Abstract
Two related actinomycetes, Glycomyces sp. strain NRRL B-16210 and Stackebrandtia nassauensis NRRL B-16338, were identified as potential phosphonic acid producers by screening for the gene encoding phosphoenolpyruvate (PEP) mutase, which is required for the biosynthesis of most phosphonates. Using a variety of analytical techniques, both strains were subsequently shown to produce phosphonate-containing exopolysaccharides (EPS), also known as phosphonoglycans. The phosphonoglycans were purified by sequential organic solvent extractions, methanol precipitation, and ultrafiltration. The EPS from the Glycomyces strain has a mass of 40 to 50 kDa and is composed of galactose, xylose, and five distinct partially O-methylated galactose residues. Per-deutero-methylation analysis indicated that galactosyl residues in the polysaccharide backbone are 3,4-linked Gal, 2,4-linked 3-MeGal, 2,3-linked Gal, 3,6-linked 2-MeGal, and 4,6-linked 2,3-diMeGal. The EPS from the Stackebrandtia strain is comprised of glucose, galactose, xylose, and four partially O-methylated galactose residues. Isotopic labeling indicated that the O-methyl groups in the Stackebrandtia phosphonoglycan arise from S-adenosylmethionine. The phosphonate moiety in both phosphonoglycans was shown to be 2-hydroxyethylphosphonate (2-HEP) by 31P nuclear magnetic resonance (NMR) and mass spectrometry following strong acid hydrolysis of the purified molecules. Partial acid hydrolysis of the purified EPS from Glycomyces yielded 2-HEP in ester linkage to the O-5 or O-6 position of a hexose and a 2-HEP mono(2,3-dihydroxypropyl)ester. Partial acid hydrolysis of Stackebrandtia EPS also revealed the presence of 2-HEP mono(2,3-dihydroxypropyl)ester. Examination of the genome sequences of the two strains revealed similar pepM-containing gene clusters that are likely to be required for phosphonoglycan synthesis.
INTRODUCTION
Phosphonic acids are a group of structurally diverse compounds similar to many common organic phosphate esters, but with direct C-P bonds in place of the more familiar C-O-P linkage. Since the discovery of 2-aminoethylphosphonate (2-AEP) as a biologically produced molecule in 1959, phosphonates have been unequivocally identified in a number of eukaryotes, bacteria, and archaea (1, 2). Recent evidence suggests that ca. 5% of all microorganisms have the genetic capacity to produce phosphonates (3). Moreover, it is likely that these molecules play a significant role in the global phosphorus cycle (3–5). Among the diverse phosphonate natural products discovered in nature, the most common is 2-AEP, which typically occurs as a component of structural macromolecules such as lipids, polysaccharides, and proteins (6, 7).
Whereas phosphate is a common modification of polysaccharides, the corresponding phosphonate group is far less usual. A phosphonate-containing polysaccharide (phosphonoglycan) was initially isolated from the plasma membrane of the soil amoeba Acanthamoeba castellanii and was later shown to be a lipophosphonoglycan comprised of neutral sugars (26%), amino sugars (3%), acid-hydrolyzable phosphate (3%), long-chain fatty acids (14%), inositol (8%), phytosphingosines (13%), and a mixture of 2-AEP and 1-hydroxy-2-AEP in a 1:1 ratio (10%) (8–10). These aminophosphonates were proposed to be involved in the linkage to the lipids, perhaps through the inositol moieties (10). In the albumin glands of the snail Megalobulimus paranaguensis, 2-AEP was found to be a component of a branched β-d-galactopyranan, in which 2-AEP is esterified to d-galactose at O-6 (11). Previato and coworkers subsequently discovered that a lipopeptidophosphonoglycan isolated from epimastigote forms of Trypanosoma cruzi contains two end units of galactose linked to a mannotetraose main chain, which is linked (1→4) to a glucosaminyl unit substituted at O-6 by ester-linked 2-AEP (12). A similar result was observed in the O-glycan of Q-mucin from jellyfish composed of three monosaccharides: N-acetylgalactosamine (GalNAc), AEP-(O→6)-GalNAc, and P-6-GalNAc (13). Among bacteria, Bacteroides fragilis NCTC 9343 produces a capsular polysaccharide complex (CPC) which is directly involved in abscess formation in animal models (14). CPC comprises at least three distinct polysaccharides, PS A, PS B, and PS C, in which a 2-AEP substituent is located at O-4 of the N-acetyl-β-d-glucopyranosyl residue in PS B (15, 16). A capsular polysaccharide has also been isolated from the outer membrane of the ruminal bacterium Fibrobacter succinogenes S85 and contains N-(2-hydroxyethyl)-2-AEP (17). Interestingly, F. succinogenes S85 lacks typical lipopolysaccharides, and a possible function for the phosphonic acids in stabilizing membranes in the presence of phosphatases and lipases was proposed (17). Perhaps the most striking example of phosphonoglycan occurrence is in the freshly laid egg masses of the freshwater snail Helisoma, where almost 85% of the phosphorus is in the form of AEP and another unknown phosphonate linked to high-molecular-weight molecules consisting mainly of carbohydrate (18).
In each of the known phosphonoglycans described above, the phosphonate moiety was either 2-AEP or a derivative. Recently, we conducted a large-scale, gene-based screen for phosphonate-producing microorganisms (3). Among the organisms we identified were Glycomyces sp. strain NRRL B-16210 and Stackebrandtia nassauensis NRRL B-16338. Here we show that both organisms produce novel phosphonoglycans containing unusual methylated sugars and both glycerol- and hexose-linked 2-hydroxyethylphosphonate (2-HEP) in place of 2-AEP.
MATERIALS AND METHODS
Bacterial strains, plasmids and culture conditions.
Strains and plasmids used in this study are listed in Table 1. Streptomyces strains were grown at 30°C on ISP2 or ISP4 agar (Difco, Sparks, MD). Glycomyces and Stackebrandtia strains were grown at 30°C on ATCC medium 172 agar (19) or ISP4 agar. To probe the source of the methoxy group of O-methylgalactose, which is part of the phosphonoglycans, 0.4 mg/ml of l-[13C-methyl]methionine and l-[2H3-methyl]methionine was used to supplement ISP4 agar plates for the growth of Glycomyces and Stackebrandtia, respectively. Escherichia coli strains were grown at 37°C on lysogeny broth (LB) supplemented with antibiotics where appropriate. Antibiotics were used at the following concentrations for plasmid maintenance: chloramphenicol, 12.5 μg/ml; ampicillin, 100 μg/ml; and apramycin, 50 μg/ml. Diaminopimelic acid (1 mM) was added to the media for the growth of E. coli WM6029.
TABLE 1.
Bacterial strains, plasmids, and oligonucleotide PCR primers used in this study
| Strain or plasmid | Relevant characteristic(s)a | Source or reference |
|---|---|---|
| Strains | ||
| E. coli WM4489 | E. coli DH10B derivative; mcrA Δ(mrr-hsdRMS-mcrBC) ϕ80(ΔlacM15) ΔlacX74 endA1 recA1 deoR Δ(ara leu)7697 araD139 galU galK nupG rpsLλattB::pAE12(PrhaB::trfA33 ΔoriR6K-cat::frt-5) | 41 |
| E. coli WM6029 | dam-3 dcm-9 metB1 galK2 galT27 lacY1 tsx-78 supE44 thi mel-1 tonA31 Δ2 (mcrC-mrr)::frt Δ(endA)::frt attλ::pAE12-Δ1(oriR6K-cat::frt5) Δ5816(dapA)::frt uidA(ΔMluI)::pir(wt) attHK::pJK1006:: Δ1/2(Δ oriR6K-cat trfA) | This study |
| Glycomyces sp. NRRL B-16210 | Phosphonoglycan producer | ARS Culture Collection |
| Stackebrandtia nassauensis NRRL B-16215 | Phosphonoglycan producer | ARS Culture Collection |
| S. lividans TK24 | Heterologous host for phosphonate production | 42 |
| S. lividans MMG559 | Derivative of S. lividans TK24 containing a putative phosphonoglycan gene cluster from Glycomyces integrated at the ϕC31 attB site; Aprr | This study |
| S. lividans MMG598 | Derivative of S. lividans TK24 containing putative phosphonoglycan gene cluster from Stackebrandtia integrated at ϕC31 attB site; Aprr | This study |
| Plasmids | ||
| pJK050 | Double-cos fosmid vector; oriV λattB loxP FRT Cmr | 21 |
| pAE4 | OriT Aprr λattP ϕC31 int ϕC31attP | 21 |
| Fosmid 4-7G | Glycomyces genomic DNA cloned into pJK050; contains putative phosphonoglycan gene cluster | This study |
| Fosmid 4-10A | Stackebrandtia genomic DNA cloned into pJK050; contains putative phosphonoglycan gene cluster | This study |
| Primers | ||
| pepMF-for | 5′-CGCCGGCGTCTGCNTNGARGAYAA-3′ | 21 |
| pepMR-rev | 5′-GGCGCGCATCATGTGRTTNGCVYA-3′ | 21 |
FRT, Flp recombination target.
DNA isolation and manipulation.
All DNA manipulations were performed by established protocols (20). Endonuclease and T4 DNA ligase were purchased from Invitrogen (Carlsbad, CA) and New England BioLabs (Ipswich, MA). The oligonucleotide PCR primers were obtained from Integrated DNA Technologies (Coralville, IA) (Table 1). Plasmids and fosmids were isolated using Qiagen (Valencia, CA) miniprep or maxiprep kits. The genomic DNA from Glycomyces and Stackebrandtia strains, extracted using UltraClean microbial DNA isolation kit (MO BIO Laboratories, Carlsbad, CA), was used as the template for PCR amplification of a 406-bp pepM fragment with degenerate primers as described previously (21). PCR amplifications were performed with GoTaq Green master mix (Promega). Correct amplifications of the pepM gene were confirmed by DNA sequencing.
Construction of Glycomyces and Stackebrandtia genomic libraries, library screening, fosmid sequencing, and sequence annotations.
Construction of genomic libraries of Glycomyces and Stackebrandtia was as described previously (21), except that E. coli WM4489 was used as the cloning host. Fosmid clones carrying the phosphonate biosynthetic gene clusters from both strains were isolated as described in reference 21. Eight pepM+ fosmids from Glycomyces were pooled and sequenced on a Roche 454 GS FLX system on a quarter of a full 454 plate after tagging and library construction using Nextera kits (Epicentre Biotechnologies, Madison, WI). Sequence assembly of 454 reads was accomplished using Newbler (22). Additional sequence-specific primers were designed to fill in remaining gaps, as needed, by traditional Sanger sequencing with the Applied Biosystems 3730xl DNA analyzer. All sequencing was performed at the Roy J. Carver Biotechnology Center at the University of Illinois at Urbana-Champaign. Potential open reading frames were initially identified using RAST (23) and BLAST (24) analysis. Start sites and additional ORFs were corrected after visual inspection of the translated sequence.
Heterologous expression of phosphonate gene clusters in Streptomyces lividans.
To add functions necessary for transfer and integration into S. lividans, pepM+ fosmids from Glycomyces and Stackebrandtia were recombined in vitro with plasmid pAE4 (21), using Gateway BP Clonase II enzyme mix (Invitrogen) following the manufacturer's instructions. Conjugal transfer of fosmids from E. coli donor strain WM6029 to S. lividans TK24 was established following the standard protocol (21). Exconjugants were purified by repeated single-colony isolations on ISP4-apramycin plates. Correct integration of the fosmid into the genome of S. lividans TK24 was verified by PCR amplification of the pepM fragment from the purified genomic DNA. To test phosphonate production, exconjugants were first inoculated into 3 ml MYG (1% malt, 0.4% yeast extract, and 1% glucose) broth and incubated at 30°C for 3 days on a roller drum. This seed culture was then plated on ISP4 agar plates (Difco) (50 μl per plate) and incubated at 30°C for an additional 10 days. The liquid fraction containing phosphonates, released from the agar as described below, was concentrated 40-fold by lyophilization, resuspended in water, and filtered through a 0.2-μm filter to remove insoluble particles.
Isolation and purification of phosphonoglycans from Glycomyces.
For isolation of the phosphonoglycans, 20 ml of ATCC 172 broth (19) was inoculated with 200 μl of the starter culture of Glycomyces sp. strain NRRL B-16210 and agitated on a rotary shaker at 200 rpm for 5 days at 30°C. The culture was then plated on agar plates containing 10 liters of ISP4 solid medium (Difco) and incubated at 30°C for an additional 10 days. The plates were frozen at −80°C, followed by subsequent thawing and squeezing, which allowed liquids to be liberated from the agar. Squeezed agar was soaked with 70% methanol for 2 h at room temperature and then subjected to a second squeezing. Liquids from two extractions were combined and concentrated 40-fold by evaporation. Hydrophobic contaminants were removed from the concentrated liquids by successive extractions with dichloromethane, ethyl acetate, and hexane, while the phosphonoglycans were retained in the aqueous phase, as determined by 31P nuclear magnetic resonance (NMR). The aqueous phase was then treated with 60% (vol/vol) methanol and incubated on ice for 30 min. Following centrifugation at 5,000 × g for 30 min, the supernatant fluid was collected and the precipitates were removed. Subsequent addition of methanol to a final concentration of 80% (vol/vol) precipitated the phosphonoglycans, which were collected by centrifugation at 5,000 × g for 30 min, air dried, and redissolved in water. The final purification step involved ultrafiltration using an Amicon Ultra-15 membrane (molecular weight cutoff [MWCO] = 50,000) (Millipore, Billerica, MA). After 10 cycles of concentration and dilution, the phosphonoglycan in the retentate was recovered and lyophilized. The total yield of the phosphonoglycan was 120 mg per liter of culture.
Isolation and purification of phosphonoglycans from Stackebrandtia.
The Stackebrandtia phosphonoglycans were purified in the same manner, except that a 30K Amicon Ultra-15 membrane (Millipore) was used for ultrafiltration. The total yield of the phosphonoglycan was 40 mg per liter of culture.
Compositional and linkage analysis by GC-MS.
Phosphonoglycans from both strains were treated with 200 mM ice-cold trifluoroacetic acid (TFA). The resulting supernatant, after high-speed centrifugation to remove precipitates, was hydrolyzed by 2 M TFA for 1 h at 110°C. The monosaccharides thus obtained were derivatized using either aldononitrile acetate or alditol acetate as described elsewhere (25, 26). Permethylation linkage analysis was performed by established methods (27). Mono-O-methylated galactose standards were synthesized by treatment of galactose with methyl iodide in aqueous acetone and were purified on a mixed-bed column of Celite/activated charcoal. Gas chromatography-mass spectrometry (GC-MS) analysis was undertaken as previously reported (26). The GC-MS analysis used an Agilent (Santa Clara, CA) 6890N gas chromatograph interfaced with an Agilent 5973N mass-selective detector configured in an electron impact (EI) mode. Multiple injections were made with a Hewlett-Packard (Santa Clara, CA) 7683 series autoinjector. The column was a Hewlett-Packard DB-5 ms column (30 m by 0.25 mm; 0.25-µm film thickness), using helium as the carrier gas. The oven temperature was ramped over a linear gradient from 150 to 300°C at 10°C per min. Injector and detector/interface temperatures were 275 and 300°C, respectively. Mass spectra were recorded in positive-ion mode over the m/z range of 60 to 550. Data analysis was done off-line using HP Chemstation.
Partial acid hydrolysis and isolation of phosphonate-containing oligosaccharides from Glycomyces and Stackebrandtia.
Partial hydrolysis of 800 mg of purified Glycomyces phosphonoglycan was performed at 100°C in 6 M HCl under reflux for 3 h. The hydrolysate was adjusted to pH 7.0 by adding 5 M NH4OH and incubated with 70 g of activated charcoal (Sigma). The aqueous supernatant was removed after centrifugation. Charcoal-bound phosphonylated oligosaccharides were eluted stepwise with increasing concentrations of methanol (25%, 50%, 75%, and 90%) and lyophilized. The resultant solid was dissolved in water, passed through an Oasis HLB extraction cartridge (Waters), and eluted with water. The water eluant was concentrated, treated with Fe3+-charged Chelex 100 resin (Sigma), and eluted with an NH4HCO3 gradient (10 to 500 mM) and 2% NH4OH, followed by lyophilization. The major phosphonate-containing fraction was further chromatographed on Bio-Gel P-2 (100 by 1.5 cm) (Bio-Rad). Late phosphonate-containing fractions eluted from the P-2 column (5 ml per tube) were pooled, concentrated, and dialyzed against water using a Micro Float-A-Lyzer dialysis device (MWCO = 500 to 1,000; Spectra/Por). The filtrate was concentrated and fractionated on a Sephadex LH-20 column (100 by 3 cm) (GE Healthcare). Collected phosphonate-containing fractions (5 ml per tube) were concentrated for further NMR, GC-MS, and liquid chromatograph-mass spectrometry (LC-MS) analyses. Partial acid hydrolysis of Stackebrandtia phosphonoglycans and isolation of phosphonate-containing fractions were performed in a similar manner.
NMR spectroscopy.
All NMR experiments except diffusion ordered spectroscopy (DOSY NMR) were performed at room temperature on a Varian Inova 600 spectrometer equipped with a 5-mm Varian 600DB AutoX probe tuned for proton at 600 MHz and phosphorus at 242.83 MHz at the University of Illinois, Urbana-Champaign. 1H DOSY spectra were acquired on a Bruker Avance spectrometer (Bruker BioSpin Corp., Billerica, MA) operating at 500.11 MHz using a standard 5-mm z-gradient BBI probe at 27°C. The deuterated solvents used in this study were from Cambridge Isotope Laboratories (Andover, MA). Spectra were collected in water supplemented with 25% to 90% D2O as a lock solvent. Chemical shifts are reported in δ (ppm), referenced to tetramethylsilane for 1H and 13C or 85% H3PO4 as an external standard for 31P chemical shifts.
Sample preparation and analysis by LC-MS.
Phosphonates were analyzed by LC-MS as described previously (28). Briefly, phosphonates in crude culture extracts were partially purified using Fe3+-charged immobilized metal affinity chromatography (IMAC), dried in a SpeedVac, and then reconstituted in 90% acetonitrile containing 10 mM ammonium bicarbonate. LC-MS analysis was performed on a custom 11T linear ion trap Fourier transform mass spectrometer (LTQ-FT; Thermo Fisher Scientific) equipped with a 1200 high-performance liquid chromatography (HPLC) system (Agilent). Samples were separated on a Zic pHILIC column (2.1 mm by 150 mm; SeQuant) using 90% acetonitrile containing 10 mM ammonium bicarbonate (B) and 10 mM ammonium bicarbonate (A) as mobile phases. The elution was performed at a 0.2-ml/min flow rate with the following gradient conditions: 0 to 5 min, 100% B; 5 to 15 min, from 100% to 40% B; 15 to 20 min, from 40% to 100% B; 20 to 35 min, 100% B. The mass-spectral analysis consisted of a full scan at a resolution of 100,000 (m/z 100 to 1,000), a source fragmentation scan (85 V, m/z 50 to 110) detected in the ion trap, and a targeted collision-induced dissociation (CID) MS-MS (MS2) scan with FT detection to obtain tandem mass spectra of target compounds. The data were analyzed manually using the Qualbrowser application of Xcalibur software (Thermo Fisher Scientific).
Elemental analysis.
CHN (carbon, hydrogen, and nitrogen) analysis was performed with a CE440 elemental analyzer (Exeter Analytical Inc.), and P (phosphorus) analysis was performed with an Optima 2000 DV optical emission spectrometer (PerkinElmer) at the Microanalytical Lab of the University of Illinois, Urbana-Champaign. Both measurements were performed in duplicate.
Nucleotide sequence accession number.
The sequence for the putative phosphonoglycan biosynthetic locus from Glycomyces sp. strain NRRL B-16210 has been deposited in GenBank under accession number KJ125437.
RESULTS
Production of phosphonoglycans by Glycomyces sp. strain NRRL B-16210 and S. nassauensis NRRL B-16338.
We recently screened a large collection of actinomycetes for the presence of the pepM gene, which encodes phosphoenolpyruvate phosphonomutase, to identify novel phosphonate producers (6). Among the pepM-positive organisms we identified were two members of the family Glycomycetaceae (29, 30): Glycomyces sp. strain NRRL B-16210 and S. nassauensis NRRL B-16338.
Spent media obtained after growth of both organisms contained substantial amounts of P-containing compounds that had chemical shifts consistent with C-P linkages in 31P NMR analyses (see Fig. S1A and B in the supplemental material). 31P NMR spectra from washed cells of Glycomyces also indicated the presence of phosphonates (see Fig. S1C in the supplemental material). Treatment of Glycomyces cells with lysozyme released significant amounts of the phosphonate into the supernatant (see Fig. S1D in the supplemental material), suggesting that the molecule was attached to the cell surface. Initial analyses of the phosphonates present in spent media showed that the molecules could be precipitated by 80% methanol and that they were retained during ultrafiltration using high-molecular-weight-cutoff filters. Taken together, these analyses suggested that the two organisms produced a phosphonate-modified exopolysaccharide.
Purification of phosphonoglycans and molecular weight determination.
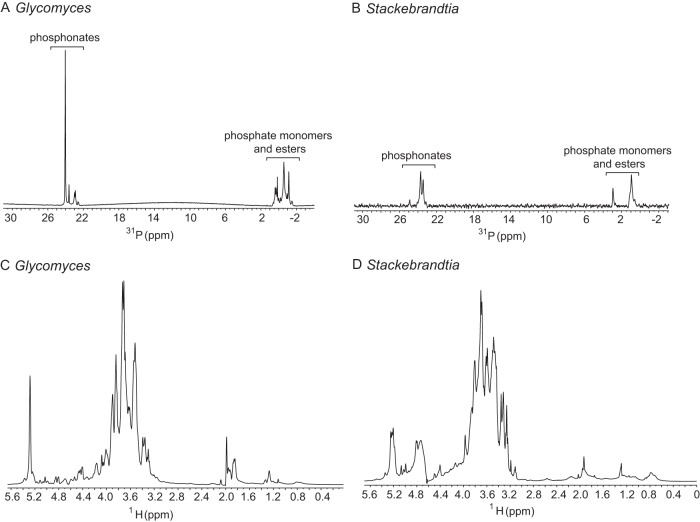
To obtain detailed structural and compositional data, we purified the phosphonoglycans from both organisms as described in Materials and Methods. In 31P NMR spectra, three to five peaks with close chemical shifts between 23 to 25 ppm were observed for both strains, suggesting that they may produce phosphonoglycans with a variety of linkages in the vicinity of the phosphonate moiety (Fig. 1A and B). Failure to separate those phosphonoglycans through purification steps, along with the fact that they had very close phosphorus chemical shifts, indicated that the structures may be very closely related. The 1H NMR spectrum of purified phosphonoglycans from Glycomyces resembled that from Stackebrandtia; at least four signals were found in the anomeric proton region (δH 4.4 to 5.3 ppm, excluding the water peak), but line broadening caused by sample viscosity precluded anomeric signal integrations. Other major signals shown in the 1H NMR spectra included the CH2P groups at δH 1.78 and 1.86 ppm for Glycomyces (and δH 1.81 and 1.94 ppm for Stackebrandtia) and other sugar ring protons in the region of δH 3.2 to 4.4 ppm (Fig. 1C and D). The proton chemical shifts of CH2P groups were determined from 1H-31P heteronuclear multiple-bond correlation (HMBC) experiments (see Fig. S2 in the supplemental material).
FIG 1.
NMR spectra of purified phosphonoglycans. (A) 31P NMR spectrum of purified phosphonoglycans from Glycomyces. (B) 31P NMR spectrum of purified phosphonoglycans from Stackebrandtia. (C) 1H NMR spectrum of purified phosphonoglycans from Glycomyces. (D) 1H NMR spectrum of purified phosphonoglycans from Stackebrandtia.
DOSY NMR, which resolves different compounds based on their different diffusion rates, provides a relatively convenient method to estimate molecular size (31, 32). Based on 1H DOSY NMR experiments and compared with known pullulan standards, predicted sizes of phosphonoglycans of Glycomyces and Stackebrandtia purified from spent media were ∼40 to 50 kDa (see Fig. S3A and B in the supplemental material), whereas sizes of cell-bound phosphonoglycans from Glycomyces were estimated to be 5 to 6 times greater (see Fig. S3C in the supplemental material). Elemental analysis (wt%) of the phosphonoglycans found the following: for Glycomyces, C, 31.60 (±0.26) (mean ± standard deviation); H, 4.31 (±0.13); P, 1.24 (±0.05); for Stackebrandtia, C, 36.12 (±0.07); H, 5.22 (±0.04); P, 0.26 (±0.03).
Compositional and per-deutero-methylation linkage analysis.
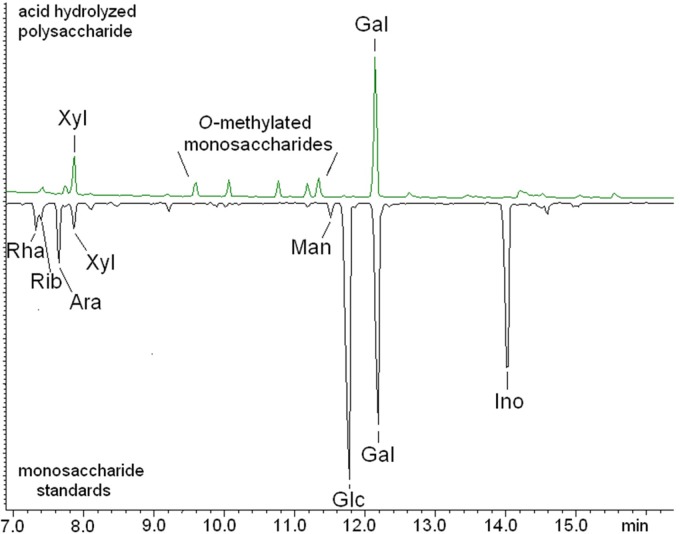
To determine the sugar composition, phosphonoglycans from the two strains were hydrolyzed, derivatized, and then analyzed by GC-MS. The major phosphonoglycan from Glycomyces was composed of galactose (70.7%), various monomethoxygalactoses (21.0%), xylose (6.4%), and 2,3-di-O-methylgalactose (1.9%) (Table 2). GC-MS monosaccharide compositional analysis of the phosphonoglycan is exemplified in Fig. 2 and in Fig. S4 in the supplemental material. The predominant monosaccharides from Stackebrandtia phosphonoglycans were glucose (73.2%) and galactose (18.2%), with different varieties of monomethoxygalactoses and xylose present as minor components (Table 2).
TABLE 2.
Glycomyces and Stackebrandtia phosphonoglycan compositional analysis by GC-MS of sugar aldononitrile acetate derivatives
| Sugar | % of total |
|
|---|---|---|
| Glycomyces | Stackebrandtia | |
| Galactose | 70.7 | 18.2 |
| Glucose | 73.2 | |
| 2-O-Methylgalactose | 7.4 | 3.9 |
| 3-O-Methylgalactose | 1.8 | <1 |
| 4-O-Methylgalactose | 3.9 | <1 |
| 6-O-Methylgalactose | 7.9 | 4.7 |
| 2,3-Di-O-methylgalactose | 1.9 | |
| Xylose | 6.4 | <1 |
FIG 2.
Overlaid gas chromatographs of aldononitrile acetate derivatives of the component monosaccharides from the major phosphonoglycan of Glycomyces. The top chromatograph is from the acid hydrolysate of the phosphonoglycan. The lower chromatograph (inverted) is from a series of monosaccharide standards. Rha, rhamnose; Rib, ribose; Ara, arabinose; Xyl, xylose; Man, mannose; Glc, glucose; Gal, galactose; Ino, inositol. Peaks due to the five partially O-methylated monosaccharides are indicated. The identities of these five peaks were confirmed by comparing with aldononitrile acetate derivatives of O-methylated galactose standards, as illustrated in Fig. S4 in the supplemental material.
Because of the presence of the various naturally occurring methoxygalactose residues in the phosphonoglycan backbone of Glycomyces, we decided to undertake permethylation linkage analysis using isotopically labeled deuterated [CD3]methyl iodide. In this case, the more standard usage of dimethyl sulfoxide as the solvent was precluded by the low solubility of the phosphonated polysaccharide and because the use of aqueous acetone plus a sodium hydroxide base catalyst was found to be more effective. Partially methylated galactose standards were prepared from methyl galactoside under the same reaction conditions. Following perdeuteromethylation and hydrolysis, aldononitrile acetate derivatives were prepared for analysis by GC-MS.
The results for the linkage analysis are summarized in Table 3. Permethylation analysis gave rise to several di-methoxygalactose residues that had one nondeuterated and one deuterated methyl group. These arose from monodeuteromethylation of the various naturally occurring monomethoxygalactose residues. The location of the deuteromethyl groups, and hence of potential linkage sites, was determined from the EI-MS fragmentation analysis. Fragmentation of O-methylated aldononitrile acetates tends to occur adjacent to methoxy groups, due to the addition stability of [R-O-Me]+ ions (33). Hence, hexose aldononitrile acetates that are substituted by a methyl group at the 6-hydroxy position are characterized by an m/z 45 fragment ion, which arises from cleavage of the hexosyl C-5–C-6 carbon-carbon bond. Replacement of this methyl group by a deuteromethyl group (CD3) increases the mass of this ion to m/z 48, and this was observed for the GC peaks at retention times of 11.95, 12.34, and 12.59 min. These primary fragment ions are degraded further by the loss of ketene (−42 Da), acetic acid (−60 Da), or acetic anhydride (−102 Da). Thus, 3,6D3-diMeGal was characterized by m/z 48 plus the two ion series 192 → 132 → 90 and 236 → 176 → 116, clearly showing that the methyl group at O-3 was nondeuterated. However, both of the methyl groups on the 2,6-diMeGal and 4,6-diMeGal derivatives were deuterated, showing that these arose from the nonmethylated galactosyl residues in the polysaccharide.
TABLE 3.
Per-deutero-methylation linkage analysis for phosphonoglycans from Glycomyces
| Derivative | Retention time (min) | % of totala | Inferred linkage |
|---|---|---|---|
| 2D3,6D3-diMeGal | 11.95 | 53.7 | 3,4-linked Gal |
| 3,6D3-diMeGal | 12.34 | 2.2 | 2,4-linked 3-MeGal |
| 4D3,6D3-diMeGal | 12.59 | 16.0 | 2,3-linked Gal |
| 2,3-diMeGal | 13.33 | 8.2 | 4,6-linked 2,3-diMeGal |
| 2,4D3-diMeGal | 13.65 | 17.4 | 3,6-linked 2-MeGal |
Fractional percentage of total dimethylated residues detected.
These per-deutero-methylation data indicated that four of the galactosyl residues in the polysaccharide backbone were 3,4-linked Gal, 2,4-linked 3-MeGal, 2,3-linked Gal, and 3,6-linked 2-MeGal. This suggested either that these residues were at branch points in the polysaccharide backbone or that another acid-labile substituent was present on these residues, other than the single methoxy group. It also showed that these residues were likely to be a part of the polysaccharide backbone, rather than terminating branch point sugars. Moreover, from the lack of deuteromethylation at the 5 position, it is reasonable to assume that these residues were predominantly present as pyranoses. Interestingly, we observed linkage types for 2-MeGal and 3-MeGal, but we did not observe a deuteromethylated derivative arising from the 4-MeGal or 6-MeGal residues. This suggested that the 4-MeGal and 6-MeGal residues present may be at a trisecting branch point, which was unusual, or that they were more heavily substituted than the 2- or 3-MeGal residues. In addition, a peak was observed with a retention time of 13.33 min, due to nondeuterated 2,3-diMeGal. This showed that the small amount of 2,3-dimethoxy-galactose in the polysaccharide backbone was 4,6-linked or, alternatively, substituted by acid-labile substituents at these positions.
GC-MS analysis of the O-methylgalactose component of phosphonoglycan isolated from Stackebrandtia cultured with isotopically enriched l-methionine.
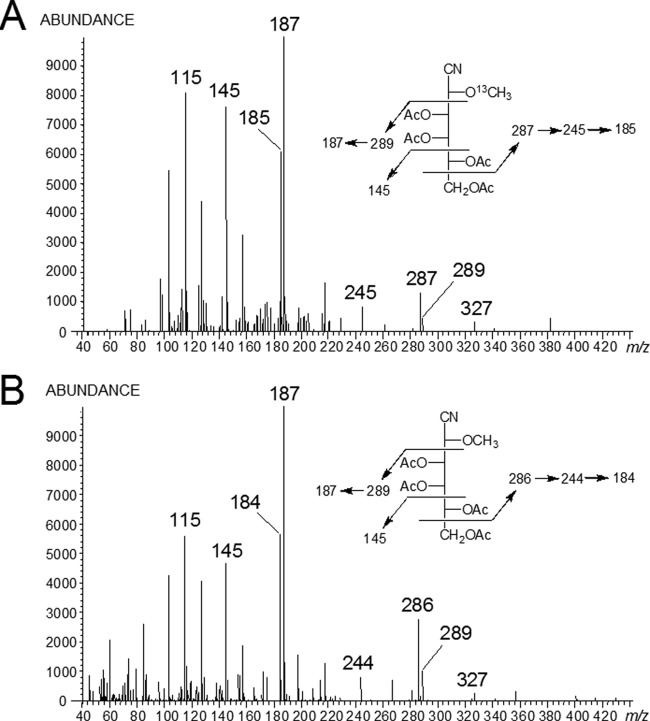
Various mono-O-methylated galactose residues were shown to be a component of both the Glycomyces and the Stackebrandtia phosphonoglycans. The biosynthetic origin of the O-methyl groups was investigated by culturing two strains with l-[13C-methyl]methionine or l-[2H3-methyl]methionine. Following culturing and extraction of the phosphonoglycans, the component monosaccharides were recovered, acid hydrolyzed, and analyzed by GC-MS as peracetylated-aldononitrile (PAAN) derivatives. For phosphonoglycans from Glycomyces, the sensitivity of the isotopic enrichment was too low to allow the unambiguous assignments of peaks, probably due to poor uptake. However, phosphonoglycans from Stackebrandtia grown in the presence of l-[13C-methyl]methionine gave rise to a GC peak at 15.2 min due to 2-O-methylgalactose PAAN and were seen to have incorporated 13C label with about 50% enrichment. As is typical for PAAN derivatives, no molecular ion was observed, but characteristic fragment ions were apparent. Electron impact mass spectrometry generated fragment ions across the C-2–C-3, C-4–C-5, and C-5–C-6 bonds of the sugar derivative. The ions arising from C-2–C-3 (m/z 289 and the daughter ion m/z 187) and C-4–C-5 (m/z 145) were not isotopically enriched and were derived from the nonmethylated part of the 2-O-methylgalactose PAAN (Fig. 3). However, the C-5–C-6 cleavage generated a series of fragments ions (m/z 287, 245, and 185) that were 1 Da greater than the equivalent fragments of the control (Fig. 3). These mass differences were due to the incorporation of 13C from l-[13C-methyl]methionine into these ions. A small enrichment was also observed when the same strain was grown on l-[2H3-methyl]methionine, with the control ion at m/z 286 being increased by 3 mass units due to the incorporation of the deuterated methyl group (data not shown). The isotopic enrichment of l-methionine suggested that the O-methyl groups on the monomethylated galactose residues arose via S-adenosylmethionine, the classic pathway for the biological formation of methyl ethers.
FIG 3.
Electron impact mass spectra of 2-O-methylgalactose PAAN derived from phosphonoglycans of Stackebrandtia, grown in the presence of l-[13C-methyl]methionine (A) or in the presence of l-methionine (control) (B). Structures and fragmentation patterns of the compound are shown in the insets. The peak labels in the spectra are observed values, whereas the numbers shown in the insets are calculated values.
Partial hydrolysis of phosphonoglycans and isolation of phosphonate-containing oligosaccharides.
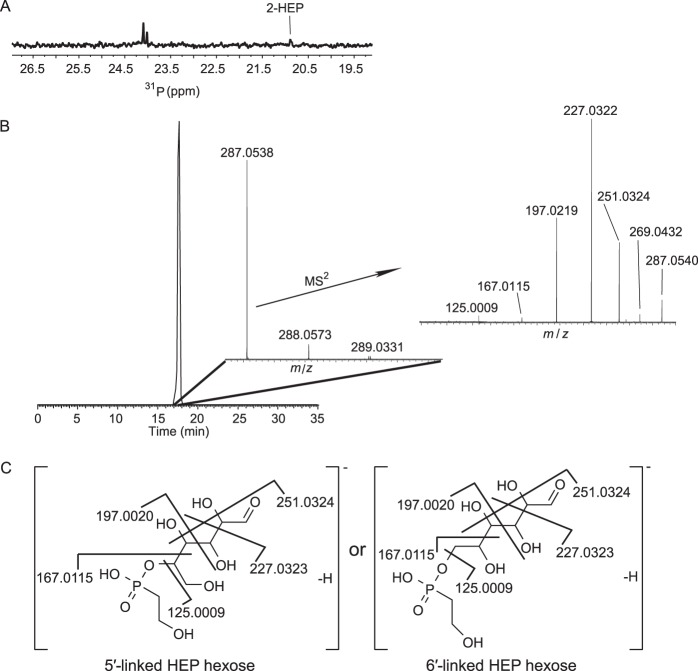
To study the linkage of the phosphonate to the polysaccharide, partial acid hydrolysis (6 M HCl, 3 h) of Glycomyces phosphonoglycans was performed at 100°C. This resulted in the release of various polymeric fragments containing 2-HEP bound oligosaccharides and 2-HEP (see Fig. S5 in the supplemental material). The hydrolysate was purified as described in Materials and Methods. Analysis of one sample after partial acid hydrolysis excluded the possibility of an ether linkage between 2-HEP and oligosaccharides, as in the 1H-13C HMBC spectrum, H-2 of 2-HEP had only one cross-peak with C-1 of 2-HEP but not with any other carbons from sugar rings (see Fig. S6 in the supplemental material). Therefore, we believe 2-HEP was most likely bound to oligosaccharides through an ester linkage. Dialysis of the hydrolysate against water with a Micro Float-A-Lyzer dialysis device (MWCO = 500 to 1,000; Spectra/Por) followed by fractionation of the filtrate on a Sephadex LH-20 column (GE Healthcare) provided a fraction which contained a mixture of two unknown phosphonates and 2-HEP (Fig. 4A). In addition to 2-HEP, high-resolution LC-MS analysis of this fraction identified the presence of the ion at m/z 287.0538, corresponding to 2-HEP linked to one hexose (Fig. 4B). Further MS2 investigation indicated that 2-HEP was possibly ester linked to hexose at either the O-5 or O-6 position, by virtue of the fragment ions at m/z 251.0324, 227.0322, 197.0219, 167.0115, and 125.0009 (Fig. 4B and C). Based on the sugar component analysis, the only hexose present in the phosphonoglycans was galactose, and it was the largest component (accounting for 70.7% of the total); we therefore assigned the hexose as galactose. Unfortunately, attempts to use GC-MS to determine the 2-HEP and galactose linkage following sodium borohydride reduction, peracetylation, and derivatization were not successful. Nonetheless, galactose more often occurs in bacterial polysaccharides in the form of pyranose. If this was the case in the Glycomyces phosphonoglycans, which was supported by the lack of deuteromethylation at the 5 position of galactose (or O-methylgalactose) as noted above (thereby implying the pyranose configuration), the O-5 of galactose would be part of the sugar ring and hence not available for substitution. Therefore, the most likely attachment to 2-HEP was through an ester bond to O-6 of galactose.
FIG 4.
Characterization of 2-HEP linked hexose after partial acid hydrolysis of Glycomyces phosphonoglycans. (A) 31P NMR spectrum of one fraction eluted from Sephadex LH-20, which contained two unknown phosphonates and 2-HEP. (B) High-resolution LC-MS analysis of the sample whose spectrum is shown in panel A. The precursor ion at m/z 287.0538 indicates HEP-linked hexose. This ion was selected for MS2. The peak labels in the spectrum are observed values. (C) Proposed structures based on fragment ions generated by MS2. The numbers are calculated monoisotopic values.
The Stackebrandtia phosphonoglycans were also subject to 6 M HCl hydrolysis at 100°C for 3 h. The hydrolysate was desalted by size exclusion chromatography on Sephadex G-25, treated with Fe3+-charged Chelex 100 resin, and dialyzed against water using a Micro Float-A-Lyzer dialysis membrane (MWCO = 500 to 1,000; Spectra/Por). The filtrate was further fractionated on Sephadex LH-20. One fraction from Sephadex LH-20 exhibited a major peak at 24.5 ppm in 31P NMR (see Fig. S7A in the supplemental material). By using the combination of NMR and LC-MS analyses, this phosphonate was determined to be 2-HEP mono(2,3-dihydroxypropyl)ester (Fig. 5; also, see Fig. S7 in the supplemental material). Assignment of most of the 1H and 13C signals to 2-HEP mono(2,3-dihydroxypropyl)ester is summarized in Table 4. High-resolution LC-MS analysis of the same sample in the negative mode detected the precursor ion at m/z 199.0382 and its fragment ions at m/z 181.0275, 169.0274, and 125.0010, suggesting the presence of 2-HEP mono(2,3-dihydroxypropyl)ester, in good agreement with NMR results (see Fig. S8 in the supplemental material). Interestingly, the same fragment ion was also detected from the above-described partially acid-hydrolyzed Glycomyces phosphonoglycan sample, albeit in lower abundance (see Fig. S9 in the supplemental material). Presumably, 2-HEP in this sample was also attached to glycerol via an ester linkage.
FIG 5.
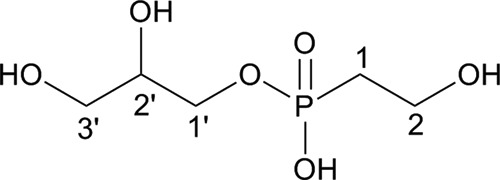
Structure of 2-HEP mono(2,3-dihydroxypropyl)ester.
TABLE 4.
1H (600 MHz) and 13C (150 MHz) spectroscopic data for 2-HEP mono(2,3-dihydroxypropyl)ester in D2O
| No. | δC | δH, multiplets (J in Hz) | 1H-1H COSY | 1H-1H TOCSY | 1H-13C HMBC |
|---|---|---|---|---|---|
| 1 | 29.3 | 1.85, m | H-2 | H-2 | C-2 |
| 2 | 56.8 | 3.67, m | H-1 | H-1 | |
| 1′ | 64.8 | 3.72, m; 3.78, m | C-2′ | ||
| 2′ | 70.7 | 3.74, m | H-3′ | ||
| 3′ | 62.0 | 3.47, dd (11.8, 5.9); 3.53, dd (11.8, 5.9) | H-2′ | H-2′ | C-1′,2′ |
Cloning, sequencing, and bioinformatic analyses of phosphonoglycan biosynthetic gene clusters.
Since genes encoding phosphonate biosynthetic pathways are usually clustered with the pepM gene (3, 6), PCR screening with pepM degenerate primers enabled the identification of fosmid clones carrying the phosphonoglycan biosynthetic gene clusters from two strains. Fosmid libraries of both strains were constructed and screened with pepM primers as described previously (21). Out of 3,072 fosmid clones screened, eight and seven fosmid clones containing overlapping fragments from the same gene locus from Glycomyces and Stackebrandtia strains, respectively, were shown to be pepM positive. The fosmids from Glycomyces were pooled, sequenced, annotated, and compared with the phosphonoglycan biosynthetic locus from Stackebrandtia (genome sequence available under GenBank accession number NC_013947) (Fig. 6 and Table 5; also, see Table S1 in the supplemental material).
FIG 6.
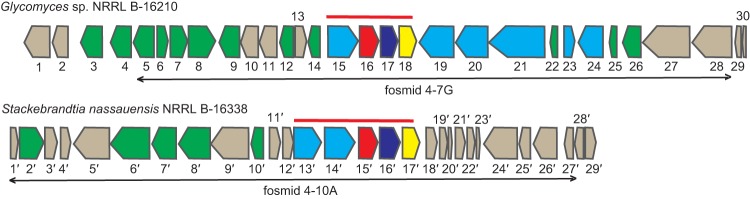
Putative phosphonoglycan biosynthetic loci of Glycomyces sp. strain NRRL B-16210 and S. nassauensis NRRL B-16338. Red lines indicate gene homologs shared by two clusters. ORFs in green encode proteins possibly involved in polysaccharide biosynthesis, whereas ORFs in blue encode proteins possibly involved in teichoic acid biosynthesis. Black arrows indicate the borders of fosmids, which are used for heterologous expression in S. lividans TK24. Annotations of ORFs from Glycomyces and Stackebrandtia putative phosphonoglycan clusters are shown in Table 5 and in Table S1 in the supplemental material.
TABLE 5.
Summary of ORFs in the putative phosphonoglycan biosynthetic locus of Glycomyces
| ORF | No. of amino acids | Homologous protein(s)a | Amino acid identity (%) |
|---|---|---|---|
| 1 | 579 | S. nassauensis hypothetical protein (YP_003514486) | 159/473 (34) |
| 2 | 307 | S. nassauensis proline dehydrogenase (YP_003514483) | 210/305 (69) |
| 3 | 441 | Salinispora hypothetical protein (WP_018799539) | 250/434 (58) |
| 4 | 404 | Actinomadura madurae hypothetical protein (WP_021595829) | 204/389 (52) |
| 5 | 496 | Streptomyces globisporus group 1 glycosyl transferase (ZP_11382042) | 287/495 (58) |
| 6 | 187 | Streptomyces griseus putative acetyltransferase (ZP_08238656) | 134/187 (72) |
| 7 | 391 | Streptomyces fulvissimus UDP-N-acetylglucosamine 2-epimerase (YP_007931007) | 256/361 (71) |
| 8 | 527 | Streptomyces globisporus group 1 glycosyl transferase (ZP_11382042) | 299/522 (57) |
| 9 | 428 | Streptomyces sp. nucleotide sugar dehydrogenase (WP_020127806) | 355/427 (83) |
| Streptomyces sp. UDP-glucose/GDP-mannose family dehydrogenase (AAM94042) | 112/370 (30) | ||
| 10 | 337 | Streptomyces somaliensis dehydrogenase (WP_010470231) | 230/332 (69) |
| Zymomonas mobilis oxidoreductase domain protein (YP_003422636) | 45/198 (23) | ||
| 11 | 368 | Micromonospora aurantiaca glutamine–scyllo-inositol transaminase (YP_003835206) | 263/358 (73) |
| Bacillus circulans l-glutamine:DOI aminotransferase (Q8G8Y2) | 120/369 (33) | ||
| 12 | 263 | Actinoplanes sp. xylose isomerase domain-containing protein (YP_007955649) | 165/257 (64) |
| 13 | 272 | S. nassauensis hypothetical protein Snas_5757 (YP_003514480) | 166/267 (62) |
| 14 | 273 | Actinoplanes sp. hypothetical protein (YP_007955491) | 131/261 (50) |
| 15 | 572 | S. nassauensis hypothetical protein Snas_5667 (YP_003514390) | 240/512 (47) |
| Staphylococcus aureus teichoic acid biosynthesis protein (WP_000714511) | 41/161 (25) | ||
| 16 | 429 | S. nassauensis PEP mutase (YP_003514388) | 354/427 (83) |
| Streptomyces fradiae PEP mutase (ACG70831) | 309/426 (73) | ||
| 17 | 375 | S. nassauensis phosphonopyruvate decarboxylase (YP_003514387) | 277/371 (75) |
| Streptomyces fradiae phosphonopyruvate decarboxylase (ACG70832) | 223/377 (59) | ||
| 18 | 387 | S. nassauensis iron-containing alcohol dehydrogenase (YP_003514386) | 239/377 (63) |
| Streptomyces fradiae group III metal-dependent alcohol dehydrogenase (ACG70833) | 200/375 (53) | ||
| 19 | 648 | S. nassauensis LicD family protein (YP_003509551) | 182/500 (36) |
| 20 | 654 | S. nassauensis LicD family protein (YP_003509551) | 190/496 (38) |
| 21 | 1,113 | Salinispora pacifica hypothetical protein (WP_018254297) | 492/1156 (43) |
| Staphylococcus epidermidis teichoic acid biosynthesis protein F (WP_012102514) | 145/399 (36) | ||
| 22 | 180 | S. nassauensis UDP-N-acetylglucosamine pyrophosphorylase-like protein (YP_003509695) | 135/178 (76) |
| 23 | 245 | Streptomyces sp. putative bifunctional ribulose 5-phosphate reductase/CDP-ribitol pyrophosphorylase (ZP_09401011) | 116/238 (49) |
| 24 | 522 | S. nassauensis hypothetical protein Snas_5667 (YP_003514390) | 216/504 (43) |
| Staphylococcus aureus teichoic acid biosynthesis protein (WP_000714511) | 39/209 (19) | ||
| 25 | 180 | S. nassauensis UDP-N-acetylglucosamine pyrophosphorylase-like protein (YP_003509695) | 135/178 (76) |
| 26 | 389 | S. nassauensis glycosyl transferase group 1 (YP_003509810) | 163/388 (42) |
| 27 | 915 | S. nassauensis DNA topoisomerase I (YP_003509305) | 646/856 (75) |
| 28 | 775 | Verrucosispora maris membrane-bound proton-translocating pyrophosphatase (YP_004402993) | 547/777 (70) |
| 29 | 173 | S. nassauensis hypothetical protein Snas_0500 (YP_003509308) | 74/150 (49) |
| 30 | 120 | S. nassauensis anti-sigma factor antagonist (YP_003509309) | 55/107 (51) |
The closest homologs were identified by BLASTP analysis (performed on 10 November 2013) of deduced amino acids of each ORF; accession numbers are in parentheses. Where possible, a homolog whose biochemical function was experimentally supported is shown as a second entry.
Comparative analyses of two phosphonoglycan gene clusters revealed significant similarities surrounding pepM. Four genes were shared in common between two clusters: open reading frame (ORF) 15 (hypothetical protein), ORF 16 (phosphoenolpyruvate [PEP] mutase), ORF 17 (PnPy decarboxylase), and ORF 18 (iron-dependent alcohol dehydrogenase) (Fig. 6). The latter three genes encode enzymes with high homology to ones involved in the synthesis of 2-HEP, which is a common intermediate in characterized phosphonate biosynthetic pathways, including dehydrophos, fosfomycin, and phosphinothricin (34). Hence, the presence of these three genes in clusters provided molecular evidence for the biosynthesis of HEP-containing phosphonoglycans. ORF 15 shared 47% and 48% sequence identity, respectively, with ORF 13′ (hypothetical protein Snas_5667) and ORF 14′ (hypothetical protein Snas_5666) from S. nassauensis (Fig. 6). Homologous proteins in other bacterial genomes have been annotated as CDP-glycerol:poly(glycerophosphate) glycerophosphotransferase. For example, tagF-encoded glycerophosphotransferase catalyzes extension of the teichoic acid main chain through sequential transfer of glycerol-phosphate units from CDP-glycerol to the linkage unit lipid in both Bacillus subtilis 168 and Staphylococcus epidermidis ATCC 14990 (35, 36). Homologs in phosphonoglycan gene clusters may perform a similar function. In addition, there were some genes whose products are involved in sugar metabolism (e.g., glycosyltransferase) and teichoic acid biosynthesis in two clusters (Table 5; also, see Table S1 in the supplemental material). Whether and how those genes are involved in phosphonoglycan biosynthesis remains to be determined. The gene responsible for O-methylation of galactose was absent in both clusters; it may be present elsewhere in the genomes.
Heterologous production in S. lividans.
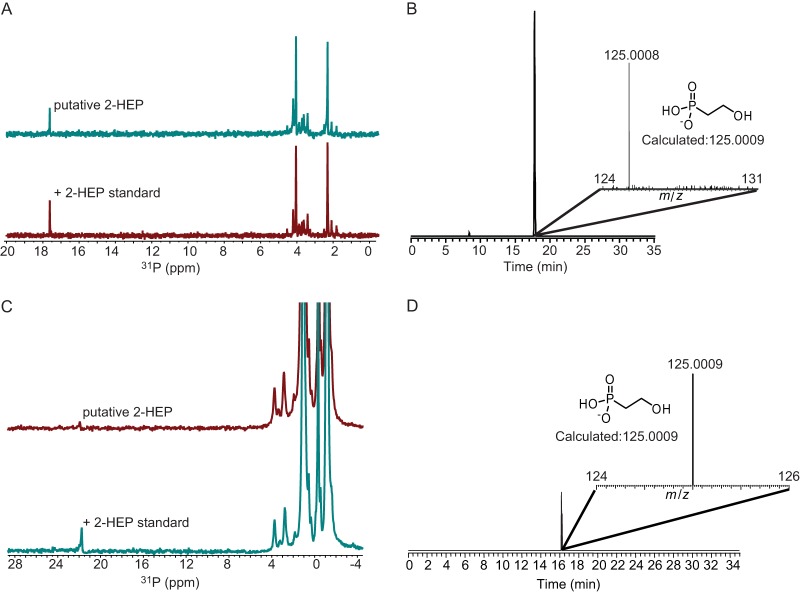
To determine genes necessary for phosphonoglycan production, we transferred pepM-containing fosmids (Fig. 6) to S. lividans TK24, which is not known to produce any phosphonates. The culture extract from one recombinant strain, S. lividans MMG559, which harbored the putative phosphonoglycan gene cluster from Glycomyces, exhibited a peak in the 31P NMR spectrum with a chemical shift of 17.7 ppm (Fig. 7A). Addition of an authentic 2-HEP standard increased the intensity of the peak at 17.7 ppm. No new peaks were observed, indicating that the phosphonate species from this recombinant strain was 2-HEP (Fig. 7A). The production of 2-HEP by S. lividans MMG559 was further confirmed by high-resolution LC-MS (Fig. 7B). Similarly, recombinant strain S. lividans MMG598, which was integrated with the putative phosphonoglycan gene cluster from Stackebrandtia, also produced 2-HEP. This was again confirmed by both spiking the sample with an authentic 2-HEP standard and high-resolution LC-MS analysis (Fig. 7C and D).
FIG 7.
Heterologous expression of putative phosphonoglycan gene clusters from Glycomyces and Stackebrandtia in S. lividans. (A) 31P NMR spectrum demonstrating heterologous expression of 2-HEP when the phosphonoglycan gene cluster is transferred from Glycomyces to S. lividans. (Top) Concentrated culture extract from S. lividans MMG559; (bottom) concentrated culture extract from S. lividans MMG559 spiked with an authentic 2-HEP standard. (B) LC-FTMS analysis showing the presence of 2-HEP from S. lividans MMG559 culture extract. (C) 31P NMR spectrum demonstrating heterologous expression of 2-HEP when the phosphonoglycan gene cluster is transferred from Stackebrandtia to S. lividans. (Top) Concentrated culture extract from S. lividans MMG598; (bottom) concentrated culture extract from S. lividans MMG598 spiked with an authentic 2-HEP standard. (D) LC-FTMS analysis showing the presence of 2-HEP from S. lividans MMG598 culture extract. The peak labels in the spectra are observed values, whereas the numbers in the insets are calculated values.
DISCUSSION
Phosphonate biosynthesis is both widespread and diverse in nature, with more than 5% of sampled microbial cells encoding putative pepM gene (3). Gene neighborhood analyses have shown that most phosphonate-biosynthetic pathways from sequenced microbial genomes are involved in the synthesis of phosphonate-containing structural components with 2-AEP as a common constituent in lipids or glycans (3). 2-HEP may be an alternative head group in some phosphonate-containing macromolecules. For example, Bacteroides eggerthii DSM 20697 has a phosphonate biosynthetic locus nearly identical to that of B. fragilis polysaccharide B complex (15) but has replaced AEP-biosynthetic genes with HEP-biosynthetic genes; thus, it has been predicted to produce a HEP-decorated capsular polysaccharide, though direct experimental evidence is lacking (3). The occurrence of a 2-HEP head group in a macromolecule has been demonstrated only from a novel biosurfactant isolated from the water blooms of Aphanizomenon flos-aquae (37). To the best of our knowledge, the structures described in the current study represent the first example of phosphonoglycans containing 2-HEP.
Currently, it is difficult to determine whether 2-HEP occurs randomly along the polysaccharide chain or whether 2-HEP is attached to certain sugars in some orderly arrangement. Detection of both glycerol-HEP and HEP-hexose by ester linkages from the same sample after partial acid hydrolysis of phosphonoglycans from Glycomyces suggests that these two molecules may be part of a more complex structure. In fact, this is reminiscent of poly (glycerol-phosphate) teichoic acids present in the cell walls of some Gram-positive bacteria (38). Three species of Glycomyces, Glycomyces tenuis, G. rutgersensis, and G. harbinensis, were reported to produce species-specific cell wall teichoic acids with different structures (39). Given that phosphonoglycans from Glycomyces are also cell bound, it is possible that they may be part of novel teichoic acid-like molecules with 2-HEP as the side chain. If this is true, occurrence of a phosphonate head group in teichoic acids would be unprecedented. No experiments have been conducted to directly test the physiological functions of phosphonate-containing macromolecules in any organism. It has been speculated that a C-P bond, in place of a C-O-P bond, in macromolecules enhances the stability to hydrolysis by hydrolases, such as phosphatases, phospholipases, and phosphodiesterases (40). Presumably, the occurrence of 2-HEP in the cell membrane may have a similar function.
The presence of various partially O-methylated galactosyl residues within a single polysaccharide structure is also highly unusual. Candidates for O-methylation of galactose are not found in both phosphonoglycan gene clusters. This may explain why heterologous expression of putative phosphonoglycan gene clusters in S. lividans produced only free 2-HEP. Furthermore, for the biosynthesis of a polysaccharide of such complexity, the machinery involved in polysaccharide assembly, export, and regulation may be specific only to the native producer. Nevertheless, given that there is only one pepM homolog and hence only one phosphonate biosynthetic locus in each genome of Glycomyces and Stackebrandtia, demonstration of 2-HEP expression in S. lividans still provides evidence linking the gene clusters with the biosynthesis of phosphonoglycans.
Supplementary Material
ACKNOWLEDGMENTS
We thank Jiangtao Gao (University of Illinois) for aid in the analysis of NMR data, Lingyang Zhu and Xudong Guan (University of Illinois) for assistance with advanced NMR experiments and Karl Vermillion for additional NMR data obtained at NCAUR, Peoria. We also thank Jaeheon Lee (University of Illinois) for assistance in LC-MS analysis. We are grateful to all colleagues in the Metcalf lab and the W. A. van der Donk lab for useful discussions.
This work was supported by the National Institutes of Health (PO1 GM077596). NMR spectra were recorded on a 600-MHz instrument purchased with support from NIH grant S10 RR028833.
Footnotes
Published ahead of print 28 February 2014
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JB.00036-14.
REFERENCES
- 1.Horiguchi M. 1984. Occurrence, identification and properties of phosphonic and phosphinic acids, p 24–52 In Hori T, Horiguchi M, Hayashi A. (ed), Biochemistry of natural C-P compounds. Japanese Association for Research on the Biochemistry of C-P Compounds, Shiga, Japan [Google Scholar]
- 2.Metcalf WW, Griffin BM, Cicchillo RM, Gao J, Janga SC, Cooke HA, Circello BT, Evans BS, Martens-Habbena W, Stahl DA, van der Donk WA. 2012. Synthesis of methylphosphonic acid by marine microbes: a source for methane in the aerobic ocean. Science 337:1104–1107. 10.1126/science.1219875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yu X, Doroghazi JR, Janga SC, Zhang JK, Circello B, Griffin BM, Labeda DP, Metcalf WW. 2013. Diversity and abundance of phosphonate biosynthetic genes in nature. Proc. Natl. Acad. Sci. U. S. A. 110:20759–20764. 10.1073/pnas.1315107110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Martinez A, Tyson GW, DeLong EF. 2010. Widespread known and novel phosphonate utilization pathways in marine bacteria revealed by functional screening and metagenomic analyses. Environ. Microbiol. 12:222–238. 10.1111/j.1462-2920.2009.02062.x [DOI] [PubMed] [Google Scholar]
- 5.McGrath JW, Chin JP, Quinn JP. 2013. Organophosphonates revealed: new insights into the microbial metabolism of ancient molecules. Nat. Rev. Microbiol. 11:412–419. 10.1038/nrmicro3011 [DOI] [PubMed] [Google Scholar]
- 6.Metcalf WW, van der Donk WA. 2009. Biosynthesis of phosphonic and phosphinic acid natural products. Annu. Rev. Biochem. 78:65–94. 10.1146/annurev.biochem.78.091707.100215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hilderbrand RL, Henderson TO. 1983. Phosphonic acids in nature, p 5–30 In Hilderbrand RL. (ed), The role of phosphonates in living systems. CRC Press, Boca Raton, FL [Google Scholar]
- 8.Korn ED, Dearborn DG, Fales HM, Sokoloski EA. 1973. Phosphonoglycan. A major polysaccharide constituent of the amoeba plasma membrane contains 2-aminoethylphosphonic acid and 1-hydroxy-2-aminoethylphosphonic acid. J. Biol. Chem. 248:2257–2259 [PubMed] [Google Scholar]
- 9.Korn ED, Dearborn DG, Wright PL. 1974. Lipophosphonoglycan of the plasma membrane of Acanthamoeba castellanii. Isolation from whole amoebae and identification of the water-soluble products of acid hydrolysis. J. Biol. Chem. 249:3335–3341 [PubMed] [Google Scholar]
- 10.Dearborn DG, Smith S, Korn ED. 1976. Lipophosphonoglycan of the plasma membrane of Acanthamoeba castellanii. Inositol and phytosphingosine content and general structural features. J. Biol. Chem. 251:2976–2982 [PubMed] [Google Scholar]
- 11.Fontana JD, Duarte JH, Gallo CBH, Iacomini M, Gorin PAJ. 1985. Occurrence of b-d-galactopyranosyl units esterified at O-6 with 2-aminoethylphosphonic acid in the D-galactan of albumin glands of the snail Megalobulimus paranaguensis. Carbohydr. Res. 143:175–183. 10.1016/S0008-6215(00)90706-7 [DOI] [Google Scholar]
- 12.Previato JO, Gorin PAJ, Mazurek M, Xavier MT, Fournet B, Wieruszesk JM, Mendoncapreviato L. 1990. Primary structure of the oligosaccharide chain of lipopeptidophosphoglycan of epimastigote forms of Trypanosoma cruzi. J. Biol. Chem. 265:2518–2526 [PubMed] [Google Scholar]
- 13.Urai M, Nakamura T, Uzawa J, Baba T, Taniguchi K, Seki H, Ushida K. 2009. Structural analysis of O-glycans of mucin from jellyfish (Aurelia aurita) containing 2-aminoethylphosphonate. Carbohydr. Res. 344:2182–2187. 10.1016/j.carres.2009.08.001 [DOI] [PubMed] [Google Scholar]
- 14.Onderdonk AB, Kasper DL, Cisneros RL, Bartlett JG. 1977. The capsular polysaccharide of Bacteroides fragilis as a virulence factor: comparison of the pathogenic potential of encapsulated and unencapsulated strains. J. Infect. Dis. 136:82–89. 10.1093/infdis/136.1.82 [DOI] [PubMed] [Google Scholar]
- 15.Baumann H, Tzianabos AO, Brisson JR, Kasper DL, Jennings HJ. 1992. Structural elucidation of two capsular polysaccharides from one strain of Bacteroides fragilis using high-resolution NMR spectroscopy. Biochemistry 31:4081–4089. 10.1021/bi00131a026 [DOI] [PubMed] [Google Scholar]
- 16.Coyne MJ, Kalka-Moll W, Tzianabos AO, Kasper DL, Comstock LE. 2000. Bacteroides fragilis NCTC9343 produces at least three distinct capsular polysaccharides: cloning, characterization, and reassignment of polysaccharide B and C biosynthesis loci. Infect. Immun. 68:6176–6181. 10.1128/IAI.68.11.6176-6181.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Vinogradov E, Egbosimba EE, Perry MB, Lam JS, Forsberg CW. 2001. Structural analysis of the carbohydrate components of the outer membrane of the lipopolysaccharide-lacking cellulolytic ruminal bacterium Fibrobacter succinogenes S85. Eur. J. Biochem. 268:3566–3576. 10.1046/j.1432-1327.2001.02264.x [DOI] [PubMed] [Google Scholar]
- 18.Miceli MV, Henderson TO, Myers TC. 1980. 2-Aminoethylphosphonic acid metabolism during embryonic development of the planorbid snail Helisoma. Science 209:1245–1247. 10.1126/science.209.4462.1245 [DOI] [PubMed] [Google Scholar]
- 19.Cote R, Daggett PM, Gantt MJ, Hay R, Hay SC, Pienta P. 1984. ATCC media handbook. ATCC, Rockville, Md [Google Scholar]
- 20.Sambrook J, Fritsch EF, Maniatis T. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Woodbury, NY [Google Scholar]
- 21.Eliot AC, Griffin BM, Thomas PM, Johannes TW, Kelleher NL, Zhao HM, Metcalf WW. 2008. Cloning, expression, and biochemical characterization of Streptomyces rubellomurinus genes required for biosynthesis of antimalarial compound FR900098. Chem. Biol. 15:765–770. 10.1016/j.chembiol.2008.07.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Margulies M, Egholm M, Altman WE, Attiya S, Bader JS, Bemben LA, Berka J, Braverman MS, Chen YJ, Chen Z, Dewell SB, Du L, Fierro JM, Gomes XV, Godwin BC, He W, Helgesen S, Ho CH, Irzyk GP, Jando SC, Alenquer ML, Jarvie TP, Jirage KB, Kim JB, Knight JR, Lanza JR, Leamon JH, Lefkowitz SM, Lei M, Li J, Lohman KL, Lu H, Makhijani VB, McDade KE, McKenna MP, Myers EW, Nickerson E, Nobile JR, Plant R, Puc BP, Ronan MT, Roth GT, Sarkis GJ, Simons JF, Simpson JW, Srinivasan M, Tartaro KR, Tomasz A, Vogt KA, Volkmer GA, Wang SH, Wang Y, Weiner MP, Yu P, Begley RF, Rothberg JM. 2005. Genome sequencing in microfabricated high-density picolitre reactors. Nature 437:376–380. 10.1038/nature03959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Aziz RK, Bartels D, Best AA, DeJongh M, Disz T, Edwards RA, Formsma K, Gerdes S, Glass EM, Kubal M, Meyer F, Olsen GJ, Olson R, Osterman AL, Overbeek RA, McNeil LK, Paarmann D, Paczian T, Parrello B, Pusch GD, Reich C, Stevens R, Vassieva O, Vonstein V, Wilke A, Zagnitko O. 2008. The RAST server: rapid annotations using subsystems technology. BMC Genomics 9:75. 10.1186/1471-2164-9-75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403–410. 10.1016/S0022-2836(05)80360-2 [DOI] [PubMed] [Google Scholar]
- 25.Jones TM, Albershem P. 1972. A gas chromatographic method for the determination of aldose and uronic acid constituents of plant cell wall polysaccharides. Plant Physiol. 49:926–936. 10.1104/pp.49.6.926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Price NPJ. 2004. Acylic sugar derivatives for GC/MS analysis of C-13-enrichment during carbohydrate metabolism. Anal. Chem. 76:6566–6574. 10.1021/ac049198m [DOI] [PubMed] [Google Scholar]
- 27.Ciucanu I, Kerek F. 1984. A simple and rapid method for the permethylation of carbohydrates. Carbohydr. Res. 131:209–217. 10.1016/0008-6215(84)85242-8 [DOI] [Google Scholar]
- 28.Evans BS, Zhao C, Gao J, Evans CM, Ju KS, Doroghazi JR, van der Donk WA, Kelleher NL, Metcalf WW. 2013. Discovery of the antibiotic phosacetamycin via a new mass spectrometry-based method for phosphonic acid detection. ACS Chem. Biol. 8:908–913. 10.1021/cb400102t [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Labeda DP, Kroppenstedt RM. 2005. Stackebrandtia nassauensis gen. nov., sp. nov. and emended description of the family Glycomycetaceae. Int. J. Syst. Evol. Microbiol. 55:1687–1691. 10.1099/ijs.0.63496-0 [DOI] [PubMed] [Google Scholar]
- 30.Labeda DP, Testa RT, Lechevalier MP, Lechevalier HA. 1985. Glycomyces, a new genus of the Actinomycetales. Int. J. Syst. Bacteriol. 35:417–421. 10.1099/00207713-35-4-417 [DOI] [Google Scholar]
- 31.Morris KF, Johnson CS. 1992. Diffusion-ordered two-dimensional nuclear magnetic resonance spectroscopy J. Am. Chem. Soc. 114:3139–3141. 10.1021/ja00034a071 [DOI] [Google Scholar]
- 32.Vermillion K, Price NPJ. 2009. Stable isotope-enhanced two- and three-dimensional diffusion ordered 13C NMR spectroscopy (SIE-DOSY 13C NMR). J. Magn. Reson. 198:209–214. 10.1016/j.jmr.2009.02.008 [DOI] [PubMed] [Google Scholar]
- 33.Chapman MF. 1987. Monosaccharides, p 1–37 In Chapman MF, Kennedy JF. (ed), Carbohydrate analysis: a practical approach. IRL Press, Oxford, United Kingdom [Google Scholar]
- 34.Shao ZY, Blodgett JAV, Circello BT, Eliot AC, Woodyer R, Li GY, van der Donk WA, Metcalf WW, Zhao HM. 2008. Biosynthesis of 2-hydroxyethylphosphonate, an unexpected intermediate common to multiple phosphonate biosynthetic pathways. J. Biol. Chem. 283:23161–23168. 10.1074/jbc.M801788200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pooley HM, Abellan FX, Karamata D. 1992. CDP-glycerol:poly(glycerophosphate) glycerophosphotransferase, which is involved in the synthesis of the major wall teichoic-acid in Bacillus subtilis 168, is encoded by tagF (rodC). J. Bacteriol. 174:646–649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fitzgerald SN, Foster TJ. 2000. Molecular analysis of the tagF gene, encoding CDP-glycerol:poly(glycerophosphate) glycerophosphotransferase of Staphylococcus epidermidis ATCC 14990. J. Bacteriol. 182:1046–1052. 10.1128/JB.182.4.1046-1052.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kaya K, Morrison LF, Codd GA, Metcalf JS, Sano T, Takagi H, Kubo T. 2006. A novel biosurfactant, 2-acyloxyethylphosphonate, isolated from waterblooms of Aphanizomenon flos-aquae. Molecules 11:539–548. 10.3390/11070539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Burger MM, Glaser L. 1964. The synthesis of teichoic acids. I. Polyglycerophosphate. J. Biol. Chem. 239:3168–3177 [PubMed] [Google Scholar]
- 39.Potekhina NyV, Streshinskaya GM, Tul'skaya EM, Shashkov AS. 2011. Cell wall teichoic acids in the taxonomy and characterization of Gram-positive bacteria. Methods Microbiol. 38:131–164. 10.1016/B978-0-12-387730-7.00006-1 [DOI] [Google Scholar]
- 40.Horiguchi M. 1984. Some physiological aspects of phosphonic and phosphinic acids, p 104–115 In Hori T, Horiguchi M, Hayashi A. (ed), Biochemistry of natural C-P compounds. Japanese Association for Research on the Biochemistry of C-P Compounds, Shiga, Japan [Google Scholar]
- 41.Circello BT, Eliot AC, Lee JH, van der Donk WA, Metcalf WW. 2010. Molecular cloning and heterologous expression of the dehydrophos biosynthetic gene cluster. Chem. Biol. 17:402–411. 10.1016/j.chembiol.2010.03.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kieser T, Bibb MJ, Buttner MJ, Chater KF, Hopwood DA. 2000. Practical Streptomyces genetics. The John Innes Foundation, Norwich, United Kingdom [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.