Abstract
Background and Aims
While molecular approaches can often accurately reconstruct species relationships, taxa that are incompletely differentiated pose a challenge even with extensive data. Such taxa are functionally differentiated, but may be genetically differentiated only at small and/or patchy regions of the genome. This issue is considered here in Poa tussock grass species that dominate grassland and herbfields in the Australian alpine zone.
Methods
Previously reported tetraploidy was confirmed in all species by sequencing seven nuclear regions and five microsatellite markers. A Bayesian approach was used to co-estimate nuclear and chloroplast gene trees with an overall dated species tree. The resulting species tree was used to examine species structure and recent hybridization, and intertaxon fertility was tested by experimental crosses.
Key Results
Species tree estimation revealed Poa gunnii, a Tasmanian endemic species, as sister to the rest of the Australian alpine Poa. The taxa have radiated in the last 0·5–1·2 million years and the non-gunnii taxa are not supported as genetically distinct. Recent hybridization following past species divergence was also not supported. Ongoing gene flow is suggested, with some broad-scale geographic structure within the group.
Conclusions
The Australian alpine Poa species are not genetically distinct despite being distinguishable phenotypically, suggesting recent adaptive divergence with ongoing intertaxon gene flow. This highlights challenges in using conventional molecular taxonomy to infer species relationships in recent, rapid radiations.
Keywords: Species tree, hybridization, polyploid, divergence, gene flow, radiation, Poa, alpine, tussock grass
INTRODUCTION
Molecular phylogenetic approaches generally assume that species relationships can be reconstructed with enough variable, neutrally evolving markers, where ‘enough’ in practice tends to mean one to tens of markers. However, it is not known how often this method will fail by way of recent differentiation, ongoing gene flow and hybridization, and/or demographic processes that obscure or slow differentiation at neutral regions. Recent work on species differentiation has reiterated that we do not know the proportion of species in this grey area of partial differentiation (Wu, 2001; Knowles and Carstens, 2007; Pinho and Hey, 2010; Feder et al., 2012; Hey and Pinho, 2012). The level of genetic differentiation that corresponds to actual speciation is taxon specific (Hey and Pinho, 2012) and, for any given group of related taxa, criteria for differentiating species may only be established following careful genetic and ecological studies. Thus well-studied taxonomic groups that exist in well-distinguished species and populations might not necessarily act as model systems for other ecologically important groups.
Grasses represent an important but challenging group to investigate from a phylogenetic perspective because of their recent evolution and the common occurrence of polyploidy and ongoing hybridization. Plastid markers have been successful in resolving some taxonomic difficulties in grasses at the tribal level (e.g. bamboos; Triplett and Clark, 2010; Zeng et al., 2010) and the genus level (Hand et al., 2010; Pirie et al., 2010), but species-level phylogenetic description has often remained elusive even when relatively variable markers such as low-copy nuclear genes have been used (e.g. Barker et al., 2007; Essi et al., 2008; Escobar et al., 2011). The grass genus Poa typifies these taxonomic and phylogenetic difficulties (Grun, 1954; Soreng, 1990; Anton and Connor, 1995; Patterson et al., 2005). It has been described as ‘bewildering’ in its complexity (Bor, 1952). Polyploidy (Darmency and Gasquez, 1997; Brysting et al., 2000; Patterson et al., 2005; Rudmann-Maurer et al., 2007; Kelley et al., 2009), hybridization (Grun, 1954; Soreng, 1990; Gillespie and Soreng, 2005) and complex breeding systems, including apomixis, cleistogamy and various forms of dioecism (Anton and Connor, 1995), occur frequently. Poa has long been considered to have a Eurasian origin, from whence it colonized North and then South America (Hartley, 1961; Anton and Connor, 1995).
Chloroplast and nuclear phylogenies favour a South American origin for Australasian Poa (Soreng, 1990; Gillespie and Soreng, 2005; Patterson et al., 2005; but see Gillespie et al., 2009), and implicate recent rather than Gondwanan dispersal as suggested some time ago (Clifford and Simon, 1981). Few Australian representatives were included in phylogenetic analyses until Gillespie et al. (2009) who surveyed representatives of many Australasian Poa species using nuclear ribosomal and chloroplast markers. All sampled Australian species (apart from P. saxicola and P. queenslandica, which were reclassified as Saxipoa saxicola and Sylvipoa queenslandica, respectively) formed a single well-supported clade, also including most New Zealand, New Guinea and some American species as well as some Eurasian species, meaning the American-origin hypothesis was neither confirmed nor rejected. In another study, both genome copies of the nuclear CDO504 region were shared with American species, but sequence data from Eurasian species were not available (Griffin et al., 2011). Within the genus, this American/Eurasian/Australasian group forms the subgenus Poa, supersection Homalopoa, with the Australasian taxa occupying the less strongly supported section Brizoides (Gillespie et al., 2009).
The Australian group has been suggested to have radiated rapidly (Gillespie and Soreng, 2005) around the end of the Pleistocene (Hoffmann et al., 2013). Taxa were first lumped as Poa australis R.Br. or Poa caespitosa G.Forst.ex Spreng. Vickery then described 34 native species (Vickery, 1970) and new taxa have been described since (Walsh, 1994; Walsh et al., 2009). Currently, 38 distinct species are recognized (Gillespie et al., 2009; Walsh et al., 2009). The Australian Poa species are characterized by hermaphroditic flowers (Anton and Connor, 1995), tussock form, shoots arising from a contracted rootstock, open leaf sheaths, flat to rolled leaf blades, and strongly laterally compressed spikelets arranged in an open, pyramidal panicle, although there are exceptions to these general characters (Vickery, 1970). Species examined so far are outcrossing (James et al., 1997; Byars et al., 2009) and allotetraploid (Griffin et al., 2011). The taxonomy of the Australian Poa species is considered imperfect (Vickery, 1970; Walsh, 1994). Many of the characters used to distinguish species (e.g. leaf colour, leaf sheath colour and leaf scabrousness) vary within species seasonally, geographically or with environmental factors such as insolation (pers. obs. and Vickery, 1970). However, there are consistent morphological differences between most species pairs (Walsh, 1994) and evidence for distinct ecological roles (Griffin and Hoffmann, 2012).
In this study, we concentrate on the Poa species of the Australian alpine and sub-alpine zone. Poa tussock grasses dominate grassland and herbfield, and play an important structural role in the alpine landscape, being early successors after fire (Walsh and McDougall, 2005; Williams et al., 2008) and reducing soil erosion (Durham, 1956) as well as inhibiting snowgum seedling establishment (Noble, 1980; Ball et al., 1997) and providing intertussock spaces for herb and forb growth. For these reasons they are key to the Australian alpine grassland, and used extensively in revegetation programmes, where single ‘species’ of local provenance tend to be used (James et al., 1997). Grasses and other alpine/sub-alpine taxa are threatened by climate change (Preston and Jones, 2006; Steffen et al., 2009), yet we lack information about the genetic population structure, hybridization propensity and adaptive potential of Australian alpine taxa that would assist prediction of climate change response (Pickering et al., 2004; Byars et al., 2007, 2009; Byars and Hoffmann, 2009; M'Baya et al., 2013).
Species relationships within the Australian Poa group have not been successfully reconstructed in previous studies. The question of whether the alpine Poa species form a monophyletic group remains unclear. There are eight taxa occurring only in the alpine and/or sub-alpine zone, seven of which we studied here. According to Vickery (1979), Poa hiemata, P. costiniana and P. fawcettiae are strictly alpine; and P. hothamensis, P. phillipsiana, P. clivicola and P. gunnii occur from the alpine to the sub-alpine zone. We excluded the Tasmanian endemic sub-alpine species Poa jugicola (Walsh et al., 2009) due to a lack of suitable herbarium specimens. A further nine taxa occur in sub-alpine to montane regions, and we studied three of these: P. ensiformis, P. helmsii and P. sieberiana var. sieberiana, which is also widespread in lowland regions. Connor and Edgar (1986) classified the alpine/sub-alpine taxa into two sub-groups, with the alpine P. helmsii and P. hothamensis clustered together in the ‘labillardieri’ sub-group; P. sieberiana, P. fawcettiae, P. petrophila and P. hiemata clustered together in the ‘sieberiana’ sub-group; and P. costiniana, P. gunnii and P. saxicola as stand-alone species. This classification was based on a few ‘innovative’ characters and on habitat preference (which differed somewhat from Vickery's initial description). It also included lowland species in each of the two sub-groups, implying that the alpine region was colonized by at least two lineages of Poa. Genetic studies have not supported this classification, but genetic inference has been limited by low variation (Gillespie et al., 2009). A recent study using one chloroplast and two nuclear regions produced sufficient genetic resolution to reclassify the former P. saxicola as Saxipoa saxicola, but was unable to delimit further species within the Australian section Brizoides. Further, a study that focused on the alpine species found gene tree incongruence, complicating species delimitation (Griffin et al., 2011).
We expected to find gene tree incongruence due to marker-specific patterns of incomplete lineage sorting if this group did indeed evolve under a recent, rapid radiation. For this reason, we used multiple gene regions and multiple individuals per species to investigate species relationships, and considered partial nuclear gene regions as well as chloroplast non-coding regions. We then aimed to distinguish between hybridization after initial species divergence, and ongoing gene flow, searching for signs of individual hybrid origin by comparing pairwise genetic distances at each marker with those of reconstructed species trees (Joly, 2012). Microsatellite markers and crossing experiments were applied to investigate gene flow in a broad-scale spatial context, and to test the possibility of gene flow between different ‘species’, respectively. We discuss our multiple lines of evidence in light of current ideas about the adaptation to alpine environments, genomic ‘stages’ of species differentiation and challenges in reconstructing species relationships.
MATERIALS AND METHODS
Sample selection and DNA extraction
Leaf samples were taken from herbarium specimens spanning the distribution range of each Australian alpine Poa species in Victoria, New South Wales and Tasmania (MEL, MELU, CANB) and from some live grasses collected in the Bogong High Plains, Victoria in 2006 (Supplementary Data Table S1). We included all five species that occur in the true alpine zone (P. costiniana, P. fawcettiae, P. hiemata, P. hothamensis var. hothamensis and P. phillipsiana), two species that occur in the sub-alpine zone only (P. clivicola and P. gunnii) and three species occuring from the sub-alpine to montane zone (P. ensiformis, P. helmsii and P. sieberiana var. sieberiana, which also occurs at lower elevation) (Walsh et al., 2009). The cosmopolitan, non-native P. annua was chosen as a related species occurring in the same geographic region that would provide an outgroup sufficiently divergent for phylogenetic inference. Specimens were restricted to those identified or confirmed by taxonomists expert in Australian Poa. By using expert-identified herbarium specimens, we avoided potential misidentification associated with field collections and obtained a broad survey of species distribution, both spatial and temporal. Herbarium specimens have been used in a range of evolutionary and ecological studies (Cozzolino et al., 2007; Gallagher et al., 2009) as well as for phylogenetic analysis (Savolainen et al., 1995; Gillespie et al., 2009). The chloroplast genome in particular can remain intact for many decades: chloroplast microsatellites have been amplified from >100-year-old specimens (e.g. Biss et al., 2003). Here we successfully amplified DNA from a specimen collected in 1893 (one P. ensiformis sample, MEL 2138127A, for which two nuclear microsatellite markers amplified successfully).
For extractions, approx. 20 mg of leaf tissue was ground to a fine powder in a 1·5 mL Eppendorf tube using a small plastic pestle and/or a Mixermill (Retsch MM300) combined with repeated freezing in liquid nitrogen. Initially, DNA extractions were performed using the CTAB (cetyltrimethylammonium bromide) method (Rogers and Bendich, 1994). All 72 specimens for the microsatellite amplification were extracted with this method (Supplementary Data Table S1); the 63 specimens used only for the gene region amplification were extracted with a DNeasy Plant Mini Kit (Qiagen).
Chloroplast non-coding region amplification
Sequences were obtained for two chloroplast regions (rpoB-trnC and rpl32-trnL) with 454 sequencing as described in Griffin et al. (2011). These were aligned with sequences obtained in a trial panel of individuals by Sanger sequencing carried out by Macrogen Inc. (Seoul, Korea) with an ABI-3730XL capillary sequencer (Applied Biosystems) on PCR products (see Supplementary Data Table S3 for primers and PCR conditions). For these extra PCR amplifications, reactions were performed using 0·4 µL of Taq (5 U μL−1, New England Biolabs), dNTPs (0·2 mm), primers (0·8 µm each primer), 1× bovine serum albumin (BSA), 5 µL of total DNA (1/20 diluted), 1× buffer (New England Biolabs) containing 2·0 mm MgCl2, and water to a total of 50 µL. Between three and 17 individuals were sequenced per species per chloroplast region (Supplementary Data Table S1).
Nuclear gene amplification
Four nuclear regions were chosen, reported to be single copy in diploid grasses: the partial gene regions waxy (also called GBSS1), trx, CDO504 and DMC1 (Supplementary Data Table S3). These had all exhibited useful infrageneric variation either in Poa (trx and CDO504, Patterson et al., 2005) or in closely related grass genera (DMC1, Petersen and Seberg, 2002; waxy, Mason-Gamer et al., 1998; Souto et al., 2006; Zhang et al., 2009). From previous work (Griffin et al., 2011) we knew each region should be present in two homeologous, non-recombining copies in the genome, due to the species' hybrid origin, so in effect the four PCRs produced data for eight independent, informative gene regions.
A combination of methods (described below) resulted in successful nuclear sequence for 1–16 individuals per homeologous gene copy per species, for a total of 90 individuals. First, reactions included 0·4 µL of Taq (5 U μL−1, New England Biolabs), dNTPs (0·2 mm), primers (0·8 µm each primer), 1× BSA, 0·5 % polyvinylpyrrolidone (PVP), 1–5 µL of total DNA (1/20 diluted), 1× buffer (New England Biolabs) containing 2·0 mm MgCl2, and water to a total of 50 µL. These conditions were generally appropriate for the fresh samples but produced poor amplification in many of the herbarium specimens. A reaction comprising 2× BIO-X-ACT™ Short Mix (Bioline) containing enzyme, buffer, dNTPs and 2·0 mm MgCl2 was used in 10–50 µL reactions along with primers (0·8 µm each primer) and template (2 µL of extracted DNA) for the herbarium samples.
According to previous findings of allotetraploidy, we expected two homeologues for each gene, which had to be separated before sequencing. To do this, PCR products were purified with an illustra™ GFX™ PCR DNA and a Gel Band purification kit (GE Healthcare). The purified PCR product was ligated into a pGEM-T Easy Vector (Promega) (total reaction volume: 5 µL) and this vector was then ligated into JM109 competent cells (Promega) following the manual, using the heat shock method. Clones with successful inserts were sequenced (without prior purification) by Macrogen Inc. Where possible, at least 20 clones were sequenced per individual as recommended by Small et al. (2004) to ensure sequencing all alleles present.
To avoid the bacterial cloning step, 454 sequencing was performed for 61 individuals using new primers designed within the nuclear and chloroplast regions. Primers and methods for DMC1, CDO504, rpoB-trnC and rpl32-trnL were described in Griffin et al. (2011); those for trx and waxy are described in Supplementary Data Table S3. Briefly, PCR products for each gene region were pooled according to individual and labelled with an individual-identifying barcode sequence. The resulting mixture was then extended by PCR amplification in order to make the ends compatible with 454 sequencing chemistry, and sequenced in a 1/4 plate run by the Australian Genome Research Facility (AGRF), Brisbane, Australia. All sequences were edited and aligned using Sequencher 4·7 (GeneCodes Corporation) or Geneious Pro v. 5 (Biomatters Ltd). PCR chimera sequences were identified and removed as described in Griffin et al. (2011) and the nucleotide diversity was calculated in MEGA v. 5·05 (Tamura et al., 2011). Tajima's D was evaluated in DNASP v. 5. (Librado and Rozas, 2009).
Multiple-gene analysis
To make simultaneous use of all sequenced DNA regions, we applied the program *BEAST (Heled and Drummond, 2010). *BEAST uses coalescent and Bayesian statistical methods to estimate species trees from multiple organellar and nuclear loci. It assumes no recombination within loci and full recombination between nuclear loci. These assumptions were verified with linkage tests in DnaSP (Librado and Rozas, 2009) and recombination tests performed with the BootScan method (Martin et al., 2005a) in RDP3 (Martin et al., 2005b).
All individuals that produced sequence for rpl32-trnL or rpoB-trnC were included in a sequence alignment for each region, coding missing individual–sequence combinations as a string of Ns. For the nuclear regions waxy, DMC1 and CDO504, separate alignments were created for each copy. For trx, only copy A was used, since amplification of copy B was poor. Alignments included sequences obtained using 454 sequencing (Griffin et al., 2011) and sequences obtained using Sanger sequencing (see Supplementary Data Table S1 for the full complement of individual–gene combinations included). Since the Australian P. annua samples failed to amplify for trx, we included two representative GenBank P. annua sequences (GenBank accession nos AY589270 and AY589271) as replacement outgroup sequences. Indels were scored as described in Griffin et al. (2011) and included in a separate data partition for each locus. Within each nuclear gene copy, each individual was represented by two sequences: either the two alleles detected (for heterozygotes) or two copies of the single allele deteted (for homozygotes). This was necessary to represent the allele distribution in the population of loci accurately, as recommended by the author of *BEAST (J. Heled, pers. comm.).
All 18 alignments (sequence and indels for each locus) were imported into BEAUTi v1·7·5 (Drummond et al., 2012) as separate data partitions. The ‘species tree ancestral reconstruction (*BEAST)’ option was enabled and each sequence was assigned to the appropriate species using the ‘Traits’ tab. Poa annua was constrained as the outgroup on the species tree, and the root age was constrained to 13 ± 1 million years (MY), the estimated age of the split between Poa I, containing P. annua, and Poa VIII, containing the Australian Poa (Hoffmann et al., 2013). The sequence and indel alignments for each locus were assigned the same tree model. Both chloroplast regions were also assigned to evolve according to the same tree model, as the chloroplast is inherited as a single, non-recombining molecule. All clock and substitution models were left unlinked. We assigned a strict molecular clock and estimated all other data partition clock rates with gamma priors (shape 2·0, scale 2·0) in relation to the rpl32-trnL clock (mean rate fixed at 1). Nucleotide substitution (‘site’) models were assigned to each data partition based on the best model out of those available in *BEAST chosen by BIC in PartitionFinder (Lanfear et al., 2012) (Table 1). All site priors were given a log-normal distribution (mean ± s.d., 1·0 ± 1·25) except transition/transversion priors (gamma distribution, shape 0·05, scale 10). The species tree was estimated using a Yule Process prior (uniform, between 0 and 10) and a piecewise constant population size model (gamma, shape = 2·0, scale = 1 × 105). The starting gene tree was randomly generated for each data partition and the ploidy type of the chloroplast tree was set to 1, while all nuclear trees had ploidy = 2. The root height was estimated automatically for each tree. The Markov chain Monte Carlo (MCMC) was run for 100 × 106 generations, logging parameters every 10 000 generations. Ten separate runs were performed with different random starts.
Table 1.
Sequence and alignment length, variation and substitution models used for each region included in the multiple gene analysis
| Region | Length range (bp) | Alignment length (bp) | Base changes (variable/parsimony-informative/parsimony-informative excluding outgroup) | % GC content | Π (nucleotide diversity) | Tajima's D | PartitionFinder best substitution model | |
|---|---|---|---|---|---|---|---|---|
| rpl32-trnL | Sequence | 426–448 | 470 | 49/23/11 | 26·0 | 0·0326 | –1·90* | TrN + I + G |
| Indels | NA/8/5 | HKY | ||||||
| rpoB-trnC | Sequence | 418–430 | 449 | 39/28/4 | 28·2 | 0·0320 | (–1·28) | HKY |
| Indels | NA/8/5 | HKY | ||||||
| CDO504 A | Sequence | 402–419 | 434 | 49/49/14 | 37·3 | 0·0039 | –1·96* | K80 |
| Indels | NA/7/2 | HKY | ||||||
| CDO504 B | Sequence | 389–433 | 467 | 78/55/16 | 49·2 | 0·0133 | –2·44** | K80 + G |
| Indels | NA/12/8 | HKY | ||||||
| DMC1 A | Sequence | 443–457 | 470 | 68/50/30 | 40·7 | 0·0103 | –2·58*** | HKY |
| Indels | NA/10/6 | HKY | ||||||
| DMC1 B | Sequence | 428–453 | 461 | 71/67/25 | 39·7 | 0·0086 | –2·54*** | TrN |
| Indels | NA/7/4 | HKY | ||||||
| trx | Sequence | 345–370 | 413 | 113/55/22 | 35·1 | 0·0174 | –2·57*** | HKY |
| Indels | NA/8/4 | HKY | ||||||
| waxy A | Sequence | 399–412 | 429 | 75/46/21 | 61·6 | 0·0207 | –2·48** | HKY |
| Indels | NA/7/2 | HKY | ||||||
| waxy B | Sequence | 379–408 | 415 | 87/46/26 | 60·4 | 0·0201 | –2·31** | K80 + G |
| Indels | NA/5/1 | HKY |
For length range, % GC content, nucleotide diversity π and Tajima's D, only the Australian native species are reported. For alignment length and nucleotide substitution model choice, the outgroup Poa annua is also included.
Asterisks indicate P-values: *0·01 < P < 0·05; **0·01 < P < 0·001; ***P < 0·001. Parentheses indicate P > 0·05.
‘NA’, only variable sites were included in the indel alignments.
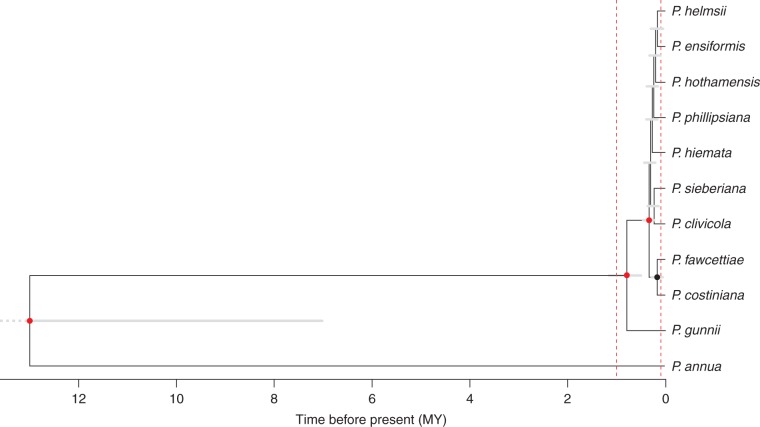
Log files were examined in Tracer v1·5 (Rambaut and Drummond, 2009) and an appropriate burn-in was identified as 20 × 106 generations (20 % of each run) based on the appearance of the posterior probability (pp) trace (Drummond et al., 2007). For each data partition and for the species trees, tree files were processed using LogCombiner v1·7·5 (Drummond et al., 2012) after removing the burn-in. TreeAnnotator 1·7·5 (Drummond et al., 2012) was then used to calculate the maximum clade credibility (MCC) tree (Fig. 1), with median node heights and a pp limit of 0. Convergence was checked by comparing the MCC tree topology for each run, and by examining the likelihood. The run with the lowest median likelihood produced a different tree topology from the other nine runs, and so was excluded. An overall MCC tree was then calculated after combining the nine highest likelihood species tree files and downsampling 10× in LogCombiner.
Fig. 1.
Maximum clade credibility (MCC) species tree estimated in *BEAST. Grey bars show 95 % HPD for node ages. 95 % HPD for root node extends to 20 MY (shown as a dashed bar). Red dots show node posterior probability (pp) ≥0·99; black dots show 0·8 ≤ pp < 0·9. Unlabelled nodes had pp < 0·8. Red dashed lines show 100 000 years and 1 million years before present.
A test of species delimitation used the software BP&P v 2·2 (Yang and Rannala, 2010), following the ‘τ-threshold’ method on a fixed, fully resolved species guide tree. This method accommodates a species phylogeny along with coalescent processes (ancestral polymorphism within species). For the root age (τ0), we used the estimate of 13 ± 1 million years ago (MYA) as per the *BEAST analysis. We assumed a generation time of 1 year and a mutation rate of 6·5 × 10−9 ± 0·7 × 10−9 mutations per site per generation (Ossowski et al., 2010). This gave a prior for τ0 of the form G(170, 2011) with mean 170/2011 = 0·085. Running the program without data verified that the threshold prior was large enough to prevent oversplitting. The other divergence time parameters were assigned the Dirichlet prior (Yang and Rannala, 2010). A gamma prior G(2,1000) was used on the population size parameters. The maximum clade credibility species tree identified in *BEAST (Fig. 1) was used as a guide tree. The heredity scalar was set to 1·0 for the nuclear loci and 0·5 for the chloroplast loci, and the mutation rate was allowed to vary among loci according to a Dirichlet prior D(2). After removal of a burn-in of 20 000 generations, 80 000 MCMC generations were run, sampling every ten generations. The analysis was run 20 times to obtain most ESS (effective sample size) values >200. For species delineation, the 95 % highest posterior density (HPD) interval of each node age was calculated in Tracer and compared with our previously chosen threshold τT age of 100 000 generations of separation.
The program jml v 1·01 compares the minimum genetic distance observed between sequences of two species at a single locus with the posterior predictive distribution of species distances between those two species simulated from a cohort of species trees (Joly, 2012). This provides a conservative test for hybridization even in the presence of incomplete lineage sorting. If the observed minimum genetic distance falls outside the posterior predictive distribution, i.e. conflicts with the species tree, either it is an instance of hybridization/introgression at that locus, or the taxa are poorly delimited taxonomically. As this software examines pairwise individual distances for each locus, it should have the power to detect rare instances of hybridization or introgression. Because jml assumes that taxa included are valid genetic entities, we reclassified sequence data as P. annua, P. gunnii or a third taxon (containing all other Australian species), following the results from both the BP&P and *BEAST analyses. Ten runs were performed using the same *BEAST parameters as previously, and, since all runs converged, species trees were combined after removal of 20 % burn-in and downsampled 100×. Median values for nucleotide substitution model parameters and relative mutation rates estimated in the combined *BEAST runs were included to inform the simulations, and a Type I error rate of α = 0·05 was used with subsequent false discovery rate (FDR) control (Benjamini and Hochberg, 1995).
Interspecies crosses
Crosses were performed with three of the high elevation alpine species, P. hiemata, P. phillipsiana and P. hothamensis. Poa hiemata plants were grown from seed collected on Mt Nelse Central, Bogong High Plains, Victoria (147°20′E, 36°50′S). Poa phillipsiana and P. hothamensis plants were grown from clonal material collected at Johnston's Hut, Bogong High Plains, Victoria (36°51S, 147°21′E). In November–December 2008, pairs of plants were chosen as they reached the appropriate flowering stage, ensuring that approximately equal numbers of crosses were performed between species pairs (between two and four crosses in each direction; Table 2). For each reciprocal cross, one flowering culm was chosen per plant several days before maturity to avoid the risk of open pollination. Any individual flowers that showed signs of maturity (anther or stigma extrusion) were removed. Each pair of flowering culms was bagged in a pollen exclusion bag (#421, Lawson Pollinating Bags, Northfield, IL, USA), which was gently shaken once a day for several weeks to encourage pollen movement, and left until the end of the flowering season. Nine single culms were bagged individually for P. hiemata to test for self-fertilization. Seeds were then air-dried and collected from each culm.
Table 2.
Results of crosses between and within Poa species
| Father |
||||
|---|---|---|---|---|
| P. hiemata | P. phillipsiana | P. hothamensis | ||
| (A) Successful crosses | ||||
| Mother | P. hiemata | 42/67 | 2/3 | 1/2 |
| P. phillipsiana | 1/3 | 2/3 | 3/3 | |
| P. hothamensis | 1/2 | 4/4 | 2/2 | |
| (B) Paternal resemblance | ||||
| Mother | P. hiemata | 1/5 | 1/1 | |
| P. phillipsiana | 5/5 | 5/7 | ||
| P. hothamensis | 1/1 | 7/16 | ||
A: number of crosses with successful germination expressed as a fraction of total crosses performed, for each cross type. B: number of offspring exhibiting the morphology of the paternal species as a fraction of the total offspring from each inter-species cross.
Two rounds of seed were sown, first using approx. 30 seeds per cross, then using all the remaining seed for all crosses in May 2009 once it was observed that many crosses completely failed to germinate. Germination was scored as successful when at least one seedling was produced from the first and/or second round of sowing. At maturity, randomly numbered plants were identified by the following morphological characters (Vickery, 1970) as one of the three parent species: (1) P. hothamensis, leaf blades flat or concave, 7–30 cm long, 1·5–5 mm wide; (2) P. phillipsiana, leaves bluish-green, blades inrolled, 7–25 cm long, 0·5–0·75 mm wide; and (3) P. hiemata, leaves green, blades closely folded and compressed, 5–25 cm long, often <0·5 mm wide.
This identification was then checked against their true parentage to determine whether they resembled the maternal or paternal parent.
Microsatellite amplification and analysis
Six microsatellite markers were trialled: CAID4, CA1F4 and GAC1 (Maurer et al., 2005); MI6-B and H01H06 (Kindiger, 2006); and AE5 (Albertini et al., 2003). These had all been used successfully in Victorian P. hiemata (Byars et al., 2009) as well as the species in which they were originally designed. Microsatellite PCRs contained Bioline PCR buffer (final concentration 1×), MgCl2 (1·5 mm), PVP (0·5 %), dNTPs (0·08 mm), 1× BSA, unlabelled forward primer (1 µm), unlabelled reverse primer (1·2 µm), forward primer radiolabelled with [33P]ATP (0·005 µm), 0·1 µL of Immolase DNA polymerase (Bioline), total DNA (2 µL of 1/20 dilution) and deionized water to a final volume of 10 µL. Polyvinylpyrrolidone is a PCR or DNA extraction additive that sequesters polyphenolic compounds that can interfere with amplification (Koonjul et al., 1999). The PCR conditions were as follows: 95 °C for 7 min; 30 cycles of 95 °C (45 s), annealing temperature (Supplementary Data Table S2) (45 s), 72 °C (60 s); final extension at 72 °C (8 min). Denaturing polyacrylamide gel electrophoresis was used to separate PCR products. Gels (5 %) were run for 2–5 h at 65 W, dried for 30 min and exposed to autoradiography film (Fuji) for 24–120 h.
Bands were scored by hand and each gel was scored at least twice to check for problematic markers or poorly defined banding patterns. Initially, marker size was measured by comparison with the fmol® λgt11 sequencing ladder (Promega). For later gels, a sub-set of four individuals was always included as a size reference. Microsatellite alleles were scored as present/absent (binary), since the copy number could not be determined. For the final analysis, results from all five successful markers were included in a single data set, excluding individuals with poor representation (only one or two successful amplifications). Individuals were compared manually to count the number of distinct genotypes, scoring genotypes as distinct if they differed by at least one band. GENALEX 6·3 (Peakall and Smouse, 2006) was used to calculate overall and pairwise species differentiation (ΦPT) by an analysis of molecular variance (AMOVA).
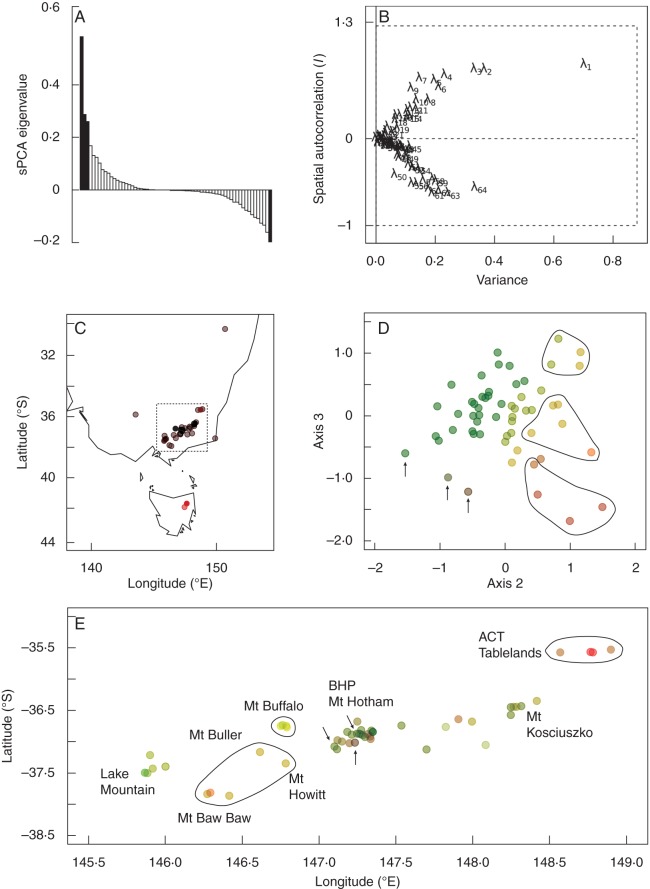
Principal component analysis (PCA) was performed on the binary data set, interpolating missing values to the overall mean frequency, using the dudi.pca command in the ade4 package (Dray and Dufour, 2004) in R v. 2·13·2 (R Development Core Team, 2011) to examine species-level and geographic patterns. For a more sensitive exploration of spatial patterns, the sPCA method (Jombart et al., 2008) was implemented using the adegenet R package. This method incorporates a matrix of geographic distance weights along with the matrix of alleles ‘x’. A PCA is performed to decompose the function C(x), which measures both the spatial structure and genetic variability. The resulting positive eigenvalues reflect global spatial patterns, while negative eigenvalues reflect local spatial patterns. Spatial and genetic variance is not additive in an sPCA, hence eigenvalues do not sum to 1. Inverse ln-transformed geographic distances were used to build the connection network. The first three positive eigenvalues and the first negative eigenvalue were retained based on the relative proportion of genetic and spatial variance they captured (Fig. 2A, B).
Fig. 2.
sPCA results for the microsatellite data set revealing spatial genetic structure across species boundaries. (A) sPCA eigenvalues. Positive eigenvalues reflect global patterns and negative eigenvalues reflect local patterns. Eigenvalues retained in the final model are highlighted in black. (B) Screeplot of sPCA eigenvalues showing their decomposition into variance and spatial autocorrelation (Moran's I) components. λ1, λ2 and λ3 are the first three positive eigenvalues from (A); λ64 is the largest negative eigenvalue. These four eigenvalues were retained in the final model. (C) Axis 1 eigenvector sample scores (represented by colour on a black to red scale) plotted on the geographic locations, revealing Tasmania–mainland differentiation. The dotted box outlines the mainland area presented in (E). (D) Axis 3 vs. Axis 2 sPCA eigenvectors. Points are coloured-coded according to their positions along both axes. (E) Geographic location of mainland samples coloured by Axis 2 and 3 sPCA eigenvector score (see D for colour scheme). Geographic regions showing some distinctiveness in eigenvector space are circled. Points with similar, outlying eigenvector scores but no overall distinctiveness of geographic region are indicated by arrows. BHP, Bogong High Plains.
RESULTS
Chloroplast and nuclear sequence results
As previously reported for DMC1 and CDO504 (Patterson et al., 2005; Griffin et al., 2011), waxy exhibited two distinct copies per individual. These were consistently separated by 13 fixed single nucleotide polymorphisms (SNPs) and two indels. For trx, a distinct second copy was identified (separated by 31 fixed SNPs and four indels), but it was present in relatively few 454 reads and could not be accurately reconstructed in all individuals. This probably indicates a primer mismatch in this copy that resulted in poor amplification. For this reason, only trx copy A was retained in further analysis. PCR recombinant sequences again formed a significant proportion of 454 reads (Griffin et al., 2011), but no evidence for recombination among the two copies was detected once the PCR recombinants were removed. New chloroplast and nuclear sequences used in this study are available in GenBank (accession nos KC154272–KC154858).
Individual gene trees co-estimated with an overall species tree in the *BEAST analysis showed little support for most nodes (Supplementary Data Figs S2–S9). Well-supported internal nodes typically contained representatives of several species [e.g. waxy copy B (Supplementary Data Fig. S9)]. The majority of Tasmanian samples populated a distinct clade in the chloroplast tree, along with some Victorian samples (Supplementary Data Fig. S2).
Multiple-gene species structure
The nine MCMC runs combined gave acceptable ESSs (>200) for all parameters except some of the species.tree.splitPopsize parameters. This reflected variation from run to run due to the high number of priors included and the low variation in the data set.
In the overall maximum clade credibility species tree (Fig. 1), Poa gunnii, which is endemic to Tasmania, was well supported as the sister group to the rest of the Australian alpine taxa with a pp of 1. No other species relationships were significantly supported, although P. fawcettiae and P. costiniana were grouped in a single clade with pp = 0·87. Node dating based on a root age of 13 ± 1 MY (Hoffmann et al., 2013) produced an estimate for the age of the Australian group of 0·510–1·19 MY (95 % HPD; mean 0·795 MY).
The species-threshold approach used in BP&P also supported P. gunnii as a distinct species from the rest of the Australian alpine taxa (Supplementary Data Fig. S1). The posterior distribution of the node age for the clade containing P. gunnii and the rest of the Australian alpine Poa using BP&P was τ = 0·246–0·515 MY (95 % HPD; mean 0·382 MY), corresponding to approx. 382 000 generations and therefore exceeding the 100 000-generation threshold τT. The node age estimates obtained with this method were younger than the *BEAST estimates since indel characters were not included. The clade containing the ten Australian alpine Poa taxa apart from P. gunnii did not exceed the 100 000-generation age threshold (95 % HPD 0·0775–0·213 MY; mean 0·147 MY). Thus, whether using the *BEAST approach to investigate structure among currently accepted species, or using the coarse threshold-delimitation method in BP&P to test species delimitation, only two Australian alpine Poa species are supported: P. gunnii and a second species subsuming the other nine taxa.
An analysis using jml on the per-locus sequence data only (no indels) detected no clear instances of hybridization among P. annua, P. gunnii and the third taxon containing the rest of the Australian alpine Poa. For the chloroplast region rpl32-trnL, minimum distances between P. annua individuals and individuals of both P. gunnii and the third taxon were smaller than expected from the posterior predictive distribution of the species trees. For the other chloroplast region, rpoB-trnC, the distances between P. gunnii and third taxon individuals were smaller than expected. However, these differences were not significant after FDR correction for multiple comparisons. No nuclear regions showed significant disagreement between the minimum observed intertaxon distance and the posterior predictive distribution.
Interspecies crosses
Two to four replicate crosses were successful for each of the nine possible crosses (Table 2). Poa hiemata produced viable seeds from self-fertilization in 8/9 crosses, suggesting that self-fertilization cannot be ruled out for this species. However, successful germination was observed for at least one cross in each species–direction combination (Table 2), pointing to the possibility of hybridization between all species pairs. At least one offspring from each type of cross fell within the morphological range of the pollen donor species, but well outside the range of the mother species under blind scoring, suggesting that the crosses had been successful (Table 2).
Microsatellite diversity and geographic patterns
Five of the six microsatellite markers were included in the analysis; GAC1 was excluded due to non-replicable banding patterns. The other markers exhibited useful genetic variation, with 13–24 alleles per marker detected overall (Supplementary Data Table S2).
More than two bands were produced in many individuals for every microsatellite marker tested (Supplementary Data Table S2). AE5 produced an average of 3·4 bands per individual (maximum 7) whereas the other four markers produced 2–2·4 on average (maximum 4), suggesting tetraploidy. Observed heterozygosity levels were correspondingly high (0·70–0·94), as expected given the fixed heterozygosity likely to be provided by multiple homeologous loci (Supplementary Data Table S2). Overall, all species showed polymorphism (18·8–58·4 % of loci polymorphic within each species) with 0–5 private alleles per species and mean unbiased diversity ranging from 0·121 to 0·201 within each species (Supplementary Data Table S2). There were 13–24 alleles detected per locus, and each individual scored for all five markers was resolved as a distinct genotype, showing that the five-marker panel provided sufficient resolution for this study.
Little overall species differentiation was detected using the microsatellite markers. Only 10 % of the total genetic variance was partitioned according to species (AMOVA P < 0·05) and the remaining 90 % was within species. A pairwise AMOVA did reveal significant differentiation between several species pairs (999 permutations, P < 0·05 after Bonferroni correction for multiple comparisons) but all ΦPT values were low. Poa phillipsiana was significantly differentiated (P < 0·05) from P. sieberiana (ΦPT = 0·15), P. fawcettiae (ΦPT = 0·084), P. ensiformis (ΦPT = 0·24) and P. hothamensis (ΦPT = 0·26); P. hothamensis was also significantly differentiated from P. costiniana (ΦPT = 0·19). Correspondingly, the PCA revealed no clear species differentiation (data not shown).
Both the PCA and the sPCA identified the Tasmania–mainland differentiation as the dominant pattern in the data set, splitting these geographic regions on Axis 1 [PCA Axis 1, 6·7 % of total variance; sPCA Axis 1, 88 % of maximum possible genetic variance (linear combination of variables) and 66 % of maximum possible variance in Moran's I]. However, the PCA revealed no other clear geographic patterns, whereas the sPCA indicated several geographic groupings. The ACT Tablelands samples, Mt Buffalo and Mt Baw Baw/Mt Howitt/Mt Buller samples all scored highly on Axis 2 of the sPCA (46 % of maximum possible genetic and 61 % of maximum possible spatial variance), but were differentiated on Axis 3 (42 % of maximum possible genetic variance and 61 % of maximum possible spatial variance), meaning that these three regions were identified as distinct overall from each other and from the rest of the samples. Within the ‘main range’ spanning Mt Hotham to Mt Kosciuszko (Fig. 2E), no clear spatial structure was revealed. No relationship was detected between either PCA or sPCA scores and altitude (examined graphically). Species structure (if any) should have been revealed in the local score axis (Axis 4), but none was observed (examined graphically and by ANOVA).
DISCUSSION
Limited species structure detected
In Poa, phylogenetic analyses have supported designation of subgeneric clades in known diploid (Soreng et al., 2010) and polyploid (Gillespie and Soreng, 2005; Gillespie et al., 2009) species, allowed identification of the parental lineages of some allopolyploids (Soreng et al., 2010) and identified relationships at the broader level of the subtribe Poinae (Gillespie et al., 2008). However, species relationships below the subgenus level have remained problematic. We aimed to increase both marker coverage and the number of individuals per species, to facilitate the detection of species structure under ongoing gene flow or incomplete lineage sorting (Small et al., 2004; Maddison and Knowles, 2006; Degnan and Rosenberg, 2009; Heled and Drummond, 2010). However, despite our use of multiple non-ribosomal nuclear and chloroplast regions, and multiple individuals per species, we obtained as little resolution as did a previous study using fewer and potentially less informative markers (Gillespie et al., 2009).
As reported by Gillespie et al. (2009) and Griffin et al. (2011), the chloroplast regions showed extensive haplotype sharing across putative species (Supplementary Data Fig. S2). Because the chloroplast genome is uniparentally inherited, chloroplast markers alone cannot reveal whether haplotype sharing stems from ongoing hybridization or incomplete lineage sorting. Low copy number nuclear gene sequences are now favoured in situations where low levels of organellar genome variation and the presence of polyploidy and hybridization complicate traditional marker use (Essi et al., 2008; Liu et al., 2008; Xu et al., 2008). A wealth of protein-coding genes are now used in family-level phylogenetic studies (Hughes et al., 2006), though in practice a small number of genes tend to be favoured once their utility has been demonstrated, for example waxy/GBSSI in grasses (Baumel et al., 2002; Ingram and Doyle, 2003; Mason-Gamer et al., 2010). Although nuclear regions are often informative, the nuclear gene trees in this study revealed no clear pattern of species divergence (Supplementary Data Figs S3–S9). To determine whether this was due to the retention of ancestral polymorphism across current species boundaries (Maddison and Knowles, 2006; Degnan and Rosenberg, 2009) as found in other plant groups including the grass genus Hordeum (Jakob and Blattner, 2006), we used two approaches. First, we applied *BEAST, a Bayesian technique to co-estimate an overall species tree with multiple gene trees (Heled and Drummond, 2010), assuming only incomplete lineage sorting. When this method was applied to the data set comprising seven nuclear and two (linked) chloroplast regions (Table 1), the species tree revealed only one well-supported relationship: Poa gunnii, a Tasmanian endemic, was well supported as a sister group (pp = 1) to a clade containing the rest of the Australian alpine species (pp = 1; Fig. 1). Secondly, we used the fixed-tree threshold approach implemented in BP&P (Yang and Rannala, 2010) to judge whether taxa were delimited as species with respect to a threshold of a separation of 100 000 generations. Again, only P. gunnii was delimited as a distinct species by this method: all other node age distribution estimates within the Australian group included, or were younger than, our threshold of 100 000 generations (Supplementary Data Fig. S1).
This general lack of species structure was also supported by the microsatellite data. The AMOVA on the microsatellite data set revealed a global differentiation (ΦPT) value of 0·099 (P < 0·05), which is low for population differentiation, let alone for species differentiation (Frankham et al., 2002). This was comparable with pairwise population differentiation values detected in P. hiemata (Byars et al., 2009). Some significant pairwise species differentiation was detected, with P. phillipsiana significantly different from four other species, yet this did not produce clear species clusters in the PCA or sPCA and was not supported in the species tree using sequence data.
The overall lack of genetic structure conflicts with the consistent morphological character differences (Walsh, 1994) between the Australian alpine Poa. In addition, distinct ecological characteristics have been reported for some species pairs, with the flat-leaved P. hothamensis exhibiting lower drought tolerance than the rolled-leaved P. hiemata (Griffin and Hoffmann, 2012). Broadly, our genetic results confirm that the alpine Poa have evolved under a recent, rapid radiation, probably as part of the larger Australian Poa radiation following long-distance dispersal to Australia (Soreng, 1990; Gillespie et al., 2009; Griffin et al., 2011). We suggest three possible and not mutually exclusive explanations for the lack of species differentiation observed. First, differentiation may have occurred too recently to be observed, even using multiple gene regions. Secondly, species relationships may be obscured by ongoing gene flow occurring during species divergence. Thirdly, previous divergence may be obscured by current hybridization. We discuss each of these possibilities below.
Informativeness of markers under a very recent radiation
Although rapid radiations have been suggested for the evolution of many Australian plant groups (reviewed in Crisp et al., 2004; Linder, 2008), a distinction can be made between more ancient radiations (from taxa that existed in Australia before its isolation >35 MYA) and recent radiations (from taxa that reached Australia only after its isolation). Australian Poaceae clearly fall into the ‘recently radiated’ category (Crisp et al., 2004). Some lineages within the Poaceae have then undergone constant, slow diversification, the endemic grass genus Austrostipa among them (Syme, 2011). However, molecular evidence points to a rapid radiation for Poa. Using the single nuclear marker ITS (internal transcribed spacer), Hoffmann et al. (2013) dated the origin of the main Australian Poa clade (containing all native Australian species investigated here) to 0·38–3·01 MYA, which corresponds to our estimated node age of 0·510–1·19 MY (95 % HPD; mean 0·795 MY). Hoffmann et al. estimated a high diversification rate of 1·1–1·5 species MY−1 and suggested that this is one of several parallel rapid radiations in multiple Poa groups around the world that have occurred in the last 2·5 MY, possibly associated with the cooler, drier climatic conditions prevalent in the Late Tertiary. Our results add support to the rapid, recent radiation scenario for the Australian Poa. In a rapid radiation, neutral regions are slow to differentiate by genome hitchhiking (Wu, 2001; Hudson and Turelli, 2003; Feder et al., 2012), and in line with this we found low variation and an excess of rare mutations in all regions examined (Table 1). As a further complicating factor, incomplete lineage sorting is likely to persist under a rapid radiation, especially where effective population sizes are large (Maddison and Knowles, 2006) as we expect in these widespread and common grasses. Genome-wide scans can reveal signatures of differentiation in such cases (e.g. Bezault et al., 2011), but it is unsurprising that the traditional molecular taxonomic methods we have used here fail to reveal differentiation (Tajima, 1983).
Ongoing gene flow during species divergence
In addition to a low base level of molecular divergence, species differentiation may be obscured by ongoing gene flow. Though small-scale, the glasshouse hybridization experiment supports the possibility that gene flow can occur: seedlings with the morphology of the pollen donor species were produced from all combinations of crosses between P. hiemata, P. phillipsiana and P. hothamensis. Australian Poa (‘P. caespitosa’) also crossed readily with North American P. arachnifera (Clausen, 1961), and various other interspecies crosses in the genus have been reported (Clausen, 1961). Microclimatic variation may limit hybridization in the natural environment by controlling flowering phenology (Byars et al., 2009). However, many of the taxa examined here are sympatric and there should be ample opportunity for hybridization at an evolutionary scale.
Previous divergence obscured by current hybridization
Species may have diverged in the past and then begun to hybridize recently, so that the genetic result of previous divergence is obscured by current hybridization. We consider this scenario unlikely in the case of the Australian alpine Poa. The jml test detected no instances of haplotype sharing between P. gunnii and other Australian alpine taxa in nuclear regions that contradicted the overall species tree relationships. The only cases where the minimum interindividual distance was smaller than the species tree posterior predictive distribution were in the chloroplast regions, which probably reflects slower chloroplast vs. nuclear genome evolution that was not completely accounted for with the mutation rate and heredity scalar parameters. This was not significant after FDR correction. Additionally, despite haplotype sharing at many loci (Supplementary Data Figs S2–S9) between P. gunnii and other Australian taxa, multiple pairs of individuals exhibited haplotype sharing in each case, suggesting a scenario of widespread genetic similarity rather than occasional hybridization. Because the distinct taxon P. gunnii may have diverged as recently as half a million years ago (Fig. 1), incomplete lineage sorting is a more likely explanation for shared haplotypes.
Distinctiveness of P. gunnii
Although most clades were not resolved, P. gunnii formed distinct clades in the CDO504 copy A, CDO504 copy B, DMC copy B and waxy copy B trees (Supplementary Data Figs S3, S4, S6, S9)], and was strongly supported as sister to the rest of the Australian alpine Poa investigated in the species tree (pp = 1; Fig. 1). Poa gunnii has been suggested to be closely related to New Zealand Poa species, especially P. colensoi (Connor and Edgar, 1986), although in its original description it was considered similar to P. costiniana (Vickery, 1970). Given the significant species differentiation observed in this study, and the previous reports that Australasian Poa share a common ancester with some Eurasian/American species (Patterson et al., 2005; Gillespie et al., 2009; Griffin et al., 2011), we suggest that P. gunnii is clearly a distinct species and may have reached Australia via a separate long-distance dispersal event. Molecular data provide evidence for common dispersal in both directions between Australia and New Zealand (Cook and Crisp, 2005; Meudt and Simpson, 2006; Meudt and Bayly, 2008; Pirie et al., 2010). Long-distance dispersal from even further afield has also occurred many times (Mummenhoff and Franze, 2007; Inda et al., 2008), including in the alpine daisy Abrotanella which shows both morphological and genetic evidence of long-distance dispersal between Australasia and South America (Wagstaff et al., 2006; Swenson et al., 2012). More extensive taxon sampling of lowland Australian Poa and congeners in New Zealand, Eurasia and America would reveal the geographic origin of the Australian Poa group and the number and dynamics of distinct introductions.
Tasmania–mainland differentiation
As well as the distinctiveness of the Tasmanian species P. gunnii revealed by the multiple gene species tree analysis, we detected mainland–Tasmanian differentiation within distinct taxa. The dominant pattern in both the PCA and the sPCA analyses was differentiation between Tasmanian and mainland samples (Fig. 2C), despite the fact that the microsatellite data set excluded the distinct Tasmanian taxon P. gunnii. This was also reported previously, based on spatial genetic autocorrelation analysis of a sub-set of the gene regions examined here (Griffin et al., 2011). This result is somewhat surprising given that Tasmania was connected to the mainland a mere 17 000 years ago (Lambeck and Chappell, 2001), and that many taxa show no significant differentiation across Bass Strait (Chapple et al., 2005; Schultz et al., 2009; Coleman et al., 2010).
Ongoing gene flow with some geographic structure on the mainland
Within the mainland samples, the sPCA method detected several regions of weak spatial–genetic clustering (Fig. 2E). The northern-most region, the Australian Capital Territory (ACT) Tablelands, was distinct on both Axes 2 and 3, though these samples were not strongly clustered. The central alpine Mt Buffalo samples showed similarity, and the Mt Buller/Mt Howitt/Mt Baw Baw region was also somewhat distinctive. A previous small-scale study in P. fawcettiae detected low but significant differentiation between Mt Hotham and Mt Kosciuszko populations (James et al., 1997). However, in our study, the high-elevation regions of the Bogong High Plains, nearby Mt Hotham, and more northerly Mt Kosciuszko, were not distinguished from each other, nor from the westerly lower elevation Lake Mountain sample. For alpine-restricted species in Australia and elsewhere, the alpine zone has been considered a chain of ‘sky islands’ – with significant population differentiation observable, for example, in the Australian lizards Cyclodomorphus praealtus (Koumoundouros et al., 2009) and Egernia species (Chapple et al., 2005), and the endangered mountain pygmy possum (Mitrovski et al., 2007), often explained by lack of suitable habitat between alpine areas. This is clearly not the case in the alpine Poa. We suggest that south-east Australian alpine Poa remains genetically well connected across its range by wind dispersal of pollen, and perhaps seeds as well. Lowland Poa species may also contribute to genetic connectivity between alpine populations. The prevailing westerly wind direction may explain the slight genetic differentiation of the ACT (northern) samples from the rest, and may also explain the Tasmanian–mainland (south–north) differentiation.
Conclusions
Current taxonomy of Australian alpine Poa was not supported by the multiple gene analysis. Poa gunnii may represent a distinct species from the rest of the alpine Poa, but the others were not genetically delimited as species. Perhaps, as Gillespie et al. (2009) suggest, these taxa are all just ‘variations of the widespread P. labillardieri–P. sieberiana complex’, despite the morphological (Walsh, 1994) and ecological (Griffin and Hoffmann, 2012) distinction between them. Conventional molecular taxonomy assumes that taxa in the process of speciation are replicably distinguishable at the genetic level. However, we still lack an understanding of where most taxon pairs lie on the scale from ‘direct selection’ to ‘post-speciation divergence’ (Pinho and Hey, 2010; Feder et al., 2012) and hence where it is especially important to support molecular taxonomy by multiple lines of evidence from ecological, physiological and traditional taxonomic studies. The Australian alpine Poa species are likely to show significant differentiation only at loci under selection generating morphological differences among the species, as well as regions linked to those loci, especially because they appear to have diversified only in the last 0·5–1·2 MY. Further differentiation may remain obscured by ongoing gene flow.
SUPPLEMENTARY DATA
ACKNOWLEDGEMENTS
We thank the staff of the Melbourne University Herbarium (MELU), the National Herbarium of Victoria (MEL) and the National Herbarium, Canberra (CANB) for permission to sample Poa specimens. Thanks also to Joseph Heled and Simon Joly for assistance with software, and to Christian Parisod and two anonymous reviewers for helpful comments on the manuscript. This work was supported by an Australian Research Council Linkage Grant with financial support from the Department of Sustainability and Environment and Parks Victoria. It was also supported by a Holsworth Wildlife Research Fellowship to P.G., an Australian Research Council Laureate Fellowship to A.H., and TERN funding through the Long Term Ecological Research Network.
LITERATURE CITED
- Albertini E, Porceddu A, Marconi G, Barcacia G, Pallottini L, Falcinelli M. Microsatellite-AFLP for genetic mapping of complex polyploids. Genome. 2003;46:824–832. doi: 10.1139/g03-058. [DOI] [PubMed] [Google Scholar]
- Anton A, Connor H. Floral biology and reproduction in Poa (Poeae: Gramineae) Australian Journal of Botany. 1995;43:577–599. [Google Scholar]
- Ball M, Egerton J, Leuning R, Cunningham R, Dunne P. Microclimate above grass adversely affects spring growth of seedling snow gum (Eucalyptus pauciflora) Plant, Cell and Environment. 1997;20:155–166. [Google Scholar]
- Barker N, Galley C, Verboom G, Mafa P, Gilbert M, Linder H. The phylogeny of the austral grass subfamily Danthonioideae: evidence from multiple data sets. Plant Systematics and Evolution. 2007;264:135–156. [Google Scholar]
- Baumel A, Ainouche M, Bayer R, Ainouche A, Misset M. Molecular phylogeny of hybridizing species from the genus Spartina Schreb. (Poaceae) Molecular Phylogenetics and Evolution. 2002;22:303–314. doi: 10.1006/mpev.2001.1064. [DOI] [PubMed] [Google Scholar]
- Benjamini Y, Hochberg Y. Controlling the False Discovery Rate: a practical and powerful approach to multiple testing. Journal of the Royal Statistical Society. Series B (Methodological) 1995;57:289–300. [Google Scholar]
- Bezault E, Mwaiko S, Seehausen O. Population genomic tests of models of adaptive radiation in Lake Victoria region cichlid fish. Evolution. 2011;65:3381–3397. doi: 10.1111/j.1558-5646.2011.01417.x. [DOI] [PubMed] [Google Scholar]
- Biss P, Freeland J, Silvertown J, McConway K, Lutman P. Successful amplification of rice chloroplast microsatellites from century-old grass samples from the Park Grass Experiment. Plant Molecular Biology Reporter. 2003;21:249–257. [Google Scholar]
- Bor N. The genus Poa L. in India. Part I. Journal of the Bombay Natural History Society. 1952;50:787–838. [Google Scholar]
- Brysting A, Holst-Jensen A, Leitch I. Genomic origin and organization of the hybrid Poa jemtlandica (Poaceae) verified by genomic in situ hybridization and chloroplast DNA sequences. Annals of Botany. 2000;85:439–445. [Google Scholar]
- Byars S, Hoffmann A. Lack of strong local adaptation in the alpine forb Craspedia lamicola in southeastern Australia. International Journal of Plant Science. 2009;170:906–917. [Google Scholar]
- Byars S, Papst W, Hoffmann A. Local adaptation and cogradient selection in the alpine plant, Poa hiemata, along a narrow altitudinal gradient. Evolution. 2007;61:2925–2941. doi: 10.1111/j.1558-5646.2007.00248.x. [DOI] [PubMed] [Google Scholar]
- Byars S, Parsons Y, Hoffmann A. Effect of altitude on the genetic structure of an Alpine grass, Poa hiemata. Annals of Botany. 2009;103:885–899. doi: 10.1093/aob/mcp018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapple D, Keogh J, Hutchinson M. Substantial genetic substructuring in southeastern and alpine Australia revealed by molecular phylogeography of the Egernia whitii (Lacertilia: Scincidae) species group. Molecular Ecology. 2005;14:1279–1292. doi: 10.1111/j.1365-294X.2005.02463.x. [DOI] [PubMed] [Google Scholar]
- Clausen J. Introgression facilitated by apomixis in polyploid Poas. Euphytica. 1961;10:87–94. [Google Scholar]
- Clifford H, Simon B. The biogeography of Australian grasses. In: Keast A, editor. Ecological biogeography of Australia. The Hague: Dr. W. Junk bv; 1981. pp. 539–554. [Google Scholar]
- Coleman R, Pettigrove V, Raadik T, Hoffmann A, Miller A, Carew M. Microsatellite markers and mtDNA indicate two distinct groups in dwarf galaxias, Galaxiella pusilla (Mack) (Pisces: Galaxiidae), a threatened freshwater fish from south-eastern Australia. Conservation Genetics. 2010;11:1911–1928. [Google Scholar]
- Connor H, Edgar E. Australasian alpine grasses: diversification and specialization. In: Barlow B, editor. Flora and fauna of alpine Australasia: ages and origins. Melbourne: CSIRO; 1986. pp. 413–434. [Google Scholar]
- Cook L, Crisp M. Directional asymmetry of long-distance dispersal and colonization could mislead reconstructions of biogeography. Journal of Biogeography. 2005;32:741–754. [Google Scholar]
- Cozzolino S, Cafasso D, Pellegrino G, Musacchio A, Widmer A. Genetic variation in time and space: the use of herbarium specimens to reconstruct patterns of genetic variation in the endangered orchid Anacamptis palustris. Conservation Genetics. 2007;8:629–639. [Google Scholar]
- Crisp M, Cook L, Steane D. Radiation of the Australian flora: what can comparisons of molecular phylogenies across multiple taxa tell us about the evolution of diversity in present-day communities? Philosophical Transactions of the Royal Society B: Biological Sciences. 2004;359:1551–1571. doi: 10.1098/rstb.2004.1528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darmency H, Gasquez J. Spontaneous hybridization of the putative ancestors of the allotetraploid Poa annua. New Phytologist. 1997;136:497–501. doi: 10.1046/j.1469-8137.1997.00772.x. [DOI] [PubMed] [Google Scholar]
- Degnan J, Rosenberg N. Gene tree discordance, phylogenetic inference and the multispecies coalescent. Trends in Ecology and Evolution. 2009;24:332–340. doi: 10.1016/j.tree.2009.01.009. [DOI] [PubMed] [Google Scholar]
- Dray S, Dufour A. The ade4 package. I. One-table methods. R News. 2004;4:5–10. [Google Scholar]
- Drummond A, Ho S, Rawlence N, Rambaut A. A rough guide to BEAST 1·4. 2007. Available at: http://beast.bio.ed.ac.uk/
- Drummond A, Suchard M, Xie D, Rambaut A. Bayesian phylogenetics with BEAUti and the BEAST 1·7. Molecular Biology and Evolution. 2012;29:1969–1973. doi: 10.1093/molbev/mss075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durham L. Soil erosion problems in the Snowy Mountains area. Journal of the Soil Conservation Service of New South Wales. 1956;12:121–131. [Google Scholar]
- Escobar JS, Scornavacca C, Cenci A, et al. Multigenic phylogeny and analysis of tree incongruences in Triticeae (Poaceae) BMC Evolutionary Biology. 2011;11:181. doi: 10.1186/1471-2148-11-181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Essi L, Longhi-Wagner H, Souza-Chies Td. Phylogenetic analysis of the Briza complex (Poaceae) Molecular Phylogenetics and Evolution. 2008;47:1018–1029. doi: 10.1016/j.ympev.2008.03.007. [DOI] [PubMed] [Google Scholar]
- Feder JL, Egan SP, Nosil P. The genomics of speciation-with-gene-flow. Trends in Genetics. 2012;28:342–350. doi: 10.1016/j.tig.2012.03.009. [DOI] [PubMed] [Google Scholar]
- Frankham R, Ballou J, Briscoe D. Introduction to conservation genetics. Cambridge: Cambridge University Press; 2002. [Google Scholar]
- Gallagher R, Hughes L, Leishman M. Phenological trends among Australian alpine species: using herbarium records to identify climate-change indicators. Australian Journal of Botany. 2009;57:1–9. [Google Scholar]
- Gillespie L, Soreng R. A phylogenetic analysis of the bluegrass genus Poa based on cpDNA restriction site data. Systematic Botany. 2005;30:84–105. [Google Scholar]
- Gillespie L, Soreng R, Bul R, Jacobs S, Refulio-Rodriguez N. Phylogenetic relationships in subtribe Poinae (Poaceae, Poeae) based on nuclear ITS and plastid trnT-trnL-trnF sequences. Botany. 2008;86:938–967. [Google Scholar]
- Gillespie L, Soreng R, Jacobs S. Phylogenetic relationships of Australian Poa (Poaceae: Poinae), including molecular evidence for two new genera, Saxipoa and Sylvipoa. Australian Systematic Botany. 2009;22:413–436. [Google Scholar]
- Griffin P, Hoffmann A. Mortality of Australian alpine grasses (Poa spp.) after drought: species differences and ecological patterns. Journal of Plant Ecology. 2012;5:121–133. [Google Scholar]
- Griffin P, Robin C, Hoffmann A. A next-generation sequencing method for overcoming the multiple gene copy problem in polyploid phylogenetics, applied to Poa grasses. BMC Biology. 2011;9:19. doi: 10.1186/1741-7007-9-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grun P. Cytogenetic studies of Poa. I. Chromosome numbers and morphology of interspecific hybrids. American Journal of Botany. 1954;41:671–678. [Google Scholar]
- Hand ML, Cogan NO, Stewart AV, Forster JW. Evolutionary history of tall fescue morphotypes inferred from molecular phylogenetics of the Lolium–Festuca species complex. BMC Evolutionary Biology. 2010;10:303. doi: 10.1186/1471-2148-10-303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartley W. Studies on the origin, evolution, and distribution of the Gramineae. IV. The genus Poa L. Australian Journal of Botany. 1961;9:152–161. [Google Scholar]
- Heled J, Drummond A. Bayesian inference of species trees from multilocus data. Molecular Biology and Evolution. 2010;27:570–580. doi: 10.1093/molbev/msp274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hey J, Pinho C. Population genetics and objectivity in species diagnosis. Evolution. 2012;66:1413–1429. doi: 10.1111/j.1558-5646.2011.01542.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann M, Schneider J, Hase P, Röser M. Rapid and recent world-wide diversification of bluegrasses (Poa, Poaceae) and related genera. PLoS One. 2013;8 doi: 10.1371/journal.pone.0060061. e60061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudson RR, Turelli M. Stochasticity overrules the ‘three-times rule’: genetic drift, genetic draft, and coalescence times for nuclear loci versus mitochondrial DNA. Evolution. 2003;57:182–190. doi: 10.1111/j.0014-3820.2003.tb00229.x. [DOI] [PubMed] [Google Scholar]
- Hughes C, Eastwood R, Bailey C. From famine to feast? Selecting nuclear DNA sequence loci for plant species-level phylogeny reconstruction. Philosophical Transactions of the Royal Society B: Biological Sciences. 2006;361:211–225. doi: 10.1098/rstb.2005.1735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inda L, Segerra-Moragues J, Müller J, Peterson P, Catalán P. Dated historical biogeography of the temperate Loliinae (Poaceae, Pooideae) grasses in the northern and southern hemispheres. Molecular Phylogenetics and Evolution. 2008;46:932–957. doi: 10.1016/j.ympev.2007.11.022. [DOI] [PubMed] [Google Scholar]
- Ingram A, Doyle J. The origin and evolution of Eragrostis tef (Poaceae) and related polyploids: evidence from nuclear waxy and plastid rps16. American Journal of Botany. 2003;90:116–122. doi: 10.3732/ajb.90.1.116. [DOI] [PubMed] [Google Scholar]
- Jakob S, Blattner F. A chloroplast genealogy of Hordeum (Poaceae): long-term persisting haplotypes, incomplete lineage sorting, regional extinction, and the consequences for phylogenetic inference. Molecular Biology and Evolution. 2006;23:1602–1612. doi: 10.1093/molbev/msl018. [DOI] [PubMed] [Google Scholar]
- James E, Isenegger D, Papst W. A study of the genetic variation of selected populations of Poa fawcettiae in the Mount Kosciusko area. 1997. National Parks and Wildlife Service of New South Wales. [Google Scholar]
- Joly S. JML: testing hybridization from species trees. Molecular Ecology Resources. 2012;12:179–184. doi: 10.1111/j.1755-0998.2011.03065.x. [DOI] [PubMed] [Google Scholar]
- Jombart T, Devillard S, Dufour A, Pontier D. Revealing cryptic spatial patterns in genetic variability by a new multivariate method. Heredity. 2008;101:92–103. doi: 10.1038/hdy.2008.34. [DOI] [PubMed] [Google Scholar]
- Kelley A, Johnson P, Waldron B, Peel M. A survey of apomixis and ploidy levels among Poa L. (Poaceae) using flow cytometry. Crop Science. 2009;49:1395–1402. [Google Scholar]
- Kindiger B. Cross-species amplification of Lolium microsatellites in Poa ssp. Grassland Science. 2006;52:105–115. [Google Scholar]
- Knowles L, Carstens B. Delimiting species without monophyletic gene trees. Systematic Biology. 2007;56:887–895. doi: 10.1080/10635150701701091. [DOI] [PubMed] [Google Scholar]
- Koonjul P, Brandt W, Farrant J, Lindsey G. Inclusion of polyvinylpyrrolidone in the polymerase chain reaction reverses the inhibitory effects of polyphenolic contamination of RNA. Nucleic Acids Research. 1999;27:915–916. doi: 10.1093/nar/27.3.915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koumoundouros T, Sumner J, Clemann N, Stuart-Fox D. Current genetic isolation and fragmentation contrasts with historical connectivity in an alpine lizard (Cyclodomorphus praealtus) threatened by climate change. Biological Conservation. 2009;142:992–1002. [Google Scholar]
- Lambeck K, Chappell J. Sea level change through the last glacial cycle. Science. 2001;292:679–686. doi: 10.1126/science.1059549. [DOI] [PubMed] [Google Scholar]
- Lanfear R, Calcott B, Ho S, Guindon S. PartitionFinder: combined selection of partitioning schemes and substitution models for phylogenetic analyses. Molecular Biology and Evolution. 2012;29:1695–1701. doi: 10.1093/molbev/mss020. [DOI] [PubMed] [Google Scholar]
- Librado P, Rozas J. DnaSP v5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics. 2009;25:1451–1452. doi: 10.1093/bioinformatics/btp187. [DOI] [PubMed] [Google Scholar]
- Linder HP. Plant species radiations: where, when, why? Philosophical Transactions of the Royal Society B: Biological Sciences. 2008;363:3097–3105. doi: 10.1098/rstb.2008.0075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z, Chen Z, Pan J, et al. Phylogenetic relationships in Leymus (Poaceae: Triticeae) revealed by the nuclear ribosomal internal transcribed spacer and chloroplast trnL-F sequences. Molecular Phylogenetics and Evolution. 2008;46:278–289. doi: 10.1016/j.ympev.2007.10.009. [DOI] [PubMed] [Google Scholar]
- Maddison W, Knowles L. Inferring phylogeny despite incomplete lineage sorting. Systematic Biology. 2006;55:21–30. doi: 10.1080/10635150500354928. [DOI] [PubMed] [Google Scholar]
- Martin D, Posada D, Crandall K, Williamson C. A modified bootscan algorithm for automated identification of recombinant sequences and recombination breakpoints. AIDS Research and Human Retroviruses. 2005a;21:98–102. doi: 10.1089/aid.2005.21.98. [DOI] [PubMed] [Google Scholar]
- Martin D, Williamson C, Posada D. RDP2: recombination detection and analysis from sequence alignments. Bioinformatics. 2005b;21:260–262. doi: 10.1093/bioinformatics/bth490. [DOI] [PubMed] [Google Scholar]
- Mason-Gamer R, Weil C, Kellogg E. Granule-bound starch synthase: structure, function, and phylogenetic utility. Molecular Biology and Evolution. 1998;15:1658–1673. doi: 10.1093/oxfordjournals.molbev.a025893. [DOI] [PubMed] [Google Scholar]
- Mason-Gamer R, Burns M, Naum M. Phylogenetic relationships among Asian Elymus (Poaceae) allotetraploids: analyses of three nuclear gene trees. Molecular Phylogenetics and Evolution. 2010;54:10–22. doi: 10.1016/j.ympev.2009.10.002. [DOI] [PubMed] [Google Scholar]
- Maurer K, Gautschi B, Weyand A, Stöcklin J, Fischer M. Isolation and characterization of microsatellite DNA markers in the grass Poa alpina L. Molecular Ecology Notes. 2005;5:719–720. [Google Scholar]
- M'Baya J, Blacket MJ, Hoffmann AA. Genetic structure of Carex species from the Australian alpine region along elevation gradients: patterns of reproduction and gene flow. International Journal of Plant Sciences. 2013;174:189–199. [Google Scholar]
- Meudt H, Bayly M. Phylogeographic patterns in the Australasian genus Chionohebe (Veronica s.l., Plantaginaceae) based on AFLP and chloroplast DNA sequences. Molecular Phylogenetics and Evolution. 2008;47:319–338. doi: 10.1016/j.ympev.2007.12.019. [DOI] [PubMed] [Google Scholar]
- Meudt H, Simpson B. The biogeography of the austral, subalpine genus Ourisia (Plantaginaceae) based on molecular phylogenetic evidence: South American origin and dispersal to New Zealand and Tasmania. Biological Journal of the Linnean Society. 2006;87:479–513. [Google Scholar]
- Mitrovski P, Heinze D, Broome L, Hoffmann A, Weeks A. High levels of variation despite genetic fragmentation in populations of the endangered mountain pygmy-possum, Burramys parvus, in alpine Australia. Molecular Ecology. 2007;16:75–87. doi: 10.1111/j.1365-294X.2006.03125.x. [DOI] [PubMed] [Google Scholar]
- Mummenhoff K, Franze A. Gone with the bird: late Tertiary and Quaternary intercontinental long-distance dispersal and allopolyploidization in plants. Systematics and Biodiversity. 2007;5:255–260. [Google Scholar]
- Noble I. Interactions between tussock grass (Poa spp.) and Eucalyptus pauciflora seedlings near treeline in south-eastern Australia. Oecologia. 1980;45:350–353. doi: 10.1007/BF00540204. [DOI] [PubMed] [Google Scholar]
- Ossowski S, Schneeberger K, Lucas-Lledó J, et al. The rate and molecular spectrum of spontaneous mutations in Arabidopsis thaliana. Science. 2010;327:92–94. doi: 10.1126/science.1180677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patterson J, Larson S, Johnson P. Genome relationships in polyploid Poa pratensis and other Poa species inferred from phylogenetic analysis of nuclear and chloroplast DNA sequences. Genome. 2005;48:76–87. doi: 10.1139/g04-102. [DOI] [PubMed] [Google Scholar]
- Peakall R, Smouse P. GENALEX 6: genetic analysis in Excel. Population genetic software for teaching and research. Molecular Ecology Notes. 2006;6:288–295. doi: 10.1093/bioinformatics/bts460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen G, Seberg O. Molecular evolution and phylogenetic application of DMC1. Molecular Phylogenetics and Evolution. 2002;22:43–50. doi: 10.1006/mpev.2001.1011. [DOI] [PubMed] [Google Scholar]
- Pickering C, Good R, Green K. Potential effects of global warming on the biota of the Australian Alps. 2004. Australian Greenhouse Office, Australian Government.
- Pinho C, Hey J. Divergence with gene flow: models and data. Annual Review of Ecology, Evolution, and Systematics. 2010;41:215–230. [Google Scholar]
- Pirie M, Lloyd K, Lee W, Linder H. Diversification of Chionochloa (Poaceae) and biogeography of the New Zealand Southern Alps. Journal of Biogeography. 2010;37:379–392. [Google Scholar]
- Preston B, Jones R. Climate change impacts on Australia and the benefits of early action to reduce global greenhouse gas emissions. Melbourne, Victoria: Commonwealth Scientific and Industrial Research Organisation (CSIRO); 2006. [Google Scholar]
- R Development Core Team. Vienna: Austria; 2011. R: a language and environment for statistical computing v 2·13·1. [Google Scholar]
- Rambaut A, Drummond A. Tracer: MCMC Trace Analysis Tool. 2009. v1·5·0 ed.
- Rogers S, Bendich A. Extraction of total cellular DNA from plants, algae and fungi. In: Gelvin S, editor. Plant molecular biology manual. Dordrecht: Kluwer Academic Press; 1994. pp. 183–190. [Google Scholar]
- Rudmann-Maurer K, Weyand A, Fischer M, Stöcklin J. Microsatellite diversity of the agriculturally important alpine grass Poa alpina in relation to land use and natural environment. Annals of Botany. 2007;100:1249–1258. doi: 10.1093/aob/mcm203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savolainen V, Cuénoud P, Spichiger R, Martinez M, Crèvecoeur M, Manen J-F. The use of herbarium specimens in DNA phylogenetics: evaluation and improvement. Plant Systematics and Evolution. 1995;197:87–98. [Google Scholar]
- Schultz M, Smith S, Horwitz P, Richardson A, Crandall K, Austin C. Evolution underground: a molecular phylogenetic investigation of Australian burrowing freshwater crayfish (Decapoda: Parastacidae) with particular focus on Engaeus Erichson. Molecular Phylogenetics and Evolution. 2009;50:580–598. doi: 10.1016/j.ympev.2008.11.025. [DOI] [PubMed] [Google Scholar]
- Small R, Cronn R, Wendel J. Use of nuclear genes for phylogeny reconstruction in plants. Australian Systematic Botany. 2004;17:145–170. [Google Scholar]
- Soreng R. Chloroplast-DNA phylogenetics and biogeography in a reticulating group: study in Poa (Poaceae) American Journal of Botany. 1990;77:1383–1400. [Google Scholar]
- Soreng R, Bull R, Gillespie L. Phylogeny and reticulation in Poa based on plastid trnTLF and nrITS sequences with attention to diploids. In: Seberg O, Peterse G, Barfod A, Davis J, editors. Diversity, phylogeny, and evolution in the monocotyledons. Aarhus, Denmark: Aarhus University Press; 2010. pp. 619–643. [Google Scholar]
- Souto DF, Catalano S, Tosto D, et al. Phylogenetic relationships of Deschampsia antarctica (Poaceae): insights from nuclear ribosomal ITS. Plant Systematics and Evolution. 2006;261:1–9. [Google Scholar]
- Steffen W, Burbidge A, Hughes L, et al. Melbourne: CSIRO Publishing; 2009. Australia's biodiversity and climate change: a strategic assessment of the vulnerability of Australia's biodiversity to climate change. A report to the Natural Resource Management Ministerial Council commissioned by the Australian Government. [Google Scholar]
- Swenson U, Nylinder S, Wagstaff SJ. Are Asteraceae 1·5 billion years old? A reply to Heads. Systematic Biology. 2012;61:522–532. doi: 10.1093/sysbio/syr121. [DOI] [PubMed] [Google Scholar]
- Syme AE. Diversification rates in the Australasian endemic grass Austrostipa: 15 million years of constant evolution. Plant Systematics and Evolution. 2011;298:221–227. [Google Scholar]
- Tajima F. Evolutionary relationship of DNA sequences in finite populations. Genetics. 1983;105:437–460. doi: 10.1093/genetics/105.2.437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Molecular Biology and Evolution. 2011;28:2731–2739. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Triplett J, Clark L. Phylogeny of the temperate bamboos (Poaceae: Bambusoideae: Bambuseae) with an emphasis on Arundinaria and allies. Systematic Botany. 2010;35:102–120. [Google Scholar]
- Vickery J. A taxonomic study of the genus Poa L. in Australia. Contributions from the N.S.W. National Herbarium. 1970;4:145–243. [Google Scholar]
- Wagstaff SJ, Breitwieser I, Swenson U. Origin and relationships of the austral genus Abrotanella (Asteraceae) inferred from DNA sequences. Taxon. 2006;55:95–106. [Google Scholar]
- Walsh N. Poaceae. In: Walsh N, Entwisle T, editors. Flora of Victoria. Vol. 2. Melbourne: Inkata Press; 1994. pp. 356–625. [Google Scholar]
- Walsh N, McDougall K. Progress in the recovery of the flora of treeless subalpine vegetation in Kosciuszko National Park after the 2003 fires. Cunninghamia. 2005;8:439–452. [Google Scholar]
- Walsh N, Weiller C, Thompson I Flora Australia Committee. Flora of Australia. Vol. 44A. Poaceae 2. Melbourne: ABRS/CSIRO; 2009. Poa L; pp. 301–338. [Google Scholar]
- Williams R, Wahren C-H, Tolsma A, et al. Large fires in Australian alpine landscapes: their part in the historical fire regime and their impacts on alpine biodiversity. International Journal of Wildland Fire. 2008;17:793–808. [Google Scholar]
- Wu CI. The genic view of the process of speciation. Journal of Evolutionary Biology. 2001;14:851–865. [Google Scholar]
- Xu X, Ke W, Yu X, Wen J, Ge S. A preliminary study on population genetic structure and phylogeography of the wild and cultivated Zizania latifolia (Poaceae) based on AdhIa sequences. Theoretical and Applied Genetics. 2008;116:835–843. doi: 10.1007/s00122-008-0717-3. [DOI] [PubMed] [Google Scholar]
- Yang Z, Rannala B. Bayesian species delimitation using multilocus sequence data. Proceedings of the National Academy of Sciences, USA. 2010;107:9264–9269. doi: 10.1073/pnas.0913022107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng C-X, Zhang Y-X, Triplett J, Yang J-B, Li D-Z. Large multi-locus plastid phylogeny of the tribe Arundinarieae (Poaceae: Bambusoideae) reveals ten major lineages and low rate of molecular divergence. Molecular Phylogenetics and Evolution. 2010;56:821–839. doi: 10.1016/j.ympev.2010.03.041. [DOI] [PubMed] [Google Scholar]
- Zhang C, Fan X, Yu H, et al. Phylogenetic relationships among the species of Elymus sensu lato in Triticeae (Poaceae) based on nuclear rDNA ITS sequences. Russian Journal of Genetics. 2009;45:696–706. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.