Abstract
Vibrio is a very diverse genus that is responsible for different human and animal diseases. The accurate identification of Vibrio at the species level is important to assess the risks related to public health and diseases caused by aquatic organisms. The ecology of Vibrio spp., together with their genetic background, represents an important key for species discrimination and evolution. Thus, analyses of population structure and ecology association are necessary for reliable characterization of bacteria and to investigate whether bacterial species are going through adaptation processes. In this study, a population of Vibrionaceae was isolated from shellfish of the Venice lagoon and analyzed in depth to study its structure and distribution in the environment. A multilocus sequence analysis (MLSA) was developed on the basis of four housekeeping genes. Both molecular and biochemical approaches were used for species characterization, and the results were compared to assess the consistency of the two methods. In addition, strain ecology and the association between genetic information and environment were investigated through statistical models. The phylogenetic and population analyses achieved good species clustering, while biochemical identification was demonstrated to be imprecise. In addition, this study provided a fine-scale overview of the distribution of Vibrio spp. in the Venice lagoon, and the results highlighted a preferential association of the species toward specific ecological variables. These findings support the use of MLSA for taxonomic studies and demonstrate the need to consider environmental information to obtain broader and more accurate bacterial characterization.
INTRODUCTION
Vibrio spp. are Gram-negative halophilic bacteria belonging to the class Gammaproteobacteria. Vibrio is one of the most studied and diverse genera of microorganisms found in aquatic ecosystems and comprises the major culturable bacteria in marine and estuarine environments (1). According to the Association of Vibrio Biologists (AViB; http://www2.ioc.fiocruz.br/vibrio/AVib/species.html), there are 99 accepted or proposed Vibrio species, although the recent description of new species has led to a constantly changing taxonomy. Vibrio spp. are frequently isolated from fish, fish products, and edible shellfish, and a large number of species are pathogenic to different hosts. Some species, such as V. cholerae, V. parahaemolyticus, and V. vulnificus, cause serious food-borne gastroenteritis in humans. Other species, such as V. anguillarum and V. salmonicida, are pathogenic for fish; V. splendidus-related species are pathogenic for bivalves, and V. harveyi and V. campbellii are pathogenic for shrimps (1, 2, 3). Recently, Austin suggested a classification of zoonotic Vibrio in two classes named “higher-risk” vibrios (V. cholerae, V. parahaemolyticus, and V. vulnificus) and “lower-risk” vibrios (V. alginolyticus, V. fluvialis, V. furnissii, V. harveyi, and V. mimicus) (4). Bivalve mollusks such as clams and mussels represent products of great economic importance and are widely distributed in the food trade. Recently, raw food has become more and more popular with consumers. The Veneto region is the main European producer of Manila clam (Venerupis philippinarum) and is also one of the main mussel-producing regions in Italy, where the pathogenic vibrios represents an emerging health problem (5, 6). The Vibrio species predominantly associated with bivalves are V. splendidus, V. alginolyticus, and V. harveyi, and the combination of these species (or some of them) is the most frequent cause of diseases affecting all life stages of bivalve mollusks (7). Moreover, V. tapetis has been also described as a serious clam pathogen (8). However, Vibrio contamination of shellfish is not easily detectable, as it does not induce any organoleptic changes. In addition, the common targets applied for detection of microbial contamination (e.g., Escherichia coli) are inappropriate for highlighting the level of Vibrionaceae in these products (9).
Although specific microbiological criteria for Vibrio species in seafood have not been adopted in the European community (regulation CE 2073), the European legislation recommended developing new reliable methods for risk assessment related to Vibrio spp., especially in shellfish. Mussels seem to be a reservoir for some pathogenic strains, for example, V. parahaemolyticus O3:K6. This species has been detected in up to 24% of mussels in Italy (10), but information on other Vibrio species in shellfish is less investigated. Thus, the accurate and reliable identification of Vibrio spp. is important to assess public health risks and to discover other potential problems linked to the presence and distribution of this genus in shellfish. Classical biochemical tests are usually applied to characterize Vibrionaceae, but their great phenotypic diversity makes microbiological identification imprecise and not always reliable (11, 12). A few official protocols were specifically designed for V. cholerae and V. parahaemolyticus isolation and identification, but they cannot be used to analyze other Vibrio species. The common biochemical commercial kits (Biolog-GN fingerprints and API 20E profiles) are not totally reliable for identifying Vibrio spp., and sometimes they are not able to distinguish Vibrio from other bacterial genera, such as Listonella, Photobacterium, and Aeromonas (13, 14). DNA-based molecular methods provide more reliable and precise results (15). Some multiplex PCR protocols are available for Vibrio spp. identification, but they are directed only at clinically important species, e.g., V. cholerae, V. parahaemolyticus, and V. vulnificus (16, 17), and sometimes V. mimicus and/or V. alginolyticus (18, 19). Next-generation sequencing methods are becoming widely used and certainly allow precise bacterial strain typing (20). However, the amount of data is often excessive for the scope. An ad hoc committee for the reevaluation of species definition in bacteriology recommended the use of multilocus sequence analysis (MLSA) as an alternative method for species delineation in bacteriology (21), and many recent studies have investigated Vibrio populations through gene sequencing (22, 23, 24). Several molecular markers, e.g., recA, pyrH, rpoA, and atpA, in single or in concatenated sequences, have been used to identify Vibrionaceae species, but these analyses have been applied mainly on type strains (1, 25, 26).
In this study, an MLSA approach was developed to identify and characterize a population of Vibrionaceae isolated from shellfish of the Venice lagoon (Italy) and to understand the natural diversity of Vibrio spp. found in the territory. The MLSA scheme has been deposited in the pubMLST database (www.pubmlst.org/vibrio) (27) and is freely accessible. The strains were analyzed using combined phylogenetic and statistical approaches to investigate the population structure in depth. The main aim of the study was to explore particular species adaptation processes and the presence of specific population niches in commercialized shellfish. In addition, a retrospective evaluation was conducted to compare molecular data with biochemical results and assess the reliability of the two approaches.
MATERIALS AND METHODS
Sample collection.
A total of 164 shellfish samples were collected in different areas of the Venice lagoon and sea. Sampling was conducted in different rearing areas and at various depths from February 2007 to December 2007. The following shellfish species were considered: Venerupis philippinarum, Ostrea edulis, Crassostrea gigas, Mytilus galloprovincialis, Ensis spp., Solen spp., Chamelea gallina, Callista chione, Cerastoderma spp., and Paracentrotus lividus. The following additional information was collected for all the shellfish samples: biological source (mollusk species), season, water temperature, risk level of the area (E. coli level according to European regulation 854/2004), area (sea or lagoon), and depth of sampling (cm). All of the detailed information is listed in Table S1 in the supplemental material.
Bacterial strains.
The complete list of the 183 strains analyzed in this study is in Table S1 in the supplemental material. Together with the field strains, we also sequenced 17 Vibrio reference and type strains of various species, and we included one Photobacterium profundum sequence and 11 Vibrio sp. sequences downloaded from GenBank.
Isolation and biochemical identification.
The collected live shellfish were immediately sent to the laboratory for microbiological analysis. The samples were scrubbed under running potable water, and dead animals were discarded. The samples were analyzed by the absence-presence method (qualitative analysis) for Vibrio spp. For each sample, an aliquot of 25 g (homogenized meat and intervalvar liquid) was enriched in alkaline saline peptone water (ASPW) with 3% NaCl and incubated at 37°C for 18 to 24 h ± 1 h. One milliliter of the enrichment broth was transferred to 9 ml of polymyxin B broth (SPB) (1% [wt/vol] peptone, 3% [wt/vol] yeast extract, 2% [wt/vol] NaCl, and 100,000 IU polymyxin B, pH 7.4) and incubated at 37°C for 18 to 24 h. Ten microliters was plated on thiosulfate-citrate-bile salts-sucrose (TCBS) agar plates (Oxoid, Hampshire, United Kingdom) and CHROMagar Vibrio plates (CHROMagar Microbiology, Paris, France) and incubated for 24 h ± 3 h at 37°C. Based on the morphology, typical and nontypical colored colonies were plated on tryptic soy agar (TSA) with 2% NaCl and incubated at 37°C for 18 to 24 h ± 1 h. Each colony was tested by Gram staining, genus tests (oxidase negative, glucose positive, lactose negative, and hydrogen sulfide negative in Kliger Iron Agar, ornithine decarboxylase [ODC] positive, lysine decarboxylase [LDC] positive, arginine dihydrolase[ADH] negative), and the species biochemical tests reported in Alsina-Blanch dichotomous keys (12).
Design of primers.
Four housekeeping genes (gyrB, pyrH, recA, and atpA) were chosen for the MLSA analysis; all of them are located on chromosome I. Most of the available partial and full-length sequences of the four Vibrio housekeeping genes were downloaded from the GenBank database and aligned by using the ClustalW program (http://www.ebi.ac.uk). Primers were designed based on the most conserved regions using Primer3 software (http://frodo.wi.mit.edu/primer3) and PriFi software for degenerate primers (28), with a length of 18 to 29 nucleotides. Primers for the amplification of atpA were obtained from a previous study (29). The complete list of genes analyzed in this study and all primers used for PCR amplification and sequencing are given in Table 1.
TABLE 1.
Primers used for amplification and sequencing of Vibrio sp. isolates
| Primer | Sequence (5′→3′) | Gene product | Trimmed amplicon length (bp) | Annealing temp (°C) | Reference |
|---|---|---|---|---|---|
| Vi_gyrBdg2F | GARGTGGTRGATAACTCWATTGATGAAGC | DNA gyrase, β subunit (GyrB) | 570 | 55 | This study |
| VigyrBR | CGGTCATGATGATGATGTTGT | ||||
| VigyrBF | GAAGGTGGTATTCAAGCGTT | ||||
| Vh_gyrB_F | CGTGAGCTTTCTTTCCTAAACTC | ||||
| VipyrHdgF | CCCTAAACCAGCGTATCAACGTATTC | Uridylate kinase (PyrH) | 501 | 55 | This study |
| VipyrHdgR | CGGATWGGCATTTTGTGGTCACGWGC | ||||
| VirecAF | TGCGCTAGGTCAAATTGAAA | Recombinase A (RecA) | 462 | 55 | This study |
| VirecAdgR | GTTTCWGGGTTACCRAACATYACACC | ||||
| Vi_atpAdg_F | ATCGGTGACCGTCARACWGGTAAAAC | ATP synthase, α subunit (AtpA) | 489 | 60 | This study |
| Vi_atpAdg_R | ATACCTGGGTCAACCGCTGG | ||||
| ViatpA-01-F | CTDAATTCHACNGAAATYAGYG | 57 | 26 | ||
| ViatpA-04-R | TTACCARGWYTGGGTTGC |
DNA extraction, PCR amplification, and sequencing.
For DNA extraction, a single colony from a fresh culture was resuspended in 100 μl nuclease-free water, vortexed at high speed for 5 s, and incubated at 94°C for 10 min. The tube was vortexed again and centrifuged for 2 min at 11,000 relative centrifugal force (RCF). The supernatant was transferred to a fresh tube and stored at −20°C.
PCR amplification was performed in a Euroclone One Advanced thermal cycler (Celbio, Milan, Italy). The final volume of the amplification reaction mixture was 20 μl, containing 1 U of GoTaq polymerase (Promega, Madison, WI), 1× GoTaq buffer, 2.5 mM MgCl2, 0.1 mM each deoxynucleoside triphosphate (dNTP), 125 nM each primer, and 5 ng of genomic DNA as the template.
For the atpA, pyrH, and recA genes, the amplification conditions comprised an initial 2-min denaturation step at 94°C followed by 35 cycles of 20 s at 94°C, 30 s at different annealing temperatures depending on the amplified target, and 30 s at 72°C, with a final extension at 72°C for 7 min. For the gyrB gene, the reaction mixture was subjected to touchdown PCR as follows: an initial step at 95°C for 2 min followed by 40 cycles of denaturation at 95°C for 10 s, annealing at changing temperature (i.e., the temperature changed from 65°C to 55°C in 0.5°C decrements during the first 20 cycles) for 30 s, and extension at 72°C for 50 s, with a final extension at 72°C for 7 min. Amplified products were analyzed by electrophoresis on 1.8% agarose–Tris-acetate-EDTA (TAE) gels, stained with SYBR Safe (Invitrogen, Carlsbad, CA), and visualized on a UV transilluminator.
Conditions for direct sequencing without any additional purification of templates were used, except for a few cases where standard PCR conditions (0.2 mM dNTPs and 250 nM both primers) were used, followed by Illustra ExoStar purification using the manufacturer's standard operating protocol (GE Healthcare Life Sciences UK Limited, United Kingdom).
Bidirectional sequencing of the four target genes was performed using the respective primer pairs used for PCR amplification as sense and antisense sequencing primers. The only exception was for the gyrB gene, where two forward primers were designed to amplify and sequence genes from some strains that were not initially amplified. The nucleotide sequences were determined using the BigDye Terminator cycle sequencing ready reaction kit with AmpliTaq DNA polymerase (Applied Biosystems, Foster City, CA), and the electrophoresis was performed on an ABI Prism 3100 Genetic Analyzer (Applied Biosystems, Foster City, CA) automated sequencer according to the manufacturer's instructions. Finally, the sequences of the amplicons were verified by BLAST search (30) to indicate whether they had similarity to the respective genes for which the primers were designed.
Phylogenetic inference and recombination analyses.
Analysis, editing, and comparison of the chromatograms obtained for the four genes were performed using FinchTV software (Geospiza). The consensus sequence for each gene fragment was determined by alignment of the forward and reverse sequences by using the ClustalW program (http://www.ebi.ac.uk). The coding sequences used for the housekeeping genes were read in frame. Allele sequences that differed from each other by one or more polymorphisms were attributed to a unique allele number in the order of discovery. Each unique allelic profile, defined by the allele numbers of the four loci, was assigned a sequence type (ST) (see Table S1 in the supplemental material). The same ST was used for the strains that shared the same allelic profile. Multiple alignments containing the concatenated sequences were straightforward and were performed according to the genomic gene order gyrB, pyrH, recA, and atpA. The concatemer length was 2,022 nucleotides. Diversity indices, such as the G+C content of each locus, number of polymorphic sites, average numbers of synonymous and nonsynonymous sites, Tajima's D, nucleotide diversity per site (π), and average number of nucleotide differences per site (θ), were calculated using DnaSP version 5.10 (31).
For the phylogenetic analysis, concatenated sequences were aligned and analyzed using MEGA v5.04 (32). Genetic distances were computed by the Kimura two-parameter model and the phylogenetic tree was constructed using the neighbor-joining method. At the same time, a phylogenetic tree was also constructed for each gene to compare the four single gene trees and the concatenated one. The taxon names of each phylogenetic cluster were attributed according to the available representative reference and type strains grouped in the same cluster. The isolates that were considered related but clearly distinct were described as “Vibrio species-like” (e.g., V. mediterranei-like), and the isolates that were not related to any reference strain were referred to by their original names (e.g., Vi20).
Evidence of recombination was investigated using the split decomposition method implemented in SplitsTree 4.10 software (33). Split networks were analyzed using the pairwise homoplasy index (PHI) test (34) to identify alleles with significant evidence of recombination. Furthermore, the phylogenetic trees obtained from each locus were compared with the concatenated sequence tree to investigate whether some genes were affected by recombination.
Population structure and habitat prediction analyses.
The Structure software was used to detect the presence of a subpopulation, depending on the distribution of distinct allele frequencies among the isolates (35). This procedure assigns a probability of ancestry for each polymorphic nucleotide for a given number of groups, K, and it estimates q, the combined probability of ancestry from each of the K groups for each individual isolate. The following parameters were used: five iterations following a burn-in period of 100,000 iterations, Markov chain Monte Carlo [MCMC] = 50,000, and a K value between 1 and 20.
An empirical parsimony algorithm (AdaptML) was used to identify populations as groups of related strains with distinct environmental distributions (24). The software employs a hidden Markov model (HMM) approach to group sequences into ecologically similar habitats, which represent distinct ecologically separated classes of strains. The AdaptML analysis results in a set of emission probabilities that describe the “projected habitats” inferred within the tree. For this purpose, the two ecological variables most correlated to the genetic clusters were included in the AdaptML input (season and shellfish species). In this case, Crassostrea gigas and Ostrea edulis were grouped together for similarity reasons. Tree figures were generated using the interactive Tree of Life web application (http://itol.embl.de) (36).
To determine whether the inferred populations were significantly different, UniFrac software was used (37). The phylogenetic tree was used as the input for the UniFrac significance test (38) for calculation of pairwise P values, for environment clustering, and for the principal-coordinate analysis.
Statistical analyses.
Principal-component analysis (PCA) and canonical correlation analysis (CCA) were performed as multivariate analyses in order to explore the sampling data and to correlate the environmental variables to the genetic subpopulations provided by Structure analysis (see “Phylogenetic inference and recombination analyses” above). Each variable was converted to a “dummy” variable linked to all the states/levels of the qualitative environmental variables. Six variables were considered: origin (shellfish species, n = 9), season (n = 4), water temperature (n = 4), risk level of the area (n = 2), sampling area (n = 2), and depth of sampling (n = 3). Detailed information on the environmental variables is reported in Table S1 in the supplemental material.
PCA was applied considering both the genetic and environmental variables (Matlab 2013 b; Mathworks). CCA was performed to highlight the strongest correlation among environmental information and each genetic group inferred by the Structure software. Taxa including fewer than four strains were removed from the data set (PLS_Toolbox 7.3.1; Eigenvector Research).
The McNemar test was used to test the difference between paired proportions (MLSA versus Alsina's test) using the Marginal Homogeneity program (v. 1.2) (http://www.john-uebersax.com/stat/mh.htm). Both the Bhapkar test (39) and the Stuart-Maxwell test (40, 41) were performed to test overall marginal homogeneity for all the categories simultaneously.
Nucleotide sequence accession numbers.
All DNA sequences were deposited in the Vibrio MLSA database (www.pubmlst.org/vibrio) (27) and in GenBank with accession numbers KF898923 to KF899349.
RESULTS
Biochemical identifications.
One hundred twenty-three shellfish samples out of 164 (75%) were found to be positive for Vibrio spp. Among them, a total of 154 strains were selected as representative of all the different colony morphologies detected through the biochemical tests. The most common species was V. parahaemolyticus (33 isolates), followed by V. alginolyticus (29 isolates) and V. vulnificus (21 isolates). Three strains were unidentified and reported as Vibrio spp. No V. cholerae was isolated in this study. Table S1 in the supplemental material reports all the results of the species assignment given by the biochemical analysis.
Genetic diversity and strain relationships.
One hundred eighty-three strains were analyzed with the MLSA approach; they included 154 Vibrio field strains, 28 Vibrio type and reference strains, and 1 Photobacterium profundum strain. Eight strains (Vi_20, Vi_51, Vi_54, Vi_60, Vi_62, Vi_73, Vi_9a, and Vi_16a) were not amplified with gyrB primers. An alternative forward primer (Vh_gyrB_F) was designed within 60 bp upstream of the Vi_gyrB_F primer in order to maintain the same final gyrB fragment length. All eight samples were amplified with this primer, and they showed an insertion of three bases.
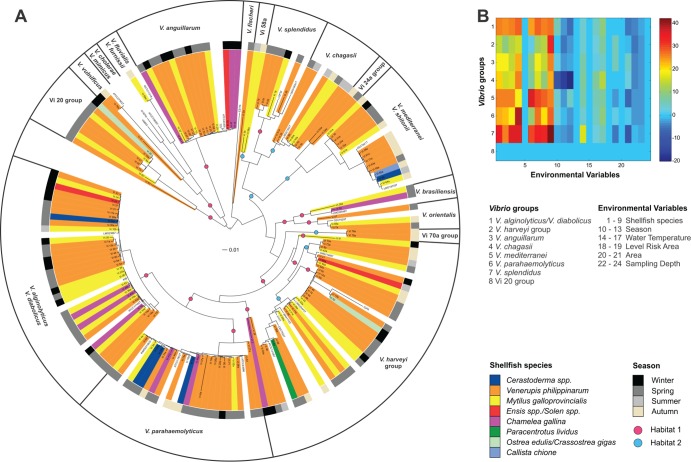
The 182 Vibrio strains analyzed in this study corresponded to 162 distinct STs (see Table S1 in the supplemental material); this high number of different alleles was expected because distinct species/taxa were processed. Only 12 STs included more than one strain: ST 33 and ST 125 each included 4 isolates; ST 3 had 3 isolates; and STs 5, 20, 30, 48, 72, 75, 78, 95, and 123 each included 2 isolates. Examination of the nucleotide variability revealed 18 times more synonymous substitutions than nonsynonymous substitutions, suggesting a predominance of neutral selection. The mean G+C contents of the four genes were very similar and varied from 47.6% (atpA) to 48.2% (pyrH). The genetic equilibrium of alleles was analyzed using Tajima's D neutrality test (42). All of the obtained D values were between −2 and 2, again supporting neutral selection of the considered genes (Table 2). The nucleotide diversity was high in all genes (ranging from 0.089 for atpA to 0.140 for pyrH), and the sequence variability among all Vibrio strains was 38.8%, which corresponded to 805 polymorphic sites (nucleotide diversity of 0.120) in the concatenated sequence. The phylogenetic tree obtained from the concatenated sequences of the four genes allowed a good discrimination of all the Vibrio species considered in this work (Fig. 1). All bootstrap values were highly supported, demonstrating a high reliability of the phylogenetic signal. The isolates that did not cluster in specific groups were not assigned a species name. The phylogenetic analysis conducted on single genes supported the distribution of the concatenated sequences, although small amounts of variation were visible on some species clustering (such as V. harveyi, V. parahaemolyticus, V. alginolyticus, and V. diabolicus) (see Fig. S1 in the supplemental material).
TABLE 2.
Nucleotide diversity observed within the 182 Vibrio strains characterized in this studya
| Locus | Fragment size (bp) | No. of alleles | G+C content | No. (%) of polymorphic sites | No. of parsimony informative sites | No. of synonymous changes | No. of nonsynonymous changes | Tajima's D | θ | π |
|---|---|---|---|---|---|---|---|---|---|---|
| gyrB | 570 | 134 | 0.481 | 235 (41.2) | 219 | 228 | 15 | 0.22409 | 0.164 | 0.135 |
| pyrH | 501 | 107 | 0.482 | 204 (40.7) | 190 | 211 | 9 | 0.17839 | 0.172 | 0.140 |
| recA | 462 | 128 | 0.478 | 195 (42.2) | 179 | 199 | 5 | 0.11590 | 0.171 | 0.139 |
| atpA | 489 | 96 | 0.477 | 171 (35.0) | 152 | 189 | 14 | −0.51451 | 0.101 | 0.089 |
| Concatenate | 2,022b | 162 | 0.480 | 805 (39.8) | 742 | 827 | 43 | 0.08054 | 0.143 | 0.120 |
π, nucleotide diversity per site; θ, average number of nucleotide differences per site.
The concatemer length of the strains belonging to the Vi20 group is 2,025 bp.
FIG 1.
(A) Phylogenetic tree of the Vibrio population analyzed in this work. The colored rectangles refer to the biological source of the strains (shellfish species). The middle ring shows the seasons of sampling. The 17 Vibrio groups inferred by Structure are reported in the outer ring. The colored circles on the tree branches represent the two habitats predicted by AdaptML. (B) CCA pattern, showing canonical correlation between variables. The plot shows the correlation between the most represented Vibrio groups inferred by Structure and the environmental categories considered in this study. The environmental variables are as follows: 1, V. philippinarum; 2, C. gallina; 3, M. galloprovincialis; 4, C. gigas; 5, O. edulis; 6, C. chione; 7, Cerastoderma spp.; 8, Ensis spp./Solen spp.; 9, P. lividus; 10, winter; 11, spring; 12, summer; 13, autumn; 14, 6 to 10°C; 15, 11 to 15°C; 16, 16 to 20°C; 17, 21 to 30°C; 18, A; 19, B; 20, lagoon; 21, sea; 22, 0 to 100 cm; 23, 101 to 200 cm; 24, >200 cm. The color bar (canonical coefficients) indicates the ranges of correlations among variables.
Evidence for recombination was investigated with the SplitsTree program using the split decomposition method separately on each locus and on the concatenated sequences of all STs. Individual genes were not significantly affected by intragenic recombination, but in all cases, parallelogram formation was evident, which is indicative of some recombination events (data not shown). However, significant recombination (P = 1.8 × 10−14) was found when the concatenated sequence of all STs were considered. In addition, the five most represented species (V. alginolyticus-V. diabolicus group, V. anguillarum-like, V. harveyi-group, V. mediterranei, and V. parahaemolyticus-like) were analyzed separately to evaluate whether some specific taxon was particularly affected by genetic transfer, and only the V. alginolyticus-V. diabolicus group showed a significant presence of recombination (P = 1.2 × 10−6). The Structure software was used to evaluate the population structure and identify the groups that differed in terms of their allele frequencies. In addition, the software allows the detection of more subtle recombination events and potential import of foreign DNA. Seventeen subpopulations were identified, as repeated analyses showed that the model probability was best supported at a K value of 17. Within the same species, most strains were homogeneous, supporting the low presence of recombination found in the previous analyses. However, some strains presented mixed colors in the corresponding column, which demonstrates the import of gene sequences from other species (see Fig. S2 in the supplemental material). The 17 groups detected by Structure corresponded in large measure with those of named Vibrio species, supporting the phylogenetic grouping obtained by the neighbor-joining analysis.
Statistical analyses and comparison of biochemical and MLSA approaches.
PCA and CCA were conducted in a restricted data set containing 134 strains (the Structure groups including fewer than four strains were removed from the analysis). Considering the data set including both the environmental and genetic variables, the first two principal components (PCs) of the PCA explained the majority of the data set variability (68%). Two main groups were separated along the scores of PC1 and PC2, and the loadings highlighted that the dominant variables involved in the clusters' definition are four descriptors: water temperature, season, shellfish species, and sampling area (data not shown). In particular, the first group of strains grouped isolates collected during cold seasons (winter/spring) and/or from Mytilus galloprovincialis, while the strains belonging to the second group were isolated mainly from clams collected in the lagoon during summer and the late part of spring. According to the Structure groups, the most informative taxa were V. alginolyticus-diabolicus and V. anguillarum, which showed the highest loading values. Moreover, V. anguillarum seemed to be more related to samples collected during cold seasons. The same data set was applied to CCA. The first six canonical components described the majority of the canonical correlations among the two data sets (Fig. 1B). According to the canonical coefficients of each variable, the shellfish species and the season were the most significant variables, followed by the water temperature. The season seemed to be anticorrelated to some Structure groups (Fig. 1B). At the same time, the shellfish species highlighted a high level of correlation to the genetic groups for the majority of the canonical components considered descriptive of the system. Taken together, the two statistical approaches suggest that the shellfish species and the season are the most informative variables in relation to the species groups. The last Structure group (Vi 20 group) did not show any significant correlation with any environmental information, probably because this cluster grouped isolates with undefined attribution and different isolation conditions. Moreover, this cluster included all the strains carrying the three-base insertion in gyrB. Six environmental variables did not show correlation with any Structure group (canonical coefficient equal to 0) (Fig. 1B).
The McNemar test was used to compare the two species identification methods used in this work (Alsina's test and MLSA). The classifications were inconsistent for the most represented taxa (i.e., those including more than three strains), except for V. mediterranei and V. anguillarum, which exhibited a good correspondence. Among the higher-risk Vibrio species, V. vulnificus and V. parahaemolyticus were overestimated by Alsina's method (Table 3).
TABLE 3.
Comparison between Alsina's test and MLSA by the McNemar testa
| Category | Frequencyd |
P | |
|---|---|---|---|
| Alsina | MLSA | ||
| V. alginolyticus-V. diabolicus group | 26 | 33 | 0.0348 |
| V. anguillarum (-like) | 10 | 15 | 0.2752 |
| V. brasiliensis (-like) | 0 | 2 | 0.5000b |
| V. harveyi group | 9 | 30 | 0.0002c |
| V. chagasii | 0 | 12 | 0.0005c |
| V. fischeri | 3 | 0 | 0.2500b |
| V. fluvialis | 17 | 0 | 0.0000c |
| V. furnissii | 0 | 1 | 10.000b |
| V. logei | 4 | 0 | 0.1250b |
| V. marinus | 1 | 0 | 10.000b |
| V. mediterranei (-like)/V. shilonii | 14 | 14 | 10.000b |
| V. mimicus | 3 | 0 | 0.2500b |
| V. nereis | 4 | 0 | 0.1250b |
| V. orientalis | 0 | 3 | 0.2500b |
| V. parahaemolyticus (-like) | 33 | 23 | 0.0039 |
| V. pelagius | 4 | 0 | 0.1250b |
| V. splendidus (ll) | 9 | 5 | 0.2482 |
| V. vulnificus (B2) | 14 | 1 | 0.0003c |
| Vibrio spp. | 3 | 15 | 0.0047 |
Tests of overall marginal homogeneity: Bhapkar chi-square, 124.716 (degrees of freedom [df] = 18, P = 0.0000); Stuart-Maxwell chi-square, 68.910 (df = 18, P = 0.0000). Bowker symmetry test chi-square, 78.867 (df = 171, P = 1.0000).
A two-tailed exact test was applied.
P < Bonferroni-adjusted significance criterion of 0.003.
Numbers of strains identified by the indicated method.
Habitat predictions and ecology.
The analysis performed through AdaptML allowed the simultaneous evaluation of ecological differentiation and genetic background of the isolates. After 100 repetitions, the predictive model identified two projected habitats as the most supported ecological diversification of the Vibrio strains (Fig. 1). Generally, the two groups were diverse in terms of seasonality: habitat 1 included strains isolated mainly during the cold seasons (winter-spring), and habitat 2 grouped the majority of the strains collected during the warm seasons (summer-autumn). There is no clear correlation between some specific taxa and the source, suggesting the wide distribution of Vibrio among different shellfish species. In addition, unweighted UniFrac was used to cluster the Vibrio strains according to shared similarities. The UniFrac significance and the environment clustering tests were employed to evaluate which environments were statistically different from each other (Fig. 2; see Table S2 in the supplemental material), while the principal-coordinate analysis showed which ecologies were most closely related to one another. The software supported the ecological differentiation based on the seasonality obtained through the AdaptML software: cold seasons were statistically different from warm seasons in terms of Vibrio species distribution. In addition, the three tests highlighted a further distinction: V. philippinarum, M. galloprovincialis, and C. gallina were found to be significantly different from the other sources and formed a distinct group (Fig. 2).
FIG 2.
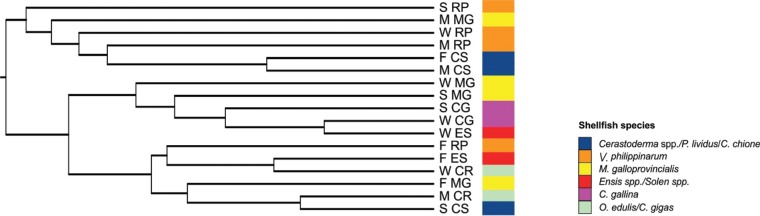
Tree obtained by the UniFrac environment clustering test. The colors refer to the biological sources of the samples. CR, Crassostrea gigas/Ostrea edulis; ES, Ensis spp./Solen spp.; CG, Chamelea gallina; CS, Cerastoderma spp./Paracentrosus lividus/Callista chione; MG, Mytilus galloprovincialis; RP, Venerupis philippinarum; F, autumn; M, summer; S, spring; W, winter.
DISCUSSION
Vibrionaceae are ubiquitous marine heterotrophic bacteria whose taxonomy has been extensively studied and revised, especially in the last years (29, 43, 44, 45, 46, 47). In particular, the new microbiological and molecular trends have recently seen the mutual analysis of phenotypic, genotypic, and environmental variables, as these features are clearly interconnected in bacterial evolution (24, 48, 49). However, the ecological processes of bacterial dynamics in the environment remain poorly understood (50). Vibrio populations from different parts of the world have been analyzed in independent studies that described species distribution in the environment (habitats and hosts) and also highlighted the strong connection between this genus and specific ecological variables (48, 50, 51). The Venice lagoon is an area of great economic importance for mollusk commerce. Little information is available concerning the distribution of Vibrionaceae in this area, and most of the studies are related to marine samples (e.g., sediments, plankton, or water) (52, 53, 54). In this work, we developed and applied an MLSA scheme based on four housekeeping genes, together with different statistical and predictive models, to characterize the Vibrio species from the Venice lagoon and to explore to what extent different ecologies represented distinct habitats for this genus. The work was based on the simultaneous analysis of the core genetic information and a set of ecological features (season, host species, risk level of the area, water temperature, and area and depth of sampling), with the aim to understand whether they were interconnected. A total of 182 Vibrio strains were analyzed. The MLSA scheme was demonstrated to be very simple and useful for discriminating Vibrio species; the distribution and clustering of the taxa achieved a high degree of discrimination that supported the results of previous studies overall (25, 43, 55). Most species were easily identified through population and phylogenetic analyses, while other species were grouped together (V. harveyi group and V. alginolyticus/V. diabolicus). This is not surprising, as other studies have already demonstrated the genetic similarity of these taxa (25, 26, 29). In particular, regarding the V. alginolyticus and V. diabolicus species, the concatenated gene sequence tree and the SplitsTree analysis revealed two subclusters, while the Structure software showed a unique group. This is also consistent with the phylogenetic analyses of the recA and atpA genes (see Fig. S1 in the supplemental material), in which the two groups seemed to be more distantly related. This result suggests that the two species might have originally been the same species and that they underwent recombination events that led to differentiation of some strains into a distinct species. Another example is the V. harveyi group: it is known that the cluster comprises four species (V. harveyi, V. campbellii, V. rotiferanus, and V. owensii) (56), but the four-gene MLSA did not permit discrimination among them. In the cases reported above, the use of a higher number of molecular markers would be useful to investigate the taxonomic positions of particular isolates or to detect pathogenic strains, as demonstrated in other studies (43, 56, 57).
However, the precise strain typing and detection of pathogens for aquaculture organisms were not the aims of this work, which were instead focused on a wider characterization of the Vibrio species of the Venice lagoon related to their risk level. Regarding this point, the MLSA developed in this study allowed easy discrimination of the high-risk Vibrio species from the low-risk species. Most of the strains isolated from mollusks belonged to the low-risk group: V. alginolyticus, V. fluvialis, V. furnissii, V. harveyi, and V. mimicus were the most frequently isolated species. Twenty-five out of 154 isolates (16.23%) clustered in the high-risk group. No V. cholerae was isolated, while both V. parahaemolyticus and V. vulnificus strains were identified and a precise clustering was achieved for both of them.
The comparative statistical analysis performed for biochemical and molecular identification demonstrated the higher reliability of the latter. In particular, the biochemical results overestimated the two high-risk Vibrio species (V. vulnificus and V. parahaemolyticus). Although they represented a small part of the total Vibrio species isolated in this study, their presence still represents worrying data on the safety of shellfish in the Venice lagoon. This highlights the need to confirm biochemical data with molecular methods to avoid false-positive results, particularly for high-risk Vibrio species.
The molecular results obtained in this work allowed characterization of the Vibrio populations that inhabit the main mollusk species reared or caught in the Venice lagoon. However, a deeper understanding of the distribution of the bacterial species in the environment also relies on additional information. These data include the phenotypic and ecological characteristics of the bacterial strains. This polyphasic approach has been commonly used in prokaryotic taxonomy in recent years (24, 48, 49, 50, 51, 58). We integrated the MLSA molecular data with a set of environmental categories to investigate whether the Vibrio populations also had an ecological trend. The most significant result was the influence of seasonality on species diversity. This is consistent with previous work conducted both on the Vibrio genus and on single species in different parts of the world (48, 50, 59). We performed several statistical and population analyses to understand whether ecological variables could be linked to particular species and which of them were the most informative. All the different approaches that we employed (PCA, UniFrac, and AdaptML) suggested a distinction between the Vibrio species isolated during the cold seasons (winter and spring) and those isolated during the warm seasons (summer and autumn), suggesting that seasons have a strong influence on the population structure and species distribution. According to AdaptML, some species, such as V. anguillarum, V. alginolyticus, V. diabolicus, and V. splendidus, were specific to cold seasons, while V. mediterranei, V. shilonii, V. chagasii, V. brasiliensis, and V. orientalis were isolated predominantly during warm seasons (Fig. 1). The V. harveyi group and V. parahaemolyticus included strains that were isolated during both cold and warm seasons, although the majority of them were found during spring. The two habitats predicted by the quantitative model AdaptML reflected this seasonality; however, there were a few exceptions, that is, strains isolated during summer/autumn seasons that were assigned to “cold season” habitats. This is the case of the underrepresented clusters, such as V. vulnificus and V. fluvialis/V. furnissii, which included just one field isolate whose informativeness was not high enough to support an independent clustering. The predictability of host association and the importance of the ecological data were tested using another method (three UniFrac tests), which supported the separation between cold and warm seasons but also pointed out a further distinction: host species diversity was detected (Fig. 2). In particular, V. philippinarum, M. galloprovincialis, and C. gallina were found to be significantly different from the other reservoirs. Nevertheless, further analyses are needed to comprehend and support this result, as the sampling scheme was not uniform with regard to the host species. As a matter of fact, the three species reported above were the most represented ones, with 91, 37, and 10 Vibrio isolates, respectively, while the remaining mollusk species bore fewer than 10 isolates each. However, it is important to report that all the low-represented mollusk species had at least one Vibrio species, supporting the wide distribution of this genus in the aquatic environment. At the same time, this may suggest that there is no selection of reservoirs by the different taxa, but this result needs to be further investigated; bacterial population structure in animals may be weak just because selection can be balanced by migration and/or adaptation to different environments (50). In fact, the ability of Vibrio species to disperse laterally among hosts is well documented (24, 50, 51).
In conclusion, this work employed different statistical and evolution analyses to shed light on the dynamics of the Vibrio genus in the mollusks of the Venice lagoon. The results highlighted the wide presence of Vibrio species in the environment, with the majority of samples bearing at least one species. Low-risk Vibrio species represented most of the species isolated, although high-risk species were also detected. This suggests the need for precise monitoring of the shellfish commerce and the importance of confirming biochemical analyses with more reliable screening techniques. The MLSA demonstrated a reliable characterization of the main Vibrio species and provided an overview of the ecology of Vibrio spp. in the Venice lagoon. The concatenated MLSA loci highlighted an evolutionary history that was ecologically coherent and supported the use of core genes for ecological studies. In addition, the combined use of different algorithms provided a clear view of the species distribution, which was clearly dependent on the temperature (seasonality). Bacterial phylogenetic diversity in this environment is structured mainly by seasons, rather than other environmental variables. This result supports the recent trend of considering an integrated approach to studying prokaryotic taxonomy and dynamics, as investigation of bacterial evolution must consider ecology.
Supplementary Material
ACKNOWLEDGMENTS
This work was funded by a Ph.D. grant from the School of Veterinary Science of the University of Padua to support the education of M.S.R. and by a Ph.D. grant from Erasmus Mundus Mobility with Asia (EMMA).
Footnotes
Published ahead of print 31 January 2014
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.04133-13.
REFERENCES
- 1.Thompson FL, Iida T, Swings J. 2004. Biodiversity of vibrios. Microbiol. Mol. Biol. Rev. 68:403–431. 10.1128/MMBR.68.3.403-431.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Austin B, Austin DA. 2007. Bacterial fish pathogens: diseases of farmed and wild fish, 4th ed. Springer Praxis, New York, NY [Google Scholar]
- 3.Le Roux F, Gay M, Lambert C, Waechter M, Poubalanne S, Chollet B, Nicolas JL, Berthe F. 2002. Comparative analysis of Vibrio splendidus-related strains isolated during Crassostrea gigas mortality events. Aquat. Living Res. 15:251–258. 10.1016/S0990-7440(02)01176-2 [DOI] [Google Scholar]
- 4.Austin B. 2010. Vibrios as causal agents of zoonoses. Vet. Microbiol. 140:310–317. 10.1016/j.vetmic.2009.03.015 [DOI] [PubMed] [Google Scholar]
- 5.Ottaviani D, Leoni F, Rocchegiani E, Santarelli S, Canonico C, Masini L, DiTrani V. 2008. First clinical report of pandemic Vibrio parahaemolyticus O3:K6 infection in Italy. J. Clin. Microbiol. 46:2144–2145. 10.1128/JCM.00683-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ottaviani D, Leoni F, Rocchegiani E, Canonico C, Potenziani S, Santarelli S, Masini L, Scuota S, Carraturo A. 2010. Vibrio parahaemolyticus-associated gastroenteritis in Italy: persistent occurrence of O3:K6 pandemic clone and emergence of O1:KUT serotype. Diagn. Microbiol. Infect. Dis. 66:452–455. 10.1016/j.diagmicrobio.2009.11.015 [DOI] [PubMed] [Google Scholar]
- 7.Beaz-Hidalgo R, Balboa S, Romalde JL, Figueras MJ. 2010. Diversity and pathogenicity of Vibrio species in cultured bivalve molluscs. Environ. Microbiol. Rep. 2:34–43. 10.1111/j.1758-2229.2010.00135.x [DOI] [PubMed] [Google Scholar]
- 8.Balboa S, Romalde JL. 2013. Multilocus sequence analysis of Vibrio tapetis, the causative agent of brown ring disease: description of Vibrio tapetis subsp. britannicus subsp. nov. Syst. Appl. Microbiol. 36:183–187. 10.1016/j.syapm.2012.12.004 [DOI] [PubMed] [Google Scholar]
- 9.Oliveira J, Cunha A, Castilho F, Romalde JL, Pereira MJ. 2011. Microbial contamination and purification of bivalve shellfish: crucial aspects in monitoring and future perspectives. Food Control 22:805–816. 10.1016/j.foodcont.2010.11.032 [DOI] [Google Scholar]
- 10.Ottaviani D, Santarelli S, Bacchiocchi S, Masini L, Ghittino C, Bacchiocchi I. 2005. Presence of pathogenic Vibrio parahaemolyticus strains in mussels from the Adriatic Sea, Italy. Food Microbiol. 22:585–590. 10.1016/j.fm.2005.01.005 [DOI] [PubMed] [Google Scholar]
- 11.Alsina M, Blanch AR. 1994. A set of keys for biochemical identification of environmental Vibrio species. J. Appl. Bacteriol. 76:79–85. 10.1111/j.1365-2672.1994.tb04419.x [DOI] [PubMed] [Google Scholar]
- 12.Alsina M, Blanch AR. 1994. Improvement and update of a set of keys for biochemical identification of Vibrio species. J. Appl. Bacteriol. 77:719–721. 10.1111/j.1365-2672.1994.tb02824.x [DOI] [PubMed] [Google Scholar]
- 13.Ottaviani D, Masini L, Bacchiocchi S. 2003. A biochemical protocol for the isolation and identification of current species of Vibrio in seafood. J. Appl. Microbiol. 95:1277–1284. 10.1046/j.1365-2672.2003.02105.x [DOI] [PubMed] [Google Scholar]
- 14.Vandenberghe J, Thompson FL, Gomez-Gil B, Swings J. 2003. Phenotypic diversity amongst Vibrio isolates from marine aquaculture systems. Aquaculture 219:9–20. 10.1016/S0044-8486(02)00312-5 [DOI] [Google Scholar]
- 15.Prakash O, Verma M, Sharma P, Kumar M, Kumari K, Singh A, Kumari H, Jit S, Gupta SK, Khanna M, Lal R. 2007. Polyphasic approach of bacterial classification—an overview of recent advances. Indian J. Microbiol. 47:98–108. 10.1007/s12088-007-0022-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bauer A, Rørvik LM. 2007. A novel multiplex PCR for the identification of Vibrio parahaemolyticus, Vibrio cholerae and Vibrio vulnificus. Lett. Appl. Microbiol. 45:371–375. 10.1111/j.1472-765X.2007.02195.x [DOI] [PubMed] [Google Scholar]
- 17.Neogi S, Chowdhury N, Asakura M, Hinenoya A, Haldar S, Saidi S, Kogure K, Lara R, Yamasaki S. 2010. A highly sensitive and specific multiplex PCR assay for simultaneous detection of Vibrio cholerae, Vibrio parahaemolyticus and Vibrio vulnificus. Lett. Appl. Microbiol. 51:293–300. 10.1111/j.1472-765X.2010.02895.x [DOI] [PubMed] [Google Scholar]
- 18.Espiñeira M, Atanassova M, Vieites JM, Santaclara FJ. 2010. Validation of a method for the detection of five species, serogroups, biotypes and virulence factors of Vibrio by multiplex PCR in fish and seafood. Food Microbiol. 27:122–131. 10.1016/j.fm.2009.09.004 [DOI] [PubMed] [Google Scholar]
- 19.Tarr CL, Patel JS, Puhr ND, Sowers EG, Bopp CA, Strockbine NA. 2007. Identification of Vibrio isolates by a multiplex PCR assay and rpoB sequence determination. J. Clin. Microbiol. 45:134–140. 10.1128/JCM.01544-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cummings PJ, Ahmed R, Durocher JA, Jessen A, Vardi T, Obom KM. 2013. Pyrosequencing for microbial identification and characterization. J. Vis. Exp. 22:e50405. 10.3791/50405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Stackebrandt E, Frederiksen W, Garrity GM, Grimont PAD, Kämpfer P, Maiden MCJ, Nesme X, Rosselló-Mora R, Swings J, Trüper HG, Vauterin L, Ward AC, Whitman WB. 2002. Report of the Ad Hoc Committee for the Re-evaluation of the Species Definition in Bacteriology. Int. J. Syst. Evol. Microbiol. 52:1043–1047. 10.1099/ijs.0.02360-0 [DOI] [PubMed] [Google Scholar]
- 22.Bisharat N, Cohen DI, Harding RM, Falush D, Crook DW, Peto T, Maiden MC. 2005. Hybrid Vibrio vulnificus. Emerg. Infect. Dis. 11:30–35. 10.3201/eid1101.040440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.González-Escalona N, Martinez-Urtaza J, Romero J, Espejo RT, Jaykus LA, DePaola A. 2008. Determination of molecular phylogenetics of Vibrio parahaemolyticus strains by multilocus sequence typing. J. Bacteriol. 190:2831–2840. 10.1128/JB.01808-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hunt DE, David LA, Gevers D, Preheim SP, Alm EJ, Polz MF. 2008. Resource partitioning and sympatric differentiation among closely related bacterioplankton. Science 320:1081–1085. 10.1126/science.1157890 [DOI] [PubMed] [Google Scholar]
- 25.Thompson FL, Gevers D, Thompson CC, Dawyndt P, Naser S, Hoste B, Munn CB, Swings J. 2005. Phylogeny and molecular identification of vibrios on the basis of multilocus sequence analysis. Appl. Environ. Microbiol. 71:5107–5115. 10.1128/AEM.71.9.5107-5115.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Thompson CC, Thompson FL, Vicente ACP, Swings J. 2007. Phylogenetic analysis of vibrios and related species by means of atpA gene sequences. Int. J. Syst. Evol. Microbiol. 57:2480–2484. 10.1099/ijs.0.65223-0 [DOI] [PubMed] [Google Scholar]
- 27.Jolley KA, Maiden M. 2010. BIGSdb: scalable analysis of bacterial genome variation at the population level. BMC Bioinformatics 11:595. 10.1186/1471-2105-11-595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fredslund J, Schauser L, Madsen LH, Sandal N, Stougaard J. 2005. PriFi: using a multiple alignment of related sequences to find primers for amplification of homologs. Nucleic Acids Res. 33:W516–W520. 10.1093/nar/gki425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Thompson FL, Gomez-Gil B, Vasconcelos ATR, Sawabe T. 2007. Multilocus sequence analysis reveals that Vibrio harveyi and V. campbelli are distinct species. Appl. Environ. Microbiol. 73:4279–4285. 10.1128/AEM.00020-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389–3402. 10.1093/nar/25.17.3389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Librado P, Rozas J. 2009. DnaSP v5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics 25:1451–1452. 10.1093/bioinformatics/btp187 [DOI] [PubMed] [Google Scholar]
- 32.Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. 2011. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 28:2731–2739. 10.1093/molbev/msr121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Huson DH, Bryant D. 2006. Application of phylogenetic networks in evolutionary studies. Mol. Biol. Evol. 23:254–267. 10.1093/molbev/msj030 [DOI] [PubMed] [Google Scholar]
- 34.Bruen TC, Philippe H, Bryant D. 2006. A simple and robust statistical test for detecting the presence of recombination. Genetics 172:2665–2681. 10.1534/genetics.105.048975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Falush D, Stephens M, Pritchard JK. 2003. Inference of population structure using multilocus genotype data: linked loci and correlated allele frequencies. Genetics 164:1567–1587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Letunic I, Bork P. 2007. Interactive Tree Of Life (iTOL): an online tool for phylogenetic tree display and annotation. Bioinformatics 23:127–128. 10.1093/bioinformatics/btl529 [DOI] [PubMed] [Google Scholar]
- 37.Lozupone C, Hamady M, Knight R. 2006. UniFrac—an online tool for comparing microbial community diversity in a phylogenetic context. BMC Bioinformatics 7:371. 10.1186/1471-2105-7-371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Martin AP. 2002. Phylogenetic approaches for describing and comparing the diversity of microbial communities. Appl. Environ. Microbiol. 68:3673–3682. 10.1128/AEM.68.8.3673-3682.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bhapkar VP. 1966. A note on the equivalence of two test criteria for hypotheses in categorical data. J. Am. Stat. Assoc. 61:228–235. 10.1080/01621459.1966.10502021 [DOI] [Google Scholar]
- 40.Stuart AA. 1955. A test for homogeneity of the marginal distributions in a two-way classification. Biometrika 42:412–416. 10.1093/biomet/42.3-4.412 [DOI] [Google Scholar]
- 41.Maxwell AE. 1970. Comparing the classification of subjects by two independent judges. Br. J. Psychiat. 116:651–655. 10.1192/bjp.116.535.651 [DOI] [PubMed] [Google Scholar]
- 42.Tajima F. 1989. Statistical methods to test for nucleotide mutation hypothesis by DNA polymorphism. Genetics 123:585–595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sawabe T, Kita-Tsukamoto K, Thompson FL. 2007. Inferring the evolutionary history of vibrios by means of multilocus sequence analysis. J. Bacteriol. 189:7932–7936. 10.1128/JB.00693-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Thompson FL, Thompson CC, Dias GM, Naka H, Dubay C, Crosa JH. 2011. The genus Listonella MacDonell and Colwell 1986 is a later heterotypic synonym of the genus Vibrio Pacini 1854 (Approved Lists 1980)—a taxonomic opinion. Int. J. Syst. Evol. Microbiol. 61:3023–3027. 10.1099/ijs.0.030015-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chimetto LA, Cleenwerck I, Moreira APB, Brocchi M, Willems A, De Vos P, Thompson FL. 2011. Vibrio variabilis sp. nov. and Vibrio maritimus sp. nov., isolated from Palythoa caribaeorum. Int. J. Syst. Evol. Microbiol. 61:3009–3015. 10.1099/ijs.0.026997-0 [DOI] [PubMed] [Google Scholar]
- 46.Yoshizawa S, Tsuruya Y, Fukui Y, Sawabe T, Yokota A, Kogure K, Higgins M, Carson J, Thompson FL. 2012. Vibrio jasicida sp. nov., a member of the Harveyi clade, from marine animals (packhorse lobster, abalone, and Atlantic salmon). Int. J. Syst. Evol. Microbiol. 62:1864–1870. 10.1099/ijs.0.025916-0 [DOI] [PubMed] [Google Scholar]
- 47.Urbanczyk H, Ast JC, Higgins MJ, Carson J, Dunlap PV. 2007. Reclassification of Vibrio fischeri, Vibrio logei, Vibrio salmonicida and Vibrio wodanis as Aliivibrio fischeri gen. nov., comb. nov., Aliivibrio logei comb. nov., Aliivibrio salmonicida comb. nov. and Aliivibrio wodanis comb. nov. Int. J. Syst. Evol. Microbiol. 57:2823–2829. 10.1099/ijs.0.65081-0 [DOI] [PubMed] [Google Scholar]
- 48.Ellis CN, Schuster BM, Striplin MJ, Jones SH, Whistler CA, Cooper VS. 2012. Influence of seasonality on the genetic diversity of Vibrio parahaemolyticus in New Hampshire shellfish waters as determined by multilocus sequence analysis. Appl. Environ. Microbiol. 78:3778–3782. 10.1128/AEM.07794-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Martino ME, Fasolato L, Montemurro F, Novelli E, Cardazzo B. Aeromonas spp.: ubiquitous or specialized bugs? Environ. Microbiol. 2013 Aug 6; doi: 10.1111/1462-2920.12215. [DOI] [PubMed] [Google Scholar]
- 50.Preheim SP, Boucher Y, Wildschutte H, David LA, Veneziano D, Alm EJ, Polz MF. 2011. Metapopulation structure of Vibrionaceae among coastal marine invertebrates. Environ. Microbiol. 13:265–275. 10.1111/j.1462-2920.2010.02328.x [DOI] [PubMed] [Google Scholar]
- 51.Preheim SP, Timberlake S, Polz MF. 2011. Merging taxonomy with ecological population prediction in a case study of Vibrionaceae. Appl. Environ. Microbiol. 77:7195–7206. 10.1128/AEM.00665-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Caburlotto G, Gennari M, Ghidini V, Tafi M, Lleo MM. 2009. Presence of T3SS2 and other virulence-related genes in tdh-negative Vibrio parahaemolyticus environmental strains isolated from marine samples in the area of the Venetian Lagoon, Italy. FEMS Microbiol. Ecol. 70:506–514. 10.1111/j.1574-6941.2009.00764.x [DOI] [PubMed] [Google Scholar]
- 53.Caburlotto G, Haley BJ, Lleò MM, Huq A, Colwell RR. 2010. Serodiversity and ecological distribution of Vibrio parahaemolyticus in the Venetian Lagoon, northeast Italy. Environ. Microbiol. Rep. 2:151–157. 10.1111/j.1758-2229.2009.00123.x [DOI] [PubMed] [Google Scholar]
- 54.Lafisca A, Pereira CS, Giaccone V, Rodrigues Ddos P. 2008. Enzymatic characterization of Vibrio alginolyticus strains isolated from bivalves harvested at Venice Lagoon (Italy) and Guanabara Bay (Brazil). Rev. Inst. Med. Trop. Sao Paulo 50:199–202. 10.1590/S0036-46652008000400002 [DOI] [PubMed] [Google Scholar]
- 55.Tall A, Hervio-Heath D, Teillon A, Boisset-Helbert C, Delesmont R, Bodilis J, Touron-Bodilis A. 2013. Diversity of Vibrio spp. isolated at ambient environmental temperature in the eastern English Channel as determined by pyrH sequencing. J. Appl. Microbiol. 114:1713–1724. 10.1111/jam.12181 [DOI] [PubMed] [Google Scholar]
- 56.Cano-Gomez A, Høj L, Owens L, Andreakis N. 2011. Multilocus sequence analysis provides basis for fast and reliable identification of Vibrio harveyi-related species and reveals previous misidentification of important marine pathogens. Syst. Appl. Microbiol. 34:561–565. 10.1016/j.syapm.2011.09.001 [DOI] [PubMed] [Google Scholar]
- 57.Hoffmann M, Monday SR, Fischer M, Brown EW. 2012. Genetic and phylogenetic evidence for misidentification of Vibrio species within the Harveyi clade. Lett. Appl. Microbiol. 54:160–165. 10.1111/j.1472-765X.2011.03183.x [DOI] [PubMed] [Google Scholar]
- 58.Mazard S, Ostrowski M, Partensky F, Scanlan DJ. 2012. Multi-locus sequence analysis, taxonomic resolution and biogeography of marine Synechococcus. Environ. Microbiol. 14:372–386. 10.1111/j.1462-2920.2011.02514.x [DOI] [PubMed] [Google Scholar]
- 59.Azandégbé A, Garnier M, Andrieux-Loyer F, Kérouel R, Philippon X, Nicolas JL. 2010. Occurrence and seasonality of Vibrio aestuarianus in sediment and Crassostrea gigas haemolymph at two oyster farms in France. Dis. Aquat. Organ. 91:213–221. 10.3354/dao02253 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.