Abstract
The most common system for synthesis of cell surface polysaccharides is the Wzx/Wzy-dependent pathway, which involves synthesis, on the cytoplasmic face of the cell membrane, of repeat units, which are then translocated to the periplasmic face by a Wzx translocase and then polymerized by Wzy to generate the polysaccharide. One such polysaccharide is O antigen, which is incorporated into lipopolysaccharide (LPS). The O antigen is extremely variable, with over 186 forms in Escherichia coli. Wzx proteins are also very diverse, but they have been thought to be specific only for the first sugar of the repeat units. However, recent studies demonstrated examples in which Wzx translocases have considerable preference for their native repeat unit, showing that specificity can extend well beyond the first sugar. These results appear to be in conflict with the early conclusions, but they involved specificity for side branch residues and could be a special case. Here we take six Wzx translocases that were critical in the earlier studies on the importance of the first sugar and assess their ability to translocate the Escherichia coli O16 and O111 repeat units. We use gene replacements to optimize maintenance of expression level and show that under these conditions the native translocases are the most effective for their native repeat unit, being, respectively, 64-fold and 4-fold more effective than the next best. We conclude that Wzx translocases are commonly adapted to their native repeat unit, which provides an explanation for the great diversity of wzx genes.
INTRODUCTION
Many eukaryotic and prokaryotic cells have on their surfaces a complex set of glycoconjugates. In bacteria, these can have important roles in pathogenesis and environmental adaptation. One notable example is lipopolysaccharide (LPS), the major structural constituent of the outer leaflet of the outer membrane in Gram-negative bacteria. In most cases, the LPS includes a polysaccharide, known as O antigen or O polysaccharide, which is comprised of repeats of a 3- to 8-sugar repeat unit, that is the outermost component of the cell wall. In many species, the O antigen is extremely variable, with variation possible in the sugars present, in their order and associated linkages, and also in the polymerization linkage between the repeat units (1). For example, there are over 186 O-antigen forms for Escherichia coli (including Shigella) (2). Shigella flexneri 2a, to be discussed below, is in effect a pathogenic form of E. coli (3); notably, the O-antigen gene clusters found in E. coli O13, O129, and O135 have very few base differences from that of S. flexneri 2a (4), so in this paper S. flexneri 2a will be treated as one of the set of E. coli strains.
Nearly all E. coli O antigens are synthesized by the Wzx/Wzy-dependent pathway (5), as shown in Fig. 1A. The genes associated with the synthesis are generally grouped to form an O-antigen gene cluster at a specific locus, which in E. coli is between the galF and gnd genes (5). The synthesis of O antigen begins on the cytoplasmic face of the inner membrane, by addition of the first sugar as a sugar phosphate to a membrane-associated undecaprenyl-phosphate (Und-P) molecule. In E. coli, the initial transferase is usually WecA, which transfers N-acetylglucosamine-phosphate (GlcNAc-P) to Und-P to give Und-PP-linked GlcNAc. A series of glycosyltransferases subsequently add the other sugars until the repeat unit is completed. The repeat units are then translocated across the inner membrane by the Wzx translocase (6) and are subsequently polymerized by the Wzy polymerase (7). This produces the O-antigen polymer that is transferred by WaaL to preformed lipid A-core, and the LPS is exported en bloc to the outer leaflet of the outer membrane by the Lpt set of proteins (8). The chain lengths of O antigens vary, and the distribution commonly exhibits a concentration around a preferred modal chain length. The distribution is controlled by the Wzz protein, the product of the wzz gene, which is a few genes downstream of the O-antigen gene cluster in E. coli (9). In the absence of a wzz gene, the O-antigen chain length distribution has a very different pattern, with the number of molecules declining steadily with increasing chain length, which was shown by Bastin et al. (10) for Salmonella enterica LT2 to fit the distribution described by Goldman and Hunt before any knowledge of Wzz (11), for the situation where the probabilities of a growing O-antigen polymer (i) being extended by addition of another repeat unit or (ii) being ligated to lipid A core were the same regardless of chain length. In the case of S. enterica LT2, the probabilities were estimated to be 0.065 for ligation and 0.935 for extension over the range of 3 to 37 repeat units. Clearly, the presence of a Wzz protein confers a preferred chain length on O-antigen chains.
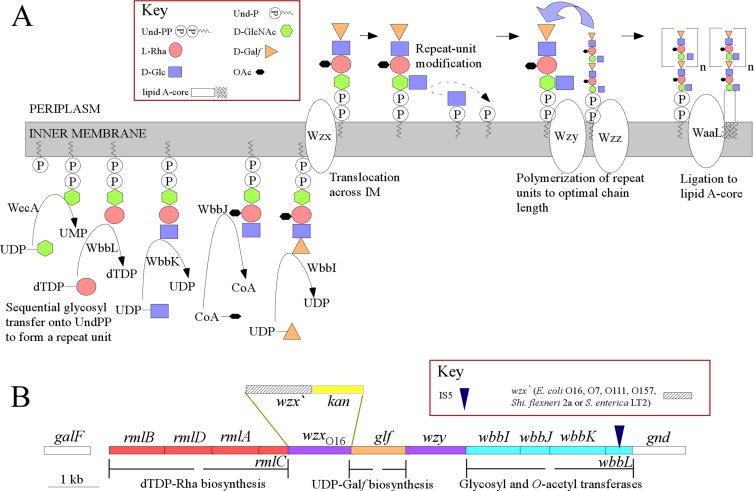
FIG 1.
(A) Biosynthesis pathway for the O16 antigen of E. coli K-12. Each sugar is represented by a symbol, color coded to correspond to the color coding of the biosynthesis genes in the gene cluster. The WbbJ O-acetyltransferase is shown here as acting on the cytoplasmic face of the inner membrane, as is known for the S. enterica group C2 O antigen with a gene-cluster-located transferase (45), but there is no evidence for the compartment for this reaction. (B) Gene cluster for the O16 O antigen of E. coli K-12 (the wbbL gene is interrupted by an IS5 element, as shown by Liu and Reeves (29). The wzx-kan cassettes used to replace the wzxO16 gene are also shown (not drawn to scale). Note that the gene encoding the glucosyltransferase for the side branch Glc residue that acts in the periplasmic face is not in the gene cluster and is not present in all E. coli O16 strains (46). Abbreviations: dTDP, thymine diphosphate; Galf, galactofuranose; Glc, glucose; GlcNAc, N-acetylglucosamine; IM, inner membrane; OAc, O-acetyl; Rha, rhamnose; Und-P, undecaprenyl phosphate; Und-PP, undecaprenyl pyrophosphate.
The Wzx/Wzy-dependent pathway is involved in the synthesis of many bacterial surface polysaccharides, but it is important to note that the translocase and polymerase proteins are not always named Wzx or Wzy, as proposed in 1996 by several groups working on O antigens and capsules (12). However, in this paper the names Wzx and Wzy cover the relevant proteins in all gene clusters that appear to code for a Wzx/Wzy-dependent pathway.
The Wzx translocases belong to the polysaccharide transport (PST) family, a member of the multidrug/oligosaccharidyl-lipid/polysaccharide (MOP) flippase superfamily, which closely resembles the multidrug and toxic compound extrusion (MATE) transporter family (13). Unfortunately, relatively little is known about these proteins, despite their vital importance in biological systems being long recognized. Wzx translocases are integral membrane proteins that contain either 12 or 14 transmembrane helices (14). An interesting and characteristic feature of Wzx translocases is their enormous sequence diversity (15). This diversity is exemplified by the finding of 20 Wzx sequence forms in 25 Acinetobacter polysaccharide gene clusters (16) and 23 Wzx sequence forms in the 37 S. enterica GlcNAc/N-acetylgalactosamine (GalNAc)-initiated O-antigen gene clusters (17).
The high level of wzx sequence diversity can be compared with the enormous structural diversity of the O-antigen repeat units that are translocated. However, Feldman et al. (18) showed that in E. coli O7 incomplete repeat units could be ligated to lipid A-core and therefore must have been translocated, showing that this Wzx translocase at least was not absolutely specific for the complete repeat unit. It was also demonstrated that several cloned Wzx translocases could complement the loss of Wzx function in E. coli K-12 strains O7 and O16 (18, 19), provided that their native repeat unit has the same first sugar, which for O7 and O16 is GlcNAc. However, translocases for non-GlcNAc-initiated repeat units showed either undetectable or only minor activity (19). It was inferred that the specificity for Wzx activity is determined largely by the identity of the first sugar.
It would be very surprising if Wzx translocases were indeed specific only for the first sugar, as there is enormous diversity in the wzx genes in species such as E. coli, in which almost all O antigens have either GlcNAc or GalNAc as the first sugar. We therefore undertook a study involving replacement of the wzx gene in the gene cluster, using a set of S. enterica serogroups that produce galactose (Gal)-initiated O antigens differing only in the presence or absence of a side branch dideoxyhexose (DDH), and showed that these Wzx translocases have strong preference for their native substrate (20). For example, the WzxD translocase encoded in the group D2 gene cluster has a strong preference for the presence of a side branch DDH residue, and the WzxE translocase for group E has a strong preference for the absence of that residue. Because the sugar involved was not in the main chain, polymerization is potentially possible and in this case highly likely, as the D2 and E gene clusters have nearly identical wzyE genes (21).
In an independent study, Wang et al. (22) reported two cases of an exopolysaccharide that could exist in two forms, with two Wzx translocases in each strain, each translocase being specific for one of the two structures. The species were Pantoea stewartii and Erwinia amylovora, and for both species the difference between the two forms was having either a glucose (Glc) or a pyruvate moiety at the end of a side branch. Clearly, some Wzx translocases have substrate specificity that extends well beyond the first sugar. However, it may be significant that in both studies the specificities are related to side branch residues, which may constitute a special case, as their absence would not necessarily block polymerization. Because translocation is potentially reversible but polymerization is not, it may be important to prevent incomplete repeat units being translocated to prevent them from being incorporated into long-chain O antigen, with selection for specificity in relation to side branch sugars.
However, it is also possible that some of the earlier data suggesting that translocases are specific only for the first sugar of the repeat unit were affected by higher-than-normal levels of Wzx expression. In this paper, we test the hypothesis that Wzx translocases are generally adapted to their native repeat unit and that this will usually make them less efficient with other structures. We used the same set of Wzx translocases that was used in the earlier studies on Wzx specificity, to assess the effect of expression level on their ability to translocate the E. coli O16 and O111 repeat units. We find that for both repeat units, only the native Wzx translocase can give normal levels of long-chain O-antigen-containing LPS when expressed from within an O-antigen gene cluster. This provides an explanation for the enormous diversity of wzx genes. However, the substrate specificity is not absolute and can be overwhelmed by high levels of Wzx expression.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions.
The strains and plasmids used in this study are described in Table 1. The E. coli strains were grown at 37°C in nutrient broth (5 g liter−1 NaCl, 5 g liter−1 yeast extract, 10 g liter−1 bacteriological peptone, pH 7.2) or on agar plates (nutrient broth with agar, 15 g liter−1). When appropriate, antibiotics were added at the following concentrations: ampicillin (25 μg ml−1), kanamycin (25 μg ml−1), and chloramphenicol (12.5 μg ml−1).
TABLE 1.
Strains and plasmids used in this study
| Strain or plasmid | Parent strain | Genotypea or description | Source or reference(s) |
|---|---|---|---|
| Strains | |||
| DH5α | E. coli K–12 strain; F− ϕ80lacZΔM15 Δ(lacZYA–argF)U169 deoR recA1 endA1 hsdR17(rk− mk+) phoA supE44 thi–1 gyrA96 relA1 λ− | Promega | |
| MG1655 | E. coli K–12 strain; F− λ− ilvG rfb–50 rph–1 | Coli Genetic Stock Centre | |
| M92 | E. coli O111:H− strain | IMVS Pathologyb and references 52 and 53 | |
| P5941 | MG1655 | MG1655, pPR2191 | This study |
| P5932 | MG1655 | MG1655, wzxO16 gene replaced by a kan gene | This study |
| P5937 | MG1655 | MG1655, wzxO16 gene replaced by wzxO16-kan | This study |
| P5933 | MG1655 | MG1655, wzxO16 gene replaced by wzxO7-kan | This study |
| P5938 | MG1655 | MG1655, wzxO16 gene replaced by wzxO111-kan | This study |
| P5935 | MG1655 | MG1655, wzxO16 gene replaced by wzxO157-kan | This study |
| P5934 | MG1655 | MG1655, wzxO16 gene replaced by wzxSf2a-kan | This study |
| P5936 | MG1655 | MG1655, wzxO16 gene replaced by wzxSeLT2-kan | This study |
| P5942 | P5932 | MG1655, wzxO16 gene replaced by a kan gene, pPR2191 | This study |
| P5946 | P5937 | MG1655, wzxO16 gene replaced by wzxO16-kan, pPR2191 | This study |
| P5944 | P5933 | MG1655, wzxO16 gene replaced by wzxO7-kan, pPR2191 | This study |
| P5947 | P5938 | MG1655, wzxO16 gene replaced by wzxO111-kan, pPR2191 | This study |
| P5952 | P5935 | MG1655, wzxO16 gene replaced by wzxO157-kan, pPR2191 | This study |
| P5943 | P5934 | MG1655, wzxO16 gene replaced by wzxSf2a-kan, pPR2191 | This study |
| P5945 | P5936 | MG1655, wzxO16 gene replaced by wzxSeLT2-kan, pPR2191 | This study |
| P5953 | P5942 | MG1655, wzxO16 gene replaced by a kan gene, pPR2191, pPR2193 | This study |
| P5954 | P5943 | MG1655, wzxO16 gene replaced by wzxSf2a-kan, pPR2191, pPR2193 | This study |
| P5955 | P5944 | MG1655, wzxO16 gene replaced by wzxO7-kan, pPR2191, pPR2193 | This study |
| P5956 | P5936 | MG1655, wzxO16 gene replaced by wzxSeLT2-kan, pPR2191, pPR2193 | This study |
| P5957 | P5947 | MG1655, wzxO16 gene replaced by wzxO111-kan, pPR2191, pPR2193 | This study |
| P5958 | P5952 | MG1655, wzxO16 gene replaced by wzxO157-kan, pPR2191, pPR2193 | This study |
| P5948 | P5938 | MG1655, wzxO16 gene replaced by wzxO111-kan, pPR2192 | This study |
| P5949 | P5937 | MG1655, wzxO16 gene replaced by wzxO16-kan, pPR2192 | This study |
| P5950 | P5933 | MG1655, wzxO16 gene replaced by wzxO7-kan, pPR2192 | This study |
| P5951 | P5934 | MG1655, wzxO16 gene replaced by wzxSf2a-kan, pPR2192 | This study |
| P5959 | P5951 | MG1655, wzxO16 gene replaced by wzxO16-kan, pPR2192, pPR2193 | This study |
| Plasmids | |||
| pKD3 | FRT-flanked cat gene, oriRγ replicon, ampicillin and chloramphenicol resistance | 23 | |
| pKD4 | FRT-flanked kan gene, oriRγ replicon, ampicillin and kanamycin resistance | 23 | |
| pKD46 | Lambda recombinase genes (α, β, γ) controlled by arabinose-inducible promoter, ParaB, temp-sensitive oriR101 replicon, ampicillin resistance | 23 | |
| pWQ552 | Low-copy-no. vector with p15A origin, with multiple cloning sites from pBAD24, tightly controllable expression by PTet, ampicillin resistance | Christ Whitfield | |
| pWQ572 | Low-copy-no. vector with p15A origin, with multiple cloning sites from pBAD24, tightly controllable expression by PTet, chloramphenicol resistance | 54 | |
| pPR2105 | pPR691 carrying the E. coli O111 gene cluster. Kanamycin resistance | 36 | |
| pPR2191 | pWQ572 (NcoI/BamHI) carrying the wbbL gene from E. coli K-12 strain WG1, which is controlled by tetracycline inducible PTet promoter, chloramphenicol resistant | This study | |
| pPR2192 | pPR691 carrying the E. coli O111 gene cluster with the wzxO111 gene replaced by a cat gene. Kanamycin and chloramphenicol resistance | This study | |
| pPR2193 | wzxO16 from E. coli K-12 cloned into pWQ552 (NcoI/PstI), controlled by tetracycline-inducible PTet promoter, ampicillin resistance | This study |
Genetic differences from the parent are underlined, including gene replacements and plasmid additions.
IMVS Pathology, Institute of Medical and Veterinary Science, Adelaide, Australia.
Cloning and transformation.
For the oligonucleotides used in cloning, see Table S1 in the supplemental material. PCR-amplified genes and plasmid vectors were separately digested with relevant restriction enzymes (New England BioLabs). Purified digested vectors were treated with calf intestinal alkaline phosphatase (New England BioLabs). Ligation was performed overnight at 16°C with T4 DNA ligase (New England BioLabs). The ligation mixtures were chemically transformed into E. coli K-12 strain DH5α.
The plasmids used in this study were introduced into the bacterial strains by electrotransformation, using the Gene Pulser Electroporation system (Bio-Rad), with settings as follows: 2.5 kV, 200 Ω, and 25 μF capacitance. Cultures were grown to an optical density at 600 nm (OD600) of ∼0.7, washed 3 times using ice-cold 10% glycerol, and concentrated in 10% ice-cold glycerol, with each 5-ml culture giving a final volume of 100 μl. The competent cells were mixed with plasmid DNA and electroporated in a 0.2-cm electroporation cuvette (Bridge).
Gene replacements.
The gene replacements within this study were performed as described previously (23) but with the modifications suggested in a recent study (24). Bacterial strains were made lambda recombination competent by incorporating pKD46. Cells for transformation were grown at 30°C, with l-arabinose (10 mM) added at an OD600 of ∼0.4, and then grown until an OD600 of ∼0.7 was reached. Cells were made electrocompetent and transformed according to the conditions described above. For the oligonucleotides used for gene replacements, see Table S2 in the supplemental material.
The wzxO16 and wzxO111 genes in E. coli K-12 MG1655 and pPR2105 were replaced by a kan and a cml cassette amplified from pKD4 and pKD3, respectively. For recombination involving wzx replacement, the wzx gene was joined to a kan gene from pKD4 by a 2-step PCR approach. Note that the wzx genes in these constructs have the same start and stop codons as the original wzxO16 gene, which should minimize the risk of variation in expression level, and that the kan element includes the P2 site from pKD4, in which a promoter is incorporated to drive the expression of genes downstream of the cassette (23), which should avoid polarity effects. The recombinants were selected using antibiotic resistance and verified by PCR. The replacement junctions were sequenced by Australian Genome Research Facility Ltd.
LPS extraction and visualization.
The E. coli strains were subcultured by 1:100 dilution from overnight cultures and grown at 37°C in nutrient broth. Note that tetracycline is used to induce the expression of genes cloned into the pWQ552 or pWQ572 vectors. Various concentrations of tetracycline were tested, of which the concentration of 50 ng ml−1 was found to give sufficient induction while not inhibiting cell growth and was used in all experiments. LPS was prepared as described previously (25), but the protocol was modified as suggested in a recent study (20). LPS samples derived from ∼2.5 × 108 cells were used per loading, but in some cases the loadings were slightly adjusted (± 11%) in referenced to the lipid A-core density of respective samples. The samples were run on 13% tricine SDS-PAGE and visualized by silver staining (26). The gels were scanned using a GS800 calibrated densitometer (Bio-Rad).
RESULTS
Reconstituting and controlling O-antigen production in E. coli K-12.
E. coli K-12 is a strain that has been widely used for genetic studies starting with Tatum using mutants to study the biosynthesis of amino acids (27). All laboratory isolates appeared to be rough (lacking O antigen), as first reported by Orskov and Orskov (28) and later found to be due to a mutation in the wbbL gene, which encodes the second transferase for the assembly of the O16 repeat unit, adding a rhamnose residue to the Und-PP-linked GlcNAc acceptor (29). This mutation, present in most K-12 strains, was due to insertion of an IS5 element in the gene (30) (Fig. 1B). We cloned the wbbL gene into plasmid pWQ572, putting wbbL under the control of the PTet promoter, which is inducible by tetracycline (see Materials and Methods), and restored O-antigen synthesis in E. coli K-12 MG1655. It is known that mutations in O-antigen genes can be deleterious due to accumulation of repeat unit intermediates (31–33), but the use of the PTet promoter allows us to control the expression of wbbL and hence the whole repeat unit. It is important to note that the WecA-facilitated transfer of GlcNAc-P to Und-P is reversible (34), so the wbbL mutation does not affect cell fitness, as the tetracycline inducer is added only for LPS extraction experiments.
Synthesis of LPS with normal levels of E. coli O16 antigen requires the native WzxO16 translocase.
The wzx genes from the E. coli O7, E. coli O111, E. coli O157, S. flexneri 2a, and S. enterica group B1 LT2 O-antigen gene clusters were introduced into E. coli K-12 MG1655 to replace the original wzxO16 gene, using the lambda red system (Fig. 1B) (see Materials and Methods). An E. coli K-12 wzxO16-kan cassette was also included as a control for the effect of the kan gene. The start codon of each inserted wzx gene is at the same site as for the original wzx. The wbbL plasmid was then transformed into MG1655 and each of the derived strains to allow induction of wbbL and thus allow controlled synthesis of the O16 repeat unit. The structures of the native O-antigen repeat unit substrates of these Wzx translocases are shown in Fig. 2.
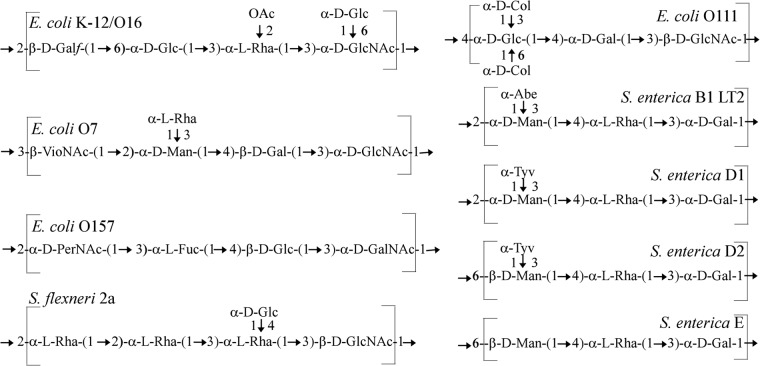
FIG 2.
Repeat unit structures of E. coli O16 (30), O7 (47), O111 (48), and O157 (49), S. flexneri 2a (50), and S. enterica group B1 (51) and D1, D2, and E (21) O antigens. The Glc side branch residues are coded by the Gtr system elsewhere in the genome and are added in the periplasmic face of the inner membrane only after translocation by Wzx translocase. Abbreviations: Galf, galactofuranose; Glc, glucose; Rha, rhamnose; GlcNAc, N-acetylglucosamine; OAc, O-acetyl; VioNAc, N-acetylviosamine; Gal, galactose; Col, colitose; PerNAc, N-acetylperosamine; Fuc, fucose; Abe, abequose; Tyv, tyvelose.
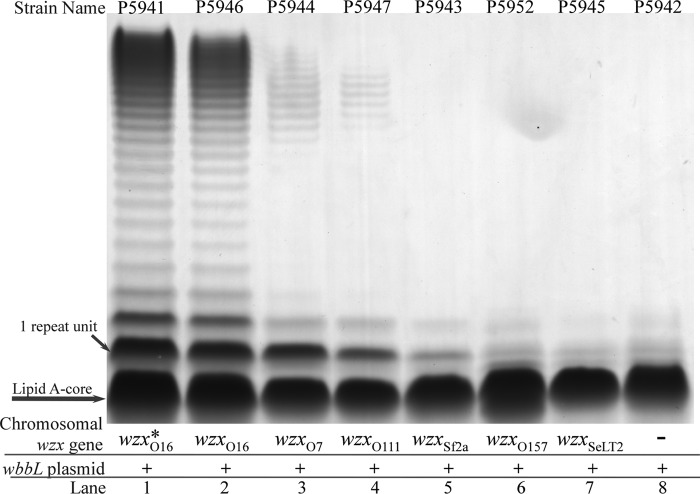
The above-described strains were grown for 16 h in nutrient broth, with tetracycline added to induce the expression of wbbL gene in the plasmid. The LPS samples were extracted and run on SDS-PAGE gels (Fig. 3) as described in Materials and Methods. The LPS extracted from strain MG1655 with the cloned wbbL gene gives a typical LPS pattern on an SDS-PAGE gel, i.e., a lipid A-core band and a ladder of LPS molecules with a modal value centered at about 18 repeat units (Fig. 3, lane 1). The MG1655 strain with the chromosomal wzxO16 gene replaced by a wzxO16-kan cassette (Fig. 3, lane 2) produces LPS with a pattern very similar to that of the strain with the native K-12 O16 gene cluster (Fig. 3, lane 1), indicating that the inclusion of the kan gene in the cassette has not had any deleterious effect. However, the ΔwzxO16 strain still produces very small amounts of LPS carrying one repeat unit (Fig. 3, lane 8). We used monospecific O16 antiserum to confirm that the band was of O16 LPS (see Fig. S1 in the supplemental material). This low level of translocation is attributed to the presence of non-O-antigen Wzx (probably for enterobacterial common antigen or colonic acid synthesis), which limits our ability to determine the translocation range of other O-antigen Wzx if it falls below this background level, so that the levels reported below for WzxO157 and WzxSeLT2 may be conservative estimates of the differences in effectiveness. We had observed a similar outcome in our previous S. enterica study, in a group D2 Δwzx mutant, in which the band has been confirmed by an immunoblot assay (20).
FIG 3.
LPS profiles of E. coli strain K-12 variants with different wzx genes in wzx-kan cassettes. All strains carry the wbbL plasmid pPR2191 to allow O16 repeat unit synthesis. The samples were prepared as described in Materials and Methods and separated in a 13% tricine SDS-PAGE gel. The LPS was visualized by silver nitrate staining. The asterisk (*) indicates that the gene cluster is the original E. coli K-12 O16 gene cluster.
MG1655 strains that have the original wzxO16 gene replaced by wzxO7-kan, wzxO111-kan, or wzxSf2a-kan, separately, produced LPS with very reduced levels of long-chain O antigen (Fig. 3, lanes 3, 4, and 5) compared with the positive-control strain that has wzxO16-kan (Fig. 3, lane 2). We assessed the amount of LPS in a band by scanning the gels, but only because diluted samples do not give an equivalent reduction in densitometry (data not shown). We did this by comparison with gels of diluted samples of the LPS of positive-control strain (see Fig. S2 in the supplemental material). The strains with wzxO7 and wzxO111 produce 64-fold less long-chain O antigen than those with the native wzxO16 translocase (Table 2), so they are seriously deficient in the synthesis of the O16 antigen. However, there is little or no reduction observed for LPS with one repeat unit (Table 2). The strain with the wzxSf2a gene gave no detectable modal chain length O antigen and about 8- to 16-fold less LPS with one repeat unit (Table 2). It is interesting that despite the severe reduction in the amount of O antigen produced, the modal chain length differed by only 2 to 3 repeat units wherever they could be detected.
TABLE 2.
Relative effectiveness for the synthesis of both E. coli O16 and O111 antigens in MG1655 strains with the wzxO16, wzxO7, wzxO111, or wzxSf2a gene within the gene cluster in respective strains
| Repeat unit | LPS form | Effectivenessa |
|||
|---|---|---|---|---|---|
| WzxO16 | WzxO7 | WzxO111 | WzxSf2a | ||
| O16 | Modal region chain | 1 | 1/64 | 1/64 | <1/64 |
| Single repeat unit | 1 | 1/1 | 1/2 | 1/8 to 1/16 | |
| Nonmodal short chain | 1 | 1/64 | 1/64 | <1/64 | |
| O111 | Modal region chain | 1/4 | 1/64 | 1 | <1/64 |
| Single repeat unit | 1/1 | 1/2 | 1 | 1/4 to 1/8 | |
| Nonmodal short chain | 1/64 | 1/64 | 1 | <1/64 | |
Effectiveness is given relative to that of the strain that encodes the native translocase and was evaluated by comparing the density of the band or group of bands for LPS from a nonnative translocase with those of serial dilutions of LPS from the positive control with the native translocase (see Fig. S2 and S4 in the supplemental material). Each set of the gels was assayed three times to obtain relative effectiveness. Three LPS forms were compared: the modal region LPS, LPS carrying a single repeat unit, and LPS carrying nonmodal short chain, i.e., 2 to 9 repeat units.
E. coli O157 and S. enterica group B1 LT2 both produce O antigens that have a first sugar other than GlcNAc. The latter has WbaP as the initial transferase, which adds Gal-P onto Und-P. However, like other GlcNAc-initiated O antigens, the E. coli O157 antigen is initialized by WecA, but the presence of a Gnu protein converts the Und-PP-linked GlcNAc to Und-PP-linked GalNAc (34, 35). Strains carrying the wzxO157 and wzxSeLT2 genes had LPS (Fig. 3, lanes 6 and 7) that was indistinguishable from that of the MG1655 ΔwzxO16 strain (Fig. 3, lane 8), putting O-antigen expression outside the detectable range.
We also added a cloned wzxO16 gene (pPR2193) to each strain used in Fig. 3, which restored the synthesis of O antigen to wild-type levels as in the ΔwzxO16 mutant (see Fig. S3 in the supplemental material). This shows that repeat unit synthesis and polymerization are not limiting and that the low levels of O antigen on the LPS discussed above are not due to any polar effect of the wzx gene replacement on expression of other genes. This confirms that the phenotypes that we observed are due to changes in the wzx genes.
We conclude that none of the five strains with an alternative wzx gene in the gene clusters can express enough O16 antigen to give a normal LPS phenotype, whereas putting the native wzxO16 gene in the same type of construct gave a normal level of expression.
Synthesis of normal levels of the E. coli O111 antigen also requires its native Wzx translocase.
We next assessed the ability of the same set of strains with the different wzx genes to express the E. coli O111 antigen. For this, we used the gene cluster constructs described above but changed the plasmids, so that the O16 repeat unit was replaced by the O111 repeat unit. These strains do not synthesize the O16 repeat unit because they do not have the wbbL plasmid, so they do not add rhamnose to Und-PP-linked GlcNAc, the first O16-specific reaction. We added the capacity to make the O111 repeat unit using plasmid pPR2192, which harbors a variant of the E. coli O111 gene cluster plasmid pPR2105 (36), in which the wzxO111 gene in the plasmid was replaced by a cat gene. The changes are such that the only O-antigen wzx genes present in these strains are those previously inserted into the chromosomal O16 gene cluster.
The last step in making the strains for this experiment was addition of the pPR2192 plasmid carrying the O111 gene cluster lacking wzx. The plasmid was successfully transformed into the strains that carry wzx genes from the E. coli O111, O16, and O7 and S. flexneri 2a gene clusters. We included strains M92 and P5938 in this experiment as the positive and negative controls. M92 is an O111 strain, and P5938 lacks the O-antigen gene cluster. The strain with the wzxO111 chromosomal gene produced LPS (Fig. 4, lane 3) similar to the LPS from M92. It should be noted that M92 has a unique modal pattern due to the presence of a wzzO111 gene in M92, in contrast with the strains used in other tracks, which had the wzzO16 gene. In contrast to the wzxO111 strain (Fig. 4, lane 3), the strain with the wzxO16 gene produced LPS with significantly smaller amounts of O antigen (Fig. 4, lane 4), while only a minor LPS ladder pattern was detected for strains with wzxO7 or wzxSf2a (the latter from S. flexneri 2a) (Fig. 4, lane 5 or 6, respectively). We again compared the modal chain length regions of these tracks with those of diluted samples of LPS from the wzxO111 strain (see Fig. S4 in the supplemental material). This showed that the strain with the wzxO16 gene is about 4-fold less effective than the strain with the wzxO111 gene for O111 antigen synthesis (Table 2), reflecting the effectiveness of the respective Wzx translocases.
FIG 4.
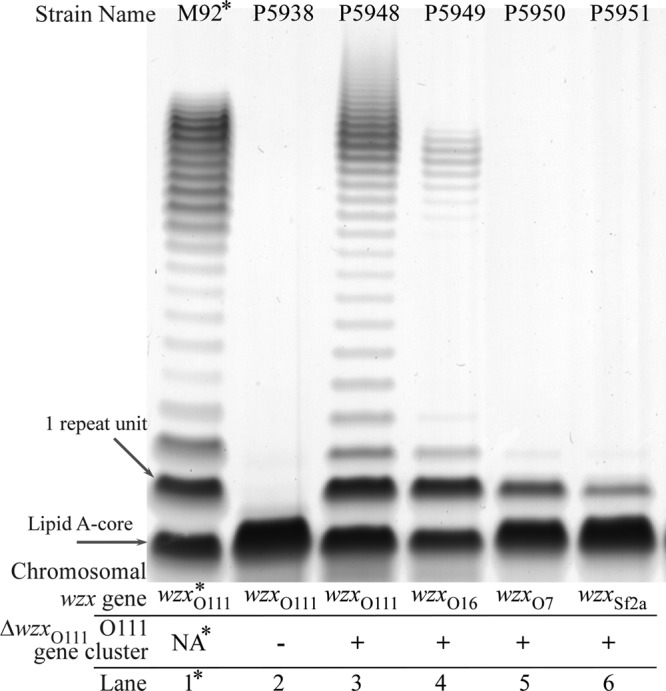
LPS profiles of E. coli K-12 variants expressing only the O111 antigen but with different wzx genes. All strains lack the wbbL plasmid pPR2191 and so are not able to synthesize the O16 repeat unit, and those in lanes 3 to 6 carry plasmid pPR2192 harboring an O111 gene cluster that lacks the wzxO111 gene. The samples were prepared, separated, and stained as described for Fig. 3. The asterisks (*) on lane 1 indicate that M92 is an E. coli O111 wild-type strain carrying the entire O111 gene cluster (including the wzxO111 gene) in its chromosome.
Similar transformation experiments to add the plasmid carrying the O111 gene cluster lacking wzx to strains with the wzxO157 gene or wzxSeLT2 gene, as well as the Δwzx strain, all failed, giving very few colonies, which did not survive after transfer to a fresh medium. These strains either have no wzx gene or have one for non-GlcNAc-initiated repeat units. We therefore interpret this as failure in translocation leading to accumulation of the Und-PP-linked repeat unit in the cytoplasmic face of the inner membrane. It should be pointed out that unlike the O16 set of experiments, where the wbbL gene is under control, there is no control over expression of the O111 repeat unit, and so the failure arises as the plasmid genes enter the cell, and it is the synthesis of repeat units that cannot be used that gives the lethality that we observed.
We conclude that WzxO111 is about 4-fold more effective than the WzxO16 form, while the other Wzx translocases make barely detectable levels on long-chain O antigen. Thus, for both O16 and O111, the native Wzx translocase is by far the most effective.
Overexpression can compensate for low translocation activity of a nonnative substrate.
We used a cloned WzxO16 translocase for translocation of the O111 repeat unit, and the gel pattern observed (Fig. 5, lane 4) resembles that of the positive-control LPS (Fig. 5, lane 2), showing that if overexpressed, WzxO16 can translocate the O111 repeat unit to produce normal levels of long-chain O antigen. The chromosomally encoded WzxO16 (Fig. 5, lane 3) showed the same reduction in effectiveness as in the previous experiment (Fig. 4, lane 4).
FIG 5.
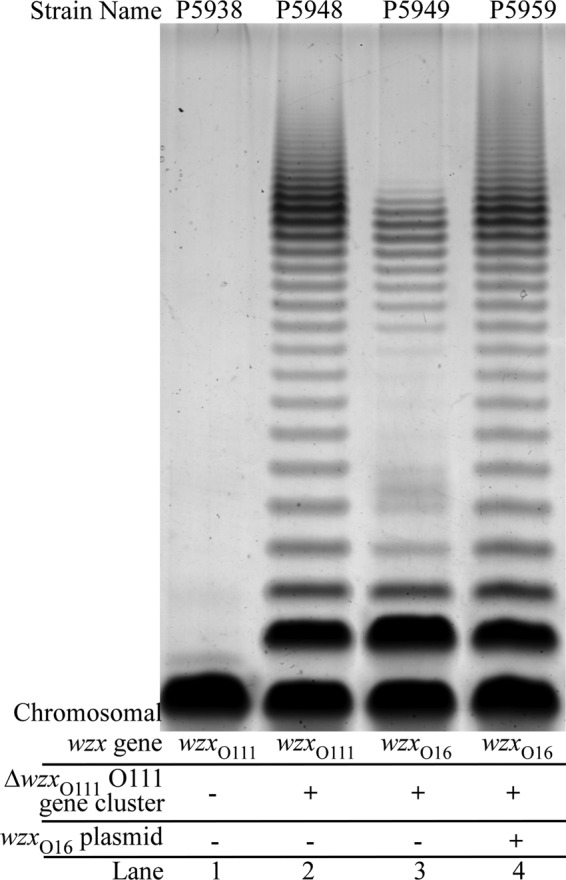
Wzx overexpression of a nonnative Wzx translocase can compensate for low translocation activity with normal expression. The samples in lanes 2 to 4 were extracted from strains carrying the pPR2192 plasmid harboring an O111 gene cluster that lacked the wzxO111 gene. An additional plasmid, pPR2193 (wzxO16), was also included in the strain used in lane 4. The samples were prepared, separated, and stained as described for Fig. 3.
We also tested the effect of overexpression in the B and D2 S. enterica strains that we used previously (20), which have Wzx translocases with a strong preference for repeat units that have a DDH side branch sugar. Using the D2 strain as an example, we took a mutant that did not have the side branch sugar and therefore made very little long-chain O antigen, and we showed that adding a clone of its native wzxD translocase gene restored normal levels of long-chain O antigen (see Fig. S5 in the supplemental material). We obtained the same results in comparable experiments with the group B strain and also with a group D1 strain (see Fig. S5 in the supplemental material).
These results show that overexpression of a wzx gene can restore ability to translocate a nonnative repeat unit and also confirm that the preference for presence of the DDH lies in the Wzx translocase and not in the Wzy polymerase, The experiment also confirmed that the reduced levels of long-chain O antigen in strains with nonnative wzx gene are not due to polar effects on expression of other genes in the gene cluster.
DISCUSSION
The native Wzx translocase is the most effective for the two repeat units studied.
We evaluated the capacity of one S. enterica and five E. coli Wzx translocases to translocate the E. coli O16 and O111 repeat units and found a very wide range in capability, with the native Wzx translocase being the most effective for both O antigens.
For the O16 repeat unit, we made a set of constructs in E. coli K-12 in which the chromosomal wzxO16 gene was replaced by a wzx gene from one of the other strains. Thus, the proteins would be regulated and expressed similarly to the native translocase. The same set of strains was then used to examine the activity of the six Wzx translocases with the O111 repeat unit. We did this by adding a clone of the O111 gene cluster that lacked the wzxO111 gene and by blocking the expression of the O16 repeat unit. This gave us the ability to examine the six Wzx translocases with the O16 and O111 repeat units in the same genetic background. In both cases, all wzx genes, including that native to the repeat unit, were in the same location in the chromosome and are expected a priori to be made in the same amount. The effectiveness of nonnative Wzx translocases for repeat units that have a GlcNAc as the first sugar ranged from barely detectable to about 1.5% for the O16 repeat unit and up to 25% for the O111 repeat unit. The wzxO16 and wzxO111 pair gives particularly strong support for specificity, as the comparison was done in both directions without changing the O-antigen gene cluster. Thus, the strain that has the wzxO16 gene within the gene cluster gave a wild-type LPS profile with its native O16 repeat unit (Fig. 3, lane 2) but gave only one-quarter the normal level of the O111 antigen (Fig. 4, lane 4). Likewise, the strain with the wzxO111 gene produced normal levels of its native O111 antigen (Fig. 4, lane 3) but gave only 1/64 the normal amount of modal chain length O16 antigen (Fig. 3, lane 4). The WzxO16 and WzxO111 translocases are clearly discriminating between the Und-PP-linked O16 and Und-PP-linked O111 repeat units. Each of the translocases gave a wild-type pattern for chain length distribution with its native repeat unit, indicating normal expression levels of both genes, but a greatly reduced level of modal chain length O antigen for the other repeat unit.
In the comparisons discussed above, we compared the relative effectiveness of nonnative Wzx translocases by estimating the amount of O antigen in the LPS. However, these estimates of translocation effectiveness will not equate to translocation efficiency if the reduced levels of translocation lead to changes in the concentration of one or more of (i) the Und-PP-linked repeat unit substrate in the cytoplasmic face of the inner membrane, (ii) the Wzx translocase, or (iii) lipid A-core. It is indeed very likely that reducing translocation would lead to accumulation of Und-PP-linked repeat units in the inner leaflet and perhaps also of lipid A-core in the outer leaflet of the membrane, due to the lack of substrate for ligation. This would increase the observed rates of translocation and ligation, respectively. It is also possible that the failure to make LPS with a normal chain length distribution would lead to increased expression of O-antigen pathway genes, including wzx, by feedback controls. All such effects are likely to increase the overall level of translocation, and our measure of translocase effectiveness would be an overestimate of the per-molecule efficiency of translocation by nonnative translocases. Therefore, the relative levels of effectiveness that we record should be treated as conservative estimates of translocation efficiency for nonnative repeat units.
The WzxO16, WzxO7, WzxO111, WzxO157, WzxSf2a, and WzxSeLT2 translocases were among those studied previously by plasmid-based complementation and were reported to be specific only for the first sugar of the repeat unit (18, 19), so we need an explanation for the different outcomes. In the earlier studies, the wzx genes were cloned into both low- and high-copy-number expression vectors, but a high-expression promoter was used for both. It may be that many of the earlier data were affected by protein overexpression, which was shown in our experiments to have a major impact on translocation of the S. enterica groups B, D1, and D2 repeat units by their respective Wzx (see Fig. S5 in the supplemental material) and the O111 repeat unit by WzxO16 (Fig. 5). It is important to note that the special status of the first sugar is not affected by our data, as overexpression of wzx genes for repeat units with different first sugars did not enable translocation of the O16 repeat unit.
Support for a complex involving at least Wzz and the Und-PP-linked O-antigen chain.
The reduction in translocation efficiency with a nonnative translocase reduces the number of Und-PP-linked repeat units for the downstream polymerization and ligation steps. The extent of the reduction depends on O-antigen chain length, and the patterns observed give insights into the competition for the diminished supply of repeat units. It is striking that the distribution of chain lengths within the major modal region is very similar whenever the bands are dense enough to be seen. For example, when WzxO7 is translocating the O16 repeat unit, there is ∼64-fold less LPS in the modal region than with the native WzxO16 translocase (Table 2), but the O-antigen chains have only 2 or 3 fewer repeat units (Fig. 3, compare lane 3 to lane 2). However, LPS molecules with only one repeat unit either are not affected or are reduced about 2-fold in the same samples, showing that the major effect of reducing the supply of repeat units is to reduce the amount, but not the length, of modal chain length LPS. The results for the O111 repeat unit set of experiments are similar (Fig. 4; see Fig. S4 in the supplemental material).
The very small variation that we observe in the Wzz-determined chain length over a 64-fold range in the proportion of modal chain length LPS supports a model proposed in 1993, in which Wzz forms a complex with Wzy, with Wzz first locking Wzy into continuous extension of the polymer and then changing to facilitate instead ligation by WaaL (10). It was also proposed that Wzz acts as a molecular clock, changing its function from facilitating chain extension to facilitating ligation after a set period of time or, alternatively, that it acts as a ruler that in some way effects the change in function when the length of the chain reaches a given length. An alternative “molecular chaperone” model involves Wzz interaction with WaaL so as to determine the molecular ratio of WaaL and Wzy, which is then proposed to determine the kinetics of ligation and polymerization (37). However, later in vivo (38) and in vitro (39) experiments showed that WaaL is not required for assembly of modal chain length Und-PP-linked O antigen. There is still no experimental evidence for either a clock-based or ruler-based determination of chain length or any direct biochemical evidence for interaction between Wzy and Wzz, but the new data on synthesis of modal chain length O antigen under limited substrate conditions provide very strong support for a complex including at least Wzz and the Und-PP-linked chain of repeat units. The existence of a complex including Wzz and the growing Und-PP-linked polymer is also supported by the ability of two or three types of “Wzz” molecules present in the same cell to each impose their specific modal chain length on a set of molecules (38, 40, 41). It is highly improbable that this could occur if Wzz was not associated with specific polymer molecules throughout the extension to modal length.
Competition between Wzz and WaaL for the Und-PP-linked repeat units when supply is limited.
The implication of the proposed Wzz-based complex for assembly of modal-length O-antigen polymer molecules is that there is competition between Wzz and WaaL for the Und-PP-linked repeat unit, as the ratio of single-repeat-unit LPS and modal chain length LPS is influenced by Und-PP-linked repeat unit availability.
In discussing the dynamics, we focus on (i) the formation of single-repeat-unit LPS, which requires only WaaL for synthesis, and (ii) synthesis of modal chain length LPS, for which O-antigen chain length is determined by Wzz, because we can record the outcome for both without ambiguity. There is also competition by Wzy for extension of short-chain-length molecules not under the control of Wzz. These are ligated as short-chain-length molecules, and as there are very few of them when repeat units are in short supply, we leave them out of the discussion and consider only the major outcomes in analysis of the competition.
We have relevant data for WzxO16, WzxO7, WzxO111, and WzxSf2a, with both O16 and O111 repeat units. When the repeat units are in short supply, it is modal chain length LPS that is most affected; for example, there are >64-fold reductions in modal chain length LPS with the WzxO7 and WzxO111 strains, and the single-repeat-unit LPS is reduced less than 2-fold (Table 2; see Fig. S2, lanes 8 and 9, in the supplemental material). This indicates that WaaL gets a higher proportion of the molecules than usual and that a much smaller proportion of the molecules enter the Wzz-controlled polymerization path to give the modal chain length LPS (Fig. 3 and 4). Indeed, only the WaaL-driven single-repeat-unit form of LPS can be seen when substrate supply is further reduced, as is the case for WzxSf2a (Fig. 3, lane 5, and Fig. 4, lane 6). It should be noted that extending the time to complete a modal length chain will not have much effect on the final outcome, as it is the relative frequencies of initiating Wzz-controlled polymerization and ligation of single repeat units that largely determine the ratio of modal length chain and single-repeat-unit LPS.
The substrate specificity of Wzx translocase provides insight into cell surface polysaccharide evolution.
The presence of long-chain cell surface polysaccharides is important in pathogenesis and colonization for many bacterial pathogens. It appears that long-chain O antigen allows evasion of the host innate immune system during the early stage of Salmonella infection, achieved by hindering both epithelial internalization and Toll-like receptor 4 recognition (42). Complement killing is also greatly inhibited by the presence of O antigen, and the wzz-determined modal chain length O antigen is most effective in this (43, 44). It should also be noted that enteropathogenic bacteria isolated from fecal samples generally exhibit long O-antigen chains on their LPS (42).
Thus, the outcome of having an ineffective Wzx translocase, and therefore inefficient translocation, is to make the organism more susceptible to host defense mechanisms, as the first effect is to reduce the proportion of long-chain LPS. If a nonnative translocase is present, the reduced efficiency means that insufficient repeat units are translocated to give a level of long-chain O antigen that would allow survival or colonization. The implication is that the evolution of a new repeat unit structure requires evolution of a translocase that is compatible with the new repeat unit. However, the fact that overexpression of Wzx can compensate for inefficiency of translocation raises the possibility that adaptation of a wzx gene to a new repeat unit structure can take place over time if expression is increased. This is in reality a likelihood, as changes in the structure occur in discrete steps, but adaptation of Wzx probably requires multiple amino acid substitutions. A further implication of this is that it is selection pressure that maintains the efficiency of the translocase against random genetic drift to less efficient forms.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by the University of Sydney Bridging Support Grant (RIMS 2012-00068).
We thank Thomas Ferenci and Ram Maharjan for the E. coli K-12 strain MG1655 and Chris Whitfield and Jerry King for the cloning vectors pWQ552 and pWQ572.
Footnotes
Published ahead of print 14 February 2014
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JB.01323-13.
REFERENCES
- 1.Wang L, Wang Q, Reeves PR. 2010. The variation of O antigens in gram-negative bacteria. Subcell. Biochem. 53:123–152. 10.1007/978-90-481-9078-2_6 [DOI] [PubMed] [Google Scholar]
- 2.Feng L, Senchenkova SN, Yang J, Shashkov AS, Tao J, Guo H, Zhao G, Knirel YA, Reeves PR, Wang L. 2004. Structural and genetic characterization of the Shigella boydii type 13 O antigen. J. Bacteriol. 186:383–392. 10.1128/JB.186.2.383-392.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pupo GM, Lan R, Reeves PR. 2000. Multiple independent origins of Shigella clones of Escherichia coli and convergent evolution of many of their characteristics. Proc. Natl. Acad. Sci. U. S. A. 97:10567–10572. 10.1073/pnas.180094797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Liu B, Knirel YA, Feng L, Perepelov AV, Senchenkova SN, Wang Q, Reeves PR, Wang L. 2008. Structure and genetics of Shigella O antigens. FEMS Microbiol. Rev. 32:627–653. 10.1111/j.1574-6976.2008.00114.x [DOI] [PubMed] [Google Scholar]
- 5.Samuel G, Reeves PR. 2003. Biosynthesis of O-antigens: genes and pathways involved in nucleotide sugar precursor synthesis and O-antigen assembly. Carbohydr. Res. 338:2503–2519. 10.1016/j.carres.2003.07.009 [DOI] [PubMed] [Google Scholar]
- 6.Liu D, Cole RA, Reeves PR. 1996. An O-antigen processing function for Wzx (RfbX): a promising candidate for O-unit flippase. J. Bacteriol. 178:2102–2107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Collins LV, Hackett J. 1991. Molecular cloning, characterization, and nucleotide sequence of the rfc gene, which encodes an O antigen polymerase of Salmonella typhimurium. J. Bacteriol. 173:2521–2529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Silhavy TJ, Kahne D, Walker S. 2010. The bacterial cell envelope. Cold Spring Harb. Perspect. Biol. 2:a000414. 10.1101/cshperspect.a000414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Batchelor RA, Alifano P, Biffali E, Hull SI, Hull RA. 1992. Nucleotide sequences of the genes regulating O-polysaccharide antigen chain length (rol) from Escherichia coli and Salmonella typhimurium: protein homology and functional complementation. J. Bacteriol. 174:5228–5236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bastin DA, Stevenson G, Brown PK, Haase A, Reeves PR. 1993. Repeat unit polysaccharides of bacteria: a model for polymerisation resembling that of ribosomes and fatty acid synthetase, with a novel mechanism for determining chain length. Mol. Microbiol. 7:725–734. 10.1111/j.1365-2958.1993.tb01163.x [DOI] [PubMed] [Google Scholar]
- 11.Goldman RC, Hunt F. 1990. Mechanism of O-antigen distribution in lipopolysaccharide. J. Bacteriol. 172:5352–5359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Reeves PR, Hobbs M, Valvano M, Skurnik M, Whitfield C, Coplin D, Kido N, Klena J, Maskell D, Raetz C, Rick P. 1996. Bacterial polysaccharide synthesis and gene nomenclature. Trends Microbiol. 4:495–503. 10.1016/S0966-842X(97)82912-5 [DOI] [PubMed] [Google Scholar]
- 13.Punta M, Coggill PC, Eberhardt RY, Mistry J, Tate J, Boursnell C, Pang N, Forslund K, Ceric G, Clements J, Heger A, Holm L, Sonnhammer EL, Eddy SR, Bateman A, Finn RD. 2012. The Pfam protein families database. Nucleic Acids Res. 40:D290–D301. 10.1093/nar/gkr1065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hvorup RN, Winnen B, Chang AB, Jiang Y, Zhou XF, Saier MH., Jr 2003. The multidrug/oligosaccharidyl-lipid/polysaccharide (MOP) exporter superfamily. Eur. J. Biochem. 270:799–813. 10.1046/j.1432-1033.2003.03418.x [DOI] [PubMed] [Google Scholar]
- 15.Cunneen MM. 2007. Ph.D. thesis Genetic and evolutionary analysis of polysaccharide flippase. The University of Sydney, Sydney, Australia [Google Scholar]
- 16.Hu D, Liu B, Dijkshoorn L, Wang L, Reeves PR. 2013. Diversity in the major polysaccharide antigen of Acinetobacter baumannii assessed by DNA sequencing, and development of a molecular serotyping scheme. PLoS One 8:e70329. 10.1371/journal.pone.0070329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liu B, Knirel YA, Feng L, Perepelov AV, Senchenkova SN, Reeves PR, Wang L. 2014. Structural diversity in Salmonella O antigens and its genetic basis. FEMS Microbiol. Rev. 38:56–89. 10.1111/1574-6976.12034 [DOI] [PubMed] [Google Scholar]
- 18.Feldman MF, Marolda CL, Monteiro MA, Perry MB, Parodi AJ, Valvano MA. 1999. The activity of a putative polyisoprenol-linked sugar translocase (Wzx) involved in Escherichia coli O antigen assembly is independent of the chemical structure of the O repeat. J. Biol. Chem. 274:35129–35138. 10.1074/jbc.274.49.35129 [DOI] [PubMed] [Google Scholar]
- 19.Marolda CL, Vicarioli J, Valvano MA. 2004. Wzx proteins involved in biosynthesis of O antigen function in association with the first sugar of the O-specific lipopolysaccharide subunit. Microbiology 150:4095–4105. 10.1099/mic.0.27456-0 [DOI] [PubMed] [Google Scholar]
- 20.Hong Y, Cunneen MM, Reeves PR. 2012. The Wzx translocases for Salmonella enterica O-antigen processing have unexpected serotype specificity. Mol. Microbiol. 84:620–630. 10.1111/j.1365-2958.2012.08048.x [DOI] [PubMed] [Google Scholar]
- 21.Reeves PR, Cunneen MM, Liu B, Wang L. 2013. Genetics and evolution of the Salmonella galactose-initiated set of O antigens. PLoS One 8:e69306. 10.1371/journal.pone.0069306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang X, Yang F, von Bodman SB. 2012. The genetic and structural basis of two distinct terminal side branch residues in stewartan and amylovoran exopolysaccharides and their potential role in host adaptation. Mol. Microbiol. 83:195–207. 10.1111/j.1365-2958.2011.07926.x [DOI] [PubMed] [Google Scholar]
- 23.Datsenko KA, Wanner BL. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. U. S. A. 97:6640–6645. 10.1073/pnas.120163297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hong Y, Duda KA, Cunneen MM, Holst O, Reeves PR. 2013. The WbaK acetyltransferase of Salmonella enterica group E gives insights into O antigen evolution. Microbiology 159:2316–2322. 10.1099/mic.0.069823-0 [DOI] [PubMed] [Google Scholar]
- 25.McGrath BC, Osborn MJ. 1991. Localisation of the terminal steps of O-antigen synthesis in Salmonella typhimurium. J. Bacteriol. 173:649–654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Brown PK, Romana LK, Reeves PR. 1991. Cloning of the rfb gene cluster of a group C2 Salmonella: comparison with the rfb regions of groups B and D. Mol. Microbiol. 5:1873–1881. 10.1111/j.1365-2958.1991.tb00811.x [DOI] [PubMed] [Google Scholar]
- 27.Gray GH, Tatum EL. 1944. X-ray induced growth factor deficiencies in bacteria. Proc. Natl. Acad. Sci. U. S. A. 30:404–410. 10.1073/pnas.30.12.404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Orskov I, Orskov F. 1960. The H antigen of the “K12” strain. A new E. coli H antigen: H48. Acta Pathol. Microbiol. Scand. 48:47. [PubMed] [Google Scholar]
- 29.Liu D, Reeves PR. 1994. Escherichia coli K12 regains its O antigen. Microbiology 140:49–57. 10.1099/13500872-140-1-49 [DOI] [PubMed] [Google Scholar]
- 30.Stevenson G, Neal B, Liu D, Hobbs M, Packer NH, Batley M, Redmond JW, Lindquist L, Reeves PR. 1994. Structure of the O-antigen of E. coli K-12 and the sequence of its rfb gene cluster. J. Bacteriol. 176:4144–4156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yuasa R, Levinthal M, Nikaido H. 1969. Biosynthesis of cell wall lipopolysaccharide in mutants of Salmonella. J. Bacteriol. 100:433–444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rick PD, Osborn MJ. 1972. Isolation of a mutant of Salmonella typhimurium dependent on D-arabinose-5-phosphate for growth and synthesis of 3-deoxy-D-mannooctulosonate. Proc. Natl. Acad. Sci. U. S. A. 69:3756–3760. 10.1073/pnas.69.12.3756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rick PD, Wolski S, Barr K, Ward S, Ramsay-Sharer L. 1988. Accumulation of a lipid-linked intermediate involved in enterobacterial common antigen synthesis in Salmonella typhimurium mutants lacking dTDP-glucose pyrophosphorylase. J. Bacteriol. 170:4008–4014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rush JS, Alaimo C, Robbiani R, Wacker M, Waechter CJ. 2010. A novel epimerase that converts GlcNAc-P-P-undecaprenol to GalNAc-P-P-undecaprenol in Escherichia coli O157. J. Biol. Chem. 285:1671–1680. 10.1074/jbc.M109.061630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cunneen MM, Liu B, Wang L, Reeves PR. 2013. Biosynthesis of UDP-GlcNAc, UndPP-GlcNAc and UDP-GlcNAcA involves three easily distinguished 4-epimerase enzymes, Gne, Gnu and GnaB. PLoS One 8:e67646. 10.1371/journal.pone.0067646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Stevenson G, Diekelmann M, Reeves PR. 2008. Determination of glycosyltransferase specificities for the Escherichia coli O111 O antigen by a generic approach. Appl. Environ. Microbiol. 74:1294–1298. 10.1128/AEM.02660-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Morona R, Van Den Bosch L, Manning P. 1995. Molecular, genetic, and topological characterization of O antigen chain regulation in Shigella flexneri. J. Bacteriol. 177:1059–1068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Daniels C, Griffiths C, Cowles B, Lam JS. 2002. Pseudomonas aeruginosa O-antigen chain length is determined before ligation to lipid A core. Environ. Microbiol. 4:883–897. 10.1046/j.1462-2920.2002.00288.x [DOI] [PubMed] [Google Scholar]
- 39.Woodward R, Yi W, Li L, Zhao G, Eguchi H, Sridhar PR, Guo H, Song JK, Motari E, Cai L, Kelleher P, Liu X, Han W, Zhang W, Ding Y, Li M, Wang PG. 2010. In vitro bacterial polysaccharide biosynthesis: defining the functions of Wzy and Wzz. Nat. Chem. Biol. 6:418–423. 10.1038/nchembio.351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Murray GL, Attridge SR, Morona R. 2003. Regulation of Salmonella typhimurium lipopolysaccharide O antigen chain length is required for virulence; identification of FepE as a second Wzz. Mol. Microbiol. 47:1395–1406. 10.1046/j.1365-2958.2003.03383.x [DOI] [PubMed] [Google Scholar]
- 41.Carter JA, Jimenez JC, Zaldivar M, Alvarez SA, Marolda CL, Valvano MA, Contreras I. 2009. The cellular level of O-antigen polymerase Wzy determines chain length regulation by WzzB and WzzpHS-2 in Shigella flexneri 2a. Microbiology 155:3260–3269. 10.1099/mic.0.028944-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Duerr CU, Zenk SF, Chassin C, Pott J, Gutle D, Hensel M, Hornef MW. 2009. O-antigen delays lipopolysaccharide recognition and impairs antibacterial host defense in murine intestinal epithelial cells. PLoS Pathog. 5:e1000567. 10.1371/journal.ppat.1000567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Murray GL, Attridge SR, Morona R. 2006. Altering the length of the lipopolysaccharide O antigen has an impact on the interaction of Salmonella enterica serovar Typhimurium with macrophages and complement. J. Bacteriol. 188:2735–2739. 10.1128/JB.188.7.2735-2739.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Murray GL, Attridge SR, Morona R. 2005. Inducible serum resistance in Salmonella typhimurium is dependent on wzz(fepE)-regulated very long O antigen chains. Microbes Infect. 7:1296–1304. 10.1016/j.micinf.2005.04.015 [DOI] [PubMed] [Google Scholar]
- 45.Liu D, Lindqvist L, Reeves PR. 1995. Transferases of O-antigen biosynthesis in Salmonella enterica: dideoxyhexosyltransferases of groups B and C2 and acetyltransferase of group C2. J. Bacteriol. 177:4084–4088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Batley M, Redmond JW, Packer NH, Liu D, Reeves PR. 1997. Relationship between the structures of the O polysaccharides from Escherichia coli O17 and O16. Carbohydr. Res. 303:313–318. 10.1016/S0008-6215(97)00177-8 [DOI] [PubMed] [Google Scholar]
- 47.L'vov VL, Shashkov AS, Dmitriev BA, Kochetrov NK. 1984. Structural studies of the O-specific side chain of the lipopolysaccharide from Escherichia coli O:7. Carbohydr. Res. 126:249–259. 10.1016/0008-6215(84)85382-3 [DOI] [PubMed] [Google Scholar]
- 48.Stenutz R, Weintraub A, Widmalm G. 2006. The structures of Escherichia coli O-polysaccharide antigens. FEMS Microbiol. Rev. 30:382–403. 10.1111/j.1574-6976.2006.00016.x [DOI] [PubMed] [Google Scholar]
- 49.Perry MB, MacLean L, Griffith DW. 1986. Structure of the O-chain polysaccharide of the phenol-phase soluble lipopolysaccharide of Escherichia coli O157:H7. Biochem. Cell Biol. 64:21–28. 10.1139/o86-004 [DOI] [PubMed] [Google Scholar]
- 50.Kenne L, Lindberg B, Petersson K, Katzenellenbogen E, Romanowska E. 1978. Structural studies of Shigella flexneri O-antigens. Eur. J. Biochem. 91:279–284. 10.1111/j.1432-1033.1978.tb20963.x [DOI] [PubMed] [Google Scholar]
- 51.Hellerqvist CG, Lindberg AA. 1971. Structural studies of the common-core polysaccharide of the cell wall lipopolysaccharide from Salmonella typhimurium. Carbohydr. Res. 16:39–48. 10.1016/S0008-6215(00)86096-6 [DOI] [Google Scholar]
- 52.Bastin DA, Reeves PR. 1995. Sequence and analysis of the O antigen gene (rfb) cluster of Escherichia coli O111. Gene 164:17–23. 10.1016/0378-1119(95)00459-J [DOI] [PubMed] [Google Scholar]
- 53.Bastin DA, Romana LK, Reeves PR. 1991. Molecular cloning and expression in Escherichia coli K-12 of the rfb gene cluster determining the O antigen of an E. coli O111 strain. Mol. Microbiol. 5:2223–2231. 10.1111/j.1365-2958.1991.tb02152.x [DOI] [PubMed] [Google Scholar]
- 54.Larue K, Ford RC, Willis LM, Whitfield C. 2011. Functional and structural characterization of polysaccharide co-polymerase proteins required for polymer export in ATP-binding cassette transporter-dependent capsule biosynthesis pathways. J. Biol. Chem. 286:16658–16668. 10.1074/jbc.M111.228221 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.