Abstract
Both host and parasite factors contribute to disease severity of malaria infection; however, the molecular mechanisms responsible for the disease and the host-parasite interactions involved remain largely unresolved. To investigate effects of parasite factors on host immune responses and pathogenesis, we measured levels of plasma cytokines/chemokines (CC) and growth rates in mice infected with two Plasmodium yoelii strains having different virulence phenotypes and in progeny from a genetic cross of the two parasites. Quantitative trait loci (QTL) analysis linked levels of many CCs, particularly IL-1β, IP-10, IFN-γ, MCP-1, and MIG, and early parasite growth rate to loci on multiple parasite chromosomes, including chromosomes 7, 9, 10, 12, and 13. Comparison of the genome sequences spanning the mapped loci revealed various candidate genes. The loci on chromosome 7 and 13 had significant (p < 0.005) additive effects on IL-1β, IL-5, and IP-10 responses, and the chromosome 9 and 12 loci had significant (p = 0.017) interaction. Infection of knockout mice showed critical roles of MCP-1 and IL-10 in parasitemia control and host mortality. These results provide important information for better understanding of malaria pathogenesis and can be used to examine the role of these factors in human malaria infection.
Keywords: genetic mapping, cytokine, chemokine, inflammation, pathogenesis, virulence
Introduction
Malaria is a deadly parasitic disease that kills ∼0.6–0.8 million people annually, mostly African children 1. The symptoms of malaria patients are highly variable, ranging from asymptomatic to lethal infections depending on the outcome of host and parasite interactions and the nature and intensity of host immune responses2, 3. Production of cytokines and chemokines (CC) is critical for controlling parasitemia and disease symptoms; however, overproduction of inflammatory related CC (IRCC) can also lead to severe disease or even host death 4-7. Host response to malaria infection can differ greatly between individuals because of variation in host genetic background and immune status 8-13. In addition to polymorphisms in the genes encoding hemoglobin, ABO blood group, HLA antigens, glucose-6-phosphate dehydrogenase (G6PD), and many other genetic loci, variation in levels of IRCC such as TNF-α, IL-1β, IFN-γ, and IL-6, and related signaling pathways have been associated with both parasite clearance and disease severity 14-18. Host responses to malaria infection are therefore greatly influenced by host genetic background 4, 8, 19, 20.
Variations in parasite traits such as cytoadherance 21, rosetting 22, and red blood cell (RBC) selectivity 23, 24 have also been shown to play a role in clinical pathology, and parasite-derived materials such as glycolipid (glycophosphatidylinositol or GPI), hemozoin pigment, TA-rich stem-loop DNA motif, DNA-protein complex, and a homolog of macrophage migration inhibition factor (MIF) have been reported to modulate host immunity and CC production 25-30. It has also been well established that laboratory strains and field isolates of Plasmodium parasites can differ in their ability to induce or suppress host innate immune responses 10, 31. The nature of parasite determinants that can differentially stimulate or modulate the host immune response and the molecular mechanisms of virulence remain unresolved; death from malaria could be due to high parasitemia and/or result from overproduction of IRCC. It is still difficult to strictly define the roles of IRCC in protection and pathogenesis during a malarial infection—particularly in human malaria, in which control or manipulation of host genetic background is not feasible 4. Variations in parasite and host genetic backgrounds as well as concurrent infections with other pathogens may also contribute to the frequent conflicting reports in studies of host-parasite interaction. Identifying parasite genes and their variants that play a role in modulating the host immune response will provide invaluable insight into the molecular mechanisms of malaria pathogenesis.
The rodent malaria parasite Plasmodium yoelii is an excellent model for studying disease-related phenotypes, because many genetically distinct parasite strains—exhibiting wide variation in blood-stage multiplication rate and virulence—are available 32, and the use of inbred mice can minimize the influence of host genetic variation on phenotype measurements. Taking advantage of the availability of P. yoelii strains with different growth characteristics and virulence, we investigated the differences in host CC response in mice infected with two rodent malaria parasites (P. yoelii nigeriensis N67and P. yoelii yoelii 17XNL) that cause different disease phenotypes and 22 progeny from a cross of the two parasites 33. Analysis of the recombinant progeny from the genetic cross revealed that differential CC production such as IL-10, IL-5, IP-10, IL-1β, KC, and MCP-1 in P. yoelii infected mice is inheritable and segregated as quantitative traits that could be linked to parasite chromosomal loci.
Results
Variation in growth rate and CC production between P. yoelii strains
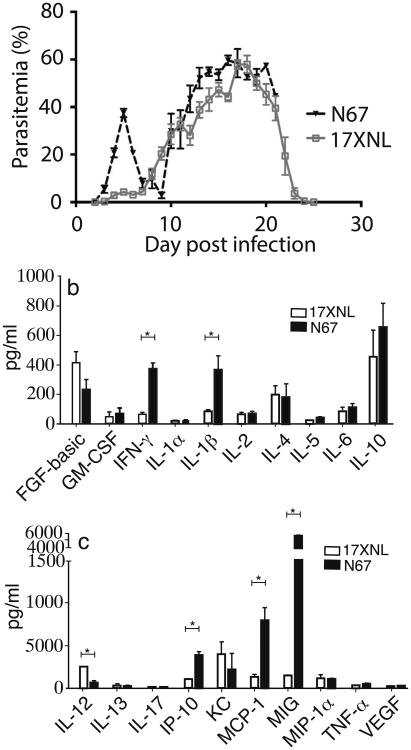
To study the mechanisms underlying malaria virulence, we compared patterns of parasitemia and CC levels in C57BL/6 mice after injection of 1 × 105 infected RBCs (iRBC) of 17XNL and N67 parasites (Figure 1a). Mice infected with 17XNL had low parasitemia (under 5%) during the first 7 days before clearing the parasite at approximately day 25. Parasitemia in mice infected with N67 peaked at 30–40% on day 5 post infection, then declined to under 10% on day 7 before returning to ∼60%. The mice died at around day 15-20 post infection. The early decline of parasitemia in the N67-infected mice suggests that the host innate response plays a critical role in controlling the parasitemia.
Fig. 1.
Variation in parasitemia and cytokine/chemokine responses in C57BL/6 mice infected with two Plasmodium yoelii parasite lines. (a) Parasitemia from mice infected with P. y. yoelii 17XNL and P. y. nigeriensis N67. (b, c) Levels of day 4 cytokines and chemokines in mice infected with 17XNL and N67. Standard deviations (SD) were determined from at least 3 mice for each group. Statistically significant (p < 0.05) differences in cytokine/chemokine levels between 17XNL and N67 are as indicated (Mann-Whitney U test).
To investigate the molecular mechanisms underlying the differences in pathology and parasitemia profiles with these parasites, we measured plasma levels of 20 CC (FGF-basic, GM-CSF, IFN-γ, IL-1α, IL-1β, IL-2, IL-4, IL-5, IL-6, IL-10, IL-12 (p40/p70), IL-13, IL-17, IP-10, KC, MCP-1, MIG, MIP-α, TNF-α, and VEGF) in mice infected with the two parasites using a commercial bead array kit (Invitrogen) (Figure 1b, 1c and Figure S1). Levels of all the CC remained low in the uninfected group during the study, whereas mice infected with the parasites had elevated levels for the majority of CC, although the CC levels of 17XNL-infected mice were generally lower. Interestingly, the levels of GM-CSF, IFN-γ, IL-1β, IL-2, IL-4, IL-5, IL-10, MCP-1, and MIG in the N67-infected mice peaked on day 4 post infection (p.i.)—one day before the parasitemia peak on day 5 p.i.—and returned to relatively low levels after day 5 (Figure S1). Comparison of the day 4 CC levels in the N67 infected mice with those infected with 17XNL parasites revealed significant differences in many IRCC levels, in particular the levels of IFN-γ, IP-10, MCP-1, MIG, IL-1β, and IL-12 (Figure 1b, 1c). We therefore focused on investigating the differences in day 4 CC levels between the two parasites.
Correlation of CC response profiles among the progeny of a genetic cross
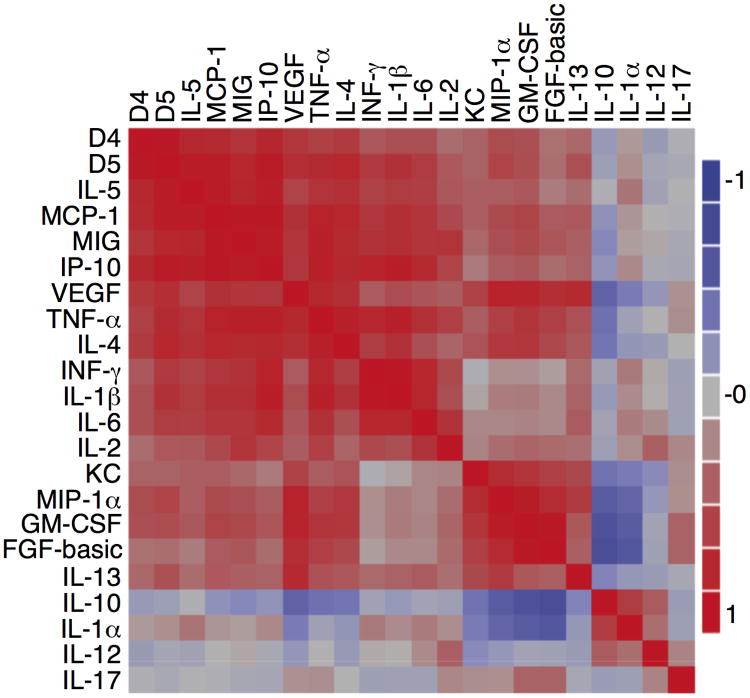
The disease outcomes and CC profiles clearly showed that host CC responses to P. yoelii infection are strain dependent, suggesting different parasites may produce some strain-specific molecules that stimulate unique CC responses. To identify the parasite genetic loci that can stimulate such differential host responses, we measured parasitemia and day 4 plasma levels of the same 20 CC from C57BL/6 mice injected with 22 progeny (1×105 iRBC/each into four mice for each progeny) from the N67×17XNL genetic cross (Table S1). Multivariate correlation analysis of the averaged CC levels revealed a high degree of correlation between the day 4/5 parasitemia and day 4 levels of MCP-1, MIG, IP-10, VEGF, TNF-α, IFN-γ, IL-1β, IL-2, IL-4, IL-5, and IL-6 (Figure 2). Another highly correlated CC cluster included KC, MIP-α, GM-CSF, FGF, IL-13, and VEGF. As a known anti-inflammatory cytokine, IL-10 was negatively correlated with most of the CC—particularly KC, MIP-1α, GM-CSF, FGF, VEGF, TNF-α, and IL-4—but positively correlated with IL-1α and IL-12. Similarly, IL-1α was negatively correlated with MIP-1α, GM-CSF, FGF, VEGF, TNF-α, and IL-4. Because the parasites are progeny from two parents, these patterns of CC responses are likely linked to differences in parasite genetic backgrounds of the parents.
Figure 2.
Correlation of cytokine/chemokine responses among the progeny of the N67×17XNL cross. The color bar indicates degree of correlation, with red indicating positive correlation and blue indicating negative correlation. Correlation analysis was performed using GraphPad Prism version 6.0 (GraphPad Software, Inc.).
Genetic loci and candidate genes linked to differential CC responses
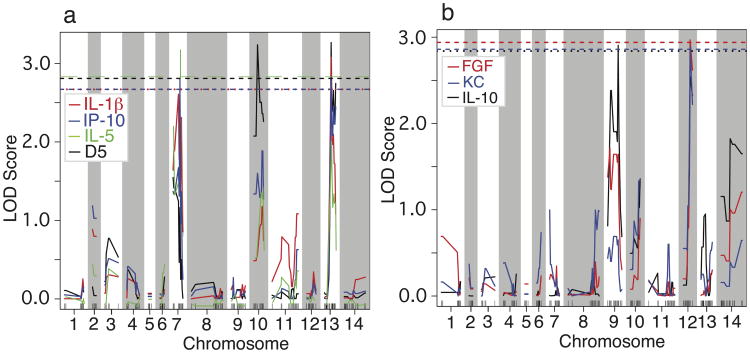
To identify genetic loci linked to the CC responses, we performed quantitative trait loci (QTL) analysis using genome-wide genotypes previously collected from 486 MSs 34 and the CC measurements from the 22 progeny (Table S1). In addition to day 4/5 parasitemia, six cytokines were significantly linked to loci on four different chromosomes (Figure 3 and Table 1). As previously reported, day 5 parasitemias were significantly linked to two loci on chromosome 10 and 13 (Figure 3A)33. Interestingly, the levels of IP-10 and IL-1β were also significantly linked to the same marker on chromosome 13 (Py2609) as well as a peak on chromosome 7 (marker Py2601-1; LOD scores close to significant cut-off level for IL-1β). Similarly, the IL-5 level was linked to the same markers on chromosome 7 and chromosome 13, but only the peak on chromosome 7 was significant. Additionally, the levels of FGF and KC were significantly linked to Py743 on chromosome 12, and the IL-10 level was significantly linked to Py148 on chromosome 9 (Figure 3B). If we lowered the cutoff criteria to a LOD score of 2.0 (suggestive linkage), we could detect two groups of linkage: the first group was the linkage of IL-1β, IL-4, IL-5, IP-10, IFN-γ, MCP-1, MIG, and TNF-α to the loci on chromosome 7 and/or chromosome 13; the second group was the linkage of KC, MIP-1α, GM-CSF, FGF, IL-10, and IL-1α to the loci on chromosome 9 and/or chromosome 12 (Table 1 and Figure S2). Note that there were two peaks on chromosome 13 for MCP-1 and TNF-α, and the highest peaks were at MS marker Py1555, not at Py2069. Therefore, the loci on chromosome 7 and 13 contributed mostly to IRCC responses and parasitemia, whereas the loci on chromosome 9 and 12 might contribute to the anti-inflammatory response and to production of growth and chemotactic factors. Finally, a peak with a LOD score of 2.5 on chromosome 2 was linked to IL-2 response (Table 1 and Figure S2).
Figure 3.
Genetic loci linked to higher cytokine and chemokine levels in response to infection of Plasmodium yoelii nigeriensis N67. (a) Loci significantly linked to day-5 parasitemia (black), IP-10 (blue), IL-1β (red), and IL-5 (green). (b) Loci significantly linked to IL-10 (black), KC (blue), and FGF (red). The dashed lines of black, blue, red, and green indicate significant LOD scores (p < 0.05) corresponding to their color curves. QTL mapping was performed using R/qtl library in R2.12.2 software (http://www.rqtl.org/) 53
Table 1.
Genetic loci/markers linked to differential levels of cytokines/chemokines in mice infected with Plasmodium yoelii yoelii 17XNL and Plasmodium yoelii nigeriensis N67.
| Phenotypes | MS. Marker | Chr | Position (cM) | LOD score |
|---|---|---|---|---|
| D5 para | Py56 | 10 | 10.7 | 3.2 |
| Py2609 | 13 | 19.4 | 3.3 | |
|
| ||||
| MCP-1 | Py2061-1 | 7 | 20 | 2.8 |
| Py1239 | 10 | 24 | 2.1 | |
| Py1555 | 13 | 30.4 | 2.5 | |
|
| ||||
| IP-10 | Py2061-1 | 7 | 20 | 2.8 |
| Py2609 | 13 | 19.4 | 2.8 | |
|
| ||||
| IFN-γ | Py1277 | 7 | 15.7 | 2.0 |
| Py2609 | 13 | 19.4 | 2.5 | |
|
| ||||
| IL-10 | Py148 | 9 | 30.6 | 2.9 |
| Py743 | 12 | 19.6 | 2.5 | |
|
| ||||
| IL-1β | Py2061-1 | 7 | 20 | 2.6 |
| Py2609 | 13 | 19.4 | 3.1 | |
|
| ||||
| IL-2 | Py652 | 2 | 0 | 2.5 |
| IL-4 | Py2061-1 | 7 | 20 | 2.2 |
| Py867 | 13 | 26.1 | 2.0 | |
|
| ||||
| IL-5 | Py627 | 7 | 20 | 3.2 |
| Py2123 | 13 | 19.4 | 2.0 | |
|
| ||||
| KC | Py743 | 12 | 19.6 | 2.9 |
|
| ||||
| TNF-α | Py1555 | 13 | 30.4 | 2.1 |
|
| ||||
| MIG | Py2061-1 | 7 | 20 | 2.4 |
|
| ||||
| IL-1α | Py743 | 12 | 19.6 | 2.1 |
|
| ||||
| MIP-1α | Py743 | 12 | 19.6 | 2.3 |
|
| ||||
| GM.CSF | Py743 | 12 | 19.6 | 2.3 |
|
| ||||
| FGF.basic | Py743 | 12 | 19.6 | 3.0 |
The LOD scores in bold were deemed significant (P<0.05) after 1000 permutations.
Phenotypes, plasma cytokine/chemokine levels or day 5 parasitemia (D5 para); MS marker, names of microsatellite markers; Chr, chromosome; Position, genetic distance in centiMorgan (cM); LOD score, logarithm of odds (LOD) scores calculated using R/qtl.
To determine the sizes of the linked loci, we first identified the MS markers immediately flanking the loci with reduced LOD scores (compared with the markers with peak LOD scores) and the progeny with crossovers that resulted in the reduction of LOD scores (Table 2). We then calculated the distances between the two flanking markers based on the assembled P. yoelii YM sequences (http://www.sanger.ac.uk/resources/downloads/protozoa/plasmodium-yoelii.html). The sizes of the loci ranged from 148 kb to 413 kb, containing 30-112 genes in each locus. The candidate genes included malarial antigens, protein phosphatases, guanylyl cyclases, protein kinases, helicases, and DNA/RNA binding proteins that may play a role in parasite ligand recognition and signaling and regulation of gene expression (Table S2).
Table 2.
Positions and sizes of genetic loci linked to host cytokine and chemokine response to Plasmodium yoelli infection.
| MS | Chr | Distance (cM) | Forward primer | Reverse primer | Physi. Posi | Length (bp) | Recomb. Progeny defining the loci | No. Can. Genes | |
|---|---|---|---|---|---|---|---|---|---|
| Py1238 | 7 | 17.8 | 5 ′ TATAATACCATGTGACATGT3 ′ | 5 ′ GCTTATACGAGATATTGTGA3 ′ | 385973 | ||||
| Py2061-1 | 7 | 20 | 5 ′ AAGGGAAGAAATTCGGCA3 ′ | 5 ′ TACTCTTTCACCTTTGCAA3 ′ | 440592 | ||||
| Py627 | 7 | 20 | 5 ′ ATACATATAAGCTGCTACAA3 ′ | 5 ′ TAATGTGTGCAAATATGTGA3 ′ | 501081 | 60489* | |||
| Py1798 | 7 | 22.2 | 5 ′ GCTTAGTGACTAAAATGTG3 ′ | 5 ′ GCATACTTGGAAAATTCCT3 ′ | 532584 | 146,611 | E890#3 | F239#4 | 38 |
| Py2179 | 9 | 28.1 | 5 ′ TGCTTTCATATTCTTCCAC3 ′ | 5 ′ TTGAAGAAGATAGGAGTAC3 ′ | 472715 | ||||
| Py1861 | 9 | 30.6 | 5 ′ GCATACACATATGTTGAGA3 ′ | 5 ′ ACGGATGGAAACATTACAA3 ′ | 402219 | ||||
| Py148 | 9 | 30.6 | 5 ′ AGATTCAAGTACAAGGTCA3 ′ | 5 ′ CTTTAGCTAGTCATCGTC3 ′ | 373266 | 28953* | |||
| Py806 | 9 | 32.7 | 5 ′ CTACTATTCTTTATTGTACC3 ′ | 5 ′ CAGATGAAGTAAAAGGCAC3 ′ | 324346 | 148,369 | F120#5 | F994#3 | 30 |
| Py189-2 | 10 | 6.4 | 5 ′ ATTGTTTGGGAATTAGCGA3 ′ | 5 ′ TGTATTCCCAGGTGTATC3 ′ | 696263 | ||||
| Py280 | 10 | 10.7 | 5 ′ ATTAAAGGCCTTATGCACA3 ′ | 5 ′ TTACATCATTAGTATTGGCA3 ′ | 790202 | ||||
| Py56 | 10 | 10.7 | 5 ′ AAGTTGAAATAACAATGCCA3 ′ | 5 ′ CAAAGTACGCAATCCTCA3 ′ | 793886 | 3684* | |||
| Py1829 | 10 | 17.6 | 5 ′ ATATGGAGAAAATTAGCGAA3 ′ | 5 ′ CTAACGCCAGACATAAAAA3 ′ | 1109682 | 413,419 | F245#l | G007#3 | 112 |
| Py1316-3 | 12 | 17.4 | 5 ′ ATTCTGCAGCGTATGATC3 ′ | 5 ′ TGTTTCTGCTCATACTAGA3 ′ | 1362004 | ||||
| Py437 | 12 | 19.6 | 5 ′ CATCACCAACTTGTATTTC3 ′ | 5 ′ CCATAATAAAGCAGTTCTC3 ′ | 1387382 | ||||
| Py743 | 12 | 19.6 | 5 ′ TAATATCTGGTCGTTATTCT3 ′ | 5 ′ ATTTAGCTATACAAATAGCC3 ′ | 1399469 | 12087* | |||
| Py2260 | 12 | 26.5 | 5 ′ TAGCAGCATATTCCTTGC3 ′ | 5 ′ ATGGCTTCTTCCAATAAGT3 ′ | 1765986 | 403,982 | E995#3 | G001#4 | 110 |
| Py2035 | 13 | 15.1 | 5 ′ TGTGTATATTGCAAAGGAC3 ′ | 5 ′ GTATACACTGTGATTCCTA3 ′ | 1121475 | ||||
| Py2123 | 13 | 19.4 | 5 ′ CATTCTTAGTCATGTCACA3 ′ | 5 ′ ATAATGGCAATAATAATGGC3 ′ | 1231545 | ||||
| Py2609 | 13 | 19.4 | 5 ′ CTCATATTTAAGTTTTTCTCT3 ′ | 5 ′ TAGGAAGACGATCTCGCT3 ′ | 1421935 | 190390* | |||
| Py1084 | 13 | 21.7 | 5 ′ GCATATCCAGTGACGCTA3 ′ | 5 ′ CAATCGATTTGGGTGCTA3 ′ | 1490422 | 368,947 | F995#4 | G001#5 | 96 |
MS, microsatellite names; Chr, chromosome; Distance (cM), genetic distance; Forward primer and Reverse primer, primers for the amplification of the mcirosatellites; Physi. Posi, physical positions of the first base of the forward primers; Length (bp), the fragment sizes of the linked loci with MS markers detecting crossovers resulting changes in LOD scores; Recomb. progeny, names of the progeny with crossovers defining the mapped loci; No. Can. Gene, numbers of candidate genes in the loci.
indicates the distance between the two markers with the highest LOD scores.
Genome sequencing and detection of polymorphisms in candidate genes
To determine the genetic variations in the genes located within the mapped loci between 17XNL and N67, we sequenced the genome of N67 parasite and obtained approximately 56 million reads that covered ∼95% of the genome with an average of 367-fold coverage (total 9,260,362,154 bp covered; sequences are being submitted to NCBI, accession number SAMN02209965). Comparison of DNA sequences encoding the candidate genes in the mapped loci revealed nearly 4,000 putative non-synonymous single nucleotide polymorphisms (nsSNPs) and many highly polymorphic genes between N67 and 17XNL (Table S2). Fine mapping using progeny from additional crosses and functional characterization using allelic exchange and other functional assays will be necessary to reduce the sizes of the loci to pinpoint and identify the candidate genes.
Interaction of genetic loci linked to CC responses
Because the levels of many cytokine responses were highly correlated and were linked to more than one locus, we investigated the additive effects and interactions of the loci contributing to the cytokine levels. The loci on chromosome 7 and 13 had significant (p < 0.005) additive effects on IL-1β, IL-5, and IP-10 responses, contributing to 58%, 59%, and 56% of the phenotype variances, respectively. In addition, a significant interaction (p = 0.017) on the IL-10 level between the loci on chromosome 9 and 12 was detected when analyzed using the fitqtl method (Table 3).
Table 3. Additive effects and interactions between loci significantly linked to responses to selected cytokines or chemokines.
| CC | Chr (position in cM) | % Var | Additive | p-Values | Interaction | p-Values |
|---|---|---|---|---|---|---|
| IP-10 | 7 (20.0) and 13 (19.4) | 58 | Yes | 0.0002 | No | 0.43 |
| IL-1β | 7(20.0) and 13(19.4) | 59 | Yes | 0.0002 | No | 0.37 |
| IL-5 | 7(20.0) and 13(19.4) | 56 | Yes | 0.0004 | No | 0.32 |
| IL-10 | 9(30.6) and 12(19.6) | 67 | Yes | 0.0003 | Yes | 0.017 |
CC, cytokines and chemokines; Chr (position in cM), chromosome and marker position in centimorgan (cM); % Var, percent variance explained by the loci; Additive, additive effect or not; Interaction, interaction between the two loci or not.
Contribution of MCP-1 and other CC to parasitemia and mouse survival
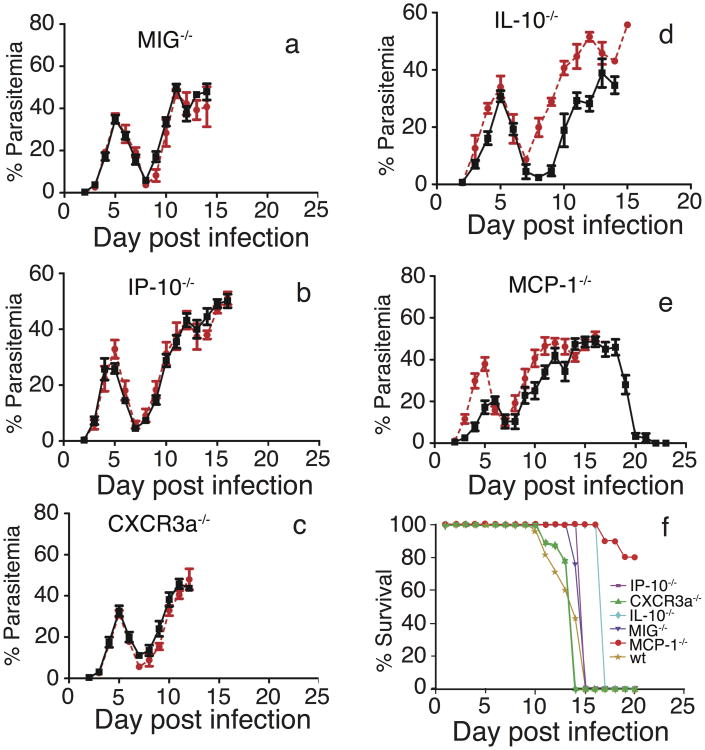
As shown above, levels of many IRCC were elevated in mice infected with the N67 parasite, suggesting that these IRCC may contribute to the disease and to host mortality. To investigate the functional roles of the IRCC during early infection, we infected wild-type (wt) C57BL/6 mice as well as mice of the same genetic background but with disrupted genes encoding selected CC significantly linked to the parasite genetic loci (MIG−/−, IP-10−/−, CXCR3a−/−, IL-10−/−, and MCP-1−/−; CXCR3a is the receptor for MIG and IP-10) with 1×105 N67 parasites and compared the dynamics of parasitemia and host survival time. Parasitemias from MIG−/−, IP-10−/−, and CXCR3a−/− mice infected with N67 were essentially the same as those of wt (Figure 4a–4c). For IL-10−/− mice infected with N67, parasitemia was approximately the same as of wt before day 7 p.i.; however, parasitemia rose more slowly in the IL-10−/− mice after day 7 p.i. (Figure 4d). Significantly, the majority (80%) of the MCP-1−/− mice were able to clear the parasites and survive N67 infection after the second parasitemia peak (Figure 4e and 4f), while the wt mice died at around day 15 p.i. Additionally, the initial parasitemia peak in the MCP-1−/− mice was also lower than that of wt at days 3–5 p.i., and there was another small dip in parasitemia at day 14 in the MCP-1−/− group. Except for the MCP-1−/− and IL-10−/− mice, all the other KO mice infected with N67 died approximately at the same time as the wt control mice (Figure 4f). These results suggest that MCP-1 or overproduction of MCP-1 may play a crucial role in immunopathology and host mortality and that IL-10 may also play some role in controlling the parasitemia.
Figure 4.
Parasitemia and mortality of wild type (wt) and cytokine/chemokine gene knockout (−/−) mice infected with Plasmodium yoelii nigeriensis N67. Mice (n = 5–8) were injected intravenously with an inoculum containing 1×105 infected red blood cells. Parasitemias were measured daily by microscopic examination of Giemsa-stained thin tail blood smears. (a–e) Red lines, wt mice; black lines, −/− mice. (a) MIG−/−; (b) IP-10−/−; (c) CXCR3a−/−; (d) IL-10−/−; (e) MCP-1−/−. (f) Mice showing severe clinical signs were humanely euthanized; the numbers of surviving mice during the course of infection were used to calculate percentage of survival.
Discussion
Host innate immune responses and IRCC in malaria infection have been implicated in disease severity 35; however, genetic variation and complex interactions between host and parasite have been major obstacles in dissecting the roles of host and parasite factors contributing to pathogenesis in human malaria. Taking advantage of the relatively homogeneous genetic background of inbred mice, here we used genetic mapping to identify the parasite genetic loci and determinants that can stimulate differential CC levels and to dissect the molecular basis of virulence during P. yoelii infection. We showed clearly that the ability to induce specific CC production was strain dependent and inheritable. Mice infected with N67 produced good levels of IRCC on day 4 p.i., one day ahead of peak parasitemia, which likely contributed to the decline of parasitemia; however, the levels of IRCC returned to baseline after day 5, and the parasitemia rebounded, leading to host death at day 15-20. It is critically important to investigate the molecular events leading to the decline of CC after day 4 p.i., when the parasites are rapidly growing in the mice infected with N67. Slow growth and low parasitemia of 17XNL, partly due to its preference for invading reticulocytes, did not stimulate strong IRCC responses or cause severe disease, allowing the host's acquired immunity to clear the parasites 3 weeks after infection. The dramatic differences in early IRCC response to infections of different parasite strains/subspecies also suggest that results obtained using a specific parasite strain may not be applied to another. It is therefore necessary to consider parasite genetic background when explaining results from studies of host immune responses and from genetic association studies searching for host genes involved in specific disease phenotypes. Moreover, variation in host genetic background can also play an important role in the host response to malaria infection 10, 36.
Our results showed that the loci on chromosomes 7 and 13 were linked to levels of IRCC such as IL-1β, IL-4, IL-5, IL-6, IP-10, IFN-γ, MCP-1, MIG, and TNF-α. On the other hand, the responses of the second CC group (IL-1α, MIP-1α, GM-CSF, FGF-basic, KC, and IL-10) were linked to loci on chromosome 9 and/or chromosome 12 that were not associated with parasitemia. Additionally, IL-10 and IL-1α were negatively associated with MIP-1α, GM-CSF, FGF-basic, and KC (Figure 2). It is likely that some molecules in the chromosome 9 and/or chromosome 12 loci can influence IL-10 and IL-1α production and at the same time suppress production of other IRCC. The results suggest that the N67 parasites have two mechanisms that can modulate host innate immune responses: one controlled by molecules on chromosomes 7 and 13 that can stimulate pro-inflammatory responses and a second controlled by molecules on chromosome 9 and 12 that can generate anti-inflammatory responses. The linkage of both higher levels of IRCC and parasitemia to the chromosome 13 locus could be due to faster growth and more parasites that in turn stimulates a stronger IRCC response. However, the growth phenotype was also significantly linked to a locus on chromosome 10, whereas many IRCC were linked to a chromosome 7 locus, suggesting differences in the genetic basis of controlling parasite growth and IRCC responses. Identifying the molecules that can modulate host immune responses, and therefore virulence, will greatly facilitate our understanding of the mechanisms of malaria pathogenesis.
We also attempted to identify the potential candidate genes linked to differential IRCC responses by sequencing the N67 genome. Comparison of the sequences of candidate genes in the mapped loci between N67 and 17XNL (genome sequence available publically) parasites identified nearly 4000 nsSNPs. It is still difficult to determine which genes contributed to the differences in the host cytokine response at this moment. The LOD scores for many of the CC were above 2.0 but did not reach significant levels (p < 0.05 after 1000 permutations or LOD = 3). This was likely due to the relatively small number of progeny used in the analysis. Cloning and genotyping more progeny from the cross will be necessary to further fine-map the loci and hopefully to increase the LOD scores. Additional progeny and crossovers in the mapped loci will also reduce the sizes of the loci and the numbers of candidate genes to be tested functionally. Unfortunately, cloning progeny in mice is relatively labor intensive; additionally, 17XNL produces very few oocysts in the mosquito midgut (fewer than 10 on average). Improvements in oocyst production of the parasite and in procedures of parasite cloning will greatly facilitate the chances of obtaining additional progeny from more genetic crosses 37.
In Toxoplasma gondii, several rhoptry kinases, a dense granule protein (GRA15), a profilin-like protein, and possibly nucleoside triphosphate hydrolases have been shown to regulate host signaling pathways and immune responses 38-41. Although no orthologs of the rhoptry kinases or GRA15 have been found in Plasmodium parasites, many molecules with putative secretion signals and functions in regulating gene expression were identified in the loci linked to levels of CC. Some examples of interesting genes include a gene encoding a rhoptry neck protein 5 (RON5) on chromosome 7 that may be involved in invasion of RBCs and interaction with host cells 42, 43; an Apetala2-like transcription factors (ApiAP2) on chromosome 9 44; a highly polymorphic carbon catabolite repressor protein (CCR4) on chromosome 12 that is a part of the gene regulatory complex 45; and the PyEBL on chromosome 13. The PyEBL has been implicated in parasite invasion of RBCs and may bind to DARC expressed on endothelial cells, triggering host innate immune responses 46. The Duffy antigen on the RBC surface is the receptor for Plasmodium vivax, Plasmodium knowlesi, and P. yoelii EBL proteins during invasion 47-49; it can also act as a receptor for chemokines of both the C-C and C-X-C families—including IL-8, MGSA, RANTES, and MCP-1—when expressed on endothelial and other cell types 50. Indeed, a rat homolog of DARC expressed on endothelial cells was shown to bind IL-8–coated microspheres, which could be inhibited by previous infusion of IL-8 as well as MCP-1 50. It is possible that PyEBL secreted or expressed on the iRBC binds to DARC on endothelial cells, interfering with host C-C or C-X-C chemokine signaling pathways. Indeed, our observations of increased survival of MCP-1−/− mice infected with N67 parasites and higher levels of IRCC suggest an association of overactivation of C-C or C-X-C chemokine signaling and disease pathogenesis. MCP-1 is a chemokine that recruits many cell types such as monocytes, memory T cells, and dendritic cells to sites of tissue injury and infection 51. A decrease in several IRCC in the brain and in activation of microglia after peripheral injection of lipopolysaccharide was observed in MCP-1−/− mice compared with wt mice 52, suggesting an important role of MCP-1 in brain inflammation. DARC is also highly expressed in the Purkinje cells of human cerebellum and may act as a receptor for MCP-1 50. The relationship of PyEBL binding to DARC, increased production of MCP-1, and disease severity requires further investigation.
In summary, the host innate immune response to P. yoelii is strain dependent. A rapid and robust response of IRCC such as seen with MCP-1 and IL-10 appeared to be necessary for controlling early parasitemia. The differences in host CC responses were linked to several parasite chromosomal loci, including the DNA segment containing Pyebl, which may play a role in modulating host innate immune response and disease severity. Elucidation of the molecular mechanism of malaria virulence and identification of parasite molecules that can modulate host immune responses will provide critical information for vaccine development and disease management.
Materials and Methods
Animals and malaria parasites
Inbred female C57BL/6 mice, aged 6–8 weeks old, and KO mice of matched genetic background were obtained from Charles River Laboratory, Jackson Laboratory, or NIAID/Taconic repository, NIH. All the experiments were performed in accordance with NIH–approved animal study protocol LMVR-11E. The P. yoelii cloned lines (17XNL and N67) and progeny from the N67×17XNL cross were described previously 33.
Infection of mice with P. yoelii parasites
An inoculum containing appropriate numbers of iRBC suspended in 100 μl phosphate buffered saline (PBS), pH 7.4, from donor mice was injected intravenously (i.v.) into experimental C57BL/6 mice or KO mice with the same genetic background. Naïve mice receiving an equivalent number of uninfected RBC served as a negative control group. Parasitemia was monitored daily by microscopic examination of Giemsa-stained thin tail blood smears. Mouse blood was collected daily to monitor parasitemia.
Cytokine and chemokine assays
Approximately 50 μl blood from individual mice was collected every other day from the tail vein into heparinized tubes and centrifuged at 10,000×g for 10 min. Supernatants were transferred to sterile cryopreserved tubes and stored at −80°C prior to analysis using mouse cytokine 20-plex kit and Luminex 200 instrument according to the manufacturer's instructions (Invitrogen). Levels of CC were exported to Excel sheets and subjected to statistical analyses (nonparametric Mann-Whitney U test and one-way ANOVA) using GraphPad Prism version 6.0 (GraphPad Software, Inc.). Differences were deemed significant at p < 0.05.
QTL analysis
QTL mapping was performed using R/qtl library in R2.12.2 software (http://www.rqtl.org/) 53 as described previously 33. Briefly, day-5 parasitemia and day-4 CC levels from the progeny of the N67×17XNL cross were natural-log transformed to obtain normally distributed datasets before QTL analysis. A 5% threshold (p = 0.05) based on 1,000 permutations using standard interval mapping (EM method) was used as the significant cutoff level. A single-QTL genome scan was performed initially, followed by a two-dimensional and finally two-QTL genome scans. The 2-dimensional, 2-QTL scan was performed to identify additional QTL and 2-locus epistatic interactions (fitqtl), if present. Correlation of the CC responses from the progeny was performed using ANOVA, and the p-values for the differences were calculated with the nonparametric Mann-Whitney U test using GraphPad Prism software.
Genome sequencing
Blood samples infected with N67 at parasitemia of 30–40% were placed into physiological citrate saline (0.85% NaCl, 1.5% sodium citrate) and passed-through three Plasmodipur filters (Europroxima) to remove host white blood cells, which was confirmed with thick smears. Genomic DNA was isolated using standard phenol-chloroform extraction after lysis of RBCs with 0.15% saponin. Whole-genome small insert libraries were prepared from the genomic DNAs as follows: 5 μg of genomic DNA was sheared to ∼450 bp using a Covaris S2 with settings: duty cycle 20%; intensity 4; cycles 200. The libraries were constructed according to the Illumina protocols using the paired-end DNA sample prep oligo-only kit (Illumina). After paired-end adapter ligation, the samples were size selected on a 2% agarose gel after staining with SybrGold. Amplification reactions were cleansed using two rounds of Agencourt AMPure beads (Beckman Coulter), and sequence data were processed using RTA1.6.32.0 and GERALD 1.15.
Genome assembly and polymorphism detection
Parameters other than default were: –no-discordant –sensitive –mp 10-R 3. Sequence information was initially processed with an Illumina pipeline using quality values Phred+64. Picard (http://picard.sourceforge.net) was used to process sequence files (add read header, sort, index, mark duplicates) and align reads. Gatk (http://www.broadinstitute.org/gatk/) was used to improve alignment in areas with indels (gaps) and to annotate variants in clusters in regions with poor coverage that were found in both samples or in one sample but could not be determined in the other sample.
Sequence reads were mapped to the assembled YM genome at the website of the Wellcome Trust Sanger Institute (accessed at http://www.genedb.org/Homepage/PyoeliiYM, accessed in August 2012). The sequence reads were trimmed with a cut-off quality-score of 30 using Btrim64 54. BWA version 0.6.1 and Bowtie2 were used to index the FASTA format reference genome and to map the quality-trimmed sequence reads to the reference genome 55, 56. The SAMtools package version 0.1.18 57 was used with the Extended Base Quality algorithm setting on a duplicate-filtered BAM file to call variants. SNPEff version 2.1 58 was used to annotate the variants using a custom library constructed from the YM genome. Due to space limitation, detailed characterization and annotation of the N67 genome will be described in later publications.
Supplementary Material
Figure S1. Cytokine and chemokine responses of mice infected with three Plasmodium yoelii parasites. Mean levels of plasma cytokines and chemokines (FGF-basic, GM-CSF, IFN-γ, IL-1α, IL-1β, IL-2, IL-4, IL-5, IL-6, IL-10, IL-12 (p40/p70), IL-13, IL-17, IP-10, KC, MCP-1, MIG, MIP-1α, TNF-α, and VGEF) were measured as described in Experimental Procedures. Error bars represent standard deviations determined from eight mice. Red, from lethal P. y. nigeriensis N67-infected mice; purple, from lethal P. y. yoelii YM-infected mice; blue, from nonlethal P. y. yoelii 17XNL-infected mice; green, naïve batch mates.
Figure S2. Genetic loci linked to levels of cytokines and chemokines during Plasmodium yoelii infection. Plots of logarithm of odds (LOD) scores for day 4 cytokines and chemokines (FGF-basic, GM-CSF, IFN-γ, IL-10, IL-1α, IL-1β, IL-2, IL-4, IL-5, IP-10, KC, MCP-1, MIG, MIP-1α and TNF-α) that have a LOD = 2.0 or higher. One dash line indicates the cutoff level of LOD = 2.0. In some plots, a second dash line indicates significant level at p < 0.05 after 1000 permutations.
Table S1. Levels of cytokines and chemokines obtained from C57BL/6 mice infected with parents (N67 and 17XNL) and 22 progeny of the genetic cross at day 4 post infection.
Table S2. Nucleotide substitutions and predicted functions of candidate genes in chromosomal loci mapped to differential cytokine/chemokine responses between Plasmodium yoelii yoelii 17XNL and P. y. nigeriensis N67.
Acknowledgments
We thank Drs. Alan Sher and David Sacks for valuable advice and comments and Brenda Rae Marshall, DPSS, NIAID, for editing. This work was supported by the Intramural Research Program of the Division of Intramural Research, National Institute of Allergy and Infectious Diseases, National Institutes of Health; and partially by “Project 111” sponsored by the State Bureau of Foreign Experts and Ministry of Education of China (B06016) and by the National Natural Science Foundation of China (#81271858, # 81201324, and #81220108019).
Because S.P., J.L., J.W., Y.Q., R.T.E., M.Z., S.C.N., M.C.H., M.Q., H.J., J.Z., K.Z., C.A.L., and X-z.S. are government employees and this is a government work, the work is in the public domain in the United States. Notwithstanding any other agreements, the NIH reserves the right to provide the work to PubMedCentral for display and use by the public, and PubMedCentral may tag or modify the work consistent with its customary practices. You can establish rights outside of the U.S. subject to a government use license.
Abbreviations
- CC
cytokines and chemokines
- IRCC
inflammatory related cytokines and chemokines
- iRBC
infected red blood cells
- MS
microsatellite
- p.i.
post infection
- PyEBL
Plasmodium yoelii erythrocyte binding ligand
- QTL
quantitative trait loci
Footnotes
Competing Interests: The authors declare that they have no competing interests.
References
- 1.WHO. Worldmalaria report 2012. 2012 http://www.who.int/malaria/publications/world_malaria_report_2012/en/index.html.
- 2.Conway DJ. Molecular epidemiology of malaria. Clin Microbiol Rev. 2007;20(1):188–204. doi: 10.1128/CMR.00021-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Miller LH, Baruch DI, Marsh K, Doumbo OK. The pathogenic basis of malaria. Nature. 2002;415(6872):673–679. doi: 10.1038/415673a. [DOI] [PubMed] [Google Scholar]
- 4.Perkins DJ, Were T, Davenport GC, Kempaiah P, Hittner JB, Ong'echa JM. Severe malarial anemia: innate immunity and pathogenesis. Int J Biol Sci. 2011;7(9):1427–1442. doi: 10.7150/ijbs.7.1427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Schofield L. Intravascular infiltrates and organ-specific inflammation in malaria pathogenesis. Immunol Cell Biol. 2007;85(2):130–137. doi: 10.1038/sj.icb.7100040. [DOI] [PubMed] [Google Scholar]
- 6.Stevenson MM, Lyanga JJ, Skamene E. Murine malaria: genetic control of resistance to Plasmodium chabaudi. Infect Immun. 1982;38(1):80–88. doi: 10.1128/iai.38.1.80-88.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Erdman LK, Finney CA, Liles WC, Kain KC. Inflammatory pathways in malaria infection: TLRs share the stage with other components of innate immunity. Mol Biochem Parasitol. 2008;162(2):105–111. doi: 10.1016/j.molbiopara.2008.08.006. [DOI] [PubMed] [Google Scholar]
- 8.Laroque A, Min-Oo G, Tam M, Radovanovic I, Stevenson MM, Gros P. Genetic control of susceptibility to infection with Plasmodium chabaudi chabaudi AS in inbred mouse strains. Genes Immun. 2012;13(2):155–163. doi: 10.1038/gene.2011.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Longley R, Smith C, Fortin A, Berghout J, McMorran B, Burgio G, et al. Host resistance to malaria: using mouse models to explore the host response. Mamm Genome. 2011;22(1-2):32–42. doi: 10.1007/s00335-010-9302-6. [DOI] [PubMed] [Google Scholar]
- 10.Walther M, Woodruff J, Edele F, Jeffries D, Tongren JE, King E, et al. Innate immune responses to human malaria: heterogeneous cytokine responses to blood-stage Plasmodium falciparum correlate with parasitological and clinical outcomes. J Immunol. 2006;177(8):5736–5745. doi: 10.4049/jimmunol.177.8.5736. [DOI] [PubMed] [Google Scholar]
- 11.Artavanis-Tsakonas K, Eleme K, McQueen KL, Cheng NW, Parham P, Davis DM, et al. Activation of a subset of human NK cells upon contact with Plasmodium falciparum-infected erythrocytes. J Immunol. 2003;171(10):5396–5405. doi: 10.4049/jimmunol.171.10.5396. [DOI] [PubMed] [Google Scholar]
- 12.Korbel DS, Newman KC, Almeida CR, Davis DM, Riley EM. Heterogeneous human NK cell responses to Plasmodium falciparum-infected erythrocytes. J Immunol. 2005;175(11):7466–7473. doi: 10.4049/jimmunol.175.11.7466. [DOI] [PubMed] [Google Scholar]
- 13.Modiano D, Petrarca V, Sirima BS, Nebie I, Diallo D, Esposito F, et al. Different response to Plasmodium falciparum malaria in west African sympatric ethnic groups. Proc Natl Acad Sci U S A. 1996;93(23):13206–13211. doi: 10.1073/pnas.93.23.13206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kremsner PG, Winkler S, Brandts C, Wildling E, Jenne L, Graninger W, et al. Prediction of accelerated cure in Plasmodium falciparum malaria by the elevated capacity of tumor necrosis factor production. Am J Trop Med Hyg. 1995;53(5):532–538. doi: 10.4269/ajtmh.1995.53.532. [DOI] [PubMed] [Google Scholar]
- 15.Lyke KE, Burges R, Cissoko Y, Sangare L, Dao M, Diarra I, et al. Serum levels of the proinflammatory cytokines interleukin-1 beta (IL-1beta), IL-6, IL-8, IL-10, tumor necrosis factor alpha, and IL-12(p70) in Malian children with severe Plasmodium falciparum malaria and matched uncomplicated malaria or healthy controls. Infect Immun. 2004;72(10):5630–5637. doi: 10.1128/IAI.72.10.5630-5637.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Driss A, Hibbert JM, Wilson NO, Iqbal SA, Adamkiewicz TV, Stiles JK. Genetic polymorphisms linked to susceptibility to malaria. Malar J. 2011;10:271. doi: 10.1186/1475-2875-10-271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ockenhouse CF, Hu WC, Kester KE, Cummings JF, Stewart A, Heppner DG, et al. Common and divergent immune response signaling pathways discovered in peripheral blood mononuclear cell gene expression patterns in presymptomatic and clinically apparent malaria. Infect Immun. 2006;74(10):5561–5573. doi: 10.1128/IAI.00408-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Franklin BS, Parroche P, Ataide MA, Lauw F, Ropert C, de Oliveira RB, et al. Malaria primes the innate immune response due to interferon-gamma induced enhancement of toll-like receptor expression and function. Proc Natl Acad Sci U S A. 2009;106(14):5789–5794. doi: 10.1073/pnas.0809742106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Omer FM, de Souza JB, Corran PH, Sultan AA, Riley EM. Activation of transforming growth factor beta by malaria parasite-derived metalloproteinases and a thrombospondin-like molecule. J Exp Med. 2003;198(12):1817–1827. doi: 10.1084/jem.20030713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Omer FM, Kurtzhals JA, Riley EM. Maintaining the immunological balance in parasitic infections: a role for TGF-beta? Parasitol Today. 2000;16(1):18–23. doi: 10.1016/s0169-4758(99)01562-8. [DOI] [PubMed] [Google Scholar]
- 21.Fairhurst RM, Wellems TE. Modulation of malaria virulence by determinants of Plasmodium falciparum erythrocyte membrane protein-1 display. Curr Opin Hematol. 2006;13(3):124–130. doi: 10.1097/01.moh.0000219655.73162.42. [DOI] [PubMed] [Google Scholar]
- 22.Doumbo OK, Thera MA, Kone AK, Raza A, Tempest LJ, Lyke KE, et al. High levels of Plasmodium falciparum rosetting in all clinical forms of severe malaria in African children. Am J Trop Med Hyg. 2009;81(6):987–993. doi: 10.4269/ajtmh.2009.09-0406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dondorp AM, Desakorn V, Pongtavornpinyo W, Sahassananda D, Silamut K, Chotivanich K, et al. Estimation of the total parasite biomass in acute falciparum malaria from plasma PfHRP2. PLoS Med. 2005;2(8):e204. doi: 10.1371/journal.pmed.0020204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chotivanich K, Udomsangpetch R, Simpson JA, Newton P, Pukrittayakamee S, Looareesuwan S, et al. Parasite multiplication potential and the severity of Falciparum malaria. J Infect Dis. 2000;181(3):1206–1209. doi: 10.1086/315353. [DOI] [PubMed] [Google Scholar]
- 25.Sharma S, DeOliveira RB, Kalantari P, Parroche P, Goutagny N, Jiang Z, et al. Innate immune recognition of an AT-rich stem-loop DNA motif in the Plasmodium falciparum genome. Immunity. 2011;35(2):194–207. doi: 10.1016/j.immuni.2011.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Coban C, Ishii KJ, Kawai T, Hemmi H, Sato S, Uematsu S, et al. Toll-like receptor 9 mediates innate immune activation by the malaria pigment hemozoin. J Exp Med. 2005;201(1):19–25. doi: 10.1084/jem.20041836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Parroche P, Lauw FN, Goutagny N, Latz E, Monks BG, Visintin A, et al. Malaria hemozoin is immunologically inert but radically enhances innate responses by presenting malaria DNA to Toll-like receptor 9. Proc Natl Acad Sci U S A. 2007;104(6):1919–1924. doi: 10.1073/pnas.0608745104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Augustijn KD, Kleemann R, Thompson J, Kooistra T, Crawford CE, Reece SE, et al. Functional characterization of the Plasmodium falciparum and P. berghei homologues of macrophage migration inhibitory factor. Infect Immun. 2007;75(3):1116–1128. doi: 10.1128/IAI.00902-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhang Y, Miura K, Li J, Tullo G, Zhu F, Hong L, et al. Macrophage migration inhibitory factor homolog from Plasmodium yoelii modulates monocyte recruitment and activation in spleen during infection. Parasitol Res. 2012;110(5):1755–1763. doi: 10.1007/s00436-011-2696-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shao D, Zhong X, Zhou YF, Han Z, Lin Y, Wang Z, et al. Structural and functional comparison of MIF ortholog from Plasmodium yoelii with MIF from its rodent host. Mol Immunol. 2010;47(4):726–737. doi: 10.1016/j.molimm.2009.10.037. [DOI] [PubMed] [Google Scholar]
- 31.Omer FM, de Souza JB, Riley EM. Differential induction of TGF-beta regulates proinflammatory cytokine production and determines the outcome of lethal and nonlethal Plasmodium yoelii infections. J Immunol. 2003;171(10):5430–5436. doi: 10.4049/jimmunol.171.10.5430. [DOI] [PubMed] [Google Scholar]
- 32.Pattaradilokrat S, Cheesman SJ, Carter R. Congenicity and genetic polymorphism in cloned lines derived from a single isolate of a rodent malaria parasite. Mol Biochem Parasitol. 2008;157(2):244–247. doi: 10.1016/j.molbiopara.2007.10.011. [DOI] [PubMed] [Google Scholar]
- 33.Li J, Pattaradilokrat S, Zhu F, Jiang H, Liu S, Hong L, et al. Linkage maps from multiple genetic crosses and loci linked to growth-related virulent phenotype in Plasmodium yoelii. Proc Natl Acad Sci U S A. 2011;108(31):E374–382. doi: 10.1073/pnas.1102261108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li J, Zhang Y, Liu S, Hong L, Sullivan M, McCutchan TF, et al. Hundreds of microsatellites for genotyping Plasmodium yoelii parasites. Mol Biochem Parasitol. 2009;166(2):153–158. doi: 10.1016/j.molbiopara.2009.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Stevenson MM, Riley EM. Innate immunity to malaria. Nat Rev Immunol. 2004;4(3):169–180. doi: 10.1038/nri1311. [DOI] [PubMed] [Google Scholar]
- 36.Torre S, van Bruggen R, Kennedy JM, Berghout J, Bongfen SE, Langat P, et al. Susceptibility to lethal cerebral malaria is regulated by epistatic interaction between chromosome 4 (Berr6) and chromosome 1 (Berr7) loci in mice. Genes Immun. 2013;14(4):249–257. doi: 10.1038/gene.2013.16. [DOI] [PubMed] [Google Scholar]
- 37.Qi Y, Zhu F, Li J, Fu Y, Pattaradilokrat S, Hong L, et al. Optimized protocols for improving the likelihood of cloning recombinant progeny from Plasmodium yoelii genetic crosses. Exp Parasitol. 2013;133(1):44–50. doi: 10.1016/j.exppara.2012.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lim DC, Cooke BM, Doerig C, Saeij JP. Toxoplasma and Plasmodium protein kinases: roles in invasion and host cell remodelling. Int J Parasitol. 2012;42(1):21–32. doi: 10.1016/j.ijpara.2011.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Melo MB, Jensen KD, Saeij JP. Toxoplasma gondii effectors are master regulators of the inflammatory response. Trends Parasitol. 2011;27(11):487–495. doi: 10.1016/j.pt.2011.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Saeij JP, Boyle JP, Coller S, Taylor S, Sibley LD, Brooke-Powell ET, et al. Polymorphic secreted kinases are key virulence factors in toxoplasmosis. Science. 2006;314(5806):1780–1783. doi: 10.1126/science.1133690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Saeij JP, Coller S, Boyle JP, Jerome ME, White MW, Boothroyd JC. Toxoplasma co-opts host gene expression by injection of a polymorphic kinase homologue. Nature. 2007;445(7125):324–327. doi: 10.1038/nature05395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cao J, Kaneko O, Thongkukiatkul A, Tachibana M, Otsuki H, Gao Q, et al. Rhoptry neck protein RON2 forms a complex with microneme protein AMA1 in Plasmodium falciparum merozoites. Parasitol Int. 2009;58(1):29–35. doi: 10.1016/j.parint.2008.09.005. [DOI] [PubMed] [Google Scholar]
- 43.Richard D, MacRaild CA, Riglar DT, Chan JA, Foley M, Baum J, et al. Interaction between Plasmodium falciparum apical membrane antigen 1 and the rhoptry neck protein complex defines a key step in the erythrocyte invasion process of malaria parasites. J Biol Chem. 2010;285(19):14815–14822. doi: 10.1074/jbc.M109.080770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Painter HJ, Campbell TL, Llinas M. The Apicomplexan AP2 family: integral factors regulating Plasmodium development. Mol Biochem Parasitol. 2011;176(1):1–7. doi: 10.1016/j.molbiopara.2010.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Balu B, Maher SP, Pance A, Chauhan C, Naumov AV, Andrews RM, et al. CCR4-associated factor 1 coordinates the expression of Plasmodium falciparum egress and invasion proteins. Eukaryot Cell. 2011;10(9):1257–1263. doi: 10.1128/EC.05099-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Culleton R, Kaneko O. Erythrocyte binding ligands in malaria parasites: intracellular trafficking and parasite virulence. Acta Trop. 2010;114(3):131–137. doi: 10.1016/j.actatropica.2009.10.025. [DOI] [PubMed] [Google Scholar]
- 47.Akimitsu N, Kim HS, Hamamoto H, Kamura K, Fukuma N, Arimitsu N, et al. Duffy antigen is important for the lethal effect of the lethal strain of Plasmodium yoelii 17XL. Parasitol Res. 2004;93(6):499–503. doi: 10.1007/s00436-004-1165-x. [DOI] [PubMed] [Google Scholar]
- 48.Swardson-Olver CJ, Dawson TC, Burnett RC, Peiper SC, Maeda N, Avery AC. Plasmodium yoelii uses the murine Duffy antigen receptor for chemokines as a receptor for normocyte invasion and an alternative receptor for reticulocyte invasion. Blood. 2002;99(8):2677–2684. doi: 10.1182/blood.v99.8.2677. [DOI] [PubMed] [Google Scholar]
- 49.Otsuki H, Kaneko O, Thongkukiatkul A, Tachibana M, Iriko H, Takeo S, et al. Single amino acid substitution in Plasmodium yoelii erythrocyte ligand determines its localization and controls parasite virulence. Proc Natl Acad Sci U S A. 2009;106(17):7167–7172. doi: 10.1073/pnas.0811313106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hadley TJ, Peiper SC. From malaria to chemokine receptor: the emerging physiologic role of the Duffy blood group antigen. Blood. 1997;89(9):3077–3091. [PubMed] [Google Scholar]
- 51.Car BD, Meloni F, Luisetti M, Semenzato G, Gialdroni-Grassi G, Walz A. Elevated IL-8 and MCP-1 in the bronchoalveolar lavage fluid of patients with idiopathic pulmonary fibrosis and pulmonary sarcoidosis. Am J Respir Crit Care Med. 1994;149(3 Pt 1):655–659. doi: 10.1164/ajrccm.149.3.8118632. [DOI] [PubMed] [Google Scholar]
- 52.Thompson WL, Karpus WJ, Van Eldik LJ. MCP-1-deficient mice show reduced neuroinflammatory responses and increased peripheral inflammatory responses to peripheral endotoxin insult. J Neuroinflammation. 2008;5:35. doi: 10.1186/1742-2094-5-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sen S, Ferdig M. QTL analysis for discovery of genes involved in drug responses. Curr Drug Targets Infect Disord. 2004;4(1):53–63. doi: 10.2174/1568005043480916. [DOI] [PubMed] [Google Scholar]
- 54.Kong Y. Btrim: a fast, lightweight adapter and quality trimming program for next-generation sequencing technologies. Genomics. 2011;98(2):152–153. doi: 10.1016/j.ygeno.2011.05.009. [DOI] [PubMed] [Google Scholar]
- 55.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25(14):1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10(3):R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25(16):2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Cingolani P, Platts A, Wang le L, Coon M, Nguyen T, Wang L, et al. A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w1118; iso-2; iso-3. Fly (Austin) 2012;6(2):80–92. doi: 10.4161/fly.19695. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Cytokine and chemokine responses of mice infected with three Plasmodium yoelii parasites. Mean levels of plasma cytokines and chemokines (FGF-basic, GM-CSF, IFN-γ, IL-1α, IL-1β, IL-2, IL-4, IL-5, IL-6, IL-10, IL-12 (p40/p70), IL-13, IL-17, IP-10, KC, MCP-1, MIG, MIP-1α, TNF-α, and VGEF) were measured as described in Experimental Procedures. Error bars represent standard deviations determined from eight mice. Red, from lethal P. y. nigeriensis N67-infected mice; purple, from lethal P. y. yoelii YM-infected mice; blue, from nonlethal P. y. yoelii 17XNL-infected mice; green, naïve batch mates.
Figure S2. Genetic loci linked to levels of cytokines and chemokines during Plasmodium yoelii infection. Plots of logarithm of odds (LOD) scores for day 4 cytokines and chemokines (FGF-basic, GM-CSF, IFN-γ, IL-10, IL-1α, IL-1β, IL-2, IL-4, IL-5, IP-10, KC, MCP-1, MIG, MIP-1α and TNF-α) that have a LOD = 2.0 or higher. One dash line indicates the cutoff level of LOD = 2.0. In some plots, a second dash line indicates significant level at p < 0.05 after 1000 permutations.
Table S1. Levels of cytokines and chemokines obtained from C57BL/6 mice infected with parents (N67 and 17XNL) and 22 progeny of the genetic cross at day 4 post infection.
Table S2. Nucleotide substitutions and predicted functions of candidate genes in chromosomal loci mapped to differential cytokine/chemokine responses between Plasmodium yoelii yoelii 17XNL and P. y. nigeriensis N67.