Abstract
Fast and accurate identification and typing of pathogens are essential for effective surveillance and outbreak detection. The current routine procedure is based on a variety of techniques, making the procedure laborious, time-consuming, and expensive. With whole-genome sequencing (WGS) becoming cheaper, it has huge potential in both diagnostics and routine surveillance. The aim of this study was to perform a real-time evaluation of WGS for routine typing and surveillance of verocytotoxin-producing Escherichia coli (VTEC). In Denmark, the Statens Serum Institut (SSI) routinely receives all suspected VTEC isolates. During a 7-week period in the fall of 2012, all incoming isolates were concurrently subjected to WGS using IonTorrent PGM. Real-time bioinformatics analysis was performed using web-tools (www.genomicepidemiology.org) for species determination, multilocus sequence type (MLST) typing, and determination of phylogenetic relationship, and a specific VirulenceFinder for detection of E. coli virulence genes was developed as part of this study. In total, 46 suspected VTEC isolates were characterized in parallel during the study. VirulenceFinder proved successful in detecting virulence genes included in routine typing, explicitly verocytotoxin 1 (vtx1), verocytotoxin 2 (vtx2), and intimin (eae), and also detected additional virulence genes. VirulenceFinder is also a robust method for assigning verocytotoxin (vtx) subtypes. A real-time clustering of isolates in agreement with the epidemiology was established from WGS, enabling discrimination between sporadic and outbreak isolates. Overall, WGS typing produced results faster and at a lower cost than the current routine. Therefore, WGS typing is a superior alternative to conventional typing strategies. This approach may also be applied to typing and surveillance of other pathogens.
INTRODUCTION
Bacterial pathogens still pose a major threat to public health, and in order to limit their spread and prevent infectious disease outbreaks, accurate and rapid diagnostics and classification of isolates are of great importance. In current routine practice, isolation and identification are mostly performed at clinical microbiological laboratories, and verification and further characterization are performed for a few selected pathogens at national, or regional, reference laboratories, using a variety of species-specific methods. Typing and surveillance of bacterial pathogens rely mainly on well-established, standardized phenotypic and molecular typing methods, such as serotyping and pulsed-field gel electrophoresis (PFGE) (1, 2). However, to obtain sufficient discrimination between isolates, it is typically necessary to combine typing results from several different typing techniques, both phenotypic and genotypic. As a result, it is laborious, time-consuming, and expensive to perform proper typing for surveillance and outbreak detection.
As the cost of whole-genome sequencing (WGS) has decreased and benchtop sequencing machines enable fast turnaround, it has become increasingly attractive for use in routine diagnostics and typing, and the approach has already been found useful in retrospective outbreak investigations (3, 4).
Although WGS provides detailed information that will, in theory, enable diagnostics and typing solely on the basis of the features in the bacterial genome, it is a challenge to define and extract the appropriate information from the large amount of sequence data that is generated. Thus, to facilitate the use of WGS data for routine diagnostics, typing, and surveillance, it is important that the sequence data can be automatically and quickly converted to clinically relevant information that can be easily interpreted by physicians and public health professionals with limited bioinformatics skills. To achieve this, the Center for Genomic Epidemiology (CGE) provides public, user-friendly web-tools for rapid handling of WGS data and extraction of relevant information, useful for diagnostics, surveillance, and outbreak investigations for the global medical society (www.genomicepidemiology.org).
In this study, as a proof of concept, we demonstrate the usefulness of WGS for routine typing, surveillance, and outbreak detection of verocytotoxin-producing Escherichia coli (VTEC) infections in Denmark. VTEC, also known as Shiga toxin-producing E. coli (STEC), is a gastrointestinal pathogen, which is typically spread by ingestion of contaminated food or water or person-to-person contact. Rapid and reliable diagnostics and detection of outbreak clusters are of utmost importance for control. VTEC infections cause bloody diarrhea and in some cases hemolytic-uremic syndrome (HUS), which is characterized by kidney failure, thrombocytopenia, and microangiopathic hemolytic anemia, and can be fatal to young children. VTEC pathogenicity is facilitated by the Shiga toxin (Stx) and a number of other virulence factors (5, 6).
We performed real-time benchtop sequencing of VTEC isolates from patients in Denmark during a 7-week period in parallel with the current routine procedure. During the period of the study, Denmark experienced a small E. coli O157:H7 outbreak with a total of 13 VTEC isolates. The VTEC outbreak strain had an infrequent toxin subtype profile (eae, vtx1a, and vtx2a) and a high proportion of HUS cases (62%), and the toxin subtype profiling proved important in the outbreak investigation and risk assessment of the VTEC strain (7).
Here, we demonstrate that this WGS-based typing approach is a superior alternative to the current routine typing of VTEC infections, rapidly producing typing results comparable to those of current routine typing and thus of great value in surveillance and outbreak detection. This approach may also be applicable to other pathogens. In addition, we here present the VirulenceFinder, a new CGE web-tool made for automatic detection and extraction of E. coli virulence genes from WGS data.
MATERIALS AND METHODS
Study design and isolates.
The use of whole-genome sequencing (WGS) for routine typing purposes of verocytotoxin-producing Escherichia coli (VTEC) isolates was examined by conducting a WGS study parallel with the routine typing performed at the Statens Serum Institut (SSI) to evaluate WGS for typing purposes with regard to typing results, time to final result, labor time, and total cost. Suspected VTEC isolates from infected patients were, as part of the routine surveillance in Denmark, sent from hospitals to the SSI for confirmation and for further phenotypic and molecular characterization. Simultaneously, the isolates were typed by the WGS approach at DTU Food (National Food Institute, Technical University of Denmark), using the IonTorrent PGM (Life Technologies, Carlsbad, CA) benchtop sequencer. All isolates included in the study were obtained from human fecal samples from patients with regular diarrhea, bloody diarrhea, or hemolytic-uremic syndrome (HUS), and epidemiological information was collected by the SSI as standard procedure. We defined the test period in the fall of 2012 to last until a total of at least 40 isolates were collected, but with a maximum duration of 12 weeks, in case fewer isolates were received than expected.
Routine typing procedures.
As part of the routine typing and surveillance at the SSI, suspected VTEC isolates were subjected to several phenotypic and molecular typing methods according to the standard procedures at the SSI. In instances where the isolates received at the SSI exhibited mixed-colony morphology, both types of isolates were subjected to typing and included in the study.
All suspected VTEC isolates were serotyped by O typing and H typing, identifying the specific cell wall antigen and flagellar antigen, respectively (8, 9). In addition, the isolates were K typed detecting capsule antigens by K1/K5 bacteriophage susceptibility (8). All isolates were additionally tested for hemolysin production (10) and β-glucuronidase activity (11) and for the production of verocytotoxin by the Vero cell assay (12).
All the suspected VTEC isolates were subjected to DNA hybridization with specific DNA probes for the E. coli attaching and effacing gene (eae) (13), bundle-forming pilus gene (bfpA) (14), enteropathogenic E. coli (EPEC) adherence factor (EAF) (15), plasmid-encoded O157 enterohemolysin gene (ehxA) (16), verotoxin 1 gene (vtx1) (17), verotoxin 2 gene (vtx2) (18), the verotoxin 2f variant gene (vtx2f) (19), and Shiga toxin-producing E. coli (STEC) autoagglutinating adhesion gene (saa) (20). In addition, subtyping of vtx1 and vtx2 was carried out for all isolates by PCR with subtype-specific primers detecting vtx1a, vtx1c, vtx1d, vtx2a, vtx2b, vtx2c, vtx2d, vtx2e, vxt2f, and vtx2g subtypes (21). Only isolates considered to be potential outbreak isolates based on the typing methods described above were additionally typed by PFGE (22).
Whole-genome sequencing.
Genomic DNA (gDNA) was purified from the isolates using the Easy-DNA extraction kit (Invitrogen, Carlsbad, CA), and DNA concentrations were determined using the Qubit dsDNA (double-stranded DNA) BR assay kit (Invitrogen). Subsequently, gDNA was fragmented by sonication on the Covaris S2 system. Specifically, 130 μl (200 ng) gDNA diluted in low-TE (Tris-EDTA) buffer was used as input, and conditions were set for generation of 200- to 300-bp fragments, using 3 cycles each of 1 min, 5°C bath temperature, frequency sweeping, 10% duty cycling, intensity of 5, and cycles/burst of 100. A total of 100 ng gDNA was introduced to the IonTorrent PGM 200-bp work flow. gDNA libraries were prepared according to the IonXpress Plus gDNA Fragment Library Preparation (Life Technologies) protocol, consisting of end repair, nick repair, ligation, and size selecting with the E-Gel SizeSelect agarose gel system and subsequent library amplification. Library concentration was determined employing a 2100 BioAnalyzer (Agilent Technologies, Santa Clara, CA) and the Agilent high-sensitivity DNA kit. Template preparation was done according to the Ion OneTouch 200 template kit (Life Technologies) protocol for 200-base-read libraries on the Ion OneTouch system (Life Technologies) and subsequent quality control of template Ion Spheres using the Qubit 2.0 fluorometer (Life Technologies). Sequencing was done on the IonTorrent PGM sequencer following the Ion PGM 200 sequencing kit (Life Technologies) protocol for either Ion 316 chips or Ion 318.
The E. coli virulence gene database.
For automatic detection of virulence genes in the suspected VTEC isolates, an E. coli FASTA database was constructed, as part of VirulenceFinder, which is a component of the publicly available web-based tools for WGS analysis hosted by the Center for Genomic Epidemiology (CGE) (http://www.genomicepidemiology.org/). The content of the E. coli virulence database was constructed on the basis of the Identibac scheme (Alere Technologies GmbH, Jena, Germany) for genotypic detection of E. coli virulence genes. All genes, and gene variants, represented in the Identibac scheme by GenBank accession numbers or identifiers (IDs) were BLASTed against the NCBI nucleotide database (https://www.ncbi.nlm.nih.gov/nuccore/), and gene variants that matched 90% on identity and size were collected. All partial genes were excluded from the database, and also gene variants belonging to genera other than Escherichia, Klebsiella, Citrobacter, Enterobacter, or Shigella were excluded. Subtypes of the vtx genes were assigned according to the previously described, sequence-based nomenclature for verocytotoxins (21). The E. coli virulence gene database contains 76 genes, and the gene content and number of gene variants can be seen in Table 1.
TABLE 1.
Gene content of the E. coli virulence database
| Gene | Descriptiona | No. of variants in the database |
|---|---|---|
| astA | Heat-stable enterotoxin 1 | 11 |
| bfpA | Major subunit of bundle-forming pili | 5 |
| cba | Colicin B | 15 |
| ccI | Cloacin | 4 |
| cdtB | Cytolethal distending toxin B | 14 |
| celb | Endonuclease colicin E2 | 10 |
| cfa_c | Colonization factor antigen I | 4 |
| cif | Type III secreted effector | 4 |
| cma | Colicin M | 19 |
| cnf1 | Cytotoxic necrotizing factor | 7 |
| cofA | Longus type IV pilus subunit | 1 |
| eae | Intimin | 45 |
| eatA | Serine protease autotransporters of Enterobacteriaceae (SPATE) | 3 |
| efa1 | EHEC factor for adherence | 11 |
| ehxA | Enterohemolysin | 12 |
| epeA | Serine protease autotransporters of Enterobacteriaceae | 1 |
| espA | Type III secretion system | 23 |
| espB | Secreted protein B | 14 |
| espC | Serine protease autotransporters of Enterobacteriaceae | 3 |
| espF | Type III secretion system | 13 |
| espI | Serine protease autotransporters of Enterobacteriaceae | 2 |
| espJ | Prophage-encoded type III secretion system effector | 2 |
| espP | Putative exoprotein precursor | 4 |
| etpD | Type II secretion protein | 3 |
| f17A | Subunit A of F17 fimbrial protein | 7 |
| f17G | Adhesin subunit of F17 fimbriae | 9 |
| fanA | Involved in biogenesis of K99/F5 fimbriae | 1 |
| fasA | Fimbrial 987P/F6 subunit | 1 |
| fedA | Fimbrial protein F107 subunit A | 3 |
| fedF | Fimbrial adhesin AC precursor | 6 |
| fim41a | Mature Fim41a/F41 protein | 2 |
| gad | Glutamate decarboxylase | 70 |
| hlyE | Avian E. coli hemolysin | 1 |
| iha | Adherence protein | 19 |
| ipaD | Invasion protein Shigella flexneri | 9 |
| ipaH9.8 | Invasion plasmid antigen | 8 |
| ireA | Siderophore receptor | 4 |
| iroN | Enterobactin siderophore receptor protein | 13 |
| iss | Increased serum survival | 14 |
| K88ab | K88/F4 protein subunit | 10 |
| katP | Plasmid-encoded catalase peroxidase | 1 |
| lngA | Longus type IV pilus | 2 |
| lpfA | Long polar fimbriae | 11 |
| ltcA | Heat-labile enterotoxin A subunit | 17 |
| mchB | Microcin H47 part of colicin H | 2 |
| mchC | MchC protein | 6 |
| mchF | ABC transporter protein MchF | 15 |
| mcmA | Microcin M part of colicin H | 4 |
| nfaE | Diffuse adherence fibrillar adhesin gene | 5 |
| nleA | Non-LEE-encoded effector A | 18 |
| nleB | Non-LEE-encoded effector B | 14 |
| nleC | Non-LEE-encoded effector C | 6 |
| perA | EPEC adherence factor | 19 |
| pet | Autotransporter enterotoxin | 1 |
| pic | Serine protease autotransporters of Enterobacteriaceae | 6 |
| prfB | P-related fimbrial regulatory gene | 22 |
| rpeA | Serine protease autotransporters of Enterobacteriaceae | 1 |
| sat | Serine protease autotransporters of Enterobacteriaceae | 6 |
| senB | Plasmid-encoded enterotoxin | 3 |
| sepA | Serine protease autotransporters of Enterobacteriaceae | 7 |
| sfaS | S-fimbrial minor subunit | 1 |
| sigA | Serine protease autotransporters of Enterobacteriaceae | 2 |
| sta1 | Heat-stabile enterotoxin ST-Ia | 2 |
| stb | Heat-stabile enterotoxin II | 3 |
| stx1A | Shiga-like toxin 1 A-subunit | 18 |
| stx1B | Shiga-like toxin 1 B-subunit | 14 |
| stx2A | Shiga toxin 2 subunit A | 114 |
| stx2B | Shiga toxin 2 subunit B | 43 |
| subA | Subtilase toxin subunit | 5 |
| saa | STEC autoagglutinating adhesin | 1 |
| tccP | Tir cytoskeleton coupling protein | 34 |
| tir | Translocated intimin receptor protein | 36 |
| toxB | Toxin B | 4 |
| tsh | Serine protease autotransporters of Enterobacteriaceae | 3 |
| vat | Serine protease autotransporters of Enterobacteriaceae | 7 |
| virF | VirF transcriptional activator | 3 |
LEE, locus of enterocyte effacement.
Using VirulenceFinder.
VirulenceFinder was constructed to enable detection of virulence genes related to E. coli in WGS data while being simple and user friendly. Sequence data can be submitted either as assembled genomes or raw reads from various sequencing technologies, since assembly of raw sequence reads is incorporated into the tool, as described previously for other CGE tools (23, 24). It is possible to select configurations for the organism of interest, and in addition, it is possible to select percent identity (%ID) threshold between the input and the best matching database gene.
The output consists of best-matching genes from BLAST analysis of the selected database, against the submitted genome, with genes set to cover a minimum of three-fifths of the length of the database genes (24). The output contains information on the virulence gene, the %ID, the length of query and database gene, the position of the hit in the contig, and the accession number of the hit. In addition, the vtx subtypes are outputted for typing purposes.
Analysis of sequence data.
Sequence data were analyzed, without further processing, using the CGE web-tools for species detection using the KmerFinder, for determination of multilocus sequence types (MLSTs) (23) and by employing VirulenceFinder for detection of E. coli virulence genes and vtx1 and vtx2 subtypes. For VirulenceFinder, the configuration was set for the E. coli database with an 85.00%ID threshold. For the MLST tool, the configuration was set to E. coli scheme 1 (23, 25).
KmerFinder is a novel program/database for rapid species identification using WGS data (36). Briefly, 1,647 (in a later update 5,029) complete bacterial genomes were downloaded from NCBI, and each k-mer (k = 16) with the prefix ATGAC was saved in a database using an in-house script. ATG was chosen to focus on coding segments, and the extending nucleotides were chosen alphabetically. Each k-mer was a key in the database, and the value was set to a list of all GenBank entries containing that k-mer. Another in-house script was used to search the database. The script finds the unique k-mers in the input file and outputs the number of times each of the GenBank entries in the database is associated with one of these k-mers.
Phylogenetic relationships were established by employing the NDtree (named NDtree for nucleotide difference tree), a newly developed method for variant calling (37), and were, in addition, examined using the SNPtree (26) CGE tool for comparison, using a minimum coverage of 10 and minimum distance between single nucleotide polymorphisms (SNPs) (prune) of 10. For SNPtree, reads were mapped using the Burrows-Wheeler alignment tool (BWA) (27), SNPs were identified and filtered using SAMtools (28), and the tree was constructed by employing Fastree (29).
For both phylogenetic approaches, sequence data were initially quality trimmed using the program AdapterRemoval (30), keeping only reads of a minimum length of 20 nucleotides (nt) and a quality score of at least 30 and without N′s. The E. coli O157:H7 strain Sakai (GenBank accession no. BA000007.2) was used as the reference for both phylogenetic methods.
For the NDtree method, the reference genome was split into 17-mers and so were all reads of at least 50 nucleotides in length. For the reads, a sliding window of size 17 was used with a step size of 1 to make all possible 17-mers, i.e., with the last 16 nucleotides of one 17-mer overlapping with the first 16 nucleotides of the next 17-mer. The 17-mers from the reads, and their reverse complement, were mapped to the reference for an ungapped alignment with a score of at least 50, using a match score of 1 and a mismatch score of −3. The significance of each base call was assessed by evaluating the number of reads with the most common nucleotide at the specific position, X, in relation to the number of reads with other nucleotides at the position, Y. A Z-score was calculated as , and Z = 3.29 was used as the threshold, and additionally, nucleotide differences were considered only when the most common nucleotide was at least 10 times more abundant than other nucleotides at the position. Positions with nonsignificant variations were assigned N, and the same for nonmapped positions of the reference genome.
Sequences were compared in pairs, nucleotide differences in positions were counted, and the tree was constructed by the unweighted-pair group method using average linkages (UPGMA) algorithm from the neighbor program in the phylip package (http://evolution.genetics.washington.edu/phylip.html). The phylogenetic trees were rooted by midpoints.
Evaluation of WGS for typing and surveillance of VTEC.
The WGS-derived typing data were, for each suspected VTEC isolate, compared to the typing data received from the routine procedure at the SSI. The phylogenies obtained from the two different methods were compared and related to the epidemiological information received from the SSI to evaluate the ability to discriminate correctly between isolates. In addition, the application of WGS for typing and surveillance of VTEC infections was assessed by comparing the WGS-based approach and the routine typing with respect to hands-on time, time for obtaining typing results, and estimated cost per isolate.
RESULTS
The study was initiated in late September 2012 and was conducted for 7 weeks before more than 40 isolates were collected. The study included all suspected VTEC isolates received at the SSI during this time period. An increased number of isolates were received during the period of the study due to the occurrence of a small O157:H7 outbreak (7), and thus, both sporadic isolates and outbreak isolates (C812-12, C818-12, C819-12, C849-12, C852-12, and C863-12) were available for evaluation of the WGS-based typing method.
During the 7 weeks, a total of 42 isolates were received at the SSI for further characterization and subtyping, and since 4 of these isolates exhibited mixed-colony morphology upon visual inspection of primary plates, a total of 46 different suspected VTEC isolates were included in the study. In addition, two of the isolates, i.e., C770-12 and C679-12A, were subjected to WGS twice for verification of the WGS method.
As part of the study, the E. coli database for VirulenceFinder was constructed for detection of important E. coli virulence genes and vtx subtypes from WGS data to enable comparison to the current routine typing results. Additionally, a newly developed method, NDtree, for determining nucleotide differences among related isolates was employed for establishing phylogenetic relationships.
WGS-based typing of the suspected VTEC isolates.
All 48 whole-genome sequences, representing the 46 isolates and 2 replicate sequences, were subjected to bioinformatics analysis using the above-mentioned web-tools. The initial WGS-based species identification led to the discovery that one isolate, C848-12, was in fact not E. coli, but Morganella morganii, and this isolate was thus not included in further analysis. The remaining isolates were all confirmed as E. coli. In addition, one isolate, C767-12, was, based on comparison of WGS typing results and routine typing results, a clear mix-up, and was thus excluded from the comparison of typing results. MLST types were successfully assigned for the remaining isolates, apart from two isolates, C887-12 and C893-12, for which the ST types could not be assigned due to problems with the de novo assembly. Table 2 shows a comparison between the most important typing results for each isolate in the study by routine typing and WGS.
TABLE 2.
Comparison of the most important typing results for each isolate in the study by routine typing and WGS typing
| Serotype | Routine typing result(s) | VirulenceFinder result(s) | MLST | Isolate |
|---|---|---|---|---|
| O5:H− | vtx1c | vtx1c | ST-447 | C750-12 |
| O26:H11 | eae, vtx1a | eae, vtx1a | ST-21 | C696-12 |
| O27:H30 | vtx2b | vtx2b | ST-753 | C770-12 (replicate) |
| O36:H− | C887-12 | |||
| O55:H12 | vtx1a | vtx1a | ST-101 | C749-12 |
| O76:H− | vtx1c | vtx1c | ST-675 | C904-12A |
| O103:H2 | eae, vtx1a | eae, vtx1a | ST-17 | C757-12 |
| O103:H− | eae, vtx1a | eae, vtx1a | ST-1967 | C813-12 |
| O rough:H2 | eae, vtx1a | eae, vtx1a | ST-17 | C850-12 |
| O115:H− | eae, vtx2a | eae, vtx2a | ST-333 | C885-12B |
| eae, vtx2a | eae, vtx2a | ST-333 | C896-12A | |
| O117:K1:H7 | vtx1a | vtx1a | ST-504 | C751-12 |
| vtx1a | vtx1a | ST-504 | C659-12 | |
| vtx1a | vtx1a | ST-504 | C760-12 | |
| O121:H19 | eae, vtx2a | eae, vtx2a | ST-655 | C821-12 |
| O128ab:H2 | vtx2b | vtx2b | ST-25 | C748-12 |
| O128abc:H− | vtx1c, vtx2b | vtx1c, vtx2b | ST-811 | C864-12 |
| eae, vtx2f | eae, vtx2f | ST-20 | C820-12 | |
| eae, vtx2f | eae, vtx2f | ST-20 | C862-12 | |
| O145:H− | eae, vtx2a | eae, vtx2a | ST-32 | C816-12 |
| eae, vtx2a | eae, vtx2a | ST-32 | C857-12 | |
| eae, vtx2a | eae, vtx2a | ST-32 | C884-12 | |
| eae, vtx2a | eae, vtx2a | ST-32 | C886-12 | |
| O146:H21 | vtx1c, vtx2b | vtx1c, vtx2b | ST-442 | C874-12 |
| eae | eae | ST-442 | C896-12B | |
| eae | eae | ST-442 | C885-12A | |
| O157:H7 (O157:H−) | eae, vtx1a, vtx2c | eae, vtx1a, vtx2c | ST-11 | C570-12 |
| eae, vtx1a, vtx2c | eae, vtx1a, vtx2c | ST-11 | C697-12A (replicate) | |
| eae, vtx1a, vtx2c | eae, vtx1a, vtx2c | ST-11 | C697-12B | |
| eae, vtx1a, vtx2c | eae, vtx1a | ST-11 | C541-12 | |
| eae, vtx1a, vtx2a | eae, vtx1a, vtx2a | ST-11 | C812-12 | |
| eae, vtx1a, vtx2a | eae, vtx1a, vtx2a | ST-11 | C818-12 | |
| eae, vtx1a, vtx2a | eae, vtx1a, vtx2a | ST-11 | C819-12 | |
| eae, vtx1a, vtx2a | eae, vtx1a, vtx2a | ST-11 | C849-12 | |
| eae, vtx1a, vtx2a | eae, vtx1a, vtx2a | ST-11 | C852-12 | |
| eae, vtx1a, vtx2a | eae, vtx1a, vtx2a | ST-11 | C863-12 | |
| eae, vtx2a | eae, vtx2a | ST-11 | C894-12 | |
| eae, vtx2c | eae, vtx2c | ST-2966 | C891-12 | |
| O165:H− | eae, vtx1a, vtx2a | eae, vtx1a, vtx2a | ST-119 | C905-12 |
| O180:H− | eae | eae | ST-301 | C641-12A |
| eae, vtx2aa | eae | ST-301 | C641-12B | |
| O181:H16 | vtx1ab | vtx1c | C893-12 | |
| OX187:H28 | vtx2dc | vtx2g | ST-200 | C892-12 |
| O rough:− | vtx2a | vtx2a | ST-678 | C895-12 |
Retyped to lack vtx2 at the SSI.
Retyped as vtx1c at the SSI.
Retyped as vtx2g at the SSI.
For all WGS analysis, apart from the SNPtree, the replicate sequences showed identical results. The complete list of typing results can be seen in Table S1 in the supplemental material, as well as the information on sequence quality and epidemiological information. The average coverage of the E. coli genome sequences ranged from 18× to 110×, and VirulenceFinder succeeded in detecting virulence genes in all the E. coli genomes. The complete list of detected virulence genes for each isolate can also be seen in Table S1.
Overall, there was high concordance between the routine typing results and the results obtained with VirulenceFinder, as can be seen in Table 3.
TABLE 3.
Concordance of results from routine typing and VirulenceFinder
| Virulence gene | No. of isolates with the virulence genea found by: |
|
|---|---|---|
| Routine typing | VirulenceFinder | |
| eae | 30 | 30 |
| vtx1 | 24 | 24 |
| vtx2 | 29 (28) | 27 |
| ehxA | 32 | 31 (32) |
| saa | 1 | 0 (1) |
| bfpA | 2 (0) | 0 |
When there was a discrepancy between the results for routine typing and VirulenceFinder, the value after retyping at the SSI or after reference mapping of raw reads is shown in parentheses.
The eae gene was detected in the same 30 isolates by routine typing and by VirulenceFinder. The routine typing detected the presence of vtx genes in 40 of the isolates, of which 11 isolates had vtx1, 16 isolates had vtx2, and the remaining 13 isolates possessed both vtx1 and vtx2. There was exact concordance between the vtx1 genes found using routine typing and VirulenceFinder. The vtx2 gene was detected in 29 isolates by routine typing, and VirulenceFinder found the vtx2 gene in 27 isolates. For one of the two isolates, C641-12B, in which VirulenceFinder did not detect vtx2, subsequent retyping at the SSI confirmed the lack of vtx2, while the presence of vtx2 was confirmed for the other isolate, C541-12. The inability of VirulenceFinder to detect the gene in this isolate was a feature of poor de novo assembly in this specific region in the gene of this isolate.
For 35 of the isolates that were hemolytic, routine typing detected ehxA, encoding enterohemolysin A, in 32 isolates, while VirulenceFinder detected ehxA in only 31 of these isolates. However, ehxA was subsequently detected in the sequence data for the last isolate, C813-12. The ehxA gene did not show up in VirulenceFinder due to problems in the de novo assembly, leading to only around half of the gene being present in the same contig of the assembled sequence data.
It was not possible to detect the plasmid-encoded virulence factor genes saa and bfpA with VirulenceFinder in the three isolates, C749-12, C862-12, and C820-12, shown by routine typing to harbor one of these genes, although the genes were included in the database. The saa gene was, however, detected in C749-12 by subsequent reference mapping of raw reads, and retyping at the SSI confirmed the absence of bfpA in the isolates. The saa gene did not show up in VirulenceFinder due to a low-coverage region in the middle of the gene that caused problems in the de novo assembly step in VirulenceFinder.
For two isolates, C893-12 and C892-12, vtx subtypes were initially assigned by routine typing as vtx1a and vtx2d, respectively. VirulenceFinder assigned the subtypes vtx1c and vtx2g. Retyping at the SSI confirmed the vtx1c and vtx2g subtypes detected by VirulenceFinder.
Phylogeny of the suspected VTEC isolates.
NDtree and SNPtree were constructed from the sequence data from the 44 confirmed E. coli isolates, as well as the two replicate sequences (Fig. 1 and Fig. 2). For NDtree, on average, 78% of the 5,498,450 bases were called in each of the strains. The isolates clustered completely according to serotype, and there was clear concordance between serotype and MLST type. For SNPtree, a total of 118,834 SNPs were called. Most isolates clustered according to serotype, although this method did not manage to cluster all three O117:K1:H7 isolates, C751-12, C760-12, and C659-12, together, and also the two replicate sequences of C770-12 were not grouped with SNPtree. This seemed to be a feature of IonTorrent data in combination with the SNPtree algorithm, causing trouble for some sequences that were very dissimilar to the employed O157 reference genome.
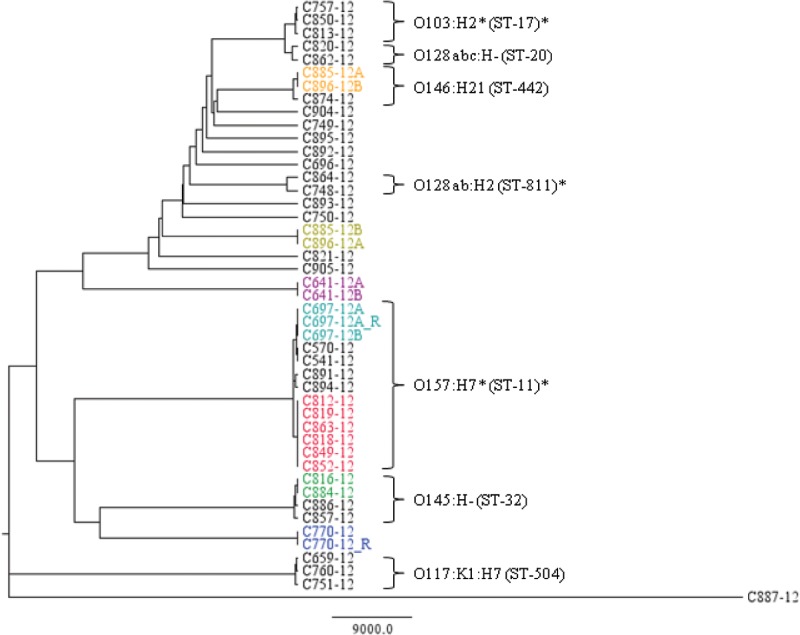
FIG 1.
Phylogeny of the isolates by the NDtree method. Isolates known to be epidemiologically related are shown in the same color, with the red group constituting the outbreak isolates. Serotypes and MLST types are shown for the main clusters. An asterisk indicates types with slight variations within the cluster.
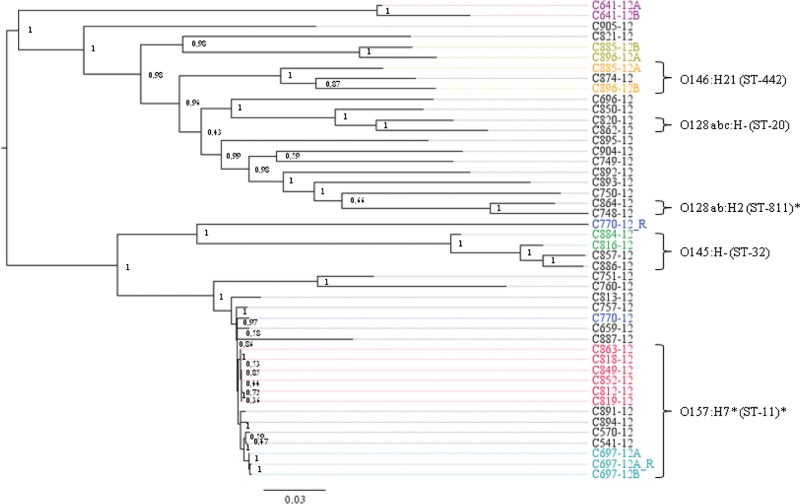
FIG 2.
Phylogeny of the isolates by the SNPtree method. Isolates known to be epidemiologically related are shown in the same color, with the red group constituting the outbreak isolates. Serotypes and MLST types are shown for the main clusters. An asterisk indicates types with slight variations within the cluster. Bootstrap values are assigned to each node.
NDtree thus seemed to cluster the isolates more correctly for this data set, although both approaches managed to group the six outbreak isolates.
The nucleotide difference (ND) and SNP divergence between the 46 genome sequences is included in Dataset S2 and Dataset S3 in the supplemental material, respectively. In addition, for further comparison of the phylogenetic approaches, a phylogenetic tree based on the NDtree matrix, but with positions called in all of the sequences, and constructed using Fastree, is included in Dataset S4.
Evaluating the NDtree phylogeny.
NDtree clustered the isolates in complete agreement with the epidemiological information obtained. The replicate sequences, C697-12A and C770-12, both had no nucleotide differences to their respective partner sequences. C641-12A and C641-12B, which were isolated from the same plate due to colony morphology differences, also exhibited no differences and were thus identical, as also concluded by routine typing. Also, C697-12A (replicate) and C697-12B were isolates from the same plate and had no nucleotide differences and were also found identical by routine typing. The C885-12 and C896-12 isolates, which in both cases exhibited mixed-colony morphology upon reception at the SSI, were from the same patient received 3 days apart. They clustered together with 2 nucleotide differences pairing isolate C885-12A together with C896-12B, and C885-12B together with C896-12A, which was also evident from the routine typing data, where the C885-12A and C896-12B isolates were both serotype O115:H−, and the C885-12B and C896-12A isolates were both serotype O146:H21.
In addition, the two isolates C816-12 and C884-12, which were not known to be epidemiologically related, but both were serotyped as O145:H−, clustered together with no nucleotide differences, and could potentially have originated from the same source.
During the 7 weeks, a VTEC outbreak occurred with high risk of HUS, with a total of 13 diagnosed cases (7). Six of these outbreak isolates were included in the study. Within the outbreak, the nucleotide differences ranged from 1 to 7, whereas the distance to the other isolates in the O157:H7 group ranged from 607 to 1,617 nucleotide differences.
Comparison of time for typing.
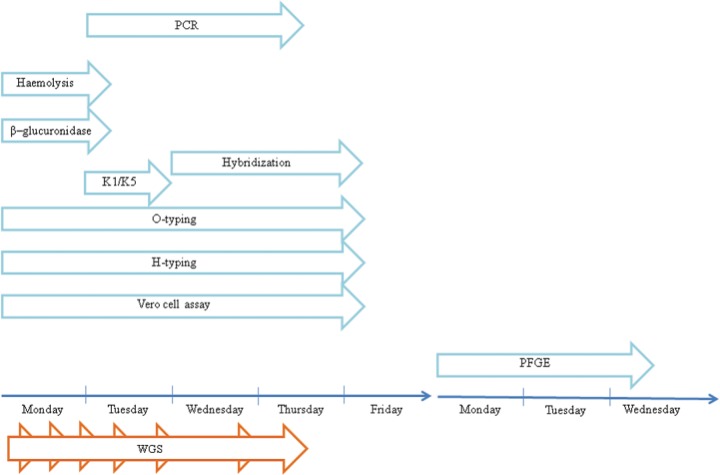
Typing of the isolates was initiated Monday each week during the study, both at the SSI for routine analysis and at DTU Food for WGS analysis, in parallel, starting from the exact same isolates. All work was performed to fit into an average working day in Denmark of maximum 7.4 h.
At the SSI, several different typing methods were employed, and although the majority were initiated Monday (hemolysis test, β-glucuronidase test, O typing, H typing, and Vero cell assay), others were initiated Tuesday (K1/K5 typing and PCR), Wednesday (hybridization), or Monday the following week (PFGE), for practical reasons as part of the standard work flow in the routine laboratory.
The time for obtaining results was dependent on the method used. Results on hemolytic activity and β-glucuronidase activity were obtained on Tuesday, K1/K5 results were ready on Wednesday, PCR results for vtx subtyping were ready on Thursday, and results from hybridizations, O typing, H typing, and Vero cell assay were ready Friday. The PFGE results were obtained on Wednesday of the second week, on day 10.
The WGS-based typing was different, with only one work flow and each step necessary for initiating the next. The first step was gDNA purification, which was initiated Monday, and took approximately 2 h (35-min hands-on time), followed by fragmentation that took around 1 h (1 h hands-on) and subsequently, library preparation (3.5 h [2.5-h hands-on]). On Tuesday, the library concentration was determined (1 h [15-min hands-on]) and the template was subsequently prepared (5 h [0.5 h hands-on]). On Wednesday, the template quality was determined (0.5 h [0.5 h hands-on]), and then sequencing was initiated, finishing after 5 h (45 min hands-on). On Thursday, the sequence data were extracted and submitted to the CGE web-tools, and the final results for all analyses were ready after 3 to 3.5 h (0.5 h hands-on).
In Fig. 3, a comparison between the time for obtaining typing results by routine typing and by the WGS approach is illustrated. The routine typing relies heavily on serotyping and vtx subtyping, and thus, sufficient results for accurate typing were not obtained before Friday. For further discrimination of suspected outbreak isolates, PFGE results were not ready before Wednesday of the following week. For the WGS approach, however, all results on MLST, vtx subtypes, and phylogenetic relationships among isolates were ready for interpretation on Thursday, saving at least half a day compared to the routine typing.
FIG 3.
Comparison of time for obtaining typing results by routine typing and WGS typing.
The time for WGS typing depended on the number of isolates to be tested. In the setup, four isolates could be run simultaneously. With a number of isolates between four and eight, an additional template preparation and sequencing were necessary. This added a total of 1 h and 45 min hands-on time to the procedure, but results were still obtained Thursday.
The total hands-on time each week for the typing procedures carried out at the SSI was approximately 14.5 h, while for the WGS, it was around 6.5 h or 8 h depending on the number of isolates.
DISCUSSION
The objective of this study was to evaluate in real time WGS for typing and surveillance of VTEC infections in Denmark by comparing the typing results, time, and cost to those of the routine typing procedure currently carried out. A set of 46 suspected VTEC isolates was employed for evaluation, with WGS-based typing being conducted in parallel to the routine typing on all suspected VTEC isolates received at the SSI during the study period.
Several recent studies have already proved the usefulness of rapid benchtop sequencing for investigations of various outbreaks (4, 31, 32). These studies have analyzed outbreaks retrospectively, and for useful implications in clinical microbiology, it is necessary to be able to conduct the relevant WGS analysis in real time, to be able to take action both regarding patient care and outbreak control. In this study, it was apparent that typing data could be extracted from WGS and analyzed in real time, and lead to faster conclusions, and more information, than routine typing.
For real-time WGS typing to work for routine surveillance, it is essential that clinical health personnel without bioinformatics skills be able to quickly extract and interpret the relevant information from the massive amount of sequence data. Different comprehensible web-tools for WGS analysis are gradually becoming available enabling extraction of relevant information from WGS data for typing purposes (23, 24, 26, 33). However, the well-established traditional typing procedures varies between pathogens, and thus, a lot of effort still needs to be put into development of more useful and user-friendly tools.
We conducted WGS typing and analysis in real time with the ongoing routine typing and surveillance of VTEC and developed VirulenceFinder for extraction of virulence genes for typing. In our study, the WGS-based typing using the CGE web-tools was able to compete with the current typing, both on typing results, time, and price. The use of VirulenceFinder enabled quick and accurate detection of eae, ehxA, and vtx genes and was in addition more robust assigning correct vtx subtypes than routine typing was. VirulenceFinder was, however, not successful in detecting saa and vtx2 in isolates C749-12 and C541-12, respectively, harboring these genes according to routine typing. This was probably a feature of poor sequence quality and low average coverage. The presence of many other important virulence genes in the isolates was detected by VirulenceFinder, giving much more information on the virulence profiles of the isolates than obtained by routine typing.
WGS-based typing also proved valuable for species detection, confirming that one of the suspected VTEC isolates was not E. coli, but instead Morganella morganii, adding to the value of performing WGS and strongly emphasizing that species confirmation should be performed as the first step of WGS typing.
The WGS-based phylogeny was very efficient for discrimination between the suspected VTEC isolates. As routine typing and surveillance did not offer any direct measure of relationships between all the isolates, but were instead based on grouping according to serotype and virulence gene profiles, and only in some cases PFGE, WGS-based typing could in this discipline offer much more information on how the isolates were related and potentially lead to faster detection of outbreaks.
The NDtree method managed to cluster the suspected VTEC isolates in complete agreement with the epidemiological information, whereas the SNPtree tool failed to cluster some of the isolates known to be identical. This was probably because the IonTorrent sequence data did not work well with SNPtree, since SNPtree is optimized for Illumina sequence data. The problem with SNPtree could be that this algorithm assigns the nucleotide for the reference genome at positions where the SNP is not accepted. This could, in theory, lead to two identical sequences not being clustered if the sequence data for one of these sequences had more sequencing errors, or perhaps low average coverage compared to the other, and was thus “mistakenly” assigned bases from the reference genome. Since SNPtree for this reason required all the query sequences to be closely related to the reference genome employed, it was not suitable for routine typing where many diverse isolates were included.
However, SNPtree did manage to cluster the main part of the isolates according to serotype, and also to identify the outbreak cluster, but since it did not seem completely reliable with the IonTorrent sequence data, NDtree, which was based on a different way to assign nucleotide differences, was employed instead, as it performed better for the data set in this study. NDtree clustered all isolates in agreement with the epidemiological data and showed no SNP differences between replicate sequences, and all isolates were clustered according to serotype. The method also perfectly clustered the outbreak isolates together and made it easy to distinguish outbreak isolates from sporadic isolates even of the same serotype. Although serotyping is an essential part of routine typing of E. coli as well as other important food-borne pathogens, for WGS-based typing, it was not necessary, since the MLST typing alone did just as good a job with clustering the isolates. This could be coincidental and due to the limited number of strains or restriction to analyze only VTEC, because in silico MLST performed on 61 whole-genome sequences of various E. coli strains rendered many of the various strains jumbled and less well resolved (34).
For the WGS analysis, many of the conventional typing results may be accessory. WGS offers ultimate resolution, and it would be valuable to develop tools to extract additional information, such as the serotype, thus enabling comparison to historical data generated by conventional serotyping. Although WGS offers ultimate resolution, it is important to note that epidemiological information is still necessary for any analysis in typing, outbreak investigation, and surveillance.
For this study, WGS typing could compete with routine typing, by employing automated extraction of WGS data for typing of VTEC. With this approach, all relevant information could be extracted from the sequence, making WGS typing faster than routine typing, and especially with regard to the workload, WGS was advantageous, since it required much less hands-on time.
At the SSI, many different E. coli isolates passed through the routine flow, being typed to different discriminatory levels by diverse methods, and it was thus impossible to determine the exact price per isolate included in the study. However, it was estimated that the overall price per isolate for each of the two approaches, routine typing and WGS typing, was around 430 euros for typing of suspected VTEC in this study, including both salaries and materials.
For routine typing, this included serotyping, PCR subtyping, DNA hybridizations, hemolysis and β-glucuronidase tests, Vero cell assay, and PFGE.
For both approaches, time could be optimized if necessary. In routine typing, PFGE could be initiated for isolates on the first day, and thus yield results Wednesday or Thursday of the first week. Similarly, in the WGS work flow, sequencing could be initiated on Tuesday, thus enabling analysis of sequencing results on Wednesday. However, for both routine and WGS typing, the procedure for typing was done according to what was believed to be most advantageous considering time, economical aspects, accessibility to equipment, etc.
Since the WGS part of this study was performed using the IonTorrent PGM, the price of routine typing and surveillance could be further decreased by performing sequencing on the Illumina MiSeq benchtop sequencer (35), as our current in-house material cost price on this sequencing platform is around 160 euros per isolate.
In conclusion, this study shows that WGS-based typing and surveillance using user-friendly web-tools are already applicable for routine purposes and that this approach can make the process even faster and cheaper. Finally, WGS delivers typing results that equal or even surpass the current typing methodologies in terms of microbiological information.
Supplementary Material
ACKNOWLEDGMENTS
This study was supported by the Center for Genomic Epidemiology (www.genomicepidemiology.org) grant 09–067103/DSF from the Danish Council for Strategic Research.
We thank Susanne Jespersen, Pia Møller Hansen, Christian Vråby Pedersen, and Christina Aaby Sørensen for excellent technical assistance.
We declare that we have no conflicts of interest.
Footnotes
Published ahead of print 26 February 2014
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JCM.03617-13.
REFERENCES
- 1.Ribot EM, Fair MA, Gautom R, Cameron DN, Hunter SB, Swaminathan B, Barrett TJ. 2006. Standardization of pulsed-field gel electrophoresis protocols for the subtyping of Escherichia coli O157:H7, Salmonella, and Shigella for PulseNet. Foodborne Pathog. Dis. 3:59–67. 10.1089/fpd.2006.3.59 [DOI] [PubMed] [Google Scholar]
- 2.Kauffmann F. 1975. Classification of bacteria. A realistic scheme with special reference to the classification of Salmonella -and Escherichia- species. Munksgaard, Copenhagen, Denmark [Google Scholar]
- 3.Mellmann A, Harmsen D, Cummings CA, Zentz EB, Leopold SR, Rico A, Prior K, Szczepanowski R, Ji Y, Zhang W, McLaughlin SF, Henkhaus JK, Leopold B, Bielaszewska M, Prager R, Brzoska PM, Moore RL, Guenther S, Rothberg JM, Karch H. 2011. Prospective genomic characterization of the German enterohemorrhagic Escherichia coli O104:H4 outbreak by rapid next generation sequencing technology. PLoS One 6:e22751. 10.1371/journal.pone.0022751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Reuter S, Harrison TG, Köser CU, Ellington MJ, Smith GP, Parkhill J, Peacock SJ, Bentley SD, Török ME. 2013. A pilot study of rapid whole-genome sequencing for the investigation of a Legionella outbreak. BMJ Open 3:1–6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Karch H, Tarr PI, Bielaszewska M. 2005. Enterohaemorrhagic Escherichia coli in human medicine. Int. J. Med. Microbiol. 295:405–418. 10.1016/j.ijmm.2005.06.009 [DOI] [PubMed] [Google Scholar]
- 6.Boerlin P, McEwen SA, Boerlin-Petzold F, Wilson JB, Johnson RP, Gyles CL. 1999. Associations between virulence factors of Shiga toxin-producing Escherichia coli and disease in humans. J. Clin. Microbiol. 37:497–503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Soborg B, Lassen SG, Müller L, Jensen T, Ethelberg S, Mølbak K, Scheutz F. 2013. A verocytotoxin-producing E. coli outbreak with a surprisingly high risk of haemolytic uraemic syndrome, Denmark, September-October 2012 Euro Surveill. 18(2):pii=20350 http://www.eurosurveillance.org/ViewArticle.aspx?ArticleId=20350 [PubMed] [Google Scholar]
- 8.Orskov F, Orskov I. 1984. Serotyping of Escherichia coli. Methods Microbiol. 14:43–112. 10.1016/S0580-9517(08)70447-1 [DOI] [Google Scholar]
- 9.Scheutz F, Cheasty T, Woodward D, Smith HR. 2004. Designation of O174 and O175 to temporary O groups OX3 and OX7, and six new E. coli O groups that include Verocytotoxin-producing E. coli (VTEC): O176, O177, O178, O179, O180 and O181. APMIS 112:569–584. 10.1111/j.1600-0463.2004.apm1120903.x [DOI] [PubMed] [Google Scholar]
- 10.Beutin L, Montenegro MA, Prada J, Zimmermann S, Stephan R. 1989. Close association of verotoxin (Shiga-like toxin) production with enterohemolysin production in strains of Escherichia coli. J. Clin. Microbiol. 27:2559–2564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lautrop H, Høiby N, Bremmelgaard A, Korsager B. 1979. Bakteriologiske undersøgelsesmetoder. FADL's Forlag, Copenhagen, Denmark [Google Scholar]
- 12.Scheutz F. 1997. Vero cytotoxin producing Escherichia coli (VTEC) isolated from Danish patients. Ph.D. thesis Statens Serum Institut, Copenhagen, Denmark [Google Scholar]
- 13.Jerse AE, Yu J, Tall BD, Kaper JB. 1990. A genetic locus of enteropathogenic Escherichia coli necessary for the production of attaching and effacing lesions on tissue culture cells. Proc. Natl. Acad. Sci. U. S. A. 87:7839–7843. 10.1073/pnas.87.20.7839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Girón JA, Donnenberg MS, Martin WC, Jarvis KG, Kaper JB, Giron JA, Donnenberg S. 1993. Distribution of the bundle-forming pilus structural gene (bfpA) among enteropathogenic Escherichia coli. J. Infect. Dis. 168:1037–1041. 10.1093/infdis/168.4.1037 [DOI] [PubMed] [Google Scholar]
- 15.Nataro JP, Baldini MM, Kaper JB, Black RE, Bravo N, Levine MM, Black E. 1985. Detection of an adherence factor of enteropathogenic Escherichia coli with a DNA probe. J. Infect. Dis. 152:560–565. 10.1093/infdis/152.3.560 [DOI] [PubMed] [Google Scholar]
- 16.Levine MM, Xu J, Kaper JB, Lior H, Prado V, Nataro J, Karch H, Wachsmuth K. 1987. A DNA probe to identify enterohemorrhagic Escherichia coli of O157:H7 and other serotypes that cause hemorrhagic colitis and hemolytic uremic syndrome. J. Infect. Dis. 156:175–182. 10.1093/infdis/156.1.175 [DOI] [PubMed] [Google Scholar]
- 17.Willshaw GA, Smith HR, Scotland SM, Rowe B. 1985. Cloning of genes determining the production of Vero cytotoxin by Escherichia coli. J. Gen. Microbiol. 131:3047–3053 [DOI] [PubMed] [Google Scholar]
- 18.Thomas A, Smith HR, Willshaw GA, Rowe B. 1991. Non-radioactively labelled polynucleotide oligonucleotide DNA probes for selectively detecting Escherichia coli strains producing Vero cytotoxins VT1, VT2 and VT2 variant. Mol. Cell Probes 5:129–135. 10.1016/0890-8508(91)90007-7 [DOI] [PubMed] [Google Scholar]
- 19.Persson S, Olsen KEP, Ethelberg S, Scheutz F. 2007. Subtyping method for Escherichia coli Shiga toxin (verocytotoxin) 2 variants and correlations to clinical manifestations. J. Clin. Microbiol. 45:2020–2024. 10.1128/JCM.02591-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Paton AW, Srimanote P, Woodrow MC, Paton JC. 2001. Characterization of Saa, a novel autoagglutinating adhesin produced by locus of enterocyte effacement-negative Shiga-toxigenic Escherichia coli strains that are virulent for humans. Infect. Immun. 69:6999–7009. 10.1128/IAI.69.11.6999-7009.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Scheutz F, Teel LD, Beutin L, Piérard D, Buvens G, Karch H, Mellmann A, Caprioli A, Tozzoli R, Morabito S, Strockbine NA, Melton-Celsa AR, Sanchez M, Persson S, O'Brien AD. 2012. Multicenter evaluation of a sequence-based protocol for subtyping Shiga toxins and standardizing Stx nomenclature. J. Clin. Microbiol. 50:2951–2963. 10.1128/JCM.00860-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.PulseNet International. 2013. Standard operating procedure for PulseNet PFGE of Escherichia coli O157:H7, Escherichia coli non-O157 (STEC), Salmonella serotypes, Shigella sonnei and Shigella flexneri. PNL05. PulseNet International, Atlanta, GA: http://www.pulsenetinternational.org/assets/PulseNet/uploads/pfge/PNL05_Ec-Sal-ShigPFGEprotocol.pdf [Google Scholar]
- 23.Larsen MV, Cosentino S, Rasmussen S, Friis C, Hasman H, Marvig RL, Jelsbak L, Sicheritz-Pontén T, Ussery DW, Aarestrup FM, Lund O. 2012. Multilocus sequence typing of total-genome-sequenced bacteria. J. Clin. Microbiol. 50:1355–1361. 10.1128/JCM.06094-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zankari E, Hasman H, Cosentino S, Vestergaard M, Rasmussen S, Lund O, Aarestrup FM, Larsen MV. 2012. Identification of acquired antimicrobial resistance genes. J. Antimicrob. Chemother. 67:2640–2644. 10.1093/jac/dks261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wirth T, Falush D, Lan R, Colles F, Mensa P, Wieler LH, Karch H, Reeves PR, Maiden MCJ, Ochman H, Achtman M. 2006. Sex and virulence in Escherichia coli: an evolutionary perspective. Mol. Microbiol. 60:1136–1151. 10.1111/j.1365-2958.2006.05172.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Leekitcharoenphon P, Kaas RS, Thomsen MCF, Friis C, Rasmussen S, Aarestrup FM. 2012. snpTree - a web-server to identify and construct SNP trees from whole genome sequence data. BMC Genomics 13(Suppl 7):S6. 10.1186/1471-2164-13-S7-S6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li H, Durbin R. 2009. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25:1754–1760. 10.1093/bioinformatics/btp324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R. 2009. The Sequence Alignment/Map format and SAMtools. Bioinformatics 25:2078–2079. 10.1093/bioinformatics/btp352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Price MN, Dehal PS, Arkin AP. 2009. FastTree: computing large minimum evolution trees with profiles instead of a distance matrix. Mol. Biol. Evol. 26:1641–1650. 10.1093/molbev/msp077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lindgreen S. 2012. AdapterRemoval: easy cleaning of next-generation sequencing reads. BMC Res. Notes 5:337. 10.1186/1756-0500-5-337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Eyre DW, Golubchik T, Gordon NC, Bowden R, Piazza P, Batty EM, Ip CLC, Wilson DJ, Didelot X, O'Connor L, Lay R, Buck D, Kearns AM, Shaw A, Paul J, Wilcox MH, Donnelly PJ, Peto TEA, Walker AS, Crook DW. 2012. A pilot study of rapid benchtop sequencing of Staphylococcus aureus and Clostridium difficile for outbreak detection and surveillance. BMJ Open 2:e001124. 10.1136/bmjopen-2012-001124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Köser CU, Holden MTG, Ellington MJ, Cartwright EJP, Brown NM, Ogilvy-Stuart AL, Hsu LY, Chewapreecha C, Croucher NJ, Harris SR, Sanders M, Enright MC, Dougan G, Bentley SD, Parkhill J, Fraser LJ, Betley JR, Schulz-Trieglaff OB, Smith GP, Peacock SJ. 2012. Rapid whole-genome sequencing for investigation of a neonatal MRSA outbreak. N. Engl. J. Med. 366:2267–2275. 10.1056/NEJMoa1109910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jolley KA, Maiden MC. 2013. Automated extraction of typing information for bacterial pathogens from whole genome sequence data: Neisseria meningitidis as an exemplar. Euro Surveill. 18(4):pii=20379.http://www.eurosurveillance.org/ViewArticle.aspx?ArticleId=20379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lukjancenko O, Wassenaar TM, Ussery DW. 2010. Comparison of 61 sequenced Escherichia coli genomes. Microb. Ecol. 60:708–720. 10.1007/s00248-010-9717-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Quail MA, Smith M, Coupland P, Otto TD, Harris SR, Connor TR, Bertoni A, Swerdlow HP, Gu Y. 2012. A tale of three next generation sequencing platforms: comparison of Ion Torrent, Pacific Biosciences and Illumina MiSeq sequencers. BMC Genomics 13:341. 10.1186/1471-2164-13-341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Larsen MV, Cosentino S, Lukjancenko O, Saputra D, Rasmussen S, Hasman H, Sicheritz-Pontén T, Aarestrup FM, Ussery DW, Lund O. 26 February 2014. Benchmarking methods for genomic taxonomy. J. Clin. Microbiol. 10.1128/JCM.02981-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Leekitcharoenphon P, Nielsen EM, Kaas RS, Lund O, Aarestrup FM. 2014. Evaluation of whole genome sequencing for outbreak detection of Salmonella enterica. PLoS One 9(2):e87991. 10.1371/journal.pone.0087991 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.