Abstract
Peptidoglycan (PG), an essential structure in the cell walls of the vast majority of bacteria, is critical for division and maintaining cell shape and hydrostatic pressure1. Bacteria comprising the Chlamydiales were thought to be one of the few exceptions. Chlamydia encodes genes for PG biosynthesis2–7 and exhibits susceptibility to "anti-PG" antibiotics8,9, yet attempts to detect PG in any chlamydial species have proven unsuccessful (the ‘chlamydial anomaly’10). We employed a novel approach to metabolically label chlamydial PG using D-amino acid dipeptide probes and click chemistry. Replicating Chlamydia trachomatis was labeled with the probes throughout its biphasic, developmental life cycle, and differential probe incorporation experiments conducted in the presence of ampicillin is consistent with the presence of chlamydial PG modifying enzymes. These findings culminate 50 years of speculation and debate concerning the chlamydial anomaly and are the strongest evidence to date that chlamydial species possess functional PG.
Chlamydia trachomatis is the leading cause of infectious blindness and sexually transmitted bacterial infection worldwide. It is a member of the Chlamydiae, a phylum consisting of obligate, intracellular, bacteria that cause a wide variety of infectious diseases in humans and animals. Their obligate intracellular nature and dimorphic life cycle has made studying Chlamydia a challenge and questions remain about even the basic processes of cell division and cell envelope maintenance in these pathogens. The infectious form of the organism, the elementary body (EB), is small (~0.3 microns) and essentially metabolically inert11. After attachment to and infection of a host cell, the EB undergoes a transition to the metabolically active reticulate body (RB), which replicates via binary fission but is incapable of attaching to or infecting new host cells. Thus, RBs must differentiate back to the EB form to complete the developmental cycle. Infected cells then lyse, releasing infectious EBs that infect new host cells.
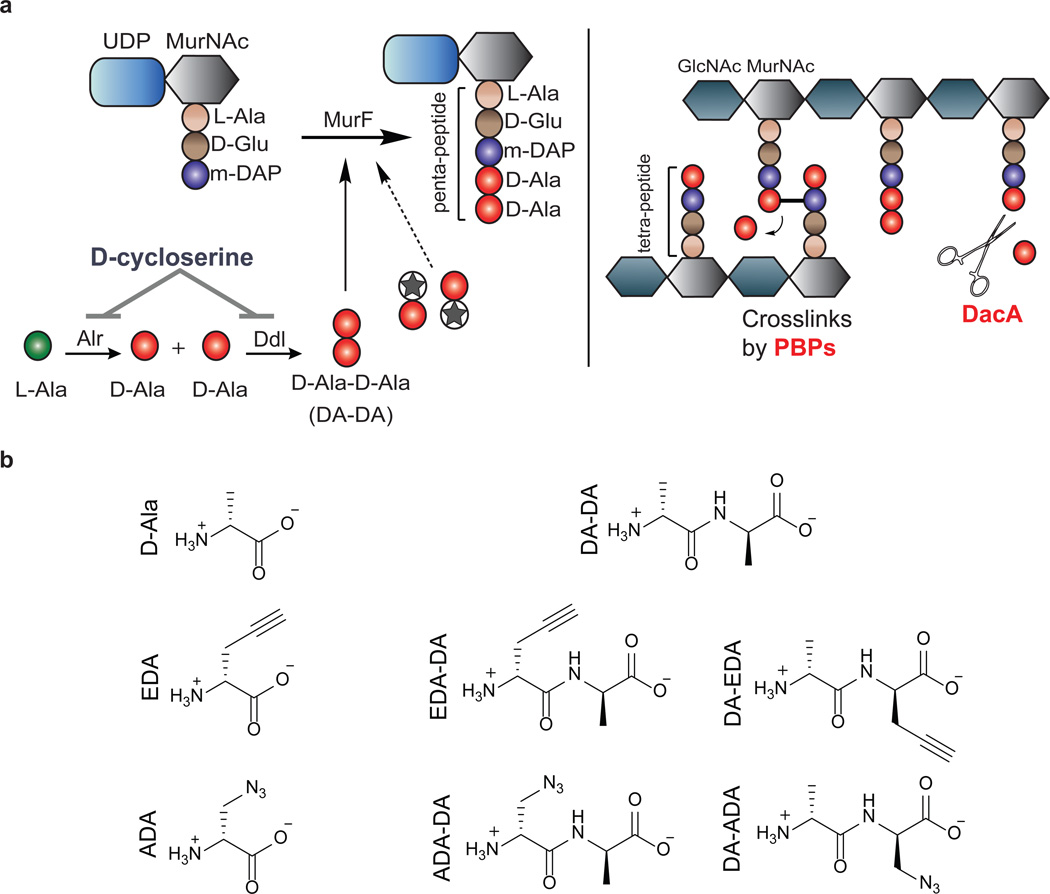
Peptidoglycan (PG) is a sugar amino acid polymer that forms a mesh-like sheet surrounding the plasma membrane of bacterial cells. In the vast majority of free-living bacteria, PG aids in cell division1, maintenance of osmotic pressure, and provides a stable anchor for transmembrane complexes and integral membrane proteins12. Bacteria maintain their cell shape largely due to the presence of this rigid yet modifiable cell wall. A single PG subunit consists of a disaccharide backbone coupled to a pentapeptide chain (Figure 1a). During cell wall synthesis, disaccharide pentapeptide monomers are linked together at their corresponding sugars, creating a sugar polymer with polypeptide stems, which are cross-linked by transpeptidation. The pentapeptide chain is assembled sequentially by a series of ligases that specifically incorporate both L- but also D-amino acids (D-glutamic acid and D-alanine) (Figure 1a). These two D-amino acids are unique to bacteria and they are not utilized by mammalian cells. Thus, the enzymes involved in their synthesis and incorporation into PG are excellent targets for antibiotics such as β-lactams and D-cycloserine.
Figure 1. Novel dipeptide PG labeling strategy.
a, Biosynthesis of the terminal PG stem peptide of Gram negative bacteria. Two D-alanines are first ligated together by D-alanine-D-alanine ligase and the dipeptide is subsequently added to the stem tripeptide by MurF, resulting in a pentapeptide. The labeling strategy relies on the inherent tolerance of the PG machinery to accept DA-DA analogs. Subsequent cross-linking between neighboring peptide stems is carried out by a series of transpeptidases (penicillin binding proteins). Upon transpeptidation, a proximal m-DAP from a neighboring peptide stem attacks the subterminal D-alanine of the PG stem. The terminal D-alanine is thus cleaved from the stem peptide, which results in a tetrapeptide. Another pathway for the loss of terminal D-alanine is D,D-carboxypeptidation catalyzed by enzymes such as DacA. b, Chemical structures of D-Ala, DA-DA, and their derivatives carrying bioorthogonal handles used in this study. Abbreviations: MurNAc, N-acetylmuramic acid; GlcNAc, N-acetylglucosamine; L-Ala, L-Alanine; D-Glu, D-Glutamic Acid; m-DAP, meso-diaminopimelic acid; D-Ala, D-Alanine; Alr, Alanine racemase; Ddl, D-alanine-D-alanine ligase; MurF, UDP-N-acetylmuramoyl-tripeptide--D-alanyl-D-alanine ligase; PBPs, penicillin-binding proteins; DA-DA, D-alanyl-D-Alanine; EDA, Ethynyl-D-alanine; ADA, Azido-D-alanine; EDA-DA, Ethynyl-D-alanyl-D-alanine; ADA-DA, Azido-D-alanyl-D-alanine.
The existence of PG in Chlamydia has long been debated. While genetic analysis and antibiotic susceptibility suggest that chlamydial PG exists8,9,13, all attempts to detect or purify PG in Chlamydia have been unsuccessful10,14–17, resulting in the ‘chlamydial anomaly’10. It has been established that the cytosolic receptor for PG, Nod1, is triggered upon infection by various chlamydial species18. Chlamydial homologs of PG biosynthetic enzymes have been extensively studied2–7 and a growing body of literature supports the functionality of a complete biosynthesis pathway. A functional chlamydial UDP-N-acetylglucosamine enolpyruvyl transferase (MurA) has been described2. The product of the MurA reaction is specifically used for the synthesis of UDP-N-acetylmuramic acid (the sugar unique to the PG disaccharide backbone), suggesting the presence of the sugar component of PG in Chlamydia. During PG biosynthesis in most bacteria, D-alanine-D-alanine (DA-DA) generated by D-alanine-D-alanine ligase (Ddl) is incorporated directly into growing PG peptide chains via the MurF enzyme (Fig. 1a). Characterization of Ddl4 and MurF6 enzymes in Chlamydia coupled with recent advances in the chemical modification of PG through single D-amino acids19,20 present an opportunity to covalently label the PG of actively growing Chlamydia.
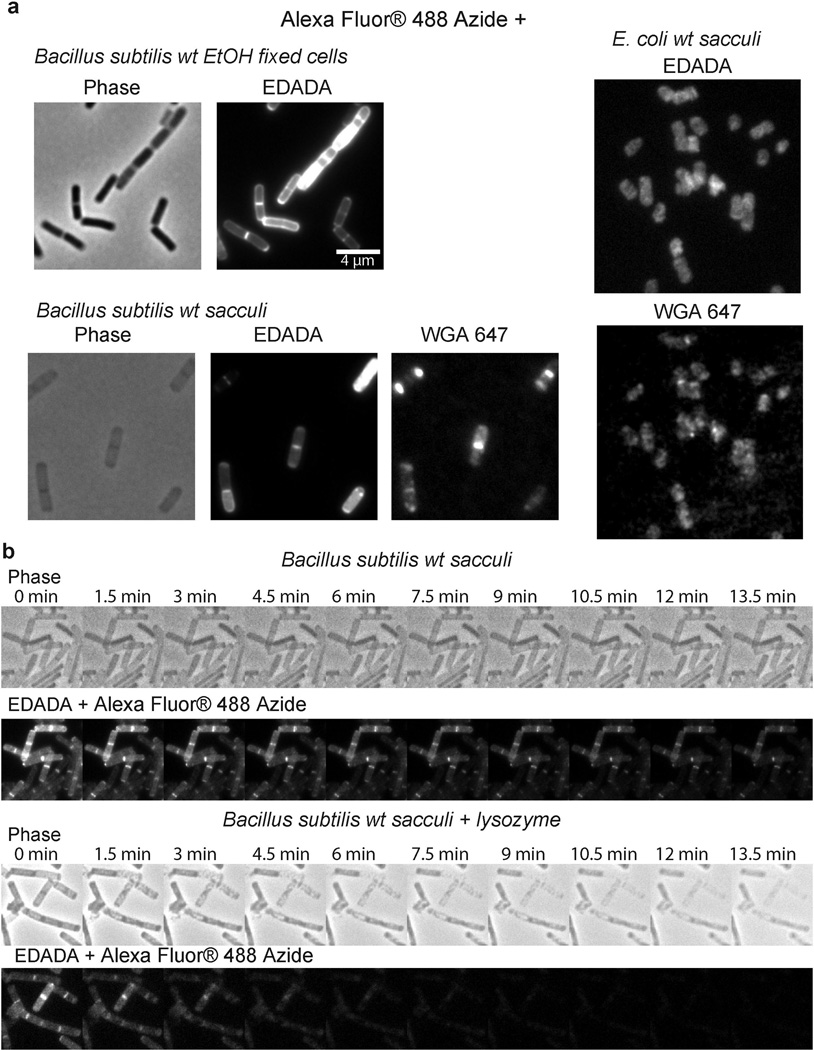
C. trachomatis can take up both D-alanine and DA-DA dipeptide4,8, however, efforts to successfully label Chlamydia employing previously characterized D-amino acid probes19,20 were unsuccessful (Extended Data Figure 1). We reasoned that this result was due to the inability of the chlamydial PG synthesis machinery to incorporate the modified single D-amino acids. Thus, we developed a novel and broadly applicable PG labeling approach that bypassed the bacterial Ddl enzyme and used DA-DA dipeptide analogs modified with alkyne- or azide- functional groups (Fig. 1). Initial studies in Escherichia coli and Bacillus subtilis established that the alkyne- and azide- analogs of DA-DA (EDA-DA, DA-EDA, ADA-DA, and DA-ADA, respectively) are capable of rescuing the growth of bacteria with depleted DA-DA dipeptide pools, while an alkyne analog of the enantiomer L-alanine-L-alanine (LA-LA) is not capable of rescuing growth (Extended Data Table 1). In rich medium, bacterial growth is unaffected by the presence of DA-DA analogs (Extended Data Figure 2). Once incorporated into a macromolecule such as PG, the functional groups of these dipeptides can be selectively captured via a click-chemistry reaction21. Labeling studies utilizing DA-DA analogs in conjunction with clickable, modified Alexa Fluor dyes confirmed D-enantiomer-specific incorporation of the modified dipeptides in diverse bacterial species (Extended Data Figure 2–3). Polarly growing Streptomyces venezuelae was grown in the presence of the previously characterized, fluorescent D-amino acid HADA19 for a few generations, and upon addition of EDA-DA for a brief period, subsequent polar-labeling confirmed that these dipeptides specifically label areas of new PG synthesis (Extended Data Figure 3e). The labeling of E. coli and B. subtilis was covalent and cell wall-specific (Extended Figure 4a) and B. subtilis label could be removed by treatment with the PG-digesting enzyme, lysozyme (Extended Figure 4b), indicating that the probes were incorporated in the PG.
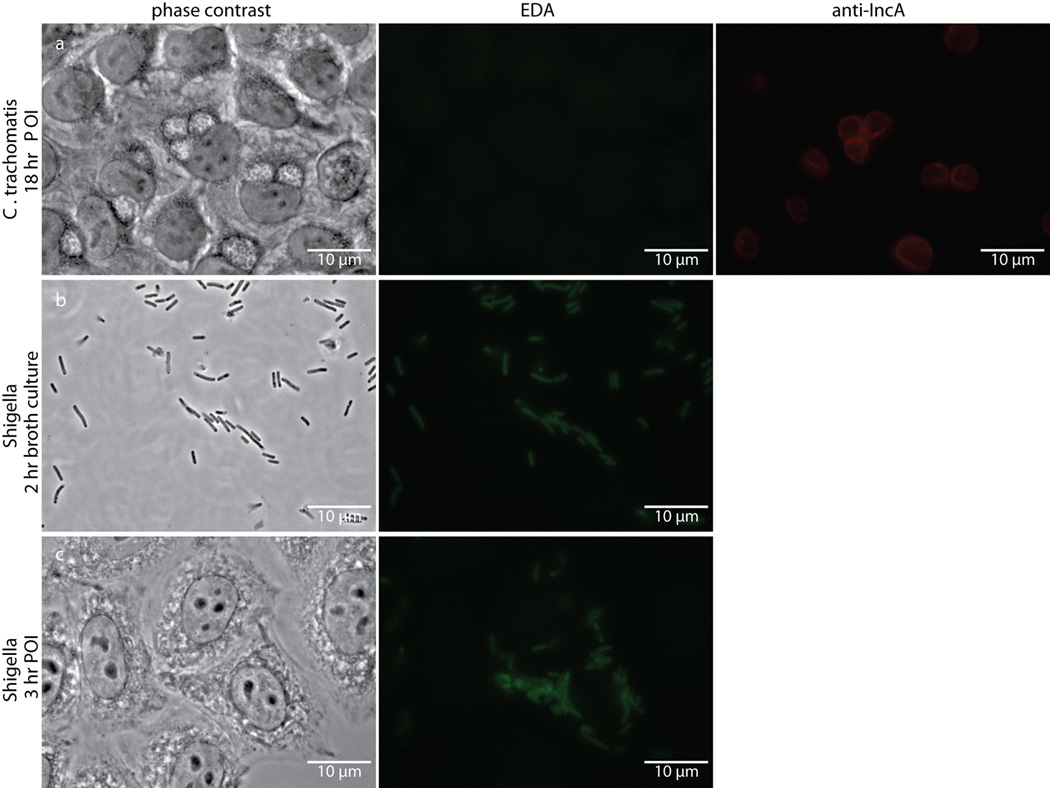
Extended Data Figure 1. Single D-amino acid probe EDA fails to label intracellular Chlamydia despite labeling intracellular Shigella flexneri.
Phase contrast and epifluorescence microscopy of (a) Chlamydia-infected L2 cells 18 hours post infection, (b) Shigella flexneri strain 2457T two hour broth cultures, and (c) Shigella-infected L2 cells three hours post infection. All were grown in the presence of 1 mM EDA. Subsequent tethering of the probe to a modified Alexa Fluor 488 (green) was achieved via click chemistry. Antibody to chlamydial inclusion protein A (IncA, red) was used to visualize chlamydial inclusions. Experiments were conducted in technical duplicates and biological triplicates, with between 4–5 fields examined (with ~3–10 inclusions viewed per field) per technical replicate.
Extended Data Table 1. Exogenous dipeptide analogs rescue growth of cells that have been depleted of DA-DA.
Growth rescue of a D-alanine-D-alanine ligase double mutant (ΔddlA ΔddlB) of E. coli was tested with varying concentrations of natural and modified dipeptides. Similarly, E. coli and B. subtilis cells were rendered auxotrophic for DA-DA by inhibiting their respective D-alanine-D-alanine ligases with D-cycloserine (DCS).. Data represent the average of at least two biological replicates conducted with technical duplicates.
| Condition |
E. coli ΔddlA, ΔddlB |
Condition | E. coli wt | B. subtilis wt |
|---|---|---|---|---|
| MM1 | − | MM | +++++ | +++++ |
| MM+ DA-DA (25 µM)2 | ++ | MM + DCS2 (20 µM for E. coli and 150 µM for B. subtilis) | − | − |
| MM+ DA-DA (200 µM)2 | +++++ | MM + DCS + DA-DA2(50 µM for E. coli and 50 µM for B.susbtilis) | ++ | ++++ |
| MM + DA-EDA (100 µM)2 | +++++ | MM + DCS + DA-EDA2(50 µM for E. coli and 800 µM for B. subtilis) | +++ | +++ |
| MM + DA-ADA (50 µM)2 | ++++ | MM + DCS + DA-ADA2(50 µM for E. coli and 400 µM for B. subtilis) | +++ | ++++ |
| MM + ELA-LA (0.8 mM)2 | − | |||
| MM + EDA–DA (0.8 mM)2 | − | |||
| MM + ADA–DA (0.8 mM)2 | − | |||
| MM+ DA-DA (25 µM)3 | + | |||
| MM + DA-DA (25 µM) + ELA–LA (0.8 mM)3 | + | |||
| MM + DA-DA (25 µM) + EDA–DA (0.8 mM)3 | ++++ | |||
| MM + DA-DA (25 µM) + ADA–DA (0.8 mM)3 | +++ |
MM = minimal medium
Initial inoculum was OD600 = 0.025
Initial inoculum was OD600 = 0.0025
+++++ indicates the highest culture density achieved within a set and − indicates no bacterial growth, with intermediate values representing the relative fractions of the highest culture density within a set.
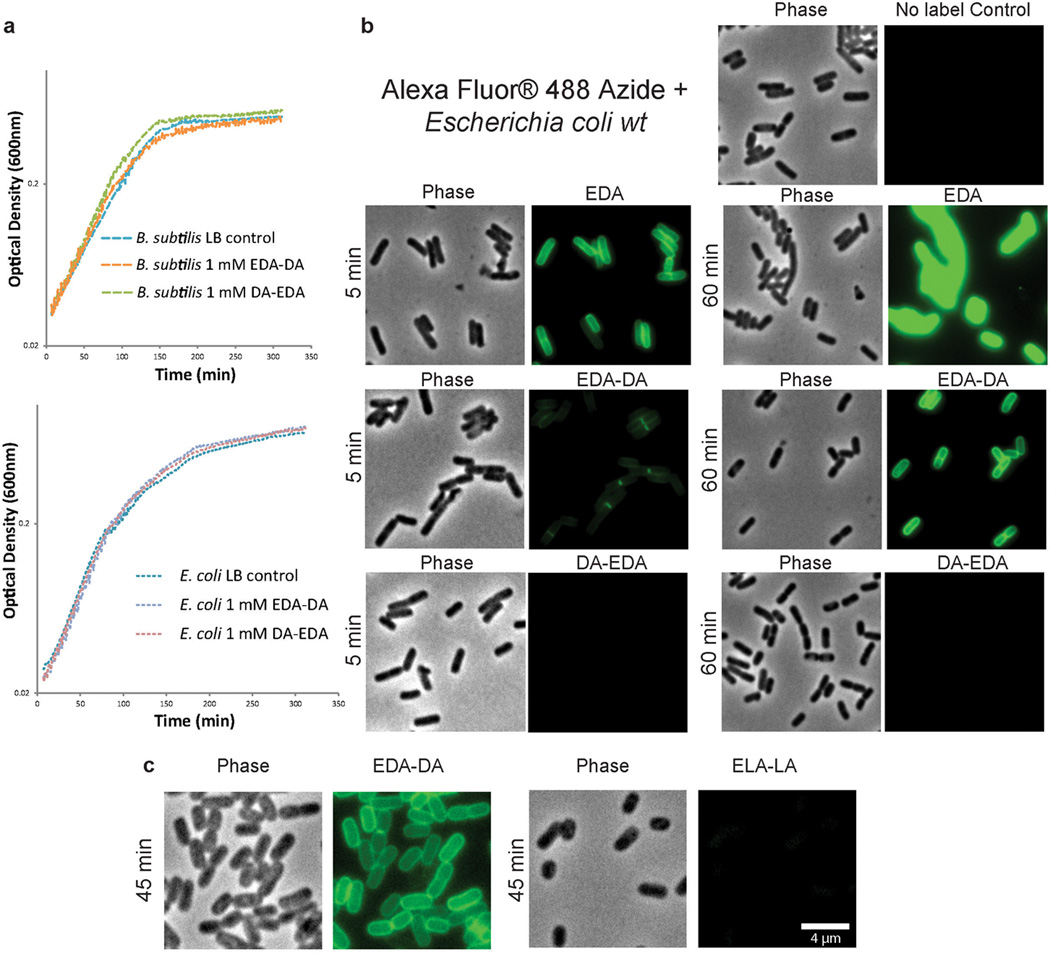
Extended Data Figure 2. D-enantiomer dipeptide probes do not affect growth in rich media, but differentially and specifically label PG of E. coli and B. subtilis.
a, Growth of wild-type E. coli and B. subtilis in the presence of experimental concentrations of EDA-DA or DA-EDA. A representative growth curve from two biological replicates, each with three technical replicates, is shown. b, Phase contrast and epifluorescence microscopy of E. coli grown with 0.5 mM alkyne containing EDA-DA, DA-EDA or as a positive control with EDA at five minutes and 60 minutes. These samples together with unlabeled controls were ‘clicked’ to Alexa Fluor 488 azide and imaged. When the alkyne is on the C-terminus (DA-EDA), the labeling is not apparent. Signal from N-terminally tagged dipeptide (EDA-DA) is significantly higher, but still lower than EDA and the patterns of labeling at the earlier time points are different. This is probably due to periplasmic incorporation of D-amino acids (e.g. EDA) by E. coli L,D-transpeptidases, which result in more efficient peripheral labeling in addition to labeling due to lipid II-dependent PG synthesis. Therefore, in bacteria that have active L,D-transpeptidases, the cytoplasmic PG labeling through dipeptide probes provides a better measure of lipid II-dependent PG synthesis than single D-amino acids. The experiment was conducted twice and images are representative of a minimum of five fields viewed per condition/time point per replicate. c, Comparison of the labeling in E. coli grown with 0.5 mM alkyne containing EDA-DA or the L-enantiomer control Ethynyl-L-alanine-L-alanine (ELA-LA) for 45 min and clicked as above shows that the labeling is D-enantiomer specific. Images are representative of a minimum of four fields viewed per replicate and the experiment was conducted twice.
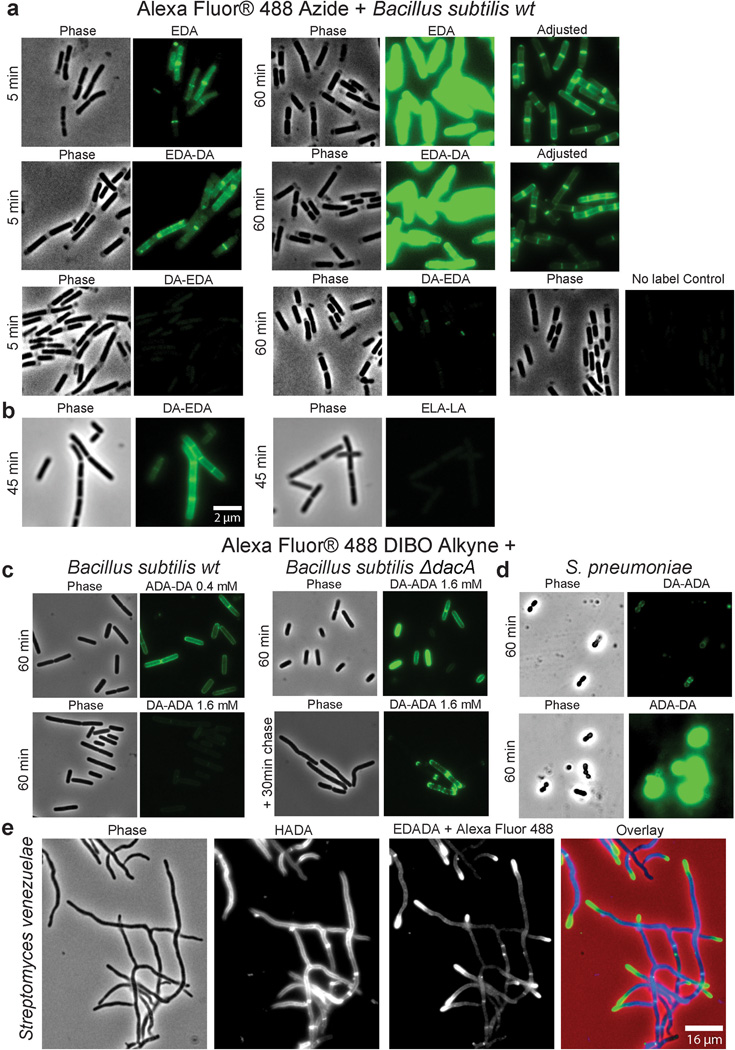
Extended Data Figure 3. Dipeptide probes differentially and specifically label PG of diverse gram-positive bacteria allowing live-cell experiments.
Phase contrast and epifluorescence microscopy of B. subtilis (a–c), Streptococcus pneumoniae (d), and Streptomyces venezuelae (e). a, Five minute and 60 minute aliquots were taken from wild-type B. subtilis grown with 0.5 mM alkyne containing EDA-DA, DA-EDA or as a positive control with EDA. These aliquots together with unlabeled controls were ‘clicked’ to Alexa Fluor 488 azide and imaged. When the alkyne is on the N-terminus (EDA-DA), labeling is comparable to EDA. On the other hand, the labeling with C-terminal tag (DA-EDA) is much fainter. b, B. subtilis grown with 0.5 mM alkyne containing EDA-DA or the L-enantiomer control ELA-LA for 45 min and clicked as above indicates that the labeling is D-enantiomer specific. The partial lysis of the cells visible in phase contrast is caused by 70% EtOH fixation. c–d, When live B. subtilis and S. pneumoniae labeled with azide containing ADA-DA and DA-ADA (0.4 mM and 1.6 mM for c and 0.5 mM for d) were clicked to Alexa Fluor 488 DIBO alkyne using a non-toxic procedure, the signals from N-terminally tagged dipeptide ADA-DA were much higher than the signal from DA-ADA labeled cells. (c) Interestingly, the signal from DA-ADA can be elevated to the ADA-DA level, if the labeling is performed in a ΔdacA, D,D-carboxpeptidase-null mutant of B. subtilis. Since copper-free click-chemistry is not toxic to cells, a pulse-chase experiment was done, which shows the trapping of old PG at the poles of the cells (lower panel). e, When polarly growing S. venezuelae cells are grown with the blue fluorescent D-amino acid HADA (2 h, 0.5 mM)19 for several generations and briefly pulsed with EDA-DA (10 min, 0.5 mM) and clicked, the signal from EDA-DA complements the signal from HADA. This result shows that dipeptide probes label the cell wall at sites of new PG synthesis. Fluorescent images (a–d) were taken and processed in the same manner for comparison. In ‘Adjusted’ images, signal intensities were lowered for comparison of labeling patterns. All experiments were conducted in biological duplicates, and images are representative of 2–5 fields viewed per condition/time point/replicate.
Extended Data Figure 4. EDA-DA labeling is specific to the PG of bacteria.
a, Alexa Fluor 488 Azide ‘clicked’ sacculi from B. subtilis and E. coli cells grown with 0.5 mM EDA-DA for several generations retained the alkyne label. The labeled cells were clicked before sacculi purification in the case of B. subtilis and after purification in the case of E. coli. Experiment was conducted in biological duplicates and images are representative of five fields viewed per replicate. b, The EDA-DA signal retained on the isolated PG can be released by PG-digesting enzymes (~ 10 mg/mL lysozyme + 200 µg/mL mutanolysin). The kinetics of signal disappearance from the lysozyme treated sacculi is much faster than the kinetics of the photo-bleaching during the time-course, indicating that the loss of signal is due to hydrolytic activity of lysozyme. Three experimental replicates were performed.
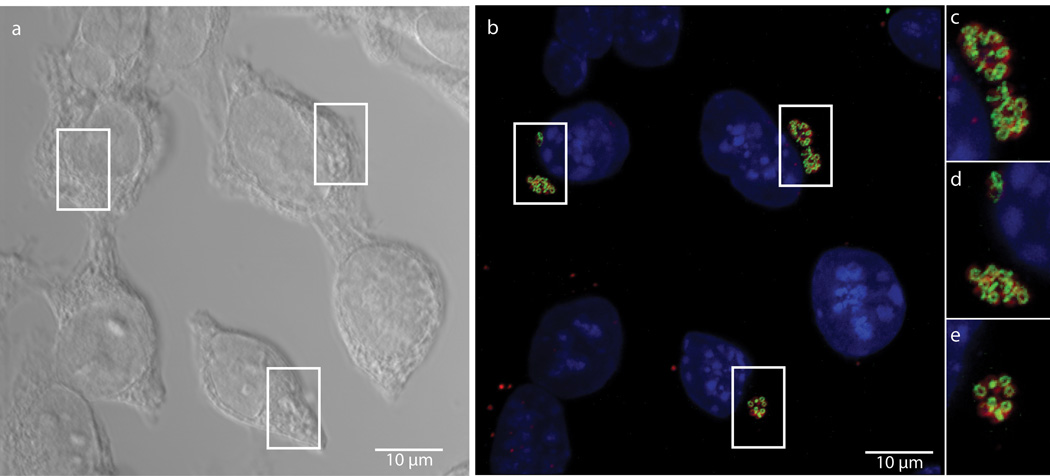
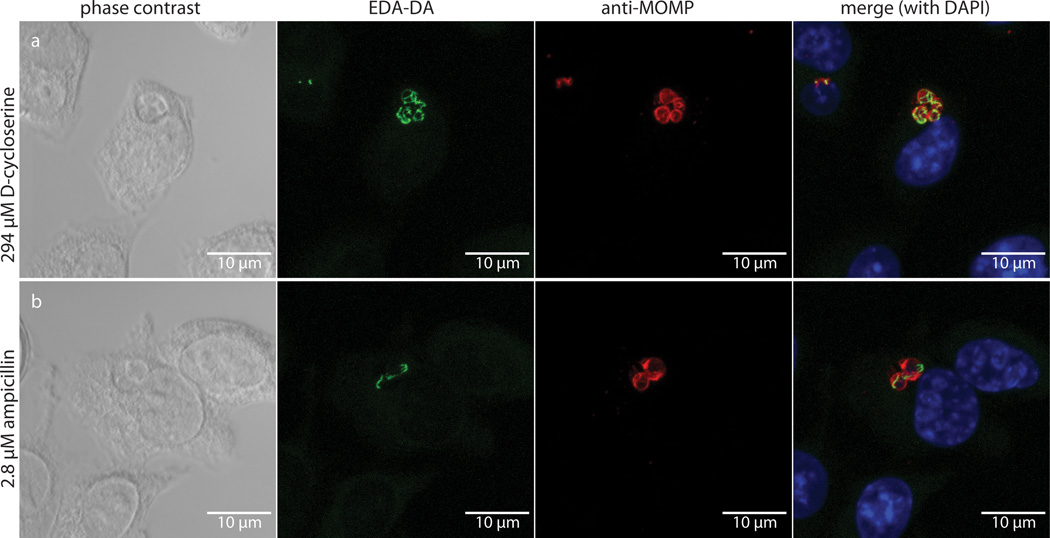
We next attempted to measure dipeptide probe uptake and incorporation in intracellular Chlamydia. L2 mouse fibroblast cells were infected for 18 hours with C. trachomatis serovar L2 strain 434/Bu in the presence of 1 mM EDA-DA. Cells were then fixed, permeabilized, and click chemistry was used to attach an Alexa fluorophore modified with a terminal azide group to the alkyne group present on the EDA-DA probe. The probe localized within chlamydial inclusions with individual bacteria clearly discernible (Fig. 2). When co-labeled with antibody to the chlamydial major outer membrane protein (MOMP), EDA-DA labeling appeared as either a ring or a single line bisecting MOMP-labeled RBs (Extended Data Figure 5). The labeling was arranged in a distinct, ring-like shape, consistent with a cellular division plane and the labeling bore a striking resemblance to images previously obtained for intracellular C. trachomatis stained with antibody generated with Ribi adjuvant22. Labeling was only present in Chlamydia-infected cells and only in the presence of probe (Extended Data Figure 6a–c). This result suggests that the majority of labeled chlamydial PG is localized to the septum of dividing RBs. However, we cannot rule out the possibility that low levels of PG exist elsewhere on the bacterium and are simply below the detection limit of fluorescence microscopy. Similar to our results with B. subtilis, we found that incubation with lysozyme for two hours was sufficient to remove EDA-DA labeling within chlamydial inclusions (Extended Data Figure 6d–e), supporting our conclusion that the dipeptide probes are incorporating into chlamydial PG.
Figure 2. Fluorescent labeling of intracellular C. trachomatis PG.
Differential interference contrast (DIC) (a) and fluorescent (b–e) microscopy of L2 cells infected for 18 hours with C. trachomatis in the presence of the dipeptide probe EDA-DA (1 mM). Subsequent binding of the probe to an azide modified Alexa Fluor 488 (green) was achieved via click chemistry. Antibody to MOMP (red) was used to label chlamydial EBs and RBs. DAPI (blue) was used for nuclear staining. Panels B–E show a merge of all three fluorescent channels. Boxes indicate location of chlamydial inclusions, and magnification of the boxes is provided in panels c–e. Fluorescent images are maximum intensity projections of deconvoluted zstacks. Three dimensional renderings are provided in the supplemental materials (Videos S1 and S2).
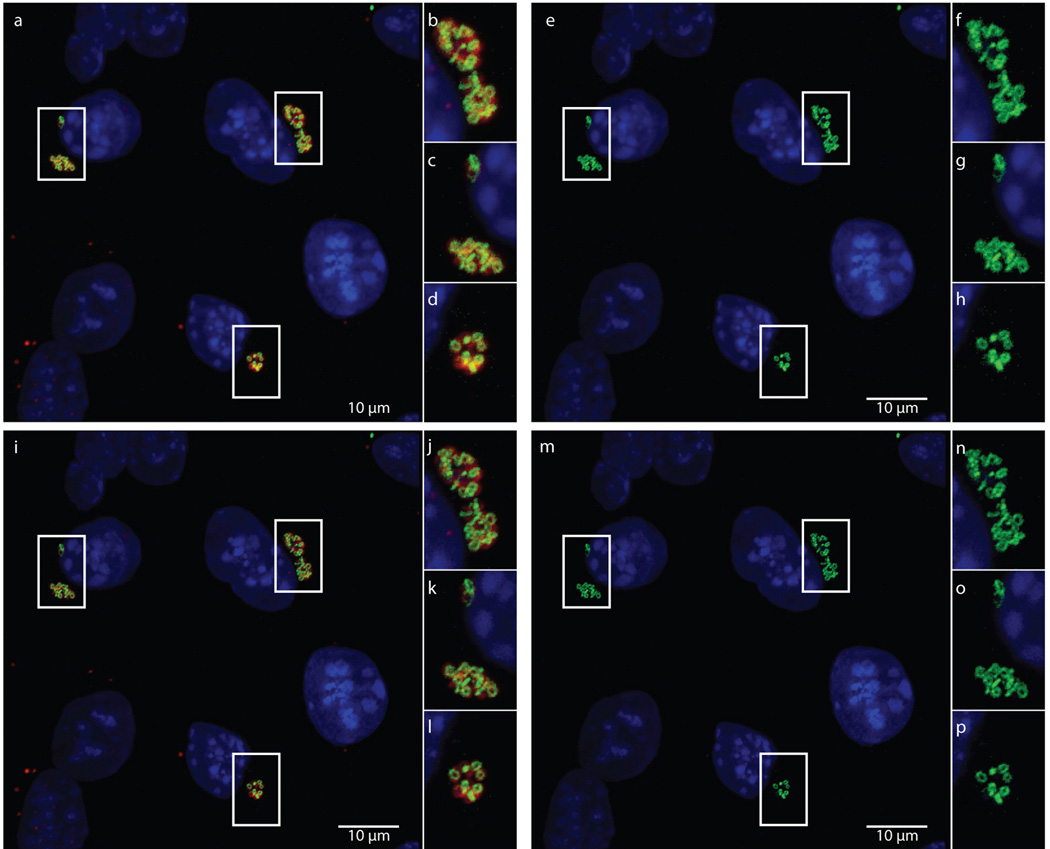
Extended Data Figure 5. Maximum intensity projections of confocal zstacks before and after deconvolution.
Raw data used for generating Figure 2, showing merged (a–d) and green (e–h) channels are compared with the same maximum intensity projections from zstacks that have undergone deconvolution, (i–l) and (m–p), respectively.
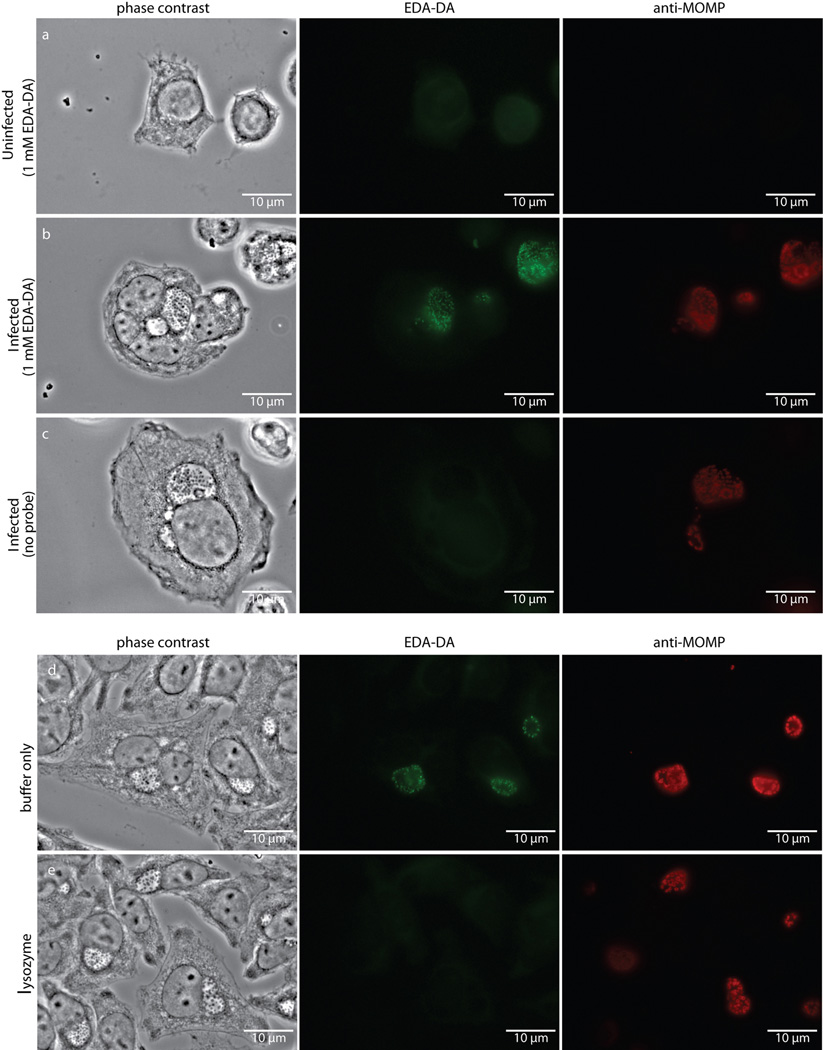
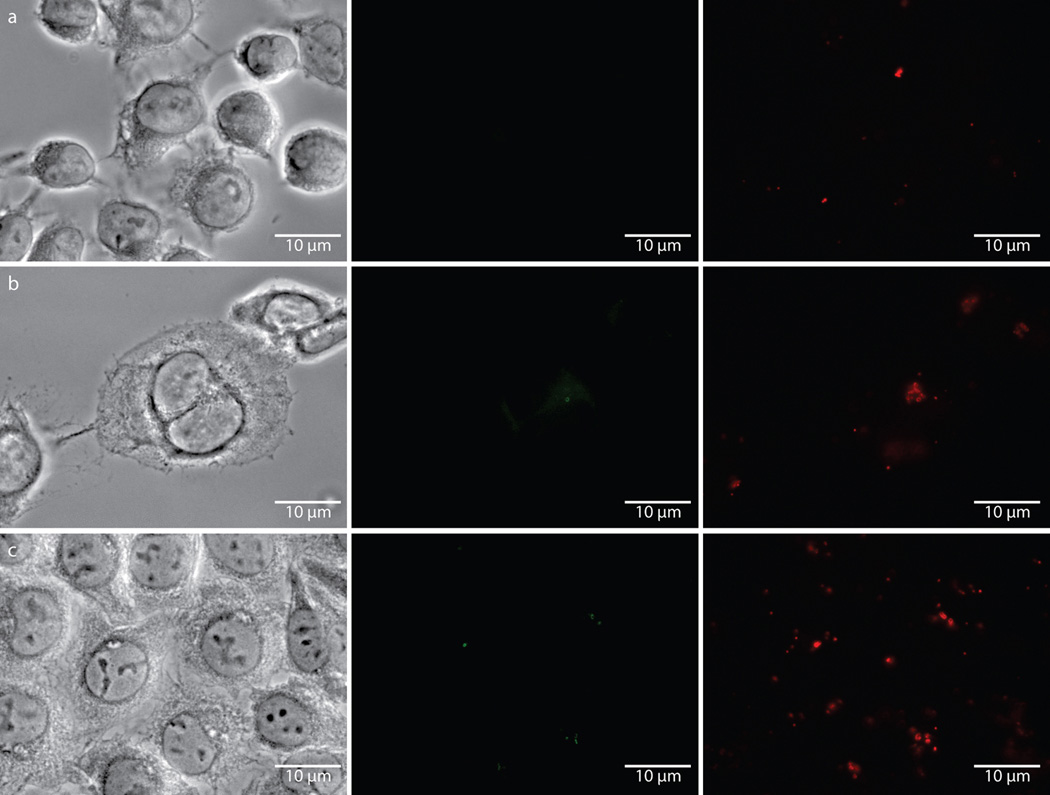
Extended Data Figure 6. Fluorescence is specific to chlamydial infected cells in the presence of the dipeptide probe EDA-DA, and lysozyme treatment is capable of removing the label from fixed bacteria.
Phase contrast and epifluorescence microscopy was conducted on (a) uninfected L2 cells grown in the presence of 1 mM EDA-DA, (b) 18 hour C. trachomatis-infected cells in the presence of 1 mM EDA-DA, and (c) 18 hour C. trachomatis-infected cells grown in the absence of probe. Subsequent binding of the probe to a modified Alexa Fluor 488 (green) was achieved via a click chemistry reaction. For lysozyme treatments, 18 hour C. trachomatis-infected cells (fixed and labeled as described above) were suspended in either (d) buffer (25 mM NaPO4 pH 6.0, 0.5 mM MgCl2) or (e) buffer and lysozyme (200 µg/mL) for two hours. Cells were subsequently washed, blocked, and counter labeled with anti-MOMP, as described previously. Images are representative of between 3–5 fields examined (with ~1–10 inclusions viewed per field) per technical replicate, each condition conducted in technical duplicates, and experiments represent a total of three biological replicates.
To further confirm that the modified probes were being taken up and incorporated into chlamydial PG, we performed plaque assays23 that allow quantification of intracellular bacterial growth and infectivity. D-cycloserine (DCS) is an inhibitor of cell wall biosynthesis that targets bacterial alanine racemase and D-alanine-D-alanine ligase24 and previous studies have shown that Chlamydia growth is inhibited by DCS at millimolar concentrations4,8,25. Growth inhibition is overcome by supplementation with exogenous D-alanine or DA-DA dipeptide4, most likely due to the exogenous single D-amino acids outcompeting DCS for the binding sites of the chlamydial ligase or, in the case of DA-DA, bypassing the need for the ligase altogether. Various D-amino acids, dipeptides, and their corresponding alkyne-modified probes were tested to determine the level of DCS rescue they conferred upon growing C. trachomatis. We found that DA-DA dipeptide and the corresponding modified dipeptides (EDA-DA and DA-EDA) were both capable of rescuing chlamydial plaquing (Table 2), indicating their successful uptake and incorporation by Chlamydia. However, while unmodified D-alanine was capable of overcoming the growth inhibitory effects of DCS, the corresponding chemically modified, single D-alanine probe (EDA) was not. These results were consistent with our inability to detect fluorescent labeling of C. trachomatis via single EDA probes (Extended Data Figure 1).
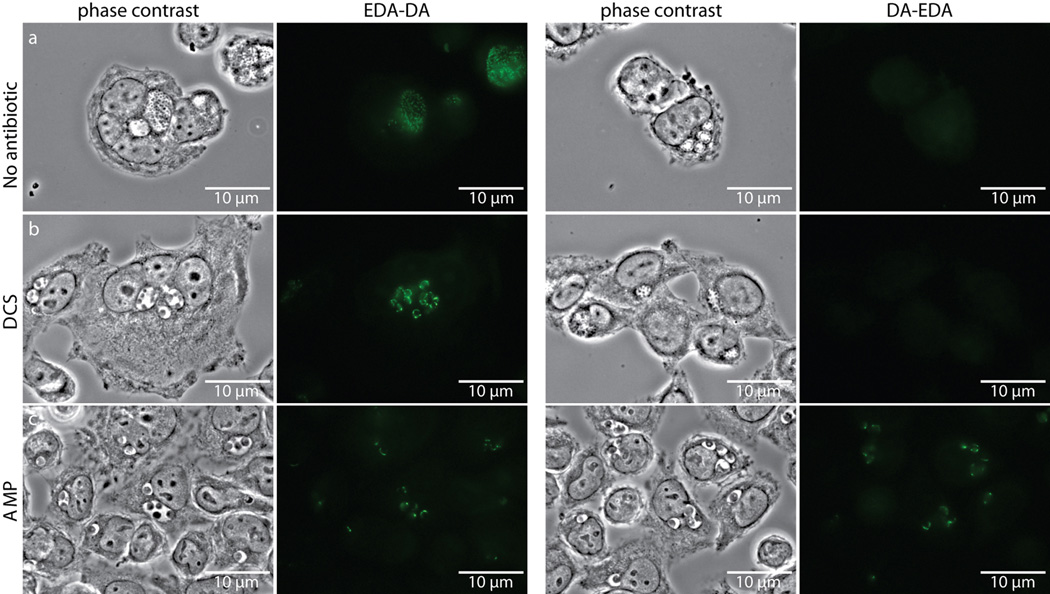
Despite rescue of chlamydial growth by both EDA-DA and DA-EDA in the DCS plaque assay, we initially were not able to label chlamydial PG with DA-EDA (Extended Data Figure 7). Similarly, we were unable to obtain labeling with DA-EDA in E. coli (Extended Data Figure 2). We reasoned that the inability to label Chlamydia with DA-EDA was due to the removal of the terminal, modified EDA amino acid from the PG pentapeptide stem during either transpeptidation or carboxypeptidation (Fig. 1a). In order to test this hypothesis and to further validate that our probes were incorporated into C. trachomatis PG, we conducted EDA-DA and DA-EDA labeling studies in the presence of two antibiotics that block PG biosynthesis: DCS, a competitive inhibitor of both alanine racemase and D-alanine-D-alanine ligase, and ampicillin, an inhibitor of PG transpeptidases/carboxypeptidases. When grown for 18 hours in the presence of either antibiotic, inclusions contained enlarged, aberrant RBs. In the presence of DCS and 1 mM EDA-DA, fluorescent PG was discernible within aberrant RBs (Fig. 3a). This result indicates that EDA-DA was capable of partly substituting for DA-DA after depletion of the bacterium's natural dipeptide pool and confirms the DCS plaquing assay results. EDA-DA labeling intensity appeared unaffected by inhibition of PG transpeptidation/carboxypeptidation with ampicillin (Extended Data Figure 7), suggesting that probe incorporation is not dependent on transpeptidation and does not occur in the periplasm in Chlamydia. When imaged via epifluorescence, labeling of aberrant bodies grown in the presence of ampicillin often appeared punctate, due to the enlarged PG ring structures that no longer exist within a single focal plane (see Extended Data Figure 8). Zstacks taken of the ampicillin-treated aberrant RBs clearly revealed labeled PG sequestered to an equatorial region where the bacterial division plane would normally form (Fig. 3b). The presence of fewer bacteria per inclusion is indicative of a pre-division block, due to the absence of transpeptidation and is consistent with the literature26. Fluorescence labeling of Chlamydia with DA-EDA was only observed when transpeptidation/carboxypeptidation was inhibited with ampicillin (Extended Data Figure 7). DA-ADA labeling of a B. subtilis D,D-carboxypeptidase mutant (ΔdacA) confirmed this finding; labeling was greatly increased compared to the parental, wild type strain and was not significantly turned over as the labeled cells were allowed to grow (Extended Data Figure 3). These observations suggest that PG modifications (through transpeptidation or carboxypeptidation) occur in vivo in Chlamydia, as inhibition of these modifications would preserve the terminal D-alanine in the stem peptide, thus allowing for labeling of PG with DA-EDA.
Extended Data Figure 7. D-cycloserine (DCS) and ampicillin (AMP) influence labeling of C. trachomatis PG by dipeptide probes EDA-DA and DA-EDA.
Phase contrast and epifluorescence microscopy of L2 cells infected with C. trachomatis 18 hours post infection. Cells were grown in the presence of either EDA-DA or DA-EDA (1 mM) and were either untreated (a), or treated with 294 µM DCS (b) or 2.8 µM AMP (c). Subsequent binding of the probe to a modified Alexa Fluor 488 (green) was achieved via click chemistry. The image used for EDA-DA labeling in the absence of antibiotics is the same image from Extended Data Figure 6b and experiments were all conducted in parallel on the same day. Images showing labeling by EDA-DA and DA-EDA in the presence or absence of DCS are representative of the vast majority of over 100 inclusions measured 18 hours post-infection. Labeling by EDA-DA in the presence of ampicillin is representative of 97% (73/75) total aberrant bodies while labeling by DA-EDA in the presence of ampicillin is representative of 95% (73/77) total aberrant bodies, as viewed by epifluorescence microscopy. Experiments were conducted in technical duplicates and represent at least three biological replicates.
Figure 3. Labeling in the presence of PG synthesis inhibitors.
DIC and fluorescent microscopy of infected L2 cells 18 hours post infection in the presence of 1 mM EDA-DA and either (a) D-cycloserine (DCS) or (b) ampicillin (AMP). Labeling was conducted as described in the Figure 2 legend. A merge of all three fluorescent channels is presented in the right hand panels. Fluorescent images are maximum intensity projections of zstacks and three dimensional renderings are provided in the supplemental materials (Videos S3 and S4).
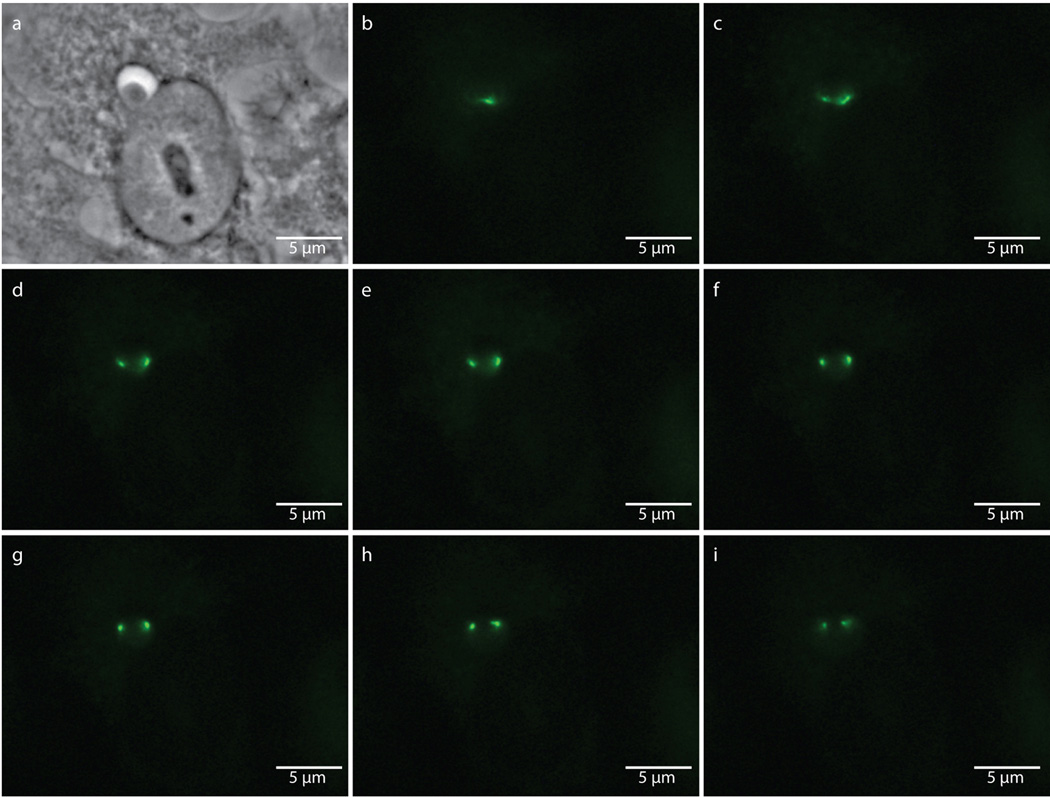
Extended Data Figure 8. Punctate labeling of aberrant bodies due to enlarged bacteria encompassing multiple focal planes.
Phase contrast (a) and epifluorescence microscopy (b–i) of an 18 hour, EDA-DA labeled, ampicillin-induced aberrant body. Images were taken through sequential focal planes in order to show how the ring-like, PG structure is maintained in aberrant bodies and can appear punctate when viewed via an epifluorescence microscope. Images are representative of between 3–5 fields viewed per technical replicate, comprising over 20 independent biological replicates, and each experiment was conducted in technical duplicates.
Multiple investigators have examined the transcriptional profile of PG biosynthesis genes throughout the chlamydial life cycle11,27–30. The current consensus is that PG biosynthesis genes are up-regulated during the transition phase between EB and RB27, indicating the need for PG prior to cell division. These results fit well with the knowledge that PG is closely involved in bacterial growth and cytokinesis1. In order to correlate these findings with actual PG production as measured via the incorporation of dipeptide probes into actively forming chlamydial PG, we conducted a time course study to determine the earliest time at which dipeptide incorporation occurs. We detected EDA-DA labeled C. trachomatis as early as 8 hours post infection (Extended Data Figure 9) and labeled bacteria were always seen in the context of developing chlamydial inclusions. Pre-incubating L2 cells with EDA-DA for 8 hours prior to infection with C. trachomatis did not result in probe incorporation at earlier time points (data not shown). This suggests that probe incorporation and new PG synthesis occur during the early stages of EB to RB transition, consistent with transcriptome data27.
Extended Data Figure 9. EDA-DA labeling of C. trachomatis is apparent as early as eight hours post infection.
L2 cells infected with C. trachomatis (a) 6, (b) 8, and (c) 10 hours post infection grown in the presence of 1 mM EDA-DA. Time points examined covered 4, 6, 8, 10, 12, 18, 24, and 40 hour infected cells. Subsequent binding of the probe to a modified Alexa Fluor 488 (green) was achieved via a click chemistry reaction. Antibody to chlamydial MOMP (red) was used to label chlamydial EBs and RBs. Experiments were conducted in technical duplicates, and the time course was conducted three independent times, with between 3–5 fields viewed per time point per technical replicate.
In conclusion, we have successfully labeled the PG of intracellular Chlamydia trachomatis using modified dipeptide probes. The development and characterization of this versatile, general, and non-toxic method for metabolically labeling PG provides a unique and powerful technique for studying bacterial PG biosynthetic pathways in a myriad of bacterial species. The strength of the click chemistry approach utilized in this work is in its specificity, but future studies will also exploit its versatility. These probes can be selectively captured by molecules other than conventional dyes, such as specified fluorophores for use in super resolution microscopy, modified gold nanoparticles for use in electron microscopy, or agarose resins for use in PG enrichment and chemical characterization. While this technique has opened the door to a wide range of studies in Chlamydia, it is readily adaptable to other model systems, thus providing a powerful investigative tool for use in the examination of bacterial PG biosynthesis, modification, and degradation pathways.
Methods
Reagents
EDA was purchased from BoaoPharma. ‘clickable’ Alexa Fluor 488 and Click-iT® Cell Reaction Buffer Kit were purchased from Invitrogen.
Cell culture conditions
L2 mouse fibroblast cells were obtained from S. Weiss (U. Penn.) cultured in T-175 flasks (BD Falcon) using Dulbecco's Modified Eagle Medium+GlutaMAX™ [Gibco®] (DMEM) supplemented with 10% heat-inactivated Fetal Bovine Serum (FBS, Hyclone) at 37°C with 5% CO2, and checked monthly for mycoplasma. When conducting chlamydial infections, cell medium was supplemented with 1× MEM Non-Essential Amino Acids Solution (Sigma) and 0.2 µg/ml cyclohexamide (Sigma).
Cell infection and bacterial growth conditions
C. trachomatis serovar L2 strain 434/Bu was provided by Harlan Caldwell (Rocky Mountain Laboratories, Hamilton, MT). Chlamydial EBs were harvested from L2 cells 40 hours post infection and stored at −80°C in sucrose phosphate glutamic acid buffer [7.5% W/V sucrose, 17 mM Na2HPO4, 3 mM NaH2PO4, 5 mM L-glutamic acid, pH 7.4] until use. Stocks were titered using an infection forming unit assay (IFU). For infections, treated glass coverslips were placed in 24 well tissue culture plates (Costar) and L2 cells were plated to a confluence of ~60–70%. Cells were washed twice with DMEM, and then infected at an MOI of between 1 and 10 with C. trachomatis. Plates were placed on a rocker for 2 hours to allow for adherence of bacterial EBs to the L2 cells, and then medium was removed and replaced with infection medium (described previously) supplemented with modified D-alanine or dipeptide probes (additional supplementation with D-cycloserine (DCS, 294 µM4) or ampicillin (2.8 µM26,31,32) where indicated). E. coli MG1655 and B. subtilis PY79 (wild type or ΔdacA19) were grown in Luria Broth (LB) [tryptone 10 g, yeast extract, 5 g NaCl 10 g] at 37°C with aeration. S. venezuelae was grown in LB at 30°C with aeration. For E. coli (wild type or ΔddlA, ΔddlB4) and B. subtilis the minimal media used were M9 + 0.2% glucose33 ± 1% LB or Spizizen’s minimal medium [composition per liter, 2 g (NH4)2SO4, 14 g K2HPO4·3H2O, 6 g KH2PO4, 1 g Na-citrate·2H2O, 0.2 g MgSO4·7H2O (plus tryptophan, final concentration of 50 µg/ml, and 0.5 % glucose added after sterilization)], respectively. S. pneumoniae IU1945 was grown at 37°C in Brain-Heart Infusion (BHI) Broth34. When appropriate, the media were supplemented with modified D-alanine and dipeptide molecules as noted in the text.
Growth curves with E. coli and B. subtilis
Exponentially growing E. coli, and B. subtilis were diluted to OD600 0.05 into wells of 24-well polystyrene plates (Falcon) containing 1 mL LB or 1 mL LB + 1 mM EDA-DA or 1 mM DA-EDA. OD600 was recorded every 45 sec for 5 h in a Molecular Devices Spectramax M5 (37°C, with 5s Automix before each measurement).
Labeling E. coli, B. subtilis, and S. pneumoniae PG
Exponentially growing cells were diluted to OD600 ~0.3 in media containing D-alanine or dipeptide analogs. Aliquots were taken after five min and 60 min. For copper catalyzed click-chemistry involving alkyne containing PG probes and Alexa Fluor 488 Azide (Invitrogen), cells were fixed in ice-cold 70% EtOH washed once with 1X PBS [NaCl 8 g/L, KCl 0.2 g/L, Na2HPO4-2H2O 1.78 g/L, KH2PO4 0.27 g/L, pH 7.4] + 0.15 % Polysorbate 20, once with 1X PBS + 1 % Bovine Serum Albumin (BSA) and the click-chemistry was performed using Click-iT® Cell Reaction Buffer Kit (Invitrogen; with 10 µM azide concentration). For live-cell experiments involving azide containing PG probes, cells were washed twice with 1X PBS and copper-free click chemistry was performed using Alexa Fluor® 488 DIBO Alkyne (Invitrogen; 50 µM DIBO alkyne concentration) at room temperature for 30 min. The cells were washed three times in 1X PBS and either directly imaged or resuspended in medium, grown for 30 min and then imaged.
Rescue experiments with E. coli and B. subtilis
For the wild-type strains, minimal DCS concentrations that inhibit growth were determined by titrating DCS in minimal medium. Growth rescue by different concentrations of DA-DA and/or its derivatives was tested using this DCS concentration (see Extended Data Table 1). For initial qualitative analysis, DCS inhibition and rescue experiments were done by inoculating 1–5 µL cells from a −80°C frozen stock. For quantitative analysis, exponentially growing cells (LB for wild-type E. coli and B. subtilis and M9 + 0.2 % glucose + 200 µM DA-DA for E. coli ΔddlA ΔddlB) were centrifuged and washed twice with M9 medium. Cells were then diluted and inoculated into wells of a 96-well plate, which contained two-fold dilutions of different DA-DA and/or its derivatives for a final concentration ranging from 6.25 µM to 800 µM. For these experiments, the inoculum was OD600 0.025, except for the partial rescue experiment with N-terminally modified dipeptides, rescue of which was more evident with a lower inoculum. The plates were incubated at 37°C with aeration and the growth was monitored. Once the cultures reached saturation, OD600 was read with a Molecular Devices Spectramax M5 and recovery was quantified by comparing average ODs of each condition to a growth control (no drug control, or M9 + 0.2 % glucose + 200 µM DA-DA for E. coli ΔddlA ΔddlB). For clarity, the average OD600 of the growth control was normalized to a score of 5 (i.e., +++++).
Purification and characterization of PG sacculi from E. coli and B. subtilis
Sacculi were purified from E. coli as described35 with the following modifications. Exponentially growing cells were diluted to OD600 0.005 in 30 mL LB containing 500 µM EDA-DA and grown to late exponential phase. Cells were rapidly chilled on ice and harvested by centrifugation at 25,000 × g for 15 min at 4°C and resuspended in 4 mL water. The suspension was added dropwise to an equal volume of boiling sodium dodecyl sulfate (SDS, 8% w/v) and boiled with stirring for 45 min. SDS insoluble material was divided into 4 × 2 mL tubes, collected with a table-top centrifuge at 18,620 g for 20 min at RT and was washed three times with 2 mL water. The pelletable material was resuspended in 1 mL 10 mM Tris-HCl pH 7.0 + 10 mM NaCl + 0.32 M immidazole + α-amylase (100 µg/mL) and incubated for 2 h at 37°C. Samples were pelleted and resuspended in 0.01 M Tris-pH 7.8 + 0.5 % SDS + 200 µg /mL pronase (type XXV from Streptomyces griseus) and incubated 2 h at 60°C. Samples were again pelleted and resuspended in 1 mL water and boiled in an equal volume of SDS (2% w/v) with stirring for 30 min. The sacculi preparations were washed a final time and the click-chemistry was performed using Click-iT® Cell Reaction Buffer Kit (Invitrogen) and Alexa Fluor 488–azide (20 µM) with 45 min incubation at RT. Sacculi were washed two more times and further stained with Wheat Germ Agglutinin Alexa Fluor® 647 Conjugate (WGA-647, 10 µg/mL, 15 min at RT) and imaged.
Unless combined with a mechanical breakage method36, the sacculi isolation method described above applied to B. subtilis results in material trapped within the intact cell wall, presumably because of the thicker gram-positive cell wall. This material is resistant to enzymatic digestion37 and causes non-specific labeling with Alexa Fluor 488–azide. Thus, the sacculi purification method for B. subtilis was modified to obtain intact sacculi that are clean of any apparent precipitates (Extended Data Figure 4). Exponentially growing cells were diluted to OD600 0.005 in 10 mL LB containing 500 µM EDA-DA and grown to late exponential phase. Cells were harvested in a table-top centrifuge at room temperature, washed once with 1 × PBS and combined into 3 × 1.7 mL tubes. The cells were resuspended in 3 × 1.5 mL ice-cold 70% EtOH and kept at −20°C for 10 – 12 min, pelleted and washed twice with an equal volume of 1 × PBS. After the last wash, click-chemistry was performed using Click-iT® Cell Reaction Buffer Kit (Invitrogen) and Alexa Fluor 488–azide (20 µM) with 45 min incubation at RT. The cells were washed once more and resuspended in 3 × 1 mL 0.01 M Tris-pH 7.8 + 0.5 % SDS + 1.5 mg/mL pronase (type XXV from Streptomyces griseus) and incubated 2 h at 60°C. The cells were pelleted and combined in 2 mL water and boiled in an equal volume of SDS (8% w/v) with stirring for 45 min. The sacculi were divided into 3 × 1.7 mL tubes and excess SDS was removed by washing three times with 1.5 mL water. Sacculi were further stained with Wheat Germ Agglutinin Alexa Fluor® 647 Conjugate (WGA-647, 10 µg/mL, 15 min at RT) and imaged.
For the lysozyme digestion experiment, a 2 % agarose pad in 25 mM NaPO4 pH 6.0, 0.5 mM MgCl2 buffer impregnated with ~ 10 mg/mL lysozyme + 200 µg/mL mutanolysin was prepared. Quickly, EDA-DA + Alexa Fluor 488-azide labeled B. subtilis sacculi were sandwiched between a 22 mm × 60 mm coverslip and a piece of the pad and imaged every 10 s for 20 min at RT. For the photo-bleach control, the same procedure was followed with the exclusion of the PG-digesting enzymes in the agarose pad.
Labeling Chlamydial PG
At designated time points post infection, infection medium was removed, coverslips were washed three times with PBS, and cells were fixed in methanol at room temperature for five minutes. Cells were again washed in PBS and further permeabilized in 0.5% TritonX for five minutes, and washed again. Cells were then blocked for one hour in 3% BSA prior to the click chemistry reaction being performed. Click-iT® Cell Reaction Buffer Kit (Invitrogen) was used to carry out the click chemistry reaction with Alexa Fluor 488-azide (10 µM). The Click-iT reaction was allowed to proceed for 1 hour, after which slides were washed with 3% BSA. For labeling major chlamydial outer membrane protein (MOMP) or inclusion protein A (IncA), coverslips were first blocked in DMEM supplemented with 10% heat-inactivated FBS for an hour. Coverslips were then incubated with monoclonal anti-MOMP antibody (LifeSpan Biosciences) or anti-IncA antibody (D. Rockey38) diluted 1:500 in DMEM (10% FBS) for one hour, washed with DMEM (10% FBS), incubated with a secondary, chicken anti-goat or anti-rabbit IgG (respectively) conjugated to Alexa Fluor 594 (Invitrogen), diluted 1:2,000 in DMEM (10% FBS). Coverslips were washed once in DMEM (10% FBS), once in PBS, then stained with DAPI (Sigma) diluted 1:80,000 in PBS, for five minutes. Coverslips were washed one final time in PBS and mounted to glass slides with SlowFade® Gold Antifade reagent (Invitrogen) for imaging.
Chlamydial lysozyme treatment assay
L2 cells were infected for 18 hours, fixed, permeabilized, and washed (as previously described), and blocked with 3% BSA for one hour. The click chemistry reaction was conducted (as described previously) and finally cells were suspended in 250 µl of 25 mM NaPO4 pH 6.0, 0.5 mM MgCl2 in the presence or absence of lysozyme (Sigma, 200 µg/mL). Cells were then rocked gently for two hours under tissue culture conditions (37°C 5% CO2). Counter labeling was then conducted as previously described and imaging was conducted via epifluorescence microscopy. The assay was also conducted prior to running the click chemistry reaction on fixed/permeabilized cells, and as the results were identical, these data were not included. Similarly, 18 hour incubation in reaction buffer or lysozyme was also conducted, with results identical to the two hour incubation.
Image acquisition and analysis
Images were acquired via epifluorescence (Olympus BX50 and IX81) or confocal (Zeiss 710) laser scanning microscopy. Image acquisition was performed with DPController (Olympus Corp.) and Zen 2009 (Carl Zeiss) software, respectively. Settings were fixed at the beginning of image acquisition. Brightness and contrast were adjusted slightly in all channels for images obtained via epifluorescence microscopy. Brightness and contrast were slightly adjusted for differential interference contrast for images taken via confocal microscopy. Image analysis was conducted with Image J. Deconvolution was used for generating the fluorescent images in Figure 2. Due to the limits of detection inherent to fluorescence microscopy utilizing click-chemistry-based probes and the statements made in the text concerning the exact localization of the label within individual bacteria (i.e., the label being present only within bisecting rings within normal and aberrant bacteria), we provide maximum intensity projections of unaltered zstacks for comparison with the deconvoluted images in Extended Data Figure 5. Deconvolution was conducted with AxioVision (Carl Zeiss) software utilizing the inverse filter setting. Figure 2 is representative of 20 inclusions viewed by confocal microscopy and over 200 inclusions viewed by epifluorescence microscopy at 18 hours post infection. Figure 3A is representative of 10 inclusions viewed by confocal microscopy and over 200 inclusions viewed by epifluorescence microscopy at 18 hours post-infection. Figure 3B is representative of 100/104 (96%) aberrant bodies induced by ampicillin treatment and viewed by confocal microscopy. All conditions were replicated technically twice and encompass at least three biological replicates of each experiment.
Phase and fluorescence microscopy of E. coli, B. subtilis and S. pneumoniae were performed with a Nikon 90i fluorescence microscope equipped with a Plan Apo 100×/1.40 Oil Ph3 DM objective and a Chroma 83700 triple filter cube. Images were captured using NIS software from Nikon and a Photometrics Cascade 1K cooled charge-coupled device camera, and were processed and analyzed using ImageJ. When a comparison was made, samples were treated the same and the same parameters were applied for collecting and post processing of the microscopy data.
Chlamydial plaque assays
Plaque assays were adapted from a previously described protocol4. Briefly, confluent monolayers of L2 mouse fibroblast cells in 24 well plates were washed twice with pre-warmed DMEM and then infected with C. trachomatis at an MOI of 1. Plates were incubated on a rocker at 37°C with 5% CO2 for two hours after which time, infection medium was removed and cells were overlayed with low-melting point agarose medium (0.25%) containing DMEM (1×), FBS (10%), nonessential amino acids (1×), cycloheximide (200 ng ml−1), and gentamicin (20 µg ml−1). Overlay medium was supplemented as indicated with DCS (30 µg ml−1) and varying concentrations of amino acids and their modified derivatives. At seven days post infection, an additional agarose overlay was added to each well. At 14 days post infection, plaque formation was visualized by staining cells for three hours with 0.5% neutral red.
Supplementary Material
Table 1. DCS Chlamydia trachomatis plaque assay in the presence of natural and modified D-amino acids.
C. trachomatis serovar L2 strain 434/Bu was grown in the plaque assay as previously described4 in the presence of D-cycloserine (DCS) and varying molar equivalent concentrations of D-alanine (D-ala), D-alanine-D-alanine (DA-DA), EDA, DA-EDA, and EDA-DA.
| DCS (µM) | Amino acid, amino acid to DCS ratio |
Plaque formation |
|---|---|---|
| 0 | − | ++++ |
| 294 | No amino acid | − |
| Dipeptides | ||
| DA-DA, 1:1 | ++ | |
| DA-DA, 10:1 | +++ | |
| DA-EDA, 1:1 | − | |
| DA-EDA, 10:1 | +++ | |
| EDA-DA, 1:1 | − | |
| EDA-DA, 10:1 | ++ | |
| Single amino acids | ||
| D-ala, 1:1 | ++++ | |
| D-ala, 1:10 | +++ | |
| EDA, 1:1 | − | |
| EDA, 10:1 | − |
++++ complete infection, bacterial growth, and lysis of the monolayer; +++ numerous large plaques but less than complete lysis of the monolayer; ++ numerous small plaques; + few (~10–20) small plaques; −, no plaque formation (no bacterial growth). Data represent the average of three biological replicates and each experiment was conducted with technical duplicates.
Acknowledgments
This work was supported by NIH grants to A.T.M. (AI044033), and Y.V.B. (GM51986). We would like to thank Dr. Dennis McDaniel and Dr. Meera Murgai for their help with image acquisition and presentation, Patricia Foster and Daniel Kearns for help in early stages of the project, Rebecca Calvo for help with strain construction, and Malcolm Winkler for providing strains and advice.
Footnotes
Supplementary Information is linked to the online version of the paper at www.nature.com/nature.
Author Contributions: G.W.L. conducted all labeling, imaging, intracellular growth-rescue, and probe characterization studies in C. trachomatis. E.K. developed the dipeptide labeling strategy, conducted all labeling, imaging, and probe characterization studies for E. coli, B. subtilis, S. pneumoniae, and S. venezuelae, purification and imaging of labeled bacterial PG, and E. coli and B. subtilis growth rescue experiments. E.H. and A.K. were involved in the synthesis of probes. M.V.N, Y.V.B., and A.T.M. were all involved in study design. G.W.L. and E.K. designed the study, analyzed the data, and wrote the manuscript. G.W.L. and E.K. contributed equally to this work. All authors discussed the results and commented on the manuscript.
We declare that the authors have no competing interests as defined by Nature Publishing Group, or other interests that might be perceived to influence the results and/or discussion reported in this article.
Disclaimer The opinions or assertions contained herein are ours and are not to be construed as official or as reflecting the views of the Department of Defense or the Uniformed Services University.
References
- 1.Egan AJ, Vollmer W. The physiology of bacterial cell division. Annals of the New York Academy of Sciences. 2013;1277:8–28. doi: 10.1111/j.1749-6632.2012.06818.x. [DOI] [PubMed] [Google Scholar]
- 2.McCoy AJ, Sandlin RC, Maurelli AT. In vitro and in vivo functional activity of Chlamydia MurA, a UDP-N-acetylglucosamine enolpyruvyl transferase involved in peptidoglycan synthesis and fosfomycin resistance. Journal of bacteriology. 2003;185:1218–1228. doi: 10.1128/JB.185.4.1218-1228.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hesse L, et al. Functional and biochemical analysis of Chlamydia trachomatis MurC, an enzyme displaying UDP-N-acetylmuramate:amino acid ligase activity. Journal of bacteriology. 2003;185:6507–6512. doi: 10.1128/JB.185.22.6507-6512.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.McCoy AJ, Maurelli AT. Characterization of Chlamydia MurC-Ddl, a fusion protein exhibiting D-alanyl-D-alanine ligase activity involved in peptidoglycan synthesis and D-cycloserine sensitivity. Molecular microbiology. 2005;57:41–52. doi: 10.1111/j.1365-2958.2005.04661.x. [DOI] [PubMed] [Google Scholar]
- 5.Patin D, Bostock J, Blanot D, Mengin-Lecreulx D, Chopra I. Functional and biochemical analysis of the Chlamydia trachomatis ligase MurE. Journal of bacteriology. 2009;191:7430–7435. doi: 10.1128/JB.01029-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Patin D, Bostock J, Chopra I, Mengin-Lecreulx D, Blanot D. Biochemical characterisation of the chlamydial MurF ligase, and possible sequence of the chlamydial peptidoglycan pentapeptide stem. Archives of microbiology. 2012;194:505–512. doi: 10.1007/s00203-011-0784-8. [DOI] [PubMed] [Google Scholar]
- 7.McCoy AJ, et al. L,L-diaminopimelate aminotransferase, a trans-kingdom enzyme shared by Chlamydia and plants for synthesis of diaminopimelate/lysine. Proceedings of the National Academy of Sciences of the United States of America. 2006;103:17909–17914. doi: 10.1073/pnas.0608643103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Moulder JW, Novosel DL, Officer JE. Inhibition of the Growth of Agents of the Psittacosis Group by D-Cycloserine and Its Specific Reversal by D-Alanine. Journal of bacteriology. 1963;85:707–711. doi: 10.1128/jb.85.3.707-711.1963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tamura A, Manire GP. Effect of penicillin on the multiplication of meningopneumonitis organisms (Chlamydia psittaci) Journal of bacteriology. 1968;96:875–880. doi: 10.1128/jb.96.4.875-880.1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Moulder JW. Why is Chlamydia sensitive to penicillin in the absence of peptidoglycan? Infectious agents and disease. 1993;2:87–99. [PubMed] [Google Scholar]
- 11.Omsland A, Sager J, Nair V, Sturdevant DE, Hackstadt T. Developmental stage-specific metabolic and transcriptional activity of Chlamydia trachomatis in an axenic medium. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:19781–19785. doi: 10.1073/pnas.1212831109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mattei PJ, Neves D, Dessen A. Bridging cell wall biosynthesis and bacterial morphogenesis. Current opinion in structural biology. 2010;20:749–755. doi: 10.1016/j.sbi.2010.09.014. [DOI] [PubMed] [Google Scholar]
- 13.Stephens RS, et al. Genome sequence of an obligate intracellular pathogen of humans: Chlamydia trachomatis. Science. 1998;282:754–759. doi: 10.1126/science.282.5389.754. [DOI] [PubMed] [Google Scholar]
- 14.Fox A, et al. Muramic acid is not detectable in Chlamydia psittaci or Chlamydia trachomatis by gas chromatography-mass spectrometry. Infection and immunity. 1990;58:835–837. doi: 10.1128/iai.58.3.835-837.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hatch TP. Disulfide cross-linked envelope proteins: the functional equivalent of peptidoglycan in chlamydiae? Journal of bacteriology. 1996;178:1–5. doi: 10.1128/jb.178.1.1-5.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chopra I, Storey C, Falla TJ, Pearce JH. Antibiotics, peptidoglycan synthesis and genomics: the chlamydial anomaly revisited. Microbiology. 1998;144(Pt 10):2673–2678. doi: 10.1099/00221287-144-10-2673. [DOI] [PubMed] [Google Scholar]
- 17.Barbour AG, Amano K, Hackstadt T, Perry L, Caldwell HD. Chlamydia trachomatis has penicillin-binding proteins but not detectable muramic acid. Journal of bacteriology. 1982;151:420–428. doi: 10.1128/jb.151.1.420-428.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Welter-Stahl L, et al. Stimulation of the cytosolic receptor for peptidoglycan, Nod1, by infection with Chlamydia trachomatis or Chlamydia muridarum. Cellular microbiology. 2006;8:1047–1057. doi: 10.1111/j.1462-5822.2006.00686.x. [DOI] [PubMed] [Google Scholar]
- 19.Kuru E, et al. In Situ probing of newly synthesized peptidoglycan in live bacteria with fluorescent D-amino acids. Angewandte Chemie. 2012;51:12519–12523. doi: 10.1002/anie.201206749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Siegrist MS, et al. (D)-amino acid chemical reporters reveal peptidoglycan dynamics of an intracellular pathogen. ACS chemical biology. 2013;8:500–505. doi: 10.1021/cb3004995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Breinbauer R, Kohn M. Azide-alkyne coupling: a powerful reaction for bioconjugate chemistry. Chembiochem : a European journal of chemical biology. 2003;4:1147–1149. doi: 10.1002/cbic.200300705. [DOI] [PubMed] [Google Scholar]
- 22.Brown WJ, Rockey DD. Identification of an antigen localized to an apparent septum within dividing chlamydiae. Infection and immunity. 2000;68:708–715. doi: 10.1128/iai.68.2.708-715.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Binet R, Maurelli AT. Frequency of spontaneous mutations that confer antibiotic resistance in Chlamydia spp. Antimicrobial agents and chemotherapy. 2005;49:2865–2873. doi: 10.1128/AAC.49.7.2865-2873.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Neuhaus FC, Hammes WP. Inhibition of cell wall biosynthesis by analogues and alanine. Pharmacology & therapeutics. 1981;14:265–319. doi: 10.1016/0163-7258(81)90030-9. [DOI] [PubMed] [Google Scholar]
- 25.Gordon FB, Quan AL. Susceptibility of Chlamydia to antibacterial drugs: test in cell cultures. Antimicrobial agents and chemotherapy. 1972;2:242–244. doi: 10.1128/aac.2.3.242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Skilton RJ, et al. Penicillin induced persistence in Chlamydia trachomatis: high quality time lapse video analysis of the developmental cycle. PloS one. 2009;4:e7723. doi: 10.1371/journal.pone.0007723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Belland RJ, et al. Genomic transcriptional profiling of the developmental cycle of Chlamydia trachomatis. Proceedings of the National Academy of Sciences of the United States of America. 2003;100:8478–8483. doi: 10.1073/pnas.1331135100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Albrecht M, Sharma CM, Reinhardt R, Vogel J, Rudel T. Deep sequencing-based discovery of the Chlamydia trachomatis transcriptome. Nucleic acids research. 2010;38:868–877. doi: 10.1093/nar/gkp1032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Albrecht M, et al. The transcriptional landscape of Chlamydia pneumoniae. Genome biology. 2011;12:R98. doi: 10.1186/gb-2011-12-10-r98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shaw EI, et al. Three temporal classes of gene expression during the Chlamydia trachomatis developmental cycle. Molecular microbiology. 2000;37:913–925. doi: 10.1046/j.1365-2958.2000.02057.x. [DOI] [PubMed] [Google Scholar]
References
- 31.Lambden PR, Pickett MA, Clarke IN. The effect of penicillin on Chlamydia trachomatis DNA replication. Microbiology. 2006;152:2573–2578. doi: 10.1099/mic.0.29032-0. [DOI] [PubMed] [Google Scholar]
- 32.Xu S, Battaglia L, Bao X, Fan H. Chloramphenicol acetyltransferase as a selection marker for chlamydial transformation. BMC research notes. 2013;6:377. doi: 10.1186/1756-0500-6-377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sambrook JFaR, D W. Molecular cloning : a laboratory manual. 3rd edn. Cold Spring Harbor Laboratory Press; 2001. [Google Scholar]
- 34.Sham LT, Barendt SM, Kopecky KE, Winkler ME. Essential PcsB putative peptidoglycan hydrolase interacts with the essential FtsXSpn cell division protein in Streptococcus pneumoniae D39. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:E1061–E1069. doi: 10.1073/pnas.1108323108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Litzinger S, et al. Muropeptide rescue in Bacillus subtilis involves sequential hydrolysis by beta-N-acetylglucosaminidase and N-acetylmuramyl-L-alanine amidase. Journal of bacteriology. 2010;192:3132–3143. doi: 10.1128/JB.01256-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.de Jonge BL, Chang YS, Gage D, Tomasz A. Peptidoglycan composition of a highly methicillin-resistant Staphylococcus aureus strain. The role of penicillin binding protein 2A. The Journal of biological chemistry. 1992;267:11248–11254. [PubMed] [Google Scholar]
- 37.Turner RD, Hurd AF, Cadby A, Hobbs JK, Foster SJ. Cell wall elongation mode in Gram-negative bacteria is determined by peptidoglycan architecture. Nature communications. 2013;4:1496. doi: 10.1038/ncomms2503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bannantine JP, Stamm WE, Suchland RJ, Rockey DD. Chlamydia trachomatis IncA is localized to the inclusion membrane and is recognized by antisera from infected humans and primates. Infection and immunity. 1998;66:6017–6021. doi: 10.1128/iai.66.12.6017-6021.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.