Abstract
Streptococcus mutans is often cited as the main bacterial pathogen in dental caries, particularly in early-childhood caries (ECC). S. mutans may not act alone; Candida albicans cells are frequently detected along with heavy infection by S. mutans in plaque biofilms from ECC-affected children. It remains to be elucidated whether this association is involved in the enhancement of biofilm virulence. We showed that the ability of these organisms together to form biofilms is enhanced in vitro and in vivo. The presence of C. albicans augments the production of exopolysaccharides (EPS), such that cospecies biofilms accrue more biomass and harbor more viable S. mutans cells than single-species biofilms. The resulting 3-dimensional biofilm architecture displays sizeable S. mutans microcolonies surrounded by fungal cells, which are enmeshed in a dense EPS-rich matrix. Using a rodent model, we explored the implications of this cross-kingdom interaction for the pathogenesis of dental caries. Coinfected animals displayed higher levels of infection and microbial carriage within plaque biofilms than animals infected with either species alone. Furthermore, coinfection synergistically enhanced biofilm virulence, leading to aggressive onset of the disease with rampant carious lesions. Our in vitro data also revealed that glucosyltransferase-derived EPS is a key mediator of cospecies biofilm development and that coexistence with C. albicans induces the expression of virulence genes in S. mutans (e.g., gtfB, fabM). We also found that Candida-derived β1,3-glucans contribute to the EPS matrix structure, while fungal mannan and β-glucan provide sites for GtfB binding and activity. Altogether, we demonstrate a novel mutualistic bacterium-fungus relationship that occurs at a clinically relevant site to amplify the severity of a ubiquitous infectious disease.
INTRODUCTION
Biofilms often contribute to and/or cause disease in humans (1). In the United States and worldwide, dental caries is the single-most common and costly biofilm-dependent oral infectious disease, which continues to compromise the health and well-being of children and adults alike (2). Furthermore, the prevalence of dental caries, particularly early-childhood caries (ECC), is increasing among preschool children (2). ECC is a hypervirulent form of the disease that is characterized by a heavy Streptococcus mutans burden (often exceeding 30% of the cultivable plaque biofilm flora) (3, 4), accompanied by protracted feeding of dietary sugars, especially sucrose (5). The child is often allowed to consume sugary beverages almost constantly from a nursing bottle. The adverse effects of sugars are enhanced by the mechanical effects of the nipple on the bottle, which restricts the access of buffering saliva to the tooth surfaces (6, 7).
Streptococcus mutans has often been regarded as one of the key etiologic agents of ECC (3, 4, 8, 9), although other organisms may also contribute to its pathogenesis (9–11). S. mutans cells can rapidly orchestrate the formation of cariogenic plaque biofilms on susceptible tooth surfaces when they are exposed frequently to dietary sucrose. Sucrose is utilized by S. mutans-derived enzymes (e.g., glucosyltransferases [Gtfs]) to produce exopolysaccharides (EPS), the prime building blocks of cariogenic biofilms (12). The Gtfs are secreted into the extracellular milieu, become constituents of the pellicle that covers teeth, and are also adsorbed to bacterial surfaces while retaining enzymatic activity (12–14). Glucan synthesis on the pellicle provides additional nonmammalian bacterial binding sites (e.g., through membrane-associated glucan-binding proteins in S. mutans), while the polymers on the surfaces of resident microorganisms increase the cohesion between organisms (12–14). As a consequence, a structured community forms, which is enmeshed in an EPS-rich matrix that is diffusion limiting (12, 15). At the same time, S. mutans and other acidogenic/aciduric organisms produce acids as by-products of sugar metabolism, creating acidic microenvironments within the biofilm that further select for the growth of these organisms (12, 15–19). Low pH values present at the biofilm-tooth interface promote the dissolution of adjacent tooth enamel, leading to the clinical onset of cavitation.
The onset and progression of carious lesions in children with ECC is rapid and aggressive, resulting in rampant destruction of the smooth surfaces of the teeth (3, 4, 8, 20, 21). The underlying biological reasons for the development of ECC remain unclear. Microbiological studies of plaque biofilms from children with ECC reveal that in addition to high levels of S. mutans, the common opportunistic fungal pathogen Candida albicans is also frequently detected; in contrast, it is detected sporadically, if at all, in the plaque of ECC-free children (22–24). Why C. albicans is found together with high levels of S. mutans in plaque biofilms and whether this bacterium-fungus association at sites of ECC infection plays a significant role in the pathogenesis of ECC remain to be elucidated.
Bacterium-fungus interactions occur commonly in humans and may influence the transition from a healthy to a diseased state within a specific host niche (25, 26). C. albicans is by far the most commonly detected fungal organism on human mucosal surfaces, and it often participates in the formation of polymicrobial biofilms on soft tissue and acrylic surfaces (26, 27). Candida coadheres with many oral commensal species, namely, viridans group streptococci (e.g., Streptococcus gordonii, Streptococcus oralis), in vitro (28–30) and enhances fungal carriage and infectivity in mucosal diseases in vivo (31). Yet Candida was initially regarded as having little to no physical adhesion with S. mutans in the absence of sucrose (30). However, when sucrose is present, the adhesive interaction between these two organisms is enhanced (32–34). Images derived from electron microscopy revealed extracellular material that had formed between cocci and yeast cells, suggesting that locally produced glucans play a role in mediating their coadherence (32, 34). We have determined that all three S. mutans Gtf exoenzymes bind to the surfaces of C. albicans cells in vitro; GtfB shows the greatest affinity (12, 35). When sucrose is available, the Gtfs adsorbed onto C. albicans cells produce large amounts of glucan on the fungal surface. These glucans formed in situ provide enhanced binding sites for S. mutans while simultaneously enhancing fungal adhesion to saliva-coated hydroxyapatite surfaces (35). Here we explore whether this sucrose-dependent cross-kingdom interaction modulates cospecies biofilm development and/or influences the infectivity and the pathogenesis of dental caries in vivo.
We hypothesize that S. mutans-C. albicans associations may enhance S. mutans infection and modulate the development of hypervirulent biofilms on tooth surfaces, which will, in turn, influence the onset and severity of dental caries in vivo. We demonstrate that the presence of C. albicans enhances the assembly of the EPS-rich matrix, such that cospecies biofilms accrue more biomass and more viable S. mutans cells than single-species biofilms in vitro. More importantly, coinfection of rats with S. mutans and C. albicans enhances the colonization and carriage of both organisms in vivo and dramatically amplifies the virulence of plaque biofilms formed on rodent dentition, leading to the development of rampant carious lesions. Furthermore, our in vitro data reveal plausible explanations for the enhanced ability of these organisms to form virulent cospecies biofilms. Altogether, we demonstrate that a novel synergistic interaction occurs between an opportunistic fungus and a bacterial pathogen on a clinically relevant site (teeth) in the mouth. Our data offer new insight into the microbiological and clinical features of ECC and may have relevance for other bacterium-fungus interactions associated with polymicrobial infections in humans (25, 27).
(Part of this paper was presented at the 91st General Session of the International Association for Dental Research, Seattle, WA, 20 to 23 March 2013 [36]).
MATERIALS AND METHODS
In vitro biofilm model.
Streptococcus mutans strain UA159 serotype c (a proven virulent cariogenic bacterial pathogen selected for genome sequencing) and Candida albicans SC5314 (a well-characterized strain whose genome has been sequenced) were used to generate single or cospecies biofilms. Biofilms were formed using our saliva-coated hydroxyapatite (sHA) disc model as described previously (15, 37). The hydroxyapatite discs (surface area, 2.7 ± 0.2 cm2; Clarkson Chromatography Products, Inc., South Williamsport, PA) coated with filter-sterilized, clarified whole saliva (15) were vertically suspended in 24-well plates using a custom-made wire disc holder, which was designed to mimic the free smooth surfaces of the teeth (15, 37). For single-species biofilms, each disc was inoculated with approximately 2 × 106 CFU of S. mutans/ml in ultrafiltered (10-kDa cutoff; Millipore, Billerica, MA) tryptone-yeast extract (UFYTE) broth containing 1% (30 mM) sucrose at 37°C under 5% CO2. For cospecies biofilms, approximately 2 × 104 CFU of C. albicans/ml (containing predominantly yeast cell forms [35]) was also added to the inoculum; the proportion of the microorganisms in the inoculum is similar to that found in saliva samples from children with ECC. During the first 18 h, the organisms were grown undisturbed so as to allow initial biofilm formation; the culture medium was then changed twice daily at 8 a.m. and 6 p.m. until the end of the experimental period (42 h). The pH of the culture medium was measured daily at each medium change.
Quantitative biofilm analysis.
The development of each of the biofilms was assessed at 18 and 42 h postinoculation using our well-established protocols optimized for biofilm imaging and quantification (15, 37, 38). The sequential assembly of the matrix was followed by incorporating an Alexa Fluor 647-labeled dextran conjugate (10 kDa; absorbance/fluorescence emission maxima, 647/668 nm; Molecular Probes, Invitrogen Corp., Carlsbad, CA) into the glucans synthesized during the assembly of the EPS matrix (37, 38). The total microbial biomass was stained with Syto 9 (485/498 nm; Molecular Probes) (15, 37). Imaging was performed using an Olympus FV 1000 two-photon laser scanning microscope (Olympus, Tokyo, Japan) equipped with a 10× (numerical aperture, 0.45) water immersion objective lens. The excitation wavelength was 810 nm, and the emission wavelength filter for Syto 9 was a 495/540 OlyMPFC1 filter, while the filter for Alexa Fluor 647 was an HQ655/40M-2P filter (37). Each biofilm was scanned at 5 positions randomly selected on the microscope stage (39), and confocal image series (512- by 512-pixel resolution) were generated by optical sectioning at each of these positions. At least 3 independent biofilm experiments were performed. The confocal images were analyzed using software for the quantitation of EPS and microbial cells within intact biofilms (15, 37). COMSTAT (available at http://www.imageanalysis.dk) was used to calculate the biomass, as well as the number and size of microcolonies (15). Furthermore, a separate set of biofilms was used for standard microbiological analysis. The biofilms were homogenized by sonication, and the number of viable cells (total number of CFU per biofilm) was determined as described elsewhere (40). It should be noted that CFU data for C. albicans have limitations given the morphology, since the cells exist in both the yeast and hyphal forms; hyphae are essentially multicellular structures that, when plated, form a single CFU, despite having a larger biomass than yeast forms.
Visualization of 3D biofilm architecture.
We examined the spatial distribution of individual microbial species and the EPS matrix within intact biofilms at 4, 6, 8, 18, and 42 h (15, 37, 38). Because Syto 9 (as well as other commercially available nucleic acid stains) labels both S. mutans and C. albicans, we used a green fluorescent protein (GFP)-expressing strain of S. mutans (constructed from S. mutans strain UA159) to resolve these species in cospecies biofilms. The GFP-expressing strain was a gift from Jose Lemos (Center for Oral Biology, University of Rochester Medical Center, Rochester, NY). Based on work from other groups (41, 42), we elected to stain the yeast using concanavalin A (ConA) lectin conjugated with tetramethylrhodamine (absorbance/fluorescence emission maxima, 555/580 nm; Molecular Probes). We used a concentration of 40 μg/ml with an incubation time of 30 min to reduce the possibility that ConA might bind S. mutans, since ConA is capable of agglutinating several serotypes of S. mutans, but not serotype c strains (43). Prior to three-color imaging, we determined that our concentration and incubation time were sufficient to label the C. albicans cells within the biofilm without cross-reacting with S. mutans (44). The EPS matrix was labeled with Alexa Fluor 647, as described above. Imaging was again performed using the Olympus FV 1000 laser scanning microscope equipped with a 25× LPlan N (numerical aperture, 1.05) water immersion objective lens. The excitation wavelength was 780 nm, and the emission wavelength filter for GFP was a 495/540 OlyMPFC1 filter, while an HQ655/40M-2P and a 598/28 OlyMPFC2 filter were used for Alexa Fluor 647 and rhodamine, respectively. In order to visualize all three fluorophores, the channels were scanned in the following pairs: (i) GFP and rhodamine and (ii) rhodamine and Alexa Fluor 647. Images at a 1,024- by 1,024-pixel resolution were collected and analyzed using software for the simultaneous visualization of EPS and each of the microbial cells within intact biofilms (15). Amira software (version 5.4.1; Visage Imaging, San Diego, CA) was used to create 3-dimensional (3D) renderings of each biofilm structural component (EPS and microorganisms) by combining the GFP and rhodamine channels from scan 1 with the Alexa Fluor 647 channel from scan 2.
Labeling of β-glucan in biofilms.
Cospecies biofilms were formed using S. mutans UA159 and C. albicans SC5314 as described above. Our protocols are optimized and limited to three-color confocal imaging. Thus, we examined the presence of C. albicans-derived β-glucan in cospecies biofilms in two ways. First, we investigated the spatial distribution of the EPS matrix, C. albicans cells, and β-glucan. The EPS matrix was labeled with Alexa Fluor 647, while C. albicans was stained with ConA conjugated with rhodamine. β-Glucan produced by C. albicans was stained using a commercially available mouse monoclonal IgG antibody to 1,3-β-glucan (Biosupplies Australia Pty. Ltd., Victoria, Australia) paired with a fluorescently labeled secondary antibody. All antibody staining steps were performed in the dark at 4°C. The primary antibody was diluted 1:20 in phosphate-buffered saline (PBS; pH 7.0) and was incubated with the biofilm for 60 min. The biofilm was then washed in fresh PBS and was blocked with 3% bovine serum albumin (Sigma-Aldrich, St. Louis, MO) for 15 min. The biofilm was again washed in fresh PBS and was incubated for 30 min with the Fab′ fragment of a goat-anti-mouse IgG antibody conjugated to Alexa Fluor 488 (absorbance/fluorescence emission maxima, 488/519 nm; Molecular Probes) at a concentration of 4 mg/ml. The biofilm was finally washed in 0.89% NaCl and was then imaged using the Olympus FV 1000 microscope equipped with a 25× LPlan N (numerical aperture, 1.05) objective as described in the preceding section. In a separate set of experiments, we determined the spatial distribution of S. mutans, C. albicans, and β-glucan within biofilms. We used our GFP-expressing strain of S. mutans, while C. albicans was labeled with ConA-tetramethylrhodamine (Molecular Probes). β-Glucan was labeled via the anti-β-glucan antibody with a secondary antibody conjugated to Alexa Fluor 647 (Molecular Probes). Images at a 1,024- by 1,024-pixel resolution were collected and were analyzed using Amira software, version 5.4.1.
Cospecies biofilm formation using gtf-defective S. mutans strains.
Knockout mutant strains of S. mutans with insertions in gtfB, gtfC, or both (ΔgtfB::kan, ΔgtfC::kan, and ΔgtfBC::kan) (15, 37) were cultured with C. albicans SC5314 to form cospecies biofilms. The gtfB-, gtfC-, and gtfBC-null mutants, constructed from the parental S. mutans UA159 strain, were kindly provided by Robert A. Burne (Department of Oral Biology, University of Florida, Gainesville, FL). The mutant strains were generated by standard allelic replacement with a nonpolar kanamycin resistance marker and were verified by DNA sequence and biochemical analysis (for Gtf production); the lack of polarity of the mutations was also verified by reverse transcription–quantitative PCR (RT-qPCR). The mutant strains were also devoid of any growth or growth rate defects (relative to the growth of UA159). Biofilms formed with the parental S. mutans strain, UA159, were compared to those formed with the ΔgtfB::kan, ΔgtfC::kan, or ΔgtfBC::kan mutant at 42 h postinoculation. The 3D architecture of cospecies biofilms was determined using confocal microscopy as described above. We used an anti-S. mutans antibody conjugated to Alexa Fluor 488 (Molecular Probes) to label the mutant strain (which does not express GFP) within biofilms as described previously (45). The antibody was provided courtesy of Robert Palmer and John Cisar at the National Institutes for Health, Bethesda, MD. In addition, we also determined (in a separate experiment) the total number of viable microbial cells in each of the cospecies biofilms (40).
Purification of biofilm RNAs.
RNA was extracted and purified using protocols optimized for biofilms formed in vitro (46). Briefly, disc sets were incubated in RNALater (Applied Biosystems/Ambion, Austin, TX), and the biofilm material was removed from the sHA discs. The RNAs were purified and were treated with DNase on a column using the Qiagen RNeasy Micro kit (Qiagen, Valencia, CA). The RNAs were then subjected to a second DNase I treatment with Turbo DNase (Applied Biosystems/Ambion) and were purified using the Qiagen RNeasy MinElute cleanup kit (Qiagen). The RNAs were quantified using the NanoDrop ND-1000 spectrophotometer (Thermo Scientific/NanoDrop, Wilmington, DE). RNA quality was evaluated using an Agilent 2100 bioanalyzer (Agilent Technologies Inc., Santa Clara, CA), and all RNAs used to prepare cDNAs were determined to have RNA integrity numbers (RIN) of 9.6 and above.
RT-qPCR analysis for selected genes.
We performed RT-qPCR to measure the expression profiles of specific genes directly associated with extracellular polysaccharide matrix development and acid stress survival, including gtfB, gtfC, gtfD, fruA, dexA, fabM, and atpD. Briefly, cDNAs were synthesized using 0.5 μg of purified RNA and the Bio-Rad iScript cDNA synthesis kit (Bio-Rad Laboratories, Inc., Hercules, CA). To check for DNA contamination, purified total RNA without reverse transcriptase served as a negative control. The resulting cDNAs were amplified with a Bio-Rad CFX96 system (Bio-Rad Laboratories, Inc., Hercules, CA) using previously published specific primers and TaqMan probes (15, 47). A standard curve was plotted for each primer set, as described elsewhere (48). The standard curves were used to transform the critical threshold cycle (CT) values to relative numbers of cDNA molecules. Comparative expression was calculated by normalizing each gene of interest to the 16S rRNA signal (48).
In vivo model of dental caries.
Animal experiments were performed as described previously with some modifications (7, 49, 50). Six litters of 8 female Sprague Dawley rats aged 15 days were purchased with their dams from Harlan Laboratories (Madison, WI). Upon arrival, animals were screened for S. mutans and C. albicans, and were determined not to be infected with either organism, by plating oral swabs on selective media: ChromAgar (VWR International LLC, Radnor, PA) for C. albicans and Mitis Salivarius Agar plus Bacitracin (MSB) for S. mutans. Half of the dams, while still nursing, were infected by mouth with an actively growing culture of S. mutans UA159, which is transmitted to the pups (49); these were maintained separately from uninfected animals. Pups in cages with S. mutans-infected dams were also directly infected with S. mutans. At weaning, pups aged 21 days were checked and confirmed for S. mutans infection, while the other half of the animals remained uninfected. At the age of 23 days, all pups were subjected to surgical hyposalivation (49). After recovery, all pups slated for infection with C. albicans were inoculated with an actively growing culture of C. albicans SC5314 at the ages of 24 and 25 days, and their infections were confirmed at 26 days. All the animals were randomly placed into 1 of the following 4 groups: (i) S. mutans infected, (ii) C. albicans infected, (iii) S. mutans plus C. albicans infected, and (iv) uninfected. Animals were screened at 26, 28, and 30 days for S. mutans and C. albicans infection. Each of the infected groups was confirmed for its respective microbial infection, while the uninfected group remained free of either S. mutans or C. albicans; no cross-contamination was observed throughout the experiment. All animals were provided the National Institutes of Health cariogenic diet 2000 (51) and 5% sucrose water ad libitum. The experiment proceeded for 2 weeks. At the end of 2 weeks, the animals were sacrificed. The jaws were aseptically dissected and were processed for microbiological analysis of each animal's plaque biofilms as described by Klein et al. (52). For microbiological analysis, the left jaws were sonicated in 5 ml of 154 mM sterile NaCl solution for plaque biofilm removal. The suspensions obtained were serially diluted and were plated on MSB or Inhibitory Mold Agar (a less costly alternative to ChromAgar, yet equally selective, as determined experimentally) to estimate the S. mutans or C. albicans population, respectively, and on blood agar to determine the total cultivable flora in the plaque biofilms (49). All the jaws were defleshed, and the teeth were prepared for caries scoring according to Larson's modification of Keyes' system (53).
Ethics statement.
All animal experiments were performed in strict accordance with the guidelines of the Animal Welfare Act of the United States, under protocols reviewed and approved by the Institutional Animal Care and Use Committee of the University of Rochester (approved protocol 2011-040).
Statistical analysis.
The data were analyzed by pairwise comparisons of multiple groups with regression models using the ranked values. Kruskal-Wallis tests, which are nonparametric and are based on ranks, were used for two-group comparisons. The significance level was set at 5%, and no adjustments were made for multiple comparisons. For the animal study, an analysis of outcome measures was done with transformed values of the measures in order to stabilize variances as detailed by Raubertas et al. (54). The data were then subjected to statistical analyses as described above. Furthermore, we also examined the possibility of a synergistic effect of coinfection on the development of carious lesions. To determine whether there is a synergistic interaction, the effects in dually infected animals were compared with the sums of the effects in singly infected animals by using estimates and statistical tests of the differences from regression models. SAS statistical software, version 9.3 (SAS Institute, Cary, NC), was used to perform the analyses.
RESULTS
Temporal development and 3D architecture of cospecies biofilms.
We examined how the EPS-mediated bacterium-fungus interactions observed in the presence of sucrose (35) influence biofilm formation on the saliva-coated hydroxyapatite (sHA) surface. We focused on the development of the biofilm architecture in 3 dimensions (3D) using our protocols optimized for the imaging and quantification of Gtf-derived EPS and microbial cells within intact biofilms (12, 15, 39). The presence of both C. albicans and S. mutans dramatically enhances the assembly of an EPS-rich matrix, leading to the development of biofilms that are larger and thicker than those formed by either species alone (Fig. 1A). Although C. albicans alone binds sporadically to an sHA surface in the presence of sucrose, it lacks the capacity to form biofilms in vitro under our experimental conditions (data not shown).
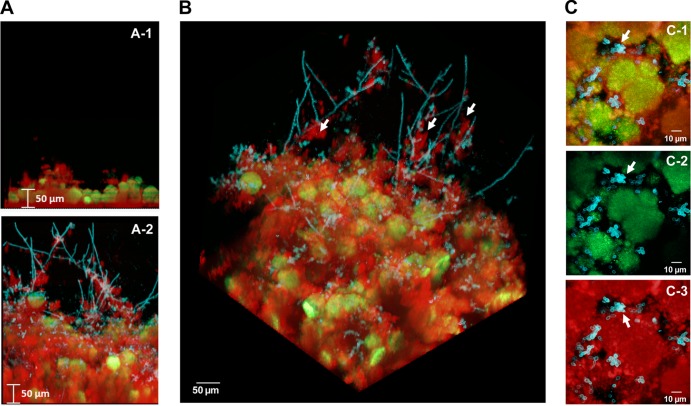
FIG 1.
Three-dimensional architecture of the cospecies biofilm. Representative images of single-species and cospecies biofilms grown for 42 h are shown. Bacterial microcolonies expressing GFP appear green, while fungal cells labeled with ConA-tetramethylrhodamine appear blue. EPS labeled with Alexa Fluor 647-dextran appear red. (A) Orthogonal views of the biofilms, illustrating the overall differences in the accumulation of biofilms between cospecies and S. mutans single-species biofilms. (B) Three-dimensional rendering of the cospecies biofilm, illustrating the complexity of its architecture. Bacterial microcolonies and yeast forms of C. albicans are enmeshed and surrounded by an EPS-rich matrix, while hyphae (indicated by white arrows) extend from the biofilm into the fluid phase and are coated with EPS. (C) Projection images of the first 20 μm (from the surface of attachment) of a cospecies biofilm, illustrating the spatial relationship between C. albicans, S. mutans, and EPS. (C-1) Merged image of all three components; (C-2) merged image of C. albicans and S. mutans; (C-3) C. albicans and EPS. The arrows indicate that there is little to no direct association between S. mutans and C. albicans (C-1 and C-2). In contrast, it is readily apparent that the fungal cells are associated with EPS (C-3), which then contacts the microcolonies (C-1).
We observed, by means of confocal microscopy, localized accumulation of EPS on the saliva-coated apatitic surface and the presence of small clusters of S. mutans cells (associated with EPS) as early as 6 h (data not shown). In contrast, C. albicans was not detected in the biofilm until 8 h postinoculation. At this time point, the initial polymeric matrix and bacterial microcolonies were detected, while yeast forms could be consistently observed throughout the biofilm (see Fig. S1A in the supplemental material). At 8 h, the appearance of pseudohyphal or hyphal forms was infrequent (see Fig. S1A). At 18 h, the EPS matrix and microbial biomass had developed further, while microcolonies of S. mutans and hyphal forms of C. albicans appeared with greater prevalence (see Fig. S1B in the supplemental material). After 42 h, the size of the biofilm had increased, revealing large microcolonies (which form as the initial microcolonies merge), abundant fungal cells (both yeast and hyphal), and the presence of an EPS-rich matrix (Fig. 1 and Table 1).
TABLE 1.
Quantitative analysis of biofilmsa
| Biofilm | Biovolume (μm3/μm2) |
Microcolonies |
|||
|---|---|---|---|---|---|
| Total | EPS | Cells | No. per total biovolume | Size (×103 μm3) | |
| At 18 h | |||||
| S. mutans alone | 35.6 ± 8.9 | 24.3 ± 7.5 | 11.5 ± 2.6 | 16.3 ± 7.7 | 3.6 ± 0.6 |
| Cospecies | 66.5 ± 21.5* | 47.2 ± 18.6* | 19.3 ± 5.8* | 33.8 ± 6.0* | 8.8 ± 5.5* |
| At 42 h | |||||
| S. mutans alone | 118.2 ± 13.4 | 84.3 ± 12.2 | 35.5 ± 5.9 | 19.4 ± 11.0 | 10.7 ± 9.3 |
| Cospecies | 251.8 ± 60.9* | 191.8 ± 60.6* | 60.2 ± 8.3* | 12.1 ± 6.8 | 30.1 ± 20.5* |
Quantitative analysis of biomass (both EPS and total microbial cells) and microcolonies within intact cells was performed using COMSTAT. The data are mean values ± standard deviations (n, ≥15) from at least 3 independent experiments. Asterisks indicate that the values for S. mutans and cospecies biofilms are significantly different from each other (P, <0.01).
The resulting 3D architecture of mature cospecies biofilms is highly intricate (Fig. 1B). Both yeast and hyphal cells were detected, along with sizable microcolonies, which were enmeshed in and surrounded by EPS. The hyphae extended out from the biofilm into the surrounding medium and were coated with EPS (Fig. 1B, white arrows). In contrast, yeast cells tended to cluster near the surface of the biofilm attachment and were closely associated with the EPS surrounding bacterial microcolonies. We were mostly unable to detect yeast and bacterial cells associated with one another without glucan as the intermediary (Fig. 1C, arrows), which is in line with a lack of cell-cell binding in the absence of sucrose, as observed previously (30, 35). Furthermore, neither yeast cells nor hyphal cells are found within the microcolony structures formed by S. mutans; rather, they are associated with the periphery. These observations could be a product of sequential assembly of the biofilm, where the colonization of yeast cells (and later differentiation into hyphae) occurs after the initial EPS is formed on sHA and the basic microcolony structure has been initiated. However, it is also possible that competitive interactions may occur locally between these organisms (55, 56), which could potentially explain their spatial relationship and physical proximity in the biofilm.
The prevalence of EPS-coated hyphae prompted us to investigate the ability of purified GtfB to bind to and produce glucan in situ on hyphal cells. We had demonstrated previously that GtfB binds to yeast cells in an active form, but we had not determined its ability to interact with hyphal cells (35). Despite the differences in size and membrane composition between yeast and hyphal cells (57), we found that the enzyme was equally efficient in producing glucans when adsorbed to either cell type (see Fig. S2 in the supplemental material); these results demonstrate that GtfB attached to C. albicans can enhance EPS-rich matrix production in the absence of bacteria. Our data also show that GtfB binds in an active form to mannan and β-1,3-glucan (see Fig. S3 and Protocol S1 in the supplemental material), which are present in the cell walls of both yeast and hyphal forms (57). Mannan is located at the outermost layer of the Candida cell wall (57). Although β-1,3-glucan is present close to the inner cell wall, it can be secreted extracellularly (58) (see below). It is possible that GtfB may bind to one or more of the carbohydrate components, such as glucose (12). However, the exact location and/or structure of the Gtf binding sites remains to be identified.
Clearly, C. albicans provides an abundance of binding sites for Gtfs derived from S. mutans; the fungus is converted to a de facto glucan producer when exposed to sucrose. Thus, the formation of copious amounts of EPS on the large surface areas of C. albicans cells facilitates the assembly of a dense and abundant EPS-rich matrix in cospecies biofilms.
Enhanced microbial carriage in cospecies biofilms.
The interactions of bacterial and fungal cells led to the development of cospecies biofilms on the sHA surface, which contained more EPS and microbial biomass than S. mutans single-species biofilms (Table 1). Furthermore, cospecies biofilms displayed more (at the early stage of 18 h) and larger (at 42 h) microcolonies than S. mutans-only biofilms (P, <0.05). Cross-sectional imaging analysis of cospecies biofilms revealed that microcolonies were composed of densely packed S. mutans cells alone, while C. albicans cells were located around the microcolonies. The enhanced microcolony development corresponded to a nearly 2.5-fold increase in EPS accumulation in cospecies biofilms relative to biofilms with S. mutans alone (Table 1). This observation agrees with earlier reports that the amount of EPS present in the biofilm directly affects the formation and size of the microcolony, since Gtf-derived glucans help to cluster the bacterial cells (15, 39).
Analyses of the viable population also revealed that there was a dramatic increase (∼6-fold increase) (Fig. 2) in the number of S. mutans cells in cospecies biofilms (versus single-species biofilms), revealing a connection between enhanced microcolony formation and carriage of S. mutans. In parallel, viable C. albicans cells were also detected in high numbers within cospecies biofilms (>107 CFU/biofilm), while the number of fungal cells in single-species biofilms was too low to count in our model. The elevated populations of S. mutans and C. albicans in the biofilms are consistent with published results for humans (22–24) and our recent microbiological analyses of plaque biofilm samples from children with ECC (data not shown). However, we recognize the limitations of C. albicans viable count data given the fungal morphology, since hyphal development would increase biomass but not necessarily CFU.
FIG 2.
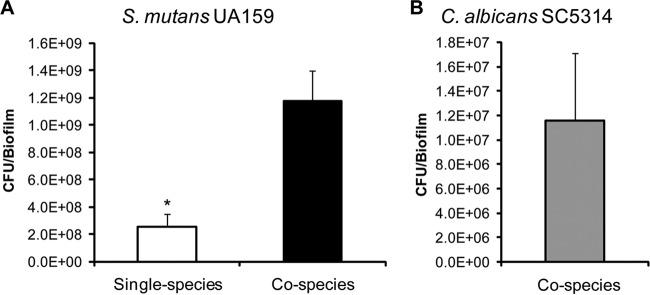
Microbial populations in the cospecies biofilm. Shown are the total viable counts (CFU) of S. mutans in single-species and cospecies biofilms (A) and of C. albicans in cospecies biofilms (B), grown for 42 h. C. albicans alone lacked the capacity to form biofilms under our experimental conditions. The data are mean values ± standard deviations (n = 16). (A) The asterisk indicates that the values for single-species and cospecies biofilms are significantly different from each other (P, <0.05). There is a dramatic increase in the number of S. mutans CFU in cospecies biofilms (∼6-fold increase). (B) There is a large number of viable C. albicans cells in cospecies biofilms (∼107 CFU/biofilm).
The pH values of the medium surrounding cospecies biofilms were highly acidic (final pH ranging from 4.5 to 4.7), as monitored during biofilm development (data not shown). However, the pH values were similar to those for single-species S. mutans biofilms at all stages of development, despite the differences in microcolony size and bacterial density, although we did not measure the pH within the biofilm.
S. mutans-C. albicans interactions enhance the virulence of plaque biofilms in vivo.
The data from our in vitro biofilm studies indicate that the infectivity and virulence of cospecies biofilms may be enhanced in vivo. Thus, we sought to determine whether the association of S. mutans and C. albicans influences the onset of dental caries by using a rodent model. We used hyposalivatory rats, which were provided a high-sucrose diet and sugared water ad libitum. The protracted feeding of sugars, coupled with the restricted access of saliva to teeth, used in our model mimics the severe conditions experienced clinically by children afflicted with ECC (3, 5, 6, 8, 20). The animals were readily infected with S. mutans, C. albicans, or both using our model, and then the impact on the development of carious lesions was assessed for each experimental condition.
Coinfection with S. mutans and C. albicans in vivo produced dramatic effects on both the level of microbial colonization and the development of carious lesions. We detected significant increases in the viable populations of both S. mutans (>3-fold increase) and C. albicans (>20-fold increase) in plaque biofilms from coinfected animals over those from animals infected with either species alone (Table 2). This observation is consistent with the data from our in vitro investigation. The uninfected animals remained free of infection by S. mutans and/or C. albicans.
TABLE 2.
Viable microbial populations in animals' plaque biofilms
| Group | Microbial population (CFU/jaw) |
||
|---|---|---|---|
| S. mutans | C. albicans | Total flora (107) | |
| Coinfected | (6.1 ± 1.3) × 106 | (2.9 ± 1.2) × 105 | 2.4 ± 0.2 |
| Infected with S. mutans UA159 alone | (1.9 ± 0.3) × 166* | — | 1.6 ± 0.1 |
| Infected with C. albicans SC5314 alone | — | (1.0 ± 0.3) × 104* | 2.7 ± 0.4 |
| Uninfected | — | — | 1.3 ± 0.2 |
Data are mean viable populations of S. mutans, C. albicans, and total flora ± standard deviations (n = 11). —, not detected. Asterisks indicate that the values for the two infection groups are significantly different from each other (P, <0.05). The values for total flora do not differ significantly for the different infection groups (P, >0.05).
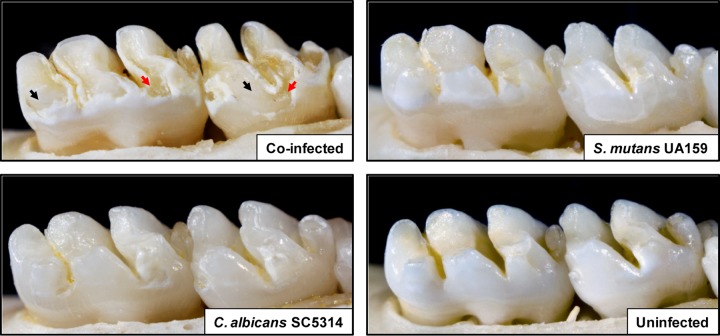
More importantly, there was a rapid onset of severe carious lesions on the smooth surfaces of teeth from coinfected animals, characterized by large areas of enamel destruction where the dentin was exposed (Fig. 3, black arrows). Areas where the dentin is eroded or missing (red arrows) indicate the most severe carious lesions. We also determined that there was a significant increase in the severity of disease at all stages of lesion development (initial, moderate, and extensive) in coinfected rats over that for animals infected with either organism alone (P, <0.05) (Fig. 4). Furthermore, there is a synergistic interaction in coinfected rats, such that the total effect on the severity of the lesions (at moderate and extensive stages) is greater than the sum of the effects of infection with each organism (single infection) (P, <0.001). The initial lesions were also more numerous in dually infected animals, but the effects were not synergistic (P, >0.05).
FIG 3.
Images of teeth from rats infected with S. mutans UA159 and/or C. albicans SC5314, or left uninfected, after 2 weeks. Photographs of lower molars in the rodent jaws are shown; jaws representing the average result have been selected. For the coinfected animal, black arrows indicate moderate to severe carious lesions where areas of the enamel are missing, exposing the underlying dentin. In some areas, the dentin is eroded or missing (red arrows), indicating the most severe carious lesions. In the S. mutans-infected animal, large areas of initial lesions were detected, although they were visibly less severe than those of coinfected animals. In the C. albicans-infected animal, small areas of demineralization and initial lesions were observed. In the uninfected animal, overt carious lesions are absent, while “white spots” (very early lesions) begin to appear in some localized areas.
FIG 4.
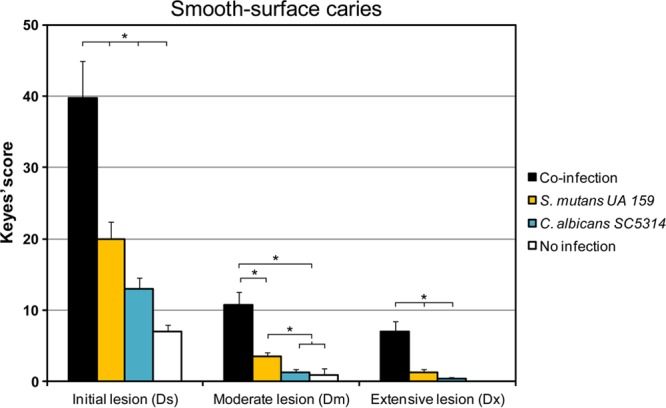
Smooth-surface carious lesions in singly infected, coinfected, or uninfected animals. Smooth-surface caries scores are presented as mean values ± standard deviations (n = 11). Scores are recorded as stages of carious lesion severity according to Larson's modification of Keyes' scoring system: Ds, initial lesion (surface enamel white, broken, and/or dry); Dm, moderate lesion (dentin exposed); Dx, extensive lesion (dentin soft or missing). Asterisks indicate that the values for different experimental groups are significantly different from each other (P, <0.05).
C. albicans alone was only moderately cariogenic, which increased the initial severity of smooth-surface caries, resulting in small carious lesions (compared to an uninfected control). Infection with S. mutans alone produced more lesions of greater severity than infection with C. albicans alone (P, <0.05). These results contrast somewhat with those reported by Klinke et al. (59), who noted that C. albicans alone did not induce the formation of smooth-surface caries. This apparent discrepancy may be related to differences in the animal model, since their animals were subjected to a lesser cariogenic challenge and were fed ampicillin, which could affect microbial colonization and biofilm development on the smooth surfaces of the teeth (59). In our study, uninfected (control) animals developed some small areas of demineralization with negligible severity.
All the animals developed sulcal-surface lesions (Fig. 5), although no differences in the number and severity of lesions were detected among the groups (P, >0.05), except for the uninfected animals, which had significantly fewer severe lesions than the coinfected and C. albicans-infected groups (P, <0.05). Although statistically significant, this difference is slight and may have limited biological significance. Nevertheless, the observation that C. albicans can induce sulcal-surface lesions in our model is consistent with a previous report from Klinke et al. (59).
FIG 5.
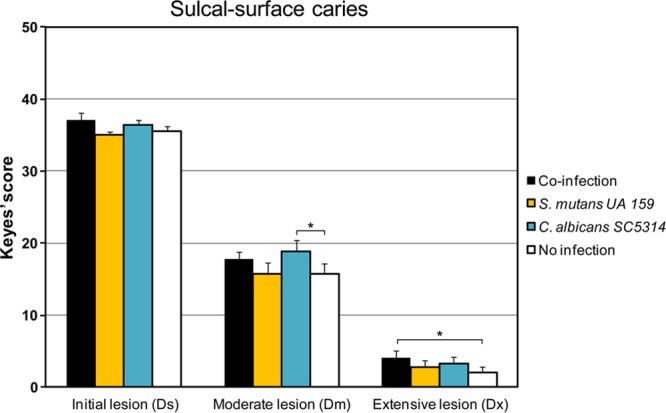
Sulcal-surface carious lesions in singly infected, coinfected, or uninfected animals. Sulcal-surface caries scores are presented as mean values ± standard deviations (n = 11). Scores are recorded as stages of carious lesion severity according to Larson's modification of Keyes' scoring system: Ds, initial lesion (surface enamel white, broken and/or dry); Dm, moderate lesion (dentin exposed); Dx, extensive lesion (dentin soft or missing). Asterisks indicate that the values for different experimental groups are significantly different from each other (P, <0.05).
Taken together, our data demonstrate that increased colonization with both species and the subsequent interactions between these species result in the establishment of hypervirulent biofilms on the smooth surfaces of the animals' dentition. We have conducted additional in vitro studies to further elucidate the possible mechanisms for the enhanced microbial carriage/coexistence and the ability of these organisms to form cospecies biofilms.
Gtf-derived EPS production modulates cospecies biofilm assembly.
Glucans formed by S. mutans GtfB (and, to a lesser extent, GtfC) on the C. albicans cell surface appear to be critical for bacterial-fungal coadhesion (35). We hypothesized that the expression of Gtfs is essential for the enhanced microbial carriage and establishment of cospecies biofilms. Therefore, we assessed the abilities of S. mutans strains lacking the gtf genes to form cospecies biofilms compared to that of the parental strain, UA159. The Δgtf mutants are well described and well characterized (with no polar mutations or growth defects) in the published literature (15, 37).
Our results reveal that all biofilms formed with mutant strains defective in one or more gtf genes harbor significantly fewer viable S. mutans and C. albicans cells than the parental strain after 42 h of growth (Fig. 6). The development of cospecies biofilms using a ΔgtfBC::kan double mutant was the most impaired; these biofilms were populated by the fewest microbial cells. Biofilms formed with either single mutant were less severely impaired. It should also be noted that the numbers of S. mutans cells present in cospecies biofilms formed with the mutants were either lower than or similar to those in single-species biofilms. This observation suggests that significant impairment of glucan production may remove the advantage of enhanced carriage conferred by cohabitating with C. albicans.
FIG 6.
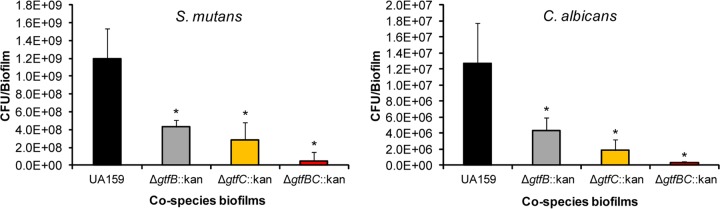
Viable counts in cospecies biofilms formed with Δgtf::kan mutant strains of S. mutans UA159. Shown are the total viable counts of S. mutans and C. albicans in 42-h cospecies biofilms formed with C. albicans SC5314 and one of the following S. mutans strains: the parental strain, UA159 (black bars), the ΔgtfB::kan mutant (gray bars), the ΔgtfC::kan mutant (orange bars), or the ΔgtfBC::kan mutant (red bars). The data are mean values ± standard deviations (n, ≥22). All cospecies biofilms formed with any of the three mutant strains contained significantly fewer viable counts of S. mutans and C. albicans than those formed in the presence of UA159 (*, P < 0.05).
The use of mutant strains of S. mutans also affected the 3D architecture of cospecies biofilms (compared to those formed with the parental strain, UA159), as shown in Fig. 7. We determined that the ΔgtfBC::kan mutant was unable to form true biofilms, and the structures formed were essentially devoid of Gtf-derived EPS; only random clusters of S. mutans cells could be observed, and minimal numbers of C. albicans cells were detected. The ΔgtfB::kan and ΔgtfC::kan mutants were also defective in their abilities to form cospecies biofilms, as evidenced by the fact that the 3D architecture of the biofilms was dramatically altered. The ΔgtfB::kan mutant formed fairly homogenous and flat biofilms that were devoid of any microcolonies and contained visibly less EPS, fewer yeast cells, and almost no hyphae. The presence of the ΔgtfC::kan mutant also altered the overall biofilm architecture, although the changes were less dramatic than those observed in cospecies biofilms with the ΔgtfB::kan mutant. Small and loosely adherent microcolonies were formed that were easily sheared from sHA discs during medium changes; these biofilms appeared to contain less EPS and fewer fungal cells than cospecies biofilms formed with the parental strain, UA159. Furthermore, these observed alterations are indeed linked to a defect in glucan synthesis, since supplementation with a purified GtfB enzyme helps to restore the cospecies biofilm phenotype/architecture in the presence of the ΔgtfB::kan mutant (see Fig. S4 in the supplemental material).
FIG 7.
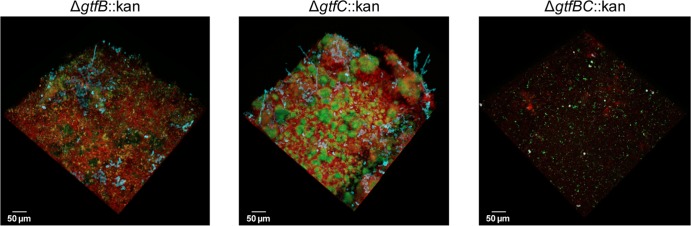
Architecture of cospecies biofilms formed with Δgtf mutants. Shown are representative images of the architectures of cospecies biofilms formed by each of the Δgtf::kan mutant strains (at 42 h). Cospecies biofilms formed by the parental strain, UA159 (image not displayed; refer to Fig. 1), were always included (as a control) for comparison. Overall, biofilms formed with the ΔgtfB::kan mutant were thin and flat; they were devoid of microcolony structures and contained few yeast cells and almost no hyphae. The presence of the ΔgtfC::kan mutant strain also altered the overall architecture of the cospecies biofilms, which contained small microcolonies, few fungal cells, and largely defective EPS-rich matrix production. The ΔgtfBC::kan mutant strain was virtually incapable of forming cospecies biofilms with C. albicans.
Although the diminished and unstable structure of the biofilms formed with each of the mutant strains prevented accurate quantification of biofilms, it is readily apparent that altering the amount and type of Gtf-derived EPS significantly impacts the architecture of cospecies biofilms.
C. albicans also contributes to the biofilm matrix.
Although S. mutans-derived EPS appears to be an articulation point between the two species, it is conceivable that C. albicans may contribute its own extracellular substances that help mediate this interaction. C. albicans alone produces matrix materials (β-glucans, chitin, β-N-acetylglucosamine) during biofilm formation on other surfaces, and these appear to confer protection from antifungal agents (58, 60–62). Results from previous biochemical studies reveal that β-glucans are likely among the major constituents of the matrices of C. albicans biofilms, although these substances largely have not been visualized within the intact matrix (63–65). The presence of β-glucans was sought within the matrices of our cospecies biofilms by means of a β-glucan-specific antibody labeled with a fluorescent secondary antibody, which was visualized via confocal imaging (66).
β-Glucan is a component of the cell wall and can also be actively secreted by C. albicans during biofilm formation on silicone or polystyrene surfaces (58, 60, 62). Confocal images of labeled β-glucan and Gtf-derived glucan (α-glucan), along with the C. albicans cells, are shown in Fig. 8. We observed that cospecies biofilms contained β-glucan (Fig. 8A) and that this glucan is found associated with C. albicans cells throughout the biofilm, as well as interspersed in areas where Gtf-derived EPS is also often present (Fig. 8A-1 and A-2). To confirm that β-glucan was accumulating within the biofilm and that the antibody was not simply labeling C. albicans cell-associated β-glucan, we also examined the spatial distribution of β-glucan, S. mutans, and C. albicans cells. We found that antibody-labeled β-glucan is closely associated with the bacterial microcolonies, and it does not appear visually to be in a 1:1 ratio with the C. albicans cells present (Fig. 8B).
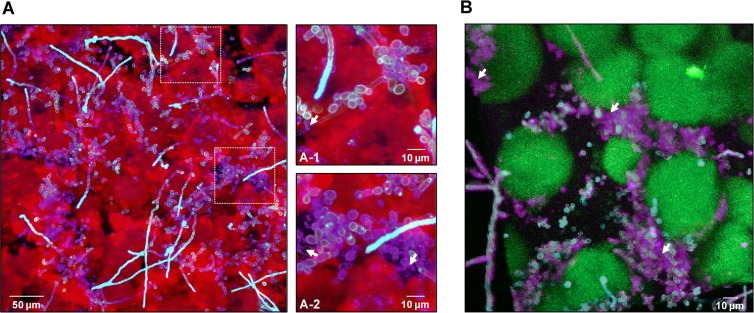
FIG 8.
Visualization and spatial distribution of β-glucan within cospecies biofilms. (A) Projection image of 42-h cospecies biofilms labeled with an anti-β-glucan antibody (purple), Alexa Fluor 647-dextran (EPS) (red), and ConA-tetramethylrhodamine (C. albicans cells) (blue). The image shows the presence of β-glucan (purple) within the biofilm, while the arrows in the closeup images of selected areas indicate punctate accumulations of β-glucan (A-1) that appear to be localized extracellularly (A-2). (B) Three-dimensional projection of a separate 42-h cospecies biofilm labeled with the anti-β-glucan antibody (purple), GFP (S. mutans cells) (green), and ConA-tetramethylrhodamine (C. albicans cells) (blue). The arrows indicate extracellular accumulations of β-glucan that appear to enmesh the C. albicans cells. Clearly, β-glucan can be found intercalated between C. albicans cells and S. mutans microcolonies, potentially having a structural role.
Furthermore, punctate accumulations of β-glucan were found in the extracellular milieu, away from the cells. Some C. albicans cells were partially labeled, while non-cell wall-associated β-glucan accumulated among the microcolonies, intercalating between the fungal cells and microcolonies (Fig. 8B). We have found that GtfB binds in an active form to β-1,3-glucan (see Fig. S3 in the supplemental material), which could explain why β-glucan is closely associated with both C. albicans cells and the Gtf-derived α-glucan produced by S. mutans. Our observations reveal that β-glucan contributes to the structural organization of the extracellular matrix in cospecies biofilms and may play a functional role yet to be elucidated.
We have also explored whether the expression of C. albicans properties associated with biofilm formation influences the development of cospecies biofilms with S. mutans. Initially, we examined C. albicans bcr1Δ/Δ and efg1Δ/Δ mutants (the bcr1 and efg1 genes are associated with the formation of single-species biofilms on catheters or acrylic surfaces in vivo [58, 67]). Analysis using the bcr1Δ/Δ or efg1Δ/Δ deletion mutant cocultured with S. mutans UA159 showed that the overall capacity to form cospecies biofilms was largely unaffected (see Fig. S5 in the supplemental material). However, we observed noticeable changes in the overall 3D architecture of cospecies biofilms with the efg1Δ/Δ mutant (from that with the parental strain), in that the mutant biofilms were devoid of hyphal cells and displayed a less-developed EPS matrix; although the CFU counts were similar, the lack of hyphae in efg1Δ/Δ cospecies biofilms likely affected the precise enumeration of the viable fungal cells. Nevertheless, S. mutans carriage in cospecies biofilms with either C. albicans mutant was similar to that in cospecies biofilms formed with the parental C. albicans strain. These observations suggest that the ability of C. albicans to form biofilms on other surfaces may not be crucial for the observed cooperativity with S. mutans in our biofilm model at least. The data emphasize the importance of S. mutans-derived Gtfs in mediating cospecies biofilm development on sHA, although the impact of other C. albicans biofilm-related properties needs additional exploration.
Coexistence with C. albicans influences gene expression in S. mutans.
S. mutans effectively responds to a dynamic and changing milieu during biofilm development (15, 16, 18, 47, 68). The presence of C. albicans clearly modifies this milieu, which can influence the transcriptomic responses of S. mutans within biofilms. Therefore, we investigated the effect of coexistence with C. albicans on the expression profile of S. mutans genes linked to EPS-rich matrix construction (gtfB, gtfC, and gtfD), glucan binding proteins and matrix degradation/remodeling (gbpB and dexA), and acid stress survival (fabM and atpD). The roles of these genes in contributing to the virulence and survival of S. mutans have been well established in vivo (12, 16). We monitored transcription in S. mutans during the formation of cospecies biofilms at 22 h and 32 h, and at 42 h, when they had reached maturity (Fig. 9).
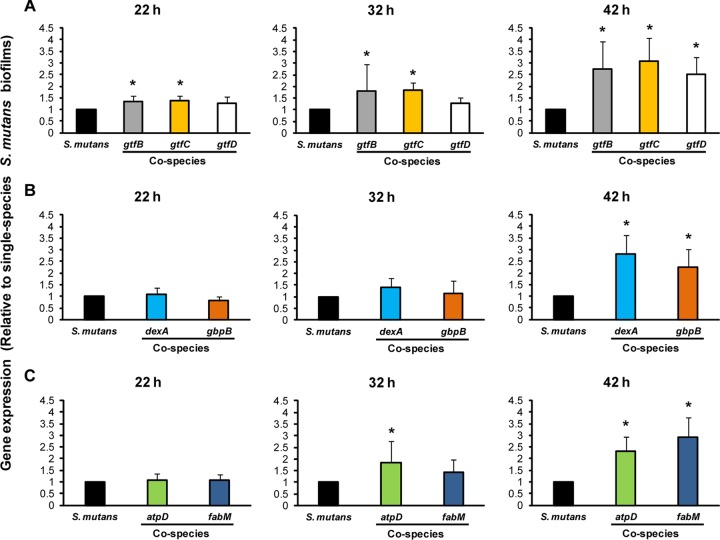
FIG 9.
Expression profiles of S. mutans UA159 genes during the development of cospecies biofilms. The expression of selected S. mutans genes associated with EPS synthesis (A), EPS degradation and binding (B), and acid stress survival (C) is shown. The data (gene expression in cospecies biofilms relative to that in single-species S. mutans biofilms, represented by the black bars) are depicted as the mean ± standard deviation (n = 8). An asterisk indicates that the expression level of a specific S. mutans gene is significantly different for single-species and cospecies biofilms (P, <0.05).
We detected significant increases in the expression of gtfB and gtfC (but not gtfD) in cospecies biofilms at 22 and 32 h, while all three gtf genes were induced at 42 h (2- to 3-fold increase in transcript abundance over that with S. mutans alone [P, <0.05]) (Fig. 9A). Although S. mutans itself enhances the expression of gtf genes during biofilm development (15, 47, 69), the presence of C. albicans appears to further increase the expression of these genes during the initial stages of cospecies biofilm formation, and these genes continue to be comparatively induced as the biofilms mature (relative to expression in single-species S. mutans biofilms). This expression pattern further supports our earlier observations about the essential roles of GtfB- and GtfC-derived polysaccharides in the assembly of S. mutans-C. albicans biofilms. In contrast, genes involved in EPS degradation (dexA) and EPS binding (gbpB) are more highly transcribed in cospecies biofilms only at 42 h (Fig. 9B). Such temporal changes in the expression of these genes may affect the composition and chemical structure of the matrix (relative to that for single-species biofilms), as well as enhancing the ability of S. mutans to bind EPS via GbpB as the cospecies biofilms reach maturity.
Genes involved in acid tolerance were also largely induced in mature cospecies biofilms (relative to expression in single-species biofilms) (P, <0.05), although atpD expression was enhanced in cospecies biofilms as early as 32 h (Fig. 9C). The fabM gene is linked to unsaturated fatty acid biosynthesis in S. mutans, while atpD is essential for the assembly of membrane-associated F-ATPase (16). Since both genes are transcriptionally upregulated when C. albicans is present, the ability of S. mutans to cope and thrive in an acidified environment may be enhanced in cospecies biofilms. Clearly, coexistence with C. albicans induces the expression of key virulence genes in S. mutans that are critical for the ability of the bacterium to persist within biofilms and to cause disease.
DISCUSSION
The results of our study provide striking evidence that S. mutans and C. albicans develop a symbiotic relationship that enhances the virulence of cospecies plaque biofilms formed on tooth surfaces, ultimately amplifying the severity of disease. Our data support and further advance the initial concept that C. albicans may be associated with the pathogenesis of early-childhood caries (ECC) (22–24, 59). More importantly, the rapid onset of disease and the enhanced number, extent, and severity of carious lesions on the free smooth surfaces of the teeth show very clearly that the presence of C. albicans and its association with S. mutans have a synergistic effect on the virulence of the disease. The presence of C. albicans with S. mutans more than doubles the number and severity of smooth-surface lesions relative to the effect of infection with either organism alone. Furthermore, our results reveal an overwhelming infection by S. mutans and C. albicans when they are growing together in the presence of sucrose. This observation certainly offers at least a partial explanation for the extent and rapidity of tooth destruction and the detection of elevated levels of both organisms seen clinically (22–24).
It may appear surprising that coinfection with C. albicans and S. mutans had a comparatively smaller effect on the number or severity of sulcal-surface carious lesions in our model. It is well recognized that the development of smooth-surface caries is highly dependent on the formation of Gtf-derived EPS (70, 71). EPS facilitates the adherence of S. mutans (and other organisms) and modulates the formation of cariogenic plaque biofilms in vivo (10), which is in line with the findings of our study. In contrast, the sulcal surfaces provide natural entrapment sites, and the retention of microorganisms is not dependent on EPS. Nevertheless, a major clinical feature of ECC is the presence of extensive lesions on the smooth surfaces, a condition that is clearly mimicked in our in vivo model.
The data from the in vitro studies may offer a further explanation for the enhanced infectivity/carriage and cooperative coexistence in the presence of sucrose and may also explain why the presence of C. albicans together with S. mutans causes the observed synergistic enhancement in virulence. The ability of C. albicans to colonize and develop cospecies biofilms with S. mutans is largely dependent on the actions of GtfB and GtfC. We propose that the Gtfs play key roles in the development of highly virulent cospecies biofilms in at least three ways: (i) they convert C. albicans cells into glucan producers, which promote the assembly of the EPS-rich matrix scaffold; (ii) they enable the fungus to colonize EPS-coated surfaces readily; and (iii) they enhance fungal-bacterial coadherence.
The surface area of C. albicans is significantly larger than that of S. mutans and provides plentiful Gtf binding sites (35). It should be noted that GtfB binds to both yeast and hyphal cell forms and remains enzymatically active. This surface-bound enzyme produces higher quantities of insoluble EPS, with more α1,6-linkages, than GtfB in solution or bound to the surfaces of S. mutans cells (35). The α1,6-linked glucosyl residues in the glucan structure in turn provide a site to which S. mutans cells adhere avidly (12, 35, 72). Thus, the enhanced surface area, together with an increased number of binding sites, offers a plausible explanation for enhanced S. mutans carriage in cospecies biofilms. These phenomena were absent when C. albicans cells were grown with S. mutans strains defective in gtfB and/or gtfC.
Gtf-derived glucans formed on the C. albicans surface enhance the ability of the fungal cells to colonize and form cospecies biofilms. Results from previous studies have shown that S. mutans-derived Gtfs (particularly GtfC) present on sHA surface rapidly form an amorphous glucan layer (13, 14, 72), which masks host-derived microbial binding sites in the salivary pellicle (72). These observations are relevant because C. albicans itself adheres poorly to the preformed EPS layer on sHA surfaces, or binds poorly to S. mutans, unless the fungal cells are first coated with Gtf-derived glucans (28, 30, 35). Our study reveals that fungal cells are detected only after the initial polymeric matrix and S. mutans microcolonies are formed on the sHA. Furthermore, the lack of gtfB and/or gtfC expression by S. mutans severely disrupts the ability of C. albicans to colonize, accumulate, and form cospecies biofilms. These findings are supported by the observation that C. albicans is detected at low numbers or not at all in the plaque of ECC-free children (22–24) and at lower number in rats infected with C. albicans alone than in coinfected rats under our experimental conditions.
Our data provide a feasible explanation for the previous reports showing that the ability of S. mutans and C. albicans to form biofilms together was promoted in the presence of sucrose (32–34), while other sugars (e.g., glucose), which are not substrates for EPS synthesis, had no effect (33). A similar mechanism may also enhance C. albicans and S. gordonii biofilm formation in vitro (73). Altogether, we demonstrate the importance of Gtfs in mediating the cooperativity between C. albicans and S. mutans. This type of interaction represents a truly unique physical interaction where a bacterially produced product adheres to, and functions on, the surface of an organism from another kingdom, transforming a relatively innocuous organism (in terms of dental caries) into a fierce stimulator of cariogenic biofilm formation.
The potential of C. albicans to contribute to the pathogenesis of caries disease has often been associated with its ability to produce and tolerate acids (22–24, 59, 75). We found that the pH values of the culture medium surrounding cospecies biofilms were highly acidic, though not significantly different from those of single-species S. mutans biofilms. There may have been differences in the pH values within the biofilm, but this measurement was beyond the scope of the present study. Although an acidic pH is undeniably the immediate cause of tooth enamel dissolution, the environment within which the acid is produced plays a crucial role in cariogenesis (12). The results of our previous studies have shown that the synthesis of Gtf-derived glucans leads to the formation of an insoluble EPS-rich matrix scaffold that acts as a diffusion-limiting barrier (15). In parallel, the metabolic activity of S. mutans clustered within the microcolony can produce copious amounts of acids that accumulate locally (15, 74). It is conceivable that the alterations in the extracellular matrix containing a dense population of bacterial cells help to prevent acid within the biofilm from diffusing outward, thus prolonging and intensifying the acid attack.
The presence of C. albicans dramatically modifies the physical environment and the 3D architecture of the biofilm. It alters the volume and the structure of the extracellular matrix by (i) increasing the amount of insoluble Gtf-derived EPS, which has been shown to have diffusion-limiting properties (15), and (ii) independently contributing to the matrix through the production of extracellular β-glucans. It is also possible that the presence of insoluble β-1,3-glucan embedded in the extracellular matrices of cospecies biofilms may help limit diffusion while contributing to stability of the 3D matrix scaffold. Furthermore, S. mutans microcolonies form more rapidly, and their size more than doubles, when the biofilms are grown in the presence of C. albicans. We have shown previously that the pH inside the microcolony becomes more acidic as the structure increases in size, due to a high density of acidogenic organisms and limited diffusion into and out of the structure (15). Thus, the elevated and localized concentration of S. mutans cells sheltered by an abundant extracellular matrix would maximize the ability of acids to demineralize teeth by retaining the acids in close proximity to the tooth surface. In this scenario, EPS may be both the point of articulation for the coexistence of S. mutans and C. albicans and a diffusion barrier that helps to maintain an acidic environment, which could explain why the transcription of S. mutans acid tolerance genes (fabM and atpD) is induced in cospecies biofilms relative to single-species biofilms. We are currently mapping the spatial distributions of pH, EPS, and microbial cells by using a fluorescent pH indicator that is incorporated into the matrix scaffold to determine the exact locations of acidic niches within undisturbed biofilms. At the same time, such changes in expression also suggest that S. mutans may be able to sense C. albicans within the surrounding biofilm milieu, in turn increasing the production of proteins involved in virulence and/or stress defense. Overall, the data indicate that the presence of C. albicans might accentuate the fitness of S. mutans, which may help to account for the enhanced virulence observed in our rodent model.
We recognize the complexity of this bacterium-fungus association. The interactions between these two organisms are multifaceted and could presumably induce additional responses in one another and/or alter the immediate environment to influence pathogenesis. Although we focus on the influence of the presence of C. albicans on S. mutans accumulation, biofilm formation, and virulence expression in this publication, it is possible that S. mutans may also provide benefits to C. albicans, such as enhanced colonization of the tooth surface. Although we have begun to investigate the consequences of this cross-kingdom interaction, much is yet unknown. Certainly, further studies are needed to investigate the changes in C. albicans virulence and matrix production.
In summary, we propose that there is a novel mutualistic relationship between a fungus and an oral bacterial pathogen that results in synergistic enhancement of the virulence of an infectious disease. The association between C. albicans and S. mutans appears to be largely mediated by a physical interaction that relies on the production of glucans, which are produced by bacterial exoenzymes (Gtfs), on yeast and hyphal cell surfaces. These interactions are essential for the assembly of an EPS-rich matrix, the formation of enlarged microcolonies containing densely packed S. mutans cells, and the development of cospecies biofilms. These findings illustrate how Gtfs can convert a moderately cariogenic organism into a major contributor to the formation of virulent plaque biofilms, ultimately modulating the pathogenesis of dental caries in a susceptible and vulnerable population. Since we have shown unequivocally that S. mutans-C. albicans association can drastically increase virulence in vivo and that our in vitro studies point to a Gtf-dependent mechanism, testing of individual Δgtf mutants with C. albicans (in the context of infection and cariogenesis) is certainly warranted. In addition, it is clear that C. albicans also contributes independently to EPS production in cospecies biofilms. However, additional factors may be at play, including signaling interactors, since the presence of C. albicans augments the expression of virulence genes in S. mutans.
Clinical implications.
We offer plausible data to support the clinical importance of the association between C. albicans and S. mutans in the pathogenesis of ECC, one of the most virulent, painful, and costly infectious diseases afflicting children. A key finding of this study is that the interactions between C. albicans and S. mutans via EPS production increase both fungal and bacterial carriage, and these organisms together synergistically enhance the virulence of plaque biofilms. Our data help to explain the high level of recovery of these organisms from the plaque of children afflicted with ECC and the overt demineralization and rampant carious lesions that typically occur on the free smooth surfaces of their primary teeth (22–24). Clearly, our data provide new perspectives for devising efficacious therapies to control ECC. For example, blocking Gtf binding to the Candida cell wall or including antifungals as part of the treatment to reduce or prevent fungal infection may be effective therapeutic approaches. Furthermore, increased knowledge concerning the identities of the molecular interactors that facilitate C. albicans-S. mutans associations and the assembly of hypervirulent biofilms may provide additional avenues for the prevention of this disease.
In addition, enhanced colonization by C. albicans and increased fungal carriage in cospecies plaque biofilms may also provide a fungal reservoir that could promote Candida infections of soft tissue and oral mucosal surfaces, as reported recently (31). In our preliminary data, we observed elevated fungal colonization of tongues from coinfected animals relative to that for animals infected with C. albicans alone, as shown in Fig. S6 in the supplemental material (albeit further quantitative analysis is warranted). Thus, our findings likely have relevance beyond teeth and the mouth, since localized bacterium-fungus interactions are associated with other polymicrobial infections and systemic complications at various sites in humans (25–27).
Supplementary Material
ACKNOWLEDGMENTS
This work was supported in part by research grants from the National Science Foundation (EFRI-1137186) and from the National Institute for Dental and Craniofacial Research (T90DE021985).
Footnotes
Published ahead of print 24 February 2014
Supplemental material for this article may be found at http://dx.doi.org/10.1128/IAI.00087-14.
REFERENCES
- 1.Hall-Stoodley L, Stoodley P. 2009. Evolving concepts in biofilm infections. Cell. Microbiol. 11:1034–1043. 10.1111/j.1462-5822.2009.01323.x [DOI] [PubMed] [Google Scholar]
- 2.Dye BA, Tan S, Smith V, Lewis BG, Barker LK, Thornton-Evans G, Eke PI, Beltran-Aguilar ED, Horowitz AM, Li CH. 2007. Trends in oral health status: United States, 1988–1994 and 1999–2004. Vital Health Stat. 11:1–92 [PubMed] [Google Scholar]
- 3.Berkowitz RJ, Turner J, Hughes C. 1984. Microbial characteristics of the human dental caries associated with prolonged bottle-feeding. Arch. Oral Biol. 29:949–951. 10.1016/0003-9969(84)90097-9 [DOI] [PubMed] [Google Scholar]
- 4.Milnes AR, Bowden GH. 1985. The microflora associated with developing lesions of nursing caries. Caries Res. 19:289–297. 10.1159/000260858 [DOI] [PubMed] [Google Scholar]
- 5.Hallett KB, O'Rourke PK. 2002. Early childhood caries and infant feeding practice. Community Dent. Health 19:237–242 [PubMed] [Google Scholar]
- 6.Chestnutt IG, Murdoch C, Robson KF. 2003. Parents and carers' choice of drinks for infants and toddlers, in areas of social and economic disadvantage. Community Dent. Health 20:139–145 [PubMed] [Google Scholar]
- 7.Bowen WH, Lawrence RA. 2005. Comparison of the cariogenicity of cola, honey, cow milk, human milk, and sucrose. Pediatrics 116:921–926. 10.1542/peds.2004-2462 [DOI] [PubMed] [Google Scholar]
- 8.Karp J, Berkowitz RJ. 2008. Clinical outcomes for severe early childhood caries. Clin. Rev. Pediatr. 4:169–173. 10.2174/157339608785855965 [DOI] [Google Scholar]
- 9.Palmer CA, Kent R, Jr, Loo CY, Hughes CV, Stutius E, Pradhan N, Dahlan M, Kanasi E, Arevalo Vasquez SS, Tanner AC. 2010. Diet and caries-associated bacteria in severe early childhood caries. J. Dent. Res. 89:1224–1229. 10.1177/0022034510376543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kanasi E, Dewhirst FE, Chalmers NI, Kent R, Jr, Moore A, Hughes CV, Pradhan N, Loo CY, Tanner AC. 2010. Clonal analysis of the microbiota of severe early childhood caries. Caries Res. 44:485–497. 10.1159/000320158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gross EL, Leys EJ, Gasparovich SR, Firestone ND, Schwartzbaum JA, Janies DA, Asnani K, Griffen AL. 2010. Bacterial 16S sequence analysis of severe caries in young permanent teeth. J. Clin. Microbiol. 48:4121–4128. 10.1128/JCM.01232-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bowen WH, Koo H. 2011. Biology of Streptococcus mutans-derived glucosyltransferases: role in extracellular matrix formation of cariogenic biofilms. Caries Res. 45:69–86. 10.1159/000324598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vacca-Smith AM, Bowen WH. 1998. Binding properties of streptococcal glucosyltransferases for hydroxyapatite, saliva-coated hydroxyapatite, and bacterial surfaces. Arch. Oral Biol. 43:103–110. 10.1016/S0003-9969(97)00111-8 [DOI] [PubMed] [Google Scholar]
- 14.Vacca-Smith AM, Venkitaraman AR, Schilling KM, Bowen WH. 1996. Characterization of glucosyltransferase of human saliva adsorbed onto hydroxyapatite surfaces. Caries Res. 30:354–360. 10.1159/000262342 [DOI] [PubMed] [Google Scholar]
- 15.Xiao J, Klein MI, Falsetta ML, Lu B, Delahunty CM, Yates JR, III, Heydorn A, Koo H. 2012. The exopolysaccharide matrix modulates the interaction between 3D architecture and virulence of a mixed-species oral biofilm. PLoS Pathog. 8:e1002623. 10.1371/journal.ppat.1002623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lemos JA, Quivey RG, Jr, Koo H, Abranches J. 2013. Streptococcus mutans: a new Gram-positive paradigm? Microbiology 159:436–445. 10.1099/mic.0.066134-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nyvad B, Crielaard W, Mira A, Takahashi N, Beighton D. 2013. Dental caries from a molecular microbiological perspective. Caries Res. 47:89–102. 10.1159/000345367 [DOI] [PubMed] [Google Scholar]
- 18.Burne RA. 1998. Oral streptococci. products of their environment. J. Dent. Res. 77:445–452. 10.1177/00220345980770030301 [DOI] [PubMed] [Google Scholar]
- 19.Marsh PD. 2003. Are dental diseases examples of ecological catastrophes? Microbiology 149:279–294. 10.1099/mic.0.26082-0 [DOI] [PubMed] [Google Scholar]
- 20.Acharya S, Tandon S. 2011. The effect of early childhood caries on the quality of life of children and their parents. Contemp. Clin. Dent. 2:98–101. 10.4103/0976-237X.83069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Skeie MS, Raadal M, Strand GV, Espelid I. 2006. The relationship between caries in the primary dentition at 5 years of age and permanent dentition at 10 years of age—a longitudinal study. Int. J. Paediatr. Dent. 16:152–160. 10.1111/j.1365-263X.2006.00720.x [DOI] [PubMed] [Google Scholar]
- 22.de Carvalho FG, Silva DS, Hebling J, Spolidorio LC, Spolidorio DM. 2006. Presence of mutans streptococci and Candida spp. in dental plaque/dentine of carious teeth and early childhood caries. Arch. Oral Biol. 51:1024–1028. 10.1016/j.archoralbio.2006.06.001 [DOI] [PubMed] [Google Scholar]
- 23.Raja M, Hannan A, Ali K. 2010. Association of oral candidal carriage with dental caries in children. Caries Res. 44:272–276. 10.1159/000314675 [DOI] [PubMed] [Google Scholar]
- 24.Yang XQ, Zhang Q, Lu LY, Yang R, Liu Y, Zou J. 2012. Genotypic distribution of Candida albicans in dental biofilm of Chinese children associated with severe early childhood caries. Arch. Oral Biol. 57:1048–1053. 10.1016/j.archoralbio.2012.05.012 [DOI] [PubMed] [Google Scholar]
- 25.Peleg AY, Hogan DA, Mylonakis E. 2010. Medically important bacterial-fungal interactions. Nat. Rev. Microbiol. 8:340–349. 10.1038/nrmicro2313 [DOI] [PubMed] [Google Scholar]
- 26.Shirtliff ME, Peters BM, Jabra-Rizk MA. 2009. Cross-kingdom interactions: Candida albicans and bacteria. FEMS Microbiol. Lett. 299:1–8. 10.1111/j.1574-6968.2009.01668.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Harriott MM, Noverr MC. 2011. Importance of Candida-bacterial polymicrobial biofilms in disease. Trends Microbiol. 19:557–563. 10.1016/j.tim.2011.07.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jenkinson HF, Douglas LJ. 2002. Candida interactions with bacterial biofilms, p 357–373 In Brogden KA, Guthmiller JM. (ed), Polymicrobial diseases. ASM Press, Washington, DC: [PubMed] [Google Scholar]
- 29.Diaz PI, Xie Z, Sobue T, Thompson A, Biyikoglu B, Ricker A, Ikonomou L, Dongari-Bagtzoglou A. 2012. Synergistic interaction between Candida albicans and commensal oral streptococci in a novel in vitro mucosal model. Infect. Immun. 80:620–632. 10.1128/IAI.05896-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jenkinson HF, Lala HC, Shepherd MG. 1990. Coaggregation of Streptococcus sanguis and other streptococci with Candida albicans. Infect. Immun. 58:1429–1436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Xu H, Sobue T, Thompson A, Xie Z, Poon K, Ricker A, Cervantes J, Diaz PI, Dongari-Bagtzoglou A. 17 September 2013. Streptococcal co-infection augments Candida pathogenicity by amplifying the mucosal inflammatory response. Cell. Microbiol. 10.1111/cmi.12216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Branting C, Sund ML, Linder LE. 1989. The influence of Streptococcus mutans on adhesion of Candida albicans to acrylic surfaces in vitro. Arch. Oral Biol. 34:347–353. 10.1016/0003-9969(89)90108-8 [DOI] [PubMed] [Google Scholar]
- 33.Pereira-Cenci T, Deng DM, Kraneveld EA, Manders EM, Del Bel Cury AA, Ten Cate JM, Crielaard W. 2008. The effect of Streptococcus mutans and Candida glabrata on Candida albicans biofilms formed on different surfaces. Arch. Oral Biol. 53:755–764. 10.1016/j.archoralbio.2008.02.015 [DOI] [PubMed] [Google Scholar]
- 34.Metwalli KH, Khan SA, Krom BP, Jabra-Rizk MA. 2013. Streptococcus mutans, Candida albicans, and the human mouth: a sticky situation. PLoS Pathog. 9:e1003616. 10.1371/journal.ppat.1003616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gregoire S, Xiao J, Silva BB, Gonzalez I, Agidi PS, Klein MI, Ambatipudi KS, Rosalen PL, Bauserman R, Waugh RE, Koo H. 2011. Role of glucosyltransferase B in interactions of Candida albicans with Streptococcus mutans and with an experimental pellicle formed on hydroxyapatite surfaces. Appl. Environ. Microbiol. 77:6357–6367. 10.1128/AEM.05203-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Falsetta ML, Gregoire S, Colonne M, Koo H. 2013. Streptococcus mutans and Candida albicans interactions during cariogenic biofilm formation. J. Dent. Res. 92(Spec Iss A):abstr 385 https://iadr.confex.com/iadr/13iags/webprogram/Paper173866.html [Google Scholar]
- 37.Koo H, Xiao J, Klein MI, Jeon JG. 2010. Exopolysaccharides produced by Streptococcus mutans glucosyltransferases modulate the establishment of microcolonies within multispecies biofilms. J. Bacteriol. 192:3024–3032. 10.1128/JB.01649-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Klein MI, Duarte S, Xiao J, Mitra S, Foster TH, Koo H. 2009. Structural and molecular basis of the role of starch and sucrose in Streptococcus mutans biofilm development. Appl. Environ. Microbiol. 75:837–841. 10.1128/AEM.01299-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Xiao J, Koo H. 2010. Structural organization and dynamics of exopolysaccharide matrix and microcolonies formation by Streptococcus mutans in biofilms. J. Appl. Microbiol. 108:2103–2113. 10.1111/j.1365-2672.2009.04616.x [DOI] [PubMed] [Google Scholar]
- 40.Koo H, Hayacibara MF, Schobel BD, Cury JA, Rosalen PL, Park YK, Vacca-Smith AM, Bowen WH. 2003. Inhibition of Streptococcus mutans biofilm accumulation and polysaccharide production by apigenin and tt-farnesol. J. Antimicrob. Chemother. 52:782–789. 10.1093/jac/dkg449 [DOI] [PubMed] [Google Scholar]
- 41.Dongari-Bagtzoglou A, Kashleva H, Dwivedi P, Diaz P, Vasilakos J. 2009. Characterization of mucosal Candida albicans biofilms. PLoS One 4:e7967. 10.1371/journal.pone.0007967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jin Y, Zhang T, Samaranayake YH, Fang HH, Yip HK, Samaranayake LP. 2005. The use of new probes and stains for improved assessment of cell viability and extracellular polymeric substances in Candida albicans biofilms. Mycopathologia 159:353–360. 10.1007/s11046-004-6987-7 [DOI] [PubMed] [Google Scholar]
- 43.Hamada S, Gill K, Slade HD. 1977. Binding of lectins to Streptococcus mutans cells and type-specific polysaccharides, and effect on adherence. Infect. Immun. 18:708–716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.London R, Schwedock J, Sage A, Valley H, Meadows J, Waddington M, Straus D. 2010. An automated system for rapid non-destructive enumeration of growing microbes. PLoS One 5:e8609. 10.1371/journal.pone.0008609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chalmers NI, Palmer RJ, Jr, Cisar JO, Kolenbrander PE. 2008. Characterization of a Streptococcus sp.-Veillonella sp. community micromanipulated from dental plaque. J. Bacteriol. 190:8145–8154. 10.1128/JB.00983-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cury JA, Koo H. 2007. Extraction and purification of total RNA from Streptococcus mutans biofilms. Anal. Biochem. 365:208–214. 10.1016/j.ab.2007.03.021 [DOI] [PubMed] [Google Scholar]
- 47.Klein MI, Xiao J, Lu B, Delahunty CM, Yates JR, III, Koo H. 2012. Streptococcus mutans protein synthesis during mixed-species biofilm development by high-throughput quantitative proteomics. PLoS One 7:e45795. 10.1371/journal.pone.0045795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Koo H, Seils J, Abranches J, Burne RA, Bowen WH, Quivey RG., Jr 2006. Influence of apigenin on gtf gene expression in Streptococcus mutans UA159. Antimicrob. Agents Chemother. 50:542–546. 10.1128/AAC.50.2.542-546.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bowen WH, Madison KM, Pearson SK. 1988. Influence of desalivation in rats on incidence of caries in intact cagemates. J. Dent. Res. 67:1316–1318. 10.1177/00220345880670101401 [DOI] [PubMed] [Google Scholar]
- 50.Koo H, Rosalen PL, Cury JA, Park YK, Ikegaki M, Sattler A. 1999. Effect of Apis mellifera propolis from two Brazilian regions on caries development in desalivated rats. Caries Res. 33:393–400. 10.1159/000016539 [DOI] [PubMed] [Google Scholar]
- 51.Keyes PH, White CL. 1959. Dental caries in the molar teeth of rats. III. Bio-assay of sodium fluoride and sodium lauroyl sarcosinate as caries-inhibitory agents. J. Am. Dent. Assoc. 58:43–55 [DOI] [PubMed] [Google Scholar]
- 52.Klein MI, Scott-Anne KM, Gregoire S, Rosalen PL, Koo H. 2012. Molecular approaches for viable bacterial population and transcriptional analyses in a rodent model of dental caries. Mol. Oral Microbiol. 27:350–361. 10.1111/j.2041-1014.2012.00647.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Larson R. 1981. Merits and modifications of scoring rat dental caries by Keyes' method, p 195–203 In Tanzer J. (ed), Animal models in cariology. IRL Press, Washington, DC [Google Scholar]
- 54.Raubertas RF, Davis BA, Bowen WH, Pearson SK, Watson GE. 1999. Litter effects on caries in rats and implications for experimental design. Caries Res. 33:164–169. 10.1159/000016511 [DOI] [PubMed] [Google Scholar]
- 55.De Sordi L, Muhlschlegel FA. 2009. Quorum sensing and fungal-bacterial interactions in Candida albicans: a communicative network regulating microbial coexistence and virulence. FEMS Yeast Res. 9:990–999. 10.1111/j.1567-1364.2009.00573.x [DOI] [PubMed] [Google Scholar]
- 56.Merritt J, Qi F. 2012. The mutacins of Streptococcus mutans: regulation and ecology. Mol. Oral Microbiol. 27:57–69. 10.1111/j.2041-1014.2011.00634.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Gow NA, van de Veerdonk FL, Brown AJ, Netea MG. 2012. Candida albicans morphogenesis and host defence: discriminating invasion from colonization. Nat. Rev. Microbiol. 10:112–122. 10.1038/nrmicro2711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Nobile CJ, Nett JE, Hernday AD, Homann OR, Deneault JS, Nantel A, Andes DR, Johnson AD, Mitchell AP. 2009. Biofilm matrix regulation by Candida albicans Zap1. PLoS Biol. 7:e1000133. 10.1371/journal.pbio.1000133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Klinke T, Guggenheim B, Klimm W, Thurnheer T. 2011. Dental caries in rats associated with Candida albicans. Caries Res. 45:100–106. 10.1159/000324809 [DOI] [PubMed] [Google Scholar]
- 60.Nett JE, Sanchez H, Cain MT, Andes DR. 2010. Genetic basis of Candida biofilm resistance due to drug-sequestering matrix glucan. J. Infect. Dis. 202:171–175. 10.1086/651200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Nett JE, Crawford K, Marchillo K, Andes DR. 2010. Role of Fks1p and matrix glucan in Candida albicans biofilm resistance to an echinocandin, pyrimidine, and polyene. Antimicrob. Agents Chemother. 54:3505–3508. 10.1128/AAC.00227-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Taff HT, Nett JE, Zarnowski R, Ross KM, Sanchez H, Cain MT, Hamaker J, Mitchell AP, Andes DR. 2012. A Candida biofilm-induced pathway for matrix glucan delivery: implications for drug resistance. PLoS Pathog. 8:e1002848. 10.1371/journal.ppat.1002848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Al-Fattani MA, Douglas LJ. 2006. Biofilm matrix of Candida albicans and Candida tropicalis: chemical composition and role in drug resistance. J. Med. Microbiol. 55:999–1008. 10.1099/jmm.0.46569-0 [DOI] [PubMed] [Google Scholar]
- 64.Baillie GS, Douglas LJ. 2000. Matrix polymers of Candida biofilms and their possible role in biofilm resistance to antifungal agents. J. Antimicrob. Chemother. 46:397–403. 10.1093/jac/46.3.397 [DOI] [PubMed] [Google Scholar]
- 65.Nett J, Lincoln L, Marchillo K, Massey R, Holoyda K, Hoff B, VanHandel M, Andes D. 2007. Putative role of β-1,3 glucans in Candida albicans biofilm resistance. Antimicrob. Agents Chemother. 51:510–520. 10.1128/AAC.01056-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Humbel BM, Konomi M, Takagi T, Kamasawa N, Ishijima SA, Osumi M. 2001. In situ localization of β-glucans in the cell wall of Schizosaccharomyces pombe. Yeast 18:433–444. 10.1002/yea.694 [DOI] [PubMed] [Google Scholar]
- 67.Nobile CJ, Fox EP, Nett JE, Sorrells TR, Mitrovich QM, Hernday AD, Tuch BB, Andes DR, Johnson AD. 2012. A recently evolved transcriptional network controls biofilm development in Candida albicans. Cell 148:126–138. 10.1016/j.cell.2011.10.048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Lemos JA, Burne RA. 2008. A model of efficiency: stress tolerance by Streptococcus mutans. Microbiology 154:3247–3255. 10.1099/mic.0.2008/023770-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Li Y, Burne RA. 2001. Regulation of the gtfBC and ftf genes of Streptococcus mutans in biofilms in response to pH and carbohydrate. Microbiology 147:2841–2848 [DOI] [PubMed] [Google Scholar]
- 70.Tanzer JM, Freedman ML, Fitzgerald RJ. 1985. Virulence of mutants defective in glucosyltransferase, dextran-mediated aggregation, or dextranase activity, p 204–211 In Mergenhagen SE, Rosan B. (ed), Molecular basis of oral microbial adhesion: proceedings of a workshop held in Philadelphia, Pennsylvania, 5–8 June 1984. American Society for Microbiology, Washington, DC [Google Scholar]
- 71.Yamashita Y, Bowen WH, Burne RA, Kuramitsu HK. 1993. Role of the Streptococcus mutans gtf genes in caries induction in the specific-pathogen-free rat model. Infect. Immun. 61:3811–3817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Schilling KM, Bowen WH. 1992. Glucans synthesized in situ in experimental salivary pellicle function as specific binding sites for Streptococcus mutans. Infect. Immun. 60:284–295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Ricker A, Vickerman M, Dongari-Bagtzoglou A. 29 January 2014. Streptococcus gordonii glucosyltransferase promotes biofilm interactions with Candida albicans. J. Oral Microbiol. 6:23419. 10.3402/jom.v6.23419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Guo L, Hu W, He X, Lux R, McLean J, Shi W. 2013. Investigating acid production by Streptococcus mutans with a surface-displayed pH-sensitive green fluorescent protein. PLoS One 8:e57182. 10.1371/journal.pone.0057182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Klinke T, Kneist S, de Soet JJ, Kuhlisch E, Mauersberger S, Forster A, Klimm W. 2009. Acid production by oral strains of Candida albicans and lactobacilli. Caries Res. 43:83–91. 10.1159/000204911 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.