Abstract
HIV undergoes high rates of mutation and recombination during reverse transcription, but it is not known whether these events occur independently or are linked mechanistically. Here we used a system of silent marker mutations in HIV and a single round of infection in primary T lymphocytes combined with a high-throughput sequencing and mathematical modeling approach to directly estimate the viral recombination and mutation rates. From >7 million nucleotides (nt) of sequences from HIV infection, we observed 4,801 recombination events and 859 substitution mutations (≈1.51 and 0.12 events per 1,000 nt, respectively). We used experimental controls to account for PCR-induced and transfection-induced recombination and sequencing error. We found that the single-cycle virus-induced mutation rate is 4.6 × 10−5 mutations per nt after correction. By sorting of our data into recombined and nonrecombined sequences, we found a significantly higher mutation rate in recombined regions (P = 0.003 by Fisher's exact test). We used a permutation approach to eliminate a number of potential confounding factors and confirm that mutation occurs around the site of recombination and is not simply colocated in the genome. By comparing mutation rates in recombined and nonrecombined regions, we found that recombination-associated mutations account for 15 to 20% of all mutations occurring during reverse transcription.
INTRODUCTION
The development of a genetically diverse quasispecies is one of the hallmarks of HIV infection. Genetic diversity underpins the ability of HIV to evolve resistance to antiretroviral therapy and to successfully evade the immune system. Viral diversity increases rapidly during initial infection (1) and is driven by mutation, recombination, and population size. Mutations are introduced primarily during reverse transcription by the error-prone reverse transcriptase (RT) enzyme (2) or by host cellular defense mechanisms (3, 4). These mutations can then be shuffled within the viral quasispecies by retroviral recombination, which relies on the copackaging of two genetically distinct RNA genomes (for a review, see references 5–9). While the dimer initiation sequence (DIS) is not critical for HIV replication in primary cells (10–12), the DIS is important to facilitate the recombination process, presumably by bringing genomic RNA sequences into close proximity during virion assembly and viral cDNA synthesis (13–17). Retroviral recombination occurs during reverse transcription when the RT enzyme switches between strands of the two copackaged RNA genomes to produce a chimeric DNA molecule (5–7, 18).
Whether retroviral mutation is linked with recombination has been a subject of debate for decades, and numerous studies have measured HIV mutation and retroviral recombination rates in vitro and in cell culture (2, 19–31). Nevertheless, the question of whether mutation and recombination are associated has not been resolved directly due to numerous inherent technical difficulties. To assess retroviral recombination, many studies utilize PCR to amplify and to clone out the engineered reporter genes within the retroviral vectors (or intersubtype HIV constructs) from the infected cells, which is followed by expression of cloned sequences in bacteria to indirectly assess the recombination and mutation rates (32–34). Others have taken advantage of retroviral vector or reporter systems, in which the expressions of reporter genes (such as fluorescent proteins, cell surface markers, and/or antibiotic resistance genes) were used to select cells containing recombined viral genomes for recombination and mutation analyses (28–31, 35–39). As coexpression of reporter gene products in a single cell is needed to identify recombination events, these systems generally use a low multiplicity of infection (MOI) to avoid dual infection introducing experimental bias (28–39). However, as these systems require expression of the gene products, natural mutation events or recombination events that lead to nonproductive expression of reporter genes will be unaccounted for. It would have been advantageous if recombination and mutation rates could be estimated directly from the newly synthesized and/or integrated viral cDNA that is independent of the MOI. Furthermore, as those studies depend on PCR to amplify specific genomic sequences for recombination and/or mutation analyses (28–30), the potential of PCR-induced recombination and mutation must be controlled for to limit overestimation of these two retroviral biological processes (40).
Using a transfection approach to generate heterozygous HIV to assess retroviral recombination, we have previously shown that HIV recombination does not randomly occur throughout the viral genome (20, 40). Based on the observed recombination events in HIV gag, we have found that HIV-1 undergoes 1.35 × 10−3 recombination events per nucleotide (REPN) per replication cycle (20). It is estimated that HIV-1 has a mutation rate that produces approximately 0.34 mutations per replication cycle (2, 19, 40, 41). Consequently, mutation and recombination associate relatively infrequently, and a large amount of sequencing data across the HIV genome is needed to determine whether recombination and mutations are linked. While reporter systems that can generate large quantities of data have been developed to measure either mutation rates or recombination rates on a nonviral sequence (30, 35, 36, 38, 42–45), these systems, however, cannot simultaneously measure both recombination and mutation rates. Furthermore, these systems measure rates within foreign nonviral gene sequences. As in vitro studies have shown that template sequence and structure are important determinants of these processes (46, 47), the most accurate measurements will be derived from direct sequencing of the viral genome (20). However, before the advent of next-generation sequencing technology, generation of the large amounts of sequencing data required to answer this question was not feasible.
We have previously described a method to quantify the rate of recombination by direct sequencing of the HIV-1 genome within infected primary T lymphocytes (20, 40). This system addresses several issues that can otherwise bias the analysis of mutation and recombination rates using conventional approaches. First, although recombination is most easily measured by using highly divergent strains of HIV (as recombination can be observed only through the mixing of genetic marker points), the rate of recombination is greatly affected by the overall homology between genomes and processivity of RT (15, 16). Thus, recombination rate measurements using highly divergent HIV strains do not reflect the recombination occurring between highly related members of a viral quasispecies found within an infected individual (48). Our system addresses this issue by measuring recombination between two closely related HIV-1 genomes that differ by silent mutations found in naturally occurring HIV sequences, and these HIV sequences display identical replication kinetics in primary T lymphocytes (20). Second, by sequencing of viral cDNAs that are produced within 24 h of infection (with the fusion inhibitor T-20 supplied at 6 h postinfection to block secondary infection), all HIV RT mutations and recombination that may lead to nonproductive infection will also be accounted for. Our previous work also showed that a low MOI is not vital for this type of recombination analysis and does not yield intervirion recombination or homologous recombination of plasmid DNA from transfection that may bias data interpretation (20). Third, we have developed experimental and analytical tools to account for artifacts that can compromise analysis. For example, the impact of multiple recombination events between two marker points (20), recombination during cotransfection of plasmids, and PCR-induced recombination (40) must be all minimized and controlled for in any analysis. Similarly, any analysis must take into account the possibility that coincident high or low rates of mutation and recombination (without the rate of mutation being specifically increased at sites of recombination) can lead to a spurious association.
In this study, we have utilized a system of marker sites introduced into HIV-1 to infect primary T lymphocytes, followed by high-throughput sequencing. We have found that the previous estimate of the nucleotide substitution rate closely matched our calculated error rate of 4.6 × 10−5 using direct sequencing and observed a significantly higher mutation rate in recombined regions. We eliminated a number of potential causes for this association to demonstrate a direct association between the processes of recombination and mutation. We show that 15 to 20% of the total mutations are associated with recombination.
MATERIALS AND METHODS
Molecular clones.
The wild-type (WT) HIV-1 plasmid used was pDRNL(AD8) (49). The marker (MK) plasmid [pDRNL(AD8)GagPolMarker] was created by introducing 15 and 35 genetic marker points by silent transition mutation in the gag and pol genes of pDRNL(AD8), respectively. pDNRL(AD8) is an R5-tropic strain of HIV and thus infects the activated/memory T-cell lymphocyte subset (50). This created a total of 46 intervals spaced an average of 53.5 nucleotides (nt) apart (range, 17 to 155 nucleotides; median, 47 nucleotides), where recombination and mutation could be simultaneously measured. The marker sites were chosen on the basis that they were (i) silent mutations in the third position, (ii) located in adjacent codons, and (iii) A→G mutations observed in the HIV sequence database (HIV Sequence Compendium [http://www.hiv.lanl.gov/]). Only four markers contained mutations in nonadjacent codons. In addition, the first and last markers in pol contained silent, but non-A→G, mutations and introduced the XhoI and NotI restriction sites, respectively, into the genome. Two mutations in adjacent codons were used so that recombination could be distinguished from mutations introduced during the PCR or sequencing reactions. We reasoned that A-to-G mutations would have the least impact on the RNA structure, as U can base pair with both A and G in RNA. Consequently, the introduction of A-to-G marker points does not grossly disrupt base pairing within RNA structure regions. Nevertheless, we did not modify regions of the HIV-1 genome with known functional secondary structure, and we preferentially modified regions of the genome containing these mutations as natural polymorphisms (HIV Sequence Compendium [http://www.hiv.lanl.gov/]). The MK plasmid was constructed, using standard molecular cloning techniques, from two regions corresponding to gag and pol that were designed electronically and synthesized chemically. The 14-day replication kinetics of marker virus was indistinguishable from that of WT virus, and no protein or known RNA sequence elements were changed.
Viruses.
Homozygous virus was produced by transfection of 293T cells with either the WT or MK pDRNLAD8 plasmid. In general, a total of 3 μg of plasmid DNA containing proviral HIV genomes was used to transfect 293T cells for the production of HIV particles. Heterozygous virus was produced by cotransfection of equal masses of WT and MK pDRNLAD8 plasmids into 293T cells. When equal amounts of two HIV plasmids are cotransfected, copackaging of RNA into virions is random (13). Consequently, we expected 50% heterozygous virions, 25% homozygous WT virions, and 25% homozygous marker virions. Transfections were carried out with polyethylenimine (PEI; Polysciences), and transfection efficiencies were measured by using a reverse transcriptase assay. While recombination has been observed during transfection under certain experimental conditions (51–53), we have previously shown that these transfection conditions do not yield observable recombination (20). Furthermore, we have also converted the virion-associated RNA to cDNA for direct sequencing, which showed that transfection-induced recombination (TIR) events had a negligible influence on our results (see below). At 36 h posttransfection, virus-containing medium was harvested, clarified by centrifugation at 1,462 × g for 30 min, and then passed through a 0.45-μm filter to remove cellular debris. Purified virus was concentrated by ultracentrifugation at 100,000 × g through a 20% sucrose cushion and stored at −80°C. Virus was treated with 90 units/ml Benzonase (Sigma) for 15 min at 37°C to remove contaminating plasmid DNA before use.
Infections.
Stimulated peripheral blood lymphocytes (PBLs) from 3 separate blood donors (patients) were infected with an equal mass of either homozygous or heterozygous virus (which contained a mix of homozygous and heterozygous viruses), as determined by an HIV-1 antigen (p24 CA) micro-enzyme-linked immunosorbent assay (ELISA) (Vironostika). In this experimental setup, the ability to measure recombination is not affected by the MOI. Using viral cDNA synthesis as a surrogate marker for successful infection, retrospective analysis showed the average MOI to be 0.5 in these peripheral blood mononuclear cells (PBMCs). Efficient removal of plasmid DNA by Benzonase treatment was confirmed for each sample by using quantitative PCR (qPCR) targeting the ampicillin gene. Only virus preparations where the infection had no or insignificant background PCR signals (<5% of wild-type infection) were used in subsequent infection and sequencing experiments (Table 1). We note that any potential carryover plasmid DNA would have the effect of reducing the observed HIV recombination and mutation. At 6 h postinfection, 10 μg/ml T-20 (Roche) was added to the cells to prevent second-round replication. At 24 h postinfection, cells were pelleted and lysed in PCR lysis buffer containing 1× PCR buffer (Roche) with 0.5% (vol/vol) Triton X-100, 0.5% (vol/vol) NP-40, and 75 μg/ml proteinase K (Roche). A total of 1 × 106 cells were lysed per 100 μl of PCR lysis buffer. Samples were incubated at 56°C for 1 h before proteinase K was inactivated at 95°C for 10 min; samples were then stored at −20°C. Lysates were diluted 10× in PCR grade water before quantification by quantitative PCR.
TABLE 1.
Removal of contaminating plasmid DNA by Benzonase treatmenta
| Sample | Donor | Sample type | No. of HIV copies | No. of ampicillin copies | % Background |
|---|---|---|---|---|---|
| 1 | JL10 | Intervirion | 13,250 | 163.3 | 1.23 |
| 2 | JL10 | Wild type | 9,559 | 121.8 | 1.27 |
| 3 | RS13 | Intervirion | 11,700 | 127.6 | 1.09 |
| 4 | RS13 | Wild type | 1,746 | 29.56 | 1.69 |
| 5 | WJ2 | Intervirion | 5,822 | 44.36 | 0.76 |
| 6 | WJ2 | Wild type | 4,148 | 51.93 | 1.25 |
Efficient removal of contaminating plasmid DNA by Benzonase treatment was confirmed by qPCR quantification of HIV and ampicillin gene sequences using specific primers and the same standard based on serial dilutions of the NL43 plasmid. HIV-specific primers used were 5′-TTAAATGGCTCTTGATAAATTTGATATGTCCATTG-3′ (P7 sense) and 5′-CCACTAACAGAAGAAGCAGAGCTAGAACTG-3′ (P7 antisense). Ampicillin-specific primers were 5′-AACTCGCCTTGATCGTTGGG-3′ (AMP sense) and 5′-TGTTGCCATTGCTACAGGCATC-3′ (AMP antisense).
Primers.
PCR primers were designed to span the 46 intervals as 14 overlapping amplicons of roughly 350 nucleotides. PCR primers were designed against regions that were identical between the WT and MK plasmids according to the manufacturer's directions (Finnzymes).
Primers used are as follows: G1 sense (5′-GGTGCGAGAGCGTCGGTATTAAG-3′), G1 antisense (5′-CTGTGTCAGCTGCTGCTTGCTG-3′), G2 sense (5′-TCCTCTATTGTGTGCATCAAAGGATAGATG-3′), G2 antisense (5′-CCACTGTGTTTAGCATGGTATTTAAATCTTGTG-3′), G3 sense (5′-CAAATGGTACATCAGGCCATATCACCTAG-3′), G3 antisense (5′-CTGCATGCACTGGATGCAATCTATC-3′), G4 sense (5′-GAAGGAGCCACCCCACAAGATTTA-3′), G4 antisense (5′-GGTTCCTTTGGTCCTTGTCTTATGTCCAG-3′), G5 sense (5′-GGAAGTGACATAGCAGGAACTACTAG-3′), G5 antisense (5′-AGTCTTACAATCTGGGTTCGCATTTTGG-3′), G6 sense (5′-AAACTCTAAGAGCCGAGCAAGCTTC-3′), G6 antisense (5′-TGCCCTTCTTTGCCACAATTGAAACAC-3′), P1 sense (5′-GCAGGAGCCGATAGACAAGGAACT-3′), P1 antisense (5′-TAAAGTGCAGCCAATCTGAGTCAACAG-3′), P2 sense (5′-AGAAATCTGCGGACATAAAGCTATAGG-3′), P2 antisense (5′-GGAGTATTGTATGGATTTTCAGGCCCAA-3′), P3 sense (5′-GTAAAATTAAAGCCAGGAATGGATGGC-3′), P3 antisense (5′-GAAAAATATGCATCGCCCACATCCAG-3′), P4 sense (5′-TGTGGGCGATGCATATTTTTCAGT-3′), P4 antisense (5′-ATGGAGTTCATAACCCATCCAAAGGAATG-3′), P5 sense (5′-CACCAGCAATATTCCAGTGTAGCATG-3′), P5 antisense (5′-CTTTAATCCCTGCATAAATCTGACTTGCC-3′), P6 sense (5′-GAACTCCATCCTGATAAATGGACAGTACAG-3′), P6 antisense (5′-TTAAATGGCTCTTGATAAATTTGATATGTCCATTG-3′), P7 sense (5′-CCACTAACAGAAGAAGCAGAGCTAGAACTG-3′), P7 antisense (5′-CAGGTGGCTTGCCAATACTCTGTC-3′), P8 sense (5′-AGGGTGCCCACACTAATGATGTGAAAC-3′), P8 antisense (5′-AGTCTTCTGATTTGTTGTGTCCGTTAGG-3′), AMP sense (5′-AACTCGCCTTGATCGTTGGG-3′), and AMP antisense (5′-TGTTGCCATTGCTACAGGCATC-3′).
Quantitative PCR.
Quantitative PCR was performed on an MX3000 instrument (Stratagene). Reverse transcription products were assessed by using the HIV-1-specific primer pair M661/M667 (54), and background plasmid levels were assessed by using primers directed against the ampicillin resistance gene (AMP) (see the primer list, above). Each PCR mixture contained 1× Brilliant II master mix (Stratagene), 400 nM each primer, and 5 μl cell lysate in a 15-μl reaction mixture volume. For viral cDNA estimation, PCR conditions were an initial denaturation step at 95°C for 15 min followed by 40 rounds of cycling at 95°C for 10 s and then 60°C for 30 s. Samples were compared to ACH2 cell standards.
PCR.
With amplicon generation for next-generation sequencing, PCR conditions were optimized to reduce the formation of artificial recombinants by using a method outlined by Smyth et al. (40). PCR mixtures were titrated to contain 2,500 copies of template DNA, 1× HF buffer (Finnzymes), 200 μM deoxynucleoside triphosphate (dNTP), 1 μM each primer, and 0.3 U of Phusion DNA polymerase (Finnzymes) in a 15-μl total reaction mixture volume. Plasmid DNA was titrated in cellular lysates from uninfected PBLs so that the DNA complexity of the control PCRs faithfully represented the experimental samples. PCRs were performed in quadruplicate, and data were pooled to guard against PCR bias. PCR cycling conditions were 98°C for 30 s followed by 29 cycles of 98°C for 10 s and 72°C for 1 min.
Transfection-induced recombination.
To assess the rate of transfection-induced recombination, RNA was extracted from heterozygous virus by using phenol chloroform-based Tri reagent (Sigma-Aldrich), according to the manufacturer's recommendations, and reverse transcribed into cDNA by using SuperScript III (SSIII) (Invitrogen Life Technologies) and gene-specific primer GAG4(4195)R (5′-ACATTTCCAACAGCCCTTTTTCCTAG-3′). To control for in vitro cell-free reverse transcription and PCR-induced recombination, control samples consisting of homozygous WT virus and homozygous MK virus were mixed in equal quantities (based on p24 values) prior to RNA extraction and were reverse transcribed in parallel with RNA extracted from heterozygous virus. Reverse transcription was performed in the presence and absence of SSIII, with the latter condition providing a control for any plasmid contamination carried over from transfection. Real-time PCR was used to estimate viral cDNA copy numbers against a standard curve based on plasmid pDRNL(AD8) by using primers GAG1(2945)F (5′-GAGATGGGTGCGAGAGCGTC-3′) and GAG1(3314)R (5′-TGTGTCAGCTGCTGCTTGCTG-3′). Twenty replicate wells containing 2,500 copies of template viral cDNA were amplified by using optimized PCR conditions to reduce the formation of artificial recombinants, as outlined by Smyth et al. (40).
454 sequencing.
PCR amplicons spanning gag and pol were pooled for each blood donor. Unique 6-nucleotide identifiers (barcodes) were then individually attached by parallel tagged sequencing to allow multiplexing of samples from different blood donors on the same sequencing run (55, 56). Single-stranded sequencing libraries were constructed from 1 μg of initial starting material by using the 454 library preparation kit (Roche) according to the manufacturer's directions. Libraries were quantified and stored at −20°C until further use. Emulsion PCR and sequencing were performed according to standard GS FLX titanium procedures. In order to avoid excessive resampling of the same DNA strand, the 454 library was constructed from PCR products derived from at least 10 times more viral cDNA molecules than the maximum number of sequencing reads allowed in a reaction. For each full 454 sequencing run, the 454 library consisted of PCR products from 4,000 PCRs, with each reaction mixture containing 2,500 templates for amplification. This results in a 454 library consisting of PCR products derived from 10 million original templates, with a maximum of 1 million sequencing reads per 454 sequencing reaction. For PCR products derived from every 2,500 copies of original template, only 10% (250 copies) of its next-generation sequencing read was used for analysis to minimize resampling of the same original sequence, which could bias the estimation of the recombination and mutation rates.
Sequence alignment.
Sequencing data were processed to remove the 6-nucleotide barcode and assigned to a sample only upon a perfect barcode match. To reduce the missense error rate of 454 sequencing, sequences were also removed if they were of poor quality (such as those containing ambiguous nucleotides [“N′s”]) or not full length. All sequencing analysis was performed by using software custom written in BioRuby (http://www.bioruby.org/). In order to count mutations and other events, the sequences were aligned against the consensus sequence of the wild-type and marker-type versions of the amplicon (plus a margin of 100 nucleotides on both ends of the amplicon) by using needle. The parameters for needle were a gap-opening penalty of 3.0 and a gap extension penalty of 0.5. Segments which contained an ambiguous marker (a mutation within the marker) were discarded from the analysis.
Measurement of recombination rate.
The conversion of raw rates of chimera formation into recombination events per nucleotide (REPN) and statistical comparisons between recombination rates were performed by using methods described previously by Schlub et al. (20). Briefly, recombination was detected by monitoring the linking of marker points from wild-type and marker-type genomes into a single genome. However, measuring the crude recombination rate by dividing the number of events by the number of nucleotides can be misleading. Therefore, our approach, which has been described in detail previously (20), takes into account a number of factors inherent in the marker system.
First, multiple recombination events can occur between two marker points. Consequently, all odd numbers of recombination events between two markers (1, 3, 5, 7, 9, …., etc.) will be counted as a single recombination event, while even numbers of recombination events between two markers (2, 4, 6, 8, 10, …., etc.) will go undetected.
Second, we infect cells using a mix of heterozygous and homozygous virions (and in the latter, recombination will go undetected). Thus, we directly estimate the proportion of heterozygous virions for each sample.
Third, the widths of our intervals vary substantially, and therefore, we must take into account different likelihoods of multiple recombination events.
Our mathematical procedure accounts for these factors to estimate the recombination rate within the experimental system. This estimated recombination rate minimizes the chi-square value:
where k is the number of intervals between markers and Ei and Oi are the expected and observed numbers, respectively, of detected recombinations for interval i. The expected number of detected recombination events in any given interval, Ei, is calculated as
where s is the number of heterozygous sequences, Li is the nucleotide length of the interval, [(Li + 1)/2] is the integer part of (Li + 1)/2, and C(Li,k) is the binomial coefficient for picking k unordered outcomes from Li possibilities.
Estimating mutation rate per recombination: method 1.
To estimate the background rate of mutation and the rate of mutation induced by recombination, we consider the numbers of point mutations in intervals with and those without recombination. Intervals derived from homozygous virions do not display recombination regardless of RT template-switching activity. Therefore, we initially limited ourselves to sequences that are known to be derived from heterozygous sequences (sequences which contain at least one observable recombination). The mutation rate in heterozygous intervals without recombination, mb, provides an estimate of the background mutation rate alone. The mutation rate per nucleotide in intervals with recombination, mr, then represents the cumulative effect of background mutation and mutation induced from the recombination event. Thus, mr = mb + p/L, where p is the mutation rate per recombination and L is the length of the interval. As mr and mb can be calculated directly from the data and L is known, p can be calculated. Despite the large data set, as mutation rates and recombination rates are relatively low, mutation rates for individual intervals may have large amounts of variation by random chance alone. Thus, mutation rate calculations will not be informative if calculated on a per-interval basis. To overcome this, we calculate mr and p from the cumulative mutation rate over all heterozygous intervals with and without recombination and in this case replace L with the average length of intervals displaying recombination.
Estimating mutation rate per recombination: method 2.
Method 1 estimates the background mutation rate per nucleotide and an additional mutation rate per recombination event. However, this method uses only a fraction of the available data (sequences known to be heterozygous). Additionally, method 1 does not compensate for recombination events occurring at a frequency of >1 per interval (although this is estimated to be minimal). To adjust for these factors, we developed a second method to measure mutation rates, as described below.
To estimate the background rate of mutation and the rate of mutation induced by recombination, we consider the numbers of point mutations in intervals with recombination (N1), heterozygous intervals without recombination (contains a recombination somewhere on the sequence) (N2), and intervals of unknown ancestry (no recombination on the remainder of the sequence) without recombination (N3). Thus, for each interval, over all sequences,
where m is the background rate of mutation; I1, I2, and I3 are the numbers of informative nucleotides in intervals with recombination, heterozygous intervals without recombination, and intervals of unknown ancestry without recombination, respectively; p is the rate of mutation per recombination; R is the probability of observing a recombination over that interval; s1 and s2 are the numbers of intervals with recombination and heterozygous intervals without recombination, respectively; and s3 and s4 are the estimated number of intervals derived from heterozygous virions that have no recombination along the entire sequence (and are thus of unknown heterozygous/homozygous ancestry) and the estimated number of homozygous intervals, respectively, and where x = P1 + 3P3 + 5P5 + …. and y = 2P2 + 4P4 + 6P6 + …., with Pi being the probability of i recombinations occurring in a single interval. Thus, the terms x/R and y/(1 − R) represent the expected numbers of recombinations per interval in heterozygous intervals with and without recombination, respectively. The term (x + y) represents the number of recombinations expected to occur in a homozygous interval (where recombination is unobservable).
To calculate a single mutation rate per nucleotide and mutation rate per recombination over all intervals, we optimize the expected values of N1, N2, and N3 to minimize the summed square error to the observed counts over all intervals. As N3 represents the majority of our data set, weighting of N1, N2, and N3 errors may be employed to amplify their effect on the mutation rate estimations. However, such weighting did not substantially change mutation rate calculations in this data set and did not affect the conclusions drawn. Optimizations were performed in Matlab (v7.1.0.124 [R14]; Mathworks Inc.) with the function fmincon.
Estimating mutation rate per recombination: subtracting controls.
As the mechanism for individual mutations and recombinations cannot be determined, we cannot remove experimentally induced mutation and/or recombination prior to the analysis and comparison of rates. Rather, we estimate rates of mutation, recombination, and mutation per recombination for the biological sample and then separately estimated these rates for the controls. We then show that the contribution of experimental mutation/recombination to the mutation rate per recombination is minimal. Specifically, by estimating the proportion of recombination events attributable to experimental factors and their corresponding mutations, we estimate that only 9 to 15% of the recombination-associated mutation rate observed following infection could be due to experimental factors. This was subtracted from the biological sample rate to estimate the recombination-associated mutation rate attributable to viral infection processes only (RT, RNA polymerase II, and, potentially, host nucleic acid-editing enzymes such as APOBEC3G). This rate is then used to calculate the mutation rate per recombination for viral infection only.
Statistical analysis.
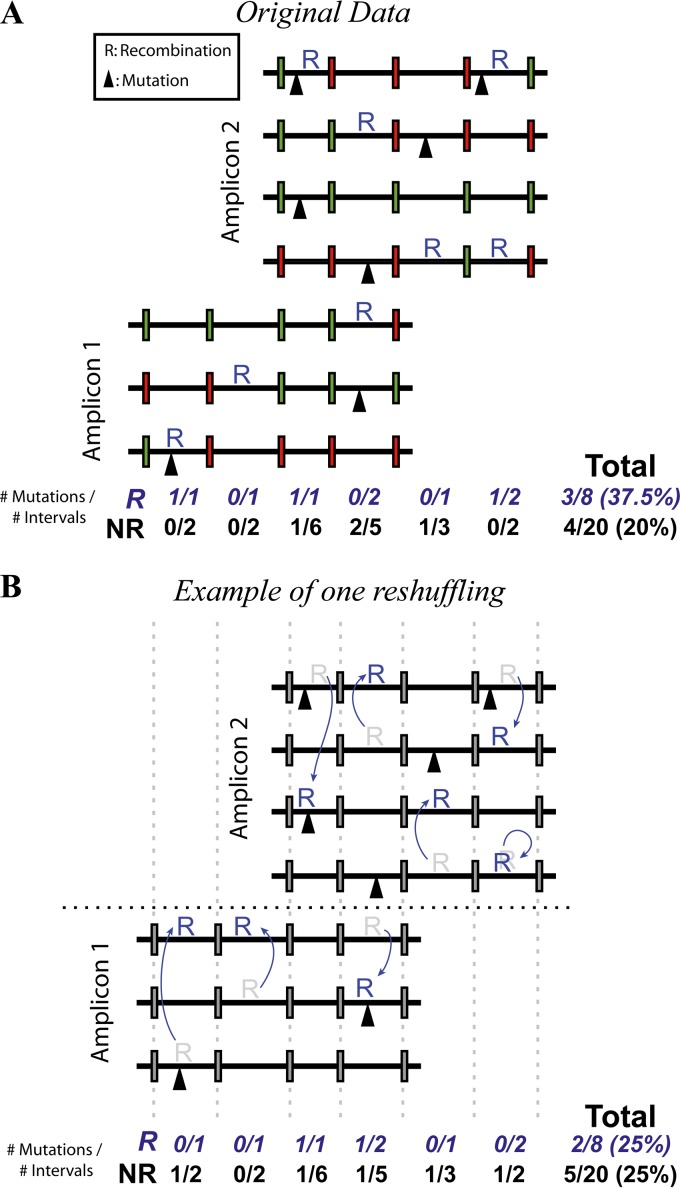
Statistical and mathematical analyses were carried out by using Graphpad Prism and Matlab, respectively. Fisher's exact tests were used to compare overall rates of mutation. To eliminate the effect of “coincident hot spots,” we developed a permutation approach to investigate the relationship between mutation and recombination (see Fig. 3A and B). In this approach, each sequence interval (the region between two marker sites) was classified as recombined (R) or nonrecombined (NR). The classification of R or NR was then randomly permuted (reshuffled) 10,000 times to generate the expected distribution of mutations within R and NR intervals if mutation and recombination were not associated. To avoid confounding variables, we reshuffled only within the same interval, same amplicon, same direction of sequencing, and same patient. By comparing the observed difference in R and NR mutation rates with those obtained from random reshuffling, we eliminated the confounding effects of coincident hot spots and Simpson's paradox.
FIG 3.
Permutation of recombination and mutation sites. An excess of mutations in recombined regions can be produced simply by having regions of the genome with high mutation and recombination rates (coincident hot spots) (Fig. 2A). To overcome this, we performed a permutation test as follows. (A) We classified each interval on each sequence to be recombined and/or mutated. (B) With each reshuffle, the recombination status of intervals was randomly permuted. To eliminate confounding factors (such as coincident hot spots), recombination status is permuted only within the same interval, amplicon, direction of sequencing, and patient. Reshuffling 10,000 times generates the distribution of mutation rates that would occur if recombination and mutation were not mechanistically associated. Statistics were calculated by comparing the original unpermuted data with the generated null distribution.
RESULTS
Measurement of recombination and mutation.
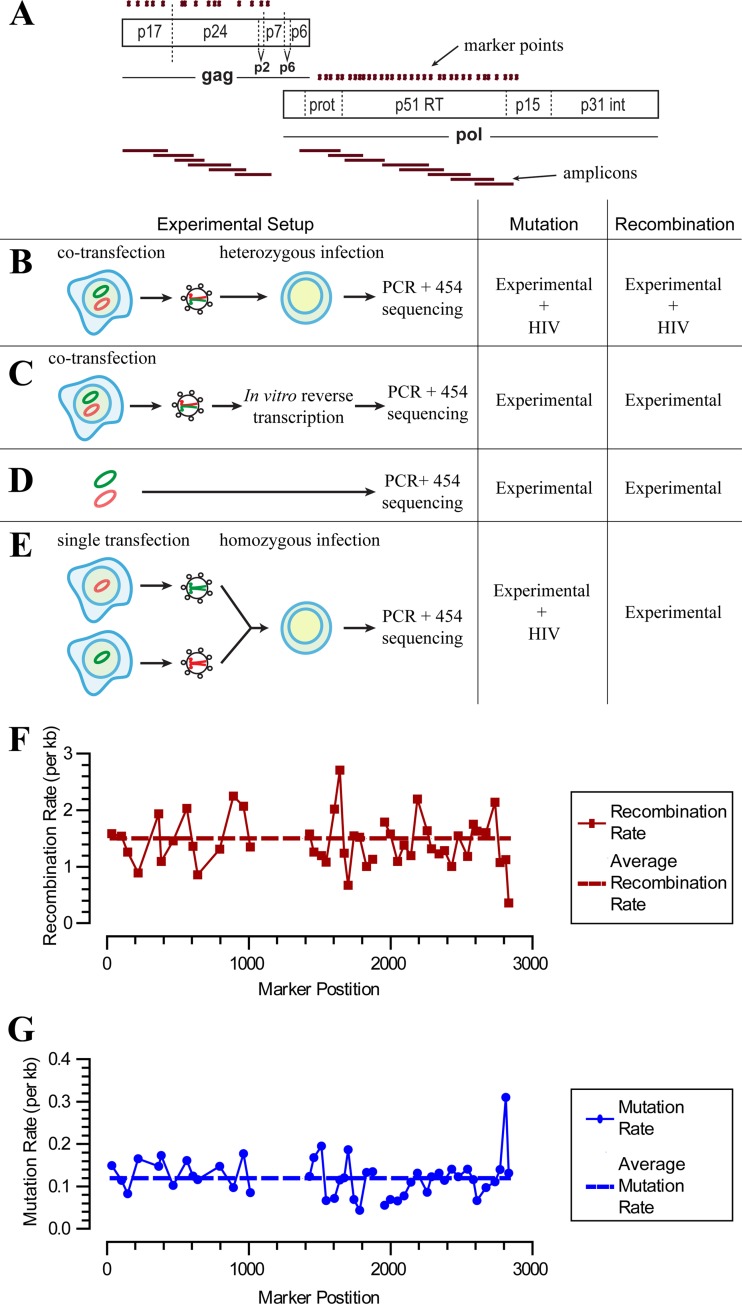
We used a system of markers introduced into the HIV gag and pol genes to simultaneously measure the rates of recombination and mutation during a single round of replication (Fig. 1A). A total of 46 intervals spaced an average of 53.5 nucleotides apart (range, 17 to 155 nucleotides; median, 47 nucleotides) were used. The positions of these mutations were chosen due to their natural polymorphism in the HIV database, and this marker HIV also has replication kinetics identical to that of our parental wild-type control. Heterozygous virus was produced by cotransfection of 293T cells with an equal mass of WT and marker (MK) plasmids, leading to a mixture of virions containing WT homozygous (25%), MK homozygous (25%), and WT-MK heterozygous (50%) copackaged RNA genomes, as described previously (13, 14, 20). The ratio of homozygous to heterozygous virions in our HIV infection stock was also internally monitored for each infection based on the frequency of nonrecombined WT and MK sequences (20). This mixture of heterozygous and homozygous viruses was used to separately infect freshly stimulated T lymphocytes from three separate blood donors. Following a single round of infection, viral DNA was extracted, PCR amplified, and sequenced (Fig. 1B). As previously reported, we have chosen to use a high-fidelity and high-processivity DNA polymerase (40), Phusion, which has a significantly higher fidelity rate than those of other polymerases, with an estimated error rate of 4.5 × 10−7 per base pair (57).
FIG 1.
Measurement of recombination and mutation rates. (A) Schematic of the marker system. A marker plasmid (MK) was generated through the introduction of silent markers into the gag and pol genes of wild-type (WT) HIV (shown as green dots). The positions of the amplicons are marked as red horizontal bars. (B) Experimental infection. MK plasmid and WT plasmid DNAs were cotransfected into 293T cells to produce a mixture of heterozygous and homozygous virus. Virus was then used to perform a single round of infection in primary T cells. DNA was subsequently extracted for PCR and high-throughput sequencing. (C to E) Controls for experimentally induced recombination and mutations. (C) The first control consisted of amplifying and sequencing of heterozygous virus reverse transcribed in vitro. This control assesses the rate of recombination occurring during cotransfection of viral plasmid. (D) The second control involved PCR amplification of a mixture of MK and WT plasmid DNAs to assess the rate of mutation and recombination during amplification and sequencing. (E) Two separate transfections of either the WT or MK plasmid were used to produce homozygous WT and MK viruses. These homozygous virus preparations were combined 50:50 and used to infect cells. Thus, any recombination during reverse transcription occurs on an identical strand (since the virus is homozygous) and is undetectable. The rate of mutation is the same as that for the experimental sample, and only experimentally (PCR) induced recombination was measured. (F and G) Recombination and mutation rates in each region of the experimental sample. The average rates that would produce this distribution are shown as dashed lines.
In addition to our experimental sample, we included a number of controls that account for sequencing error and PCR-induced recombination and mutation (Fig. 1D and E and Table 2). The DNA control consisted of PCR amplification and subsequent sequencing of plasmid DNA (Fig. 1D and Table 2). This sample allowed us to control for the rates of recombination and sequencing error during PCR amplification and 454 sequencing. Plasmid DNA was titrated in cellular lysates from uninfected PBLs so that the DNA complexity of the control PCRs faithfully represented the experimental samples. The second control involved infecting cells with a mixture of homozygous (containing identical copackaged RNA genomes) WT and MK viruses, before PCR amplification and sequencing (Fig. 1E and Table 2). In this case, recombination during HIV reverse transcription is expected to occur at a normal rate but effectively be “silent,” as the homozygous virus simply recombines onto an identical RNA strand. However, recombinant chimeras between WT and MK DNA strands may still occur during subsequent PCR amplification after cells are lysed. Thus, the level of PCR-induced recombination in this sample should occur at exactly the same rate as that during heterozygous virus infection. This second recombination control also directly monitors the levels of intervirion recombination events, demonstrating that the MOIs used in these infections did not bias the observed recombination rate (Table 2). Finally, this control incorporates the possibility of multiple rounds of infection, which are not expected to occur due to the addition of fusion inhibitors 6 h after infection and the termination of infection at 24 h postinfection.
TABLE 2.
Summary of mutation and recombination ratesa
| Sample | Total no. of nucleotides | No. of recombination events | Recombination rate/1,000 nt | No. of point mutations | Mutation rate/1,000 nt | No. of indels | Indel rate/1,000 nt |
|---|---|---|---|---|---|---|---|
| DNA control | 4,931,016 | 37 | 0.024 | 366 | 0.074 | 33,008 | 6.70 |
| Homozygous control | 15,407,974 | 338 | 0.050 | 1,830 | 0.119 | 98,281 | 6.38 |
| Infection (total) | 7,180,712 | 4,801 | 1.51 | 859 | 0.120 | 47,316 | 6.59 |
| Total R sequences | 297,908 | 4,801 | NA | 54 | 0.181 | 2,082 | 6.99 |
| Total NR sequences | 6,882,804 | 0 | NA | 805 | 0.117 | 45,234 | 6.57 |
| NR sequences on NR strands | 6,312,179 | 0 | NA | 762 | 0.121 | 41,659 | 6.60 |
| NR sequences on R strands | 570,625 | 0 | NA | 43 | 0.075 | 3,575 | 6.27 |
For each sample, the number of informative nucleotides from recombined or nonrecombined intervals as well as the numbers of recombination events, point mutations, and indels (frameshift errors) are indicated. We included controls for measuring the magnitude of recombination and mutation resulting from PCR and 454 sequencing: the DNA control, where plasmid DNA was amplified and spiked with cellular lysates to control for any impact of these on amplification efficiency/fidelity, measuring PCR recombination and mutation by PCR and 454 sequencing, and the homozygous control, where two homozygous virus infections (one MK and one WT) were combined after lysis and before PCR amplification so that recombination during reverse transcription is silent and recombination during PCR is observable, measuring PCR recombination and mutation of viral cDNA by 454 sequencing. The experimental infection samples are further broken up into recombined and nonrecombined sequences. The latter are also broken up into nonrecombined sequences on recombined strands and on nonrecombined strands. NA, not applicable.
In response to reviewer concerns, a further round of controls was performed to assess whether transfection-induced recombination (TIR) might be occurring during the initial stage of transfection of the plasmids. TIR might occur when the WT and MK plasmids are cotransfected and undergo recombination in vitro. To exclude this possibility, we sequenced the virus produced by transfected cells (Fig. 1C and Table 3). Because this involved a reverse transcription step performed on viral RNA, it is possible that any recombination events observed in the viral sequences may have arisen from reverse transcription rather than TIR. Therefore, we included controls where WT and MK plasmids were transfected separately into cells, and the resultant homozygous viruses were combined only for the stage of reverse transcription. These assays showed that transfection of the two different plasmids did not result in more recombination than seen with reverse transcription of homozygous virions, indicating very little or no TIR (approximately 0.006 × 10−3 REPN, <0.5% of the total recombination rate) (Table 3). Moreover, the total level of recombination seen was so low that even if all of these recombination events were caused by TIR (and not by the reverse transcription step), they would not account for any significant proportion of recombination events seen in the data.
TABLE 3.
Summary of mutation and recombination for the transfection controla
| Sample | Total no. of nucleotides | No. of recombination events | Recombination rate/1,000 nt |
|---|---|---|---|
| Transfection-induced recombination control | 22,600,749 | 58 | 0.006 |
| SuperScript III control | 3,261,995 | 4 | 0.005 |
| DNA control | 24,893,615 | 31 | 0.004 |
| Heterozygous infection | 6,558,479 | 4,243 | 1.59 |
To exclude the possibility that recombination during transfection of plasmids is biasing our results, we sequenced virus produced by transfected cells. This rate measures the cumulative effect of transfection-induced recombination and the reverse transcription step using SuperScript III (see Materials and Methods). To determine the level of reverse transcription recombination using SuperScript III, we sequenced virus produced by cells separately transfected with either MK plasmids or WT plasmids only, which were mixed before reverse transcription. We also repeated the PCR control (DNA control) for this assay and the experimental sample.
Under each experimental condition, six pairs of overlapping gag PCR amplicons and eight pairs of overlapping pol PCR amplicons were used to amplify the corresponding viral sequences. These PCR amplicons cover all the marker points in gag and pol and have an approximate length of 350 nucleotides, which is optimal for 454 pyrosequencing. Each PCR amplicon contains between 5 and 7 marker points and, hence, 4 to 6 intervals each for our analysis.
Following sequencing and alignment, we obtained on average 3,364 sequences per interval (range, 1,290 to 6,889) for the experimental sample. Each marker position was then classified as WT, MK, or ambiguous (if it was not identical to either a WT or MK sequence). Each interval between markers was then classified as recombined (R) if adjacent markers were WT and MK, nonrecombined (NR) if adjacent markers were identical, or ambiguous if either adjacent marker could not be classified. We calculated the recombination rate using a method that takes into account the proportion of homozygous sequences (where virally induced recombination cannot be detected) as well as the possibility of multiple undetectable recombination events between marker sets (described in detail in reference 20). From this, we estimated the overall recombination rate in our experimental samples (1.51 per 1,000 nt) and in our controls (Fig. 1F and Table 2).
We next studied the rate of mutation in these samples (Fig. 1G and Table 1). To avoid resampling of the viral cDNA, which could lead to an inaccurate estimation of the mutation rate in the HIV genome, 10-fold excesses of viral cDNA were used as the templates for 454 sequencing; i.e., >10 million distinct viral cDNA templates were used for a complete run (1 million reads) of 454 sequencing. In the experimental sample, from a total of 7,180,712 informative nucleotides, we observed 859 mutations, giving an overall mutation rate in of 0.120 per 1,000 nt. This mutation rate represents the cumulative effect of experimentally induced “background” PCR and sequencing errors and the viral RT-induced mutation. We estimated the background rate by amplifying and sequencing a plasmid DNA with a 50:50 mix of the WT and MK sequences in the presence of cell lysates from uninfected PBLs. Here our sample consisted of 4,931,016 nt, from which we observed 366 mutations, giving a mutation rate of 0.0742 per 1,000 nt (Table 2). We note that our background rate of substitution mutations from PCR amplification and sequencing is significantly lower than some other estimates, and this difference likely reflects the high-fidelity nature of Phusion DNA polymerase in our analyses. In addition, removal of low-quality sequencing reads (i.e., those containing ambiguous nucleotides [“n”] or that were not full-length amplicons) also kept the rate low. The mutation rate attributable to viral factors (including RT, RNA polymerase II, and, potentially, host nucleic acid-editing enzymes [e.g., APOBEC3G]) is the difference between the error rate in the biological sample and that for the plasmid DNA control. This rate is 0.120 − 0.0742 = 0.0458 per 1,000 nt. This is similar to the rate of 1.4 × 10−5 to 4 × 10−5 previously estimated for HIV-1 (2, 19, 41).
Higher mutation rate in recombined intervals.
In order to test whether recombination was associated with mutation, we compared the mutation rates in intervals where recombination is and is not observed. We found a significantly higher mutation rate in recombined intervals than in nonrecombined intervals (0.181/1,000 nt versus 0.117/1,000 nt; P = 0.003 by Fisher's exact test). However, since our procedure involved PCR amplification, and we have previously demonstrated that PCR-induced recombination occurs at a low but significant rate (20), it is possible that PCR-induced recombination during sample preparation was responsible for the observed association between recombination and mutation. To exclude this possibility, we analyzed control infections where we infected cells with a 50:50 mix of homozygous WT and MK viruses. Because of the low rate of PCR-induced recombination, we sequenced twice as many samples, leading to a total of 15,407,974 informative nucleotides from control samples. As previously described (40), the rate of PCR-induced recombination in this sample (0.050 per 1,000 nt) was much lower than the cumulative effect of viral RT-induced and PCR-induced recombination measured for heterozygous infection (1.51 per 1,000 nt). PCR-induced recombination was associated with a higher mutation rate (mutation rate of 0.537 per 1,000 nt in R sequences versus 0.118 per 1,000 nt in NR sequences; P < 0.0001 by Fisher's exact test). Although the mutation rate was increased with PCR-induced recombination, the overall rate of PCR-induced recombination was much too low to account for the observed difference in the heterozygous HIV infection samples; that is, correcting for the sample size, we expect that around 3% of recombination events are due to PCR error and that in our total sample, these PCR-recombined regions would contain approximately 5 mutations (including both background and recombination-associated mutations). Thus, <10% (5/54) of the total mutations in the recombined regions could be attributed to PCR-induced recombination. Similarly, transfection-induced recombination accounted for <0.5% of mutations when we sequenced the virion genomes directly (Table 3) and is too low to contribute to the observed rates.
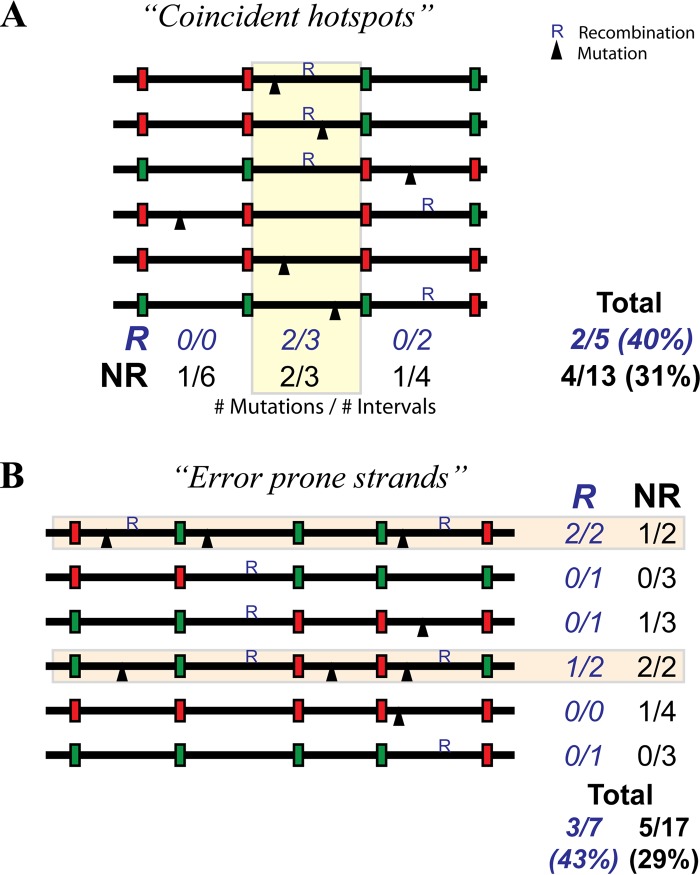
Although we observed an overall higher rate of mutation in recombined regions, this could have occurred for a number of potential reasons. First, the apparent association between mutation and recombination could have been artificially created by pooling data over the various intervals and sequences from various amplicons. This could occur in a scenario where coincident recombination and mutation hot spots led to higher incidences of both mutation and recombination at particular segments, even though recombined intervals had the same mutation rate as nonrecombined intervals within the segment (Fig. 2A, yellow box). This would be an example of the so-called “Simpson's paradox” (58). A second potential confounder is the effect of “error-prone cells” or “error-prone strands” driving the observed association; that is, if some cells, RT enzymes, or viral RNA strands produce a particularly high rate of both mutation and recombination in any particular set of amplicons, mutation and recombination will appear to be associated, even if it is only because the mutation rate is higher all along the error-prone strand and not higher in all recombined intervals on the strand in general (Fig. 2B, light orange boxes).
FIG 2.
Association between recombination and mutation. An experimental association between mutation and recombination may be observed due to either coincident hot spots (A) or error-prone strands (B), even though there is no real mechanistic association. (A) A coincident hot spot scenario can occur if there is a region of the genome with both high mutation and high recombination rates (yellow box). In this scenario, overall, we see that 2 of the 5 recombined segments (R) (40%) have mutations, and only 4 of the 13 (31%) nonrecombined segments (NR) have a mutation. However, this apparent association is due simply to the high mutation and recombination rates in the yellow region. There is no higher mutation rate in R segments when individual regions are analyzed separately; that is, there are no mutations in the recombined regions outside the hot spot and an equal rate of recombination in R and NR within the hot spot (i.e., 2/3 segments have mutations for both R and NR). (B) The presence of error-prone strands (light orange) with high mutation and recombination rates can also lead to an erroneous association. Here we observed that 3/7 (43%) recombined regions contain a mutation and that 5/17 (29%) nonrecombined regions contain a mutation. However, there are no mutations in recombined regions outside the error-prone strands, and in the error-prone strands, there are a total of 3/4 regions mutated in both the recombined and nonrecombined regions.
Mutation is associated with recombination.
To eliminate the effect of coincident hot spots, we developed a permutation test (Fig. 3A and B; see also Materials and Methods). In this approach, recombination classification (R or NR) for each sequence interval (the region between two markers) was randomly permuted to generate the expected distribution of mutations within R and NR intervals if mutation and recombination were not associated. By comparing the empirical difference in R and NR mutation rates with those obtained from permutation, the probability of randomly observing an association between recombination and mutation, assuming no underlying association, can be eliminated with the confounding effects of coincident hot spots, and Simpson's paradox can be eliminated. From 10,000 reshuffling runs, we observed only 51 runs where the difference in mutation rates was as great as that observed experimentally. Thus, the association between mutation and recombination is significant (P = 0.0102 by two-tailed test) and is not affected by the confounding effects of coincident hot spots.
In the case of error-prone strands, the confounding effect of high mutation and high recombination rates of a subset of sequences would lead to a spurious association between recombination and mutation. In this scenario, we expect that sequences with a high recombination rate would also have a high mutation rate (Fig. 2B). Therefore, the mutation rate should be on average higher in recombined strands (in both recombined and nonrecombined intervals) than it is in nonrecombined strands. To investigate this, we classified all NR intervals into those on a strand with recombination elsewhere (NRR) and those on a strand with no recombination (NRNR). If the association between mutation and recombination is due to error-prone strands, we expect that the mutation rate of NRR would be higher than that of NRNR. Alternatively, if the association between recombination and mutation is not due to error-prone strands, we expect that the mutation rate of NRR would be less than or equal to that of NRNR. The case of the mutation rate of NRR being less than that of NRNR can occur because NRR intervals are truly nonrecombined (they are known to be derived from a heterozygous infection, as they have an observed recombination elsewhere), whereas NRNR will be a mix of heterozygous intervals without recombination, homozygous intervals without recombination, and homozygous intervals with undetectable recombination. The homozygous intervals with undetectable recombination will increase the mutation rate in these intervals so that NRR is less than NRNR. Upon analysis, we found that the mutation rate in nonrecombined intervals in strands with recombination was lower than the rate on strands without recombination (NRR < NRNR; P < 0.004 by using the reshuffling approach). This indicates that the association between recombination and mutation was not a product of confounding effects such as error-prone strands.
This analysis of mutation rates on NRR and NRNR provides a useful independent verification that neither PCR-induced recombination nor transfection-induced recombination was a factor driving the association between mutation and recombination; that is, both PCR-induced and transfection-induced recombinations occur without consideration of whether the virions were homozygous or heterozygous. Hence, if the association was due to either of these mechanisms, we would expect the rate of mutation in NRR and NRNR to be the same.
Pattern of mutation in recombined and nonrecombined sequences.
Given the increased mutation rates in recombined intervals, we sought to investigate whether the pattern of mutation in R intervals was significantly different from that in NR intervals. Because many NR intervals have come from homozygous virions and thus have undetected recombination, we compared mutations in nonrecombined intervals from recombined sequences (NRR, where any odd number of recombination events should have been detectable). We found no substantial differences in the patterns of mutation observed in NRR and recombined sequences (Table 4).
TABLE 4.
Mutation pattern of recombined and nonrecombined intervalsa
| Original nucleotide | No. of nucleotides | % (no.) of mutated sequences (10−5) | % (no.) of nucleotide mutations out of total mutations |
|||
|---|---|---|---|---|---|---|
| A | C | G | T | |||
| Recombined interval | ||||||
| A | 111,789 | 8.1 (9) | 0 | 3.7 (2) | 13.0 (7) | 0.0 (0) |
| C | 56,088 | 12.5 (7) | 3.7 (2) | 0 | 1.9 (1) | 7.4 (4) |
| G | 66,473 | 45.1 (30) | 46.3 (25) | 3.7 (2) | 0 | 5.6 (3) |
| T | 63,558 | 12.6 (8) | 5.6 (3) | 5.6 (3) | 3.7 (2) | 0 |
| Nonrecombined interval from recombined sequence | ||||||
| A | 220,054 | 5.0 (11) | 0 | 4.7 (2) | 20.9 (9) | 0.0 (0) |
| C | 101,042 | 4.9 (5) | 0.0 (0) | 0 | 2.3 (1) | 9.3 (4) |
| G | 124,497 | 16.9 (21) | 41.9 (18) | 0.0 (0) | 0 | 7.0 (3) |
| T | 125,032 | 4.8 (6) | 0.0 (0) | 11.6 (5) | 2.3 (1) | 0 |
| Plasmid DNA control | ||||||
| A | 1,892,627 | 4.9 (99) | 0 | 1.1 (4) | 20.8 (76) | 4.4 (16) |
| C | 883,997 | 12.4 (117) | 5.7 (21) | 0 | 2.2 (8) | 20.8 (76) |
| G | 1,074,185 | 11.4 (131) | 25.4 (93) | 0.5 (2) | 0 | 6 (22) |
| T | 1,080,207 | 4.5 (52) | 1.9 (7) | 10.7 (39) | 0.5 (2) | 0 |
The frequencies of substitution mutations in A, C, G, and T as well as the nucleotide to which the nucleotide is mutated are indicated for recombined intervals and for nonrecombined intervals from recombined sequences. These mutation patterns are the cumulative product of experimental factors (such as sequencing error) and viral factors. To give some indication of the contribution of sequencing error to the pattern seen, the frequency of substitution mutations in the plasmid DNA control is shown.
Recombination and frameshift mutations.
The above-described results have analyzed the rate of substitution mutations. However, it has also been suggested that recombination may be associated with insertions and deletions (indels) (59). To investigate this, we performed the same analysis as that described above to investigate whether the indel rate was higher in R or NR intervals (Table 2). Although we observed a trend toward higher rates of indels in recombined sequences (6.989 versus 6.572 indels per 1,000 nt), this was not significant by using a two-tailed permutation test (P = 0.31). One reason for the inability to demonstrate a consistent significant difference in indel rates is the high background indel rate due to the difficulties in identifying homopolymer lengths during 454 sequencing (60). Indeed, the rate of indels of the DNA control is higher than that of the experimental sequences (6.694 versus 6.589 indels per 1,000 nt; P = 0.028 by Fischer's exact test). The 454 sequencing indels in the DNA sample are preferentially clustered around longer homopolymer tracts. Thus, we filtered the indel detection algorithm to exclude indels arising from homopolymer tracts of ≥3 nt. This reduced the indel rate in both R and NR sequences, although the difference was still not significant (P = 0.21 by two-tailed permutation test).
One issue with comparing all intervals with observed recombination with all intervals with no observed recombination is that the latter intervals include homozygous sequences in which silent recombination (and the associated rate of mutation) occurred on a homozygous virion and so was not observable. Thus, including these “silently recombined” sequences in the nonrecombined group will push up the observed mutation rate. In order to try to exclude this, we can limit our analysis to nonrecombined regions that were observed on recombined sequences (NRR); that is, because recombination was observed elsewhere on a sequence, we know that the sequence came from a heterozygous virion, and thus, silent recombination was not possible. The indel rate in R was significantly higher than the indel rate in NRR (6.989 versus 6.265 indels per 1,000 nt; P = 0.0464 by two-tailed permutation test). Additionally, when the data were filtered to eliminate indels from homopolymers (a limitation of 454 pyrosequencing) of ≥3 nt, the recombination rate in R was significantly higher than that in NRR (2.02 versus 1.55; P = 0.003 by two-tailed permutation test). While the rate of indels is significantly higher in R than in NRR sequences, the high background rate of indels complicates the analysis.
DISCUSSION
Genetic diversity in HIV arises primarily during reverse transcription via the introduction of mutations that are then shuffled between viral genomes by recombination. It is generally thought that recombination and mutation are not associated, although previous studies addressing this question have led to conflicting results (28–30). In this study, we have employed a novel HIV marker system and a high-throughput pyrosequencing system to simultaneously measure mutation and recombination rates directly on an authentic HIV-1 genome. Our comprehensive analysis demonstrates that recombination is associated with point mutations in HIV infection of primary T cells.
Three previous studies have attempted to evaluate the potential linkages between recombination and mutation in retroviruses (28–30). In the first two studies, 65,000 and 4,100 bp were sequenced in a spleen necrosis virus (SNV) vector and HIV genomes (with 2 and 0.1 anticipated mutation events), yet 0 and 2 mutations were detected, respectively, leading the authors of those studies to conclude that any association was absent or uncertain (28, 29). In a third study, a larger analysis was carried out in which five recombined sequences were observed to contain substitution mutations, leading those authors to conclude that around 6% of recombination resulted in mutation (30). However, those authors pointed out that the background mutation rate of that study was four times higher than expected. Moreover, the estimated rate of recombination-associated mutation would lead to a higher mutation rate in HIV than theoretically possible, even if it was the only source of mutation and excluding other mechanisms, such as APOBEC-mediated mutations; that is, if there is a 6% chance of mutation per recombination, and HIV undergoes 5 to 15 recombination events per genome (20, 35, 45), this would lead to 0.3 to 0.9 mutation-associated recombinations per genome. However, only ∼0.3 to 0.4 mutations are usually observed (2, 19), suggesting that this rate is higher than what is compatible with the observed mutation rates. Those authors speculate that this high level of mutation might result from the two rounds of reverse transcription and subsequent PCR (30). With these higher-than-expected mutation rates and the potential confounding recombination events (30), it is difficult to utilize the data set to determine the potential linkage between retroviral mutation and recombination.
In the current study, we analyzed over 27 million nt of sequencing data (including 7 million nt of a viral genome from a heterozygous HIV infection) and observed 859 mutations and 4,801 recombination events during HIV infection. Using this system, we found an overall mutation rate of 0.0458 mutations per 1,000 nt per replication cycle and a recombination rate of 1.51 events per 1,000 nt per replication cycle, which are similar to the rates previously reported for HIV-1. Furthermore, using appropriate controls and statistical analyses, we demonstrated that this association between mutation and recombination represents a biological association rather than experimental or statistical artifacts.
The association between recombination and mutation can be explained in three ways: mutation increases the chance of recombination, recombination increases the chance of mutation, or the chance of individual mutation and recombination events can be influenced simultaneously by some other factor. Our analysis cannot identify the direction of causality. However, if the direction is such that mutation increases the chance of recombination, the effect of each mutation on the overall recombination rate will be minimal, as the mutation rate is approximately 100-fold lower than the recombination rate. Conversely, as recombination occurs much more frequently than mutation, if recombination influences mutation, this may have a significant impact on the overall mutation rate and mutation hot spots.
Assuming the scenario where recombination increases the probability of mutation, we calculated what combination of background mutation rate per nucleotide and additional mutation rate per recombination event best fits the experimental data, thus estimating the proportion of mutations that are attributable to recombination. Using the overall rate of recombination and the rates of mutation in R, NRNR, and NRR sequences, we used two mathematical methods to calculate the recombination-induced mutation rate. After subtracting the recombination-associated mutation rates from sequencing and PCR, we found that each viral recombination has a 0.5 to 0.6% chance of inducing a mutation in the same interval. This corresponds to viral recombination inducing approximately 0.07 to 0.09 mutations per genome (of 9,600 bp in length), representing between 15% and 20% of all virus-associated mutations.
One plausible explanation for this observation is that recombination is mutagenic and, thus, that mutations are introduced into the viral genome at the site of recombination (24, 26, 27, 61). Indeed, early in vitro investigations showed that HIV-1 RT frequently adds nontemplated nucleotides at the 3′ ends of nascent DNA during reverse transcription and that these nontemplated nucleotides are misincorporated upon strand transfer (25, 27). In support of this mechanism, the mutation rate for murine leukemia virus (MLV), a related retrovirus, was reported to be 1,000-fold higher at the site of first-strand transfer than in other regions of the genome (62). However, in vitro recombination and first-strand transfer occur at template ends, and it is not known whether this corresponds to recombination at internal positions within the genome analyzed in that study. It was reported that a nontemplated addition is highly specific for purines (A > G ≫ T ≫ C), yet there was no difference in the mutation spectra observed between recombined and nonrecombined intervals. This suggests that this mechanism of mutagenic recombination does not take place or that the mutation spectrum during a natural infection cycle is different from that observed in vitro. Another mechanism of mutation at recombination sites is referred to as “slippage synthesis” (24). This occurs due to the misalignment of the 3′ end of the nascent DNA onto the acceptor RNA template. This type of recombination-induced mutagenesis is expected to be highly dependent on local sequence characteristics of the template. Indeed, one in vitro study demonstrated that while one recombination location was associated with a 30% mutation rate, another recombination location was not associated with mutations (21). Differences in template sequence may explain why some studies support the notion that recombination causes mutation (21, 24, 25) whereas other studies do not (63). One important advantage of our system is that mutation and recombination rates are measured directly on the HIV-1 genome, meaning that these results are not biased by foreign gene sequences.
Another explanation for the association between mutation and recombination is that mutation increases the likelihood of template switching, rather than template switching being mutagenic (22, 23, 64). HIV-1 RT is capable of extending mismatched template primers, but this extension is associated with pausing of DNA synthesis (22). The most widely accepted model for retroviral recombination, the dynamic copy choice model, suggests that the rate of recombination is determined by the dynamic steady state between DNA polymerase and RNase H activities (65). This model predicts that increasing the RNase H-to-polymerase ratio by stalling DNA synthesis will increase the local rate of recombination. In agreement with this model, synthesis from a mismatched primer increased strand transfer by 50% compared to a complementary primer, and this was associated with a significant pause in synthesis (22, 23). Furthermore, in an in vitro template-switching assay, a lower frequency of mutations was observed on the donor template when in the presence of an acceptor template, implying that mutation induces template switching onto the acceptor (64).
In this study, the direction of causality is unclear. Indeed, it is plausible that recombination-induced mutation and mutation-induced recombination are both responsible for the observed association. Regardless of the causality, our study demonstrates a direct linkage between recombination and mutation in driving overall viral evolution. In the event that the association is due only to recombination-induced mutation, our calculations show that up to 20% of mutations result from recombination. It is unclear whether the observed association between mutation and recombination provides an evolutionary advantage to the virus or is simply a result of the mechanisms of transcription of the error-prone RT. Given the importance of recombination and mutation to the evolution of drug resistance and immune escape, dissecting parameters and molecular determinants that regulate these events will be vital to define the process of HIV evolution. Such information is likely to be invaluable for understanding the emergence of immune escape and antiviral drug-resistant HIV in the course of HIV infection and AIDS pathogenesis.
Footnotes
Published ahead of print 22 January 2014
REFERENCES
- 1.Maldarelli F, Kearney M, Palmer S, Stephens R, Mican J, Polis MA, Davey RT, Kovacs J, Shao W, Rock-Kress D, Metcalf JA, Rehm C, Greer SE, Lucey DL, Danley K, Alter H, Mellors JW, Coffin JM. 2013. HIV populations are large and accumulate high genetic diversity in a nonlinear fashion. J. Virol. 87:10313–10323. 10.1128/JVI.01225-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mansky LM, Temin HM. 1995. Lower in vivo mutation rate of human immunodeficiency virus type 1 than that predicted from the fidelity of purified reverse transcriptase. J. Virol. 69:5087–5094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mangeat B, Turelli P, Caron G, Friedli M, Perrin L, Trono D. 2003. Broad antiretroviral defence by human APOBEC3G through lethal editing of nascent reverse transcripts. Nature 424:99–103. 10.1038/nature01709 [DOI] [PubMed] [Google Scholar]
- 4.Zhang H, Yang B, Pomerantz RJ, Zhang C, Arunachalam SC, Gao L. 2003. The cytidine deaminase CEM15 induces hypermutation in newly synthesized HIV-1 DNA. Nature 424:94–98. 10.1038/nature01707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Johnson SF, Telesnitsky A. 2010. Retroviral RNA dimerization and packaging: the what, how, when, where, and why. PLoS Pathog. 6:e1001007. 10.1371/journal.ppat.1001007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Moore MD, Hu WS. 2009. HIV-1 RNA dimerization: it takes two to tango. AIDS Rev. 11:91–102 http://www.aidsreviews.com/files/2009_11_2_091-102.pdf [PMC free article] [PubMed] [Google Scholar]
- 7.Negroni M, Buc H. 2001. Mechanisms of retroviral recombination. Annu. Rev. Genet. 35:275–302. 10.1146/annurev.genet.35.102401.090551 [DOI] [PubMed] [Google Scholar]
- 8.D'Souza V, Summers MF. 2005. How retroviruses select their genomes. Nat. Rev. Microbiol. 3:643–655. 10.1038/nrmicro1210 [DOI] [PubMed] [Google Scholar]
- 9.Paillart JC, Shehu-Xhilaga M, Marquet R, Mak J. 2004. Dimerization of retroviral RNA genomes: an inseparable pair. Nat. Rev. Microbiol. 2:461–472. 10.1038/nrmicro903 [DOI] [PubMed] [Google Scholar]
- 10.Hill MK, Shehu-Xhilaga M, Campbell SM, Poumbourios P, Crowe SM, Mak J. 2003. The dimer initiation sequence stem-loop of human immunodeficiency virus type 1 is dispensable for viral replication in peripheral blood mononuclear cells. J. Virol. 77:8329–8335. 10.1128/JVI.77.15.8329-8335.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jones KL, Sonza S, Mak J. 2008. Primary T-lymphocytes rescue the replication of HIV-1 DIS RNA mutants in part by facilitating reverse transcription. Nucleic Acids Res. 36:1578–1588. 10.1093/nar/gkm1149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Huthoff H, Das AT, Vink M, Klaver B, Zorgdrager F, Cornelissen M, Berkhout B. 2004. A human immunodeficiency virus type 1-infected individual with low viral load harbors a virus variant that exhibits an in vitro RNA dimerization defect. J. Virol. 78:4907–4913. 10.1128/JVI.78.9.4907-4913.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen J, Nikolaitchik O, Singh J, Wright A, Bencsics CE, Coffin JM, Ni N, Lockett S, Pathak VK, Hu WS. 2009. High efficiency of HIV-1 genomic RNA packaging and heterozygote formation revealed by single virion analysis. Proc. Natl. Acad. Sci. U. S. A. 106:13535–13540. 10.1073/pnas.0906822106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Moore MD, Nikolaitchik OA, Chen J, Hammarskjold ML, Rekosh D, Hu WS. 2009. Probing the HIV-1 genomic RNA trafficking pathway and dimerization by genetic recombination and single virion analyses. PLoS Pathog. 5:e1000627. 10.1371/journal.ppat.1000627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Balakrishnan M, Fay PJ, Bambara RA. 2001. The kissing hairpin sequence promotes recombination within the HIV-I 5′ leader region. J. Biol. Chem. 276:36482–36492. 10.1074/jbc.M102860200 [DOI] [PubMed] [Google Scholar]
- 16.Balakrishnan M, Roques BP, Fay PJ, Bambara RA. 2003. Template dimerization promotes an acceptor invasion-induced transfer mechanism during human immunodeficiency virus type 1 minus-strand synthesis. J. Virol. 77:4710–4721. 10.1128/JVI.77.8.4710-4721.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dykes C, Balakrishnan M, Planelles V, Zhu Y, Bambara RA, Demeter LM. 2004. Identification of a preferred region for recombination and mutation in HIV-1 gag. Virology 326:262–279. 10.1016/j.virol.2004.02.033 [DOI] [PubMed] [Google Scholar]
- 18.Basu VP, Song M, Gao L, Rigby ST, Hanson MN, Bambara RA. 2008. Strand transfer events during HIV-1 reverse transcription. Virus Res. 134:19–38. 10.1016/j.virusres.2007.12.017 [DOI] [PubMed] [Google Scholar]
- 19.Mansky LM. 1996. Forward mutation rate of human immunodeficiency virus type 1 in a T lymphoid cell line. AIDS Res. Hum. Retroviruses 12:307–314. 10.1089/aid.1996.12.307 [DOI] [PubMed] [Google Scholar]
- 20.Schlub TE, Smyth RP, Grimm AJ, Mak J, Davenport MP. 2010. Accurately measuring recombination between closely related HIV-1 genomes. PLoS Comput. Biol. 6:e1000766. 10.1371/journal.pcbi.1000766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wu W, Palaniappan C, Bambara RA, Fay PJ. 1996. Differences in mutagenesis during minus strand, plus strand and strand transfer (recombination) synthesis of the HIV-1 nef gene in vitro. Nucleic Acids Res. 24:1710–1718. 10.1093/nar/24.9.1710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Palaniappan C, Wisniewski M, Wu W, Fay PJ, Bambara RA. 1996. Misincorporation by HIV-1 reverse transcriptase promotes recombination via strand transfer synthesis. J. Biol. Chem. 271:22331–22338. 10.1074/jbc.271.37.22331 [DOI] [PubMed] [Google Scholar]
- 23.Diaz L, DeStefano JJ. 1996. Strand transfer is enhanced by mismatched nucleotides at the 3′ primer terminus: a possible link between HIV reverse transcriptase fidelity and recombination. Nucleic Acids Res. 24:3086–3092. 10.1093/nar/24.15.3086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wu W, Blumberg BM, Fay PJ, Bambara RA. 1995. Strand transfer mediated by human immunodeficiency virus reverse transcriptase in vitro is promoted by pausing and results in miscorporation. J. Biol. Chem. 270:325–332. 10.1074/jbc.270.1.325 [DOI] [PubMed] [Google Scholar]
- 25.Peliska JA, Benkovic SJ. 1994. Fidelity of in vitro DNA strand transfer reactions catalyzed by HIV-1 reverse transcriptase. Biochemistry 33:3890–3895. 10.1021/bi00179a014 [DOI] [PubMed] [Google Scholar]
- 26.Peliska JA, Benkovic SJ. 1992. Mechanism of DNA strand transfer reactions catalyzed by HIV-1 reverse transcriptase. Science 258:1112–1118. 10.1126/science.1279806 [DOI] [PubMed] [Google Scholar]
- 27.Patel PH, Preston BD. 1994. Marked infidelity of human immunodeficiency virus type 1 reverse transcriptase at RNA and DNA template ends. Proc. Natl. Acad. Sci. U. S. A. 91:549–553. 10.1073/pnas.91.2.549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhuang J, Jetzt AE, Sun G, Yu H, Klarmann G, Ron Y, Preston BD, Dougherty JP. 2002. Human immunodeficiency virus type 1 recombination: rate, fidelity, and putative hot spots. J. Virol. 76:11273–11282. 10.1128/JVI.76.22.11273-11282.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bircher LA, Rigano JC, Ponferrada VG, Wooley DP. 2002. High fidelity of homologous retroviral recombination in cell culture. Arch. Virol. 147:1665–1683. 10.1007/s00705-002-0843-1 [DOI] [PubMed] [Google Scholar]
- 30.Chin MP, Lee SK, Chen J, Nikolaitchik OA, Powell DA, Fivash MJ, Jr, Hu WS. 2008. Long-range recombination gradient between HIV-1 subtypes B and C variants caused by sequence differences in the dimerization initiation signal region. J. Mol. Biol. 377:1324–1333. 10.1016/j.jmb.2008.02.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jetzt AE, Yu H, Klarmann GJ, Ron Y, Preston BD, Dougherty JP. 2000. High rate of recombination throughout the human immunodeficiency virus type 1 genome. J. Virol. 74:1234–1240. 10.1128/JVI.74.3.1234-1240.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Simon-Loriere E, Galetto R, Hamoudi M, Archer J, Lefeuvre P, Martin DP, Robertson DL, Negroni M. 2009. Molecular mechanisms of recombination restriction in the envelope gene of the human immunodeficiency virus. PLoS Pathog. 5:e1000418. 10.1371/journal.ppat.1000418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Galetto R, Moumen A, Giacomoni V, Veron M, Charneau P, Negroni M. 2004. The structure of HIV-1 genomic RNA in the gp120 gene determines a recombination hot spot in vivo. J. Biol. Chem. 279:36625–36632. 10.1074/jbc.M405476200 [DOI] [PubMed] [Google Scholar]
- 34.Moumen A, Polomack L, Unge T, Veron M, Buc H, Negroni M. 2003. Evidence for a mechanism of recombination during reverse transcription dependent on the structure of the acceptor RNA. J. Biol. Chem. 278:15973–15982. 10.1074/jbc.M212306200 [DOI] [PubMed] [Google Scholar]
- 35.Levy DN, Aldrovandi GM, Kutsch O, Shaw GM. 2004. Dynamics of HIV-1 recombination in its natural target cells. Proc. Natl. Acad. Sci. U. S. A. 101:4204–4209. 10.1073/pnas.0306764101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chin MP, Chen J, Nikolaitchik OA, Hu WS. 2007. Molecular determinants of HIV-1 intersubtype recombination potential. Virology 363:437–446. 10.1016/j.virol.2007.01.034 [DOI] [PubMed] [Google Scholar]
- 37.Motomura K, Chen J, Hu WS. 2008. Genetic recombination between human immunodeficiency virus type 1 (HIV-1) and HIV-2, two distinct human lentiviruses. J. Virol. 82:1923–1933. 10.1128/JVI.01937-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chen J, Rhodes TD, Hu WS. 2005. Comparison of the genetic recombination rates of human immunodeficiency virus type 1 in macrophages and T cells. J. Virol. 79:9337–9340. 10.1128/JVI.79.14.9337-9340.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chen J, Powell D, Hu WS. 2006. High frequency of genetic recombination is a common feature of primate lentivirus replication. J. Virol. 80:9651–9658. 10.1128/JVI.00936-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Smyth RP, Schlub TE, Grimm A, Venturi V, Chopra A, Mallal S, Davenport MP, Mak J. 2010. Reducing chimera formation during PCR amplification to ensure accurate genotyping. Gene 469:45–51. 10.1016/j.gene.2010.08.009 [DOI] [PubMed] [Google Scholar]
- 41.Abram ME, Ferris AL, Shao W, Alvord WG, Hughes SH. 2010. Nature, position, and frequency of mutations made in a single cycle of HIV-1 replication. J. Virol. 84:9864–9878. 10.1128/JVI.00915-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chen J, Dang Q, Unutmaz D, Pathak VK, Maldarelli F, Powell D, Hu WS. 2005. Mechanisms of nonrandom human immunodeficiency virus type 1 infection and double infection: preference in virus entry is important but is not the sole factor. J. Virol. 79:4140–4149. 10.1128/JVI.79.7.4140-4149.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chin MP, Rhodes TD, Chen J, Fu W, Hu WS. 2005. Identification of a major restriction in HIV-1 intersubtype recombination. Proc. Natl. Acad. Sci. U. S. A. 102:9002–9007. 10.1073/pnas.0502522102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rhodes T, Wargo H, Hu WS. 2003. High rates of human immunodeficiency virus type 1 recombination: near-random segregation of markers one kilobase apart in one round of viral replication. J. Virol. 77:11193–11200. 10.1128/JVI.77.20.11193-11200.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rhodes TD, Nikolaitchik O, Chen J, Powell D, Hu WS. 2005. Genetic recombination of human immunodeficiency virus type 1 in one round of viral replication: effects of genetic distance, target cells, accessory genes, and lack of high negative interference in crossover events. J. Virol. 79:1666–1677. 10.1128/JVI.79.3.1666-1677.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Moumen A, Polomack L, Roques B, Buc H, Negroni M. 2001. The HIV-1 repeated sequence R as a robust hot-spot for copy choice recombination. Nucleic Acids Res. 29:3814–3821. 10.1093/nar/29.18.3814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Harrison GP, Mayo MS, Hunter E, Lever AM. 1998. Pausing of reverse transcriptase on retroviral RNA templates is influenced by secondary structures both 5′ and 3′ of the catalytic site. Nucleic Acids Res. 26:3433–3442. 10.1093/nar/26.14.3433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Keele BF, Giorgi EE, Salazar-Gonzalez JF, Decker JM, Pham KT, Salazar MG, Sun C, Grayson T, Wang S, Li H, Wei X, Jiang C, Kirchherr JL, Gao F, Anderson JA, Ping LH, Swanstrom R, Tomaras GD, Blattner WA, Goepfert PA, Kilby JM, Saag MS, Delwart EL, Busch MP, Cohen MS, Montefiori DC, Haynes BF, Gaschen B, Athreya GS, Lee HY, Wood N, Seoighe C, Perelson AS, Bhattacharya T, Korber BT, Hahn BH, Shaw GM. 2008. Identification and characterization of transmitted and early founder virus envelopes in primary HIV-1 infection. Proc. Natl. Acad. Sci. U. S. A. 105:7552–7557. 10.1073/pnas.0802203105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Englund G, Theodore TS, Freed EO, Engelman A, Martin MA. 1995. Integration is required for productive infection of monocyte-derived macrophages by human immunodeficiency virus type 1. J. Virol. 69:3216–3219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bleul CC, Wu L, Hoxie JA, Springer TA, Mackay CR. 1997. The HIV coreceptors CXCR4 and CCR5 are differentially expressed and regulated on human T lymphocytes. Proc. Natl. Acad. Sci. U. S. A. 94:1925–1930. 10.1073/pnas.94.5.1925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rauth S, Song KY, Ayares D, Wallace L, Moore PD, Kucherlapati R. 1986. Transfection and homologous recombination involving single-stranded DNA substrates in mammalian cells and nuclear extracts. Proc. Natl. Acad. Sci. U. S. A. 83:5587–5591. 10.1073/pnas.83.15.5587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sprengel R, Varmus HE, Ganem D. 1987. Homologous recombination between hepadnaviral genomes following in vivo DNA transfection: implications for studies of viral infectivity. Virology 159:454–456. 10.1016/0042-6822(87)90486-7 [DOI] [PubMed] [Google Scholar]
- 53.Wake CT, Vernaleone F, Wilson JH. 1985. Topological requirements for homologous recombination among DNA molecules transfected into mammalian cells. Mol. Cell. Biol. 5:2080–2089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zack JA, Arrigo SJ, Weitsman SR, Go AS, Haislip A, Chen IS. 1990. HIV-1 entry into quiescent primary lymphocytes: molecular analysis reveals a labile, latent viral structure. Cell 61:213–222. 10.1016/0092-8674(90)90802-L [DOI] [PubMed] [Google Scholar]
- 55.Meyer M, Stenzel U, Myles S, Prufer K, Hofreiter M. 2007. Targeted high-throughput sequencing of tagged nucleic acid samples. Nucleic Acids Res. 35:e97. 10.1093/nar/gkm566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hoffmann C, Minkah N, Leipzig J, Wang G, Arens MQ, Tebas P, Bushman FD. 2007. DNA bar coding and pyrosequencing to identify rare HIV drug resistance mutations. Nucleic Acids Res. 35:e91. 10.1093/nar/gkm435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kinde I, Wu J, Papadopoulos N, Kinzler KW, Vogelstein B. 2011. Detection and quantification of rare mutations with massively parallel sequencing. Proc. Natl. Acad. Sci. U. S. A. 108:9530–9535. 10.1073/pnas.1105422108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Simpson EH. 1951. The interpretation of interaction in contingency tables. J. R. Stat. Soc. B 13:238–241 [Google Scholar]
- 59.Baird HA, Galetto R, Gao Y, Simon-Loriere E, Abreha M, Archer J, Fan J, Robertson DL, Arts EJ, Negroni M. 2006. Sequence determinants of breakpoint location during HIV-1 intersubtype recombination. Nucleic Acids Res. 34:5203–5216. 10.1093/nar/gkl669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Margulies M, Egholm M, Altman WE, Attiya S, Bader JS, Bemben LA, Berka J, Braverman MS, Chen YJ, Chen Z, Dewell SB, Du L, Fierro JM, Gomes XV, Godwin BC, He W, Helgesen S, Ho CH, Irzyk GP, Jando SC, Alenquer ML, Jarvie TP, Jirage KB, Kim JB, Knight JR, Lanza JR, Leamon JH, Lefkowitz SM, Lei M, Li J, Lohman KL, Lu H, Makhijani VB, McDade KE, McKenna MP, Myers EW, Nickerson E, Nobile JR, Plant R, Puc BP, Ronan MT, Roth GT, Sarkis GJ, Simons JF, Simpson JW, Srinivasan M, Tartaro KR, Tomasz A, Vogt KA, Volkmer GA, Wang SH, Wang Y, Weiner MP, Yu P, Begley RF, Rothberg JM. 2005. Genome sequencing in microfabricated high-density picolitre reactors. Nature 437:376–380. 10.1038/nature03959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.DeStefano JJ, Raja A, Cristofaro JV. 2000. In vitro strand transfer from broken RNAs results in mismatch but not frameshift mutations. Virology 276:7–15. 10.1006/viro.2000.0533 [DOI] [PubMed] [Google Scholar]
- 62.Kulpa D, Topping R, Telesnitsky A. 1997. Determination of the site of first strand transfer during Moloney murine leukemia virus reverse transcription and identification of strand transfer-associated reverse transcriptase errors. EMBO J. 16:856–865. 10.1093/emboj/16.4.856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.DeStefano J, Ghosh J, Prasad B, Raja A. 1998. High fidelity of internal strand transfer catalyzed by human immunodeficiency virus reverse transcriptase. J. Biol. Chem. 273:1483–1489. 10.1074/jbc.273.3.1483 [DOI] [PubMed] [Google Scholar]
- 64.Diaz L, Cristofaro JV, DeStefano JJ. 2000. Human immunodeficiency virus reverse transcriptase base misincorporations can promote strand transfer. Arch. Virol. 145:1117–1131. 10.1007/s007050070113 [DOI] [PubMed] [Google Scholar]
- 65.Nikolenko GN, Svarovskaia ES, Delviks KA, Pathak VK. 2004. Antiretroviral drug resistance mutations in human immunodeficiency virus type 1 reverse transcriptase increase template-switching frequency. J. Virol. 78:8761–8770. 10.1128/JVI.78.16.8761-8770.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]