Abstract
Mycoplasma genitalium is a sexually transmitted organism commonly treated with azithromycin. However, macrolide resistance has been reported and is associated with point mutations in the 23S rRNA gene. To evaluate the prevalence of macrolide resistance in M. genitalium isolates from clinical specimens from France, we first used a previously reported high-resolution melting assay. Because susceptible and resistant M. genitalium isolates were hardly discriminated in M. genitalium-positive clinical specimens, we developed a new molecular assay for the rapid detection of macrolide resistance. An assay using real-time PCR based on fluorescence resonance energy transfer (FRET) coupled with melting curve analysis was designed. The assay was first validated on characterized macrolide-resistant M. genitalium isolates and then applied to 202 urogenital M. genitalium-positive specimens collected from 178 patients from France in 2011 and 2012. Resistant genotypes were confirmed by 23S rRNA gene sequencing. Among the 202 M. genitalium-positive specimens, 155 were amplified, demonstrating a sensitivity of 76.7%. A substitution in the 23S rRNA gene was found in 14.2% of the patient samples. Nine and six patients had M. genitalium isolates with a substitution at positions 2059 and 2058, respectively. In four cases, a mixed population of wild-type and mutated M. genitalium isolates was observed. The prevalence of M. genitalium macrolide resistance has been stable in France since its detection in 2006. Our FRET PCR assay is able to discriminate between wild-type and resistant genotypes directly from clinical specimens. This assay will allow clinicians to shorten the time to the initiation of effective disease treatment.
INTRODUCTION
Mycoplasma genitalium is a sexually transmitted organism associated with 15 to 20% of cases of acute and chronic nongonococcal urethritis (NGU) in men (1) and is implicated in female cervicitis and pelvic inflammatory disease (2). It is a fastidious microorganism that is extremely difficult to culture from clinical samples and requires cell culture on Vero cells (3). Thus, nucleic acid amplification testing is the only method available for routine M. genitalium detection. Several in-house PCR assays and a few commercially available simplex and multiplex detection kits that target the MgPa adhesin gene (mgpB) or the 16S rRNA gene have been developed recently (4–6).
The lack of a cell wall makes M. genitalium intrinsically resistant to a number of antibiotics such as beta-lactams (4). However, M. genitalium is intrinsically susceptible to tetracyclines, macrolides, and fluoroquinolones. Tetracyclines are not adequate for the treatment of M. genitalium infections because of the high rate of therapeutic failure (1, 4). Azithromycin regimens have been commonly used for the treatment of M. genitalium infections (7), but the failure of azithromycin treatment has been reported in many cases of M. genitalium-positive NGU (8, 9). This can be due to the selection of resistant mutants during treatment with a single 1-g dose of azithromycin and/or the presence of preexisting resistant isolates (10, 11). In this situation, moxifloxacin is usually used as a second-line treatment (4).
Macrolide-resistant M. genitalium isolates have been detected in patients from Australia, Sweden, Norway, and the United States, and mutations in domain V of the 23S rRNA gene have been detected in specimens from Greenland, Japan, France, and an increasing number of other countries (8, 9, 12–14). The resistance mutations consist mainly of adenine-to-guanine mutations in the 23S rRNA gene at position 2058 or 2059 (Escherichia coli numbering) (9, 12, 15, 16).
Macrolide resistance requires epidemiological surveillance with molecular tools that detect 23S rRNA mutations directly from clinical samples. Such tools already exist for the phylogenetically closely related respiratory pathogen Mycoplasma pneumoniae, which has a similar pattern of resistance to macrolides and for which rapid real-time assays have been developed for the direct detection of macrolide resistance in respiratory specimens (17–19).
To evaluate the prevalence of macrolide resistance in M. genitalium isolates from clinical specimens from France in 2011 and 2012, we first used a recently reported high-resolution melting (HRM) assay (10). Because susceptible and resistant M. genitalium isolates were hardly discriminated in M. genitalium-positive clinical specimens, we developed a new real-time PCR based on fluorescence resonance energy transfer (FRET) coupled with melting curve analysis for the rapid detection of macrolide resistance directly in clinical specimens.
MATERIALS AND METHODS
Bacterial strains.
Genomic DNAs from the susceptible M. genitalium reference strain G37 (ATCC 33530) and from 3 macrolide-resistant clinical isolates from Denmark (9), M6270 (20), M6271, and M6302, with characterized A2059G, A2058G, and A2058C mutations (E. coli numbering) in the 23S rRNA gene, respectively, were used as controls for validation of each real-time PCR assay.
Clinical samples.
Ten urogenital M. genitalium-positive specimens with genotypes characterized by PCR and sequencing (6 wild-type isolates, 3 with the A2058G mutation, and 1 with the A2059G mutation) were used to validate the HRM and FRET assays.
Between January 2011 and December 2012, 202 urogenital specimens were collected retrospectively from 178 patients (30 endocervical swabs and 117 vaginal swabs from 139 women and 37 first-void urine samples and 18 urethral swabs from 39 men). All the specimens were confirmed to be positive for M. genitalium at Pellegrin Hospital (Bordeaux, France) by using an in-house TaqMan PCR assay targeting the MgPa adhesin gene (21). DNA extraction was performed with MagNa Pure LC DNA isolation kit I (Roche Diagnostics, Meylan, France), according to the manufacturer's instructions. These specimens and their DNA extracts were stored at −80°C.
HRM analysis.
To evaluate the prevalence of macrolide resistance in M. genitalium isolates from clinical specimens from France, a recently reported HRM assay using primers Mg23S1992F and Mg23S2138R (10) was applied to the characterized isolates and specimens. The assay was run on a LightCycler 480 real-time thermocycler (Roche Diagnostics, Meylan, France) according to a previously reported protocol (10). Three different enzyme mixes were tested: LightCycler 480 High Resolution Melting Master mix (Roche), Kapa HRM Fast PCR mix (Kapa Biosystems, Nanterre, France), and MeltDoctor HRM Master mix (Applied Biosystems, Courtaboeuf, France).
For data analysis, the HRM curves were derived by selecting two normalization regions, one occurring prior to the melting of the double-stranded product and one following complete separation of the two strands. The normalized and temperature-shifted difference plot was generated by selecting the melting profile of reference strain G37 as the susceptible genotype.
FRET real-time PCR.
We designed a real-time PCR based on FRET technology coupled with melting curve analysis. The assay included the amplification of a fragment of the M. genitalium 23S rRNA gene with the simultaneous hybridization of two probes and analysis of melting curves. Primers specific for the M. genitalium 23S rRNA gene were designed after the alignment of reported 23S rRNA sequences from other human mycoplasmas, such as Mycoplasma pneumoniae, M. hominis, M. penetrans, Ureaplasma parvum, and U. urealyticum. A Basic Local Alignment Search Tool (BLAST) analysis of each primer and probe was performed, and all organisms that had a BLAST hit were included in the analytical specificity assay (see below).
Primers F1-Mg and R1-Mg (Table 1) amplified a 266-bp fragment encompassing nucleotides at positions 2058 and 2059. Two hybridization probes were designed for the detection of point mutations associated with macrolide resistance in the 23S rRNA gene. The probes included a sensor probe, 5′ labeled with LC-Red 640, which hybridized to the region containing the mutation sites and a fluorescein 3′-labeled anchor probe that hybridized 3 bases upstream of the former probe (Table 1). The probes were obtained from Eurofins MWG Operon (Ebersberg, Germany). The PCR and hybridization reactions were carried out in glass capillaries by using a LightCycler 1.5 thermocycler (Roche). To determine the accurate volume of DNA extract from specimens for PCR, optimization tests were performed. Ten DNA extracts from M. genitalium-positive specimens with detection cycle thresholds ranging from 24 to 39 were tested in triplicate with volumes ranging from 2 to 8 μl. The manufacturer's instructions for the LightCycler FastStart DNA Master hybridization kit probes recommend not using >8 μl of DNA per 20-μl PCR mixture because of the potential inhibitory effects. Because extract volumes of 5, 6, 7, and 8 μl led to the same satisfactory results, a volume of 5 μl of DNA extract was retained for PCR. Twenty microliters of a PCR mixture containing 5 μl of template DNA from clinical specimens (or 2 μl of template DNA from isolates), 1.6 μl of 25 mM MgCl2, 1 μl of the two primers (5 μM each), 2 μl of the two probes (2 μM each), and 2 μl of FastStart DNA Master hybridization probes (Roche) was prepared. The cycling conditions consisted of an initial denaturation cycle at 95°C for 10 min, followed by 50 amplification cycles (with a transition rate of 20°C/s) consisting of 95°C for 10 s, annealing at 58°C for 20 s, and extension at 72°C for 20 s. After amplification, a melting step was performed, consisting of 95°C for 1 s, 35°C for 40 s, and a slow increase in the temperature to 85°C at a rate of 0.1°C/s with the continuous acquisition of fluorescence; a final cooling step was performed for 30 s at 40°C. Data were analyzed with LightCycler software version 3.5 (Roche). The “polynomial” option was selected as the calculation method for melting curve analysis, and melting temperature (Tm) values were determined by using the “manual Tm” function. The assay was validated first on wild-type strain G37 and the characterized macrolide-resistant M. genitalium isolates and then on the 10 characterized M. genitalium-positive urogenital specimens. For the control strain and isolates, PCR was performed 10 times, and the Tm values are presented as means ± standard deviations (SD).
TABLE 1.
Primers and probes used for FRET real-time PCR
| Primer or probe | Sequence (5′→3′) | Amplicon size (bp) |
|---|---|---|
| Primers | ||
| F1-Mg | GAAGGAGGTTAGCAATTTATTGC | 266 |
| R1-Mg | TTCTCTACATGGTGGTGTTTTG | |
| Probes | ||
| Anchor-probe | CGGGTGAAGACACCCGTTAGGC-fluorescein | |
| Sensor-probe | LC-Red 640–ACGGGACGGAAAGACC-phosphate |
Sequencing analysis.
The fragment of domain V of the 23S rRNA gene encompassing positions 2058, 2059, and 2062 was amplified with primers F1-Mg and R1-Mg (Table 1) and sequenced with the same primers in both strands for clinical specimens that were determined to have a resistant genotype by using the FRET real-time PCR assay.
Specificity and limit of detection.
To assess the specificity of the assay, 20 specimens with negative results for M. genitalium determined by an in-house real-time PCR (21) but with positive results for pathogens involved in urogenital infections or commensals of the urogenital tract (Chlamydia trachomatis, Neisseria gonorrhoeae, M. hominis, U. urealyticum, Candida albicans, Enterococcus faecalis, E. coli, Gardnerella vaginalis, Corynebacterium sp., Lactobacillus sp., Pseudomonas aeruginosa, Staphylococcus aureus, coagulase-negative staphylococci, Streptococcus agalactiae, Bacteroides fragilis, Prevotella bivia, and Clostridium perfringens) as well as DNA extracts from Mycoplasma strains with a BLAST hit for the probes (M. pneumoniae, M. penetrans, M. hominis, M. amphoriforme, and M. gallisepticum) were tested.
To evaluate the limit of detection of the FRET assay, M. genitalium G37 DNA was extracted and amplified with the F1-Mg/R1-Mg primer set. The amplified product was purified by using the Wizard PCR Preps DNA purification system (Promega, Charbonnière, France) and subsequently cloned into the pGEM-T Easy plasmid vector (Promega), according to the manufacturer's instructions. Plasmid pGEM-T MgF1-R1 was transformed into E. coli DH5α cells and purified from E. coli transformants by using the Wizard Plus SV Minipreps DNA purification system (Promega). The purity and concentration of plasmid DNA were determined by optical density measurements (NanoDrop 1000). The detection limit of the assay was assessed by using 10-fold serial dilutions of plasmid pGEM-T MgF1-R1.
Statistical analysis.
Comparisons were made by using Fisher's exact test and the paired t test to determine if there were any gender differences in the observed macrolide resistance and if the sensitivity of the assay varies according to the specimen type, respectively. A P value of <0.05 was considered statistically significant.
RESULTS
HRM analysis.
The recently reported HRM assay was used to detect macrolide resistance-associated mutations in clinical specimens (10). For this purpose, we tested 10 M. genitalium-positive urogenital specimens and four M. genitalium isolates with characterized genotypes. Three different enzyme mixes were successively used, as the first results were not satisfactory (see below).
In terms of sensitivity, the HRM assay was able to amplify all of the clinical specimens with a high cycle threshold (close to 39 [data not shown]), regardless of the master mix used. However, wild-type and mutated M. genitalium isolates were inadequately discriminated.
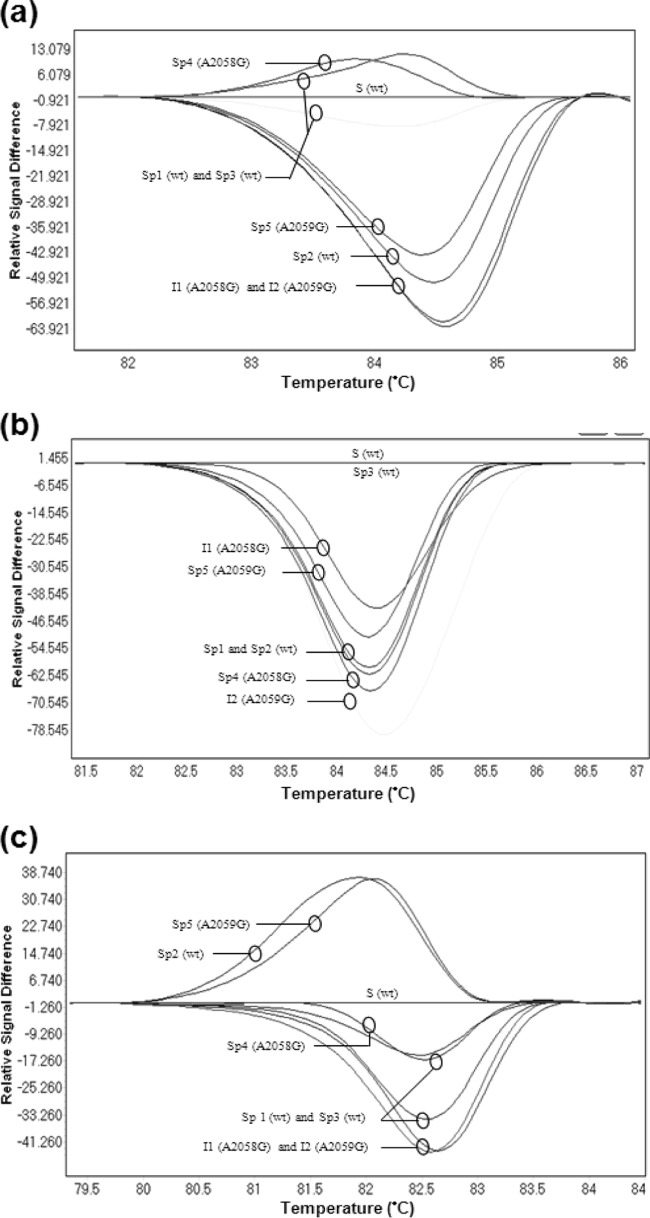
Using the Roche LightCycler 480 High Resolution Melting Master mix, the Tms of wild-type and resistant isolates and specimens differed by <0.5°C; thus, the differentiation of both genotypes was not achieved. Moreover, analysis based on normalized and shifted melting curves or normalized and temperature-shifted difference plots did not allow for differentiation between wild-type and mutated specimens (Fig. 1a). The melting curves of resistant isolates with the A2058G, A2059G, or A2058C mutation overlapped without differentiation, and the results were not reproducible when the PCRs were repeated. In addition, HRM results varied according to the M. genitalium DNA concentration. Different profiles were observed when different concentrations of wild-type strain DNA were tested with the Roche and Applied Biosystems enzyme mixes. Indeed, the wild-type curves sometimes overlapped the curves of mutated isolates (see Fig. S1 in the supplemental material).
FIG 1.
Normalized and temperature-shifted difference plots obtained with the real-time high-resolution melting assay using three distinct enzyme mixes for three characterized isolates and five clinical specimens. The curve of wild-type (wt) M. genitalium strain G37 was selected as a reference. (a) LightCycler 480 High Resolution Melting Master mix (Roche); (b) Kapa HRM Fast PCR mix (Kapa Biosystems); (c) MeltDoctor HRM Master mix (Applied Biosystems). S, strain G37 (wild type); I1, resistant isolate harboring an A2058G mutation; I2, resistant isolate harboring an A2059G mutation; Sp1, Sp2, and Sp3, clinical specimens with the wild-type genotype; Sp4, clinical specimen with the A2058G mutation; Sp5, clinical specimen with the A2059G mutation.
Using the Kapa HRM Fast PCR mix, it was possible to differentiate wild-type strain G37 from the mutated isolates and to distinguish the types of mutations among the mutated isolates. However, when clinical specimens were tested, the Tms were very similar, and no significant difference in the melting profiles and plots was observed between the different genotypes (Fig. 1b).
Finally, we tested the MeltDoctor HRM Master mix (Applied Biosystems), which was previously used by Twin et al. (10). Results comparable to those of the Roche HRM mix were obtained. The profiles of wild-type and mutated specimens were not separated (Fig. 1c; see also Fig. S1 in the supplemental material).
Detection of a point mutation associated with macrolide resistance using the FRET assay.
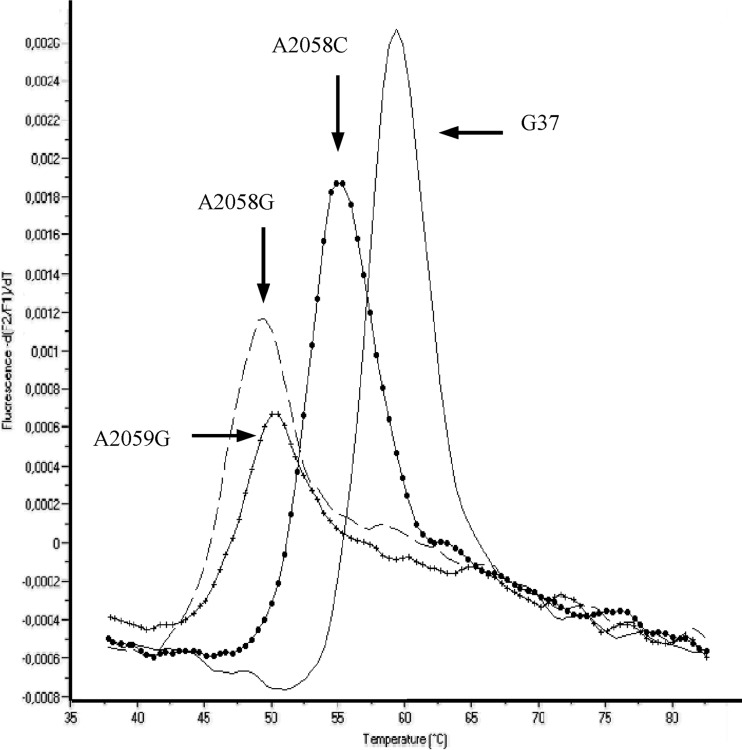
A FRET PCR assay was developed to detect point mutations conferring resistance to macrolides in the M. genitalium 23S rRNA gene. This assay was first evaluated with genetically characterized M. genitalium strains and was able to discriminate wild-type from resistant genotypes (Fig. 2). The Tm for the mutated strains was lower than the Tm for the wild-type strains because of the existence of a nucleotide mismatch between the gene sequence and the hybridization probe. The results were reproducible, and the same profile and Tm were obtained for the wild-type strain when the concentration of DNA varied from 50 to 5 × 109 copies.
FIG 2.
Melting curve analysis using the FRET assay for M. genitalium wild-type strain G37 (continuous line) and mutated isolates harboring the A2058C (filled circles), A2058G (large dashes), and A2059G (crosses) mutations. Wild-type and mutated isolates were run in the same PCR.
Melting curve analysis of DNA from the characterized strains produced four different curves, with mean Tms of 59.5°C ± 0.56°C for the wild-type strain, 55°C ± 0.5°C for the isolate harboring the A2058C mutation, 50.2°C ± 0.45°C for the isolate harboring the A2059G mutation, and 49°C ± 0.46°C for the isolate harboring the A2058G mutation (Fig. 2).
The A2058G and A2059G mutant-related Tms were very similar, and thus, these two resistant genotypes were hardly differentiated from one another. Consequently, the exact position of the mutation had to be determined by sequencing.
Specificity and limit of detection.
No amplification was observed with the 20 specimens that were positive for known pathogens involved in urogenital infections or urogenital commensals or with the DNA extracts from different human Mycoplasma strains (see Materials and Methods). The assay showed a limit of detection of 50 copies/μl.
Clinical specimen testing.
After validation using characterized macrolide-resistant strains, the FRET real-time PCR was optimized to be applied directly to clinical specimens by increasing the amount of template DNA. Five microliters of DNA from characterized clinical specimens was necessary for amplification, instead of the 2 μl that was used for the genomic DNA from isolates. All 10 clinical specimens with characterized genotypes were found to have the expected profiles by using the real-time PCR assay.
A total of 202 M. genitalium PCR-positive clinical specimens from 178 patients were collected in Bordeaux, France, in 2011 and 2012. Among them, 155 samples from 134 patients were amplified with the FRET assay, demonstrating a sensitivity of 76.7% (155/202). The sensitivity of our assay did not vary according to the specimen type (P = 0.22 by the Student t test). A resistant genotype was detected for 23 specimens from 19 patients, and sequencing analysis confirmed the presence of the mutations in all cases (Table 2). Eleven and eight clinical specimens harbored the A2059G and A2058G mutations, respectively. In four cases, a mixed population was observed (Table 2). Two specimens had a mixed population of wild-type and A2059G mutant bacteria; one specimen had wild-type and A2058G mutant bacteria. For these three specimens, the amplification produced 2 melting peaks, one with a Tm corresponding to the wild-type strain and one with a Tm corresponding to the mutation. In specimen Mg-67096, the FRET assay showed a unique melting peak with a Tm at 49.8°C. Analysis of the M. genitalium 23S rRNA sequence from this specimen showed a mixture of A and G nucleotides at positions 2058 and 2059, suggesting the presence of two distinct mutated M. genitalium populations in this clinical specimen.
TABLE 2.
Characteristics of patients with macrolide-resistant M. genitalium infectiona
| Patient | Patient age (yr) | Specimen designation | Date of collection (day/mo/yr) | Source of specimen | Genotype(s) detected by: |
|
|---|---|---|---|---|---|---|
| FRET real-time PCR | Sequence analysis | |||||
| 1 | 20 | Mg-27932 | 03/01/2011 | Vaginal swab | A2058G or A2059G | A2058G |
| Mg-35761 | 06/03/2011 | Endocervical swab | A2058G or A2059G | A2058G | ||
| 2 | 33 | Mg-28521 | 03/07/2011 | First-void urine | Wild type | ND |
| Mg-32157 | 04/18/2011 | First-void urine | A2058G or A2059G | A2058G | ||
| 3 | 35 | Mg-36534 | 06/14/2011 | Urethral swab | A2058G or A2059G | A2058G |
| Mg-36593 | 06/14/2011 | First-void urine | A2058G or A2059G | A2058G | ||
| 4 | 49 | Mg-42600 | 09/02/2011 | First-void urine | A2058G or A2059G | A2059G |
| Mg-47150 | 10/28/2011 | First-void urine | A2058G or A2059G | A2059G | ||
| 5 | 50 | Mg-42613 | 09/02/2011 | Vaginal swab | A2058G or A2059G | A2059G |
| Mg-47142 | 10/28/2011 | Endocervical swab | A2058G or A2059G | A2059G | ||
| 6 | 22 | Mg-44723 | 09/29/2011 | Vaginal swab | A2058G or A2059G | A2058G |
| 7 | 25 | Mg-46414 | 10/19/2011 | Endocervical swab | A2058G or A2059G | A2059G |
| 8 | 29 | Mg-49903 | 12/02/2011 | Urethral swab | A2058G or A2059G | A2059G |
| 9 | NA | Mg-52242 | 12/30/2011 | Urethral swab | A2058G or A2059G | A2058G |
| 10 | 47 | Mg-66652 | 06/08/2012 | First-void urine | A2058G or A2059G | A2059G |
| 11 | 49 | Mg-70716 | 07/23/2011 | First-void urine | Wild type and A2058G or A2059G | A2058A/Gb |
| 12 | 26 | Mg-58433 | 03/05/2012 | Vaginal swab | A2058G or A2059G | A2059G |
| 13 | 24 | Mg-74722 | 09/07/2012 | Vaginal swab | Wild type and A2058G or A2059G | A2059A/Gb |
| 14 | 29 | Mg-84127 | 12/14/2012 | Vaginal swab | A2058G or A2059G | A2059G |
| 15 | 23 | Mg-68123 | 06/22/2012 | Vaginal swab | A2058G or A2059G | A2059G |
| 16 | 28 | Mg-70412 | 07/18/2012 | Vaginal swab | A2058G or A2059G | A2058G |
| 17 | 41 | Mg-81092 | 11/14/2012 | Urethral swab | Wild type | ND |
| Mg-83545 | 12/10/2012 | Urethral swab | A2058G or A2059G | A2059G | ||
| 18 | 19 | Mg-70762 | 07/23/2012 | Endocervical swab | Wild type and A2058G or A2059G | A2059A/Gb |
| 19 | 17 | Mg-67096 | 06/13/2012 | Vaginal swab | A2058G or A2059G | A2058A/G and A2059A/Gb |
NA, not available; ND, not determined.
A2058A/G or A2059A/G indicates a simultaneous finding of both the macrolide-sensitive (2058A or 2059A) and the macrolide-resistant (2058G or 2059G) genotypes.
Overall, a substitution in the 23S rRNA gene was found in 14.2% (19/134) of the M. genitalium-positive patients, with rates of 14.5% in 2011 and 13.8% in 2012. Moreover, no gender difference was found between the 11 women and the 8 men harboring a strain with macrolide resistance (P = 0.08 by Fisher's exact test).
DISCUSSION
Macrolides are generally considered the first-line treatment for M. genitalium infections. However, resistance to macrolides seems to be increasing worldwide (4, 8, 10, 14, 22). Susceptibility testing is rarely achieved because of the difficulty of culturing clinical isolates of M. genitalium (23). For this reason, the development of rapid assays for the detection of macrolide resistance directly in clinical specimens is necessary, especially to guide the clinician in the choice of a second-line treatment in the event of therapeutic failure.
A real-time PCR assay based on HRM analysis was reported previously to detect resistance-associated mutations at positions 2058 and 2059 in region V of the 23S rRNA gene (10). The HRM assay is a rapid molecular method for the detection of single nucleotide changes and is suitable for direct application on clinical specimens (24). We first attempted to use the previously reported HRM assay to evaluate the prevalence of macrolide resistance in M. genitalium in clinical specimens from France in 2011 and 2012. However, wild-type and mutated M. genitalium isolates could not be discriminated despite the use of three different enzyme mixes. The reason for the differences between the mixes is unknown. The difference may be attributed to the chemistries of the different dyes used in the mixes. Because there is no information about the dyes in the kits that were used, this hypothesis cannot be confirmed. Moreover, HRM results varied according to the M. genitalium DNA concentration. Consequently, this method was not suitable for the routine detection of macrolide resistance-associated mutations in clinical specimens in which the DNA concentration is highly variable. This lack of reproducibility due to variations in the initial nucleic acid template concentrations was previously reported for HRM techniques (24). Nevertheless, it remains unclear why the Australian researchers were successful with HRM-PCR and we were not.
Given the limitations of the HRM assay, we designed a real-time PCR assay based on FRET technology coupled with melting curve analysis. Our assay was successfully validated on genetically characterized M. genitalium isolates as well as on M. genitalium-positive clinical specimens and was able to separate wild-type from resistant genotypes.
The FRET assay proved to be specific because no amplification was observed with a range of clinical specimens containing bacteria that are involved in urogenital infection, but the assay was less sensitive than the in-house real-time PCR performed at the Bordeaux University Hospital laboratory for the routine detection of M. genitalium in urogenital specimens (21). Among the 202 specimens with a positive M. genitalium real-time PCR result, 155 (76.7%) were amplified. Our assay appears to have a sensitivity equivalent to that of a FRET assay that was used for the detection of macrolide resistance-associated mutations in M. pneumoniae-positive respiratory tract clinical samples, which had a global sensitivity of 72.6% (18). Finally, our FRET assay should be used only for resistance detection purposes on clinical samples for which M. genitalium in-house real-time PCR would be positive.
The presence of macrolide-resistant M. genitalium was first reported by Bradshaw et al. in 2006 (13). A troubling fact is that macrolide resistance in M. genitalium seems to be spreading throughout the world. Studies reported the detection of macrolide resistance in Japan (16), Australia (9, 10, 13, 22), Northern Europe (9), France (8), and, recently, Greenland (14). In our study, 19/134 patients (14.2%) had a substitution in the 23S rRNA gene, with prevalence rates of 14.5% in 2011 and 13.8% in 2012. Macrolide resistance-associated mutations have been detected in France since 2006 at a rate of 13.8%, ranging from 10% to 15.4% of patients per year between 2006 and 2010 (8). Thus, in France, the prevalence of macrolide resistance-associated mutations in M. genitalium seems to be stable.
High-level resistance to macrolides is associated with point mutations in domain V of the 23S rRNA gene at positions 2058 and 2059 in several bacterial species (25). In our study, a resistant genotype was detected in 19 patients, 9 with an A-to-G substitution at position 2059 and 6 with an A-to-G substitution at position 2058. A mixed population was observed for the remaining four patients. In a previous study covering the period from 2006 to 2010 in France, we also reported a higher number of substitutions at position 2059 than at position 2058 (9 at position 2059 versus 2 at position 2058) (8). This predominance of the mutation at position 2059 was also reported in Australia (10) and in New Zealand (12). Jensen et al. reported an identical number of mutations at positions A2058 and A2059 in Denmark (9), while in Greenland, mutations at position 2058 were predominant. In M. pneumoniae, a close relative of M. genitalium, most resistant strains harbor the A2058 mutation (26).
Macrolide resistance in M. genitalium is associated with therapeutic failure of 1 g azithromycin in about 85% of patients infected by a mutant strain (4), and selection of macrolide-resistant mutants during azithromycin treatment has been reported by several studies (9, 12, 15, 27). In our study, we also observed the selection of macrolide resistance for 2 patients, patient 2 in 2011 and patient 17 in 2012. In both cases, there was no macrolide resistance-associated mutation before azithromycin treatment, but a substitution in the 23S rRNA gene (A2058G mutation for patient 2 and A2059G mutation for patient 17) was observed after treatment (Table 2). Patient 2 received an extended 5-day course of azithromycin as a first-line treatment. Because symptoms persisted, a new urogenital specimen was collected, and another 5-day regimen of azithromycin was administered. Patient 2 was M. genitalium negative after the second therapy, although he was infected by an isolate harboring the A2058G mutation. This could be due to a low organism load in the follow-up specimen, or this is more likely due to spontaneous clearance. For patient 7, a single dose of 1 g azithromycin was given as primary treatment, followed by an extended 5-day course of azithromycin after the persistence of symptoms. A posttreatment sample harbored an isolate that carried an A2059G mutation. Patient 17 was cured by treatment with 400 mg moxifloxacin daily for 10 days. Moxifloxacin is known to cure patients who experience azithromycin failure (1, 9, 28). At present, the treatment of M. genitalium infections is not standardized (4, 29, 30). The common first-line treatment is azithromycin. In the case of azithromycin failure, moxifloxacin is the most commonly used second-line antibiotic. However, the side effects and the risk of selection of resistant strains are limiting factors for the frequent use of moxifloxacin (29, 31). Given the high prevalence of macrolide resistance, exceeding 40%, described in recent works from Australia and the United Kingdom (22, 32), it may be reasonable to screen all M. genitalium-positive specimens for macrolide resistance. This strategy was adopted at the Bordeaux University Hospital (Bordeaux, France) using our FRET assay. Such an approach might not be cost-effective; however, the early detection of macrolide resistance will guide clinicians in the use of moxifloxacin only in the case of macrolide resistance and thus eventually decrease the global therapeutic cost of M. genitalium infections.
In conclusion, we have developed a FRET real-time assay that is able to discriminate wild-type from macrolide-resistant M. genitalium isolates directly from urogenital specimens that are M. genitalium positive by PCR. According to the results of this assay with French clinical specimens, we observed that macrolide resistance in M. genitalium has been stable in France since its detection in 2006, with a rate of 14%. Our method allows the rapid detection of macrolide resistance-associated mutations and will allow clinicians to shorten the time to the initiation of effective treatment.
Supplementary Material
ACKNOWLEDGMENTS
We have no conflicts of interest to declare.
This work was supported by internal funding. A.T. benefited from a postdoctoral fellowship from the AXA Research Fund.
Footnotes
Published ahead of print 26 February 2014
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JCM.03318-13.
REFERENCES
- 1.Taylor-Robinson D, Jensen JS. 2011. Mycoplasma genitalium: from chrysalis to multicolored butterfly. Clin. Microbiol. Rev. 24:498–514. 10.1128/CMR.00006-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Haggerty CL. 2008. Evidence for a role of Mycoplasma genitalium in pelvic inflammatory disease. Curr. Opin. Infect. Dis. 21:65–69. 10.1097/QCO.0b013e3282f3d9ac [DOI] [PubMed] [Google Scholar]
- 3.Jensen JS. 2004. Mycoplasma genitalium: the aetiological agent of urethritis and other sexually transmitted diseases. J. Eur. Acad. Dermatol. Venereol. 18:1–11. 10.1111/j.1468-3083.2004.00923.x [DOI] [PubMed] [Google Scholar]
- 4.Cazanave C, Manhart LE, Bébéar C. 2012. Mycoplasma genitalium, an emerging sexually transmitted pathogen. Med. Mal. Infect. 42:381–392. 10.1016/j.medmal.2012.05.006 [DOI] [PubMed] [Google Scholar]
- 5.Le Roy C, Le Hen I, Clerc M, Arfel V, Normandin F, Bébéar C, de Barbeyrac B. 2012. The first performance report for the Bio-Rad Dx CT/NG/MG assay for simultaneous detection of Chlamydia trachomatis, Neisseria gonorrhoeae and Mycoplasma genitalium in urogenital samples. J. Microbiol. Methods 89:193–197. 10.1016/j.mimet.2012.03.009 [DOI] [PubMed] [Google Scholar]
- 6.Wroblewski JK, Manhart LE, Dickey KA, Hudspeth MK, Totten PA. 2006. Comparison of transcription-mediated amplification and PCR assay results for various genital specimen types for detection of Mycoplasma genitalium. J. Clin. Microbiol. 44:3306–3312. 10.1128/JCM.00553-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mena LA, Mroczkowski TF, Nsuami M, Martin DH. 2009. A randomized comparison of azithromycin and doxycycline for the treatment of Mycoplasma genitalium-positive urethritis in men. Clin. Infect. Dis. 48:1649–1654. 10.1086/599033 [DOI] [PubMed] [Google Scholar]
- 8.Chrisment D, Charron A, Cazanave C, Pereyre S, Bébéar C. 2012. Detection of macrolide resistance in Mycoplasma genitalium in France. J. Antimicrob. Chemother. 67:2598–2601. 10.1093/jac/dks263 [DOI] [PubMed] [Google Scholar]
- 9.Jensen JS, Bradshaw CS, Tabrizi SN, Fairley CK, Hamasuna R. 2008. Azithromycin treatment failure in Mycoplasma genitalium-positive patients with nongonococcal urethritis is associated with induced macrolide resistance. Clin. Infect. Dis. 47:1546–1553. 10.1086/593188 [DOI] [PubMed] [Google Scholar]
- 10.Twin J, Jensen JS, Bradshaw CS, Garland SM, Fairley CK, Min LY, Tabrizi SN. 2012. Transmission and selection of macrolide resistant Mycoplasma genitalium infections detected by rapid high resolution melt analysis. PLoS One 7:e35593. 10.1371/journal.pone.0035593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jensen JS. 2012. Protocol for the detection of Mycoplasma genitalium by PCR from clinical specimens and subsequent detection of macrolide resistance-mediating mutations in region V of the 23S rRNA gene. Methods Mol. Biol. 903:129–139. 10.1007/978-1-61779-937-2_8 [DOI] [PubMed] [Google Scholar]
- 12.Yew HS, Anderson T, Coughlan E, Werno A. 2011. Induced macrolide resistance in Mycoplasma genitalium isolates from patients with recurrent nongonococcal urethritis. J. Clin. Microbiol. 49:1695–1696. 10.1128/JCM.02475-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bradshaw CS, Jensen JS, Tabrizi SN, Read TR, Garland SM, Hopkins CA, Moss LM, Fairley CK. 2006. Azithromycin failure in Mycoplasma genitalium urethritis. Emerg. Infect. Dis. 12:1149–1152. 10.3201/eid1207.051558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gesink DC, Mulvad G, Montgomery-Andersen R, Poppel U, Montgomery-Andersen S, Binzer A, Vernich L, Frosst G, Stenz F, Rink E, Olsen OR, Koch A, Jensen JS. 2012. Mycoplasma genitalium presence, resistance and epidemiology in Greenland. Int. J. Circumpolar Health 71:1–8. 10.3402/ijch.v71i0.18203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ito S, Shimada Y, Yamaguchi Y, Yasuda M, Yokoi S, Nakano M, Ishiko H, Deguchi T. 2011. Selection of Mycoplasma genitalium strains harbouring macrolide resistance-associated 23S rRNA mutations by treatment with a single 1 g dose of azithromycin. Sex. Transm. Infect. 87:412–414. 10.1136/sextrans-2011-050035 [DOI] [PubMed] [Google Scholar]
- 16.Shimada Y, Deguchi T, Nakane K, Yasuda M, Yokoi S, Ito S, Nakano M, Ishiko H. 2011. Macrolide resistance-associated 23S rRNA mutation in Mycoplasma genitalium, Japan. Emerg. Infect. Dis. 17:1148–1150. 10.3201/eid1706.101055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wolff BJ, Thacker WL, Schwartz SB, Winchell JM. 2008. Detection of macrolide resistance in Mycoplasma pneumoniae by real-time PCR and high-resolution melt analysis. Antimicrob. Agents Chemother. 52:3542–3549. 10.1128/AAC.00582-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Peuchant O, Menard A, Renaudin H, Morozumi M, Ubukata K, Bébéar CM, Pereyre S. 2009. Increased macrolide resistance of Mycoplasma pneumoniae in France directly detected in clinical specimens by real-time PCR and melting curve analysis. J. Antimicrob. Chemother. 64:52–58. 10.1093/jac/dkp160 [DOI] [PubMed] [Google Scholar]
- 19.Spuesens EB, Meijer A, Bierschenk D, Hoogenboezem T, Donker GA, Hartwig NG, Koopmans MP, Vink C, van Rossum AM. 2012. Macrolide resistance determination and molecular typing of Mycoplasma pneumoniae in respiratory specimens collected between 1997 and 2008 in The Netherlands. J. Clin. Microbiol. 50:1999–2004. 10.1128/JCM.00400-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hjorth SV, Bjornelius E, Lidbrink P, Falk L, Dohn B, Berthelsen L, Ma L, Martin DH, Jensen JS. 2006. Sequence-based typing of Mycoplasma genitalium reveals sexual transmission. J. Clin. Microbiol. 44:2078–2083. 10.1128/JCM.00003-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jensen JS, Bjornelius E, Dohn B, Lidbrink P. 2004. Use of TaqMan 5′ nuclease real-time PCR for quantitative detection of Mycoplasma genitalium DNA in males with and without urethritis who were attendees at a sexually transmitted disease clinic. J. Clin. Microbiol. 42:683–692. 10.1128/JCM.42.2.683-692.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tagg KA, Jeoffreys NJ, Couldwell DL, Donald JA, Gilbert GL. 2013. Fluoroquinolone and macrolide resistance-associated mutations in Mycoplasma genitalium. J. Clin. Microbiol. 51:2245–2249. 10.1128/JCM.00495-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hamasuna R. 2013. Identification of treatment strategies for Mycoplasma genitalium-related urethritis in male patients by culturing and antimicrobial susceptibility testing. J. Infect. Chemother. 19:1–11. 10.1007/s10156-012-0487-3 [DOI] [PubMed] [Google Scholar]
- 24.Tong SY, Giffard PM. 2012. Microbiological applications of high-resolution melting analysis. J. Clin. Microbiol. 50:3418–3421. 10.1128/JCM.01709-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kannan K, Mankin AS. 2011. Macrolide antibiotics in the ribosome exit tunnel: species-specific binding and action. Ann. N. Y. Acad. Sci. 1241:33–47. 10.1111/j.1749-6632.2011.06315.x [DOI] [PubMed] [Google Scholar]
- 26.Bébéar C, Pereyre S, Peuchant O. 2011. Mycoplasma pneumoniae: susceptibility and resistance to antibiotics. Future Microbiol. 6:423–431. 10.2217/fmb.11.18 [DOI] [PubMed] [Google Scholar]
- 27.Walker J, Fairley CK, Bradshaw CS, Tabrizi SN, Twin J, Chen MY, Taylor N, Donovan B, Kaldor JM, McNamee K, Urban E, Walker S, Currie M, Birden H, Bowden FJ, Gunn J, Pirotta M, Gurrin L, Harindra V, Garland SM, Hocking JS. 2013. Mycoplasma genitalium incidence, organism load, and treatment failure in a cohort of young Australian women. Clin. Infect. Dis. 56:1094–1100. 10.1093/cid/cis1210 [DOI] [PubMed] [Google Scholar]
- 28.Jernberg E, Moghaddam A, Moi H. 2008. Azithromycin and moxifloxacin for microbiological cure of Mycoplasma genitalium infection: an open study. Int. J. STD AIDS 19:676–679. 10.1258/ijsa.2008.008038 [DOI] [PubMed] [Google Scholar]
- 29.Manhart LE, Broad JM, Golden MR. 2011. Mycoplasma genitalium: should we treat and how? Clin. Infect. Dis. 53(Suppl 3):S129–S142. 10.1093/cid/cir702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Weinstein SA, Stiles BG. 2011. A review of the epidemiology, diagnosis and evidence-based management of Mycoplasma genitalium. Sex. Health 8:143–158. 10.1071/SH10065 [DOI] [PubMed] [Google Scholar]
- 31.Anagrius C, Lore B, Jensen JS. 2013. Treatment of Mycoplasma genitalium. Observations from a Swedish STD clinic. PLoS One 8:e61481. 10.1371/journal.pone.0061481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pond MJ, Nori AV, Witney AA, Lopeman RC, Butcher PD, Sadiq ST. 2014. High prevalence of antibiotic-resistant Mycoplasma genitalium in nongonococcal urethritis: the need for routine testing and the inadequacy of current treatment options. Clin. Infect. Dis. 58:631–637. 10.1093/cid/cit752 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.