Abstract
The accumulation and aggregation of misfolded proteins is the primary hallmark for more than 45 human degenerative diseases. These devastating disorders include Alzheimer’s, Parkinson’s, Huntington’s, and amyotrophic lateral sclerosis. Over 15 degenerative diseases are associated with the aggregation of misfolded proteins specifically in the nucleus of cells. However, how the cell safeguards the nucleus from misfolded proteins is not entirely clear. In this review, we discuss what is currently known about the cellular mechanisms that maintain protein homeostasis in the nucleus and protect the nucleus from misfolded protein accumulation and aggregation. In particular, we focus on the chaperones found to localize to the nucleus during stress, the ubiquitin–proteasome components enriched in the nucleus, the signaling systems that might be present in the nucleus to coordinate folding and degradation, and the sites of misfolded protein deposition associated with the nucleus.
Keywords: Nucleus, Chaperone, Ubiquitin-protein ligase, Ubiquitin, Proteasome, Unfolded protein response, Misfolded protein, Aggregation, Inclusion, Aggresome, JUNQ
Introduction
Proteins are principal molecular machines in the cell—catalyzing enzymatic reactions, building key cellular structures, and mediating information transfer from DNA to RNA to protein. The centrality of proteins to cellular physiology means that the cell must produce and maintain a cohort of functional proteins that are folded into correct conformations, assembled into appropriate complexes, localized to proper subcellular compartments, regulated by important signals, and sustained at suitable concentrations. The cell’s collective ability to do this has been generally referred to as protein homeostasis (or proteostasis [1]).
Of considerable importance in protein homeostasis is the folding state of proteins. The birth of every protein begins as a nascent peptide produced by the ribosomal translation of mRNA. Nascent peptides must subsequently be folded into specific three-dimensional conformations to confer final protein function and interactions with other proteins, membranes, DNA, RNA, or small molecules. And while the initial folding of a protein is essential for its function, so too is the maintenance of a protein’s functional structure throughout the lifetime of the protein. The initial folding of a protein and the continued maintenance of its folded structure are vulnerable to misfolding events that can be triggered by genetic mutations, synthesis errors, or post-synthesis damage caused by physical or chemical stresses. When protein folding goes awry, not only is protein function lost, misfolded proteins can acquire aggregation-prone states that are toxic to the cell.
The deleterious nature of protein misfolding and subsequent aggregation is best viewed through the lens of human health. It has been revealed that misfolded protein aggregation underlies the pathology for many devastating human disorders such as Alzheimer’s disease, Parkinson’s disease, Huntington’s disease, amyotrophic lateral sclerosis (ALS), and prion disorders like Creutzfeldt–Jakob [2]. Presently, it is not clear how misfolding and aggregation cause cellular toxicity in each pathology. In the simplest sense, the misfolded proteins produced could overwhelm the cell’s capacity to maintain general protein homeostasis. Alternatively, the misfolded proteins could gain a toxic function that affects specific cellular pathways. One important feature of most protein aggregation diseases is that they are degenerative and typically manifest their symptoms later in life [3], suggesting that one or more age-dependent factors is required for the pathogenesis of these diseases. It is also thought that aging itself might result from the increasing accumulation and aggregation of misfolded proteins with age [4]. How misfolded proteins contribute to the aging process and how aging contributes to protein aggregation diseases is currently a topic of intense interest. The precise mechanism(s) by which a misfolded protein affects a cell will depend upon the nature of the misfolded protein, the cellular compartment in which it is expressed, the state of the cellular milieu during its expression, and the particular cell type in which it is expressed. The failures in protein folding that lead to protein aggregation diseases underscores the need to understand the ways in which the cell normally manages misfolded proteins to prevent toxic misfolded protein aggregation.
General cellular protein quality control
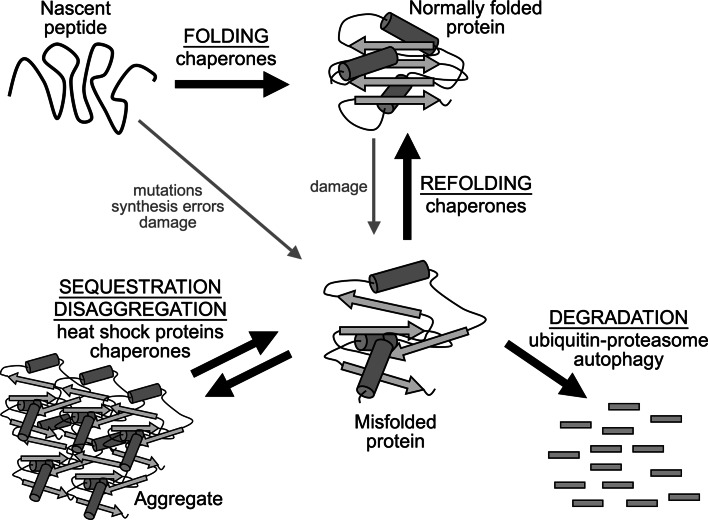
Due to the deleterious nature of protein misfolding, the eukaryotic cell has evolved a battery of compartment-specific protein quality control (PQC) systems to handle misfolded proteins as they arise within a specific organelle. In general, cellular PQC management of misfolded proteins can be divided into three functions: the repair, sequestration, and degradation of misfolded proteins (Fig. 1). The repair and sequestration aspects of PQC primarily involve protein chaperones that aid in the folding of nascent proteins, refolding of misfolded proteins, disassembling of aggregates, binding of misfolded proteins to prevent their aggregation, and sequestering of misfolded protein aggregates into cellular inclusions [5–7]. The destruction aspect of PQC typically employs ubiquitin-protein ligases that recognize and ubiquitinate misfolded proteins for proteasomal degradation [8]. Autophagy is another key route for the destruction of misfolded proteins [9]. For the most part, each cellular compartment possesses its own distinct PQC systems to manage misfolded proteins as they arise locally. Compartment-specific repair, sequestration, and destruction systems are also thought to function together in a “triage” hierarchy [10, 11], which would provide each cellular compartment the ability to determine the best action towards a misfolded protein depending on whether it can be salvaged. For example, if chaperones cannot unfold/refold a misfolded protein, they would direct the misfolded protein to degradation machinery for its destruction.
Fig. 1.
Primary PQC systems in the eukaryotic cell. Misfolded proteins can be generated through mutations, synthesis errors, and damage through physical or chemical stress during and after nascent peptide folding. Stages of the main PQC system action are indicated
In addition to the compartment-specific primary PQC systems that directly manage misfolded proteins, the cell also possesses compartment-specific secondary PQC systems that sense the burden of misfolded proteins and adjust production of the primary PQC components in an organelle accordingly. For example, the ER unfolded protein response modulates the expression of the primary ER PQC systems to accommodate the burden of misfolded proteins in the ER during ER stress [12]. Similarly, the mitochondrial unfolded protein response regulates the expression of the primary mitochondrial PQC systems as misfolded proteins accumulate in mitochondria [13]. These secondary PQC systems are critical because the misfolded protein load in a cellular compartment often changes with varying external environmental or internal metabolic conditions, and the primary PQC repair, sequestration, and degradation systems must be coordinately modulated to meet these changing demands.
The nucleus and protein quality control
More than 15 human diseases are associated with misfolded protein aggregation and inclusion formation of misfolded proteins in the nucleus [14]. These include the known polyQ-expansion diseases: Huntington’s disease, six different spinal cerebellar ataxias, spinal-bulbar muscular atrophy (Kennedy’s disease), and dentatorubral-pallidoluysian atrophy [15]. One polyA-expansion disease, oculopharyngeal muscular dystrophy, is also associated with aggregation of proteins in the nucleus [16]. Nuclear inclusions are also observed in diseases caused by mutations that are not tract-expanding such as neuronal intranuclear inclusion disease (NIID), neuronal intermediate filament inclusion disease (NIFID), multiple system atrophy (MSA), neuroferritinopathy, and inclusion body myopathy with early onset Paget's disease and frontotemporal dementia (IBMPFD) [14].
The existence of nuclear aggregation diseases indicates that the nuclear environment is susceptible to the toxicity of misfolded proteins, but it is not clear why. Do misfolded proteins disrupt chromatin organization, DNA replication, DNA repair, transcription, ribosome biogenesis, nuclear import/export, or nuclear structure? What are the PQC systems in the nucleus that protect it from toxic misfolded proteins? What do they recognize as abnormal, and are they incapable of targeting or processing certain types of misfolded proteins? If they are capable, are lesions in the nuclear PQC systems required for nuclear aggregation diseases to develop? Alternatively, are there age-dependent decrements in nuclear PQC function that contribute to the toxic accumulation and aggregation of misfolded proteins? Before these questions can be answered, it is important to understand how proteins arrive in the nucleus and the environment to which they are exposed while in the nucleus.
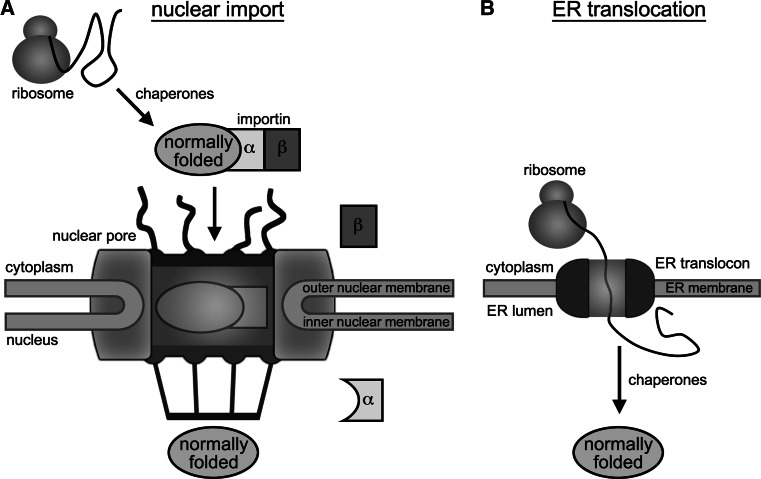
Nuclear proteins are first synthesized in the cytoplasm (Fig. 2). Thus, unlike the cytoplasm, the nucleus is not a primary site of protein synthesis and does not have to contend with a high burden of nascent proteins that must be folded immediately after translation by ribosomes. We do note, however, there are reports suggesting that some translation might occur in the nucleus [17, 18], perhaps in part to mediate nonsense-mediated decay of defectively produced mRNAs [19, 20]. If nuclear translation does occur to facilitate nonsense-mediated decay, the nucleus would need to manage the translation products generated, especially those produced from defective mRNAs. At this time, the existence and functional purpose for nuclear translation remains controversial [21–24], but could be an important parameter to consider in the context of protein misfolding in the nucleus.
Fig. 2.
Nuclear import versus ER translocation. Nuclear proteins are transported through the nuclear pore in intact, folded states, whereas ER proteins are transported as nascent peptides across the ER translocon and folded in the ER lumen
After synthesis in the cytoplasm, nuclear proteins enter the nucleus through either passive diffusion (only if they are below the ~40-kDa passive diffusion limit of the nuclear pore) or active transport by karyopherins (importins) [25] (Fig. 2). During active transport, the nuclear pore can expand from 9 to ~39 nm [26], allowing it to accommodate very large single-subunit proteins and multi-subunit protein complexes that can reach megadaltons in size [25]. Import of proteins into the nucleus is therefore very different from import of proteins into the ER or mitochondria, in which nascent proteins targeted to these compartments are trafficked across the ER membrane or outer mitochondrial membrane in an unfolded state by threading through a membrane translocon [27, 28] (Fig. 2). Given the ability of nuclear pores to expand greatly during active transport, it is generally thought that most resident nuclear proteins are in a properly folded state and assembled into appropriate subcomplexes prior to their transport into the nucleus [25, 26]. Based on the ability of nuclear pores to accommodate folded proteins, it is reasonable to suspect that PQC in the nucleus will be predominantly involved in managing nuclear proteins that have become damaged and misfolded after transiting the nuclear pore.
However, recent evidence suggests that the nucleus might also be a site of PQC for misfolded cytoplasmic proteins. While it would be predicted that misfolded proteins arising de novo in the cytoplasm should be handled by cytoplasmic PQC mechanisms, there is now a growing body of literature demonstrating that some misfolded cytoplasmic proteins in yeast are trafficked to the nucleus for PQC degradation [29–34]. It is currently unknown how these misfolded cytoplasmic proteins enter the nucleus. In some cases, the misfolded protein’s size is at or below the ~40-kDa passive diffusion limit of the yeast nuclear pore, such as ∆2GFP (~27 kDa) and Ste6*C (~28 kDa) [32, 33]. Thus, they could enter the nucleus via passive diffusion. In other cases, the misfolded protein’s size exceeds the passive diffusion limit, such as ∆ssPrA (~43 kDa) and CPY†-GFP (~85 kDa) [30, 32]. An active import mechanism would be required for nuclear localization of these proteins. Why misfolded cytoplasmic proteins would be actively imported into the nucleus remains a mystery. Perhaps it is a function of protein synthesis, where the cytoplasm must manage the folding of nascent peptides and the nucleus does not. As a consequence, the nucleus could have evolved to harbor the most aggressive PQC degradation systems aimed at destroying any protein that is not in a properly folded state. In fact, the proteasome is enriched in the nucleus [35], indicating that the nucleus likely has robust degradative capabilities. A mechanism that sends grossly misfolded cytoplasmic proteins to the nucleus could have been evolutionarily selected to partition PQC degradation from nascent PQC folding.
While there appears to be a directed action towards sending some misfolded cytoplasmic proteins to the nucleus in yeast, this has yet to be established in mammalian cells. However, it has been shown that nuclear pores break down in mammalian cells as a consequence of aging and become more permissive to larger cytoplasmic proteins, such as tubulin, leaking into the nucleus [36]. Nuclear pore breakdown during the course of aging, subsequently leading to increased access of cytoplasmic proteins to the nucleus, has the potential to challenge nuclear PQC mechanisms as the cell ages.
The observations showing that cytoplasmic proteins gain access to the nucleus have important implications in terms nuclear protein aggregation diseases. Purposeful trafficking or accidental leakage of misfolded proteins into the nucleus could have dire consequences for the health of the cell if the imported misfolded proteins are not managed appropriately within the confines of the nucleus. For example, if the misfolded cytoplasmic proteins reach sufficient levels in the nucleus, they could overwhelm nuclear PQC systems leading to a general increase in the burden of misfolded proteins in the nucleus. Alternatively, the misfolded cytoplasmic proteins themselves could confer a specific toxicity in the nucleus. This latter scenario might be the case for Huntington’s disease, which is caused by aggregation of a polyQ-expanded, truncated form of the huntingtin protein [37]. Huntingtin, in its full-length form, is primarily localized to the cytoplasm and associated with secretory vesicles in neurons [38]. However, polyQ-expanded, truncated huntingtin localizes to nuclear inclusions [39], and is particularly toxic in the nucleus [40, 41]. How common a trend it is for misfolded cytoplasmic proteins to mislocalize to the nucleus in nuclear protein aggregation diseases remains to be determined. Many misfolded proteins causally linked to nuclear protein aggregation diseases are normally nuclear localized such as the nuclear transcriptional corepressor atrophin-1 in dentatorubral-pallidoluysian atrophy [42], the nuclear transcription factor androgen receptor in spinal-bulbar muscular atrophy (Kennedy’s disease) [43], and the nuclear mRNA polyadenine-binding protein PABPN1 in oculopharyngeal muscular dystrophy [44].
Once in the nucleus, proteins face a different environment than the cytoplasm in terms of the molecules they encounter and the compartments to which they partition. Molecular crowding is similar between the nucleoplasm and cytoplasm [45, 46], indicating that overall movement and molecular collisions will be fairly equivalent. However, the nucleus is not homogenous and is divided into a number of distinct subcompartments. Thus, it is possible that the unique nature of each subcompartment poses different threats for misfolding and necessitates PQC mechanisms discrete from each other.
The most generally recognized feature of the nucleus is that it harbors the cell’s genetic material packaged into chromatin. There are potential dangers associated with chromatin related to protein misfolding. Most notably, the processes that regulate chromatin structure could contribute to protein misfolding in the nucleus. For example, access of transcription factors, histone-modifying enzymes, and RNA and DNA polymerases to compacted chromatin requires powerful ATP-dependent chromatin remodelers [47], which could unfold proteins during the chromatin-remodeling process. Key protein-dependent chromatin processes that could be impaired by protein misfolding and failures in nuclear PQC include DNA stability and repair, DNA replication, chromosome segregation, transcription, and gene silencing.
Another important subcompartment in the nucleus is the nucleolus, which encompasses the cell’s ribosomal DNA and is the center of ribosome biogenesis and assembly [48]. The process of ribosome biogenesis could burden PQC systems in the nucleus due to the dozens of ribosomal proteins in exceptionally high concentrations that must be correctly incorporated into immature ribosome subunits as they are built in the nucleolus. Any stoichiometric imbalances in the ribosomal proteins or the dozens of complexes that build the subunits could significantly add to the PQC burden within the nucleus. Furthermore, protein misfolding or failures of PQC in the nucleolus could negatively impact production of the cell’s cohort of ribosomes, which in turn could disrupt global protein homeostasis in the cell.
There are other, well-defined nuclear subcompartments where protein misfolding could affect the viability of the cell. Cajal bodies are implicated in many RNA-related functions including snRNP biogenesis, mRNA processing, and telomere maintenance [49]. PML bodies are punctate structures associated with transcriptional activation, DNA-damage repair, resistance to viral infection, and apoptosis [50]. Nuclear lamins provide an underlying architecture for the nucleus and are involved in an array of nuclear functions including transcription, DNA replication, and DNA repair [51]. Alteration of nuclear lamin structure is known to cause various premature aging diseases [52], which could be exacerbated if lamin structure is perturbed by protein misfolding.
In addition to considering that the mode of nuclear PQC might be distinct between nuclear subcompartments, it is also worth contemplating whether nuclear PQC varies between different cell types. The a priori assumption would be that nuclear PQC would be the same between cell types because the same nuclear functions occur in all nondividing somatic cells: e.g., DNA transcription, mRNA processing, ribosome biogenesis, nuclear transport, and maintenance of nuclear architecture. However, there does appear to be cell-type sensitivity for misfolded protein toxicity in nuclear protein aggregation diseases as neurons are primarily affected [14]. The neuronal cell bias could exist because nuclear protein aggregation simply has not yet been identified in the etiology of other diseases affecting non-neuronal cell types. On the other hand, this bias could have arisen because neurons are one of the longest-lived cell types and may be more affected by decrements in nuclear protein homeostasis as they age compared to other cell types. Even among the nuclear protein aggregation diseases, not all neurons are equally affected despite the fact that the deleterious misfolded protein is often widely expressed in many different cell types. For example, it is striatal medium spiny neurons that are the most vulnerable to polyQ-expanded huntingtin in Huntington’s disease [53], whereas it is lower motor neurons in the spinal cord and brainstem that are the most vulnerable to polyQ-expanded androgen receptor in spinal bulbar muscular atrophy (SBMA) [43]. It is possible that selective vulnerability of distinct neuronal cells is due to a cell-specific effect of the misfolded protein, such as cell-specific transcriptional dysregulation [54]. Alternatively, cell-specific sensitivity to the particular misfolded protein could arise due to differences in the nuclear PQC capabilities between cell types.
At this point, we do not understand the full scope of PQC capabilities in the nucleus. Thus, we do not know if distinct nuclear PQC systems generally operate throughout the nucleus or are specific to individual subcompartments in the nucleus. We also do not understand if there are differences in the nature or robustness of nuclear PQC between different cell types that could lead to selective vulnerability. In the following sections, we discuss what is currently known about PQC systems that protect the nuclear environment.
Nuclear PQC: chaperones
Chaperones are central components of cellular PQC and are pivotal in the proper folding of nascent polypeptides, the assembly or disassembly of macromolecular complexes, the transport of proteins across membranes, the unfolding/refolding of misfolded proteins, and the disassociation of protein aggregates [55]. In addition, there is significant crosstalk between chaperones and PQC degradation systems such as the ubiquitin–proteasome system [56]. To understand the primary, direct role for chaperones in nuclear PQC, we think it is important to separate chaperone functions in the folding of newly synthesized nuclear proteins in the cytoplasm from PQC functions explicitly in the nucleus. Thus, we will limit our discussion to those chaperones that have demonstrated nuclear localization. Since we will not provide a detailed review of how these chaperones function mechanistically in terms of protein folding, refolding, and disaggregation, we refer the reader to the recent excellent review on chaperone function by Hartl and colleagues [55].
The main evidence demonstrating a role for chaperones in maintaining nuclear protein homeostasis comes from studies examining the localization of chaperones under stress conditions known to cause protein misfolding. For example, a number of Hsp70 chaperones undergo considerable relocalization to the nucleus during heat shock in mammalian cells [57–63]. The budding yeast Hsp70 chaperone Ssa4 becomes enriched in the nucleus after ethanol or starvation stress [64–66]. Similarly, yeast Hsp104 also undergoes significant nuclear relocalization during heat shock [67–69]. In mammalian and yeast cells, the small heat shock protein Hsp26 has notable relocalization to the nucleus under heat shock conditions [70, 71]. Our understanding of nuclear PQC and the factors that function in the nuclear environment is still fledgling. As such, we think it is likely that other chaperones relocalize to the nucleus during protein misfolding stress. However, these are the best examples to date for chaperones involved in nuclear PQC.
Enhanced nuclear localization of chaperones under protein misfolding stress conditions, such as heat shock or oxidative stress, is quite interesting due to the fact that protein misfolding stress conditions inhibit normal nuclear protein import [72–77]. Inhibition of normal nuclear import during protein misfolding stress happens, in large part, by prevention of nuclear export of the karyopherin proteins (importins) that are required for active nuclear transport [72–77]. Because the karyopherins are not recycled to the cytoplasm, they can no longer function in nuclear import. Nuclear export blockade of karyopherins has been posited to be due to disruption of the Ran gradient essential for recycling of the karyopherins [72–75, 78]. Alternatively, it could be due to the misfolding of the karyopherins under stress conditions. Whatever the mechanism(s), normal nuclear import is decreased during stress conditions, yet nuclear localization of chaperones is increased [62, 65]. To facilitate increased chaperone levels in the nucleus during misfolded protein stress conditions, the cell appears to use two mechanisms: reduced chaperone export from the nucleus [57, 65, 79], and enhanced import of chaperones into the nucleus using specialized nuclear transport carriers that are unaffected by protein stress [62]. The overall effect of reduced normal nuclear import and increased chaperone nuclear localization is likely to be an important combined physiological response to protein misfolding in the nucleus. It would be advantageous for the cell to prevent new nuclear proteins from entering the nucleus until the burden of misfolded proteins in the nucleus generated under the current stress is resolved.
In addition to relocalizing to the nucleus under conditions of protein misfolding stress, chaperones have also been shown to colocalize with nuclear inclusions formed by misfolded proteins associated with human disease. For example, nuclear inclusions formed by polyQ expansions in huntingtin (htt), androgen receptor, ataxin-1, ataxin-3, and ataxin-7 show considerable colocalization with Hsp70, Hsp40, and/or Hsp110 chaperones [80–88]. Nuclear inclusions formed by polyA expansions in PABPN1 and ataxin-3 also demonstrate colocalization with Hsp70 and Hsp40 chaperones [86, 89–92]. Nuclear inclusions formed by other misfolded proteins have also been found to include Hsp70 and Hsp40 chaperones [93–95]. The functional purpose of chaperones in nuclear inclusions is not yet clear. Perhaps they help to form the inclusion, thereby reducing the amount of free misfolded species within the nucleus. Alternatively, they may act to disaggregate the inclusion to facilitate refolding or degradation. Whatever their particular mode of action in the inclusion, the fact that chaperones localize to nuclear inclusions is another key piece of evidence for chaperone function in nuclear PQC.
As mentioned earlier in the review, recent studies have shown that a number of misfolded cytoplasmic proteins enter the nucleus and are subject to nuclear PQC degradation [29–33]. In budding yeast, the Hsp70 chaperones Ssa1/Ssa2, the Hsp110 chaperone Sse1, the Hsp40 chaperones Ydj1 and Sis1 are involved in the nuclear PQC degradation of this class of misfolded protein [29–33]. How these chaperones function in nuclear PQC degradation is not clear at this time. Interestingly, both Sse1 and Sis1 have been implicated in the nuclear transport of certain misfolded cytoplasmic proteins [30–32], which suggests that at least one function for these chaperones may be to facilitate the trafficking of misfolded cytoplasmic proteins into the nucleus.
Nuclear PQC: ubiquitin-protein ligases
The ubiquitin–proteasome system is a principal means for the destruction of misfolded proteins in the cell [8]. As mentioned above, the proteasome is enriched in the nucleus [35], supporting the idea that ubiquitin-mediated proteasomal degradation is a primary route for misfolded protein elimination in the nucleus. The small signaling protein ubiquitin is covalently attached to target substrates via a conserved enzymatic cascade [96]. Initially, a ubiquitin-activating enzyme, or E1, binds ubiquitin in a high-energy thiol-ester bond. A ubiquitin-conjugating enzyme, or E2, then accepts ubiquitin from the activating enzyme with a similar high-energy thiol-ester bond. Lastly, a ubiquitin-protein ligase, or E3, acts to transfer the ubiquitin molecule to the substrate. The E3 is typically thought to confer substrate specificity via intrinsic interaction domains or recruitment of ancillary substrate-recognition factors [97]. Identification of the E3s that function in PQC degradation in the nucleus and characterization of how they function are current topics of investigation. Here we focus our discussion on E3s with demonstrated nuclear localization and defined roles in the degradation of misfolded proteins.
In the budding yeast Saccharomyces cerevisiae, the major E3 that is involved in the PQC degradation of misfolded proteins in the nucleus is San1 [98–102]. In support of San1 mediating nuclear PQC, San1 is predominantly localized to the nucleus [99], and cannot recognize misfolded protein substrates if they are redirected to the cytoplasm [103]. Furthermore, San1 is highly selective for misfolded proteins and does not target normally folded proteins for degradation [99, 103]. Substrate recognition by San1 occurs through the binding of misfolded proteins to regions in San1 located N- and C-terminal to its RING domain [103]. These N- and C-terminal regions are predicted to contain short, linear, substrate-binding modules separated by highly disordered linker sequences [103]. This distinct configuration is thought to provide San1 with a conformationally plastic structure that allows San1 the ability to adapt itself to the wide variety of shapes its misfolded substrates are likely to adopt [103]. Within its misfolded protein substrates, San1 generally recognizes exposed hydrophobicity that is at the threshold that can cause aggregation of the misfolded protein [104, 105]. San1 homologs exist in a variety of other fungi [103, 106], with a fission yeast Schizosaccharomyces pombe homolog also shown to function in the PQC degradation of misfolded nuclear proteins [107].
Although no mammalian homolog of San1 has been identified to date, the mammalian E3s UHRF-2, PML IV, and E6-AP have been suggested to function in nuclear PQC degradation [87, 93, 108, 109]. The nuclear PQC degradation function of UHRF-2 is proposed from its involvement in the nuclear degradation of polyQ-expanded, truncated huntingtin [108]. The nuclear PQC degradation function for PML IV is posited from its role in the degradation of a nuclear protein with an expanded polyQ tract [87]. In addition, PML IV colocalizes with nuclear inclusions formed by polyQ-expanded proteins [87, 93]. Lastly, E6-AP has been implicated in nuclear PQC by its involvement in the degradation of polyQ-expanded, truncated huntingtin and its localization to huntingtin nuclear inclusions [109]. Because these E3s have only been shown to target polyQ-expanded proteins, further studies on other misfolded proteins are required to support bona fide global functions for these E3s in nuclear PQC degradation.
In addition to San1 in budding yeast, the yeast E3 Ubr1 is also involved in the PQC degradation of misfolded proteins that have been shown to be nuclear localized [29–34]. This function is likely conserved as Ubr1 homologs exist in mammalian cells [110], and mammalian UBR1 has been implicated in controlling the levels of Hsp90 client proteins [111]. The protein degradation functions of Ubr1 were initially characterized in terms of its ability to select proteins containing particular destabilizing N-terminal residues [112, 113], with Ubr1 as the major recognition component of the N-end rule pathway [114]. By contrast, how Ubr1 selects misfolded proteins for PQC degradation is not yet known. In particular, it is not clear what feature of misfolding Ubr1 recognizes. The N-terminal residue of misfolded proteins does not appear to be the recognized feature [30], indicating that the PQC recognition aspect of Ubr1 is independent of its N-end rule recognition function. Despite that, it is worth considering the N-end rule function of Ubr1 in the context of nuclear PQC. For example, it is conceivable that improper post-translational processing of a nuclear protein’s N-terminus could affect its folding or associations [115, 116]. Also, nuclear proteins that have undergone regulated proteolytic cleavage have been shown to expose destabilizing N-end rule residues in the truncated C-terminal fragments that are recognized by Ubr1 [117–119]. Persistence of these truncated proteins in the absence of Ubr1 function could exert dominant-negative effects in their normal physiological pathways [117–119], or allow misfolded fragments to aggregate and potentially lead to neurodegeneration [120].
In considering the nuclear PQC functions of yeast Ubr1 and San1, there is some overlap between the E3s in the misfolded proteins that they target for degradation [29–34]. However, the majority of misfolded San1 substrates are solely targeted by San1 [99, 103–105] and some misfolded Ubr1 substrates are solely targeted by Ubr1 [30, 121], demonstrating that Ubr1 and San1 do have at least a partial separation of function in PQC degradation. One explanation is that some misfolded proteins display both the feature of misfolding recognized by San1 and the feature of misfolding recognized by Ubr1 thus leading to their degradation by both pathways. On the other hand, some substrates might display only the feature of misfolding recognized by either San1 or Ubr1 and are thus subject to degradation solely by either pathway. Until the feature of misfolding recognized by Ubr1 is discovered as has been for San1 [104, 105], this question will remain unresolved.
Alternatively, it is possible that the separation in PQC degradation functions for Ubr1 and San1 are due to the PQC degradation role of Ubr1 being limited to the cytoplasm [30], and the PQC degradation role of San1 limited to the nucleus [99]. If compartmentalization of San1 and Ubr1 underlies their partial separation of function, then the Ubr1-dependency for a misfolded nuclear protein’s PQC degradation might consequently depend upon the time the misfolded protein spends in the cytoplasm prior to its nuclear import. The longer the misfolded protein dwells in the cytoplasm, the more its PQC degradation will be Ubr1 dependent. Currently, the localization for budding yeast Ubr1 has not yet been published, but fission yeast Ubr1 is localized to both the cytoplasm and nucleus [122]. Additional work will be needed to clarify the exact location for the PQC degradation functions of budding yeast Ubr1.
Lastly, the budding yeast protein Doa10 was originally characterized as an ER-membrane localized E3 that ubiquitinates proteins in the ER for proteasomal degradation [123–126]. However, the ER membrane and the nuclear envelope are continuous [127], and it was found that Doa10 does indeed localize to the inner nuclear membrane where it is required to mediate the ubiquitination and degradation of the transcription factor Matα2 [128]. Although the degradation of Matα2 regulates mating-type switching in yeast, Doa10 has also been shown to mediate the degradation of misfolded proteins that are predicted or known to localize to the nucleus [126, 129, 130]. Like San1, Doa10 appears to recognize exposed hydrophobicity [130], though the molecular rules for substrate hydrophobicity appear to be different than those for San1 recognition because there is little overlap between Doa10 and San1 substrates. Perhaps San1 and Doa10 have a division of labor in terms of the type of exposed hydrophobicity they recognize, and this broadens the capabilities of the nucleus for recognizing and destroying misfolded proteins. Alternatively, it could be that San1 mediates the degradation of misfolded proteins that are nucleoplasmic in nature whereas Doa10 mediates the degradation of misfolded proteins that localize to the surface of the inner nuclear membrane. Future comparative studies will be necessary to understand the potential interplay and functional separation of these two nuclear PQC E3s. Doa10 does have a mammalian homolog called TEB4 [131], though a nuclear PQC role for TEB4 has yet to be established.
Nuclear PQC: protein SUMOylation
While ubiquitination of misfolded proteins is a key posttranslational mechanism used in nuclear PQC to facilitate misfolded protein degradation, another posttranslational mechanism potentially used in nuclear PQC is modification of proteins by the small ubiquitin-like modifier (SUMO). Structurally similar to ubiquitin, SUMO has been shown to modify many cellular proteins, particularly during stresses that can cause protein misfolding [132, 133]. For example, after heat shock or oxidative stress, there is a general increase in the SUMOylation of predominantly nuclear proteins in yeast, plants, and mammalian cells [134–137]. The majority of these nuclear SUMOylation targets are involved in transcription, chromatin regulation, RNA processing, and metabolism [134–137]. In many cases, it is reasonable to suspect that the stress-induced SUMOylation of nuclear proteins will alter their function in order to mediate important physiological responses to stress.
It is also important to consider that SUMOylation could be a direct factor involved in maintaining nuclear protein homeostasis during stress. For example, a subset of plant proteins targeted for stress-induced SUMOylation also undergo increased ubiquitination [134]. In addition, proteasome impairment in yeast has been shown to increase the global levels of high molecular weight SUMO conjugates that are also ubiquitinated [138]. Furthermore, proteasome inhibition of mammalian cells has been found to increase protein SUMOylation [138, 139], to an extent similar to what is observed during heat shock [139]. Interestingly, the proteasome inhibition-dependent increase in SUMOylation in mammalian cells requires active protein synthesis [139], suggesting that SUMOylation could potentially act as a degradation signal for nascent misfolded proteins. The same study also found that SUMO conjugates accumulate within insoluble inclusions during proteasome inhibition [139].
The relationship between SUMO, protein solubility, and protein aggregation is intriguing and complex. SUMO is known to be an extremely soluble protein. In fact, as an artificial fusion tag, it is a highly effective tool for enhancing the solubility of difficult-to-express recombinant proteins [140, 141]. In recent years, SUMO has been found to modify a number of aggregation-prone proteins associated with neurodegenerative diseases [142]. In some cases, SUMO may function as a potential solubilizing factor. For example, SUMOylation of α-synuclein, a major component of Lewy body inclusions [143], has been shown to prevent α-synuclein fibril formation in vitro and reduced SUMOylation of α-synuclein enhances its neurotoxicity in vivo [144]. By contrast, increased SUMOylation of nuclear polyQ-expanded ataxin-1 during oxidative stress enhances ataxin-1 insolubility and inclusion formation in vivo [145]. The modulation of huntingtin by SUMO is also complex. SUMOylation of polyQ-expanded huntingtin reduces its inclusion formation in mammalian cells but enhances neurodegeneration in flies [146]. The role of SUMO in protein aggregation diseases is clearly not straightforward and will likely depend upon the particular nature of the misfolded protein. From the combined studies, however, it is emerging that SUMO plays a broader role in PQC than simply direct regulation of protein activity during stress. The studies are beginning to reveal a distinct axis in PQC in which SUMO likely functions in modulating misfolded protein solubility and stability.
Nuclear PQC: signaling pathways
Eukaryotic cells dramatically alter their transcriptome to adapt to stress conditions that result in an increased burden of misfolded proteins. Most cellular compartments have been shown to possess the means to sense the levels of misfolded proteins in the compartment and signal to the nucleus to increase the transcription of the compartment’s primary PQC protection components. As previously mentioned, accumulation of misfolded proteins in the ER can lead to the ER unfolded protein response, which results in the increased transcription of ER PQC chaperones and E3s [12]. The mitochondria also possess a similar unfolded response that elevates the expression of mitochondrial PQC chaperones and proteases as misfolded proteins accumulate in the mitochondria [13]. In the cytoplasm, accumulation of misfolded proteins can lead to induction of the heat shock response, which elevates the expression of cytoplasmic chaperones [147–149].
While unfolded protein responses have been well characterized in other cellular compartments, none have yet been described for the nucleus. Nevertheless, we think that one does exist. In our previous studies characterizing the role of the E3 San1 in nuclear PQC degradation, we found that there was increased expression of several protein chaperone and stress genes in san1∆ cells [99]. The chaperone genes include those that encode for Ssa4, Hsp104, and Hsp26, which have been shown to become enriched in the nucleus during protein misfolding stress [64–69, 71]. Because the deletion of SAN1 would lead to the accumulation of misfolded proteins specifically in the nucleus, we think that the increased expression of these chaperone genes in san1∆ cells is due to a secondary PQC system within the nucleus that senses the level of protein misfolding and adjusts expression of the primary nuclear PQC systems accordingly. However, it has yet to be established that increased expression of these chaperones in turn leads to their increased presence in the nucleus, which is an important parameter if it is a nucleus-specific response. Future work will be required to clarify the consequences of chaperone up-regulation in the absence of San1. Should the increased expression of chaperones in san1∆ cells constitute a nuclear unfolded protein response, it will be important to discover the misfolded protein sensors and transcription factors of this response to determine how it operates in the nucleus.
Nuclear PQC: inclusion sites
Despite the formidable chaperone and degradative PQC capabilities deployed by the eukaryotic cell to manage misfolded proteins, the burden of misfolded proteins can become quite high under conditions that exacerbate protein misfolding, such as chemical or physical stresses, nutritional deficiencies, and aging [3, 4, 150]. As misfolded proteins accumulate and overwhelm the primary PQC systems, they can progressively oligomerize and aggregate in the cell [151]. It is now well understood that the etiology for dozens of degenerative human diseases is linked to the oligomerization and aggregation of misfolded proteins [2, 151]. These diseases are typically characterized by the age-dependent accumulation of pathogenic misfolded proteins into visible cellular inclusions [2, 151]. Because of their association with degenerative human diseases, inclusions were long thought to be the pathological manifestation of misfolded protein aggregation.
Recent work, however, has indicated that inclusion formation is not a passive result of aggregation and may not be a pathological process [152–156]. Rather, it appears that organization of misfolded proteins into specific subcellular domains is a key physiological aspect of PQC that assists in the efficient triage and processing of misfolded proteins by concentrating them into dedicated PQC regions. Inclusions also appear to serve as a means to sequester small soluble oligomers of misfolded proteins, which are currently thought to be the species that cause cellular toxicity [151]. Prominent cellular inclusions where misfolded proteins are known to be directed during high levels of protein misfolding stress are the perinuclear aggresome [157], the perinuclear JUNQ [153], and the perivacuolar IPOD [153]. For the purposes of this review, we will focus on cytoprotective inclusions associated with the nucleus—the JUNQ and the aggresome.
In budding yeast, a key inclusion associated with the nucleus is the JUNQ (for juxtanuclear quality control) [153]. The JUNQ is formed within a proliferation of the nuclear envelope and contains misfolded proteins that are intended for ubiquitin–proteasome degradation [153]. The misfolded proteins localized within the JUNQ are fairly mobile [153], but under severe stress or in the presence of toxic aggregates the mobility of misfolded proteins in the JUNQ can decrease [158, 159]. Consistent with the notion that the JUNQ is an active site for PQC management of misfolded proteins, Hsp70 chaperones and proteasome subunits colocalize with the JUNQ [158]. Similar to the misfolded proteins that compose the JUNQ, chaperones that associate with the JUNQ also exchange freely with the cytoplasm [153, 158], but the rate of chaperone exchange with the cytoplasm decreases during stress [158]. Additionally, compartmentalization of misfolded proteins in the JUNQ plays a key PQC role in the asymmetric inheritance of misfolded proteins, as localization to the JUNQ prevents misfolded proteins from being passed to the daughter cell from the mother cell during mitosis [155]. Altogether, it is thought that the JUNQ provides a storage and processing platform for misfolded proteins intended for proteasomal degradation and perhaps refolding.
The accumulation of misfolded proteins in compartments like the JUNQ depends upon the type of misfolding, the nature of the stress and its duration. For example, ubiquitinated misfolded proteins in budding yeast are sent to the JUNQ, but those that fail to be ubiquitinated are sent to the perivacuolar IPOD (for insoluble protein deposit) instead of the JUNQ [153]. By contrast, detergent insoluble amyloidogenic proteins are sent directly to the IPOD and not the JUNQ [153]. Despite their differences, both the JUNQ and IPOD appear to be “final destination” compartments for proteins that are committed to degradation or sequestration, respectively. Recently, it was found that, upon exposure to protein misfolding stress, a conditionally misfolded protein localizes to transient intermediary inclusions called Stress Foci (SF) [155], which have also been called Q Bodies (QB) [152]. Under prolonged exposure to protein misfolding stress, SF/QB subsequently coalesce into the JUNQ and IPOD [152, 155]. This suggests that SF/QB are important early sites for PQC triage, although the mechanism that determines which misfolded proteins eventually partition to the JUNQ or the IPOD remains to be determined.
Studies in mammalian cells have identified several types of inclusions in different subcellular compartments, indicating that similar sequestration mechanisms for managing misfolded proteins and aggregation exist in mammalian cells [157–160]. The best-characterized mammalian inclusion is the perinuclear aggresome, which sequesters misfolded cytoplasmic proteins at the microtubule-organizing center (MTOC) adjacent to the nucleus [157]. The aggresome has several properties similar to the JUNQ. Overexpression of misfolded proteins or inhibition of the proteasome leads to accumulation of misfolded proteins in the aggresome [157]. It has been shown that Hsp70 and Hsp40 chaperones as well as proteasome subunits associate with the aggresome [80, 161, 162], suggesting that the aggresome is a site of active PQC like the JUNQ. Until recently, most mammalian inclusions were called aggresomes. However, it is becoming clear that, similar to yeast, mammalian cells can direct misfolded proteins to distinct inclusion structures that have features in common with the yeast JUNQ and IPOD [158, 160].
In terms of the nuclear interior, we now know that more than 15 human degenerative diseases are marked by visible inclusions within the nuclei of affected cells [14]. In many cases, chaperones colocalize with the nuclear inclusions [80–92]. Thus, the nuclear inclusions have at least one characteristic of the perinuclear aggresome and JUNQ. A key open question is whether the nuclear inclusions formed by distinct misfolded proteins are the same nuclear structure or if they are different in each specific case. If they are different, is the difference due to aggregation within a specific subcompartment of the nucleus? This may be the case in the nucleolus. It was recently reported that an inclusion body containing polyadenylated RNA, cyclins, and transcription factors forms within the nucleolus after proteasome inhibition [163]. Currently, it is not clear if the nucleolar inclusion is the result of misfolded protein aggregation that occurs when proteasomal degradation is impaired, or if it is another nuclear body formed when particular proteasome substrates cannot be degraded. Additional work will be required to cement the nucleolar inclusion as an aggresome-like structure involved in the sequestration of misfolded proteins. Broadening the outlook to the entire nucleus, more focus will need to be applied to understand the sequestration mechanisms that protect the nucleus from toxic misfolded protein aggregation.
Concluding remarks
Over the last two decades, our understanding of cellular PQC systems that protect the cell from the toxic aggregation of misfolded proteins has grown by leaps and bounds. Studies examining the general biochemical mechanisms for PQC chaperones and E3s have revealed important insights into their modes of action. The focus on the compartmentalization of PQC systems has provided a window into how organelles specifically manage misfolded proteins as they arise on site. Because the nucleus houses our genetic material and performs essential cellular functions, we think greater attention should be paid to understand how the cell protects this organelle from deleterious alterations in protein homeostasis. We think this is especially important when viewed through the lens of the many degenerative human diseases caused by protein aggregation in the nucleus and the loss of protein homeostasis as cells age.
Acknowledgments
We tried to cite all primary literature pertaining to nuclear PQC. We apologize to any colleagues if we unintentionally missed their studies. This work was supported by an NIH/NIA grant R01AG031136 (R.G.G.), an Ellison Medical Foundation New Scholar Award in Aging (R.G.G), an Israel Science Foundation Grant ISF 843/11 (D.K), a German–Israel Foundation Grant GIF I-1201-242.13/2012 (D.K.), an ERC-StG2013 337713 DarkSide grant (D.K.), and an American Federation for Aging Research grant (D.K.).
References
- 1.Balch WE, Morimoto RI, Dillin A, Kelly JW. Adapting proteostasis for disease intervention. Science. 2008;319:916–919. doi: 10.1126/science.1141448. [DOI] [PubMed] [Google Scholar]
- 2.Wang SS, Wu JW, Yamamoto S, Liu HS. Diseases of protein aggregation and the hunt for potential pharmacological agents. Biotechnol J. 2008;3:165–192. doi: 10.1002/biot.200700065. [DOI] [PubMed] [Google Scholar]
- 3.Kikis EA, Gidalevitz T, Morimoto RI. Protein homeostasis in models of aging and age-related conformational disease. Adv Exp Med Biol. 2010;694:138–159. doi: 10.1007/978-1-4419-7002-2_11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Taylor RC, Dillin A. Aging as an event of proteostasis collapse. Cold Spring Harb Perspect Biol. 2011;3:a004440. doi: 10.1101/cshperspect.a004440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Voisine C, Pedersen JS, Morimoto RI. Chaperone networks: tipping the balance in protein folding diseases. Neurobiol Dis. 2010;40:12–20. doi: 10.1016/j.nbd.2010.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen B, Retzlaff M, Roos T, Frydman J. Cellular strategies of protein quality control. Cold Spring Harb Perspect Biol. 2011;3:a004374. doi: 10.1101/cshperspect.a004374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hartl FU, Bracher A, Hayer-Hartl M. Molecular chaperones in protein folding and proteostasis. Nature. 2011;475:324–332. doi: 10.1038/nature10317. [DOI] [PubMed] [Google Scholar]
- 8.Fredrickson EK, Gardner RG. Selective destruction of abnormal proteins by ubiquitin-mediated protein quality control degradation. Semin Cell Dev Biol. 2012;23:530–537. doi: 10.1016/j.semcdb.2011.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sridhar S, Botbol Y, Macian F, Cuervo AM. Autophagy and disease: always two sides to a problem. J Pathol. 2012;226:255–273. doi: 10.1002/path.3025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Arndt V, Rogon C, Hohfeld J. To be, or not to be–molecular chaperones in protein degradation. Cell Mol Life Sci. 2007;64:2525–2541. doi: 10.1007/s00018-007-7188-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wickner S, Maurizi MR, Gottesman S. Posttranslational quality control: folding, refolding, and degrading proteins. Science. 1999;286:1888–1893. doi: 10.1126/science.286.5446.1888. [DOI] [PubMed] [Google Scholar]
- 12.Gardner BM, Pincus D, Gotthardt K, Gallagher CM, Walter P. Endoplasmic reticulum stress sensing in the unfolded protein response. Cold Spring Harb Perspect Biol. 2013;5:a013169. doi: 10.1101/cshperspect.a013169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Haynes CM, Fiorese CJ, Lin YF. Evaluating and responding to mitochondrial dysfunction: the mitochondrial unfolded-protein response and beyond. Trends Cell Biol. 2013;23:311–318. doi: 10.1016/j.tcb.2013.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Woulfe J. Nuclear bodies in neurodegenerative disease. Biochim Biophys Acta. 2008;1783:2195–2206. doi: 10.1016/j.bbamcr.2008.05.005. [DOI] [PubMed] [Google Scholar]
- 15.Ross CA. Polyglutamine pathogenesis: emergence of unifying mechanisms for Huntington’s disease and related disorders. Neuron. 2002;35:819–822. doi: 10.1016/s0896-6273(02)00872-3. [DOI] [PubMed] [Google Scholar]
- 16.Amiel J, Trochet D, Clement-Ziza M, Munnich A, Lyonnet S. Polyalanine expansions in human. Hum Mol Genet. 2004;13:R235–R243. doi: 10.1093/hmg/ddh251. [DOI] [PubMed] [Google Scholar]
- 17.David A, Dolan BP, Hickman HD, Knowlton JJ, Clavarino G, Pierre P, Bennink JR, Yewdell JW. Nuclear translation visualized by ribosome-bound nascent chain puromycylation. J Cell Biol. 2012;197:45–57. doi: 10.1083/jcb.201112145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Iborra FJ, Jackson DA, Cook PR. Coupled transcription and translation within nuclei of mammalian cells. Science. 2001;293:1139–1142. doi: 10.1126/science.1061216. [DOI] [PubMed] [Google Scholar]
- 19.Iborra FJ, Escargueil AE, Kwek KY, Akoulitchev A, Cook PR. Molecular cross-talk between the transcription, translation, and nonsense-mediated decay machineries. J Cell Sci. 2004;117:899–906. doi: 10.1242/jcs.00933. [DOI] [PubMed] [Google Scholar]
- 20.Iborra FJ, Jackson DA, Cook PR. The case for nuclear translation. J Cell Sci. 2004;117:5713–5720. doi: 10.1242/jcs.01538. [DOI] [PubMed] [Google Scholar]
- 21.Dahlberg JE, Lund E, Goodwin EB. Nuclear translation: what is the evidence? RNA. 2003;9:1–8. doi: 10.1261/rna.2121703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pederson T. The persistent plausibility of protein synthesis in the nucleus: process, palimpsest or pitfall? Curr Opin Cell Biol. 2013;25:520–521. doi: 10.1016/j.ceb.2013.06.006. [DOI] [PubMed] [Google Scholar]
- 23.Nathanson L, Xia T, Deutscher MP. Nuclear protein synthesis: a re-evaluation. RNA. 2003;9:9–13. doi: 10.1261/rna.2990203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dahlberg J, Lund E. Nuclear translation or nuclear peptidyl transferase? Nucleus. 2012;3:320–321. doi: 10.4161/nucl.20754. [DOI] [PubMed] [Google Scholar]
- 25.Grossman E, Medalia O, Zwerger M. Functional architecture of the nuclear pore complex. Annu Rev Biophys. 2012;41:557–584. doi: 10.1146/annurev-biophys-050511-102328. [DOI] [PubMed] [Google Scholar]
- 26.Pante N, Kann M. Nuclear pore complex is able to transport macromolecules with diameters of about 39 nm. Mol Biol Cell. 2002;13:425–434. doi: 10.1091/mbc.01-06-0308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dudek J, Rehling P, van der Laan M. Mitochondrial protein import: common principles and physiological networks. Biochim Biophys Acta. 2013;1833:274–285. doi: 10.1016/j.bbamcr.2012.05.028. [DOI] [PubMed] [Google Scholar]
- 28.Johnson N, Powis K, High S. Post-translational translocation into the endoplasmic reticulum. Biochim Biophys Acta. 2012;1833:2403–2409. doi: 10.1016/j.bbamcr.2012.12.008. [DOI] [PubMed] [Google Scholar]
- 29.Guerriero CJ, Weiberth KF, Brodsky JL. Hsp70 targets a cytoplasmic quality control substrate to the San1p ubiquitin ligase. J Biol Chem. 2013;288:18506–18520. doi: 10.1074/jbc.M113.475905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Heck JW, Cheung SK, Hampton RY. Cytoplasmic protein quality control degradation mediated by parallel actions of the E3 ubiquitin ligases Ubr1 and San1. Proc Natl Acad Sci USA. 2010;107:1106–1111. doi: 10.1073/pnas.0910591107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Park SH, Kukushkin Y, Gupta R, Chen T, Konagai A, Hipp MS, Hayer-Hartl M, Hartl FU. PolyQ proteins interfere with nuclear degradation of cytosolic proteins by sequestering the Sis1p chaperone. Cell. 2013;154:134–145. doi: 10.1016/j.cell.2013.06.003. [DOI] [PubMed] [Google Scholar]
- 32.Prasad R, Kawaguchi S, Ng DT. A nucleus-based quality control mechanism for cytosolic proteins. Mol Biol Cell. 2010;21:2117–2127. doi: 10.1091/mbc.E10-02-0111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Prasad R, Kawaguchi S, Ng DT. Biosynthetic mode can determine the mechanism of protein quality control. Biochem Biophys Res Commun. 2012;425:689–695. doi: 10.1016/j.bbrc.2012.07.080. [DOI] [PubMed] [Google Scholar]
- 34.Summers DW, Wolfe KJ, Ren HY, Cyr DM. The Type II Hsp40 Sis1 cooperates with Hsp70 and the E3 ligase Ubr1 to promote degradation of terminally misfolded cytosolic protein. PLoS ONE. 2013;8:e52099. doi: 10.1371/journal.pone.0052099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wojcik C, DeMartino GN. Intracellular localization of proteasomes. Int J Biochem Cell Biol. 2003;35:579–589. doi: 10.1016/s1357-2725(02)00380-1. [DOI] [PubMed] [Google Scholar]
- 36.D’Angelo MA, Raices M, Panowski SH, Hetzer MW. Age-dependent deterioration of nuclear pore complexes causes a loss of nuclear integrity in postmitotic cells. Cell. 2009;136:284–295. doi: 10.1016/j.cell.2008.11.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Arrasate M, Finkbeiner S. Protein aggregates in Huntington’s disease. Exp Neurol. 2012;238:1–11. doi: 10.1016/j.expneurol.2011.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.DiFiglia M, Sapp E, Chase K, Schwarz C, Meloni A, Young C, Martin E, Vonsattel JP, Carraway R, Reeves SA, et al. Huntingtin is a cytoplasmic protein associated with vesicles in human and rat brain neurons. Neuron. 1995;14:1075–1081. doi: 10.1016/0896-6273(95)90346-1. [DOI] [PubMed] [Google Scholar]
- 39.DiFiglia M, Sapp E, Chase KO, Davies SW, Bates GP, Vonsattel JP, Aronin N. Aggregation of huntingtin in neuronal intranuclear inclusions and dystrophic neurites in brain. Science. 1997;277:1990–1993. doi: 10.1126/science.277.5334.1990. [DOI] [PubMed] [Google Scholar]
- 40.Peters MF, Nucifora FC, Jr, Kushi J, Seaman HC, Cooper JK, Herring WJ, Dawson VL, Dawson TM, Ross CA. Nuclear targeting of mutant Huntingtin increases toxicity. Mol Cell Neurosci. 1999;14:121–128. doi: 10.1006/mcne.1999.0773. [DOI] [PubMed] [Google Scholar]
- 41.Schilling G, Savonenko AV, Klevytska A, Morton JL, Tucker SM, Poirier M, Gale A, Chan N, Gonzales V, Slunt HH, Coonfield ML, Jenkins NA, Copeland NG, Ross CA, Borchelt DR. Nuclear-targeting of mutant huntingtin fragments produces Huntington’s disease-like phenotypes in transgenic mice. Hum Mol Genet. 2004;13:1599–1610. doi: 10.1093/hmg/ddh175. [DOI] [PubMed] [Google Scholar]
- 42.Shen Y, Peterson AS. Atrophins’ emerging roles in development and neurodegenerative disease. Cell Mol Life Sci. 2009;66:437–446. doi: 10.1007/s00018-008-8403-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Katsuno M, Tanaka F, Adachi H, Banno H, Suzuki K, Watanabe H, Sobue G. Pathogenesis and therapy of spinal and bulbar muscular atrophy (SBMA) Prog Neurobiol. 2012;99:246–256. doi: 10.1016/j.pneurobio.2012.05.007. [DOI] [PubMed] [Google Scholar]
- 44.Brais B. Oculopharyngeal muscular dystrophy: a polyalanine myopathy. Curr Neurol Neurosci Rep. 2009;9:76–82. doi: 10.1007/s11910-009-0012-y. [DOI] [PubMed] [Google Scholar]
- 45.Guigas G, Kalla C, Weiss M. The degree of macromolecular crowding in the cytoplasm and nucleoplasm of mammalian cells is conserved. FEBS Lett. 2007;581:5094–5098. doi: 10.1016/j.febslet.2007.09.054. [DOI] [PubMed] [Google Scholar]
- 46.Guigas G, Kalla C, Weiss M. Probing the nanoscale viscoelasticity of intracellular fluids in living cells. Biophys J . 2007;93:316–323. doi: 10.1529/biophysj.106.099267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Clapier CR, Cairns BR. The biology of chromatin remodeling complexes. Annu Rev Biochem. 2009;78:273–304. doi: 10.1146/annurev.biochem.77.062706.153223. [DOI] [PubMed] [Google Scholar]
- 48.Lempiainen H, Shore D. Growth control and ribosome biogenesis. Curr Opin Cell Biol. 2009;21:855–863. doi: 10.1016/j.ceb.2009.09.002. [DOI] [PubMed] [Google Scholar]
- 49.Morris GE. The Cajal body. Biochim Biophys Acta. 2008;1783:2108–2115. doi: 10.1016/j.bbamcr.2008.07.016. [DOI] [PubMed] [Google Scholar]
- 50.Lallemand-Breitenbach V, de The H. PML nuclear bodies. Cold Spring Harb Perspect Biol. 2010;2:a000661. doi: 10.1101/cshperspect.a000661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Dittmer TA, Misteli T. The lamin protein family. Genome Biol. 2011;12:222. doi: 10.1186/gb-2011-12-5-222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Schreiber KH, Kennedy BK. When lamins go bad: nuclear structure and disease. Cell. 2013;152:1365–1375. doi: 10.1016/j.cell.2013.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ehrlich ME. Huntington’s disease and the striatal medium spiny neuron: cell-autonomous and non-cell-autonomous mechanisms of disease. Neurotherapeutics. 2012;9:270–284. doi: 10.1007/s13311-012-0112-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Desplats PA, Kass KE, Gilmartin T, Stanwood GD, Woodward EL, Head SR, Sutcliffe JG, Thomas EA. Selective deficits in the expression of striatal-enriched mRNAs in Huntington’s disease. J Neurochem. 2006;96:743–757. doi: 10.1111/j.1471-4159.2005.03588.x. [DOI] [PubMed] [Google Scholar]
- 55.Kim YE, Hipp MS, Bracher A, Hayer-Hartl M, Ulrich Hartl F. Molecular chaperone functions in protein folding and proteostasis. Annu Rev Biochem. 2013;82:323–355. doi: 10.1146/annurev-biochem-060208-092442. [DOI] [PubMed] [Google Scholar]
- 56.Kriegenburg F, Ellgaard L, Hartmann-Petersen R. Molecular chaperones in targeting misfolded proteins for ubiquitin-dependent degradation. FEBS J. 2012;279:532–542. doi: 10.1111/j.1742-4658.2011.08456.x. [DOI] [PubMed] [Google Scholar]
- 57.Kodiha M, Chu A, Lazrak O, Stochaj U. Stress inhibits nucleocytoplasmic shuttling of heat shock protein hsc70. Am J Physiol Cell Physiol. 2005;289:C1034–C1041. doi: 10.1152/ajpcell.00590.2004. [DOI] [PubMed] [Google Scholar]
- 58.Welch WJ, Feramisco JR. Nuclear and nucleolar localization of the 72,000-dalton heat shock protein in heat-shocked mammalian cells. J Biol Chem. 1984;259:4501–4513. [PubMed] [Google Scholar]
- 59.Velazquez JM, Lindquist S. hsp70: nuclear concentration during environmental stress and cytoplasmic storage during recovery. Cell. 1984;36:655–662. doi: 10.1016/0092-8674(84)90345-3. [DOI] [PubMed] [Google Scholar]
- 60.Pelham HR. Hsp70 accelerates the recovery of nucleolar morphology after heat shock. EMBO J. 1984;3:3095–3100. doi: 10.1002/j.1460-2075.1984.tb02264.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lamian V, Small GM, Feldherr CM. Evidence for the existence of a novel mechanism for the nuclear import of Hsc70. Exp Cell Res. 1996;228:84–91. doi: 10.1006/excr.1996.0302. [DOI] [PubMed] [Google Scholar]
- 62.Kose S, Furuta M, Imamoto N. Hikeshi, a nuclear import carrier for Hsp70s, protects cells from heat shock-induced nuclear damage. Cell. 2012;149:578–589. doi: 10.1016/j.cell.2012.02.058. [DOI] [PubMed] [Google Scholar]
- 63.Imamoto N, Kose S. Heat-shock stress activates a novel nuclear import pathway mediated by Hikeshi. Nucleus. 2012;3:422–428. doi: 10.4161/nucl.21713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Quan X, Rassadi R, Rabie B, Matusiewicz N, Stochaj U. Regulated nuclear accumulation of the yeast hsp70 Ssa4p in ethanol-stressed cells is mediated by the N-terminal domain, requires the nuclear carrier Nmd5p and protein kinase C. FASEB J. 2004;18:899–901. doi: 10.1096/fj.03-0947fje. [DOI] [PubMed] [Google Scholar]
- 65.Quan X, Tsoulos P, Kuritzky A, Zhang R, Stochaj U. The carrier Msn5p/Kap142p promotes nuclear export of the hsp70 Ssa4p and relocates in response to stress. Mol Microbiol. 2006;62:592–609. doi: 10.1111/j.1365-2958.2006.05395.x. [DOI] [PubMed] [Google Scholar]
- 66.Chughtai ZS, Rassadi R, Matusiewicz N, Stochaj U. Starvation promotes nuclear accumulation of the hsp70 Ssa4p in yeast cells. J Biol Chem. 2001;276:20261–20266. doi: 10.1074/jbc.M100364200. [DOI] [PubMed] [Google Scholar]
- 67.Kawai R, Fujita K, Iwahashi H, Komatsu Y. Direct evidence for the intracellular localization of Hsp104 in Saccharomyces cerevisiae by immunoelectron microscopy. Cell Stress Chaperones. 1999;4:46–53. doi: 10.1054/csac.1999.0076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Tkach JM, Glover JR. Nucleocytoplasmic trafficking of the molecular chaperone Hsp104 in unstressed and heat-shocked cells. Traffic. 2008;9:39–56. doi: 10.1111/j.1600-0854.2007.00666.x. [DOI] [PubMed] [Google Scholar]
- 69.Parsell DA, Kowal AS, Singer MA, Lindquist S. Protein disaggregation mediated by heat-shock protein Hsp104. Nature. 1994;372:475–478. doi: 10.1038/372475a0. [DOI] [PubMed] [Google Scholar]
- 70.Willsie JK, Clegg JS. Small heat shock protein p26 associates with nuclear lamins and HSP70 in nuclei and nuclear matrix fractions from stressed cells. J Cell Biochem. 2002;84:601–614. [PubMed] [Google Scholar]
- 71.Rossi JM, Lindquist S. The intracellular location of yeast heat-shock protein 26 varies with metabolism. J Cell Biol. 1989;108:425–439. doi: 10.1083/jcb.108.2.425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Kodiha M, Chu A, Matusiewicz N, Stochaj U. Multiple mechanisms promote the inhibition of classical nuclear import upon exposure to severe oxidative stress. Cell Death Differ. 2004;11:862–874. doi: 10.1038/sj.cdd.4401432. [DOI] [PubMed] [Google Scholar]
- 73.Czubryt MP, Austria JA, Pierce GN. Hydrogen peroxide inhibition of nuclear protein import is mediated by the mitogen-activated protein kinase, ERK2. J Cell Biol. 2000;148:7–16. doi: 10.1083/jcb.148.1.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Miyamoto Y, Saiwaki T, Yamashita J, Yasuda Y, Kotera I, Shibata S, Shigeta M, Hiraoka Y, Haraguchi T, Yoneda Y. Cellular stresses induce the nuclear accumulation of importin alpha and cause a conventional nuclear import block. J Cell Biol. 2004;165:617–623. doi: 10.1083/jcb.200312008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Furuta M, Kose S, Koike M, Shimi T, Hiraoka Y, Yoneda Y, Haraguchi T, Imamoto N. Heat-shock induced nuclear retention and recycling inhibition of importin alpha. Genes Cells. 2004;9:429–441. doi: 10.1111/j.1356-9597.2004.00734.x. [DOI] [PubMed] [Google Scholar]
- 76.Kodiha M, Tran D, Qian C, Morogan A, Presley JF, Brown CM, Stochaj U. Oxidative stress mislocalizes and retains transport factor importin-alpha and nucleoporins Nup153 and Nup88 in nuclei where they generate high molecular mass complexes. Biochim Biophys Acta. 2008;1783:405–418. doi: 10.1016/j.bbamcr.2007.10.022. [DOI] [PubMed] [Google Scholar]
- 77.Yasuda Y, Miyamoto Y, Saiwaki T, Yoneda Y. Mechanism of the stress-induced collapse of the Ran distribution. Exp Cell Res. 2006;312:512–520. doi: 10.1016/j.yexcr.2005.11.017. [DOI] [PubMed] [Google Scholar]
- 78.Kelley JB, Paschal BM. Hyperosmotic stress signaling to the nucleus disrupts the Ran gradient and the production of RanGTP. Mol Biol Cell. 2007;18:4365–4376. doi: 10.1091/mbc.E07-01-0089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Crampton N, Kodiha M, Shrivastava S, Umar R, Stochaj U. Oxidative stress inhibits nuclear protein export by multiple mechanisms that target FG nucleoporins and Crm1. Mol Biol Cell. 2009;20:5106–5116. doi: 10.1091/mbc.E09-05-0397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Waelter S, Boeddrich A, Lurz R, Scherzinger E, Lueder G, Lehrach H, Wanker EE. Accumulation of mutant huntingtin fragments in aggresome-like inclusion bodies as a result of insufficient protein degradation. Mol Biol Cell. 2001;12:1393–1407. doi: 10.1091/mbc.12.5.1393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Abel A, Walcott J, Woods J, Duda J, Merry DE. Expression of expanded repeat androgen receptor produces neurologic disease in transgenic mice. Hum Mol Genet. 2001;10:107–116. doi: 10.1093/hmg/10.2.107. [DOI] [PubMed] [Google Scholar]
- 82.Jana NR, Tanaka M, Wang G, Nukina N. Polyglutamine length-dependent interaction of Hsp40 and Hsp70 family chaperones with truncated N-terminal huntingtin: their role in suppression of aggregation and cellular toxicity. Hum Mol Genet. 2000;9:2009–2018. doi: 10.1093/hmg/9.13.2009. [DOI] [PubMed] [Google Scholar]
- 83.Bailey CK, Andriola IF, Kampinga HH, Merry DE. Molecular chaperones enhance the degradation of expanded polyglutamine repeat androgen receptor in a cellular model of spinal and bulbar muscular atrophy. Hum Mol Genet. 2002;11:515–523. doi: 10.1093/hmg/11.5.515. [DOI] [PubMed] [Google Scholar]
- 84.Adachi H, Katsuno M, Minamiyama M, Sang C, Pagoulatos G, Angelidis C, Kusakabe M, Yoshiki A, Kobayashi Y, Doyu M, Sobue G. Heat shock protein 70 chaperone overexpression ameliorates phenotypes of the spinal and bulbar muscular atrophy transgenic mouse model by reducing nuclear-localized mutant androgen receptor protein. J Neurosci. 2003;23:2203–2211. doi: 10.1523/JNEUROSCI.23-06-02203.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Ishihara K, Yamagishi N, Saito Y, Adachi H, Kobayashi Y, Sobue G, Ohtsuka K, Hatayama T. Hsp105alpha suppresses the aggregation of truncated androgen receptor with expanded CAG repeats and cell toxicity. J Biol Chem. 2003;278:25143–25150. doi: 10.1074/jbc.M302975200. [DOI] [PubMed] [Google Scholar]
- 86.Latouche M, Fragner P, Martin E, El Hachimi KH, Zander C, Sittler A, Ruberg M, Brice A, Stevanin G. Polyglutamine and polyalanine expansions in ataxin7 result in different types of aggregation and levels of toxicity. Mol Cell Neurosci. 2006;31:438–445. doi: 10.1016/j.mcn.2005.10.013. [DOI] [PubMed] [Google Scholar]
- 87.Janer A, Martin E, Muriel MP, Latouche M, Fujigasaki H, Ruberg M, Brice A, Trottier Y, Sittler A. PML clastosomes prevent nuclear accumulation of mutant ataxin-7 and other polyglutamine proteins. J Cell Biol. 2006;174:65–76. doi: 10.1083/jcb.200511045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Seidel K, Meister M, Dugbartey GJ, Zijlstra MP, Vinet J, Brunt ER, van Leeuwen FW, Rub U, Kampinga HH, den Dunnen WF. Cellular protein quality control and the evolution of aggregates in spinocerebellar ataxia type 3 (SCA3) Neuropathol Appl Neurobiol. 2012;38:548–558. doi: 10.1111/j.1365-2990.2011.01220.x. [DOI] [PubMed] [Google Scholar]
- 89.Bao YP, Cook LJ, O’Donovan D, Uyama E, Rubinsztein DC. Mammalian, yeast, bacterial, and chemical chaperones reduce aggregate formation and death in a cell model of oculopharyngeal muscular dystrophy. J Biol Chem. 2002;277:12263–12269. doi: 10.1074/jbc.M109633200. [DOI] [PubMed] [Google Scholar]
- 90.Corbeil-Girard LP, Klein AF, Sasseville AM, Lavoie H, Dicaire MJ, Saint-Denis A, Page M, Duranceau A, Codere F, Bouchard JP, Karpati G, Rouleau GA, Massie B, Langelier Y, Brais B. PABPN1 overexpression leads to upregulation of genes encoding nuclear proteins that are sequestered in oculopharyngeal muscular dystrophy nuclear inclusions. Neurobiol Dis. 2005;18:551–567. doi: 10.1016/j.nbd.2004.10.019. [DOI] [PubMed] [Google Scholar]
- 91.Chartier A, Benoit B, Simonelig M. A Drosophila model of oculopharyngeal muscular dystrophy reveals intrinsic toxicity of PABPN1. EMBO J. 2006;25:2253–2262. doi: 10.1038/sj.emboj.7601117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Tavanez JP, Bengoechea R, Berciano MT, Lafarga M, Carmo-Fonseca M, Enguita FJ. Hsp70 chaperones and type I PRMTs are sequestered at intranuclear inclusions caused by polyalanine expansions in PABPN1. PLoS ONE. 2009;4:e6418. doi: 10.1371/journal.pone.0006418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Fu L, Gao YS, Tousson A, Shah A, Chen TL, Vertel BM, Sztul E. Nuclear aggresomes form by fusion of PML-associated aggregates. Mol Biol Cell. 2005;16:4905–4917. doi: 10.1091/mbc.E05-01-0019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Fu L, Gao YS, Sztul E. Transcriptional repression and cell death induced by nuclear aggregates of non-polyglutamine protein. Neurobiol Dis. 2005;20:656–665. doi: 10.1016/j.nbd.2005.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Moran DM, Shen H, Maki CG. Puromycin-based vectors promote a ROS-dependent recruitment of PML to nuclear inclusions enriched with HSP70 and Proteasomes. BMC Cell Biol. 2009;10:32. doi: 10.1186/1471-2121-10-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Ciechanover A. Intracellular protein degradation: from a vague idea through the lysosome and the ubiquitin-proteasome system and onto human diseases and drug targeting. Neurodegener Dis. 2012;10:7–22. doi: 10.1159/000334283. [DOI] [PubMed] [Google Scholar]
- 97.Finley D, Ulrich HD, Sommer T, Kaiser P. The ubiquitin-proteasome system of Saccharomyces cerevisiae . Genetics. 2012;192:319–360. doi: 10.1534/genetics.112.140467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Dasgupta A, Ramsey KL, Smith JS, Auble DT. Sir antagonist 1 (San1) is a ubiquitin ligase. J Biol Chem. 2004;279:26830–26838. doi: 10.1074/jbc.M400894200. [DOI] [PubMed] [Google Scholar]
- 99.Gardner RG, Nelson ZW, Gottschling DE. Degradation-mediated protein quality control in the nucleus. Cell. 2005;120:803–815. doi: 10.1016/j.cell.2005.01.016. [DOI] [PubMed] [Google Scholar]
- 100.Evans DR, Brewster NK, Xu Q, Rowley A, Altheim BA, Johnston GC, Singer RA. The yeast protein complex containing cdc68 and pob3 mediates core-promoter repression through the cdc68 N-terminal domain. Genetics. 1998;150:1393–1405. doi: 10.1093/genetics/150.4.1393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Estruch F, Peiro-Chova L, Gomez-Navarro N, Durban J, Hodge C, Del Olmo M, Cole CN. A genetic screen in Saccharomyces cerevisiae identifies new genes that interact with me67-5, a temperature-sensitive allele of the gene encoding the mRNA export receptor. Mol Genet Genomics. 2009;281:125–134. doi: 10.1007/s00438-008-0402-x. [DOI] [PubMed] [Google Scholar]
- 102.Lewis MJ, Pelham HR. Inefficient quality control of thermosensitive proteins on the plasma membrane. PLoS ONE. 2009;4:e5038. doi: 10.1371/journal.pone.0005038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Rosenbaum JC, Fredrickson EK, Oeser ML, Garrett-Engele CM, Locke MN, Richardson LA, Nelson ZW, Hetrick ED, Milac TI, Gottschling DE, Gardner RG. Disorder targets misorder in nuclear quality control degradation: a disordered ubiquitin ligase directly recognizes its misfolded substrates. Mol Cell. 2011;41:93–106. doi: 10.1016/j.molcel.2010.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Fredrickson EK, Gallagher PS, Clowes Candadai SV, Gardner RG. Substrate recognition in nuclear protein quality control degradation is governed by exposed hydrophobicity that correlates with aggregation and insolubility. J Biol Chem. 2013;288:6130–6139. doi: 10.1074/jbc.M112.406710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Fredrickson EK, Rosenbaum JC, Locke MN, Milac TI, Gardner RG. Exposed hydrophobicity is a key determinant of nuclear quality control degradation. Mol Biol Cell. 2011;22:2384–2395. doi: 10.1091/mbc.E11-03-0256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Fredrickson EK, Clowes Candadai SV, Tam CH, Gardner RG. Means of self-preservation: how an intrinsically disordered ubiquitin-protein ligase averts self-destruction. Mol Biol Cell. 2013;24:1041–1052. doi: 10.1091/mbc.E12-11-0811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Matsuo Y, Kishimoto H, Tanae K, Kitamura K, Katayama S, Kawamukai M. Nuclear protein quality is regulated by the ubiquitin-proteasome system through the activity of Ubc4 and San1 in fission yeast. J Biol Chem. 2011;286:13775–13790. doi: 10.1074/jbc.M110.169953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Iwata A, Nagashima Y, Matsumoto L, Suzuki T, Yamanaka T, Date H, Deoka K, Nukina N, Tsuji S. Intra-nuclear degradation of polyglutamine aggregates by the ubiquitin proteasome system. J Biol Chem. 2009;284:9796–9803. doi: 10.1074/jbc.M809739200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Mishra A, Dikshit P, Purkayastha S, Sharma J, Nukina N, Jana NR. E6-AP promotes misfolded polyglutamine proteins for proteasomal degradation and suppresses polyglutamine protein aggregation and toxicity. J Biol Chem. 2008;283:7648–7656. doi: 10.1074/jbc.M706620200. [DOI] [PubMed] [Google Scholar]
- 110.Tasaki T, Sriram SM, Park KS, Kwon YT. The N-end rule pathway. Annu Rev Biochem. 2012;81:261–289. doi: 10.1146/annurev-biochem-051710-093308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Sultana R, Theodoraki MA, Caplan AJ. UBR1 promotes protein kinase quality control and sensitizes cells to Hsp90 inhibition. Exp Cell Res. 2012;318:53–60. doi: 10.1016/j.yexcr.2011.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Bartel B, Wunning I, Varshavsky A. The recognition component of the N-end rule pathway. EMBO J. 1990;9:3179–3189. doi: 10.1002/j.1460-2075.1990.tb07516.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Xia Z, Webster A, Du F, Piatkov K, Ghislain M, Varshavsky A. Substrate-binding sites of UBR1, the ubiquitin ligase of the N-end rule pathway. J Biol Chem. 2008;283:24011–24028. doi: 10.1074/jbc.M802583200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Varshavsky A. The N-end rule pathway and regulation by proteolysis. Protein Sci. 2011;20:1298–1345. doi: 10.1002/pro.666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Shemorry A, Hwang CS, Varshavsky A. Control of protein quality and stoichiometries by N-terminal acetylation and the N-end rule pathway. Mol Cell. 2013;50:540–551. doi: 10.1016/j.molcel.2013.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Hwang CS, Shemorry A, Varshavsky A. N-terminal acetylation of cellular proteins creates specific degradation signals. Science. 2010;327:973–977. doi: 10.1126/science.1183147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Piatkov KI, Brower CS, Varshavsky A. The N-end rule pathway counteracts cell death by destroying proapoptotic protein fragments. Proc Natl Acad Sci USA. 2012;109:E1839–E1847. doi: 10.1073/pnas.1207786109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Piatkov KI, Colnaghi L, Bekes M, Varshavsky A, Huang TT. The auto-generated fragment of the Usp1 deubiquitylase is a physiological substrate of the N-end rule pathway. Mol Cell. 2012;48:926–933. doi: 10.1016/j.molcel.2012.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Rao H, Uhlmann F, Nasmyth K, Varshavsky A. Degradation of a cohesin subunit by the N-end rule pathway is essential for chromosome stability. Nature. 2001;410:955–959. doi: 10.1038/35073627. [DOI] [PubMed] [Google Scholar]
- 120.Brower CS, Piatkov KI, Varshavsky A. Neurodegeneration-associated protein fragments as short-lived substrates of the N-end rule pathway. Mol Cell. 2013;50:161–171. doi: 10.1016/j.molcel.2013.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Eisele F, Wolf DH. Degradation of misfolded protein in the cytoplasm is mediated by the ubiquitin ligase Ubr1. FEBS Lett. 2008;582:4143–4146. doi: 10.1016/j.febslet.2008.11.015. [DOI] [PubMed] [Google Scholar]
- 122.Kitamura K, Taki M, Tanaka N, Yamashita I. Fission yeast Ubr1 ubiquitin ligase influences the oxidative stress response via degradation of active Pap1 bZIP transcription factor in the nucleus. Mol Microbiol. 2011;80:739–755. doi: 10.1111/j.1365-2958.2011.07605.x. [DOI] [PubMed] [Google Scholar]
- 123.Swanson R, Locher M, Hochstrasser M. A conserved ubiquitin ligase of the nuclear envelope/endoplasmic reticulum that functions in both ER-associated and Matalpha2 repressor degradation. Genes Dev. 2001;15:2660–2674. doi: 10.1101/gad.933301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Huyer G, Piluek WF, Fansler Z, Kreft SG, Hochstrasser M, Brodsky JL, Michaelis S. Distinct machinery is required in Saccharomyces cerevisiae for the endoplasmic reticulum-associated degradation of a multispanning membrane protein and a soluble luminal protein. J Biol Chem. 2004;279:38369–38378. doi: 10.1074/jbc.M402468200. [DOI] [PubMed] [Google Scholar]
- 125.Carvalho P, Goder V, Rapoport TA. Distinct ubiquitin-ligase complexes define convergent pathways for the degradation of ER proteins. Cell. 2006;126:361–373. doi: 10.1016/j.cell.2006.05.043. [DOI] [PubMed] [Google Scholar]
- 126.Ravid T, Kreft SG, Hochstrasser M. Membrane and soluble substrates of the Doa10 ubiquitin ligase are degraded by distinct pathways. EMBO J. 2006;25:533–543. doi: 10.1038/sj.emboj.7600946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.English AR, Voeltz GK. Endoplasmic reticulum structure and interconnections with other organelles. Cold Spring Harb Perspect Biol. 2013;5:a013227. doi: 10.1101/cshperspect.a013227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Deng M, Hochstrasser M. Spatially regulated ubiquitin ligation by an ER/nuclear membrane ligase. Nature. 2006;443:827–831. doi: 10.1038/nature05170. [DOI] [PubMed] [Google Scholar]
- 129.Alfassy OS, Cohen I, Reiss Y, Tirosh B, Ravid T. Placing a disrupted degradation motif at the C terminus of proteasome substrates attenuates degradation without impairing ubiquitylation. J Biol Chem. 2013;288:12645–12653. doi: 10.1074/jbc.M113.453027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Furth N, Gertman O, Shiber A, Alfassy OS, Cohen I, Rosenberg MM, Doron NK, Friedler A, Ravid T. Exposure of bipartite hydrophobic signal triggers nuclear quality control of Ndc10 at the endoplasmic reticulum/nuclear envelope. Mol Biol Cell. 2011;22:4726–4739. doi: 10.1091/mbc.E11-05-0463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Kreft SG, Wang L, Hochstrasser M. Membrane topology of the yeast endoplasmic reticulum-localized ubiquitin ligase Doa10 and comparison with its human ortholog TEB4 (MARCH-VI) J Biol Chem. 2006;281:4646–4653. doi: 10.1074/jbc.M512215200. [DOI] [PubMed] [Google Scholar]
- 132.Castro PH, Tavares RM, Bejarano ER, Azevedo H. SUMO, a heavyweight player in plant abiotic stress responses. Cell Mol Life Sci. 2012;69:3269–3283. doi: 10.1007/s00018-012-1094-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Tempe D, Piechaczyk M, Bossis G. SUMO under stress. Biochem Soc Trans. 2008;36:874–878. doi: 10.1042/BST0360874. [DOI] [PubMed] [Google Scholar]
- 134.Miller MJ, Scalf M, Rytz TC, Hubler SL, Smith LM, Vierstra RD. Quantitative proteomics reveals factors regulating RNA biology as dynamic targets of stress-induced SUMOylation in Arabidopsis . Mol Cell Proteomics. 2013;12:449–463. doi: 10.1074/mcp.M112.025056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Miller MJ, Barrett-Wilt GA, Hua Z, Vierstra RD. Proteomic analyses identify a diverse array of nuclear processes affected by small ubiquitin-like modifier conjugation in Arabidopsis . Proc Natl Acad Sci USA. 2010;107:16512–16517. doi: 10.1073/pnas.1004181107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Golebiowski F, Matic I, Tatham MH, Cole C, Yin Y, Nakamura A, Cox J, Barton GJ, Mann M, Hay RT (2009) System-wide changes to SUMO modifications in response to heat shock. Sci Signal 2: ra24 [DOI] [PubMed]
- 137.Zhou W, Ryan JJ, Zhou H. Global analyses of sumoylated proteins in Saccharomyces cerevisiae. Induction of protein sumoylation by cellular stresses. J Biol Chem. 2004;279:32262–32268. doi: 10.1074/jbc.M404173200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Uzunova K, Gottsche K, Miteva M, Weisshaar SR, Glanemann C, Schnellhardt M, Niessen M, Scheel H, Hofmann K, Johnson ES, Praefcke GJ, Dohmen RJ. Ubiquitin-dependent proteolytic control of SUMO conjugates. J Biol Chem. 2007;282:34167–34175. doi: 10.1074/jbc.M706505200. [DOI] [PubMed] [Google Scholar]
- 139.Tatham MH, Matic I, Mann M, Hay RT (2011) Comparative proteomic analysis identifies a role for SUMO in protein quality control. Sci Signal 4: rs4 [DOI] [PubMed]
- 140.Marblestone JG, Edavettal SC, Lim Y, Lim P, Zuo X, Butt TR. Comparison of SUMO fusion technology with traditional gene fusion systems: enhanced expression and solubility with SUMO. Protein Sci. 2006;15:182–189. doi: 10.1110/ps.051812706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Malakhov MP, Mattern MR, Malakhova OA, Drinker M, Weeks SD, Butt TR. SUMO fusions and SUMO-specific protease for efficient expression and purification of proteins. J Struct Funct Genomics. 2004;5:75–86. doi: 10.1023/B:JSFG.0000029237.70316.52. [DOI] [PubMed] [Google Scholar]
- 142.Krumova P, Weishaupt JH. Sumoylation in neurodegenerative diseases. Cell Mol Life Sci. 2013;70:2123–2138. doi: 10.1007/s00018-012-1158-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Goedert M, Spillantini MG, Del Tredici K, Braak H. 100 years of Lewy pathology. Nat Rev Neurol. 2013;9:13–24. doi: 10.1038/nrneurol.2012.242. [DOI] [PubMed] [Google Scholar]
- 144.Krumova P, Meulmeester E, Garrido M, Tirard M, Hsiao HH, Bossis G, Urlaub H, Zweckstetter M, Kugler S, Melchior F, Bahr M, Weishaupt JH. Sumoylation inhibits alpha-synuclein aggregation and toxicity. J Cell Biol. 2011;194:49–60. doi: 10.1083/jcb.201010117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Ryu J, Cho S, Park BC, Lee do H. Oxidative stress-enhanced SUMOylation and aggregation of ataxin-1: implication of JNK pathway. Biochem Biophys Res Commun. 2010;393:280–285. doi: 10.1016/j.bbrc.2010.01.122. [DOI] [PubMed] [Google Scholar]
- 146.Steffan JS, Agrawal N, Pallos J, Rockabrand E, Trotman LC, Slepko N, Illes K, Lukacsovich T, Zhu YZ, Cattaneo E, Pandolfi PP, Thompson LM, Marsh JL. SUMO modification of Huntingtin and Huntington’s disease pathology. Science. 2004;304:100–104. doi: 10.1126/science.1092194. [DOI] [PubMed] [Google Scholar]
- 147.Mittler R, Finka A, Goloubinoff P. How do plants feel the heat? Trends Biochem Sci. 2012;37:118–125. doi: 10.1016/j.tibs.2011.11.007. [DOI] [PubMed] [Google Scholar]
- 148.Morano KA, Grant CM, Moye-Rowley WS. The response to heat shock and oxidative stress in Saccharomyces cerevisiae . Genetics. 2012;190:1157–1195. doi: 10.1534/genetics.111.128033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Morimoto RI. The heat shock response: systems biology of proteotoxic stress in aging and disease. Cold Spring Harb Symp Quant Biol. 2011;76:91–99. doi: 10.1101/sqb.2012.76.010637. [DOI] [PubMed] [Google Scholar]
- 150.Goldberg AL. Protein degradation and protection against misfolded or damaged proteins. Nature. 2003;426:895–899. doi: 10.1038/nature02263. [DOI] [PubMed] [Google Scholar]
- 151.Caughey B, Lansbury PT. Protofibrils, pores, fibrils, and neurodegeneration: separating the responsible protein aggregates from the innocent bystanders. Annu Rev Neurosci. 2003;26:267–298. doi: 10.1146/annurev.neuro.26.010302.081142. [DOI] [PubMed] [Google Scholar]
- 152.Escusa-Toret S, Vonk WI, Frydman J. Spatial sequestration of misfolded proteins by a dynamic chaperone pathway enhances cellular fitness during stress. Nat Cell Biol. 2013;15:1231–1243. doi: 10.1038/ncb2838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Kaganovich D, Kopito R, Frydman J. Misfolded proteins partition between two distinct quality control compartments. Nature. 2008;454:1088–1095. doi: 10.1038/nature07195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Malinovska L, Kroschwald S, Munder MC, Richter D, Alberti S. Molecular chaperones and stress-inducible protein-sorting factors coordinate the spatiotemporal distribution of protein aggregates. Mol Biol Cell. 2012;23:3041–3056. doi: 10.1091/mbc.E12-03-0194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Spokoini R, Moldavski O, Nahmias Y, England JL, Schuldiner M, Kaganovich D. Confinement to organelle-associated inclusion structures mediates asymmetric inheritance of aggregated protein in budding yeast. Cell Rep. 2012;2:738–747. doi: 10.1016/j.celrep.2012.08.024. [DOI] [PubMed] [Google Scholar]
- 156.Tyedmers J, Treusch S, Dong J, McCaffery JM, Bevis B, Lindquist S. Prion induction involves an ancient system for the sequestration of aggregated proteins and heritable changes in prion fragmentation. Proc Natl Acad Sci USA. 2010;107:8633–8638. doi: 10.1073/pnas.1003895107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Johnston JA, Ward CL, Kopito RR. Aggresomes: a cellular response to misfolded proteins. J Cell Biol. 1998;143:1883–1898. doi: 10.1083/jcb.143.7.1883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Weisberg SJ, Lyakhovetsky R, Werdiger AC, Gitler AD, Soen Y, Kaganovich D. Compartmentalization of superoxide dismutase 1 (SOD1G93A) aggregates determines their toxicity. Proc Natl Acad Sci USA. 2012;109:15811–15816. doi: 10.1073/pnas.1205829109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Zhang X, Qian SB. Chaperone-mediated hierarchical control in targeting misfolded proteins to aggresomes. Mol Biol Cell. 2011;22:3277–3288. doi: 10.1091/mbc.E11-05-0388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Zaarur N, Meriin AB, Gabai VL, Sherman MY. Triggering aggresome formation. Dissecting aggresome-targeting and aggregation signals in synphilin 1. J Biol Chem. 2008;283:27575–27584. doi: 10.1074/jbc.M802216200. [DOI] [PubMed] [Google Scholar]
- 161.Garcia-Mata R, Bebok Z, Sorscher EJ, Sztul ES. Characterization and dynamics of aggresome formation by a cytosolic GFP-chimera. J Cell Biol. 1999;146:1239–1254. doi: 10.1083/jcb.146.6.1239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Wigley WC, Fabunmi RP, Lee MG, Marino CR, Muallem S, DeMartino GN, Thomas PJ. Dynamic association of proteasomal machinery with the centrosome. J Cell Biol. 1999;145:481–490. doi: 10.1083/jcb.145.3.481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Latonen L, Moore HM, Bai B, Jaamaa S, Laiho M. Proteasome inhibitors induce nucleolar aggregation of proteasome target proteins and polyadenylated RNA by altering ubiquitin availability. Oncogene. 2011;30:790–805. doi: 10.1038/onc.2010.469. [DOI] [PubMed] [Google Scholar]