Abstract
Background
Fetal hemoglobin (HbF) is the major modifier of the clinical course of sickle cell anemia. Its levels are highly heritable and its interpersonal variability is modulated in part by three quantitative trait loci (QTL) that effect HbF gene expression. Genome-wide association studies (GWAS) have identified single nucleotide polymorphisms (SNPs) in these QTLs that are highly associated with HbF but explain only 10 to 12% of the variance of HbF. Combining SNPs into a genetic risk score (GRS) can help to explain a larger amount of the variability of HbF level but the challenge of this approach is to select the optimal number of SNPs to be included in the GRS.
Methods and Results
We develop a collection of 14 models with GRS composed of different numbers of SNPs, and use the ensemble of these models to predict HbF in sickle cell anemia patients. The models were trained in 841 sickle cell anemia patients and were tested in three independent cohorts. The ensemble of 14 models explained 23.4% of the variability in HbF in the discovery cohort, while the correlation between predicted and observed HbF in the 3 independent cohorts ranged between 0.28 and 0.44. The models included SNPs in BCL11A, the HBS1L-MYB intergenic region and the site of the HBB gene cluster, QTL previously associated with HbF.
Conclusions
An ensemble of 14 genetic risk models can predict HbF levels with accuracy between 0.28 and 0.44 and the approach may prove useful in other applications.
Keywords: sickle cell disease, hemoglobin, genetics, association studies, risk prediction, risk factor
Introduction
HbF is the major modifier of the clinical features of sickle cell anemia (homozygosity for HBB glu6val) and β thalassemia. HbF inhibits sickle hemoglobin (HbS) polymerization and compensates for the deficit of normal HbA in β thalassemia1. If it were possible to know at birth the HbF level likely to be present after stabilization of this measurement at about age 5 years2, then a better patient-specific prognosis might be given and HbF-inducing treatments better tailored to the individual. The γ-globin chain of HbF is encoded by the linked HBG1 and HBG2 genes. Levels of HbF in adults are highly heritable and the production of HbF is genetically regulated by several quantitative trait loci that modulate HBG1 and HBG2 expression.3–6 Genome wide association studies (GWAS) in sickle cell anemia have identified single nucleotide polymorphisms (SNPs) in BCL11A, the HBS1L-MYB intergenic region and elements linked to the HBB gene cluster that jointly explain 10–12% of the variability of HbF.2, 7 However, it is possible that SNPs that are significantly associated with HbF levels but do not reach genome-wide significance may explain additional variability and be used to predict HbF levels. This is in part due to the difference in the goals of the analysis of GWAS and phenotype prediction; in order to increase the amount of variability explained in a phenotype one may need to use SNPs that fall below the genome-wide significance threshold.8
One approach to genetic risk prediction uses a summary of the risk alleles in the form of a genetic risk score (GRS) as a covariate of the model.9–12 A GRS can summarize a large amount of genetic information into a single covariate, but the challenge is to identify the optimal number of SNPs to be included in the score. To overcome this challenge, we present a novel method of creating an ensemble of models with the GRS composed of a different number of SNPs to produce more robust predictions. This method extends the approach introduced in Sebastiani et al 2012 for building genetic risk prediction models from case control studies to predict quantitative traits.13 We show that an ensemble of 14 models with GRSs comprising 1 to 14 SNPs that were trained in a set of 841 sickle cell anemia patients from the Cooperative Study of Sickle Cell Disease (CSSCD) can predict HbF in three different cohorts of African Americans with sickle cell anemia with correlation between observed and predicted values between 0.28 and 0.44.
Methods
Participants
HbF levels were measured in 841 African American subjects from the CSSCD (NCT00005277) homozygous for the HbS gene or with HbS-β0 thalassemia.14 The validation cohorts included 181 patients from the Pulmonary Hypertension and Sickle Cell Disease with Sildenafil Therapy (Walk-PHaSST) Study (NCT00492531), 77 patients from a study of pulmonary hypertension in children with sickle cell disease (PUSH NCT 00495638), and 127 sickle cell anemia patients from the Comprehensive Sickle Cell Centers Collaborative Data (C-data) project. Subjects for all cohorts were selected based on the following criteria: age >5 years for the HbF measurement, no hydroxyurea use, and no recent transfusion. In addition, patients in the validation cohorts had hemoglobin phenotypes similar to that of the discovery set. The demographics of these studies have been described.14–16 Some characteristics of the sickle cell anemia patients are described in Table 1. All studies were approved by the institutional review board (IRB) of each participating institution.
Table 1.
Patient characteristics. Summary statistics of patient characteristics in the CSSCD cohort, and the PUSH, WALK-PHaSST and C-Data replication cohorts. For each cohort statistics (mean and standard deviate or frequencies) are reported for all patients included in the analysis. The last row reports the frequency and proportion of individuals with gene deletion alpha thalassemia (at least one alpha gene deletion).
| CSSCD (N=841) Mean (StD) | PUSH (N=77) Mean (StD) | Walk-PHaSST (N=181) Mean (StD) | C-Data (N=127) Mean (StD) | |
|---|---|---|---|---|
| Age (years) | 17.19(10.69) | 12.49 (4.69) | 36.35 (12.54) | 13–17 |
| Gender (% male) | 53.7% | 50.6% | 51.9% | 55.9% |
| HbF (%) | 6.65 (5.50) | 9.81 (8.23) | 6.07 (5.60) | 7.59(5.09) |
| Hemoglobin (d/mL) | 8.42 (1.33) | 8.62 (1.25) | 8.50 (1.69) | NA |
| α thalassemia (% yes) | 143 (31.4%) | 21 (27.2%) | 46 (25.4%) | NA |
HbF Values
HbF was measured by alkali denaturation in the CSSCD or by high-pressure liquid chromatography. Within the range of observed HbF, both alkali denaturation and high-pressure liquid chromatography give similar results. For the CSSCD subjects, longitudinal HbF values were gathered from phases 1 through 3 of the study, only steady-state values were used (ie, measurements not taken during an acute event) and summarized by the median of the longitudinal values.2 Because HbF is known to decrease in early years of life, we only used HbF measurements at age 5 years or older. The cubic root transformation of HbF was used in all statistical analyses, to remove asymmetry.
Genotyping
DNA from the CSSCD, PUSH, C-Data and Walk-PHaSST samples that formed the discovery and replication cohorts were genotyped at Boston University using Illumina Human610-Quad SNP arrays with approximately 600,000 SNPs. Samples were processed according the manufacturer's protocol and BeadStudio Software was used to make genotype calls utilizing the Illumina pre-defined clusters. Samples with less than a 95% call rate were removed and SNPs with a call rate <97.5% were re-clustered. After re-clustering, SNPs with call rates >97.5%, cluster separation score > 0.25, excess heterozygosity between −0.10 and 0.10, and minor allele frequency > 5% were retained in the analysis. We used the genome-wide identity by descent analysis in PLINK to discover unknown relatedness.17 Pairs with IBD measurements greater than 0.2 were deemed to be related and related subjects within individual or different studies were removed. We also removed samples with inconsistent gender findings defined by heterozygosity of the X chromosome and gender recorded in the database.
Statistical Analysis
Genotype data from the CSSCD were used as the training data set to develop different GRSs. Initially, the association of each SNP was tested using a linear regression model adjusted for gender using the additive coding in PLINK17, and the p-values for each association were computed. No significant associations were found between HbF and the first ten principal components (PCs) computed using EIGENSOFT.18 Lack of inflation of the associations was confirmed by the genomic inflation factor 1.003.18 SNPs were sorted by increasing p-values, starting from the most significant SNP associated with HbF (rs766432, p-value=2.61×10−21), and the list of SNPs was pruned by removing those SNPs in high linkage disequilibrium (r2 > 0.8). If two SNPs were found to be in high linkage disequilibrium, the SNP with lowest p-value was kept and the other SNP was removed. This process was repeated until no SNPs in high linkage disequilibrium remained which left 500,325 SNPs. This pruned and sorted list of SNPs was used to generate a sequence of unweighted GRS for each subject in the CSSCD by cumulatively adding the number of risk alleles (an allele that causes a decrease in HbF) for each SNP. The first GRS included only the most significant SNP, the second GRS was generated by adding the second SNP from the sorted list of SNPs to the first GRS and so on using the iterative formula:
where n is the number of SNPs, and Risk Allelei,j is the number of risk alleles carried by individual i for the jth SNP. A sequence of weighted GRS was also generated by weighting each risk allele as:
where tj is the t-statistic to test the association of the jth SNP with HbF. This analysis was repeated for the first 10,000 SNPs (p-value< .02185) and generated 10,000 GRS, for each of the subjects in the CSSCD. Each of these GRS was included as covariate in a linear regression model and the regression coefficients of the resultant 10,000 linear regression models were estimated using Least Squares methods in the CSSCD data in the R package. The fitted regression models were used to predict HbF using the formula:
where is the predicted HbF value for individual i using the nth GRS, and , are the estimate of the regression coefficients. Cumulative ensembles of the predictions13, 19–21 were computed using the formula:
The predictive value of the genetic risk models and their ensembles was evaluated in the CSSCD, and in the three independent cohorts. The proportion of variability explained in the ensemble of GRS models in the CSSCD set was computed as the squared Pearson correlation between the predicted and observed values while the predictive accuracy in the independent sets was evaluated by computing the Pearson correlation between the observed and predicted values of HbF. The number of SNPs with GRS that maximized the correlation between observed and predicted values in the three independent cohorts was selected as the optimal number of SNPs. To evaluate the predictive value of genetic data relative to non-genetic risk factors, a non-genetic prediction model based on age, gender and the presence of alpha thalassemia was estimated and the genetic risk models were also adjusted by age, gender and alpha thalassemia. The accuracy of these additional models was assessed by the correlation between the predicted and observed HbF values.
Results
Table 1 shows patient characteristics of the four studies. The percent male and HbF concentration were approximately the same across the Walk-PhaSST, C-Data and CSSCD cohorts. Average age varied across the four cohorts; the CSSCD studied both children and adults, C-Data and PUSH were skewed toward pediatric age patients while the Walk-PHaSST cohort consisted mainly of adults. Patients in the PUSH study were younger and had significantly higher HbF levels (p-value=0.0016).
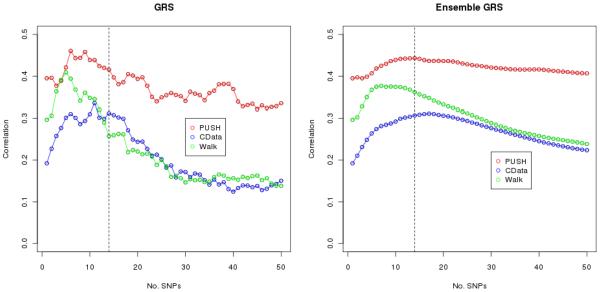
Figure 1 shows the correlation between the observed and predicted HbF values using genetic risk models with unweighted GRS (left panel) and the ensemble of these GRS models (right panel) for the top 50 SNPs, in each of the validation cohorts. The correlation ranged between 0.2 and 0.4 for prediction with only 1 SNP; it peaked at 0.45 for prediction with GRSs that include 10 to 15 SNPs in the PUSH data, with both the standard genetic risk models and their ensembles, and declines for larger numbers of SNPs in the models. Inclusion of more than 50 SNPs decreased the correlation even further. While the inclusion of new SNPs in the GRS had substantial effects on the predictive accuracy of the genetic risk model, as shown by the up and down pattern from one model to the next (left panel of Figure 1), the accuracy of the ensemble of these GRS models was more stable. The results using the weighted GRS were similar (Supplementary Figure 1).
Figure 1.
Correlation between observed and predicted values for increasing number of SNPs in the unweighted GRS. Plot of the correlation between the observed and predicted HbF in the three independent cohorts versus the number of SNPs in the top 50 unweighted GRS models (left panel) and ensemble of unweighted GRS models (right panel). The vertical bars are at No. SNP=14.
An ensemble of the first 14 GRS models had the highest average correlation among all three data sets and explained 23.4% of the variability in HbF in the CSSCD cohort. The correlation between observed and predicted HbF using the ensemble of 14 GRS models was 0.44, 0.28 and 0.39 in the PUSH, Walk-PHaSST and C-Data cohorts, respectively. Of these 14 SNPs, 5 were located in BCL11A; other SNPs were located in the olfactory receptor region on chromosome 11p15 and the site of the HBB gene cluster, and in the HBS1L-MYB interval on chromosome 6q and were found previously to be associated with HbF.2, 22, 23 Table 2 shows details of these 14 SNPs.
Table 2.
Summary of SNPs in prediction model. GWAS results of the 14 SNPs in the best GRS prediction model in the CSSCD cohort. β=estimate of genetic effect from additive model; SE= standard error.
| SNP | Gene | Coding Allele | β | SE | pvalue |
|---|---|---|---|---|---|
| rs766432 | Chr 2; BCL11A | C | 0.25 | 0.02 | 1.22E-22 |
| rs1O195871 | Chr 2; BCL11A | A | 0.21 | 0.02 | 3.30E-18 |
| rs6706648 | Chr 2; BCL11A | A | −0.19 | 0.02 | 1.30E-16 |
| rs6709302 | Chr 2; BCL11A | A | −0.14 | 0.02 | 3.69E-08 |
| rs9494145 | Chr 6; intergenic | G | 0.24 | 0.05 | 1.80E-07 |
| rs6732518 | Chr 2; BCL11A | G | 0.13 | 0.02 | 1.86E-07 |
| rs6446085 | Chr 3; FHIT | A | −0.11 | 0.02 | 9.43E-07 |
| rs10152034 | Chr 14; intergenic | A | −0.11 | 0.03 | 1.21E-05 |
| rs17114175 | Chr 14; intergenic | C | −0.16 | 0.02 | 1.59E-06 |
| rs2855039 | Chr 11; HBG1, HBG2 | G | 0.18 | 0.04 | 2.24E-06 |
| rs2239580 | Chr 14; COCH | A | −0.13 | 0.03 | 4.46E-06 |
| rs5006883 | Chr 11; OR51B5,OR51B6 | G | 0.17 | 0.04 | 5.32E-06 |
| rs9525079 | Chr 13; UGGT2 | G | 0.13 | 0.03 | 6.28E-06 |
| rs416586 | Chr 11; OR51A | G | 0.13 | 0.02 | 6.37E-06 |
| rs11794652 | Chr 9; FUBP3 | A | −0.10 | 0.02 | 6.51E-06 |
| rs12469604 | Chr 2; intergenic | A | 0.19 | 0.04 | 8.31E-06 |
| rs6932510 | Chr 6; RPS6KA2 | A | 0.16 | 0.04 | 8.52E-06 |
| rs7113817 | Chr 11; intergenic | A | 0.17 | 0.04 | 9.88E-06 |
| rs1O837814 | Chr 11; OR51B2,OR51B3P | A | 0.14 | 0.03 | 1.39E-05 |
| rs2021966 | Chr 6; intergenic | G | 0.10 | 0.02 | 1.79E-05 |
Adding non-genetic risk factors age and gender to the GRS models did not increase the amount of variability explained of HbF in the Howard and Walk-PHaSST cohorts. The non-genetic prediction model which included information on age, alpha thalassemia and gender only explained 6.8% of the variability in HbF in the CSSCD cohort.
It is noticeable that the genetic risk models had consistently higher predictive accuracy in the PUSH cohort in comparison with the C-Data and Walk-PHaSST cohorts. The age distribution of the CSSCD and PUSH cohorts was skewed toward children and young adolescents while the age distribution of Walk-PHaSST cohort was skewed toward adult patients (Supplementary Figure 2). Exact age of patients was not available for the C-Data cohort.
Discussion
In African Americans with sickle cell anemia, HbF level does not stabilize until the age of 5 years.2 Early prediction of stable adult HbF levels might help foresee some complications of sickle cell anemia and aid in its clinical management by guiding the decision of how vigorously to pursue HbF induction therapies. Our goal was to identify methods that could combine SNPs to provide a better prediction of HbF levels than single SNP analysis. We developed an ensemble of GRS that predicted HbF in 3 independent cohorts. In an ensemble of GRS models, as few as 14 SNPs explained a larger fraction of the variability in HbF and better predicted HbF levels when compared with single SNP analysis, or a single GRS model. Even though the ensemble of GRS explains 23.4% of the variability in HbF in the CSSCD cohort, it does not explain the totality of the variability in HbF that is due to heritability, estimated to be between 60% and 90%.5, 24 This missing heritability could be due to gene-gene interactions, epigenetic factors or multiple rare variants with small effects that GWAS are poorly designed to detect.25 Our GRS included SNPs with a MAF > 5%; however, as sickle cell anemia is a rare disease it is possible that some major genetic modifiers are rare variants with a lower allele frequency. Next generation sequencing might discover rare alleles that could be incorporated into a GRS to increase prediction accuracy. Increasing the number of genetic variants in the GRS may increase the total explained variability of HbF26; however, this could lead to overfitting and as one continues to add more genetic variants to the GRS the prediction accuracy in independent cohorts will decrease (as shown in Figure 1). The prediction accuracy of our weighted GRS model was similar to that of our unweighted GRS model. We hypothesize that this is due to the fact that weighted GRS models perform better when there is a difference in the genetic effects; however, Table 2 shows that the regression coefficients and standard errors from the GWAS of our top SNPs are all very similar.
An interesting result of our analysis is the systematically higher correlation between predicted and observed HbF values in the PUSH study. This result might suggest that the genetic models predict more accurately in children and young adults. Blood counts decline with advancing age in sickle cell disease and could reflect decades of bone marrow damage due to sickle vasoocclusion with relative bone marrow “failure”.27–29 Perhaps the higher correlation between the predicted and observed HbF levels in the younger patients of the PUSH cohort is a result of gene x environment interaction, where the genetic elements regulating HbF production are not impeded by the erythroid bone marrow injury associated with aging in the older cohorts. However, the difference in prediction accuracy could also be due to unobservable patient characteristics not accounted for in the model. For example, the CSSCD was conducted before the establishment of hydroxyurea as standard therapy for sickle cell anemia patients, and there may be survivor effects in more contemporary cohorts that are not accounted for in older cohorts. Testing these results in additional contemporary cohorts of sickle cell anemia will be necessary to explain the result.
The 14 SNP model included SNPs in the BCL11A region, the HBS1L-MYB intergenic region and SNPs in the olfactory receptor gene cluster 5' to the HBB gene complex. BCL11A down regulates HBG expression 30, 31, the HBS1L-MYB intergenic region might affect erythropoiesis and modulate BCL11A expression while the olfactory receptor region might control expression with the HBB gene-like cluster.32
There are alternate methods of genetic prediction that we did not explore in this paper including combining SNP information into a haplotype-based analysis,33, 34 multivariate regression models and machine learning type approaches such as support vector machines and cross validation (CV)35, 36, principal component analysis37, and Bayesian networks38–42. In our analysis, we used traditional regression based models and ensemble of these models: we trained the models in the discovery cohorts and examined the prediction results in independent cohorts to determine which model predicts most accurately. We investigated using 10 fold cross validation (CV) method to select the best number of SNPs in the discovery set. However, we noted in a large simulation study that using 10 fold cross validation tends to underestimate the true number of SNPs (Supplementary Information; Supplementary Figure 3), and noted that an ensemble of regression models appear to be more robust for prediction. This result is consistent with published literature.43. Since the 3 test sets were used to determine the optimal number of SNPs, replication of the results in additional independent sets is necessary to confirm the results. However, the substantial agreement of prediction of the ensemble of 14 models in the 3 sets is reassuring that this result is robust.
The ensemble of GRS models explained 23.4% of the variability in HbF in the CSSCD data and the correlation between predicted and observed HbF values in the 3 independent sets ranged from 0.28 to 0.44. These numbers are higher than results reported in the literature when GRS have been used as a predictive tool. For example, a study of 3,575 subjects from the Doetinchem Cohort Study computed a GRS to predict plasma total cholesterol levels which are highly genetically determined with a heritability estimated to be 40 to 60%.44, 45 Using 12 SNPs they were able to explain 6.9% of the total variability in total cholesterol levels.46 Participants in the Atherosclerosis Risk in Communities cohort of 10,745 individuals were used to construct a GRS of obesity in order to predict BMI.47, 48 The obesity GRS showed a correlation with BMI of r=0.12 for the unweighted GRS model and r=0.13 for the weighted model. Our ensemble of GRS models of HbF can explain more phenotype variability and had a higher predictive accuracy in comparison with single SNP analyses.
One of the major goals of GWAS was to identify genetic variants that are associated with disease or measures of disease severity to be used in personalized medicine. Many studies have shown the importance of including genetic variants beyond those that meet the genome-wide association threshold of 5×10−08 but many of these SNPs associations may be false positives and lower the accuracy of a prediction model.49, 50 Our study shows that the use of an ensemble of genetic risk models is robust to inclusion of false positive associations and the approach may prove useful in other applications.
Supplementary Material
Acknowledgments
Funding Sources: This work was supported by National Institutes of Health Grants R21HL114237 (PS), R01 HL87681 (MHS), RC2 L101212 (MHS), 5T32 HL007501 (JNM), 2R25 HL003679-8 (VRG), R01 HL079912 (VRG), 2M01 RR10284-10 (VRG), R01HL098032 (MTG), R01HL096973 (MTG), P01HL103455 (MTG), the Institute for Transfusion Medicine and the Hemophilia Center of Western Pennsylvania (MTG), U54 HL70583. The C-Data Project of the Comprehensive Sickle Cell Centers, U54HL70583 and NIH intramural funding 1 ZIA HL005116 and 1 ZIA HL006016
Footnotes
Conflict-of Interest Disclosure: The authors declare no competing financial interests.
References
- 1.Steinberg MH, Nagel RL, Forget BG, Higgs DR, Weatherall DJ. Genetic Modulation of Sickle Cell Disease and Thalassemia. In: Steinberg MH, Forget BG, Higgs DR, Weatherall DJ, editors. Disorders of hemoglobin: Genetics, Pathophysiology and Clinical Management. Cambridge University Press; Cambridge: 2009. pp. 638–657. [Google Scholar]
- 2.Solovieff N, Milton JN, Hartley SW, Sherva R, Sebastiani P, Dworkis DA, et al. Fetal hemoglobin in sickle cell anemia: genome-wide association studies suggest a regulatory region in the 5' olfactory receptor gene cluster. Blood. 2010;115:1815–1822. doi: 10.1182/blood-2009-08-239517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Steinberg MH, Sebastiani P. Genetic modifiers of sickle cell disease. Am J Hematol. 2012;87:795–803. doi: 10.1002/ajh.23232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Steinberg MH, Hsu H, Nagel RL, Milner PF, Adams JG, Benjamin L, et al. Gender and haplotype effects upon hematological manifestations of adult sickle cell anemia. Am J Hematol. 1995;48:175–181. doi: 10.1002/ajh.2830480307. [DOI] [PubMed] [Google Scholar]
- 5.Garner C, Tatu T, Reittie JE, Littlewood T, Darley J, Cervino S, et al. Genetic influences on F cells and other hematologic variables: a twin heritability study. Blood. 2000;95:342–346. [PubMed] [Google Scholar]
- 6.Menzel S, Thein SL. Genetic architecture of hemoglobin F control. Curr Opin Hematol. 2009;16:179–186. doi: 10.1097/moh.0b013e328329d07a. [DOI] [PubMed] [Google Scholar]
- 7.Bae HT, Baldwin CT, Sebastiani P, Telen MJ, Ashley-Koch A, Garrett M, et al. Meta-analysis of 2040 sickle cell anemia patients: BCL11A and HBS1L-MYB are the major modifiers of HbF in African Americans. Blood. 2012;120:1961–1962. doi: 10.1182/blood-2012-06-432849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gibson G. Rare and common variants: twenty arguments. Nat Rev Genet. 2011;13:135–145. doi: 10.1038/nrg3118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Meigs JB, Shrader P, Sullivan LM, McAteer JB, Fox CS, Dupuis J, et al. Genotype score in addition to common risk factors for prediction of type 2 diabetes. N Engl J Med. 2008;359:2208–2219. doi: 10.1056/NEJMoa0804742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Purcell SM, Wray NR, Stone JL, Visscher PM, O'Donovan MC, Sullivan PF, et al. Common polygenic variation contributes to risk of schizophrenia and bipolar disorder. Nature. 2009;460:748–752. doi: 10.1038/nature08185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Paynter NP, Chasman DI, Pare G, Buring JE, Cook NR, Miletich JP, et al. Association between a literature-based genetic risk score and cardiovascular events in women. JAMA. 2010;303:631–637. doi: 10.1001/jama.2010.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sebastiani P, Solovieff N, Sun JX. Naive Bayesian Classifier and Genetic Risk Score for Genetic Risk Prediction of a Categorical Trait: Not so Different after all! Front Genet. 2012;3:26. doi: 10.3389/fgene.2012.00026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sebastiani P, Solovieff N, Dewan AT, Walsh KM, Puca A, Hartley SW, et al. Genetic signatures of exceptional longevity in humans. PLoS ONE. 2012;7:e29848. doi: 10.1371/journal.pone.0029848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gaston M, Rosse WF. The cooperative study of sickle cell disease: review of study design and objectives. Am J Pediatr Hematol Oncol. 1982;4:197–201. [PubMed] [Google Scholar]
- 15.Dham N, Ensing G, Minniti C, Campbell A, Arteta M, Rana S, et al. Prospective echocardiography assessment of pulmonary hypertension and its potential etiologies in children with sickle cell disease. Am J Cardiol. 2009;104::713–720. doi: 10.1016/j.amjcard.2009.04.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Machado RF, Barst RJ, Yovetich NA, Hassell KL, Kato GJ, Gordeuk VR, et al. Hospitalization for pain in patients with sickle cell disease treated with sildenafil for elevated TRV and low exercise capacity. Blood. 2011;118:855–864. doi: 10.1182/blood-2010-09-306167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, et al. PLINK: A Tool Set for Whole-Genome Association and Population-Based Linkage Analyses. Am J Hum Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Price AL, Patterson NJ, Plenge RM, Weinblatt ME, Shadick NA, Reich D. Principal components analysis corrects for stratification in genome-wide association studies. Nat Genet. 2006;38:904–9. doi: 10.1038/ng1847. [DOI] [PubMed] [Google Scholar]
- 19.Breiman L. Bagging Predictors. Machine Learning. 1996;24:123–140. [Google Scholar]
- 20.Mevik B-H, Segtnan VH, Næs T. Ensemble methods and partial least squares regression. J Chemom. 2004;18:498–507. [Google Scholar]
- 21.Rokach L. Ensembled-based classifiers. Art Intell Review. 2010;33:1–39. [Google Scholar]
- 22.Sedgewick AE, Timofeev N, Sebastiani P, So JC, Ma ES, Chan LC, et al. BCL11A is a major HbF quantitative trait locus in three different populations with beta-hemoglobinopathies. Blood Cells Mol Dis. 2008;41:255–258. doi: 10.1016/j.bcmd.2008.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Uda M, Galanello R, Sanna S, Lettre G, Sankaran VG, Chen W, et al. Genome-wide association study shows BCL11A associated with persistent fetal hemoglobin and amelioration of the phenotype of beta-thalassemia. Proc Natl Acad Sci U S A. 2008;105:1620–1625. doi: 10.1073/pnas.0711566105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pilia G, Chen WM, Scuteri A, Orru M, Albai G, Dei M, et al. Heritability of cardiovascular and personality traits in 6,148 Sardinians. PLoS Genet. 2006;2:e132. doi: 10.1371/journal.pgen.0020132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hindorff LA, Sethupathy P, Junkins HA, Ramos EM, Mehta JP, Collins FS, et al. Potential etiologic and functional implications of genome-wide association loci for human diseases and traits. Proc Natl Acad Sci U S A. 2009;106:9362–9367. doi: 10.1073/pnas.0903103106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Galarneau G, Palmer CD, Sankaran VG, Orkin SH, Hirschhorn JN, Lettre G. Fine-mapping at three loci known to affect fetal hemoglobin levels explains additional genetic variation. Nat Genet. 2010;42:1049–1051. doi: 10.1038/ng.707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.West MS, Wethers D, Smith J, Steinberg M. Laboratory profile of sickle cell disease: a cross-sectional analysis. The Cooperative Study of Sickle Cell Disease. J Clin Epidemiol. 1992;45:893–909. doi: 10.1016/0895-4356(92)90073-v. [DOI] [PubMed] [Google Scholar]
- 28.Morris J, Dunn D, Beckford M, Grandison Y, Mason K, Higgs D, et al. The haematology of homozygous sickle cell disease after the age of 40 years. Br J Haematol. 1991;77:382–385. doi: 10.1111/j.1365-2141.1991.tb08588.x. [DOI] [PubMed] [Google Scholar]
- 29.Hayes RJ, Beckford M, Grandison Y, Mason K, Serjeant BE, Serjeant GR. The haematology of steady state homozygous sickle cell disease: frequency distributions, variation with age and sex, longitudinal observations. Br J Haematol. 1985;59:369–382. doi: 10.1111/j.1365-2141.1985.tb03002.x. [DOI] [PubMed] [Google Scholar]
- 30.Sankaran VG, Menne TF, Xu J, Akie TE, Lettre G, Van Handel B, et al. Human fetal hemoglobin expression is regulated by the developmental stage-specific repressor BCL11A. Science. 2008;322:1839–1842. doi: 10.1126/science.1165409. [DOI] [PubMed] [Google Scholar]
- 31.Chen Z, Luo HY, Steinberg MH, Chui DH. BCL11A represses HBG transcription in K562 cells. Blood Cells Mol Dis. 2009;42:144–149. doi: 10.1016/j.bcmd.2008.12.003. [DOI] [PubMed] [Google Scholar]
- 32.Feingold EA, Penny LA, Nienhuis AW, Forget BG. An olfactory receptor gene is located in the extended human beta-globin gene cluster and is expressed in erythroid cells. Genomics. 1999;61:15–23. doi: 10.1006/geno.1999.5935. [DOI] [PubMed] [Google Scholar]
- 33.Hughes MF, Saarela O, Stritzke J, Kee F, Silander K, Klopp N, et al. Genetic Markers Enhance Coronary Risk Prediction in Men: The MORGAM Prospective Cohorts. PLoS ONE. 2012;7:e40922. doi: 10.1371/journal.pone.0040922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Onuki R, Shibuya T, Kanehisa M. New kernel methods for phenotype prediction from genotype data. Genome Inform. 2010;22:132–141. [PubMed] [Google Scholar]
- 35.Wei Z, Wang K, Qu HQ, Zhang H, Bradfield J, Kim C, et al. From disease association to risk assessment: an optimistic view from genome-wide association studies on type 1 diabetes. PLoS Genet. 2009;5:e1000678. doi: 10.1371/journal.pgen.1000678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wu C, Walsh K, DeWan A, Hoh J, Wang Z. Disease risk prediction with rare and common variants. BMC Proceedings. 2011;5:S61. doi: 10.1186/1753-6561-5-S9-S61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Moore JH, Gilbert JC, Tsai CT, Chiang FT, Holden T, Barney N, et al. A flexible computational framework for detecting, characterizing, and interpreting statistical patterns of epistasis in genetic studies of human disease susceptibility. J Theor Biol. 2006;241:252–261. doi: 10.1016/j.jtbi.2005.11.036. [DOI] [PubMed] [Google Scholar]
- 38.Rodin AS, Boerwinkle E. Mining genetic epidemiology data with Bayesian networks I: Bayesian networks and example application (plasma apoE levels) Bioinformatics. 2005;21:3273–3278. doi: 10.1093/bioinformatics/bti505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sebastiani P, Ramoni MF, Nolan V, Baldwin CT. Steinberg MH. Genetic dissection and prognostic modeling of overt stroke in sickle cell anemia. Nat Genet. 2005;37:435–440. doi: 10.1038/ng1533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jiang X, Barmada MM, Cooper GF, Becich MJ. A Bayesian method for evaluating and discovering disease loci associations. PLoS ONE. 2011;6:e22075. doi: 10.1371/journal.pone.0022075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kang J, Zheng W, Li L, Lee J, Yan X, Zhao H. Use of Bayesian networks to dissect the complexity of genetic disease: application to the Genetic Analysis Workshop 17 simulated data. BMC Proceedings. 2011;5:S37. doi: 10.1186/1753-6561-5-S9-S37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sebastiani P, Perls TT. Prediction models that include genetic data. Circ Cardiovasc Genet. 2010;3:1–2. doi: 10.1161/CIRCGENETICS.110.937862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hartley SW, Monti S, Liu CT, Steinberg MH, Sebastiani P. Bayesian methods for multivariate modeling of pleiotropic SNP associations and genetic risk prediction. Front Genet. 2012;3:176. doi: 10.3389/fgene.2012.00176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lusis AJ, Fogelman AM, Fonarow GC. Genetic basis of atherosclerosis: part I: new genes and pathways. Circulation. 2004;110:1868–1873. doi: 10.1161/01.CIR.0000143041.58692.CC. [DOI] [PubMed] [Google Scholar]
- 45.Verschuren WM, Blokstra A, Picavet HS, Smit HA. Cohort profile: the Doetinchem Cohort Study. Int J Epidemiol. 2008;37:1236–1241. doi: 10.1093/ije/dym292. [DOI] [PubMed] [Google Scholar]
- 46.Lu Y, Feskens EJ, Boer JM, Imholz S, Verschuren WM, Wijmenga C, et al. Exploring genetic determinants of plasma total cholesterol levels and their predictive value in a longitudinal study. Atherosclerosis. 2010;213:200–205. doi: 10.1016/j.atherosclerosis.2010.08.053. [DOI] [PubMed] [Google Scholar]
- 47.Belsky DW, Moffitt TE, Sugden K, Williams B, Houts R, McCarthy J, et al. Development and evaluation of a genetic risk score for obesity. Biodemography Soc Biol. 2013;59:85–100. doi: 10.1080/19485565.2013.774628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Folsom AR, Chambless LE, Ballantyne CM, Coresh J, Heiss G, Wu KK, et al. An assessment of incremental coronary risk prediction using C-reactive protein and other novel risk markers: the atherosclerosis risk in communities study. Arch Intern Med. 2006;166:1368–1373. doi: 10.1001/archinte.166.13.1368. [DOI] [PubMed] [Google Scholar]
- 49.Kooperberg C, LeBlanc M. Obenchain V. Risk prediction using genome-wide association studies. Genet Epidemiol. 2009;34:643–652. doi: 10.1002/gepi.20509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yang J, Manolio TA, Pasquale LR, Boerwinkle E, Caporaso N, Cunningham JM, et al. Genome partitioning of genetic variation for complex traits using common SNPs. Nat Genet. 2010;43:519–525. doi: 10.1038/ng.823. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.