Abstract
Single-locus blaOXA-51-like sequence-based typing (SBT) was evaluated for its ability to determine correctly sequence types (STs) in Acinetobacter baumannii clinical isolates, in comparison with the Pasteur's multilocus sequence typing (MLST) reference method and 3-locus sequence typing (3-LST). The comparative study was performed in 585 multidrug-resistant (MDR) A. baumannii clinical isolates recovered from 21 hospitals located throughout Greece, Italy, Lebanon, and Turkey. The isolates belonged to nine clonal complexes (CCs) that correspond to 12 distinct sequence types (STs) and to one singleton ST. These clonal lineages predominate worldwide among nosocomial MDR A. baumannii strains. The most common clone was CC2 (ST2 and ST45; n = 278 isolates) followed by CC1 (ST1 and ST20; n = 155), CC25 (n = 65), ST78 (n = 62), CC15 (ST15 and ST84; n = 9), CC10 (n = 4), CC3 (n = 4), CC6 (n = 3), CC54 (n = 3), and CC83 (n = 2). Using the blaOXA-51-like SBT method, all 585 isolates of the study were typed and assigned correctly to the nine CCs and the singleton ST78. The 3-LST method was not able to classify isolates belonging to CC6, CC10, CC54, and CC83, which are not yet characterized in its database. The low-cost and convenient blaOXA-51-like SBT method, compared with 3-LST and MLST, discriminated all epidemic and sporadic lineages of our collection and could be effectively applied to type rapidly A. baumannii strains.
INTRODUCTION
Outbreaks due to multidrug-resistant (MDR) Acinetobacter baumannii clinical strains are increasingly documented worldwide (1). Currently, A. baumannii strains causing hospital outbreaks usually belong to a limited number of clonal lineages, among which those applying to the international clones I and II are prevailing (1–3). This development increasingly necessitates monitoring the epidemic evolution of A. baumannii strains by grouping them to specific clonal lineages. For this purpose, several genotyping methods exist, such as 3-locus sequence typing (3-LST) (3), ribotyping (4), infrequent-restriction-site analysis (5), repetitive sequence-based PCR (rep-PCR) (6), amplified fragment length polymorphism (7), electrospray ionization mass spectrometry (8), pulsed-field gel electrophoresis (PFGE) (9), and multilocus sequence typing (MLST) (10, 11).
Among the methods most commonly used for tracking the worldwide clonal dissemination of A. baumannii, as the reference, the method considered the “gold standard” is MLST, of which two schemes exist, PubMLST (11) and Pasteur MLST (10). Both schemes are based on sequences of seven housekeeping genes and are used for global but also for local epidemiological studies. Concerning MLST, it still remains to be answered whether several loci are required to obtain robust results and if the selection of housekeeping genes is sufficiently reliable to reveal population structures. For instance, the PubMLST gyrB and gpi genes present recombination, resulting in failure to type some isolates (12). Another method that has been widely applied and produced accurate results for the classification of worldwide clones is the 3-LST scheme (3). It involves amplification and sequencing of the ompA, csuE, and blaOXA-51-like genes that are under selective pressure and assigns A. baumannii strains to seven different sequence groups (SGs), SG1 to SG7, corresponding to international clonal lineages identified by other methods (2, 3, 13, 14). Based on 3-LST, two multiplex PCRs were also designed to selectively amplify the ompA, csuE, and blaOXA-51-like alleles that correspond to SG1, SG2, SG3, and SG6 (3, 14) without being followed by sequencing, thus serving as a convenient preliminary method to study local epidemiology. The increasing need for molecular surveillance of multidrug-resistant A. baumannii in the current era of financial restrictions prompts investigation for reliable typing approaches requiring reduced time, labor, and costs.
One of the simplest approaches for the identification (15) and typing (12) of A. baumannii involves amplification and sequencing of the blaOXA-51-like gene, which is unique to A. baumannii, although the same gene has occasionally been detected in Acinetobacter nosocomialis and Acinetobacter genomic species “close to 13TU” (16). The potential of the identification of the specific blaOXA-51-like gene to correctly assign A. baumannii isolates to international clones as single-locus-based typing (SBT) has been scarcely evaluated previously on international collections, in comparison only with PubMLST (12) and rep-PCR (17). In the present study, the performance of this simple, low-cost, and rapid SBT method was evaluated in a large international collection of A. baumannii strains in parallel with the widely used Institute Pasteur's MLST and 3-LST schemes.
MATERIALS AND METHODS
Bacterial collection.
A collection that included 585 A. baumannii clinical isolates recovered between 2000 and 2010 from four different countries (Italy, Greece, Turkey, and Lebanon), representing most common worldwide lineages, was tested. The isolates included in this collection were recovered during outbreaks that occurred in 21 unrelated hospitals in the four countries. Representative isolates of this collection were molecularly characterized previously (13, 14, 18, 19). Although not all of the study isolates were selected because of a multidrug resistance (MDR) phenotype, almost all of them actually exhibited MDR and >80% were carbapenem resistant.
Identification of isolates as A. baumannii was initially performed by PCR and sequencing of the blaOXA-51-like gene (15) and was confirmed by subsequent assignment of isolates by MLST to clonal lineages specific for A. baumannii.
SBT, 3-LST, and MLST.
All isolates were tested using the 3-LST protocol developed by the United Kingdom Health Protection Agency (HPA) (3), involving initially two multiplex PCRs identifying only SG1, SG2, SG3, and SG6 and, subsequently, sequencing, confirming the grouping and discriminating SG1, SG2, SG3, SG4, SG5, SG6, and SG7 (available at the Health Protection Agency website [http://www.hpa-bioinformatics.org.uk/AB/]). The sequencing of solely the blaOXA51-like gene using primers encompassing the total gene (forward primer, 5′-ATGAACATTAAAGCACTCTTAC-3′; reverse primer, 5′-CTATAAAATACCTAATTGTTCT-3′ [825-bp amplicon]) was applied as a separate approach for typing the isolates. These primers include the start and stop codons, and a few OXA-51-like variants that differ from the common alleles at the 5′ and 3′ proximities could be misidentified. Nevertheless, the primers were able to amplify blaOXA-51-like alleles of all 585 strains included in our collection. Furthermore, all isolates were tested by the MLST scheme developed by the Pasteur Institute (10), which served as the gold standard method against which the other approaches were evaluated. This scheme involves PCR amplification and sequencing of seven housekeeping genes (fusA, gltA, pyrG, recA, cpn60, rpoB, and rplB).
E burst.
To determine the clonal complexes (CCs) where isolates belonged, e-BURST analysis was performed using the eBURST software program (http://eburst.mlst.net/).
Phylogeny trees.
Neighbor-joining phylogenetic trees based on blaOXA-51-like genes and concatenated alleles of the seven housekeeping genes of Pasteur's MLST scheme were constructed using the phylogeny.fr platform (20) available at http://www.phylogeny.fr/.
RESULTS
STs of the studied isolates using the Pasteur's MLST scheme.
Table 1 represents the origin, the typing results, and the classification of the isolates using the three different typing schemes. Overall, by using the reference Pasteur's MLST scheme, 13 STs (ST1, ST2, ST3, ST15, ST20, ST25, ST45, ST54, ST78, ST82, ST83, ST84, and ST85) were identified among the 585 A. baumannii isolates of the study.
TABLE 1.
Characteristics of the 585 study isolatesa
| Index strainb | Country | blaOXA-51-like allele (no. of isolates) | Pasteur's MLST |
3-LST group | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CC | cpn60 | fusA | gltA | pyrG | recA | rpiB | rpoB | ST (no. of isolates) | ||||
| AB3990 | Italy | blaOXA-66 (278) | CC2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | ST2 (265) | 1 |
| AB3 | Greece | 2 | 2 | 6 | 2 | 2 | 2 | 2 | ST45 (13) | |||
| AB700 | Italy | blaOXA-69 (155) | CC1 | 1 | 1 | 1 | 1 | 5 | 1 | 1 | ST1 (124) | 2 |
| AB2979 | Italy | 3 | 1 | 1 | 1 | 5 | 1 | 1 | ST20 (31) | |||
| AB3237 | Lebanon | blaOXA-71 (4) | CC3 | 3 | 3 | 2 | 2 | 3 | 1 | 3 | ST3 (4) | 3 |
| AB3890 | Greece | blaOXA-64 (65) | CC25 | 3 | 3 | 2 | 4 | 7 | 2 | 4 | ST25 (65) | 4 |
| AB3909 | Italy | blaOXA-90 (62) | Singleton | 25 | 3 | 6 | 2 | 28 | 1 | 29 | ST78 (62) | 6 |
| AB6 | Greece | blaOXA-94 (3) | CC6 | 5 | 2 | 4 | 1 | 3 | 3 | 4 | ST85 (3) | NA |
| AB17 | Greece | blaOXA-365c (3) | CC54 | 12 | 3 | 18 | 2 | 17 | 4 | 5 | ST54 (3) | NA |
| AB3868 | Turkey | blaOXA-51 (9) | CC15 | 6 | 6 | 8 | 2 | 3 | 5 | 4 | ST15 (6) | 5 |
| AB3871 | Turkey | 6 | 6 | 8 | 2 | 3 | 5 | 30 | ST84 (3) | |||
| AB2977 | Italy | blaOXA-128 (4) | CC10 | 28 | 3 | 2 | 1 | 4 | 4 | 4 | ST82 (4) | NA |
| AB3866 | Turkey | blaOXA-86 (2) | CC83 | 26 | 4 | 2 | 2 | 9 | 1 | 4 | ST83 (2) | NA |
| OIFC032 | Germany | blaOXA-100d | CC32 | 1 | 1 | 2 | 2 | 3 | 4 | 4 | ST32 | NA |
| AB_TG27343 | USA | blaOXA-65e | CC79 | 26 | 2 | 2 | 2 | 29 | 4 | 5 | ST79 | NA |
SBT results.
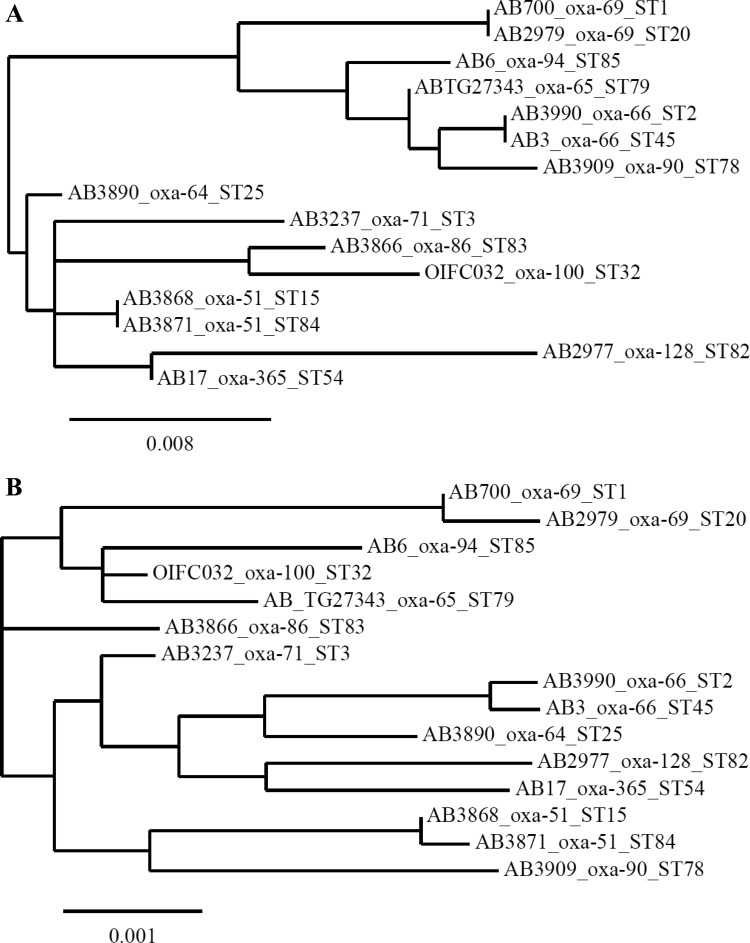
In comparison with the MLST, SBT correctly identified isolates belonging to all major lineages (Fig. 1). In particular, the two major international clonal lineages corresponding to CC1 and CC2 each carried a single blaOXA-51-like variant. All isolates carrying identical blaOXA-69 genes belonged to CC1, which included the founder ST of CC1 (ST1) and its single-locus variant (SLV) ST20. CC2 isolates, consisting in our collection of ST2 and the SLV ST45, in all cases carried identical blaOXA-66 genes. Isolates belonging to CC3, CC6, CC10, CC15 (ST15 and ST84), CC25, CC54, ST78, and CC83 all carried a unique blaOXA-51-like variant (Table 1). Finally, isolates of CC32 and CC79, although not available in the present collection, could be effectively typed by SBT, as was shown using data obtained from the GenBank; CC32 isolates carry blaOXA-100 (GenBank accession number AM231720), and CC79 isolates carry blaOXA-65 (accession numbers AY750908 and JQ412185) (Table 1). These data clearly showed that CC1 and CC2 predominated in this international collection.
FIG 1.
Neighbor-joining phylogenetic trees based on blaOXA-51like genes (A) and concatenated alleles of the seven housekeeping genes of Pasteur's MLST scheme (B). The dendrograms show the amounts of genetic change based on multiple alignments and were generated using phylogeny.fr software (17). The bar at the bottom of each figure shows the amount of genetic change corresponding to the length of each branch.
The relatively limited number of OXA-51-like alleles that were identified among the study isolates prompted a GenBank search of all publicly available whole-genome sequences of A. baumannii. The results of this search are presented in the Table S1 in the supplemental material. In particular, of the 104 strains available that belonged to ST2, 98 harbored OXA-66 or the single-amino-acid variant OXA-82; the OXA-66 allele had identical nucleotide sequences in all strains, and OXA-82 had identical sequences in all but 2 strains that harbored alleles differing by 1 nucleotide (nt). Also, 2 strains and 1 strain assigned to ST2 harbored the single-amino-acid variants OXA-109 and OXA-254, respectively, and 3 strains harbored OXA-113 alleles that differed from OXA-66 in six amino acid residues, thus suggesting that limited variability of the blaOXA-51-like gene existed in international clonal linage 2. Despite this variability, SBT was able to correctly type and assign to international clonal lineage 2 all ST2 strains available in GenBank. Furthermore, all 28 ST1 strains harbored identical OXA-69 alleles, 6/8 ST3 strains harbored OXA-71 and 2/8 harbored an allele (OXA-312) with 1-nt difference, 1/2 CC10 strains harbored OXA-68 and the other harbored an allele (OXA-128) with 1-nt difference, 7/7 CC25 strains harbored identical OXA-64 alleles, 3/3 CC32 strains harbored OXA-100, and 3/3 CC79 strains harbored identical OXA-65 alleles.
3-LST results.
The 3-LST scheme, although overall it provided data relative to three genes under selective pressure, failed to classify isolates belonging to CC10, CC54, CC83, ST85, CC32, and CC79, as alleles corresponding to these lineages are not yet assigned in its database. Twelve isolates, identified as belonging to ST54, ST82, ST83, and ST85, are considered to belong to microepidemic lineages and are not assigned to any SG. As for the simple and rapid 3-LST multiplex PCR, it discriminated only SG1 (CC2), SG2 (CC1), SG3 (CC3), and SG6 (ST78).
DISCUSSION
Several methods have been applied during the last 2 decades to identify the clonal lineages of A. baumannii isolates. Among them, the most common ones have been 3-LST (3) and MLST (10, 11). These methods are appropriately discriminatory but can be considered either time-consuming and labor-intensive or costly. Amplification and sequencing of the blaOXA-51-like gene have been used successfully for A. baumannii identification and within the 3-LST scheme, respectively. It has also been observed during 3-LST analysis (3) that blaOXA-51-like variants are conserved within most SGs and often correlate well with the MLSTs, thus deserving investigation for the potential of blaOXA-51-like SBT to be utilized as an independent typing scheme. Until now, this possibility was tested in parallel only with the PubMLST scheme (12), against which the blaOXA-51-like SBT was shown to identify accurately isolates belonging to the three major epidemic lineages. In another study, the blaOXA-51-like SBT compared with rep-PCR identified international clonal lineages I to III and worldwide clonal lineages 4 to 8 (17). In the current report, the performance of blaOXA-51-like SBT was compared with that of the Pasteur's MLST scheme, which does not exhibit recombination in its target loci in contrast with PubMLST, and also with that of 3-LST, including both the initial multiplex PCR and the sequencing analysis. The total blaOXA-51-like gene was sequenced, as the cost is the same and the nucleotide yield is bigger, enabling the identification of novel blaOXA-51-like variants, such as the blaOXA-365 allele corresponding to ST54. This comparison was applied to a large, diverse collection of 585 A. baumannii strains from several countries, including most common lineages. This collection was also tested by PFGE, but these results were not included in the current presentation because PFGE is not suitable for international population studies.
In the tested collection, blaOXA-51-like SBT identified successfully the common lineages CC1, CC2, CC3, CC10, CC15, and CC25. It was also shown in the publicly available databases that each of the remaining rather common clones CC32 and CC79 contains a specific blaOXA-51-like allele (blaOXA-100 and blaOXA-65, respectively) and can be efficiently identified by the blaOXA-51-like SBT. A single blaOXA-51-like allele was found to correspond to two different STs (blaOXA-69 to ST1 and ST20 and blaOXA-66 to ST2 and ST45) that, however, were included in the same CC, not affecting prompt identification of the respective international lineages. In all cases, the same blaOXA-51-like allele was detected in strains belonging to the same CC and no case was observed where different blaOXA-51-like alleles were detected in the same ST. In contrast, the application of blaOXA-51-like SBT in comparison with PubMLST (12) resulted in the detection of the same blaOXA-51-like allele(s) in unrelated STs (blaOXA-51 in CC4 and ST20) and of distinct alleles in a single lineage (blaOXA-69 and blaOXA-112 in international clone I; blaOXA-82 and blaOXA-83 in ST22; blaOXA-107 and blaOXA-110 in ST49; and blaOXA-51 and blaOXA-68 in ST20). The correlation of the blaOXA-51-like gene with PubMLST was better when alleles exhibiting recombination (gpi and gyrB) were excluded from the analysis (12).
In this study, testing 585 multidrug-resistant clinical isolates, only 13 STs were identified, while at least 450 STs were found to be currently distinguishable by the Pasteur's MLST. However, it is well known that the high diversity of the general population of A. baumannii at the strain level especially involves susceptible strains (10). Also, although this large collection includes also epidemiologically related isolates, these isolates came from numerous unrelated hospitals in four different countries, indicating the circulation of limited clones in the hospital epidemiological niche and supporting the observed bottleneck effect for MDR A. baumannii (10). Furthermore, the present analysis, which detected identical alleles in multiple A. baumannii isolates from each lineage, underlines the intrastrain stability of typing characteristics using the blaOXA-51-like SBT method.
The reliability of the blaOXA-51-like SBT method is further validated by the analysis of A. baumannii whole-genome sequences (WGS) available in GenBank. Despite a slight variability of the blaOXA-51-like gene found in A. baumannii strains belonging to international clone 2 and also within the predominant international clones 1 and 3 (see Table S1 in the supplemental material), this SBT is able to correctly type and assign to the major international clonal lineages A. baumannii WGS available in GenBank. These data indicate that the vast majority of the predominant lineages of A. baumannii could be effectively identified by this SBT and clearly support our view of the clinical applicability of blaOXA-51-like SBT as a simple first-line typing approach.
As for 3-LST, although this scheme analyzes three loci and provides more comprehensive data from an evolutionary point of view than the blaOXA-51-like SBT scheme, it has not yet been applied in collections large enough to identify all common lineages. Its database currently includes only SG2 (CC1 according to Pasteur's scheme), SG1 (CC2), SG3 (CC3), SG4 (CC25), SG5 (CC15), SG6 (ST78), and SG7, which were almost all identified in our collection. In that respect, blaOXA-51-like SBT identified more STs defined by MLST than 3-LST. However, it is evident that a future application of 3-LST in larger collections representing all lineages might further discriminate strains. We should note here for 3-LST that two unrelated strains were previously shown to carry the blaOXA-51-like gene that corresponds to CC1 but also a csuE allele that was characteristic of CC2 (21). However, both of these strains actually belonged to CC1 as shown by MLST and it was concluded that they had acquired the csuE gene from other A. baumannii strains. In fact, one of these strains, A388, carried OXA-92 (12), which differs from OXA-69 by only one nucleotide (22), indicating that it would be correctly grouped by OXA-51-like SBT as CC1. It is thus indicated that we should treat with caution results that include the csuE allele, such as those from 3-LST, and that OXA-51-like alleles are possibly more conserved and may correlate better with MLST than does correlate the 3-LST scheme. Finally, as could be anticipated, simple multiplex PCR of the 3-LST scheme, when applied as an independent method, identified fewer SGs than the other typing approaches tested.
Concluding remarks.
Overall, blaOXA-51-like SBT, compared with Pasteur's MSLT and 3-LST, was shown to identify accurately isolates belonging to all major A. baumannii lineages that were available in our collection. It can be assumed that this SBT, being evidently easier, faster, and cheaper than MLST, could be applied efficiently for the provisional molecular typing of A. baumannii.
Supplementary Material
Footnotes
Published ahead of print 12 March 2014
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JCM.03565-13.
REFERENCES
- 1.Peleg AY, Seifert H, Paterson DL. 2008. Acinetobacter baumannii: emergence of a successful pathogen. Clin. Microbiol. Rev. 21:538–582. 10.1128/CMR.00058-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zarrilli R, Pournaras S, Giannouli M, Tsakris A. 2013. Global evolution of multidrug-resistant Acinetobacter baumannii clonal lineages. Int. J. Antimicrob. Agents 41:11–19. 10.1016/j.ijantimicag.2012.09.008 [DOI] [PubMed] [Google Scholar]
- 3.Turton JF, Gabriel SN, Valderrey S, Kaufmann ME, Pitt TL. 2007. Use of sequence-based typing and multiplex PCR to identify clonal lineages of outbreak strains of Acinetobacter baumannii. Clin. Microbiol. Infect. 13:807–815. 10.1111/j.1469-0691.2007.01759.x [DOI] [PubMed] [Google Scholar]
- 4.Brisse S, Milatovic D, Fluit AC, Kusters K, Toelstra A, Verhoef J, Schmitz FJ. 2000. Molecular surveillance of European quinolone-resistant clinical isolates of Pseudomonas aeruginosa and Acinetobacter spp. using automated ribotyping. J. Clin. Microbiol. 38:3636–3645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yoo JH, Choi JH, Shin WS, Huh DH, Cho YK, Kim KM, Kim MY, Kang MW. 1999. Application of infrequent-restriction-site PCR to clinical isolates of Acinetobacter baumannii and Serratia marcescens. J. Clin. Microbiol. 37:3108–3112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Huys G, Cnockaert M, Vaneechoutte M, Woodford N, Nemec A, Dijkshoorn L, Swings J. 2005. Distribution of tetracycline resistance genes in genotypically related and unrelated multiresistant Acinetobacter baumannii strains from different European hospitals. Res. Microbiol. 156:348–355. 10.1016/j.resmic.2004.10.008 [DOI] [PubMed] [Google Scholar]
- 7.Spence RP, van der Reijden TJ, Dijkshoorn L, Towner KJ. 2004. Comparison of Acinetobacter baumannii isolates from United Kingdom hospitals with predominant northern European genotypes by amplified-fragment length polymorphism analysis. J. Clin. Microbiol. 42:832–834. 10.1128/JCM.42.2.832-834.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ecker JA, Massire C, Hall TA, Ranken R, Pennella TT, Agasino Ivy C, Blyn LB, Hofstadler SA, Endy TP, Scott PT, Lindler L, Hamilton T, Gaddy C, Snow K, Pe M, Fishbain J, Craft D, Deye G, Riddell S, Milstrey E, Petruccelli B, Brisse S, Harpin V, Schink A, Ecker DJ, Sampath R, Eshoo MW. 2006. Identification of Acinetobacter species and genotyping of Acinetobacter baumannii by multilocus PCR and mass spectrometry. J. Clin. Microbiol. 44:2921–2932. 10.1128/JCM.00619-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Seifert H, Dolzani L, Bressan R, van der Reijden T, van Strijen B, Stefanik D, Heersma H, Dijkshoorn LJ. 2005. Standardization and interlaboratory reproducibility assessment of pulsed-field gel electrophoresis-generated fingerprints of Acinetobacter baumannii. J. Clin. Microbiol. 43:4328–4335. 10.1128/JCM.43.9.4328-4335.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Diancourt L, Passet V, Nemec A, Dijkshoorn L, Brisse S. 2010. The population structure of Acinetobacter baumannii: expanding multiresistant clones from an ancestral susceptible genetic pool. PLoS One 5:e10034. 10.1371/journal.pone.0010034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bartual SG, Seifert H, Hippler C, Luzon MA, Wisplinghoff H, Rodríguez-Valera F. 2005. Development of a multilocus sequence typing scheme for characterization of clinical isolates of Acinetobacter baumannii. J. Clin. Microbiol. 43:4382–4390. 10.1128/JCM.43.9.4382-4390.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hamouda A, Evans BA, Towner KJ, Amyes SG. 2010. Characterization of epidemiologically unrelated Acinetobacter baumannii isolates from four continents by use of multilocus sequence typing, pulsed-field gel electrophoresis, and sequence-based typing of blaOXA-51-like genes. J. Clin. Microbiol. 48:2476–2483. 10.1128/JCM.02431-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Giannouli M, Tomasone F, Agodi A, Vahaboglu H, Daoud Z, Triassi M, Tsakris A, Zarrilli R. 2009. Molecular epidemiology of carbapenem-resistant Acinetobacter baumannii strains in intensive care units of multiple Mediterranean hospitals. J. Antimicrob. Chemother. 63:828–830. 10.1093/jac/dkp032 [DOI] [PubMed] [Google Scholar]
- 14.Giannouli M, Cuccurullo S, Crivaro V, Di Popolo A, Bernardo M, Tomasone F, Amato G, Brisse S, Triassi M, Utili R, Zarrilli R. 2010. Molecular epidemiology of multidrug-resistant Acinetobacter baumannii in a tertiary care hospital in Naples, Italy, shows the emergence of a novel epidemic clone. J. Clin. Microbiol. 48:1223–1230. 10.1128/JCM.02263-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Turton JF, Woodford N, Glover J, Yarde S, Kaufmann ME, Pitt TL. 2006. Identification of Acinetobacter baumannii by detection of the blaOXA-51-like carbapenemase gene intrinsic to this species. J. Clin. Microbiol. 44:2974–2976. 10.1128/JCM.01021-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lee YT, Kuo SC, Chiang MC, Yang SP, Chen CP, Chen TL, Fung CP. 2012. Emergence of carbapenem-resistant non-baumannii species of Acinetobacter harboring a blaOXA-51-like gene that is intrinsic to Acinetobacter baumannii. Antimicrob. Agents Chemother. 56:1124–1127. 10.1128/AAC.00622-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zander E, Nemec A, Seifert H, Higgins PG. 2012. Association between β-lactamase-encoding bla(OXA-51) variants and DiversiLab rep-PCR-based typing of Acinetobacter baumannii isolates. J. Clin. Microbiol. 50:1900–1904. 10.1128/JCM.06462-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Di Popolo A, Giannouli M, Triassi M, Brisse S, Zarrilli R. 2011. Molecular epidemiology of multidrug-resistant Acinetobacter baumannii strains in four Mediterranean countries with a multilocus sequence typing scheme. Clin. Microbiol. Infect. 17:197–201. 10.1111/j.1469-0691.2010.03254.x [DOI] [PubMed] [Google Scholar]
- 19.Gogou V, Pournaras S, Giannouli M, Voulgari E, Piperaki E-T, Zarrilli R, Tsakris A. 2011. Evolution of multidrug-resistant Acinetobacter baumannii clonal lineages: a 10 years study in Greece (2000–2009). J. Antimicrob. Chemother. 66:2767–2772. 10.1093/jac/dkr390 [DOI] [PubMed] [Google Scholar]
- 20.Post V, Hamidian M, Hall RM. 2012. Antibiotic-resistant Acinetobacter baumannii variants belonging to global clone 1. J. Antimicrob. Chemother. 67:1039–1040. 10.1093/jac/dkr586 [DOI] [PubMed] [Google Scholar]
- 21.Dereeper A, Guignon V, Blanc G, Audic S, Buffet S, Chevenet F, Dufayard JF, Guindon S, Lefort V, Lescot M, Claverie JM, Gascuel O. 2008. Phylogeny.fr: robust phylogenetic analysis for the non-specialist. Nucleic Acids Res. 36 (Web Server Issue):W465–W469. 10.1093/nar/gkn180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tsakris A, Ikonomidis A, Spanakis N, Pournaras S, Bethimouti K. 2007. Identification of a novel bla(OXA-51) variant, bla(OXA-92), from a clinical isolate of Acinetobacter baumannii. Clin. Microbiol. Infect. 13:348–349. 10.1111/j.1469-0691.2006.01598.x [DOI] [PubMed] [Google Scholar]
- 23.Sahl JW, Gillece JD, Schupp JM, Waddell VG, Driebe EM, Engelthaler DM, Keim P. 2013. Evolution of a pathogen: a comparative genomics analysis identifies a genetic pathway to pathogenesis in Acinetobacter. PLoS One 8:e54287. 10.1371/journal.pone.0054287 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.