Abstract
Self-assembled monolayers (SAMs) of alkanethiolates on gold can be used to carefully probe immobilized biomolecule interactions with cell-surface receptors. However, due to a lack of experimental throughput associated with labor-intensive production, specialized fabrication apparatus, and other practical challenges, alkanethiolate SAMs have not had widespread use by biological researchers. In this Minireview, we investigate a range of techniques that could enhance the throughput of SAM-based approaches by patterning substrates with arrays of different conditions. Here we highlight microfluidic, photochemical, localized removal, and backfilling techniques to locally pattern SAM substrates with biomolecules and also describe how these approaches have been applied in SAM-based screening systems. Furthermore we provide perspectives on several crucial barriers that need to be overcome to enable widespread use of SAM chemistry in biological applications.
Keywords: alkanethiolates, arrays, patterning, peptides, self-assembled monolayers
1. Introduction
In vivo, cells interact with a complex mixture of insoluble extracellular matrix components that supports cell adhesion and also confers specific biochemical signals. Multiple extracellular cues arising from insoluble factors, soluble factors, and cell-cell interactions are coordinated to tightly regulate cell behavior. The complexity of these extracellular cues presents a significant challenge for understanding basic biological processes or designing biomaterials to influence cell behavior. In view of this complexity, screening platforms that deconstruct the influence of specific biomolecules on cell behavior are useful tools, and can enable fundamental biological discoveries and associated biomedical applications. In vitro, the majority of mammalian cells are adherent to, and constantly interact with, their insoluble microenvironment. As a result, the interface between cells and surfaces is a major source of biochemical signals. Therefore, chemically defined substrates, in which biological interactions with surfaces are rigorously controlled, can be used to systematically probe the effects of individual biomolecules on cell behavior by presenting them to cells in a defined manner.
Self-assembled monolayers (SAMs) of alkanethiolates on gold are ideal substrates with which to screen for the effects of specific biomolecules on cell behavior, as they provide biological interfaces that regulate protein–surface interactions and, consequently, cell–surface interactions.[1,2] Specifically, this can be achieved by using SAMs formed using alkanethiolates bearing two key components: 1) oligo(ethylene glycol) (OEG) moieties that prevent nonspecific protein interactions; and 2) controlled densities of biomolecules, such as peptide ligands, to regulate substrate interactions with integrins, [2] growth factor receptors [3-5] or glycosaminoglycans [3,6-8] (Figure 1). In this manner, any downstream changes in cell phenotype can be directly correlated to the density and composition of biomolecules displayed on the surface. This offers significant advantages over canonical cell culture surfaces, in which the type and density of biomolecules present in an adsorbed protein layer are difficult to determine or predict, [9] and thus difficult to correlate to downstream changes in cell phenotype. SAMs have been widely used to characterize the effects of cell adhesion ligands on cell behavior, such as cell attachment, spreading, proliferation, and migration. [2,10-12] For example, Roberts and co-workers demonstrated that the density of the integrin-binding cell-adhesion ligand Arg-Gly-Asp (RGD) presented on an otherwise bioinert SAM substrate dictated attachment and spreading of bovine endothelial cells. [10] Additionally, SAMs have been used as biological tools to investigate more detailed cell signaling mechanisms associated with cell adhesion. Kato and co-workers used well-defined SAM substrates to examine the effects of linear versus cyclic versions of RGD on integrin-mediated activation of the intracellular enzyme focal adhesion kinase (FAK). Immobilized cyclic RGD promoted increased FAK activation as well as increased downstream mitogen-activated protein (MAP) kinase activation when compared to linear RGD. [13] In addition to investigations aimed at probing cell adhesion, we have recently used SAMs to regulate growth factor signaling. SAMs presenting a vascular endothelial growth factor (VEGF) receptor-binding peptide regulated human umbilical vein endothelial cell (HUVEC) response to soluble VEGF. [4] Additionally, SAMs presenting a heparin-binding peptide could locally sequester heparin and subsequently bind growth factors to regulate cell activity. [6,8] For example, sequestered heparin promoted fibroblast growth factor-mediated increases in HUVEC proliferation, [8] and also amplified human mesenchymal stem cell (hMSC) proliferation and osteogenesis. [6] Taken together, these results and others [1,2] demonstrate that chemically defined SAMs can be used as a versatile tool in order to understand and manipulate cell behavior.
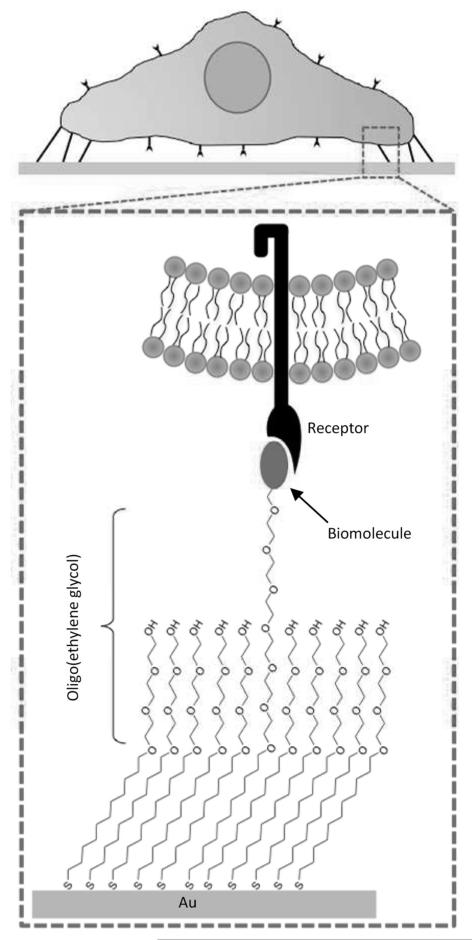
Figure 1.
Generating defined culture substrates using alkanethiolate SAM arrays. SAMs can be designed to present covalently immobilized biomolecules to cells while minimizing the effects of nonspecific protein adsorption by using oligo(ethylene glycol) moieties that limit protein-surface interaction.
While chemically defined SAM-based approaches have provided unique insights into several biological processes, SAMs have yet to become a commonly used tool for biological studies. One potential reason for this lack of utilization is that SAM fabrication can be labor intensive. In a typical experiment investigating SAMs presenting a range of different biomolecules or biomolecule densities, each condition and replicate requires an individual gold substrate. In most approaches, substrates are manually handled before and after each step of an experiment that can include 1) gold substrate cleaning 2) SAM formation, 3) biomolecule conjugation, 4) cell seeding, 5) and immunostaining or other analysis of cellular behavior. To perform a SAM-based experiment with throughput comparable to a standard 96-well plate, a researcher may have to perform close to 1000 handling steps before performing any type of data acquisition or analysis. While SAMs are an excellent model substrate for interrogating the effects of an immobilized biomolecule on cell behavior, to address the complexities of biology we need to generate SAM platforms that can rapidly interrogate a wide range of conditions. Furthermore, to have widespread use in biological investigations, these substrates must be accessible to researchers with very little training in organic chemistry.
Array technologies in the areas of DNA, RNA, and protein detection have revolutionized our ability to interrogate cell behavior. At the center of these approaches is the ability to spatially pattern specific biomolecules on a single substrate to screen for the presence of a target analyte. Recently, SAM patterning approaches have also been developed to spatially localize biomolecules such as peptides to create spatially and chemically defined cell culture substrates. In the interest of generating SAM-based tools for investigating biology, we focus this Minireview on patterning approaches that have been applied to SAMs and discuss how these approaches can be used to generate SAM array platforms. We also highlight several current array platforms that have been used in screening applications to identify novel biological phenomena, and individual sections are organized according to the type of array patterning approach employed in each case. Here, we limit our discussion to bioinert alkanethiolate SAMs, which are designed to limit nonspecific protein adsorption and thereby generate chemically defined substrates. For more information regarding methods of patterning SAMs formed using other types of alkanethiolates the reader is referred to other reviews. [1,2,14]
2. Microcontact Printing
One of the most widespread methods of generating patterned SAMs is to stamp alkanethiolates onto a gold surface by using microcontact printing. [15,16] In this approach, molecules are “inked” onto a flexible elastomeric stamp, which is then used to transfer a pattern of molecules onto a gold substrate. [15] In most cases, hydrophobic alkanethiolates (such as HS –C10–CH3) are stamped onto a gold surface to create a pattern of hydrophobic features. The remaining areas of bare gold are then “backfilled” with a second alkanethiolate species, such as HS–C11–EG3–OH, generate a bioinert SAM layer surrounding the stamped hydrophobic domains. These substrates are then bathed in a solution of extracellular matrix proteins that spontaneously adsorb to the hydrophobic regions to create patterned islands for cell attachment. Microcontact printed islands for cell attachment have been used to study the effects of cell spreading, [17-19] cell–cell contact, [20,21] and cell shape [22] on cell behaviors, including proliferation [17,18,20,21] and differentiation. [19,22] However, while microcontact printing often uses bioinert alkanethiolates for regions surrounding patterned features, OEG-terminated alkanethiolates are not easily microcontact printed due to difficulties in inking and transfer of OEG-alkanethiolates from an elastomeric stamp to a gold surface. While microcontact printing is extremely useful for creating patterns of adsorbed proteins, it is less useful for approaches aiming to generate patterns of specific biomolecules on a bioinert background, with precise control over biomolecule identity and density.
3. Microfluidics
Microfluidics offers a simple method to handle small volumes of reaction buffer and spatially control biomolecule conjugation. Microfluidics approaches typically rely on the use of elastomeric stamps with microscale features (tens of mm) that form channels when passively adhered to a SAM substrate. Localized biomolecule conjugation can then be achieved by flowing reaction solutions through the channels exposing them to reactive terminal moieties presented by the underlying SAM. Microfluidics has been used to pattern biomolecules on SAMs formed using alkanethiolates with several different terminal groups including carboxylate, [6] maleimide, [23] hydroquinone, [24] and aldehyde. [25] Recently we used microfluidics to pattern regions of SAMs with different peptides to spatially control hMSC behavior (Figure 2). [6] In this approach, we patterned immobilization of a cell adhesion peptide (RGD) in combination with a heparin-binding peptide (HEPpep) that sequesters heparin from fetal bovine serum and consequently localizes heparin-binding growth factors such as basic fibroblast growth factor (bFGF) on the SAM surface. To generate patterns of peptide, we formed a SAM substrate containing carboxylate-terminated OEG-alkanethiolates, which were then converted to active ester groups using an N-hydroxysuccinimide (NHS) group. Then, an array of microfluidic channels was adhered to the NHS surface and solutions containing different mixtures of RGD and HEPpep—or mutated versions of either peptide—were flowed through channels to pattern the surface with a range of immobilized peptide compositions (Figure 2A and B). [6] When this surface was seeded with hMSCs, we found that regions containing HEPpep significantly accelerated hMSC proliferation compared to regions presenting a mutant “control” peptide (Figure 2C). In addition to locally controlling the composition in discrete regions on a SAM, microfluidics has also been used to pattern SAMs with immobilized ligand gradients by leveraging the laminar flow that is characteristic of microscale channels. [23] Furthermore, while microfluidics offer a convenient method for manipulating liquids, liquid handling approaches such as the use of robotic microspotters also offer methods to print patterns of biomolecules onto SAM substrates that could be leveraged to rapidly generate arrays of conditions on a SAM surface. [26] Taken together, microfluidics and liquid handling approaches offer a straightforward method for controlling the liquid–surface interface that enables spatial control over biomolecule conjugation that can be used to generate SAM arrays.
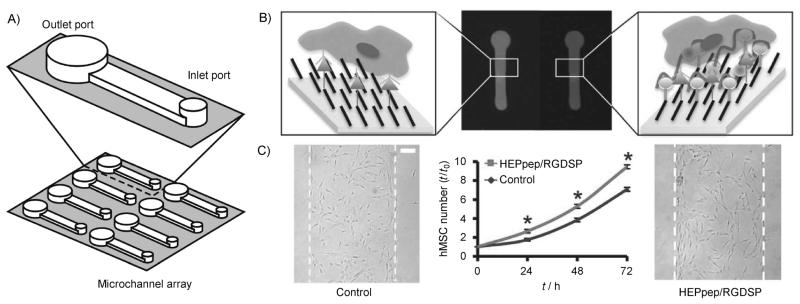
Figure 2.
Pattering SAMs using microfluidics. A) Microfluidic channels were adhered to a SAM substrate and used to locally conjugate a cell adhesion ligand (RGDSP) and a heparin-binding peptide (HEPpep). B) Schematic representation and fluorescent images of peptides patterned using microfluidics with control peptide (left) and HEPpep (right). C) hMSC number over time and bright-field photomicrographs at t= 72 h of hMSCs in regions presenting 2% RGDSP and 2% control peptide (left image), or 2% RGDSP and 2% HEPpep (right image). Scale bar=100 μm; an asterisk indicates significant difference compared to “control” at p<0.05. Adapted from ref. [6] with permission. Copyright: The Royal Society of Chemistry, 2011.
4. Photochemistry
Photochemistry, in combination with micro-patterned photomasks, can be used to pattern SAM substrates with microscale features. One patterning strategy that has been used with OEG-alkanethiolates has involved protecting a reactive terminal moiety with a photolabile molecule, which is then selectively deprotected to locally immobilize biomolecules on a SAM substrate. [27-31] For example, OEG-alkanethiolate SAMs presenting nitroveratryloxycarbonyl (NVOC)-protected hydroquinone groups can be selectively deprotected using 365 nm UV light and then used to covalently immobilize cyclopentadiene- or oxyamine-bearing molecules by a Diels–Alder reaction (Figure 3). [27-30] Using this strategy, Dillmore and co-workers used a photomask to generate SAMs patterned with 15 μm diameter spots and demonstrated that Swiss 3T3 fibroblast attachment was confined to regions presenting the cell adhesion peptide RGD. [29] Additionally, graded photomasks can be used to generate gradients of immobilized biomolecules. [27-30] In particular, this approach has been used to study the effects of adhesion ligand gradients on cell polarization, and have demonstrated that Swiss 3T3 fibroblasts align in directions of increasing adhesion ligand density. [28] Furthermore, the intrinsic electrochemical properties of covalent linkages formed by using hydroquinone groups can be used to release immobilized biomolecules in response to specific voltages, generating SAM substrates with both spatial and temporal control over cell attachment. [27-30] In summary, photochemical deprotection offers a controllable strategy for biomolecule immobilization, and when this strategy is combined with tools commonly used in DNA array fabrication (e.g. “maskless array” technology [32]), it could be used to rapidly generate arrays of biomolecules on a single SAM substrate.
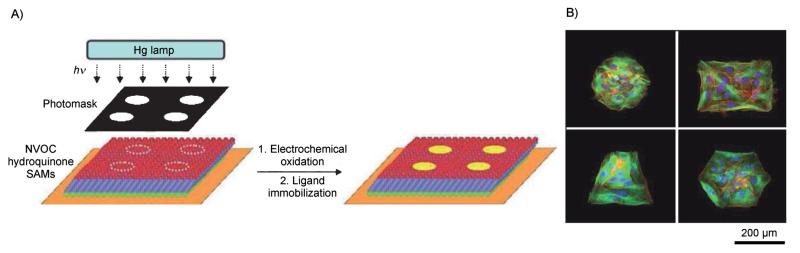
Figure 3.
Photopatterning SAM substrates. A) A photochemical strategy for generating patterns of immobilized ligands onto an electrochemically active monolayer. UV illumination of the NVOC-protected hydroquinone through a photomask reveals the hydroquinone in select regions on the monolayer. Electrochemical oxidation of the hydroquinone monolayer to the quinine permits selective immobilization of ligands to the patterned surface. B) Representative fluorescent images of Swiss 3T3 Fibroblasts on various photo patterns of RGD ligands. Patterned cells were stained for actin (red), microtubules (green), and nuclei (blue). Adapted from ref. [28] with permission. Copyright: The Royal Society of Chemistry, 2008.
5. Localized SAM Removal and Replacement
In addition to approaches to locally control biomolecule conjugation on the surface of a fully formed SAM substrate (as described in the previous sections), SAM patterning can also be achieved by locally modifying the composition of preformed alkanethiolates in the SAM. One method involves locally destroying regions of a fully formed SAM, then reforming new SAMs in the destroyed regions. This methodology has been performed using photochemical [7,33-35] as well as microfluidic [36] approaches to locally remove/destroy and replace regions of SAMs, to generate arrays with spot-to-spot control over alkanethiolate composition and resultant immobilized biomolecule composition. For example, starting with a SAM composed of fluorinated alkanethiolates, prolonged exposure to UV light through a photomask can be used to locally destroy regions of the initial SAM. After this step, the “solvophobic” nature of the fluorinated SAM allows for localized spotting of alkanethiolate solutions to form new SAMs with different alkanethiolate content in each spot. [35] In a similar approach, we recently developed a SAM array system using a combination of microfluidic liquid handling, NaBH4-mediated thiolate reduction, and aqueous SAM re-formation conditions to locally remove and replace regions of a monolayer and create an array of cell-adhesive regions (Figure 4A). In this fabrication approach, bioinert HS–C11EG3–OH SAM was preformed on a gold substrate and a device containing an array of microchannels was passively adhered to the SAM surface. Then, a solution of NaBH4 was flowed through microchannels to locally remove regions of SAM, followed by solutions of different alkanethiolates to locally form new SAMs with varied ratios of carboxylic acid- and hydroxyl-terminated OEG-alkanethiolates. Carbodiimide chemistry was then used to couple the N terminus of peptides to carboxylic acid groups present in the newly formed SAMs. After peptide coupling, the device was removed, and cells were seeded on the resulting patterned SAM substrate. This approach allowed us to rapidly fabricate SAMs patterned with a range of peptide densities and identities in multiple different pattern geometries that are commonly used in microfluidic device applications. Furthermore, hMSCs cultured on patterned SAMs exhibited significant behavioral dependencies on cell adhesion ligand density, including spreading and focal adhesion formation (Figure 4B-D). Taken together, SAM removal-and-replacement approaches enable control over local alkanethiolate composition and, in turn, the fabrication of patterned SAM substrates with local control over biomolecule density and identity.
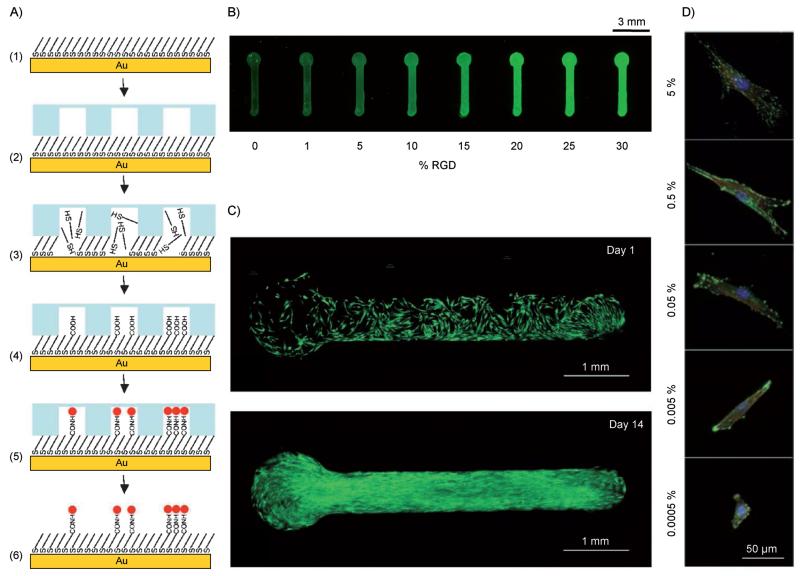
Figure 4.
Patterning using localized SAM removal and replacement. Schematic representation of a localized SAM replacement approach using borohydride chemistry to remove regions of a preformed SAM. A) The approach involves the following: 1) HS–C11EG3OH alkanethiolate SAM; 2) apply microfluidic device; 3) flow aq. 0.5m NaBH 4 through channels to locally remove SAM; 4) locally form a new SAM with a mixture of aqueous HS–C11–EG3–OH and HS–C11–EG6–COOH alkanethiolates to create features with varied carboxylic acid densities; 5) covalently conjugate primary amine-bearing peptide to COOH-terminated alkanethiolates. B) Using this approach, biomolecule immobilization was monitored by coupling a fluorescently tagged peptide to patterned SAM containing discrete features with varied mol% HS–C11 –EG6–COOH alkanethiolate. C) hMSC attachment and growth over 14 days was confined to RGD presenting regions (live hMSCs were stained with calcein AM, green) and D) MSCs attaching to patterned regions presenting varied percentages of RGD exhibited significant differences in cytoskeletal organization (vinculin: green, actin: red, and nuclei: blue). Adapted from ref. [36] with permission. Copyright: The American Chemical Society, 2009.
6. Backfilling
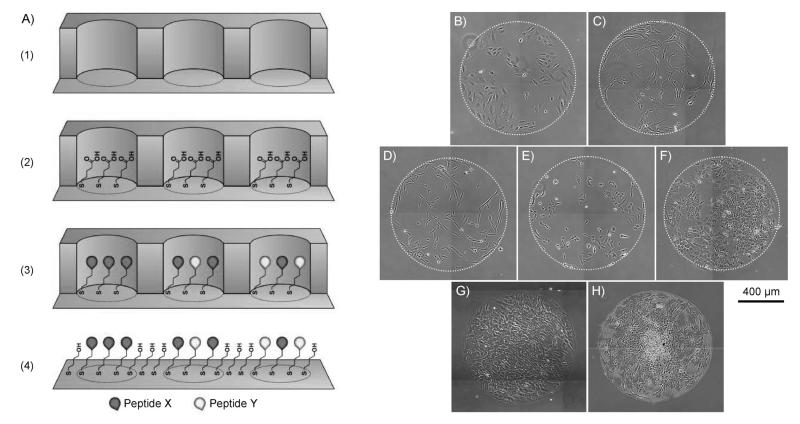
Similar to microcontact printing approaches described in Section 2, bare gold regions of a substrate initially patterned with one alkanethiolate species can be “backfilled” with a second alkanethiolate species to generate surfaces with patterned alkanethiolate composition. However, since microcontact printing does not provide an efficient way to transfer OEG-alkanethiolates to a gold substrate, other methods are used to create an initial pattern of alkanethiolates. [29] Initial patterns of alkanethiolates can be generated using microfluidics, [37-40] simple stencils, [3,4,41] or robotic microspotting techniques [43] to locally control where alkanethiolate solutions contact and subsequently form SAMs on a gold surface. In these approaches, bare regions of gold remaining after initial SAM formation are backfilled with a second, typically bioinert and unreactive alkanethiolate species, such as HS–C11–EG3–OH. Recently, our lab has leveraged the simplicity of elastomeric stencils in combination with alkanethiolate backfilling to generate patterned surfaces to investigate the effects of the adhesion ligand RGD [42] and other peptides [3,4] on the behavior of several different cell types (Figure 5). Here, a simple elastomeric stencil was used to form an array of wells on a gold substrate, and a SAM was formed in each well. SAMs presenting varied peptide composition could be formed using a standard pipette and commercially available alkanethiolates and peptides (Figure 5A). After these steps, backfilling was performed by immersing the gold substrate and the attached elastomeric stencil into a solution of bioinert alkanethiolates and removing the stencil to expose regions of bare gold. Using this approach, one can generate well-defined patterned SAMs with arrays of different peptide conditions in less than two hours. Furthermore, we have used these patterned substrates to study a range of different cell types, including HUVECs, hMSCs, human fibrosarcoma cells (HT-1080s), human dermal fibroblasts (hDFs), and human embryonic stem cells (hESCs) (Figure 5B-F) and cells are maintained within a patterned spot for at least 20 days during extended cell culture (Figure 5G-H). Taken together, approaches that combine liquid handling with SAM backfilling can be used to rapidly pattern SAMs for a range of biological applications.
Figure 5.
SAM patterning using backfilling. [42] A) Schematic representation of one type of backfilling patterning approach: 1) Adhere elastomeric stencil to gold substrate to generate a microwell array superstructure, 2) locally form a SAM in each well with alkanethiolate mixtures containing carboxylic acid-terminated and hydroxyl-terminated OEG-alkanethiolates, 3) covalently conjugate peptides to array spots by carbodiimide condensation of peptide N-terminal primary amine and SAM carboxylic acid terminal moities, and 4) remove mask and backfill remaining bare gold areas with an inert alkanethiolate such as HS–C11–EG3–OH. SAMs patterned with GRGDSP using this backfilling approach have been used to culture a range of human cell types including B) umbilical vein endothelial cells, C) mesenchymal stem cells, D) dermal fibroblasts, E) fibrosarcoma cells (HT-1080s), and F) embryonic stem cells. Furthermore, we have demonstrated that hMSCs seeded on SAM arrays at G) 50000 cellscm −2 remain confined to the patterned spot over H) 20 days.
7. Screening by Using SAM Arrays
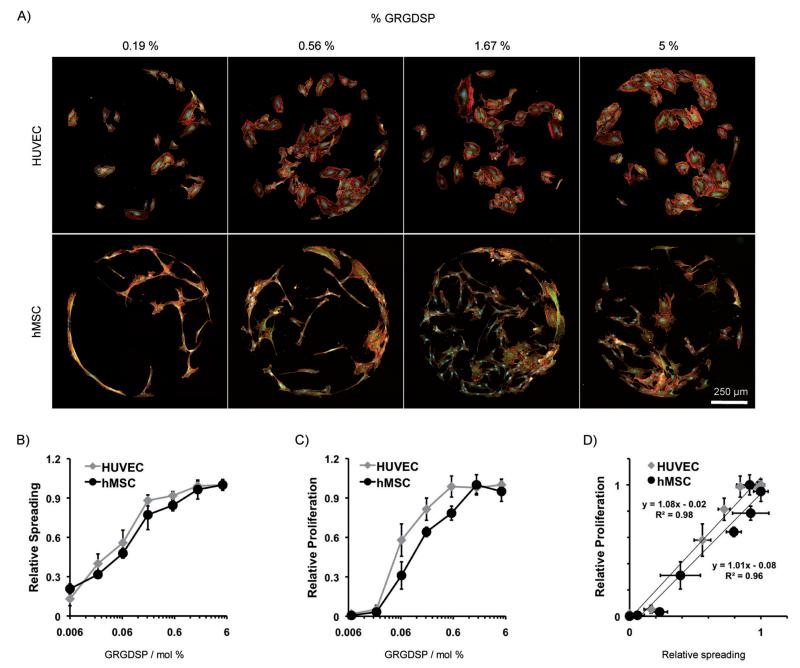
Local control over biomolecule immobilization by SAM patterning has been used to generate arrays that present a wide range of conditions on a single SAM substrate. Here, we discuss key examples in which SAM arrays were used to screen a wide range of peptide conditions. We particularly discuss studies that have identified biological phenomena that would be otherwise impossible to identify with the limited throughput of standard SAM cell culture substrates. Several studies have used SAM arrays to identify novel peptide sequences for maintenance of hESC pluripotency. Kiessling and co-workers have generated SAM arrays using a localized removal-and-replacement approach, and used the arrays to screen for the effects of a panel of known integrin-binding peptides, [33] and a panel of glycosaminoglycan-binding peptides. [7]These studies identified synthetic substrates that promoted hESC proliferation and maintained pluripotency, similar to levels observed when hESCs are grown on Matrigel. Similar SAM arrays were also used in combination with phage display [34] to identify novel peptides that supported hESC proliferation and pluripotency. In addition to using SAM arrays to identify novel synthetic substrates for stem cell culture, we have recently used SAM arrays to directly compare the influence of a cell adhesion ligand on two different cell types. We used a backfilling approach (described in Section 6, Figure 5A) to generate arrays presenting the peptide GRGDSP in densities over three orders of magnitude and seeded parallel arrays with HUVECs and hMSCs (Figure 6). [42] These arrays identified unexpected similarities in cell attachment, spreading, and proliferation for HUVECs and hMSCs and detailed comparison between cell behaviors identified an unexpected 1:1 correlation between spreading and proliferation for both cell types (Figure 6B-D). Additionally, when arrays were integrated with time-lapse microscopy, we observed profoundly different migratory behavior between cell types. Applying patterning techniques to generate chemically defined SAM arrays enables the discovery of novel substrates for targeted cell behaviors, such as maintenance of pluripotency, as well as platforms for fundamental comparisons of cell behaviors across distinct lineages.
Figure 6.
Screening using SAM arrays. [42] SAM arrays presenting a range of GRGDSP peptide densities were used to compare the effects of cell adhesion between two different cell types. A) HUVECs and hMSCs cultured for 72 h on arrays were stained for focal adhesion complexes and cytoskeletal organization (vinculin: green, actin: red, and nuclei: blue). To compare cell behavior, relative HUVEC and hMSC B) spreading and C) proliferation were calculated at 72 h on GRGDSP densities spanning three orders of magnitude. D) Comparison of spreading and proliferation revealed a 1:1 correlation in both cell types. Error bars represent standard error of the mean.
8. Future Perspectives: SAM Array Tools for Cell Biology
A series of recent studies, including many of the studies referenced herein, have demonstrated the significance of SAM arrays in cell biology. SAM arrays can present ligands to cells in a well-defined manner, which allows for precise control over ligand identity and density. In turn, SAMs enable investigators to probe for the effects of a specific molecule of interest, while avoiding the confounding factors that exist in standard cell culture. In addition, SAM arrays afford a series of additional capabilities that may enable new discoveries in cell biology over the next decade and beyond. First, the chemically defined nature of SAM arrays enables investigators to probe for “context dependence” of a particular biological signal. For example, our recent work indicates that heparin-binding ligands impacted stem cell proliferation when presented in conjunction with a relatively high RGD density, but not in conjunction with lower RGD density. [3] In the same study, we found that a bone morphogenic protein receptor-binding peptide exhibited the most pronounced effects on stem cell behavior in conjunction with lower RGD densities compared to higher RGD density. These types of context dependencies are critical for manipulation of stem cell behavior, and also suggest important mechanisms by which distinct signaling pathways interact. Second, SAM arrays are co-planar and translucent, which enables simultaneous time-lapse microscopy of hundreds of individual SAM array spots. This capability allows investigators to efficiently study dynamic processes such as cell migration, stem cell differentiation, and early tissue development in real time. Finally, the raw materials needed to form alkanethiolate SAMs for cell biology applications are now readily available from commercial sources, so that biologists and bioengineers with no experience in synthetic chemistry can readily form alkanethiolate SAMs. In summary, it is clear that SAM arrays are a significant tool, and are now positioned to have unique impact on discoveries in cell biology.
Despite their significance and potential, emerging SAM-based array substrates will bring with them some critical challenges, which will need to be addressed before they are readily used by a broad range of biologists and bioengineers. One critical issue is SAM stability in cell culture, which is highly dependent on the quality of the gold substrate as well as the purity of alkanethiolate molecules and solvents used during monolayer formation. Our experience indicates that SAM stability can be significantly enhanced by instituting well-defined quality control protocols (e.g., NMR and infrared spectroscopy, scanning electron microscopy) to validate the properties of raw materials. Another critical challenge involves the development of methods by which researchers with little to no training in surface chemistry (e.g., biologists) can easily create SAM arrays. While the raw materials for SAM preparation are readily commercially available, the methods for SAM array processing have generally required a high degree of expertise or significant organic synthesis. Our most recent SAM patterning approaches have attempted to address this challenge by relying on commercially available raw materials and simple, universally understood tools, such as elastomeric stencils and standard pipetting. [3,4,42] Taken together, as chemists and engineers continue to develop simple patterning approaches to generate SAM arrays, we envision that SAMs will become more accessible and have widespread impact in cell biology.
Acknowledgements
The authors would like to thank Dr. Jay Warrick at the University of Wisconsin–Madison for his assistance preparing the schematic of microfluidic channels in Figure 2.
References
- [1].Hudalla GA, Murphy WL. Soft Matter. 2011;7:9561–9571. doi: 10.1039/C1SM05596H. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Mrksich M. Acta Biomater. 2009;5:832–841. doi: 10.1016/j.actbio.2009.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Koepsel JT, Brown PT, Loveland SG, Li W-J, Murphy WL. J. Mat. Chem. 2012 doi: 10.1039/C2JM32242K. DOI: 10.1039/C2JM32242K. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Koepsel JT, Nguyen EH, Murphy WL. Integr. Biol. 2012 doi: 10.1039/c2ib20055d. DOI: 10.1039/C2IB20055D. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Li L, Klim JR, Derda R, Courtney AH, Kiessling LL. Proc. Natl. Acad. Sci. USA. 2011;108:11745–11750. doi: 10.1073/pnas.1101454108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Hudalla GA, Kouris NA, Koepsel JT, Ogle BM, Murphy WL. Integr. Biol. 2011;3:832–842. doi: 10.1039/c1ib00021g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Klim JR, Li L, Wrighton PJ, Piekarczyk MS, Kiessling LL. Nat. Methods. 2010;7:989–994. doi: 10.1038/nmeth.1532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Hudalla GA, Koepsel JT, Murphy WL. Adv. Mater. 2011;23:5415–5418. doi: 10.1002/adma.201103046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Vroman L, Adams AL. Surf. Sci. 1969;16:438–446. [Google Scholar]
- [10].Roberts C, Chen CS, Mrksich M, Martichonok V, Ingber DE, Whitesides GM. J. Am. Chem. Soc. 1998;120:6548–6555. [Google Scholar]
- [11].Houseman BT, Mrksich M. Biomaterials. 2001;22:943–955. doi: 10.1016/s0142-9612(00)00259-3. [DOI] [PubMed] [Google Scholar]
- [12].Kato M, Mrksich M. J. Am. Chem. Soc. 2004;126:6504–6505. doi: 10.1021/ja039058e. [DOI] [PubMed] [Google Scholar]
- [13].Kato M, Mrksich M. Biochemistry. 2004;43:2699–2707. doi: 10.1021/bi0352670. [DOI] [PubMed] [Google Scholar]
- [14].Smith RK, Lewis PA, Weiss PS. Prog. Surf. Sci. 2004;75:1–68. [Google Scholar]
- [15].Alom Ruiz S, Chen CS. Soft Matter. 2007;3:168–177. doi: 10.1039/b613349e. [DOI] [PubMed] [Google Scholar]
- [16].Raghavan S, Chen CS. Adv. Mater. 2004;16:1303–1313. [Google Scholar]
- [17].Huang S, Chen CS, Ingber DE. Mol. Biol. Cell. 1998;9:3179–3193. doi: 10.1091/mbc.9.11.3179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Chen CS, Mrksich M, Huang S, Whitesides GM, Ingber DE. Science. 1997;276:1425–1428. doi: 10.1126/science.276.5317.1425. [DOI] [PubMed] [Google Scholar]
- [19].Gao L, McBeath R, Chen CS. Stem Cells. 2010;28:564–572. doi: 10.1002/stem.308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Nelson CM, Chen CS. FEBS Lett. 2002;514:238–242. doi: 10.1016/s0014-5793(02)02370-0. [DOI] [PubMed] [Google Scholar]
- [21].Nelson CM, Chen CS. J. Cell Sci. 2003;116:3571–3581. doi: 10.1242/jcs.00680. [DOI] [PubMed] [Google Scholar]
- [22].Kilian KA, Bugarija B, Lahn BT, Mrksich M. Proc. Natl. Acad. Sci. USA. 2010;107:4872–4877. doi: 10.1073/pnas.0903269107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Petty RT, Li HW, Maduram JH, Ismagilov R, Mrksich M. J. Am. Chem. Soc. 2007;129:8966–8967. doi: 10.1021/ja0735709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Westcott NP, Yousaf MN. Langmuir. 2008;24:2261–2265. doi: 10.1021/la703744v. [DOI] [PubMed] [Google Scholar]
- [25].Westcott NP, Pulsipher A, Lamb BM, Yousaf MN. Langmuir. 2008;24:9237–9240. doi: 10.1021/la802208v. [DOI] [PubMed] [Google Scholar]
- [26].Chan EW, Yousaf MN. J. Am. Chem. Soc. 2006;128:15542–15546. doi: 10.1021/ja065828l. [DOI] [PubMed] [Google Scholar]
- [27].Chan EW, Park S, Yousaf MN. Angew. Chem. 2008;120:6363–6367. doi: 10.1002/anie.200800166. [DOI] [PubMed] [Google Scholar]; Angew. Chem. Int. Ed. 2008;47:6267–6271. doi: 10.1002/anie.200800166. [DOI] [PubMed] [Google Scholar]
- [28].Chan EW, Yousaf MN. Mol. Biosyst. 2008;4:746–753. doi: 10.1039/b801394b. [DOI] [PubMed] [Google Scholar]
- [29].Dillmore WS, Yousaf MN, Mrksich M. Langmuir. 2004;20:7223–7231. doi: 10.1021/la049826v. [DOI] [PubMed] [Google Scholar]
- [30].Lee EJ, Chan EW, Yousaf MN. ChemBioChem. 2009;10:1648–1653. doi: 10.1002/cbic.200900277. [DOI] [PubMed] [Google Scholar]
- [31].Park S, Yousaf MN. Langmuir. 2008;24:6201–6207. doi: 10.1021/la8005663. [DOI] [PubMed] [Google Scholar]
- [32].Singh-Gasson S, Green RD, Yue Y, Nelson C, Blattner F, Sussman MR, Cerrina F. Nat. Biotechnol. 1999;17:974–978. doi: 10.1038/13664. [DOI] [PubMed] [Google Scholar]
- [33].Derda R, Li LY, Orner BP, Lewis RL, Thomson JA, Kiessling LL. ACS Chem. Biol. 2007;2:347–355. doi: 10.1021/cb700032u. [DOI] [PubMed] [Google Scholar]
- [34].Derda R, Musah S, Orner BP, Klim JR, Li L, Kiessling LL. J. Am. Chem. Soc. 2010;132:1289–1295. doi: 10.1021/ja906089g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Orner BP, Derda R, Lewis RL, Thomson JA, Kiessling LL. J. Am. Chem. Soc. 2004;126:10808–10809. doi: 10.1021/ja0474291. [DOI] [PubMed] [Google Scholar]
- [36].Koepsel JT, Murphy WL. Langmuir. 2009;25:12825–12834. doi: 10.1021/la901938e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Lamb BM, Park S, Yousaf MN. Langmuir. 2010;26:12817–12823. doi: 10.1021/la1022642. [DOI] [PubMed] [Google Scholar]
- [38].Lamb BM, Westcott NP, Yousaf MN. ChemBioChem. 2008;9:2220–2224. doi: 10.1002/cbic.200800400. [DOI] [PubMed] [Google Scholar]
- [39].Lamb BM, Westcott NP, Yousaf MN. ChemBioChem. 2008;9:2628–2632. doi: 10.1002/cbic.200800473. [DOI] [PubMed] [Google Scholar]
- [40].Pulsipher A, Yousaf MN. Langmuir. 2010;26:4130–4135. doi: 10.1021/la903297d. [DOI] [PubMed] [Google Scholar]
- [41].Yeo WS, Mrksich M. Langmuir. 2006;22:10816–10820. doi: 10.1021/la061212y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Koepsel JT, Loveland SG, Le NN, Schwartz MP, Murphy WL. 2012 unpublished results. [Google Scholar]
- [43].Luo W, Chan EW, Yousaf MN. J. Am. Chem. Soc. 2010;132:2614–2621. doi: 10.1021/ja907187f. [DOI] [PubMed] [Google Scholar]