent of American foulbrood of honeybees. Appl. Environ. Microbiol. 75:3344–3347. 10.1128/AEM.02839-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Matheson A, Reid M. 1992. Strategies for the prevention and control of American foulbrood. Am. Bee J. 132:399–402 [Google Scholar]
- 11.Brødsgaard CJ, Hansen H, Ritter W. 2000. Progress of Paenibacillus larvae larvae infection in individually inoculated honey bee larvae reared singly in vitro, in micro colonies, or in full-size colonies. J. Apicult. Res. 39:19–27 http://www.ibra.org.uk/articles/20091216_3 [Google Scholar]
- 12.Bailey L, Ball BV. 1991. American foulbrood, p 36–41 In Bailey L, Ball BV. (ed), Honey bee pathology. Academic Press, London, United Kingdom [Google Scholar]
- 13.Yue D, Nordhoff M, Wieler LH, Genersch E. 2008. Fluorescence in situ hybridization (FISH) analysis of the interactions between honeybee larvae and Paenibacillus larvae, the causative agent of American foulbrood of honeybees (Apis mellifera). Environ. Microbiol. 10:1612–1620. 10.1111/j.1462-2920.2008.01579.x [DOI] [PubMed] [Google Scholar]
- 14.Sturtevant AP. 1932. Relation of commercial honey to the spread of American foulbrood. J. Agric. Res. 45:257–285 [Google Scholar]
- 15.Lindström A, Korpela S, Fries I. 2008. Horizontal transmission of Paenibacillus larvae spores between honey bee (Apis mellifera) colonies through robbing. Apidologie 39:515–522. 10.1051/apido:2008032 [DOI] [Google Scholar]
- 16.Rieg S, Bauer TM, Peyerl-Hoffmann G, Held J, Ritter W, Wagner D, Kern WV, Serr A. 2010. Paenibacillus larvae bacteremia in injection drug users. Emerg. Infect. Dis. 16:487–489. 10.3201/eid1603.091457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Beatty PH, Jensen SE. 2002. Paenibacillus polymyxa produces fusaricidin-type antifungal antibiotics active against Leptosphaeria maculans, the causative agent of blackleg disease of canola. Can. J. Microbiol. 48:159–169. 10.1139/w02-002 [DOI] [PubMed] [Google Scholar]
- 18.Qian C-D, Wu X-C, Teng Y, Zhao W-P, Li O, Fang S-G, Huang Z-H, Gao H-C. 2012. Battacin (octapeptin B5), a new cyclic lipopeptide antibiotic from Paenibacillus tianmuensis active against multidrug-resistant Gram-negative bacteria. Antimicrob. Agents Chemother. 56:1458–1465. 10.1128/AAC.05580-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wu X-C, Qian C-D, Fang H-H, Wen Y-P, Zhou J-Y, Zhan Z-J, Ding R, Li O, Gao H. 2011. Paenimacrolidin, a novel macrolide antibiotic from Paenibacillus sp. F6-B70 active against methicillin-resistant Staphylococcus aureus. Microb. Biotechnol. 4:491–502. 10.1111/j.1751-7915.2010.00201.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Anandaraj B, Vellaichamy A, Kachman M, Selvamanikandan A, Pegu S, Murugan V. 2009. Co-production of two new peptide antibiotics by a bacterial isolate Paenibacillus alvei NP75. Biochem. Biophys. Res. Commun. 379:179–185. 10.1016/j.bbrc.2008.12.007 [DOI] [PubMed] [Google Scholar]
- 21.Ding R, Wu X-C, Qian C-D, Teng Y, Li O, Zhan Z-J, Zhao Y-H. 2011. Isolation and identification of lipopeptide antibiotics from Paenibacillus elgii B69 with inhibitory activity against methicillin-resistant Staphylococcus aureus. J. Microbiol. 49:942–949. 10.1007/s12275-011-1153-7 [DOI] [PubMed] [Google Scholar]
- 22.Wen Y, Wu X, Teng Y, Qian C, Zhan Z, Zhao Y, Li O. 2011. Identification and analysis of the gene cluster involved in biosynthesis of paenibactin, a catecholate siderophore produced by Paenibacillus elgii B69. Environ. Microbiol. 13:2726–2737. 10.1111/j.1462-2920.2011.02542.x [DOI] [PubMed] [Google Scholar]
- 23.Shaheen M, Li J, Ross AC, Vederas JC, Jensen SE. 2011. Paenibacillus polymyxa PKB1 produces variants of polymyxin B-type antibiotics. Chem. Biol. 18:1640–1648. 10.1016/j.chembiol.2011.09.017 [DOI] [PubMed] [Google Scholar]
- 24.Li J, Jensen SE. 2008. Nonribosomal biosynthesis of fusaricidins by Paenibacillus polymyxa PKB1 involves direct activation of a d-amino acid. Chem. Biol. 15:118–127. 10.1016/j.chembiol.2007.12.014 [DOI] [PubMed] [Google Scholar]
- 25.Bode HB. 2009. Entomopathogenic bacteria as a source of secondary metabolites. Curr. Opin. Chem. Biol. 13:224–230. 10.1016/j.cbpa.2009.02.037 [DOI] [PubMed] [Google Scholar]
- 26.Brachmann AO, Bode HB. 2013. Identification and bioanalysis of natural products from insect symbionts and pathogens. In Scheper T, Belkin S, Doran PM, Endo I, Gu MB, Hu WS, Mattiasson B, Nielsen J, Stephanopoulos GN, Ulber R, Zeng A-P, Zhong J-J, Zhou W. (ed), Advances in biochemical engineering/biotechnology. Springer, Berlin, Germany: [DOI] [PubMed] [Google Scholar]
- 27.Gliński Z, Jarosz J. 1992. Does Bacillus larvae produce an antibacterial substance in infected honey bee larvae? Apidologie 23:193–201. 10.1051/apido:19920301 [DOI] [Google Scholar]
- 28.Antúnez K, Anido M, Evans JD, Zunino P. 2010. Secreted and immunogenic proteins produced by the honeybee bacterial pathogen, Paenibacillus larvae. Vet. Microbiol. 141:385–389. 10.1016/j.vetmic.2009.09.006 [DOI] [PubMed] [Google Scholar]
- 29.Fünfhaus A, Poppinga L, Genersch E. 2013. Identification and characterization of two novel toxins expressed by the lethal honey bee pathogen Paenibacillus larvae, the causative agent of American foulbrood. Environ. Microbiol. 15:2951–2965. 10.1111/1462-2920.12229 [DOI] [PubMed] [Google Scholar]
- 30.Thanabalu T, Hindley J, Jackson-Yap J, Berry C. 1991. Cloning, sequencing, and expression of a gene encoding a 100-kilodalton mosquitocidal toxin from Bacillus sphaericus SSII-1. J. Bacteriol. 173:2776–2785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schirmer J, Just I, Aktories K. 2002. The ADP-ribosylating mosquitocidal toxin from Bacillus sphaericus: proteolytic activation, enzyme activity, and cytotoxic effects. J. Biol. Chem. 277:11941–11948. 10.1074/jbc.M108463200 [DOI] [PubMed] [Google Scholar]
- 32.Peng Y-SC, Mussen E, Fong A, Montague MA, Tyler T. 1992. Effects of chlortetracycline of honey bee worker larvae reared in vitro. J. Invertebr. Pathol. 60:127–133. 10.1016/0022-2011(92)90085-I [DOI] [Google Scholar]
- 33.Genersch E, Otten C. 2003. The use of repetitive element PCR fingerprinting (rep-PCR) for genetic subtyping of German field isolates of Paenibacillus larvae subsp. larvae. Apidologie 34:195–206. 10.1051/apido:2003025 [DOI] [Google Scholar]
- 34.Neuendorf S. 2004. Biochemical characterization of different genotypes of Paenibacillus larvae subsp. larvae, a honey bee bacterial pathogen. Microbiology 150:2381–2390. 10.1099/mic.0.27125-0 [DOI] [PubMed] [Google Scholar]
- 35.Qin X, Evans JD, Aronstein KA, Murray KD, Weinstock GM. 2006. Genome sequences of the honey bee pathogens Paenibacillus larvae and Ascosphaera apis. Insect Mol. Biol. 15:715–718. 10.1111/j.1365-2583.2006.00694.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nordström S, Fries I. 1995. A comparison of media and cultural conditions for identification of Bacillus larvae in honey. J. Apicult. Res. 34:97–103 [Google Scholar]
- 37.Forsgren E, Olofsson TC, Vásquez A, Fries I. 2009. Novel lactic acid bacteria inhibiting Paenibacillus larvae in honey bee larvae. Apidologie 41:99–108 [Google Scholar]
- 38.Govan VA, Allsopp MH, Davison S. 1999. A PCR detection method for rapid identification of Paenibacillus larvae. Appl. Environ. Microbiol. 65:2243–2245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Reimer D, Pos KM, Thines M, Grün P, Bode HB. 2011. A natural prodrug activation mechanism in nonribosomal peptide synthesis. Nat. Chem. Biol. 7:888–890. 10.1038/nchembio.688 [DOI] [PubMed] [Google Scholar]
- 40.Jaskolla TW, Lehmann WD, Karas M. 2008. 4-Chloro-α-cyanocinnamic acid is an advanced, rationally designed MALDI matrix. Proc. Natl. Acad. Sci. U. S. A. 105:12200–12205. 10.1073/pnas.0803056105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jaskolla T, Fuchs B, Karas M, Schiller J. 2009. The new matrix 4-chloro-α-cyanocinnamic acid allows the detection of phosphatidylethanolamine chloramines by MALDI-TOF mass spectrometry. J. Am. Soc. Mass Spectrom. 20:867–874. 10.1016/j.jasms.2008.12.028 [DOI] [PubMed] [Google Scholar]
- 42.Medema MH, Blin K, Cimermancic P, de Jager V, Zakrzewski P, Fischbach MA, Weber T, Takano E, Breitling R. 2011. antiSMASH: rapid identification, annotation and analysis of secondary metabolite biosynthesis gene clusters in bacterial and fungal genome sequences. Nucleic Acids Res. 39:W339–W346. 10.1093/nar/gkr466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Blin K, Medema MH, Kazempour D, Fischbach MA, Breitling R, Takano E, Weber T. 2013. antiSMASH 2.0—a versatile platform for genome mining of secondary metabolite producers. Nucleic Acids Res. 41:W204–W212. 10.1093/nar/gkt449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bachmann BO, Ravel J. 2009. Methods for in silico prediction of microbial polyketide and nonribosomal peptide biosynthetic pathways from DNA sequence data. Methods Enzymol. 458:181–217. 10.1016/S0076-6879(09)04808-3 [DOI] [PubMed] [Google Scholar]
- 45.Lindström A. 2008. Distribution of Paenibacillus larvae spores among adult honey bees (Apis mellifera) and the relationship with clinical symptoms of American foulbrood. Microb. Ecol. 56:253–259. 10.1007/s00248-007-9342-y [DOI] [PubMed] [Google Scholar]
- 46.Brachmann AO, Joyce SA, Jenke-Kodama H, Schwar G, Clarke DJ, Bode HB. 2007. A type II polyketide synthase is responsible for anthraquinone biosynthesis in Photorhabdus luminescens. ChemBioChem 8:1721–1728. 10.1002/cbic.200700300 [DOI] [PubMed] [Google Scholar]
- 47.Grundmann F, Kaiser M, Kurz M, Schiell M, Batzer A, Bode HB. 2013. Structure determination of the bioactive depsipeptide xenobactin from Xenorhabdus sp. PB30.3. RSC Adv. 3:22072–22077. 10.1039/c3ra44721a [DOI] [Google Scholar]
- 48.Schneider P, Bouhired S, Hoffmeister D. 2008. Characterization of the atromentin biosynthesis genes and enzymes in the homobasidiomycete Tapinella panuoides. Fungal Genet. Biol. 45:1487–1496. 10.1016/j.fgb.2008.08.009 [DOI] [PubMed] [Google Scholar]
- 49.Forseth RR, Amaike S, Schwenk D, Affeldt KJ, Hoffmeister D, Schroeder FC, Keller NP. 2012. Homologous NRPS-like gene clusters mediate redundant small-molecule biosynthesis in Aspergillus flavus. Angew. Chem. Int. Ed. Engl. 52:1590–1594. 10.1002/anie.201207456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Reimer D, Bode HB. 2014. A natural prodrug activation mechanism in the biosynthesis of nonribosomal peptides. Nat. Prod. Rep. 31:154–159. 10.1039/c3np70081j [DOI] [PubMed] [Google Scholar]
- 51.Dubois D, Baron O, Cougnoux A, Delmas J, Pradel N, Boury M, Bouchon B, Bringer MA, Nougayrede JP, Oswald E, Bonnet R. 2011. ClbP is the prototype of a peptidase subgroup involved in the biosynthesis of nonribosomal peptides. J. Biol. Chem. 286:35562–35570. 10.1074/jbc.M111.221960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Brotherton CA, Balskus EP. 2013. A prodrug resistance mechanism is involved in colibactin biosynthesis and cytotoxicity. J. Am. Chem. Soc. 135:3359–3362. 10.1021/ja312154m [DOI] [PubMed] [Google Scholar]
- 53.Koumoutsi A, Chen XH, Henne A, Liesegang H, Hitzeroth G, Franke P, Vater J, Borriss R. 2004. Structural and functional characterization of gene clusters directing nonribosomal synthesis of bioactive cyclic lipopeptides in Bacillus amyloliquefaciens strain FZB42. J. Bacteriol. 186:1084–1096. 10.1128/JB.186.4.1084-1096.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Duitman EH, Hamoen LW, Rembold M, Venema G, Seitz H, Saenger W, Bernhard F, Reinhardt R, Schmidt M, Ullrich C, Stein T, Leenders F, Vater J. 1999. The mycosubtilin synthetase of Bacillus subtilis ATCC6633: a multifunctional hybrid between a peptide synthetase, an amino transferase, and a fatty acid synthase. Proc. Natl. Acad. Sci. U. S. A. 96:13294–13299. 10.1073/pnas.96.23.13294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Arnison PG, Bibb MJ, Bierbaum G, Bowers AA, Bugni TS, Bulaj G, Camarero JA, Campopiano DJ, Challis GL, Clardy J, Cotter PD, Craik DJ, Dawson M, Dittmann E, Donadio S, Dorrestein PC, Entian K-D, Fischbach MA, Garavelli JS, Göransson U, Gruber CW, Haft DH, Hemscheidt TK, Hertweck C, Hill C, Horswill AR, Jaspars M, Kelly WL, Klinman JP, Kuipers OP, Link AJ, Liu W, Marahiel MA, Mitchell DA, Moll GN, Moore BS, Müller R, Nair SK, Nes IF, Norris GE, Olivera BM, Onaka H, Patchett ML, Piel J, Reaney MJT, Rebuffat S, Ross RP, Sahl H-G, Schmidt EW, Selsted ME, et al. 2013. Ribosomally synthesized and post-translationally modified peptide natural products: overview and recommendations for a universal nomenclature. Nat. Prod. Rep. 30:108. 10.1039/c2np20085f [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Antúnez K, Arredondo D, Anido M, Zunino P. 2011. Metalloprotease production by Paenibacillus larvae during the infection of honeybee larvae. Microbiology 157:1474–1480. 10.1099/mic.0.044321-0 [DOI] [PubMed] [Google Scholar]
- 57.Antúnez K, Anido M, Schlapp G, Evans JD, Zunino P. 2009. Characterization of secreted proteases of Paenibacillus larvae, potential virulence factors involved in honeybee larval infection. J. Invertebr. Pathol. 102:129–132. 10.1016/j.jip.2009.07.010 [DOI] [PubMed] [Google Scholar]
- 58.Antúnez K, Anido M, Arredondo D, Evans JD. 2011. Paenibacillus larvae enolase as a virulence factor in honeybee larvae infection. Vet. Microbiol. 147:83–89. 10.1016/j.vetmic.2010.06.004 [DOI] [PubMed] [Google Scholar]
- 59.Kersten RD, Yang Y-L, Xu Y, Cimermancic P, Nam S-J, Fenical W, Fischbach MA, Moore BS, Dorrestein PC. 2011. A mass spectrometry-guided genome mining approach for natural product peptidogenomics. Nat. Chem. Biol. 7:794–802. 10.1038/nchembio.684 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.