ABSTRACT
Candida albicans is a fungus that colonizes oral cavity surfaces, the gut, and the genital tract. Streptococcus gordonii is a ubiquitous oral bacterium that has been shown to form biofilm communities with C. albicans. Formation of dual-species S. gordonii-C. albicans biofilm communities involves interaction of the S. gordonii SspB protein with the Als3 protein on the hyphal filament surface of C. albicans. Mannoproteins comprise a major component of the C. albicans cell wall, and in this study we sought to determine if mannosylation in cell wall biogenesis of C. albicans was necessary for hyphal adhesin functions associated with interkingdom biofilm development. A C. albicans mnt1Δ mnt2Δ mutant, with deleted α-1,2-mannosyltransferase genes and thus defective in O-mannosylation, was abrogated in biofilm formation under various growth conditions and produced hyphal filaments that were not recognized by S. gordonii. Cell wall proteomes of hypha-forming mnt1Δ mnt2Δ mutant cells showed growth medium-dependent alterations, compared to findings for the wild type, in a range of protein components, including Als1, Als3, Rbt1, Scw1, and Sap9. Hyphal filaments formed by mnt1Δ mnt2Δ mutant cells, unlike wild-type hyphae, did not interact with C. albicans Als3 or Hwp1 partner cell wall proteins or with S. gordonii SspB partner adhesin, suggesting defective functionality of adhesins on the mnt1Δ mnt2Δ mutant. These observations imply that early stage O-mannosylation is critical for activation of hyphal adhesin functions required for biofilm formation, recognition by bacteria such as S. gordonii, and microbial community development.
IMPORTANCE
In the human mouth, microorganisms form communities known as biofilms that adhere to the surfaces present. Candida albicans is a fungus that is often found within these biofilms. We have focused on the mechanisms by which C. albicans becomes incorporated into communities containing bacteria, such as Streptococcus. We find that impairment of early stage addition of mannose sugars to C. albicans hyphal filament proteins deleteriously affects their subsequent performance in mediating formation of polymicrobial biofilms. Our analyses provide new understanding of the way that microbial communities develop, and of potential means to control C. albicans infections.
INTRODUCTION
Candida albicans is an opportunistic fungal pathogen found in the microbiota of the gut, oral cavity, and genital tract (1). In healthy individuals, C. albicans growth is kept in check by a combination of the resident microbiota and both the innate and acquired immune systems. Overgrowth of C. albicans, associated with administration of broad-spectrum antibiotics or immune dysfunction, may lead to superficial infections, such as oropharyngeal candidiasis (thrush) and vulvovaginal candidiasis (vaginal candidiasis). Systemic infections have become increasingly prevalent because more individuals are immunocompromised. C. albicans biofilm infections are common in patients with urinary or intravascular catheters or artificial joints or voice boxes (2). Indeed, C. albicans is associated with >90% of human oral fungal diseases and is becoming an increasingly serious problem in hospital infections (3).
C. albicans forms biofilms on a range of surfaces in vivo (4–6) and is well adapted to conditions in the oral cavity. Biofilm formation occurs in three or four phases, starting first with the deposition of yeast-form cells onto the substratum and then the formation of hyphae (7). The latter are critical for biofilm formation because mutants that do not form hyphae under biofilm-promoting conditions are unable to form robust biofilms (8). Biofilm formation is under genetic regulation by the transcription factors Bcr1 (9, 10), Efg1 (11), and Ace2 (12), among others. In particular, Bcr1 is known to control expression of cell surface proteins, such as Als3 and Hwp1 (13), that are involved in adherence of hyphal filaments to each other (14) in the building of biofilms. A number of techniques may be utilized to grow C. albicans biofilms in vitro on surfaces of acrylic, rubber disks, catheter strips, or glass coverslips (15). Static biofilm models provide good in vitro data but do not take into account conditions in vivo, where C. albicans biofilms growing on medical implants, such as catheters, prosthetic joints and heart valves, would be exposed to flow forces and replenishment of nutrients from body fluids. In addition, biofilms formed under flow have been shown to develop architectures different from those of static biofilms grown under the same conditions (16).
The cell surface of C. albicans is the major point of contact between fungus and host, along with its constituent microbiota, and provides adhesive and immunomodulatory functions (17). The outer cell wall layer is comprised principally of mannoproteins which are embedded in a polysaccharide skeleton provided by β-(1,3)- and β-(1,6)-linked glucan chains and covalently linked chitin (18). The protein and carbohydrate components of the outer wall layers have both been implicated in adhesion to host surfaces (19) and in biofilm formation (20). It is predicted that ~115 proteins encoded by the C. albicans genome are glycosylphosphatidylinositol (GPI)-modified proteins (21). Among these are adhesins, such as Hwp1, Rbt1, Eap1, and the Als family of proteins (22), some of which have established roles in biofilm formation (13, 23, 24) and in interactions with host tissues (25). Glycosylation of these and other cell wall proteins is thought to be important for their functions (26). The N-linked glycans carry a conserved core structure and highly branched outer mannose chains (27). The O-linked glycans are, conversely, linear oligosaccharides of one to five α-1,2-linked mannose residues attached to serine or threonine residues (28). The N- and O-glycosylations are completed in the Golgi apparatus, mediated by a set of multifunctional mannosyltransferases (29). Mnt1 and Mnt2 are partially redundant α-1,2-mannosyltransferases that catalyze the addition of the second and third mannose residues in an O-linked mannose pentamer (30). The Mnt1 enzyme may also add the fourth and fifth mannose residues (31). C. albicans mutants deficient in the Mnt1 and Mnt2 proteins are modified in cell wall structure (30, 32), diminished in adherence to human buccal epithelial cells (28), and attenuated in virulence (33). In addition, mnt1Δ, mnt2Δ, and mnt1Δ mnt2Δ mutants form hyphae that are hypersensitive to killing by Pseudomonas aeruginosa (34).
In polymicrobial infections (35), C. albicans has been found in close association with P. aeruginosa (36), Staphylococcus aureus (37), and oral streptococci in denture- and mucosa-related diseases (38). The interactions of oral streptococci, such as Streptococcus gordonii, with C. albicans are proposed to facilitate oral carriage and persistence of C. albicans in mixed-species biofilms on natural or prosthetic surfaces (39, 40). The cell surface protein SspB expressed on S. gordonii is responsible at least in part for mediating adherence to C. albicans hyphal filaments (41, 42). S. gordonii appears to promote hyphal filament formation by C. albicans (42, 43), in contrast to P. aeruginosa and Salmonella enterica serovar Typhimurium, which kill hyphae (44, 45). The aims of this work were to determine if C. albicans cell wall O-mannosylation was necessary for interactions of hyphae with S. gordonii and for formation of dual-species biofilms.
RESULTS
Mannosyltransferase Mnt1 is necessary for C. albicans to bind streptococci.
Previous work has demonstrated that S. gordonii cells adhered to hyphae formed by C. albicans SC5314 (43). In the present study, C. albicans CAI4+Clp10 has been designated wild type, this being a Ura3+ reintegrant generated from C. albicans CAI4 to control for any effects of ura3 gene deletion and transposition in mutagenesis (46). The strain behaved identically to SC5314, from which it was initially derived (see Table S1 in the supplemental material). Hyphal filament formation by the wild-type strain was induced in planktonic phase under our standard conditions with glucose (YPT [1× Difco yeast nitrogen base, 20 mM NaH2PO4–H3PO4 buffer, pH 7.0, and 0.1% Bacto-tryptone]-Glc). Hyphae were shown to avidly bind S. gordonii DL1 cells along their lengths (Fig. 1A). A homozygous mnt1Δ mutant, deficient in production of α-1,2-mannosyltransferase (30), which adds the second mannose residue to growing O-linked glycan chains (30), showed reduced binding of S. gordonii (Fig. 1B). Hyphae formed by a homozygous mnt2Δ mutant were not noticeably impaired in streptococcal cell attachment (Fig. 1C). In the homozygous double mutant strain UB1933 mnt1Δ mnt2Δ, hyphal filaments were abrogated in binding S. gordonii (Fig. 1D). Reintroduction of MNT1 into the double mutant resulted in restoration of S. gordonii attachment (Fig. 1E), while reintroduction of MNT2 did not (Fig. 1F). These results suggested that expression of MNT1 was essential in order to provide hyphal cell wall receptors for adherence of S. gordonii to C. albicans.
FIG 1 .
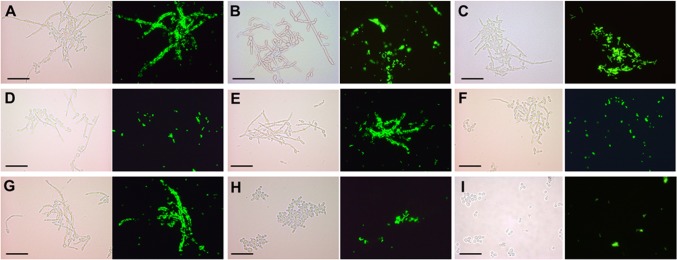
Interactions of S. gordonii DL1 cells with C. albicans wild-type or glycosylation-deficient mutants induced to form hyphae in YPT-glucose medium in suspension culture (planktonic conditions). Bacteria were fluorescently labeled with fluorescein isothiocyanate (FITC) and incubated with hypha-forming cells of C. albicans in YPT-Glc medium for 1 h at 37°C with gentle agitation. Samples (10 µl) were then removed and placed on glass microscope slides, coverslips were applied, and cells were visualized with a Leica microscope under phase contrast or fluorescence at magnification ×400. The panels each show a light microscopic image and the corresponding fluorescence image. (A) C. albicans wild type; (B) mnt1Δ mutant; (C) mnt2Δ strain; (D) mnt1Δ mnt2Δ strain; (E) mnt1Δ mnt2Δ + MNT1 strain; (F) mnt1Δ mnt2Δ + MNT2 strain; (G) mnn4Δ strain; (H) och1Δ strain; (I) Saccharomyces cerevisiae BY4742 (negative control). Scale bar = 50 µm.
We also tested mnn4Δ and och1Δ N-linked glycosylation-impaired mutants (47, 48) for the ability to recognize S. gordonii. Deficiency in Mnn4, which normally regulates mannosyl phosphorylation of N- and O-linked glycans, had no effect on S. gordonii binding to hyphae (Fig. 1G). The mutant strain UB1939 och1Δ (48), deficient in production of α-1,6-mannosyltransferase, which initiates N-glycan outer chain branch addition, did not support adherence of S. gordonii (Fig. 1H). However, this mutant had a highly pleiotropic phenotype, including being impaired in growth rate and hypha production. Under the conditions of these experiments, Saccharomyces cerevisiae cells did not bind S. gordonii (Fig. 1I).
Effect of growth medium on development of biofilms.
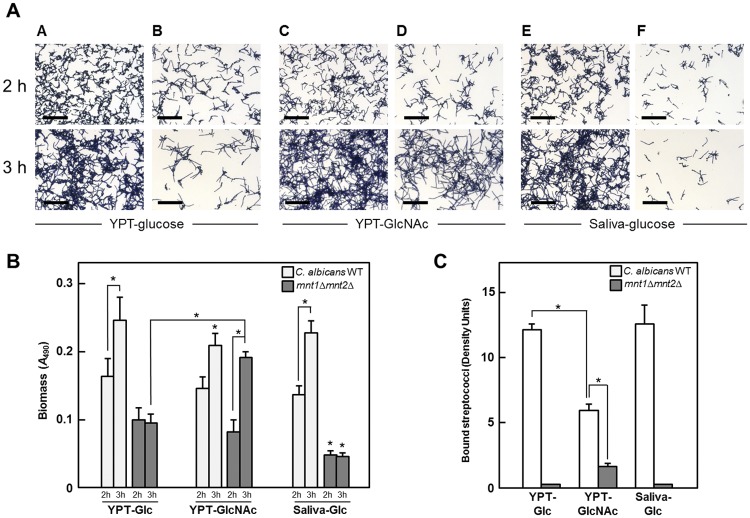
To investigate in more detail the role of O-mannosylation in biofilm-forming and S. gordonii-binding properties of C. albicans, we compared phenotypes of the wild type and the mnt1Δ mnt2Δ double mutant under different growth conditions. In biofilm studies with YPT-Glc medium, a high proportion of deposited C. albicans wild-type cells had formed hyphal filaments at 2 h (Fig. 2A). After a 3-h incubation, an intensified network of hyphae was apparent, with a concomitant increase in biofilm biomass (Fig. 2B). Although at 2 h, fewer mnt1Δ mnt2Δ mutant cells were associated with the substratum (Fig. 2A, column B), a proportion of cells similar to that for the wild type had formed hyphae. However, after 2 h, the mutant cells appeared to become arrested in biofilm formation (Fig. 2A, column B), and no further significant increase in biomass had occurred at 3 h (Fig. 2B). When N-acetyl-d-glucosamine (GlcNAc), a known inducer of hypha formation (49), was substituted for glucose, the previous inability of the mnt1Δ mnt2Δ mutant to further develop a biofilm and increased biomass over the 2- to 3-h period was reversed (Fig. 2A, columns C and D, and 2B). The numbers of cells forming hyphae at 3 h in YPT-GlcNAc were similar (65% ± 6.0%) for the wild type and double mutant (Fig. 2A). Fewer S. gordonii cells bound to wild-type hyphae in YPT-GlcNAc than in YPT-Glc (Fig. 2C). This we attributed to a deficiency in metabolism of GlcNAc by S. gordonii. We also utilized 10% saliva containing 0.01% glucose (saliva-Glc) as a growth medium, and in this medium, the C. albicans wild type showed patterns of biofilm formation and biomass production similar to those with YPT-Glc (Fig. 2A, column E, and 2B). With saliva-Glc medium, the mnt1Δ mnt2Δ mutant formed only sparse biofilms (Fig. 2A, column F) and showed no increase in biomass after 2 h (Fig. 2B).
FIG 2 .
Effects of different hyphal filament induction media on biofilm formation and intermicrobial adherence by C. albicans wild type and mnt1Δ mnt2Δ mutant strains. (A) Biofilms were grown on saliva-coated coverslips (as described in Materials and Methods) for 2 h or 3 h in YPT-Glc, YPT-GlcNAc, or saliva-Glc medium, as indicated, and the cells were then stained with crystal violet and visualized by light microscopy. Biofilms of the wild-type strain showed increased density and extent of hyphal filament formation over 2 to 3 h (columns A, C, and E). Between 2 h and 3 h, the biofilms formed by the mnt1Δ mnt2Δ mutant in YPT-Glc (column B) and saliva-Glc (column F) ceased to develop further, while biofilms in YPT-GlcNAc continued to grow (column D). Bar = 50 µm. (B) corresponding biofilm biomass values for panel A images (*, P < 0.05; n = 3). (C) Extent of FITC-labeled S. gordonii DL1 cells binding to hyphal filaments formed by wild-type or mnt1Δ mnt2Δ cells after a 3-h incubation in suspension cultures in the three different induction media. Density units of bound streptococci were calculated from image capture software measurements, as described in Materials and Methods (*, P < 0.05; n = 3).
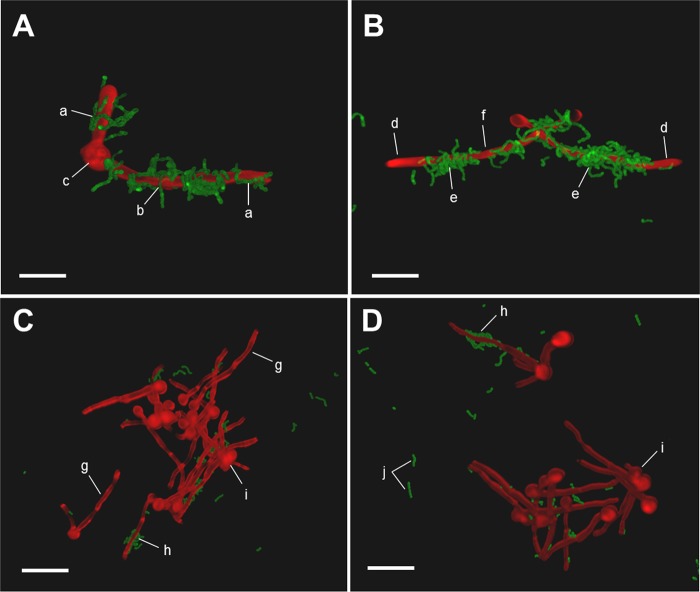
Although the presence of GlcNAc rescued the mnt1Δ mnt2Δ mutant from deficiency in biofilm production in YPT-Glc medium, the ability of these double mutant hyphae to bind S. gordonii was not restored to wild-type levels (Fig. 2C). Confocal scanning laser microscopy (CSLM) images showed that streptococci in pairs or short chains interacted avidly with hyphae formed by wild-type C. albicans (Fig. 3A and B), accumulating as localized clusters of cells but tending not to bind to the hyphal tip regions (Fig. 3B). Conversely, S. gordonii cells bound only sparsely to hyphae produced by the mnt1Δ mnt2Δ mutant (Fig. 3C), although some hyphal filaments seemed to support localized S. gordonii attachment (Fig. 3D).
FIG 3 .
CSLM images of S. gordonii cells interacting with hyphal filaments formed by C. albicans wild-type or mnt1Δ mnt2Δ mutant strains in YPT-GlcNAc suspension culture. S. gordonii DL1 cells (green) expressing GFP (Table 1) were incubated for 1 h at 37°C with filament-forming C. albicans cells stained with calcofluor white (images recolored red). C. albicans wild type (A and B) or mnt1Δ mnt2Δ mutant (C and D) is shown. (A) short chains of streptococci may be seen aligning along the hyphal filaments (a) or bound to the filament by one end of a streptococcal cell chain (b) but not associated with yeast morphology mother cells (c). (B) the tips of hyphal filaments and their adjacent regions were not usually seen to bind S. gordonii cells (d); some parts of filaments were heavily populated by streptococcal communities (e), while other regions were not bound by streptococci (f). (C and D) Hyphal filaments of the mnt1Δ mnt2Δ mutant were sometimes unevenly stained with calcofluor white (g) and were generally not recognized by S. gordonii (g), but some individual filaments among the population showed low levels of bacterial cell binding (h). The mnt1Δ mnt2Δ mutant mother cells often emanated multiple hyphal filaments from the one cell (i). Nonadhered streptococci (j) were clearly visible in the mnt1Δ mnt2Δ strain preparations. Bar = 20 µm.
Effect of mnt1Δ and mnt2Δ mutations on formation of monospecies or dual-species biofilms.
In monospecies biofilms, C. albicans wild-type cells formed a confluent mat of blastospores and hyphae after 6 h at 37°C in YPT-Glc or YPT-GlcNAc growth conditions (Fig. 4). When saliva-Glc was utilized to grow C. albicans biofilms, aggregates or clusters of cells were visible and consequently overall coverage of the substratum was lower (Fig. 4). Saliva-Glc medium did not support such extensive hyphal filament and biofilm formation. The mnt1Δ mnt2Δ mutant cells were unable to form biofilms in YPT-Glc or saliva-Glc medium (Fig. 4). However, the deleterious effects of the mnt1Δ and mnt2Δ mutations on hypha formation and biofilm formation in YPT-Glc and saliva-Glc media were not evident in YPT-GlcNAc medium (Fig. 4). Although the mnt1Δ mnt2Δ mutant biofilms still showed reduced coverage compared to findings for the wild type (Fig. 4), there was extensive hyphal filament formation (more detail can be seen in Video S1 and Video S2 in the supplemental material).
FIG 4 .
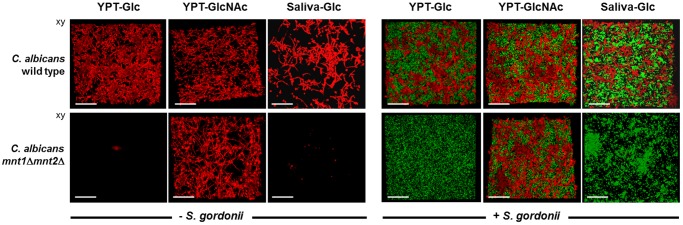
CSLM images of biofilms formed by C. albicans wild type or mnt1Δ mnt2Δ mutant strains in the absence (monospecies) or presence (dual species) of S. gordonii DL1. Monospecies or dual-species biofilms were prepared as described in Materials and Methods and were grown for 6 h at 37°C in the three different growth media indicated. The mnt1Δ mnt2Δ mutant was unable to form monospecies or dual-species biofilms in YPT-Glc or saliva-Glc but produced robust monospecies and dual-species biofilms in YPT-GlcNAc. Bar = 50 µm.
C. albicans wild-type cells formed dual-species biofilms with S. gordonii under the three different growth conditions, with more abundant growth and hypha formation in YPT-GlcNAc (Fig. 4). In YPT-Glc medium, S. gordonii cells formed an evenly distributed biofilm over the substratum surface between the deposited C. albicans cells, and hyphae appeared integrated within the streptococcal cell community (Fig. 4). More extensive C. albicans hypha formation in YPT-GlcNAc medium was accompanied by hyphae appearing to emanate from the S. gordonii community. In saliva-Glc medium, both C. albicans and S. gordonii formed patches of growth (Fig. 4) associated with saliva-mediated aggregation of the microorganisms. In dual-species biofilm experiments, the mnt1Δ mnt2Δ mutant was unable to form biofilms in YPT-Glc or saliva-Glc in the presence of S. gordonii (Fig. 4). In YPT-GlcNAc medium, a dual-species biofilm with S. gordonii was formed (Fig. 4), similar in architecture to the monospecies biofilm. Taken collectively with the results shown in Fig. 2, it is evident that the mnt1Δ mnt2Δ deletions result in the following: (i) reduced levels of adhesion to a salivary pellicle substratum, (ii) a depressed rate of hyphal filament extension, and (iii) a curtailed ability (in YPT-Glc or saliva-Glc medium) to form a robust biofilm.
Architecture of monospecies or dual-species biofilms.
We then investigated in finer detail the architecture of C. albicans and S. gordonii in mono- or dual-species biofilms. In YPT-Glc biofilms, C. albicans was present as a mixture of blastospores and filaments integrated within and on top of a dense layer of streptococci (see Fig. S1A in the supplemental material). A vertical section (see Fig. S1B) showed C. albicans attached to regions of the pellicle substratum and to the streptococcal layer. In the presence of YPT-GlcNAc, C. albicans formed a more robust and filamentous biofilm (see Fig. S1, center panels) either alone (see panel E) or with S. gordonii (see panel D). The S. gordonii component of the dual-species biofilm was much less compact (panel C), and S. gordonii cells were more clearly interdigitated with the extending hyphal filaments. In saliva-Glc medium, a highly integrated dual-species biofilm was produced (see Fig. S1), with streptococci and hyphal filaments extending up to 115 µm from the substratum. Both of the microbial components grew more luxuriantly under these conditions, which was especially apparent when comparing C. albicans dual- and monospecies biofilms (see Fig. S1D and E).
Biofilm formation under flow.
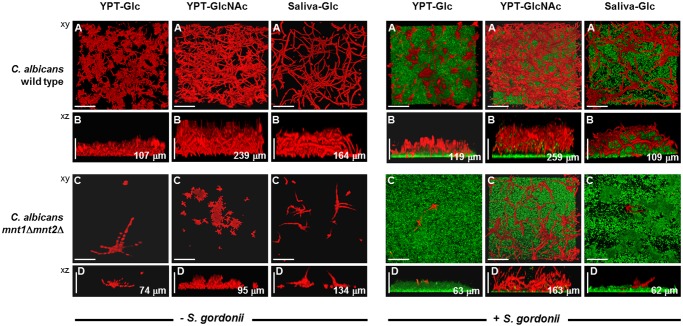
Under flow conditions, which might more closely mimic those in the human oral cavity, there were some remarkable changes in morphology and composition of single- or dual-species biofilms. C. albicans monospecies biofilms that formed in YPT-Glc under flow contained more hyphae than when grown under nonflow conditions, and hyphae extended further from the surface (107 µm) (Fig. 5B) than under nonflow (73-80 µm) (see Fig. S1D and E in the supplemental material). C. albicans wild-type biofilms formed in YPT-GlcNAc produced hyphal filaments that protruded far out into the environment (to 239 µm), giving a brush-like appearance (Fig. 5B; see also Video S3). In saliva-Glc medium, extensive hypha formation was also observed, but the hyphae tended to lie flatter across the surface of the biofilm (Fig. 5B). Conversely, the mnt1Δ mnt2Δ mutant was unable to form robust biofilms under flow conditions, even in YPT-GlcNAc (Fig. 5C and D; see also Video S4).
FIG 5 .
CSLM images of mono- or dual-species biofilms formed under flow conditions by the C. albicans wild type or mnt1Δ mnt2Δ mutant in three different growth media as indicated. Biofilms were prepared as described in Materials and Methods and grown for 6 h at 37°C at a medium flow rate of 6 ml/h. In the left-side group of 12 panels, monospecies biofilms of C. albicans wild-type and mnt1Δ mnt2Δ mutant strains are shown; the right-side group of 12 panels shows dual-species biofilms with S. gordonii. The image at the top of each group of 6 is a representative xy stack of the biofilm assembled from the top down (A and C). The lower image in each column (B and D) is the corresponding xz image showing organization and thickness (depth) of the biofilm. Red, C. albicans; green, S. gordonii. The significant structural and architectural differences associated with the different growth conditions employed are described in the text. The values shown in μm are average thickness (depth) measurements calculated from across the sections of the biofilms shown. Bar = 100 µm.
In dual-species biofilms, patterns of hypha formation by C. albicans wild-type cells were observed that were similar to those of monospecies biofilms (see Video S5 in the supplemental material). Hyphae formed by the C. albicans wild type in saliva-Glc were less protruding and lay flatter over the S. gordonii (Fig. 5B). The presence of S. gordonii did not promote incorporation of mnt1Δ mnt2Δ mutant cells into the flow biofilm except in YPT-GlcNAc medium (Fig. 5). In this medium, the presence of S. gordonii stimulated hypha formation by the mnt1Δ mnt2Δ mutant (Fig. 5C; see also Video S6), suggesting that metabolic signaling between these two strains was occurring.
These interactions under flow are shown in greater detail in Fig. S2 in the supplemental material, in which the dual fluorescence channels are separated. Hyphae produced by the C. albicans wild type in YPT-Glc medium grew within, and protruded through the S. gordonii biofilm (see Fig. S2B in the supplemental material). In YPT-GlcNAc, hyphal filaments extended into the environment by at least 250 µm and streptococcal cells were seen to be associated with the hyphae through the depth of the biofilm (see Fig. S2B and C and Video S5). In saliva-Glc medium, hyphal filaments tended to grow within and across the surface of the S. gordonii biofilm (see Fig. S2B, C, and D), the biofilms being more compact than the corresponding monospecies C. albicans flow biofilm (see Fig. S2E). The mnt1Δ mnt2Δ mutant formed a dual-species flow biofilm with S. gordonii in YPT-GlcNAc medium (see Fig. S2). However, the S. gordonii component of the biofilm appeared to be reduced compared to that for the wild type (see Video S6 for greater detail). Taken collectively, these results show that Mnt1/Mnt2 deficiency can severely restrict C. albicans biofilm development but that this restriction can be at least partly negated in the presence of GlcNAc. Importantly, C. albicans wild-type monospecies biofilms or dual-species biofilms with S. gordonii appeared to be more substantive and vigorous when formed under flow conditions. This suggests that suppressing factors under nonflow conditions or shear forces under flow may play a role in modulating biofilm development.
Expression of C. albicans Als3.
We have shown that a major adhesin for S. gordonii on the hyphal cell surface is Als3 (42), a filament-specific glycophosphatidylinositol (GPI)-modified cell wall protein with multiple adhesin functions (50, 51). Therefore, a possible explanation for the inability of mnt1Δ mnt2Δ mutant hyphae to bind S. gordonii was that expression of Als3 was affected. To test this, we reacted hyphal filaments formed by wild-type or mnt1Δ mnt2Δ mutant cells after a 3-h incubation in YPT-GlcNAc with a monoclonal antibody specific to an epitope within the N-terminal region of Als3 (52). Immunofluorescence microscopy showed that hyphal filaments of the wild type expressed Als3 evenly along their lengths (Fig. 6A and B). However, hyphal filaments formed by the mnt1Δ mnt2Δ mutant cells also reacted with the antibody along their lengths (Fig. 6C and D), with no evidence for any differences in distribution or intensity of antibody reactivity compared to findings for the wild type.
FIG 6 .
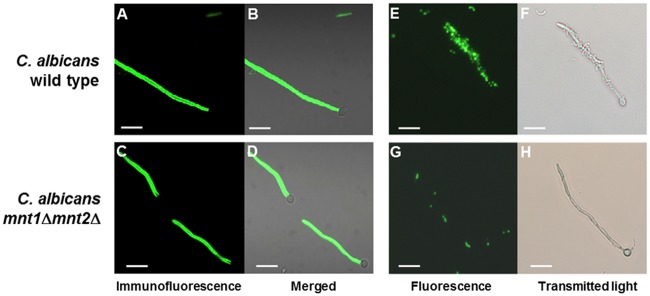
Hypha-forming cells of the C. albicans wild type or mnt1Δ mnt2Δ mutant immunolabeled with monoclonal antibody to the hyphal cell wall protein Als3 or reacted with L. lactis cells expressing SspB. C. albicans cells were induced to form hyphae in YPT-GlcNAc medium suspension culture, as described in Materials and Methods, for 3 h at 37°C, harvested by centrifugation, and then incubated with anti-Als3 MAb 3-A5 (52). (A to D) Antibody binding was detected with FITC-conjugated goat anti-mouse F(ab)2 fragment-specific antibody, and wet mounts were observed microscopically by phase contrast and fluorescence (52). (E and F) Light and corresponding fluorescence microscopic images of FITC-labeled L. lactis cells expressing SspB binding to hyphae. (A, B, E, and F) C. albicans wild type. (C, D, G, and H) mnt1Δ mnt2Δ mutant. Bar = 20 µm.
To investigate the functionality of Als3, hypha-forming cells of the wild type or mutant were incubated with fluorescently labeled cells of Lactococcus lactis expressing the S. gordonii adhesin SspB. This streptococcal cell wall-anchored protein has been shown to interact with Als3 (42). L. lactis SspB+ cells adhered to wild-type hyphae (Fig. 6E and F) but not to hyphae produced by the mnt1Δ mnt2Δ mutant (Fig. 6G and H). L. lactis MG1363 control cells did not bind to hyphae, as previously shown (42). Thus, it was concluded that although Als3 was present on the surface of mnt1Δ mnt2Δ mutant hyphae, as detected by antibody binding, the protein was not expressed in a conformation that was recognized by the S. gordonii SspB protein.
C. albicans WT and mnt1Δ mnt2Δ cell wall proteomes.
To determine if there were significant differences in the expression of cell wall proteins (CWPs) by C. albicans wild-type and mutant strains under the different growth conditions, cell wall preparations (see Materials and Methods) were subjected to trypsin digestion and the resulting peptides were identified by matrix-assisted laser desorption ionization–time of flight (MALDI-TOF) mass spectrometry. The experiments allowed us to reproducibly identify 23 CWPs (Table 1). Thirteen of these were produced by both strains under each of the hypha-forming conditions, while 10 CWPs were not expressed under all conditions. In YPT-Glc medium, there were significant (P < 0.05) reductions in abundances of the proteins Als1, Als3, Phr1, Rbt1, Sap9, Utr2, and Scw1 for the mnt1Δ mnt2Δ mutant compared with those for the wild type (Table 1). The apparent reductions for the mnt1Δ mnt2Δ mutant of most other proteins, except for Rbt5 and Ecm33.3, were not statistically significant. Thus, deficiency in Mnt1 and Mnt2 affected incorporation of glycan-modifying enzymes, e.g., glycosidases and adhesins, into the cell wall when hyphal induction was performed in YPT-Glc medium.
TABLE 1 .
Cell wall proteomes for the C. albicans wild type or mnt1Δ mnt2Δ mutant undergoing hyphal filament formation in three different media
| Protein | Description or function | Glucose (Glc) medium emPAI valuea |
GlcNAc medium emPAI value |
Saliva-Glc medium emPAI value |
|||
|---|---|---|---|---|---|---|---|
| Wild type | mnt1Δ mnt2Δ | Wild type | mnt1Δ mnt2Δ | Wild type | mnt1Δ mnt2Δ | ||
| Als1 | GPI-modified yeast cell wall and hyphal filament base adhesin | 0.18 ± 0.03 | —c | 0.22 ± 0.08 | 0.05 ± 0.09 | 0.09 ± 0.05 | 0.10 ± 0.05 |
| Als3 | GPI-modificed hyphal cell wall adhesin and invasin | 0.34 ± 0.09 | 0.03 ± 0.06 | 0.44 ± 0.08 | 0.41 ± 0.02 | 0.13 ± 0.08 | 0.02 ± 0.02 |
| Als10 | GPI-modified, related to Als3 | 0.03 ± 0.01 | — | — | — | — | — |
| Pga4/Gas1 | GPI-modified cell surface β-1,3-glucanosyltransferase | 0.48 ± 0.17 | 0.12 ± 0.22 | 0.29 ± 0.09 | 0.19 ± 0.09 | 0.47 ± 0.09 | 0.29 ± 0 |
| Phr1 | Cell wall glycosidase, glucan remodeling, pH regulated | 0.31 ± 0.21 | — | 0.17 ± 0.06 | 0.22 ± 0.19 | 1.07 ± 0.17 | 0.51 ± 0.17 |
| Phr2 | Cell wall glycosidase, pH regulated | 0.09 ± 0.03 | 0.06 ± 0 | 0.06 ± 0b | 0.08 ± 0.09 | 0.31 ± 0.11 | 0.22 ± 0.03 |
| Rbt1 | GPI-modified hyphal cell wall adhesin similar to Hwp1 | 0.15 ± 0.03 | 0.04 ± 0 | 0.19 ± 0.05 | 0.09 ± 0.05 | 0.01 ± 0.02b | 0.04 ± 0b |
| Rbt5 | GPI-modified hyphal cell wall protein | 0.14 ± 0 | 0.14 ± 0 | 0.19 ± 0.09b | 0.14 ± 0b | 0.14 ± 0b | 0.14 ± 0b |
| Ssr1 | GPI-modified glucan-associ- ated role in cell wall structure |
0.38 ± 0.20 | 0.20 ± 0.09 | 0.38 ± 0.20 | 0.26 ± 0.09 | 0.26 ± 0.09 | 0.31 ± 0 |
| Sap9 | GPI-modified aspartyl protein- ase, adhesion and cell sur- face integrity |
0.04 ± 0.06 | — | 0.04 ± 0.07 | — | 0.20 ± 0.03b | 0.04 ± 0.03 |
| Rhd3/Pga29 | GPI-modified yeast cell wall protein, induced by high iron | 0.58 ± 0.36 | 0.11 ± 0.20b | 0.22 ± 0.10 | 0.05 ± 0.09b | 0.17 ± 0.17 | 0.17 ± 0.17 |
| Utr2 | GPI-modified cell wall glycosidase, role in adhesion | 0.13 ± 0.06 | 0.01 ± 0.02b | 0.02 ± 0.03b | 0.04 ± 0.03b | 0.30 ± 0.09 | 0.20 ± 0.08 |
| Hyr1 | GPI-modified hyphal cell wall protein, induced by macrophages (Bcr1 acti- vated) |
0.01 ± 0.02 | — | 0.05 ± 0.06 | 0.33 ± 0.09 | — | — |
| Scw1 | Cell surface O-glycosylated mannoprotein, glucan metabolism and adhesion | 0.72 ± 0.30 | 0.24 ± 0.12 | 0.20 ± 0.10 | 0.15 ± 0.14 | 0.20 ± 0.11 | 0.05 ± 0.05 |
| Cht2 | GPI-modified chitinase, re- quired for filamentous growth |
0.55 ± 0.05 | 0.48 ± 0.04 | 0.34 ± 0.04 | 0.50 ± 0.17 | 0.32 ± 0.08 | 0.02 ± 0.03b |
| Ecm33.3 | GPI-modified hypha-specific protein, adhesion | 0.53 ± 0.17 | 0.74 ± 0.31 | 0.38 ± 0.06 | 0.55 ± 0.34 | 0.77 ± 0.07 | 1.00 ± 0.17 |
| Crh11 | GPI-modified transglycosylase | 0.55 ± 0.28 | 0.33 ± 0.18 | 0.35 ± 0.10 | 0.38 ± 0.20 | 0.72 ± 0.12 | 0.90 ± 0.32 |
| Sod4 | Superoxide dismutase (Cu/Zn) | 0.19 ± 0.09 | 0.05 ± 0.08b | — | — | — | — |
| Sod5 | Superoxide dismutase (Cu/Zn) | 0.19 ± 0.09 | 0.05 ± 0.08b | 0.25 ± 0.09 | 0.32 ± 0.12 | 0.30 ± 0 | 0.36 ± 0.10 |
| Pir1 | 1,3-β-Glucan-linked cell wall protein, highly glycosylated, full virulence | — | 0.03 ± 0.05b | 0.03 ± 0.05b | — | 0.12 ± 0.06 | 0.09 ± 0b |
| Pga24/YWp1 | Secreted yeast form specific protein, dispersal in vivo | 0.04 ± 0.01 | 0.02 ± 0.35b | — | 0.02 ± 0.03b | 0.04 ± 0.03 | 0.06 ± 0b |
| Pga31 | GPI-modified cell wall protein | — | 0.03 ± 0.05b | — | 0.03 ± 0.05b | — | 0.19 ± 0.10 |
| Ihd1/Pga36 | GPI-modified protein, hypha induced | — | — | — | 0.03 ± 0.05b | — | — |
Conversely, in YPT-GlcNAc medium, there were no significant decreases in cell wall protein abundance between the wild type and the mutant except for Sap9 (aspartyl proteinase), which was absent from mnt1Δ mnt2Δ cell walls (Table 1). The presence of GlcNAc appeared to restore wild-type levels of Als3, Phr1, and Rbt1 in the mnt1Δ mnt2Δ mutant cell walls, while Hyr1 was significantly (P < 0.05) increased compared with that for the wild type. However, in saliva-Glc medium, there were contrasting differences, with the Pga4/Gas1, Phr1, Phr2, Sap9, Utr2, Ecm33.3, and Crh11 proteins being more abundant than in YPT-GlcNAc, and a significant reduction (P < 0.05) in levels of Als3, Pga4/Gas1, Phr1, Sap9, and Cht2 was seen for the mutant (Table 1). The apparent increases in Ecm33.3 and Crh11 for the mnt1Δ mnt2Δ mutant were not statistically significant within the data set. In summary, the mnt1Δ and mnt2Δ deletions can have pleiotropic effects on hyphal cell wall protein composition depending upon the medium and conditions employed to induce hypha formation. Since the cell wall proteome data indicated that Sap9 was the only protein consistently reduced in abundance from cell wall preparations of the mnt1Δ mnt2Δ mutant, the possibility that SAP9 expression might be necessary for activating receptors, e.g., Als3 for S. gordonii, was considered. To test this, we examined a sap9Δ mutant (53) for ability to form biofilms and to support adhesion of S. gordonii. The incidence of hyphal filament formation by the sap9Δ mutant was similar to that for the C. albicans wild type, and S. gordonii cells bound at similar levels to filaments formed by the wild type or the sap9Δ mutant (data not shown). Therefore, absence of Sap9 from the cell wall of mnt1Δ mnt2Δ hyphae could not account directly for the deficiency in binding S. gordonii cells.
Loss of adhesin function by mnt1Δ mnt2Δ mutant hyphae.
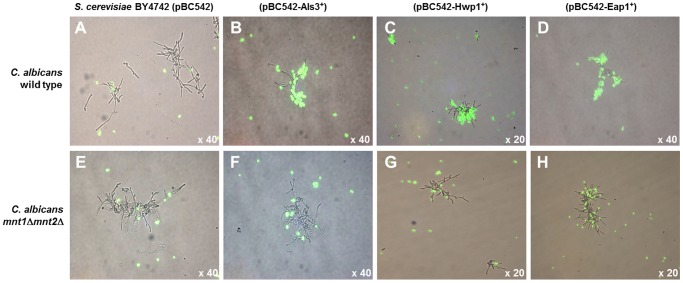
The proteome studies supported the observations shown in Fig. 6 that the hyphal cell wall Als3 receptor for S. gordonii (42) in the mnt1Δ mnt2Δ mutant was present in normal amounts in YPT-GlcNAc (Table 1). We hypothesized, therefore, that the mnt1Δ mnt2Δ mutations affected cell wall protein structure such that S. gordonii receptors, e.g., Als3, were not presented in a functional conformation. Since the hyphal surface proteins Als3 and Hwp1 have been shown to interact with one another (14), we further tested our hypothesis by determining the abilities of Als3 and Hwp1 to interact with hyphae produced by the wild type or the mnt1Δ mnt2Δ mutant. Saccharomyces cerevisiae cells expressing Als3 on their surfaces (24) (Fig. 7B) were able to interact with hyphae formed by the C. albicans wild type but not with hyphae formed by the mnt1Δ mnt2Δ mutant (Fig. 7F). Wild-type S. cerevisiae containing empty vector pBC542 (54) did not interact with hyphae from either strain of C. albicans (Fig. 7A and E). S. cerevisiae cells expressing Hwp1 (Fig. 7C) also bound to hyphae produced by the C. albicans wild type but not to those formed by the mnt1Δ mnt2Δ mutant (Fig. 7G and H). S. cerevisiae cells expressing adhesin Eap1 (55) showed partial recognition of mnt1Δ mnt2Δ mutant hyphae (Fig. 7D). These results suggested that while the hyphal filaments of the mnt1Δ mnt2Δ mutant appeared to express Als3, this protein was not presented in a conformation that enabled homotypic (Als3-Als3) or heterotypic (Hwp1-Als3) interactions to occur, as they did with wild-type hyphal filaments. The observations provide direct evidence that mannosylation mediated by Mnt1/Mnt2 is essential for determining hyphal cell wall protein functions and hence interkingdom biofilm formation with S. gordonii.
FIG 7 .
Interactions of C. albicans wild type or mnt1Δ mnt2Δ mutants with S. cerevisiae strains expressing C. albicans cell wall proteins. S. cerevisiae cultures were grown to mid-exponential phase in CSM medium, and the cells were FITC labeled and then mixed with C. albicans cells that had been induced to form hyphal filaments for 3 h at 37°C in YPT-GlcNAc medium (see Materials and Methods). After a 2-h incubation of cultures at 30°C with gentle agitation, samples (10 µl) were removed and placed on glass microscope slides, coverslips were applied, and the cells were visualized using a Leica microscope under phase contrast or fluorescence. The images show phase micrographs merged with respective green fluorescence images of S. cerevisiae cells. (A to D) C. albicans wild type; (E to H) C. albicans mnt1Δ mnt2Δ mutant. S. cerevisiae BY4742 empty vector pBC542 control (A and E), S. cerevisiae expressing Als3 (B and F), S. cerevisiae expressing Hwp1 (C and G), or S. cerevisiae expressing Eap1 (D and H) is shown. Magnification, ×20 or ×40, as indicated.
DISCUSSION
A number of studies have demonstrated that mannosylation reactions in C. albicans are required for adherence, invasion of host tissues, and virulence (30, 33). The marked reduction in adherence of mnt1Δ mnt2Δ mutants to a number of substrata observed previously suggested that O-linked glycosylation might indeed be specifically required for adhesion (56). It has not been precisely clear why mannosylation reactions necessarily affect these properties. One possibility is that deficiency in mannosylation directly affects the synthesis, expression, or activities of cell wall proteins and adhesins crucial for invasion and virulence. Oligosaccharide chains are believed to confer stabilizing properties upon the extended regions of fungal extracellular glycoprotein adhesins (57). Thus, mannosylation could be key to formation of a fully functional C. albicans cell surface, especially since there is now good evidence that a number of CWP families, e.g., Als and Rbt, crucial to biofilm formation and virulence, carry extensive amino acid residue repeat blocks in their C-terminal regions that are heavily O-glycosylated (58). In addition to being attenuated in adherence and invasion, C. albicans glycosylation mutants induced lower levels of cytokine production by human peripheral blood monocytes (29). Disruption of the mannosylation processes seems therefore to have a range of damaging effects on the fungus and interactions with the host (28). Despite these observations, the potential for mannan synthesis to be an antifungal target has generally received less attention.
Previously, it was suggested that deletion of MNT1 or MNT2 in C. albicans might not result in marked changes in the tertiary structure of cell surface mannoproteins, because protein stabilization is thought to be achieved by addition of the first O-linked mannose rather than subsequent residues (30). However, our results suggest that the subsequent (second and third) steps in mannosylation are important. Biofilm development and interactions of C. albicans with streptococci were dependent upon expression of MNT1 and MNT2, encoding the enzymes adding the second and third mannose residues to the growing mannan chain. Interestingly, the phenotypic effects of the mnt1Δ mnt2Δ double deletion on biofilm development were environmentally sensitive. When induced to form hyphal filaments by nitrogen starvation (YPT-Glc) or with salivary glycoproteins, the double mutant produced a mixture of pseudohyphae and true hyphae. However, when induced with GlcNAc, the double mutant formed hyphae similar to wild-type hyphae in structure and proteome.
GlcNAc induces two sets of responses in C. albicans. One causes it to switch from budding to hyphal growth, activated by stimulation of adenylyl cyclase and increased cAMP signaling (59). The second activates a pathway independently of cAMP to induce expression of genes necessary to catabolize GlcNAc (60), but this is not required for hypha induction (61). The hyphal-induction signal from GlcNAc is transmitted through Ngt1 and then to the hyphal gene transcription regulator Efg1 (62). Conversely, the induction signals of low nitrogen or salivary (serous) proteins are received through Mep2 or Ras1, with activation of Cyr1 (adenylyl cyclase). Possibly the latter hyphal induction pathway is more sensitive to the effects of MNT1/MNT2 deletions, such as cell wall stress (63), than the Ngt1 induction pathway.
GlcNAc is not involved directly in O-mannosylation, but GlcNAc treatment induces chitin synthesis and could potentially be converted to other monosaccharide cell wall building blocks via its conversion to fructose-6-phosphate. Therefore, the effect of GlcNAc in effectively complementing the mnt1Δ mnt2Δ mutant phenotypes may be multifactorial. Despite GlcNAc enhancing biofilm and hypha formation by the mnt1Δ mnt2Δ mutant, the hyphae were still deficient in binding S. gordonii and in interacting with the C. albicans cell wall adhesins Als3 and Hwp1, expressed on the surface of S. cerevisiae. This provides biochemical and physiological demonstration that without the functions of MNT1 and MNT2, the intermicrobial cell adhesins are not presented in a way that promotes dual-species community development. Nevertheless, there must be sufficient levels of interaction between C. albicans mnt1Δ mnt2Δ cells in GlcNAc to enable monospecies biofilm formation.
The ability of oral streptococci to interact with C. albicans may be relevant to longer-term carriage or persistence of C. albicans in the oral cavity (29). Evidence suggests that streptococci and C. albicans exhibit growth synergy, implying mutual benefit in cocolonization (43, 64). In this article, dual-species communities formed under flow or nonflow conditions, and S. gordonii cells could be seen within the biofilms closely associated with hyphal filaments (see Video S5 in the supplemental material). In biofilms formed under medium flow, hyphal filaments extended further into the environment (see Video S3) than they did under nonflow conditions, where the biofilms were structurally more compact (see Video S1). The stimulation of biofilm development under flow could result from better provision of nutrients, removal of inhibitory metabolic end products, shear-force-induced gene expression, or diffusion away from the biofilm of compounds, such as farnesol (65), that inhibit hypha formation.
To determine the effects of mnt1Δ mnt2Δ mutation on the production of hyphal cell wall proteins, we prepared cell walls from hypha-forming cells and subjected them to trypsin digestion and proteome analyses. In C. albicans cell walls, the more abundant covalently attached proteins are GPI modified, whereas the least abundant are attached via an alkali-labile linkage (66). Our experiments identified peptides from proteins present in the cell wall through covalent linkage. Proteins missing from the cell wall extracts would be either not expressed (transcription) or not properly incorporated (covalently linked) into the cell wall or lacking amenable trypsin cleavage sites. In the mnt1Δ mnt2Δ mutant grown in YPT-Glc, there were significant reductions in a wide range of CWPs that would be expected to have major effects on phenotypes such as adhesion, biofilm formation, and invasion of host cells (9, 50, 51). In the presence of GlcNAc, however, the mnt1Δ mnt2Δ mutant showed a cell wall proteome much more similar to that of the wild type. Peptides from the GPI-modified hypha-specific adhesin Hwp1 were not detected by liquid chromatography-tandem mass spectrometry (LC-MS/MS), as also reported by others (67). During hypha formation there is enrichment of carbohydrate-active or cell wall-remodeling enzymes Cht2, Crh11, Mp65, Pga4, Phr1, Phr2, and Utr2 (49, 67). We have identified six of these proteins in our studies. Actual relative amounts will be dependent upon the methods employed in cell wall purification and MS analysis and most importantly the precise conditions utilized for growth and hypha formation.
A protein consistently deficient in the mnt1Δ mnt2Δ cell wall proteome was Sap9, one of a family of 10 aspartyl proteinases (Sap1 to 10) that are differentially expressed in C. albicans (68). The main effects of SAP9 deletion are reduced chitinase activity and increased adhesion to epithelial cells (53). It has been suggested that Sap9 is able to trim cell surface proteins, on C. albicans or the host, to modulate receptor-ligand interactions (69–71). In a model for Als3-mediated interactions in vivo, soluble or membrane-bound Saps are hypothesized to partly digest proteins on the host cell surface, thus providing additional ligands for Als3 to interact with (71). However, it was concluded that this protease activity did not function in providing receptor availability for binding of S. gordonii.
Initial adherence in C. albicans biofilm formation is mediated at least in part by Als1 (25), and ALS1 expression is under control of the transcriptional regulator Bcr1. Other adhesin targets of Bcr1 are Als3 and Hwp1, which mediate cell-cell interactions in biofilms (14). The cell wall protein Eap1 is required for biofilm formation in vitro and in vivo (55) and may be needed for initial layer formation on specific substrata (24). It has been suggested that the Eap1 and Als proteins may play a role in environmental sensing as well as directly as adhesins (72). Genes encoding transglycosidases involved in maintenance of cell wall integrity, such as CRH11 (Table 1), are coregulated with other hyphal or virulence genes, while BGL2, PHR1, and XOG1 encode enzymes that are crucial for delivery of β-1,3-glucan to the biofilm matrix and for accumulation of matrix biomass (73). Levels of a number of these factors were altered in the mnt1Δ mnt2Δ mutant cell wall proteomes from cells forming hyphae in YPT-Glc and saliva-Glc medium, possibly disabling biofilm formation. The observation that between 2 h and 3 h biofilm development by the mnt1Δ mnt2Δ mutant effectively ceased is similar to that made for an als3Δ mutant (42). This might be consistent with the 80 to 90% reduction in Als3 in the cell wall proteome of the double mutant under YPT-Glc or saliva-Glc medium conditions. On the other hand, GlcNAc-induced hyphae formed by the mnt1Δ mnt2Δ mutant had a full complement of adhesins (Table 1). We conclude that in the presence of GlcNAc, the functional expression of adhesins on the C. albicans cell surface for binding S. gordonii but not for initial biofilm formation is affected by the block in mannosylation.
Evidence that adhesins expressed on the surface of GlcNAc-induced hyphae produced by the mnt1Δ mnt2Δ mutant were not properly functional was provided by the experiments utilizing S. cerevisiae strains expressing the adhesin Als3, Hwp1, or Eap1. Strains expressing these proteins bound avidly to C. albicans wild-type hyphae but feebly to hyphae produced by the mnt1Δ mnt2Δ mutant. This suggested that the mnt1Δ mnt2Δ mutant hyphae did not present a correct configuration of adhesins to interact with these CWP adhesins. Such effects would be extremely difficult to detect by current imaging techniques and are implicated by probing for functionality, such as ability to interact with known ligands. Of note is that some hyphae in all populations (wild type or mutant) appeared not to bind S. gordonii cells, thus indicating potential heterogeneity of hyphal adhesin expression or activation within hyphal filament populations.
A current model for the broad and strong adhesive activity of Als3 suggests that Als3 molecules are able to cluster on the cell surface and bind to each other through rapidly dissociable hydrophobic interactions (74). A long glycosylated C-terminal region stalk elevates the N-terminal region and allows flexibility needed for the adhesins to form amyloids. The Als3 ligand binding cleft is located within the N-terminal region (71). We suggest that the initial SspB-Als3 interaction unfolds or extends the Als3 protein such that further interactions are substantiated, possibly between amyloid-like regions of the proteins. The Als3 protein contains at least one amyloid sequence (32) and SspB carries three potential amyloid sequences within the central region and C-terminal regions of the polypeptide (VVYTYT, IWFAF, and TTSFVLN), as predicted by the TANGO computer algorithm (75), that could have amyloidigenic activity.
Amyloid interactions between adhesins may provide cohesive strength to C. albicans Als proteins. Formation of Als5 adhesin nanodomains on the cell surface was observed in response to mechanical stimuli, which probably caused the T region (Thr-rich N-terminally located sheet) to partially unfold and expose the amyloid-forming sequences (76). The formation of adhesin clusters could thus explain why Als proteins mediate strong adherence (51). These various in vitro observations taken collectively with our results in this article provide the basis for a model (see Fig. S3 in the supplemental material) through which we envisage O-mannosylation to play a critical role in development of CWP adhesin functions. In this model, O-mannosylation catalyzed by the enzymes Mnt1 and Mnt2 initially acts by stabilizing adhesin structure and then allowing for subsequent unfolding and extension. The apical N-terminal domains of Als3 protrude from the hyphal cell wall surface and are recognized by S. gordonii cells. Binding causes force-induced unfolding of Als3, which would be enhanced in biofilms under flow conditions (as shown in Results). The adhesive (pulling) forces then elicit clustering of Als3 protein molecules to form nanodomains over the cell surface, thus promoting multiple force interactions of Als3 proteins with bacteria. This would be consistent with observations that streptococci can be seen on some hyphae to show localized attachment and accumulations, suggesting that they are interacting with clusters of adhesins. In Fig. S3, Sap9 aspartyl proteinase is shown as modulating the activities of these nanodomains by proteolytically disrupting the intermolecular forces. This would disaggregate hyphal filaments, leading to increased biofilm permeability and more flexibility for recognition of additional binding partner molecules. Dispersal of intermicrobial cell contacts might also be regulated, at least in part, via Sap9-catalyzed hydrolysis of intramolecular peptide bonds of proteins engaged in intermolecular adhesion events. To date, the evidence for adhesin activation has been derived from biophysical in vitro analyses (76), but our data provide direct experimental evidence at the physiological level for the hypothesis that Als-mediated adhesion largely depends upon conformational modifications of existing adhesins and that this is dependent upon early-stage O-mannosylation reactions. These reactions are clearly worthy of further consideration as potential targets for control of C. albicans colonization and virulence.
MATERIALS AND METHODS
Microbial growth conditions.
The microbial strains utilized in this study are listed in Table S1 in the supplemental material. C. albicans strains, all derived from CAI4 (77), were cultivated on Sabouraud dextrose agar (Lab M) aerobically at 37°C. S. gordonii strains were cultivated anaerobically at 37°C on BHYN agar (per liter: 37 g brain heart infusion broth, 5 g yeast extract, 5 g Neopeptone, and 15 g agar), which was supplemented with erythromycin (5 µg/ml) for S. gordonii expressing green fluorescent protein (GFP). Lactococcus lactis strains were grown anaerobically at 30°C on M17-glucose agar (Oxoid) containing 5 µg erythromycin/ml where appropriate. Saccharomyces cerevisiae strains (24, 78) were cultivated aerobically at 30°C on CSM medium (per liter: 6.7 g Difco yeast nitrogen base, 0.77 g complete supplement mixture [CSM] drop-out [CSM-Ura] [Formedium, Hunstanton, United Kingdom], 20 g glucose, and 30 g agar). Suspension cultures of C. albicans were grown in YPD medium (1% yeast extract, 2% peptone, and 2% glucose) in conical flasks at 37°C with shaking (200 rpm). S. gordonii strains were grown in BHY medium (brain heart infusion, containing 5 g/liter yeast extract), stationary, at 37°C. YPT medium (1× Difco yeast nitrogen base, 20 mM NaH2PO4–H3PO4 buffer, pH 7.0, and 0.1% Bacto-tryptone) supplemented with 0.4% glucose (YPT-Glc) or 0.4% N-acetyl-d-glucosamine (YPT-GlcNAc) was utilized to support growth of S. gordonii and C. albicans in planktonic cultures or biofilms.
Generation of S. gordonii expressing GFP.
Plasmid pAR4 (rpsT-gfp-mut3b*pJAR2), kindly provided by Patrick Piggot (Temple University School of Medicine), was extracted from Streptococcus mutans UA159 (79) by a modified alkaline-lysis technique (41). The plasmid was purified using a Qiaprep spin miniprep kit (Qiagen). S. gordonii DL1 cells were made competent for transformation as previously described (80), and transformants containing pAR4 were selected on BHYN agar containing 5 µg erythromycin/ml (see Table S1 in the supplemental material).
Preparation of saliva.
Collection of saliva from human subjects was approved by the National Research Ethics Committee South Central Oxford C) (no. 08/H0606/87+5). All samples were collected from at least 6 adult subjects, who provided written informed consent. The samples were pooled, treated with 0.25 M dithiothreitol on ice for 10 min, and centrifuged (8,000 × g for 10 min). The supernatant was diluted to 10% with sterile water, filter sterilized (0.45-µm-pore-size membrane), and stored at −70°C in 10-ml portions. Saliva (10%) containing 0.1% glucose was utilized as a microbial growth medium (saliva-Glc).
Preparation of microbial cells.
C. albicans strains were grown for 16 h in YPD. Cells were harvested by centrifugation (5,000 × g for 5 min), washed twice in YPT, and suspended at an optical density at 600 nm (OD600) of 1.0 (approximately 1 × 107 cells/ml). S. gordonii or L. lactis cells were grown for 16 h in 10 ml YPT-Glc, harvested by centrifugation (5,000 × g for 7 min), and washed twice with YPT (no glucose). Bacteria were suspended in 0.05 M Na2CO3–0.1 M NaCl containing 1.5 mM fluorescein isothiocyanate (FITC) and incubated in the dark at 20°C for 30 min with gentle agitation. The bacterial cells were then washed three times with YPT and suspended at an OD600 of 0.5 (2 × 108 cells/ml) in YPT.
C. albicans interactions with S. gordonii in planktonic phase.
Portions (0.2 ml, 2 × 106 cells) of C. albicans cell suspension in YPT were added to glass tubes containing warm YPT-Glc, YPT-GlcNAc, or saliva-Glc medium (1.8 ml). The cultures were incubated for 37°C for 3 h with shaking at 220 rpm; S. gordonii (or in some experiments L. lactis) cell suspension (1 ml, 2 × 108 cells) labeled with FITC or expressing GFP was then added, and incubation was continued at 37°C for 1 h. Samples of suspension (10 µl) were applied to glass microscope slides and visualized by light or fluorescence microscopy (Leica DMLB) or with a Leica SP5-AOBS confocal laser scanning microscope attached to a Leica DM I6000 inverted epifluorescence microscope. The Volocity software program was utilized to determine density of fluorescence associated with multiple hyphal filaments over at least 10 randomly selected fields of view (expressed as density units per 50 hyphae).
Planktonic-phase interactions of S. cerevisiae with C. albicans.
S. cerevisiae BY4742 strains expressing C. albicans cell wall proteins (see Table S1 in the supplemental material) were inoculated into warm CSM (10 ml) and incubated for 16 h at 30°C with shaking at 220 rpm. Portions (5 ml) were centrifuged (5,000 × g, 5 min), washed twice with YPT, and fluorescently labeled with FITC as described above. The labeled cells were washed thoroughly with carbonate buffer and twice with YPT before being suspended in YPT-Glc at an OD600 of 1.0 (1 × 107 cells/ml). A 1-ml suspension of each strain was transferred to a glass tube containing 1 ml C. albicans cells preinduced to form hyphae for 3 h in YPT-GlcNAc and incubated at 30°C for 2 h with shaking at 220 rpm. Portions (10 µl) of the cell suspensions were visualized by transmitted light or fluorescence microscopy.
Monospecies and dual-species biofilms of C. albicans and S. gordonii.
Sterile 19-mm glass coverslips were incubated with 10% filter-sterilized saliva at 4°C for 16 h. Individual coverslips were then transferred to 12-well tissue culture plates containing 1.9 ml YPT-Glc, YPT-GlcNAc, or saliva-Glc, and 0.1 ml C. albicans cell suspension (1 × 106 cells) was added. The plates were incubated at 37°C for up to 6 h with gentle motion (50 rpm) in a humid environment. Coverslips were removed at intervals, rinsed gently with phosphate-buffered saline (PBS), dried, and stained with 0.5% crystal violet solution for light microscopy or estimation of biomass after releasing the stain with 10% acetic acid and measuring the OD595 (43).
For visualization of S. gordonii interactions with C. albicans hyphae in early biofilms, coverslips were incubated with C. albicans as described above for 3 h, removed, and placed in wells containing fresh YPT-Glc medium (0.5 ml). Suspensions (0.5 ml) of FITC-labeled S. gordonii were added, and the cultures were incubated for a further 1 h. For comparative monospecies biofilms of C. albicans, culture medium alone (0.5 ml) was added. Coverslips were then removed, gently rinsed once with PBS, inverted onto clean glass microscope slides, and examined by transmitted light or fluorescence microscopy as described above.
Preparation of biofilms for CSLM.
Plastic culture dishes (35-mm diameter; Mat Tek) with 14-mm No. 1.0 coverslip base glass windows were incubated with 2 ml 10% saliva at 4°C for 16 h. The saliva was aspirated, and 1.8-ml growth medium (YPT-Glc, YPT-GlcNAc, or saliva-Glc) was added to each dish, followed by 0.2 ml C. albicans cell suspension (2 × 106 cells). Dishes were incubated in a humid environment at 37°C for 1 h with gentle motion at 50 rpm. All subsequent incubation steps were carried out under these growth conditions. The culture suspensions were aspirated and replaced with the appropriate medium, and the dishes were incubated for 2 h. For dual-species biofilms, the C. albicans suspension was gently aspirated, and 0.2 ml S. gordonii UB2549 (expressing GFPmut3b*) suspension in appropriate medium was added. The dishes were incubated for 30 min, the S. gordonii culture suspension was aspirated, and 2 ml appropriate growth medium was added for further incubation at 37°C for 4 h. The cell suspensions were then aspirated, the dishes were washed gently with sterile deionized water, and C. albicans cells were stained with calcofluor white (0.2 µg/ml) just prior to visualizing by confocal scanning laser microscopy (CSLM) with a Leica SP5-AOBS confocal microscope attached to a Leica DM I6000 inverted epifluorescence microscope. Volocity software was utilized to prepare three-dimensional (3D) images and to calculate biofilm heights (in μm). For monospecies biofilms, exactly the same protocols were applied, minus bacteria or C. albicans, substituted with sterile growth medium.
Flow-cell biofilms.
Flow cell units, consisting of two parallel chambers sealed with a glass coverslip, were prepared as described by Palmer (81). The growth medium input line was connected, medium was drawn through the cells with a syringe, and the cells were injected with 0.5 ml 10% human saliva and incubated at 4°C for 16 h to coat the inside surfaces of the chambers with salivary glycoproteins. The effluent line to a peristaltic pump was then connected, and appropriate growth medium was drawn through the flow cell for 15 min at a flow rate of 6 ml/h. C. albicans cell suspension in growth medium (0.2 ml) was injected into the flow cell chamber and incubated statically at 37°C for 1 h. The growth medium (containing 1 µg/ml calcofluor white) was then drawn through the flow cell chambers at a rate of 6 ml/h for up to 16 h. For dual-species biofilms with S. gordonii, the medium flow was stopped after 2 h, and S. gordonii UB2549 (expressing GFPmut3b*) cell suspension (0.2 ml) was injected into the chamber and incubated without medium flow at 37°C for 30 min. Flow of growth medium was then recommenced and continued for 4 h at a rate of 6 ml/h. Biofilms were visualized by CSLM.
Immunolabeling of C. albicans hyphae.
Suspension cultures of C. albicans were grown in YPD medium for 16 h at 37°C with shaking (200 rpm). Cells were harvested by centrifugation, washed with YPT medium, suspended at an OD600 of 0.5 in YPT-GlcNAc medium, and incubated with shaking for 3 h at 37°C. Cells were harvested by centrifugation, washed, blocked with goat serum, and then reacted with MAb 3-5A monoclonal antibody to the protein Als3, as previously described (52). Antibody binding was detected with FITC-labeled goat anti-mouse IgG F(ab) fragment-specific antibody, and wet mounts were visualized by light microscopy using an Olympus BX50 microscope.
C. albicans cell wall purification.
Cell wall protein samples were prepared by a modification of the method previously described (82). C. albicans cells (200-ml cultures) were grown at 37°C for 6 h with shaking to an OD600 of 0.7 to 0.9. The cells were harvested by centrifugation (5,000 × g, 5 min), washed with 10 mM Tris-HCl, pH 7.5, at 4°C, suspended in 0.2 ml Tris-HCl buffer, and mixed with 0.5 g cold silica beads (Biospec). Protease inhibitor cocktail (Sigma) and 1 mM phenylmethylsulfonyl fluoride (PMSF) were added, and the cells were disrupted by shaking using a FastPrep cell breakage machine (FastPrep-24 bead beater; MP Biosciences). The contents of the tubes were washed out into cold 50-ml tubes with 1 M NaCl, the beads were allowed to settle, and the supernatant was removed (repeated twice). Cell wall suspensions were centrifuged (3,000 × g, 5 min, 4°C), and the pellets were washed 5 times with 1 M NaCl and once with double-distilled water (ddH2O). The crude cell walls were then twice extracted with SDS-mercaptoethanol buffer (50 mM Tris, 2% SDS, 0.3 M β-mercaptoethanol, and 1 mM EDTA [pH 8.0]) at 100°C for 10 min to remove noncovalently bound proteins. Cell walls were then washed thoroughly by alternate suspension in ddH2O and centrifugation (5 times) and freeze-dried.
Preparation of cell wall peptides.
Freeze-dried cell wall sample (1 to 2 mg) was mixed with 0.5 M ammonium bicarbonate in water containing 3 mM dithiothreitol and heated at 60°C for 20 min. Iodoacetamide (30 µl of 55 mM stock solution) was then added, and the suspension was incubated at 25°C for 10 min in the dark. Trypsin (30 µl of 20 mg/ml stock) was added, and the suspension was incubated at 37°C for 14 h and centrifuged at 14,000 × g for 10 min. The supernatant was freeze-dried and extracted with 10% formic acid, and peptides were purified using ZipTip mC18 pipette tips (Millipore) and dissolved in 0.1% formic acid.
Proteomic analysis.
Samples (3 µl) were injected into an LC-MS system which comprised an UltiMate 3000 LC instrument (Dionex Ltd., United Kingdom) fitted with a PepSwift monolithic poly(styrene-co-divinylbenzene) (PS-DVB) column (200-µm inside diameter [i.d.] by 5 cm; Dionex) coupled to an HCTultra ion trap mass spectrometer (Bruker Daltonik GmbH, Bremen, Germany) fitted with a low-flow nebulizer in the electrospray ionization (ESI) source and controlled by HyStar software (version 4.0; Bruker Daltonik). Peptides were separated at a flow rate of 2 µl/min using a linear gradient of 0 to 40% acetonitrile-water-formic acid (80:20:0.04) (solvent B) in water-acetonitrile-formic acid (97:3:0.05) (solvent A) over 40 min, followed by a 1-min column wash in 90% solvent B and a 12-min equilibration step in solvent A. MS/MS data (scan range, m/z 100 to 2,200; averages = 2) were acquired in positive data-dependent AutoMS(n) mode using the esquireControl software program (version 6.2; Bruker Daltonik). Up to three precursor ions were selected from the MS scan (range, m/z 300 to 1,500; averages = 3) in each AutoMS(n) cycle. Precursors were actively excluded after being selected twice within a 1-min window, and singly charged ions were also excluded. Peptide peaks were detected (maximum of 9,999 compounds above an intensity threshold of 50,000) and deconvoluted automatically using DataAnalysis software (version 3.4; Bruker Daltonik). Mass lists in the form of Mascot Generic Format (*. mgf) files were created automatically and used as inputs to Mascot MS/MS ion searches via a local Mascot server (version 2.2; Matrix Science, London, United Kingdom) with a database built from the <CALBI_prot.txt > file (date stamp, 5 February 2002; 6,166 sequences) downloaded from CandidaDB (ftp://ftp.pasteur.fr/pub/GenomeDB/CandidaDB/FlatFiles). Search parameters used were the following: enzyme = trypsin; fixed modifications = carbamidomethyl (C); variable modifications = oxidation (M); mass values = monoisotopic; peptide mass tolerance = 1.5 Da; fragment mass tolerance = 0.5 Da; max missed cleavages = 1; instrument type = ESI-TRAP. Search results were displayed by Mascot after selection of the following parameters: standard scoring; require bold red; ion score or expect cutoff = 0.05. An open-source web application (emPAI Calc) was utilized for estimation of protein abundance (83, 84).
SUPPLEMENTAL MATERIAL
CSLM images of dual-species biofilms formed by the C. albicans wild type with S. gordonii DL1 in three different growth media as indicated. The image at the top of each column (A) is a representative xy stack of the biofilm assembled from the top down. The four images underneath (B, C, D, and E) are xz images showing organization and thickness (depth) of the dual-species biofilms from the xy image immediately above (B), the green S. gordonii component only (red subtracted) (C), the red C. albicans component only (green subtracted) (D), or (bottom image) a representative monospecies C. albicans wild-type biofilm grown under the same conditions (E). The significant structural and architectural differences associated with the different growth conditions employed are described in the text. The values shown in μm are average thickness (depth) measurements calculated from across the sections of the biofilms shown. Bar = 80 µm. Download
CSLM images of dual-species biofilms formed under flow conditions by the C. albicans wild type or mnt1Δ mnt2Δ mutant with S. gordonii DL1. The image at the top of each column (A) is a representative xy stack of the biofilm assembled from the top down. The four images underneath (B, C, D, and E) are xz images showing organization and thickness (depth) of dual-species biofilms from the xy image immediately above (B), the green S. gordonii component only (red subtracted) (C), the red C. albicans component only (green subtracted) (D), or (bottom image) a representative monospecies C. albicans wild-type or mnt1Δ mnt2Δ mutant biofilm grown under the same conditions (E). Note that YPT-GlcNAc medium data only are shown for the mnt1Δ mnt2Δ mutant because the other media did not support biofilm formation (see Fig. 5). The values shown in μm are average thickness (depth) measurements calculated from across the sections of the biofilms shown. Bar = 80 µm. Download
Diagrammatic model for cell wall protein adhesin-mediated interactions of C. albicans with S. gordonii. Mannosylation catalyzed by the enzymes Mnt1/Mnt2 is proposed to be essential for formation of a functional adhesin on the cell surface. The initial mannosylation acts by stabilizing adhesin structure in the first instance, necessary for allowing subsequent unfolding and extension. The Als3 adhesin is depicted as an example. The apical N-terminal domains (red) of Als3 protrude from the hyphal cell wall surface and are recognized by S. gordonii cells (71). Binding causes force-induced unfolding of Als3, and evidence presented in this article suggests that this may be enhanced under conditions of flow (shear). The adhesive (pulling) forces then elicit clustering of Als3 protein molecules to form nanodomains over the cell surface, as has been suggested previously (32), thus promoting multiple force interactions of Als3 proteins with bacteria, forming aggregates. It is suggested that the T domains (orange) containing amyloidogenic sequences are important for hydrophobic clustering and the formation of nanoadesomes (74). In this model, the Sap9 aspartyl proteinase is shown as modulating the activities of these nanodomains by disrupting the intermolecular forces. This would tend to enhance disaggregation of filaments, leading to increased biofilm permeability and more flexibility for recognition of additional binding partner molecules. Dispersal of cells from intermicrobial cell contacts might also be regulated via Sap9 hydrolysis of intramolecular peptide bonds of proteins engaged in intermolecular adhesion events. Download
Strains used in this study
3D video clip of biofilm structure formed by C. albicans wild type in YPT-GlcNAc Download
3D video clip of biofilm structure formed by C. albicans mnt1Δ mnt2Δ mutant Download
3D video clip of biofilm structure formed by C. albicans wild type in YPT-GlcNAc, under flow Download
3D video clip of biofilm structure formed by C. albicans mnt1Δ mnt2Δ mutant, under flow Download
3D video clip of dual-species biofilm structure formed by C. albicans wild type with S. gordonii DL1 in YPT-GlcNAc, under flow Download
3D video clip of dual-species biofilm structure formed by C. albicans mnt1Δ mnt2Δ mutant with S. gordonii in YPT-GlcNAc, under flow Download
ACKNOWLEDGMENTS
We are most grateful to Neil Gow, Steven Bates, and Bernhard Hube for the provision of strains and general advice, to Rob Palmer and Paul Kolenbrander for flow-cell design and training, to Patrick Piggot for supplying plasmids prior to publication, to Ciara Keene and Richard Silverman for strain construction and laboratory assistance, to Lois Hoyer and X. Zhao for providing antibody and performing immunofluorescence microscopy, and to David Stead (Aberdeen Proteomics) for proteomics analysis. We thank the Medical Research Council and Wolfson Foundation for establishing the Bioimaging Facility at the University of Bristol. We also acknowledge the importance of the Candida Genome Database (http://www.candidagenome.org) for our studies.
This work was supported by NIH/NIDCR grant R01 DE016690.
Footnotes
Citation Dutton LC, Nobbs AH, Jepson K, Jepson MA, Vickerman MM, Aqeel Alawfi S, Munro CA, Lamont RJ, Jenkinson HF. 2014. O-Mannosylation in Candida albicans enables development of interkingdom biofilm communities. mBio 5(2):e00911-14. doi:10.1128/mBio.00911-14.
REFERENCES
- 1. Odds FW. 1988. Candida and candidosis, 2nd ed. Baillière Tindall, London, United Kingdom [Google Scholar]
- 2. Ramage G, Martínez JP, López-Ribot JL. 2006. Candida biofilms on implanted biomaterials: a clinically significant problem. FEMS Yeast Res. 6:979–986. 10.1111/j.1567-1364.2006.00117.x [DOI] [PubMed] [Google Scholar]
- 3. Azie N, Neofytos D, Pfaller M, Meier-Kriesche HU, Quan SP, Horn D. 2012. The PATH (prospective antifungal therapy) Alliance® registry and invasive fungal infections: update 2012. Diagn. Microbiol. Infect. Dis. 73:293–300. 10.1016/j.diagmicrobio.2012.06.012 [DOI] [PubMed] [Google Scholar]
- 4. Paulitsch AH, Willinger B, Zsalatz B, Stabentheiner E, Marth E, Buzina W. 2009. In-vivo Candida biofilms in scanning electron microscopy. Med. Mycol. 47:690–696. 10.3109/13693780802635237 [DOI] [PubMed] [Google Scholar]
- 5. Busscher HJ, Rinastiti M, Siswomihardjo W, van der Mei HC. 2010. Biofilm formation on dental restorative and implant materials. J. Dent. Res. 89:657–665. 10.1177/0022034510368644 [DOI] [PubMed] [Google Scholar]
- 6. Harriott MM, Lilly EA, Rodriguez TE, Fidel PL, Jr, Noverr MC. 2010. Candida albicans forms biofilms on the vaginal mucosa. Microbiology 156:3635–3644. 10.1099/mic.0.039354-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Chandra J, Kuhn DM, Mukherjee PK, Hoyer LL, McCormick T, Ghannoum MA. 2001. Biofilm formation by the fungal pathogen Candida albicans: development, architecture, and drug resistance. J. Bacteriol. 183:5385–5394. 10.1128/JB.183.18.5385-5394.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Banerjee M, Uppuluri P, Zhao XR, Carlisle PL, Vipulanandan G, Villar CC, López-Ribot JL, Kadosh D. 2013. Expression of UME6, a key regulator of Candida albicans hyphal development, enhances biofilm formation via Hgc1- and Sun41-dependent mechanisms. Eukaryot. Cell 12:224–232. 10.1128/EC.00163-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Dwivedi P, Thompson A, Xie Z, Kashleva H, Ganguly S, Mitchell AP, Dongari-Bagtzoglou A. 2011. Role of Bcr1-activated genes hwp1 and hyr1 in Candida albicans oral mucosal biofilms and neutrophil evasion. PLoS One 6:e16218. 10.1371/journal.pone.0016218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Fanning S, Xu W, Solis N, Woolford CA, Filler SG, Mitchell AP. 2012. Divergent targets of Candida albicans biofilm regulator Bcr1 in vitro and in vivo. Eukaryot. Cell 11:896–904. 10.1128/EC.00103-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Ramage G, VandeWalle K, López-Ribot JL, Wickes BL. 2002. The filamentation pathway controlled by the Efg1 regulator protein is required for normal biofilm formation and development in Candida albicans. FEMS Microbiol. Lett. 214:95–100. 10.1111/j.1574-6968.2002.tb11330.x [DOI] [PubMed] [Google Scholar]
- 12. Mulhern SM, Logue ME, Butler G. 2006. Candida albicans transcription factor Ace2 regulates metabolism and is required for filamentation in hypoxic conditions. Eukaryot. Cell 5:2001–2013. 10.1128/EC.00155-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Nobile CJ, Andes DR, Nett JE, Smith FJ, Yue F, Phan QT, Edwards JE, Filler SG, Mitchell AP. 2006. Critical role of Bcr1-dependent adhesins in C. albicans biofilm formation in vitro and in vivo. PLoS Pathog. 2:e63. 10.1371/journal.ppat.0020063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Nobile CJ, Schneider HA, Nett JE, Sheppard DC, Filler SG, Andes DR, Mitchell AP. 2008. Complementary adhesin function in C. albicans biofilm formation. Curr. Biol. 18:1017–1024. 10.1016/j.cub.2008.09.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Douglas LJ. 2003. Candida biofilms and their role in infection. Trends Microbiol. 11:30–36. 10.1016/S0966-842X(02)00002-1 [DOI] [PubMed] [Google Scholar]
- 16. Uppuluri P, Chaturvedi AK, Lopez-Ribot JL. 2009. Design of a simple model of Candida albicans biofilms formed under conditions of flow: development, architecture, and drug resistance. Mycopathologia 168:101–109. 10.1007/s11046-009-9205-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Gow NA, Hube B. 2012. Importance of the Candida albicans cell wall during commensalism and infection. Curr. Opin. Microbiol. 15:406–412. 10.1016/j.mib.2012.04.005 [DOI] [PubMed] [Google Scholar]
- 18. Gow NA, van de Veerdonk FL, Brown AJ, Netea MG. 2011. Candida albicans morphogenesis and host defence: discriminating invasion from colonization. Nat. Rev. Microbiol. 10:112–122. 10.1038/nrmicro2711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Herrero AB, Uccelletti D, Hirschberg CB, Dominguez A, Abeijon C. 2002. The Golgi GDPase of the fungal pathogen Candida albicans affects morphogenesis, glycosylation, and cell wall properties. Eukaryot. Cell 1:420–431. 10.1128/EC.1.3.420-431.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Peltroche-Llacsahuanga H, Goyard S, d’Enfert C, Prill SK, Ernst JF. 2006. Protein O-mannosyltransferase isoforms regulate biofilm formation in Candida albicans. Antimicrob. Agents Chemother. 50:3488-3491. 10.1128/AAC.00606-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Plaine A, Walker L, Da Costa G, Mora-Montes HM, McKinnon A, Gow NA, Gaillardin C, Munro CA, Richard ML. 2008. Functional analysis of Candida albicans GPI-anchored proteins: roles in cell wall integrity and caspofungin sensitivity. Fungal Genet. Biol. 45:1404–1414. 10.1016/j.fgb.2008.08.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Hoyer LL, Green CB, Oh SH, Zhao X. 2008. Discovering the secrets of the Candida albicans agglutinin-like sequence (ALS) gene family—a sticky pursuit. Med. Mycol. 46:1-15. 10.1080/13693780701435317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Garcia MC, Lee JT, Ramsook CB, Alsteens D, Dufrêne YF, Lipke PN. 2011. A role for amyloid in cell aggregation and biofilm formation. PLoS One 6:e17632. 10.1371/journal.pone.0017632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Nobbs AH, Vickerman MM, Jenkinson HF. 2010. Heterologous expression of Candida albicans cell wall-associated adhesins in Saccharomyces cerevisiae reveals differential specificities in adherence and biofilm formation and in binding oral Streptococcus gordonii. Eukaryot. Cell 9:1622–1634. 10.1128/EC.00103-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Wachtler B, Citiulo F, Jablonowski N, Forster S, Dalle F, Schaller M, Wilson D, Hube B. 2012. Candida albicans-epithelial cell interactions: dissecting the roles of active penetration, induced endocytosis and host factors on the infection process. PLoS One 7:e36952. 10.1371/journal.pone.0036952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Richard M, Ibata-Ombetta S, Dromer F, Bordon-Pallier F, Jouault T, Gaillardin C. 2002. Complete glycosylphosphatidylinositol anchors are required in Candida albicans for full morphogenesis, virulence and resistance to macrophages. Mol. Microbiol. 44:841–853. 10.1046/j.1365-2958.2002.02926.x [DOI] [PubMed] [Google Scholar]
- 27. Cutler JE. 2001. N-glycosylation of yeast, with emphasis on Candida albicans. Med. Mycol. 39:75–86. 10.1080/714030993 [DOI] [PubMed] [Google Scholar]
- 28. Buurman ET, Westwater C, Hube B, Brown AJ, Odds FC, Gow NA. 1998. Molecular analysis of CaMnt1p, a mannosyl transferase important for adhesion and virulence of Candida albicans. Proc. Natl. Acad. Sci. U. S. A. 95:7670–7675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Mora-Montes HM, Bates S, Netea MG, Castillo L, Brand A, Buurman ET, Díaz-Jiménez DF, Jan Kullberg B, Brown AJ, Odds FC, Gow NA. 2010. A multifunctional mannosyltransferase family in Candida albicans determines cell wall mannan structure and host-fungus interactions. J. Biol. Chem. 285:12087–12095. 10.1074/jbc.M109.081513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Munro CA, Bates S, Buurman ET, Hughes HB, MacCallum DM, Bertram G, Atrih A, Ferguson MA, Bain JM, Brand A, Hamilton S, Westwater C, Thomson LM, Brown AJ, Odds FC, Gow NA. 2005. Mnt1p and Mnt2p of Candida albicans are partially redundant alpha-1,2-mannosyltransferases that participate in O-linked mannosylation and are required for adhesion and virulence. J. Biol. Chem. 280:1051–1060. 10.1074/jbc.M411413200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Díaz-Jiménez DF, Mora-Montes HM, Hernández-Cervantes A, Luna-Arias JP, Gow NA, Flores-Carreón A. 2012. Biochemical characterization of recombinant Candida albicans mannosyltransferases Mnt1, Mnt2 and Mnt5 reveals new functions in O- and N-mannan biosynthesis. Biochem. Biophys. Res. Commun. 419:77–82. 10.1016/j.bbrc.2012.01.131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Beaussart A, Alsteens D, El-Kirat-Chatel S, Lipke PN, Kucharíková S, Van Dijck P, Dufrêne YF. 2012. Single-molecule imaging and functional analysis of Als adhesins and mannans during Candida albicans morphogenesis. ACS Nano 6:10950–10964. 10.1021/nn304505s [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Castillo L, MacCallum DM, Brown AJ, Gow NA, Odds FC. 2011. Differential regulation of kidney and spleen cytokine responses in mice challenged with pathology-standardized doses of Candida albicans mannosylation mutants. Infect. Immun. 79:146–152. 10.1128/IAI.01004-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Brand A, Barnes JD, Mackenzie KS, Odds FC, Gow NA. 2008. Cell wall glycans and soluble factors determine the interactions between the hyphae of Candida albicans and Pseudomonas aeruginosa. FEMS Microbiol. Lett. 287:48–55. 10.1111/j.1574-6968.2008.01301.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Harriott MM, Noverr MC. 2011. Importance of Candida-bacterial polymicrobial biofilms in disease. Trends Microbiol. 19:557–563. 10.1016/j.tim.2011.07.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Delhaes L, Monchy S, Fréalle E, Hubans C, Salleron J, Leroy S, Prevotat A, Wallet F, Wallaert B, Dei-Cas E, Sime-Ngando T, Chabé M, Viscogliosi E. 2012. The airway microbiota in cystic fibrosis: a complex fungal and bacterial community—implications for therapeutic management. PLoS One 7:e36313. 10.1371/journal.pone.0036313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Klotz SA, Chasin BS, Powell B, Gaur NK, Lipke PN. 2007. Polymicrobial bloodstream infections involving Candida species: analysis of patients and review of the literature. Diagn. Microbiol. Infect. Dis. 59:401–406. 10.1016/j.diagmicrobio.2007.07.001 [DOI] [PubMed] [Google Scholar]
- 38. Campos MS, Marchini L, Bernardes LA, Paulino LC, Nobrega FG. 2008. Biofilm microbial communities of denture stomatitis. Oral Microbiol. Immunol. 23:419–424. 10.1111/j.1399-302X.2008.00445.x [DOI] [PubMed] [Google Scholar]
- 39. Wright CJ, Burns LH, Jack AA, Back CR, Dutton LC, Nobbs AH, Lamont RJ, Jenkinson HF. 2013. Microbial interactions in building of communities. Mol. Oral Microbiol. 28:83–101. 10.1111/omi.12012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Diaz PI, Xie Z, Sobue T, Thompson A, Biyikoglu B, Ricker A, Ikonomou L, Dongari-Bagtzoglou A. 2012. Synergistic interaction between Candida albicans and commensal oral streptococci in a novel in vitro mucosal model. Infect. Immun. 80:620–632. 10.1128/IAI.05896-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Holmes AR, Gilbert C, Wells JM, Jenkinson HF. 1998. Binding properties of Streptococcus gordonii SspA and SspB (antigen I/II family) polypeptides expressed on the cell surface of Lactococcus lactis MG1363. Infect. Immun. 66:4633–4639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Silverman RJ, Nobbs AH, Vickerman MM, Barbour ME, Jenkinson HF. 2010. Interaction of Candida albicans cell wall Als3 protein with Streptococcus gordonii SspB adhesin promotes development of mixed-species communities. Infect. Immun. 78:4644–4652. 10.1128/IAI.00685-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Bamford CV, d’Mello A, Nobbs AH, Dutton LC, Vickerman MM, Jenkinson HF. 2009. Streptococcus gordonii modulates Candida albicans biofilm formation through intergeneric communication. Infect. Immun. 77:3696–3704. 10.1128/IAI.00438-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Hogan DA, Vik A, Kolter R. 2004. A Pseudomonas aeruginosa quorum-sensing molecule influences Candida albicans morphology. Mol. Microbiol. 54:1212–1223. 10.1111/j.1365-2958.2004.04349.x [DOI] [PubMed] [Google Scholar]
- 45. Kim Y, Mylonakis E. 2011. Killing of Candida albicans filaments by Salmonella enterica serovar Typhimurium is mediated by sopB effectors, parts of a type III secretion system. Eukaryot. Cell 10:782–790. 10.1128/EC.00014-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Brand A, MacCallum DM, Brown AJ, Gow NA, Odds FC. 2004. Ectopic expression of URA3 can influence the virulence phenotypes and proteome of Candida albicans but can be overcome by targeted reintegration of URA3 at the RPS10 locus. Eukaryot. Cell 3:900–909. 10.1128/EC.3.4.900-909.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Hobson RP, Munro CA, Bates S, MacCallum DM, Cutler JE, Heinsbroek SE, Brown GD, Odds FC, Gow NA. 2004. Loss of cell wall mannosylphosphate in Candida albicans does not influence macrophage recognition. J. Biol. Chem. 279:39628–39635. 10.1074/jbc.M405003200 [DOI] [PubMed] [Google Scholar]
- 48. Bates S, Hughes HB, Munro CA, Thomas WP, MacCallum DM, Bertram G, Atrih A, Ferguson MA, Brown AJ, Odds FC, Gow NA. 2006. Outer chain N-glycans are required for cell wall integrity and virulence of Candida albicans. J. Biol. Chem. 281:90–98. 10.1074/jbc.M510360200 [DOI] [PubMed] [Google Scholar]
- 49. Simonetti N, Strippoli V, Cassone A. 1974. Yeast-mycelial conversion induced by N-acetyl-D-glucosamine in Candida albicans. Nature 250:344–346 [DOI] [PubMed] [Google Scholar]
- 50. Almeida RS, Brunke S, Albrecht A, Thewes S, Laue M, Edwards JE, Filler SG, Hube B. 2008. The hyphal-associated adhesin and invasin Als3 of Candida albicans mediates iron acquisition from host ferritin. PLoS Pathog. 4:e1000217. 10.1371/journal.ppat.1000217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. De Groot PW, Bader O, de Boer AD, Weig M, Chauhan N. 2013. Adhesins in human fungal pathogens: glue with plenty of stick. Eukaryot. Cell 12:470–481. 10.1128/EC.00364-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Coleman DA, Oh SH, Zhao X, Zhao H, Hutchins JT, Vernachio JH, Patti JM, Hoyer LL. 2009. Monoclonal antibodies specific for Candida albicans Als3 that immunolabel fungal cells in vitro and in vivo and block adhesion to host surfaces. J. Microbiol. Methods 78:71–78. 10.1016/j.mimet.2009.05.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Albrecht A, Felk A, Pichova I, Naglik JR, Schaller M, de Groot P, Maccallum D, Odds FC, Schäfer W, Klis F, Monod M, Hube B. 2006. Glycosylphosphatidylinositol-anchored proteases of Candida albicans target proteins necessary for both cellular processes and host-pathogen interactions. J. Biol. Chem. 281:688–694. 10.1074/jbc.M509297200 [DOI] [PubMed] [Google Scholar]
- 54. Zupancic ML, Frieman M, Smith D, Alvarez RA, Cummings RD, Cormack BP. 2008. Glycan microarray analysis of Candida glabrata adhesin ligand specificity. Mol. Microbiol. 68:547–559. 10.1111/j.1365-2958.2008.06184.x [DOI] [PubMed] [Google Scholar]
- 55. Li F, Svarovsky MJ, Karlsson AJ, Wagner JP, Marchillo K, Oshel P, Andes D, Palecek SP. 2007. Eap1p, an adhesin that mediates Candida albicans biofilm formation in vitro and in vivo. Eukaryot. Cell 6:931–939. 10.1128/EC.00049-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Timpel C, Zink S, Strahl-Bolsinger S, Schröppel K, Ernst J. 2000. Morphogenesis, adhesive properties, and antifungal resistance depend on the Pmt6 protein mannosyltransferase in the fungal pathogen Candida albicans. J. Bacteriol. 182:3063–3071. 10.1128/JB.182.11.3063-3071.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Frieman MB, McCaffery JM, Cormack BP. 2002. Modular domain structure in the Candida glabrata adhesin Eap1p, a β1,6 glucan-cross-linked cell wall protein. Mol. Microbiol. 46:479–492. 10.1046/j.1365-2958.2002.03166.x [DOI] [PubMed] [Google Scholar]
- 58. Frank AT, Ramsook CB, Otoo HN, Tan C, Soybelman G, Rauceo JM, Gaur NK, Klotz SA, Lipke PN. 2010. Structure and function of glycosylated tandem repeats from Candida albicans Als adhesins. Eukaryot. Cell 9:405–414. 10.1128/EC.00235-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Gunasekera A, Alvarez FJ, Douglas LM, Wang HX, Rosebrock AP, Konopka JB. 2010. Identification of GIG1, a GlcNAc-induced gene in Candida albicans needed for normal sensitivity to the chitin synthase inhibitor nikkomycin Z. Eukaryot. Cell. 9:1476–1483. 10.1128/EC.00178-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Kumar MJ, Jamaluddin MS, Natarajan K, Kaur D, Datta A. 2000. The inducible N-acetylglucosamine catabolic pathway gene cluster in Candida albicans: discrete N-acetylglucosamine-inducible factors interact at the promoter of NAG1. Proc. Natl. Acad. Sci. U. S. A. 97:14218–14223. 10.1073/pnas.250452997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Naseem S, Gunasekera A, Araya E, Konopka JB. 2011. N-acetylglucosamine (GlcNAc) induction of hyphal morphogenesis and transcriptional responses in Candida albicans are not dependent on its metabolism. J. Biol. Chem. 286:28671-28580. 10.1074/jbc.M111.249854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Sudbery PE. 2011. Growth of Candida albicans hyphae. Nat. Rev. Microbiol. 9:737–748. 10.1038/nrmicro2636 [DOI] [PubMed] [Google Scholar]
- 63. Heilmann CJ, Sorgo AG, Mohammadi S, Sosinska GJ, de Koster CG, Brul S, de Koning LJ, Klis FM. 2013. Surface stress induces a conserved cell wall stress response in the pathogenic fungus Candida albicans. Eukaryot. Cell 12:254–264. 10.1128/EC.00278-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Dongari-Bagtzoglou A, Kashleva H, Dwivedi P, Diaz P, Vasilakos J. 2009. Characterization of mucosal Candida albicans biofilms. PLoS One 4:e7967. 10.1371/journal.pone.0007967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Ramage G, Saville SP, Wickes BL, López-Ribot JL. 2002. Inhibition of Candida albicans biofilm formation by farnesol, a quorum-sensing molecule. Appl. Environ. Microbiol. 68:5459–5463. 10.1128/AEM.68.11.5459-5463.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Vialás V, Perumal P, Gutierrez D, Ximénez-Embún P, Nombela C, Gil C, Chaffin WL. 2012. Cell surface shaving of Candida albicans biofilms, hyphae, and yeast form cells. Proteomics 12:2331–2339. 10.1002/pmic.201100588 [DOI] [PubMed] [Google Scholar]
- 67. Sosinska GJ, de Koning LJ, de Groot PW, Manders EM, Dekker HL, Hellingwerf KJ, de Koster CG, Klis FM. 2011. Mass spectrometric quantification of the adaptations in the wall proteome of Candida albicans in response to ambient pH. Microbiology 157:136–146. 10.1099/mic.0.044206-0 [DOI] [PubMed] [Google Scholar]
- 68. Naglik JR, Challacombe SJ, Hube B. 2003. Candida albicans secreted aspartyl proteinases in virulence and pathogenesis. Microbiol. Mol. Biol. Rev. 67:400–428. 10.1128/MMBR.67.3.400-428.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Schild L, Heyken A, de Groot PW, Hiller E, Mock M, de Koster C, Horn U, Rupp S, Hube B. 2011. Proteolytic cleavage of covalently linked cell wall proteins by Candida albicans Sap9 and Sap10. Eukaryot. Cell 10:98–109. 10.1128/EC.00210-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Naglik J, Albrecht A, Bader O, Hube B. 2004. Candida albicans proteinases and host/pathogen interactions. Cell. Microbiol. 6:915–926. 10.1111/j.1462-5822.2004.00439.x [DOI] [PubMed] [Google Scholar]
- 71. Salgado PS, Yan R, Taylor JD, Burchell L, Jones R, Hoyer LL, Matthews SJ, Simpson PJ, Cota E. 2011. Structural basis for the broad specificity to host-cell ligands by the pathogenic fungus Candida albicans. Proc. Natl. Acad. Sci. U. S. A. 108:15775–15779. 10.1073/pnas.1103496108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Fox SJ, Shelton BT, Kruppa MD. 2013. Characterization of genetic determinants that modulate Candida albicans filamentation in the presence of bacteria. PLoS One 8:e71939. 10.1371/journal.pone.0071939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Taff HT, Nett JE, Zarnowski R, Ross KM, Sanchez H, Cain MT, Hamaker J, Mitchell AP, Andes DR. 2012. A Candida biofilm-induced pathway for matrix glucan delivery: implications for drug resistance. PLoS Pathog. 8:e1002848. 10.1371/journal.ppat.1002848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Lipke PN, Garcia MC, Alsteens D, Ramsook CB, Klotz SA, Dufrêne YF. 2012. Strengthening relationships: amyloids create adhesion nanodomains in yeasts. Trends Microbiol. 20:59–65. 10.1016/j.tim.2011.10.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Fernandez-Escamilla AM, Rousseau F, Schymkowitz J, Serrano L. 2004. Prediction of sequence-dependent and mutational effects on the aggregation of peptides and proteins. Nat. Biotechnol. 22:1302–1306. 10.1038/nbt1012 [DOI] [PubMed] [Google Scholar]
- 76. Alsteens D, Garcia MC, Lipke PN, Dufrêne YF. 2010. Force-induced formation and propagation of adhesion nanodomains in living fungal cells. Proc. Natl. Acad. Sci. U. S. A. 107:20744–20749. 10.1073/pnas.1013893107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Fonzi WA, Irwin MY. 1993. Isogenic strain construction and gene mapping in Candida albicans. Genetics 134:717–728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Brachmann CB, Davies A, Cost GJ, Caputo E, Li J, Hieter P, Boeke JD. 1998. Designer deletion strains derived from Saccharomyces cerevisiae S288C: a useful set of strains and plasmids for PCR-mediated gene disruption and other applications. Yeast 14:115–132. [DOI] [PubMed] [Google Scholar]
- 79. Busuioc M, Buttaro BA, Piggot PJ. 2010. The pdh operon is expressed in a subpopulation of stationary-phase bacteria and is important for survival of sugar-starved Streptococcus mutans. J. Bacteriol. 192:4395–4402. 10.1128/JB.00574-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. McNab R, Jenkinson HF. 1998. Altered adherence properties of a Streptococcus gordonii hppA (oligopeptide permease) mutant result from transcriptional effects on cshA adhesin gene expression. Microbiology 144:127–136. 10.1099/00221287-144-1-127 [DOI] [PubMed] [Google Scholar]
- 81. Palmer RJ., Jr. 1999. Microscopy flowcells: perfusion chambers for real-time study of biofilms. Methods Enzymol. 310:160–166 [DOI] [PubMed] [Google Scholar]
- 82. de Groot PW, de Boer AD, Cunningham J, Dekker HL, de Jong L, Hellingwerf KJ, de Koster C, Klis FM. 2004. Proteomic analysis of Candida albicans cell walls reveals covalently bound carbohydrate-active enzymes and adhesins. Eukaryot. Cell 3:955–965. 10.1128/EC.3.4.955-965.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Ishihama Y, Oda Y, Tabata T, Sato T, Nagasu T, Rappsilber J, Mann M. 2005. Exponentially modified protein abundance index (emPAI) for estimation of absolute protein amount in proteomics by the number of sequenced peptides per protein. Mol. Cell. Proteomics 4:1265–1272. 10.1074/mcp.M500061-MCP200 [DOI] [PubMed] [Google Scholar]
- 84. Shinoda K, Tomita M, Ishihama Y. 2010. emPAI Calc—for the estimation of protein abundance from large-scale identification data by liquid chromatography-tandem mass spectrometry. Bioinformatics 26:576–577. 10.1093/bioinformatics/btp700 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
CSLM images of dual-species biofilms formed by the C. albicans wild type with S. gordonii DL1 in three different growth media as indicated. The image at the top of each column (A) is a representative xy stack of the biofilm assembled from the top down. The four images underneath (B, C, D, and E) are xz images showing organization and thickness (depth) of the dual-species biofilms from the xy image immediately above (B), the green S. gordonii component only (red subtracted) (C), the red C. albicans component only (green subtracted) (D), or (bottom image) a representative monospecies C. albicans wild-type biofilm grown under the same conditions (E). The significant structural and architectural differences associated with the different growth conditions employed are described in the text. The values shown in μm are average thickness (depth) measurements calculated from across the sections of the biofilms shown. Bar = 80 µm. Download
CSLM images of dual-species biofilms formed under flow conditions by the C. albicans wild type or mnt1Δ mnt2Δ mutant with S. gordonii DL1. The image at the top of each column (A) is a representative xy stack of the biofilm assembled from the top down. The four images underneath (B, C, D, and E) are xz images showing organization and thickness (depth) of dual-species biofilms from the xy image immediately above (B), the green S. gordonii component only (red subtracted) (C), the red C. albicans component only (green subtracted) (D), or (bottom image) a representative monospecies C. albicans wild-type or mnt1Δ mnt2Δ mutant biofilm grown under the same conditions (E). Note that YPT-GlcNAc medium data only are shown for the mnt1Δ mnt2Δ mutant because the other media did not support biofilm formation (see Fig. 5). The values shown in μm are average thickness (depth) measurements calculated from across the sections of the biofilms shown. Bar = 80 µm. Download
Diagrammatic model for cell wall protein adhesin-mediated interactions of C. albicans with S. gordonii. Mannosylation catalyzed by the enzymes Mnt1/Mnt2 is proposed to be essential for formation of a functional adhesin on the cell surface. The initial mannosylation acts by stabilizing adhesin structure in the first instance, necessary for allowing subsequent unfolding and extension. The Als3 adhesin is depicted as an example. The apical N-terminal domains (red) of Als3 protrude from the hyphal cell wall surface and are recognized by S. gordonii cells (71). Binding causes force-induced unfolding of Als3, and evidence presented in this article suggests that this may be enhanced under conditions of flow (shear). The adhesive (pulling) forces then elicit clustering of Als3 protein molecules to form nanodomains over the cell surface, as has been suggested previously (32), thus promoting multiple force interactions of Als3 proteins with bacteria, forming aggregates. It is suggested that the T domains (orange) containing amyloidogenic sequences are important for hydrophobic clustering and the formation of nanoadesomes (74). In this model, the Sap9 aspartyl proteinase is shown as modulating the activities of these nanodomains by disrupting the intermolecular forces. This would tend to enhance disaggregation of filaments, leading to increased biofilm permeability and more flexibility for recognition of additional binding partner molecules. Dispersal of cells from intermicrobial cell contacts might also be regulated via Sap9 hydrolysis of intramolecular peptide bonds of proteins engaged in intermolecular adhesion events. Download
Strains used in this study
3D video clip of biofilm structure formed by C. albicans wild type in YPT-GlcNAc Download
3D video clip of biofilm structure formed by C. albicans mnt1Δ mnt2Δ mutant Download
3D video clip of biofilm structure formed by C. albicans wild type in YPT-GlcNAc, under flow Download
3D video clip of biofilm structure formed by C. albicans mnt1Δ mnt2Δ mutant, under flow Download
3D video clip of dual-species biofilm structure formed by C. albicans wild type with S. gordonii DL1 in YPT-GlcNAc, under flow Download
3D video clip of dual-species biofilm structure formed by C. albicans mnt1Δ mnt2Δ mutant with S. gordonii in YPT-GlcNAc, under flow Download