Abstract
Alzheimer’s disease (AD) is the most common neurodegenerative disorder affecting the elderly people. AD is characterized by progressive and gradual decline in cognitive function and memory loss. While familial early-onset AD is usually associated with gene mutations, the etiology of sporadic late-onset form of AD is largely unknown. It has been reported that environmental factors and epigenetic alterations significantly contribute to the process of AD. Our previous studies have documented that chronic hypoxia is one of the environmental factors that may trigger the AD development and aggravate the disease progression. In this review, we will summarize the pathological effects of chronic hypoxia on the onset and development of AD and put forward the possible molecule mechanisms underlying the chronic hypoxia mediated AD pathogenesis. Finally, we propose that epigenetic regulations may represent new opportunity for the therapeutic intervention of this disease.
Keywords: Alzheimer’s disease, Chronic hypoxia, Epigenetic modification, DNA methylation, Histone acetylation
Introduction
Alzheimer’s disease (AD) is the most common neurodegenerative disorder in the elderly people. The latest epidemiology study of AD reported that the number of people with the disease in China is significantly increased from 1.93 million in 1990 to 5.69 million in 2010 [1], representing a major health problem and a heavy financial burden on individuals and society as a whole. Clinically, AD is characterized by progressive and gradual decline in cognitive function, accompanying with severe memory loss and, ultimately, decreasing physical functions and death. The clinical manifestations of AD are associated with the specific pathological changes in the brain of the patient. The pathological hallmarks of the disease are extracellular neuritic plaques, intracellular neurofibrillary tangles (NFTs), synaptic loss and neuron degeneration. Aberrant hyperphosphorylation of tau protein is the main constituent of NFTs. Neuritic plaques are composed of abnormal cleavaged β-amyloid (Aβ) peptide and are surrounded by reactive astrocytes and activated microglia [2,3].
AD can be classified into two forms: early-onset familial AD (FAD) and sporadic late-onset form of AD (LOAD). Most cases of AD are sporadic, with only 5% of the total number of AD exhibiting a clear genetic inheritance [4]. Gene mutations in the amyloid precursor protein (APP), presenilin1 (PS1) and presenilin2 (PS2) have been documented as causatives for FAD, and the Apolipoprotein E4 (ApoE4) allele has been associated with LOAD [5]. Several new genetic findings derived from genome-wide association studies (GWAS) have been identified as new susceptibility loci. These include clusterin (CLU), phosphatidylinositol binding clathrin assembly protein (PICALM), complement receptor 1 (CR1), bridging integrator (BIN1), microtubule affinity-regulating kinase4 (MARK4), the ATP cassette transporter (ABCA7), a membrane-spanning 4-domains, subfamily A (MS4A6A), CD33, a CD2-associated protein (CD2AP), and ephrin A1 (EPHA1) [6,7]. While gene mutations undoubtedly play an important role in the etiology of AD, more and more studies support that the environmental risk factors contribute greatly to the disease onset and progression [8-10]. It is believed that most cases of AD arise through interaction between genetic and environmental factors. Gene-environment interaction refers to a certain environmental exposure in the presence of a susceptibility gene [8]. It is thought that various environmental exposures can contribute to the risk of AD, such as aging, diet and nutrition, educational level, exposure to metals, pesticides, diabetes, brain trauma and etc. [11-14]. Among all the environmental exposures, chronic hypoxia has been extensively studied recently [15]. It has been shown that chronic hypoxia may trigger the AD development and may also aggravate the disease progression [15]. Hereby we summarize current findings of the pathological effects of chronic hypoxia on the onset and development of AD and put forward the potential molecule mechanisms of epigenetic modification underlying the hypoxia mediated AD pathogenesis.
Chronic hypoxia is a risk factor for dementia and Alzheimer’s disease
Chronic hypoxia is a reduction of oxygen supply, which is a direct consequence of hypoperfusion and restricts the function of organs, tissues or cells. Efficient oxygen delivery to brain tissues is essential for normal brain function and cells survival. Clinically, chronic hypoxia is a common pathophysiological event and contributes significantly to the progression of widespread diseases including fever, chronic obstructive pulmonary disease, obstructive sleep apnea syndrome, stroke, cancer, neurodegenerative disorders and etc. [16]. It has been shown that individuals who have suffered severe hypoxia or ischemia are more susceptible to developing AD [17-19]. Researchers reported that Aβ deposited in the brain after severe head injury [12,20], and cardiac arrest induced a time-dependent increase level of Aβ in the serum of heart attack patients [21]. A recent clinical prospective study by Yaffe and colleagues, demonstrated that older women with sleep-disordered breathing and hypoxia had a significantly increased risk of developing cognitive impairment and dementia [22]. Their findings suggested that chronic hypoxia but not sleep fragmentation or duration was associated with the higher risk of mild cognitive impairment or dementia. Another prospective study named Health, Aging, and Body Composition (Health ABC) that took a follow-up observation over the 11 years has shown that anemia was associated with an increased risk of developing dementia among older adults [23].
Consistent results have been published in transgenic mouse models or cell models showing the impact of chronic hypoxia on AD. It is found that hypoxia increased both mRNA and protein levels of APP in cells and elevated the level of Aβ in culture medium significantly [24]. Our previous study has reported that repeated hypoxia treatment could lead to more and larger senile plaque formations and more Aβ42 production in aged APPSwe + PS1A246E double transgenic mice [25]. Interestingly, similar neuropathology was observed in the study of prenatal hypoxia in AD development in adult mice [26]. It has been found that prenatal chronic intermittent hypoxia significantly contributed to memory and cognition deficit in adult mice [26]. Moreover, prenatal hypoxia increased senile plaque formation and Aβ production in adult mice [26]. Another study also showed that hypoxia treatment impaired spatial learning and memory of AD transgenic mice, and caused ER stress and neuronal apoptosis [27]. These effects were associated with abnormal calpain activation because suppression of m-calpain expression could attenuate the hypoxia-induced ER stress and apoptosis [27].
Chronic hypoxia may facilitate AD pathogenesis by increasing Aβ generation and decreasing Aβ degradation
Abnormal cleavaged β-amyloid peptide is the main constituent of neuritic plaques, which derives from sequential cleavage of APP by β-secretase and γ-secretase [28]. In the brain, the clearance of Aβ is mainly degraded by neprilysin (NEP) and insulin-degrading enzyme (IDE) [29]. Several studies including ours have shown that chronic hypoxia may elevate the production of Aβ and reduce the degradation of Aβ [25,26,30-33]. Chronic hypoxia in the human neuroblastoma SH-SY5Y cells caused reduced expression ADAM10, part of the α-secretases and increased the expression of β-site APP cleaving enzyme 1 (BACE1) [34,35]. Our previous study has demonstrated that chronic hypoxia increased Aβ generation by altering β- and γ-cleavage of APP [25]. The ratio of C99/C83 was elevated by chronic hypoxia in the brain of AD transgenic mice and the expression level of APH-1a was enhanced under hypoxia condition, which in turn would lead to increase in Aβ production [25]. Others have also shown that hypoxia could up-regulate BACE1 and APH-1a at both transcriptional and translational levels in vitro and in vivo[30,31,33]. It is widely accepted that hypoxia-inducible factor-1 (HIF-1) is the master regulator of the cellular response to chronic hypoxia [36]. Further investigations have revealed a functional hypoxia responsive element in the BACE1 gene promoter, to which HIF-1α can bind and result in increased activity of β-secretase under hypoxia conditions [30,33]. Similar HIF-1α binding site AP4 was identified in the promoter of APH-1a [31]. The binding of AP4 and HIF-1 to the promoter under hypoxia conditions may significantly affect the expression of APH-1a and lead to increased activity of γ-secretase [31]. All these findings suggest that hypoxia may increase the generation of Aβ in vivo and in vitro.
It is known that NEP plays a key role in the degradation of Aβ in the brain and it is one of the most important Aβ-degrading enzymes [29,37]. Evidence showed that NEP mRNA, protein and activity levels were declined not only in AD but also in the normal aging in the brains [38-42]. Fisk et al. demonstrated that hypoxia reduced NEP expression at the protein and mRNA levels as well as its activity [43,44]. We previously tested the expression levels of NEP in the cortex of AD transgenic mice, and found that the protein level of NEP started to reduce at the age of 6-month old in the brain of AD mice in comparison with age-matched wide-type mice [26]. Interestingly, the results also documented that chronic hypoxia induced significantly down regulation of NEP protein level in both transgenic mice and wide-type mice [26]. The declined level of NEP would reduce the clearance of Aβ. Given to the aggravated pathogenesis of AD and the over-load of Aβ burden under hypoxia condition, the chronic hypoxia-induced down regulation of NEP was implied to be a significant event in AD.
Chronic hypoxia may aggravate Aβ burden through epigenetic modifications on genes associated with Aβ metabolism
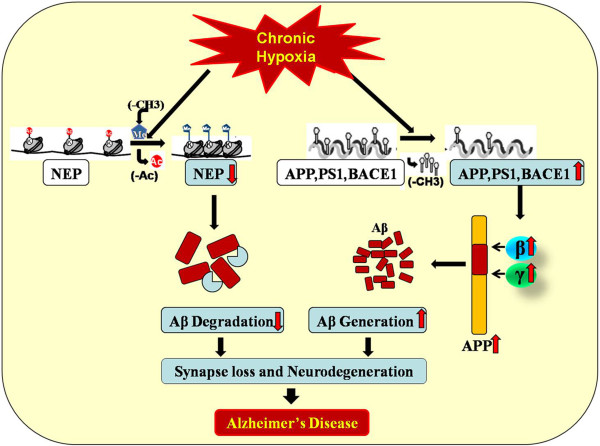
It has been indicated that environmental factors and epigenetic mechanisms are likely to contribute to the etiology of LOAD [8-10]. Epigenetic mechanisms modify heritable and non-heritable traits without altering the underlying DNA code, mediated through the reversible modifications of DNA and histones [45]. Epigenetic processes play an important role in normal physical functions of cells and the body, so aberrant epigenetic modification are hypothesized to contribute to a majority of pathologies [46]. Abnormal PS1 methylation patterns have previously been associated with hypomethylation in promoter [47]. DNA demethylation has a strong correlation with transcriptional activation [48]. Hypomethylation in promoter CpG islands of other AD-associated genes such as APP and BACE1 has also been reported, which in turn may lead to abnormal up regulation of these genes and over-production of Aβ [49,50]. In the study of epigenetic differences in monozygotic twins discordant for AD, a significantly reduction of DNA methylation was observed in the temporal cortex neuronal nuclei of AD twin [51]. It was shown that hypoxia can reduce global DNA methylation in cancer cell lines in vivo and in vitro[52]. Importantly, hypoxia could cause long-lasting change in DNA methylation change in promoter regions, some of which could be highly correlated with transcriptional modulation in a number of genes involved in neural growth and development [53]. Chen et al. found that Aβ could reduce global DNA methylation and increase NEP promoter methylation and further suppress the NEP expression in mRNA and protein levels [54]. However, the methylation status of the NEP promoters did not regulate its expression in vitro[55], and chronic hypoxia did not affect the methylation patterns of NEP gene promoters in mouse primary cortical and hippocampal neurons [32]. Interestingly, chronic hypoxia could cause significant down regulation of NEP by up-regulating G9a histone methyltransferase and histone deacetylase 1 (HDAC1), which resulted in increased expression of H3K9me2 and decreased expression of H3-Ace respectively [32]. In addition, methylation inhibitor 5-Aza, HDAC inhibitor valproic acid VA, and siRNA-mediated knockdown of G9a or HDAC1 could reverse the expression of NEP [32]. Others also reported that the NEP promoter could be repressed by HDACs and the expression of NEP was repressed in neuronal cells via the competitive binding of HDACs to its promoter [55]. All these findings suggest that chronic hypoxia aggravates AD by epigenetic modulations, mainly focusing on DNA methylaion and histones modifications, which are of tremendous importance to the onset and progression of the disease (Figure 1).
Figure 1.
The effect of chronic hypoxia on AD. Hypoxia increases the β and γ cleavage of APP by demethylation in promoter CpG islands of APP, PS1, and BACE1. In addition, chronic hypoxia reduces the expression of NEP by up-regulating histone methyltransferase and histone deacetylase. All these contribute to the deposition of Aβ and AD pathogenesis.
Epigenetic regulation targeting Aβ burden may help relieve the chronic hypoxia-mediated disease process
Currently, the treatment of AD focuses mainly on improving symptoms, targeting cholinergic and glutamatergic neuron transmission, with no therapeutics that can cure the disease [56]. Researchers around the world have worked hard to search for novel therapeutics for AD, which include the anti-Aβ (drugs to reduce its production, prevent its aggregation and promote its clearance), anti-tau (drugs to prevent its aggregation or phosphorylation), neurotrophins, and others [56]. Epigenetic regulation is a strong candidate therapeutic, since epigenetic modifications are reversible while genetic mutations are not. Drugs which can modulate DNA methylation and remodel the structure of chromatin through post-translational modifications of histones are promising potential candidates [57-60]. Recently, several drugs have been reported to be effective on AD mice models by modulating DNA methylation and histone acetylation separately, although none have yet entered clinical development [58,59,61,62]. Sodium valproic acid is one of these disease-modifying drugs, which may not only attenuate AD pathogenesis in transgenic AD mice, but also show effect in hypoxia-induced AD in the model. Valproic acid is a widely used antiepileptic drug, which has recently been found to have neuroprotection [63,64], and histone deacetylase inhibitory property may offer potential therapeutic option for AD [65]. It has been reported that valproic acid could inhibit Aβ production, reduce neuritic plaque formation, and improve behavioral deficits in AD mice [66]. Further study has demonstrated that valproic acid could attenuate the prenatal hypoxia-induced AD neuropathology, improved learning deficits and decrease Aβ42 levels in AD transgenic mice by up regulation of NEP [67]. Moreover, valproic acid could also restore memory deficit in adult offspring caused by prenatal hypoxia via elevating NEP expression and activity [68]. Hence, epigenetic drugs that increase histone acetylation by inhibiting HDAC could be useful in the disease treatment. Strategies targeting Aβ burden by epigenetic regulations, may represent new opportunities for therapeutic intervention to AD.
Conclusion
Epigenetic effects can present throughout life and environmental factors can influence the individuals repeatedly along their whole life. A wide range of clinical diseases may be associated with chronic hypoxia including obstructive sleep apnea, cerebrovascular diseases, systemic hypertension, cardiovascular disease, chronic obstructive pulmonary disease, pulmonary hypertension, congestive heart failure and others [69]. Given to the possibility that chronic hypoxia is associated with AD, more attention should be paid to this pathological event and the underlying mechanisms. In addition, we hypothesize that prevent chronic hypoxic condition may be helpful for the reduction of AD. Unlike genetic mutations in FAD, epigenetic alterations are reversible, which are easier to modulate the process. Therefore, understanding the mechanisms of epigenetic alteration during AD pathogenesis caused by environmental factors, is essential for searching new treatment strategy for the disease.
Abbreviations
AD: Alzheimer’s disease; NFTs: Neurofibrillary tangles; Aβ: beta-amyloid; FAD: Familial AD; LOAD: Late-onset form of AD; APP: Amyloid-precursor protein; PS: Presenilin; Apo E: Apolipoprotein E; GWAS: Genome-wide association studies; CLU: Clusterin; PICALM: Phosphatidylinositol binding clathrin assembly protein; CR1: Complement receptor 1; BIN1: Bridging integrator; MARK4: Microtubule affinity-regulating kinase4; ABCA7: The ATP cassette transporter; MS4A6A: A membrane-spanning 4-domains, subfamily A; CD2AP: A CD2-associated protein; EPHA1: Ephrin A1; Health ABC: Health, Aging, and Body Composition; NEP: Neprilysin; IDE: Insulin-degrading enzyme; HIF-1: Hypoxia-inducible factor-1; BACE: Beta-site APP-cleaving enzyme; CBZ: Carbamazepine.
Competing interests
All authors declare no competing financial interests.
Authors’ contributions
HL reviewed the literature and has written the initial manuscript draft; WL reviewed and critiqued the manuscript. Both authors read and approved the final manuscript.
Contributor Information
Hui Liu, Email: liuhui@sjtu.edu.cn.
Weidong Le, Email: wdle@sibs.ac.cn.
Acknowledgement
This study was funded by research grants from the National Nature Science Foundation (NO. 81000541and NO. 81171201) and by the National Basic Research Program (NO. 2011CB510003).
References
- Chan KY, Wang W, Wu JJ, Liu L, Theodoratou E, Car J, Middleton L, Russ TC, Deary IJ, Campbell H, Wang W, Rudan I. Epidemiology of Alzheimer’s disease and other forms of dementia in China, 1990–2010: a systematic review and analysis. Lancet. 2013;381:2016–2023. doi: 10.1016/S0140-6736(13)60221-4. [DOI] [PubMed] [Google Scholar]
- Pimplikar SW, Nixon RA, Robakis NK, Shen J, Tsai LH. Amyloid-independent mechanisms in Alzheimer’s disease pathogenesis. J Neurosci. 2010;30:14946–14954. doi: 10.1523/JNEUROSCI.4305-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swerdlow RH. Pathogenesis of Alzheimer’s disease. Clin Interv Aging. 2007;2:347–359. [PMC free article] [PubMed] [Google Scholar]
- Goedert M, Spillantini MG. A century of Alzheimer’s disease. Science. 2006;314:777–781. doi: 10.1126/science.1132814. [DOI] [PubMed] [Google Scholar]
- Bertram L, Lill CM, Tanzi RE. The genetics of Alzheimer disease: back to the future. Neuron. 2010;68:270–281. doi: 10.1016/j.neuron.2010.10.013. [DOI] [PubMed] [Google Scholar]
- Harold D, Abraham R, Hollingworth P, Sims R, Gerrish A, Hamshere ML, Pahwa JS, Moskvina V, Dowzell K, Williams A. Genome-wide association study identifies variants at CLU and PICALM associated with Alzheimer’s disease. Nat Genet. 2009;41:1088–1093. doi: 10.1038/ng.440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollingworth P, Harold D, Sims R, Gerrish A, Lambert J-C, Carrasquillo MM, Abraham R, Hamshere ML, Pahwa JS, Moskvina V. Common variants at ABCA7, MS4A6A/MS4A4E, EPHA1, CD33 and CD2AP are associated with Alzheimer’s disease. Nat Genet. 2011;43:429–435. doi: 10.1038/ng.803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chouliaras L, Sierksma AS, Kenis G, Prickaerts J, Lemmens MA, Brasnjevic I, van Donkelaar EL, Martinez-Martinez P, Losen M, De Baets MH, Kholod N, Van Leeuwen F, Hof PR, Van Os J, Steinbusch HWM, Van den Hove DLA, Rutten BPF. Gene-environment interaction research and transgenic mouse models of Alzheimer’s disease. Int J Alzheimers Dis. 2010;2010 doi: 10.4061/2010/859101. doi: 10.4061/2010/859101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lahiri DK, Maloney B, Zawia NH. The LEARn model: an epigenetic explanation for idiopathic neurobiological diseases. Mol Psychiatry. 2009;14:992–1003. doi: 10.1038/mp.2009.82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lahiri DK, Zawia NH, Greig NH, Sambamurti K, Maloney B. Early-life events may trigger biochemical pathways for Alzheimer’s disease: the “LEARn” model. Biogerontology. 2008;9:375–379. doi: 10.1007/s10522-008-9162-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrillo MC, Blackwell A, Hampel H, Lindborg J, Sperling R, Schenk D, Sevigny JJ, Ferris S, Bennett DA, Craft S, Timothy H, Klunk W. Early risk assessment for Alzheimer’s disease. Alzheimers Dement. 2009;5:182–196. doi: 10.1016/j.jalz.2009.01.019. [DOI] [PubMed] [Google Scholar]
- Fleminger S, Oliver D, Lovestone S, Rabe-Hesketh S, Giora A. Head injury as a risk factor for Alzheimer’s disease: the evidence 10 years on; a partial replication. J Neurol Neurosurg Psychiatry. 2003;74:857–862. doi: 10.1136/jnnp.74.7.857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sivanandam TM, Thakur MK. Traumatic brain injury: a risk factor for Alzheimer’s disease. Neurosci Biobehav Rev. 2012;36:1376–1381. doi: 10.1016/j.neubiorev.2012.02.013. [DOI] [PubMed] [Google Scholar]
- Seshadri S, Beiser A, Selhub J, Jacques PF, Rosenberg IH, D’Agostino RB, Wilson PW, Wolf PA. Plasma homocysteine as a risk factor for dementia and Alzheimer’s disease. N Engl J Med. 2002;346:476–483. doi: 10.1056/NEJMoa011613. [DOI] [PubMed] [Google Scholar]
- Zhang X, Le W. Pathological role of hypoxia in Alzheimer’s disease. Exp Neurol. 2010;223:299–303. doi: 10.1016/j.expneurol.2009.07.033. [DOI] [PubMed] [Google Scholar]
- Michiels C. Physiological and pathological responses to hypoxia. Am J Pathol. 2004;164:1875–1882. doi: 10.1016/S0002-9440(10)63747-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivan CS, Seshadri S, Beiser A, Au R, Kase CS, Kelly-Hayes M, Wolf PA. Dementia after stroke the Framingham study. Stroke. 2004;35:1264–1268. doi: 10.1161/01.STR.0000127810.92616.78. [DOI] [PubMed] [Google Scholar]
- Savva GM, Stephan BC. Epidemiological studies of the effect of stroke on incident dementia a systematic review. Stroke. 2010;41:e41–e46. doi: 10.1161/STROKEAHA.109.559880. [DOI] [PubMed] [Google Scholar]
- Ukraintseva S, Sloan F, Arbeev K, Yashin A. Increasing rates of dementia at time of declining mortality from stroke. Stroke. 2006;37:1155–1159. doi: 10.1161/01.STR.0000217971.88034.e9. [DOI] [PubMed] [Google Scholar]
- Roberts GW, Gentleman SM, Lynch A, Murray L, Landon M, Graham DI. Beta amyloid protein deposition in the brain after severe head injury: implications for the pathogenesis of Alzheimer’s disease. J Neurol Neurosurg Psychiatry. 1994;57:419–425. doi: 10.1136/jnnp.57.4.419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zetterberg H, Mörtberg E, Song L, Chang L, Provuncher GK, Patel PP, Ferrell E, Fournier DR, Kan CW, Campbell TG. Hypoxia due to cardiac arrest induces a time-dependent increase in serum amyloid β levels in humans. PLoS One. 2011;6:e28263. doi: 10.1371/journal.pone.0028263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kristine Yaffe AML, Harrison SL, Redline S, Spira AP, Ensrud KE, Ancoli-Israel S, Stone KL. Sleep-disordered breathing, hypoxia, and risk of mild cognitive impairment and dementia in older women. JAMA. 2011;306:613–619. doi: 10.1001/jama.2011.1115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong CH, Falvey C, Harris TB, Simonsick EM, Satterfield S, Ferrucci L, Metti AL, Patel KV, Yaffe K. Anemia and risk of dementia in older adults: findings from the Health ABC study. Neurology. 2013;81:528–533. doi: 10.1212/WNL.0b013e31829e701d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Dong Z, Liu B, Zhuo Y, Sun X, Yang Z, Ge J, Tan Z. Hypoxia induces beta-amyloid in association with death of RGC-5 cells in culture. Biochem Biophys Res Commun. 2011;410:40–44. doi: 10.1016/j.bbrc.2011.05.101. [DOI] [PubMed] [Google Scholar]
- Li L, Zhang X, Yang D, Luo G, Chen S, Le W. Hypoxia increases Abeta generation by altering beta- and gamma-cleavage of APP. Neurobiol Aging. 2009;30:1091–1098. doi: 10.1016/j.neurobiolaging.2007.10.011. [DOI] [PubMed] [Google Scholar]
- Zhang X, Li L, Zhang X, Xie W, Li L, Yang D, Heng X, Du Y, Doody RS, Le W. Prenatal hypoxia may aggravate the cognitive impairment and Alzheimer’s disease neuropathology in APPSwe/PS1A246E transgenic mice. Neurobiol Aging. 2013;34:663–678. doi: 10.1016/j.neurobiolaging.2012.06.012. [DOI] [PubMed] [Google Scholar]
- Wang CY, Xie JW, Wang T, Xu Y, Cai JH, Wang X, Zhao BL, An L, Wang ZY. Hypoxia-triggered m-calpain activation evokes endoplasmic reticulum stress and neuropathogenesis in a transgenic mouse model of Alzheimer’s disease. CNS Neurosci Ther. 2013;19:820–833. doi: 10.1111/cns.12151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearson HA, Peers C. Physiological roles for amyloid beta peptides. J Physiol. 2006;575:5–10. doi: 10.1113/jphysiol.2006.111203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malito E, Hulse RE, Tang W-J. Amyloid β-degrading cryptidases: insulin degrading enzyme, presequence peptidase, and neprilysin. Cell Mol Life Sci. 2008;65:2574–2585. doi: 10.1007/s00018-008-8112-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun X, He G, Qing H, Zhou W, Dobie F, Cai F, Staufenbiel M, Huang LE, Song W. Hypoxia facilitates Alzheimer’s disease pathogenesis by up-regulating BACE1 gene expression. Proc Natl Acad Sci U S A. 2006;103:18727–18732. doi: 10.1073/pnas.0606298103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang R, Zhang YW, Zhang X, Liu R, Zhang X, Hong S, Xia K, Xia J, Zhang Z, Xu H. Transcriptional regulation of APH-1A and increased gamma-secretase cleavage of APP and Notch by HIF-1 and hypoxia. FASEB J. 2006;20:1275–1277. doi: 10.1096/fj.06-5839fje. [DOI] [PubMed] [Google Scholar]
- Wang Z, Yang D, Zhang X, Li T, Li J, Tang Y, Le W. Hypoxia-induced down-regulation of neprilysin by histone modification in mouse primary cortical and hippocampal neurons. PLoS One. 2011;6:e19229. doi: 10.1371/journal.pone.0019229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Zhou K, Wang R, Cui J, Lipton SA, Liao FF, Xu H, Zhang YW. Hypoxia-inducible factor 1alpha (HIF-1alpha)-mediated hypoxia increases BACE1 expression and beta-amyloid generation. J Biol Chem. 2007;282:10873–10880. doi: 10.1074/jbc.M608856200. [DOI] [PubMed] [Google Scholar]
- Marshall AJ, Rattray M, Vaughan PF. Chronic hypoxia in the human neuroblastoma SH-SY5Y causes reduced expression of the putative α-secretases, ADAM10 and TACE, without altering their mRNA levels. Brain Res. 2006;1099:18–24. doi: 10.1016/j.brainres.2006.05.008. [DOI] [PubMed] [Google Scholar]
- Webster NJ, Green KN, Peers C, Vaughan PF. Altered processing of amyloid precursor protein in the human neuroblastoma SH SY5Y by chronic hypoxia. J Neurochem. 2002;83:1262–1271. doi: 10.1046/j.1471-4159.2002.01236.x. [DOI] [PubMed] [Google Scholar]
- Iyer NV, Kotch LE, Agani F, Leung SW, Laughner E, Wenger RH, Gassmann M, Gearhart JD, Lawler AM, Aimee YY. Cellular and developmental control of O2 homeostasis by hypoxia-inducible factor 1α. Genes Dev. 1998;12:149–162. doi: 10.1101/gad.12.2.149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farris W, Schütz SG, Cirrito JR, Shankar GM, Sun X, George A, Leissring MA, Walsh DM, Qiu WQ, Holtzman DM. Loss of neprilysin function promotes amyloid plaque formation and causes cerebral amyloid angiopathy. Am J Pathol. 2007;171:241–251. doi: 10.2353/ajpath.2007.070105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Apelt J, Ach K, Schliebs R. Aging-related down-regulation of neprilysin, a putative β-amyloid-degrading enzyme, in transgenic Tg2576 Alzheimer-like mouse brain is accompanied by an astroglial upregulation in the vicinity of β-amyloid plaques. Neurosci Lett. 2003;339:183–186. doi: 10.1016/S0304-3940(03)00030-2. [DOI] [PubMed] [Google Scholar]
- Caccamo A, Oddo S, Sugarman MC, Akbari Y, LaFerla FM. Age-and region-dependent alterations in Aβ-degrading enzymes: implications for Aβ-induced disorders. Neurobiol Aging. 2005;26:645–654. doi: 10.1016/j.neurobiolaging.2004.06.013. [DOI] [PubMed] [Google Scholar]
- Wang D-S, Lipton RB, Katz MJ, Davies P, Buschke H, Kuslansky G, Verghese J, Younkin SG, Eckman C, Dickson DW. Decreased neprilysin immunoreactivity in Alzheimer disease, but not in pathological aging. J Neuropathol Exp Neurol. 2005;64:378–385. doi: 10.1093/jnen/64.5.378. [DOI] [PubMed] [Google Scholar]
- Russo R, Borghi R, Markesbery W, Tabaton M, Piccini A. Neprylisin decreases uniformly in Alzheimer’s disease and in normal aging. FEBS Lett. 2005;579:6027–6030. doi: 10.1016/j.febslet.2005.09.054. [DOI] [PubMed] [Google Scholar]
- Carpentier M, Robitaille Y, DesGroseillers L, Boileau G, Marcinkiewicz M. Declining expression of neprilysin in Alzheimer disease vasculature: possible involvement in cerebral amyloid angiopathy. J Neuropathol Exp Neurol. 2002;61:849–856. doi: 10.1093/jnen/61.10.849. [DOI] [PubMed] [Google Scholar]
- Fisk L, Nalivaeva NN, Boyle JP, Peers CS, Turner AJ. Effects of hypoxia and oxidative stress on expression of neprilysin in human neuroblastoma cells and rat cortical neurones and astrocytes. Neurochem Res. 2007;32:1741–1748. doi: 10.1007/s11064-007-9349-2. [DOI] [PubMed] [Google Scholar]
- Nalivaevaa NN, Fisk L, Kochkina EG, Plesneva SA, Zhuravin IA, Babusikova E, Dobrota D, Turner AJ. Effect of hypoxia/ischemia and hypoxic preconditioning/reperfusion on expression of some amyloid degrading enzymes. Ann N Y Acad Sci. 2004;1035:21–33. doi: 10.1196/annals.1332.002. [DOI] [PubMed] [Google Scholar]
- Jaenisch R, Bird A. Epigenetic regulation of gene expression: how the genome integrates intrinsic and environmental signals. Nat Genet. 2003;33:245–254. doi: 10.1038/ng1089. [DOI] [PubMed] [Google Scholar]
- Rodenhiser D, Mann M. Epigenetics and human disease: translating basic biology into clinical applications. Can Med Assoc J. 2006;174:341–348. doi: 10.1503/cmaj.050774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S-C, Oelze B, Schumacher A. Age-specific epigenetic drift in late-onset Alzheimer’s disease. PLoS One. 2008;3:e2698. doi: 10.1371/journal.pone.0002698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones PA. Functions of DNA methylation: islands, start sites, gene bodies and beyond. Nat Rev Genet. 2012;13:484–492. doi: 10.1038/nrg3230. [DOI] [PubMed] [Google Scholar]
- Fuso A, Seminara L, Cavallaro RA, D’Anselmi F, Scarpa S. S-adenosylmethionine/homocysteine cycle alterations modify DNA methylation status with consequent deregulation of PS1 and BACE and beta-amyloid production. Mol Cell Neurosci. 2005;28:195–204. doi: 10.1016/j.mcn.2004.09.007. [DOI] [PubMed] [Google Scholar]
- Roger L, West JML, Maroun LE. Hypomethylation of the amyloid precursor protein gene in the brain of an Alzheimer’s disease patient. J Mol Neurosci. 1995;6:141–146. doi: 10.1007/BF02736773. [DOI] [PubMed] [Google Scholar]
- Mastroeni D, McKee A, Grover A, Rogers J, Coleman PD. Epigenetic differences in cortical neurons from a pair of monozygotic twins discordant for Alzheimer’s disease. PLoS One. 2009;4:e6617. doi: 10.1371/journal.pone.0006617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shahrzad S, Bertrand K, Minhas K, Coomber B. Induction of DNA hypomethylation by tumor hypoxia. Epigenetics. 2007;2:119–125. doi: 10.4161/epi.2.2.4613. [DOI] [PubMed] [Google Scholar]
- Hartley I, Elkhoury FF, Shin JH, Xie B, Gu X, Gao Y, Zhou D, Haddad GG. Long-lasting changes in DNA methylation following short-term hypoxic exposure in primary hippocampal neuronal cultures. PLoS One. 2013;8:e77859. doi: 10.1371/journal.pone.0077859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen K-L, Wang SS-S, Yang Y-Y, Yuan R-Y, Chen R-M, Hu C-J. The epigenetic effects of amyloid-β < sub > 1–40</sub > on global DNA and neprilysin genes in murine cerebral endothelial cells. Biochem Biophys Res Commun. 2009;378:57–61. doi: 10.1016/j.bbrc.2008.10.173. [DOI] [PubMed] [Google Scholar]
- Belyaev ND, Nalivaeva NN, Makova NZ, Turner AJ. Neprilysin gene expression requires binding of the amyloid precursor protein intracellular domain to its promoter: implications for Alzheimer disease. EMBO Rep. 2008;10:94–100. doi: 10.1038/embor.2008.222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mangialasche F, Solomon A, Winblad B, Mecocci P, Kivipelto M. Alzheimer’s disease: clinical trials and drug development. Lancet Neurol. 2010;9:702–716. doi: 10.1016/S1474-4422(10)70119-8. [DOI] [PubMed] [Google Scholar]
- Lee JH. Mini review: epigenetic modification is linked to alzheimers disease: is it a maker or a marker? BMB Rep. 2010;43:649–655. doi: 10.5483/BMBRep.2010.43.10.649. [DOI] [PubMed] [Google Scholar]
- Adwan L, Zawia NH. Epigenetics: a novel therapeutic approach for the treatment of Alzheimer’s disease. Pharmacol Ther. 2013;139(1):41–50. doi: 10.1016/j.pharmthera.2013.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caraci F, Leggio GM, Drago F, Salomone S. Epigenetic drugs for Alzheimer’s disease: hopes and challenges. Br J Clin Pharmacol. 2012;75:1154–1155. doi: 10.1111/j.1365-2125.2012.04443.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Yu J-T, Tan M-S, Jiang T, Tan L. Epigenetic mechanisms in Alzheimer’s disease: Implications for pathogenesis and therapy. Ageing Res Rev. 2013;12(4):1024–41. doi: 10.1016/j.arr.2013.05.003. [DOI] [PubMed] [Google Scholar]
- Cuadrado-Tejedor M, Oyarzabal J, Lucas MP, Franco R, García-Osta A. Epigenetic drugs in Alzheimer’s disease. Biomolecular Concepts. 2013;4:433–445. doi: 10.1515/bmc-2013-0012. [DOI] [PubMed] [Google Scholar]
- Peedicayil J. Epigenetic drugs for Alzheimer’s disease. Br J Clin Pharmacol. 2012;75:1152–1153. doi: 10.1111/j.1365-2125.2012.04444.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monti B, Polazzi E, Contestabile A. Biochemical, molecular and epigenetic mechanisms of valproic acid neuroprotection. Curr Mol Pharmacol. 2009;2:95–109. doi: 10.2174/1874467210902010095. [DOI] [PubMed] [Google Scholar]
- Nalivaeva NN, Belyaev ND, Turner AJ. Sodium valproate: an old drug with new roles. Trends Pharmacol Sci. 2009;30:509–514. doi: 10.1016/j.tips.2009.07.002. [DOI] [PubMed] [Google Scholar]
- Zhang X-Z, Li X-J, Zhang H-Y. Valproic acid as a promising agent to combat Alzheimer’s disease. Brain Res Bull. 2010;81:3–6. doi: 10.1016/j.brainresbull.2009.09.003. [DOI] [PubMed] [Google Scholar]
- Qing H, He G, Ly PT, Fox CJ, Staufenbiel M, Cai F, Zhang Z, Wei S, Sun X, Chen C-H. Valproic acid inhibits Aβ production, neuritic plaque formation, and behavioral deficits in Alzheimer’s disease mouse models. J Exp Med. 2008;205:2781–2789. doi: 10.1084/jem.20081588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Zhang XJ, Li T, Li J, Tang Y, Le W. Valproic acid reduces neuritic plaque formation and improves learning deficits in APPSwe/PS1A246E transgenic mice via preventing the prenatal hypoxia induced down regulation of neprilysin. CNS Neurosci Ther. 2013;20:209–217. doi: 10.1111/cns.12186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nalivaeva NN, Belyaev ND, Lewis DI, Pickles AR, Makova NZ, Bagrova DI, Dubrovskaya NM, Plesneva SA, Zhuravin IA, Turner AJ. Effect of sodium valproate administration on brain neprilysin expression and memory in rats. J Mol Neurosci. 2012;46:569–577. doi: 10.1007/s12031-011-9644-x. [DOI] [PubMed] [Google Scholar]
- Daulatzai MA. Death by a thousand cuts in Alzheimer’s disease: hypoxia—the prodrome. Neurotox Res. 2013;24:216–243. doi: 10.1007/s12640-013-9379-2. [DOI] [PubMed] [Google Scholar]