ABSTRACT
Many murine leukemia viruses (MLVs) are partially resistant to restriction by mouse APOBEC3 (mA3) and essentially fully resistant to induction of G-to-A mutations by mA3. In contrast, Vif-deficient HIV-1 (ΔVif HIV-1) is profoundly restricted by mA3, and the restriction includes high levels of G-to-A mutation. Human APOBEC3G (hA3G), unlike mA3, is fully active against MLVs. We produced a glutathione S-transferase–mA3 fusion protein in insect cells and demonstrated that it possesses cytidine deaminase activity, as expected. This activity is localized within the N-terminal domain of this 2-domain protein; the C-terminal domain is enzymatically inactive but required for mA3 encapsidation into retrovirus particles. We found that a specific arginine residue and several aromatic residues, as well as the zinc-coordinating cysteines in the C-terminal domain, are necessary for mA3 packaging; a structural model of this domain suggests that these residues line a potential nucleic acid-binding interface. Mutation of a few potential phosphorylation sites in mA3 drastically reduces its antiviral activity by impairing either deaminase activity or its encapsidation. mA3 deaminates short single-stranded DNA oligonucleotides preferentially toward their 3′ ends, whereas hA3G exhibits the opposite polarity. However, when packaged into infectious ΔVif HIV-1 virions, both mA3 and hA3G preferentially induce deaminations toward the 5′ end of minus-strand viral DNA, presumably because of the sequence of events during reverse transcription in vivo. Despite the fact that mA3 in MLV particles does not induce detectable deaminations upon infection, its deaminase activity is easily detected in virus lysates. We still do not understand how MLV resists mA3-induced G-to-A mutation.
IMPORTANCE One way that mammalian cells defend themselves against infection by retroviruses is with APOBEC3 proteins. These proteins convert cytidine bases to uridine bases in retroviral DNA. However, mouse APOBEC3 protein blocks infection by murine leukemia viruses without catalyzing this base change, and the mechanism of inhibition is not understood in this case. We have produced recombinant mouse APOBEC3 protein for the first time and characterized it here in a number of ways. Our mutational studies shed light on the mechanism by which mouse APOBEC3 protein is incorporated into retrovirus particles. While mouse APOBEC3 does not catalyze base changes in murine leukemia virus DNA, it can be recovered from these virus particles in enzymatically active form; it is still not clear why it fails to induce base changes when these viruses infect new cells.
INTRODUCTION
Mammals possess several innate mechanisms for defense against retroviruses. One of these restriction factors is the APOBEC3 system. APOBEC3 proteins are cytidine deaminases (1). In many cases, they are encapsidated into assembling virions; when these virions infect a new cell and copy their RNA into DNA, the APOBEC3 protein from the virion binds to the single-stranded DNA (ssDNA; the initial DNA product) and deaminates deoxycytidine residues into deoxyuridine residues. Copying this minus-strand DNA into the complementary coding strand yields replacement of guanine residues with adenine residues, often at remarkably high frequencies. This hypermutation of the viral genome is often a major contributor to the block in infection induced by APOBEC3 proteins (2). Humans have seven APOBEC3 family members, of which human APOBEC3G (hA3G) is the most active against HIV-1. In turn, HIV-1 encodes a small protein, Vif, which binds hA3G in the virus-producing cell and, by causing its degradation, prevents hA3G incorporation into nascent virions (3–5). The actions of hA3G on many gammaretroviruses, including murine leukemia viruses (MLVs), are similar to its effects on Vif-deficient HIV-1 (ΔVif HIV-1) (6, 7).
Rodents encode only a single APOBEC3 family member. We and others have previously analyzed the restriction of retroviruses by mouse APOBEC3 (mA3), which shares ∼33% sequence identity with hA3G (8, 9). mA3 is similar to hA3G in its potency toward ΔVif HIV-1 and in inducing very high levels of G-to-A mutations in ΔVif HIV-1. However, its ability to restrict many murine retroviruses is far lower than that of hA3G. Moreover, the limited restriction of many MLVs by mA3 is not accompanied by elevated levels of mutations in the viral genome; in fact, in these cases the mA3 block to MLV infection apparently occurs at or before the initiation of reverse transcription (8–10). This restriction mechanism is not yet understood.
In order to enhance our understanding of mA3 and its actions on MLVs, we have now produced it in enzymatically active form, using a baculovirus expression system. Our experiments with mutants of mA3 have revealed several characteristics of the enzyme and its biological actions. We also tested MLV particles carrying mA3 for the presence of active deaminase; the results show that the resistance of MLV to mA3-induced hypermutation does not involve irreversible inactivation of the enzyme.
MATERIALS AND METHODS
Cloning.
The clone that expresses mA3 in insect cells was constructed as follows. The mA3-coding region, together with a C-terminal hemagglutinin (HA) epitope tag, was amplified from the plasmid used previously for mammalian cell expression (9); this mA3 gene, which lacks exon 5, was originally from C57BL/6 mice, and this plasmid was a gift from Nathaniel Landau (New York University) (11). The mA3 gene was then placed in the pDest-603 Gateway destination vector using the Gateway cloning technology (Invitrogen) according to the manufacturer's instructions. The resulting baculovirus-based expression construct encodes a protein with an N-terminal histidine tag, followed by glutathione S-transferase (GST), a tobacco etch virus protease cleavage site, mA3, and finally, the HA tag. This clone also expresses green fluorescent protein, so that the growth of the baculovirus vector in infected insect cells can be easily monitored. It was grown initially by transformation of DH10 Bac competent Escherichia coli (Invitrogen), and the recombinant bacmid clones were screened by blue/white selection and confirmed by PCR amplification of the mA3 sequence. All mutations were generated by standard PCR-based mutagenesis, and all mutants were fully confirmed by sequencing.
Experiments in mammalian cells.
Except where noted, experiments with mA3 and hA3G were performed in 293T human cells and used reagents and methods described previously (9, 12). Viruses were produced by transient transfection of 293T cells with 5 μg of a plasmid expressing hA3G or mA3 (or empty control plasmid) together with either 10 μg of our infectious Moloney MLV clone and 3 μg of pBabeLuc or 3 μg of the ΔVif HIV-1 packaging vector, 1 μg of the HIV-1 luciferase-expressing transfer vector, and 0.25 μg of the vesicular stomatitis virus G-protein expression vector (9, 12). The infectivity of the virus preparations was assessed by measuring luciferase activity, as described previously (9). Target cells for viral infections were 293 cells stably expressing the mouse cationic acid transporter (MCAT), the Moloney MLV receptor. Relative specific infectivity is the ratio of luciferase activity to reverse transcriptase (RT) activity divided by the corresponding ratio for virus produced with no mA3 and multiplied by 100. Each experiment was repeated at least once, and all measurements in each experiment were performed in triplicate.
Expression and purification of mA3 from Sf9 insect cells.
Sf9 insect cell lines were cultured at 27°C in Insect-XPRESS medium (Lonza). The cells were transfected with the recombinant bacmids using the Cellfectin reagent (Invitrogen), and the baculovirus was propagated in Sf9 cells as described by the manufacturer (Invitrogen Bac-to-Bac expression system). Sf9 cells expressing wild-type (WT) or mutant mA3 were lysed by sonication in buffer A (50 mM Tris HCl, pH 7.5, 500 mM NaCl, 0.1% NP-40, 1% glycerol, 2 mM β-mercaptoethanol [β-ME], Complete protease inhibitor [Roche]). The lysate was clarified by centrifugation at 20,000 × g, and the clarified extract was loaded onto a nickel-nitrilotriacetic acid (Ni-NTA) column (Novagen). The protein was eluted in buffer B (50 mM Tris HCl, pH 7.5, 500 mM NaCl, 2 mM β-ME) supplemented with 200 mM imidazole, followed by dialysis in buffer B.
Cytidine deaminase assay.
Single-stranded DNA substrates, labeled at their 5′ ends with Alexa Fluor 488 through an N-hydroxysuccinimide ester linkage and with TCC or TTC target sites embedded in AT-rich sequences, were obtained from Integrated DNA Technologies and are listed in Table 1. One substrate, also from Integrated DNA Technologies, was labeled internally with fluorescein. For assays of deamination activity, substrates at 0.25 to 0.5 pmol/μl were incubated with protein in 10 mM Tris (pH 8.0), 50 mM NaCl, 1 mM dithiothreitol (DTT) for 2 h at 37°C. Each reaction mixture in an experiment contained a similar amount of the His-GST-mA3 fusion protein. The reaction mixture was then treated with uracil DNA glycosylase (UDG; Fermentas) in a buffer supplied by the manufacturer and then with NaOH, as described by Iwatani et al. (13). The DNA was then fractionated in denaturing polyacrylamide gels and scanned on a Typhoon phosphorimager (GE Healthcare).
TABLE 1.
Sequences of oligodeoxynucleotides used in in vitro cytidine deaminase assays
| Oligodeoxynucleotide | Sequence |
|---|---|
| Sequence with 3 target sites | 5′-Alexa 488-ATAATAAATTAAATTAATAATCCCTATTTCCTTAATCAATTATTTAAT-3′ |
| Sequence with 1 target site | 5′-Alexa 488-ATAATA AATTAAATTAATATATAATTCAATATTTAAATAAATTATATT-3′ |
| Control | 5′-Alexa 488-ATAATAAATTAAATTAATAATUUUTATTTUUTTAATUAATTATTTAAT-3′ |
| 9D | 5′-Alexa 488-ATAATAATAATAATAATAATAATAATAATAATAATAATAATAATAATAATATCCATAATAATA-3′ |
| 18D | 5′-Alexa 488-ATAATAATAATAATAATAATAATAATAATAATAATAATAATATCCATAATAATAATAATAATA-3′ |
| 27D | 5′-Alexa 488-ATAATAATAATAATAATAATAATAATAATAATATCCATAATAATAATAATAATAATAATAATA-3′ |
| 36D | 5′-Alexa 488-ATAATAATAATAATAATAATAATATCCATAATAATAATAATAATAATAATAATAATAATAATA-3′ |
| 45D | 5′-Alexa 488-ATAATAATAATAATATCCATAATAATAATAATAATAATAATAATAATAATAATAATAATAATA-3′ |
| 51D | 5′-Alexa 488-ATAATAATATCCATAATAATAATAATAATAATAATAATAATAATAATAATAATAATAATAATA-3′ |
| Int-fluor | 5′-ATAATAATAATAATATCCATAATAATAATAATA-fluorescein-ATAATAATAATAATATCCATAATAATAATA ATAATAATAATAATAATA-3′ |
Deaminase activity in virus particles was assayed as follows. Virus was pelleted from culture fluids by centrifugation at 25,000 rpm for 1 h in an SW28 rotor (Beckman Coulter) through a 20% (wt/vol) sucrose cushion in phosphate-buffered saline. The pellet was resuspended in lysis buffer (50 mM Tris, pH 8.0, 40 mM KCl, 50 mM NaCl, 5 mM EDTA, 10 mM DTT, 0.1% Triton X-100) (14). These viral lysates were then assayed for deaminase activity as described above.
RT assays.
In all experiments, the viral infectivity (as measured by luciferase activity) was normalized to the RT activity of the virus preparation. For RT assays, virus particles were precipitated from the culture fluid with 30% polyethylene glycol (molecular weight, 8000; Sigma-Aldrich). The precipitate was resuspended in viral resuspension buffer (50 mM Tris HCl, pH 7.5, 100 mM NaCl, 1 mM EDTA, 5% fetal bovine serum). These preparations were diluted into RT cocktail [for MLV, 62.5 mM Tris, pH 8.0, 25 mM KCl, 0.6 mM MnCl2, 10 mM DTT, 5 μg/ml oligo(dT), 50 μg/ml poly(rA), 0.25% NP-40, 3.75 Ci/mmol [α-32P]dTTP; for HIV, 62.5 mM Tris, pH 8.0, 125 mM NaCl, 7.5 mM MgCl2, 12.5 mM DTT, 5 μg/ml oligo(dT), 50 μg/ml poly(rA), 0.25% NP-40, 3.75 Ci/mmol [α-32P]dTTP] and incubated for 4 h at 37°C. The reaction mixtures were then assayed by spotting on DEAE-cellulose filters; the filters were washed and counted in a scintillation counter. Values were in the linear range with respect to virus concentration (data not shown).
Immunoblotting.
APOBEC3 proteins and viral proteins were detected by immunoblotting using antisera against the HA epitope and p30CA and p24CA, as described previously (9, 15).
Modeling of N- and C-terminal domains of mA3.
The Phyre2 server (16) was employed for homologous modeling. The top four models of the N- or C-terminal domain were constructed on the basis of human APOBEC2 (hA2; PDB accession no. 2NYT), human APOBEC3A (PDB accession no. 2M65), human APOBEC3C (PDB accession no. 3VOW), and the C-terminal domain of hA3G (PDB accession no. 2KBO) structures with 100% confidence. Among the four templates, those with PDB accession no. 2KBO and 2M65 are nuclear magnetic resonance solution structures, whereas those with PDB accession no. 2NYT and 3VOW are crystal structures. Nevertheless, the four models for each domain are very similar. We mainly used the model derived from hA2 in designing mutants of mA3 for analysis. hA2 has ∼30% sequence identity with either the N- or the C-terminal domain of mA3.
Polarity in vivo.
A clone termed pNLdVdELuc, derived from the pNL4-3 molecular clone of HIV-1 but lacking vpr and containing firefly luciferase in place of env (a kind gift of Alok Mulky and Vineet KewalRamani, NCI) (17), was further modified by introduction of two stop codons in the vif gene. Virus was produced by cotransfection of 293T cells with this clone together with a plasmid expressing the vesicular stomatitis virus G protein and either the mammalian mA3 or hA3G expression vector. Fresh cultures of 293T cells were infected with filtered culture fluids from the transfected cells. Twenty-four hours later, DNA was extracted from the infected cells as described by Sanchez-Martinez et al. (10). The DNA was amplified with primers ∼4 kb apart on either the 5′ or the 3′ side of the central polypurine tract, specifically, either with 5′-TGGAAGGGCTAATTCACTCCCAAAGAAGAC-3′ and 5′-CCTGGGCTACAGTCTACTTGTCCATGC-3′, representing nucleotides (nt) 1 to 30 and the complement of nt 4405 to 4379 of pNLdVdELuc, respectively, or with 5′-CAGGGACAGCAGAGATCCAGTTTGGAAAGG-3′ and 5′-GGCTAAGATCTACAGCTGCCTTGTAAGTC, representing nt 4519 to 4548 and the complement of nt 9380 to 9352, respectively. The resulting ∼4-kb PCR products were cloned into pCR2.1 TOPO (Invitrogen), and ∼1 kb at the 5′ and 3′ ends of these 4-kb inserts was sequenced using the M13 forward and reverse primers as sequencing primers. Sequences were aligned using the EBI Clustal W program, and G-to-A mutations were scored with the help of the HYPERMUT program (http://www.hiv.lanl.gov/content/sequence/HYPERMUT/hypermut.html).
RESULTS
Protein expression and purification.
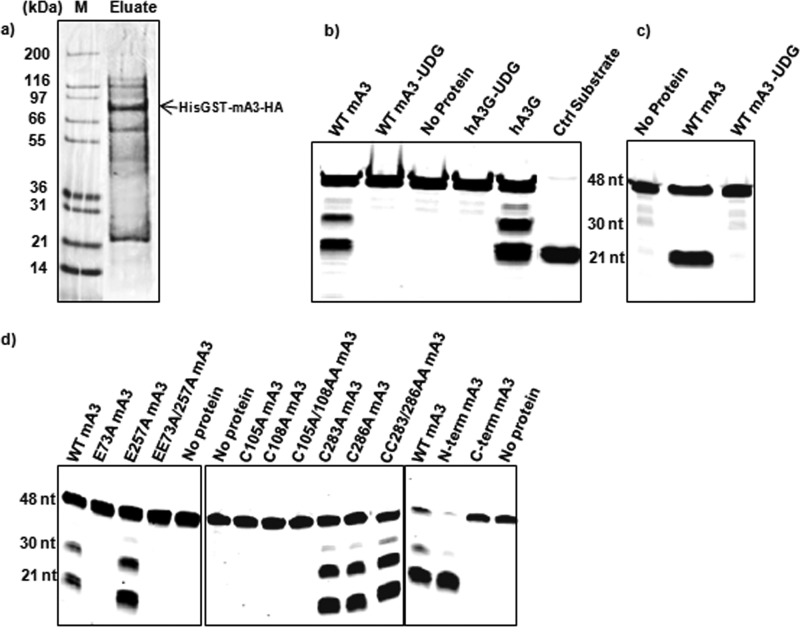
As described in Materials and Methods, we used Sf9 cells to produce a fusion protein with an N-terminal His tag, followed by GST and by the HA-tagged mA3. This protein was purified from the Sf9 lysates by Ni-NTA chromatography. An SDS-polyacrylamide gel of a typical preparation stained with Coomassie blue is shown in Fig. 1a. It is evident that the fusion protein is the predominant protein in this preparation but that it is not completely pure; one major species copurifying with mA3 is an ∼25-kDa protein. As this protein reacts with anti-GST antiserum (data not shown), it appears to arise by spontaneous cleavage of the expressed fusion protein. Smaller quantities of other proteins are also visible in the gel. Although the desired protein was only partially purified by this procedure, it possesses cytidine deaminase activity (see below), and preparations of this type have been used in all of the biochemical experiments described herein.
FIG 1.
Properties of recombinant mA3. (a) SDS-polyacrylamide gel of a typical His-GST-mA3-HA protein preparation used in cytidine deaminase assays. The gel was stained with Coomassie brilliant blue. Lane M, molecular mass markers. (b) Cytidine deaminase assay performed on a 48-base oligodeoxynucleotide with 3 target sites (Table 1) as described in Materials and Methods. −UDG, the reaction mix was not treated with UDG before NaOH was added; Ctrl, control. (c) Cytidine deaminase assay using an oligodeoxynucleotide with 1 target site. (d) Cytidine deaminase assay on mutant and wild-type proteins using the oligodeoxynucleotide with 3 target sites. N-term, N-terminal; C-term, C-terminal.
Deaminase activity.
Recombinant proteins were assayed for cytidine deaminase activity as described in Materials and Methods. Except where otherwise noted, the substrate oligodeoxynucleotides were labeled at their 5′ ends with Alexa Fluor 488 and contained both TCC and TTC sequences embedded in an AT-rich sequence (Table 1). As hA3G will deaminate the final C in the sequence (T/C)CC, a GST-hA3G fusion protein was used as a control in our assays. To assay the partially purified protein for deaminase activity, we incubated mA3 and hA3G proteins with a substrate oligodeoxynucleotide containing the sequences TCCC and TTCC and then treated them with UDG and alkali; oligodeoxynucleotides containing the 5′ label were then visualized on a denaturing polyacrylamide gel. As expected, mA3, like hA3G, deaminated the oligodeoxynucleotide substrate at two or three sites, since two bands were seen if the reaction mixture was treated with UDG and alkali (Fig. 1b) (two of the sites are contained in the 4-base sequence TTCC, and products arising by cleavage at the first and second C residues in this sequence would not be resolved in this gel). A control protein containing only the His-tagged GST moiety was also prepared; no deaminase activity was detected in this protein preparation (data not shown).
As both of the target sequences in the substrate used in Fig. 1b contain TCC, the results did not reveal whether our recombinant mA3 could also deaminate TTC, as mA3 does in vivo. We therefore tested it with a second substrate oligodeoxynucleotide, in which the only cytidine residue was in the context TTC. As shown in Fig. 1c, this substrate was also deaminated by our mA3. Thus, the results show that the recombinant mA3 can deaminate both TCC and TTC sequences.
Mutational analysis of mA3 protein and domain arrangement.
mA3, like hA3G and many other APOBEC3 family members, has two catalytic domains, each with the hallmark H-X-E active site, followed 23 to 28 residues later by the zinc finger motif P-C-X2-4--C (18, 19). In the case of hA3G, only the C-terminal catalytic domain actually possesses catalytic activity; it is not known why the N-terminal domain is not an active deaminase (14). It has been reported, on the basis of experiments in mammalian cells, that in mA3 only the N-terminal domain exhibits deaminase activity (20). To further characterize the deaminase activity of mA3, we tested a series of mA3 mutants in our in vitro enzymatic assay. As shown in Fig. 1d, replacement of the active-site glutamate and the zinc finger cysteines in the N-terminal domain, i.e., E73, C105, or C108, with alanine abolished the deaminase activity of mA3. In contrast, the corresponding changes in the C-terminal domain, i.e., E257A, C283A, or C286A, had no detectable effect on the deaminase activity of the protein. In fact, the N-terminal domain is evidently the sole locus of activity, as removal of the entire C-terminal domain, i.e., residues 200 to 396, did not reduce the activity, while no activity could be detected in the isolated C-terminal domain (Fig. 1d).
Encapsidation: role of C-terminal domain.
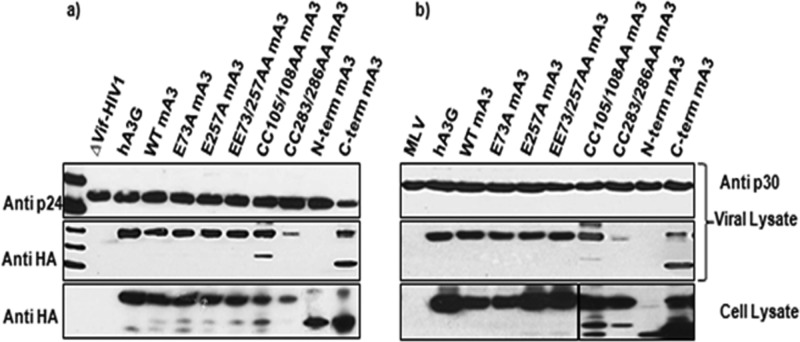
The enzymatic assay results showed that the C-terminal domain is not required for deamination. In hA3G, the domain that lacks deaminase activity is needed for packaging of the protein into virions (14). Therefore, it was of interest to determine whether the C-terminal domain of mA3 contributes to packaging. Cells were transfected with proviral clones together with constructs expressing either the N- or the C-terminal domain of mA3. Virus was then collected and assayed for the presence of the mA3 fragments by immunoblotting for the HA tag, present at the C terminus of each mA3 fragment. As shown in Fig. 2a and b, the C-terminal domain of mA3 is packaged into either ΔVif HIV-1 or MLV particles, even in the complete absence of the N-terminal domain. In contrast, the N-terminal domain is not encapsidated, although it is readily detectable in the virus-producing cells. (A higher-molecular-mass band is also visible in the lane with the C-terminal domain; this is presumably a dimer of the C-terminal domain but was not further characterized).
FIG 2.
Encapsidation of wild-type and mutant mA3s. 293T cells were cotransfected with mA3 expression plasmids and the ΔVif HIV-1 packaging vector (12) (a) or a full-length MLV proviral clone (b). Viruses and cells were harvested and analyzed for CA proteins and the HA tag on mA3 as described previously (9, 15).
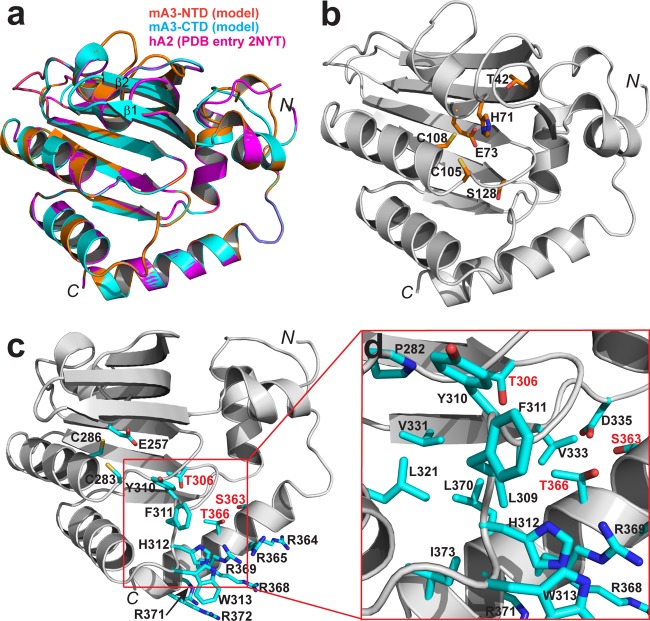
In hopes of obtaining some insight into mA3 activities, we constructed models of the mA3 N- and C-terminal domains. The models were based on homology with hA2, which has ∼30% sequence identity with each mA3 domain. The resulting structural models are shown in Fig. 3. As in the hA2 structure, the models for both mA3 domains show a typical deaminase fold, comprising a compact core structure of 6 helices and 6 β strands in arranged in an α1-β1-β2-β3-α2-β4-α3-β5-α4-β6-α5-α6 configuration, with β1, β2, and β3 being in an antiparallel orientation and β4, β5, and β6 being in parallel orientation (Fig. 3). Differences between the two models and between them and hA2 are confined to the length of two β strands (β1 and β2) and the conformations of loops between secondary structure elements (Fig. 3a). The active-site His-Ala-Glu and zinc finger cysteines are contributed by helices 2 and 3, respectively, in both the N- and C-terminal domains (Fig. 3b and c, respectively). The model-based sequence alignment shows that the two domains are approximately 32% identical in sequence; the differences are most conspicuous on the surfaces, while a hydrophobic core in the interior of the protein is more conserved (the core of the C-terminal domain is depicted in Fig. 3d).
FIG 3.
Structural model of mA3. The N-terminal and C-terminal domains of mA3 (mA3-NTD and mA3-CTD, respectively) were modeled as described in Materials and Methods and are shown superimposed with the template structure hA2 (PDB accession no. 2NYT) (a) and separately in panels b and c, respectively. (d) A zoomed-in view of the mA3-CTD, showing the environment of side chains T306, S363, and T366. Polypeptide chains are illustrated as ribbon diagrams in gray. Helices are shown as spirals, strands are shown as arrows, and loops are shown as tubes. Side chains of key residues are highlighted as stick models in atomic color scheme (N in blue, C in cyan, O in red, and S in yellow). This figure was prepared with the PyMOL molecular graphics system (version 1.5.0.5; Schrödinger, LLC).
It has been shown that encapsidation of hA3G is dependent upon several motifs in its N-terminal domain, including the zinc-coordinating cysteine residues (14); a few basic residues, i.e., R24 and R30, with some contribution from R136 as well (21); and a stretch of aromatic residues, i.e., YYFW (residues 124 to 127) (22, 23); human APOBEC3F and human APOBEC3H encapsidation are similarly dependent upon a cluster of aromatic residues (24, 25). Several lines of evidence suggest that encapsidation of hA3G involves binding to packaged RNA (26), and it has also been proposed that hA3G dimerization is important in hA3G packaging (21, 27, 28). We noted a stretch of aromatic residues in the mA3 C-terminal domain, i.e., Y310-F-H-W313, and, nearby in the model structure, a stretch of nine residues, six of which are arginines (R364-R-T-Q-R-R-L-R-R372). In fact, the zinc-binding C283 and C286, together with the YFHW stretch and arginine residues 365 and 369, could plausibly line a nucleic acid-binding surface in the structure (Fig. 3). Neither the aromatic stretch nor the arginine-rich cluster is present in the N-terminal domain of mA3.
We tested the contributions of each of these motifs to mA3 encapsidation in both MLV and ΔVif HIV-1. Virus particles were produced in transiently transfected 293T cells in the presence of WT or mutant mA3s. As shown in Fig. 2, replacement of the zinc-coordinating cysteines in the C-terminal domain with alanines almost completely prevented encapsidation, while the corresponding change in the N-terminal domain had no evident effect upon packaging. Similar results were reported earlier by Hakata and Landau (20). In contrast, replacement of the active-site glutamate residue in either domain with alanine did not interfere with packaging.
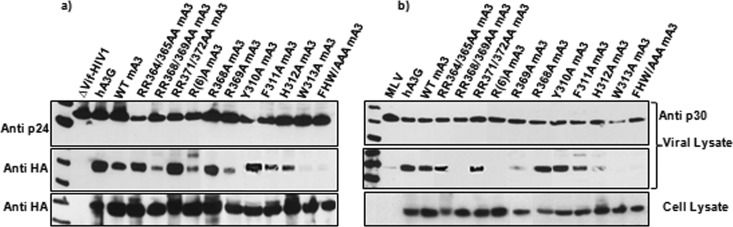
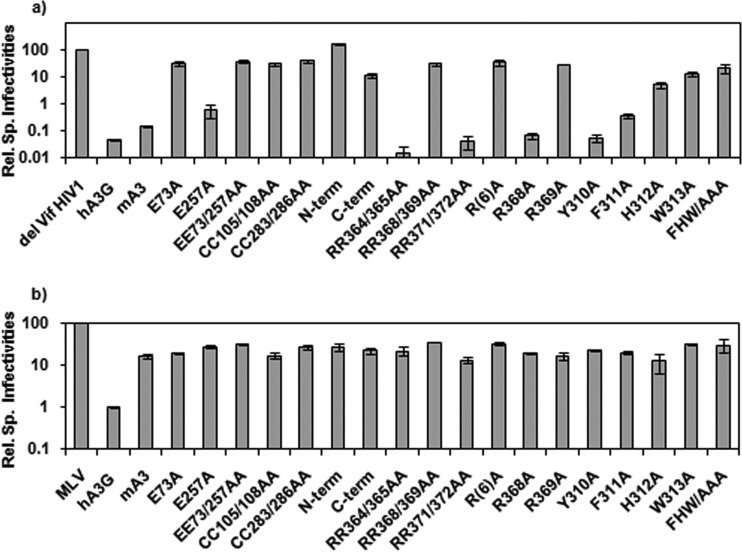
Pairs of arginine residues between R364 and R372 were replaced with alanines. As shown in Fig. 4, RR368/369AA was packaged poorly. Further experiments showed that the R369A point mutant was almost completely defective with respect to encapsidation. When the aromatic stretch was analyzed, it was found that the Y310A mutant retained its ability to be packaged, while packaging of the F311A and H312A mutants was diminished, and there was no detectable encapsidation of W313A mA3 (Fig. 4). The triple mutant 311FHW313/AAA also could not be detected in virus particles. Taken together, these results show that packaging of mA3 into either MLV or ΔVif HIV-1 particles depends upon R369, F311, H312, and W313, as well as upon C283 and/or C286.
FIG 4.
Encapsidation of mA3s with mutations in the C-terminal domain. 293T cells were cotransfected with mA3 expression plasmids and the ΔVif HIV-1 packaging vector (a) or the MLV proviral clone (b). Viruses and cells were analyzed by immunoblotting as described previously (9, 15).
Anti HIV-1 activity of mA3.
We also tested these mutant mA3s for their ability to restrict ΔVif HIV-1. Virus particles were produced in 293T cells as described above, along with retroviral vectors encoding luciferase. The resulting virus preparations were assayed for their ability to induce luciferase activity in permissive target cells, and the resulting infectivity values were normalized to the reverse transcriptase activity. As shown in Fig. 5a, WT mA3 decreased the specific infectivity of ΔVif HIV-1 by nearly 1,000-fold. In contrast, mutants with the following mutations lost virtually all their activity against the virus: E73A, EE73/257AA, CC105/108AA, CC283/286AA, RR368/369AA, R(6)A, R369A, H312A, and W313A. In contrast, mutants with the E257A, RR364/365AA, RR371/372AA, R368A, and Y310A mutations retained their anti-HIV-1 activity, while the F311A mutant was partially active. Although the C-terminal domain was packaged into virions (Fig. 2), it had little antiviral activity.
FIG 5.
Antiviral activity of mutant mA3s. ΔVif HIV-1 (a) or MLV (b) carrying luciferase vectors were produced and assayed as described in Materials and Methods. Infectivities (as measured by the ability to induce luciferase in 293-MCAT cells) were normalized to RT activities, and this ratio for the controls lacking mA3 was set at 100%. Results are presented as relative specific (Rel. Sp.) infectivities. Error bars represent standard deviations.
Anti-MLV activity of mA3.
We also tested the mutant mA3s for their activity against MLV (Fig. 5b). Unfortunately, because WT mA3 does not depress the infectivity of MLV very far, the antiviral activities of the mutants were difficult to assess. For example, in this experiment the MLV-specific infectivity was reduced only ∼8-fold relative to that for the APOBEC3-free control virus by WT mA3 but only ∼3-fold by the EE73/253AA mutant. This 3-fold decrease could, in principle, represent the residual antiviral activity of the mutant mA3, but alternatively, it might simply reflect some nonspecific inhibition due to the presence of mA3. As is evident in Fig. 5b, almost all of the mutants produced MLVs with specific infectivities intermediate between the infectivity of the no-APOBEC3 control and that of the virus with WT mA3. In no case was a specific infectivity greater than that of the no-APOBEC3 control ever observed.
Mutagenesis of putative phosphorylation sites in mA3.
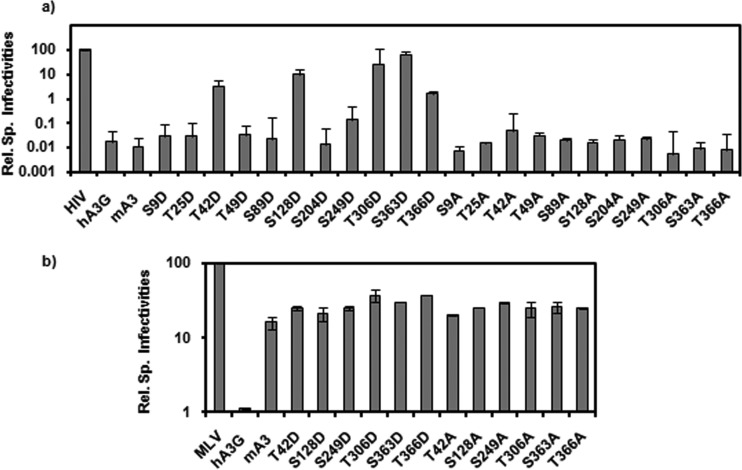
Several reports have raised the possibility that the antiviral activity of hA3G might be regulated by phosphorylation, although this remains controversial (29–31). It was therefore of interest to explore the possibility of phosphorylation of mA3. Inspection of the mA3 sequence with the help of the Prosite (http://prosite.expasy.org/) and Motif Scan (http://myhits.isb-sib.ch/cgi-bin/motif_scan) databases suggested that residues S9, T25, S89, S128, S249, T306, S363, and T366 are possible sites for phosphorylation by protein kinase C; T42 is a possible target site for protein kinase A; and T49, S89, and S204 are possible substrates for casein kinase II phosphorylation. To test the significance of these residues for mA3 activities, each of these residues was mutated to alanine, blocking any phosphorylation at that site in the protein, and to aspartate, mimicking phosphorylation.
The ability of these mutants to restrict ΔVif HIV-1 infectivity was tested as described above. As shown in Fig. 6a, most of the mutations had no significant effect on the anti-HIV-1 activity of mA3, but the T42D, S128D, T306D, S363D, and T366D mutants had all lost most or all of this activity. We also tested a subset of these mA3 mutants against MLV, with the results shown in Fig. 6b; again, the dynamic range of these anti-MLV assays was so small that it is difficult to draw any definitive conclusions from these data.
FIG 6.
Antiviral activity of mA3s with mutations altering putative phosphorylation sites. mA3 mutants were assayed for activity against ΔVif HIV-1 (a) or MLV (b) as described in the legend to Fig. 5.
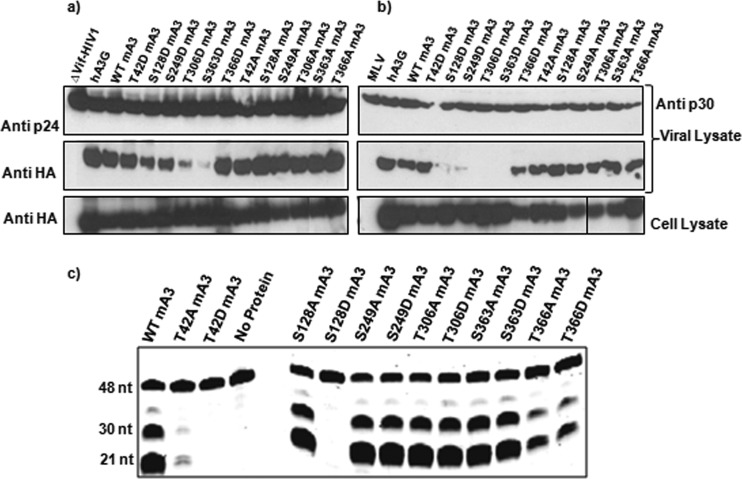
The loss in antiviral activity in these phosphomimetic mutants might reflect a loss in enzymatic activity or a defect in encapsidation. We tested both of these possibilities. We found (Fig. 7a and b) that little if any T306D or S363D mA3 was packaged into ΔVif HIV-1 or MLV virions, while both S128D and S249D mA3s were packaged somewhat less efficiently than wild-type mA3. It is notable that S249D mA3 retained highly significant anti-HIV-1 activity, despite the reduction in the efficiency with which it was packaged. Finally, we expressed some of these mA3 mutants in insect cells and tested the semipurified proteins for cytidine deaminase activity. As shown in Fig. 7c, no enzymatic activity could be detected in the T42D or S128D protein. The activity of T42A mA3 was also diminished relative to that of the wild-type protein; however, we also observed that this mutant mA3 was somewhat unstable in solution (data not shown).
FIG 7.
Encapsidation and cytidine deaminase activity of mA3s with mutations altering putative phosphorylation sites. mA3 mutants were tested for encapsidation into ΔVif HIV-1 (a) or MLV (b) particles, as described previously (9, 15). (c) Mutant and wild-type His-GST-mA3-HA preparations were tested using the oligodeoxynucleotide with 3 target sites.
Polarity of mA3 deamination in vitro and in vivo.
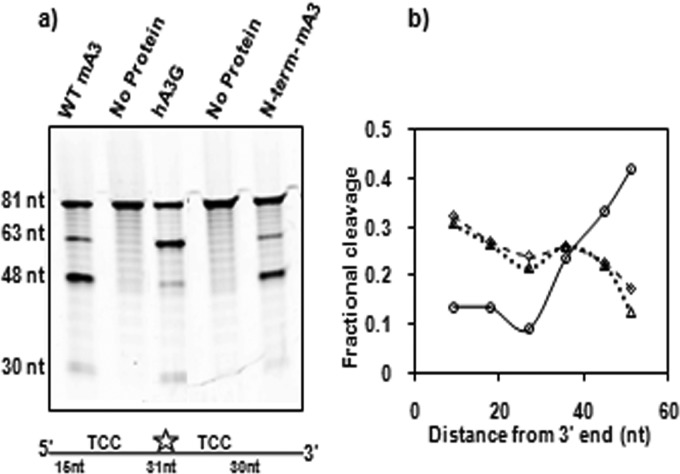
Results from deaminase, encapsidation, and ΔVif HIV-1 infectivity assays clearly demonstrated that mA3 has a domain organization opposite that of hA3G. As originally reported by Chelico et al., hA3G displays a 3′-to-5′ directional bias when deaminating single-stranded oligodeoxynucleotides in vitro (32). It was of interest to investigate whether mA3, with its reversed domain arrangement, exhibited a polarity opposite that of hA3G. We tested this possibility with an oligonucleotide substrate containing 2 TCC sites spaced at different distances from either end. This 81-nucleotide substrate, designated Int-fluor in Table 1, was internally labeled with fluorescein and was composed exclusively of ATA repeats, except for a TCC sequence 15 bases from the 5′ end and another TCC sequence 30 bases from the 3′ end. This asymmetric arrangement of the two target sites meant that cleavage at the 5′ site yields an ∼60-base labeled fragment, while cleavage at the 3′ site produces an ∼45-base labeled fragment. As shown in Fig. 8a, mA3 strongly favored deamination at the 3′ target site, while, as expected, hA3G preferentially deaminated the 5′ site. In fact, the isolated N-terminal domain of mA3 retained the 5′-to-3′ bias of mA3 (Fig. 8a). As a second experimental check of these polarities, we performed an in vitro deamination assay with a series of 63-base ssDNA substrates that were labeled at their 5′ ends; each substrate contained a single TCC site positioned at different distances from the 3′ end (Table 1). As shown in Fig. 8b, purified hA3G showed increased deamination of TCC closer to the 5′ end, as expected. On the other hand, this effect was reversed with mA3: sites closer to the 3′ end were preferentially deaminated (Fig. 8b), confirming the finding that mA3 exhibits a 5′-to-3′ deamination polarity in vitro.
FIG 8.
Polarity of cytidine deamination on oligodeoxynucleotides in vitro. (a) His-GST-mA3-HA or its hA3G equivalent or the protein containing the N-terminal domain of mA3 rather than full-length mA3 was tested with the internally labeled substrate oligodeoxynucleotide (Table 1). (b) Sixty-three-base substrate oligodeoxynucleotides with TCC sites placed at different distances from their 3′ ends (Table 1) were incubated with His-GST-hA3G-HA (solid line) or His-GST-mA3-HA or the protein containing the N-terminal domain of mA3 (dashed and dotted lines, respectively). Cleavage resulting from deamination of each oligodeoxynucleotide was measured.
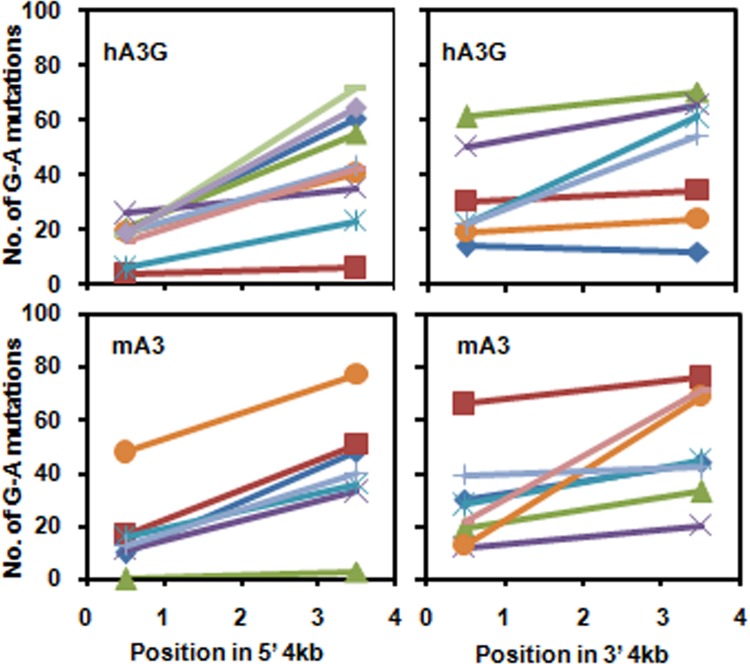
When an HIV-1 particle copies its RNA into DNA in a newly infected cell in the presence of hA3G, the frequency of hA3G-induced G-to-A mutations increases toward the 3′ end of the proviral DNA (33); in fact, this polarity is observed both in the 5′ half and in the 3′ half of the DNA (34, 35). As discussed below, it has been suggested that this dual gradient reflects the complex sequence of events during reverse transcription and the fact that plus-strand DNA synthesis is initiated at two points in the genome. As the functional domains in mA3 are reversed relative to those in hA3G and as the polarity of deamination on short DNAs in vitro is opposite that seen with hA3G, it was of interest to determine whether deaminations with the same polarity as those induced by hA3G would be induced by mA3 in vivo. We therefore infected cells with ΔVif HIV-1 bearing either hA3G or mA3. DNA was isolated from the infected cells, and PCR was performed to amplify the 5′ or the 3′ half of the viral genome. Individual amplification products were then cloned and the 5′ and 3′ kilobase of each clone was sequenced and scored for G-to-A mutations. With the virus bearing hA3G, we found the expected increase in mutation frequency toward the 3′ end of each fragment (Fig. 9), confirming the observations of others (34, 35). We also observed the same trend with the mA3-containing virus (Fig. 9). Thus, although the polarity of deamination by mA3 on short oligonucleotides in vitro differs between hA3G and mA3, they share the same long-range polarity of deamination in vivo.
FIG 9.
Polarity of G-to-A mutation during reverse transcription in vivo. Virus containing mA3 or hA3G was produced from the pNLdVdELuc clone lacking an intact vif open reading frame as described in Materials and Methods. Cells were infected with these viruses, and the DNA produced during infection was analyzed as described in Materials and Methods. Each line in the graphs shows the number of G-to-A mutations in the 5′ ∼1 kb and the 3′ ∼1 kb for a single ∼4-kb PCR product, representing ∼4 kb of DNA on either side of the central polypurine tract produced by a single virus particle.
Deaminase activity of mA3 encapsidated in MLV.
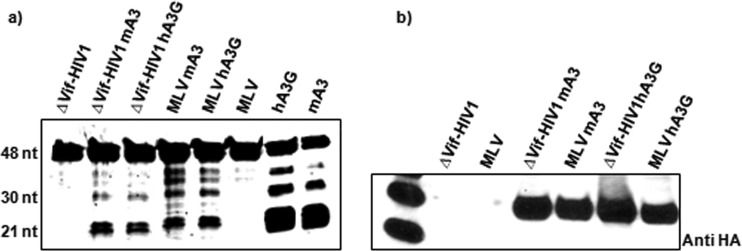
MLV genomes are not deaminated by mA3 in vivo (8, 9). In order to test the possibility that mA3 encapsidated in MLV is somehow inactivated and, hence, incapable of deamination, we assayed viral lysates for the deaminase activity of mA3. MLV or ΔVif HIV-1 virions containing either mA3 or hA3G were lysed as described in Materials and Methods. The viral lysates were then incubated with the DNA substrate containing target deamination sites. As seen in Fig. 10a, mA3 in MLV particles is still enzymatically active, like mA3 in ΔVif HIV-1 and hA3G in MLV or ΔVif HIV-1. Immunoblotting showed that similar levels of mA3 and hA3G were present in the four virus preparations (Fig. 10b). Thus, mA3 retains its deaminase activity within MLV virions; why it fails to deaminate viral DNA following infection is still not known.
FIG 10.
Cytidine deaminase activity in viral lysates. Viruses were produced by transfection of 293T cells with the ΔVif HIV-1 transfer vector (12) or the MLV proviral clone with or without the hA3G or mA3 expression vector (9). Viruses were lysed as described in Materials and Methods and assayed for cytidine deaminase activity, using the DNA substrate with three target sites. Positive control lanes contained recombinant GST-APOBEC3 proteins (a). Viruses were also tested for their mA3 or hA3G content by immunoblotting with anti-HA antiserum (b).
DISCUSSION
The results presented here provide new information regarding the biochemical and biological properties of mA3. Briefly, our biochemical and biological studies confirmed the original finding of Hakata and Landau (20) that the overall domain organization of mA3 is the opposite of that of hA3G: in mA3, the N-terminal domain possesses deaminase activity, while the C-terminal domain is necessary for encapsidation into retrovirus particles. With the help of a homology-based structural model, we identified several aromatic and basic residues in the C-terminal domain that are required for packaging; these residues may all participate in binding viral RNA during virion assembly. These residues are conserved in the mA3s of laboratory mice, despite considerable mA3 polymorphism (19).
mA3 is very potent in the restriction of ΔVif HIV-1. Thus, the effects of mutations upon this restriction can be very large, and so it is obvious from the properties of the mutant mA3s that both deaminase activity and encapsidation are necessary for full restriction of ΔVif HIV-1. However, because the effect of wild-type mA3 upon infectivity of MLV is so much less than that on ΔVif HIV-1, the anti-MLV properties of the mutant mA3s are more difficult to assess. Many mutations in mA3 appeared to reduce, but not eliminate, its anti-MLV activity, but these effects are so small that they might, in some cases, reflect nonspecific phenomena.
It has been suggested that the antiviral activity of hA3G is regulated by phosphorylation (29–31). Although we have no data addressing this possibility directly, we did examine the properties of a number of mA3 mutants in which individual serines or threonines, possibly sites of phosphorylation, were replaced by alanine or aspartate residues. We found that the T42D, S128D, T306D, S363D, and T366D mutants are very deficient in anti-HIV-1 activity. The T42D and S128D mutants are encapsidated efficiently but do not exhibit detectable cytidine deaminase activity in vitro; in our structural model, both T42 and S128 are near the catalytic site in the N-terminal domain of mA3 (Fig. 3b). It seems possible that placement of the negatively charged aspartate side chain at either of these positions inhibits the placement of the nucleic acid substrate in the active site of the enzyme. It is also interesting to note that S128 has undergone positive selection during mouse evolution (19). In contrast, the T306D and S363D mutants exhibit highly active deaminase activities in vitro but are poorly encapsidated. The T366D mutant is packaged efficiently and is enzymatically active, although perhaps less so than wild-type mA3. Our mA3 model (Fig. 3c and d) suggests that the T306 and T366 side chains contact the hydrophobic core of the C-terminal domain; replacement of either of these residues with aspartate would disrupt the core. S363 is very near D335, and an aspartate at position 363 would alter the protein structure by its repulsive interaction with D335. Thus, we would predict that the T306D, S363D, and T366D mutants might interfere with encapsidation by their disruptive effects on the C-terminal domain of mA3.
hA3G has previously been shown to exhibit polarity in its deamination of ssDNA oligonucleotides in vitro, with the frequency of deaminations increasing toward the 5′ end of the substrate (32). It has been suggested that this property reflects the asymmetric charge distribution in hA3G, in which the N-terminal domain has a strong positive charge (pI 9.30), while the C-terminal domain is slightly acidic (pI 5.67) (36). It was thus of interest to test mA3 for its polarity, since as noted above its overall domain arrangement is the reverse of that of hA3G and also since (unlike hA3G) both of its domains are positively charged (pIs of the N- and C-terminal domains, 9.15 and 8.74, respectively). We found (Fig. 8) that the frequency of deamination by mA3 increases toward the 3′ end of an oligodeoxynucleotide that has the reverse polarity relative to that of hA3G. As the overall charge distribution in mA3 is not asymmetric, as it is in hA3G, these results would seem to argue against the role of charge asymmetry in APOBEC3 polarity in vitro.
hA3G also has a strong directional bias in deamination of viral genomes in vivo, where the G-to-A mutation frequency increases significantly toward the 3′ end of the viral genome. As the substrate for deamination in vivo is minus-strand DNA, this represents a bias toward the 5′ end of the substrate. Tests with ΔVif HIV-1 (Fig. 9) showed that mA3 exhibits the same bias as hA3G in vivo; this result is fully consistent with the hypothesis that the 5′ end of the minus strand undergoes a higher frequency of cytidine deamination than the 3′ end because it is single stranded longer during reverse transcription in vivo. There are actually independent gradients in the 5′ and 3′ halves of the HIV-1 genome with mA3 (Fig. 9), just as previously observed with hA3G (34, 35); this is presumably because in HIV-1, synthesis of plus-strand DNA, which progressively covers the minus-strand DNA and protects it from deamination, is initiated from the central polypurine tract in the middle of the genomic RNA, as well as from the polypurine tract near the 3′ end of the RNA (37).
It is striking that the restriction of MLV by mA3 does not involve detectable levels of G-to-A mutation. One conceivable explanation for the lack of mutations could be that the deaminase activity of mA3 is somehow inactivated by encapsidation within an MLV particle. We tested this possibility by lysing MLV virions with mild detergent and found (Fig. 10), contrary to this hypothesis, that these virions contained enzymatically active deaminase. Indeed, similar levels of deaminase activity were recovered from MLV and ΔVif HIV-1 particles containing equivalent amounts of mA3; these levels were also similar to those of hA3G in MLV or ΔVif HIV-1 lysates. Thus, when a population of cells is infected with MLV bearing mA3, DNA synthesis is prevented in a fraction of the virions, while the remainder proceed through the infectious process without hindrance. In contrast, if the virions are HIV-1 rather than MLV, some are blocked before or during DNA synthesis (38–40), perhaps by the roadblock mechanism (41); however, the DNA produced by the remaining particles suffers high levels of mA3-induced G-to-A mutation (10, 11). The action of mA3 on ΔVif HIV-1 thus resembles that of hA3G. The dual effects of hA3G appear to be regulated by a switch between monomers or dimers, which induce G-to-A mutation, and larger oligomers, which act as roadblocks because they dissociate only slowly from ssDNA (42); it is possible that mA3 undergoes similar transitions in ΔVif HIV-1. In contrast, perhaps the structure of MLV particles limits the interconversion of these hypothetical forms of mA3. Recent evidence indicates that the glyco-Gag of MLV affects virus structure in a way that interferes with mA3 action (43, 44). In any case, further work will be required to elucidate the mechanisms of mA3 restriction of MLV and of the resistance of MLV to mA3-induced hypermutation.
Jern et al. (45) have carefully analyzed the sequences of endogenous MLVs, i.e., MLV genomes present in the genomic DNA of mice as a result of ancient infections of germline cells. Notably, some of these sequences show all the hallmarks of mA3-induced G-to-A mutations (including the increase in frequency toward the 3′ end); thus, some of the MLVs whose genomes are now embedded in mouse DNA were evidently not resistant to mA3-induced deamination when they infected the mouse germ line, unlike the recently derived laboratory strains of MLV used here. We have previously reported that a chimeric MLV whose gag gene is from one of these endogenous genomes, while the remainder of its genome is from Moloney MLV, is no more sensitive to mA3-induced deamination than Moloney MLV (10). Thus, some region of the MLV genome other than gag must be responsible for this striking difference between endogenous MLVs and Moloney MLV. It should be noted that the G-to-A mutations are found in members of the polytropic and modified polytropic families of endogenous MLVs, but not in the xenotropic MLV family; this difference correlates well with the presence of glyco-Gag in xenotropic, but not in polytropic or modified polytropic MLVs (45, 46).
Several investigators have compared the replication of MLV in mice lacking a functional mA3 gene to its replication in wild-type mice (47–49). These comparisons show clearly that endogenous mA3 naturally restricts MLV in mice. However, this restriction of laboratory isolates of MLV by naturally expressed mA3 is not accompanied by detectable G-to-A mutation in mice (a low frequency of such mutations has been reported for the endogenous MLV AKV [50]). In other words, the restriction that we have studied under highly nonphysiological circumstances, using transient transfection of 293T human cells with plasmid clones expressing mA3 and the MLV genome, appears to proceed by the same mechanism as that exerted by natural, endogenous mA3 in mice. The restriction by mA3 of the betaretrovirus mouse mammary tumor virus also resembles its restriction of MLV (a gammaretrovirus) in many important respects (51–54). These observations underscore once again the dramatic difference between the mechanism(s) by which hA3G restricts retroviruses and the mechanism of murine beta- and gammaretrovirus restriction by mA3.
ACKNOWLEDGMENTS
We are grateful to Tiyun Wu and Judith Levin for the kind gift of the GST-hA3G protein, Alok Mulky and Vineet KewalRamani for the pNLdVdELuc clone, Jennifer Miller for help with the Typhoon imager, Doaminic Esposito for copious advice as well as many reagents for Gateway cloning and baculovirus expression, Sid Datta for many helpful suggestions, and Jane Mirro and Demetria Harvin for superb technical assistance.
This work was supported by the Intramural Research Program of the National Institutes of Health, National Cancer Institute, Center for Cancer Research.
Footnotes
Published ahead of print 22 January 2014
REFERENCES
- 1.Jarmuz A, Chester A, Bayliss J, Gisbourne J, Dunham I, Scott J, Navaratnam N. 2002. An anthropoid-specific locus of orphan C to U RNA-editing enzymes on chromosome 22. Genomics 79:285–296. 10.1006/geno.2002.6718 [DOI] [PubMed] [Google Scholar]
- 2.Harris RS, Liddament MT. 2004. Retroviral restriction by APOBEC proteins. Nat. Rev. Immunol. 4:868–877. 10.1038/nri1489 [DOI] [PubMed] [Google Scholar]
- 3.Sheehy AM, Gaddis NC, Choi JD, Malim MH. 2002. Isolation of a human gene that inhibits HIV-1 infection and is suppressed by the viral Vif protein. Nature 418:646–650. 10.1038/nature00939 [DOI] [PubMed] [Google Scholar]
- 4.Mehle A, Strack B, Ancuta P, Zhang C, McPike M, Gabuzda D. 2004. Vif overcomes the innate antiviral activity of APOBEC3G by promoting its degradation in the ubiquitin-proteasome pathway. J. Biol. Chem. 279:7792–7798. 10.1074/jbc.M313093200 [DOI] [PubMed] [Google Scholar]
- 5.Yu X, Yu Y, Liu B, Luo K, Kong W, Mao P, Yu XF. 2003. Induction of APOBEC3G ubiquitination and degradation by an HIV-1 Vif-Cul5-SCF complex. Science 302:1056–1060. 10.1126/science.1089591 [DOI] [PubMed] [Google Scholar]
- 6.Harris RS, Bishop KN, Sheehy AM, Craig HM, Petersen-Mahrt SK, Watt IN, Neuberger MS, Malim MH. 2003. DNA deamination mediates innate immunity to retroviral infection. Cell 113:803–809. 10.1016/S0092-8674(03)00423-9 [DOI] [PubMed] [Google Scholar]
- 7.Mangeat B, Turelli P, Caron G, Friedli M, Perrin L, Trono D. 2003. Broad antiretroviral defence by human APOBEC3G through lethal editing of nascent reverse transcripts. Nature 424:99–103. 10.1038/nature01709 [DOI] [PubMed] [Google Scholar]
- 8.Browne EP, Littman DR. 2008. Species-specific restriction of apobec3-mediated hypermutation. J. Virol. 82:1305–1313. 10.1128/JVI.01371-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rulli SJ, Jr, Mirro J, Hill SA, Lloyd P, Gorelick RJ, Coffin JM, Derse D, Rein A. 2008. Interactions of murine APOBEC3 and human APOBEC3G with murine leukemia viruses. J. Virol. 82:6566–6575. 10.1128/JVI.01357-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sanchez-Martinez S, Aloia AL, Harvin D, Mirro J, Gorelick RJ, Jern P, Coffin JM, Rein A. 2012. Studies on the restriction of murine leukemia viruses by mouse APOBEC3. PLoS One 7:e38190. 10.1371/journal.pone.0038190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mariani R, Chen D, Schrofelbauer B, Navarro F, Konig R, Bollman B, Munk C, Nymark-McMahon H, Landau NR. 2003. Species-specific exclusion of APOBEC3G from HIV-1 virions by Vif. Cell 114:21–31. 10.1016/S0092-8674(03)00515-4 [DOI] [PubMed] [Google Scholar]
- 12.Derse D, Hill SA, Lloyd PA, Chung H, Morse BA. 2001. Examining human T-lymphotropic virus type 1 infection and replication by cell-free infection with recombinant virus vectors. J. Virol. 75:8461–8468. 10.1128/JVI.75.18.8461-8468.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Iwatani Y, Takeuchi H, Strebel K, Levin JG. 2006. Biochemical activities of highly purified, catalytically active human APOBEC3G: correlation with antiviral effect. J. Virol. 80:5992–6002. 10.1128/JVI.02680-05 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Navarro F, Bollman B, Chen H, Konig R, Yu Q, Chiles K, Landau NR. 2005. Complementary function of the two catalytic domains of APOBEC3G. Virology 333:374–386. 10.1016/j.virol.2005.01.011 [DOI] [PubMed] [Google Scholar]
- 15.Crist RM, Datta SA, Stephen AG, Soheilian F, Mirro J, Fisher RJ, Nagashima K, Rein A. 2009. Assembly properties of human immunodeficiency virus type 1 Gag-leucine zipper chimeras: implications for retrovirus assembly. J. Virol. 83:2216–2225. 10.1128/JVI.02031-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kelley LA, Sternberg MJ. 2009. Protein structure prediction on the Web: a case study using the Phyre server. Nat. Protoc. 4:363–371. 10.1038/nprot.2009.2 [DOI] [PubMed] [Google Scholar]
- 17.Datta SA, Temeselew LG, Crist RM, Soheilian F, Kamata A, Mirro J, Harvin D, Nagashima K, Cachau RE, Rein A. 2011. On the role of the SP1 domain in HIV-1 particle assembly: a molecular switch? J. Virol. 85:4111–4121. 10.1128/JVI.00006-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.LaRue RS, Andresdottir V, Blanchard Y, Conticello SG, Derse D, Emerman M, Greene WC, Jonsson SR, Landau NR, Lochelt M, Malik HS, Malim MH, Munk C, O'Brien SJ, Pathak VK, Strebel K, Wain-Hobson S, Yu XF, Yuhki N, Harris RS. 2009. Guidelines for naming nonprimate APOBEC3 genes and proteins. J. Virol. 83:494–497. 10.1128/JVI.01976-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sanville B, Dolan MA, Wollenberg K, Yan Y, Martin C, Yeung ML, Strebel K, Buckler-White A, Kozak CA. 2010. Adaptive evolution of Mus Apobec3 includes retroviral insertion and positive selection at two clusters of residues flanking the substrate groove. PLoS Pathog. 6:e1000974. 10.1371/journal.ppat.1000974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hakata Y, Landau NR. 2006. Reversed functional organization of mouse and human APOBEC3 cytidine deaminase domains. J. Biol. Chem. 281:36624–36631. 10.1074/jbc.M604980200 [DOI] [PubMed] [Google Scholar]
- 21.Huthoff H, Autore F, Gallois-Montbrun S, Fraternali F, Malim MH. 2009. RNA-dependent oligomerization of APOBEC3G is required for restriction of HIV-1. PLoS Pathog. 5:e1000330. 10.1371/journal.ppat.1000330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Huthoff H, Malim MH. 2007. Identification of amino acid residues in APOBEC3G required for regulation by human immunodeficiency virus type 1 Vif and virion encapsidation. J. Virol. 81:3807–3815. 10.1128/JVI.02795-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang T, Tian C, Zhang W, Luo K, Sarkis PT, Yu L, Liu B, Yu Y, Yu XF. 2007. 7SL RNA mediates virion packaging of the antiviral cytidine deaminase APOBEC3G. J. Virol. 81:13112–13124. 10.1128/JVI.00892-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang T, Tian C, Zhang W, Sarkis PT, Yu XF. 2008. Interaction with 7SL RNA but not with HIV-1 genomic RNA or P bodies is required for APOBEC3F virion packaging. J. Mol. Biol. 375:1098–1112. 10.1016/j.jmb.2007.11.017 [DOI] [PubMed] [Google Scholar]
- 25.Zhen A, Du J, Zhou X, Xiong Y, Yu XF. 2012. Reduced APOBEC3H variant anti-viral activities are associated with altered RNA binding activities. PLoS One 7:e38771. 10.1371/journal.pone.0038771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Svarovskaia ES, Xu H, Mbisa JL, Barr R, Gorelick RJ, Ono A, Freed EO, Hu WS, Pathak VK. 2004. Human apolipoprotein B mRNA-editing enzyme-catalytic polypeptide-like 3G (APOBEC3G) is incorporated into HIV-1 virions through interactions with viral and nonviral RNAs. J. Biol. Chem. 279:35822–35828. 10.1074/jbc.M405761200 [DOI] [PubMed] [Google Scholar]
- 27.Bulliard Y, Turelli P, Rohrig UF, Zoete V, Mangeat B, Michielin O, Trono D. 2009. Functional analysis and structural modeling of human APOBEC3G reveal the role of evolutionarily conserved elements in the inhibition of human immunodeficiency virus type 1 infection and Alu transposition. J. Virol. 83:12611–12621. 10.1128/JVI.01491-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Friew YN, Boyko V, Hu WS, Pathak VK. 2009. Intracellular interactions between APOBEC3G, RNA, and HIV-1 Gag: APOBEC3G multimerization is dependent on its association with RNA. Retrovirology 6:56. 10.1186/1742-4690-6-56 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Demorest ZL, Li M, Harris RS. 2011. Phosphorylation directly regulates the intrinsic DNA cytidine deaminase activity of activation-induced deaminase and APOBEC3G protein. J. Biol. Chem. 286:26568–26575. 10.1074/jbc.M111.235721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kopietz F, Jaguva Vasudevan AA, Kramer M, Muckenfuss H, Sanzenbacher R, Cichutek K, Flory E, Munk C. 2012. Interaction of human immunodeficiency virus type 1 Vif with APOBEC3G is not dependent on serine/threonine phosphorylation status. J. Gen. Virol. 93:2425–2430. 10.1099/vir.0.043273-0 [DOI] [PubMed] [Google Scholar]
- 31.Shirakawa K, Takaori-Kondo A, Yokoyama M, Izumi T, Matsui M, Io K, Sato T, Sato H, Uchiyama T. 2008. Phosphorylation of APOBEC3G by protein kinase A regulates its interaction with HIV-1 Vif. Nat. Struct. Mol. Biol. 15:1184. 10.1038/nsmb.1497 [DOI] [PubMed] [Google Scholar]
- 32.Chelico L, Pham P, Calabrese P, Goodman MF. 2006. APOBEC3G DNA deaminase acts processively 3′ → 5′ on single-stranded DNA. Nat. Struct. Mol. Biol. 13:392–399. 10.1038/nsmb1086 [DOI] [PubMed] [Google Scholar]
- 33.Yu Q, Konig R, Pillai S, Chiles K, Kearney M, Palmer S, Richman D, Coffin JM, Landau NR. 2004. Single-strand specificity of APOBEC3G accounts for minus-strand deamination of the HIV genome. Nat. Struct. Mol. Biol. 11:435–442. 10.1038/nsmb758 [DOI] [PubMed] [Google Scholar]
- 34.Suspène R, Rusniok C, Vartanian J-P, Wain-Hobson S. 2006. Twin gradients in APOBEC3 edited HIV-1 DNA reflect the dynamics of lentiviral replication. Nucleic Acids Res. 34:4677–4684. 10.1093/nar/gkl555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hu C, Saenz DT, Fadel HJ, Walker W, Peretz M, Poeschla EM. 2010. The HIV-1 central polypurine tract functions as a second line of defense against APOBEC3G/F. J. Virol. 84:11981–11993. 10.1128/JVI.00723-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chelico L, Prochnow C, Erie DA, Chen XS, Goodman MF. 2010. Structural model for deoxycytidine deamination mechanisms of the HIV-1 inactivation enzyme APOBEC3G. J. Biol. Chem. 285:16195–16205. 10.1074/jbc.M110.107987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Charneau P, Alizon M, Clavel F. 1992. A second origin of DNA plus-strand synthesis is required for optimal human immunodeficiency virus replication. J. Virol. 66:2814–2820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bishop KN, Holmes RK, Malim MH. 2006. Antiviral potency of APOBEC proteins does not correlate with cytidine deamination. J. Virol. 80:8450–8458. 10.1128/JVI.00839-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Holmes RK, Malim MH, Bishop KN. 2007. APOBEC-mediated viral restriction: not simply editing? Trends Biochem. Sci. 32:118–128. 10.1016/j.tibs.2007.01.004 [DOI] [PubMed] [Google Scholar]
- 40.Mbisa JL, Barr R, Thomas JA, Vandegraaff N, Dorweiler IJ, Svarovskaia ES, Brown WL, Mansky LM, Gorelick RJ, Harris RS, Engelman A, Pathak VK. 2007. Human immunodeficiency virus type 1 cDNAs produced in the presence of APOBEC3G exhibit defects in plus-strand DNA transfer and integration. J. Virol. 81:7099–7110. 10.1128/JVI.00272-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Iwatani Y, Chan DS, Wang F, Maynard KS, Sugiura W, Gronenborn AM, Rouzina I, Williams MC, Musier-Forsyth K, Levin JG. 2007. Deaminase-independent inhibition of HIV-1 reverse transcription by APOBEC3G. Nucleic Acids Res. 35:7096–7108. 10.1093/nar/gkm750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chaurasiya KR, McCauley MJ, Wang W, Qualley DF, Wu T, Kitamura S, Geertsema H, Chan DS, Hertz A, Iwatani Y, Levin JG, Musier-Forsyth K, Rouzina I, Williams MC. 2014. Oligomerization transforms human APOBEC3G from an efficient enzyme to a slowly dissociating nucleic acid-binding protein. Nat. Chem. 6:28–33. 10.1038/nchem.1795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kolokithas A, Rosenke K, Malik F, Hendrick D, Swanson L, Santiago ML, Portis JL, Hasenkrug KJ, Evans LH. 2010. The glycosylated Gag protein of a murine leukemia virus inhibits the antiretroviral function of APOBEC3. J. Virol. 84:10933–10936. 10.1128/JVI.01023-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Stavrou S, Nitta T, Kotla S, Ha D, Nagashima K, Rein AR, Fan H, Ross SR. 2013. Murine leukemia virus glycosylated Gag blocks apolipoprotein B editing complex 3 and cytosolic sensor access to the reverse transcription complex. Proc. Natl. Acad. Sci. U. S. A. 110:9078–9083. 10.1073/pnas.1217399110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Jern P, Stoye JP, Coffin JM. 2007. Role of APOBEC3 in genetic diversity among endogenous murine leukemia viruses. PLoS Genet. 3:2014–2022. 10.1371/journal.pgen.0030183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nitta T, Lee S, Ha D, Arias M, Kozak CA, Fan H. 2012. Moloney murine leukemia virus glyco-gag facilitates xenotropic murine leukemia virus-related virus replication through human APOBEC3-independent mechanisms. Retrovirology 9:58. 10.1186/1742-4690-9-58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Low A, Okeoma CM, Lovsin N, de las Heras M, Taylor TH, Peterlin BM, Ross SR, Fan H. 2009. Enhanced replication and pathogenesis of Moloney murine leukemia virus in mice defective in the murine APOBEC3 gene. Virology 385:455–463. 10.1016/j.virol.2008.11.051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Santiago ML, Montano M, Benitez R, Messer RJ, Yonemoto W, Chesebro B, Hasenkrug KJ, Greene WC. 2008. Apobec3 encodes Rfv3, a gene influencing neutralizing antibody control of retrovirus infection. Science 321:1343–1346. 10.1126/science.1161121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Takeda E, Tsuji-Kawahara S, Sakamoto M, Langlois M-A, Neuberger MS, Rada C, Miyazawa M. 2008. Mouse APOBEC3 restricts friend leukemia virus infection and pathogenesis in vivo. J. Virol. 82:10998–11008. 10.1128/JVI.01311-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Langlois MA, Kemmerich K, Rada C, Neuberger MS. 2009. The Akv murine leukemia virus is restricted and hypermutated by mouse Apobec3. J. Virol. 83:11550–11559. 10.1128/JVI.01430-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.MacMillan AL, Kohli RM, Ross SR. 2013. APOBEC3 inhibition of mouse mammary tumor virus infection: the role of cytidine deamination versus inhibition of reverse transcription. J. Virol. 87:4808–4817. 10.1128/JVI.00112-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Okeoma CM, Huegel AL, Lingappa J, Feldman MD, Ross SR. 2010. APOBEC3 proteins expressed in mammary epithelial cells are packaged into retroviruses and can restrict transmission of milk-borne virions. Cell Host Microbe 8:534–543. 10.1016/j.chom.2010.11.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Okeoma CM, Lovsin N, Peterlin BM, Ross SR. 2007. APOBEC3 inhibits mouse mammary tumour virus replication in vivo. Nature 445:927–930. 10.1038/nature05540 [DOI] [PubMed] [Google Scholar]
- 54.Okeoma CM, Petersen J, Ross SR. 2009. Expression of murine APOBEC3 alleles in different mouse strains and their effect on mouse mammary tumor virus infection. J. Virol. 83:3029–3038. 10.1128/JVI.02536-08 [DOI] [PMC free article] [PubMed] [Google Scholar]