Abstract
In the biosphere, many species live in close proximity and can thus interact in many different ways. Such interactions are dynamic and fall along a continuum between antagonism and cooperation. Because interspecies interactions are the key to understanding biological communities, it is important to know how species interactions arise and evolve. Here, we show that the feedback between ecological and evolutionary processes has a fundamental role in the emergence and dynamics of species interaction. Using a two-species artificial community, we demonstrate that ecological processes and rapid evolution interact to influence the dynamics of the symbiosis between a eukaryote (Saccharomyces cerevisiae) and a bacterium (Rhizobium etli). The simplicity of our experimental design enables an explicit statement of causality. The niche-constructing activities of the fungus were the key ecological process: it allowed the establishment of a commensal relationship that switched to ammensalism and provided the selective conditions necessary for the adaptive evolution of the bacteria. In this latter state, the bacterial population radiates into more than five genotypes that vary with respect to nutrient transport, metabolic strategies and global regulation. Evolutionary diversification of the bacterial populations has strong effects on the community; the nature of interaction subsequently switches from ammensalism to antagonism where bacteria promote yeast extinction. Our results demonstrate the importance of the evolution-to-ecology pathway in the persistence of interactions and the stability of communities. Thus, eco-evolutionary dynamics have the potential to transform the structure and functioning of ecosystems. Our results suggest that these dynamics should be considered to improve our understanding of beneficial and detrimental host–microbe interactions.
Keywords: fungal–bacterial interactions, eco-evolutionary feedbacks, adaptive evolution, niche construction, Saccharomyces cerevisiae, Rhizobium etli
Introduction
In nature, species are bound together by a web of complex interactions that are central to the structure, composition and function of any community (Thompson, 1994). Such interactions are dynamic and fall along a continuum between antagonism and cooperation (Ewald, 1987; Hayashi, 2006; Relman, 2008; Sachs et al., 2011). Despite the central importance of species interactions to the diversification and organization of life, we still know little about how these relations arise and evolve (Thompson, 1999). Many studies describing interspecies interactions have focused on how ecological processes, for example, environmental changes, affect the nature of the interaction (Kim et al., 2008; Seyedsayamdost et al., 2011; Duffy et al., 2012). For example, the nature and strength of the interactions between ants and certain plants depend upon environmental factors such as nutrient or light availability, herbivore pressure or relationships with other organisms (Palmer et al., 2008). Other studies have shown how the evolutionary process can affect the nature of interspecies interactions (Yoshida et al., 2003; Harmon et al., 2009; Hillesland and Stahl, 2010; Gómez and Buckling, 2011; Lawrence et al., 2012). Using two unrelated soil-inhabiting bacteria, Hansen et al. (2007) showed that simple mutations in the genome of one species caused it to adapt to the presence of the other, forming an intimate and specialized association. A recent analysis of the symbiotic interaction between a bacteria and a nematode demonstrated that a genetic change in the bacteria reversibly switched the interaction from mutualism to parasitism (Somvanshi et al., 2012).
Eco-evolutionary theory suggests that ecological and evolutionary process often occur on similar time scales, and that they codetermine the dynamic behavior of ecological communities (Post and Palkovacs, 2009; Schoener, 2011). For example, in two-species community variation in antipredator defense among algal genotypes can strongly influence rotifer growth rates and densities, which feed back to influence gene frequencies in algal populations (Meyer et al., 2006).
A recent study with a single species showed that feedback between population and evolutionary dynamics determines the fate of social microbial populations (Sanchez and Gore, 2013). In contrast, fewer studies on interspecific interactions include eco-evolutionary dynamics as one of the working hypotheses to know how species interactions arise and evolve, even though its potential importance is clear (Thompson, 1999; Post and Palkovacs, 2009; Schoener, 2011).
Artificial microbial communities constructed from species that have no known history of previous interaction provide a platform to address many important questions about the origin of natural interactions (Hansen et al., 2007; Kim et al., 2008; Hillesland and Stahl, 2010). Natural interactions, which have evolved over millions of years, have received the most attention and have been examined extensively, for example, the Rhizobium-legume symbioses. Here, we take a different vantage point by investigating what happens when two species with no known history of previous interaction meet for the first time. To this end, we use a two-species artificial community composed of a fungus (Saccharomyces cerevisiae) and a bacterium (Rhizobium etli). S. cerevisiae is a widely-studied eukaryote for which there are a large number of genomic tools that make it ideal for work in areas such as evolution and ecology of species interactions (Replansky et al., 2008). R. etli is a nitrogen-fixing bacterium that has evolved to form intimate, intracellular associations with legumes (Denison, 2000; González et al., 2003). This soil bacterium has been widely studied during its interaction with the plant and in pure cultures, but less is known about how this plant symbiont interacts with other microbial species in free life.
To study how species interactions arise and evolve, R. etli CE3 and S. cerevisiae (a common lab strain) were grown together in rich medium for 9 days under well-mixed culture conditions. Measurements of the fitness of each species in coculture and monoculture were used to determine the nature of the interaction throughout the incubation period. Coculturing the bacteria with ∼4700 yeast mutants allowed us to establish a causal relationship between changes in the environment caused by yeast and its effect on the nature of the interaction. At the beginning of the interaction, yeast simultaneously secreted an inhibitor (orotic acid, OA) of bacterial growth and growth promoters (C4-dicarboxylates) that blocked the effect of the OA, and it allowed the establishment of a commensal relationship. When the growth promoters became depleted (ecological change), the interaction shifted from commensalism to ammensalism.
We conducted proteomic, genome-wide transcriptomic and biochemical analysis of the population of cells exposed to OA, and showed that the OA-sensitive phenotype is caused by an imbalance between purine and pyrimidine nucleotides. Interestingly, we found that during ammensalism OA-resistant (OAR) bacterial variants arose that allowed bacterial growth, which modified the environment and created competition for nutrients with the yeast. Thus, the rapid evolution prevents extinction of the bacteria (evolutionary rescue) and switched the interaction from ammensalism to antagonism.
Here, we show for the first time, to our knowledge, how the metabolic perturbation of an essential pathway may be the cause of a rapid evolutionary response that consequently changes the course of a biological interaction. We provide experimental evidence that the interplay between ecological (niche construction and metabolic relationships between species) and evolutionary processes (adaptative evolution) has a fundamental role in the emergence and dynamics of species interaction.
Finally, our results show that the R. etli–S. cerevisiae community can be a useful experimental model for discovering new evolutionary and physiological aspects of fungi and bacteria that cannot be found in pure culture or in the interaction with their natural partners.
Materials and methods
Strains, plasmids and culture conditions
The yeast, bacterial strains and plasmids used in the study are listed in Supplementary Table S1. S. cerevisiae was routinely grown in YPD (1% yeast extract, 2% bacto peptone and 2% dextrose) medium at 30 °C. R. etli was grown at 30 °C in PY (0.5% peptone, 0.3% yeast extract and 7 mM CaCl2) medium. Escherichia coli was grown at 37 °C in Luria–Bertani medium. Antibiotics were supplied to each medium to maintain selection for plasmids or to select recombinant strains at the following concentrations: nalidixic acid (20 μg ml−1), streptomycin (200 μg ml−1), tetracycline (5–10 μg ml−1), spectinomycin (200 μg ml−1), kanamycin (30 μg ml−1) and gentamicin (30 μg ml−1). Triparental conjugations and site-directed mutagenesis of R. etli were performed as described previously (D'hooghe et al., 1995). DNA preparation and recombinant DNA techniques were performed according to standard procedures (Sambrook and Russell, 2001).
Cocultures
R. etli CE3 and S. cerevisiae Σ1278h were cultured together in 150 ml flasks, each containing 50 ml of PY-D medium (0.5% peptone, 0.3% yeast extract, 10 mM dextrose and 7 mM CaCl2). We inoculated with ∼4 × 106 bacterial cells per ml and ∼5 × 105 yeast cells per ml in each flask at the start of an experiment. R. etli CE3 and yeast monocultures were grown in PY-D medium and PY-DOA100 (PY-D medium with 100 μg ml−1 OA). Each flask was inoculated with ∼4 × 106 bacterial cells per ml. The cultures were incubated at 30 °C and shaken at 200 r.p.m. Samples of the cultures were periodically preserved at −80 °C in 20% glycerol. Each sample was tested for the presence of contamination by plating a sample (50 μl) on an Luria–Bertani agar plate.
Bacterial densities were measured by plating diluted cultures onto PY-DOA100 agar plates and counting the number of colony-forming units (CFUs) after 72 h of incubation at 30 °C. The colonies of OAR variants were larger in size and easily distinguished from the parental strain. Isolates were stored at −80 °C in 20% glycerol for use in subsequent assays.
Phenotypic analysis of OAR isolates
We measured the total number of distinct phenotypes of OAR variants based on a sample of 94 randomly chosen OAR colonies from each sampling time. The bacterial isolates were arranged in 96-well microtiter plates containing 100 μl of PY-D medium with 100 μg ml−1 OA. The diversity of the population was determined by evaluating the growth of the clones in different culture media, assaying melanin production and performing a genetic complementation test.
The isolates were grown on PY-DOA plates for 48 h. Next, the cells were transferred to 96-well microtiter plates containing 100 μl of H2O per well using a prong. Then, 100 μl of liquid medium per well was inoculated with 1 μl of the bacterial cell suspension and grown in a static incubator at 30 °C for 24 h. The growth yields were measured via optical density (OD) at 600 nm using a microplate reader (3550-UV, Bio-Rad). OD600 data (mean±s.d.) were obtained from at least two independent cultures of each strain.
The growth of the isolates was evaluated in the following culture media:
PY-DOA100, PY-D medium with 100 μg ml−1 OA; PY-DFOA50, PY-D medium with 50 μg ml−1 5-fluoroorotic acid; MM-S, minimal medium containing 10 mM succinate (Encarnación et al., 1995); MM-S+Uridine, minimal medium containing 10 mM succinate supplemented with 20 μg ml−1 uridine; MM-D, minimal medium containing 10 mM dextrose and MM-D+Uridine, minimal medium containing 10 mM dextrose supplemented with 20 μg ml−1 uridine.
Melanin production was assayed on PY-D plates containing 30 μg ml−1 tyrosine and 10 μg ml−1 CuSO4, as described previously (Hawkins and Johnston, 1988). The bacteria were incubated at 30 °C for 72 h, and the presence or absence of black pigment was monitored visually. The rpoN1 mutant strains were unable to produce melanin, whereas no differences between the wild-type strains and rpoNd mutants were observed (Supplementary Figure S10). Strains that did not produce melanin were PCR screened for the presence of the tyrosinase gene (melA), which is plasmid encoded and can be lost (González et al., 2003).
Genotyping assays to estimate allele frequencies
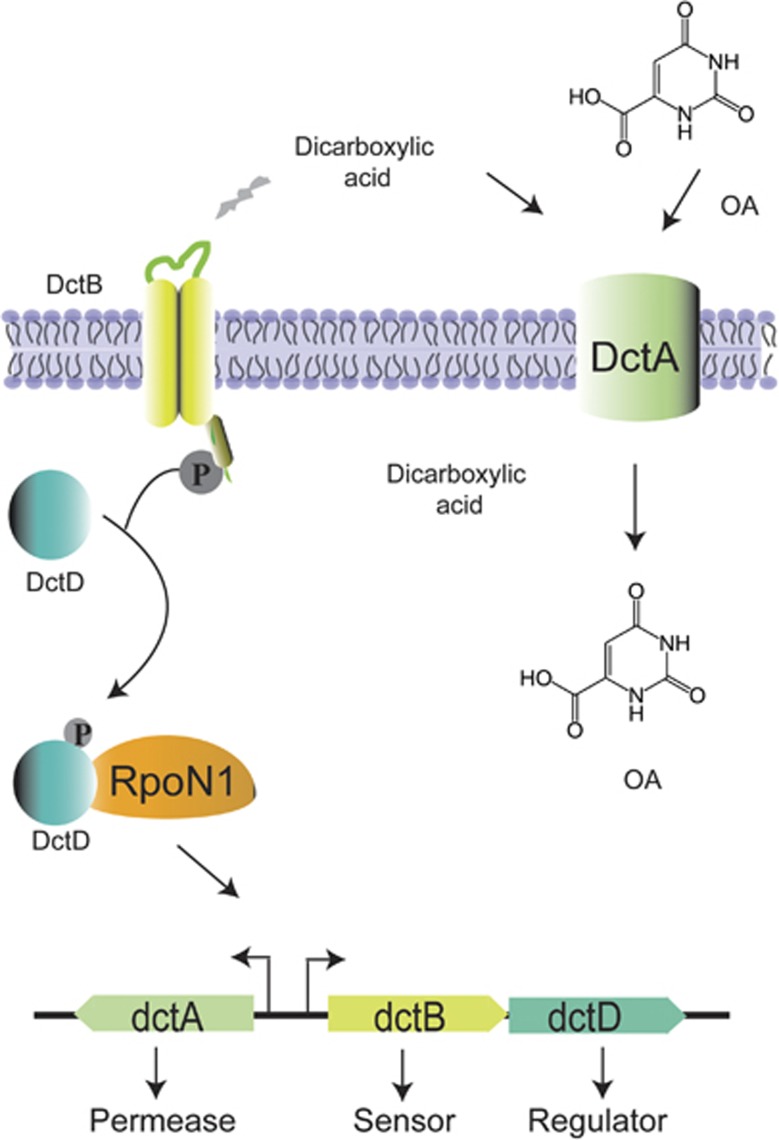
We constructed plasmids (Supplementary Table S1) containing each of the following wild-type genes: dctA, dctB, dctB-dctD operon, rpoN1, pyrE and pyrF. R. etli CE3 chromosomal DNA was used as the template for PCR to amplify each gene with its endogenous promoter (see the list of primers in Supplementary Table S2). The amplified genes were cloned into pBBR1MCS5. The resulting pAT plasmids were transformed into E. coli DH5α. All plasmids were transferred to the OAR isolates by triparental mating (D'hooghe et al., 1995). The plasmids pAT-dctA and pAT-dctB-dctD were transferred to the dct-group. The resultant strains did not transport succinate, produce melanin or grow in MM-D.
The plasmid pAT-rpoN1 was transferred to isolates that did not produce melanin and were unable to grow on MM-S and MM-S+uridine media. The uracil auxotrophs isolates were transformed with plasmids AT-pyrE and AT-pyrF. The growth of the transformed strains was evaluated in PY-DFOA, MM-S and MM-D media. As a control, the vector pBBR1MCS-5 was also transferred to the OAR isolates, but no change in phenotype was observed. The complementation of OAR isolates with a wild-type gene allowed them to growing MM-S and MM-D but not in PY-DFOA.
Gene sequencing
We sequenced the entirety of the dctA, dctB, dctD, rpoN1, pyrE and pyrF genes from the OAR clones isolated in the evolution experiments. The PCR products were amplified using primer-F and primer-R (Supplementary Table S3). The primers in Supplementary Table S9 were used in separate Sanger sequencing reactions per template to cover the lengths of the genes.
Measurement of bacterial diversity
Diversity was measured by determining the characteristics of 94 OAR isolates. Diversity was calculated as the complement of Simpson's index (1−λ) (Simpson, 1949):
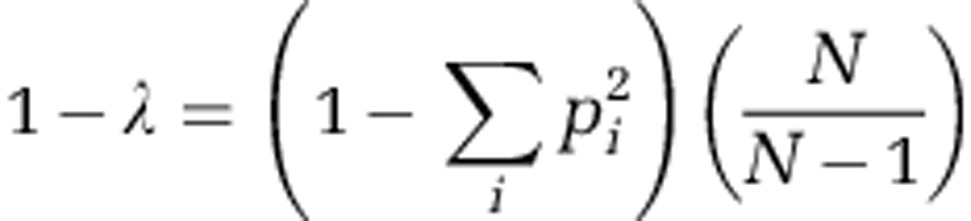 |
where pi is the proportion of the i phenotype and N is the total number of colonies sampled.
Fitness measures
For competitive fitness assays, the medium initially contained ∼4 × 106 CFU per ml of both ancestral R. etli CE3 and OAR variants (∼1:1) and additionally ∼5 × 105 CFU per ml of yeast when competing in the presence thereof. The mixed culture was allowed to grow for 48 h at 30 °C and was shaken at 200 r.p.m. After the desired number of daily growth cycles, a sample of the mixture was again diluted in Tween solution (0.01% Tween 80 and 10 mM MgSO4) and plated onto PY-DOA100 agar to estimate population densities of bacterial cells of each type. To estimate the yeast population density, the sample was centrifuged (4 min at 3000 × g), washed and deflocculated using 300 mM EDTA, diluted and plated onto YPD agar (R. etli does not grow on this medium) to determine the number of CFU (Smukalla et al., 2008). The Malthusian parameter (Lenski et al., 1991) m, which is the average rate of increase, was calculated for both competitors: m=ln(Nf/Ni), where Nf is the number of individuals present at the final count and Ni is the number of individuals present at the initial count. Relative fitness was calculated as the selection rate constant: rij=mi−mj, resulting in a fitness of zero when competing organisms are equally fit.
Statistical analysis
Analysis of variance and Tukey's Multiple Comparison Tests were used to identify significant differences between treatments of interest.
Yeast deletion screen
Yeast knock-out strains (YKO library, Open Biosystems, Huntsville, AL, USA) were tested against R. etli CE3 to identify the genes involved in OA production. The library contains ∼4700 null mutations in nonessential genes. Individual deletion strains (4 μl) were transferred from frozen stocks to YPD (containing 200 μg ml−1 G418) medium plates using a prong and were grown at 30 °C for 2 days. Following this, 96 mutants were spotted (the distance between strains was 1.5 cm) over 24 × 24 cm PY-D (25 mM dextrose) agar plates inoculated with ∼3 × 107 cells ml−1 R. etli CE3. The yeast and bacteria were co-incubated at 30 °C for 24 h, and the presence or absence of an inhibition zone was monitored visually (Supplementary Figure S1). Furthermore, the mutants that did not promote bacterial growth within the inhibition zone were isolated (Figure 1d). We identified three strains that did not produce inhibition zones (Supplementary Figure S3B). These strains were then retested, and OA production was quantified by HPLC as described in Supplementary Materials and Methods.
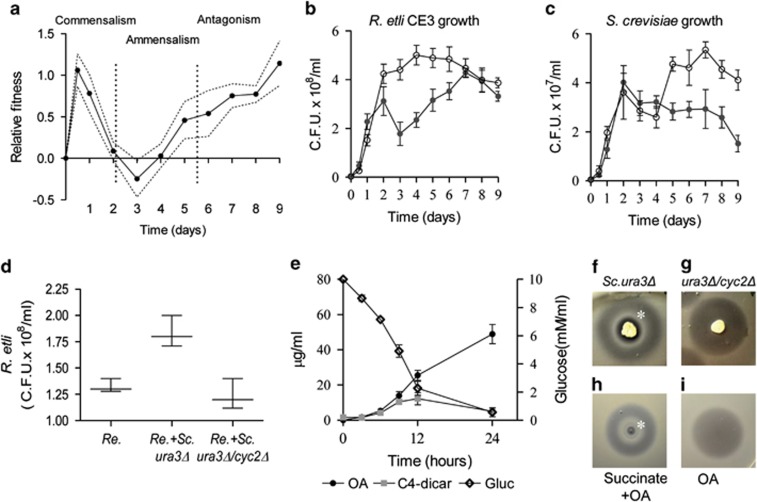
Figure 1.
Temporal dynamics of the fungal–bacterial interaction and niche construction by yeast. (a) The fitness of R. etli CE3 relative to S. cerevisiae Σ1278h ura3Δ. Fitness is given as the difference in Malthusian parameters (m). A fitness of zero indicates that the two species are equally fit. Dotted lines indicate ±s.e.m. (n=3). (b, c) Individual growth curves of R. etli and S. cerevisiae in coculture (filled symbols) and pure culture (open symbols). (d–i) C4-dicarboxylate production by yeast is required for the establishment of commensalism. (d) Population density of R. etli at 24 h in monoculture (Re.), coculture with S. cerevisiae ura3Δ (Re.+Sc.ura3Δ) and coculture with the S. cerevisiae cyc2 mutant (Re.+Sc. ura3Δ/cyc2Δ), in the BY4741 background. (e) The environmental changes in coculture through the course of the evolution experiment. Left y-axis: concentrations of OA and C4-dicarboxylates (plotted as the sum of succinate, fumarate and α-ketoglutarate). Right y-axis: concentrations of Glucose. (f, g) The experiments demonstrate the capacity of yeast strains to modify the environment and determine the community structure on solid media. (f) R. etli CE3 grow as a commensal organism inside the inhibition zone (asterisk) surrounding the ura3Δ S. cerevisiae colony. (d) cyc2Δ/ura3Δ S. cerevisiae cells, which do not accumulate C4-dicarboxylates, do not promote bacterial growth. (h, i) Experiments to mimic community structure generated by yeast strains. (h) OA and succinate (50 μg each) were placed together on the plate. Succinate promoted bacterial growth inside the inhibition zone (asterisk). (i) Bacterial growth inhibition by OA (50 μg). All panels show the mean±s.e.m. (n=3).
Transcriptome analysis of R. etli
R. etli CE3 was grown overnight in PY medium. The cells were collected by centrifugation and washed twice with water. These cells were used to inoculate 100 ml MM-D medium in a 250-ml Erlenmeyer flask at an OD600 of 0.2. After 4 h incubation at 30 °C with constant shaking (200 r.p.m.), the cultures were supplemented with 100 μl of H2O (negative control) or 50 μg ml−1 of OA. After 30 min incubation at 30 °C with constant shaking (200 r.p.m.), the cells were collected by centrifugation at 5520 × g for 5 min, flash-frozen and stored in liquid nitrogen. The total RNA was extracted using an RNeasy mini kit (Qiagen, Hilden, Germany) following the manufacturer's instructions. Microarray experiments were conducted using three independently isolated RNA preparations from independent cultures. Microarray preparation, synthesis of fluorescent cDNA probes, hybridization with R. etli CE3 cDNA microarray and signal analysis were strictly conducted as reported previously (Salazar et al., 2010).
We used the mean Z-score of the three replicates to produce a preordered ranked gene list data set, which was then tested for enrichment by Gene Set Enrichment Analysis (GSEA v. 2.0, Broad Institute, Cambridge, MA, USA) (Subramanian et al., 2005). Gene sets were generated and modified with biochemical pathway data publicly available from the Kyoto Encyclopedia of Genes and Genomes (KEGG; www.genome.jp/kegg).
To study the extent of the effects of OA on R. etli CE3, the transcriptome microarray data were first organized into biological process clusters according to the KEGG. We tested the KEGG gene sets of the R. etli organism microarray data for enrichment by GSEA. The overall results of the R. etli CE3 enrichment are depicted in Supplementary Table S8. Several gene sets were differentially enriched (P<0.05, FDR<0.25) in the R. etli CE3 under OA stress. Eleven gene sets were negatively enriched (repressed), and one was positively enriched (activated).
Results
Niche construction by yeast allows the establishment of a commensal relationship with the bacteria
The budding yeast S. cerevisiae (strain Σ1278h ura3Δ) and the nitrogen-fixing bacterium R. etli (strain CE3) are microorganisms that do not have a known history of previous interaction. In our experiments, we cultured the species together in rich medium with shaking for 9 days. The absolute fitness as measured by the average rate of population increase, or Malthusian parameter, was calculated for both species in the cocultures. To identify changes in the nature of their interactions, we analyzed the fitness of bacteria relative to yeast throughout the incubation period.
We found that the interaction was dynamic, switching from commensalism to ammensalism and to antagonism (Figure 1a). In the early stages of the interaction, the presence of the yeast improved bacterial growth; a higher yield was observed compared with their growth in pure culture (Figure 1b). In contrast, during the first four days of coculture the bacterium does not reduce or promote the growth of the yeast, that is, yeast growth is similar in the cocultures and pure cultures (Figure 1c). The capacity of yeast to increase bacterial growth yield was associated with the production of C4-carboxylates, because respiration-deficient yeast that excreted low quantities of C4-carboxylates (Dibrov et al., 1997) were less efficient at promoting bacterial growth (Figures 1d, and g, Supplementary Figure S2). At this stage, the relationship between these two species approximated that of a commensal partner (R. etli) and a host (S. cerevisiae). Thus, the niche-constructing activities of yeast were the key ecological process that allowed the establishment of a commensal relationship.
An ecological change transforms the interaction from commensalism to ammensalism
After 24 h of coculture, environmental changes shifted the interaction from commensalism to ammensalism (Figure 1a). Ammensalism is the ecological interaction in which an individual harms other without obtaining any benefit. We found that this change in the interaction was caused by a metabolite excreted by the yeast that inhibits the growth of the bacterium (Figures 1f–i, Supplementary Figure S1). To confirm this, we isolated the bacterial growth inhibitor (we designated it PΣA) from S. cerevisiae Σ1278h ura3Δ culture supernatants (see Supplementary Methods, Supplementary Figure S1D). The identity of PΣA was confirmed by chromatographic and analytical methods, such as infrared spectroscopy, mass spectrometry, 1H NMR, 13C NMR and gHSQC. Infrared spectroscopy data (see Supplementary Methods and Supplementary Table S4) and the mass spectrum of PΣA were identical (Supplementary Figure S3) to the standard OA spectrum. The complete structure of PΣA was solved by interpreting one- and two-dimensional NMR spectra (1H and 13C NMR and gHSQC; see Supplementary Methods and Supplementary Table S5). Furthermore, PΣA had biological activity identical to OA (Supplementary Figure S1D). OA is an intermediate in the uridine-5′-phosphate biosynthetic pathway. OA accumulates in the medium of our strain of yeast because it lacks the URA3 gene, which encodes orotidine-5′-phosphate decarboxylase (Figure 1d and Supplementary Figure S1). Importantly, yeast mutants unable to synthesize OA did not inhibit the growth of the bacterium, indicating that OA was responsible for the inhibition observed in the interaction (Supplementary Figure S1B).
We found that OA at concentrations greater than 15 μg ml−1 (±s.d. 3) inhibited the growth of the bacterium. In the cocultures, OA reached inhibitory concentrations (25.5 μg ml−1, ±s.d. 5) by 12 h, but its effect on the growth of the bacteria was observed only after 24 h and correlated with the decrease in the concentration of C4-dicarboxylates derived from glucose fermentation (Figure 1d). Previous reports have demonstrated that the production of C4-dicarboxylates by yeast increases over time and is dependent on the rate of glucose consumption (Romano and Kolter, 2005). We hypothesized that C4-dicarboxylates secreted by yeast blocked OA transport into the bacterium and allowed the bacteria to grow as commensals. We found that in liquid cultures, succinate, malate and fumarate blocked the effect of OA (Supplementary Figure S4A). Previous studies have shown that in Sinorhizobium meliloti, dicarboxylates compete with OA for the DctA permease and block OA transport (Yurgel and Kahn, 2005). Figure 1f shows that on culture plates, diffusion of C4-dicarboxylates secreted by yeast colonies blocked OA transport into the bacterium allowing it to grow within the inhibition zone. Using yeast knock-out strains (YKO library), we identified 120 yeast mutants that did not promote the establishment of a commensal relationship (Supplementary Table S6). The largest class of mutants were affected in mitochondrial functions and these mutants produced low amounts of organic acids (Supplementary Figure S2) (Dibrov et al., 1997). Taken together, these results demonstrate that yeast has a strong impact on the community and therefore had the potential for niche construction (Laland et al., 1999; Post and Palkovacs, 2009). The mechanism used by yeast for niche construction was the simultaneous secretion of bacterial growth promoters (C4-dicarboxylates from glucose fermentation) and a bacterial growth inhibitor (OA) (Figures 1e–i and Supplementary Figure S1). Thus, the interaction switched from commensalism to ammensalism when glucose was consumed and the concentration of C4-dicarboxylates decreased to levels below those required to block the transport of OA (Figure 1e).
The OA-sensitive phenotype is caused by an imbalance between purine and pyrimidine nucleotides
When individuals of two species interact, they can adjust their phenotypes in response to their respective partner, be they antagonists or mutualists. Single genotypes can change their chemistry, physiology, development, morphology or behavior in response to environmental cues (Hansen et al., 2007; Seyedsayamdost et al., 2011; Lawrence et al., 2012; Somvanshi et al., 2012). Here, we analyze the physiological changes of the bacteria caused by the factor (OA) that changed the interaction from commensalism to ammensalism.
Growth inhibition of R. etli and other organisms by exogenous OA has not been previously reported. We conducted experiments to determine the possible mechanism by which OA exerts its growth-inhibitory effect on R. etli CE3. We conducted proteomic and genome-wide transcriptomic analysis of population of cells exposed to OA.
The results of the transcriptome analysis indicated that cells exposed to OA displayed considerable differences in gene expression compared with nonexposed cells (Supplementary Table S7 and S8). The observed changes fall into several broad categories. First, the decreased expression of genes involved in protein synthesis (including ribosomal genes), replication and energy production indicates that the cells arrest or at least slow their growth, which agrees with the observed growth arrest induced by OA (Supplementary Figure S4). The lack of growth might be associated with the limited availability of purines, as indicated by an upregulation of genes required for de novo purine nucleotide biosynthesis and genes involved in 5′-phosphoribosyl-1′-pyrophosphate (PRPP) biosynthesis. PRPP is an essential intermediate involved in many biosynthetic pathways, such as biosynthesis of purine nucleotides, pyrimidine nucleotides, NAD+, NADP+, histidine and tryptophan. PRPP is also consumed by purine and pyrimidine salvage pathways when purine or pyrimidine derivatives are added exogenously (Shimosaka et al., 1984). PRPP and OA are substrates for the orotate phosphoribosyl transferase (OPTase), which catalyzes the reversible phosphoribosyl transfer from PRPP to OA, forming pyrophosphate and orotidine 5′-monophosphate (OMP) (Figure 2). The upregulation of a key enzyme (ribose-phosphate pyrophosphokinase, PrsAch) for PRPP synthesis suggests that cells exposed to OA suffer PRPP deprivation. We hypothesized that the PRPP pools were depleted as a result of an increase in the synthesis of OMP stimulated by exogenous OA. PRPP depletion would result in a marked decrease in purine synthesis, which would lead to an imbalance between purine and pyrimidine nucleotides (Shimosaka et al., 1984), causing arrest of DNA synthesis (Sheikh et al., 1993) and a lack of energetic nucleotides (ADP, ATP) (Reaves et al., 2013).
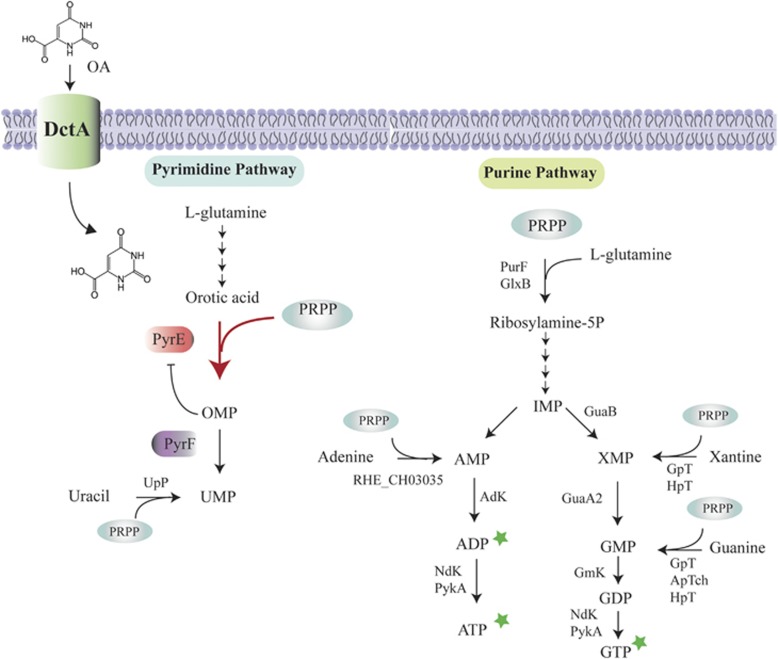
Figure 2.
A proposed mechanism for the inhibitory effect of OA on the growth of R. etli CE3. OA is transported into cells by DctA permease (Reid and Poole, 1998; Yurgel and Kahn, 2005). Inside the cell, OA causes an increase in the rate of de novo pyrimidine synthesis at the orotate phosphoribosyl transferase (PyrE) step (thick arrow). Because this step consumes PRPP (5′-phosphoribosyl-1′-pyrophosphate), we propose that the intracellular level of this metabolite was diminished by the exogenously supplied OA and caused retardation of the de novo purine synthesis. This notion is supported by the reversal of the inhibitory effect of OA by the simultaneous addition of purine derivatives (stars). We found that mutants lacking orotidine-5'-phosphate decarboxylase activity (PyrE) did not consume PRPP and were not affected by OA (Supplementary Figure S5). Furthermore, mutants lacking orotidine5′-phosphate decarboxylase activity (PyrF) were OA-resistant (Supplementary Figure S5). Our hypothesis is that pyrF mutants accumulate orotidine5′-monophosphate (OMP) and that this metabolite inhibits the activity of PyrE and therefore reduces the consumption of PRPP. The image shows the simplified purine and pyrimidine pathways and indicates the PRPP-requiring reactions. Only the intermediates relevant to this study are shown.
Test if low purine ribonucleotide levels cause growth arrest in cells exposed to OA, we supplemented cultures with purines and analyzed the growth after treatment with OA (Supplementary Figure S4B). First, the addition of adenine and guanine did not reverse the growth inhibition caused by OA. These results were consistent with our proposal because purine nucleotide synthesis starting from adenine and guanine requires PRPP (Figure 2). Supplementation with guanine nucleotides or adenine nucleotides partially restored the growth of the bacteria. In contrast, simultaneous supplementation with GTP and ATP completely blocks the effects of OA (Supplementary Figure S4B). These results suggest that the level of intracellular PRPP in the cells exposed to OA was not enough to support de novo purine synthesis in R. etli CE3.
The notion that PRPP pools were depleted by OPTase activity was also supported by isolating OAR variants lacking OPTase activity (pyrE−) (Figure 3). Furthermore, mutants (pyrF−) lacking orotidine-5′-phosphate decarboxylase were resistant to OA (Supplementary Figure S5A). In addition, pyrE and pyrF mutants excrete high concentrations of OA and do not display growth defects (Supplementary Figure S6). We suggest that pyrF mutants accumulate OMP, which inhibits the OPTase activity; consequently, PRPP and OA are not consumed by this step (Figure 2).
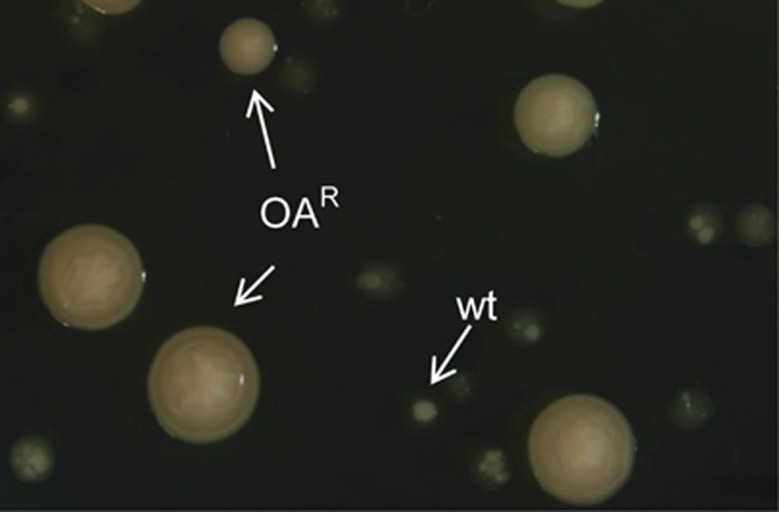
Figure 3.
The characteristic phenotypes of the ancestral and the derived variants of R. etli CE3 on PYD100 agar (96 h of growth). OAR cells develop large colonies and wild-type (wt) cells and translucent small colonies. The inset shows a wt colony with opaque sectors that arise after 3 days of incubation. These sectors are OAR variants that evolved in the colony.
Proteomic analysis revealed that OA affects the expression of the two main glutamine synthetases (GSI and GSII) of R. etli. The GSII (encoded by glnII) was downregulated, and GSI was predominantly non-adenylylated (Supplementary Figure S7). We demonstrated that OA decreased GSI adenylylation, thereby, increasing its specific activity. We propose that ATP depletion and an increased UTP may be the cause of the deadenylylation of GSI induced by OA because ATP is required for the adenylation reaction and UTP stimulates GSI deadenylylation via AT/PII-UMP (Jiang et al., 1998). These results show that increasing GSI activity and downregulating glnII transcription, OA may alter ammonium assimilation in R. etli.
The niche construction and rapid evolution interact to alter the nature of the interaction
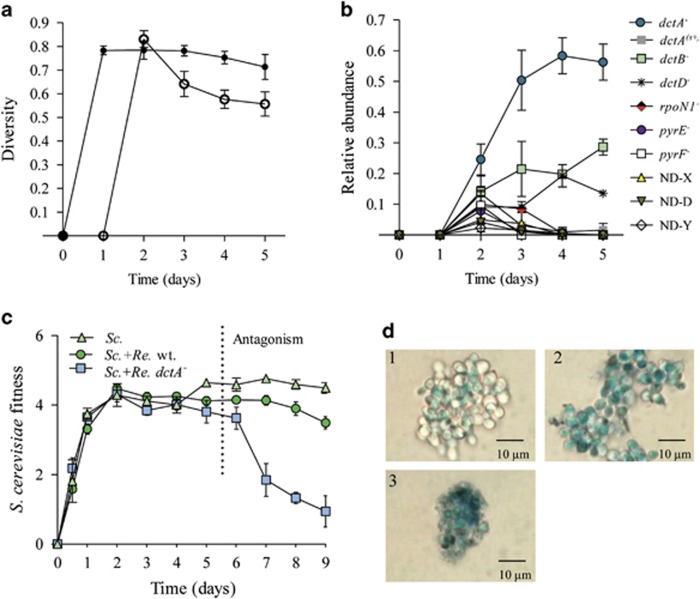
Eco-evolutionary feedbacks require that populations alter their environment (niche construction) and that those changes in the environment feed back to influence the subsequent evolution of the population (Post and Palkovacs, 2009). We show that during ammensalism bacterial growth was affected by the environmental changes generated by yeast (niche construction) (Figures 1e–g). Interestingly, we observed that the growth of the bacteria resumed after 3 days of coculture (Figure 1b), and yeast fitness then gradually declined (Figure 1c). To determine the cause of this change in the community, bacterial cells from 1- to 9-day-old cultures were grown on agar plates with OA (100 μg ml−1). We determined that after 24 h in the cocultures, OAR bacterial variants arose (Figures 3 and 4a). In cocultures, the OAR population increased in frequency and displaced the ancestral type (Figure 4b).
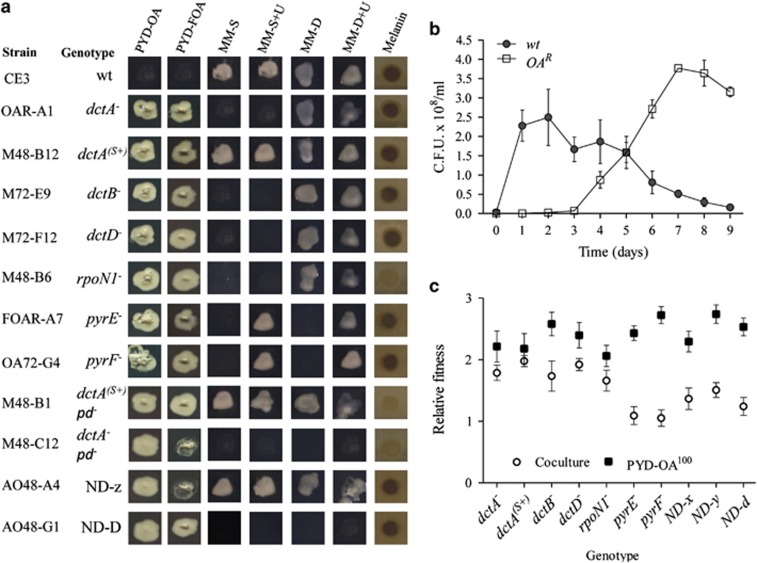
Figure 4.
Adaptive radiation of R. etli CE3 in ammensal–enemy interaction. (a) The traits and genotypes of the representative OAR variants that evolved in coculture. Each column shows the growth on different culture media and melanin production. Culture media are as follows: PYD-OA (complete medium with 100 μg ml−1 OA), PYD-FOA (complete medium with 50 μg ml−1 FOA), MM-S (succinate minimal medium), MM-S+U (succinate minimal medium with uridine), MM-D (dextrose minimal medium) and MM-D+U (dextrose minimal medium with uridine). The genotypes were determined by Sanger sequencing (Table 1). The dctA(s+) mutants retained some capacity to transport succinate but not OA and FOA. M48-B1 and M48-C12 strains lacked the symbiotic plasmid (pd). This plasmid contains the genes necessary for melanin synthesis; therefore, these strains do not produce the pigment. ND-z and ND-D indicate that the genetic change was not identified. (b) The rapid fixation of OAR variants in coculture. The population dynamics of wt and OAR cell populations. (c) The competitive fitness of OAR variants. The fitness was determined relative to ancestral R. etli CE3 (initial ratio 1:1) in the presence of S. cerevisiae Σ1278h (open symbols) and in monoculture with 100 μg ml−1 OA (filled symbols). Competitive fitness was determined in monoculture and coculture at 24 and 48 h, respectively. All panels show the mean±s.e.m. (n=3).
The phenotype of the variants was heritable and was more competitive than the ancestral type in the presence of yeast and when cultured with OA (Figure 4c). To investigate the genetic changes underlying this adaptive evolution, the mutations responsible for the OA resistance of 28 isolates were determined (Table 1). Spontaneous mutations that led to loss-of-function in components of the dicarboxylate uptake system (dctA, dctB, dctD), sigma factor (rpoN1) and pyrimidine auxotrophs (pyrE and pyrF) were identified (Figure 4a, Table 1). In rhizobia, C4-dicarboxylates trigger the activation of the DctB/DctD two-component system, which enables the binding of RNA polymerase by the RpoN1 sigma factor and transcription of the dctA gene (Figure 5) (Reid and Poole, 1998). We demonstrated that inactivation individually of each of these genes prevents uptake of OA and FOA (5-fluoro-orotic acid, a toxic analog of OA) (Reid and Poole, 1998; Yurgel and Kahn, 2005). Thus, these mutated bacteria can grow even when exposed to high concentrations of these metabolites (Figure 4, Supplementary Figure S5A, Supplementary Table S9). These results show that the niche-constructing activities of yeast change the nature of the interaction and were sufficient to produce selection and subsequent evolution of the bacteria.
Table 1. Mutations in evolved OAR isolates from the fungal–bacterial community.
| Isolate | Gene |
Mutation |
|
|---|---|---|---|
| Change in nucleotide sequencea | Change in amino-acid sequence | ||
| OAR-A1 | DctA | Δ375–527 | In-frame deletion of 50 aa (Δ126–176) |
| M48-B12 | 1145 T>G | 382 M>R | |
| M48-A8 | C insertion at 92 | Truncation; retains first 31 native aa+10 nonsense aa added | |
| M48-B8 | T insertion at 88 | Truncation; retains first 29 native aa+12 nonsense aa added | |
| M72-F8 | G insertion at 96 | Truncation; retains first 33 native aa+8 nonsense aa added | |
| M72-F10 | G insertion at 92 | Truncation; retains first 30 native aa+11 nonsense aa added | |
| M72-G12 | ΔT, 84 | Truncation; retains first 94 native aa+11 nonsense aa added | |
| M72-E9 | DctB | ΔT, 380 | Truncation; retains first 126 native aa+103 nonsense aa added |
| M72-F12 | A insertion at 162 | Truncation; retains first 54 native aa+53 nonsense aa added | |
| M72-G1 | ΔG, 627 | Truncation; retains first 218 native aa+20 nonsense aa added | |
| M72-G6 | 352 G>T | 118V>F | |
| M72-H4 | ΔT, 380 | Truncation; retains first 218 native aa+20 nonsense aa added | |
| M72-H10 | 352 G>T | 118V>F | |
| M72-E3 | dctD | G insertion at 246 | Truncation; retains first 82 native aa+18 nonsense aa added |
| M72-F12 | ΔA, 227 | Truncation; retains first 75 native aa+3 nonsense aa added | |
| M48-B6 | rpoN1 | ΔT, 1409 | Truncation; retains first 469 native aa+9 nonsense aa added |
| M72-E8 | C insertion at 116 | Truncation; retains first 190 native aa+2 nonsense aa added | |
| M72-E10 | ΔA, 1507 | Truncation; retains first 502 native aa+40 nonsense aa added | |
| OA48-B11 | 34 A>T ΔA, 1507 | 12 S>C retains first 502 native aa+40 nonsense aa added | |
| FOAR-A6 | pyrE | G insertion at 130, 368 T>C | Truncation; retains first 43 native aa+64 nonsense aa added |
| FOAR-A7 | Δ74,75 | Truncation; retains first 25 native aa+81 nonsense aa added | |
| FOAR-C8 | Δ74,75 | Truncation; retains first 25 native aa+81 nonsense aa added | |
| FOAR-C11 | Δ74,75 | Truncation; retains first 25 native aa+81 nonsense aa added | |
| FOAR-C12 | Δ74,75 | Truncation; retains first 25 native aa+81 nonsense aa added | |
| FOAR-B6 | A insertion at 124 | Truncation; retains first 41 native aa+66 nonsense aa added | |
| FOAR-A9 | pyrF | 312 C>A 313 T>G 320 T>G | 105 S>A 107 L>R 108 C>G |
| FOAR-D5 | Δ15 | Truncation; retains first 4 native aa+2 nonsense aa added | |
| OA72-G4 | Δ248 | Truncation; retains first 82 native aa+3 nonsense aa added | |
| OA72-H7 | A insertion at 39 | Truncation; retains first 13 native aa+20 nonsense aa added | |
Abbreviations: aa, amino acids; OAR, OA-resistant. aNumbering of sequences is from the first nucleotide in the start codon.
Figure 5.
Transport of OA via the C4-dicarboxylic transport (Dct) system. In Rhizobium, the transport of L-malate, fumarate and succinate occurs via the Dct system (Reid and Poole, 1998; Yurgel and Kahn, 2005). This system consists of three genes: dctA, which codes for the transport protein, and two divergently transcribed genes, dctB and dctD, which activate the transcription of dctA in response to the presence of dicarboxylates (Reid and Poole, 1998). DctB and DctD are well characterized as a two-component sensor-regulator pair. In free-living cultures, mutations in any of the three dct genes result in the loss of the capacity to transport and grow on C4-dicarboxylates. Transcription from dctA is RpoN1 sigma factor-dependent (Meyer et al., 2001), which binds to a site located 93-bp upstream from the dctA start codon in R. leguminosarum and a similar position in R. etli (Ronson et al., 1987). In addition to dicarboxylates, DctA can transport compounds that are not dicarboxylates, such as OA and FOA, a toxic analog of OA (Yurgel and Kahn, 2005).
Interspecies competition for resources affects genotypic composition and reduces the diversity of bacterial populations
To understand the changes in the community generated by these evolutionary processes, we evaluated the genotypic diversity of OAR variants through time. We found that bacterial populations evolving in coculture rapidly diversified into a variety of genotypes (Figure 4), resulting in high levels of sympatric diversity (Figure 6a). However, in the cocultures this diversity declined over time. In contrast, in pure culture with OA as a selective agent, the OAR diversity did not change significantly over time (Figure 6a). Furthermore, we noted that the relative abundances of genotypes were significantly different with and without yeast. In the cocultures, the populations of pyrimidine auxotrophs (pyrE and pyrF mutants) and rpoN1 mutants declined rapidly and were displaced by dct mutants (Figure 6b).
Figure 6.
The ecological consequences of rapid adaptative evolution. (a) The diversity over time for OAR populations evolving with S. cerevisiae Σ1278h ura3 mutant (open symbols) and in monoculture with 100 μg ml−1 OA (filled symbols). Diversity was calculated as the complement of Simpson's index (1−λ). (b) The relative abundance of different OAR genotypes along the symbiosis continuum. (c) Yeast fitness declined after the bacteria evolved. Yeast fitness in monoculture (Sc.), coculture with R. etli (Sc.+Re.) and coculture with R. etli dctA− (Sc.+Re. dctA−). Fitness was measured using a growth rate or Malthusian parameter (m) based on colony counts. (d) The assay of yeast strain viability confirmed that yeast cells lost viability after 5 and 8 days of coculture with R. etli wt and R. etli dctA−, respectively. Cell viability was assayed using methylene blue after 8 days of culturing: stained cells were dead, and live cells remained white. Representative light microscopy images: (1) S. cerevisiae monoculture, (2) S. cerevisiae-R. etli coculture and (3) S. cerevisiae-R. etli dctA− coculture. All panels show the mean±s.e.m. (n=3).
To investigate the causes of the reduction of diversity in the cocultures, competitive fitness of OAR variants was determined relative to a dctA mutant in coculture with yeast and in culture with OA. We found that the pyrE and pyrF mutants were less fit than the dctA mutants in the presence of yeast (competition environment) (Supplementary Figure S8). In contrast, this difference was not observed in medium supplemented with pyrimidines (Supplementary Figure S8). Given that the yeast strain used in this study was a pyrimidine auxotroph, we suggest that in the cocultures yeast consumed pyrimidines from the medium and, therefore, the populations of pyrE and pyrF mutants could not proliferate and compete with other OAR variants. These results suggest that interspecies interactions may affect diversification within populations by opposing diversifying selection that arises from resource competition (Buckling and Rainey, 2002). Furthermore, these experiments illustrate how the niche construction affects the genetic composition of the community and the change of abundance of multiple populations.
Evolutionary diversification of the bacterial populations switched the interaction from ammensalism to antagonism
Previous studies have found that rapid evolution can affect ecosystem function (Harmon et al., 2009). To investigate the possible effects of rapid evolution in the interaction, we monitored changes in yeast fitness throughout the cocultivation with R. etli wild type. We found that the yeast fitness declines (Figure 6c) when the population of OAR variants reaches its maximum density (Figure 4). In contrast, the fitness of yeast did not decrease in pure culture compared with its fitness in coculture (Figure 6c). Furthermore, the yeast fitness was quickly affected in coculture with a dctA mutant (Figure 6c).
We found that alkaline pH and starvation were factors contributing to the loss of yeast viability in cocultures (Supplementary Figure S9). This is consistent with previous reports, which demonstrated that alkalinization of the external environment affects the availability of different nutrients, such as phosphate, copper and iron (Serrano et al., 2004). In addition, it has been demonstrated that iron deficiency increases oxidative stress, which significantly affects yeast lifespan (Almeida et al., 2008).
Thus, evolutionary diversification of the bacterial populations switched the interaction from ammensalism to antagonism where bacteria promote yeast extinction (Figures 1 and 6c). Because rapid evolution allowed R. etli to significantly modify the environment, we suggest that adaptive evolution can give rise to an ecosystem engineer (Jones et al., 1994).
Discussion
Many studies have documented that ecological change affects evolution and that evolutionary dynamics can also affect ecology (Yoshida et al., 2003; Meyer et al., 2006; Post and Palkovacs, 2009; Schoener, 2011; Sanchez and Gore, 2013). Despite the potential for a directional influence of evolution on ecology, we still do not know the importance of the evolution-to-ecology pathway in the persistence of interactions and the stability of communities (Thompson, 1999). Studies have documented rapid evolutionary change that affects the interspecific interactions and genotypic structure within natural communities (Thompson, 1998; Whitham et al., 2006).
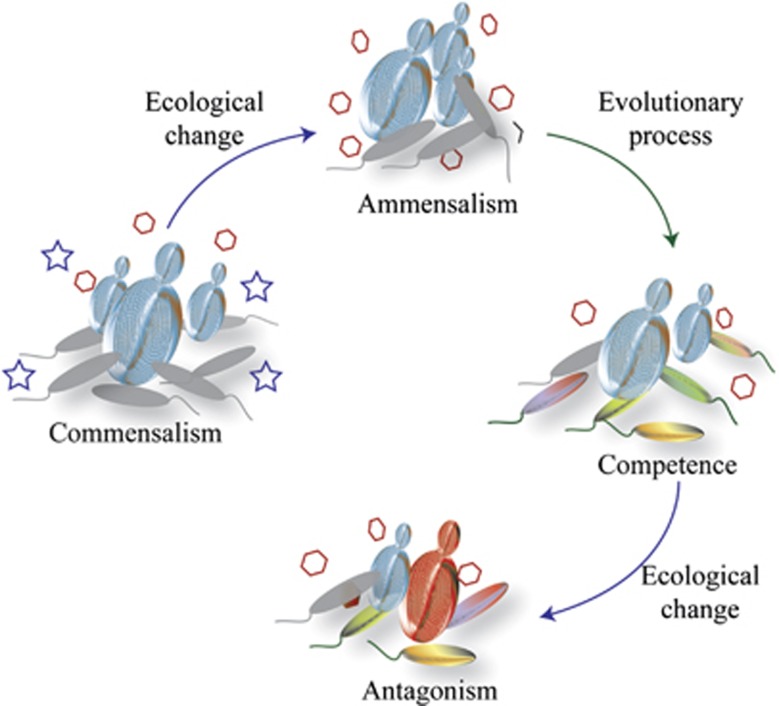
Here, we show that in a two-species community of microorganisms that do not have a known history of previous interaction, the feedback between ecological (niche construction) and evolutionary processes (rapid evolution) has a fundamental role in the emergence and dynamics of species interaction. We further developed a highly simplified, conceptual model to delineate how the evolutionary dynamics emerge and alter the nature of fungus–bacterium interaction (see Figure 7). First, at the beginning of the interaction, yeast simultaneously secreted an inhibitor of bacterial growth (OA) and growth promoters (C4-dicarboxylates) that blocked the effect of the inhibitor and allowed the establishment of a commensal relationship. When the growth promoters became depleted (ecological change), the interaction shifted from commensalism to ammensalism.
Figure 7.
An abstract model delineates how ecological change and evolutionary processes drive the fungal–bacterial interaction. In our system, at the beginning of the interaction, yeast simultaneously secreted an inhibitor of bacterial growth (OA, hexagons) and growth promoters (C4-dicarboxylates, stars) that blocked the effect of the inhibitor and it allowed the establishment of a commensal relationship. When the growth promoters became depleted (ecological change), the interaction shifted from commensalism to ammensalism. During ammensalism, the bacterial population radiated into more than five phenotypes, with multiple variations in nutrient transport (dct system mutations), global regulation (rpoN1 mutations) and metabolic strategies (pyrE and pyrF mutations). The interaction between species affected the diversity and determined the phenotypic composition of the bacteria population. Adaptive evolution allowed the bacteria growth, which modified the environment and created competition for nutrients with the yeast. The ecological consequence of evolutionary diversification of the bacteria is a new change in the community: the interaction switch from ammensalism to antagonism.
The niche construction theory explicitly recognizes the ability of organisms to shape the biotic and abiotic attributes of their environments and the potential for those changes to influence subsequent adaptive evolution (Laland et al., 1999; Post and Palkovacs, 2009; Schoener, 2011). We demonstrated that the strong effect of yeast on the environment caused the subsequent evolution of the bacterial population, therefore, yeast had the potential for niche construction. We were able to establish the genetic and physiological basis of adaptations to ammensalism and thus provide an explicit statement of causality between ecological change and evolutionary dynamics.
A population that is exposed to a stress may adapt through natural selection and thereby avoid extinction (Bell and Collins, 2008). This process of ‘evolutionary rescue' has been described for single populations in the laboratory (Bell and Gonzalez, 2011) but has not been studied in species interactions. Our study demonstrates for the first time, to our knowledge that evolutionary rescue, an eco-evolutionary outcome of our system, has the potential to change the course of a biological interaction.
Harmon et al. (2009), showed that adaptive radiation has considerable effects on the composition and abundance of species at lower trophic levels. These consequences are then reflected in changes in ecosystem features that in turn are likely to affect the course of adaptive radiation. Here, we observed that adaptive evolution allowed the bacteria to grow and modify the environment, competing for nutrients and alkalinizing the culture medium. These changes caused the loss of yeast viability (Figures 3c and d). Thus, evolutionary diversification of the bacterial populations switched the interaction from ammensalism to antagonism.
Our results show that the feedback in the community emerges from niche construction and from the ability of the bacteria to evolve in response to selection caused by changes in the environment. Thus, eco-evolutionary dynamics have the potential to transform the structure and functioning of the communities.
Despite the abundance and diversity of fungal–bacterial interactions in nature (Azam and Malfatti, 2007) and the human body (Wargo and Hogan, 2006), very little is known about the contribution of the ecological and evolutionary process underlying these interactions and their importance to human health (Robinson et al., 2010). Our results show that eco-evolutionary approaches will provide a deeper understanding of host–microbial interactions, including disease dynamics, and of the structuring of ecological communities in general.
Acknowledgments
We thank Prof Jaime Mora for his support and comments. We also thank Victoria Labastida (CIQ-UAEM) and Mariano Martínez (Instituto de Química-UNAM) for their support in solving the structure of orotic acid and Miguel Elizalde for technical assistance. Yeast knock-out strains were provide by Gabriel del Rio Guerra, and CE3 rpoN mutant strains by Ma de Lourdes Girard. We are grateful to the Biomedical Sciences PhD Program of the Universidad Nacional Autónoma de México. AAD was a recipient of a PhD Studentship from the CONACyT. Part of this work was supported by DGAPA-PAPIIT grant IN-206113. Finally, the authors appreciate the valuable comments and suggestions from two anonymous referees during the review process.
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on The ISME Journal website (http://www.nature.com/ismej)
Accession number
The microarray data discussed in this work have been deposited in Gene Expression Omnibus (GEO). www.ncbi.nlm.nih.gov/geo/query/acc.cgi?token=nnibzqiwueyacha&acc=GSE46013.
Supplementary Material
References
- Almeida T, Marques M, Mojzita D, Amorim MA, Silva RD, Almeida B, et al. Isc1p plays a key role in hydrogen peroxide resistance and chronological lifespan through modulation of iron levels and apoptosis. Mol Biol Cell. 2008;19:865–876. doi: 10.1091/mbc.E07-06-0604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azam F, Malfatti F. Microbial structuring of marine ecosystems. Nat Rev Microbiol. 2007;5:782–791. doi: 10.1038/nrmicro1747. [DOI] [PubMed] [Google Scholar]
- Bell G, Collins S. Adaptation, extinction and global change. Evol Appl. 2008;1:3–16. doi: 10.1111/j.1752-4571.2007.00011.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell G, Gonzalez A. Adaptation and evolutionary rescue in metapopulations experiencing environmental deterioration. Science. 2011;332:1327–1330. doi: 10.1126/science.1203105. [DOI] [PubMed] [Google Scholar]
- Buckling A, Rainey PB. The role of parasites in sympatric and allopatric host diversification. Nature. 2002;420:496–499. doi: 10.1038/nature01164. [DOI] [PubMed] [Google Scholar]
- D'hooghe I, Michiels J, Vlassak K, Verreth C, Waelkens F, Vanderleyden J. Structural and functional analysis of the fixLJ genes of Rhizobium leguminosarum biovar phaseoli CNPAF512. Mol Gen Genet. 1995;249:117–126. doi: 10.1007/BF00290243. [DOI] [PubMed] [Google Scholar]
- Denison RF. Legume sanctions and the evolution of symbiotic cooperation by Rhizobia. Am Nat. 2000;156:567–576. doi: 10.1086/316994. [DOI] [PubMed] [Google Scholar]
- Dibrov E, Robinson KM, Lemire BD. The COQ5 gene encodes a yeast mitochondrial protein necessary for ubiquinone biosynthesis and the assembly of the respiratory chain. J Biol Chem. 1997;272:9175–9181. doi: 10.1074/jbc.272.14.9175. [DOI] [PubMed] [Google Scholar]
- Duffy MA, Ochs JH, Penczykowski RM, Civitello DJ, Klausmeier CA, Hall SR. Ecological context influences epidemic size and parasite-driven evolution. Science. 2012;335:1636–1638. doi: 10.1126/science.1215429. [DOI] [PubMed] [Google Scholar]
- Encarnación S, Dunn M, Willms K, Mora J, Dunn M, Willms K, et al. Fermentative and aerobic metabolism in Rhizobium etli. J Bacteriol. 1995;177:3058–3066. doi: 10.1128/jb.177.11.3058-3066.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ewald PW. Transmission modes and evolution of the parasitism-mutualism continuum. Ann N Y Acad Sci. 1987;503:295–306. doi: 10.1111/j.1749-6632.1987.tb40616.x. [DOI] [PubMed] [Google Scholar]
- Gómez P, Buckling A. Bacteria-phage antagonistic coevolution in soil. Science. 2011;332:106–109. doi: 10.1126/science.1198767. [DOI] [PubMed] [Google Scholar]
- González V, Bustos P, Ramírez-Romero M, Medrano-Soto A, Salgado H, Hernández-González I, et al. The mosaic structure of the symbiotic plasmid of Rhizobium etli CFN42 and its relation to other symbiotic genome compartments. Genome Biol. 2003;4:R36. doi: 10.1186/gb-2003-4-6-r36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen SK, Rainey PB, Haagensen JAJ, Molin S. Evolution of species interactions in a biofilm community. Nature. 2007;445:533–536. doi: 10.1038/nature05514. [DOI] [PubMed] [Google Scholar]
- Harmon LJ, Matthews B, Des RochesS, Chase JM, Shurin JB, Schluter D. Evolutionary diversification in stickleback affects ecosystem functioning. Nature. 2009;458:1167–1170. doi: 10.1038/nature07974. [DOI] [PubMed] [Google Scholar]
- Hawkins FK, Johnston AW. Transcription of a Rhizobium leguminosarum biovar phaseoli gene needed for melanin synthesis is activated by nifA of Rhizobium and Klebsiella pneumoniae. Mol Microbiol. 1988;2:331–337. doi: 10.1111/j.1365-2958.1988.tb00036.x. [DOI] [PubMed] [Google Scholar]
- Hayashi T. Breaking the barrier between commensalism and pathogenicity. Science. 2006;313:772–773. doi: 10.1126/science.1131752. [DOI] [PubMed] [Google Scholar]
- Hillesland KL, Stahl DA. Rapid evolution of stability and productivity at the origin of a microbial mutualism. Proc Natl Acad Sci USA. 2010;107:2124–2129. doi: 10.1073/pnas.0908456107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang P, Peliska JA, Ninfa AJ. The regulation of Escherichia coli glutamine synthetase revisited: role of 2-ketoglutarate in the regulation of glutamine synthetase adenylylation state. Biochemistry. 1998;37:12802–12810. doi: 10.1021/bi980666u. [DOI] [PubMed] [Google Scholar]
- Jones CG, Lawton JH, Shachak M. Organisms as ecosystem engineers. Oikos. 1994;69:373–386. [Google Scholar]
- Kim HJ, Boedicker JQ, Choi JW, Ismagilov RF. Defined spatial structure stabilizes a synthetic multispecies bacterial community. Proc Natl Acad Sci. USA. 2008;105:18188–18193. doi: 10.1073/pnas.0807935105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laland KN, Odling-Smee FJ, Feldman MW. Evolutionary consequences of niche construction and their implications for ecology. Proc Natl Acad Sci USA. 1999;96:10242–10247. doi: 10.1073/pnas.96.18.10242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrence D, Fiegna F, Behrends V, Bundy JG, Phillimore AB, Bell T, et al. Species interactions alter evolutionary responses to a novel environment. PLoS biol. 2012;10:e1001330. doi: 10.1371/journal.pbio.1001330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenski RE, Rose MR, Simpson SC, Tadler SC. Long-term experimental evolution in Escherichia coli. I. Adaptation and divergence during 2,000 generations. Am Nat. 1991;138:1315–1341. [Google Scholar]
- Meyer JR, Ellner SP, Hairston NG, Jones LE, Yoshida T. Prey evolution on the time scale of predator–prey dynamics revealed by allele-specific quantitative PCR. Proc Natl Acad Sci USA. 2006;103:10690–10695. doi: 10.1073/pnas.0600434103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer MG, Park S, Zeringue L, Staley M, McKinstry M, Kaufman RI, et al. A dimeric two-component receiver domain inhibits the sigma54-dependent ATPase in DctD. FASEB J. 2001;15:1326–1328. doi: 10.1096/fj.00-0516fje. [DOI] [PubMed] [Google Scholar]
- Palmer TM, Stanton ML, Young TP, Goheen JR, Pringle RM, Karban R. Breakdown of an ant-plant mutualism follows the loss of large herbivores from an African savanna. Science. 2008;319:192–195. doi: 10.1126/science.1151579. [DOI] [PubMed] [Google Scholar]
- Post DM, Palkovacs EP. Eco-evolutionary feedbacks in community and ecosystem ecology: interactions between the ecological theatre and the evolutionary play. Phil Trans R Soc B. 2009;364:1629–1640. doi: 10.1098/rstb.2009.0012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reaves ML, Young BD, Hosios AM, Xu Y-F, Rabinowitz JD. Pyrimidine homeostasis is accomplished by directed overflow metabolism. Nature. 2013;500:237–241. doi: 10.1038/nature12445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reid CJ, Poole PS. Roles of DctA and DctB in signal detection by the dicarboxylic acid transport system of Rhizobium leguminosarum. J Bacteriol. 1998;180:2660–2669. doi: 10.1128/jb.180.10.2660-2669.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Relman DA. ‘Til death do us part': coming to terms with symbiotic relationships. Nat Rev Microbiol. 2008;6:721–724. doi: 10.1038/nrmicro1990. [DOI] [PubMed] [Google Scholar]
- Replansky T, Koufopanou V, Greig D, Bell G. Saccharomyces sensu stricto as a model system for evolution and ecology. Trends Ecol Evol. 2008;23:494–501. doi: 10.1016/j.tree.2008.05.005. [DOI] [PubMed] [Google Scholar]
- Robinson CJ, Bohannan BJ, Young VB. From structure to function: the ecology of host-associated microbial communities. Microbiol Mol Biol Rev. 2010;74:453–476. doi: 10.1128/MMBR.00014-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romano JD, Kolter R. Pseudomonas-Saccharomyces interactions: influence of fungal metabolism on bacterial physiology and survival. J Bacteriol. 2005;187:940–948. doi: 10.1128/JB.187.3.940-948.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ronson CW, Nixon BT, Albright LM, Ausubel FM. Rhizobium meliloti ntrA (rpoN) gene is required for diverse metabolic functions. J Bacteriol. 1987;169:2424–2431. doi: 10.1128/jb.169.6.2424-2431.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sachs JL, Skophammer RG, Regus JU. Evolutionary transitions in bacterial symbiosis. Proc Natl Acad Sci USA. 2011;108:10800–10807. doi: 10.1073/pnas.1100304108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salazar E, Díaz-Mejía JJ, Moreno-Hagelsieb G, Martínez-Batallar G, Mora Y, Mora J, et al. Characterization of the NifA-RpoN regulon in Rhizobium etli in free life and in symbiosis with Phaseolus vulgaris. Appl Environ Microbiol. 2010;76:4510–4520. doi: 10.1128/AEM.02007-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J, Russell DW.2001Molecular Cloning3rd edn. Cold Spring Harbor Laboratory Press: New York [Google Scholar]
- Sanchez A, Gore J. Feedback between population and evolutionary dynamics determines the fate of social microbial populations. PLoS Biol. 2013;11:e1001547. doi: 10.1371/journal.pbio.1001547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoener TW. The newest synthesis: understanding the interplay of evolutionary and ecological dynamics. Science. 2011;331:426–429. doi: 10.1126/science.1193954. [DOI] [PubMed] [Google Scholar]
- Serrano R, Bernal D, Simón E, Ariño J. Copper and iron are the limiting factors for growth of the yeast Saccharomyces cerevisiae in an alkaline environment. J Biol Chem. 2004;279:19698–19704. doi: 10.1074/jbc.M313746200. [DOI] [PubMed] [Google Scholar]
- Seyedsayamdost MR, Case RJ, Kolter R, Clardy J. The Jekyll-and-Hyde chemistry of Phaeobacter gallaeciensis. Nat Chem. 2011;3:331–335. doi: 10.1038/nchem.1002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheikh A, Yusuf A, Laconi E, Rao PM, Rajalakshmi S, Sarma DSR. Effect of orotic acid on in vivo DNA synthesis in hepatocytes of normal rat liver and in hepatic foci/nodules. Carcinogenesis. 1993;14:907–912. doi: 10.1093/carcin/14.5.907. [DOI] [PubMed] [Google Scholar]
- Shimosaka M, Fukuda Y, Murata K, Kimura A. Purine-mediated growth inhibition caused by a pyrE mutation in Escherichia coli K-12. J Bacteriol. 1984;160:1101–1104. doi: 10.1128/jb.160.3.1101-1104.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson EH. Measurement of diversity. Nature. 1949;163:688. [Google Scholar]
- Smukalla S, Caldara M, Pochet N, Beauvais A, Guadagnini S, Yan C, et al. FLO1 is a variable green beard gene that drives biofilm-like cooperation in budding yeast. Cell. 2008;135:726–737. doi: 10.1016/j.cell.2008.09.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Somvanshi VS, Sloup RE, Crawford JM, Martin AR, Heidt AJ, Kim K, et al. A single promoter inversion switches Photorhabdus between pathogenic and mutualistic states. Science. 2012;337:88–93. doi: 10.1126/science.1216641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson JN. Rapid evolution as an ecological process. Trends Ecol Evol. 1998;13:329–332. doi: 10.1016/s0169-5347(98)01378-0. [DOI] [PubMed] [Google Scholar]
- Thompson JN. The evolution of species interactions. Science. 1999;284:2116–2118. doi: 10.1126/science.284.5423.2116. [DOI] [PubMed] [Google Scholar]
- Thompson JN. The Coevolutionary Process. University of Chicago Press: Chicago, IL; 1994. [Google Scholar]
- Wargo MJ, Hogan DA. Fungal-bacterial interactions: a mixed bag of mingling microbes. Curr Opin Microbiol. 2006;9:359–364. doi: 10.1016/j.mib.2006.06.001. [DOI] [PubMed] [Google Scholar]
- Whitham TG, Bailey JK, Schweitzer JA, Shuster SM, Bangert RK, LeRoy CJ, et al. A framework for community and ecosystem genetics: from genes to ecosystems. Nat Rev Genet. 2006;7:510–523. doi: 10.1038/nrg1877. [DOI] [PubMed] [Google Scholar]
- Yoshida T, Jones LE, Ellner SP, Fussmann GF, Hairston NG. Rapid evolution drives ecological dynamics in a predator-prey system. Nature. 2003;424:303–306. doi: 10.1038/nature01767. [DOI] [PubMed] [Google Scholar]
- Yurgel SN, Kahn ML. Sinorhizobium meliloti dctA mutants with partial ability to transport dicarboxylic acids. J Bacteriol. 2005;187:1161–1172. doi: 10.1128/JB.187.3.1161-1172.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.