Abstract
We previously identified a decapeptide from the model plant Arabidopsis thaliana, OSIP108, which is induced upon fungal pathogen infection. In this study, we demonstrated that OSIP108 interferes with biofilm formation of the fungal pathogen Candida albicans without affecting the viability or growth of C. albicans cells. OSIP108 displayed no cytotoxicity against various human cell lines. Furthermore, OSIP108 enhanced the activity of the antifungal agents amphotericin B and caspofungin in vitro and in vivo in a Caenorhabditis elegans-C. albicans biofilm infection model. These data point to the potential use of OSIP108 in combination therapy with conventional antifungal agents. In a first attempt to unravel its mode of action, we screened a library of 137 homozygous C. albicans mutants, affected in genes encoding cell wall proteins or transcription factors important for biofilm formation, for altered OSIP108 sensitivity. We identified 9 OSIP108-tolerant C. albicans mutants that were defective in either components important for cell wall integrity or the yeast-to-hypha transition. In line with these findings, we demonstrated that OSIP108 activates the C. albicans cell wall integrity pathway and that its antibiofilm activity can be blocked by compounds inhibiting the yeast-to-hypha transition. Furthermore, we found that OSIP108 is predominantly localized at the C. albicans cell surface. These data point to interference of OSIP108 with cell wall-related processes of C. albicans, resulting in impaired biofilm formation.
INTRODUCTION
Yeasts of the genus Candida are opportunistic human fungal pathogens causing life-threatening systemic infections, particularly in immunocompromised patients. Candida spp. are recognized as the fourth most common cause of bloodstream infections in the United States (1), with high attributable mortality rates up to 40% (2). Although Candida glabrata and Candida krusei are increasingly being isolated and have an increased resistance to commonly used antifungals, Candida albicans remains the most common fungal pathogen (3–5). In natural environments like the human body, Candida spp. occur preferably in biofilms. The latter are well-structured populations of microbial cells attached to a surface and embedded in a self-produced polymer matrix (6, 7). Biofilms have a great significance for public health, as they are critical in the development of clinical infections and are frequently refractory to conventional antimicrobial agents (8, 9). From the currently available antifungal agents, liposomal formulations of amphotericin B (AMB) and the echinocandins such as caspofungin (CAS) can effectively treat C. albicans biofilms (10). As those two classes of antifungal agents have limitations due to toxicity, resistance, and bioavailability issues, we are clearly in need of new antibiofilm agents and therapies. In combination with improved assays for faster and more sensitive diagnosis of biofilm involvement in fungal infections (11), another need in current antimycotic therapies, such novel antibiofilm compounds would significantly improve the efficacy of current treatments of biofilm-associated fungal infections.
Candida species often form biofilms on medical devices, such as urinary and vascular catheters, (dental) implants, prostheses, and heart valves (12, 13). Besides their role as a constant source of cells leading to chronic infections due to their increased resistance to antifungal treatment, those biofilms may also cause device failure. Treatment often requires the removal of the device, which is associated with high costs and patient discomfort (13). Besides systemic administration of antibiofilm molecules to cure biofilm-associated infections, such molecules can be used as a coating on medical devices, thereby preventing proper biofilm formation of microbial pathogens on the device and thus resulting in a reduced risk for the development of biofilm-associated device infections. Current antibiofilm coatings of medical devices are mainly based on the use of silver ions, which are toxic upon accumulation (14), or on the release of standard antibiotics/antimycotics for which biofilms display increased resistance.
The study of plants as an alternative to other forms of drug discovery has attracted great attention because, according to the World Health Organization, these would be the best sources for obtaining a wide variety of drugs and could benefit a large population (15). We previously identified a decapeptide of Arabidopsis thaliana, OSIP108, which is induced in plants upon infection by the fungal pathogen Botrytis cinerea (16). In this study, we investigated the potential antifungal effect of OSIP108 against different fungi, including the human pathogen C. albicans. Furthermore, as microbial biofilm formation was recently found to be a prerequisite for infection in various hosts, we also investigated the effect of OSIP108 on biofilm formation of C. albicans. We subsequently performed a preliminary structure-activity relationship study of OSIP108 consisting of a scrambled version and cyclic derivatives of OSIP108 and investigated possible cytotoxic effects of OSIP108 against human osteoblasts, mesenchymal stem cells, and endothelial cells. In addition, we investigated the effects of OSIP108 on the activities of commonly used antifungal agents such as AMB and CAS against mature biofilms in vitro and in vivo in a Caenorhabditis elegans-C. albicans infection model. Finally, to gain more insight into the antibiofilm activity of OSIP108, we screened a library of 137 C. albicans homozygous mutants affected in cell wall proteins or transcription factors for both resistance and hypersensitivity to OSIP108, aiming at identification of OSIP108 targets and tolerance mechanisms to OSIP108 in C. albicans, respectively.
MATERIALS AND METHODS
Strains and chemicals.
The strains C. albicans SC5314, CAI4, CAF2 (17), DAY286 (18), B2630, and B63195 (19); C. glabrata BG2 (20); Candida dubliniensis NCPF 3949 (21); and C. krusei IHEM 6104 (Belgian Coordinated Collections Of Microorganisms [BCCM]/IHEM, Brussels, Belgium) were used in this study. Homozygous deletion mutants derived from C. albicans BWP17 (22) with the isogenic wild-type (WT) strain DAY286 were ordered from the Fungal Genetics Stock Center (FGSC) (23) (University of Missouri, Kansas City, MO) and sent by M. L. Richard (24). The following homozygous deletion mutants of (i) cell wall proteins (Δpga62, Δpde2, Δhoc1, Δorf19.10953, Δgal10, Δecm17, Δrot11, Δmpt5, Δorf19.12732, Δorf19.1277, Δiff11, Δwsc2, Δmnn4, Δpga7, Δmnn10, Δhwp1, Δrbt1, Δsnf7, Δcis2, ΔFig1, Δnup, Δeap1, Δecm29, Δirs4, Δmsb2, Δspr3, Δecm3, Δpga44, Δmp65, Δcyc3, Δccw14, Δdfg5, Δvsp28, Δspr1, Δorf19.2296, Δorf.19.2332, Δorf19.2336, Δsap8, Δorf19.2476, Δorf19.251, Δecm4, Δahp1, Δpga62, Δecm14, Δorf19.2996, Δecm33, Δbmh1, Δpra1, Δdot4, Δhwp2, Δorf19.3434, Δsun41, Δrax2, Δsap10, Δorf19.3869, Δcht2, Δcrh12, Δadh1, Δecm331, Δbni4, Δktr4, Δcsh1, Δals9, Δbgl2, Δpga6, Δecm21, Δecm25, Δhyr1, Δorf19.4981, Δpga21, Δorf19.5401, Δorf19.5412, Δcdc10, Δphr3, Δrbt5, Δpga10, Δwsc1, Δcmp1, Δatc1, Δsdh2, Δplb2, Δssr1, Δcsa1, Δrbe1, Δwsc4, Δbgl22, Δkre6, Δmnn9, Δiff4, Δlrg1, Δcda2, Δcht1, Δcht3, Δchs5, Δsap98, Δkre62, Δpga17, Δpga55, Δpga33, Δpga17, Δpga40, Δpga50, Δpga27, Δpga3 [sod5], Δpga2 [sod4], Δpga9 [sod6], Δpga45, Δpga42, Δpga43, Δmid1, Δpga8 [hwp2], Δywp1, Δpga5, Δpga23, Δpga37, Δpga4, Δpga57, Δpga6, Δpga30, Δpga36, Δpga39, Δpga32, Δsap9, and Δpga31) and (ii) transcription factors involved in hyphal morphogenesis (Δrim101, Δcph1, Δtec1, Δczf1, Δcph2, Δtup1, and Δrfg1) were tested. Candida strains were grown routinely on yeast-peptone-dextrose (YPD) (1% yeast extract, 2% peptone, and 2% glucose) agar plates at 30°C for 2 days. RPMI 1640 medium (pH 7.0) with l-glutamine and without sodium bicarbonate was purchased from Sigma and buffered with MOPS (morpholinepropanesulfonic acid) (Sigma, St. Louis, MO, USA). Amphotericin B (AMB), cholesterol, ampicillin, and kanamycin were purchased from Sigma (St. Louis, MO, USA). Caspofungin (CAS) (Cancidas) was purchased from Merck (Beeston Nottingham, United Kingdom). The plant defensin RsAFP2 was isolated from radish seeds (Raphanus sativus) as described previously (25).
Peptide synthesis.
The following peptides (and corresponding sequences) were used: OSIP108 (MLCVLQGLRE), S-OSIP108 (scrambled-OSIP108 with sequence ELRLVCMGQL), d-OSIP108 (MLCVLQGLRE [all d-amino acids]), [CYC1]OSIP108 (head-to-tail cyclized-MLCVLQGLRE), [CYC2]OSIP108 (head-to-tail cyclized-MLCVLQGLREGG), and [CYC3]OSIP108 (disulfide cyclized-MLCVLQGLREC [C3 to C11 cyclization]). OSIP108, S-OSIP108, d-OSIP108, and [Cyc3]OSIP108 (MLCVLQLGREC) were synthesized via Fmoc-protected solid-phase peptide chemistry using standard methods (26). Briefly, the peptides were assembled on a 2-chlorotritylchloride resin (100 to 200 mesh) from Peptide International (Louisville, KY) and cleaved off the resin after complete chain assembly with trifluoroacetic acid (TFA)/triisopropylsilane/water (96:2:2) for 2.5 h. [Cyc3]OSIP108 was oxidized in a folding buffer made of acetic acid/dimethyl sulfoxide (DMSO)/water (5:10:85) at room temperature for 48 h at a final peptide concentration of 0.25 mg/ml. The other two cyclic OSIP108 peptides, [Cyc1]OSIP108 (cyclo-MLCVLQGLRE) and [Cyc2]OSIP108 (cyclo-MLCVLQGLREGG), were synthesized using Boc-protected solid-phase peptide chemistry, with thioester-mediated native chemical ligation to achieve the head-to-tail cyclization (27). Briefly, peptides were assembled on a Boc-Gly-PAM resin (100 to 200 mesh) from Peptide International (Louisville, KY). After coupling of the thioester linker and complete assembly of the peptide chain, the linear peptide precursors were cleaved off the resin. The head-to-tail cyclization was carried out using native chemical ligation in 0.1 M ammonium bicarbonate buffer (pH 8.3), containing 2 mM Tris(2-carboxyethyl)phosphine (TCEP), overnight at room temperature with a final peptide concentration of 0.25 mg/ml.
Liquid chromatography and mass spectrometry.
All OSIP108 analogues were purified using a Shimadzu Prominence high-pressure liquid chromatography (HPLC) system (Shimadzu Corp., Kyoto, Japan), including an LC-20AT pump and SPD-20A UV-visible light (UV-VIS) detector, and using a reversed-phase Jupiter C18, 250- by 21.20-mm, 5-μm column (Phenomenex, Inc., Torrance, CA). Final purity of the compounds was checked on a liquid chromatography-mass spectrometry (LC-MS) Shimadzu Prominence system equipped with double LC-20AD micropumps, an SPD-20A UV/VIS detector, and a Shimadzu electrospray ionization (ESI)-MS 2020 mass spectrometer. Peptides were injected onto a Jupiter C18, 150- by 2.00-mm, 5-μm (Phenomenex, Inc.) column. After 5 min in isocratic 1% solvent B, a gradient of 2% per minute from 1 to 81% was applied before washing and equilibrating the column back to 1% B (solvent A, 0.05% formic acid [FA] in H2O, and solvent B, 0.045% FA, 10% H2O, and 90% acetonitrile [AcN]). All synthetic peptides were of high purity (>97%) and had the correct mass (see Fig. S1 and Table S1 in the supplemental material).
Circular dichroism.
Circular dichroism (CD) spectroscopy is commonly used to evaluate peptide secondary structure (28) and is also useful to confirm enantiomeric relationships (29). Thus it was used here to confirm the mirror-image nature of the d- and l-OSIP108 isomers. Briefly, each individual enantiomer and a 50:50 mixture of both were prepared at a concentration of 50 μM in water. A 0.1-cm path-length cuvette was filled with the desired solution and placed into a Jasco J-810 CD spectropolarimeter. The spectrum was recorded from 185 to 260 nm at a speed of 50 nm per minute. The solvent signal was subtracted and the CD spectrum transformed to mean residue molar ellipticity [θ]MR.
MIC assay.
The MIC-2, which is the minimum concentration that reduces growth by 50% compared to the growth control, was determined according to the CLSI M27-A3 protocol for antifungal susceptibility testing of yeasts in RPMI 1640 medium with a 1% DMSO background (30). The concentration range used for the OSIP108 peptide was 0.39 to 200 μM.
Antibiofilm assay.
The inhibitory potential of OSIP108 and its analogues on Candida biofilm formation was assessed using the CellTiter Blue (CTB) quantification method (31) or the 2,3-bis(2-methoxy-4-nitro-5-sulfophenyl)-5-[(phenylamino)carbonyl]-2H-tetrazolium hydroxide (XTT) assay (32). To this end, an overnight culture of Candida in YPD was washed with phosphate-buffered saline (PBS) and a cell suspension of 106 cells/ml was prepared in RPMI 1640 medium (pH 7.0). Two-fold dilution series of the peptides (dissolved in 100% DMSO) were prepared in RPMI 1640. Five-microliter volumes of these series were added with 95 μl of inoculum to each well of a round-bottomed polystyrene 96-well microtiter plate (TPP; Trasadingen, Switzerland), resulting in a background of 0.5% DMSO. After 24 h of incubation, biofilms were washed and quantified with CTB as described previously (33). Activities of OSIP108 against biofilms of C. glabrata, C. dubliniensis, and C. krusei were assessed with the XTT assay as described previously (33), since those Candida spp. did not convert CTB within 1 h. The activity of OSIP108 and its derivatives against Candida biofilm formation is expressed as the biofilm-inhibiting concentration 2 (BIC-2), which is the minimum concentration of the peptides that reduces biofilm formation by 50% compared to the growth control. RPMI 1640 medium for OSIP108 antibiofilm tests against C. albicans CAI4 was supplemented with 50 μg/ml uridine (URA) as CAI4 is URA deficient.
To determine activities of OSIP108 against mature biofilms of C. albicans, we used a slightly adapted protocol. One hundred microliters of the above-described cell suspension (106 cells/ml in RPMI 1640) was added to the wells of a round-bottomed polystyrene 96-well microtiter plate, and after 1 h of adhesion at 37°C, wells were washed with PBS. Subsequently, 100 μl fresh RPMI 1640 medium was added to each well. Upon 24 h of incubation, biofilms were washed with PBS, and 2-fold dilution series of OSIP108 prepared in RPMI 1640 were added to the biofilms (0.5% DMSO background). After 24 h of treatment, biofilms were washed with PBS and quantified with CTB. Antibiofilm activity of OSIP108 against mature C. albicans biofilms is expressed as the biofilm-eradicating concentration 2 (BEC-2), which is the minimum concentration that eradicates 50% of the biofilm cells compared to the growth control.
Assessment of cytotoxicity.
Cell viability of three human primary cell cultures, namely, osteoblasts, mesenchymal stromal cells, and microvascular endothelial cells, was tested with trypan blue staining according to the ISO 10993-5 standard. Briefly, cells were seeded at 2 × 104 cells/cm2 to 96-well plates, and then were exposed in four replicates to (i) medium for osteoblasts and mesenchymal stromal cells (advanced Dulbecco modified Eagle medium [DMEM], 10% fetal bovine serum [FBS], 1× GlutaMAX, and 0.05 mg/ml gentamicin) or for microvascular endothelial cells (medium 131 with microvascular growth supplement and 0.05 mg/ml gentamicin [all from Gibco, Carlsbad, CA] with 0.5% DMSO background), (ii) medium with 0.05% phenol (cytotoxic control), and (iii) medium with OSIP108 (12.5 μM). After 6 days of culture, the cell culture medium was removed from 96 wells and one-third of trypan blue in DMEM medium was added to the culture for 3 min. Dead (blue) and live (transparent) cells in two visual fields were counted in each of the four wells.
Tube formation assay.
Human aortic endothelial cells were cultured in M200 medium with low serum growth supplement (all from Gibco, Carlsbad, CA). For the tube formation assay, the surface of a 24-well plate was coated with 250 μl of phenol-red free Matrigel (Becton, Dickinson, Bedford, MA) per well and cells were seeded at 2 × 104 cells/cm2. The medium was supplemented with DMSO (0.5%) and phenol (0.05%) or OSIP108 (12.5 μM). Eight hours after seeding of the cells, three random phase-contrast digital images in each of the three wells per treatment were taken with a digital camera (DS-Fi1; Nikon, Tokyo, Japan) using a 10× objective lens. Tube-like structures were analyzed with ImageJ software (http://rsbweb.nih.gov/ij/), and the average tube length per field was calculated.
Osteogenic differentiation.
To test the effect of OSIP108 on osteogenic differentiation potential, osteoblasts and mesenchymal stromal cells were cultured in following culture media: (i) osteogenic medium (osteogenic differentiation Bulletkit [Lonza, Basel, Switzerland] and 0.05 mg/ml gentamicin) with 0.5% DMSO background and (ii) osteogenic medium with 12.5 μM OSIP108. Osteoblast and mesenchymal stromal cell cultures were harvested after 3 or 5 weeks for the calcium and DNA assay.
Calcium and DNA assay.
Calcium deposition of osteoblasts and mesenchymal stromal cells was measured with the Stanbio calcium CPC LiquiColor test (Stanbio Laboratory, Boerne, TX). Briefly, cell cultures were extracted by 5% trichloroacetic acid (500 μl per sample). o-Cresolphthalein complex was added, and the calcium content was determined spectrophotometrically at 550 nm.
To determine the DNA content, cells were washed with PBS, and 200 μl of digestion buffer (10 mM Tris, 1 mM EDTA, 0.1% Triton X-100, and 0.1 mg/ml proteinase K) was added. Samples were incubated in digestion buffer overnight at 56°C. The supernatants were drawn off, and PicoGreen dye (Molecular Probes, Eugene, OR) was added to the supernatants in a 1:1 ratio and read in a fluorescent plate reader (excitation 485 nm and emission 528 nm). DNA values were used to normalize calcium content. Four wells per condition were examined, and two samples from each well were taken for each assay.
Statistical analysis.
Statistical significance of the results from the above-described cell assays was tested with one-way ANOVA followed by Tukey's test. Differences were considered statistically significant at P values of <0.05.
Checkerboard antibiofilm assay.
To determine synergistic interactions between OSIP108 and the antifungals AMB or CAS against C. albicans biofilms, checkerboard analysis was used and fractional inhibitory concentration index (FICI) values were calculated. The FICI was calculated by the formula FICI = [C(BEC-2A)/BEC-2A] + [C(BEC-2B)/BEC-2B], in which C(BEC-2A) and C(BEC-2B) are the BEC-2s of the antifungal drugs in combination, and BEC-2A and BEC-2B are the BEC-2s of antifungal drugs A and B alone. The interaction was defined as synergistic if the FICI was ≤0.5, indifferent if it was between 0.5 and 4, and antagonistic if it was >4.0 (34). Biofilms were grown to maturity as described above. After 24 h of biofilm formation at 37°C, biofilms were washed with 100 μl PBS, after which 100 μl of a combination of OSIP108 (3.125 to 100 μM) with AmB or CAS (0.01 to 5 μM), 2-fold diluted in RPMI 1640 medium across columns and rows of the microtiter plate, respectively, was added to each well (final DMSO background 0.6%). After 24 h of treatment at 37°C, metabolic activity was quantified with the CTB method as described previously (33). The FICI value for the OSIP108-AMB and OSIP108-CAS interactions of 3 independent experiments is shown.
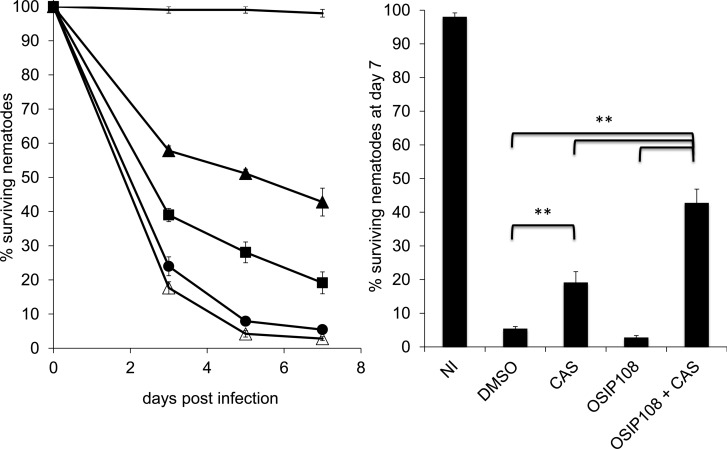
Worm infection assay.
In vivo experiments using the C. elegans-C. albicans biofilm infection model were performed as described previously (33). The glp4/sek1 deletion strain of C. elegans was used to avoid the production of progeny, which is undesirable in this assay as (i) it makes it difficult to score the viability of the infected parents and (ii) Candida-infected wild-type nematodes exhibit significant matricidal death (over 30%) (35). To investigate potential synergy between OSIP108 and CAS in vivo, worms, infected with C. albicans, were treated with 100 μM OSIP108, 0.095 μM CAS, and 100 μM OSIP108 plus 0.095 μM CAS or 0.6% DMSO (negative control). We used this specific concentration of CAS as it has only a modest effect on the survival of the infected C. elegans, namely, 20% of the worms survived after 7 days. As a control, the survival of noninfected worms was also monitored. Worm survival was expressed as a percentage of their viability at day zero. Data shown represent the mean and standard error of the mean of a representative experiment out of 3 independent experiments with at least 3 replicates/condition. Results were analyzed for statistical significance by unpaired Student's t test. Values were considered to be statistically significant when the P value was <0.05.
Screening of homozygous deletion mutants.
Homozygous C. albicans cell wall protein deletion mutants (n = 130) and homozygous deletion mutants of transcription factors (n = 7) involved in hyphal morphogenesis derived from C. albicans BWP17 with isogenic WT DAY286 were screened for their ability to form biofilms in the presence of OSIP108. Biofilms were grown and treated as mentioned above in the presence of 25 μM or 6.25 μM OSIP108 or 0.5% DMSO (control) for 24 h at 37°C. Biofilm formation was quantified with the CTB assay, and a sensitivity factor (SF) was calculated as SF = average % fluorescence mutant at 25 or 6.25 μM OSIP108 relative to control treatment mutant (0.5% DMSO)/average % fluorescence WT DAY286 at 25 or 6.25 μM OSIP108 relative to control treatment WT (0.5% DMSO). Subsequently, resistant or hypersensitive mutants were selected (SF, ≥2 or ≤0.5) for further susceptibility testing toward OSIP108 in a broader concentration range (0.78 to 100 μM) to determine the antibiofilm effect of OSIP108 against the selected mutants compared to the isogenic WT DAY286.
FITC-OSIP108 localization study.
Cellular localization of OSIP108 was investigated using an N-terminal-labeled fluorescein isothiocyanate (FITC)-OSIP108 (FITC-MLCVLQGLRE; ChinaPeptides, Shanghai, China). C. albicans biofilms were grown in the presence of 50 μM FITC-OSIP108 as described above. FITC labeling of the peptide did not affect antibiofilm activity. Biofilm cells were collected and washed 3 times with PBS and visualized using a Zeiss Axio Imager Z1 fluorescence microscope using the FITC channel (excitation wavelength, 450 to 490 nm; emission wavelength, 500 to 550 nm; exposure time, 1.41 s).
Mkc1p phosphorylation assay.
A single colony of WT SC5314 C. albicans was inoculated into 10 ml YPD and grown 8 h at 30°C. Cells were diluted 1:125 into 50 ml RPMI 1640 and grown with shaking at 25°C overnight. Cells were treated with OSIP108 (100 μM) or DMSO (0.5%) as the control treatment for 1 h. Cells were collected over ice, and total protein was extracted as described previously (36). A total of 60 μg (phospho-Mkc1p) or 20 μg (actin) total protein was loaded per sample on an 8.5% (phospho-Mkc1p) or 10% (actin) SDS-PAGE gel. Gels were transferred onto 0.2-μm polyvinylidene difluoride (PVDF) membranes using standard protocols and probed with anti-dually phosphorylated p42/44 mitogen-activated protein kinase (MAPK) rabbit monoclonal antibody (CS-4380; Cell Signaling) or with rabbit anti-actin (A5060; Sigma). Horseradish peroxidase (HRP)-conjugated goat anti-rabbit (Invitrogen) was used as a secondary antibody, and signal was detected using Pierce ECL Western-blotting substrate (Thermo Scientific number 32209) as directed. The signal was visualized and quantified with a Syngene GBox Chemi imager and GeneSys software. The experiment was repeated three times, and a representative blot is shown.
Yeast-to-hypha transition assay using solid-phase cytometry.
To determine the effect of OSIP108 on the yeast-to-hypha transition of C. albicans, the fraction of hyphae was determined using solid-phase cytometry as described previously (37). Biofilms were grown as described above in the presence of OSIP108 (12.5 and 50 μM), S-OSIP108 (12.5 and 50 μM), and the DMSO control (0.5%). As the active OSIP108 concentrations used allow only the formation of loosely attached biofilms, the total percentage of hyphal mass was determined per well after 24 h of biofilm formation without washing. Biofilms were detached by sonication for 10 min and thorough pipetting up and down. Biofilm cells were diluted 1,000 times in particle-free 0.9% (wt/vol) NaCl, and further processing of the diluted suspensions and analysis of the fluorescent signals was performed as previously described (37).
RESULTS AND DISCUSSION
OSIP108 interferes with C. albicans biofilm formation without affecting yeast viability.
We assessed the effect of OSIP108 on planktonic C. albicans cells and on the biofilm formation process of this pathogen. OSIP108 displayed no antifungal activity against C. albicans (MIC-2, >200 μM), and the addition of up to 200 μM OSIP108 to C. albicans cells did not affect their viability after 24 h of incubation. In contrast, OSIP108 strongly reduced C. albicans biofilm formation when added during the adhesion and biofilm formation phases. Biofilms formed in the presence of OSIP108 (6.25 to 100 μM) were very easily detached, pointing to a strong interference of OSIP108 with the C. albicans biofilm structure. The BIC-2 of OSIP108 was 5.3 μM (Fig. 1; Table 1). However, OSIP108 did not eradicate 24-h-old C. albicans biofilms, as the BEC-2 of OSIP108 was >800 μM. In order to compare the antibiofilm potential of OSIP108 with that of conventional antifungal agents, we also determined the antibiofilm activity of amphotericin B (AMB) and caspofungin (CAS) in our setup. The BIC-2 values of AMB and CAS were 0.4 and 0.06 μM, respectively, while the BEC-2 values were 0.72 and 0.52 μM, respectively. It is clear that the antibiofilm activity of OSIP108 is lower than that of the conventional antifungal agents. However, in contrast to these antifungal agents, OSIP108 has no fungicidal or antifungal activity, which is beneficial in terms of the development of resistance. Next, we tested OSIP108's ability to inhibit biofilm formation against other C. albicans strains and Candida spp. (Table 1). C. albicans strains CAI4, CAF2, and DAY286 are laboratory strains derived from SC5314 (17). C. albicans B2630 and B63195 are azole-sensitive strains (19). OSIP108 displayed antibiofilm activity against all C. albicans strains tested (BIC-2, 4.5 to 10.8 μM) (Table 2) but not against other pathogenic Candida spp., such as C. glabrata, C. dubliniensis, and C. krusei (BIC-2, >100 μM) (Table 1). Together, these data demonstrate that the activity of OSIP108 is biofilm specific, as OSIP108 displayed no antifungal/fungicidal activity, but was restricted to interference with the biofilm formation of C. albicans strains. Though the lack of activity against other Candida spp. is not favorable for the development of OSIP108 as a broad-spectrum antibiofilm agent, it remains a valuable peptide in tackling biofilm-associated infections caused by C. albicans, the predominant fungal pathogen in humans (3, 4).
FIG 1.
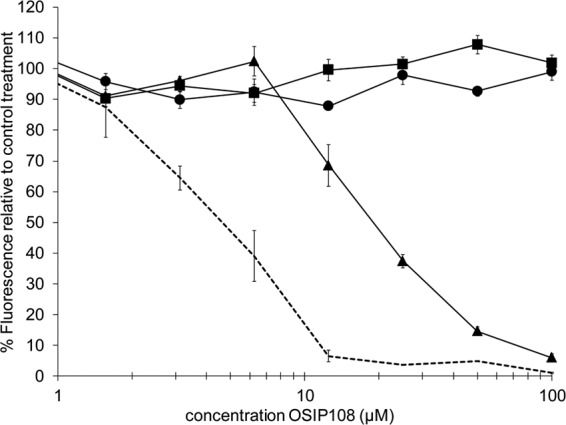
OSIP108 antibiofilm activity with and without the yeast-to-hypha inhibitor RsAFP2. C. albicans SC5314 biofilms were grown in the presence of OSIP108 alone (0.78 to 100 μM; dotted line) or in the presence of the plant defensin RsAFP2 (solid lines) at concentrations of 25 μg/ml (triangles), 50 μg/ml (squares), or 100 μg/ml (circles) for 24 h at 37°C in RPMI medium. After 24 h of biofilm formation, biofilms were quantified with the CTB assay. Graph shows the percentage of fluorescence relative to the control treatment (0.5% DMSO) and a representative of two independent biological experiments that consisted of triplicate measurements.
TABLE 1.
Activity of OSIP108 against biofilm formation of different Candida strains
| Strain | OSIP108 BIC-2 (μM) |
|---|---|
| C. albicans CAI4 | 4.5 |
| C. albicans SC5314 | 5.3 |
| C. albicans DAY286 | 7.0 |
| C. albicans CAF2 | 9.0 |
| C. albicans B2630 | 10.8 |
| C. albicans B63195 | 10.8 |
| C. glabrata BG2 | >100 |
| C. dubliniensis NCPF 3949 | >100 |
| C. krusei | >100 |
TABLE 2.
Activity of OSIP108 and analogues against biofilm formation of C. albicans
| OSIP108 or analogue | Peptide | BIC-2 (μM) | Fold change |
|---|---|---|---|
| OSIP108 | MLCVLQGLRE | 5.3 | |
| S-OSIP108 | ELRLVCMGQL | >100 | >20 |
| d-OSIP108 | MLCVLQGLRE (all d-amino acids) | 13.7 | 2.6 |
| [CYC1]OSIP108 | Cyclo-MLCVLQGLRE | >100 | >20 |
| [CYC2]OSIP108 | Cyclo-MLCVLQGLREGG | 36.0 | 6.8 |
| [CYC3]OSIP108 | Cyclo-MLCVLQGLREC | >100 | >20 |
The use of (antimicrobial) peptides from various natural sources as new antibiofilm agents as alternatives to antibiotics/antimycotics has been increasingly studied during the last few years. For example, antibiofilm activity was reported for LL-37 (the major human cationic host defense peptide) against several Gram-negative and Gram-positive bacteria (38, 39), as well as against C. albicans, where it interferes with adhesion (40, 41). Moreover, the antibiofilm activity against C. albicans of synthetic antimicrobial peptides such as KSL-W and KABT-AMP has been reported (42, 43). In contrast to OSIP108, these synthetic peptides do not act in a biofilm-specific way, as they display rather general fungicidal activity.
Structure-activity relationship study of OSIP108.
We undertook a preliminary structure-activity relationship (SAR) study of OSIP108 to identify elements of the structure important for its antibiofilm activity. A scrambled version of OSIP108 (S-OSIP108), containing the same amino acids as OSIP108 but in a random order, was tested (Table 2) but displayed no antibiofilm activity (BIC-2, >100 μM), pointing to the importance of the order of the amino acids for the antibiofilm activity of OSIP108. Moreover, a d-stereoisomer of OSIP108, in which all the amino acids are in the d-configuration (d-OSIP108), was synthesized and tested to assess the role of stereochemistry on the activity (Table 2). The d-OSIP108 was confirmed by CD spectroscopy to have equal and opposite mean residue ellipticity ([θ]MR) and thereby mirror image symmetry to its OSIP108 enantiomer (see Fig. S2 in the supplemental material). The optical isomerization was further demonstrated in the CD spectrum of a 50:50 mixture of l-OSIP108 and d-OSIP108, which counterbalanced each other and showed a signal close to zero (see Fig. S2). Interestingly, the BIC-2 of d-OSIP108 (13.7 μM) was only 2.6-fold higher than that of the native OSIP108 peptide (5.3 μM). If OSIP108 acts exclusively via a stereospecific receptor, a more pronounced increase in BIC-2 (i.e., a greater loss of activity) would have been expected, while if it acts exclusively via a nonchiral mechanism (e.g., via membrane lipid disruption), a minimal change in activity would have been expected. The observed BIC-2 suggests that OSIP108 might have several alternative mechanisms contributing to its mode of action. Moreover, the fact that d-OSP108 retains antibiofilm activity is beneficial in terms of possible applications of OSIP108 as antibiofilm therapy, as peptides in the d-configuration are less sensitive to endogenous proteases (44).
We subsequently examined the role of conformational restriction on the activity of OSIP108. As cyclization is a well-established way to stabilize peptides for therapeutic applications (45), we synthesized three cyclic derivatives using either head-to-tail or disulfide-based cyclization. The antibiofilm properties of the cyclic derivatives of OSIP108 ([CYC1]OSIP108, [CYC2]OSIP108, and [CYC3]OSIP108) were tested (Table 2). [CYC1]OSIP108 has the same sequence as the native OSIP108, except that the N and C termini are joined head-to-tail to cyclize the peptide. Cyclization of the [CYC2]OSIP108 peptide was similarly performed, but two glycine residues were added to alter the steric restraints associated with head-to-tail cyclization of the peptide. In the [CYC3]OSIP108 peptide, an extra cysteine residue was added to form a disulfide bridge with the cysteine at position 3 (Cys3), to form a disulfide-cyclized peptide. For [CYC1]OSIP108 and [CYC3]OSIP108, the antibiofilm activity was totally abolished (BIC-2, >100 μM). For [CYC2]OSIP108, the BIC-2 (36 μM) was increased 6.8-fold compared to that of the native OSIP108 peptide (BIC-2, 5.3 μM). These data demonstrate that cyclization reduces the antibiofilm activity of OSIP108.
As well as being cyclic, the [CYC3]OSIP108 analogue sequesters the native Cys3 side chain in a disulfide bond, rather than as a free thiol, and this could be a factor in reducing its activity. Furthermore, we considered the possibility that this Cys might be involved in the formation of a disulfide-linked dimer in OSIP108 but found no evidence of such a dimer in any of the chromatographic or mass spectrometry data (data not shown).
OSIP108 does not affect viability or functionality of osteoblasts, mesenchymal stromal cells, or endothelial cells.
As an application of OSIP108 could imply its coating on medical devices, we assessed the viability and functionality of various human cell types such as osteoblasts, mesenchymal stromal cells, and endothelial cells in the presence of OSIP108. OSIP108 (12.5 μM) did not significantly decrease viability of osteoblasts, mesenchymal stromal cells, and endothelial cells compared to the DMSO control (data not shown). Phenol (0.05%) was used as a positive control and significantly affected the viabilities of all cell types. In addition, OSIP108 (12.5 μM) had no significant effect (P > 0.05) on the osteogenic potential of osteoblasts compared to the DMSO control treatment, as measured by calcium deposition (data not shown). OSIP108 (12.5 μM) significantly affected the osteogenic potential of mesenchymal stromal cells compared to the DMSO control treatment (0.161 ± 0.003 ng Ca/μg DNA for OSIP108-treated cells versus 0.167 ± 0.0045 ng Ca/μg DNA for the DMSO control treatment). However, as this difference is very small and no significant difference in calcium deposition between the OSIP108-treated sample and the mesenchymal stromal cells cultured in osteogenic medium (without DMSO) (0.162 ng ± 0.006 Ca/μg DNA) was observed, we can conclude that there is no significant negative effect of OSIP108 on the osteogenic potential of this cell type. Furthermore, OSIP108 did not reduce the potential of endothelial cells to form tubular structures (Fig. 2). No significant differences in average tube length of endothelial cells were observed in the presence of 12.5 μM OSIP108 (126 ± 18 pixels for OSIP108-treated endothelial cells compared to 117 ± 5 pixels for the DMSO control treatment), in contrast to 0.05% phenol, which significantly (P < 0.05) affected the average tube length (87 ± 8 pixels).
FIG 2.
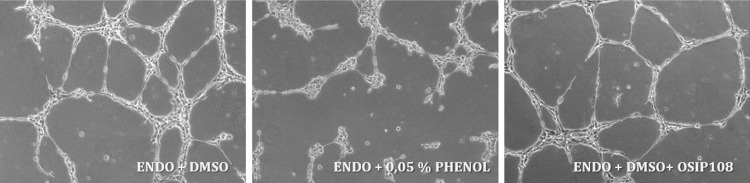
OSIP108 does not affect average tube length formed by endothelial cells. Human aortic endothelial cells were cultured in M200 medium with low serum growth supplement (ENDO) in the presence of 12.5 μM OSIP108, 0.5% DMSO (control treatment), or 0.05% phenol (positive control). Eight hours after seeding, digital images for each treatment were taken.
OSIP108 enhances the activity of amphotericin B and caspofungin in vitro and in vivo in a C. albicans worm infection assay.
As OSIP108 is not antifungal or fungicidal, and hence has a different mode of antibiofilm action than conventional antifungal agents like AMB and CAS, we investigated a potential synergy between OSIP108 and AMB or CAS against mature C. albicans biofilms using checkerboard analysis. OSIP108 (12.5 to 100 μM) significantly reduced the BEC-2 of AMB and CAS against mature C. albicans biofilms (P < 0.05). Moreover, in this concentration range (12.5 to 100 μM), OSIP108 acted synergistically with AMB and CAS against mature C. albicans biofilms, as indicated by their corresponding FICI coefficients being <0.5. The highest reductions in the BEC-2 values of AMB and CAS were observed in combination with 100 μM OSIP108, resulting in 4- to 5-fold reductions of the BEC-2 values of AMB and CAS (from 0.72 μM to 0.17 μM for AMB and from 0.52 μM to 0.11 μM for CAS). FICI coefficients for these OSIP108-AMB and OSIP108-CAS interactions were <0.36 and <0.33, respectively.
To translate the in vitro findings regarding the OSIP108 antibiofilm activity to an in vivo infection model, we used the Caenorhabditis elegans-C. albicans infection assay, which is regarded as a good infection model for studying biofilm-associated infections (35, 46–50). Application of OSIP108 (25 to 100 μM) alone did not significantly cure C. elegans worms infected with C. albicans compared to the control treatment (data not shown). We further investigated the in vivo synergy with different types of antifungal agents using the OSIP108-CAS combination. Treatment of the infected worms with a combination of 100 μM OSIP108 and 0.095 μM CAS significantly (P < 0.05) increased the survival of the worms compared to treatment with 100 μM OSIP108 or 0.095 μM CAS alone or control treatment (0.6% DMSO) after 3, 5, and 7 days postinfection (Fig. 3). Seven days postinfection, 42.78% ± 4.07% of the worms survived when treated with the combination of OSIP108 and CAS. In contrast, treatment with CAS or OSIP108 alone resulted in only 19.15% ± 3.21% or 2.81% ± 0.57% surviving worms, respectively, whereas 5.45% ± 0.57% of the worms treated with 0.6% DMSO (control treatment) survived 7 days postinfection (Fig. 3). These data indicate that OSIP108 also enhances the activity of CAS in vivo in the C. elegans infection model and suggest that OSIP108 might be a valuable adjuvant for conventional antifungal agents like AMB and CAS in systemic combination therapy against C. albicans biofilm-associated infections. Further studies using appropriate models (e.g., a well-established in vivo biofilm-associated rat catheter model [51]) are required to confirm this synergy. Although the primary application of OSIP108 could be envisaged in systemic combination therapy, another potential application concerns its use as a coating of specific medical devices, such as dental implants, dentures, and voice prostheses, as C. albicans is the most prevalent fungus in the oral cavity (52) and is the most frequently isolated Candida spp. on voice prostheses in laryngectomized patients (53). Regarding to the latter, infection risk of these voice prostheses is very high (50 to 100%) compared to the risk with other implantable biomaterials (13). We reason that biofilms growing on OSIP108-coated medical devices such as voice prostheses might be more easily eradicated by conventional antifungal agents. In this regard, it is of great importance that the OSIP108-containing coatings are not cytotoxic for the surrounding tissue. As described above, OSIP108 had no cytotoxic effects on various human cell lines.
FIG 3.
OSIP108 enhances the activity of caspofungin in a C. elegans-C. albicans biofilm infection model. Δglp4/Δsek1 C. elegans worms were infected with C. albicans by feeding them on a YPD plate containing C. albicans for 2 h at 25°C. Worms were subsequently treated with 100 μM OSIP108 (open triangles), 0.095 μM CAS (filled squares), or 100 μM OSIP108 + 0.095 μM CAS (filled triangles). Untreated worms (filled circles) and noninfected worms (solid line) served as controls with a DMSO background of 0.6%. Worms were counted regularly for 7 days postinfection. Worm survival is expressed as a percentage of their viability at day 0. Data presented are the mean and SEM for one representative experiment out of three independent biological experiments, each consisting of at least three repeats/condition. NI, noninfected worms; **, P < 0.01.
C. albicans mutants defective in components important for cell wall integrity or hyphal morphogenesis show increased tolerance to OSIP108.
Important steps in the biofilm formation process of C. albicans include the adhesion of cells to various surfaces via cell wall proteins and the yeast-to-hypha transition (54). Furthermore, the importance of the yeast-to-hypha transition and the presence of both morphological forms for proper C. albicans biofilm formation and structure were demonstrated by Baillie and Douglas (55). In a first attempt to obtain more insight into the antibiofilm mode of action of OSIP108 against C. albicans biofilm formation, we screened a set of homozygous C. albicans deletion mutants, affected in genes encoding cell wall proteins or transcription factors involved in hyphal morphogenesis, for altered sensitivity to OSIP108 during biofilm formation. We identified 9 mutants that displayed at least a 5-fold increased tolerance and 1 mutant that displayed a 5-fold increased sensitivity to the antibiofilm activity of OSIP108. Targeted genes of these mutants were divided in two main functional groups, genes encoding proteins that are important to maintain cell wall integrity (CWI) and structure and genes encoding proteins involved in hyphal morphogenesis.
The first group of OSIP108-tolerant mutants was affected in genes encoding proteins that function in cell wall integrity (CWI) or cell wall structure (WSC1, SAP10, and CMP1). For example, Wsc1 is the orthologue of S. cerevisiae Wsc2/Wsc3, sensor-transducer of the CWI pathway (56–58), and SAP10 encodes the secreted aspartyl protease 10, which is important in adhesion, virulence, and cell surface integrity in C. albicans (59). CMP1 encodes the catalytic subunit of calcineurin and is required for virulence and tolerance to several stresses (60, 61). The second group of mutants consisted of 2 mutants with a homozygous deletion of transcription factors involved in hyphal morphogenesis, namely, Cph2 and Tec1 (62, 63), and 2 mutants affected in hypha-specific genes (HWP1, RAX2). The Δtec1 deletion mutant was the only mutant characterized by an increased sensitivity to OSIP108. In addition, three homozygous deletion mutants (Δorf19.2476, Δpga45, and Δsod6) not directly related to these two functional groups were identified as tolerant to the antibiofilm activity of OSIP108. Based on these results taken together, we can conclude that mutants defective in components important for CWI or hyphal morphogenesis showed increased tolerance to OSIP108.
OSIP108 dominantly localizes to the surface of C. albicans cells.
Based on our findings of a possible cell wall-related interaction of OSIP108, we further investigated its localization using an FITC-labeled OSIP108 peptide. We found FITC-OSIP108 (50 μM) to be dominantly associated with the cell surface of C. albicans (Fig. 4). The addition of the FITC group to the N-terminal side of OSIP108 did not alter the antibiofilm activity compared with that of the native OSIP108 peptide (data not shown). From these data, it seems that OSIP108 exerts its antibiofilm effect by associating with the C. albicans cell surface.
FIG 4.
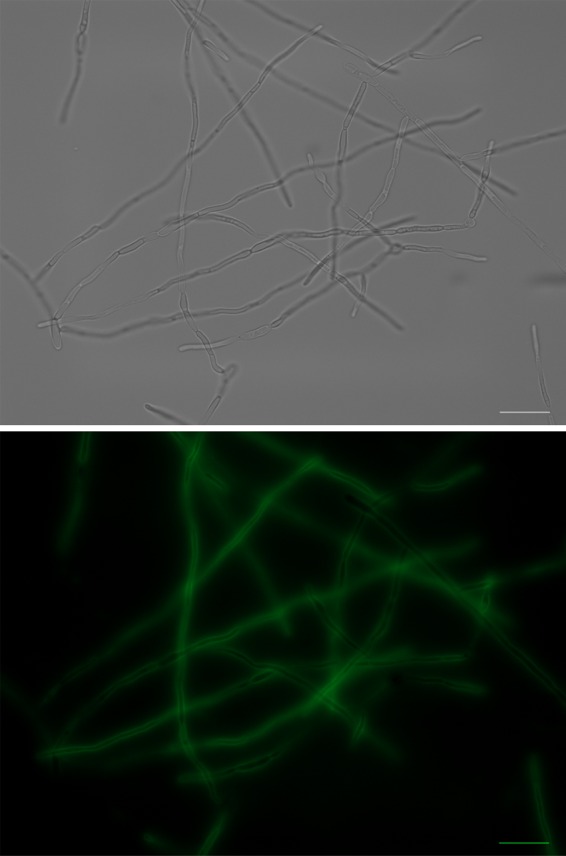
Localization of FITC-labeled OSIP108 on C. albicans biofilm cells. C. albicans biofilms were grown for 24 h in the presence of 50 μM FITC-OSIP108. Biofilm cells were collected, and the localization of FITC-OSIP108 was visualized using a fluorescence microscope (×63 magnification). Scale bar represents 20 μM.
OSIP108 activates the cell wall integrity pathway of C. albicans via Mkc1p activation.
As the deletion mutant of the sensor-transducer of the C. albicans CWI pathway (Δwsc1) and deletion mutants of other genes important for cell surface integrity (Δsap10 and Δcmp1) showed increased tolerance to OSIP108, we investigated a potential activation of the CWI pathway by OSIP108 (Fig. 5). The C. albicans CWI pathway contains the mitogen-activated protein (MAP) kinase Mkc1p, which is dually phosphorylated upon activation of this pathway (36). We observed a 2.7-fold increase in dually phosphorylated Mkc1p, in response to OSIP108 compared to the control treatment (0.5% DMSO), pointing to an activation of the CWI pathway by OSIP108. Together with the above-described FITC localization data, these data demonstrate that cell wall-related processes seem important for the antibiofilm activity of OSIP108.
FIG 5.
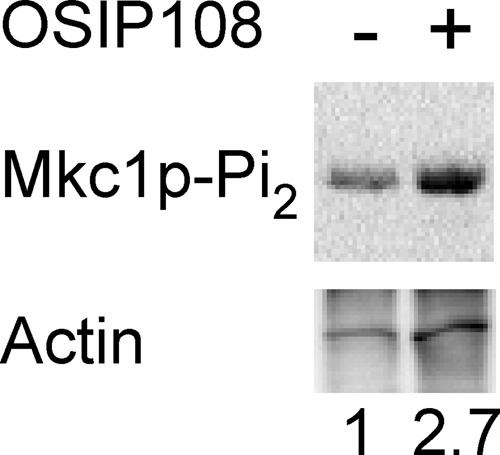
OSIP108 activates the cell wall integrity pathway of C. albicans. WT (SC5314) C. albicans cells growing in RPMI at 25°C were treated for 1 h with 100 μM OSIP108 (+) or 0.5% DMSO (−) as control treatment. Total protein was extracted, and equal amounts of protein were loaded in each lane of an SDS-PAGE gel and Western blotted to detect dually phosphorylated Mkc1p (top) or actin (bottom). Numbers at the bottom indicate amount of dually phosphorylated Mkc1p normalized to actin levels and shown relative to the DMSO control. Blot presented is a representative blot out of three biological experiments.
The yeast-to-hypha inhibitor RsAFP2 blocks the antibiofilm activity of OSIP108.
As some deletion mutants targeted in transcription factors related to hyphal morphogenesis showed an altered susceptibility toward OSIP108 (Δcph2 and Δtec1), we first investigated a potential effect of OSIP108 on the yeast-to-hypha transition of C. albicans. To this end, we determined the percentage of hyphal mass in the presence of OSIP108 or the nonactive S-OSIP108 relative to the control treatment (0.5% DMSO). No significant differences in hyphal mass were observed between OSIP108, S-OSIP108, and DMSO treatments, indicating that OSIP108 does not interfere with the yeast-to-hypha transition of C. albicans (data not shown). Next, we investigated the effect of the plant defensin RsAFP2, which reportedly blocks the yeast-to-hypha transition in C. albicans (64), on the antibiofilm activity of OSIP108 (Fig. 1). Coincubation of OSIP108 with 25 μg/ml RsAFP2 increased the BIC-2 of OSIP108 from 5 μM to 19 μM. Moreover, coincubation of OSIP108 with 50 and 100 μg/ml RsAFP2 totally abolished its antibiofilm activity (BIC-2, >100 μM). Likewise, another synthetic yeast-to-hypha inhibitor, conjugated linoleic acid (CLA) (65), had an antagonistic effect on the antibiofilm activity of OSIP108. Although coincubation of 100 μM CLA with a concentration series of OSIP108 did not change the BIC-2 value of OSIP108, a 7-fold increase in the OSIP108 concentration necessary to reduce biofilm formation by 80% was observed in the presence of 100 μM CLA (data not shown). It is important to note that RsAFP2, besides its action on the yeast-to-hypha transition of C. albicans, also induces cell wall stress, septin mislocalization, and accumulation of ceramides in C. albicans (64). Hence, it cannot be ruled out that the antagonistic activity of RsAFP2 on the antibiofilm activity of OSIP108, is not solely due to its effect on the yeast-to-hypha transition. However, the fact that another yeast-to-hypha inhibitor, CLA, also antagonizes the antibiofilm activity of OSIP108 points to the importance of this morphological transition switch of C. albicans for OSIP108 antibiofilm activity. In line with these findings, fungal pathogens that do not form true hyphae in their biofilms, like C. glabrata and C. krusei (66, 67), were resistant to OSIP108. However, C. dubliniensis, which is closely related to C. albicans and forms true hyphae in biofilms (68), was also resistant to OSIP108.
Conclusions.
In this study, we found that the plant-derived decapeptide OSIP108 interferes with the biofilm formation process of the human fungal pathogen C. albicans. This antibiofilm activity is biofilm specific, as OSIP108 does not affect the growth and viability of planktonic C. albicans cultures. Using in vitro checkerboard analysis and an in vivo C. elegans-C. albicans biofilm infection model, we showed that OSIP108 synergistically enhances the activity of conventional antifungal agents such as AMB and CAS, pointing to a potential application of OSIP108 in systemic combination therapies. Since OSIP108 shows no toxicity against several cell types, like osteoblasts, mesenchymal stem cells, and endothelial cells, OSIP108 might be an interesting peptide to coat specific medical devices, like voice prostheses, as C. albicans biofilm formation is a significant clinical problem associated with this medical device. Initial steps in the elucidation of the mode of action revealed the importance of the yeast-to-hypha transition of C. albicans for OSIP108's antibiofilm activity, as well as of cell wall-associated processes, as indicated by OSIP108's dominant localization to the C. albicans cell surface and the activation of its cell wall integrity pathway.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by the European Commission's Seventh Framework Programme (FP7/2007-2013) under the grant agreement COATIM (project number 278425), ”Fonds Wetenschappelijk Onderzoek (FWO)-Vlaanderen” (G.A062.10N, G.0414.09, W0.026.11N, and K220313N); “Agentschap voor Innovatie door Wetenschap en Technologie (IWT)-Vlaanderen” (SBO grant 120005); KU Leuven (knowledge platform IOF/KP/11/007); and “Bijzonder Onderzoeksfonds KU Leuven” (GOA/2008/11). Furthermore, this work was supported by the “Industrial Research Fund” KU Leuven (to K.T.), FWO-Vlaanderen (12A7213N) (to B.D.C.), IWT Flanders (IWT101095) (to N.D.), a National Health and Medical Research Council Professorial Fellowship (APP1026501 and APP1028509) (to D.J.C.), and the National Institute of Allergy and Infectious Diseases (R01AI081794) (to C.A.K.).
We thank M. L. Richard for sending the homozygous mutants of the GPI-anchored cell wall proteins and Violeta Morcuende Ventura for her technical assistance in the mutant screening.
Footnotes
Published ahead of print 24 February 2014
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AAC.01274-13.
REFERENCES
- 1.Wisplinghoff H, Bischoff T, Tallent SM, Seifert H, Wenzel RP, Edmond MB. 2004. Nosocomial bloodstream infections in US hospitals: analysis of 24,179 cases from a prospective nationwide surveillance study. Clin. Infect. Dis. 39:309–317. 10.1086/421946 [DOI] [PubMed] [Google Scholar]
- 2.Lai CC, Tan CK, Huang YT, Shao PL, Hsueh PR. 2008. Current challenges in the management of invasive fungal infections. J. Infect. Chemother. 14:77–85. 10.1007/s10156-007-0595-7 [DOI] [PubMed] [Google Scholar]
- 3.Marr KA. 2004. Invasive Candida infections: the changing epidemiology. Oncology 18:9–14 [PubMed] [Google Scholar]
- 4.Pfaller MA, Diekema DJ. 2010. Epidemiology of invasive mycoses in North America. Crit. Rev. Microbiol. 36:1–53. 10.3109/10408410903241444 [DOI] [PubMed] [Google Scholar]
- 5.Pfaller MA. 2012. Antifungal drug resistance: mechanisms, epidemiology, and consequences for treatment. Am. J. Med. 125(1 Suppl):S3–S13. 10.1016/j.amjmed.2011.11.001 [DOI] [PubMed] [Google Scholar]
- 6.Costerton JW, Stewart PS, Greenberg EP. 1999. Bacterial biofilms: a common cause of persistent infections. Science 284:1318–1322. 10.1126/science.284.5418.1318 [DOI] [PubMed] [Google Scholar]
- 7.Douglas LJ. 2003. Candida biofilms and their role in infection. Trends Microbiol. 11:30–36. 10.1016/S0966-842X(02)00002-1 [DOI] [PubMed] [Google Scholar]
- 8.Fanning S, Mitchell AP. 2012. Fungal biofilms. PLoS Pathog. 8:e1002585. 10.1371/journal.ppat.1002585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ramage G, Rajendran R, Sherry L, Williams C. 2012. Fungal biofilm resistance. Int. J. Microbiol. 2012:528521. 10.1155/2012/528521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kuhn DM, George T, Chandra J, Mukherjee PK, Ghannoum MA. 2002. Antifungal susceptibility of Candida biofilms: unique efficacy of amphotericin B lipid formulations and echinocandins. Antimicrob. Agents Chemother. 46:1773–1780. 10.1128/AAC.46.6.1773-1780.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Brown GD, Denning DW, Levitz SM. 2012. Tackling human fungal infections. Science 336:647. 10.1126/science.1222236 [DOI] [PubMed] [Google Scholar]
- 12.Kojic EM, Darouiche RO. 2004. Candida infections of medical devices. Clin. Microbiol. Rev. 17:255–267. 10.1128/CMR.17.2.255-267.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ramage G, Martinez JP, Lopez-Ribot JL. 2006. Candida biofilms on implanted biomaterials: a clinically significant problem. FEMS Yeast Res. 6:979–986. 10.1111/j.1567-1364.2006.00117.x [DOI] [PubMed] [Google Scholar]
- 14.Danscher G, Locht LJ. 2010. In vivo liberation of silver ions from metallic silver surfaces. Histochem. Cell Biol. 133:359–366. 10.1007/s00418-009-0670-5 [DOI] [PubMed] [Google Scholar]
- 15.Sardi JC, Scorzoni L, Bernardi T, Fusco-Almeida AM, Mendes Giannini MJ. 2013. Candida species: current epidemiology, pathogenicity, biofilm formation, natural antifungal products and new therapeutic options. J. Med. Microbiol. 62:10–24. 10.1099/jmm.0.045054-0 [DOI] [PubMed] [Google Scholar]
- 16.De Coninck B, Carron D, Tavormina P, Willem L, Craik DJ, Vos C, Thevissen K, Mathys J, Cammue BP. 2013. Mining the genome of Arabidopsis thaliana as a basis for the identification of novel bioactive peptides involved in oxidative stress tolerance. J. Exp. Bot. 64:5297–5307. 10.1093/jxb/ert295 [DOI] [PubMed] [Google Scholar]
- 17.Fonzi WA, Irwin MY. 1993. Isogenic strain construction and gene mapping in Candida albicans. Genetics 134:717–728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Davis D, Edwards JE, Jr, Mitchell AP, Ibrahim AS. 2000. Candida albicans RIM101 pH response pathway is required for host-pathogen interactions. Infect. Immun. 68:5953–5959. 10.1128/IAI.68.10.5953-5959.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.de Wit K, Paulussen C, Matheeussen A, van Rossem K, Cos P, Maes L. 2010. In vitro profiling of pramiconazole and in vivo evaluation in Microsporum canis dermatitis and Candida albicans vaginitis laboratory models. Antimicrob. Agents Chemother. 54:4927–4929. 10.1128/AAC.00730-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kaur R, Ma B, Cormack BP. 2007. A family of glycosylphosphatidylinositol-linked aspartyl proteases is required for virulence of Candida glabrata. Proc. Natl. Acad. Sci. U. S. A. 104:7628–7633. 10.1073/pnas.0611195104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sullivan DJ, Westerneng TJ, Haynes KA, Bennett DE, Coleman DC. 1995. Candida dubliniensis sp. nov.: phenotypic and molecular characterization of a novel species associated with oral candidosis in HIV-infected individuals. Microbiology 141(Pt 7):1507–1521 [DOI] [PubMed] [Google Scholar]
- 22.Wilson RB, Davis D, Mitchell AP. 1999. Rapid hypothesis testing with Candida albicans through gene disruption with short homology regions. J. Bacteriol. 181:1868–1874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.McCluskey K, Wiest A, Plamann M. 2010. The Fungal Genetics Stock Center: a repository for 50 years of fungal genetics research. J. Biosci. 35:119–126. 10.1007/s12038-010-0014-6 [DOI] [PubMed] [Google Scholar]
- 24.Plaine A, Walker L, Da Costa G, Mora-Montes HM, McKinnon A, Gow NA, Gaillardin C, Munro CA, Richard ML. 2008. Functional analysis of Candida albicans GPI-anchored proteins: roles in cell wall integrity and caspofungin sensitivity. Fungal Genet. Biol. 45:1404–1414. 10.1016/j.fgb.2008.08.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Terras FR, Schoofs HM, De Bolle MF, Van Leuven F, Rees SB, Vanderleyden J, Cammue BP, Broekaert WF. 1992. Analysis of two novel classes of plant antifungal proteins from radish (Raphanus sativus L.) seeds. J. Biol. Chem. 267:15301–15309 [PubMed] [Google Scholar]
- 26.Halai R, Clark RJ, Nevin ST, Jensen JE, Adams DJ, Craik DJ. 2009. Scanning mutagenesis of alpha-conotoxin Vc1.1 reveals residues crucial for activity at the alpha9alpha10 nicotinic acetylcholine receptor. J. Biol. Chem. 284:20275–20284. 10.1074/jbc.M109.015339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Simonsen SM, Sando L, Rosengren KJ, Wang CK, Colgrave ML, Daly NL, Craik DJ. 2008. Alanine scanning mutagenesis of the prototypic cyclotide reveals a cluster of residues essential for bioactivity. J. Biol. Chem. 283:9805–9813. 10.1074/jbc.M709303200 [DOI] [PubMed] [Google Scholar]
- 28.Greenfield NJ. 2006. Using circular dichroism spectra to estimate protein secondary structure. Nat. Protoc. 1:2876–2890. 10.1038/nprot.2006.202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sando L, Henriques ST, Foley F, Simonsen SM, Daly NL, Hall KN, Gustafson KR, Aguilar MI, Craik DJ. 2011. A synthetic mirror image of kalata B1 reveals that cyclotide activity is independent of a protein receptor. Chembiochem 12:2456–2462. 10.1002/cbic.201100450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Clinical and Laboratory Standards Institute. 2008. Reference method for broth dilution antifungal susceptibility testing of yeast, approved standard. CLSI document M27-A3 Clinical and Laboratory Standards Institute, Wayne, PA [Google Scholar]
- 31.O'Brien J, Wilson I, Orton T, Pognan F. 2000. Investigation of the Alamar Blue (resazurin) fluorescent dye for the assessment of mammalian cell cytotoxicity. Eur. J. Biochem. 267:5421–5426. 10.1046/j.1432-1327.2000.01606.x [DOI] [PubMed] [Google Scholar]
- 32.Tellier R, Krajden M, Grigoriew GA, Campbell I. 1992. Innovative endpoint determination system for antifungal susceptibility testing of yeasts. Antimicrob. Agents Chemother. 36:1619–1625. 10.1128/AAC.36.8.1619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Delattin N, De Brucker K, Vandamme K, Meert E, Marchand A, Chaltin P, Cammue BP, Thevissen K. 27 November 2013. Repurposing as a means to increase the activity of amphotericin B and caspofungin against C. albicans biofilms. J. Antimicrob. Chemother. 10.1093/jac/dkt449 [DOI] [PubMed] [Google Scholar]
- 34.Odds FC. 2003. Synergy, antagonism, and what the chequerboard puts between them. J. Antimicrob. Chemother. 52:1. 10.1093/jac/dkg301 [DOI] [PubMed] [Google Scholar]
- 35.Breger J, Fuchs BB, Aperis G, Moy TI, Ausubel FM, Mylonakis E. 2007. Antifungal chemical compounds identified using a C. elegans pathogenicity assay. PLoS Pathog. 3:e18. 10.1371/journal.ppat.0030018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kumamoto CA. 2005. A contact-activated kinase signals Candida albicans invasive growth and biofilm development. Proc. Natl. Acad. Sci. U. S. A. 102:5576–5581. 10.1073/pnas.0407097102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nailis H, Vandenbroucke R, Tilleman K, Deforce D, Nelis H, Coenye T. 2009. Monitoring ALS1 and ALS3 gene expression during in vitro Candida albicans biofilm formation under continuous flow conditions. Mycopathologia 167:9–17. 10.1007/s11046-008-9148-6 [DOI] [PubMed] [Google Scholar]
- 38.Overhage J, Campisano A, Bains M, Torfs EC, Rehm BH, Hancock RE. 2008. Human host defense peptide LL-37 prevents bacterial biofilm formation. Infect. Immun. 76:4176–4182. 10.1128/IAI.00318-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hell E, Giske CG, Nelson A, Romling U, Marchini G. 2010. Human cathelicidin peptide LL37 inhibits both attachment capability and biofilm formation of Staphylococcus epidermidis. Lett. Appl. Microbiol. 50:211–215. 10.1111/j.1472-765X.2009.02778.x [DOI] [PubMed] [Google Scholar]
- 40.Tsai PW, Yang CY, Chang HT, Lan CY. 2011. Human antimicrobial peptide LL-37 inhibits adhesion of Candida albicans by interacting with yeast cell-wall carbohydrates. PLoS One 6:e17755. 10.1371/journal.pone.0017755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chang HT, Tsai PW, Huang HH, Liu YS, Chien TS, Lan CY. 2012. LL37 and hBD-3 elevate the beta-1,3-exoglucanase activity of Candida albicans Xog1p, resulting in reduced fungal adhesion to plastic. Biochem. J. 441:963–970. 10.1042/BJ20111454 [DOI] [PubMed] [Google Scholar]
- 42.Theberge S, Semlali A, Alamri A, Leung KP, Rouabhia M. 2013. C. albicans growth, transition, biofilm formation, and gene expression modulation by antimicrobial decapeptide KSL-W. BMC Microbiol. 13:246. 10.1186/1471-2180-13-246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Thankappan B, Jeyarajan S, Hiroaki S, Anbarasu K, Natarajaseenivasan K, Fujii N. 2013. Antimicrobial and antibiofilm activity of designed and synthesized antimicrobial peptide, KABT-AMP. Appl. Biochem. Biotechnol. 170:1184–1193. 10.1007/s12010-013-0258-3 [DOI] [PubMed] [Google Scholar]
- 44.Hamamoto K, Kida Y, Zhang Y, Shimizu T, Kuwano K. 2002. Antimicrobial activity and stability to proteolysis of small linear cationic peptides with D-amino acid substitutions. Microbiol. Immunol. 46:741–749. 10.1111/j.1348-0421.2002.tb02759.x [DOI] [PubMed] [Google Scholar]
- 45.Clark RJ, Jensen J, Nevin ST, Callaghan BP, Adams DJ, Craik DJ. 2010. The engineering of an orally active conotoxin for the treatment of neuropathic pain. Angew. Chem. Int. Ed. Engl. 49:6545–6548. 10.1002/anie.201000620 [DOI] [PubMed] [Google Scholar]
- 46.Darby C, Hsu JW, Ghori N, Falkow S. 2002. Caenorhabditis elegans: plague bacteria biofilm blocks food intake. Nature 417:243–244. 10.1038/417243a [DOI] [PubMed] [Google Scholar]
- 47.Joshua GW, Karlyshev AV, Smith MP, Isherwood KE, Titball RW, Wren BW. 2003. A Caenorhabditis elegans model of Yersinia infection: biofilm formation on a biotic surface. Microbiology 149:3221–3229. 10.1099/mic.0.26475-0 [DOI] [PubMed] [Google Scholar]
- 48.Tan L, Darby C. 2004. A movable surface: formation of Yersinia sp. biofilms on motile Caenorhabditis elegans. J. Bacteriol. 186:5087–5092. 10.1128/JB.186.15.5087-5092.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Begun J, Gaiani JM, Rohde H, Mack D, Calderwood SB, Ausubel FM, Sifri CD. 2007. Staphylococcal biofilm exopolysaccharide protects against Caenorhabditis elegans immune defenses. PLoS Pathog. 3:e57. 10.1371/journal.ppat.0030057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Edwards S, Kjellerup BV. 2012. Exploring the applications of invertebrate host-pathogen models for in vivo biofilm infections. FEMS Immunol. Med. Microbiol. 65:205–214. 10.1111/j.1574-695X.2012.00975.x [DOI] [PubMed] [Google Scholar]
- 51.Ricicova M, Kucharikova S, Tournu H, Hendrix J, Bujdakova H, Van Eldere J, Lagrou K, Van Dijck P. 2010. Candida albicans biofilm formation in a new in vivo rat model. Microbiology 156:909–919. 10.1099/mic.0.033530-0 [DOI] [PubMed] [Google Scholar]
- 52.Burgers R, Hahnel S, Reichert TE, Rosentritt M, Behr M, Gerlach T, Handel G, Gosau M. 2010. Adhesion of Candida albicans to various dental implant surfaces and the influence of salivary pellicle proteins. Acta Biomater. 6:2307–2313. 10.1016/j.actbio.2009.11.003 [DOI] [PubMed] [Google Scholar]
- 53.De Prijck K, De Smet N, Coenye T, Schacht E, Nelis HJ. 2010. Prevention of Candida albicans biofilm formation by covalently bound dimethylaminoethylmethacrylate and polyethylenimine. Mycopathologia 170:213–221. 10.1007/s11046-010-9316-3 [DOI] [PubMed] [Google Scholar]
- 54.Finkel JS, Mitchell AP. 2011. Genetic control of Candida albicans biofilm development. Nat. Rev. Microbiol. 9:109–118. 10.1038/nrmicro2475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Baillie GS, Douglas LJ. 1999. Role of dimorphism in the development of Candida albicans biofilms. J. Med. Microbiol. 48:671–679. 10.1099/00222615-48-7-671 [DOI] [PubMed] [Google Scholar]
- 56.Verna J, Lodder A, Lee K, Vagts A, Ballester R. 1997. A family of genes required for maintenance of cell wall integrity and for the stress response in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. U. S. A. 94:13804–13809. 10.1073/pnas.94.25.13804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zu T, Verna J, Ballester R. 2001. Mutations in WSC genes for putative stress receptors result in sensitivity to multiple stress conditions and impairment of Rlm1-dependent gene expression in Saccharomyces cerevisiae. Mol. Genet. Genomics 266:142–155. 10.1007/s004380100537 [DOI] [PubMed] [Google Scholar]
- 58.Norice CT, Smith FJ, Jr, Solis N, Filler SG, Mitchell AP. 2007. Requirement for Candida albicans Sun41 in biofilm formation and virulence. Eukaryot. Cell 6:2046–2055. 10.1128/EC.00314-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Albrecht A, Felk A, Pichova I, Naglik JR, Schaller M, de Groot P, Maccallum D, Odds FC, Schafer W, Klis F, Monod M, Hube B. 2006. Glycosylphosphatidylinositol-anchored proteases of Candida albicans target proteins necessary for both cellular processes and host-pathogen interactions. J. Biol. Chem. 281:688–694. 10.1074/jbc.M509297200 [DOI] [PubMed] [Google Scholar]
- 60.Sanglard D, Ischer F, Marchetti O, Entenza J, Bille J. 2003. Calcineurin A of Candida albicans: involvement in antifungal tolerance, cell morphogenesis and virulence. Mol. Microbiol. 48:959–976. 10.1046/j.1365-2958.2003.03495.x [DOI] [PubMed] [Google Scholar]
- 61.Bader T, Bodendorfer B, Schroppel K, Morschhauser J. 2003. Calcineurin is essential for virulence in Candida albicans. Infect. Immun. 71:5344–5354. 10.1128/IAI.71.9.5344-5354.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lane S, Zhou S, Pan T, Dai Q, Liu H. 2001. The basic helix-loop-helix transcription factor Cph2 regulates hyphal development in Candida albicans partly via TEC1. Mol. Cell. Biol. 21:6418–6428. 10.1128/MCB.21.19.6418-6428.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Liu H. 2001. Transcriptional control of dimorphism in Candida albicans. Curr. Opin. Microbiol. 4:728–735. 10.1016/S1369-5274(01)00275-2 [DOI] [PubMed] [Google Scholar]
- 64.Thevissen K, de Mello Tavares P, Xu D, Blankenship J, Vandenbosch D, Idkowiak-Baldys J, Govaert G, Bink A, Rozental S, de Groot PW, Davis TR, Kumamoto CA, Vargas G, Nimrichter L, Coenye T, Mitchell A, Roemer T, Hannun YA, Cammue BP. 2012. The plant defensin RsAFP2 induces cell wall stress, septin mislocalization and accumulation of ceramides in Candida albicans. Mol. Microbiol. 84:166–180. 10.1111/j.1365-2958.2012.08017.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Shareck J, Nantel A, Belhumeur P. 2011. Conjugated linoleic acid inhibits hyphal growth in Candida albicans by modulating Ras1p cellular levels and downregulating TEC1 expression. Eukaryot. Cell 10:565–577. 10.1128/EC.00305-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Silva S, Negri M, Henriques M, Oliveira R, Williams DW, Azeredo J. 2011. Adherence and biofilm formation of non-Candida albicans Candida species. Trends Microbiol. 19:241–247. 10.1016/j.tim.2011.02.003 [DOI] [PubMed] [Google Scholar]
- 67.Ramage G, Mowat E, Jones B, Williams C, Lopez-Ribot J. 2009. Our current understanding of fungal biofilms. Crit. Rev. Microbiol. 35:340–355. 10.3109/10408410903241436 [DOI] [PubMed] [Google Scholar]
- 68.Ramage G, Vande Walle K, Wickes BL, Lopez-Ribot JL. 2001. Biofilm formation by Candida dubliniensis. J. Clin. Microbiol. 39:3234–3240. 10.1128/JCM.39.9.3234-3240.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.