Abstract
Shigella flexneri is the major cause of shigellosis in developing countries. A new S. flexneri serotype, Xv, appeared in 2000 and replaced serotype 2a as the most prevalent serotype in China. Serotype Xv is a variant of serotype X, with phosphoethanolamine modification of its O antigen mediated by a plasmid that contained the opt gene. Serotype Xv isolates belong to sequence type 91 (ST91). In this study, whole-genome sequencing of 59 S. flexneri isolates of 14 serotypes (serotypes 1 to 4, Y, Yv, X, and Xv) indicated that ST91 arose around 1993 by acquiring multidrug resistance (MDR) and spread across China within a decade. A comparative analysis of the chromosome and opt-carrying plasmid pSFXv_2 revealed independent origins of 3 serotype Xv clusters in China, with different divergence times. Using 18 cluster-dividing single-nucleotide polymorphisms (SNPs), SNP typing divided 380 isolates from 3 provinces (Henan, Gansu, and Anhui) into 5 SNP genotypes (SGs). One SG predominated in each province, but substantial interregional spread of SGs was also evident. These findings suggest that MDR is the key selective pressure for the emergence of the S. flexneri epidemic clone and that Shigella epidemics in China were caused by a combination of local expansion and interregional spread of serotype Xv.
INTRODUCTION
Among the four species of Shigella, Shigella flexneri is the most common cause of bacillary dysentery in developing countries (1). Worldwide, there are 164.7 million cases of shigellosis annually with 1.1 million deaths, mostly of children less than 5 years of age (1). S. flexneri is divided into at least 19 serotypes (serotypes 1a, 1b, 1c, 1d, 2a, 2b, 3a, 3b, 4a, 4av, 4b, 5a, 5b, X, Xv, Y, Yv, F6, and 7b), based on the combinations of antigenic determinants present on the O antigen of the cell envelope lipopolysaccharide (LPS) (2–14). Serotypes 1c, 4av, 7b, 1d, Yv, and Xv were new serotypes reported in recent years (3–14), among which serotypes 1c and Xv have caused epidemic-level disease (5, 7, 9). Serotype Xv first appeared in 2000 in Henan province and subsequently replaced serotype 2a to become the predominant serotype in Henan between 2002 and 2006. Serotype Xv was also the most prevalent serotype in Gansu (67%) and Anhui (54%) in 2007 (9). A novel plasmid (pSFXv_2) carrying the opt gene encoding the O antigen phosphoethanolamine (PEtN) transferase is responsible for serotype conversion from X to Xv (10).
Our previous study indicated that S. flexneri epidemics in China have been caused by a sequence type 91 (ST91) clone, and the majority of the ST91 isolates belong to serotype Xv (9). Genome sequencing of a representative ST91 strain, 2002017, revealed that it gained 2 unique multidrug resistance (MDR) islands, i.e., the Shigella resistance locus (SRL) and Tn7 (9). The SRL, which was first identified in the Japanese S. flexneri 2a strain YSH6000, carries genes for resistance against streptomycin, ampicillin, chloramphenicol, and tetracycline and also carries an iron acquisition system (9, 15–17). The SRL in 2002017 carries an additional set of tetracycline resistance genes but does not contain the iron acquisition system present in YSH6000. Tn7 is a composite transposon conferring resistance to trimethoprim, streptothricin, and streptomycin/spectinomycin, all of which were recommended antibiotics used to treat dysentery in the early 1990s (9). To elucidate the temporal and geographic dynamics of S. flexneri epidemics in China, we sequenced 59 strains, predominantly of ST91 and serotype Xv, across a 10-year time period and found that acquisition of multidrug resistance was the key factor for the expansion of ST91 across China in the early 1990s; we also found that the novel serotype Xv emerged more than once in different regions independently.
MATERIALS AND METHODS
Bacterial strains and DNA extraction.
Fifty-nine strains, which were isolated in the Henan (38 strains), Gansu (8 strains), Anhui (7 strains), Shanxi (3 strains), Guizhou (2 strains), and Sichuan (1 strain) provinces of China between 1997 and 2006, were selected for Illumina paired-end sequencing (18). The 59 isolates covered 14 serotypes, including Xv (25 isolates), X (9 isolates), 1a (7 isolates), 1b (1 isolate), 1d (1 isolate), 2a (5 isolates), 2b (1 isolate), 3a (3 isolates), 3b (1 isolate), 4a (1 isolate), 4av (1 isolate), 4b (1 isolate), Y (2 isolates), and Yv (1 isolate). The majority of the strains belong to ST91, within which serotype Xv strains dominate; therefore, more serotype Xv isolates were selected. The selection also reflected pulsed-field gel electrophoresis (PFGE) diversity, representing 30 pulsotypes. The strain details are presented in Table S1 in the supplemental material. Multilocus sequence typing (MLST) using 15 housekeeping genes was performed based on the protocol obtained from the EcMLST website (http://www.shigatox.net/ecmlst).
Bacterial isolates were grown on brain heart infusion agar at 37°C. Genomic DNA was extracted using a Wizard genomic DNA purification kit (Promega Corp., Madison, WI), as described in the manufacturer's manual. The plasmid purification minikit from Qiagen (Hilden, Germany) was used to isolate the plasmids, according to the manufacturer's instructions.
Sequencing and bioinformatics analysis.
The whole-genome sequencing of the 59 isolates was performed using an Illumina Genome Analyzer IIx system (San Diego, CA), with mate-pair and pair-end sequencing technology (18). The Illumina reads were mapped to the 2002017 reference genome using Burrows-Wheeler alignment (BWA) with default parameters (19). Repeat regions and phage sequences were excluded from single-nucleotide polymorphism (SNP) analysis. SAMtools (20) was used to calculate per-position coverage and to determine the nucleotide for each position. High-quality potential SNPs were identified where a position was covered by >10 reads with >80% covering reads exhibiting the same SNP. All SNPs were recorded by position in reference to the 2002017 genome. Potential genome-wide SNPs were obtained.
Recombination was detected using the method of Feng et al. (21). The raw sequence data were also assembled de novo using SOAPdenovo (release 1.04) (22). Genomic indels were analyzed by methods described previously (23). Identification of insertional sequence (IS) sites was performed using the method of Smith (24).
Detection of plasmid SNPs was performed by mapping sequenced reads to the reference plasmid sequences of pSFXv_2 from 2002017 and pSFyv_2 from HN006 (10). The opt gene was detected by PCR amplification, as described by Sun et al. (10).
Phylogenetic analysis.
Phylogenetic trees were constructed by the maximum likelihood (ML) method (25). Determination of the time of divergence of a branch and substitution rates was performed using the BEAST v1.7.5 package (26). The best-fit evolutionary model for the data set was found to be the general time reversible (GTR) model with a gamma-distribution of among-site rate heterogeneity and a proportion of invariant sites. A relaxed (uncorrelated exponential) molecular clock and an extended Bayesian Skyline tree prior were selected for the analysis. Two independent runs were performed with sampling every 1,000 generations, and the output was analyzed with the Tracer module (v1.7.5).
Antibiotic resistance analysis.
Antimicrobial susceptibility testing was performed using the disk diffusion method, following the standard protocol recommended by the Clinical and Laboratory Standards Institute (CLSI) (27). Resistance genes commonly present in Shigella spp. and Enterobacteriaceae, including the SRL, Tn7, gyrA, dfrA5, blaCTX-M, blaTEM, blaSHV, blaCMY, qnr, qepA, and aac(6′)-Ib-cr, were selected as references (9, 28–31) (see Table S1 in the supplemental material). The antibiotic resistance genes were searched using BLASTn, with an E value of 1 × 10−15 as the cutoff value for a significant match, based on the data generated by whole-genome sequencing.
Detection of SNPs by Sequenom MassArray.
SNP typing was performed with a Sequenom MassArray detection system (San Diego, CA), as described by Jaremko et al. (32). Eighteen cluster-specific SNPs (see results below) obtained from whole-genome sequencing were used to type a large set of isolates. The distribution and nature of the 18 SNPs and primers used are listed in Table 1 and Table S2 in the supplemental material. The initial PCR amplification was carried out with 10 ng of genomic DNA in a 5-μl PCR mixture, and base-extension reactions were performed in a 9-μl PCR extension reaction mixture, according to the manufacturer's instructions. Excess deoxynucleoside triphosphates (dNTPs) were removed by incubation with shrimp alkaline phosphatase for 40 min at 37°C. The final base-extension products were treated with SpectroCLEAN resin (Sequenom, San Diego, CA) to remove salts. The final reaction solution was dispensed into a 384-format SpectroCHIP microarray (Sequenom, San Diego, CA) for mass spectrometry to detect SNPs.
TABLE 1.
Cluster-specific SNPs
| Cluster | SNP genotypea | SNP | Location | Reference nucleotideb | SNP | Nature of SNPc | Amino acid change | Gene (locus tag) | Gene functiond |
|---|---|---|---|---|---|---|---|---|---|
| 1 | SG2 | 1 | 660,521 | C | A | ns | E/D | nagE | PTS N-acetylglucosamine-specific EIICBA |
| 2e | 2,093,579 | T | A | ns | L/Q | hchA | Chaperone protein hchA | ||
| 3 | 3,385,278 | T | C | nc | |||||
| 4 | 3,420,397 | A | C | s | A/A | yhdP | Putative membrane protein | ||
| 5 | 4,408,641 | T | C | s | Y/Y | yjiE | Putative LysR-type transcriptional regulator | ||
| 2 | SG3 and SG4 | 6 | 188,894 | A | G | s | Q/Q | bamA | Outer membrane protein assembly factor YaeT precursor |
| 7 | 1,026,507 | T | G | ns | T/P | yccW | Putative SAM-dependent methyltransferase | ||
| 8e | 1,083,507 | G | A | ns | R/H | SFxv_1115 | Putative P4-family integrase | ||
| 9 | 1,307,817 | C | A | ns | V/F | ychF | GTP-dependent nucleic acid-binding protein EngD | ||
| 10 | 2,077,539 | G | T | nc | |||||
| 11e | 2,470,089 | C | T | s | A/A | SFxv_2626 | Hypothetical protein | ||
| 12e | 2,667,545 | G | A | s | V/V | hisS | Histidyl-tRNA synthetase | ||
| 13 | 3,107,434 | G | A | nc | |||||
| 14 | 3,301,086 | A | G | nc | |||||
| 15 | 3,504,262 | G | A | ns | G/S | cysG | Uroporphyrinogen III methylase | ||
| 16 | 3,804,107 | T | A | ns | T/S | SFxv_3993 | Lipopolysaccharide 1,2-glucosyltransferase | ||
| 17 | 3,817,753 | T | C | ns | K/R | pyrE | Orotate phosphoribosyltransferase | ||
| 18 | 4,201,242 | G | A | ns | C/Y | pflC | Glycyl-radical enzyme-activating protein family | ||
| 19 | 4,202,145 | G | A | ns | V/I | frwD | PTS fructose-like IIB component 2 | ||
| 20 | 4,456,749 | T | C | nc | |||||
| 3 | SG5 | 21 | 2,821,789 | C | A | nc | |||
| 22 | 3,058,327 | G | A | ns | S/F | tktA | Transketolase 1 isozyme | ||
| 3 (extended) | 23 | 267,157 | G | T | ns | L/F | SFxv_0262 | Hypothetical protein | |
| 24 | 1,409,095 | T | C | nc | |||||
| 25 | 2,969,268 | T | A | nc | |||||
| 26 | 4,511,008 | C | T | s | T/T | dipZ | Thiol-disulfide interchange protein | ||
| 27 | 4,600,805 | T | C | nc |
See Table S4 in the supplemental material.
Reference strain 2002017.
ns, nonsynonymous SNP (nsSNP); s, synonymous SNP (sSNP); nc, noncoding SNP (ncSNP).
PTS, phosphotransferase system; SAM, S-adenosylmethionine.
These SNPs failed in Sequenom MassArray SNP typing.
opt variation analysis.
The opt gene was amplified using a primer pair (lpt-O-3 primers) described by Sun et al. (10). The primer pair of opt-zn-U (TCTGTGAGTTCACCTGACTT) and opt-zn-L (CAACCATACCGCAGCTACAT) was used for opt gene sequencing.
Nucleotide sequence accession numbers.
This whole-genome shotgun project has been deposited in GenBank under accession no. AZOG00000000 to AZQM00000000 (Bioproject identification number PRJNA230538) (see Table S1 in the supplemental material). The versions described in this paper are versions AZOG00000000.1 to AZQM00000000.1.
RESULTS AND DISCUSSION
Whole-genome sequence analysis of 59 S. flexneri isolates.
In order to elucidate the genomic basis of the epidemic spread of S. flexneri serotype Xv and the endemicity of shigellosis in China, we sequenced 59 S. flexneri isolates, of which 50 were ST91 and 25 were serotype Xv, from different regions of China, covering 10 years from 1997 to 2006 (Fig. 1A). The genomes were sequenced using Illumina 100-bp paired-end sequencing technology, with an average of 121-fold coverage. Reads were mapped to the reference sequence 2002017 to produce 4.48 Mb per genome, on average. Five published complete S. flexneri genomes (2002017 [Xv; GenBank accession no. CP001383.1], Sf301 [2a; GenBank accession no. AE005674.2], 2457T [2a; GenBank accession no. AE014073.1], Sf8401 [5b; GenBank accession no. CP000266.1], and M90T [5a; GenBank accession no. CM001474.1]) were included for comparison (9, 33–36). A final set of 1,790 chromosomal SNPs in the core genome regions, excluding SNPs in repetitive or mobile sequences, were identified from a total of 64 genomes. Fifty-two homologous recombination regions with high frequencies of substitutions that were assumed to be introduced by horizontal transfer were detected. The recombinant regions containing 108 homoplastic SNPs were removed from the phylogenetic analysis described below.
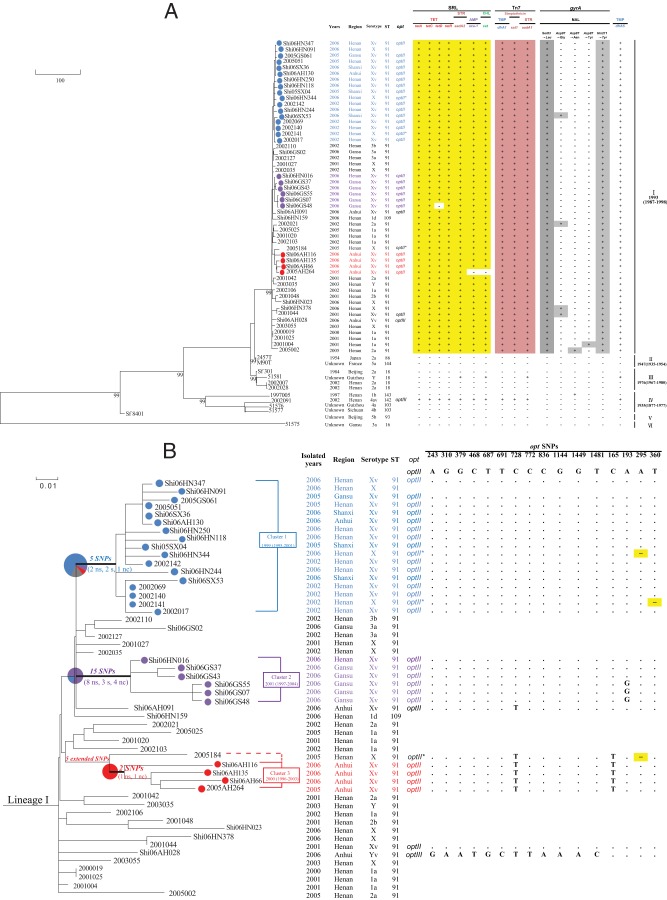
FIG 1.
Genomic relationships of S. flexneri isolates. (A) Phylogenetic tree of 64 S. flexneri isolates based on 1,790 SNPs constructed by the maximum likelihood method. Maximum likelihood bootstrap values that supported major lineages are indicated below the branches. The 6 lineages are marked on the right. The distribution of antibiotic resistance genomic contents is shown for the Shigella resistance locus (SRL) island, Tn7, gyrA mutations, and dfrA5. +, presence of a given gene or mutation; −, absence of a given gene or mutation. Isolate details (year, region, serotype, ST, and opt type) are shown. opt* indicates that the isolate carries a defective optII. The median and estimated 95% highest posterior density (HPD) of divergence time are given for the major lineages. Blue, cluster 1; purple, cluster 2; red, cluster 3. TET, tetracycline; STR, streptomycin; AMP, ampicillin; CHL, chloramphenicol; TMP, trimethoprim; NAL, nalidixic acid. (B) Phylogenetic tree of lineage I, to show opt gene variations. The phylogenetic tree was from A. The 3 clusters of Xv isolates are marked with boxed cluster names and divergence times. Blue, cluster 1; purple, cluster 2; red, cluster 3. The number above the branch that defines a cluster is the number of cluster-specific SNPs. ns, nonsynonymous SNP (nsSNP); s, synonymous SNP (sSNP); nc, noncoding SNP (ncSNP). Isolate details (year, region, serotype, ST, and opt type) are shown. opt* indicates that the isolate carries a defective optII. The opt SNPs are listed on the right, with the consensus bases in the top row. Dots, bases identical to the consensus bases. −, single-base deletion, resulting in a stop codon at the position shown.
We analyzed the 220-kb virulence plasmid pINV for SNPs. The presence of SNPs in the IS regions was noted, but no SNPs were found in the other regions of the plasmid genome of the 59 isolates. We also searched for potential new IS sites and found only one novel IS site, located in gene SFxv_1148 (encoding TnpA transposase), in 15 of the 59 isolates examined (see Table S1 in the supplemental material).
The genomes were assembled de novo and examined for any strain-specific genes. With exclusion of serotype-specific phage sequences, the average number of strain-specific genes was 12, with values ranging from 65 strain-specific genes for the most diverse isolate 51575 to none for 4 Xv isolates (see Table S1 in the supplemental material).
Phylogenetic relationship of the 64 isolates.
The 59 genomes together with 5 published genome sequences of S. flexneri were used to construct a strain phylogeny using the ML method. The topology shows that the 64 strains can be divided into 6 lineages, each with 99% maximum likelihood bootstrap support (Fig. 1). The internal nodes separating the lineages are well supported by SNPs and other genomic changes. All ST91 isolates and the sole ST109 isolate (Shi06HN159) grouped together as lineage I. There are 34 SNPs, 9 small indels, and one 1,727-bp deletion (SFxv_3884, encoding cellulose synthase subunit BcsC) marking the internal node for lineage I. Among the 34 SNPs, 12, 13, and 9 were noncoding SNPs (ncSNPs), synonymous SNPs (sSNPs), and nonsynonymous SNPs (nsSNPs), respectively. Lineage II contained strains 2457T (serotype 2a, ST86) and M90T (serotype 5a, ST144), which were isolated in Japan (in 1954) and France (year of isolation unknown), respectively; both strains have been extensively used in laboratory experiments (34, 36–38). Lineage III included 4 ST18 isolates, with 2 indels and 9 SNPs supporting the lineage. It is interesting to note that 3 of the 4 isolates were serotype 2a. Two isolates were isolated in the early 2000s, when ST91 began rising, suggesting that this lineage may have been prevalent before it was replaced by lineage I. Lineage IV contained 4 isolates belonging to 3 different STs (2 isolates in ST103, 1 isolate in ST142, and 1 isolate in ST143), with 12 indels, 36 SNPs, and a 69-bp deletion in open reading frame (ORF) SFxv_2673 (IS4) supporting the node. In a MLST analysis of 15 genes, ST142 (strain 2002091) differed from ST103 by only 1 allele in cyaA, while ST143 (strain 1997005) differed from ST103 by 2 alleles in clpX and fadD. Both lineage V and lineage VI contained a single isolate from the early 1980s (year of isolation unknown), and the lineage VI isolate in particular was very divergent from the other 5 lineages, with 786 SNPs and 468 indels on the external branch. Our phylogenetic analysis suggested that the nonlineage I S. flexneri strains had diverged largely before ST91 spread in China; all recent isolates were very closely related.
Multiple origins of serotype Xv.
Lineage I exclusively contained epidemic ST91, which carries serotype Xv, a new serotype that was noted in Henan in 2000 and subsequently replaced 2a to become predominant in Henan, Anhui, Gansu, and other provinces (9). Notably, lineage I contained 25 Xv isolates (Henan, 10 isolates; Anhui, 6 isolates; Gansu, 6 isolates; Shanxi, 3 isolates), 23 of which were grouped into 3 distinct phylogenetic clusters (Fig. 1B). Only 2 isolates (2001014 and Shi06AH091) fell outside the 3 main clusters. These Xv isolates form distinctive and extremely tight clusters on branches separate from those of other serotype isolates, suggesting multiple origins of the Xv serotype. The 3 serotype Xv clusters are supported by multiple SNPs, with 5, 15, and 2 SNPs supporting clusters 1, 2 and 3, respectively (Table 1). Four SNPs are shared by clusters 1 and 2, showing a possible common origin. However, cluster 3 seems to have arisen independently, with 2 unique SNPs and a 544-bp deletion at base position 339,247. Cluster 1 contained 17 isolates, 12 of which were from Henan (including the reference strain 2002017), 3 from Shanxi, 1 from Gansu, and 1 from Anhui, covering 14 serotype Xv and 3 serotype X isolates. Thus, cluster 1 is predominantly a Henan cluster. Cluster 2 contained 6 serotype Xv isolates, 5 of which were from Gansu and 1 from Henan. Cluster 3 contained 4 serotype Xv isolates, all of which were from Anhui. Note that the 3 cluster 3 isolates from 2006 were not known to be epidemiologically linked. Thus, there seems to be a regional prevalence of different serotype Xv clusters.
Substitution rate and divergence time estimates for lineage I and serotype Xv clusters.
The 58 isolates with known isolation years from 1954 to 2006 were used for BEAST analysis. With a total of 1,790 SNPs, a BEAST tree was constructed to visualize the overall relationships of the strains, revealing a relationship between root-to-tip branch length and the known isolation dates for the sequenced isolates. We estimated a substitution rate of 8.66 × 10−4 substitutions per site per year at 1,790 chromosomal SNP loci (95% highest posterior density [HPD], 4.17 × 10−4 to 1.34 × 10−3), corresponding to the accumulation of approximately 1.5 SNPs per chromosome per year. The estimated time to the most recent common ancestor (MRCA) for lineage I is 12.25 years (95% HPD, 7.9 to 18.48 years) from 2006; therefore, this lineage seems to have arisen in 1993 (1987 to 1998). The 3 Xv clusters within lineage I seem to have arisen successively. Cluster 1 arose in 1999 (7.53 years [95% HPD, 5.15 to 10.49 years] from 2006) and was the oldest cluster, cluster 2 arose in 2001 (5.42 years [95% HPD, 2.12 to 8.77 years] from 2006) and was the youngest cluster, and cluster 3 arose in 2000 (6.52 years [95% HPD, 3.16 to 10.53 years] from 2006).
Coevolution between chromosome and plasmid pSFXv_2.
The opt gene, on a 6.85-kb plasmid, encodes the O antigen PEtN transferase, which is responsible for attaching a PEtN at the second (RhaII) or third (RhaIII) rhamnose, or both, of the tetrasaccharide repeat unit on the O antigen for the variant serotypes, Xv, Yv, and 4av strains (10, 14). There are 2 known opt alleles, optII and optIII, which are carried by plasmids pSFXv_2 and pSFyv_2, respectively, and preferentially mediate modifications of RhaII and RhaIII, respectively (10, 14). We determined the presence of opt by mapping Illumina reads to the pSFXv_2 sequence and further confirmed the sequence by sequencing the amplified opt PCR products. All Xv strains carried optII, and 3 variants, differing from the optII prototype by 1 or 2 bases, were observed. A single Yv strain carried an optIII form with a single base change from the prototype optIII. All cluster 1 Xv strains carried an opt form identical to the prototype optII. However, the optII from 3 isolates from cluster 2 (Shi06GS55, Shi06GS07, and Shi06GS48), which are grouped together on the SNP tree, contained a single base change. The opt alleles of the other 2 Gansu isolates (Shi06GS37 and Shi06GS43) and 1 Henan isolate (Shi06HN016) in cluster 2 were identical to the optII prototype. The optII allele in cluster 3 differed from the prototype optII by 2 bases. Of the 2 Xv strains outside the main clusters, the optII allele of Shi06AH091, isolated in Anhui, shared 1 base change with cluster 3, while the optII allele of 2001044, isolated in Henan, was identical to the prototype optII. Three nonfunctional opt alleles were also observed, with one each in 3 serotype X strains. The 3 serotype X isolates (2002141, Shi06HN344, and 2005184) were found to carry the pSFXv_2 plasmid. A single-base deletion rendered the opt gene nonfunctional in all 3 cases. The 2002141 optII contained a single-base deletion at base 360, while Shi06HN344 and 2005184 contained a single-base deletion at base 295, resulting in a stop codon that abolished the function of gene. The single-base deletions in Shi06HN344 and 2005184 occurred independently, since the 2 isolates were not clustered together and 2005184 shared 2 base changes with cluster 3 isolates (Fig. 1B).
We further extended our SNP analysis to the whole pSFXv_2 plasmid (see Table S3 in the supplemental material). pSFXv_2 is 6,850 bp in length and encodes a mobilization protein, a replication initiation protein, a lipoprotein, and 7 proteins of unknown function in addition to the O antigen PEtN transferase (10). By mapping raw reads to the reference pSFXv_2 plasmid, we found that all serotype Xv isolates and 3 serotype X isolates carried pSFXv_2, while all other strains did not contain the plasmid. The pSFXv_2 plasmids of cluster 1 and cluster 2 isolates are highly homologous. The 12 serotype Xv isolates and 1 serotype X isolate (2002141) in cluster 1 share 7 base changes with cluster 2. One serotype Xv isolate (Shi06HN347) and 1 serotype X isolate (Shi06HN344) in cluster 1 share only 5 of these 7 bases. There is an additional base change in the opt gene in the 3 isolates from cluster 2. Cluster 3 was dissimilar from the other 2 clusters, with 15 unique base changes. There was good concordance between pSFXv_2 variants and chromosomal clusters, consistent with coevolution of the plasmid and chromosome. The pSFXv_2 variation further confirms that cluster 3 arose independently.
Acquisition of drug resistance genetic elements accompanying lineage I expansion.
Our previous analysis of serotype Xv isolates showed that all were resistant to ampicillin and nalidixic acid, 96% were also resistant to chloramphenicol and tetracycline, and 89.7% were resistant to trimethoprim-sulfamethoxazole (9). In serotype Xv isolate 2002017, the SRL resistance island carries MDR genes for resistance to tetracycline (tetA, tetR, tetC, and tetD), streptomycin (aadA2), ampicillin (oxa-1), and chloramphenicol (cat), while Tn7 harbors MDR elements for resistance to trimethoprim (dfrA1, encoding dihydrofolate reductase), streptothricin (sat1, encoding streptothricin acetyltransferase), and streptomycin/spectinomycin (aadA1, encoding aminoglycoside adenyltransferase) (9). All lineage I isolates carried the full SRL resistance island, except that 2 isolates had partial deletions. Shi06GS48 did not contain the tetD gene, whereas 2005AH264 lacked the oxa-1 and cat genes. All lineage I isolates carried Tn7. There was no sequence variation in the SRL or Tn7 among the lineage I isolates. No other lineages carried the SRL except for a lineage IV isolate, 2002091, which also carried a SRL and Tn7 identical to those of 2002017.
Mutations in gyrA are known to confer quinolone resistance (39). All lineage I isolates harbored 2 gyrA point mutations (248 harbored C→T, resulting in Ser83→Leu, and 631 C→T, resulting in His211→Tyr). Ser83→Leu is a known quinolone resistance mutation (40), whereas His211→Tyr is a novel mutation, located in the quinolone resistance-determining region. The 2 nonsynonymous mutations are markers for lineage I and possibly conferred an advantage to the lineage with quinolone resistance, although it is yet to be determined experimentally whether the His211→Tyr mutation confers quinolone resistance. Additionally, 4 ST91 isolates (Shi06HN378, Shi06SX53, 2001044, and 2002021) harbored an Asp87→Gly mutation in gyrA, 1 ST91 isolate (2001004) harbored an Asp87→Tyr mutation, and 1 ST91 isolate (2005002) had an Asp87→Asn mutation. Only 1 nonlineage I isolate (1997005) had an Asp87→Asn mutation, which clearly is a mutation parallel to that in the lineage I isolate.
There are other signals of antibiotic resistance-driven selection. dfrA5 confers resistance to trimethoprim. Cotrimoxazole, a combination of trimethoprim and sulfamethoxazole, is known to have been used widely in China in the 1990s, although it is no longer recommended for empirical treatment (41). All lineage I isolates (except 2005AH264) carried dfrA5, which is identical to that in 2002017. Outside lineage I, 1 lineage IV isolate (2002091) and 1 lineage III isolate (51581) also carried dfrA5. Additionally, 3 isolates (Shi06AH130, Shi06HN244, and 2005AH264) carried blaCTX-M-14, another 3 isolates (Shi06HN250, Shi06HN118, and 2001042) carried blaTEM-1, and Shi06HN250 carried blaCTX-M-3. Shi06HN250, Shi06SX36, and Shi05SX04 carried qnrS. Strains 2001042 and Shi06HN250 also carried qepA and aac(6′)-Ib-cr, respectively. Acquisition of these genes occurred sporadically and independently. However, acquisition of the SRL, Tn7, and two gyrA mutations are likely to have occurred once in lineage I, as they are present in all lineage I isolates and are associated with lineage I expansion during the 1990s.
Regional dynamics of serotype Xv clusters.
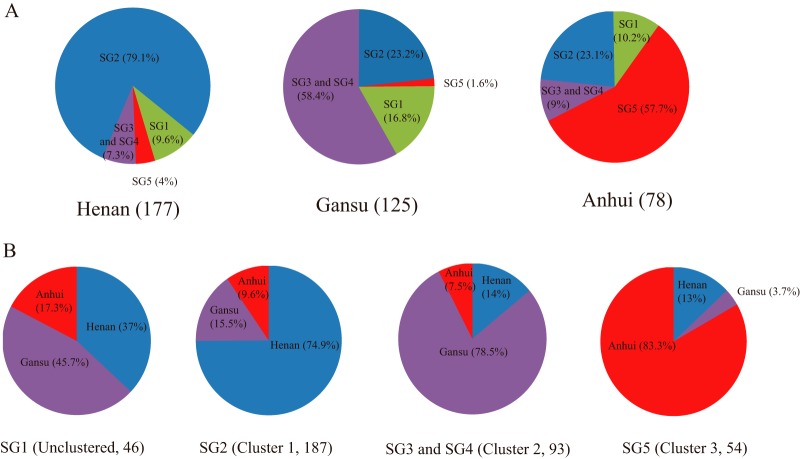
The overwhelming regional clustering of serotype Xv isolates as 3 genome clusters based on the whole-genome sequences prompted us to determine the regional dynamics of the 3 clusters using a much larger sample, by SNP typing of cluster-specific SNPs. There are 18 cluster-specific SNPs, including 4 SNPs from cluster 1, 12 from cluster 2, and 2 from cluster 3 (Fig. 1B). We typed all 380 serotype Xv isolates in our collection from Henan, Gansu, and Anhui provinces using the 18 cluster-specific SNPs. Of these 380 isolates, 177 were isolated from Henan, 125 from Gansu, and 78 from Anhui. The 18 SNPs divided the 380 isolates into 5 SNP genotypes (SGs). SG1 contained 46 isolates, including 17 from Henan, 21 from Gansu, and 8 from Anhui. SG1 grouped together all isolates that did not belong to any of the 3 clusters. SG2 and SG5 corresponded to genome clusters 1 and 3, respectively. Cluster 2 was divided into SG3 and SG4, because one of the cluster 2-specific SNPs (SNP 14) varied within cluster 2 and divided cluster 2 into 2 SGs (see Table S4 in the supplemental material).
Overall, SNP cluster 1 (SG2) is predominant, with 49.2% of all isolates (187/380 isolates) belonging to this SG. SG2 and SG5 are predominantly Henan and Anhui SGs, while SG3 and SG4 are predominantly Gansu SGs. Thus, the regional clustering seen for the genome-sequenced strains is confirmed with this large set of isolates. However, there is also a substantial interregional flow of strains. Henan isolates included 79.1% cluster 1 (SG2), 7.3% cluster 2 (SG3 and SG4), and 4% cluster 3 (SG5) isolates. Gansu isolates included 58.4% cluster 2 (SG3 and SG4), 23.2% cluster 1 (SG2), and 1.6% cluster 3 (SG5) isolates, while Anhui isolates included 57.7% cluster 3 (SG5), 23.1% cluster 1 (SG2), and 9% cluster 2 (SG3 and SG4) isolates (Fig. 2; also see Table S4 in the supplemental material).
FIG 2.
Frequency and geographic distribution of the SNP genotypes of the 380 S. flexneri isolates in China, typed using cluster-specific SNPs. (A) Frequencies of the 5 SGs in different provinces, shown in pie charts. The numbers in parentheses after the province names are the numbers of isolates from the given province. SG3 and SG4 were grouped together to be consistent with genome clusters. (B) Geographic distribution of different SGs (genome clusters). The frequencies of the 5 SGs in different geographic regions (provinces) are shown in pie charts. For consistency with genome clusters, SG3 and SG4 are combined in one group. The genome clusters are indicated in parentheses after the SGs. The numbers in parentheses are the numbers of isolates from the given SG(s).
Since strain 2005184 carries a defective optII (Fig. 1), serotype Xv in cluster 3 must have arisen before 2005184 diverged. This led us to hypothesize that some of the unclustered SG1 isolates may belong to this lineage. We used the 5 unique SNPs present in the branch to 2005184 (Table 1; also see Table S2 in the supplemental material) to type the 46 SG1 isolates, and we found that 10 isolates, including 3 from Henan and 7 from Anhui, belonged to this lineage. Thus, cluster 3 could be extended to include 2005184 and could be defined using the 5 unique SNPs.
Conclusions.
Whole-genome sequencing of 59 S. flexneri isolates revealed that, by acquiring MDR islands (SRL and Tn7) and quinolone resistance gyrA mutations, ST91 arose in the early 1990s, rapidly expanded, and spread across China within a decade. The acquisition of a novel plasmid by S. flexneri, leading to the emergence of a new serotype, occurred more than once. Both the pSFXv_2 plasmid and the opt gene variation suggest independent gain of the plasmid by different clusters, with clear evidence that cluster 3 arose independently as an Xv serotype. There were 3 main clusters of serotype Xv isolates, which were localized in their respective regions, further supporting local origins of serotype Xv. However, there was also a substantial interregional flow of the clusters. In particular, cluster 1, which was predominant in Henan and emerged earliest, spread to other regions with much greater proportions. Therefore, S. flexneri epidemics represent a dynamic play of local strains and interregional spread of different strains. Our results provide significant insights into the evolution of S. flexneri.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by grants from the National Natural Science Foundation of China (grant no. 81271788, 81290340, and 81290345), the National Basic Research Priorities Program (grant no. 2011CB504901), the Project of State Key Laboratory for Infectious Disease Prevention and Control (grant no. 2011SKLID203 and 2008SKLID106), and the National Key Program for Infectious Diseases of China (grant no. 2013ZX10004221 and 2013ZX10004216-001-002).
Footnotes
Published ahead of print 22 January 2014
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JCM.02669-13.
REFERENCES
- 1.Kotloff KL, Winickoff JP, Ivanoff B, Clemens JD, Swerdlow DL, Sansonetti PJ, Adak GK, Levine MM. 1999. Global burden of Shigella infections: implications for vaccine development and implementation of control strategies. Bull. World Health Organ. 77:651–666 [PMC free article] [PubMed] [Google Scholar]
- 2.Simmons DA, Romanowska E. 1987. Structure and biology of Shigella flexneri O antigens. J. Med. Microbiol. 23:289–302. 10.1099/00222615-23-4-289 [DOI] [PubMed] [Google Scholar]
- 3.Carlin NI, Rahman M, Sack DA, Zaman A, Kay B, Lindberg AA. 1989. Use of monoclonal antibodies to type Shigella flexneri in Bangladesh. J. Clin. Microbiol. 27:1163–1166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.El-Gendy A, El-Ghorab N, Lane EM, Elyazeed RA, Carlin NI, Mitry MM, Kay BA, Savarino SJ, Peruski LF., Jr 1999. Identification of Shigella flexneri subserotype 1c in rural Egypt. J. Clin. Microbiol. 37:873–874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Talukder KA, Dutta DK, Safa A, Ansaruzzaman M, Hassan F, Alam K, Islam KM, Carlin NI, Nair GB, Sack DA. 2001. Altering trends in the dominance of Shigella flexneri serotypes and emergence of serologically atypical S. flexneri strains in Dhaka, Bangladesh. J. Clin. Microbiol. 39:3757–3759. 10.1128/JCM.39.10.3757-3759.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Talukder KA, Islam Z, Islam MA, Dutta DK, Safa A, Ansaruzzaman M, Faruque AS, Shahed SN, Nair GB, Sack DA. 2003. Phenotypic and genotypic characterization of provisional serotype Shigella flexneri 1c and clonal relationships with 1a and 1b strains isolated in Bangladesh. J. Clin. Microbiol. 41:110–117. 10.1128/JCM.41.1.110-117.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Stagg RM, Cam PD, Verma NK. 2008. Identification of newly recognized serotype 1c as the most prevalent Shigella flexneri serotype in northern rural Vietnam. Epidemiol. Infect. 136:1134–1140. 10.1017/S0950268807009600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Foster RA, Carlin NI, Majcher M, Tabor H, Ng LK, Widmalm G. 2011. Structural elucidation of the O-antigen of the Shigella flexneri provisional serotype 88-893: structural and serological similarities with S. flexneri provisional serotype Y394 (1c). Carbohydr Res. 346:872–876. 10.1016/j.carres.2011.02.013 [DOI] [PubMed] [Google Scholar]
- 9.Ye C, Lan R, Xia S, Zhang J, Sun Q, Zhang S, Jing H, Wang L, Li Z, Zhou Z, Zhao A, Cui Z, Cao J, Jin D, Huang L, Wang Y, Luo X, Bai X, Wang P, Xu Q, Xu J. 2010. Emergence of a new multidrug-resistant serotype X variant in an epidemic clone of Shigella flexneri. J. Clin. Microbiol. 48:419–426. 10.1128/JCM.00614-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sun Q, Knirel YA, Lan R, Wang J, Senchenkova SN, Jin D, Shashkov AS, Xia S, Perepelov AV, Chen Q, Wang Y, Wang H, Xu J. 2012. A novel plasmid-encoded serotype conversion mechanism through addition of phosphoethanolamine to the O-antigen of Shigella flexneri. PLoS One 7:e46095. 10.1371/journal.pone.0046095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sun Q, Lan R, Wang Y, Wang J, Luo X, Zhang S, Li P, Ye C, Jing H, Xu J. 2011. Genesis of a novel Shigella flexneri serotype by sequential infection of serotype-converting bacteriophages SfX and SfI. BMC Microbiol. 11:269. 10.1186/1471-2180-11-269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Luo X, Sun Q, Lan R, Wang J, Li Z, Xia S, Zhang J, Wang Y, Jin D, Yuan X, Yu B, Cui Z, Xu J. 2012. Emergence of a novel Shigella flexneri serotype 1d in China. Diagn. Microbiol. Infect. Dis. 74:316–319. 10.1016/j.diagmicrobio.2012.06.022 [DOI] [PubMed] [Google Scholar]
- 13.Sun Q, Lan R, Wang J, Xia S, Wang Y, Jin D, Yu B, Knirel YA, Xu J. 2013. Identification and characterization of a novel Shigella flexneri serotype Yv in China. PLoS One 8:e70238. 10.1371/journal.pone.0070238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Knirel YA, Lan R, Senchenkova SN, Wang J, Shashkov AS, Wang Y, Perepelov AV, Xiong Y, Xu J, Sun Q. 2013. O-antigen structure of Shigella flexneri serotype Yv and effect of the lpt-O gene variation on phosphoethanolamine modification of S. flexneri O-antigens. Glycobiology 23:475–485. 10.1093/glycob/cws222 [DOI] [PubMed] [Google Scholar]
- 15.Rajakumar K, Bulach D, Davies J, Ambrose L, Sasakawa C, Adler B. 1997. Identification of a chromosomal Shigella flexneri multi-antibiotic resistance locus which shares sequence and organizational similarity with the resistance region of the plasmid NR1. Plasmid 37:159–168. 10.1006/plas.1997.1280 [DOI] [PubMed] [Google Scholar]
- 16.Turner SA, Luck SN, Sakellaris H, Rajakumar K, Adler B. 2001. Nested deletions of the SRL pathogenicity island of Shigella flexneri 2a. J. Bacteriol. 183:5535–5543. 10.1128/JB.183.19.5535-5543.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Turner SA, Luck SN, Sakellaris H, Rajakumar K, Adler B. 2003. Molecular epidemiology of the SRL pathogenicity island. Antimicrob. Agents Chemother. 47:727–734. 10.1128/AAC.47.2.727-734.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bentley DR, Balasubramanian S, Swerdlow HP, Smith GP, Milton J, Brown CG, Hall KP, Evers DJ, Barnes CL, Bignell HR, Boutell JM, Bryant J, Carter RJ, Keira Cheetham R, Cox AJ, Ellis DJ, Flatbush MR, Gormley NA, Humphray SJ, Irving LJ, Karbelashvili MS, Kirk SM, Li H, Liu X, Maisinger KS, Murray LJ, Obradovic B, Ost T, Parkinson ML, Pratt MR, Rasolonjatovo IM, Reed MT, Rigatti R, Rodighiero C, Ross MT, Sabot A, Sankar SV, Scally A, Schroth GP, Smith ME, Smith VP, Spiridou A, Torrance PE, Tzonev SS, Vermaas EH, Walter K, Wu X, Zhang L, Alam MD, Anastasi C, et al. 2008. Accurate whole human genome sequencing using reversible terminator chemistry. Nature 456:53–59. 10.1038/nature07517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li H, Durbin R. 2010. Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics 26:589–595. 10.1093/bioinformatics/btp698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R. 2009. The Sequence Alignment/Map format and SAMtools. Bioinformatics 25:2078–2079. 10.1093/bioinformatics/btp352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Feng L, Reeves PR, Lan R, Ren Y, Gao C, Zhou Z, Cheng J, Wang W, Wang J, Qian W, Li D, Wang L. 2008. A recalibrated molecular clock and independent origins for the cholera pandemic clones. PLoS One 3:e4053. 10.1371/journal.pone.0004053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li R, Zhu H, Ruan J, Qian W, Fang X, Shi Z, Li Y, Li S, Shan G, Kristiansen K, Yang H, Wang J. 2010. De novo assembly of human genomes with massively parallel short read sequencing. Genome Res. 20:265–272. 10.1101/gr.097261.109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Reeves PR, Liu B, Zhou Z, Li D, Guo D, Ren Y, Clabots C, Lan R, Johnson JR, Wang L. 2011. Rates of mutation and host transmission for an Escherichia coli clone over 3 years. PLoS One 6:e26907. 10.1371/journal.pone.0026907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Smith HE. 2011. Identifying insertion mutations by whole-genome sequencing. Biotechniques 50:96–97. 10.2144/000113600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Posada D. 2008. jModelTest: phylogenetic model averaging. Mol. Biol. Evol. 25:1253–1256. 10.1093/molbev/msn083 [DOI] [PubMed] [Google Scholar]
- 26.Drummond AJ, Rambaut A. 2007. BEAST: Bayesian evolutionary analysis by sampling trees. BMC Evol. Biol. 7:214. 10.1186/1471-2148-7-214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Clinical and Laboratory Standards Institute. 2011. Performance standards for antimicrobial susceptibility testing; 21st informational supplement. M100-S21. Clinical and Laboratory Standards Institute, Wayne, PA [Google Scholar]
- 28.Ahmed AM, Furuta K, Shimomura K, Kasama Y, Shimamoto T. 2006. Genetic characterization of multidrug resistance in Shigella spp. from Japan. J. Med. Microbiol. 55:1685–1691. 10.1099/jmm.0.46725-0 [DOI] [PubMed] [Google Scholar]
- 29.Avsaroglu MD, Helmuth R, Junker E, Hertwig S, Schroeter A, Akcelik M, Bozoglu F, Guerra B. 2007. Plasmid-mediated quinolone resistance conferred by qnrS1 in Salmonella enterica serovar Virchow isolated from Turkish food of avian origin. J. Antimicrob. Chemother. 60:1146–1150. 10.1093/jac/dkm352 [DOI] [PubMed] [Google Scholar]
- 30.Nagano Y, Nagano N, Wachino J, Ishikawa K, Arakawa Y. 2009. Novel chimeric β-lactamase CTX-M-64, a hybrid of CTX-M-15-like and CTX-M-14 β-lactamases, found in a Shigella sonnei strain resistant to various oxyimino-cephalosporins, including ceftazidime. Antimicrob. Agents Chemother. 53:69–74. 10.1128/AAC.00227-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kim JY, Kim SH, Jeon SM, Park MS, Rhie HG, Lee BK. 2008. Resistance to fluoroquinolones by the combination of target site mutations and enhanced expression of genes for efflux pumps in Shigella flexneri and Shigella sonnei strains isolated in Korea. Clin. Microbiol. Infect. 14:760–765. 10.1111/j.1469-0691.2008.02033.x [DOI] [PubMed] [Google Scholar]
- 32.Jaremko M, Justenhoven C, Abraham BK, Schroth W, Fritz P, Brod S, Vollmert C, Illig T, Brauch H. 2005. MALDI-TOF MS and TaqMan assisted SNP genotyping of DNA isolated from formalin-fixed and paraffin-embedded tissues (FFPET). Hum. Mutat. 25:232–238. 10.1002/humu.20141 [DOI] [PubMed] [Google Scholar]
- 33.Jin Q, Yuan Z, Xu J, Wang Y, Shen Y, Lu W, Wang J, Liu H, Yang J, Yang F, Zhang X, Zhang J, Yang G, Wu H, Qu D, Dong J, Sun L, Xue Y, Zhao A, Gao Y, Zhu J, Kan B, Ding K, Chen S, Cheng H, Yao Z, He B, Chen R, Ma D, Qiang B, Wen Y, Hou Y, Yu J. 2002. Genome sequence of Shigella flexneri 2a: insights into pathogenicity through comparison with genomes of Escherichia coli K12 and O157. Nucleic Acids Res. 30:4432–4441. 10.1093/nar/gkf566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wei J, Goldberg MB, Burland V, Venkatesan MM, Deng W, Fournier G, Mayhew GF, Plunkett G, III, Rose DJ, Darling A, Mau B, Perna NT, Payne SM, Runyen-Janecky LJ, Zhou S, Schwartz DC, Blattner FR. 2003. Complete genome sequence and comparative genomics of Shigella flexneri serotype 2a strain 2457T. Infect. Immun. 71:2775–2786. 10.1128/IAI.71.5.2775-2786.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nie H, Yang F, Zhang X, Yang J, Chen L, Wang J, Xiong Z, Peng J, Sun L, Dong J, Xue Y, Xu X, Chen S, Yao Z, Shen Y, Jin Q. 2006. Complete genome sequence of Shigella flexneri 5b and comparison with Shigella flexneri 2a. BMC Genomics 7:173. 10.1186/1471-2164-7-173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Onodera NT, Ryu J, Durbic T, Nislow C, Archibald JM, Rohde JR. 2012. Genome sequence of Shigella flexneri serotype 5a strain M90T Sm. J. Bacteriol. 194:3022. 10.1128/JB.00393-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Allaoui A, Mounier J, Prevost MC, Sansonetti PJ, Parsot C. 1992. icsB: a Shigella flexneri virulence gene necessary for the lysis of protrusions during intercellular spread. Mol. Microbiol. 6:1605–1616. 10.1111/j.1365-2958.1992.tb00885.x [DOI] [PubMed] [Google Scholar]
- 38.Formal SB, Dammin GJ, Labrec EH, Schneider H. 1958. Experimental Shigella infections: characteristics of a fatal infection produced in guinea pigs. J. Bacteriol. 75:604–610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fu Y, Zhang W, Wang H, Zhao S, Chen Y, Meng F, Zhang Y, Xu H, Chen X, Zhang F. 2013. Specific patterns of gyrA mutations determine the resistance difference to ciprofloxacin and levofloxacin in Klebsiella pneumoniae and Escherichia coli. BMC Infect. Dis. 13:8. 10.1186/1471-2334-13-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jeon YL, Nam YS, Lim G, Cho SY, Kim YT, Jang JH, Kim J, Park M, Lee HJ. 2012. Quinolone-resistant Shigella flexneri isolated in a patient who travelled to India. Ann. Lab. Med. 32:366–369. 10.3343/alm.2012.32.5.366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Xia S, Xu B, Huang L, Zhao JY, Ran L, Zhang J, Chen H, Pulsrikarn C, Pornruangwong S, Aarestrup FM, Hendriksen RS. 2011. Prevalence and characterization of human Shigella infections in Henan Province, China, in 2006. J. Clin. Microbiol. 49:232–242. 10.1128/JCM.01508-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.