Abstract
In recent years, the term personalized medicine has received more and more attention in the field of healthcare. The increasing use of this term is closely related to the astonishing advancement in DNA sequencing technologies and other high-throughput biotechnologies. A large amount of personal genomic data can be generated by these technologies in a short time. Consequently, the needs for managing, analyzing, and interpreting these personal genomic data to facilitate personalized care are escalated. In this article, we discuss the challenges for implementing genomics-based personalized medicine in healthcare, current solutions to these challenges, and the roles of health information management (HIM) professionals in genomics-based personalized medicine.
Introduction
Personalized medicine utilizes personal medical information to tailor strategies and medications for diagnosing and treating diseases in order to maintain people's health.1 In this medical practice, physicians combine results from all available patient data (such as symptoms, traditional medical test results, medical history and family history, and certain personal genomic information) so that they can make accurate diagnoses and determine personalized treatment strategies accordingly.2 Health information management (HIM) professionals will play a critical role in this personalized healthcare practice because they are responsible for managing patient data.
With the improvement of high-throughput biotechnologies and the rapid decrease in DNA sequencing cost and time, genomics-based personalized medicine has already been practiced in a number of places, including the Geisinger Genomic Medicine Institute, Scripps Health, Cleveland Clinic, Vanderbilt University Medical Center, and the Medical College of Wisconsin, to name just a few. Doctors have ordered whole-genome sequencing for their patients, and other healthcare professionals have performed genomic data analysis to identify genetic reasons for certain diseases that cannot be determined by conventional approaches.3 Some hospitals, such as Children's Mercy Hospital in Kansas City4 and the University of Pittsburgh Medical Center,5, 6, 7 have started to consider or have already taken the first few steps toward genomics-based personalized medicine.
In December 2013, the US Food and Drug Administration (FDA) granted marketing authorization for the first high-throughput (next-generation) genomic sequencer, Illumina's MiSeqDx. This marketing authorization is an important step for utilizing genomic information in a healthcare setting because it allows the development and use of many new genome-based tests.8 The FDA and the National Institute for Standards and Technology have collaborated to create genomic reference materials for performance assessment, such as the whole human genome DNA and the best possible sequence interpretation of such genomes. The first human genome reference materials are expected to be available for public use around the end of 2014.9
Once genomic information is widely used in healthcare, it will be connected to the existing electronic health record (EHR) systems in certain ways, for instance, as one component of the data set or as an external link to a stand-alone genomic information database. In either case, HIM professionals will be responsible for managing and protecting these genomic data sets and extracting useful information from them. HIM professionals already have many of the needed skills and knowledge for managing genomic information, such as knowledge of database management, sensitive data encryption and decryption, statistical analysis, regulations on patient data, and procedures and policies for handling legal and ethical issues related to patient records. In this article, we specifically discuss some unique features of genomic information that bring up corresponding challenges for HIM professionals. This article is intended to inform HIM professionals about some highly relevant issues in this particular field so that they can be better prepared in their career. The topics discussed in this article are not a complete list of all the relevant challenges.
The first challenge is from the recently developed high-throughput biotechnologies, such as next-generation DNA sequencing technologies10 for whole-genome sequencing, microarray for gene expression patterns, RNA-seq11, 12 for identifying all versions of genes and their expression patterns, ChIP-seq13, 14 for determining all DNA segments that regulate expression of genes (regulatory elements) in a genome, and MDB-seq15 for collecting genome-wide DNA methylation profiles (DNA methylation can change the expression of genes). These technologies can generate huge amounts of data in a short period of time. At the same time, the accuracy of data from these technologies is problematic.
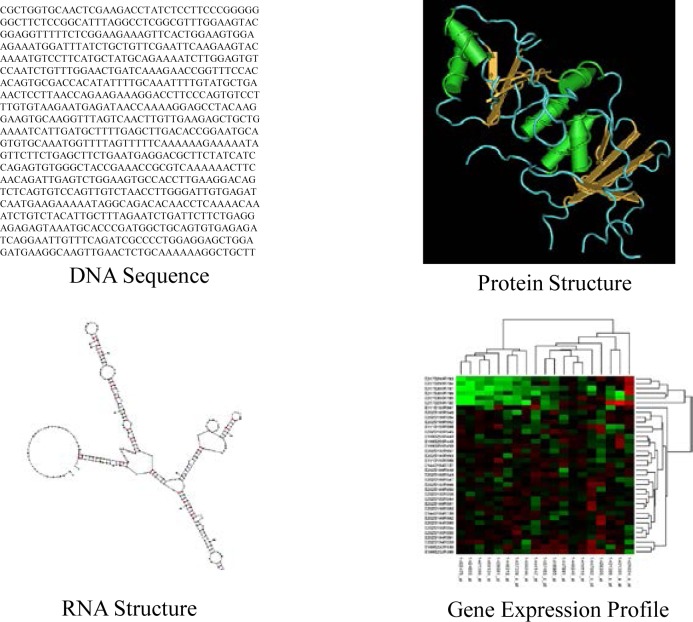
The next challenge is from genomic data analysis. Extracting valuable information from large but low-quality data sets is a difficult task. The situation is even worse when the data sets themselves are complex. Genomic data sets include different types of data, such as DNA sequences, RNA sequences and structures, protein sequences and structures, and gene expression profiles. Figure 1 illustrates these four types of genomic data. Each data type has its own characteristics, and none of the data sets is simple to analyze, especially when its volume is large. For instance, analyzing a DNA sequence with 300 nucleotide bases is not difficult, but identifying a short, informative DNA segment from 3 billion nucleotide bases (which is the length of a human genome) is a nontrivial task.
Figure 1.
Four Types of Genomic Data
For healthcare purposes, one critical piece of information is which genes and mutations are involved in the development of a disease. Decades ago, scientists already could determine associations between mutations in some genes and certain diseases. Recent technological advances have made it possible to determine the role of specific genetic variations in genetic diseases in large-scale genome-wide analyses. However, the newly generated research results are overwhelming (hundreds of papers have reported genetic variations associated with one disease, and many of them have reported completely different variations) and sometimes even conflict with each other; therefore, applying them in clinical practice is another challenge in genomics-based personalized medicine and HIM.
Since the cost of sequencing a human genome has sharply dropped to an affordable level (several thousand dollars per genome) and DNA sequencing may become a routine task in hospitals in the near future, doctors will want to apply the genomic information in clinical practice. One expensive and challenging task for obtaining clinically useful information is analyzing these large-scale sequences and other genomic data files, connecting the analysis results with research results in the literature,16 and linking the results to the EHR in a meaningful way.
Even if the ideal solutions to all of these technical challenges were found, unique and serious challenges remain in genomics-based personalized medicine: the security, privacy, legal, social, and ethical issues related to personal genomics. Many tough questions must be answered in this area before physicians can apply genomic information in clinical practice. Recent discussion on the release of the HeLa genome,17, 18 the most widely used human cell line in medical research, highlights the sensitivity of personal genomic data.19 After all, genomic data are different from traditional medical data. The release of an individual's genomic data can leak information about the descendants for many generations in the same family, while traditional medical data are, in most cases, only about the patient himself or herself. Therefore, public release of the HeLa genome has many privacy, social, and ethical issues and may impact the life of the Lacks family members even though the donor of the HeLa cell has been deceased for many years. HIM professionals need to consider this issue when they release personal genomic information to their patients. The information is not just about the patient but also is related to all the family members of the patient.
Apparently, these challenges cannot be solved by one group of healthcare professionals in one specific field. Individuals from many different fields must work together to solve these problems. In the traditional clinical practice, HIM professionals use the latest information technologies to provide timely, accurate, and complete medical data to physicians so that they can provide high-quality healthcare services to patients. In the genomics-based personalized medicine practice, HIM professionals will be responsible for managing complex genomic data, protecting highly sensitive personal genomic data, and extracting valuable information from these data sets.20 Although these tasks are not necessarily more challenging than what they are currently performing, HIM professionals do need to have more preparation to enhance their skills and knowledge in this new field. The purpose of this article is to help HIM professionals become aware of the unique challenges of genomic information and the current status of the profession in handling those challenges.
In the remainder of this article, we discuss each of these challenges in more detail, present the current solutions to these challenges, discuss other relevant issues, and offer our recommendations for healthcare and HIM professionals.
Challenges in Genomics-based Personalized Medicine
As mentioned previously, practicing genomics-based personalized medicine involves many challenges. Here we divide these challenges into five categories: (1) the overwhelming volume and poor quality of genomic data from high-throughput technologies; (2) the difficulties in genomic data analysis; (3) information security and privacy issues in managing personal genomic data; and (4) legal, social, and ethical issues related to personal genomic information. We will discuss each of them and focus on issues relevant to HIM professionals.
Challenges Related to Genomic Data from High-Throughput Technologies
The Human Genome Project (HGP)21, 22, 23 was conducted from 1990 to 2003 for the purpose of sequencing and understanding the human genome. The total cost of the HGP was about $3 billion, and one human genome was sequenced. In recent years, next-generation sequencing (NGS) technologies have dramatically reduced the DNA sequencing cost. The price dropped sharply from $3 billion to $100 million in 2007, then to roughly $1.5 million in 2008,24 and to roughly $5,000 in 2012.25 By the end of 2013, one could sequence a human genome in 24 hours for a cost of several thousand dollars.26 The commercialization and wide application of more advanced DNA sequencing technologies currently in development will further reduce the sequencing cost and increase the speed. Several other high-throughput technologies, such as RNA-seq, ChIP-seq, and MDB-seq, were developed to collect other types of genomic data. These technologies are based on sequencing and therefore share the advantages of NGS technologies (high speed and low cost). They also share the same problems, that is, they can generate a huge amount of raw data in a short time (terabytes in a few days), and they contain multiple sources of sequencing errors, such as errors in determining the nucleotide bases from the signal reader (base-calling), problems resulting from putting short DNA segments from sequencers together (DNA assembly), mistakes resulting from detecting genetic variations in the sequenced genome, and sequence differences obtained from different sequencing platforms because of different methods, chemical properties, and base-calling algorithms.27 The reason for these problems is that genome sequencing technologies are still in active development. Each of these NGS technologies also has its own advantages and disadvantages in aspects such as sequence accuracy, yield, and cost per million bases. Table 1 presents a quick summary of the advantages and disadvantages of these NGS technologies.
Table 1.
Advantages and Disadvantages of Next-Generation Sequencing Technologies
| Single-Molecule Real-Time Sequencing (Pacific Biosciences) | Ion Semiconductor (Ion Torrent) | Pyrosequencing (Roche/454) | Sequencing by Synthesis (Illumina) | Sequencing by Ligation (Applied Biosystems SOLiD) | |
|---|---|---|---|---|---|
| Advantages | Fast and informative data | Low cost and fast run | Fast | Simple, scalable, high yield | Low cost |
| Disadvantages | Low yield of high-quality sequences | High rate of sequencing errors | Expensive equipment | Expensive equipment | Slow |
Organizing and managing these raw data sets can be a big challenge. Fortunately, this task is typically handled by professional computer scientists. HIM professionals manage the processed genomic data (a few gigabytes per genome), which are significantly smaller than the raw data sets (several hundred gigabytes or a few terabytes per genome). The more important issue for HIM professionals when they want to apply personal genomic information in personalized medicine is the accuracy of the data obtained from these raw data sets.28 The “garbage in, garbage out” principle applies here. In other words, if the genomic data from these high-throughput technologies are not accurate enough, the results from the analysis of the data will not be valuable, either. Therefore, when they perform genomic data analysis, HIM professionals need to keep in mind that current high-throughput technologies generate multiple types of errors.29
One other issue is the management of complex genomic data sets. Multiple types of genomic data (e.g., DNA sequences, RNA sequences and structures, protein sequences and structures, gene expression profiles, regulatory motifs, gene-gene interactions, DNA-protein interactions, genetic pathways, metagenomics profiles, DNA methylation profiles) are generated from different technologies and platforms. HIM professionals should have a proper way to manage these data types so that they can be conveniently available for clinical practice. For instance, genome sequences can be saved as large plain text files; genetic variations and associated genes and diseases are better organized into a well-designed database for fast query; and RNA and protein structures, as well as gene expression and DNA methylation profiles, can be visualized for an overall picture. In the past, HIM professionals did not need to handle these data sets, and therefore most are not familiar with these data types and the corresponding data management programs. However, with proper training, HIM professionals should be able to confidently deal with these data sets in a short time because they are already familiar with database operations, information retrieval techniques, and the use of external software programs for patient data analysis. The necessary training may include the enhancement of knowledge of genomic terms and tutorials about genomic data processing programs.
Challenges in Personal Genomic Data Analysis
In parallel to the astonishing advancement of high-throughput technologies is the significant progress researchers have made in understanding disease at the molecular level in the past decade. Before the HGP, it was quite difficult and time-consuming to sequence even a small genome, and therefore scientists could only focus on analyzing one gene or a few genes in their research projects. Consequently, researchers believed that most genetic diseases are caused by mutations in one gene or a few genes. However, post-HGP research has indicated that this understanding was not correct. Further investigations performed on the human genome and other species’ genomes have demonstrated the complexity of these genomes. The associations between disease and genomic information have been extended from one single gene to multiple genes in one or multiple chromosomes; from protein-coding regions of a gene to intragenic regions (introns), to all sorts of regulatory elements, to intergenic regions; from protein-coding genes to other types of RNA (for instance, tRNA, microRNA); from genetics to epigenetics; from single nucleotide polymorphisms (SNPs, or point mutations) to short insertions and deletions (indels, or insertion or deletion of DNA segments), to copy number variations (CNVs, or different numbers of copies of a large chunk of DNA sequence) and other structural variations; from individual genes to gene-gene interactions and gene-environment interactions; and from one single or a few genes per study to genome-wide association studies (GWAS). Table 2 shows the research targets before and after the HGP. With the development of more advanced biotechnologies, more powerful computational facilities, and more sophisticated data integration and analysis algorithms, researchers will likely discover many other previously unexpected factors involved in the development of disease and add to the current understanding of genetic reasons for disease in the next few years.
Table 2.
Research Targets in Genetic Variation and Disease Association Studies Before and After the Human Genome Project
| Before the Human Genome Project | After the Human Genome Project |
|---|---|
| Mutations in one or a few genes | Genetic variations in many genes |
| Protein-coding genes | Protein-coding genes and RNA |
| Protein-coding regions in a gene | Intragenic regions (introns); regulatory elements; intergenic regions |
| Mainly point mutations | SNPs; short insertions and deletions; copy number variations; other structural variations |
| Individual genes | Gene-gene interactions; gene-environment interactions |
| One single gene or a few genes per study | Tens of thousands of genes or the whole genome in one study |
From the great advancement in research, scientists have a better idea of the association between genetic variations in genomes and the patients’ risk of developing certain diseases, and patients’ possible responses to some drug therapies. On the other hand, these extensive research results also make it challenging to analyze personal genomic data for healthcare purposes. For example, in the past, when doctors wanted to determine the genetic reason for sickle cell anemia in a patient, they would only check one point mutation in the hemoglobin beta gene (HBB), a mutation that leads to a change of the shape of the red blood cell.30, 31, 32, 33 Therefore, the data analysis procedure could be quite simple. Today, with the availability of extensive genomic data sets, researchers know that genetic variations in multiple genes might be associated with sickle cell anemia.34 The task of determining the precise genetic reason for one patient's condition can be much more difficult because not just more genetic variations but also the expression of those genes,35 the effect of regulatory elements,36 and the interactions among all or some of those genes37 must be examined. Another example is genes associated with hypertension (which affects roughly 80 million Americans).38 For hypertension, a search done in November 2013 in the Phenopedia system39 indicates that 1,565 genes are reported to be associated with the development of this disease, and this number is growing with further investigations.40 To make the situation worse, some research results from different research groups are not consistent41 and directly conflict with each other.42
Highly skilled genomic data analysts, well-designed databases and accurate research results, sophisticated algorithms and software programs, and sufficient computational power are needed for genomic data analysis. At this moment, these resources are typically not available to most physicians. Even if one can easily access excellent genomic data analysis programs (which are available for certain types of data sets) and powerful workstations, it is still challenging for most healthcare professionals to select the best program and the correct parameters for that particular data set because they typically do not have the required training in this field. Almost all current HIM curricula include some relevant courses such as general biology, anatomy and pathophysiology, computer programming, database design, and statistics. However, only a few programs offer courses in bioinformatics or computational genomics, which will be needed as the fields of genomics and personalized medicine progress.
Challenges in Personal Genomic Information Security and Privacy
As mentioned previously, personal genomic data are highly sensitive and need to be protected properly because each record contains not just the health information about one particular patient, but potentially information about a large group of people who have a blood relation with the person who takes the genetic test. This impact can last for generations because the genomic information will be passed to these people's descendants, as in the HeLa genome release case mentioned in the introduction to this article. In addition, by using a clever algorithm and data collected from public databases and social media, it is already possible to uniquely identify the owner of a de-identified genome sequence stored in public databases.43, 44 The privacy of the individual and his or her relatives may be threatened, and the confidentiality of the personal genomic data is lost. The threat to the individual's offspring could be even more serious because research in genomics will likely enable the discovery of more information from a human genome in the future. Therefore, a stronger and more sophisticated security measure may be needed for personal genomic data protection, and this system should be set up before the wide application of genomic information in clinical practice.
Challenges in Legal, Social, and Ethical Aspects
A number of legal, social, and ethical issues are linked with the application of personal genomic information in personalized medicine. For instance, certain knowledge about currently incurable diseases gained from personal genomic data may lead to a social impact among families and friends and/or employment discrimination in workplaces.45 Some people may be socially isolated by their friends, lose their jobs, or be unable to obtain medical insurance because their genetic test results are not kept confidential. Even if the results are kept confidential, they may still generate serious psychological shock to patients themselves. For instance, some patients may choose to take extreme actions to avoid the possible suffering from those diseases. For example, research results have indicated that cancer and AIDS patients have a statistically higher risk of committing suicide, especially shortly after diagnosis.46, 47, 48 In these extreme cases, proper patient education and counseling are critically important, and HIM professionals may conduct some of these tasks when working as patient advocates.
Challenges to consider include the following:
Should a genetic test result be made available if there are currently no treatment options for the corresponding disorder? If genetic tests are available for these disorders, should doctors or patients share the positive results with the patients’ family members? How can families resolve conflicts when some members want to be tested for a genetic disorder and others do not?49
Should parents have the right to order genetic tests for their minor children, especially if the concerned disease is adult onset? Do children of a certain age, for instance, between 14 and 18 years, have the right to make their own decisions on this issue? How can the parents be prevented from treating these children differently after they know the positive results from these tests?50
Who has the right to access stored personal genomic information? Who owns or controls the information: patients, primary doctors, or an independent third party?51, 52 Who is responsible for providing genetic counseling services to patients?
How can the government protect individuals from insurance and employment discrimination because of the breach of personal genomic information? Are the current regulations sufficient for the required protection?53
The next section describes some current solutions to the aforementioned challenges.
Current Solutions to the Challenges
Genomic Data Management
NGS technologies can produce an enormous amount of sequencing data in a short time, and therefore storing and managing these huge data sets can be challenging. One straightforward suggestion is to apply a compression algorithm to reduce the sizes of these sequences.54 A basic fact about the human genome is that genome sequences of two unrelated individuals are highly similar (roughly 99 percent identical). Therefore, directly storing hundreds and thousands of human genome sequences in a database would be highly redundant. One alternative approach is to keep only one reference genome (3 billion bases) and record all the differences between other human genomes and this reference genome. This approach can significantly reduce the size of stored data. Christley et al. combined these two approaches (compression and storage of differences only) and reduced the information about one human genome from 3 gigabytes to about 4 megabytes.55 In other words, the obtained data set is 750 times smaller than its original size. This approach also has problems, however. For instance, the information in the data set depends on the reference genome. Once the reference genome is updated, all the information in the stored data set needs to be updated as well. When the database contains a huge number of patient genomic records, this information update process may take significant time. Table 3 summarizes the advantages and disadvantages of these approaches.
Table 3.
Advantages and Disadvantages of DNA Sequence Storage Strategies
| Data Storage Strategy | Advantages | Disadvantages |
|---|---|---|
| Saving as a plain text file | Easy to retrieve and analyze the sequence | Large file size (3 GB/genome) |
| Saving as a compressed file | Smaller file size (roughly 1.5 to 2 GB/genome) | Time required (can be hours) to perform compression and decompression before data analysis |
| Saving only differences between the genome and the reference genome and compressing the file | Very small file size (4 MB/genome) | Time required to rebuild the needed DNA sequence; need to update all the files when the reference sequence is changed |
Some information systems are specifically dedicated to the management of large-scale biological information. For instance, openBIS is a distributed information system that can be used for managing DNA sequences generated by NGS technologies.56 This system has two major components:57 one is an application server mainly used to manage metadata (systematic descriptions of data) of the whole database, and the other is a data storage server that stores high-volume data such as genome sequences. A web interface allows users to retrieve their information. The architecture of openBIS is shown in Figure 2.
Figure 2.
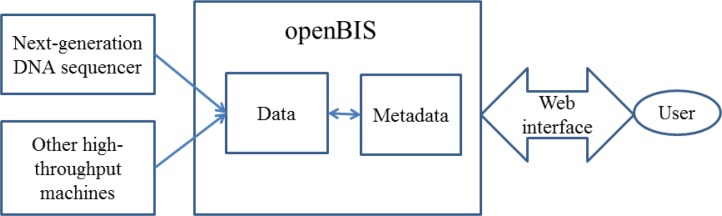
Architecture of the openBIS System for Genomic Data Management
For challenging task of integrating different types of genomic data and organizing them in a way that is convenient for clinicians to use in their practices, the current solution is just to store different genomic data sets in different databases and provide links between certain items in those databases. The genomic databases created by the National Center for Biotechnology Information (NCBI, http://www.ncbi.nlm.nih.gov) are an example of this solution.58 These databases can be very useful; however, clinicians and other healthcare providers such as HIM professionals face a serious challenge to fully utilize or correctly combine useful data items from multiple databases because this information integration task requires extensive knowledge of genetics and genomics to accomplish. An integrated database for clinical use should organize all relevant information for each disease in one place and present the information in an easy-to-understand format. When the information is needed, it can be retrieved immediately in a single step, instead of requiring healthcare providers to come up with a data analysis procedure to query and combine pieces of information from multiple places. In other words, an integrated and comprehensive genomic information management system for clinical use is desirable.
Personal Genomic Data Analysis
Because of the complexity of genomic data, many people have realized that it is necessary to enhance education in genetics and genomics59 for future healthcare providers. Such education is apparently beneficial, and the corresponding author of this article (L.Z.) has made significant efforts to educate HIM students in genomic data analysis.60 Cincinnati Children's Hospital Medical Center hosted a training course called Applying Genomics in Nursing Practice. This course aimed to help the participant nurses become familiar with genetics terms, identify the relevance of genetics and genomics to nursing practice, and identify patterns of inheritance displayed in pedigrees.61 On the other hand, expecting every physician to become an expert in human genetics and genomics and keep track of the research progress in these fields would not be reasonable. It is also not reasonable to expect a physician to search the scientific literature to find out all the factors related to one common genetic disease so that he or she can order the correct genetic test or genomic analysis for the patient. After all, the research literature may contain many papers about one common disease, and results in those papers do not necessarily agree with one another. Physicians would face significant difficulty in determining which results are applicable to their specific patients. For similar reasons, it would be equally challenging for physicians to use the literature to determine the correct drug and dosage for their patients. This scenario is quite different from the literature searches physicians need to do occasionally on rare diseases. In that case, the difficulty is to identify the small number of papers about the rare disease among a huge number of irrelevant articles. In genomics-based personalized care, physicians deal with common diseases (such as cancer, obesity, cardiovascular disease, diabetes) in a different way, and the difficulty is to identify the most suitable information for each patient from a large number of highly relevant articles. Therefore, the available genomic research results should be preprocessed and organized in a certain way before they are presented to physicians for diagnosis and treatment purposes.
Here is one proposed approach for organizing genetic variants determined in genome-wide association studies. In this approach, all genetic variants are categorized into three groups:62
Group 1 for deleterious variants that are clinically actionable and would be reported to patients;
Group 2 for variants that are possibly deleterious, are not clinically actionable, and may be reported to patients upon request with certain restrictions; and
Group 3 for variants that are nondeleterious at present, are not clinically actionable, and would not be reported to patients.
This model helps both patients and clinicians to determine the best treatment options currently available. If variants are reported to patients, they would also be included in the patients’ EHR so that clinicians can easily access them when needed and make correct healthcare decisions. Data in these groups apparently need to be updated regularly because new investigations may reveal new information. With some necessary training, HIM professionals would be able to perform this job.
Although most clinical sites, especially small healthcare facilities, do not have highly sophisticated genomic data analysts, skilled programmers, or high-performance computers for large-scale data analysis, they may use resources from large sequencing centers and cloud-based computing platforms to have those tasks done with low costs. Large genome-sequencing centers have well-trained genomic data analysts and extensive experience in processing large-scale genomic data. Those cloud-based computing facilities (e.g., Amazon Web Services) can configure their high-performance computers according to the requests from their customers and conduct intensive computation tasks. The customers do not need to hire dedicated software engineers or purchase and maintain expensive high-performance computers. Some genomic data analysis pipelines are already available for this approach. One example is the Atlas2-Cloud pipeline (http://sourceforge.net/projects/atlas2cloud). This pipeline has been successfully implemented into Amazon Web Services. With their extensive skills in data management and data analysis, HIM professionals are readily capable of preparing the genomic data sets for this type of analysis, configuring the needed data analyses, and organizing the obtained analysis results from the remote high-performance computers.
At the same time, HIM professionals are not totally free from the need to perform personal genomic data analysis themselves. They need to have certain skills to perform some quick and relatively simple genomic data analyses locally. After all, not all genomic data analyses need high-performance computers. In many cases, simple BLAST63, 64, 65, 66 searches for sequence similarities or queries in public databases may be sufficient. HIM professionals therefore need to be trained in using some common genomic data analysis tools for extracting information from personal genomic data. They also should be familiar with frequently used genomic databases. For instance, HIM professionals should be able to use BLAST to identify similar sequences in a database or compare two given sequences and interpret the results, view a patient's chromosome or one particular gene in a genome browser such as the UCSC Genome Browser,67 search the details of one genetic variation in the dbSNP database,68 and find information about a genetic disease in the OMIM database.69 HIM professionals also need to be able to follow physicians’ orders to make certain genomic data available for further analysis and present the analysis results in a way that is convenient for physicians to use. For all of these tasks, HIM professionals only need a short period of training, mainly to refresh their knowledge of genomics terms and become familiar with the necessary software programs, because they already have extensive skills in database query, data analysis, and presenting search results so that they can be used by other healthcare providers.
Personal genomic data or results from genomic data analysis will become an integral part of electronic health records. Therefore, one essential component of HIM training is to educate HIM students on how to manage, protect, and apply genomic data in clinical settings.70, 71 HIM professionals may also need to build knowledge-based decision making systems72 for genomics-based personalized medicine practices so that clinicians can extract the most needed information from complex genomic data.73, 74 The Office of the National Coordinator for Health Information Technology (ONC) is currently working on integrating genomic information into health information technology systems in order to take advantage of the full potential of genomic information in healthcare.75
Security and Privacy Issues
Polices and laws play a key role in protecting individuals’ genetic information. The Health Insurance Portability and Accountability Act (HIPAA) ensures the basic protection of the privacy of information stored in patients’ records.76 On January 25, 2013, the Office for Civil Rights of the US Department of Health and Human Services published a final rule containing modifications to the privacy, security, breach notification, and enforcement rules in HIPAA and the Health Information Technology for Economic and Clinical Health (HITECH) Act. One of these modifications emphasizes that personal genetic information is protected health information and prohibits the use and disclosure of genetic information by any health plans for underwriting purposes.77
Besides these policies and laws, many technical applications aim to protect personal genomic information. For instance, Interpretome78 is a client-side genome interpretation system developed to analyze personal genomic data on the customer's local machine. The personal genomic data and the analysis results would not leave the customer's machine, thereby protecting this sensitive information. Another example is the GenePING system.79 This system provides secure storage for genome data sets and enables the sharing of personal genetic variations and gene expressions by applying the Advanced Encryption Standard (AES) encryption algorithm. As long as the AES keys are protected properly, these genomic data sets are secure.
Legal, Social, and Ethical Issues
The Genetic Information Nondiscrimination Act (GINA) is the law specifically created to protect individuals from discrimination based on their genetic test results.80 GINA extended the medical privacy and confidentiality rules to the disclosure of genetic information81 before the modifications of HIPAA and the HITECH Act. GINA represents a landmark in the field of personal genomics because it removes one important concern when patients consider taking genetic tests. It is also beneficial for biomedical research because it removes similar concerns for participants of genetic research. On the other hand, just as with any other law, GINA alone cannot cover all possible situations related to discrimination based on genetic information. For example, GINA does not apply to employers with fewer than 15 employees; it also does not extend to the US military (http://www.ginahelp.org/).
Currently, people in different roles handle the legal and ethical issues related to genetic and genomic data according to some general practices. Physicians suggest genetic tests if they are certain that the tests are necessary for correct diagnosis.82 Physicians may also order genetic tests according to patients’ requests provided that there is supporting evidence for the patient's case.83 In both cases, physicians should be familiar with all available genetic tests and the limitations or consequences of these tests. Physicians will rely on HIM professionals to collect this information and present it to them in an easy-to-access manner.
Ethical issues in pediatrics is one of the big concerns when applying genetic tests.84 The American Academy of Pediatrics (AAP) encourages pediatricians to help parents and their children to understand all relevant information about genetic tests, discuss the importance of each genetic test, and protect the privacy and confidentiality of test results according to the existing laws. Pediatricians should also refer children and their parents to genetic counselors and order genetic tests only if such testing is best choice for the patient and their parents after they understand the benefits and risks of the tests. The AAP offers several recommendations related to genetic tests for children. For instance, newborn screening tests should be evaluated for both risks and benefits periodically to ensure their efficiency, informed consent should be obtained from parents of minor children who take genetic tests,85 and genetic tests for adult-onset conditions generally should be postponed until patients reach adulthood and can make their own critical decisions.86 Of course, if the disease would be very severe and would be a life-or-death matter, or if there are interventions that may be effective in childhood, parents may insist on having their children tested. In some cases, there is a family history of a disease; for example, girls in families with a history of breast cancer may have anxiety about their BRCA gene test results.87 Bioethics experts are still debating the proper ways of handling these issues.
The federal government can play a key role in some of the legal, social, and ethical issues in the practice of genomics-based personalized medicine. For instance, the federal government can create guidance for genomic databases and standard quality improvement metrics for genetic tests, provide information to the general public about the benefits and risks of genetic tests, establish regulations to control the price of patented genomic tests or treatment approaches, and develop policies and procedures to handle genetic discrimination cases.88 With the availability of these government rules and regulations, people would have confidence to take advantage of genomics-based personalized medicine.
In general, HIM professionals can adopt two strategies to overcome the challenges posed by legal, social, and ethical issues in genomics-based personalized medicine. One is to ensure that the practice of HIM complies with the federal policies and regulations regarding personal genomic data.89 The other strategy is to educate people about the proper way of accessing their genomic information while maintaining the confidentiality and privacy of the data.90 Other specific approaches HIM professionals can take to overcome these challenges include the following:
Know the federal, state, and local regulations that surround genomic data.
Conduct education and training sessions for healthcare employees on the laws that surround genomic data and its use, privacy, and security.
Conduct seminars for patients and serve as patient advocates for the use and proper maintenance of genomic data.
Conduct scenarios of different ethical dilemmas that include issues around genomic data for all healthcare employees. The more these issues are discussed, the better they will be handled when questions arise.
Research how genomic data should be handled within the EHR.
Practice the proper techniques for handling genomic data within the EHR or other clinical databases.
Write policies and procedures about how to handle, store, release, and process genomic data for employees, patients, outside healthcare providers, third-party payers, the Joint Commission, departments of health, and so forth.
Discussion
The previous two sections described several types of challenges in genomics-based personalized medicine and some current solutions or practices for dealing with these challenges. In this section, we briefly mention a few other issues related to genomics-based personalized medicine.
As mentioned previously, the analysis of genomic data is critically important in genomics-based personalized medicine. HIM professionals can play a critical role in the development of personal genomic information management and analysis systems for clinical use because HIM professionals have firsthand knowledge about how to organize, manage, and report health information needed by other healthcare providers, and they also have a clear idea of the typical workflow of healthcare facilities. All of these skills of HIM professionals are critically important for creating a successful clinical information management system.
HIM professionals can also contribute to patient education when patients are considering taking a genetic test or providing their personal genomic information to physicians for diagnosis or treatment purposes. Although it is becoming increasingly clear that personal genomics will have a potential role in improving the diagnosis and treatment of diseases, patients typically do not receive much education in this field, and therefore it is difficult for them to make informed decisions. HIM professionals can serve as patient advocates and explain the benefits and risks related to genetic tests and genomic data analysis.91 This explanation is not formal genetic counseling but simply provides general information about those tests and analyses.
Reimbursement is an important issue that should be carefully considered in order to facilitate wide implementation of genomics-based personalized medicine. The coverage and analysis officer of the Centers for Medicare and Medicaid Services (CMS) once said that “whole genome sequencing itself is probably something that CMS would never cover.”92 Insurance coverage could be problematic even though the cost of genome sequencing has dropped quickly in recent years.93 To solve this problem, HIM professionals may conduct robust cost-effectiveness analysis for the purpose of receiving reimbursement for genome sequencing. This analysis may take into account factors such as the clinical benefits and economic savings over the long term as compared to an alternative diagnosis and treatment approach.94 In fact, the cost of human genome sequencing may drop to point that the price is comparable to other typical clinical lab tests when the sequencing service is performed in a high-volume manner. George Church, a professor of genetics at Harvard Medical School, said that “overall, the cost of sequencing is expected to be recovered over a lifetime through the avoidance of unnecessary diagnostics and therapeutics and time spent in waiting rooms and hospitals.”95 The much more expensive service is actually the genomic data analysis. Hospitals, labs, and physicians may choose to put this service into the category of data interpretation, just as they do for professional image reading services (ultrasound, MRI, CT images).96 The National Human Genome Research Institute (NHGRI) assists all types of payers to evaluate emerging genetic tests for reimbursement and promotes research into the health benefits and cost-effectiveness of genetic testing.97
In April 2011, the NHGRI held a conference titled “Genomics and Health Information Technology Systems: Exploring the Issues.” This conference aimed to explore the research and policy issues related to the integration of genomics into health information systems.98 Having more conferences and discussions like this one would be beneficial so that genomics-based personalized medicine can be eventually applied widely in healthcare.
Management of genomic databases, conducting data analytics to mine genomic data for healthcare providers’ use, and securing the privacy and confidentiality of the genomic data sets are just some of the critical skills HIM professionals can apply to genomic data sets. Other skills include choosing an EHR system that best aligns with genomic databases for use by healthcare professionals; determining the types of genomic data sets that can be merged with clinical data within the EHR; using computer-assisted coding software that can search through the EHR and genomic data and automatically assign diagnosis and procedure codes that can then be used for research, public health statistics, and reimbursement; and developing and enforcing policies and procedures related to genomic data that is housed within the EHR or in separate clinical databases. The HIM professional's role within the genomic data and personalized medicine arena will continue to expand in the future. HIM professionals should embrace this role through education and training, clinical practice, and group discussion and inquiry. HIM professionals will need to explore the new roles they may take on as the genetic informatics specialist role continues to grow and change. This role may include building stronger ties with patients by providing patient advocacy as well as patient-centered care and patient-centered information. It may also include exploring new ideas for unique security measures that will keep genomic information safe but still accessible when needed. Whatever role the HIM professional chooses to take on, it will prove to be important not only for each individual patient and his or her information but for the betterment of the HIM field in general.
In other words, working on genomic data is a natural extension of HIM professionals’ typical job functions, such as managing data, performing data analysis using software programs, protecting patients’ data security and privacy, and following federal regulations to handle social, ethical, and legal issues related to personal data. Although challenges exist and the current solutions are not perfect, HIM professionals will just need relevant training to handle the new type of data. This training is within the reach of HIM students, as the authors of this article have demonstrated in courses and course modules.99, 100
Summary and Recommendations
Genomics-based personalized medicine has the potential to provide truly individualized and high-quality healthcare. To realize this goal, however, a number of challenges must be addressed, and partial solutions are currently available to conquer some of them. To fully clear the way for widespread adoption of genomics-based personalized medicine, joint efforts from people in different roles and with different skills will be required. Education, basic research, software development, policy construction, and law making will be needed to remove the obstacles related to personal genomic data and their applications in clinical care.
HIM professionals in particular will need to enhance their skills in data analysis and receive basic training in genomics. They should be familiar with frequently used genomic databases, have the skills to use some genomic data analysis tools, and be capable of extracting desired information from large-scale genomic data sets with certain programs. HIM professionals may also play an important role in educating the general public and provide assistance in personal genomic information management and the use of genomic data analysis software.
Some general recommendations to professionals in other fields are also in order. Policy makers need to investigate the consequences of applying genomic information in the clinic and create policies, laws, and regulations to protect individuals’ security and privacy and help them to avoid other possible damaging social, ethical, and legal consequences. All healthcare providers have the responsibility to educate the general public about the benefits of personalized medicine and the consequences of genetic tests. Software developers need to work closely with healthcare providers to determine how to incorporate genomic information into medical record systems for clinical decision support.101, 102 It might be beneficial to develop a nationwide information infrastructure with specific standards implemented to guide clinicians in the use of genomics in their practices.103, 104 Clinicians also need to improve their knowledge of genomics so that they can order the proper genetic tests and genomic analyses, understand the results from genetic tests and genomic analysis, and individualize treatment options for each patient.
Contributor Information
Amal Alzu'bi, The Department of Health Information Management at the University of Pittsburgh in Pittsburgh, PA.
Leming Zhou, The Department of Health Information Management at the University of Pittsburgh in Pittsburgh, PA.
Valerie Watzlaf, The Department of Health Information Management at the University of Pittsburgh in Pittsburgh, PA.
Notes
- 1.Kawamoto K., et al. A National Clinical Decision Support Infrastructure to Enable the Widespread and Consistent Practice of Genomic and Personalized Medicine. BMC Medical Informatics and Decision Making. 2009;9:17. doi: 10.1186/1472-6947-9-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Abrahams E., et al. The Personalized Medicine Coalition: Goals and Strategies. American Journal of Pharmacogenomics. 2005;5(6):345–55. doi: 10.2165/00129785-200505060-00002. [DOI] [PubMed] [Google Scholar]
- 3.Kerschner J. E. Clinical Implementation of Whole Genome Sequencing a Valuable Step toward Personalized Care. WMJ. 2013;112(5):224–25. [PubMed] [Google Scholar]
- 4.Karow J. “Children's Mercy Hospital Explores Rapid WGS for Newborn Screening, Disease Diagnosis.” Genomeweb. September 25, 2013. Available at http://www.genomeweb.com/sequencing/childrens-mercy-hospital-explores-rapid-wgs-newborn-screening-disease-diagnosis (accessed February 24, 2014).
- 5.University of Pittsburgh Medical Center (UPMC) “Division of Anatomic Pathology.” 2013. Available at http://path.upmc.edu/divisions/anatomic.htm (accessed December 22, 2013).
- 6.“UPMC Launches $100M Personalized Care Initiative.” HIMSS Media. October 1, 2012. Available at http://www.healthcareitnews.com/news/upmc-launches-100m-personalized-care-initiative (accessed December 22, 2013).
- 7.Khorey L. “UPMC's $100M Bid to Change Medicine.” HIMSS Media. October 2, 2012. Available at http://www.healthcareitnews.com/news/4-facets-upmcs-100m-bid-change-medicine (accessed December 22, 2013).
- 8.Collins F. S., Hamburg M. A. First FDA Authorization for Next-Generation Sequencer. New England Journal of Medicine. 2013;369(25):2369–71. doi: 10.1056/NEJMp1314561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ibid.
- 10.Mardis E. R. Next-Generation Sequencing Platforms. Annual Review of Analytical Chemistry. 2013;6(1):287–303. doi: 10.1146/annurev-anchem-062012-092628. [DOI] [PubMed] [Google Scholar]
- 11.Zhao Q., et al. Tracing the Transcriptomic Changes in Synthetic Trigenomic Allohexaploids of Brassica Using an RNA-seq Approach. PLoS ONE. 2013;8(7):e68883. doi: 10.1371/journal.pone.0068883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pepke S., et al. Computation for ChIP-seq and RNA-seq Studies. Nature Methods. 2009;6(11, suppl.):S22–S32. doi: 10.1038/nmeth.1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ibid.
- 14.Furey T. S. ChIP-seq and Beyond: New and Improved Methodologies to Detect and Characterize Protein-DNA Interactions. Nature Reviews Genetics. 2012;13(12):840–52. doi: 10.1038/nrg3306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li N., et al. Whole Genome DNA Methylation Analysis Based on High Throughput Sequencing Technology. Methods. 2010;52(3):203–12. doi: 10.1016/j.ymeth.2010.04.009. [DOI] [PubMed] [Google Scholar]
- 16.Ashley E. A., et al. Clinical Evaluation Incorporating a Personal Genome. Lancet. 2010;375(9725):1525–35. doi: 10.1016/S0140-6736(10)60452-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Adey A., et al. The Haplotype-resolved Genome and Epigenome of the Aneuploid HeLa Cancer Cell Line. Nature. 2013;500(7461):207–11. doi: 10.1038/nature12064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Landry J. J., et al. The Genomic and Transcriptomic Landscape of a HeLa Cell Line. G3. 2013;3(8):1213–24. doi: 10.1534/g3.113.005777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hudson K. L., Collins F. S. Family Matters. Nature. 2013;500(7461):141–42. doi: 10.1038/500141a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bailey J., Rudman W. “The Expanding Role of the HIM Professional: Where Research and HIM Roles Intersect.” Perspectives in Health Information Management (2004). [PMC free article] [PubMed]
- 21.Collins F. S., et al. The Human Genome Project: Lessons from Large-Scale Biology. Science. 2003;300(5617):286–90. doi: 10.1126/science.1084564. [DOI] [PubMed] [Google Scholar]
- 22.Wiechers I. R., et al. The Emergence of Commercial Genomics: Analysis of the Rise of a Biotechnology Subsector During the Human Genome Project, 1990 to 2004. Genome Medicine. 2013;5(9):83. doi: 10.1186/gm487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hood L., Rowen L. The Human Genome Project: Big Science Transforms Biology and Medicine. Genome Medicine. 2013;5(9):79. doi: 10.1186/gm483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wadman M. James Watson's Genome Sequenced at High Speed. Nature. 2008;452(788) doi: 10.1038/452788b. [DOI] [PubMed] [Google Scholar]
- 25.Bonetta L. Whole-Genome Sequencing Breaks the Cost Barrier. Cell. 2010;141(6):917–19. doi: 10.1016/j.cell.2010.05.034. [DOI] [PubMed] [Google Scholar]
- 26.Collins F. S., Hamburg M. A. “First FDA Authorization for Next-Generation Sequencer.” [DOI] [PMC free article] [PubMed]
- 27.Mardis E. R. “Next-Generation Sequencing Platforms.” [DOI] [PubMed]
- 28.Koboldt D. C., et al. Challenges of Sequencing Human Genomes. Briefings in Bioinformatics. 2010;11(5):484–98. doi: 10.1093/bib/bbq016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kircher M., Kelso J. High-Throughput DNA Sequencing—Concepts and Limitations. Bioessays. 2010;32(6):524–36. doi: 10.1002/bies.200900181. [DOI] [PubMed] [Google Scholar]
- 30.Chen S. Y., et al. The Genomic Analysis of Erythrocyte MicroRNA Expression in Sickle Cell Diseases. PLoS ONE. 2008;3(6):e2360. doi: 10.1371/journal.pone.0002360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Romana M., et al. Thrombosis-associated Gene Variants in Sickle Cell Anemia. Thrombosis and Haemostasis. 2002;87(2):356–58. [PubMed] [Google Scholar]
- 32.Bender M. A., Hobbs W. Sickle Cell Disease. In: Pagon R. A., editor. GeneReviews. Seattle: University of Washington; 1993. [Google Scholar]
- 33.Singh J., et al. Dental and Periodontal Health Status of Beta Thalassemia Major and Sickle Cell Anemic Patients: A Comparative Study. Journal of International Oral Health. 2013;5(5):53–58. [PMC free article] [PubMed] [Google Scholar]
- 34.Bender M. A., Hobbs W. “Sickle Cell Disease.”
- 35.Williams R. B., et al. The Influence of Genetic Variation on Gene Expression. Genome Research. 2007;17(12):1707–16. doi: 10.1101/gr.6981507. [DOI] [PubMed] [Google Scholar]
- 36.Vernot B., et al. Personal and Population Genomics of Human Regulatory Variation. Genome Research. 2012;22(9):1689–97. doi: 10.1101/gr.134890.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kumar R., et al. Interactions between the FTO and GNB3 Genes Contribute to Varied Clinical Phenotypes in Hypertension. PLoS ONE. 2013;8(5):e63934. doi: 10.1371/journal.pone.0063934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Roger V. L., et al. Heart Disease and Stroke Statistics—2012 Update: A Report from the American Heart Association. Circulation. 2012;125(1):e2–e220. doi: 10.1161/CIR.0b013e31823ac046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yu W., et al. Phenopedia and Genopedia: Disease-centered and Gene-centered Views of the Evolving Knowledge of Human Genetic Associations. Bioinformatics. 2010;26(1):145–46. doi: 10.1093/bioinformatics/btp618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Larsson E., et al. Hypertension and Genetic Variation in Endothelial-Specific Genes. PLoS ONE. 2013;8(4):e62035. doi: 10.1371/journal.pone.0062035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Collins F. S., et al. A Vision for the Future of Genomics Research. Nature. 2003;422(6934):835–47. doi: 10.1038/nature01626. [DOI] [PubMed] [Google Scholar]
- 42.Foster M. W., et al. The Role of Community Review in Evaluating the Risks of Human Genetic Variation Research. American Journal of Human Genetics. 1999;64(6):1719–27. doi: 10.1086/302415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Claerhout B., DeMoor G. J. Privacy Protection for Clinical and Genomic Data: The Use of Privacy-enhancing Techniques in Medicine. International Journal of Medical Informatics. 2005;74(2–4):257–65. doi: 10.1016/j.ijmedinf.2004.03.008. [DOI] [PubMed] [Google Scholar]
- 44.Hayden E. C. Privacy Protections: The Genome Hacker. Nature. 2013;497(7448):172–74. doi: 10.1038/497172a. [DOI] [PubMed] [Google Scholar]
- 45.Vogenberg F. R., et al. Personalized Medicine: Part 2. Ethical, Legal, and Regulatory Issues. Pharmacy and Therapeutics. 2010;35(11):624–42. [PMC free article] [PubMed] [Google Scholar]
- 46.Hem E., et al. Suicide Risk in Cancer Patients from 1960 to 1999. Journal of Clinical Oncology. 2004;22(20):4209–16. doi: 10.1200/JCO.2004.02.052. [DOI] [PubMed] [Google Scholar]
- 47.Mishara B. L. Suicide, Euthanasia and AIDS. Crisis. 1998;19(2):87–96. doi: 10.1027/0227-5910.19.2.87. [DOI] [PubMed] [Google Scholar]
- 48.Urban D., et al. Suicide in Lung Cancer: Who Is at Risk? Chest. 2013;144(4):1245–52. doi: 10.1378/chest.12-2986. [DOI] [PubMed] [Google Scholar]
- 49.Fulda K. G., Lykens K. Ethical Issues in Predictive Genetic Testing: A Public Health Perspective. Journal of Medical Ethics. 2006;32(3):143–47. doi: 10.1136/jme.2004.010272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pelias M. K. Genetic Testing of Children for Adult-Onset Diseases: Is Testing in the Child's Best Interests? Mount Sinai Journal of Medicine. 2006;73(3):605–8. [PubMed] [Google Scholar]
- 51.Gullapalli R. R., et al. Next Generation Sequencing in Clinical Medicine: Challenges and Lessons for Pathology and Biomedical Informatics. Journal of Pathology Informatics. 2012;3:40. doi: 10.4103/2153-3539.103013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.McGuire A. L., et al. Research Ethics and the Challenge of Whole-Genome Sequencing. Nature Reviews Genetics. 2008;9(2):152–56. doi: 10.1038/nrg2302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Clayton E. W. Ethical, Legal, and Social Implications of Genomic Medicine. New England Journal of Medicine. 2003;349(6):562–69. doi: 10.1056/NEJMra012577. [DOI] [PubMed] [Google Scholar]
- 54.Chen X., et al. DNACompress: Fast and Effective DNA Sequence Compression. Bioinformatics. 2002;18(12):1696–98. doi: 10.1093/bioinformatics/18.12.1696. [DOI] [PubMed] [Google Scholar]
- 55.Christley S., et al. Human Genomes as Email Attachments. Bioinformatics. 2009;25(2):274–75. doi: 10.1093/bioinformatics/btn582. [DOI] [PubMed] [Google Scholar]
- 56.Bauch A., et al. OpenBIS: A Flexible Framework for Managing and Analyzing Complex Data in Biology Research. BMC Bioinformatics. 2011;12:468. doi: 10.1186/1471-2105-12-468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wruck W., et al. Data Management Strategies for Multinational Large-Scale Systems Biology Projects. Briefings in Bioinformatics. 2014;15(1):65–78. doi: 10.1093/bib/bbs064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.NCBI Resource Coordinators Database Resources of the National Center for Biotechnology Information. Nucleic Acids Research. 2014;42(D1):D7–D17. doi: 10.1093/nar/gkt1146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.McCarthy J. J., et al. Genomic Medicine: A Decade of Successes, Challenges, and Opportunities. Science Translational Medicine. 2013;5(189):189sr4. doi: 10.1126/scitranslmed.3005785. [DOI] [PubMed] [Google Scholar]
- 60.Zhou L., et al. “Flexible Approaches for Teaching Computational Genomics in a Health Information Management Program.” Perspectives in Health Information Management (2013). [PMC free article] [PubMed]
- 61.Cincinnati Children's Hospital Medical Center “Applying Genomics in Nursing Practice.” 2013. Available at http://www.cincinnatichildrens.org/education/clinical/nursing/genetics/cont/applying-genomics/ (accessed December 23, 2013).
- 62.Berg J. S., et al. Deploying Whole Genome Sequencing in Clinical Practice and Public Health: Meeting the Challenge One Bin at a Time. Genetics in Medicine. 2011;13(6):499–504. doi: 10.1097/GIM.0b013e318220aaba. [DOI] [PubMed] [Google Scholar]
- 63.Altschul S. F., et al. Basic Local Alignment Search Tool. Journal of Molecular Biology. 1990;215(3):403–10. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 64.Newell P. D., et al. A Small-Group Activity Introducing the Use and Interpretation of BLAST. Journal of Microbiology & Biology Education. 2013;14(2):238–43. doi: 10.1128/jmbe.v14i2.637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Boratyn G. M., et al. “BLAST: A More Efficient Report with Usability Improvements.” Nucleic Acids Research 41, Web Server issue (2013): W29–W33. [DOI] [PMC free article] [PubMed]
- 66.Neumann R. S., et al. “BLAST Output Visualization in the New Sequencing Era.” Briefings in Bioinformatics (2013). [DOI] [PubMed]
- 67.Karolchik D., et al. “The UCSC Genome Browser.” Current Protocols in Bioinformatics (2012): Unit 1.4. [DOI] [PubMed]
- 68.Bhagwat M. “Searching NCBI's dbSNP Database.” Current Protocols in Bioinformatics (2010): Unit 1.19. [DOI] [PMC free article] [PubMed]
- 69.Li Z., et al. An Examination of the OMIM Database for Associating Mutation to a Consensus Reference Sequence. Protein & Cell. 2012;3(3):198–203. doi: 10.1007/s13238-012-2037-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Zhou L., et al. “Flexible Approaches for Teaching Computational Genomics in a Health Information Management Program.” [PMC free article] [PubMed]
- 71.Ludwig B., et al. “Adding a Genomic Healthcare Component to a Health Information Management Curriculum.” Perspectives in Health Information Management (2010). [PMC free article] [PubMed]
- 72.Watt S., et al. Clinical Genomics Information Management Software Linking Cancer Genome Sequence and Clinical Decisions. Genomics. 2013;102(3):140–47. doi: 10.1016/j.ygeno.2013.04.007. [DOI] [PubMed] [Google Scholar]
- 73.Gregory F. W., et al. Genomics and HIM: Three Areas of Increasing Intersection. Journal of AHIMA. 2009;80(11):52–53. [PubMed] [Google Scholar]
- 74.Khoury M. J., et al. The Scientific Foundation for Personal Genomics: Recommendations from a National Institutes of Health–Centers for Disease Control and Prevention Multidisciplinary Workshop. Genetics in Medicine. 2009;11(8):559–67. doi: 10.1097/GIM.0b013e3181b13a6c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Vest J. R., Gamm L. D. Health Information Exchange: Persistent Challenges and New Strategies. Journal of the American Medical Informatics Association. 2010;17(3):288–94. doi: 10.1136/jamia.2010.003673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Lindberg D. A. Biomedical Informatics: Precious Scientific Resource and Public Policy Dilemma. Transactions of the American Clinical and Climatological Association. 2003;114:113–20. [PMC free article] [PubMed] [Google Scholar]
- 77.Broccolo B. M., et al. OCR Issues Final Modifications to the HIPAA Privacy, Security, Breach Notification and Enforcement Rules to Implement the HITECH Act. McDermott Will & Emery, 2013.
- 78.Karczewski K. J., et al. Interpretome: A Freely Available, Modular, and Secure Personal Genome Interpretation Engine. Pacific Symposium on Biocomputing. 2012;17:339–50. [PMC free article] [PubMed] [Google Scholar]
- 79.Adida B., Kohane I. S. GenePING: Secure, Scalable Management of Personal Genomic Data. BMC Genomics. 2006;7:93. doi: 10.1186/1471-2164-7-93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Feldman E. A. The Genetic Information Nondiscrimination Act (GINA): Public Policy and Medical Practice in the Age of Personalized Medicine. Journal of General Internal Medicine. 2012;27(6):743–46. doi: 10.1007/s11606-012-1988-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Glinskii V. G., Glinsky G. V. Emerging Genomic Technologies and the Concept of Personalized Medicine. Cell Cycle. 2008;7(15):2278–85. doi: 10.4161/cc.6476. [DOI] [PubMed] [Google Scholar]
- 82.Bonham V. L., et al. Patient Physical Characteristics and Primary Care Physician Decision Making in Preconception Genetic Screening. Public Health Genomics. 2010;13(6):336–44. doi: 10.1159/000262328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.White D. B., et al. Too Many Referrals of Low-Risk Women for BRCA1/2 Genetic Services by Family Physicians. Cancer Epidemiology, Biomarkers & Prevention. 2008;17(11):2980–86. doi: 10.1158/1055-9965.EPI-07-2879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Burke W., Diekema D. S. Ethical Issues Arising from the Participation of Children in Genetic Research. Journal of Pediatrics. 2006;149(1, suppl.):S34–S38. doi: 10.1016/j.jpeds.2006.04.049. [DOI] [PubMed] [Google Scholar]
- 85.American Academy of Pediatrics Committee on Bioethics Ethical Issues with Genetic Testing in Pediatrics. Pediatrics. 2001;107(6):1451–55. doi: 10.1542/peds.107.6.1451. [DOI] [PubMed] [Google Scholar]
- 86.Caga-anan E. C. F., et al. Testing Children for Adult-Onset Genetic Diseases. Pediatrics. 2012;129(1):163–67. doi: 10.1542/peds.2010-3743. [DOI] [PubMed] [Google Scholar]
- 87.Ibid.
- 88.Vogenberg F. R., et al. “Personalized Medicine: Part 2. Ethical, Legal, and Regulatory Issues.” [PMC free article] [PubMed]
- 89.Zeng X., et al. “Redefining the Roles of Health Information Management Professionals in Health Information Technology.” Perspectives in Health Information Management (2009). [PMC free article] [PubMed]
- 90.Ibid.
- 91.Karczewski K. J. The Future of Genomic Medicine Is Here. Genome Biology. 2013;14:304. doi: 10.1186/gb-2013-14-3-304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Gullapalli R. R., et al. “Next Generation Sequencing in Clinical Medicine: Challenges and Lessons for Pathology and Biomedical Informatics.” [DOI] [PMC free article] [PubMed]
- 93.Wetterstrand K. A. “DNA Sequencing Costs: Data from the NHGRI Genome Sequencing Program (GSP).” 2013. Available at http://www.genome.gov/sequencingcosts
- 94.Higashi M. K., Veenstra D. L. Managed Care in the Genomics Era: Assessing the Cost Effectiveness of Genetic Tests. American Journal of Managed Care. 2003;9(7):493–500. [PubMed] [Google Scholar]
- 95.Church G. Improving Genome Understanding. Nature. 2013;502(7470):143. doi: 10.1038/502143a. [DOI] [PubMed] [Google Scholar]
- 96.Green R. C., et al. Clinical Genome Sequencing. In: Ginsburg G. S., Willard H. F., editors. Genomic and Personalized Medicine. London: Academic Press; 2013. pp. 102–122. [Google Scholar]
- 97.National Human Genome Research Institute “Coverage and Reimbursement of Genetic Tests.” 2012. Available at http://www.genome.gov/19016729 (accessed December 11, 2013).
- 98.Genomics and Health Information Technology Systems: Exploring the Issues. Bethesda, MD: National Human Genome Research Institute; 2011. [Google Scholar]
- 99.Zhou L., et al. “Flexible Approaches for Teaching Computational Genomics in a Health Information Management Program.” [PMC free article] [PubMed]
- 100.Ludwig B., et al. “Adding a Genomic Healthcare Component to a Health Information Management Curriculum.” [PMC free article] [PubMed]
- 101.Dressler L. G. Integrating Personalized Genomic Medicine into Routine Clinical Care: Addressing the Social and Policy Issues of Pharmacogenomic Testing. North Carolina Medical Journal. 2013;74(6):509–13. [PubMed] [Google Scholar]
- 102.National Research Council . Integrating Large-Scale Genomic Information into Clinical Practice: Workshop Summary. Washington, DC: National Academies Press; 2012. [PubMed] [Google Scholar]
- 103.Kawamoto K., Lobach D. F., Willard H. F., Ginsburg G. S. “A National Clinical Decision Support Infrastructure to Enable the Widespread and Consistent Practice of Genomic and Personalized Medicine.” [DOI] [PMC free article] [PubMed]
- 104.Buntin M. B., et al. Health Information Technology: Laying the Infrastructure for National Health Reform. Health Affairs. 2010;29(6):1214–19. doi: 10.1377/hlthaff.2010.0503. [DOI] [PubMed] [Google Scholar]