ABSTRACT
HIV-1 membranes contain gp120-gp41 trimers. Binding of gp120 to CD4 and a coreceptor (CCR5 or CXCR4) reduces the constraint on metastable gp41, enabling a series of conformational changes that cause membrane fusion. An analytic difficulty occurs because these steps occur slowly and asynchronously within cohorts of adsorbed virions. We previously isolated HIV-1JRCSF variants that efficiently use CCR5 mutants severely damaged in the tyrosine-sulfated amino terminus or extracellular loop 2. Surprisingly, both independent adaptations included gp120 mutations S298N, F313L, and N403S, supporting other evidence that they function by weakening gp120's grip on gp41 rather than by altering gp120 binding to specific CCR5 sites. Although several natural HIV-1 isolates reportedly use CCR5(Δ18) (CCR5 with a deletion of 18 N-terminal amino acids, including the tyrosine-sulfated region) when the soluble tyrosine-sulfated peptide is present, we show that HIV-1JRCSF with the adaptive mutations [HIV-1JRCSF(Ad)] functions approximately 100 times more efficiently and that coreceptor activation is reversible, enabling synchronous efficient entry control under physiological conditions. This system revealed that three-stranded gp41 folding intermediates susceptible to the inhibitor enfuvirtide form slowly and asynchronously on cell surface virions but resolve rapidly, with virions generally forming only one target. Adsorbed virions asynchronously and transiently become competent for entry at 37°C but are inactivated if the CCR5 peptide is absent during their window of opportunity. This competency is conferred by endocytosis, which results in inactivation if the peptide is absent. For both wild-type and adapted HIV-1 isolates, early gp41 refolding steps obligatorily occur on cell surfaces, whereas the final step(s) is endosomal. This system powerfully dissects HIV-1 entry and inhibitor mechanisms.
IMPORTANCE We present a powerful means to reversibly and efficiently activate or terminate HIV-1 entry by adding or removing a tyrosine-sulfated CCR5 peptide from the culture medium. This system uses stable cell clones and a variant of HIV-1JRCSF with three adaptive mutations. It enabled us to show that CCR5 coreceptor activation is rapidly reversible and to dissect aspects of entry that had previously been relatively intractable. Our analyses elucidate enfuvirtide (T-20) function and suggest that HIV-1 virions form only one nonredundant membrane fusion complex on cell surfaces. Additionally, we obtained novel and conclusive evidence that HIV-1 entry occurs in an assembly line manner, with some steps obligatorily occurring on cell surfaces and with final membrane fusion occurring in endosomes. Our results were confirmed for wild-type HIV-1. Thus, our paper provides major methodological and mechanistic insights about HIV-1 infection.
INTRODUCTION
Human immunodeficiency virus type 1 (HIV-1) envelope glycoproteins are trimeric heterodimers containing a surface subunit gp120 that binds receptors and a transmembrane subunit gp41 that mediates membrane fusion. In native virions, gp120 holds gp41 in a metastable conformation. Binding of gp120 to CD4 and then to a coreceptor (CCR5 or CXCR4) reduces this constraint, enabling gp41 to refold into a fusion-active conformation at an accelerated rate. After CD4 binding, the gp41 trimers extend heptad repeat 1 regions in a harpoon-like manner to form a three-stranded coil (3SC) that embeds gp41 amino termini into the cell membrane. Membrane fusion is driven by subsequent folding of gp41 heptad repeat 2 regions in an antiparallel orientation around the 3SC to form the more energetically stable six-helix bundle (6HB) (1). This process, which probably also occurs in stages (1, 2), pulls HIV-1 close to the cell surface and is irreversibly blocked by the 36-amino-acid peptide enfuvirtide (T-20) that mimics heptad repeat 2 and tightly binds into external grooves of the 3SC (1). Sensitivity to T-20 depends on its concentration and on the lifetime of the susceptible 3SC intermediate, which is influenced by CCR5 mutations and concentrations and by the sequences of gp120 and gp41 (3–7). These influences suggest that gp120 and CCR5 remain present during these refolding steps, in agreement with the idea that gp120 controls the magnitude of the activation energy barrier that constrains gp41 in native virions as well as the degree to which that barrier is reduced by CD4 and coreceptors. Optimal control by gp120 is important because premature gp41 refolding inactivates virions and damages virus-producing cells (8–11). Nevertheless, because successful infection depends upon winning a race between entry and competing inactivating processes, excessive constraint by gp120 slows gp41 refolding and reduces infectivity (4, 7, 12, 13). Although we experimentally define 3SC resolution to be escape from T-20 susceptibility, it should be understood that this loss of reactivity may occur substantially before 6HB formation has been completed (1, 2).
HIV-1 mutants resistant to small-molecule CCR5 antagonists have adaptive mutations in gp120 variable region V3 (14). Similarly, adaptations to other entry limitations and shifts to CXCR4 are principally determined by V3, and V3 mutations also alter sensitivities to T-20 (3–6, 15). Although V3 interacts directly with coreceptors (15–18), the mechanisms by which this modulates gp41 and controls infection are substantially unknown.
The tyrosine sulfate-containing amino terminus and extracellular loop 2 (ECL2) regions of CCR5 are most critical for its coreceptor activity. Nevertheless, we independently isolated HIV-1JRCSF variants that efficiently use CCR5 with a deletion of 18 N-terminal amino acids, including the tyrosine-sulfated region [CCR5(Δ18)], and others that efficiently use CCR5s with damaging mutations in ECL2 (5, 6, 19). All adapted viruses had gp120 mutations in V3, and some also had single mutations in V2 and V4. Surprisingly, the mutations that enable efficient use of CCR5s that lack amino termini or that are severely damaged in ECL2 overlapped, with S298N and F313L in V3 and N403S in V4 making critical contributions in both cases. The N403S mutation, which has a major adaptive effect, eliminates an N-linked glycan (6). During the latter investigations, we made panels of HeLa-CD4 cell clones that express wild-type or mutant CCR5s in discrete quantities. The results of studies using these clonal panels support the idea that these critical adaptive mutations function by reducing gp120's hold on gp41 rather than by strengthening gp120 interactions with specific CCR5 sites. Accordingly, the adapted variants form larger syncytia in infected CCR5-expressing cultures, efficiently use lower concentrations of damaged or wild-type CCR5s, and infect faster, as indicated by resistance to inactivation by T-20 (5, 6, 19). The adaptive mutations reduce the activation energy barrier that limits gp41 refolding (5), thus enabling the virions to efficiently use low concentrations of severely damaged CCR5s (5, 19).
It was previously shown by Farzan and coworkers that several natural HIV-1 isolates can infect canine thymocytic cells transfected with CCR5(Δ18) only if the soluble tyrosine sulfate-containing amino-terminal peptide is added to the medium (20). Tyrosine sulfate residues also occur in CXCR4 and in the neutralizing monoclonal antibody (MAb) 412d that associates with the coreceptor-binding region of gp120 (18, 21). The structure of gp120 complexed with 412d showed that the tyrosine sulfates bind to the conserved base of the V3 loop and to the fourth conserved region, and nuclear magnetic resonance analyses confirmed that the tyrosine-sulfated amino terminus of CCR5 also binds to this site (18). Considered together, these investigations suggest that the tyrosine sulfate-containing amino-terminal peptide, which we term Nt, can reconstitute the coreceptor function of CCR5(Δ18) to enable entry of HIV-1 isolates, including HIV-1YU2, and that several tyrosine-sulfated monoclonal antibodies bind to this site and inhibit HIV-1 infections.
Here we investigate the Nt peptide-dependent entry process using our variant HIV-1JRCSF that contains the adaptive mutations S298N, F313L, and N403S [HIV-1JRCSF(Ad)]. We demonstrate that Nt-dependent infection of HIV-1JRCSF(Ad) into stable HeLa-CD4/CCR5(Δ18) cell clones is approximately 100 times more efficient than infections by wild-type HIV-1JRCSF and previously tested HIV-1 isolates. Indeed, HIV-1JRCSF(Ad) infects under these conditions with an efficiency similar to wild-type HIV-1JRCSF and other highly infectious HIV-1 isolates when they utilize intact CCR5. As a consequence of this high efficiency, we were able to show that Nt-dependent CCR5(Δ18) coreceptor activation is rapidly reversible, thereby enabling synchronous activation and inactivation of HIV-1JRCSF(Ad) entry by adding or removing Nt from the culture medium under physiological conditions. By controlling Nt additions, we obtained important information about the mechanisms and limiting steps of HIV-1 entry and about entry inhibition by T-20. Our additional evidence strongly suggests that results obtained with this system are true for wild-type HIV-1 entry mediated by intact CCR5. This efficient split CCR5 system provides a powerful means to investigate factors that influence HIV-1 entry, including cellular components and neutralizing antibodies.
MATERIALS AND METHODS
Cells and viruses.
HeLa-CD4 cell clones expressing wild-type CCR5 (JC.53) or large amounts (∼6.3 × 104 molecules/cell) of CCR5 lacking 18 residues of the tyrosine-sulfated N terminus [HeLa-CD4/CCR5(Δ18)] were previously described (6, 22). All HeLa-derived cell clones were maintained in Dulbecco modified Eagle medium (DMEM)–10% fetal bovine serum (FBS) with 100 U/ml penicillin and 100 μg/ml streptomycin (complete medium). HEK293T cells were grown in high-glucose DMEM–10% FBS supplemented with 100 U/ml penicillin and 100 μg/ml streptomycin. Some experiments required HeLa-CD4 cells expressing large and small amounts of CCR5(Δ18), and these new clones were generated by retroviral transduction of HeLa-CD4 cells (cell clone HI-J, which expresses a large amount of CD4 [23]). Briefly, this entailed cotransfection of plasmids encoding CCR5(Δ18) [pFB-neo-CCR5(Δ18)], Moloney murine leukemia virus gag-pol (pCeB; from François-Loïc Cosset [24]), and the vesicular stomatitis virus envelope G protein (pME-VSV-G; from Richard E. Sutton [25]) into HEK293T cells. Supernatants from transfected cells were harvested, filtered, and used to transduce HI-J cells. Individual clones were isolated by limiting dilution, and the CCR5(Δ18) expression levels were determined by flow cytometry using the anti-CCR5 MAb 2D7 (BD Biosciences, San Jose, CA) and a Dako Qifikit (Dako North America, Inc., Carpinteria, CA) according to the manufacturers' instructions and as previously described (26). Approximate CCR5(Δ18) expression levels were high at 5.6 × 104 molecules/cell and low at 1.7 × 104 molecules/cell. These clones were used to investigate the effects of coreceptor expression levels on Nt-activated infections and T-20 sensitivities. All other experiments used clones that expressed large amounts (5.6 × 104 to 6.3 × 104 molecules/cell) or, where noted, moderate amounts (2.7 ×104 molecules/cell) of CCR5(Δ18).
Using a sequential adaptation scheme described previously (5), we generated HIV-1JRCSF(Ad) [formerly called CCR5(HHMH)-Ad], which efficiently replicated in HeLa-CD4/CCR5(HHMH) cells expressing small amounts of this human-mouse chimeric coreceptor that possesses an NIH/Swiss mouse ECL2 (27). In addition, in some studies we used HIV-1JRCSF adapted to use a damaged CCR5 with the ECL2 G163R mutation [CCR5(G163R)] (19, 28). The HIV-1JRCSF and HIV-1YU2 isolates were obtained as molecular clones from the AIDS Reagent Program, Division of AIDS, NIAID, NIH. Irvin Chen and Yoshio Koyanagi contributed pYK-JRCSF, and pYU2 was contributed by Beatrice Hahn and George Shaw. HIV-1JRCSF stocks were produced by transfecting HEK293T cells, with subsequent expansion in JC.53 cells. Virus-containing supernatants were harvested at 2 and 3 days postinfection. HIV-1JRCSF variants able to use mutant CCR5s were propagated in HeLa-CD4 cells expressing the appropriate mutant coreceptors. HIV-1YU2 stocks were generated by transfection of HEK293T cells, and supernatants harvested at 72 h were used in these studies. All virus-containing supernatants were harvested, filtered, aliquoted, and stored at −80°C. The titers of the virus stocks were determined on JC.53 cells as described previously (4). Pseudotyped viruses were produced by cotransfection of HEK293T cells with HIV-gpt (29) and HIV-1 Env expression vectors encoding gp120 with three (S298N, F313L, N403S) or four (S298N, F313L, N403S, A428T) adaptive mutations (5).
Plasmids.
The pFB-neo-CCR5(Δ18) plasmid was constructed by excising the 1.1-kb CCR5(Δ18)-coding sequence from pcDNA3.0-CCR5(Δ18) (19) by double digestion with BamHI and XhoI restriction endonucleases and then ligation with the pFB-neo plasmid (Stratagene, La Jolla, CA) digested with the same enzymes. Plasmids encoding a chimeric protein comprising the CCR5 tyrosine-sulfated N-terminal portion fused to the human IgG Fc region (CCR5-IgG) (21, 30) and tyrosylprotein sulfotransferase 2 (TPST-2) (31) were generous gifts from Michael Farzan.
CCR5-based peptides and proteins.
We used a soluble 22-residue (MDYQVSSPIYDINYYTSEPSQK) peptide (named Nt), based on the tyrosine-sulfated N terminus of CCR5, with tyrosine residues 10 and 14 sulfated (bold) and cysteine 20 replaced by serine (underlined). The peptide was synthesized to 95% purity by the American Peptide Company (Sunnyvale, CA), and the lyophilized peptide was resuspended in 100 mM sodium bicarbonate (pH 9.0) to a concentration of 20 mg/ml (∼7.2 mM). We produced CCR5-IgG in HEK293T cells cotransfected with plasmids encoding CCR5-IgG and TPST-2 using the PolyFect transfection reagent (Qiagen, Valencia, CA) or the linear polyethylenimine reagent (32). We purified CCR5-IgG from medium collected 72 and 96 h after transfection using protein A-Sepharose resin according to the method of Dorfman et al. (30). We quantitated purified CCR5-IgG by taking A280 readings using a NanoDrop spectrophotometer (ND-1000; Thermo Fisher Scientific, Waltham, MA) according to the manufacturer's instructions, and also by subjecting CCR5-IgG to SDS-PAGE and comparing the Coomassie-stained bands to the bands for bovine serum albumin standards loaded on the same gel.
Infectivity assays using the Nt system. (i) General conditions.
All infectivity assays were performed using target cells that had been seeded at 3 × 103 cells per well in a 96-well plate 24 h prior to the assay. On the day of the assay, cells were treated with 8 μg/ml DEAE-dextran for 20 min at 37°C and rinsed once before adding the viruses. Viruses were mixed with various concentrations of Nt or CCR5-IgG (see below) before the mixture was added to HeLa-CD4/CCR5(Δ18) or JC.53 cells. In some experiments, we assessed the effect of clathrin-mediated endocytosis (CME) inhibitors on Nt-activated infections (see below). Infected cultures were incubated in a humidified atmosphere containing 5% CO2 for 72 h at 37°C, at which time the cultures were fixed and stained for p24 Gag proteins using supernatant from an HIV-1 p24 hybridoma obtained from the NIH AIDS Reagent Program (HIV-1 p24 hybridoma [183-H12-5C], contributed by Bruce Chesebro) (33, 34). In some assays, Nt activation of HIV-1JRCSF(Ad) infections was analyzed in HeLa-CD4 target cells expressing large or small amounts of CCR5(Δ18). The T-20 sensitivities of Nt-activated infections were investigated using the same clones by infecting cells with HIV-1JRCSF(Ad) in the presence of 100 μM Nt and serial 10-fold dilutions of T-20. T-20 sensitivities were also tested in HIV-gpt viruses pseudotyped with adapted envelopes, using JC.53 cells as targets. T-20 was obtained from the NIH AIDS Reagent Program and was contributed by Roche.
(ii) Inhibition and activation of infection by CCR5-based peptides and proteins.
Activation of infection by Nt was investigated for HIV-1JRCSF(Ad), wild-type HIV-1JRCSF, and variants adapted to use CCR5(G163R) or CCR5(HHMH), as well as the primary isolate HIV-1YU2. Infections were initiated by combining equal volumes of 2× concentrations of Nt and virus and then adding this mixture to HeLa-CD4/CCR5(Δ18) cells. Investigation of entry activation by CCR5-IgG compared to that by Nt was done using HIV-1JRCSF(Ad) and HeLa-CD4/CCR5(Δ18) target cells. Inhibition of entry by CCR5-IgG and Nt was compared for wild-type HIV-1JRCSF and HIV-1JRCSF(Ad), using JC.53 cells as targets.
(iii) Kinetic assays.
We investigated the kinetics of Nt-activated infections by binding HIV-1JRCSF(Ad) onto HeLa-CD4/CCR5(Δ18) target cells by spinoculation (12). After spinoculation, the cultures were placed on ice and the unbound viruses were washed away, 100 μM Nt was added to each well, and the cultures were moved to 37°C. Upon warming the cultures, the progress of infection was monitored by the addition of fully inhibiting TAK779 (final concentration, 15 μM) at different times (TAK779 was obtained from the NIH AIDS Reagent Program and was contributed by the Division of AIDS, NIAID [35]). After the last time point at 20 to 24 h, the cultures were incubated for an additional 48 h at 37°C and then fixed and stained for HIV-1 p24 Gag. We assessed the reversibility of Nt activity by washing out Nt after 120- or 180-min incubations at 37°C and continued incubation at 37°C in the absence of Nt. The infectivity of the virions remaining on cell surfaces was determined by binding virions to cells at 4°C, washing, and adding 100 μM Nt after different times of incubation at 37°C. We investigated the formation of the 3SC during a 180-min incubation period at 37°C by binding virions to cells in the cold, washing, and then incubating them for 180 min at 37°C in the presence of 100 μM Nt and completely inhibiting 1 μg/ml T-20. Then, the cultures were washed to remove the T-20, Nt was added back, and subsequent Nt-dependent infections were measured.
(iv) Assay for cell surface lifetimes of 3SCs.
We examined the cell surface lifetimes of 3SCs by ascertaining virus T-20 sensitivities during different time windows determined by the presence of Nt. This was accomplished by spinoculating viruses onto HeLa-CD4/CCR5(Δ18) cells at 4°C, washing away unbound virus, and then infecting cells in the presence of 100 μM Nt plus serial 10-fold T-20 dilutions. Infections were terminated after 15 min or 2 h at 37°C by washing out the Nt and adding inhibiting TAK779. Control cultures were incubated with Nt and T-20 dilutions until the experiment was terminated at 72 h. Additional experiments using wild-type HIV-1JRCSF and JC.53 cells in the absence of Nt were performed identically, with the time window during which T-20 sensitivity was tested created by adding inhibiting TAK779 after 15 min or 2 h of incubation at 37°C.
Inhibition of Nt-activated HIV-1JRCSF(Ad) infections by compounds that block CME.
We employed two inhibitors of CME: dynasore, which inhibits the scission of endocytic vesicles by dynamin (36), and chlorpromazine, which inhibits AP2-dependent clathrin recycling (37). We purchased dynasore and chlorpromazine from Sigma-Aldrich (St. Louis, MO). Dynasore was suspended in dimethyl sulfoxide (DMSO) to make a 20 mM stock, while chlorpromazine was suspended in 100% ethanol to a concentration of 2.5 mM, and aliquots of each were stored at −20°C. For all of our studies, we used dynasore and chlorpromazine at final working concentrations of 80 μM and 2.5 μM, respectively. To study the effects of these inhibitors on HIV-1JRCSF(Ad) infection of HeLa-CD4/CCR5(Δ18) cells in the presence of Nt, we first washed cells five times in serum-free (SF) DMEM and then treated cells with 8 μg/ml DEAE-dextran plus inhibitors or vehicle (0.4% DMSO or 0.1% ethanol in DMEM) for 30 min at 37°C. Cultures were then washed, viruses were adhered to cells by centrifugation at 1,000 × g at 4°C for 30 min, cells were washed on ice, 100 μM Nt plus inhibitors or vehicle in SF DMEM was added to the cells, and the cultures were incubated for 1 h at 37°C. All washes and preparations of virus and Nt contained inhibitors or vehicle, as appropriate. After 1 h at 37°C, the cultures were washed 3 times with complete medium, and further infections were prevented by the addition of fully inhibiting TAK779. The cytotoxic effects of dynasore and chlorpromazine were investigated by adsorbing HIV-1JRCSF(Ad) onto cells, adding Nt, allowing infections to proceed in the absence of inhibitors for 1 h at 37°C, and then washing the cultures and adding fully inhibiting TAK779 plus CME inhibitors for 1 h. The cultures were then washed, and incubations were continued in the absence of CME inhibitors and in the presence of TAK779. Additional experiments were performed identically using wild-type HIV-1JRCSF and JC.53 cells in the absence of Nt.
Visualizing HIV-1 uptake by target cells.
HIV-1JRCSF(Ad) uptake and the effect of the endocytosis inhibitor dynasore were investigated using HIV-1 p24 Gag immunostaining and deconvolution microscopy (12, 38). At 24 h before the assay, glass coverslips (diameter, 13 mm; VWR International, Radnor, PA) were placed in the wells of 24-well plates and HeLa-CD4/CCR5(Δ18) cells were seeded at 3.0 × 104 cells per well. On the day of the assay, cells were washed 5 times in SF DMEM and then incubated with 80 μM dynasore or vehicle (0.4% DMSO in SF DMEM) for 30 min at 37°C. DEAE-dextran was not used because it interfered with the release of cell surface virions by pronase. All subsequent incubations and washes were performed using SF DMEM plus dynasore or vehicle, as appropriate. HIV-1JRCSF(Ad) was then added to the cells on ice and spinoculated by centrifugation at 1,000 × g for 30 min at 4°C. After spinoculation, cultures were placed on ice, the unbound viruses were washed away, 100 μM Nt plus dynasore or vehicle was added, and the cultures were incubated at 37°C for 1 or 4 h. After incubation at 37°C, cells were placed on ice, digested for 20 min with 1.0 mg/ml pronase (Sigma-Aldrich, St. Louis, MO) to remove cell surface virions, and then fixed in 3.7% formaldehyde–2% sucrose in phosphate-buffered saline (PBS) for 30 min at room temperature. Control cultures were placed on ice after spinoculation, rinsed, immediately subjected to pronase digestion without incubation at 37°C, and then fixed. Fixed cells were permeabilized with 0.5% NP-40–10% sucrose in PBS and stained for HIV-1 Gag proteins using the 183-H12-5C p24 monoclonal antibody and goat antimouse Alexa 594 (Invitrogen Life Technologies Corp., Grand Island, NY), as described previously (38). The fixed cells on coverslips were mounted on glass slides and viewed with a Zeiss Axiovert 200M deconvolution microscope, and z-stacks were acquired using the ×40 objective and AxioVision software (release 4.6.3; Carl Zeiss). z-stacks were deconvoluted, and the optical sections were compressed so that all virions could be viewed. Immunofluorescence figures were created in Photoshop software (version 8.0), and contrast and brightness were adjusted equally in all panels. Additionally, wild-type HIV-1JRCSF uptake into JC.53 cells was investigated using the same procedures.
Endocytosis inhibition by dynasore and reversal of inhibition were confirmed by investigating the cellular uptake of transferrin-Alexa 488 (Invitrogen Life Technologies Corp., Grand Island, NY). To accomplish this, we washed HeLa-CD4/CCR5(Δ18) cells 5 times in SF DMEM and then pretreated the cells with dynasore or vehicle alone for 30 min. In some wells, dynasore was washed away and the cells were allowed to recover for 20 min. We then added transferrin-Alexa 488 (final concentration, 12.5 μg/ml) to all of the wells for 5 min at 37°C. The cultures were then washed, excess unlabeled transferrin (final concentration, 625 μg/ml; EMD Millipore Corp. Billerica, MA) was added, and the cells were incubated for an additional 5 min at 37°C. The cultures were then placed at 4°C, washed in SF DMEM, digested with pronase to remove surface-bound transferrin, washed once in PBS, and then fixed in 3.7% formaldehyde. The fixed cells on coverslips were mounted on glass slides and viewed with a Zeiss Axiovert deconvolution microscope, and z-stacks were acquired using the ×40 objective and AxioVision software (release 4.6.3; Carl Zeiss). The images (see Fig. 7G) were constructed from single deconvoluted optical sections using Photoshop software. The contrast was adjusted equally in all panels.
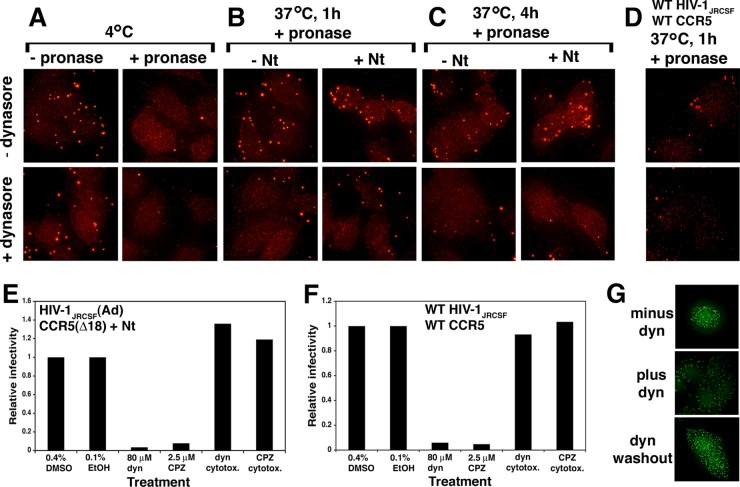
FIG 7.
Nt-activated infections and uptake of HIV-1JRCSF(Ad) by HeLa-CD4/CCR5(Δ18) cells are blocked by inhibitors of clathrin-mediated endocytosis. (A to C) Inhibition of virus uptake by dynasore. Virions were adsorbed at 4°C to cells preincubated with serum-free medium containing 80 μM dynasore (bottom) or 0.4% DMSO alone (without dynasore [−dynasore]; top). After these treatments, cultures were either treated with pronase or not treated on ice and fixed immediately (A) or incubated at 37°C with or without 100 μM Nt and with or without dynasore for 1 (B) or 4 (C) h and then digested with pronase and fixed. Cells in fixed cultures were permeabilized and stained for p24 Gag using a p24 monoclonal antibody and goat antimouse Alexa 594, and images were obtained using deconvolution microscopy. Results of a representative experiment of six total experiments performed are shown. (D) Dynasore inhibition of wild-type HIV-1JRCSF uptake by cells expressing wild-type CCR5 (JC.53). The same experiment described for panels A to C was performed without Nt. Cultures that were fixed after 1 h at 37°C are shown. Conditions in which fixation of cultures occurred immediately after virus binding at 4°C or after 4 h of incubation at 37°C yielded results identical to those shown in panels A and C. (E) Dynasore and chlorpromazine inhibition of Nt-activated infections. Cells were pretreated with the inhibitors dynasore (dyn) or chlorpromazine (CPZ) or with vehicle (0.4% DMSO or 0.1% ethanol [EtOH]) and then adsorbed with HIV-1JRCSF(Ad) with or without inhibitors at 4°C. Cultures were then placed at 37°C for 1 h, at which time the inhibitors were washed away and inhibiting TAK779 was added. Controls revealed that the inhibitors were not cytotoxic under the conditions of our assays. Results of a single representative experiment (executed in duplicate) of two independent experiments performed are shown. (F) The same experiment described for panel E performed using wild-type HIV-1JRCSF and JC.53 cells. Relative infectivity values were generated by normalizing the titers obtained under each condition to the titers obtained in the presence of vehicles. (G) Dynasore inhibition is reversible. Transferrin-Alexa 488 uptake was assessed in HeLa-CD4/CCR5(Δ18) cells treated with 80 μM dynasore (middle), in cells treated with 0.4% DMSO alone (top), or in cells treated with dynasore and then washed free of inhibitor (bottom).
Molecular modeling.
The gp120 crystal structure (PDB accession number 2B4C) (39), including the V3 loop and adaptive gp120 mutations important for growth on HeLa-CD4/CCR5(HHMH) cells, was modeled using the PyMOL molecular graphics program (version 1.2r1; Schrödinger).
Kinetics of virus inactivation by T-20.
Adsorbed virions that successfully infect cells in the absence of T-20 are irreversibly inactivated by T-20 in a manner that correlates with T-20 concentrations and the cell surface lifetimes of the susceptible fusion complex intermediates on the virus (VS) (4, 5, 7, 40). Here, we derived a quantitative kinetic model that closely fit our data and has important implications concerning the viral entry process. Although infectious virions form VS at different times, each VS forms and decays by the same processes under the conditions of our assays, and this allows us to consider them as a unit present initially. We assume that the rate of VS inactivation is first order in T-20 concentration, occurring at the rate ki[T-20][VS], where ki is the inactivation rate constant, and that Vs intermediates escape T-20 by resolving at the rate kr[VS], where kr is the resolution rate constant. The total rate at which VS leaves the cell surfaces is the sum of these two rates. Thus,
| (1) |
where [VS] is the concentration of susceptible intermediates remaining at time t.
Integration gives
| (2) |
where [VS,0] is the initial concentration of VS.
The rate at which susceptible intermediates escape inactivation by T-20 is therefore
| (3) |
where [Ve] is the concentration of susceptible intermediates that escape inactivation. Integrating until all infectious virions have left cell surfaces gives
| (4) |
where F is the fraction of susceptible intermediates that eventually escape successfully in the presence of the constant given concentration of T-20, regardless of the moment when any specific susceptible intermediate forms. Conceivably, infectious cell surface virions could form redundant T-20-susceptible fusion complexes, in which case a virion would be inactivated only if all of its fusion complexes react with T-20. Consequently, the probability that a virus will be inactivated by a given concentration of T-20 would be (1 − F)N, where N is the number of redundant VS complexes on the virion. The fraction of virions that successfully infect (f) would therefore be
| (5) |
Thus, virions with redundant fusion complexes are predicted to be more resistant to T-20 than virions that form only one complex. If all infectious virions form only one susceptible fusion complex, graphs of f versus log[T-20] are expected to resemble simple titration curves, with ƒ values being approximately 0.9 when [T-20] is equal to 0.1 time the 50% inhibitory concentration (IC50) and 0.1 when [T-20] is equal to 10 times the IC50. In contrast, if virions can form redundant fusion complexes, a distribution of fusion complex numbers would be expected and graphs of f versus log[T-20] would decline over a broader concentration range in several steplike waves. Similar broadening would be expected in assays using uncloned cells expressing different amounts of CD4 and/or coreceptors (4, 5, 26). According to equation 5, the IC50s for virions with one susceptible fusion complex are directly proportional to kr, occurring when [T-20] is equal to kr/ki. Many factors, including increases in coreceptor and Nt concentrations and adaptive gp120 mutations, increase HIV-1 infectivities by accelerating entry (4, 5, 22), suggesting that these factors reduce T-20 sensitivities by increasing kr. Data analysis, graphing, and curve fitting employed KaleidaGraph software (version 3.6.2) by Synergy Software (Reading, PA).
RESULTS
Efficient peptide-dependent use of CCR5(Δ18) by HIV-1JRCSF(Ad).
HIV-1JRCSF(Ad) was isolated by iteratively selecting HIV-1JRCSF for replication in HeLa-CD4 cell clones that stably express CCR5s with damaging mutations in ECL2 (5). Selection using mildly compromised CCR5(G163R) yielded an adapted variant with the gp120 V3 loop mutation F313I (19). Subsequent selection using a HeLa-CD4/CCR5(HHMH) clone that expresses a high concentration of this severely damaged human-mouse CCR5 chimera yielded an adapted variant with gp120 mutations F313L, N403S, and A428T. Final selection in a cell clone with a low concentration of the same coreceptor yielded the most highly adapted variant HIV-1JRCSF(Ad) with V3 mutations S298N and F313L and the V4 mutation N403S that eliminates an N-glycan (6). The positions of these three adaptive mutations in the gp120 sequence and in the crystal structure of the CD4-gp120 complex are shown in Fig. 1A and B, respectively. Importantly, an HIV-1JRCSF variant independently selected for efficient use of CCR5(Δ18) also contained S298N, F313L, and N403S, in addition to the other mutations indicated in the Fig. 1 legend (6). These results support other evidence that these common adaptive mutations weaken gp120's grip on gp41, enabling membrane fusion to accelerate and become more efficient (5), consistent with indications that the V3 loop region constrains gp41 in the unliganded quaternary native structure (41–45). Consistent with the idea that the adaptive mutations destabilize the Env trimers, HIV-1JRCSF(Ad) forms larger syncytia in infected cultures (5) and is much more sensitive than wild-type HIV-1JRCSF to inactivation by soluble CD4 (results not shown).
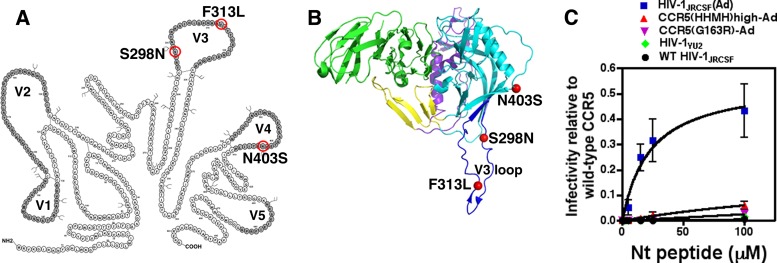
FIG 1.
HIV-1JRCSF(Ad), adapted to growth in HeLa-CD4/CCR5(HHMH) cells, has gp120 mutations that enable Nt-activated infections of HeLa-CD4/CCR5(Δ18) cells. (A) The gp120 mutations S298N, F313L, and N403S shared by virus isolates adapted to use CCR5(HHMH) or CCR5(Δ18) are circled in red, and the variable loops are in dark gray. HIV-1 adapted to growth in HeLa-CD4/CCR5(Δ18) cells possessed additional gp120 V3 mutations, one of which, N300Y, contributed specifically to CCR5(Δ18) use. Two other V3 mutations, I307M and T315P, while also contributing to CCR5(HHMH) use, facilitated Nt-independent infections of HeLa-CD4/CCR5(Δ18) cells and were therefore not investigated in this study. (B) Crystal structure (PDB accession number 2B4C) (39) of HIV-1 gp120 bound to the membrane distal two domains of CD4, with mutations conferring efficient activation by the Nt peptide represented by red spheres. Dark blue, V3; cyan, outer domain; purple, inner domain; yellow, bridging sheet; green, CD4. The image is oriented with the virus membrane at the top and the target cell membrane at the bottom. (C) HIV-1 adapted to use low levels of CCR5(HHMH) expression [HIV-1JRCSF(Ad)] demonstrates efficient infection of CCR5(Δ18) cells in the presence of Nt, while isolates less stringently adapted [i.e., CCR5(HHMH)high-Ad and CCR5(G163R)-Ad] show much less activation by Nt. The wild-type (WT) parental isolate HIV-1JRCSF and patient isolate HIV-1YU2 are poorly activated by Nt. Infectivity relative to that of wild-type CCR5 was generated by dividing the titers obtained for every virus isolate at each Nt concentration by the titers measured on HeLa-CD4/CCR5 clone JC.53 cells expressing large amounts of wild-type CCR5. Data points are averages of two independent experiments performed in duplicate, and error bars are the ranges. The plot was generated using GraphPad Prism software (version 4.0a).
Initial reports concerning the peptide-dependent split CCR5 system employed an HIV-1 vector pseudotyped with envelope glycoproteins derived from natural HIV-1 isolates (HIV-1ADA and HIV-1YU2) and Cf2Th-CD4 canine thymocytes transiently transfected with a CCR5(Δ18) expression vector (20). Because cells in transiently transfected cultures express large and variable amounts of CCR5(Δ18), the efficiencies of the peptide-dependent infections were difficult to evaluate. To more precisely analyze this issue, we used a HeLa-CD4/CCR5(Δ18) cell clone that expresses a uniformly large amount of CCR5(Δ18), and we measured infections of replication-competent wild-type and adapted viruses in media with different concentrations of the Nt peptide (Fig. 1C). The titer of each virus preparation was normalized relative to its titer in the HeLa-CD4/CCR5 cell clone JC.53 that is efficiently infected by all these viruses and contains the same amount of CD4 and a large optimal amount of wild-type CCR5 (22). The infectivity of HIV-1JRCSF(Ad) in this split CCR5 system is approximately 100 times greater than that of wild-type HIV-1JRCSF, and HIV-1JRCSF(Ad) is much more infectious than the partially adapted viruses mentioned above. In addition, HIV-1JRCSF(Ad) is approximately 100 times more infectious under these assay conditions than HIV-1YU2, which was originally considered to use split CCR5 relatively efficiently (20). At saturating concentrations of Nt, HIV-1JRCSF(Ad) infected this HeLa-CD4/CCR5(Δ18) cell clone approximately 44% (∼0.44 ± 0.09) as efficiently as it infected JC.53 cells with intact CCR5 (Fig. 1C).
The sequences of the V3 and V4 regions of HIV-1JRCSF, HIV-1JRCSF(Ad), and HIV-1YU2 are compared in Fig. 2A. Importantly, 298N and 313L occur not only in HIV-1JRCSF(Ad) but also in HIV-1YU2 and in a large percentage of natural HIV-1 isolates, as summarized in Fig. 2B. Although HIV-1YU2 differs from HIV-1JRCSF(Ad) in containing a consensus site for N-linked glycosylation at the V4 position corresponding to N403 (Fig. 2A), approximately 42% of natural HIV-1 isolates lack N-glycans at this site (Fig. 2B). Although we have not studied these polymorphisms in the context of other HIV-1 isolates, they occurred together in approximately 4% of the HIV-1 Env sequences that we examined (see the Fig. 2 legend). Thus, the three adaptive mutations in HIV-1JRCSF(Ad) occur very frequently in natural HIV-1 isolates.
FIG 2.
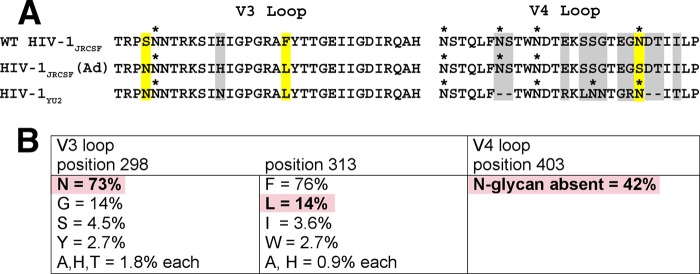
Adaptive mutations present in HIV-1JRCSF(Ad) occur commonly in many HIV-1 isolates. (A) Protein sequence alignment of V3 and V4 loops from wild-type HIV-1JRCSF, HIV-1JRCSF(Ad), and HIV-1YU2. HIV-1YU2 has V3 loop 298N and 313L residues that are also present in HIV-1JRCSF(Ad) but, unlike HIV-1JRCSF(Ad), possesses an N-glycan at position 403 within the heavily glycosylated V4 loop. Adaptive mutations are outlined in yellow, and glycosylation sites are marked with an asterisk. Other HIV-1YU2 residues that are different from those in HIV-1JRCSF(Ad) but are not known to contribute to entry activation by Nt are outlined in gray. The HXB2 standard numbering for HIV-1JRCSF(Ad) adaptive mutations is 300N, 317L, and 411S. The alignment was generated using MacVector software (version 7.2.3). (B) HIV-1JRCSF(Ad) adaptive mutations occur commonly in natural HIV-1 isolates. The frequencies of adaptive mutations in V3 and V4 loop sequences from 112 HIV-1 isolates, including isolates of clades A to D, F to H, J, and K, were analyzed using sequence data from the HIV Sequence Compendium 2013. Three clade D isolates (D.CM.10.DEMD10CM009, D.KE.97.ML415 2, and D.TD.99.MN011) and one clade B isolate (B.ES.09.P2149 3), representing 3.6% of total isolates analyzed, possessed all three mutations. Due to the hypervariability of the V4 loop, the location of position 403 within diverse isolates relative to its location within HIV-1JRCSF is approximate. Additionally, approximately 6% of the HIV-1 isolates surveyed did not possess an N-glycan within 2 amino acids of position 403. Pink boxes, adaptive mutations present in HIV-1JRCSF(Ad) that also occur in diverse HIV-1 isolates.
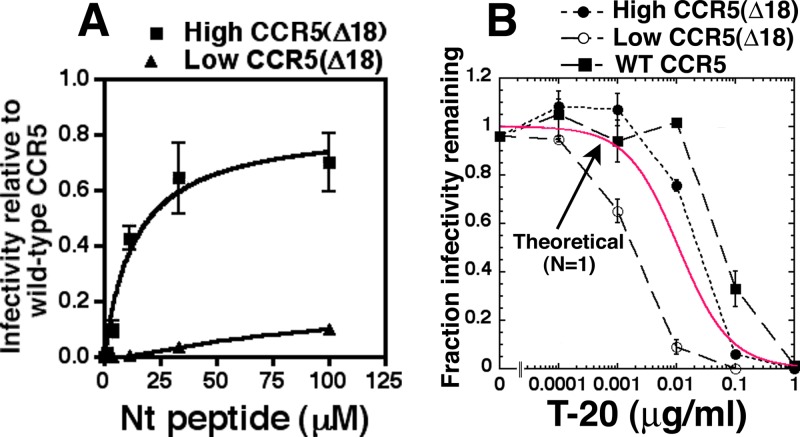
The efficiency of HIV-1JRCSF(Ad) infection is highly dependent on CCR5(Δ18) expression in cell clones, with much lower efficiencies occurring when we used a HeLa-CD4/CCR5(Δ18) clone with a small amount of CCR5(Δ18) (Fig. 3A). Moreover, this lower entry efficiency closely correlated with increased sensitivity to T-20, consistent with other evidence that inefficient infections caused by low coreceptor concentrations or by damaged coreceptors are associated with the slow resolution of 3SC conformational intermediates and correspondingly more prolonged exposures and greater sensitivities to T-20 (Fig. 3B) (4–7). Importantly, all the T-20 sensitivity curves in Fig. 3B resemble simple titration curves, with fractional infectivities of approximately 0.9 when T-20 concentrations are 1/10 the IC50s and approximately 0.1 when T-20 concentrations are 10 times greater than the IC50s. Thus, although the IC50s differ in the cell clones used, their shapes appear identical, within experimental error, to each other and to the theoretical curve for N equal to 1 shown in red (Fig. 3B). As further analyzed below, this suggests that many infectious virions form only one nonredundant T-20-susceptible fusion complex under the conditions of our assays. Moreover, based on our kinetic model, we infer that the 3SC resolution rate constant is approximately 10 times greater on the cells with more CCR5(Δ18) and is further increased about 3-fold when HIV-1JRCSF(Ad) employs wild-type CCR5 (Fig. 3B). It is also notable that the titers approach 0 at high concentrations of the membrane-impermeant T-20 peptide, suggesting that all infectious virions expose 3SCs while on cell surfaces. Therefore, if endocytosis is a prerequisite for infection, as has been proposed (46), all or nearly all virions that successfully infect cells must form their 3SCs before endocytosis. However, 3SCs cannot be a limiting requirement or signal for endocytosis because the cell surface lifetimes of 3SCs differed in the cell clones analyzed (Fig. 3B). These considerations suggest that a coreceptor-dependent process that follows 3SC formation generally or always precedes movement of infectious virions off cell surfaces (see Discussion). The possibility that endocytosis is required for entry and the kinetics of 3SC resolution are analyzed further below.
FIG 3.
Nt-dependent infectivity of HIV-1JRCSF(Ad) in cells expressing large and small amounts of CCR5(Δ18). (A) Nt activation of infection in HeLa-CD4 cells expressing high or low levels of CCR5(Δ18). Relative infectivity values were generated by normalizing the titers obtained at each Nt peptide concentration to the titers obtained on JC.53 cells expressing large amounts of wild-type CCR5. Data are averages of two independent experiments performed in duplicate, and error bars are the ranges. The plots were generated using GraphPad Prism software. (B) T-20 inhibition curves on cells with high and low levels of CCR5(Δ18) and on JC.53 cells with large amounts of wild-type CCR5. Infections were performed in the presence of 100 μM Nt and 10-fold dilutions of T-20. For cells with wild-type CCR5, T-20 dose-response curves were performed in the absence of Nt. Values of the fraction of infectivity remaining were calculated by dividing the titers obtained in the presence of T-20 by the average of the titers obtained in the absence of T-20 inhibition. Data are averages of 2 independent experiments performed in duplicate, and error bars are the ranges. A theoretical curve (in red) was generated using equation 5, assuming a single fusion complex (N = 1), and the graph was generated in KaleidaGraph (version 3.6.2).
Activation versus inhibition of entry by tyrosine-sulfated peptides and proteins.
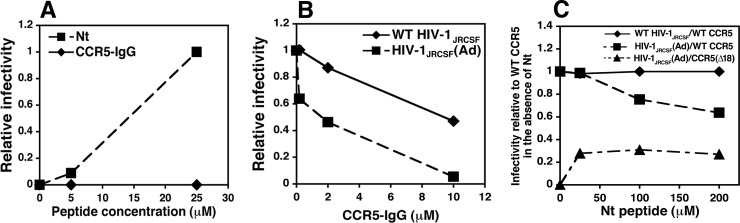
Although HIV-1JRCSF infection of HeLa-CD4/CCR5(Δ18) cells is induced by Nt, no entry occurs in identical concentrations of a CCR5-IgG chimera that has the same tyrosine-sulfated CCR5 amino terminus (Fig. 4A) (21, 30). Consistent with a previous report (30), CCR5-IgG inhibits infections mediated by wild-type CCR5 (Fig. 4B). However, HIV-1JRCSF(Ad) is inhibited much more efficiently by CCR5-IgG than unadapted HIV-1JRCSF is (Fig. 4B). Similarly, Nt inactivates HIV-1JRCSF(Ad) entry mediated by wild-type CCR5 much more efficiently than it does HIV-1JRCSF (Fig. 4C). However, as reported previously for other HIV-1 isolates by Farzan and collaborators (17, 30, 47), CCR5-IgG is a much more potent inhibitor per μM than Nt (compare the inactivation curves in Fig. 4B and C).
FIG 4.
Inhibition and activation of HIV-1 infections by CCR5-derived proteins and peptides. (A) Comparison of Nt versus CCR5-IgG activation of HIV-1JRCSF(Ad) infections of HeLa-CD4/CCR5(Δ18) cells. Infectivity values were normalized relative to the infectivity obtained at 25 μM Nt. The results of a single experiment performed in duplicate are shown. A parallel study using an antibody-derived tyrosine-sulfated CCR5-mimetic IgG (30, 69) also showed no activation. (B) Comparison of CCR5-IgG inhibition of wild-type HIV-1JRCSF and HIV-1JRCSF(Ad) on JC.53 cells expressing wild-type CCR5. Relative infectivity values were generated by dividing the titers obtained at each concentration by the titers obtained in the absence of CCR5-IgG. The results of a representative experiment of two performed in duplicate are shown. (C) Nt inhibition of wild-type HIV-1JRCSF and HIV-1JRCSF(Ad) on JC.53 cells. Relative infectivity values were calculated by normalizing the titers obtained at each peptide concentration to the titers obtained on cells expressing wild-type CCR5 in the absence of Nt. Nt effectiveness was confirmed by its ability to activate HIV-1JRCSF(Ad) infections of HeLa-CD4/CCR5(Δ18) cells. The results of a representative experiment of two performed in duplicate are shown. All graphs were created with Microsoft Excel software (version 11.5.5).
Activation of CCR5(Δ18) by Nt is rapidly reversible.
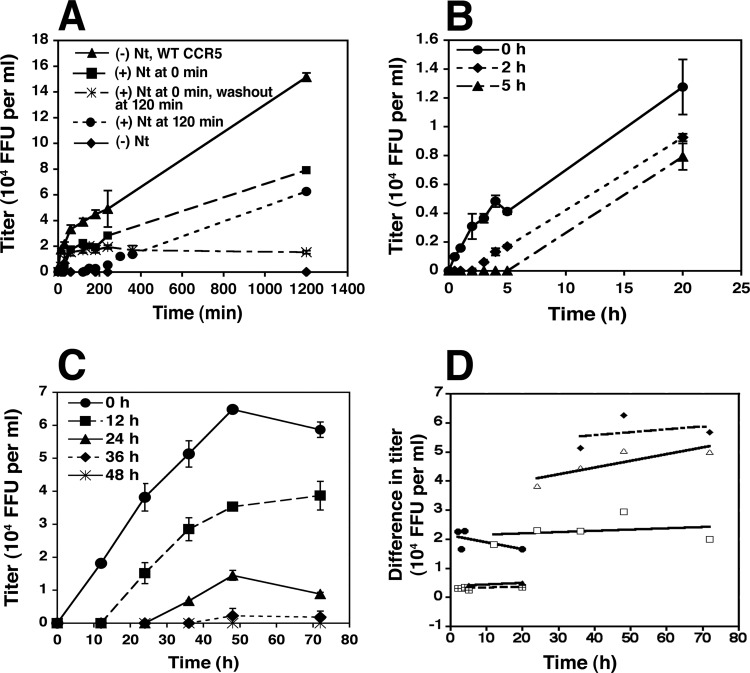
As recently demonstrated, HIV-1 virions adsorb irreversibly onto cells pretreated with polycations, such as DEAE-dextran or Polybrene, whereas they rapidly dissociate from untreated cells, thus explaining how these polycations greatly increase viral titers in infectivity assays (12). Moreover, because dissociation in the absence of polycations is much faster than successful entry, dissociation often dominates entry kinetic assays, making it appear, incorrectly, that entry is substantially completed within 1 to 2 h after a cohort of virions is initially adsorbed onto cells (12). To reduce this problem and to more faithfully analyze the entry kinetics of a complete cohort of simultaneously adsorbed virions, we spinoculated virions at 4°C onto cells pretreated with DEAE-dextran, and we terminated entry by adding a fully inhibiting concentration of the CCR5 antagonist TAK779 at different times after warming cultures to 37°C.
Figure 5A shows a representative analysis of HIV-1JRCSF(Ad) entry kinetics into cells expressing either wild-type CCR5 or CCR5(Δ18). Consistent with the results in Fig. 1, entry into the latter cells was approximately 50% as efficient and was completely dependent on the presence of Nt. Moreover, removing the peptide after 2 h caused the cessation of entry, suggesting that Nt-dependent CCR5(Δ18) activation is rapidly reversible. In addition, when Nt addition to the cultures was delayed until 2 h after warming, viral entry ensued, although it did not catch up to the level of infection obtained when Nt was added earlier. Indeed, the rates of infection after 2 h were identical regardless of whether Nt was added at the time of warming or was delayed. Thus, the titers in these two cultures (curves with solid squares and solid circles) increased in parallel throughout the remainder of the analysis. These data are inconsistent with the idea that infectious virions accumulate at a stage of arrested entry when Nt is absent and that they can subsequently enter after Nt addition. These conclusions were strongly supported by withholding Nt for different times after cultures with adsorbed HIV-1JRCSF(Ad) were warmed to 37°C (Fig. 5B and C). Regardless of the delay before Nt addition, entry reproducibly ensued at the same rate as ongoing entry into cultures given Nt at earlier times. To quantitatively analyze this issue, we measured the titer differences between cultures given Nt initially and after delays, plotted these differences as a function of time after warming, and fit the data points by least squares to lines using KaleidaGraph (Fig. 5D). These lines all appeared flat and had an average slope ± standard deviation equal to 0.00345 × 104 ± 0.014 × 104 focus-forming units (FFU)/ml-h, strongly suggesting that HIV-1JRCSF(Ad) virions leave cell surfaces at the same rate and therefore by the same mechanism regardless of whether they have interacted with Nt and are destined to infect the cells or whether they lack Nt and are destined for eventual degradation. Considered as a whole, the results in Fig. 5 further imply that each of the adsorbed virions has a transient window of opportunity to infect, that this opportunity window occurs independently of Nt, and that the virions become inactivated if Nt is absent at that time. However, each virion in the cohort experiences this window of opportunity at a different time, suggesting that this critical process is influenced by stochastic factors.
FIG 5.
Activation of HIV-1JRCSF(Ad) infection by Nt is reversible and can occur many hours after viruses have been adsorbed onto HeLa-CD4/CCR5(Δ18) cells. (A) Nt reversibility. HIV-1JRCSF(Ad) was adsorbed onto cells at 4°C, and 100 μM Nt was added upon warming to 37°C (squares) or after 2 h at 37°C (circles). In some cultures, Nt was washed out after 2 h of incubation at 37°C (asterisks). Kinetics of infection were also measured in HeLa-CD4 cells with wild-type CCR5 (triangles) or in CCR5(Δ18) cells in the absence of Nt (diamonds). This experiment was performed with HeLa-CD4 cells expressing moderate amounts of CCR5(Δ18) and gave similar results to those obtained in experiments performed with cells expressing large or small amounts of CCR5(Δ18). Results of a representative experiment of eight performed in duplicate are shown. Error bars are the ranges. (B and C) Nt activates HIV-1JRCSF(Ad) infections many hours after virus adsorption onto target cells. Viruses were bound to HeLa-CD4/CCR5(Δ18) cells at 4°C, and 100 μM Nt was added after 0, 2, or 5 h (B) or 0, 12, 24, 36, or 48 h (C) of incubation at 37°C. Error bars are the ranges. Titer values (104 focus-forming units [FFU] per ml) were plotted versus time using Microsoft Excel software. (D) Quantitative analysis of infectivity data from panels A, B, and C. The graph was obtained by plotting titer differences between cultures that had Nt added initially and after delays versus time after warming.
The fact that stably adsorbed virions remain viable and continue to infect cells for prolonged periods (e.g., Fig. 5C) initially seemed surprising, although virions stably adsorbed onto follicular dendritic cells also have long life spans (48). The factors that influence this prolonged viability are not understood.
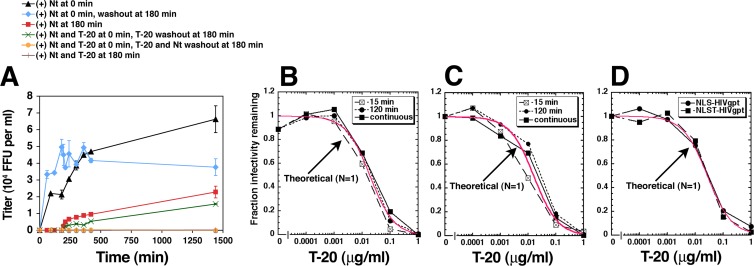
Simultaneously adsorbed virions form 3SCs at widely different times but resolve them rapidly.
Figure 6A shows an entry kinetic experiment in which we added or removed Nt from HeLa-CD4/CCR5(Δ18) cultures after 3 h at 37°C. Consistent with the results in Fig. 5, adding or removing Nt activated or terminated entry, respectively. To learn whether 3SCs form rapidly after warming the cultures or slowly throughout the incubation period, some cultures were incubated with Nt plus a fully inhibiting concentration of T-20 for 3 h before rinsing the cells and adding fresh medium containing Nt alone. The Nt-dependent infections that ensued after removing T-20 were slower at early time points than they were in the control cultures that had not been exposed to T-20 (e.g., compare the red and green curves in Fig. 6A). This difference was highly reproducible (n = 3). This and related experiments suggest that infectious virions generally form T-20-susceptible 3SCs substantially before they complete the Nt-dependent cell surface step(s) required for infection, in agreement with evidence that T-20-reactive 3SCs can be induced by CD4 prior to HIV-1 interactions with coreceptors (49). Most importantly, it is also notable that a large proportion of the infectious virions on HeLa-CD4/CCR5(Δ18) cell surfaces remained viable throughout this 3-h cell exposure to T-20, indicating that infectious virions slowly and asynchronously form T-20-susceptible 3SCs during their residencies on cell surfaces and that many virions had not yet formed 3SCs by 3 h at 37°C.
FIG 6.
Formation of three-stranded coils and their cell surface lifetimes during Nt-activated infections of HeLa-CD4/CCR5(Δ18) cells. (A) 3SC formation. HIV-1JRCSF(Ad) was adsorbed onto cells at 4°C, and the cultures were incubated at 37°C for 180 min in the absence of any peptides or in the presence of 100 μM Nt (red curve) and 1 μg/ml T-20 (green curve), at which time these cultures were washed and 100 μM Nt was added back. In control cultures, virions were bound to cells at 4°C, Nt was added upon warming to 37°C (0-min time point), and the cultures were either incubated until the last time point at 1,440 min (black curve) or Nt was washed away after 180 min of incubation at 37°C (blue curve). Not shown are data from cultures incubated at 37°C for 180 min with T-20 alone followed by washes and Nt addition, which gave results that were not reproducibly different from those shown on the green curve. T-20 inhibitory activity was confirmed by additional incubations of viruses with T-20 plus Nt (orange and brown curves). The results of a representative experiment of three performed in duplicate are shown. Error bars are the ranges, and the graph was created in Microsoft Excel software. (B) 3SC lifetimes of HIV-1JRCSF(Ad). The T-20 sensitivities of HIV-1JRCSF(Ad) exposed to Nt for different times were used to ascertain the lifetime of the 3SC. Virions were adsorbed to HeLa-CD4/CCR5(Δ18) cells at 4°C, and then 100 μM Nt was added in the presence of serial 10-fold T-20 dilutions and the cultures were incubated at 37°C for 15 or 120 min, at which time the cultures were washed and infections were terminated by addition of inhibiting TAK779. Control cultures (continuous) were incubated with Nt and T-20 until the experiment was terminated at 72 h. The results of a representative experiment of three performed in triplicate are shown. (C) An experiment identical to that whose results are shown in panel B performed using wild-type HIV-1JRCSF with JC.53 cells that express wild-type CCR5 and in the absence of Nt. (D) T-20 dose-response curves generated using HIV-gpt viruses pseudotyped with HIV-1JRCSF envelopes containing 3 (NLS; S298N, F313L, N403S) or 4 (NLST; S298N, F313L, N403S, A428T) adaptive mutations. The experiment was performed in the absence of Nt using JC.53 as target cells. For panels B, C, and D, values of the fraction of infectivity remaining were calculated by dividing the titers in the presence of T-20 by the average of the titers obtained in the absence of T-20 inhibition. Theoretical curves (in red) were generated using equation 5, assuming a single fusion complex (N = 1). The graphs were generated in KaleidaGraph.
To determine whether 3SC conformational intermediates have long or short cell surface lifetimes and to more thoroughly analyze the fusion complex redundancies of the virions, we analyzed the T-20 sensitivities of infections that occurred in HeLa-CD4/CCR5(Δ18) cultures exposed to Nt for different times (Fig. 6B). Our approach was based on evidence that the T-20-susceptible 3SC intermediates have lifetimes distributed around a mean that depends on the coreceptor structure and concentration (e.g., see Fig. 3) (3–7). Consequently, when exposures to Nt become shorter than the average natural lifetimes of the T-20-susceptible fusion complex intermediates, only virions that resolve their 3SCs more rapidly than the mean would successfully infect the cells. Because these 3SCs would be exposed to T-20 for only relatively short periods, they would be inactivated less efficiently and would be expected to have correspondingly large increases in IC50s (see equation 4 in Materials and Methods). Moreover, because susceptible fusion complex intermediates form slowly and asynchronously over a prolonged period of many hours (Fig. 6A), individual virions would be unlikely to form redundant fusion complexes during short periods. Consequently, during brief entry windows, infections would become more reliant on one fusion complex. Importantly, IC50s were not significantly altered by reducing the Nt exposure time to as short as 15 min (Fig. 6B), implying that the average T-20-susceptible 3SC lifetime under these conditions is shorter than 15 min. Moreover, these susceptibility curves and those in Fig. 3B do not significantly differ from the theoretical curves (shown in red) generated using equation 5 for N equal to 1, implying that the T-20-susceptible fusion complexes resolve with a uniform first-order rate constant and further suggesting that infectious virions generally form only one nonredundant fusion complex that can be inactivated by a single T-20 molecule. Statistical analyses confirmed the compatibility of these data with equation 5 for N equal to 1, with chi-square values for the three assays in Fig. 6B being less than 0.023 and correlation coefficient (R) values being greater than 0.98. A related analysis was done using wild-type HIV-1JRCSF in JC.53 cells that express intact CCR5, in this case, limiting the entry windows by adding the CCR5 antagonist TAK779 after different times of exposure to T-20 (Fig. 6C). Wild-type HIV-1JRCSF adsorbed onto optimally susceptible JC.53 cells also resolved its 3SCs rapidly in a manner consistent within experimental error with kinetics of N equal to 1 (chi-square values, less than 0.017; R values, greater than 0.99). In addition, we analyzed the T-20 susceptibilities of HIV-gpt virions pseudotyped with adaptive HIV-1JRCSF Envs containing the gp120 mutations S298N, F313L, and N403S or with the additional mutation A428T (Fig. 6D). This experiment was done because cells in transiently transfected cultures express widely differing amounts of Env, which would be expected to cause correspondingly large differences in the Env contents of the released virions. Presumably, virions with large amounts of Env might form redundant fusion complexes more readily than virions with limiting amounts. These T-20 susceptibility curves did not suggest virion heterogeneity and were fully consistent with kinetics of N equal to 1 (red curve in Fig. 6D) (chi-square values, less than 0.014; R values, greater than 0.99). Despite these results, we believe that fusion complex redundancy requires further investigation. For example, we cannot exclude the possibility that a small proportion of virions in our preparations form redundant fusion complexes. Additionally, we consider it possible that HIV-1 virions have the potential to form redundant fusion complexes but accomplish this inefficiently under the conditions of our cell culture assays. Considered together, these analyses suggest that T-20-susceptible fusion complex intermediates form slowly and asynchronously in a cohort of simultaneously adsorbed HIV-1JRCSF(Ad) virions (Fig. 6A), that a critical Nt-dependent process subsequently occurs (Fig. 6A), and that they resolve rapidly (Fig. 6B) at a uniform rate that strongly depends on the cell surface coreceptor and its concentration (Fig. 3B and 6B). Moreover, our data suggest that adsorbed wild-type HIV-1JRCSF, HIV-1JRCSF(Ad), and pseudotyped HIV-1JRCSF(Ad) virions generally form only one nonredundant fusion complex under the conditions of our assays (Fig. 3B and 6B to D).
Role of endocytosis in cell entry of HIV-1.
Our data in Fig. 5 strongly suggest that HIV-1JRCSF(Ad) virions on cell surfaces have a transient window of opportunity to successfully infect and that this opportunity occurs at different times for each virion in a cohort that was simultaneously adsorbed onto the cells. Moreover, in HeLa-CD4/CCR5(Δ18) cultures lacking Nt, this opportunity window ultimately results in virus inactivation rather than in successful entry. This implies that the opportunity window is caused by a cellular process rather than by a specific step in gp41 refolding. Because endocytosis has previously been implicated in the entry process (46, 50–52), we hypothesized that this opportunity window might correspond to endocytosis, which would enable HIV-1JRCSF(Ad) infection if Nt were present and ultimately result in lysosomal degradation in the absence of Nt. To test this, we adsorbed virions onto HeLa-CD4/CCR5(Δ18) cells at 4°C and then either treated the cultures immediately with pronase or incubated them at 37°C for 1 to 4 h in the presence or absence of dynasore before pronase treatment. We then visualized the virions by immunofluorescence in a deconvolution microscope using a monoclonal antibody to p24 Gag (12, 34, 38).
Pronase efficiently removed virions that had been spinoculated onto cell surfaces at 4°C (Fig. 7A). After 1 to 4 h at 37°C, however, many HIV-1JRCSF(Ad) virions that initially attached to the cells had become resistant to pronase when dynasore was absent, whereas substantially fewer were pronase resistant when endocytosis was inhibited by dynasore (Fig. 7B and C). Although it seems possible that cell surface virions might become resistant to pronase by forming 3SC anchors or hemifusing with the cell membrane (46, 53), it is very unlikely that the formation of these linkages would require dynamin. Indeed, cytoplasmic dynamin, which facilitates endocytosis by forming a collar around the necks of coated pits (54), would be topologically excluded from cell surface 3SCs and hemifused virions. Consequently, we infer that most of the pronase-resistant virions after 1- to 4-h incubations at 37°C had been endocytosed by a dynamin-dependent process. Importantly, this endocytic process occurred independently of whether Nt was present or absent during the 37°C incubations of HeLa-CD4/CCR5(Δ18) cultures (Fig. 7B and C). Infectivity assays confirmed that dynasore completely inhibited infection in the cultures incubated at 37°C for 1 h before removing it and adding an excess of TAK779 and that this loss of infectivity was not caused by the cytotoxic effects of dynasore (Fig. 7E). In agreement with these interpretations, 1-h exposures to chlorpromazine, which blocks AP2-dependent clathrin recycling, also strongly inhibited HIV-1 infections (Fig. 7E). Control analyses demonstrated that this loss of infectivity was not caused by the cytotoxic effects of chlorpromazine.
The results presented above imply that a limiting step in infectious HIV-1JRCSF(Ad) entry involves a cellular process of dynamin- and AP2-dependent endocytosis rather than a viral process and that endocytosis ultimately results in inactivation of virions that had not interacted with a functional coreceptor on cell surfaces (see Discussion). Further evidence for a limiting cellular process was obtained by exposing cultures with stably adsorbed HIV-1JRCSF(Ad) to dynasore or chlorpromazine for 1 h. After their removal, HIV-1 infections ensued at a steady rate consistent with the onset of a constitutive cellular process rather than with a burst that would have been expected if virions had accumulated in an arrested preentry state during exposure to these inhibitors (results not shown). The recovery of HIV-1 entry after dynasore removal was similar to the recovery of transferrin endocytosis, which is mediated by clathrin-coated pits (Fig. 7G) (55).
It has previously been suggested that endocytosis may be a default pathway for infectious HIV-1 entry into cells and that it predominates only when the virions are unable to fuse rapidly on cell surfaces due to cellular or viral deficiencies (51, 52, 56). We initially considered this unlikely in our assays because HIV-1JRCSF(Ad) employs the split CCR5 system with a high efficiency similar to that of intact CCR5 usage by HIV-1JRCSF and other natural HIV-1 isolates (e.g., Fig. 1C) and because the adaptive mutations in HIV-1JRCSF(Ad) occur very frequently in natural HIV-1 isolates (Fig. 2). Nevertheless, we tested this by repeating our deconvolution microscopy and infectivity assays using wild-type HIV-1JRCSF and JC.53 cells, which express optimal amounts of CD4 and intact CCR5. As shown in Fig. 7F, HIV-1JRCSF infections were strongly inhibited by dynasore and chlorpromazine. Accordingly, dynasore inhibited the endocytosis of wild-type HIV-1JRCSF virion particles, as seen by deconvolution microscopy (Fig. 7D).
Based on the report of Guo et al. (57), we tested the effects of spinoculation on the viral entry pathway. Virion particle endocytosis was strongly inhibited by dynasore independently of spinoculation. Moreover, the infectivities of wild-type virus on JC.53 cells and of HIV-1JRCSF(Ad) in the Nt-dependent system were inhibited in the absence of spinoculation by at least 80 to 97% (results not shown).
DISCUSSION
The adaptive gp120 mutations S298N and F313L in V3 and N403S in V4 of HIV-1JRCSF(Ad) weaken gp120's constraining hold on the metastable gp41 subunit, thereby enabling low concentrations of damaged CCR5s to efficiently induce the gp41 conformational changes required for entry. The critical adaptive mutation N403S eliminates an N-linked glycan. All observed mutations that eliminate this NDT consensus site for N-linked glycosylation (i.e., N403K, N403S, T405N, and T405A) have similar adaptive effects (6). Multiple lines of evidence support our conclusion that these V3 and V4 adaptive mutations facilitate viral use of damaged CCR5s, including CCR5(Δ18) plus Nt peptide, primarily by reducing gp120's hold on gp41 (5, 6), implying that they specifically destabilize the gp41 native conformation. Accordingly, HIV-1JRCSF(Ad) uses this Nt-dependent infection pathway approximately 100 times more efficiently than HIV-1JRCSF and previously tested wild-type HIV-1 isolates (Fig. 1C) (20) and with an efficiency similar to the efficiencies of intact CCR5 usage by wild-type HIV-1JRCSF and other commonly employed virus isolates. Although the tyrosine-sulfated Nt peptide activates CCR5(Δ18), our results substantiate previous evidence that a chimeric protein, CCR5-IgG, functions only as an entry inhibitor (Fig. 4) (30). Consistent with these conclusions, HIV-1JRCSF(Ad) is more fusogenic than the wild-type virus (5) and is more susceptible to inactivation by soluble CD4 (results not shown) and by CCR5-IgG (Fig. 4).
Using this highly efficient system, we found that Nt-dependent CCR5(Δ18) activation is rapidly reversible, allowing coreceptor activity to be synchronously controlled under physiological conditions. This enabled us to activate entry during brief exposures to Nt and to investigate aspects of entry that were previously obscure. For example, we found that T-20-susceptible 3SCs form slowly and asynchronously within cohorts of virions that had been simultaneously adsorbed onto the cells (Fig. 6A). Moreover, by reducing the durations of Nt exposure, we demonstrated that the T-20-susceptible 3SC intermediates have short lifetimes (Fig. 6B) and that they resolve at a uniform first-order rate that strongly depends on the structure and concentration of the cell surface coreceptor (Fig. 3B). Thus, 3SCs form asynchronously and slowly by a stochastic process but resolve rapidly at a uniform rate that depends on the cell clone employed. Our data also suggest that highly infectious HIV-1JRCSF(Ad) and wild-type HIV-1JRCF virions generally form single rather than redundant fusion complexes with CD4 and coreceptors under the conditions of our assays and that a single T-20 peptide suffices to inactivate the entire entry ensemble (Fig. 6B to D). These interpretations are compatible with recent evidence that gp120/gp41 trimers coalesce into a single cluster on the surfaces of mature HIV-1 virions (58), thereby focusing the virus's entry resources at a single point of cellular contact. Our analyses also provide novel evidence that a critical stochastic step in the viral entry pathway involves endocytosis, which segregates the virions from subsequently added extracellular Nt. We conclude that this reversible peptide-dependent system is a powerful means to investigate pathways of HIV-1 entry and mechanisms of entry inhibitor function. We recently used a variant of this system to show that the neutralizing monoclonal antibody 2G12 binds and neutralizes HIV-1 reversibly by slowing the CCR5-dependent entry steps (38).
Our results strongly suggest that each cell surface virion within a simultaneously adsorbed cohort experiences a transient window of opportunity to successfully infect the cells (Fig. 5) and that this opportunity occurs at different times for each virion. If this window opens while Nt is absent, the virus eventually becomes inactivated, whereas infection can proceed if Nt is present during this time. Consequently, virions leave cell surfaces at the same rate and, consequently, by the same mechanism, regardless of whether Nt is present or absent in the culture medium (Fig. 5). Because membrane fusion requires coreceptor activity, this strongly implies that the pathway for HIV-1 infection does not involve fusion on cell surfaces. Our analyses of this unexpected result strongly suggest that this opportunity is conferred by dynamin- and AP2-dependent endocytosis (Fig. 7), resulting in infection if Nt is present in the endocytosed virion complex or in eventual inactivation if Nt is absent from these endosomes. While these conclusions are compatible with important evidence for endocytic HIV-1 entry of Melikyan and coworkers (46) and others (56, 59), our evidence relies on different methods and additionally indicates that endocytosis ultimately results in virus inactivation if it occurs before coreceptors prime the virions (see below). Our system also employs highly infectious replication-competent viruses.
A unique implication of our investigation is that several steps of CD4 and coreceptor-induced gp41 refolding obligatorily occur before infectious HIV-1 virions leave cell surfaces. For example, a substantial proportion of viable virions on cell surfaces do not form T-20-susceptible 3SC intermediates until many hours after warming the cultures to 37°C (Fig. 6A), yet all the virions become inactivated if high concentrations of the membrane-impermeant T-20 peptide are maintained in the medium (Fig. 3B and 6B to D). Because virion endocytosis occurs randomly and independently of CD4 or coreceptors (Fig. 7A to D) (60) and therefore presumably occurs independently of gp41 refolding, this strongly suggests that any virions endocytosed before they form T-20-susceptible intermediates cannot complete the infection process. Although incorporation of the soluble T-20 peptide into the endosomes could potentially explain these results, this interpretation is incompatible with evidence that this inhibition occurs on cell surfaces (11, 46). Moreover, our data suggest that all or nearly all virions that successfully infect cells resolve their T-20-susceptible 3SCs while on cell surfaces (Fig. 6). This conclusion is principally based on our finding that 3SCs resolve at a uniform rate (consistent with equation 5 for N equal to 1) that depends on the structure and cell surface concentration of the coreceptor (Fig. 3) and that adaptive gp120 mutations dramatically accelerate 3SC resolution (5, 6). Moreover, infectious virions appear to form T-20-susceptible 3SCs substantially before the Nt-dependent cell surface entry steps are completed (Fig. 6A). On the contrary, if escape from T-20 was caused by virion endocytosis rather than by gp41 conformational changes, the lifetimes of T-20-susceptible 3SCs would not be expected to depend on cell surface coreceptor concentrations or on gp120 mutations. Moreover, an endocytic mechanism of escape from T-20 would necessitate that T-20 not enter endosomes, supporting our conclusion presented above that virion infections cannot succeed unless 3SCs previously formed on cell surfaces. Although further investigations of these issues are needed, we conclude that at least one gp41 refolding step required for infection obligatorily occurs on cell surfaces and that final entry occurs exclusively from endosomes. If endocytosis occurs in the absence of Nt or before the requisite cell surface steps are completed, the virus becomes inactivated, presumably by subsequent degradation in lysosomes. Spatially segregated entry steps have previously been reported for avian leukosis and Jaagsiekte sheep oncoretroviruses, in which cases a receptor-induced cell surface priming step is a prerequisite for successful membrane fusion in endosomes (61, 62), and similar cell surface priming is required by other viruses that infect from endosomes (63–65). Thus, HIV-1 entry occurs in an assembly line manner, with requisite steps occurring at different locations. The factors that restrict or favor these HIV-1 entry steps at these distinct locales remain to be determined.
It is notable in this context that adsorbed HIV-1 virions infect efficiently only when gp41 refolding steps occur rapidly (12). This implies that infection occurs in kinetic competition with processes that inactivate virions (38), in agreement with our evidence that HIV-1JRCSF(Ad) infects inefficiently and slowly when the amounts of Nt or coreceptor are low and with our finding that the adaptive mutations increase infection efficiencies and accelerate resolution of 3SCs (Fig. 3) (5). We previously demonstrated that rapid virion dissociation from cells is a predominant competitive process in the absence of polycations and that pretreatment of cultures with DEAE-dextran or other polycations greatly increases viral infectivities by stabilizing virion adsorption (12). Even when adsorption is stabilized, however, infection remains in kinetic competition with an inactivating process(es), occurring slowly and inefficiently when coreceptor concentrations are suboptimal (Fig. 3). Our new evidence strongly suggests that endocytosis leads to inactivation of HIV-1 virions that have not formed T-20-susceptible 3SC intermediates or interacted with active coreceptors at the time of engulfment (Fig. 7), thus implicating endocytosis as a process that is necessary for infection but is also inactivating for virions that undergo cell surface gp41 refolding steps too slowly. This potentially explains the evidence that HIV-1 isolates that infect cells inefficiently and slowly are substantially inactivated in lysosomes but can be partially rescued by adding weak bases that prevent endosome fusion with lysosomes (51, 52). In contrast, because highly infectious HIV-1 isolates were unaffected by these weak bases, it was suggested that they fuse rapidly on cell surfaces and thereby escape lethal endocytosis. We believe that those results are compatible with an obligatory endosomal pathway for infection, with rapid fusion within endosomes competing with delivery to lysosomes. Similarly, the monoclonal antibody 2G12 neutralizes HIV-1 by slowing the coreceptor-dependent steps of entry (38), and coreceptor antagonists slow entry by reducing cell surface concentrations of functional coreceptors (38). This evidence indicates that HIV-1 infection occurs in kinetic competition with processes that inactivate virions and demonstrates that potent entry inhibitors and neutralizing antibodies can reduce HIV-1 infectivities simply by slowing rather than preventing gp41 refolding.
Although wild-type HIV-1JRCSF entry into JC.53 cells could not be dissected using the Nt system, our experiments suggest that it uses the same pathway and mechanisms as Nt-dependent HIV-1JRCSF(Ad) infection of cells expressing CCR5(Δ18). First, HIV-1JRCSF(Ad) employs the Nt-dependent entry system with a high efficiency similar to the efficiencies of intact CCR5 use by HIV-1JRCSF and other commonly studied highly infectious HIV-1 isolates (Fig. 1A, 3A, and 5A). This implies that HIV-1JRCSF(Ad) use of CCR5(Δ18) plus Nt peptide does not proceed on a default pathway employed by weakly infectious viruses (51, 52, 56). The adaptive mutations S298N and F313L and N-glycan loss in V4 at position 403 in HIV-1JRCSF(Ad) are also very common in natural HIV-1 isolates (Fig. 2), consistent with the hypothesis that they modulate natural HIV-1 functions in vivo. Our results also suggest that both wild-type HIV-1JRCSF and HIV-1JRCSF(Ad) generally form nonredundant (N = 1) fusion complexes on the surfaces of susceptible cells under the conditions of our assays and that their T-20-susceptible 3SC intermediates resolve rapidly at uniform rates within 15 min (compare Fig. 6B and D with 6C). Finally, infections by both viruses require clathrin-mediated endocytosis, as indicated by strong inhibition by dynasore and chlorpromazine and by deconvolution microscopy (Fig. 7D and F).
Our conclusion that distinct steps of HIV-1 entry obligatorily occur on cell surfaces and within endosomes implies that this spatial segregation is advantageous for the virus. Generally, it has become accepted that fusion within endosomes is helpful for many viruses because it enables virion cores to efficiently move through the cytoskeletal meshwork into the cell interior (59). We suggest that CD4- and coreceptor-dependent cell surface steps are useful for HIV-1 because they induce 3SCs and hemifusion intermediates that anchor virions onto cells, thereby preventing virion dissociation and committing the virus to cells that have suitable receptors (12). Consequently, these cell surface priming steps would enable virions to graze among cells until they encounter selected targets. Although slow virus diffusion (66) and rapid dissociation (12) can severely limit adsorption in cell cultures, viral grazing between different cells is much more likely in compact tissues in vivo. Indeed, HIV-1 virions congregate in vivo at functionally important sites of strong adhesion, such as follicular dendritic cells and gut lymphocytes (67, 68). The fact that avian leukosis and Jaagsiekte sheep retroviruses also require separate cell surface and endosomal entry steps (61, 62) suggests that this two-step assembly line entry mechanism may occur more widely.
ACKNOWLEDGMENTS
We thank S. Kozak and L. Schwanemann for technical assistance. These experiments were planned and analyzed by E.J.P. and D.K. E.J.P. did all the experiments and collaborated with M.M.G. to produce and perform the CCR5-IgG analyses in Fig. 4. The paper was written by D.K. and E.J.P.
We thank Michael Chapman for his thoughtful and thorough review of the manuscript.
This work was supported by grant R01 CA67358 from the National Institutes of Health.
Footnotes
Published ahead of print 29 January 2014
REFERENCES
- 1.Eckert DM, Kim PS. 2001. Mechanisms of viral membrane fusion and its inhibition. Annu. Rev. Biochem. 70:777–810. 10.1146/annurev.biochem.70.1.777 [DOI] [PubMed] [Google Scholar]
- 2.Melikyan GB, Markosyan RM, Hemmati H, Delmedico MK, Lambert DM, Cohen FS. 2000. Evidence that the transition of HIV-1 gp41 into a six-helix bundle, not the bundle configuration, induces membrane fusion. J. Cell Biol. 151:413–423. 10.1083/jcb.151.2.413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Derdeyn CA, Decker JM, Sfakianos JN, Zhang Z, O'Brien WA, Ratner L, Shaw GM, Hunter E. 2001. Sensitivity of human immunodeficiency virus type 1 to fusion inhibitors targeted to the gp41 first heptad repeat involves distinct regions of gp41 and is consistently modulated by gp120 interactions with the coreceptor. J. Virol. 75:8605–8614. 10.1128/JVI.75.18.8605-8614.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Platt EJ, Durnin JP, Kabat D. 2005. Kinetic factors control efficiencies of cell entry, efficacies of entry inhibitors, and mechanisms of adaptation of human immunodeficiency virus. J. Virol. 79:4347–4356. 10.1128/JVI.79.7.4347-4356.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Platt EJ, Durnin JP, Shinde U, Kabat D. 2007. An allosteric rheostat in HIV-1 gp120 reduces CCR5 stoichiometry required for membrane fusion and overcomes diverse entry limitations. J. Mol. Biol. 374:64–79. 10.1016/j.jmb.2007.09.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Platt EJ, Shea DM, Rose PP, Kabat D. 2005. Variants of human immunodeficiency virus type 1 that efficiently use CCR5 lacking the tyrosine-sulfated amino terminus have adaptive mutations in gp120, including loss of a functional N-glycan. J. Virol. 79:4357–4368. 10.1128/JVI.79.7.4357-4368.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Reeves JD, Gallo SA, Ahmad N, Miamidian JL, Harvey PE, Sharron M, Pohlmann S, Sfakianos JN, Derdeyn CA, Blumenthal R, Hunter E, Doms RW. 2002. Sensitivity of HIV-1 to entry inhibitors correlates with envelope/coreceptor affinity, receptor density, and fusion kinetics. Proc. Natl. Acad. Sci. U. S. A. 99:16249–16254. 10.1073/pnas.252469399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cao J, Park IW, Cooper A, Sodroski J. 1996. Molecular determinants of acute single-cell lysis by human immunodeficiency virus type 1. J. Virol. 70:1340–1354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Moore JP, McKeating JA, Weiss RA, Sattentau QJ. 1990. Dissociation of gp120 from HIV-1 virions induced by soluble CD4. Science 250:1139–1142. 10.1126/science.2251501 [DOI] [PubMed] [Google Scholar]
- 10.Sattentau QJ, Moore JP. 1991. Conformational changes induced in the human immunodeficiency virus envelope glycoprotein by soluble CD4 binding. J. Exp. Med. 174:407–415. 10.1084/jem.174.2.407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Madani N, Hubicki AM, Perdigoto AL, Springer M, Sodroski J. 2007. Inhibition of human immunodeficiency virus envelope glycoprotein-mediated single cell lysis by low-molecular-weight antagonists of viral entry. J. Virol. 81:532–538. 10.1128/JVI.01079-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Platt EJ, Kozak SL, Durnin JP, Hope TJ, Kabat D. 2010. Rapid dissociation of HIV-1 from cultured cells severely limits infectivity assays, causes the inactivation ascribed to entry inhibitors, and masks the inherently high level of infectivity of virions. J. Virol. 84:3106–3110. 10.1128/JVI.01958-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Reeves JD, Lee FH, Miamidian JL, Jabara CB, Juntilla MM, Doms RW. 2005. Enfuvirtide resistance mutations: impact on human immunodeficiency virus envelope function, entry inhibitor sensitivity, and virus neutralization. J. Virol. 79:4991–4999. 10.1128/JVI.79.8.4991-4999.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kuhmann SE, Pugach P, Kunstman KJ, Taylor J, Stanfield RL, Snyder A, Strizki JM, Riley J, Baroudy BM, Wilson IA, Korber BT, Wolinsky SM, Moore JP. 2004. Genetic and phenotypic analyses of human immunodeficiency virus type 1 escape from a small-molecule CCR5 inhibitor. J. Virol. 78:2790–2807. 10.1128/JVI.78.6.2790-2807.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hartley O, Klasse PJ, Sattentau QJ, Moore JP. 2005. V3: HIV's switch-hitter. AIDS Res. Hum. Retroviruses 21:171–189. 10.1089/aid.2005.21.171 [DOI] [PubMed] [Google Scholar]
- 16.Trkola A, Dragic T, Arthos J, Binley JM, Olson WC, Allaway GP, Cheng-Mayer C, Robinson J, Maddon PJ, Moore JP. 1996. CD4-dependent, antibody-sensitive interactions between HIV-1 and its co-receptor CCR-5. Nature 384:184–187. 10.1038/384184a0 [DOI] [PubMed] [Google Scholar]
- 17.Farzan M, Vasilieva N, Schnitzler CE, Chung S, Robinson J, Gerard NP, Gerard C, Choe H, Sodroski J. 2000. A tyrosine-sulfated peptide based on the N terminus of CCR5 interacts with a CD4-enhanced epitope of the HIV-1 gp120 envelope glycoprotein and inhibits HIV-1 entry. J. Biol. Chem. 275:33516–33521. 10.1074/jbc.M007228200 [DOI] [PubMed] [Google Scholar]
- 18.Huang CC, Lam SN, Acharya P, Tang M, Xiang SH, Hussan SS, Stanfield RL, Robinson J, Sodroski J, Wilson IA, Wyatt R, Bewley CA, Kwong PD. 2007. Structures of the CCR5 N terminus and of a tyrosine-sulfated antibody with HIV-1 gp120 and CD4. Science 317:1930–1934. 10.1126/science.1145373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Platt EJ, Kuhmann SE, Rose PP, Kabat D. 2001. Adaptive mutations in the V3 loop of gp120 enhance fusogenicity of human immunodeficiency virus type 1 and enable use of a CCR5 coreceptor that lacks the amino-terminal sulfated region. J. Virol. 75:12266–12278. 10.1128/JVI.75.24.12266-12278.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Farzan M, Chung S, Li W, Vasilieva N, Wright PL, Schnitzler CE, Marchione RJ, Gerard C, Gerard NP, Sodroski J, Choe H. 2002. Tyrosine-sulfated peptides functionally reconstitute a CCR5 variant lacking a critical amino-terminal region. J. Biol. Chem. 277:40397–40402. 10.1074/jbc.M206784200 [DOI] [PubMed] [Google Scholar]
- 21.Farzan M, Mirzabekov T, Kolchinsky P, Wyatt R, Cayabyab M, Gerard NP, Gerard C, Sodroski J, Choe H. 1999. Tyrosine sulfation of the amino terminus of CCR5 facilitates HIV-1 entry. Cell 96:667–676. 10.1016/S0092-8674(00)80577-2 [DOI] [PubMed] [Google Scholar]
- 22.Platt EJ, Wehrly K, Kuhmann SE, Chesebro B, Kabat D. 1998. Effects of CCR5 and CD4 cell surface concentrations on infections by macrophagetropic isolates of human immunodeficiency virus type 1. J. Virol. 72:2855–2864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kabat D, Kozak SL, Wehrly K, Chesebro B. 1994. Differences in CD4 dependence for infectivity of laboratory-adapted and primary patient isolates of human immunodeficiency virus type 1. J. Virol. 68:2570–2577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cosset FL, Takeuchi Y, Battini JL, Weiss RA, Collins MK. 1995. High-titer packaging cells producing recombinant retroviruses resistant to human serum. J. Virol. 69:7430–7436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Baliga CS, van Maanen M, Chastain M, Sutton RE. 2006. Vaccination of mice with replication-defective human immunodeficiency virus induces cellular and humoral immunity and protects against vaccinia virus-gag challenge. Mol. Ther. 14:432–441. 10.1016/j.ymthe.2006.02.021 [DOI] [PubMed] [Google Scholar]
- 26.Kuhmann SE, Platt EJ, Kozak SL, Kabat D. 2000. Cooperation of multiple CCR5 coreceptors is required for infections by human immunodeficiency virus type 1. J. Virol. 74:7005–7015. 10.1128/JVI.74.15.7005-7015.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kuhmann SE, Platt EJ, Kozak SL, Kabat D. 1997. Polymorphisms in the CCR5 genes of African green monkeys and mice implicate specific amino acids in infections by simian and human immunodeficiency viruses. J. Virol. 71:8642–8656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Siciliano SJ, Kuhmann SE, Weng Y, Madani N, Springer MS, Lineberger JE, Danzeisen R, Miller MD, Kavanaugh MP, DeMartino JA, Kabat D. 1999. A critical site in the core of the CCR5 chemokine receptor required for binding and infectivity of human immunodeficiency virus type 1. J. Biol. Chem. 274:1905–1913. 10.1074/jbc.274.4.1905 [DOI] [PubMed] [Google Scholar]
- 29.Page KA, Landau NR, Littman DR. 1990. Construction and use of a human immunodeficiency virus vector for analysis of virus infectivity. J. Virol. 64:5270–5276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dorfman T, Moore MJ, Guth AC, Choe H, Farzan M. 2006. A tyrosine-sulfated peptide derived from the heavy-chain CDR3 region of an HIV-1-neutralizing antibody binds gp120 and inhibits HIV-1 infection. J. Biol. Chem. 281:28529–28535. 10.1074/jbc.M602732200 [DOI] [PubMed] [Google Scholar]
- 31.Choe H, Li W, Wright PL, Vasilieva N, Venturi M, Huang CC, Grundner C, Dorfman T, Zwick MB, Wang L, Rosenberg ES, Kwong PD, Burton DR, Robinson JE, Sodroski JG, Farzan M. 2003. Tyrosine sulfation of human antibodies contributes to recognition of the CCR5 binding region of HIV-1 gp120. Cell 114:161–170. 10.1016/S0092-8674(03)00508-7 [DOI] [PubMed] [Google Scholar]
- 32.Kirschner M, Monrose V, Paluch M, Techodamrongsin N, Rethwilm A, Moore JP. 2006. The production of cleaved, trimeric human immunodeficiency virus type 1 (HIV-1) envelope glycoprotein vaccine antigens and infectious pseudoviruses using linear polyethylenimine as a transfection reagent. Protein Expr. Purif. 48:61–68. 10.1016/j.pep.2006.02.017 [DOI] [PubMed] [Google Scholar]
- 33.Chesebro B, Wehrly K. 1988. Development of a sensitive quantitative focal assay for human immunodeficiency virus infectivity. J. Virol. 62:3779–3788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chesebro B, Wehrly K, Nishio J, Perryman S. 1992. Macrophage-tropic human immunodeficiency virus isolates from different patients exhibit unusual V3 envelope sequence homogeneity in comparison with T-cell-tropic isolates: definition of critical amino acids involved in cell tropism. J. Virol. 66:6547–6554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Baba M, Nishimura O, Kanzaki N, Okamoto M, Sawada H, Iizawa Y, Shiraishi M, Aramaki Y, Okonogi K, Ogawa Y, Meguro K, Fujino M. 1999. A small-molecule, nonpeptide CCR5 antagonist with highly potent and selective anti-HIV-1 activity. Proc. Natl. Acad. Sci. U. S. A. 96:5698–5703. 10.1073/pnas.96.10.5698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Macia E, Ehrlich M, Massol R, Boucrot E, Brunner C, Kirchhausen T. 2006. Dynasore, a cell-permeable inhibitor of dynamin. Dev. Cell 10:839–850. 10.1016/j.devcel.2006.04.002 [DOI] [PubMed] [Google Scholar]
- 37.Wang LH, Rothberg KG, Anderson RG. 1993. Mis-assembly of clathrin lattices on endosomes reveals a regulatory switch for coated pit formation. J. Cell Biol. 123:1107–1117. 10.1083/jcb.123.5.1107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Platt EJ, Gomes MM, Kabat D. 2012. Kinetic mechanism for HIV-1 neutralization by antibody 2G12 entails reversible glycan binding that slows cell entry. Proc. Natl. Acad. Sci. U. S. A. 109:7829–7834. 10.1073/pnas.1109728109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Huang CC, Tang M, Zhang MY, Majeed S, Montabana E, Stanfield RL, Dimitrov DS, Korber B, Sodroski J, Wilson IA, Wyatt R, Kwong PD. 2005. Structure of a V3-containing HIV-1 gp120 core. Science 310:1025–1028. 10.1126/science.1118398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Miyauchi K, Kozlov MM, Melikyan GB. 2009. Early steps of HIV-1 fusion define the sensitivity to inhibitory peptides that block 6-helix bundle formation. PLoS Pathog. 5:e1000585. 10.1371/journal.ppat.1000585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kwon YD, Finzi A, Wu X, Dogo-Isonagie C, Lee LK, Moore LR, Schmidt SD, Stuckey J, Yang Y, Zhou T, Zhu J, Vicic DA, Debnath AK, Shapiro L, Bewley CA, Mascola JR, Sodroski JG, Kwong PD. 2012. Unliganded HIV-1 gp120 core structures assume the CD4-bound conformation with regulation by quaternary interactions and variable loops. Proc. Natl. Acad. Sci. U. S. A. 109:5663–5668. 10.1073/pnas.1112391109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Xiang SH, Finzi A, Pacheco B, Alexander K, Yuan W, Rizzuto C, Huang CC, Kwong PD, Sodroski J. 2010. A V3 loop-dependent gp120 element disrupted by CD4 binding stabilizes the human immunodeficiency virus envelope glycoprotein trimer. J. Virol. 84:3147–3161. 10.1128/JVI.02587-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mao Y, Wang L, Gu C, Herschhorn A, Xiang SH, Haim H, Yang X, Sodroski J. 2012. Subunit organization of the membrane-bound HIV-1 envelope glycoprotein trimer. Nat. Struct. Mol. Biol. 19:893–899. 10.1038/nsmb.2351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Julien JP, Cupo A, Sok D, Stanfield RL, Lyumkis D, Deller MC, Klasse PJ, Burton DR, Sanders RW, Moore JP, Ward AB, Wilson IA. 2013. Crystal structure of a soluble cleaved HIV-1 envelope trimer. Science 342:1477–1483. 10.1126/science.1245625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lyumkis D, Julien JP, de Val N, Cupo A, Potter CS, Klasse PJ, Burton DR, Sanders RW, Moore JP, Carragher B, Wilson IA, Ward AB. 2013. Cryo-EM structure of a fully glycosylated soluble cleaved HIV-1 envelope trimer. Science 342:1484–1490. 10.1126/science.1245627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Miyauchi K, Kim Y, Latinovic O, Morozov V, Melikyan GB. 2009. HIV enters cells via endocytosis and dynamin-dependent fusion with endosomes. Cell 137:433–444. 10.1016/j.cell.2009.02.046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chiang JJ, Gardner MR, Quinlan BD, Dorfman T, Choe H, Farzan M. 2012. Enhanced recognition and neutralization of HIV-1 by antibody-derived CCR5-mimetic peptide variants. J. Virol. 86:12417–12421. 10.1128/JVI.00967-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Smith-Franklin BA, Keele BF, Tew JG, Gartner S, Szakal AK, Estes JD, Thacker TC, Burton GF. 2002. Follicular dendritic cells and the persistence of HIV infectivity: the role of antibodies and Fcgamma receptors. J. Immunol. 168:2408–2414 http://www.jimmunol.org/content/168/5/2408 [DOI] [PubMed] [Google Scholar]
- 49.Furuta RA, Wild CT, Weng Y, Weiss CD. 1998. Capture of an early fusion-active conformation of HIV-1 gp41. Nat. Struct. Biol. 5:276–279. 10.1038/nsb0498-276 [DOI] [PubMed] [Google Scholar]
- 50.Daecke J, Fackler OT, Dittmar MT, Krausslich HG. 2005. Involvement of clathrin-mediated endocytosis in human immunodeficiency virus type 1 entry. J. Virol. 79:1581–1594. 10.1128/JVI.79.3.1581-1594.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Fredericksen BL, Wei BL, Yao J, Luo T, Garcia JV. 2002. Inhibition of endosomal/lysosomal degradation increases the infectivity of human immunodeficiency virus. J. Virol. 76:11440–11446. 10.1128/JVI.76.22.11440-11446.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Schaeffer E, Soros VB, Greene WC. 2004. Compensatory link between fusion and endocytosis of human immunodeficiency virus type 1 in human CD4 T lymphocytes. J. Virol. 78:1375–1383. 10.1128/JVI.78.3.1375-1383.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.de la Vega M, Marin M, Kondo N, Miyauchi K, Kim Y, Epand RF, Epand RM, Melikyan GB. 2011. Inhibition of HIV-1 endocytosis allows lipid mixing at the plasma membrane, but not complete fusion. Retrovirology 8:99. 10.1186/1742-4690-8-99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Schmid SL, Frolov VA. 2011. Dynamin: functional design of a membrane fission catalyst. Annu. Rev. Cell Dev. Biol. 27:79–105. 10.1146/annurev-cellbio-100109-104016 [DOI] [PubMed] [Google Scholar]
- 55.Mayle KM, Le AM, Kamei DT. 2012. The intracellular trafficking pathway of transferrin. Biochim. Biophys. Acta 1820:264–281. 10.1016/j.bbagen.2011.09.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Permanyer M, Ballana E, Este JA. 2010. Endocytosis of HIV: anything goes. Trends Microbiol. 18:543–551. 10.1016/j.tim.2010.09.003 [DOI] [PubMed] [Google Scholar]
- 57.Guo J, Wang W, Yu D, Wu Y. 2011. Spinoculation triggers dynamic actin and cofilin activity that facilitates HIV-1 infection of transformed and resting CD4 T cells. J. Virol. 85:9824–9833. 10.1128/JVI.05170-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Chojnacki J, Staudt T, Glass B, Bingen P, Engelhardt J, Anders M, Schneider J, Muller B, Hell SW, Krausslich HG. 2012. Maturation-dependent HIV-1 surface protein redistribution revealed by fluorescence nanoscopy. Science 338:524–528. 10.1126/science.1226359 [DOI] [PubMed] [Google Scholar]
- 59.Mercer J, Schelhaas M, Helenius A. 2010. Virus entry by endocytosis. Annu. Rev. Biochem. 79:803–833. 10.1146/annurev-biochem-060208-104626 [DOI] [PubMed] [Google Scholar]
- 60.Marechal V, Clavel F, Heard JM, Schwartz O. 1998. Cytosolic Gag p24 as an index of productive entry of human immunodeficiency virus type 1. J. Virol. 72:2208–2212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Cote M, Zheng YM, Liu SL. 2009. Receptor binding and low pH coactivate oncogenic retrovirus envelope-mediated fusion. J. Virol. 83:11447–11455. 10.1128/JVI.00748-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Mothes W, Boerger AL, Narayan S, Cunningham JM, Young JA. 2000. Retroviral entry mediated by receptor priming and low pH triggering of an envelope glycoprotein. Cell 103:679–689. 10.1016/S0092-8674(00)00170-7 [DOI] [PubMed] [Google Scholar]
- 63.Coyne CB, Bergelson JM. 2006. Virus-induced Abl and Fyn kinase signals permit coxsackievirus entry through epithelial tight junctions. Cell 124:119–131. 10.1016/j.cell.2005.10.035 [DOI] [PubMed] [Google Scholar]
- 64.Helle F, Dubuisson J. 2008. Hepatitis C virus entry into host cells. Cell. Mol. Life Sci. 65:100–112. 10.1007/s00018-007-7291-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Nemerow GR. 2000. Cell receptors involved in adenovirus entry. Virology 274:1–4. 10.1006/viro.2000.0468 [DOI] [PubMed] [Google Scholar]
- 66.Allison AC, Valentine RC. 1960. Virus particle adsorption. III. Adsorption of viruses by cell monolayers and effects of some variables on adsorption. Biochim. Biophys. Acta 40:400–410 [DOI] [PubMed] [Google Scholar]
- 67.Arthos J, Cicala C, Martinelli E, Macleod K, Van Ryk D, Wei D, Xiao Z, Veenstra TD, Conrad TP, Lempicki RA, McLaughlin S, Pascuccio M, Gopaul R, McNally J, Cruz CC, Censoplano N, Chung E, Reitano KN, Kottilil S, Goode DJ, Fauci AS. 2008. HIV-1 envelope protein binds to and signals through integrin alpha4beta7, the gut mucosal homing receptor for peripheral T cells. Nat. Immunol. 9:301–309. 10.1038/ni1566 [DOI] [PubMed] [Google Scholar]
- 68.Geijtenbeek TB, Kwon DS, Torensma R, van Vliet SJ, van Duijnhoven GC, Middel J, Cornelissen IL, Nottet HS, KewalRamani VN, Littman DR, Figdor CG, van Kooyk Y. 2000. DC-SIGN, a dendritic cell-specific HIV-1-binding protein that enhances trans-infection of T cells. Cell 100:587–597. 10.1016/S0092-8674(00)80694-7 [DOI] [PubMed] [Google Scholar]
- 69.Kwong JA, Dorfman T, Quinlan BD, Chiang JJ, Ahmed AA, Choe H, Farzan M. 2011. A tyrosine-sulfated CCR5-mimetic peptide promotes conformational transitions in the HIV-1 envelope glycoprotein. J. Virol. 85:7563–7571. 10.1128/JVI.00630-11 [DOI] [PMC free article] [PubMed] [Google Scholar]