Abstract
Infection of intestinal epithelial cells is dependent on the Salmonella enterica serovar Typhimurium pathogenicity island 1 (Spi1)-encoded type III injectisome system and flagellar motility. Thus, the expression of virulence and flagellar genes is subject to tight regulatory control mechanisms in order to ensure the correct spatiotemporal production of the respective gene products. In this work, we reveal a new level of cross-regulation between the Spi1 and flagellar regulatory systems. Transposon mutagenesis identified a class of mutants that prevented flhDC autorepression by overexpressing HilD. HilD, HilC, RtsA, and HilA comprise a positive regulatory circuit for the expression of the Spi1 genes. Here, we report a novel transcriptional cross talk between the Spi1 and flagellar regulons where HilD transcriptionally activates flhDC gene expression by binding to nucleotides −68 to −24 upstream from the P5 transcriptional start site. We additionally show that, in contrast to the results of a previous report, HilA does not affect flagellar gene expression. Finally, we discuss a model of the cross-regulation network between Spi1 and the flagellar system and propose a regulatory mechanism via the Spi1 master regulator HilD that would prime flagellar genes for rapid reactivation during host infection.
INTRODUCTION
The enteropathogenic Gram-negative bacteria of the genus Salmonella are responsible for the food-borne illness gastroenteritis, localized infection of the small intestine, and systemic enteric (typhoid) fevers. Symptoms of Salmonella infection include diarrhea, abdominal cramps, and fever (1). The effector-driven manipulations of the vertebrate host cells are dependent on two virulence-associated type III secretion systems (vT3SS; injectisome) (2–4) encoded in Salmonella enterica serovar Typhimurium pathogenicity island 1 (Spi1) (5) and Spi2 (6).
The Spi1 and Spi2 virulence systems are responsible for different processes related to Salmonella pathogenesis that occur at different time points during infection. Spi1 is needed for the invasion of the intestinal epithelium leading to gastroenteritis (7, 8), while Spi2 plays a role during trafficking to the basolateral side of epithelial cells (9) and during later Salmonella replication and survival within macrophages (10, 11). Both systems are regulated in a spatial and temporal manner to ensure the production of gene products at the correct points during infection.
The Spi1 genes are highly regulated by a set of DNA-binding proteins, including the AraC-like regulators HilD, HilC, and RtsA. In a feed-forward loop, each of those regulators can activate the hilD, hilC, and rtsA genes, as well as the gene encoding the transcriptional Spi1 activator HilA (12). HilD is a dominant regulator of hilA transcription, while HilC and RtsA amplify hilA gene expression (12, 13).
The needle-like injectisome system is evolutionarily related to the bacterial flagellum (14). Bioinformatic and structural analysis demonstrated that the two share many similar features (15); however, differences exist regarding the purpose of protein secretion in the two systems. In case of the flagellum, secreted substrate proteins are mainly needed for flagellum assembly. The flagellar type III secretion system (fT3SS) exports substrate subunits that assemble into a functional flagellum and regulatory factors that control the assembly process. Completed flagella are used by the bacterium to move in liquid environments and across hydrated surfaces by rotation of the rigid, helical flagellar filaments. The virulence-associated vT3SS of Spi1 is essential for both the assembly of the injectisome needle-like structure (16, 17) and the secretion of effector proteins into host cells, where they can alter cellular processes to facilitate the infection process as described above.
Flagellar gene expression is under spatiotemporal control by a transcriptional hierarchy of three promoter classes. On top of the cascade is the flagellar master operon, flhDC, which is under the control of a σ70-dependent flagellar class 1 promoter. A functional FlhD4C2 complex is required for subsequent flagellar class 2 promoter transcription. Flagellar class 2 gene products are required for the structure and assembly of a flagellar hook-basal body (HBB). One class 2 gene product, σ28 (encoded by the fliA gene), is a sigma transcription factor that directs RNA polymerase to transcribe the flagellar class 3 promoters. The products of flagellar class 3 transcription are needed after HBB completion (18–20) to form the filament, motor force generators, and chemosensory components.
The flagellar master operon is under the control of a variety of different factors that either positively or negatively influence flhDC expression. Both global positive regulators, such as the nucleoid proteins Fis and H-NS or the cyclic AMP (cAMP)-catabolite activator protein (CAP) complex (21–23), and negative regulators, such as RflM, the Spi1 regulator RtsB, or SlyA, bind within and act upon the flhDC promoter (24, 25). The regulator RtsB is encoded in an operon with rtsA and functions as a repressor of flagellar class 1 gene expression. The rtsAB operon is transcriptionally activated by HilD, HilC, and RtsA. In addition, a regulatory feedback loop acting on flhDC transcription via RflM has been reported (26, 27). The inhibitory effect of RflM on flhDC expression has been shown to be dependent on the RcsCDB system (28), a positive regulator of Spi2 and other genes associated with bacterial cell growth in macrophages (29).
Significant amounts of biosynthetic resources and energy are required in order to synthesize, assemble, and rotate a functional flagellum (30, 31). For that reason, the expression of the flagellar regulon is controlled in response to various factors and according to the bacterium's motility needs. Under low-nutrient conditions, flagellar gene expression is either induced (Escherichia coli) or repressed (Salmonella) (32–34). Importantly, flagellar gene expression and assembly are repressed during early biofilm formation, during survival inside macrophages, during early infection (2 h postinfection [p.i.]) of epithelial cells, and within the mesenteric lymph nodes and the spleen (34–39). However, the expression of flagellar biosynthesis genes was upregulated during late infection of epithelial cells (4 h and 6 h p.i.) and fliC::gfp transcription was reported in the Peyer's patches (7 days p.i.), providing evidence for de novo synthesis of flagellin during the infection process (38, 39). Regarding survival within host cells, undermining the host cell's defense system and preventing the immune system from recognizing the surface-exposed filament appears to be associated with pathogenesis (40).
Salmonella bacteria can grow and survive in multiple different niches, and this requires a precise coordination of gene expression with environmental sensing. Therefore, Spi1, Spi2, and the flagellar regulon share common regulatory components that coregulate the many genes of each system. Cross talk between the flagellar and Spi1 regulon exists through FlhDC-dependent fliZ gene transcription. FliZ is expressed from flagellar class 2 and 3 promoters and functions as a regulator of HilD protein activity. Elevated levels of HilD protein activate hilA gene expression (41–44). In addition, FliZ positively regulates flagellar class 2 gene transcription via the repression of YdiV, an anti-FlhD4C2 factor (34, 45–47). YdiV binds to FlhD and prevents the FlhD4C2 complex from binding to flagellar class 2 promoters and targets FlhDC for ClpXP-dependent proteolytic degradation (34, 48). Stewart et al. reported that ΔydiV mutant strains were unable to fully repress flagellin production and thus caused increased caspase-1-dependent pyroptosis as a defense mechanism of Salmonella-infected macrophages (49). At the same time, an increased rate of macrophage killing was reported for ydiV-deficient Salmonella (50).
Controversial results have been reported regarding the regulation of the flagellar system via the Spi1 master regulator HilA. Thijs et al. showed direct binding of HilA to the flhDC promoter region and downregulation of flhDC expression under invasive conditions (51). However, earlier studies showed that HilA did not affect flhD-lux transcriptional fusions when bacteria were grown in motility agar (52). An interconnecting cross talk can also be found between the Spi1 and Spi2 regulons at the level of HilD (53). During stationary growth in lysogeny broth (LB) in vitro, the onset of HilD-dependent Spi2 gene activation occured at a later time point than the HilD-dependent activation of the Spi1 system. In experiments under conditions resembling the intracellular environment, HilD was not required for activation of the Spi2 regulon. These data of Bustamante et al. (53) suggest that the activation of the Spi2 system is purposely regulated via two distinct pathways, which come into play depending on the environmental factors within a given niche. Cross talk between the Spi1, Spi2, and flagellar regulatory systems is therefore likely of great importance to Salmonella for spatiotemporal coordination of motility and virulence.
MATERIALS AND METHODS
Bacterial strains, plasmids, and media.
Detailed information about bacterial strains and plasmids used in this study is listed in Table S1 in the supplemental material. Salmonella Typhimurium strain LT2 or ATCC 14028 was cultured in lysogeny broth (LB) (54) that was supplemented with kanamycin (50 μg ml−1), tetracycline (Tc; 15 μg ml−1), or anhydrotetracycline (1 μg ml−1) if needed. Gene expression from the arabinose promoter was induced by the addition of 0.2% l-arabinose. The Salmonella Typhimurium generalized transducing phage P22 HT105/1 int-201 was used in all crosses (55). Cultures of the virulent Salmonella Typhimurium strain ATCC 14028 were grown under Spi1-inducing conditions (high osmolarity and low oxygen) as described previously (26).
Isolation of random T-POP insertions.
Transposon T-POP insertions in strain TH15941 [ΔaraBAD1007::flhD+C+ flhC5213::MudJ fliA5886(R91C L207P) (changes of R to C at position 91 and L to P at position 207 are encoded by fliA5886)] were isolated as described previously (26). Briefly, TH15941 expresses the flhD+C+ operon from the chromosomal araBAD promoter (ParaBAD) and carries a chromosomal flhC-lac transcriptional reporter fusion (flhC5213::MudJ), as well as a fliA null allele that is defective in DNA binding (56). TH15941 becomes Lac deficient in the presence of arabinose (Ara+ Lac−) due to induction of flhD+C+ transcription from ParaBAD and autorepression of flhC-lac reporter transcription by FlhD4C2. T-POP insertions were introduced into TH15941 carrying plasmid pNK2881, and approximately 30,000 random T-POP insertions were screened for the loss of FlhD4C2-mediated repression of flhDC in the presence of tetracycline.
RNA isolation and quantitative real-time PCR.
RNA isolation was performed for three independent biological replicates using the RNeasy minikit (Qiagen). For removal of genomic DNA, RNA was treated with DNase I for 30 min at 37°C using the DNA-Free RNA Kit (Zymo Research). Subsequently, RNA samples were reverse transcribed according to the RevertAid first strand cDNA synthesis kit (Fermentas). Quantitative real-time PCRs were performed using the EvaGreen quantitative PCR (qPCR) master mix (Bio-Rad) on a CFX96 real-time PCR instrument (Bio-Rad). Relative changes in mRNA levels were analyzed according to the method of Pfaffl (57) and normalized against the transcription levels of reference genes rpoB, rpoD, gyrB, and gmk.
β-Galactosidase assays.
β-Galactosidase activity was measured as described previously using at least three independent biological replicates (26). Cultures were supplemented with 0.2% l-arabinose and 1 μg ml−1 anhydrotetracycline if needed. ST14028 experiments were performed under Spi1-inducing (high-salt and low-oxygen) and regular LB conditions.
Luminescence assay.
Luminescence measurements were made using a PerkinElmer 2030 microplate reader. Overnight cultures were diluted 1:100 in LB supplemented with 0.2% arabinose and grown in a microtiter plate for 3 h at 37°C. An amount of 25 μg ml−1 kanamycin was added to all cultures in order to retain the flhDC promoter duplication. Luminescence was measured for 3 s, and absorbance at 595 nm was measured before and after the luminescence readout for 0.1 s each time. The luminescence was normalized to the average optical density. Within one experiment, all samples were grown on the same plate. For each strain, at least four biological replicates were measured per plate. All samples were normalized against the wild-type control.
Purification of HilD protein.
For HilD protein purification, HilD was fused to an Ulp1-cleavable His6-SUMO tag (58). Protein expression was induced for 6 h by the addition of 0.2 mM isopropyl-β-d-thiogalactopyranoside (IPTG) during growth at 18°C with shaking at 120 rpm. Soluble protein was purified using Ni-nitrilotriacetic acid (NTA) agarose (Protino; Macherey-Nagel) under native conditions. Purified His6-SUMO-HilD protein was dialyzed overnight and afterwards incubated with recombinantly produced His6-Ulp1 protease overnight at 4°C. His6-Ulp1 protease, cleaved His6-SUMO, and uncut His6-SUMO-HilD were removed by binding to Ni-NTA agarose.
EMSA.
DNA fragments were amplified from Salmonella Typhimurium LT2 genomic DNA by PCR using a 20-bp overhang in the 5′ region that allowed 5′ biotinylation in a subsequent PCR using a biotinylated primer (5′-biotin-GATCATGCTGACACGTACGG-3′). The amplified PCR products were designated fragment 1 (5′-GATCATGCTGACACGTACGGTCACATATTTTCTAAAATCGCC-3′ and 5′-GAAGCAAAAAGGTCAAATGC-3′), fragment 2 (5′-GATCATGCTGACACGTACGGCGTTATTTTAACAGAGAGAAAC-3′ and 5′-CATACAACGGAGCGGGAC-3′), fragment 3 (5′-GATCATGCTGACACGTACGGGCTAAAAGTTAAATCAAATGAGC-3′ and 5′-GTCAACACCAAATTCTTTTTTG-3′), fragment 3′ (5′-GATCATGCTGACACGTACGGATTCTTATGTAAAGAATCGTGGC-3′ and 5′-ATTTTAGAAACGCTTTTATTTTACC-3′), flhDC coding (5′-GATCATGCTGACACGTACGGGGAGTTGATTAATCTTGGCG-3′ and 5′-GACACTGCTCAAGATAAAGC-3′), and gyrA (5′-GATCATGCTGACACGTACGGATGAGCGACCTTGCGAGAG-3′ and 5′-GCGCACGGCCAACAATGACC-3′). Gel shift assays were carried out using the LightShift chemiluminescent electrophoretic mobility shift assay (EMSA) kit (Thermo Scientific) according to the manufacturer's protocol. Increasing amounts of purified HilD were incubated with 0.01 pmol of each of the biotinylated DNA fragments and 50 ng μl−1 unspecific competitor DNA [poly(dI-dC)] (Sigma) for 20 min at room temperature (RT). A 250-fold molar excess of the unlabeled DNA fragment was added to the highest protein concentration to demonstrate specific binding. Biotin-labeled DNA was detected after polyacrylamide gel electrophoresis, transfer to a nylon membrane, and UV cross-linking using chemiluminescence detection.
DNase I footprint.
The flhDC DNA fragment comprising the putative HilD binding site was PCR amplified from Salmonella Typhimurium LT2 genomic DNA using 5′ digoxigenin (DIG)-labeled primers (5′-GATCATGCTGACACGTACGGATTCTTATGTAAAGAATCGTGGC-3′ and 5′-DIG-GTCAACACCAAATTCTTTTTTG-3′). DNase I (AppliChem) was added to the binding reaction mixtures containing 100 ng DIG-labeled DNA fragments and increasing amounts of HilD after 20 min at RT. The reaction was stopped by adding stop solution (10 mM EDTA, 10 μg ml−1 yeast tRNA), and DNA was recovered using phenol-chloroform extraction. Sequencing reactions were carried out using the USB Thermo Sequenase cycle sequencing kit (Affimetrix). Samples were separated on 6% Tris-borate-EDTA (TBE)–7 M urea polyacrylamide gels. DNA was transferred to a Nytran nylon membrane (GE Healthcare) and, after UV cross-linking, was detected using CDP-Star (Roche) and the anti-digoxigenin-AP (alkaline phosphatase) antibody (Fab fragments; Roche).
RESULTS
HilD links Spi1 and flagellar gene regulation.
We have recently identified RflM (formerly EcnR) as a negative regulator of flhDC transcription that is activated by FlhDC in an FlhDC-RflM feedback loop. Transposon T-POP insertions in the rflM gene resulted in loss of FlhDC autorepression (26). In this screen, a class of T-POP insertions were also isolated that lost FlhDC autorepression but only in the presence of tetracycline (Tc). The T-POP transposon will transcribe genes adjacent to the site of insertion by induction of a Tc-inducible promoter, PtetA, within the T-POP element. We hypothesized that transcription from the T-POP-encoded PtetA into genes adjacent to the site of T-POP insertion resulted in Tc-dependent expression of an flhDC activator. DNA sequence analysis revealed that this class of transposons had inserted upstream from the hilD coding region. The hilD-linked insertions required induction of PtetA within the T-POP element for loss of FlhDC autorepression. We envisioned at least two mechanisms by which activation of HilD could bypass the autoregulatory effect of FlhDC overexpression. HilD could act to either repress rflM, the inhibitor of flhDC transcription, or activate flhDC expression and overcome the inhibition of flhDC via RflM. HilD is a known activator of hilA transcription; HilA then activates the transcription of genes encoding the Spi1 type III secretion apparatus and Spi1 effector proteins through InvF (24, 59, 60). HilD could also act directly on the rflM promoter region to repress rflM transcription or activate transcription of another protein that represses rflM. We first analyzed the potential inhibition of rflM gene transcription by HilD using quantitative real-time PCR. As shown in Fig. S1A in the supplemental material, HilD induction, surprisingly, resulted in about a 50% increase in rflM transcript levels. This suggested a direct activation of the rflM activator FlhDC by HilD. This was confirmed by analyzing the β-galactosidase activity of transcriptional rflM fusions, which showed an increase in rflM-lac transcription upon HilD overexpression (see Fig. S1B, lanes 2 and 3). Upon deletion of flhDC, the activating effect of HilD was neutralized and rflM transcription was abolished (see Fig. S1B, lane 4).
We next tested the possibility that HilD might act directly on flhDC transcription. As shown by the results in Fig. 1A, HilD overexpression had a strong positive effect on flhDC mRNA levels as analyzed by quantitative real-time PCR. This activation of flhDC expression was confirmed using an flhC-lac reporter fusion. As shown by the results in Fig. 1B, flhC-lac expression increased approximately 3-fold upon HilD overexpression in an attenuated Salmonella LT2 background under normal LB growth conditions (Fig. 1B, bar 2). In results comparable to those for the LT2 strain, flhC transcription upon HilD overexpression also increased in the virulent Salmonella background ST14028s grown under Spi1-inducing conditions (Fig. 1B, bars 5 and 6). The induction of Spi1 gene transcription by HilD additionally resulted in a growth defect, as described previously (61). However, deletion of the Spi1 region of the chromosome [Δ(invH-sprB)] (Fig. 1B, Spi1−) relieved the growth-inhibitory effect of HilD overproduction (not shown). The retarded growth under HilD-overproducing conditions had an effect on our growth-based β-galactosidase assays (Fig. 1B, compare bar 5, Spi1+, to bar 6, Spi1−), and the HilD-activating effect on flhDC operon transcription was therefore more pronounced in strains harboring the invH-sprB deletion (Spi1−).
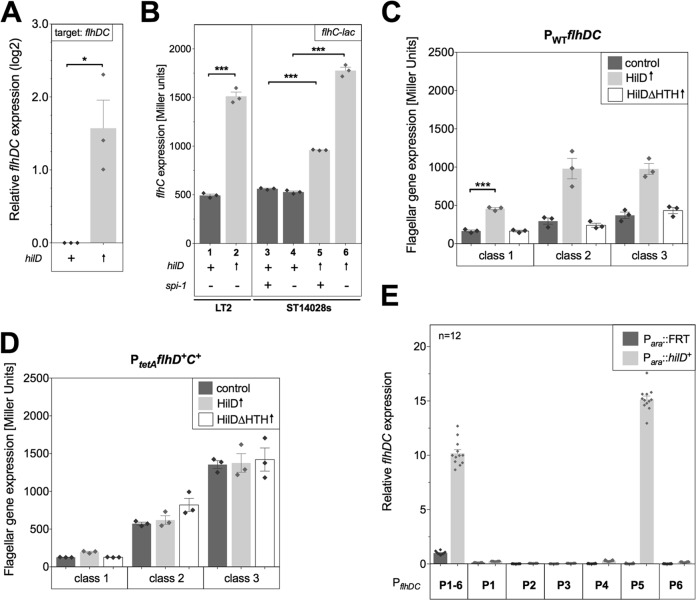
FIG 1.
HilD activates flhDC transcription via P5 of flhDC. (A) Relative flhDC expression (log2) compared to wild-type mRNA levels of strain TH6701 (Para::tetRA). Transcript levels were compared by quantitative real-time PCR. The effect of arabinose-induced overexpression of HilD (TH16339) on flhDC expression was monitored using total mRNA of three independent biological samples grown in arabinose-containing medium. (B) The level of transcription of an flhC-lac fusion was quantified under HilD-overproducing conditions. The activity from the lac reporter was measured at mid-log phase. For Salmonella LT2, transcription levels of EM4 (Para::hilD+ flhC::MudJ ΔinvH-sprB::FCF [FLP recombination target {FRT}-chloramphenicol acetyltransferase-FRT]) were compared to those of the control strain EM97 (Para::tetRA flhC::MudJ ΔinvH-sprB::FCF). Both strains had Salmonella pathogenicity island 1 (Spi1) deleted to ensure comparable growth of the cultures, as described in the text. The pathogenic strain ATCC 14028s was grown under Spi1-inducing conditions as further described in Materials and Methods. The ATCC 14028s mutants analyzed were EM665 (Para::FRT flhC::MudJ), EM674 (Para::FRT ΔinvH-sprB::FCF flhC::MudJ), EM640 (Para::hilD+ flhC::MudJ), and EM667 (Para::hilD+ ΔinvH-sprB::FCF flhC::MudJ). (C) Effects of overproduced HilD and of HilD with its DNA-binding domain deleted (hilDΔHTH) on flagellar genes expressed from class 1 (flhC-lac), class 2 (fliL-lac), and class 3 (fljB-lac) promoters. Strains TH13751 (Para::FCF flhC::MudJ), TH13752 (Para::FCF fliL::MudJ), TH14571 (Para::FCF fljB::MudJ), TH16386 (Para::hilD+ flhC::MudJ), TH16385 (Para::hilD+ fliL::MudJ), TH16423 (Para::hilD+ fljB::MudJ), EM886 (Para::hilDΔHTH+ flhC::MudJ), EM887 (Para::hilDΔHTH+ fliL::MudJ), and EM885 (Para::hilDΔHTH+ fljB::MudJ) were analyzed by β-galactosidase assay. (D) Effect of overproduced HilD on flagellar gene expression under artificially induced flhDC expression conditions. The strains analyzed were TH13659 (Para::FCF PtetA::flhD+C+ flhC::MudJ), TH13919 (Para::FCF PtetA::flhD+C+ fliL::MudJ), TH14845 (Para::FCF PtetA::flhD+C+ fljB::MudJ), EM804 (Para::hilD+ PtetA::flhD+C+ flhC::MudJ), EM802 (Para::hilD+ PtetA::flhD+C+ fliL::MudJ), EM801 (Para::hilD+ PtetA::flhD+C+ fljB::MudJ), EM858 (Para::hilDΔHTH+ PtetA::flhD+C+ flhC::MudJ), EM868 (Para::hilDΔHTH+ PtetA::flhD+C+ fliL::MudJ), and EM869 (Para::hilDΔHTH+ PtetA::flhD+C+ fljB::MudJ). (E) flhDC operon transcription from individual flhDC promoters was analyzed by introducing a (luxDCABE-Km)-flhDC promoter fusion. In these constructs, the entire flhDC promoter region was fused to a luxCDBAE-kanamycin cassette. We generated six individual strains in which the −10 boxes of five of the six known transcriptional flhDC start sites (63) were mutated with GTTGGT (72). The individual strains, retaining only one functional, wild-type −10 box, were labeled P1, P2, P3, P4, P5, and P6, respectively. In order to retain a functional flhDC operon, a duplication of PflhDC-flhD+C+ follows the PflhDC-luxCDBAE cassette. To hold the duplication, 25 μg/ml kanamycin was added to the LB growth medium. For all strains, a ΔinvH-sprB (ΔSpi1) background was used. Luminescence is shown relative to that of the strain with genotype Para::FRT PflhDC(P1-6)-luxCDBAE-Km-PflhD+C+. Two independent runs with six replicates each were combined. (A to E) Error bars represent the standard errors of the means, and asterisks indicate the gene expression levels that differed significantly (*, P < 0.05, or ***, P < 0.001). Data were analyzed by the Student t test. Biological replicates are shown as individual data points (diamonds), and the relevant genotypes are indicated as follows: +, a chromosomal wild-type copy of the gene is present; ↑, the gene is chromosomally overexpressed from an arabinose-inducible promoter; −, the respective gene was deleted from the strain.
In order to further characterize the effect of HilD on flagellar gene expression, we analyzed transcriptional lac fusions to individual flagellar class 1, class 2, and class 3 promoters. Upon overexpression of HilD from the arabinose promoter (Fig. 1C, HilD↑), we observed a 2-fold increase in flhC-lac (class 1) transcription and a 3-fold increase in both fliL-lac (class 2) and fljB-lac (class 3) expression (Fig. 1C). Mutant strains overexpressing a DNA-binding-deficient variant of HilD missing its helix-turn-helix binding motif (Fig. 1C, HilDΔHTH↑) were lacking the HilD effect and showed flagellar gene expression levels comparable to those of the wild-type control.
We next uncoupled the expression of the flagellar master regulator flhDC from any transcriptional regulation using a tetracycline-inducible flhDC promoter (62). Under PtetA-flhD+C+ conditions, neither HilD nor HilDΔHTH overexpression showed any effect on flagellar gene expression (Fig. 1D). These results suggested that HilD acted by transcriptional activation of the class 1 flhDC promoter.
In a complementary experiment, we analyzed flagellar gene expression levels in a background overexpressing the Spi1 master regulator HilA. HilA was suggested to be a regulator of flhDC transcription (51). As shown in Fig. S2 in the supplemental material, we could rule out an effect of HilA on flagellar gene transcription and thereby confirmed that the observed HilD effect on flhDC transcription was not an indirect effect via HilA.
The flhDC promoter region was reported to consist of six transcriptional start sites upstream from the flhD gene and annotated P1 through P6 (63). To determine which flhDC transcriptional start site was activated by HilD, we subsequently measured HilD-mediated expression of lux fusions to individual flhDC promoters. Upon induction of HilD, flhDC expression was detected for the construct with P1 through P6, as well as for the P5 promoter fusion (Fig. 1E). We therefore concluded that HilD acts solely on the P5 promoter to activate flhDC transcription.
Many regulatory proteins that act on flhDC transcription bind to a region close to the P1 promoter, like RcsB (+5 to +19 nucleotides from the P1 transcription start site) or RtsB (−4 to +106 nucleotides from the P1 transcription start site) (24, 29). We tested possible dominant effects of simultaneous overproduction of both HilD (acting as activator of P5 transcription) and RtsB (acting as repressor of P1 transcription) on flhDC operon transcription. As shown by the results in Fig. S3A in the supplemental material, RtsB overproduction decreased flhD-lac levels, whereas HilD had an activating effect. We did not observe a change in flhD-lac expression compared to that in the wild type when we overproduced both RtsB and HilD concurrently. This suggested that the repressor RtsB and the activator HilD acted simultaneously on different promoters of flhDC. We further explored this possibility by analyzing the expression of lux fusions to individual flhDC promoters under conditions where either HilD or RtsB was overexpressed (see Fig. S3B). As demonstrated by the results described above, the expression of HilD activated flhDC expression from the P5 promoter, whereas the expression of RtsB had no effect on flhDC transcription from the P5 promoter. In contrast, RtsB overexpression showed a significant repression of flhDC expression from the P1 promoter, demonstrating that RtsB and HilD can act independently as repressors and activators of different flhDC promoters.
To further test the idea that HilD directly activates flhDC by binding to the flhDC promoter region, we performed electrophoretic mobility shift assays using purified HilD protein and various promoter fragments of flhDC. DNA fragments comprising the P1 promoter (fragment 1; nucleotides −271 to −71 upstream from the flhD coding region), the P2, P3, P4, and P6 promoters (fragment 2; nucleotides −431 to −231 upstream from the flhD coding region), the P5 promoter (fragment 3; nucleotides −588 to −388 upstream from the flhD coding region and fragment 3′ nucleotides −668 to −462 upstream from the flhD coding region), and a control sequence outside the flhDC promoter region within the coding sequence of flhDC (designated flhDC coding; nucleotides + 403 to +672 downstream from the flhD start codon) were analyzed according to the schematic in Fig. 2A. As shown by the results in Fig. 2B, we observed binding of increasing concentrations of purified HilD protein to DNA fragments comprising the P5 promoter (fragment 3 and 3′). Increasing amounts of HilD decreased the amount of free DNA, while the amount of HilD-bound DNA increased. Control DNAs (gyrA and flhDC coding) did not bind to purified HilD, and neither did the flhDC promoter fragments 1 and 2.
FIG 2.
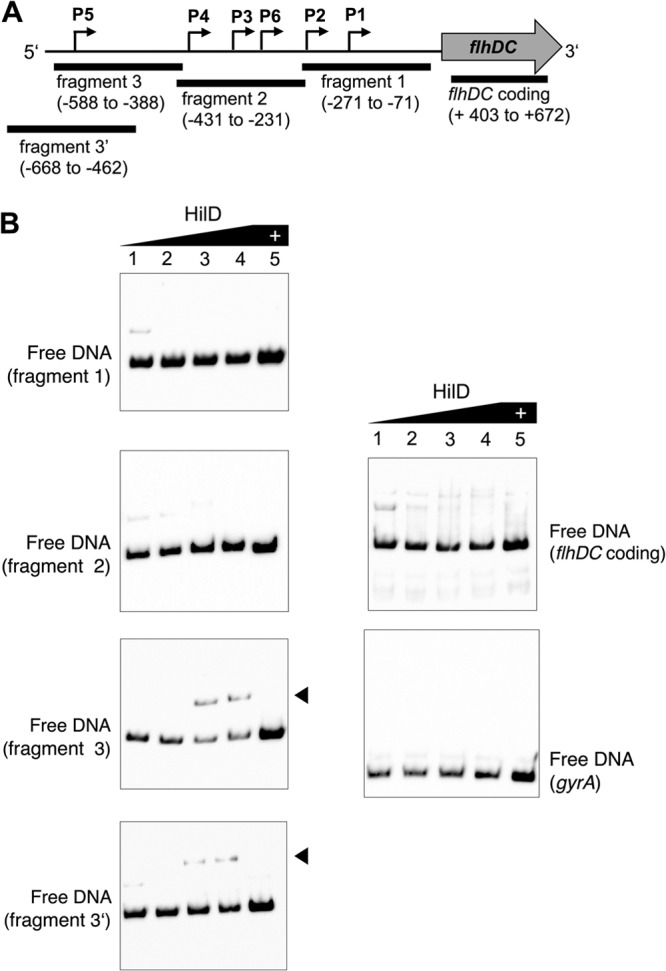
EMSA of the binding of HilD to flhDC promoter fragments. (A) Four DNA fragments covering the flhDC promoter were analyzed for specific binding of purified HilD protein in electrophoretic mobility shift assays (EMSA) as described in Materials and Methods. The region of the flhDC promoter covered by each fragment is indicated relative to the flhD start codon. The fragment designated flhDC coding is outside the promoter region. (B) Gel shift analysis of HilD binding to the flhDC promoter region. A concentration of 0.01 pmol of each of the 5′-biotinylated flhDC promoter fragments was incubated with increasing concentrations of purified HilD protein (lane 1, 0 pmol; lane 2, 1.06 pmol; lane 3, 4.23 pmol; lane 4, 8.45 pmol; lane 5, 8.45 pmol). An excess amount of unlabeled competitor DNA (lane 5) was added to the highest protein concentration to demonstrate specific binding. Arrowheads indicate HilD-DNA complexes. DNA fragments of the flhDC coding region and gyrA (E. coli) served as negative controls.
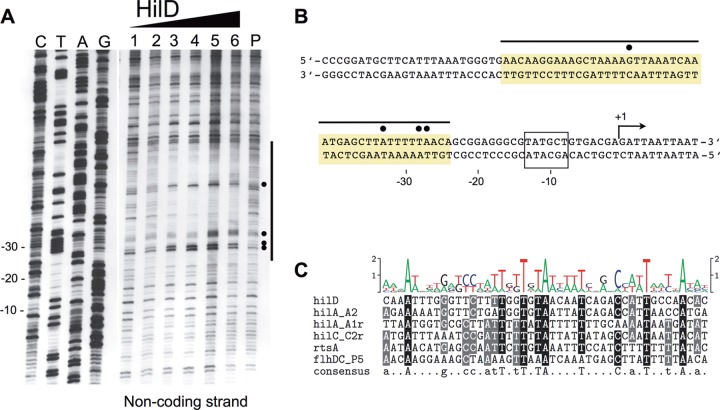
We next performed DNase I footprinting to precisely map the HilD binding site in the flhDC P5 promoter region. An flhDC promoter fragment comprising nucleotides −668 to −388 upstream from the flhDC coding region was incubated with increasing concentrations of purified HilD protein and, after partial digestion with DNase I, the resulting fragments were subjected to denaturing gel electrophoresis (Fig. 3A). We identified a region from −68 to −24 nucleotides upstream from the transcriptional start site of the P5 flhDC promoter that was protected from DNase I digestion in the presence of HilD. In addition, the presence of HilD resulted in an enhancement of DNase cleavage at nucleotides −51, −33, −28, and −27 upstream from the P5 transcriptional start, which indicates a potential DNA bend (Fig. 3A and B).
FIG 3.
DNase I footprinting demonstrates HilD binding to the flhDC P5 promoter region. (A) DNase I footprinting of an flhDC P5 promoter DNA fragment. A DNA fragment covering a region of nucleotides from position −668 to −388 upstream from the flhD start codon was DIG labeled on the noncoding strand and incubated alone (lane P) and with increasing amounts of purified HilD protein (lane 1, 4.23 pmol; lane 2, 8.45 pmol; lane 3, 12.68 pmol; lane 4, 16.9 pmol; lane 5, 21.13 pmol; lane 6, 42.25 pmol) and digested with DNase I before being loaded on a sequencing gel. The vertical line indicates the region protected from DNase I digestion. Lanes C, T, A, and G show the specific nucleotides of the noncoding strand. Exposed nucleotides are highlighted by dots. (B) Partial nucleotide sequence of the P5 promoter of flhDC that is relevant for HilD binding. A horizontal line marks the protected region, and the four most sensitive nucleotides are highlighted by dots. The transcriptional start site (marked as +1) and the −10 element of the P5 flhDC promoter are indicated. (C) Comparison of HilD binding sites in the flhDC, rtsA, hilC, hilD, and hilA promoters. The alignment of HilD binding sites is as defined by DNase I footprinting in the present and previous studies (60, 64). Shading indicates the predominant (black) and conserved (gray) nucleotides. The consensus motif logo of the experimentally determined HilD binding sites is shown at the top and was generated using WebLogo (71). The proposed consensus is displayed at the bottom; uppercase letters indicate predominant nucleotides (>80% conserved), and lowercase letters indicate conserved nucleotides (>60% conserved).
Comparison of known HilD binding sites from the rtsA, hilC, hilD, and hilA promoters (64) with the identified P5 PflhDC binding site revealed several nucleotides that were conserved between these sites (Fig. 3C, consensus; uppercase letters indicate predominant nucleotides [>80% conserved], and lowercase letters indicate conserved nucleotides [>60% conserved]). Similar to the data of Olekhnovich and Kadner (64), the 45-bp consensus sequence harbors two direct repeats of CNatTNtTNTA (uppercase and lowercase are as defined for Fig. 3C).
DISCUSSION
In a previous study, we investigated the feedback regulation occurring at the level of the flagellar master regulator flhDC. Using a genetic screen, we identified RflM as a negative regulator of flhDC transcription and demonstrated that the FlhDC complex activated rflM expression in a regulatory feedback loop (26). Another class of transposon insertions that we obtained from our screen for regulators affecting flhDC autorepression resulted in overexpression of the hilD gene. This suggested a role of the Spi1 activator HilD in regulation of the flagellar master operon flhDC. In the present study, we show that HilD acts as a direct activator of flhDC expression via activation of the P5 transcriptional start site, thus revealing a novel transcriptional cross talk between the flagellar and virulence regulons in Salmonella. Importantly, a recent study by Kroeger et al. (65) analyzed the Salmonella transcriptome under 22 different infection-relevant environmental conditions. The RNA-sequencing data of this study reveal specific activation of the flhDC P5 promoter only under Spi1-inducing conditions. In addition, the P5 transcriptional start site and the upstream HilD binding site appear not to be present in Escherichia coli, indicating that HilD-dependent activation of flhDC evolved concurrently after horizontal gene transfer of Spi1 in Salmonella.
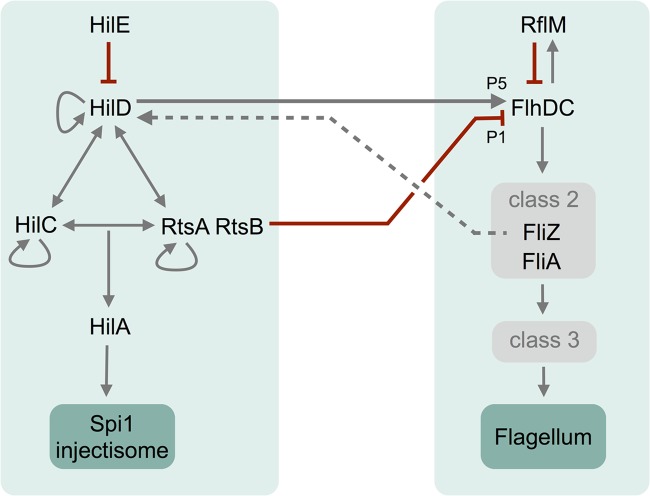
The transcriptional cross talk between the flagellar and Spi1 virulence regulons is complex, with several regulatory feedback mechanisms implemented on various levels (Fig. 4). The negative regulator of flagellar class 1 gene expression, RflM, is activated by the FlhDC protein complex, which additionally activates the expression of genes transcribed from flagellar class 2 promoters, including fliZ. FliZ protein is a posttranscriptional regulator of HilD protein. HilD acts in a positive-feedback loop together with HilC and RtsA to activate HilA and as an activator of flagellar class 1 gene expression through activation of the flhDC P5 promoter, as described in this work. RtsB is another Spi1-related regulator that is encoded in an operon together with rtsA and functions as a repressor of flagellar class 1 gene expression through repression of flhDC transcription at the level of the P1 promoter.
FIG 4.
Schematic model of cross talk between the Spi1 and flagellar regulons. A schematic model of the Spi1 and flagellar regulatory pathways with cross talk at the HilD/RtsB level is shown. For simplification, only protein names are given. Red blunt lines indicate inhibitory effects on gene transcription. The dashed line between class 2 product FliZ and HilD represents posttranscriptional activation, as described in the text.
What might be the physiological relevance of this complex cross-regulation network? At the moment, this is unclear, but for the initiation of infection, motility plays a crucial role. Therefore, we speculate that the transcriptional link between the flagellar and virulence systems is of importance during a specific step in the Salmonella infection cycle, as outlined below.
The Spi1 and Spi2 virulence-associated genes are transcribed at different stages during Salmonella infection. In the insect pathogen Xenorhabdus nematophila, hemolysin production and a full virulence phenotype have been reported to be dependent on the flagellar protein FliZ, and mutants with mutations of fliAZ or flhDC had attenuated virulence (66). For the food-borne pathogen Salmonella, FliZ was previously described to posttranslationally modify the Spi1 regulator HilD, forwarding a positive effect to HilA, the transcriptional activator of Spi1 structural genes (41–44). However, recent data suggested that only a few of the known mechanisms involved in Spi1 regulation are in fact FliZ dependent (67). Transcriptional regulation between the Spi1 system, the related flagella, and the type I fimbria systems have been subject to intensive research within the last few years. However, the detailed mechanisms are not well understood. Saini et al. (68) recently described the cross-regulation between the three systems to constitute a gene expression hierarchy that has the expression of flagellar genes at the top of a complex cascade. Fimbrial genes are repressed during flagellar gene expression and vice versa. At the same time, flagellar gene expression increased the expression of Spi1 genes (68). However, it was shown that overproduction of the Spi1-related protein RtsB completely abolished flgA promoter activity, which can be explained by repression of the flagellar master regulatory operon flhDC. Saini et al. (68) argued that the loss of flagellar gene expression and motility, which is downregulated during intracellular growth, would correspond to the bacterium's need to stay nonmotile after successful invasion. In this paper, however, we identified the Spi1 regulator HilD as a positive regulator of flagellar class 1 gene expression.
The activation of the flagellar system via HilD and simultaneous inactivation via RtsB argue for a dual or even multiprocess interplay between flagellar and Spi1 regulation. Thus, depending on the environmental niche and spatiotemporal stage of infection, the Spi1 regulator HilD could activate flhDC transcription through direct binding to the flhDC promoter or repress flhDC transcription via activation of RtsB. We postulate that during the early stages of epithelial cell infection, Spi1 gene expression is induced and motility is downregulated but flagellar genes are in a state ready to be immediately upregulated at some later time during infection by activation via HilD. The secretion of bacterial Spi1 effectors results in the internalization of Salmonella cells and formation of Salmonella-containing vacuoles. At this point, motility is probably no longer required or could hinder the infection process. In epithelial cells, flagellar proteins were downregulated during early infection (2 h p.i.), while the simultaneous expression of Spi1 and flagellar genes 4 to 6 h postinfection (resembling the late stages of infection) has been reported (39). Earlier results by Cummings et al. showed heterogeneous FliC expression by 60% of the bacterial population in Peyer's patches 7 days p.i. (38). However, as soon as eukaryotic host cells burst or lyse, the bacteria released seek further host cells for infection. At this stage, it might be advantageous for the bacteria to have flagellar gene expression ready to allow for a fast switch back into a motile state that could be mediated by activation of the flagellar regulon via HilD. This argument is also supported by the results of Sano et al. (69), who observed the requirement for flagella in order for Salmonella to exit host macrophages. Accordingly, flagellum-negative cells were unable to escape from host cells. During the first 2 h of macrophage infection, intracellular Salmonella cells were nonflagellated, whereas flagellum reexpression was observed 4 h later, an effect also previously reported in Legionella pneumophila (70). Together, these results argue in favor of a HilD-mediated de novo synthesis of flagella during some later step in the infection process.
While many questions remain, analysis of the temporal course of gene expression will be crucial to understand the interplay between motility and virulence in detail. Further research will need to focus on the aspects of gene regulation during different stages of the infection process and will need to give consideration to other cross-connected players, like the link to Spi2 and fimbria genes. Together, these studies will give a detailed picture of the steady state and the spatiotemporal interplay between the flagellar and virulence-associated regulons during Salmonella infection.
Supplementary Material
ACKNOWLEDGMENTS
We thank James M. Slauch for providing the ΔrtsAB::FRT allele, Beat Schwaller for his permission for using the luminometer, and Chakib Mouslim for advice and assistance with strain construction.
H.M.S. gratefully acknowledges scholarship support from the Boehringer Ingelheim Fonds. C.K. and J.A.D. acknowledge support by the President's Initiative and Networking Funds of the Helmholtz Association of German Research Centers (HGF) under contract number VH-GS-202. This work was supported by grant FN-7626 subsidy no. 31003A_132947/1 from the Swiss National Science Foundation and grant GM056141 from the National Institutes of Health to K.T.H. and by funding from the Helmholtz Association under the Helmholtz Young Investigator grant no. VH-NG-932 and the People Programme (Marie Curie Actions) of the European Unions' Seventh Framework Programme (FP7/2007-2013) under REA grant agreements no. 300718 and 334030 to M.E.
The authors declare that they have no conflict of interest.
Footnotes
Published ahead of print 31 January 2014
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JB.01438-13.
REFERENCES
- 1.Rubin RH, Weinstein L. 1977. Salmonellosis: microbiologic, pathogenic, and clinical features. Stratton Intercontinental Medical Book Corp., New York, NY [Google Scholar]
- 2.Cornelis GR, Van Gijsegem F. 2000. Assembly and function of type III secretory systems. Annu. Rev. Microbiol. 54:735–774. 10.1146/annurev.micro.54.1.735 [DOI] [PubMed] [Google Scholar]
- 3.Galán J, Collmer A. 1999. Type III secretion machines: bacterial devices for protein delivery into host cells. Science 284:1322–1328. 10.1126/science.284.5418.1322 [DOI] [PubMed] [Google Scholar]
- 4.Hueck C. 1998. Type III protein secretion systems in bacterial pathogens of animals and plants. Microbiol. Mol. Biol. Rev. 62:379–433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Galan JE. 1999. Interaction of Salmonella with host cells through the centisome 63 type III secretion system. Curr. Opin. Microbiol. 2:46–50. 10.1016/S1369-5274(99)80008-3 [DOI] [PubMed] [Google Scholar]
- 6.Hensel M. 2000. Salmonella pathogenicity island 2. Mol. Microbiol. 36:1015–1023. 10.1046/j.1365-2958.2000.01935.x [DOI] [PubMed] [Google Scholar]
- 7.Hapfelmeier S, Ehrbar K, Stecher B, Barthel M, Kremer M, Hardt WD. 2004. Role of the Salmonella pathogenicity island 1 effector proteins SipA, SopB, SopE, and SopE2 in Salmonella enterica subspecies 1 serovar Typhimurium colitis in streptomycin-pretreated mice. Infect. Immun. 72:795–809. 10.1128/IAI.72.2.795-809.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Muller AJ, Hoffmann C, Galle M, Van Den Broeke A, Heikenwalder M, Falter L, Misselwitz B, Kremer M, Beyaert R, Hardt WD. 2009. The S. Typhimurium effector SopE induces caspase-1 activation in stromal cells to initiate gut inflammation. Cell Host Microbe 6:125–136. 10.1016/j.chom.2009.07.007 [DOI] [PubMed] [Google Scholar]
- 9.Muller AJ, Kaiser P, Dittmar KE, Weber TC, Haueter S, Endt K, Songhet P, Zellweger C, Kremer M, Fehling HJ, Hardt WD. 2012. Salmonella gut invasion involves TTSS-2-dependent epithelial traversal, basolateral exit, and uptake by epithelium-sampling lamina propria phagocytes. Cell Host Microbe 11:19–32. 10.1016/j.chom.2011.11.013 [DOI] [PubMed] [Google Scholar]
- 10.Cirillo DM, Valdivia RH, Monack DM, Falkow S. 1998. Macrophage-dependent induction of the Salmonella pathogenicity island 2 type III secretion system and its role in intracellular survival. Mol. Microbiol. 30:175–188. 10.1046/j.1365-2958.1998.01048.x [DOI] [PubMed] [Google Scholar]
- 11.Hensel M, Shea JE, Waterman SR, Mundy R, Nikolaus T, Banks G, Vazquez-Torres A, Gleeson C, Fang FC, Holden DW. 1998. Genes encoding putative effector proteins of the type III secretion system of Salmonella pathogenicity island 2 are required for bacterial virulence and proliferation in macrophages. Mol. Microbiol. 30:163–174. 10.1046/j.1365-2958.1998.01047.x [DOI] [PubMed] [Google Scholar]
- 12.Ellermeier CD, Ellermeier JR, Slauch JM. 2005. HilD, HilC and RtsA constitute a feed forward loop that controls expression of the SPI1 type three secretion system regulator hilA in Salmonella enterica serovar Typhimurium. Mol. Microbiol. 57:691–705. 10.1111/j.1365-2958.2005.04737.x [DOI] [PubMed] [Google Scholar]
- 13.Saini S, Ellermeier JR, Slauch JM, Rao CV. 2010. The role of coupled positive feedback in the expression of the SPI1 type three secretion system in Salmonella. PLoS Pathog. 6:e1001025. 10.1371/journal.ppat.1001025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gophna U, Ron EZ, Graur D. 2003. Bacterial type III secretion systems are ancient and evolved by multiple horizontal-transfer events. Gene 312:151–163. 10.1016/S0378-1119(03)00612-7 [DOI] [PubMed] [Google Scholar]
- 15.Van Gijsegem F, Gough C, Zischek C, Niqueux E, Arlat M, Genin S, Barberis P, German S, Castello P, Boucher C. 1995. The hrp gene locus of Pseudomonas solanacearum, which controls the production of a type III secretion system, encodes eight proteins related to components of the bacterial flagellar biogenesis complex. Mol. Microbiol. 15:1095–1114. 10.1111/j.1365-2958.1995.tb02284.x [DOI] [PubMed] [Google Scholar]
- 16.Kubori T, Matsushima Y, Nakamura D, Uralil J, Lara-Tejero M, Sukhan A, Galán JE, Aizawa SI. 1998. Supramolecular structure of the Salmonella typhimurium type III protein secretion system. Science 280:602–605. 10.1126/science.280.5363.602 [DOI] [PubMed] [Google Scholar]
- 17.Kubori T, Sukhan A, Aizawa SI, Galan JE. 2000. Molecular characterization and assembly of the needle complex of the Salmonella typhimurium type III protein secretion system. Proc. Natl. Acad. Sci. U. S. A. 97:10225–10230. 10.1073/pnas.170128997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hughes KT, Gillen KL, Semon MJ, Karlinsey JE. 1993. Sensing structural intermediates in bacterial flagellar assembly by export of a negative regulator. Science 262:1277–1280. 10.1126/science.8235660 [DOI] [PubMed] [Google Scholar]
- 19.Chadsey MS, Karlinsey JE, Hughes KT. 1998. The flagellar anti-sigma factor FlgM actively dissociates Salmonella typhimurium sigma28 RNA polymerase holoenzyme. Genes Dev. 12:3123–3136. 10.1101/gad.12.19.3123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chadsey MS, Hughes KT. 2001. A multipartite interaction between Salmonella transcription factor sigma28 and its anti-sigma factor FlgM: implications for sigma28 holoenzyme destabilization through stepwise binding. J. Mol. Biol. 306:915–929. 10.1006/jmbi.2001.4438 [DOI] [PubMed] [Google Scholar]
- 21.Kelly A, Goldberg MD, Carroll RK, Danino V, Hinton JC, Dorman CJ. 2004. A global role for Fis in the transcriptional control of metabolism and type III secretion in Salmonella enterica serovar Typhimurium. Microbiology 150:2037–2053. 10.1099/mic.0.27209-0 [DOI] [PubMed] [Google Scholar]
- 22.Soutourina O, Kolb A, Krin E, Laurent-Winter C, Rimsky S, Danchin A, Bertin P. 1999. Multiple control of flagellum biosynthesis in Escherichia coli: role of H-NS protein and the cyclic AMP-catabolite activator protein complex in transcription of the flhDC master operon. J. Bacteriol. 181:7500–7508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Stojiljkovic I, Baumler AJ, Hantke K. 1994. Fur regulon in gram-negative bacteria. Identification and characterization of new iron-regulated Escherichia coli genes by a fur titration assay. J. Mol. Biol. 236:531–545 [DOI] [PubMed] [Google Scholar]
- 24.Ellermeier CD, Slauch JM. 2003. RtsA and RtsB coordinately regulate expression of the invasion and flagellar genes in Salmonella enterica serovar Typhimurium. J. Bacteriol. 185:5096–5108. 10.1128/JB.185.17.5096-5108.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Erhardt M, Hughes KT. 2010. C-ring requirement in flagellar type III secretion is bypassed by FlhDC upregulation. Mol. Microbiol. 75:376–393. 10.1111/j.1365-2958.2009.06973.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Singer HM, Erhardt M, Hughes KT. 2013. RflM functions as transcriptional repressor in the autogenous control of the Salmonella flagellar master operon flhDC. J. Bacteriol. 195:4274–4282. 10.1128/JB.00728-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kutsukake K. 1997. Autogenous and global control of the flagellar master operon, flhD, in Salmonella typhimurium. Mol. Gen. Genet. 254:440–448. 10.1007/s004380050437 [DOI] [PubMed] [Google Scholar]
- 28.Wozniak C, Lee C, Hughes K. 2009. T-POP array identifies EcnR and PefI-SrgD as novel regulators of flagellar gene expression. J. Bacteriol. 191:1498–1508. 10.1128/JB.01177-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang Q, Zhao Y, McClelland M, Harshey RM. 2007. The RcsCDB signaling system and swarming motility in Salmonella enterica serovar Typhimurium: dual regulation of flagellar and SPI-2 virulence genes. J. Bacteriol. 189:8447–8457. 10.1128/JB.01198-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Macnab RM. 1996. Flagella and motility, p 123–145 In Neidhardt FC, Curtiss R, III, Ingraham JL, Lin ECC, Low KB, Magasanik B, Reznikoff WS, Riley M, Schaechter M, Umbarger HE. (ed), Escherichia coli and Salmonella: cellular and molecular biology, 2nd ed, vol 1 ASM Press, Washington, DC [Google Scholar]
- 31.Fontaine F, Stewart EJ, Lindner AB, Taddei F. 2008. Mutations in two global regulators lower individual mortality in Escherichia coli. Mol. Microbiol. 67:2–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yokota T, Gots JS. 1970. Requirement of adenosine 3′,5′-cyclic phosphate for flagella formation in Escherichia coli and Salmonella typhimurium. J. Bacteriol. 103:513–516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhao K, Liu M, Burgess RR. 2007. Adaptation in bacterial flagellar and motility systems: from regulon members to ‘foraging'-like behavior in E. coli. Nucleic Acids Res. 35:4441–4452. 10.1093/nar/gkm456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wada T, Morizane T, Abo T, Tominaga A, Inoue-Tanaka K, Kutsukake K. 2011. EAL domain protein YdiV acts as an anti-FlhD4C2 factor responsible for nutritional control of the flagellar regulon in Salmonella enterica serovar Typhimurium. J. Bacteriol. 193:1600–1611. 10.1128/JB.01494-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Prigent-Combaret C, Vidal O, Dorel C, Lejeune P. 1999. Abiotic surface sensing and biofilm-dependent regulation of gene expression in Escherichia coli. J. Bacteriol. 181:5993–6002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Beloin C, Ghigo JM. 2005. Finding gene-expression patterns in bacterial biofilms. Trends Microbiol. 13:16–19. 10.1016/j.tim.2004.11.008 [DOI] [PubMed] [Google Scholar]
- 37.Eriksson S, Lucchini S, Thompson A, Rhen M, Hinton JC. 2003. Unravelling the biology of macrophage infection by gene expression profiling of intracellular Salmonella enterica. Mol. Microbiol. 47:103–118. 10.1046/j.1365-2958.2003.03313.x [DOI] [PubMed] [Google Scholar]
- 38.Cummings LA, Wilkerson WD, Bergsbaken T, Cookson BT. 2006. In vivo, fliC expression by Salmonella enterica serovar Typhimurium is heterogeneous, regulated by ClpX, and anatomically restricted. Mol. Microbiol. 61:795–809. 10.1111/j.1365-2958.2006.05271.x [DOI] [PubMed] [Google Scholar]
- 39.Hautefort I, Thompson A, Eriksson-Ygberg S, Parker ML, Lucchini S, Danino V, Bongaerts RJ, Ahmad N, Rhen M, Hinton JC. 2008. During infection of epithelial cells Salmonella enterica serovar Typhimurium undergoes a time-dependent transcriptional adaptation that results in simultaneous expression of three type 3 secretion systems. Cell. Microbiol. 10:958–984. 10.1111/j.1462-5822.2007.01099.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cummings LA, Barrett SL, Wilkerson WD, Fellnerova I, Cookson BT. 2005. FliC-specific CD4+ T cell responses are restricted by bacterial regulation of antigen expression. J. Immunol. 174:7929–7938 [DOI] [PubMed] [Google Scholar]
- 41.Lucas RL, Lee CA. 2001. Roles of hilC and hilD in regulation of hilA expression in Salmonella enterica serovar Typhimurium. J. Bacteriol. 183:2733–2745. 10.1128/JB.183.9.2733-2745.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Iyoda S. 2001. A flagellar gene fliZ regulates the expression of invasion genes and virulence phenotype in Salmonella enterica serovar Typhimurium. Microb. Pathog. 30:81–90. 10.1006/mpat.2000.0409 [DOI] [PubMed] [Google Scholar]
- 43.Kage H, Takaya A, Ohya M, Yamamoto T. 2008. Coordinated regulation of expression of Salmonella pathogenicity island 1 and flagellar type III secretion systems by ATP-dependent ClpXP protease. J. Bacteriol. 190:2470–2478. 10.1128/JB.01385-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chubiz JEC, Golubeva YA, Lin D, Miller LD, Slauch JM. 2010. FliZ regulates expression of the Salmonella pathogenicity island 1 invasion locus by controlling HilD protein activity in Salmonella enterica serovar Typhimurium. J. Bacteriol. 192:6261–6270. 10.1128/JB.00635-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kutsukake K, Ikebe T, Yamamoto S. 1999. Two novel regulatory genes, fliT and fliZ, in the flagellar regulon of Salmonella. Genes Genet. Syst. 74:287–292. 10.1266/ggs.74.287 [DOI] [PubMed] [Google Scholar]
- 46.Saini S, Koirala S, Floess E, Mears PJ, Chemla YR, Golding I, Aldridge C, Aldridge PD, Rao CV. 2010. FliZ induces a kinetic switch in flagellar gene expression. J. Bacteriol. 192:6477–6481. 10.1128/JB.00751-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wada T, Tanabe Y, Kutsukake K. 2011. FliZ acts as a repressor of the ydiV gene, which encodes an anti-FlhD4C2 factor of the flagellar regulon in Salmonella enterica serovar Typhimurium. J. Bacteriol. 193:5191–5198. 10.1128/JB.05441-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Takaya A, Erhardt M, Karata K, Winterberg K, Yamamoto T, Hughes KT. 2012. YdiV: a dual function protein that targets FlhDC for ClpXP-dependent degradation by promoting release of DNA-bound FlhDC complex. Mol. Microbiol. 83:1268–1284. 10.1111/j.1365-2958.2012.08007.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Stewart MK, Cummings LA, Johnson ML, Berezow AB, Cookson BT. 2011. Regulation of phenotypic heterogeneity permits Salmonella evasion of the host caspase-1 inflammatory response. Proc. Natl. Acad. Sci. U. S. A. 108:20742–20747. 10.1073/pnas.1108963108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hisert KB, MacCoss M, Shiloh MU, Darwin KH, Singh S, Jones RA, Ehrt S, Zhang Z, Gaffney BL, Gandotra S, Holden DW, Murray D, Nathan C. 2005. A glutamate-alanine-leucine (EAL) domain protein of Salmonella controls bacterial survival in mice, antioxidant defence and killing of macrophages: role of cyclic diGMP. Mol. Microbiol. 56:1234–1245. 10.1111/j.1365-2958.2005.04632.x [DOI] [PubMed] [Google Scholar]
- 51.Thijs IM, De Keersmaecker SC, Fadda A, Engelen K, Zhao H, McClelland M, Marchal K, Vanderleyden J. 2007. Delineation of the Salmonella enterica serovar Typhimurium HilA regulon through genome-wide location and transcript analysis. J. Bacteriol. 189:4587–4596. 10.1128/JB.00178-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Teplitski M, Goodier RI, Ahmer BMM. 2003. Pathways leading from BarA/SirA to motility and virulence gene expression in Salmonella. J. Bacteriol. 185:7257–7265. 10.1128/JB.185.24.7257-7265.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bustamante VH, Martinez LC, Santana FJ, Knodler LA, Steele-Mortimer O, Puente JL. 2008. HilD-mediated transcriptional cross-talk between SPI-1 and SPI-2. Proc. Natl. Acad. Sci. U. S. A. 105:14591–14596. 10.1073/pnas.0801205105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bertani G. 2004. Lysogeny at mid-twentieth century: P1, P2, and other experimental systems. J. Bacteriol. 186:595–600. 10.1128/JB.186.3.595-600.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sanderson KE, Roth JR. 1983. Linkage map of Salmonella typhimurium, edition VI. Microbiol. Rev. 47:410–453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Aldridge PD, Karlinsey JE, Aldridge C, Birchall C, Thompson D, Yagasaki J, Hughes KT. 2006. The flagellar-specific transcription factor, sigma28, is the type III secretion chaperone for the flagellar-specific anti-sigma28 factor FlgM. Genes Dev. 20:2315–2326. 10.1101/gad.380406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Pfaffl MW. 2001. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 29:e45. 10.1093/nar/29.9.e45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Andreasson C, Fiaux J, Rampelt H, Mayer MP, Bukau B. 2008. Hsp110 is a nucleotide-activated exchange factor for Hsp70. J. Biol. Chem. 283:8877–8884. 10.1074/jbc.M710063200 [DOI] [PubMed] [Google Scholar]
- 59.Schechter LM, Lee CA. 2001. AraC/XylS family members, HilC and HilD, directly bind and derepress the Salmonella typhimurium hilA promoter. Mol. Microbiol. 40:1289–1299. 10.1046/j.1365-2958.2001.02462.x [DOI] [PubMed] [Google Scholar]
- 60.Olekhnovich IN, Kadner RJ. 2002. DNA-binding activities of the HilC and HilD virulence regulatory proteins of Salmonella enterica serovar Typhimurium. J. Bacteriol. 184:4148–4160. 10.1128/JB.184.15.4148-4160.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sturm A, Heinemann M, Arnoldini M, Benecke A, Ackermann M, Benz M, Dormann J, Hardt W-D. 2011. The cost of virulence: retarded growth of Salmonella Typhimurium cells expressing type III secretion system 1. PLoS Pathog. 7:e1002143. 10.1371/journal.ppat.1002143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Karlinsey JE, Tanaka S, Bettenworth V, Yamaguchi S, Boos W, Aizawa SI, Hughes KT. 2000. Completion of the hook-basal body complex of the Salmonella typhimurium flagellum is coupled to FlgM secretion and fliC transcription. Mol. Microbiol. 37:1220–1231. 10.1046/j.1365-2958.2000.02081.x [DOI] [PubMed] [Google Scholar]
- 63.Yanagihara S, Iyoda S, Ohnishi K, Iino T, Kutsukake K. 1999. Structure and transcriptional control of the flagellar master operon of Salmonella typhimurium. Genes Genet. Syst. 74:105–111. 10.1266/ggs.74.105 [DOI] [PubMed] [Google Scholar]
- 64.Olekhnovich IN, Kadner RJ. 2007. Role of nucleoid-associated proteins Hha and H-NS in expression of Salmonella enterica activators HilD, HilC, and RtsA required for cell invasion. J. Bacteriol. 189:6882–6890. 10.1128/JB.00905-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kroger C, Colgan A, Srikumar S, Handler K, Sivasankaran SK, Hammarlof DL, Canals R, Grissom JE, Conway T, Hokamp K, Hinton JC. 2013. An infection-relevant transcriptomic compendium for Salmonella enterica serovar Typhimurium. Cell Host Microbe 14:683–695. 10.1016/j.chom.2013.11.010 [DOI] [PubMed] [Google Scholar]
- 66.Lanois A, Jubelin G, Givaudan A. 2008. FliZ, a flagellar regulator, is at the crossroads between motility, haemolysin expression and virulence in the insect pathogenic bacterium Xenorhabdus. Mol. Microbiol. 68:516–533. 10.1111/j.1365-2958.2008.06168.x [DOI] [PubMed] [Google Scholar]
- 67.Golubeva YA, Sadik AY, Ellermeier JR, Slauch JM. 2012. Integrating global regulatory input into the salmonella pathogenicity island 1 type III secretion system. Genetics 190:70–90. 10.1534/genetics.111.132779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Saini S, Slauch JM, Aldridge PD, Rao CV. 2010. Role of cross talk in regulating the dynamic expression of the flagellar Salmonella pathogenicity island 1 and type 1 fimbrial genes J. Bacteriol. 192:5767–5777. 10.1128/JB.00624-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Sano G, Takada Y, Goto S, Maruyama K, Shindo Y, Oka K, Matsui H, Matsuo K. 2007. Flagella facilitate escape of Salmonella from oncotic macrophages. J. Bacteriol. 189:8224–8232. 10.1128/JB.00898-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Byrne B, Swanson MS. 1998. Expression of Legionella pneumophila virulence traits in response to growth conditions. Infect. Immun. 66:3029–3034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Crooks GE, Hon G, Chandonia JM, Brenner SE. 2004. WebLogo: a sequence logo generator. Genome Res. 14:1188–1190. 10.1101/gr.849004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Mouslim C, Hughes KT. The effect of cell growth phase on the regulatory cross-talk between flagellar and Spi1 virulence gene expression. PLOS Pathog., in press [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.