Abstract
Tuberculosis remains a major worldwide epidemic because of its sole etiological agent, Mycobacterium tuberculosis. Ethionamide (ETH) is one of the major antitubercular drugs used to treat infections with multidrug-resistant M. tuberculosis strains. ETH is a prodrug that requires activation within the mycobacterial cell; its bioactivation involves the ethA-ethR locus, which encodes the monooxygenase EthA, while EthR is a transcriptional regulator that binds to the intergenic promoter region of the ethA-ethR locus. While most studies have focused on the role of EthA-EthR in ETH bioactivation, its physiological role in mycobacteria has remained elusive, although a role in bacterial cell detoxification has been proposed. Moreover, the importance of EthA-EthR in vivo has never been reported on. Here we constructed and characterized an EthA-EthR-deficient mutant of Mycobacterium bovis BCG. Our results indicate that absence of the ethA-ethR locus led to greater persistence of M. bovis BCG in the mouse model of mycobacterial infection, which correlated with greater adherence to mammalian cells. Furthermore, analysis of cell wall lipid composition by thin-layer chromatography and mass spectrometry revealed differences between the ethA-ethR KO mutant and the parental strain in the relative amounts of α- and keto-mycolates. Therefore, we propose here that M. bovis BCG ethA-ethR is involved in the cell wall-bound mycolate profile, which impacts mycobacterial adherence properties and in vivo persistence. This study thus provides some experimental clues to the possible physiological role of ethA-ethR and proposes that this locus is a novel factor involved in the modulation of mycobacterial virulence.
INTRODUCTION
Despite persistent efforts by public health officials, tuberculosis (TB) continues to remain a major worldwide epidemic. The situation appears to be deteriorating. With an already existing pool of 2 billion latently infected individuals, there has also been an increase in the number of new cases per year, up from 7 million in the 1990s to a staggering number of almost 9 million new cases per year today (1–3). While first-line drugs such as isoniazid (INH) and rifampin (RIF) have historically been successful in the treatment of TB infection (4), today, poor compliance with prolonged regimens, in conjunction with the acquired immunodeficiency disease pandemic, has compounded the problem and fueled the emergence of multidrug-resistant (MDR) and extremely drug-resistant (XDR) Mycobacterium tuberculosis strains (5–7). MDR TB cases currently represent nearly 5% of the world's annual TB burden (8), necessitating treatment with second-line drugs that are less effective and/or are poorly tolerated because of increased toxicity (9).
Ethionamide (ETH) has been clinically used to treat humans for more than 35 years and represents one of the most efficient second-line anti-TB drugs (10). In fact, ETH was shown to be as potent as INH in a mouse model of TB infection when the drugs are combined with streptomycin (11). However, the potential of ETH has yet to be fully exploited clinically because of its serious side effects, which include hepatotoxicity and gastrointestinal disturbances (12). The side effects associated with ETH have been a major hurdle to ensuring patient compliance, thus further fueling the emergence of resistant mycobacteria.
As a structural analog of INH, ETH inhibits the molecular target InhA, a NADH-specific enoyl-acyl carrier protein reductase involved in mycolic acid (MA) synthesis (13, 14). ETH is a prodrug that is enzymatically activated by the mycobacterial cell itself. In 2000, two laboratories concurrently and independently reported the identification in the M. tuberculosis genome of ethA, whose product, EthA, was proposed to be responsible for the bioactivation of ETH (15, 16). A number of studies have ensued, demonstrating that EthA is a common bioactivator of two other thiocarbamide-containing anti-TB drugs, thiacetazone and isoxyl, thus supporting the broad substrate specificity of this enzyme (17, 18).
EthA is a member of the Bayer-Villiger monooxygenase (BVMO) family and catalyzes the NADPH- and O2-dependent monooxygenation of ETH to its corresponding S-oxide, ETH-SO, and is also capable of further oxidizing ETH-SO to its final cytotoxic species (19). On the basis of observations made in recombinant EthA-expressing Escherichia coli (19, 20), it was suggested that EthA is a membrane-associated protein in M. tuberculosis as well and that in its absence, ETH is either quickly expelled or unable to penetrate the mycobacterial cell (21, 22). However, while EthA has been shown to be able to accept a wide range of ketones as substrates (20), the exact nature of the physiological substrate remains unknown. Transcriptome analysis of ethA has revealed downregulation of the gene under starvation (23) and its upregulation under low-iron conditions (24), suggesting a role for EthA in the pathogen's virulence.
At the genetic level, ethA expression is negatively regulated by the transcriptional regulator EthR (15, 16) with ethA and ethR open reading frames (ORFs) organized in a divergent operon (25). Interestingly, the X-ray crystal structure of EthR homodimers revealed the presence of a small ligand identified as hexadecyl octanoate (HexOc) (26). EthR-HexOc complexes were shown to prevent the binding of EthR to its target promoter region, thereby leading to derepression of ethA-ethR transcription and suggesting that regulation of the ethA-ethR locus is more complex than initially thought. More recently, the identification of small synthetic inhibitors of EthR that boost the anti-TB activity of ETH in vivo was reported, which may prompt reconsideration of ETH as a possible first-line anti-TB drug (27–29).
To date, while most studies have focused on dissecting the role of EthA and its transcriptional repressor EthR in ETH bioactivation, few attempts have been made to understand the function and physiological role of the ethA-ethR locus in Mycobacterium species. Since the presence of an EthA-encoding gene ortholog could be found in all of the mycobacterial genomes (16), it is anticipated that EthA serves an important function in mycobacteria. However, it does not appear to be essential since ETH-resistant M. tuberculosis clinical isolates can be found with mutations in ethA that likely impair its physiological function as well (15). The presence of several genes encoding BVMO-like compounds together with an abundant number of other oxidizing enzymes, such as P450 cytochromes, has led to the idea that such high oxidative potential in mycobacteria may help the pathogen resist oxidative stress in vivo (15). In this context, it was proposed that EthA and other BVMOs may play a role in detoxifying the bacterial cell by removing toxic ketones (20). Moreover, the importance of EthA in vivo during host infection has never been reported. This work describes the construction and phenotypic characterization of an ethA-ethR deletion mutant of Mycobacterium bovis BCG.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
Wild-type (WT) M. bovis BCG (Pasteur strain ATCC 35734) and derivative strains were grown at 37°C in Middlebrook liquid 7H9 medium (Difco) or on 7H11 agar supplemented with ADS (0.5% bovine serum albumin fraction V, 0.2% dextrose, 0.85% saline) enrichment, 0.05% Tween 80, and 0.2% glycerol with appropriate antibiotics (80 μg/ml hygromycin [Roche], 20 μg/ml kanamycin [Sigma]).
Construction of M. bovis BCG ethA-ethR knockout (KO) and complemented strains.
The ethA-ethR locus was deleted by double homologous recombination as described previously (30). Briefly, two primer pairs (5′-TTC TCG AGG TCC TGG CAT GAT GGG ACC G-3′ plus 5′-TTA AGC TTG ACA TCC GGC TCA TCC GGC-3′ and 5′-TTC TTA AGG TGC CGG AAG CCC GCG TG-3′ plus 5′-TTT CTA GAG GCC GCG AGC CGG ACC TG-3′) were used to amplify the DNA regions flanking the ethA-ethR locus in M. bovis BCG via PCR. The bases in bold are extra nucleotides added for cloning convenience. The PCR-amplified regions (each approximately 800 bp long) were sequenced and cloned directionally into vector pYUB854 such that the hygromycin resistance cassette (hyg) lies between the flanking regions. The lacZ ORF and promoter region from the pGoal17 plasmid (31) were then cloned into the unique PacI site of the pYUB construct. M. bovis BCG was electroporated (2,500 mV, 800 Ω, 25 µF) with 2 μg of recombinant plasmid and plated onto hygromycin-containing 7H11 medium supplemented with 40 μg/ml 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal), and incubated at 37°C. White, hygromycin-resistant clones were selected after 16 days of incubation and screened by PCR with a set of internal ethA-ethR primers (5′-TCC AGC GGT TTT CCG CGG TC-3′and 5′-TCC CGG TGC GCC ACA TGT TC-3′). Deletion of the ethA-ethR locus was further confirmed by Southern and Western blot analyses (see below).
To complement the ethA-ethR KO mutant, the full-length 2.2-kb ethA-ethR locus was PCR amplified, cloned into the multiple cloning site of the single-copy integrative vector pMV306 (32), and introduced into the genome of ethA-ethR KO M. bovis BCG via electroporation as described above. The resulting transformants were plated onto kanamycin-containing 7H11 agar and incubated at 37°C. After 16 days of incubation, kanamycin-resistant colonies were PCR screened with the internal ethA-ethR primers mentioned above.
Southern blot analysis.
From 1 to 3 μg of genomic DNA was digested with SacI (Promega), separated on a 1.5% agarose gel, and treated in accordance with standard protocols (33). DNA was transferred onto a Millipore Immobilon-Ny+ Transfer Membrane and UV cross-linked. The 415-bp digoxigenin (DIG)-labeled probe was amplified with a set of primers that binds approximately 1.5 kb downstream of ethR (5′-TGA GTT TAG TTG GGA CCT AGG CC-3′ and 5′-CTA GAG TCA CAT CAG AAA CAT TTG A-3′) according to the manufacturer's instructions (DIG labeling kit; Roche). Hybridization and signal detection were performed with a detection kit (Roche) according to the manufacturer's protocols. EasyHyb (Roche) was used as the prehybridization and hybridization solutions, and CSPD (Roche) was used as the detection substrate for chemical luminescence.
Western blot analysis.
Whole-cell bacterial lysates were prepared by the addition of protein sample buffer (8% sodium dodecyl sulfate [SDS], 20% β-mercaptoethanol, 20% glycerol, 0.04% bromophenol blue). Lysates were subjected to SDS-PAGE and electrotransferred onto polyvinylidene difluoride membranes. Blocking and incubation with antibodies were performed with 5% nonfat dry milk (Bio-Rad) and 0.1% Tween 20 in Tris-buffered saline. Immunoblotting was performed with primary rabbit polyclonal antibodies raised against an EthA epitope (GenicBio) and horseradish peroxidase-conjugated goat anti-rabbit secondary antibodies (Sigma). Detection was performed by enhanced chemiluminescence assay. Molecular sizes were determined by using prestained molecular weight marker SDS-7B (Sigma).
Determination of MIC and MBC.
The MIC was determined by a visual method as reported previously (34). ETH (Sigma) and INH (Sigma) were dissolved in 90% dimethyl sulfoxide for stock solutions. Ninety-six-well flat-bottom clear plates were prepared with 2-fold serially diluted concentrations of INH and ETH (0.02 to 5 and 0.3 to 80 μM, respectively), one drug per row, and the last row was filled with 7H9 medium only to serve as drug-free controls. M. bovis BCG strains were cultured to log phase and diluted in 7H9 medium to obtain an optical density at 600 nm (OD600) of 0.04. A 100-μl volume of this prepared inoculum was added to each test well. The plates were then sealed in an air-tight box on water-soaked paper towels for 5 days at 37°C. On the 5th day, the plates were read visually by detecting the presence of a bacterial pellet at the bottom of the well when it was placed against a black background. Visible pellets were then scored against pellets in drug-free medium for size. The visual MIC corresponds to the lowest drug concentration that correlates with the absence of a visible bacterial pellet at the bottom of the well as detected by the unaided eye. After MIC determination, 50-μl volumes of concentrations equal to 1, 2, and 4 times the MIC from the assay plates were plated onto 7H11 agar plates to determine the minimum bactericidal concentration (MBC). The MBC was defined as the drug concentration that reduced the number of CFU by 99% compared to the drug-free control after 5 days of incubation.
Mouse infection.
All animal experiments were carried out upon approval and under the guidelines of the Institutional Animal Care and Use Committee, National University of Singapore. Six- to 8-week-old female BALB/c mice were kept under specific-pathogen-free conditions in individual ventilated cages. For intranasal infection, sedated mice were intranasally administered 5 × 106 CFU of parental, ethA-ethR KO, or complemented M. bovis BCG in 20 μl of sterile phosphate-buffered saline (PBS) supplemented with 0.05% Tween 80 (PBST) (Sigma). Intravenous infection was performed retroorbitally with 5 × 106 CFU of the parental, mutant, or complemented strain in 200 μl of sterile PBST. At the times indicated, four mice per group were euthanized and individual lungs, spleens, and livers were harvested and homogenized. Appropriate dilutions were plated onto 7H11 agar for colony counting.
Ex vivo infection and adherence assays.
THP-1 (ATCC TIB-202), A549 (ATCC CCL-185), and Huh-7 (Health Science Research Resources Bank JCRB0403) cells were maintained and stored according to the American Type Culture Collection guidelines. THP1 cells were differentiated into adherent macrophages by seeding 5 × 104 monocytes per well (in 24-well plates) with 0.04 μg/ml phorbol 12-myristate 13-acetate (Sigma) 24 to 26 h prior to infection. Murine bone marrow-derived macrophages (BMMOs) were isolated from adult BALB/c mice and maintained in DMEM (Gibco) supplemented with 10% fetal calf serum, 5% horse serum, and 1.5 g/liter sodium pyruvate (Invitrogen). Infection assays were performed by coincubating mycobacteria and mammalian cells for 45 min at a multiplicity of infection (MOI) of 1 (THP-1 and BMMOs), 2 (Huh-7), or 3 (A549) in 24-well plates. At the time points indicated, the cell monolayers were washed thrice with PBS to remove extracellular bacteria and subsequently lysed with 0.1% Triton X-100 (Sigma) to release the intracellular bacteria. Appropriate dilutions of the cell lysates in 7H9 medium were plated onto 7H11 agar for colony counting. Bacterial uptake percentages were calculated by normalizing the day 0 bacterial load to the respective inoculum, and survival percentages were calculated by expressing day 2, 5, and 7 counts as percentages of the initial day 0 load. For adherence assays, 45 min of coincubation of mycobacterial and mammalian cells was performed at 4°C or in the presence of 10 μg/ml cytochalasin D (CCD) as described before (35). Extracellular bacteria were removed, and cell monolayers were then thoroughly washed thrice with PBS to remove nonadherent bacteria and lysed with 0.1% Triton X-100 to release them, and appropriate dilutions of the lysates were plated onto 7H11 agar for colony counting.
Analysis of total, extractable, and cell wall-bound lipids.
Total lipid extraction from bacterial cells and preparation of fatty acid and MA methyl esters (MAMEs) from extractable lipids and delipidated cells followed procedures described earlier (36, 37). Briefly, M. bovis BCG strains were grown at 37°C in 7H9 medium supplemented with ADS, 0.2% glycerol, and 0.05% Tyloxapol (Sigma) for 20 days, harvested, washed twice with ultrapure water (Gibco), resuspended in chloroform-methanol (2:1, vol/vol), and incubated at 4°C overnight to allow inactivation of the cells. Briefly, the whole mixture was subsequently dried and subjected to a series of extractions with CHCl3-CH3OH (1:2) and two times with CHCl3-CH3OH (2:1). Each extraction was performed overnight at room temperature. The extracts obtained by centrifugation of the mixture at 1,800 × g were collected, combined, dried, and subjected to biphasic Folch washing as described previously (38). The upper phase was removed and discarded, and the bottom phase was dried under a stream of N2 and dissolved in CHCl3-CH3OH (2:1) at a ratio of 100 μl of solvent to 300 mg of wet weight. Thin-layer chromatography (TLC) of the lipid extracts was performed on silica gel plates (Merck) in different solvent systems (CHCl3-CH3OH-H2O [20:4:0.5], CHCl3-CH3OH-NH4OH-H2O [65:25:0.5:4], n-hexane–ethyl acetate [95:5]) to reveal the total lipid species profiles. For further MAME analysis, MAMEs prepared from whole cells, extractable lipids, and delipidated cells were run in three different solvent systems (n-hexane–ethyl acetate [95:5], petroleum ether-acetone [90:10], dichloromethane) by using silver (Ag)-impregnated plates to reveal additional types of MAMEs. Lipids were visualized by spraying with cupric sulfate (10% in an 8% phosphoric acid solution) or α-naphthol (0.5% α-naphthol in 5% sulfuric acid in ethanol) and heating.
Mass spectrometry for MA lipid analysis.
Mycobacterial cells at mid-log phase were harvested in Teflon-fluorinated ethylene propylene tubes (Nalgene, Rochester, NY), and total lipids were extracted as described previously (39). Briefly, cell pellets were resuspended in chloroform-methanol (2:1, vol/vol), incubated overnight at 4°C under constant shaking at 200 rpm, and separated into organic and aqueous phases by adding deionized water. The white intermediate layer (delipidated cells) was used for subsequent alkaline hydrolysis. To release esterified MAs from the cell wall, the defatted cells obtained by chloroform-methanol extraction were washed once with deionized water and dried. For alkaline hydrolysis, 1 ml of 1 M KOH-methanol was added for 2 h of incubation at 80°C at 600 rpm and the resulting extracts were cooled to room temperature before acidification to pH 4.5 with HCl. Liberated MAs were extracted twice with 1 ml of diethyl ether. The ether phase was washed once with deionized water and dried. Samples were analyzed via a QTRAP 4000 mass spectrometer as described previously (39).
Statistical analysis.
Unless stated otherwise, bars in the figures represent means plus standard deviations (SD) and averages were compared by using a bidirectional unpaired Student t test with a 5% significance level (P ≤ 0.05).
RESULTS
Construction of an M. bovis BCG ethA-ethR KO mutant strain.
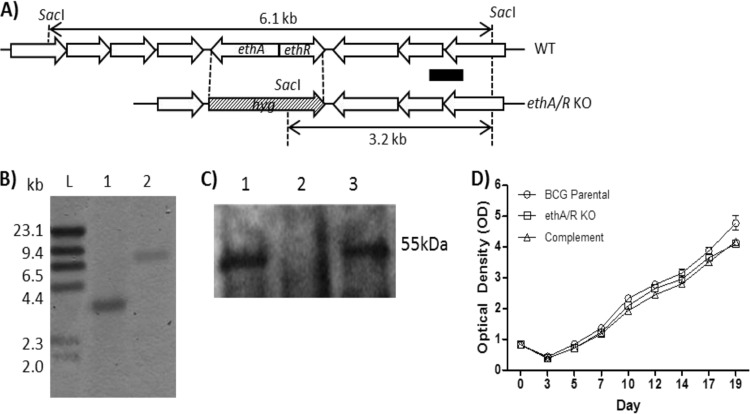
An M. bovis BCG mutant strain with the ethA-ethR locus deleted was constructed by homologous recombination (Fig. 1A). Deletion of the ethA-ethR locus was verified by Southern blotting (Fig. 1B) and Western blot analysis with anti-EthA polyclonal immune serum (Fig. 1C). A complemented strain was also constructed by reintroducing the ethA-ethR locus back into the genome of the ethA-ethR KO mutant with the promoterless integrative plasmid pMV306 (40). Similar parental, ethA-ethR KO mutant, and complemented strain in vitro growth kinetic profiles in liquid culture medium were observed (Fig. 1D), indicating that deletion of the ethA-ethR locus did not impair the general fitness of the mycobacteria.
FIG 1.
Construction of an ethA-ethR KO BCG mutant. (A) Chromosomal organization of ethA-ethR in the M. bovis BCG WT and ethA-ethR KO mutant strains. The arrows depict the lengths and directions of ethA, ethR, and the neighboring genes. The black bar corresponds to the probe used for Southern blot analysis. (B) Southern blot analysis of chromosomal DNA. Lanes: L, DNA molecular size ladder; 1, ethA-ethR KO BCG; 2, WT BCG. (C) Western blot analysis of whole-cell lysate with anti-EthA polyclonal antibodies. Lanes: 1, WT BCG; 2, ethA-ethR KO BCG; 3, complemented ethA-ethR KO BCG strain. (D) Growth kinetics of the WT, KO, and complemented strains in 7H11 medium.
Furthermore, since ethA-ethR is necessary for ETH bioactivation, removal of this locus is expected to lead to an ETH resistance phenotype (15, 16). Consistently, the ETH MIC and MBC for the ethA-ethR KO strain were significantly higher than those for the parental and complemented strains, while the MIC and MBC of INH were similar for all three strains (Table 1).
TABLE 1.
MICs and MBCs of ETH and INH for the parental, ethA-ethR KO, and complemented M. bovis BCG strainsa
| Strain | MIC (µm) of: |
MBC (µm) of: |
||
|---|---|---|---|---|
| ETH | INH | ETH | INH | |
| Parent | 40 | 0.6 | 20 | <0.6 |
| ethA-ethR KO | >80 | 0.6 | >80 | <0.6 |
| Complemented ethA-ethR KO | 40 | 0.6 | 20–40 | <0.6 |
MICs determined by a visual method as reported previously (34). The visual MIC is the lowest drug concentration that completely inhibits the growth of mycobacteria as detected by the unaided eye after 5 days of exposure in a range of INH concentrations of 0.02 to 5 μM and ETH concentrations of 0.3 to 80 μM. After MIC determination, 50 μl of a concentration equal to 1, 2, or 4 times the MIC from the assay plates was plated at appropriate dilutions on 7H11 agar plates to determine the MBC99, the lowest drug concentration required to kill 99% of existing mycobacteria. Plates were incubated at 37°C, and CFU were counted after 14 to 16 days.
The M. bovis BCG ethA-ethR KO strain displays increased virulence in the mouse model.
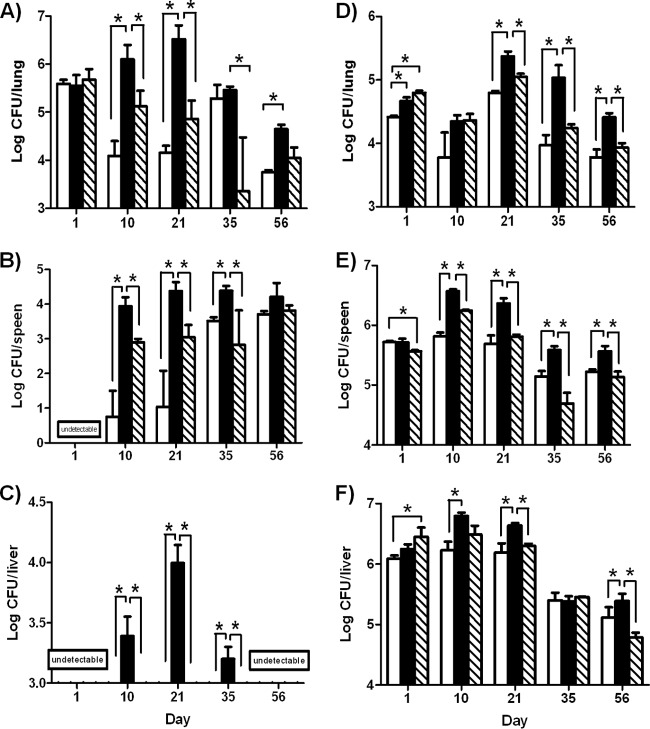
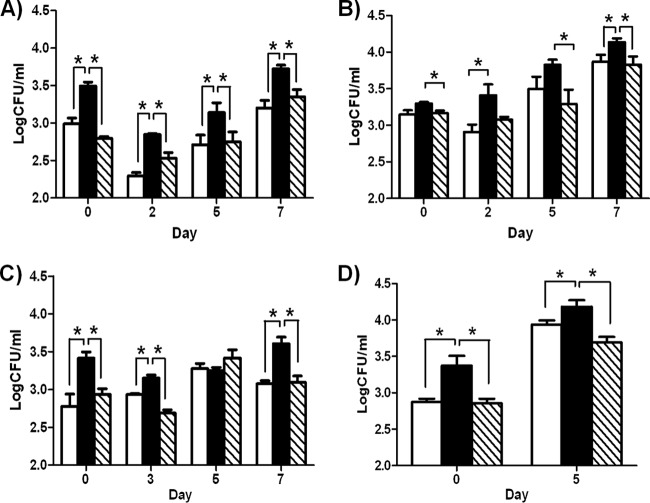
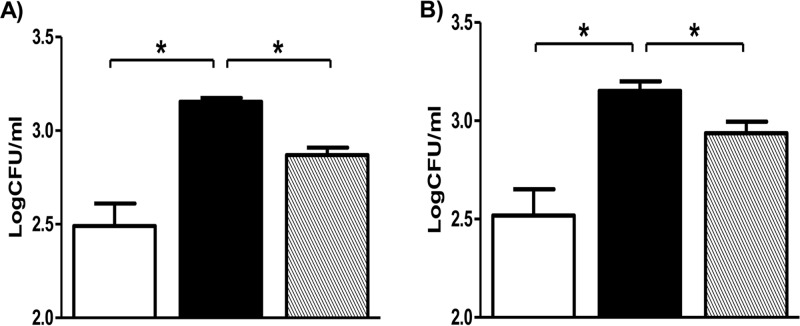
To study the role of the ethA-ethR locus during infection, we monitored the infection profiles of the BCG ethA-ethR KO mutant and its parental and complemented counterparts in the mouse model. Upon nasal administration of comparable inoculums of each strain (data not shown), the bacterial loads in the lungs, spleens, and livers of infected animals were monitored over time. Comparable counts were obtained in the lungs at day 1 postinfection (Fig. 2A). Interestingly, the number of colonies recovered from the animals infected with the ethA-ethR KO strain was several orders of magnitude higher in all of the organs examined than the number of colonies obtained in mice infected with the parental and complemented strains (Fig. 2A to C). The difference between the bacterial loads of the parental and mutant strains was particularly striking in the liver, where only the ethA-ethR KO mutant strain could establish infection and multiply transiently (Fig. 2C). Thus, the infection profiles obtained upon nasal infection indicate that absence of the ethA-ethR locus from M. bovis BCG leads to a more virulent phenotype in vivo.
FIG 2.
Infection profile of the ethA-ethR KO mutant in mice. Adult BALB/c mice were intranasally (A to C) or intravenously (D to F) infected with approximately 5 × 106 CFU of the WT (open bars), ethA-ethR KO (black bars), or complemented (striped bars) M. bovis BCG strain. The bacterial loads in the lungs (A, D), spleens (B, E), and livers (C, F) of the infected mice were monitored. The results are expressed in log10 CFU/ml as the average of four mice per group per time point ± the SD. *, P < 0.05.
The enhanced-virulence phenotype of the ethA-ethR KO mutant seen upon its nasal administration could be due to enhanced colonization or persistence ability and/or to an increased ability of the bacteria to disseminate from the lungs to other systemic organs. To test both hypotheses, infection was performed via the intravenous route, thereby bypassing the extrapulmonary dissemination step. Although less pronounced than for the nasal route of infection, significantly greater bacterial loads were again generally recovered from ethA-ethR KO-infected mice than from mice infected with the WT and complemented strains (Fig. 2D to F).
Together, these results indicate that the ethA-ethR KO mutant strain displays a greater intrinsic ability than the WT strain to persist in murine organs. This suggests that whereas the ethA-ethR locus is not critical at the initial stage of infection (day 1 postinfection), it may play a modulatory role in mycobacterial colonization of and persistence in the lungs, spleen, and liver.
M. bovis BCG ethA-ethR KO mutant displays a greater ability to adhere to mammalian cells.
To further investigate the greater ability of the ethA-ethR KO mutant to colonize mouse organs, its infection profile in various mammalian cells was determined and compared to that of the parental and complemented strains. Human macrophages (THP1), murine BMMOs, human liver cells (Huh7), and human pulmonary epithelial cells (A549) were infected at an MOI of 1, and at the time points indicated, the infected cells were lysed and appropriate dilutions were plated for colony counting. Bacterial inoculums were plated as well and revealed that comparable amounts of bacteria from each strain were added to the cell monolayers (data not shown). Significantly higher counts of KO mutant strain bacteria were obtained across the various cell lines tested and at all of the time points analyzed than of the parental and complemented strain bacteria (Fig. 3). These data thus suggested that the ethA-ethR KO mutant strain displays a greater intrinsic ability to infect mammalian cells. Furthermore and importantly, this phenotype could be observed as early as the first time point postinfection, which corresponds to the initial 45 min of coincubation of bacterial and mammalian cells (Fig. 3). This observation suggested that ethA-ethR KO may display greater adherence properties than its WT counterpart, thereby allowing higher bacterial uptake within host cells. Further analysis of the data obtained with human macrophages and murine BMMOs was performed. First, in order to take into account the variations between the inoculums of the strains (WT, KO, and complemented), the counts obtained at the day 0 time point were expressed as percentages of the respective inoculums. The results clearly indicated a greater percentage of the ethA-ethR KO strain than of the WT and complemented strains, further supporting the greater adherence/uptake of KO bacteria by macrophages (Fig. 4, right panels). Second, in order to evaluate the intracellular survival of each bacterial strain, discounting the initial uptake differences observed, the bacterial counts obtained for days 2, 5, and 7 postinfection were expressed in reference to the respective day 0 counts. The profiles obtained indicated that while there was no significant difference among the KO, WT, and complemented strains in intracellular survival in human macrophages (Fig. 4A, left panel), higher counts of the KO mutant in murine macrophages were obtained at days 2 and 7 postinfection (Fig. 4B, left panel). Together, these observations thus supported the idea that ethA-ethR KO bacteria display a greater ability to adhere to mammalian cells in general and a greater ability to survive intracellularly in murine macrophages but not in human macrophages.
FIG 3.
Infection profile of ethA-ethR KO BCG in mammalian cells. THP-1 human macrophages (A), murine BMMOs (B), Huh-7 human hepatocyte cells (C), and A549 human pulmonary epithelial cells (D) were infected with the WT (open bars), ethA-ethR KO (black bars), or complemented (striped bars) strain at an MOI of 1 (THP-1 cells and BMMOs), 2 (Huh-7 cells), or 3 (A549). At the time points indicated, the cells were washed and lysed and appropriate dilutions were plated for colony counting. The results are expressed in log10 CFU/ml and represent the averages of quadruplicates ± the SD. *, P < 0.05.
FIG 4.
Uptake (left panel) and intracellular survival (right panel) profiles of ethA-ethR KO BCG in macrophages. THP-1 human macrophages (A) and murine BMMOs (B) were infected with the WT (open bars), ethA-ethR KO (black bars), or complemented (striped bars) strain at an MOI of 1 as described in the legend to Fig. 3. (Left panels) Uptake percentages were calculated by expressing day 0 bacterial loads as a percentage of the respective inoculum. (Right panels) Intracellular survival percentages were calculated by expressing counts obtained at the time points indicated as percentages of the day 0 load. The results are expressed in log10 CFU/ml and represent the averages of quadruplicates ± the SD. *, P < 0.05.
To further test the hypothesis of the greater ability of the EthA-EthR KO mutant to adhere to the surface of macrophages, an adherence assay was performed in which mycobacteria were coincubated with BMMOs at 4°C or in the presence of CCD; both conditions are known to prevent cellular uptake (35, 41). Significantly higher counts of the ethA-ethR KO mutant bacteria than of the parental and complemented strain bacteria were obtained (Fig. 5). These data thus support the idea that absence of the ethA-ethR locus confers on mycobacteria a greater ability to adhere to mammalian cells.
FIG 5.
Assays of ethA-ethR KO BCG adherence to macrophages. THP-1 macrophages were infected with the WT (open bars), ethA-ethR KO (black bars), or complemented (striped bars) BCG strain at an MOI of 1. After 45 min of coincubation at 4°C (A) or in the presence of CCD (B), the cells were washed and lysed. Appropriate dilutions of the lysates were plated for colony counting. The results are expressed in log10 CFU/ml as the averages of quadruplicates ± the SD. *, P < 0.05.
EthA affects cell wall MA composition.
The physiological role of membrane-associated EthA is unknown. However, given that BVMOs have been suggested to be involved in MA synthesis and/or degradation (20), we thus hypothesized that the absence of EthA from the ethA-ethR KO mutant may lead to some qualitative and/or quantitative differences in cell wall lipid composition, in particular, MAs, that may account for the greater adherence of the ethA-ethR KO mutant to mammalian cells.
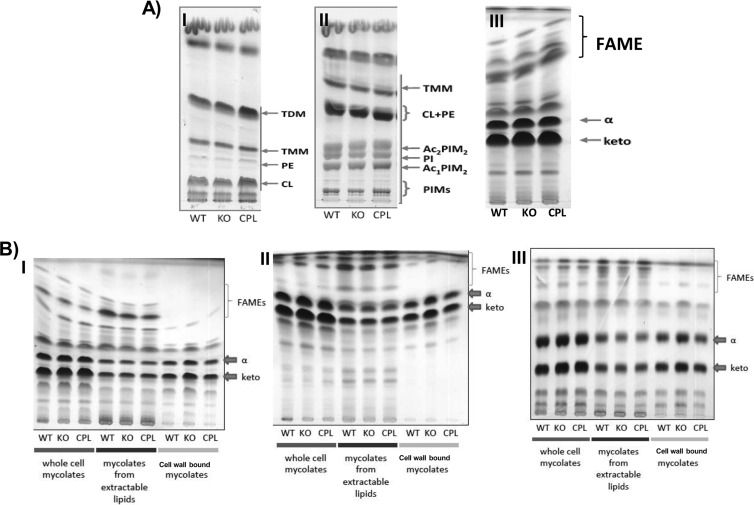
The fatty acid methyl ester (FAME) and MAME compositions of the whole-cell MAs prepared from the WT, ethA-ethR KO, and complemented strains were thus analyzed by TLC. No visible differences in the FAME profiles of the whole-cell lipid esters were observed (Fig. 6A, panel III). Similarly, no significant changes were observed in the total lipid species, including trehalose di-MAs and mono-MAs, phosphatidylethanolamine, phosphatidylinositol, cardiolipin, diacylated and monoacylated phosphatidylinositol dimannosides, or higher phosphatidylinositol mannosides (Fig. 6A, panels I and II). In contrast, a slightly greater signal intensity was consistently seen with both α- and keto-MAME species in the ethA-ethR KO mutant (Fig. 6A, panel III). To further investigate the slightly greater amounts of MAMEs seen in the KO mutant, TLC analysis of whole-cell MAs, MAs prepared from extractable lipids, or cell wall-bound MAs in different solvent systems was performed. Interestingly, the results consistently and convincingly showed greater amounts of cell wall-bound MAMEs in the KO strain than of the WT and complemented strains, whereas no visible difference in the amounts of MAMEs prepared from extractable lipids was observed (Fig. 6B).
FIG 6.
TLC analysis of the lipid composition of the BCG ethA-ethR KO mutant. (A) TLC of whole-cell MAs in different solvents: I, CHCl3-CH3OH-H2O (20:4:0.5); II, CHCl3-CH3OH-NH4OH-H2O (65:25:0.5:4); III, n-hexane–ethyl acetate (95:5). (B) TLC analysis of FAME and MAME prepared from whole cells, extractable lipids, or cell wall in different solvents: I, n-hexane–ethyl acetate (95:5); II, petroleum ether-acetone (90:10); III, dichloromethane on Ag-impregnated plates. The WT, ethA-ethR KO, and complemented (CPL) BCG strains were grown at 37°C in liquid 7H9 medium for 20 days. The bacteria were harvested and processed for total, extractable, and cell wall-bound lipid extraction. Abbreviations: TDM, trehalose di-MA; TMM, trehalose mono-MA; PE, phosphatidylethanolamine; CL, cardiolipin; PI, phosphatidylinositol; Ac2PIM2, diacylated phosphatidylinositol dimannoside; Ac1PIM1, monoacylated phosphatidylinositol dimannoside; PIMs, higher phosphatidylinositol mannosides; α- and keto, α- and keto-MAMEs, respectively.
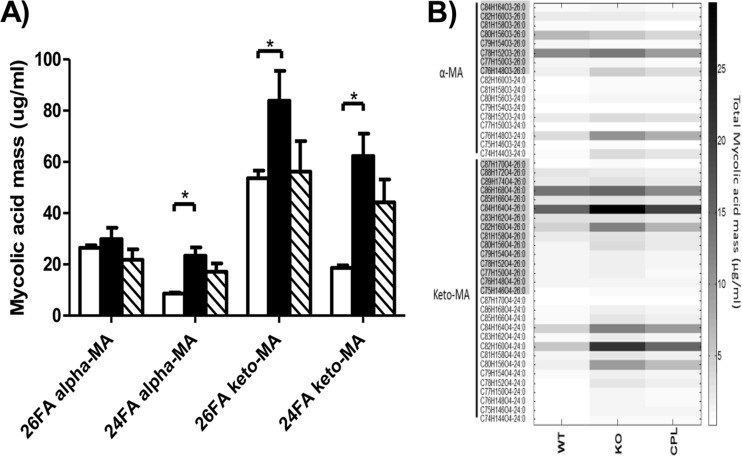
To further analyze the qualitative difference in α- and keto-MAs seen by TLC in the cell wall of the ethA-ethR KO mutant, electrospray ionization-based multiple-reaction monitoring mass spectrometry was performed. This method allows qualitative and relative analyses of various species and subspecies of MAs (39). The total amount of MAs was greater by 83% in the ethA-ethR KO mutant than in the parental strain, and reintroduction of the ethA-ethR locus into the complemented strain reduced the accumulation of MAs back to levels comparable to that in the parental strain (Fig. 7A). In-depth analysis of the various MA subspecies from the ethA-ethR KO mutant revealed significant increases in the overall amounts of the C24:0 α-MA (170% increase over the parental strain), C26:0 keto-MA (56% increase), and C24:0 keto-MA (235% increase) subspecies (Fig. 7A). The quantitative differences of individual C24:0 and C26:0 α- and keto-MA subspecies among the parental, KO, and complemented strains have been heat mapped (Fig. 7B). Together, these data indicated the existence of substantial alterations in the MA profile of the ethA-ethR KO mutant compared to its parental and complemented counterparts, implying that EthA-EthR may be involved in the metabolism of the MA species and subspecies composition in the mycobacterial cell wall.
FIG 7.
Mass spectrometry analysis of MAs. MAs were extracted from mid-log-phase liquid 7H9 medium cultures. Samples were analyzed via a QTRAP 4000 mass spectrometer. (A) Individual sums of C26 α-, C24 α-, C26 keto-, and C24 keto-MA profiles in the WT (open bars), ethA-ethR KO (black bars), and complemented (striped bars) BCG strains. Results are expressed as the averages of quintuplicates ± the SD. *, P < 0.05. (B) Heat map representation of the MA profiles of the WT (n = 5), ethA-ethR KO (n = 5), and complemented (CPL; n = 5) BCG strains. 26FA alpha-MA, C26 α-unit-containing MA; 24FA alpha-MA, C24 α-unit-containing MA; 26FA MeO-MA, C26 methoxy-unit-containing MA; 24FA alpha-MA, C24 methoxy-unit-containing MA; 26FA keto-MA, C26 keto-unit-containing MA; 24FA keto-MA, C24 keto-unit-containing MA. The carbon number indicates the chain length of the product ion.
DISCUSSION
The function of EthA in the mycobacterial cell has never been experimentally addressed, although it was previously proposed to be involved in cell detoxification through toxic ketone removal and/or in MA metabolism (20). Moreover, the importance of this factor in vivo during macrophage or host infection has never been reported on.
In this work, we constructed and characterized an EthA-EthR-deficient M. bovis BCG mutant with the primary aim of studying the physiological role of EthA in pathogenic mycobacteria. As there is no experimental evidence that EthR directly modulates the expression of genes other than ethA and itself (25), we reasoned that the observed phenotypic differences between the parental and ethA-ethR KO strains are very likely attributable to the lack of EthA monooxygenase activity and not to EthR-mediated repression of other unknown target genes, although we cannot completely rule out this remote possibility. Furthermore, while previous studies provided only indirect genetic evidence of the two players involved in ETH bioactivation through overexpression of either ethA or ethR or through ethR deletion in M. bovis BCG (15, 16), we demonstrate here for the first time that deletion of the entire ethA-ethR locus from M. bovis BCG led to ETH resistance, thus further confirming the crucial role of this locus in ETH bioactivation.
Deletion of the ethA-ethR locus from M. bovis BCG resulted in the recovery of greater bacterial loads from mouse organs upon nasal infection, thus supporting a role for the ethA-ethR locus in modulating mycobacterial virulence. Consistently, greater in vitro adherence to mammalian cells was also observed with the ethA-ethR KO mutant, strongly supporting the idea that the greater adherence ability of the ethA-ethR KO mycobacteria translated into greater persistence in murine organs. Interestingly, genetic studies have shown that 40 to 50% of the ETH-resistant clinical isolates harbor mutations in the ethA gene while the rest bear mutations in other genes, such as inhA, for example (42, 43). The possibility that the ethA-mutated, ETH-resistant isolates display greater in vitro adherence properties and enhanced in vivo persistence ability would (at least partially) explain why the ethA locus is the most commonly mutated gene among existing ETH-resistant clinical isolates, as this would confer a selective advantage on these mutants.
TLC analysis revealed greater amounts of cell wall-bound MAs in the ethA-ethR KO mutant than in the parental and complemented strains. In-depth quantitative analysis of the cell wall-bound MAs via mass spectrometry further unveiled differences between the parental and ethA-ethR KO strains in the composition of α- and keto-MAs. MAs not only constitute the major mycobacterial hydrophobic barrier responsible for drug resistance and oxidative stress but have also been shown to play an active role in host-pathogen interactions through host receptor binding (44) and immunomodulatory properties (40, 45). In addition, by controlling the fluidity and hence the outer permeability barrier of mycobacteria, MAs directly control nutrient intake during mycobacterial growth in host tissues (46, 47). Surprisingly, even the most subtle changes in the MA structure have been shown to have profound effects on the physiology and virulence of mycobacteria (47). We thus propose here that the overall greater amounts of keto- and α-MAs in the ethA-ethR KO strain may account for its observed greater ability to adhere to mammalian cells. This working hypothesis is supported by a previous study where the absence of keto-MAs was found to lead to profound alterations in envelope permeability and to an attenuated phenotype in mice (46, 48). Conversely, here, a significant increase in the abundance of both α- and keto-MAs correlated with a hypervirulence phenotype in mice. While the cell wall permeability of the ethA-ethR KO mutant strain has yet to be investigated, our data support the idea that the altered MA profile has potentially modified the ability of the mycobacterial cell wall to interact with the mammalian cell surface.
Fraaije and colleagues previously proposed that the oxidative activity of EthA and other mycobacterial BVMOs may help the pathogen survive oxidative stress conditions encountered in vivo (20). The authors also speculated that EthA may contribute to detoxification activity through the removal of toxic ketones in mycobacteria. However, we found that EthA deletion neither impaired the in vitro general fitness of the mycobacteria nor attenuated the infection capabilities of the mutant in macrophages but instead enhanced its adherence properties and in vivo persistence. Alternatively, BVMOs in general have been shown to be involved in specific metabolic processes through the conversion of relatively hydrophobic substances such as MAs, although our knowledge of MA metabolism still remains somewhat fragmentary (49). The altered MA profile in the ethA-ethR KO strain implies dysregulation of either the MA synthesis pathway or the MA degradation pathway, resulting in greater accumulation of long-chain C24:0 and C26:0 α- and keto-MAs. Furthermore, as keto-MA overproduction appears to be more pronounced in the ethA-ethR KO mutant than the change in α-MA levels, one could also speculate that EthA plays a metabolic role by oxidizing keto-MAs to yield wax ester MAs, which have been shown to be the result of a Baeyer-Villiger reaction on the keto group of keto-MAs (50, 51).
In conclusion, the work presented here suggests that the ethA-ethR locus is involved in the composition of cell wall MAs in M. bovis BCG, specifically, the relative amounts of α- and keto-MAs, which impact the ability of mycobacteria to adhere to mammalian cells ex vivo and their ability to colonize their host.
ACKNOWLEDGMENTS
We gratefully thank Pablo Bifani, Manjunatha Ujjini (Novartis Institute for Tropical Diseases, Singapore), Thomas Dick (National University of Singapore), and Alain Baulard (Institut Pasteur, Lille, France) for their critical and constructive comments throughout this work.
This work was supported by the National Medical Research Council of Singapore (IRG grant IRG07nov088 to S.A.) and by the Slovak Research and Development Agency (contract APVV-0441-10). We have no conflict of interest to declare.
Footnotes
Published ahead of print 24 February 2014
REFERENCES
- 1.WHO. 2011. Global tuberculosis control: WHO report 2011 World Health Organization, Geneva, Switzerland [Google Scholar]
- 2.Comas I, Gagneux S. 2009. The past and future of tuberculosis research. PLoS Pathog. 5:e1000600. 10.1371/journal.ppat.1000600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Farmer P, Bayona J, Becerra M, Furin J, Henry C, Hiatt H, Kim JY, Mitnick C, Nardell E, Shin S. 1998. The dilemma of MDR TB in the global era. Int. J. Tuberc. Lung Dis. 2:869–876 [PubMed] [Google Scholar]
- 4.Espinal MA, Laszlo A, Simonsen L, Boulahbal F, Kim SJ, Reniero A, Hoffner S, Rieder HL, Binkin N, Dye C, Williams R, Raviglione MC. 2001. Global trends in resistance to antituberculosis drugs. World Health Organization-International Union against Tuberculosis and Lung Disease Working Group on Anti-Tuberculosis Drug Resistance Surveillance. N. Engl. J. Med. 344:1294–1303. 10.1056/NEJM200104263441706 [DOI] [PubMed] [Google Scholar]
- 5.Barnes PF, Bloch AB, Davidson PT, Snider DE., Jr 1991. Tuberculosis in patients with human immunodeficiency virus infection. N. Engl. J. Med. 324:1644–1650. 10.1056/NEJM199106063242307 [DOI] [PubMed] [Google Scholar]
- 6.Snider DE, Jr, Roper WL. 1992. The new tuberculosis. N. Engl. J. Med. 326:703–705. 10.1056/NEJM199203053261011 [DOI] [PubMed] [Google Scholar]
- 7.Prasad R. 2012. Multidrug and extensively drug-resistant tuberculosis management: evidences and controversies. Lung India 29:154–159. 10.4103/0970-2113.95321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wells CD, Cegielski JP, Nelson LJ, Laserson KF, Holtz TH, Finlay A, Castro KG, Weyer K. 2007. HIV infection and multidrug-resistant tuberculosis: the perfect storm. J. Infect. Dis. 196(Suppl 1):S86–S107. 10.1086/518665 [DOI] [PubMed] [Google Scholar]
- 9.Cox H, Kebede Y, Allamuratova S, Ismailov G, Davletmuratova Z, Byrnes G, Stone C, Niemann S, Rusch-Gerdes S, Blok L, Doshetov D. 2006. Tuberculosis recurrence and mortality after successful treatment: impact of drug resistance. PLoS Med. 3:e384. 10.1371/journal.pmed.0030384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Crofton J, Chaulet P, Maher D, Grosset J, Harris W, Norman H, Iseman M, Watt B. 1997. Guidelines for the management of multidrug-resistant tuberculosis. World Health Organization, Geneva, Switzerland [Google Scholar]
- 11.Schwartz WS. 1966. Comparison of ethionamide with isoniazid in original treatment cases of pulmonary tuberculosis. XIV. A report of the Veterans Administration-Armed Forces cooperative study. Am. Rev. Respir. Dis. 93:685–692 [DOI] [PubMed] [Google Scholar]
- 12.Jenner PJ, Smith SE. 1987. Plasma levels of ethionamide and prothionamide in a volunteer following intravenous and oral dosages. Lepr. Rev. 58:31–37 [DOI] [PubMed] [Google Scholar]
- 13.Quémard A, Laneelle G, Lacave C. 1992. Mycolic acid synthesis: a target for ethionamide in mycobacteria? Antimicrob. Agents Chemother. 36:1316–1321. 10.1128/AAC.36.6.1316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Banerjee A, Dubnau E, Quémard A, Balasubramanian V, Um KS, Wilson T, Collins D, de Lisle G, Jacobs WR., Jr 1994. inhA, a gene encoding a target for isoniazid and ethionamide in Mycobacterium tuberculosis. Science 263:227–230. 10.1126/science.8284673 [DOI] [PubMed] [Google Scholar]
- 15.DeBarber AE, Mdluli K, Bosman M, Bekker LG, Barry CE., III 2000. Ethionamide activation and sensitivity in multidrug-resistant Mycobacterium tuberculosis. Proc. Natl. Acad. Sci. U. S. A. 97:9677–9682. 10.1073/pnas.97.17.9677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Baulard AR, Betts JC, Engohang-Ndong J, Quan S, McAdam RA, Brennan PJ, Locht C, Besra GS. 2000. Activation of the pro-drug ethionamide is regulated in mycobacteria. J. Biol. Chem. 275:28326–28331. 10.1074/jbc.M003744200 [DOI] [PubMed] [Google Scholar]
- 17.Dover LG, Alahari A, Gratraud P, Gomes JM, Bhowruth V, Reynolds RC, Besra GS, Kremer L. 2007. EthA, a common activator of thiocarbamide-containing drugs acting on different mycobacterial targets. Antimicrob. Agents Chemother. 51:1055–1063. 10.1128/AAC.01063-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Korduláková J, Janin YL, Liav A, Barilone N, Dos Vultos T, Rauzier J, Brennan PJ, Gicquel B, Jackson M. 2007. Isoxyl activation is required for bacteriostatic activity against Mycobacterium tuberculosis. Antimicrob. Agents Chemother. 51:3824–3829. 10.1128/AAC.00433-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Vannelli TA, Dykman A, Ortiz de Montellano PR. 2002. The antituberculosis drug ethionamide is activated by a flavoprotein monooxygenase. J. Biol. Chem. 277:12824–12829. 10.1074/jbc.M110751200 [DOI] [PubMed] [Google Scholar]
- 20.Fraaije MW, Kamerbeek NM, Heidekamp AJ, Fortin R, Janssen DB. 2004. The prodrug activator EtaA from Mycobacterium tuberculosis is a Baeyer-Villiger monooxygenase. J. Biol. Chem. 279:3354–3360. 10.1074/jbc.M307770200 [DOI] [PubMed] [Google Scholar]
- 21.Hanoulle X, Wieruszeski JM, Rousselot-Pailley P, Landrieu I, Baulard AR, Lippens G. 2005. Monitoring of the ethionamide pro-drug activation in mycobacteria by (1)H high resolution magic angle spinning NMR. Biochem. Biophys. Res. Commun. 331:452–458. 10.1016/j.bbrc.2005.03.197 [DOI] [PubMed] [Google Scholar]
- 22.Hanoulle X, Wieruszeski JM, Rousselot-Pailley P, Landrieu I, Locht C, Lippens G, Baulard AR. 2006. Selective intracellular accumulation of the major metabolite issued from the activation of the prodrug ethionamide in mycobacteria. J. Antimicrob. Chemother. 58:768–772. 10.1093/jac/dkl332 [DOI] [PubMed] [Google Scholar]
- 23.Betts JC, Lukey PT, Robb LC, McAdam RA, Duncan K. 2002. Evaluation of a nutrient starvation model of Mycobacterium tuberculosis persistence by gene and protein expression profiling. Mol. Microbiol. 43:717–731. 10.1046/j.1365-2958.2002.02779.x [DOI] [PubMed] [Google Scholar]
- 24.Rodriguez GM, Voskuil MI, Gold B, Schoolnik GK, Smith I. 2002. ideR, an essential gene in Mycobacterium tuberculosis: role of IdeR in iron-dependent gene expression, iron metabolism, and oxidative stress response. Infect. Immun. 70:3371–3381. 10.1128/IAI.70.7.3371-3381.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Engohang-Ndong J, Baillat D, Aumercier M, Bellefontaine F, Besra GS, Locht C, Baulard AR. 2004. EthR, a repressor of the TetR/CamR family implicated in ethionamide resistance in mycobacteria, octamerizes cooperatively on its operator. Mol. Microbiol. 51:175–188. 10.1046/j.1365-2958.2003.03809.x [DOI] [PubMed] [Google Scholar]
- 26.Frenois F, Baulard AR, Villeret V. 2006. Insights into mechanisms of induction and ligands recognition in the transcriptional repressor EthR from Mycobacterium tuberculosis. Tuberculosis (Edinb.) 86:110–114. 10.1016/j.tube.2005.07.005 [DOI] [PubMed] [Google Scholar]
- 27.Willand N, Dirié B, Carette X, Bifani P, Singhal A, Desroses M, Leroux F, Willery E, Mathys V, Déprez-Poulain R, Delcroix G, Frénois F, Aumercier M, Locht C, Villeret V, Déprez B, Baulard AR. 2009. Synthetic EthR inhibitors boost antituberculous activity of ethionamide. Nat. Med. 15:537–544. 10.1038/nm.1950 [DOI] [PubMed] [Google Scholar]
- 28.Flipo M, Desroses M, Lecat-Guillet N, Dirié B, Carette X, Leroux F, Piveteau C, Demirkaya F, Lens Z, Rucktooa P, Villeret V, Christophe T, Jeon HK, Locht C, Brodin P, Déprez B, Baulard AR, Willand N. 2011. Ethionamide boosters: synthesis, biological activity, and structure-activity relationships of a series of 1,2,4-oxadiazole EthR inhibitors. J. Med. Chem. 54:2994–3010. 10.1021/jm200076a [DOI] [PubMed] [Google Scholar]
- 29.Flipo M, Desroses M, Lecat-Guillet N, Villemagne B, Blondiaux N, Leroux F, Piveteau C, Mathys V, Flament MP, Siepmann J, Villeret V, Wohlkönig A, Wintjens R, Soror SH, Christophe T, Jeon HK, Locht C, Brodin P, Déprez B, Baulard AR, Willand N. 2012. Ethionamide boosters. 2. Combining bioisosteric replacement and structure-based drug design to solve pharmacokinetic issues in a series of potent 1,2,4-oxadiazole EthR inhibitors. J. Med. Chem. 55:68–83. 10.1021/jm200825u [DOI] [PubMed] [Google Scholar]
- 30.Bardarov S, Bardarov S, Jr, Pavelka MS, Jr, Sambandamurthy V, Larsen M, Tufariello J, Chan J, Hatfull G, Jacobs WR., Jr 2002. Specialized transduction: an efficient method for generating marked and unmarked targeted gene disruptions in Mycobacterium tuberculosis, M. bovis BCG and M. smegmatis. Microbiology 148:3007–3017 [DOI] [PubMed] [Google Scholar]
- 31.Parish T, Stoker NG. 2000. Use of a flexible cassette method to generate a double unmarked Mycobacterium tuberculosis tlyA plcABC mutant by gene replacement. Microbiology 146(Part 8):1969–1975 [DOI] [PubMed] [Google Scholar]
- 32.Stover CK, de la Cruz VF, Fuerst TR, Burlein JE, Benson LA, Bennett LT, Bansal GP, Young JF, Lee MH, Hatfull GF. 1991. New use of BCG for recombinant vaccines. Nature 351:456–460. 10.1038/351456a0 [DOI] [PubMed] [Google Scholar]
- 33.Sambrook J, Russell DW. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 34.Nozawa RT, Yokota T. 1983. Rapid drug susceptibility testing of mycobacteria in tissue culture medium. Antimicrob. Agents Chemother. 24:268–272. 10.1128/AAC.24.2.268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Balaji KN, Boom WH. 1998. Processing of Mycobacterium tuberculosis bacilli by human monocytes for CD4+ alphabeta and gammadelta T cells: role of particulate antigen. Infect. Immun. 66:98–106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Grzegorzewicz AE, Pham H, Gundi VA, Scherman MS, North EJ, Hess T, Jones V, Gruppo V, Born SE, Korduláková J, Chavadi SS, Morisseau C, Lenaerts AJ, Lee RE, McNeil MR, Jackson M. 2012. Inhibition of mycolic acid transport across the Mycobacterium tuberculosis plasma membrane. Nat. Chem. Biol. 8:334–341. 10.1038/nchembio.794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Stadthagen G, Korduláková J, Griffin R, Constant P, Bottová I, Barilone N, Gicquel B, Daffé M, Jackson M. 2005. p-Hydroxybenzoic acid synthesis in Mycobacterium tuberculosis. J. Biol. Chem. 280:40699–40706. 10.1074/jbc.M508332200 [DOI] [PubMed] [Google Scholar]
- 38.Folch J, Lees M, Sloane Stanley GH. 1957. A simple method for the isolation and purification of total lipides from animal tissues. J. Biol. Chem. 226:497–509 [PubMed] [Google Scholar]
- 39.Shui G, Bendt AK, Pethe K, Dick T, Wenk MR. 2007. Sensitive profiling of chemically diverse bioactive lipids. J. Lipid Res. 48:1976–1984. 10.1194/jlr.M700060-JLR200 [DOI] [PubMed] [Google Scholar]
- 40.Vander Beken S, Al Dulayymi JR, Naessens T, Koza G, Maza-Iglesias M, Rowles R, Theunissen C, De Medts J, Lanckacker E, Baird MS, Grooten J. 2011. Molecular structure of the Mycobacterium tuberculosis virulence factor, mycolic acid, determines the elicited inflammatory pattern. Eur. J. Immunol. 41:450–460. 10.1002/eji.201040719 [DOI] [PubMed] [Google Scholar]
- 41.Neo Y, Li R, Howe J, Hoo R, Pant A, Ho S, Alonso S. 2010. Evidence for an intact polysaccharide capsule in Bordetella pertussis. Microbes Infect. 12:238–245. 10.1016/j.micinf.2009.12.002 [DOI] [PubMed] [Google Scholar]
- 42.Morlock GP, Metchock B, Sikes D, Crawford JT, Cooksey RC. 2003. ethA, inhA, and katG loci of ethionamide-resistant clinical Mycobacterium tuberculosis isolates. Antimicrob. Agents Chemother. 47:3799–3805. 10.1128/AAC.47.12.3799-3805.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Boonaiam S, Chaiprasert A, Prammananan T, Leechawengwongs M. 2010. Genotypic analysis of genes associated with isoniazid and ethionamide resistance in MDR TB isolates from Thailand. Clin. Microbiol. Infect. 16:396–399. 10.1111/j.1469-0691.2009.02838.x [DOI] [PubMed] [Google Scholar]
- 44.Nuzzo I, Galdiero M, Bentivoglio C, Galdiero R, Romano Carratelli C. 2002. Apoptosis modulation by mycolic acid, tuberculostearic acid and trehalose 6,6′-dimycolate. J. Infect. 44:229–235. 10.1053/jinf.2002.1001 [DOI] [PubMed] [Google Scholar]
- 45.Riley LW. 2006. Of mice, men, and elephants: Mycobacterium tuberculosis cell envelope lipids and pathogenesis. J. Clin. Invest. 116:1475–1478. 10.1172/JCI28734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Dubnau E, Chan J, Raynaud C, Mohan VP, Lanéelle MA, Yu K, Quémard A, Smith I, Daffé M. 2000. Oxygenated mycolic acids are necessary for virulence of Mycobacterium tuberculosis in mice. Mol. Microbiol. 36:630–637. 10.1046/j.1365-2958.2000.01882.x [DOI] [PubMed] [Google Scholar]
- 47.Liu J, Barry CE, Besra GS, Nikaido H. 1996. Mycolic acid structure determines the fluidity of the mycobacterial cell wall. J. Biol. Chem. 271:29545–29551. 10.1074/jbc.271.47.29545 [DOI] [PubMed] [Google Scholar]
- 48.Glickman MS, Cox JS, Jacobs WR. 2000. A novel mycolic acid cyclopropane synthetase is required for cording, persistence, and virulence of Mycobacterium tuberculosis. Mol. Cell 5:717–727. 10.1016/S1097-2765(00)80250-6 [DOI] [PubMed] [Google Scholar]
- 49.Asselineau C, Asselineau J, Lanéelle G, Lanéelle MA. 2002. The biosynthesis of mycolic acids by mycobacteria: current and alternative hypotheses. Prog. Lipid Res. 41:501–523. 10.1016/S0163-7827(02)00008-5 [DOI] [PubMed] [Google Scholar]
- 50.Toriyama S. 1982. Biosynthesis and regulation of cell wall mycolic acid in mycobacteria. Kekkaku 57:279–294 (In Japanese.) [PubMed] [Google Scholar]
- 51.Etemadi AH, Lederer E. 1965. On the structure of the alpha-mycolic acids of the human test strain of Mycobacterium tuberculosis. Bull. Soc. Chim. Fr. 9:2640–2645 (In French.) [PubMed] [Google Scholar]