EB1 enables spindle microtubules to regulate the phosphorylation of kinetochores through recruitment of Aurora B kinase.
Abstract
The Aurora B kinase coordinates kinetochore–microtubule attachments with spindle checkpoint signaling on each mitotic chromosome. We find that EB1, a microtubule plus end–tracking protein, is required to enrich Aurora B at inner centromeres in a microtubule-dependent manner. This regulates phosphorylation of both kinetochore and chromatin substrates. EB1 regulates the histone phosphorylation marks (histone H2A phospho-Thr120 and histone H3 phospho-Thr3) that localize Aurora B. The chromosomal passenger complex containing Aurora B can be found on a subset of spindle microtubules that exist near prometaphase kinetochores, known as preformed K-fibers (kinetochore fibers). Our data suggest that EB1 enables the spindle microtubules to regulate the phosphorylation of kinetochores through recruitment of the Aurora B kinase.
Introduction
The chromosomal passenger complex (CPC) contains a catalytic subunit Aurora B kinase and three regulatory proteins, inner centromere protein (INCENP), Survivin, and Borealin/Dasra (Borealin). It is a central regulator of mitotic events (Ruchaud et al., 2007). It has distinct roles in different stages of mitosis. In prophase, Aurora B phosphorylation is found along the length of mitotic chromosomes where it releases cohesion and phosphorylates histone H3 on Ser10 to release the heterochromatin protein 1 (Hsu et al., 2000; Losada et al., 2002; Fischle et al., 2005; Hirota et al., 2005). From late in prophase until the onset of anaphase, Aurora B concentrates at inner centromeres, where it regulates inner centromere substrates, kinetochore microtubule attachments, and the spindle checkpoint signal (Kallio et al., 2002; Lampson et al., 2004; Lan et al., 2004; Cimini et al., 2006; Knowlton et al., 2006; Ruchaud et al., 2007; Liu et al., 2009). Although it is well accepted that the movement of the CPC to distinct locations is critical for its ability to carry out multiple functions in different stages of mitosis (Terada, 2001; Wheatley et al., 2001), it is not known how Aurora B kinase is differentially regulated during mitotic progression.
Aurora B kinase regulates spindle checkpoint signaling and the release of improper kinetochore attachments (Kallio et al., 2002; Lan et al., 2004; Cimini et al., 2006; Knowlton et al., 2006; Liu et al., 2009). These events must be measured independently on each chromosome. How Aurora B kinase can integrate local information about microtubule attachment status to regulate these chromosome autonomous events is a critical unanswered question.
The concentration of the CPC at inner centromeres is mediated by posttranslational modifications of histones. The survivin subunit binds the histone H3 tails that are phosphorylated at Thr3 by haspin kinase (Kelly et al., 2010; Wang et al., 2010; Yamagishi et al., 2010; Jeyaprakash et al., 2011; Du et al., 2012; Niedzialkowska et al., 2012). The CPC also interacts with Shugoshin (Sgo1), which is recruited to histone H2A, which is phosphorylated on Thr120 by Bub1 (Yamagishi et al., 2010). It is not clear whether the presence of these histone marks at inner centromeres is sufficient for CPC localization, and there are many reports of additional requirements, including survivin phosphorylation (Wheatley et al., 2004; Tsukahara et al., 2010; Chu et al., 2011) and regulation by microtubules, TD-60, exportin binding, and nuclear pore proteins (Mollinari et al., 2003; Knauer et al., 2006; Rosasco-Nitcher et al., 2008; Platani et al., 2009; Tseng et al., 2010).
EB1 is a microtubule plus end–tracking protein that interacts with growing tips of microtubules (Morrison et al., 1998), and its yeast homologue Bim1p forms a stable association with Ipl-1/Aurora B to regulate anaphase spindle morphology (Zimniak et al., 2009). Furthermore, EB1 has been shown to coimmunoprecipitate with Aurora B and regulate its activity by inhibiting PP2A-mediated dephosphorylation (Sun et al., 2008). However, the biological significance of this interaction remains unclear.
The microtubule-associated protein TPX2 and microtubules have been shown to activate the related kinase, Aurora A, in mitosis. High RanGTP on condensed chromosomes releases TPX2 from an importin-bound inactive state, which then binds Aurora A and protects the T loop from dephosphorylation by PP1 in a microtubule-dependent manner (Bayliss et al., 2003; Tsai et al., 2003). Microtubules also regulate Aurora B activity (Rosasco-Nitcher et al., 2008; Tseng et al., 2010). The CPC binds microtubules in two distinct regions. INCENP has a central coiled-coil region that can bind microtubules and regulate spindle checkpoint signaling (Mackay et al., 1998; Tseng et al., 2010), and Aurora B bound to a C-terminal region of INCENP distinct from this microtubule-binding domain (Rosasco-Nitcher et al., 2008). Although Aurora B kinase can autoactivate in vitro, it requires some active kinase to initiate this reaction (Kelly et al., 2007; Rosasco-Nitcher et al., 2008). Fully inactive Xenopus laevis Aurora B can be activated in vitro by microtubules and a cofactor, TD60/RCC2 (Rosasco-Nitcher et al., 2008), and spindle formation in Xenopus requires both chromatin and microtubule-binding activities (Tseng et al., 2010). Moreover, anaphase cells treated with nocodazole for 8 min are not phosphorylated on an activating site on INCENP (Fuller et al., 2008). However, microtubules are absent in prophase nuclei that have histone H3 phosphorylated on Ser10. It is also unclear whether the CPC is regulated by microtubules in prometaphase. The CPC proteins are primarily localized to the inner centromeres at this time, although pools on microtubules have been reported (Tseng et al., 2010). Moreover, Aurora B kinase activity can be measured in mitotic cells arrested by the spindle poison nocodazole.
Here, we demonstrate that the CPC at the inner centromere is substantially enriched by microtubules near the kinetochore by a novel pathway that requires the EB1 plus end–tracking protein. There is a similar EB1/microtubule-dependent increase in phosphorylation of Aurora B substrates at kinetochores and chromosome arms. The regulation by EB1/microtubules is upstream or interdependent of the histone phosphorylation pathways that localize the CPC. We show that microtubules in preformed K-fiber (kinetochore fiber; pre–K-fiber) bundles contain Aurora B and can enrich Aurora B at inner centromeres. These findings establish a new prometaphase pathway regulating Aurora B localization that requires EB1/microtubules and provides mechanisms for the spindle to regulate CPC activity and kinetochores.
Results
EB1 regulates histone phosphorylations to recruit the CPC to centromeres and phosphorylate kinetochore substrates
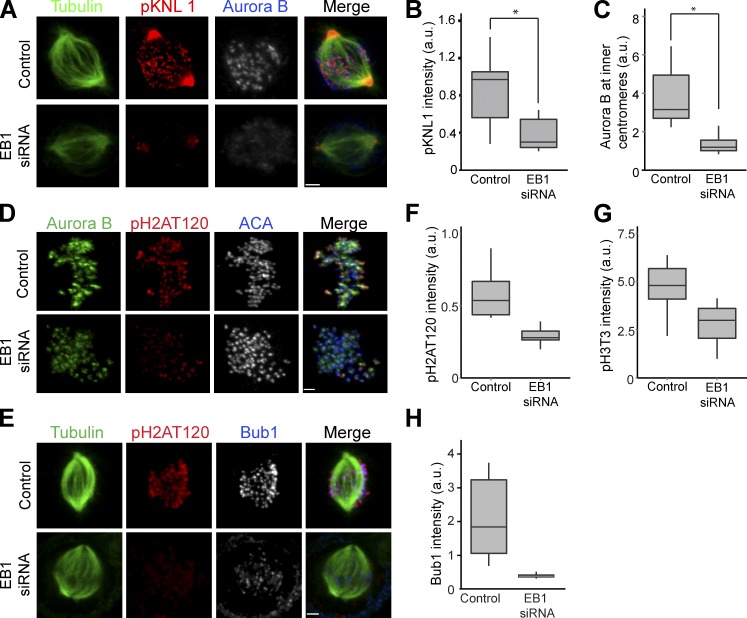
We asked whether Aurora B phosphorylation of kinetochore substrates in prometaphase required EB1. HeLa cells were depleted of EB1 using either a coding sequence–targeted siRNA (EB1 siRNA) or a combination of two EB1 siRNAs targeted to 3′-UTR (siEB13UTR; Fig. S1 A), and KNL1 phosphorylation by Aurora B was measured using a phospho-KNL1 antibody. The antibody recognized phospho-KNL1 at kinetochores but also cross-reacted with a centrosome protein as previously shown (Welburn et al., 2010). We specifically quantified kinetochores from early prometaphase cells because metaphase-aligned chromosomes show reduced KNL1 phosphorylation (Welburn et al., 2010). It was significantly reduced in cells depleted of EB1 with either set of siRNAs (Fig. 1, A and B; and Fig. S2 A). KNL1 protein levels were not reduced in EB1-depleted HeLa cells (Fig. 2 E). Surprisingly, inner centromeric Aurora B levels were also reduced in EB1-depleted prometaphase cells (Fig. 1, A and C; and Fig. S2, A and B). There was a similar drop in two other CPC proteins, Borealin/Dasra (Borealin) and INCENP, at the inner centromeres, suggesting that EB1 is required to recruit the whole CPC complex (Fig. S2, D–F). Aurora B, INCENP, and Survivin protein levels in EB1-depleted cells were similar to control HeLa cells, so the depletion from centromeres was not caused by destabilization of CPC proteins (Fig. S1 E).
Figure 1.
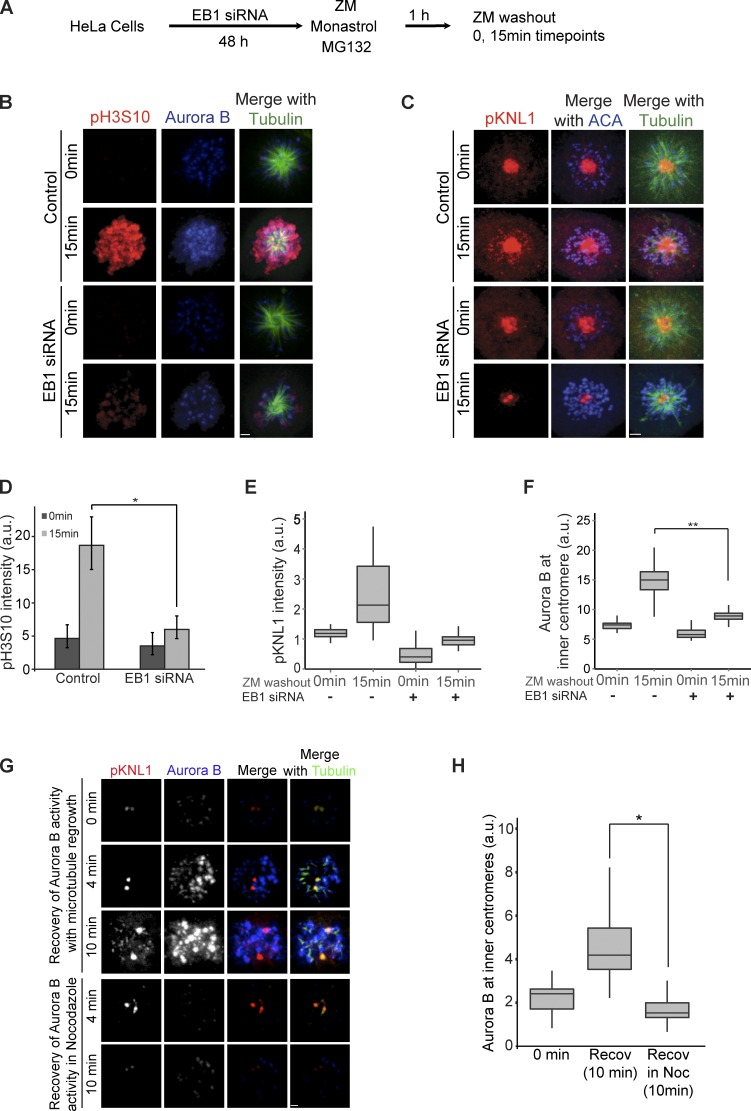
EB1 localizes Aurora B to centromeres to phosphorylate kinetochores. (A) HeLa cells depleted of EB1 were immunostained with antiphospho-KNL1(Ser60) (pKNL1) and Aurora B antibodies. Bar, 2 µm. (B) Quantification of immunostaining intensities of pKNL1 shown in A. *, P = 2.0 × 10−127. (C) Centromeric Aurora B levels measured in control and EB1-depleted cells (n > 300 centromeres). *, P = 1.2 × 10−105. (D) EB1 depletion reduces Bub1-mediated phosphorylation of histone H2A (pH2AT120). Bar, 1.6 µm. (E) EB1 depletion reduces Bub1 at kinetochores. Bar, 2.2 µm. (F–H) Quantification of pH2AT120 (F), phospho–histone H3Thr3 (pH3T3; G), and Bub1 (H) levels in control and EB1-depleted HeLa cells (see Fig. S1 B for pH3T3 staining examples). The height of the boxes represents the interquartile range (IQR). The central horizontal lines depict the median. The top whiskers represent the 75th percentile + 1.5× IQR, and the bottom whiskers represent the 25th percentile − 1.5× IQR. ACA, anticentromere antigen; a.u., arbitrary unit.
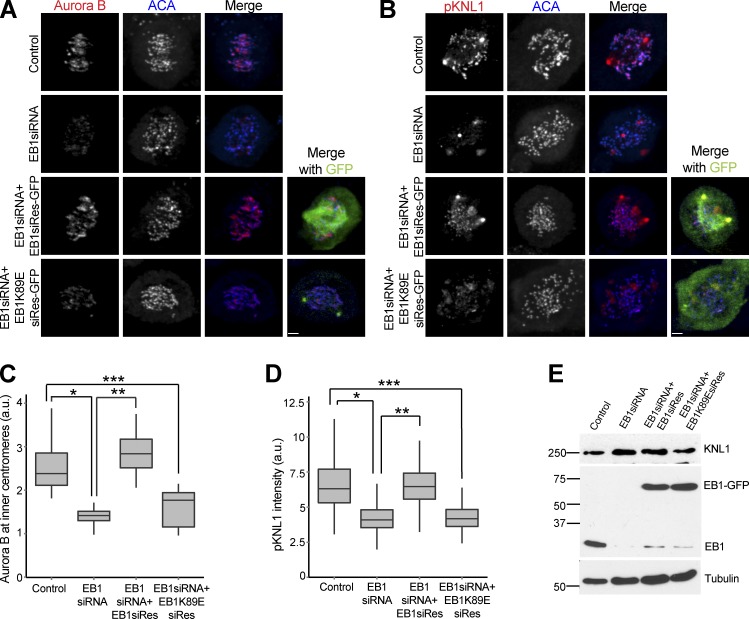
Figure 2.
EB1 localizes Aurora B at the centromere in a microtubule-dependent manner. (A) Loss of centromeric Aurora B is rescued by expressing siRNA-resistant EB1 (EB1siRes) but not the EB1K89E mutant (EB1K89EsiRes). Bar, 2.2 µm. (B) Loss of KNL1(Ser60) phosphorylation was rescued by expressing siRNA-resistant EB1 but not the EB1K89E mutant. Bar, 2.3 µm. (C) Quantification of immunostaining Aurora B intensities in A. *, P = 3.78 × 10−69; **, P = 2.54 × 10−75; ***, P = 1.33 × 10−35. (D) Quantification of immunostaining phospho-KNL1(S60) intensities in B. *, P = 5.23 × 10−42; **, P = 1.34 × 10−42; ***, P = 1.02 × 10−39. (E) Western blot of HeLa lysates showing endogenous and GFP-tagged EB1 and KNL1 levels (in kilodaltons). (Additional rescue experiments are shown in Fig. S2, A–C.) The height of the boxes represents the IQR. The central horizontal lines depict the median. The top whiskers represent the 75th percentile + 1.5× IQR, and the bottom whiskers represent the 25th percentile − 1.5× IQR. a.u., arbitrary unit.
EB1 depletion also reduced both of the histone marks that recruit Aurora B to inner centromeres. Cells depleted of EB1 had reduced levels of histone H2A phospho-Thr120 (pH2AT120 in the figures) and histoneH3 phospho-Thr3 (pH3T3 in the figures) as measured by immunofluorescence with phosphospecific antibodies (Fig. 1, D, F, and G; and Fig. S1 B). Bub1 kinase levels were also reduced at the kinetochores of EB1-depleted cells (Fig. 1, E and H). HEK293T cells also showed reduced phospho-KNL1, Aurora B, Bub1, and phospho–histone H2A Thr120 levels after EB1 depletion (Fig. S1, C and D). We conclude that EB1 is required to generate the phosphohistone marks that recruit the CPC to phosphorylate kinetochores.
EB1 localizes Aurora B to the centromeres in a microtubule-dependent manner
We rescued EB1 depletion phenotypes by multiple methods to ensure that they were not caused by off-target effects. Both the reduction of Aurora B at inner centromeres and the reduced activity at kinetochores were rescued by transfecting a plasmid expressing EB1 mutated to escape siRNA targeting (Fig. 2, A–E, EB1siRes). Moreover, the drop in Aurora B levels by transfection of a 3′-UTR–targeted siRNAs was rescued in a HeLa cell line transfected with or engineered with an integrated copy of EB1–localization and affinity purification (LAP) that lacked the 3′-UTR (Fig. S2, A and B). The protein levels of Aurora B and Bub1 were similar to control cell lysates by Western blotting (Fig. S2 C).
EB1 is a plus end–tracking protein, and most of its activities have been associated with microtubules (Morrison et al., 1998). We rescued EB1 depletion with a plasmid expressing a microtubule-binding mutant of EB1 (EB1K89EsiRes; Hayashi and Ikura, 2003) that was similarly modified to escape siRNA targeting to determine whether EB1 used a nonmicrotubule-associated activity to localize the CPC. The EB1K89E mutant failed to rescue the reduction in Aurora B levels at the centromeres (Fig. 2, A and C) and phosphorylation of KNL1 in EB1-depleted cells (Fig. 2, B and D). We conclude that EB1 enriches the CPC at inner centromeres in a microtubule-dependent manner.
Microtubules stimulate the recruitment of Aurora B to inner centromeres
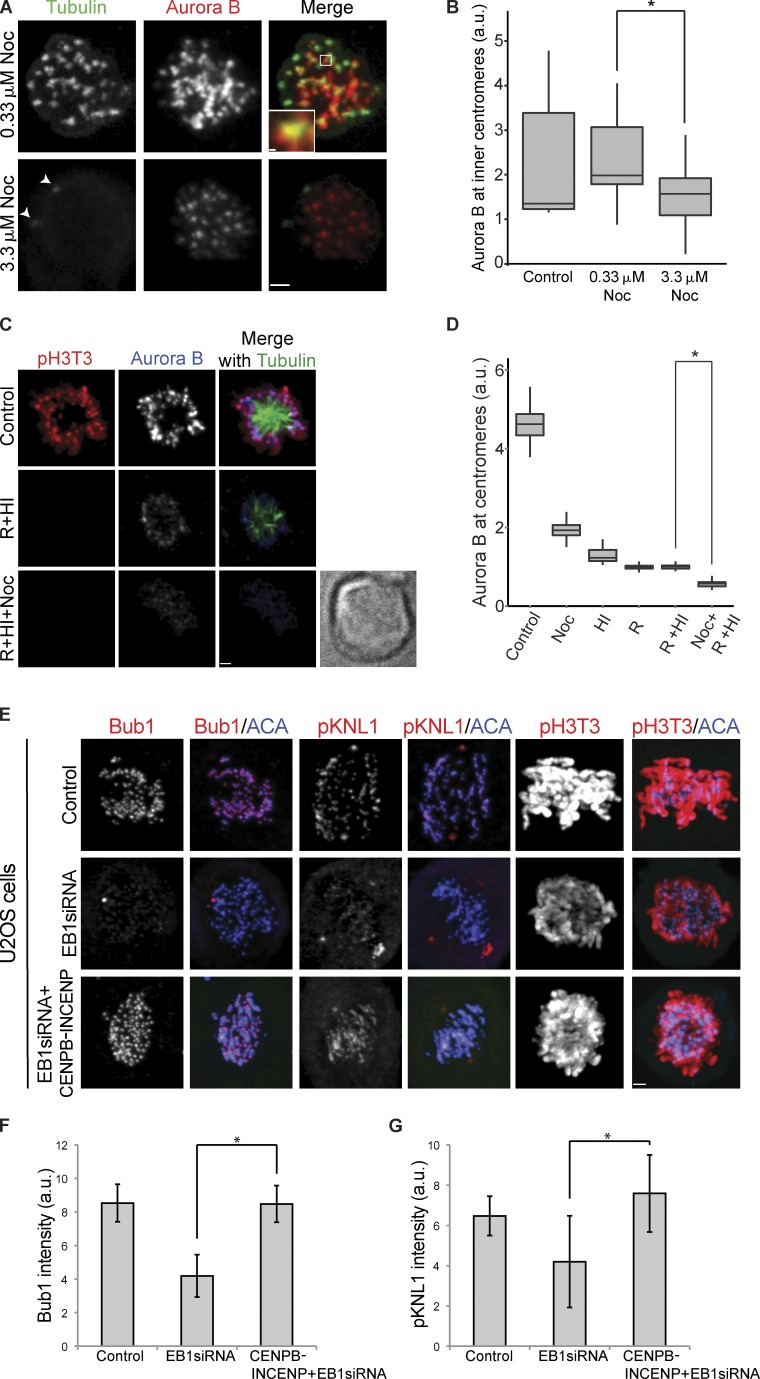
We decided to reexamine the effects of depolymerizing microtubules on CPC localization using the drug nocodazole. It is important to mention here that treating HeLa cells with different concentrations of nocodazole has radically different effects on the state of microtubules at the kinetochores. 0.33 µM nocodazole has been traditionally used to generate the spindle checkpoint arrest (Hauf et al., 2003), but at this concentration, most kinetochores have microtubule foci surrounding them (Fig. 3 A; Brito et al., 2008; Matson et al., 2012). These microtubules are absent in 3.3 µM nocodazole (Fig. 3 A; Brito et al., 2008; Matson et al., 2012).
Figure 3.
Relationship of the EB1/microtubules and the histone phosphorylation pathways in CPC localization. (A) HeLa cells treated with 0.33 and 3.3 µM nocodazole for 7 h were fixed and stained with tubulin and Aurora B antibodies. The inset is a projection of six z sections. White arrowheads in 3.3 µM nocodazole (Noc; tubulin images) point to centrosomes. Bars: (main images) 1.8 µm; (inset) 0.22 µm. Settings that allow visualization of spindle microtubules obscure the microtubule foci in 0.33 µM nocodazole, so we have not shown control cells treated with DMSO. (B) Box and whisker plot of centromeric Aurora B levels measured in HeLa cells after the indicated treatments. The lines within the boxes represent the medians. Mean inner centromeric Aurora B levels from the same experiment shown in Fig. S3 A. *, P = 0.01182. (C) HeLa cells were treated with 10 µM reversine and 1 µM 5-iodotubericidine (HI) separately or in combination (reversine [R] + HI) in the presence of MG132. (Reversine only is shown in Fig. S3 B, and HI only with the same control cells is shown in Fig. S3 C.) To determine whether microtubules can recruit Aurora B in the absence of phosphohistone marks, HeLa cells were treated with or without 3.3 µM nocodazole along with the reversine, HI, and MG132 (reversine + HI + nocodazole). Control cells were treated with MG132 only. Cells were fixed after 30 min and stained with anti-pH3T3, anti-tubulin, and anti–Aurora B antibodies. Bright-field image (reversine + HI + nocodazole treatment) to show that there is a cell that lacks detectable staining. Bar, 1.7 µm. (D) Centromeric Aurora B levels were measured at indicated treatment conditions. In this experiment, 1 µM reversine was used. *, P = 6.65 × 10−127. (E) Expression of CENPB-INCENP fusion protein in U2OS-TR cells rescued the reduction of Bub1, phospho-KNL1, and pH3T3 levels after EB1 depletion. A stable U2OS-TR line was either mock treated (control) or EB1 siRNA treated with or without CENPB-INCENP induction. Bar, 1.8 µm. (F) Quantification of Bub1 levels. *, P = 1.05 × 10−262. (G) Quantification of phospho-KNL1(Ser60) levels. *, P = 8.95 × 10−31. Error bars show standard deviations. The height of the boxes represents the IQR. The central horizontal lines depict the median. The top whiskers represent the 75th percentile + 1.5× IQR, and the bottom whiskers represent the 25th percentile − 1.5× IQR. ACA, anticentromere antigen; a.u., arbitrary units; res, resistant.
We measured Aurora B levels at centromeres in HeLa cells treated with either 0.33 or 3.3 µM nocodazole for 7 h. After treatment with 0.33 µM nocodazole, the amount of Aurora B at the centromeres was similar to DMSO-treated controls, whereas cells treated with 3.3 µM nocodazole had significantly reduced levels of Aurora B (Fig. 3, A and B; and Fig. S3 A). This suggests that the microtubule foci that surround kinetochores in 0.33 µM nocodazole are sufficient to recruit additional Aurora B.
We hypothesized that EB1 and microtubules are in the same pathway. We quantified INCENP and Borealin levels in HeLa cells that were EB1 depleted, treated with 3.3 µM nocodazole, or had both treatments (Fig. S2, E and F). The effects of EB1 depletion and microtubule depolymerization were not additive, and cells that received both treatments had similar reduction of centromeric CPC as individual treatments. Together, these data suggest that EB1 and microtubules are part of the same pathway that enriches centromeric CPC.
Microtubules cooperate with the histone phosphorylation pathways to recruit Aurora B to inner centromeres
There is a positive feedback loop by which Aurora B targets haspin kinase, to target the CPC (Wang et al., 2011b). Moreover, Aurora B regulates MPS1, which is an activator of Bub1, suggesting a second positive feedback loop in which Aurora B localizes Sgo1 to localize the CPC (Saurin et al., 2011; van der Waal et al., 2012). We designed an assay to compare the relative contributions of microtubules and histone phosphorylation in regulating centromeric Aurora B levels. We used 5-iodotubercidin, a small molecule inhibitor to haspin kinase (De Antoni et al., 2012; Wang et al., 2012), referred henceforth as haspin inhibitor (HI), and reversine, an Mps1 inhibitor that inhibits Bub1 recruitment to kinetochores (Santaguida et al., 2010, 2011; van der Waal et al., 2012), to reduce the phosphohistone marks required to localize Aurora B to centromeres. HeLa cells were treated with nocodazole, reversine, or HI separately or in combination for 30 min, along with MG132 to prevent mitotic exit. Both reversine and HI treatment reduced the histone phosphorylation of its associated pathway to levels below the level of detection (Fig. S3, B and C). Individual drug treatments caused a severe drop in Aurora B levels (Fig. 3 D). There was no additional effect of combining the kinase inhibitors together (Fig. 3 D, reversine + HI). This suggests that both haspin and MPS1 are in the same pathway or that the pathways have a common component, which we suggest is Aurora B. Adding nocodazole along with reversine and HI (Fig. 3 D, reversine + HI + nocodazole) resulted in a significant drop in Aurora B levels compared with reversine and HI (P = 6.6 × 10−127; Fig. 3, C and D). Inhibition of MPS1 by 1 µM reversine has been previously shown to not affect Aurora B levels (Santaguida et al., 2010). However, we note that these experiments were performed in nocodazole, in which we observe reduced levels of Aurora B. In agreement with the previous study, we find Aurora B levels in nocodazole were not substantially reduced upon addition of reversine (unpublished data). We conclude that microtubules can recruit Aurora B to inner centromeres independent of the histone phosphorylation pathways, but the histone phosphorylation pathways are required to obtain the full enrichment of centromeric Aurora B.
These data suggest that EB1 and microtubules are either upstream or work in combination with histone phosphorylation pathways that recruit CPC. We asked whether targeting Aurora B to centromeres suppressed the reduction of the histone phosphorylation pathways observed after EB1 depletion. We depleted EB1 from U2OS-TR cells containing an integrated transgene encoding CENPB1–158 fused to INCENP47–920 (CENPB-INCENP) driven by a doxycycline-inducible promoter (Liu et al., 2009). CENPB1–158 binds α-satellite DNA sequences at the centromere so that Aurora B is targeted independent of the normal pathways. INCENP47–920 cannot bind Survivin and Borealin but does bind Aurora B. EB1 depletion in U2OS-TR cells also reduced phospho-KNL1 and Bub1 at kinetochores and phospho–histone H3Thr3 levels when the CENPB-INCENP was not expressed. However, expressing CENPB-INCENP in EB1-depleted cells rescued both Bub1 and phospho-KNL1 levels at the kinetochore bypassing the requirement of EB1 (Fig. 3, E–G). We also observed recovery of phospho-H3Thr3 levels on CENPB-INCENP expression (Fig. 3 E). We conclude that the EB1–microtubule pathway works either upstream or is interdependent with the histone H2A and H3 phosphorylation pathways to recruit Aurora B to the centromeres.
EB1 interacts with Aurora B at the centromeres in prometaphase
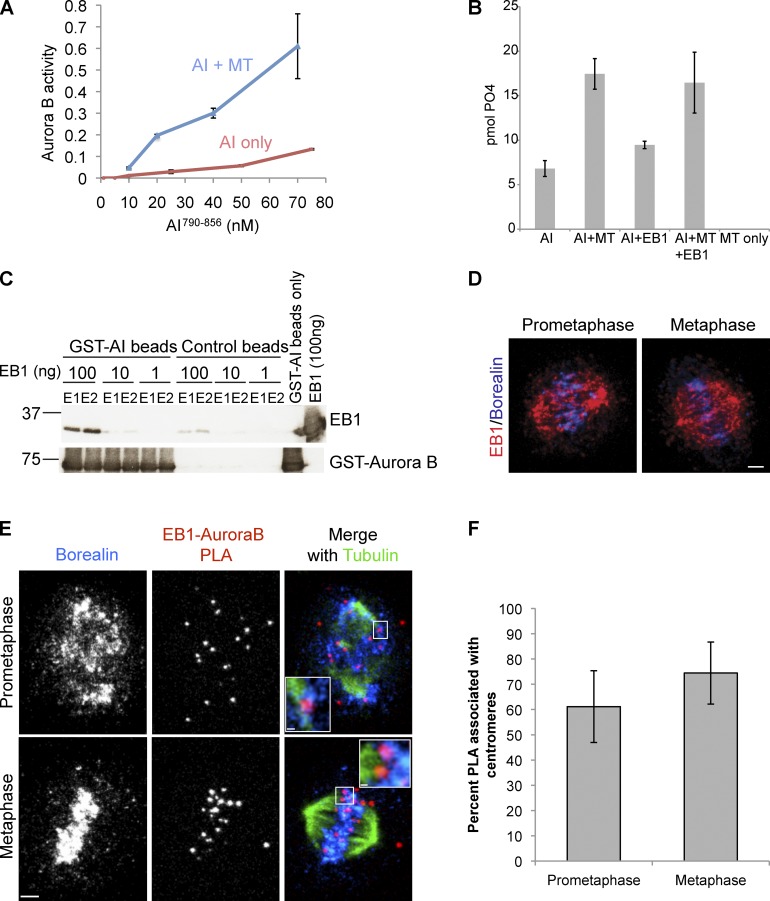
Recombinant Xenopus Aurora B bound to a C-terminal fragment of Xenopus INCENP (AI790–856) that includes the IN-box purified from Escherichia coli has a basal amount of activity (Sessa et al., 2005). The addition of microtubules stimulated Aurora B kinase activity in vitro between four- to sixfold on a myelin basic protein (MBP) substrate over a range of kinase concentrations (Fig. 4 A). However, the addition of EB1 did not further stimulate kinase activity in the presence or absence of microtubules (Fig. 4 B). We conclude that microtubules can stimulate active kinase in vitro.
Figure 4.
EB1 is in close proximity to Aurora B at centromeres. (A) Microtubules stimulate Aurora B bound to INCENP790–856 (AI790–856) activity on MBP in vitro. The assay was performed with or without 3 µM taxol-stabilized microtubules (MT). (B) EB1 does not stimulate Aurora B kinase activity alone or in combination with microtubules. AI790–-856 in vitro kinase assay, using MBP as a substrate, showing the effect of adding 3 µM taxol-stabilized microtubules and 100 ng EB1 separately or in combination. Kinase activity of 20 nM AI790–856 was assayed similarly as in A. (C) Direct interaction between EB1 and the catalytic subunit of the CPC. Recombinant Xenopus GST–Aurora B bound to a fragment of INCENP790–856 (GST-AI) on glutathione–Sepharose 4 beads was incubated with the indicated concentration of recombinant xEB1, washed, and eluted with glutathione, and the two peak fractions of the elutions (E1 and E2) were quantified by immunoblotting. GST beads were used as a control. Molecular markers are given in kilodaltons. (D) Immunostaining of EB1 and Borealin in HeLa cells. (The single channel split of images is shown in Fig. S4 A.) Bar, 2.6 µm. (E) HeLa cells immunostained for Borealin and tubulin and the close proximity of EB1 and Aurora B shown by PLA. Insets illustrate PLA at individual centromeres. Prometaphase inset is a projection of four z sections, and metaphase inset is a projection of seven z sections (PLA controls are shown in Fig. S4, C and C′). Bars: (main images) 2.4 µm; (prometaphase inset) 0.39 µm; (metaphase inset) 0.35 µm. (F) Quantification showing percentage of occurrence of PLA spots proximal to centromeres from the PLA experiment shown in C. Error bars show standard deviations.
EB1 and Aurora B can coimmunoprecipitate in HeLa cells (Sun et al., 2008). To confirm that Aurora B can directly interact with EB1, we purified full-length Xenopus EB1 and Xenopus Aurora B bound to a C-terminal fragment of Xenopus INCENP (AI790–856) that includes the IN-box from E. coli (Sessa et al., 2005). AI790–856 bound to beads could pull down EB1 in a concentration-dependent manner (Fig. 4 C).
To identify the subcellular location of the interaction between EB1 and Aurora B, we performed a proximity ligation in situ assay (PLA) using anti-EB1 and anti–Aurora B antibodies. PLA is an antibody-based technique that allows the visualization of two proteins only if they are in close proximity (∼35-nm maximum) with high sensitivity (Söderberg et al., 2006). Standard immunofluorescence staining with anti-EB1 and anti-Borealin (CPC member) antibodies showed spindle and inner centromere localization as previously shown (Fig. 4 D and Fig. S4 A). However, most EB1–Aurora B interactions identified by PLA were only found near inner centromeres in prometaphase cells (Fig. 4, E and F). This was confirmed by costaining for Borealin in these cells (Fig. 4 E, prometaphase inset, which shows a stack of four z sections). Similarly, EB1–Aurora B interactions were proximal to microtubule–inner centromere (Borealin) junctions in metaphase-aligned chromosomes (Fig. 4 E, metaphase inset, which was a stack of seven z sections). Consistent with the fact that there are no microtubules within the prophase nuclei, we don’t see any EB1–Aurora B interactions in prophase cells (Fig. S4 B). To verify the specificity of the interaction, we performed PLA reactions lacking either the EB1 or the Aurora B primary antibodies, and very little PLA signal was produced (Fig. S4, C and C′). We conclude that subsets of EB1 and Aurora B interact near or at inner centromeres, which is consistent with the regulation of Aurora B localization by EB1.
Aurora B interacts with K-fibers, pre–K-fibers, and astral microtubules in mitosis
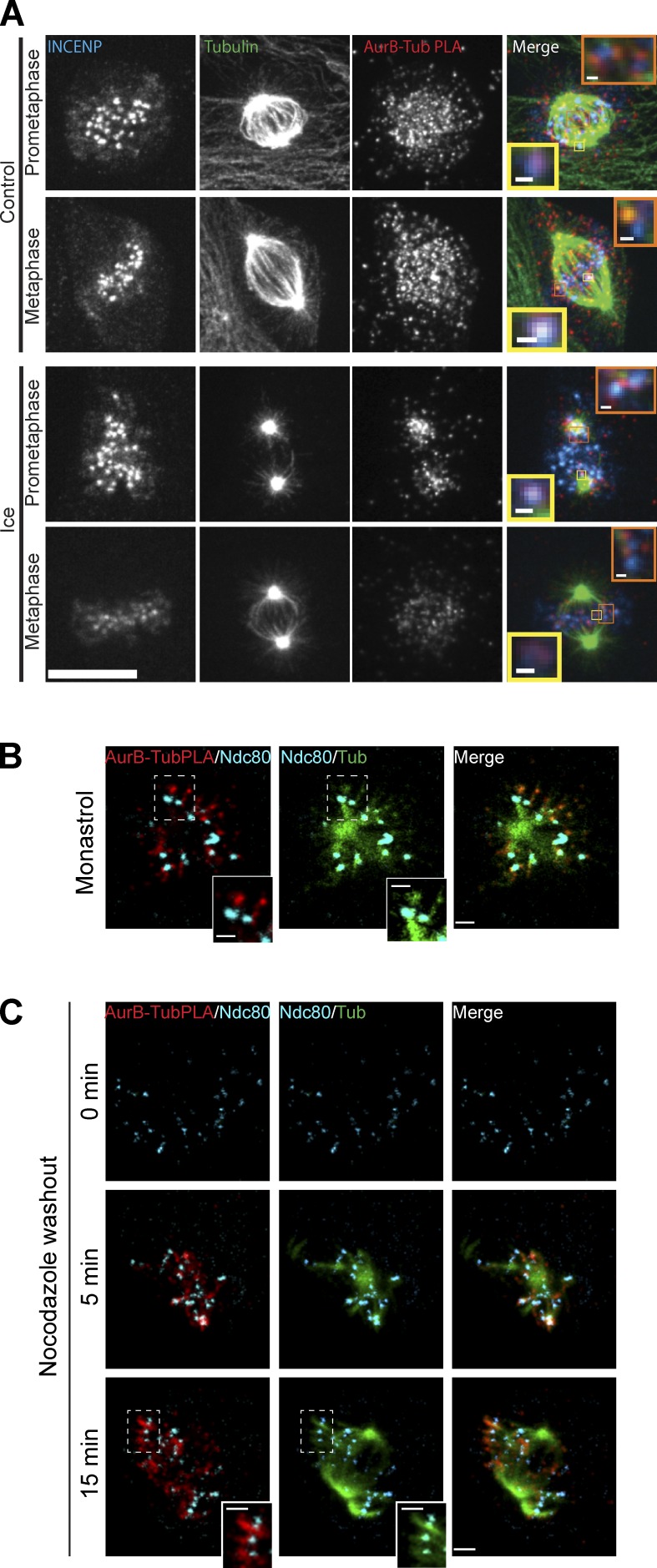
PLA was performed to identify the subcellular locations where Aurora B was in close proximity to tubulin. We used Xenopus S3 cells for two reasons. First, these cells possess a normal karyotype and remain very flat in mitosis to provide outstanding imaging. Second, our Xenopus Aurora B antibodies are highly specific and provide very reproducible signals in the PLA assay. We performed PLA for Aurora B–tubulin, and then, the cells were further processed by standard immunofluorescence with antibodies directly conjugated with fluorophores to tubulin and INCENP to generate fiducial marks on the spindle. The PLA signal in prometaphase cells could be detected at inner centromeres (Fig. 5 A, inset yellow boxes; and Video 1) and adjacent to inner centromeres on microtubules (Fig. 5 A, inset orange boxes). This is similar to the EB1–Aurora B interactions seen by PLA and is consistent with our observation that EB1/microtubules localize Aurora B to inner centromeres. In addition, we detected an additional PLA signal throughout the spindle. Cells were subjected to a brief ice treatment to destabilize nonkinetochore-associated microtubules (Fig. 5 A, ice). We observed a significant drop in PLA signal in ice-treated prometaphase cells, including most kinetochores that were not situated near centrosomes (Fig. 5 A, prometaphase). Some kinetochore/centromere PLA signals persisted in metaphase, suggesting that Aurora B can interact with K-fibers. However, most of the metaphase signals throughout the spindle were cold sensitive, suggesting that Aurora B can interact with astral microtubules. To verify the specificity of the PLA signals, we performed parallel assays in which the anti–Aurora B antibody was omitted and the PLA signal was greatly reduced (Fig. S5 A).
Figure 5.
Aurora B interacts with a distinct class of spindle microtubules. (A) Xenopus S3 cells immunostained for INCENP and tubulin (Tub) and the close proximity of tubulin and Aurora B (AurB) are shown by PLA. Insets are single z sections of Aurora B–tubulin interactions at the centromere (yellow boxes) and at the kinetochores (orange boxes). The bottom shows the effect of ice treatment on S3 cells before fixation. Bars: (main images) 11 µm; (insets) 0.51 µm. (B) Monastrol-treated Xenopus S3 cells immunostained for Ndc80 and tubulin and the close proximity of tubulin and Aurora B shown by PLA—projection of four z sections. Bars: (main images) 1.6 μm; (insets) 1 µm. (C) Nocodazole washout experiments were performed to show Aurora B specifically enriched on pre–K-fibers. Xenopus S3 cells were fixed and stained 5 min (projection of seven z sections) and 15 min (projection of nine z sections) after nocodazole washout. Bars: (main images) 2.3 μm; (insets) 1.4 µm. B and C insets are zoomed in views of the areas highlighted by the boxes showing Aurora B–tubulin PLA spots on pre–K-fibers. Aurora B–tubulin PLA signals are shown with Ndc80 and tubulin costaining. (Individual z slices of whole cell projections are shown in Videos 1–4.)
We hypothesized that the interactions between EB1–Aurora B and tubulin–Aurora B that we visualize at prometaphase kinetochores were with pre–K-fibers. This hypothesis is strongly supported by the observation that inner centromeric Aurora B levels are similar in cells treated with 0.33 µM nocodazole- and DMSO-treated prometaphase cells (Fig. 3 A). In addition, after cold treatment, the bulk of Aurora B tubulin interactions next to prometaphase kinetochores were lost. Pre–K-fibers are small bundles of microtubules that protrude from kinetochores before they make mature attachments and can mediate lateral attachments between dynein on kinetochores and microtubules (Khodjakov et al., 2003). Pre–K-fibers are difficult to distinguish in a whole spindle, but they are readily visualized in monastrol and after cells are washed out of nocodazole.
Xenopus S3 cells were incubated in monastrol, and cells were stained for xNdc80 to visualize kinetochores, tubulin to visualize the spindle, and PLA to detect where Aurora B was in close proximity to microtubules. Fig. 5 B shows four z sections through a monopolar spindle with centrosomes in the center. Chromosomes tend to have a distinct orientation in monastrol, where the kinetochore facing the pole forms K-fibers with the centrosome, whereas its sister generates pre–K-fibers extending away from the central pole axis (Khodjakov et al., 2003). We find the pre–K-fiber microtubule bundles that extend out of kinetochores directed away from the central pole have PLA signals, indicating an interaction between Aurora B and tubulin (Fig. 5 B and Video 2). The PLA signals on the kinetochore–microtubule bundles pointing away from the center are almost always stronger than those on bundles toward the center, arguing that Aurora B has specificity for pre–K-fibers over K-fibers.
We similarly stained cells that were washed out of nocodazole into fresh media for 5 and 15 min. We observed prominent PLA densities between Aurora B and tubulin emanating from most kinetochores generating pre–K-fibers 5 min after washout (Fig. 5 C, 5 min; and Video 3). Also note that the PLA signals were lost in cells treated with nocodazole (Fig. 5 C, 0 min), confirming that our PLA assay only detects when Aurora B is close to microtubules and not simply free tubulin. After 15 min, many chromosomes had aligned; however, the chromosomes that were not aligned had prominent pre–K-fibers (Fig. 5 C, 15 min; and Video 4). Fig. 5 C insets show clear bundles of microtubules next to these kinetochores that have bright Aurora B–tubulin PLA densities. These data suggest that Aurora B binds to pre–K-fibers.
EB1-dependent localization of Aurora B to centromeres in prometaphase is required to phosphorylate kinetochore and chromatin substrates
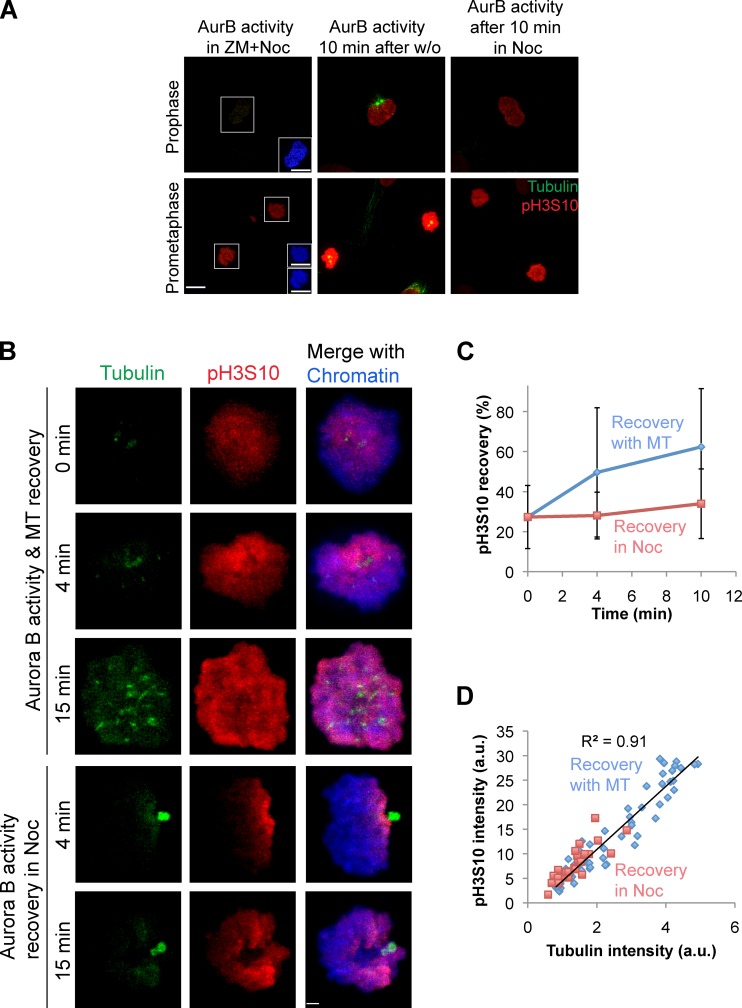
We postulated that one reason that it has been difficult to measure the effects of microtubules on Aurora B localization and phosphorylation in the past is that Aurora B activity in prophase nuclei is independent of microtubules. Aurora B phosphorylates histone H3 on Ser10 (pH3S10 in the figures) on the arms of mitotic chromosomes in late G2/prophase (Hendzel et al., 1997), and this phosphorylation persists through early anaphase. Indeed, depletion of EB1 had little effect on phospho–histone H3Ser10 levels (unpublished data), and we postulate that EB1 is not required for prophase activity of Aurora B. To separate Aurora B phosphorylation in prophase and prometaphase, we applied a recently developed assay that specifically measures the generation of Aurora B phosphorylation in prometaphase cells (Wang et al., 2011a).
EB1-depleted cells were treated with a reversible Aurora B kinase inhibitor, ZM447439 (ZM), and phosphorylation was measured on histone H3Ser10 by phosphospecific antibodies after washing out the drug. The small molecule monastrol was used to generate monopolar spindles, thereby limiting the variables of tension forces generated by the mitotic spindle, and cells were arrested in mitosis by the addition of the proteasome inhibitor MG132 for 1 h (Fig. 6 A, experimental outline). Cells depleted of EB1 were unable to phosphorylate histone H3 on Ser10 15 min after ZM washout (Fig. 6, B and D). Aurora B and phospho-KNL1 levels were similarly affected (Fig. 6, B, C, E, and F). We conclude that EB1 contributes to the phosphorylation of both kinetochore and chromatin substrates by Aurora B in prometaphase.
Figure 6.
EB1 and microtubules are required for Aurora B to phosphorylate kinetochores and chromatin substrates in prometaphase. (A) Experimental outline of B and C. (B) HeLa cells treated with ZM, MG132, and monastrol were washed out of ZM to monitor recovery of phospho–histone H3Ser10 (pH3S10) after 15 min. EB1-depleted cells are compared with control cells treated similarly and fixed immediately (ZM washout, 0 min) or replaced in ZM-free media and fixed after 15 min (ZM washout, 15 min). Bar, 1.6 µm. (C) Spreading of Aurora activity from centromeres to kinetochores (pKNL1) requires EB1. Note that centrosomal staining is an artifact, and kinetochore staining represents phospho-KNL1. Bar, 1.7 µm. (D) Quantification of the mean total pH3S10 recovery in B. *, P = 2.2 × 10−5. Error bars show standard deviations. (E) Quantification of kinetochore phosphorylation in C. (F) Centromeric Aurora B levels from B. **, P = 5.3 × 10−157. (G) Microtubules are required for the recovery of inner centromeric Aurora B and KNL1 phosphorylation after ZM washout. Note that the phospho-KNL1 has nonspecific staining of centrosomes (Welburn et al., 2010). Bar, 1.4 μm. (H) Quantification of inner centromeric Aurora B intensities in G (n > 160). Recov, recover; Noc, nocodazole. *, P = 4.19 × 10−91. (Scheme for the experiment is shown in Fig. S5 B.) The height of the boxes represents the IQR. The central horizontal lines depict the median. The top whiskers represent the 75th percentile + 1.5× IQR, and the bottom whiskers represent the 25th percentile − 1.5× IQR. a.u., arbitrary units.
We modified the ZM washout assay (Wang et al., 2011a) to determine whether spreading Aurora B activity to kinetochores was microtubule dependent in prometaphase (Fig. S5 B, experimental outline). Aurora B levels were reduced in HeLa cells after treating them with ZM (1 h) and 3.3 µM nocodazole (10 min; Fig. 6, G and H, 0 min). Centromeric Aurora B levels increased over time in cells that were shifted to ZM-nocodazole–free media (Fig. 6, G and H, recovery 10 min), whereas Aurora B levels did not increase after ZM washout in cells held in nocodazole (Fig. 6, G and H, recovery in nocodazole 10 min). Microtubules were also required for cells to phosphorylate kinetochores after ZM washout as measured by staining with phospho-KNL1 antibodies (Fig. 6 G; Welburn et al., 2010). We conclude that microtubules are required to both localize Aurora B to inner centromeres and for Aurora B–dependent phosphorylation of kinetochore substrates in prometaphase.
Aurora B activity on chromatin is regulated by microtubules in prometaphase
We compared the microtubule dependence of Aurora B activity on noncentromeric chromatin in prophase and prometaphase by washing cells out of ZM in the presence or absence of nocodazole (Fig. S5 B, assay scheme). The presence or absence of microtubules made little to no difference to the prophase cells (Fig. 7 A). In contrast, the absence of microtubules decreased the amount of chromatin phosphorylation in prometaphase cells. We conclude that prometaphase, but not prophase, Aurora B activity levels are dependent on microtubules.
Figure 7.
Aurora B activity on chromatin is regulated by microtubules in prometaphase but not in prophase. (A) Phospho–histone H3Ser10 (pH3S10) and tubulin immunostaining of prophase and prometaphase cells at the indicated conditions. Insets show chromatin staining. AurB, Aurora B; w/o, washout. (Assay scheme is shown in Fig. S5 B.) Bars, 10 µm. (B) HeLa cells in prometaphase immunostained for tubulin, phospho–histone H3Ser10 (pH3S10), and chromatin at the indicated conditions. Bar, 2.2 µm. (C) Relative pH3S10 intensity plotted as a function of time after ZM washout in the presence or absence of nocodazole. Error bars show standard deviations. (D) Correlation between total cellular pH3S10 intensity from all time points and total tubulin intensity measured per cell (R2 = 0.91). Cellular pH3S10 intensities at 0 min and recovery phases in the absence of nocodazole (Noc) are represented as blue diamonds, and intensities from cells in nocodazole are shown in red squares (n = 58; data shown are one representative experiment of three repeats). MT, microtubule; a.u., arbitrary units.
We further examined the microtubule stimulation of Aurora B kinase activity on prometaphase chromatin. We observed both a spatial and quantitative correlation between the microtubules and Aurora B activity (Fig. 7 B). Before washing out the ZM, there was weak phosphorylation of phospho-H3Ser10, and this was spatially restricted to areas of chromatin that were adjacent to microtubule foci that are likely centrosomes (Fig. 7 B, 0 min). After washing out both ZM and nocodazole, we observed robust spreading of Ser10 phosphorylation on histone H3 throughout chromatin over time. Spreading did not happen in the cells that remained in nocodazole (Fig. 7, B and C).
Microtubule regrowth was not homogenous: in some cells, we could detect microtubules only at centrosomes, and in other cells, we could detect microtubules at both centrosomes and at foci, which are likely pre–K-fibers. We plotted the amount of phospho-H3Ser10 activity in each cell as a function of the amount of polymerized tubulin in the cell. The single cell intensities fell on a diagonal signifying correlation, and the R2 value was 0.91 (Fig. 7 D). This tight correlation held true for each of the time points (Fig. S5 C). This suggests a surprising connection between the microtubules of the mitotic spindle and histone phosphorylation throughout chromatin.
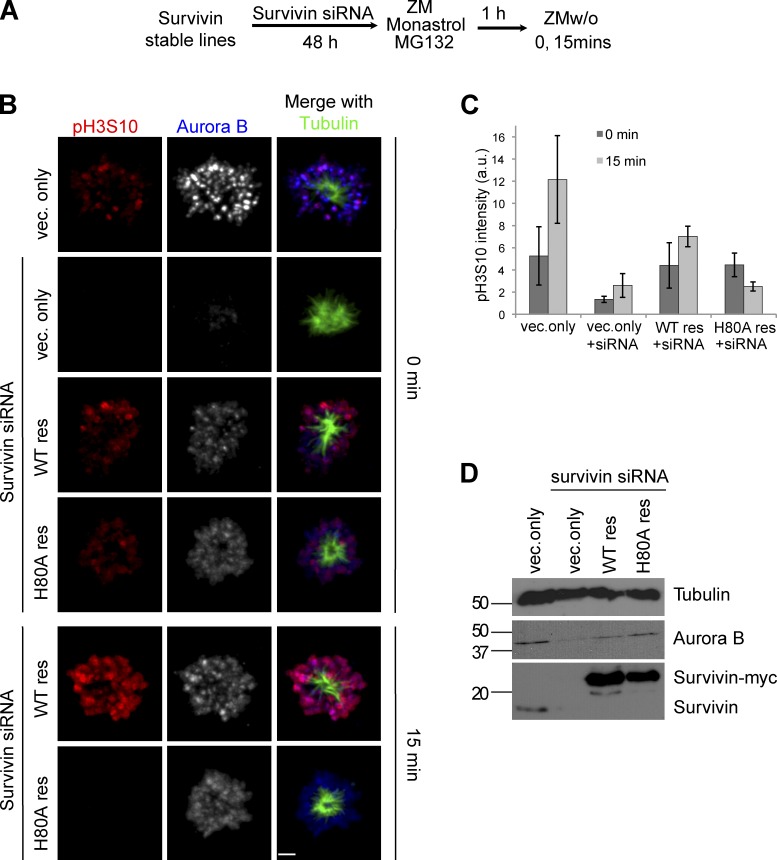
We have shown that a major role of EB1 and microtubules is to localize the CPC to centromeres. To measure the importance of localizing the CPC to inner centromeres in the ZM washout assay (Wang et al., 2011a), we replaced the endogenous survivin subunit with the survivin H80A mutant, which is unable to bind histone H3 phosphorylated on Thr3 by Haspin (Niedzialkowska et al., 2012). Aurora B kinase was reactivated by removal of ZM, and the recovery of phospho–histone H3Ser10 activity was followed by immunofluorescence over the next 15 min (Fig. 8 A, scheme). Phospho–histone H3Ser10 staining was apparent on chromatin 15 min after washing out ZM in cells expressing wild-type survivin (Fig. 8, B and C, 15 min, wild type resistant). In contrast, cells expressing survivinH80A were unable to phosphorylate chromatin (Fig. 8, B and C, H80A resistant). As expected, Aurora B did not localize to centromeres in cells expressing the survivinH80A (Fig. 8 B). We verified the Aurora B and Survivin protein levels in each condition by Western blotting (Fig. 8 D). We conclude that preventing the localization of the CPC to inner centromeres phenocopies the loss of spreading of Aurora B kinase activity on chromosome arms seen after depletion of either microtubules or EB1. Together, our data suggest that EB1/microtubules localize the CPC to prometaphase centromeres to phosphorylate both kinetochores and chromatin.
Figure 8.
CPC activity spreads from centromeres to chromosome arms after nuclear envelope breakdown. (A) Assay scheme. (B) Cells that have endogenous survivin replaced with the survivinH80A mutant are unable to phosphorylate histone H3Ser10 in the ZM washout assay. HeLa cells stably expressing vector only, survivin-myc (siRNA resistant [res]), and survivinH80A-myc (siRNA resistant) were treated with survivin siRNA for 48 h followed by 1 h in ZM, MG132, and monastrol. Bar, 2.7 µm. (C) Mean total phospho–histone H3Ser10 intensity at 0 and 15 min in mock-treated (vector [vec.] only) and survivin siRNA-treated vector only (vector only + siRNA) and survivin-myc (wild type [WT] resistant)– and survivinH80A-myc (H80A resistant)–expressing cells. Error bars show standard deviations. a.u., arbitrary units. (D) Western blot showing Aurora B, survivin-myc, and endogenous survivin levels. Molecular mass markers are given in kilodaltons.
Discussion
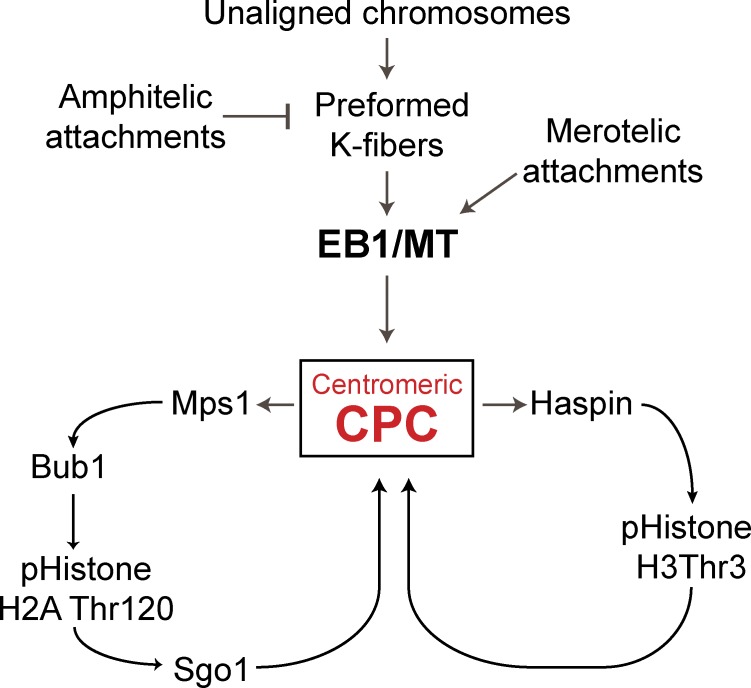
We have demonstrated that EB1 and microtubules play important roles in the recruitment of the CPC to inner centromeres after, but not before, nuclear envelope breakdown. We show that EB1 and microtubules are in the same pathway. In the absence of EB1 or microtubules, we still find pools of Aurora B at inner centromeres, but the levels are highly enriched by EB1 and microtubules. EB1 and microtubules are also essential for spreading Aurora B activity from inner centromeres to both kinetochores and to chromosome arms. EB1 has been previously shown to coimmunoprecipitate with the CPC and to block PP2A activity against Aurora B (Sun et al., 2008). Our data are consistent with these observations. Although we can detect a weak direct interaction of EB1 with Aurora B, we don’t see any direct effect on kinase activity (Fig. 4, B and C). Thus, we suggest that one role of EB1 is to inhibit PP2A to prolong Aurora B activity so that it can concentrate at centromeres by triggering the feedback loops and then spread to distant substrates.
Aurora B phosphorylates substrates at the centromere in cells treated with nocodazole. How then can we argue that microtubules regulate Aurora B? We find that Aurora B levels at the inner centromeres drop significantly in 3.3 µM nocodazole (Fig. 3, A and B), but there is still a pool that localizes in a microtubule-independent manner. This pool is dependent on the histone phosphorylation pathways and is further reduced by simultaneous inhibition of Aurora B activity by ZM (Fig. 6 G), suggesting that there are at least two pathways that localize Aurora B. Our data suggest that EB1/microtubule stimulation is upstream and activates the histone phosphorylation loops to generate increased levels of CPC at centromeres. These redundancies and positive feedback loops complicate the study of the CPC, and it is critical to knock out one pathway to study the regulation of a second pathway.
How do EB1 and microtubules localize the CPC? We show direct stimulation of kinase activity by microtubules and direct interaction of Aurora B and EB1. Moreover, EB1(K89E) microtubule binding mutant is unable to enrich CPC at the inner centromeres in EB1-depleted cells. The loss of EB1 also reduces phospho–histone H3Thr3, Bub1 at kinetochores, and Bub1 phosphorylation of histone H2A, which are all required to localize the CPC to centromeres (Kelly et al., 2010; Wang et al., 2010; Yamagishi et al., 2010; Jeyaprakash et al., 2011; Du et al., 2012; Niedzialkowska et al., 2012). The simplest model is that EB1 on microtubule plus ends enrich Aurora B at inner centromeres. This triggers positive feedback loops that regulate the histone kinases (Fig. 3 and Fig. 9; Wang et al., 2011b; van der Waal et al., 2012). The Mps1 kinase is also stimulated by microtubules and enhances phospho–histone H2A levels to rapidly recruit additional Aurora B to centromeres (Stucke et al., 2004; van der Waal et al., 2012). We postulate that the EB1–microtubule pathways modulate the amount of CPC at centromeres, whereas the histone phosphorylation pathways ensure that the CPC can only be stimulated at inner centromeres where the two histone marks intersect.
Figure 9.
Cooperation of histone phosphorylation and microtubules in centromeric localization of Aurora B. Model depicting the role of EB1 and microtubules (MT) in regulating inner centromeric Aurora B and its activity. Arrows do not imply direct interaction.
There are distinct types of microtubules in the spindle, and our data suggest that the major pool of microtubules that activate the CPC recruitment in prometaphase are the pre–K-fibers that are nucleated by kinetochores (Mitchison and Kirschner, 1985; Khodjakov et al., 2003; Platani et al., 2009). This is supported by our observation that there are higher levels of Aurora B at the microtubule foci near kinetochores in cells treated with 0.33 µM, but not 3.3 µM, nocodazole (Fig. 3, A and B; and Fig. S3 A). In addition, PLA suggests that Aurora B–tubulin interactions and Aurora B–EB1 interactions are highest at centromeres in early prometaphase cells and that Aurora B can be found on pre–K-fibers (Fig. 4 E and Fig. 5, B and C). These conclusions are also supported by earlier work that showed that the CPC requires both chromosomes and microtubules for proper function and organizes pre–K-fibers (Tulu et al., 2006; Tseng et al., 2010). Finally, when kinetochores nucleate microtubules during nocodazole washout, the amount of Aurora B and its activity are higher at kinetochores with preformed K-fibers than adjacent kinetochores without microtubule bundles in the same cell. We found a subpool of Aurora B on astral microtubules and a second pool on K-fibers of metaphase-aligned chromosomes. The function of the astral microtubule pool is still unclear, but we speculate that it is involved in spreading Aurora B kinase activity from inner centromeres to distant substrates such as chromosome arms. Consistent with this idea, it has been shown that a microtubule-targeted Förster resonance energy transfer sensor was phosphorylated by Aurora B on the metaphase spindle (Tseng et al., 2010).
Microtubules/EB1 allow the CPC to communicate with the spindle
Microtubule/EB1 regulation of Aurora B levels provides a mechanism for spindle status to regulate inner centromere signaling. We will outline three examples from the literature that could be explained by this regulation.
EB1 is the only protein that localizes specifically to the sister kinetochores that have the growing microtubule ends (antipoleward; Tirnauer et al., 2002). This finding, combined with our data, suggests that Aurora B activity would be higher on the antipoleward sister kinetochores. Consistent with this idea, most PLA signals are typically found near one of the two sister kinetochores (Fig. 4, E and F). Differential phosphorylation on the poleward and antipoleward sides could provide mechanisms to coordinate the two sisters to allow proper chromosome movements (Tirnauer et al., 2002; Liu et al., 2009; Dumont et al., 2012).
Aurora B is recruited specifically to merotelic attachment points, and Aurora B activity is required to resolve merotelic attachments (Cimini et al., 2006; Knowlton et al., 2006). Our findings can in part explain these phenomena. A defining feature of a merotelic attachment is the presence of microtubules proximal to the inner centromeres. We suggest that the presence of microtubules recruits additional Aurora B to resolve merotelic attachments. In addition, a merotelic attachment brings kinetochores close to inner centromeres to allow efficient phosphorylation of kinetochore substrates. such as Ndc80 and Ska, which release microtubule attachments (Cheeseman et al., 2006; DeLuca et al., 2006; Welburn et al., 2010).
Centromeric Aurora B levels decrease as chromosomes become aligned at the metaphase plate in nontransformed cells (Lan et al., 2004; Salimian et al., 2011). We suggest that microtubule stimulation of Aurora B recruitment could also underlie this phenomenon. The nucleation of microtubules by kinetochores is regulated by RanGTP, and mature kinetochore attachments recruit a RanGAP (Joseph et al., 2004; Orjalo et al., 2006; Mishra et al., 2010). However, brief treatments with nocodazole actually increase CPC recruitment (Salimian et al., 2011). Although this experiment initially seems to contradict our findings, we note that nocodazole takes at least 10 min to fully depolymerize microtubules. Thus, brief nocodazole treatment may decrease the stability of K-fibers and increase the pre–K-fibers that stimulate Aurora B localization (Fig. 3 A; Brito et al., 2008; Salimian et al., 2011).
We have also identified conditions that reveal global regulation of the histone code by microtubules. Specifically, the amount of histone H3Ser10 phosphorylation is both spatially and quantitatively correlated with the amount of microtubules after washout of ZM and nocodazole (Fig. 7 D). Although still speculative, this suggests a previously unappreciated role of microtubules and EB1 in regulating a histone modification throughout chromatin. It will be important to determine whether the mitotic spindle structure influences gene transcription and/or the maintenance of chromatin state in the following interphase.
Materials and methods
Kinase activity assay
Recombinant AI790–856 was obtained from pGEX6P2-Aurora B-INCENP790–856–transformed BL21 bacterial expression system using a GST tag on Aurora B by affinity purification. PreScission Protease–cleaved product was run over a Superdex 200 column, and 0.3 mg/ml AI790–856 was purified to homogeneity (previously described in Sessa et al., 2005). This was diluted to the required concentrations and incubated with either 3 µM taxol-stabilized microtubules (prepared as in Desai et al., 1999) or BRB80 (80 mM Pipes, 1 mM MgCl2, and 1 mM EGTA, pH 6.8, with KOH) for 15 min. Xenopus EB1 was purified from the BL21 bacterial expression system from a pET28c-EB1 vector. Assays were performed in kinase buffer (20 mM Tris-HCl, pH 7.5, 1 mM MgCl2, 25 mM KCl, 1 mM DTT, and 100 µM ATP/1 µM γ-[32P]ATP mix) with MBP (Invitrogen) as a substrate. Activity was initiated by incubation with microtubules/BRB80 and stopped after 2 min by adding SDS-PAGE sample buffer. Samples were separated on 15% SDS-PAGE gel, stained with Coomassie blue, dried on Whatman paper together with aliquots of γ-[32P]ATP, and exposed to Phosphor Screen (Molecular Dynamics) overnight. Phosphor Screens were scanned on a phosphor scanner (Storm 860; Molecular Dynamics), and resulting images were processed and quantified using ImageQuant (Molecular Dynamics) to calculate the amount of PO43− incorporated on MBP. Error bars represent standard deviations.
Cell lines and plasmids
Wild-type or K89E mutant EB1 was cloned into the DLAP destination vector, which is derived from the pcDNA5.0/FRT vector (Invitrogen) with C-terminal dual tags of S peptide and GFP (pDLAP-EB1). These clones were modified by site-directed mutagenesis to make them resistant to the coding sequence–targeted siRNA (pDLAP-EB1siRes and EB1K89EsiRes). Flp-In HeLa T-REx cells (Tighe et al., 2008) were cotransfected with pDLAP-EB1 and pOG44. Stable lines expressing (wild type) EB1-LAP were subsequently created by selection with hygromycin. U2OS-TR stable cell line with doxycycline-inducible CENPB-INCENP fusion protein was a gift from S. Lens (University Medical Center Utrecht, 3584 CG Utrecht, Netherlands; Saurin et al., 2011).
Tissue culture and transfection
HeLa T-REx cells (Invitrogen) were grown and passaged in DMEM supplemented with 10% FBS. Xenopus S3 cells were cultured in 66% L-15 media (Sigma-Aldrich) supplemented with 10% FBS, 100 IU/ml penicillin, 100 µg/ml streptomycin, and 1 nM sodium pyruvate at 18°C. HeLa cells were plated at 25% confluency onto poly-l-lysine–coated 18-mm coverslips in a 12-well dish (Corning) overnight. siRNA transfection for EB1 and Mad2 knockdown was performed using Lipofectamine RNAiMAX (Invitrogen) according to the manufacturer’s protocol, and survivin knockdown was performed using Oligofectamine (Invitrogen) as previously described (Niedzialkowska et al., 2012). HeLa cells were treated with EB1 siRNAs (Custom siRNA, 5′-AAGUGAAAUUCCAAGCUAAGCUU-3′; Custom 3′UTR siRNAs, 5′-GAATGCTGGAGAGATGTTATG-3′ and 5′-GCACTAATCTCTTTGGAGA-3′; Thermo Fisher Scientific) at 10 nM and a SMARTpool of siRNA oligonucleotides against Mad2 (L-003271-00-0005; Thermo Fisher Scientific) with a final concentration of 20 nM, either separately or in combination for 48 h. pDLAP-EB1 (wild type or siRNA resistant) was transiently transfected with Lipofectamine 2000 (Invitrogen) following the manufacturer’s protocol, 24 h after siRNA transfection, for rescue experiments. Cells grown in a 12-well dish were transfected with 250 ng (500 ng for a 6-well dish) plasmid and fixed for immunofluorescence after 24 h.
Immunoblotting
HeLa cells from two wells of a 6-well dish were scraped and spun down at 1,500 rpm to generate cell lysates for Western blotting. Pellets were washed with Dulbecco’s PBS (Invitrogen) and resuspended in 2× SDS sample buffer, sonicated, and run on SDS-polyacrylamide gel. Antibodies used were as follows: anti-survivin (Cell Signaling Technology), anti-tubulin DM1-α (Sigma-Aldrich), anti-AIM1 (BD), anti-EB1 (BD), anti-Mad2 (Bethyl Laboratories, Inc.), and anti-Bub1 (Abcam).
ZM washout assays
HeLa cells were plated at 75% confluency onto poly-l-lysine–coated 18-mm coverslips in a 12-well dish (Corning) overnight. Cells were treated with 2 µM ZM (Enzo Life Sciences), 42 µM MG132 (Tocris Biosciences), and 100 µM monastrol (Tocris Bioscience) for 1 h in DMEM + 10% FBS at 37°C. Cells were either fixed or, to assay recovery of Aurora B activity after ZM washout, washed with Dulbecco’s PBS for three times and replaced in fresh DMEM + 10% FBS with 42 µM MG132 and 100 µM monastrol.
ZM + nocodazole washout assay.
After 60 min in ZM/MG132/monastrol, nocodazole was added to a final concentration of 3.3 µM and kept at 37°C for 10 min. Cells were either fixed at this point or shifted to fresh DMEM + 10% FBS with 42 µM MG132 and 100 µM monastrol with or without 3.3 µM nocodazole.
PLA
Xenopus S3 or HeLa cells were grown on acid-washed, poly-lysine–coated coverslips affixed with a silicone gasket (Grace Bio-Labs, Inc.). To destabilize microtubules (Fig. 5 A), cells were washed into ice-cold growth media and incubated in an ice water bath for 5 min immediately before fixation. Untreated control cells were fixed simultaneously.
100 µM monastrol treatment was performed for 1 h and fixed with 4% paraformaldehyde in PHEM at RT for 20 min. Coverslips were washed three times with TBS + 0.05% Tween and stored at 4°C until ready for use in PLA.
Nocodazole washout.
Xenopus S3 cells were treated with 2 µM nocodazole or an equivalent dilution of DMSO for 1 h. Cells were either left in DMSO and 2 µM nocodazole or washed three times with fresh media and released for 5, 10, or 15 min before fixation with 4% paraformaldehyde. Coverslips were washed three times with TBS + 0.05% Tween and stored at 4°C until ready for use in PLA.
PLA was performed after fixation using Duolink In Situ PLA probe anti–Mouse MINUS and anti–Rabbit PLUS and Duolink II Detection Reagents orange (Olink Bioscience). The assay was performed following the manufacturer’s recommended protocol using the provided blocking solution and antibody diluent. Samples were incubated in primary antibodies overnight at 4°C. The following primary antibodies were used at the indicated dilutions: anti–xlAurora B (1:400; P.T. Stukenberg), anti–β tubulin AA2 (1:500; University of Virginia Lymphocyte Culture Center), anti-hEB1 (1:500; BD), and anti–hAurora B (1 µg/ml; Abcam).
Immunofluorescence
HeLa cells were fixed with 100% methanol for EB1 immunostaining and PLA experiments (anti-EB1 antibody). All other immunostaining and PLA experiments were performed after cofixing cells with 4% paraformaldehyde, PHEM (60 mM Pipes, 25 mM Hepes, 10 mM EGTA, and 4 mM MgCl2, pH 6.9), and 0.5% Triton X-100 for 20 min. Immunofluorescence staining was performed in 3% BSA/TBS–0.05% Tween 20 using these antibodies anti-tubulin DM1-α, antiphospho-H3Ser10 (EMD Millipore), antiphospho–histone H3T3 (EMD Millipore), antiphospho-H2AT120 (Active Motif), phospho-KNL1 (S24 and S60; I.M. Cheeseman, Whitehead Institute for Biomedical Research, Cambridge, MA; Welburn et al., 2010), anti-AIM1 (Aurora B), anticentromere antigen (Antibodies, Inc.), anti-hEB1, and TO-PRO-3 (Invitrogen).
After the PLA procedure, cells were incubated in 10% normal mouse serum and 10% normal rabbit serum (Jackson ImmunoResearch Laboratories, Inc.) in 3% BSA/TBS + 0.05% Tween 20 for 30 min at RT to block any open binding sites on the PLA probes. FITC-conjugated anti–α-tubulin, DM1-α, and Alexa Fluor 647–conjugated polyclonal antibodies (xlNdc80, xlINCENP, or hBorealin) were used for costaining with PLA reactions. Alexa Fluor 647 polyclonal antibody conjugations were prepared using an Alexa Fluor 647 labeling kit following the manufacturer’s recommended protocol (Invitrogen).
Fluorescence microscopy, image acquisition, and processing
Images were captured using an inverted microscope (Axiovert 200; Carl Zeiss) fitted with a confocal scanner using a krypton/argon laser (PerkinElmer), an electron multiplying charge-coupled device camera (C9100-50; Hamamatsu Photonics), a motor (NanoScanZ; Prior Scientific), and a 63 or 100× oil Plan Apochromat objective. An acousto-optic tunable filter was used for detection of light at 488, 568, and 647 nm. Photographs were taken as z series with 0.4-µm z steps at RT. All aspects of image acquisition and processing were controlled by Volocity 5.5 (PerkinElmer). Images from the same experiment were captured using identical acquisition settings, and the contrast enhancement tool (Volocity 5.5 or ImageJ [National Institutes of Health]) was used to scale the images to the same black and white values.
Total sum intensity in each cell was determined by drawing regions of interest (ROIs) around individual cells with the freehand or lasso tool. To measure intensities specifically at kinetochores or centromeres, a volume-thresholding algorithm was made for each channel. ROIs picked up by the algorithm were manually confirmed as kinetochores or centromeres by comparing with anticentromere antigen. Here, it is important to mention that the algorithm picked voxel volumes above a certain intensity. This resulted in some underestimation of the severity of loss of signal after treatments for signals that were not detectable above the background. This was considered reasonable, as the changes were still highly significant. Background intensity per micrometer cubed was calculated for each image by drawing an ROI and dividing the sum intensity of the ROI by its volume. This value was multiplied by the volume of the ROI drawn by freehand tool or picked up by the algorithm and subtracted from the sum intensity measurement for the cell/kinetochore to find the corrected sum intensity. Whenever we used the volume thresholding algorithm, we divided the corrected total sum intensity per voxel volume by the voxel volume (intensity/micrometer cubed) and referred to it as arbitrary units. This was performed to avoid error in estimation of absolute centromeric intensities in cases in which the voxel failed to distinguish between two or more closely situated centromeres. Standard error measurements are standard deviations computed in Excel (Microsoft). We performed F-test of equality of variance to determine scedasticity followed by Student’s t test to estimate significant difference. Error bars represent standard deviations unless mentioned otherwise.
Volocity 5.5 volume thresholding algorithm
The thresholding algorithm was as follows: (a) Find Objects Using Intensity with channel X and range (lower to upper) in arbitrary units; (b) Exclude Objects by Size greater than n1 micrometers cubed; (c) Exclude Objects by Size less than n2 micrometers cubed.
Online supplemental material
Fig. S1 shows immunoblots measuring EB1 depletion in HeLa and HEK293T cells. Fig. S2 shows rescue of centromeric Aurora B levels after EB1 depletion by alternative approaches. Fig. S3 shows mean centromeric Aurora B levels measured at different concentrations of nocodazole and controls demonstrating that reversine and 5′-iodotubericidine (HI) were active in HeLa cells. Fig. S4 shows immunostaining of EB1 and Borealin in the HeLa cell. Fig. S4 shows controls for the PLA experiments and additional images of EB1 and Aurora B PLA in prophase, prometaphase, and telophase cells. Fig. S5 shows that microtubules regulate spreading Aurora B activity from centromeres to chromosome arms. Video 1 shows sequential z sections of a prometaphase cell. Video 2 shows sequential z sections of a monastrol-treated cell. Video 3 shows sequential z sections of a prometaphase cell fixed and stained 5 min after nocodazole washout. Video 4 shows sequential z sections of a prometaphase cell fixed and stained 15 min after nocodazole washout. The R code for the box and whisker plot graphs is also provided online as a Word (Microsoft) file. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201307119/DC1.
Supplementary Material
Acknowledgments
We would like to thank Iain M. Cheeseman for the gift of the phospho-KNL1 antibody, Stefan Bekiranov and Stephen Hoang for help with statistical analysis, Sussane Lens for the U2OS-TR stable cell line with doxycycline-inducible CENPB-INCENP fusion protein, and D.J. Burke, D.R. Foltz, M. Mitchell Smith, and Michael A. Lampson for critical review of the manuscript.
This work was supported by grants from the National Institutes of Health (GM063045) to P.T. Stukenberg and in part by a gift provided to the University of Virginia by the Altria Group, and B. Banerjee was in part supported by the Robert R. Wagner Fellowship Fund.
The authors declare no competing financial interests.
Author contributions: B. Banerjee, C.A. Kestner, and P.T. Stukenberg designed and executed the experiments, and B. Banerjee and P.T. Stukenberg wrote the manuscript.
Footnotes
Abbreviations used in this paper:
- CPC
- chromosomal passenger complex
- HI
- haspin inhibitor
- INCENP
- inner centromere protein
- IQR
- interquartile range
- LAP
- localization and affinity purification
- MBP
- myelin basic protein
- PLA
- proximity ligation in situ assay
- ROI
- region of interest
References
- Bayliss R., Sardon T., Vernos I., Conti E. 2003. Structural basis of Aurora-A activation by TPX2 at the mitotic spindle. Mol. Cell. 12:851–862 10.1016/S1097-2765(03)00392-7 [DOI] [PubMed] [Google Scholar]
- Brito D.A., Yang Z., Rieder C.L. 2008. Microtubules do not promote mitotic slippage when the spindle assembly checkpoint cannot be satisfied. J. Cell Biol. 182:623–629 10.1083/jcb.200805072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheeseman I.M., Chappie J.S., Wilson-Kubalek E.M., Desai A. 2006. The conserved KMN network constitutes the core microtubule-binding site of the kinetochore. Cell. 127:983–997 10.1016/j.cell.2006.09.039 [DOI] [PubMed] [Google Scholar]
- Chu Y., Yao P.Y., Wang W., Wang D., Wang Z., Zhang L., Huang Y., Ke Y., Ding X., Yao X. 2011. Aurora B kinase activation requires survivin priming phosphorylation by PLK1. J. Mol. Cell Biol. 3:260–267 10.1093/jmcb/mjq037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cimini D., Wan X., Hirel C.B., Salmon E.D. 2006. Aurora kinase promotes turnover of kinetochore microtubules to reduce chromosome segregation errors. Curr. Biol. 16:1711–1718 10.1016/j.cub.2006.07.022 [DOI] [PubMed] [Google Scholar]
- De Antoni A., Maffini S., Knapp S., Musacchio A., Santaguida S. 2012. A small-molecule inhibitor of Haspin alters the kinetochore functions of Aurora B. J. Cell Biol. 199:269–284 10.1083/jcb.201205119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeLuca J.G., Gall W.E., Ciferri C., Cimini D., Musacchio A., Salmon E.D. 2006. Kinetochore microtubule dynamics and attachment stability are regulated by Hec1. Cell. 127:969–982 10.1016/j.cell.2006.09.047 [DOI] [PubMed] [Google Scholar]
- Desai A., Murray A., Mitchison T.J., Walczak C.E. 1999. The use of Xenopus egg extracts to study mitotic spindle assembly and function in vitro. Methods Cell Biol. 61:385–412 10.1016/S0091-679X(08)61991-3 [DOI] [PubMed] [Google Scholar]
- Du J., Kelly A.E., Funabiki H., Patel D.J. 2012. Structural basis for recognition of H3T3ph and Smac/DIABLO N-terminal peptides by human Survivin. Structure. 20:185–195 10.1016/j.str.2011.12.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dumont S., Salmon E.D., Mitchison T.J. 2012. Deformations within moving kinetochores reveal different sites of active and passive force generation. Science. 337:355–358 10.1126/science.1221886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischle W., Tseng B.S., Dormann H.L., Ueberheide B.M., Garcia B.A., Shabanowitz J., Hunt D.F., Funabiki H., Allis C.D. 2005. Regulation of HP1-chromatin binding by histone H3 methylation and phosphorylation. Nature. 438:1116–1122 10.1038/nature04219 [DOI] [PubMed] [Google Scholar]
- Fuller B.G., Lampson M.A., Foley E.A., Rosasco-Nitcher S., Le K.V., Tobelmann P., Brautigan D.L., Stukenberg P.T., Kapoor T.M. 2008. Midzone activation of aurora B in anaphase produces an intracellular phosphorylation gradient. Nature. 453:1132–1136 10.1038/nature06923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hauf S., Cole R.W., LaTerra S., Zimmer C., Schnapp G., Walter R., Heckel A., van Meel J., Rieder C.L., Peters J.-M. 2003. The small molecule Hesperadin reveals a role for Aurora B in correcting kinetochore–microtubule attachment and in maintaining the spindle assembly checkpoint. J. Cell Biol. 161:281–294 10.1083/jcb.200208092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi I., Ikura M. 2003. Crystal structure of the amino-terminal microtubule-binding domain of end-binding protein 1 (EB1). J. Biol. Chem. 278:36430–36434 10.1074/jbc.M305773200 [DOI] [PubMed] [Google Scholar]
- Hendzel M.J., Wei Y., Mancini M.A., Van Hooser A., Ranalli T., Brinkley B.R., Bazett-Jones D.P., Allis C.D. 1997. Mitosis-specific phosphorylation of histone H3 initiates primarily within pericentromeric heterochromatin during G2 and spreads in an ordered fashion coincident with mitotic chromosome condensation. Chromosoma. 106:348–360 10.1007/s004120050256 [DOI] [PubMed] [Google Scholar]
- Hirota T., Lipp J.J., Toh B.-H., Peters J.-M. 2005. Histone H3 serine 10 phosphorylation by Aurora B causes HP1 dissociation from heterochromatin. Nature. 438:1176–1180 10.1038/nature04254 [DOI] [PubMed] [Google Scholar]
- Hsu J.Y., Sun Z.W., Li X., Reuben M., Tatchell K., Bishop D.K., Grushcow J.M., Brame C.J., Caldwell J.A., Hunt D.F., et al. 2000. Mitotic phosphorylation of histone H3 is governed by Ipl1/aurora kinase and Glc7/PP1 phosphatase in budding yeast and nematodes. Cell. 102:279–291 10.1016/S0092-8674(00)00034-9 [DOI] [PubMed] [Google Scholar]
- Jeyaprakash A.A., Basquin C., Jayachandran U., Conti E. 2011. Structural basis for the recognition of phosphorylated histone h3 by the survivin subunit of the chromosomal passenger complex. Structure. 19:1625–1634 10.1016/j.str.2011.09.002 [DOI] [PubMed] [Google Scholar]
- Joseph J., Liu S.-T., Jablonski S.A., Yen T.J., Dasso M. 2004. The RanGAP1-RanBP2 complex is essential for microtubule-kinetochore interactions in vivo. Curr. Biol. 14:611–617 10.1016/j.cub.2004.03.031 [DOI] [PubMed] [Google Scholar]
- Kallio M.J., McCleland M.L., Stukenberg P.T., Gorbsky G.J. 2002. Inhibition of aurora B kinase blocks chromosome segregation, overrides the spindle checkpoint, and perturbs microtubule dynamics in mitosis. Curr. Biol. 12:900–905 10.1016/S0960-9822(02)00887-4 [DOI] [PubMed] [Google Scholar]
- Kelly A.E., Sampath S.C., Maniar T.A., Woo E.M., Chait B.T., Funabiki H. 2007. Chromosomal enrichment and activation of the aurora B pathway are coupled to spatially regulate spindle assembly. Dev. Cell. 12:31–43 10.1016/j.devcel.2006.11.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly A.E., Ghenoiu C., Xue J.Z., Zierhut C., Kimura H., Funabiki H. 2010. Survivin reads phosphorylated histone H3 threonine 3 to activate the mitotic kinase Aurora B. Science. 330:235–239 10.1126/science.1189505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khodjakov A., Copenagle L., Gordon M.B., Compton D.A., Kapoor T.M. 2003. Minus-end capture of preformed kinetochore fibers contributes to spindle morphogenesis. J. Cell Biol. 160:671–683 10.1083/jcb.200208143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knauer S.K., Bier C., Habtemichael N., Stauber R.H. 2006. The Survivin-Crm1 interaction is essential for chromosomal passenger complex localization and function. EMBO Rep. 7:1259–1265 10.1038/sj.embor.7400824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knowlton A.L., Lan W., Stukenberg P.T. 2006. Aurora B is enriched at merotelic attachment sites, where it regulates MCAK. Curr. Biol. 16:1705–1710 10.1016/j.cub.2006.07.057 [DOI] [PubMed] [Google Scholar]
- Lampson M.A., Renduchitala K., Khodjakov A., Kapoor T.M. 2004. Correcting improper chromosome-spindle attachments during cell division. Nat. Cell Biol. 6:232–237 10.1038/ncb1102 [DOI] [PubMed] [Google Scholar]
- Lan W., Zhang X., Kline-Smith S.L., Rosasco S.E., Barrett-Wilt G.A., Shabanowitz J., Hunt D.F., Walczak C.E., Stukenberg P.T. 2004. Aurora B phosphorylates centromeric MCAK and regulates its localization and microtubule depolymerization activity. Curr. Biol. 14:273–286 10.1016/j.cub.2004.01.055 [DOI] [PubMed] [Google Scholar]
- Liu D., Vader G., Vromans M.J.M., Lampson M.A., Lens S.M.A. 2009. Sensing chromosome bi-orientation by spatial separation of aurora B kinase from kinetochore substrates. Science. 323:1350–1353 10.1126/science.1167000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Losada A., Hirano M., Hirano T. 2002. Cohesin release is required for sister chromatid resolution, but not for condensin-mediated compaction, at the onset of mitosis. Genes Dev. 16:3004–3016 10.1101/gad.249202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackay A.M., Ainsztein A.M., Eckley D.M., Earnshaw W.C. 1998. A dominant mutant of inner centromere protein (INCENP), a chromosomal protein, disrupts prometaphase congression and cytokinesis. J. Cell Biol. 140:991–1002 10.1083/jcb.140.5.991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matson D.R., Demirel P.B., Stukenberg P.T., Burke D.J. 2012. A conserved role for COMA/CENP-H/I/N kinetochore proteins in the spindle checkpoint. Genes Dev. 26:542–547 10.1101/gad.184184.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishra R.K., Chakraborty P., Arnaoutov A., Fontoura B.M.A., Dasso M. 2010. The Nup107-160 complex and gamma-TuRC regulate microtubule polymerization at kinetochores. Nat. Cell Biol. 12:164–169 10.1038/ncb2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchison T.J., Kirschner M.W. 1985. Properties of the kinetochore in vitro. I. Microtubule nucleation and tubulin binding. J. Cell Biol. 101:755–765 10.1083/jcb.101.3.755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mollinari C., Reynaud C., Martineau-Thuillier S., Monier S., Kieffer S., Garin J., Andreassen P.R., Boulet A., Goud B., Kleman J.-P., Margolis R.L. 2003. The mammalian passenger protein TD-60 is an RCC1 family member with an essential role in prometaphase to metaphase progression. Dev. Cell. 5:295–307 10.1016/S1534-5807(03)00205-3 [DOI] [PubMed] [Google Scholar]
- Morrison E.E., Wardleworth B.N., Askham J.M., Markham A.F., Meredith D.M. 1998. EB1, a protein which interacts with the APC tumour suppressor, is associated with the microtubule cytoskeleton throughout the cell cycle. Oncogene. 17:3471–3477 10.1038/sj.onc.1202247 [DOI] [PubMed] [Google Scholar]
- Niedzialkowska E., Wang F., Porebski P.J., Minor W., Higgins J.M.G., Stukenberg P.T. 2012. Molecular basis for phosphospecific recognition of histone H3 tails by Survivin paralogues at inner centromeres. Mol. Biol. Cell. 23:1457–1466 10.1091/mbc.E11-11-0904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orjalo A.V., Arnaoutov A., Shen Z., Boyarchuk Y., Zeitlin S.G., Fontoura B., Briggs S., Dasso M., Forbes D.J. 2006. The Nup107-160 nucleoporin complex is required for correct bipolar spindle assembly. Mol. Biol. Cell. 17:3806–3818 10.1091/mbc.E05-11-1061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Platani M., Santarella-Mellwig R., Posch M., Walczak R., Swedlow J.R., Mattaj I.W. 2009. The Nup107-160 nucleoporin complex promotes mitotic events via control of the localization state of the chromosome passenger complex. Mol. Biol. Cell. 20:5260–5275 10.1091/mbc.E09-05-0377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosasco-Nitcher S.E., Lan W., Khorasanizadeh S., Stukenberg P.T. 2008. Centromeric Aurora-B activation requires TD-60, microtubules, and substrate priming phosphorylation. Science. 319:469–472 10.1126/science.1148980 [DOI] [PubMed] [Google Scholar]
- Ruchaud S., Carmena M., Earnshaw W.C. 2007. Chromosomal passengers: conducting cell division. Nat. Rev. Mol. Cell Biol. 8:798–812 10.1038/nrm2257 [DOI] [PubMed] [Google Scholar]
- Salimian K.J., Ballister E.R., Smoak E.M., Wood S., Panchenko T., Lampson M.A., Black B.E. 2011. Feedback control in sensing chromosome biorientation by the Aurora B kinase. Curr. Biol. 21:1158–1165 10.1016/j.cub.2011.06.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santaguida S., Tighe A., D’Alise A.M., Taylor S.S., Musacchio A. 2010. Dissecting the role of MPS1 in chromosome biorientation and the spindle checkpoint through the small molecule inhibitor reversine. J. Cell Biol. 190:73–87 10.1083/jcb.201001036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santaguida S., Vernieri C., Villa F., Ciliberto A., Musacchio A. 2011. Evidence that Aurora B is implicated in spindle checkpoint signalling independently of error correction. EMBO J. 30:1508–1519 10.1038/emboj.2011.70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saurin A.T., van der Waal M.S., Medema R.H., Lens S.M.A., Kops G.J.P.L. 2011. Aurora B potentiates Mps1 activation to ensure rapid checkpoint establishment at the onset of mitosis. Nat. Commun. 2:316 10.1038/ncomms1319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sessa F., Mapelli M., Ciferri C., Tarricone C., Areces L.B., Schneider T.R., Stukenberg P.T., Musacchio A. 2005. Mechanism of Aurora B activation by INCENP and inhibition by hesperadin. Mol. Cell. 18:379–391 10.1016/j.molcel.2005.03.031 [DOI] [PubMed] [Google Scholar]
- Söderberg O., Gullberg M., Jarvius M., Ridderstråle K., Leuchowius K.-J., Jarvius J., Wester K., Hydbring P., Bahram F., Larsson L.-G., Landegren U. 2006. Direct observation of individual endogenous protein complexes in situ by proximity ligation. Nat. Methods. 3:995–1000 10.1038/nmeth947 [DOI] [PubMed] [Google Scholar]
- Stucke V.M., Baumann C., Nigg E.A. 2004. Kinetochore localization and microtubule interaction of the human spindle checkpoint kinase Mps1. Chromosoma. 113:1–15 10.1007/s00412-004-0288-2 [DOI] [PubMed] [Google Scholar]
- Sun L., Gao J., Dong X., Liu M., Li D., Shi X., Dong J.-T., Lu X., Liu C., Zhou J. 2008. EB1 promotes Aurora-B kinase activity through blocking its inactivation by protein phosphatase 2A. Proc. Natl. Acad. Sci. USA. 105:7153–7158 10.1073/pnas.0710018105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terada Y. 2001. Role of chromosomal passenger complex in chromosome segregation and cytokinesis. Cell Struct. Funct. 26:653–657 10.1247/csf.26.653 [DOI] [PubMed] [Google Scholar]
- Tighe A., Staples O., Taylor S. 2008. Mps1 kinase activity restrains anaphase during an unperturbed mitosis and targets Mad2 to kinetochores. J. Cell Biol. 181:893–901 10.1083/jcb.200712028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tirnauer J.S., Canman J.C., Salmon E.D., Mitchison T.J. 2002. EB1 targets to kinetochores with attached, polymerizing microtubules. Mol. Biol. Cell. 13:4308–4316 10.1091/mbc.E02-04-0236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai M.-Y., Wiese C., Cao K., Martin O., Donovan P., Ruderman J., Prigent C., Zheng Y. 2003. A Ran signalling pathway mediated by the mitotic kinase Aurora A in spindle assembly. Nat. Cell Biol. 5:242–248 10.1038/ncb936 [DOI] [PubMed] [Google Scholar]
- Tseng B.S., Tan L., Kapoor T.M., Funabiki H. 2010. Dual detection of chromosomes and microtubules by the chromosomal passenger complex drives spindle assembly. Dev. Cell. 18:903–912 10.1016/j.devcel.2010.05.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsukahara T., Tanno Y., Watanabe Y. 2010. Phosphorylation of the CPC by Cdk1 promotes chromosome bi-orientation. Nature. 467:719–723 10.1038/nature09390 [DOI] [PubMed] [Google Scholar]
- Tulu U.S., Fagerstrom C., Ferenz N.P., Wadsworth P. 2006. Molecular requirements for kinetochore-associated microtubule formation in mammalian cells. Curr. Biol. 16:536–541 10.1016/j.cub.2006.01.060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Waal M.S., Saurin A.T., Vromans M.J.M., Vleugel M., Wurzenberger C., Gerlich D.W., Medema R.H., Kops G.J.P.L., Lens S.M.A. 2012. Mps1 promotes rapid centromere accumulation of Aurora B. EMBO Rep. 13:847–854 10.1038/embor.2012.93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang E., Ballister E.R., Lampson M.A. 2011a. Aurora B dynamics at centromeres create a diffusion-based phosphorylation gradient. J. Cell Biol. 194:539–549 10.1083/jcb.201103044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang F., Dai J., Daum J.R., Niedzialkowska E., Banerjee B., Stukenberg P.T., Gorbsky G.J., Higgins J.M. 2010. Histone H3 Thr-3 phosphorylation by Haspin positions Aurora B at centromeres in mitosis. Science. 330:231–235 10.1126/science.1189435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang F., Ulyanova N.P., van der Waal M.S., Patnaik D., Lens S.M.A., Higgins J.M.G. 2011b. A positive feedback loop involving Haspin and Aurora B promotes CPC accumulation at centromeres in mitosis. Curr. Biol. 21:1061–1069 10.1016/j.cub.2011.05.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang F., Ulyanova N.P., Daum J.R., Patnaik D., Kateneva A.V., Gorbsky G.J., Higgins J.M.G. 2012. Haspin inhibitors reveal centromeric functions of Aurora B in chromosome segregation. J. Cell Biol. 199:251–268 10.1083/jcb.201205106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welburn J.P.I., Vleugel M., Liu D., Yates J.R., III, Lampson M.A., Fukagawa T., Cheeseman I.M. 2010. Aurora B phosphorylates spatially distinct targets to differentially regulate the kinetochore-microtubule interface. Mol. Cell. 38:383–392 10.1016/j.molcel.2010.02.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wheatley S.P., Carvalho A., Vagnarelli P., Earnshaw W.C. 2001. INCENP is required for proper targeting of Survivin to the centromeres and the anaphase spindle during mitosis. Curr. Biol. 11:886–890 10.1016/S0960-9822(01)00238-X [DOI] [PubMed] [Google Scholar]
- Wheatley S.P., Henzing A.J., Dodson H., Khaled W., Earnshaw W.C. 2004. Aurora-B phosphorylation in vitro identifies a residue of survivin that is essential for its localization and binding to inner centromere protein (INCENP) in vivo. J. Biol. Chem. 279:5655–5660 10.1074/jbc.M311299200 [DOI] [PubMed] [Google Scholar]
- Yamagishi Y., Honda T., Tanno Y., Watanabe Y. 2010. Two histone marks establish the inner centromere and chromosome bi-orientation. Science. 330:239–243 10.1126/science.1194498 [DOI] [PubMed] [Google Scholar]
- Zimniak T., Stengl K., Mechtler K., Westermann S. 2009. Phosphoregulation of the budding yeast EB1 homologue Bim1p by Aurora/Ipl1p. J. Cell Biol. 186:379–391 10.1083/jcb.200901036 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.