Abstract
Fanconi anemia (FA) is a rare genetic disorder caused by defects in a DNA damage repair system, the FA pathway. FA patients frequently develop squamous cell carcinoma (SCC) at sites that are associated with HPV-driven cancer including the female reproductive tract. To assess experimentally whether FA deficiency increases susceptibility to HPV-associated cervical/vaginal cancer, we monitored cancer incidence in the female lower reproductive tract of FA-deficient mice expressing HPV16 oncogenes, E6 and/or E7. FA deficiency specifically increased the incidence of cancers in mice expressing E7; but, this effect was not observed in mice just expressing E6. We also observed that E7, but not E6, induced DNA damage as scored by induction of γ-H2AX and 53BP1 nuclear-foci, and this induction was heightened in FA-deficient tissue. Finally, we discovered that this induction of DNA damage responses was recapitulated in mice deficient in expression of ‘pocket’ proteins, pRb, p107, and p130, which are established targets of E7. Our findings support the hypothesis that E7 induces cancer by causing DNA damage at least in part through the inactivation of pocket proteins. This hypothesis explains why a deficiency in DNA damage repair would increase susceptibility to E7-driven cancer.
Keywords: HPV16 E7, HPV-positive cancer, Fanconi Anemia
Introduction
Fanconi Anemia (FA) is an extremely rare heterogeneous and recessive genetic disorder. FA patients frequently display developmental abnormalities, defects and malignancies in hematopoietic cells that arise in early childhood, and shortened lifespans (1). To date 15 cellular genes (fanc genes) have been identified for which homozygous mutations are associated with FA. fanc genes encode proteins that act together to carry out repair of damaged DNA. The majority of the fanc gene products (FancA, B, C, E, F, G, L, and M) form a protein complex called the FA core complex. FancL protein in the core complex is an E3 ubiquitin ligase that mono-ubiquitinates FancD2 in the response of DNA damage (2, 3). This modified form of FancD2 is recruited to sites of DNA damage where it interacts with other proteins known to be involved in the DNA repair, such as BRCA1, FancD1/BRCA2, FancN/PALB2 and Rad51 leading to DNA repair (4-6). Disruption of this FA pathway leads to increased sensitivity to DNA cross-linkers, chromosomal instability, spontaneous sister chromatid exchange and cell cycle perturbations. The FA pathway acts together with the homologous recombination repair system (HR) to maintain genomic stability. Depletion of FancD2 inhibits the HR response, indicating that HR acts downstream of FancD2 (6). Increased genomic instability is believed to contribute to the increased susceptibility of FA patients to cancer.
In the general population, 99% of cervical cancer, and more than 50% of anal, vaginal, vulva, and penile cancers are associated with mucosotropic, high-risk HPVs. Among these HPVs, HPV16 is the genotype associated with most of these cancers (7, 8). HPV16 expresses three oncogenes, E5, E6, and E7. From our prior in vivo studies for HPV-associated cancers in mice, we learned that E7's manipulation of the cell cycle through its inactivation of cell cycle regulators including pRb, other pRb family proteins (p107 and p130), and the cdk inhibitor, p21, contributes to its promoting the development of cancers of the head/neck and/or anogenital regions (9-12). Inactivation of pRb by E7 is thought to contribute to alterations in differentiation, DNA damage responses, centrosome synthesis, and tumorigenesis (13-15). In addition, E6, through its inactivation of the tumor suppressor p53 and interactions with PDZ domain proteins, accelerates the incidence of E7-dependent cancers in mice (16). The mechanism for E5's role in carcinogenesis in the context of mouse models remains unclear but might be mediated through its activation of EGFR (17).
Persistent infection with oncogenic HPVs is necessary but not sufficient for the development of HPV-associated cancers. Recent studies identified a single-nucleotide polymorphism (SNP) in fancA, one of the 15 fanc genes, which is associated with an increased risk of cervical intraepithelial neoplasia grade 3 (CIN3) and cervical cancer (18) consistent with the fact that FA patients are at increased risk of anogenital cancers (1); however, another group failed to find this relationship with different SNPs for fancA (19). Thus a role of the FA pathway in anogenital cancers remains unclear. A role of the FA pathway in mitigating HPV-induced anogenital cancers, however, is predicted based upon a number of laboratory observations: 1) in cells expressing HPV16 E7, there is an accumulation of DNA breaks, as monitored by the comet assay, and an increased frequency of cells harboring γ-H2AX foci (20); 2) expression of HPV16 E7 causes an induction in the level of mono-ubiquitinated FancD2 protein, a marker for the activation of the FA pathway (21); 3) in FA-deficient cells, E7's induction of chromosomal abnormalities such as breaks and fusions is enhanced (21) as is HPV's induction of hyperplasia in differentiating keratinocytes (22), and 4) we previously observed increased susceptibility of HPV16 E7 transgenic mice to head and neck cancers on a FA-deficient background (23). Furthermore, It has been reported that about a half of SCCs in the patients in the International Fanconi Anemia Registry (IFAR) are located in the anogenital tract. The majority of FA patients with anogenital SCCs have a prior history of HPV-associated condylomas (1). Whether HPV is etiologically related to the development of anogenital SCCs in these patients remains unclear due to the very small number of FA patients.
We hypothesize that disruption of the FA pathway increases the frequency of HPV-associated cancers in the lower reproductive tract of female FA patients. To test this hypothesis, we utilized our animal model for HPV16-associated cancers of the lower reproductive tract. In contrast to our earlier study on the influence of FA-deficiency on head and neck cancers, which was limited to studies with HPV16 E7 transgenic mice, we included in our current study an evaluation of the influence of FA-deficiency on lower female reproductive tract cancers driven not only by HPV16 E7 oncogene, but also the HPV16 E6 oncogene, and the combination of the two. We found that deficiency in the FA pathway (i.e. in fancD2-null mice) significantly increased the frequency of HPV16 E7, but surprisingly not HPV16 E6, -induced cancers in the female lower reproductive tract. FancD2 loss independently and additively with HPV16 oncogenes led to an increase in proliferation of epithelium of cervix, but this did not correlate with the selective increase in cancers in E7-expressing mice. In contrast, an increased frequency of cells harboring markers for DNA damage responses in FA deficient, E7 transgenic and E6/E7 bi-transgenic mice, but not in non-transgenic and E6 transgenic mice, correlating with the selective increased susceptibility of E7-expressing mice to cancers on the FA deficient background. Finally, we discovered that this induction of DNA damage responses by E7 could be recapitulated in mice deficient in expression of pRb family pocket proteins, pRb, p107, and p130, well-known targets of E7. These results support the hypothesis that the FA pathway attenuates the oncogenic potential of HPV16 E7 specifically and this correlates with a suppression of DNA damage responses induced by E7, an activity that at least in part is mediated by its inactivation of the pRb family of pocket proteins.
Results
Disruption of the FA DNA damage repair pathway increases the incidence of HPV-associated cancers in the female lower reproductive tract
To address whether disruption of the FA pathway elevates susceptibility to HPV-associated cancers in the female lower reproductive tract, we utilized HPV16 oncogene-transgenic mice that direct expression of the viral oncogenes to stratified squamous epithelia including that of the cervix and vagina. HPV16 E6 transgenic (K14E6) and HPV16 E7 transgenic (K14E7) mice were crossed to fancD2 null mice deficient in the FA pathway to generate 8 different genotypes of mice; non-transgenic (NTG), K14E6, K14E7, and HPV16 E6/E7 bi-transgenic (K14E6E7) mice on the fancD2-sufficient and -deficient backgrounds. Six to seven week old virgin female mice of each genotype were treated with 17β-estradiol, which cooperates with HPV oncogenes to induce cancers in the female reproductive tract (24) for a period of 6 months. At this endpoint, the lower reproductive tracts were harvested, fixed, paraffin-embedded, sectioned and every tenth 5 μm section stained with H&E and histopathologically scored for the worst neoplastic disease (25).
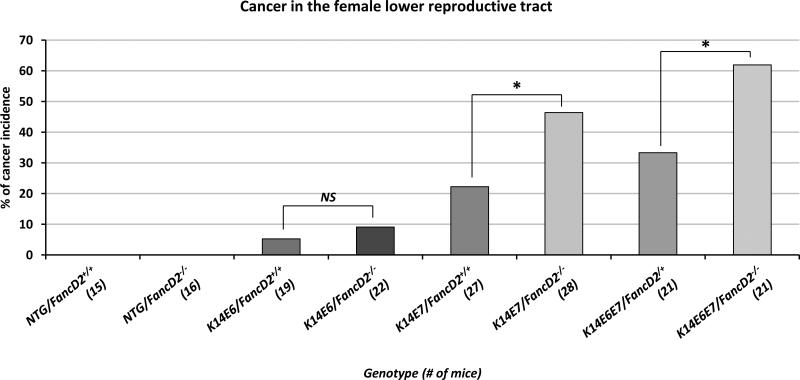
On the fancD2-sufficient background, E7 expression significantly increased the incidence of cancer in the female lower reproductive tract of K14E7 and K14E6E7 mice compared with NTG mice (NTG/FancD2+/+ vs. K14E7/FancD2+/+; P=0.03 and NTG/FancD2+/+ vs. K14E6E7/FancD2+/+; P=0.01 in Fig. 1). E6 expression failed to increase the incidence of cancer compared with NTG mice (NTG/FancD2+/+ vs. K14E6/FancD2+/+; P=0.30 in Fig. 1). These observations are consistent with our previous observations (24). Interestingly, fancD2 deficiency caused a significant increase in the cancer incidences of the reproductive tract of K14E7 and K14E6E7 mice (K14E7/FancD2+/+ vs. K14E7/FancD2−/−; P=0.03 and K14E6E7/FancD2+/+ vs. K14E6E7/FancD2−/−; P=0.04 in Fig. 1), but not in NTG and K14E6 mice. To discriminate any tissue specificity of the impact of FA pathway deficiency on HPV-associated carcinogenesis within the lower female reproductive tract, we analyzed the data obtained from the cervix and vagina separately. The incidence of cancers in K14E7 mice was significantly increased by fancD2 deficiency in both the cervix and the vagina. In K14E6E7 mice, fancD2 deficiency also caused an increased numbers of cervical or vaginal cancers, but the increases were not statistically significant (Supplementary Fig. 1). Our findings support the hypothesis that the FA pathway selectively suppresses the oncogenic activity of HPV16 E7 in causing cancers of the female lower reproductive tract.
Figure 1. HPV-associated cancer incidence in the lower reproductive tract.
Mice were treated with 17β-estradiol which is predominant estrogen for a period of 6 months to achieve a state of persistent estrus. After the estrogen treatment, the female lower reproductive tract was harvested, fixed and sectioned. Every tenth, 5 μm section was stained with hematoxylin and eosin (H&E) and histopathologically evaluated the worst lesion scored as the final diagnosis. Cervical and/or vaginal cancers were scored in each genotype of mice. The cancer incidence in K14E7/FancD2+/+ is significantly higher than it in NTG/FancD2+/+ (P=0.03) or K14E6/FancD2+/+ mice (P=0.07). fancD2 deficiency significantly increased cancer incidence in K14E7 and K14E6E7 mice. Asterisk (*) means significant difference (P<0.05). NS means no significant difference. All statistical analyses were performed using a one-sided Barnard's exact test.
Deficiency in the FA pathway triggers neoplastic disease and worsens the grade of E7-associated neoplastic disease, but not E6-associated neoplastic disease
HPV16 oncogene-transgenic mice develop a progressive disease ranging from hyperplasia and cervical (or vaginal) intraepithelial neoplasia grade 1-3, CIN1-3 (or VAIN1-3) to cervical (or vaginal) cancer similar to that seen in women (24). We therefore assessed whether FA deficiency led to a worse grade of neoplastic disease. Although none of NTG mice developed cancer on either the fancD2-sufficient or -deficient background, fancD2 deficiency alone did lead to a significantly increased severity of disease in the lower reproductive tract (NTG/FancD2+/+ vs. NTG/FancD2−/−; P=0.02 in Table 1), indicating that the FA deficiency alone can induce neoplastic cells in cervical or vaginal region, but is not sufficient to develop frank cancer in estrogen-treated mice. Consistent with the increased induction of cancers in K14E7 and K14E6E7 mice on the fancD2-deficient background, the severity of disease in the lower reproductive tract in these mice was significantly worse in mice deficient for fancD2 (K14E7/FancD2+/+ vs. K14E7/FancD2−/−; P=0.02 and K14E6E7/FancD2+/+ vs. K14E6E7/FancD2−/−; P=0.02 in Table 1). We also scored severity of disease in cervical and vaginal tissues separately. In most of our mice, the cervix displayed more severe disease than the vagina; nevertheless, in both tissues, fancD2 deficiency led to more severe disease in K14E7, and K14E6E7 mice, though not in K14E6 mice (Supplementary Table 1). This increased severity of disease (Table 1 and Supplementary Table 1) and the increased incidence of cancer (Fig. 1) in mice expressing E7 that are nulligenic for fancD2 again support the hypothesis that the FA pathway specifically inhibits the oncogenic potential of HPV16 E7 in the female lower reproductive tract. Moreover, FA deficiency itself can cause neoplastic lesions in arise in our in vivo model system (Table 1).
Table 1.
Histopathological analysis of disease grade in female lower reproductive tract from mice treated with 17β-estradiol for 6 months.
| Genotype | Group size, n | Severity of disease, n (%) |
||||
|---|---|---|---|---|---|---|
| Hyperplasia | CIN1/VIN1 | CIN2/VIN2 | CIN3/VIN3 | Cancer | ||
| NTG/FancD2+/+ | 15 | 14 (93) | 1 (7) | - | - | - |
| NTG/FancD2−/− | 16 | 10 (63) | 6 (37) | - | - | - |
| K14E6/FancD2+/+ | 19 | 4 (21) | 14 (74) | - | - | 1 (5) |
| K14E6/FancD2−/− | 22 | 5 (23) | 10 (46) | 4 (18) | 1 (5) | 2 (9) |
| K14E7/FancD2+/+ | 27 | - | 1 (4) | 8 (30) | 12 (44) | 6 (22) |
| K14E7/FancD2−/− | 28 | - | - | 4 (14) | 11 (39) | 13 (46) |
| K14E6E7/FancD2+/+ | 21 | - | - | 7 (33) | 7 (33) | 7 (33) |
| K14E6E7/FancD2−/− | 21 | - | - | 2 (10) | 6 (29) | 13 (62) |
Note: fancD2 deficiency significantly worsens the grade of neoplastic disease in NTG, K14E7, and K14E6E7 mice, but not in K14E6 mice (NTG/FancD2+/+ vs. NTG/FancD2−/−; P=0.02, K14E6/FancD2+/+ vs. K14E6/FancD2−/−; P=0.13, K14E7/FancD2+/+ vs. K14E7/FancD2−/−; P=0.02, and K14E6E7/FancD2+/+ vs. K14E6E7/FancD2−/−; P=0.02). All statistical analyses were performed by two-sided Wilcoxon rank sum test.
Expression levels of two biomarkers, MCM7 and p16, are up-regulated in E7-expressing carcinomas regardless of fancD2 gene status
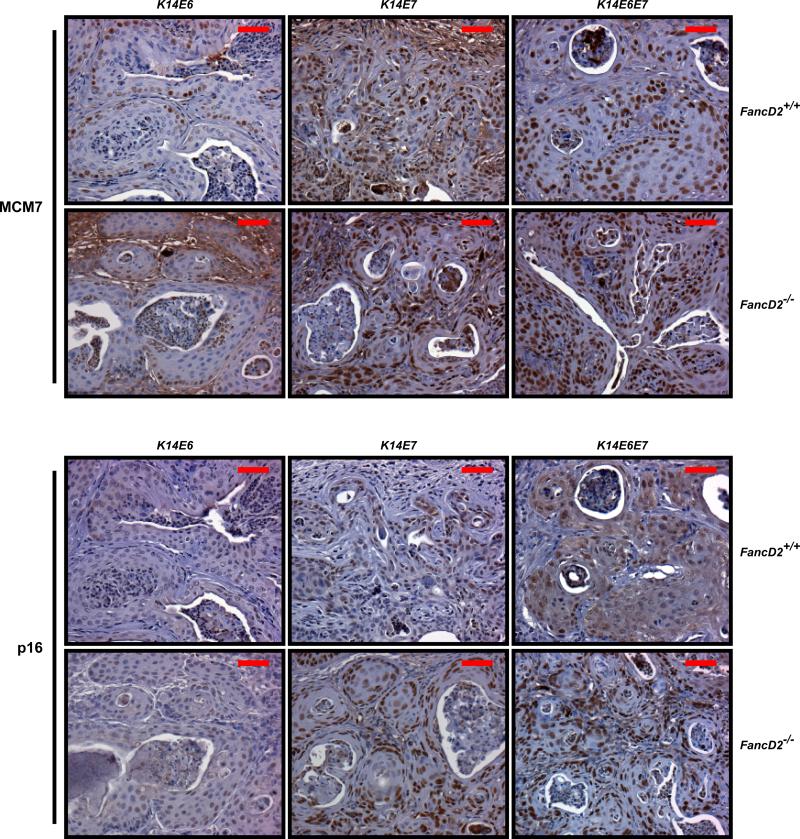
Of the HPV16 oncogenes, E7 plays a dominant role in the induction of MCM7 and p16, which are well-known biomarkers for HPV-positive cervical cancer in humans and mice (26-30). To determine if their expression levels are altered by FA deficiency, MCM7 and p16 specific immunohistochemistry was carried out (Fig. 2). On the fancD2-sufficient background, these two biomarkers were highly up-regulated in the nuclei of all of the cervical carcinomas arising in K14E7 and K14E6E7 mice. In the cervical carcinomas arising in K14E6, K14E7, and K14E6E7 mice, fancD2 deficiency did not cause any change in the expression pattern of either biomarkers compared to that observed in the same transgenic mice on the fancD2-sufficient background (Fig. 2). These findings lead us to conclude that these two biomarkers will be valuable for diagnosing HPV status in cancers from FA patients.
Figure 2. Expression of two biomarkers, MCM7 and p16, in cervical carcinomas derived from K14E6, K14E7, K14E6E7 mice on both fancD2-sufficient and -deficient backgrounds.
Shown are representative immunohistochemistry with anti-MCM7 and anti-p16. Brown-stained nuclear cells are positive for the both markers. Hematoxylin (Blue) is used for nuclear counter staining. Scale bar, 50 μm. *K14E6/FancD2+/+ tumor was a vaginal cancer due to no cervical cancer.
Proliferation is up-regulated independently and additively by HPV16 oncogenes and FA deficiency
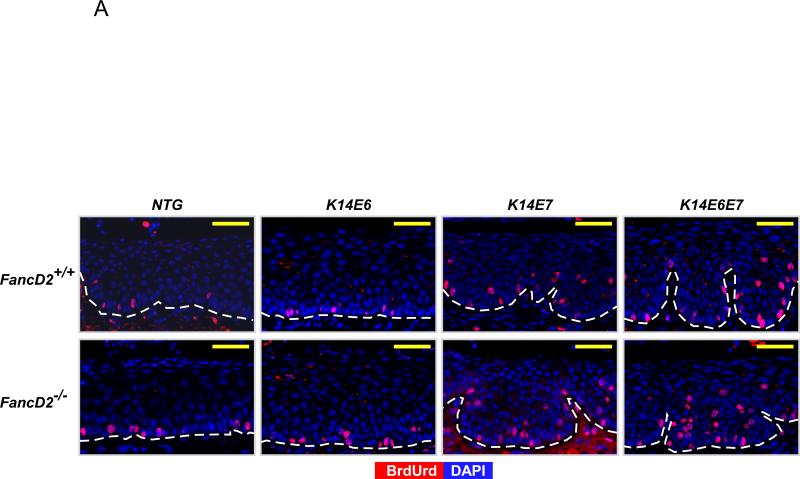
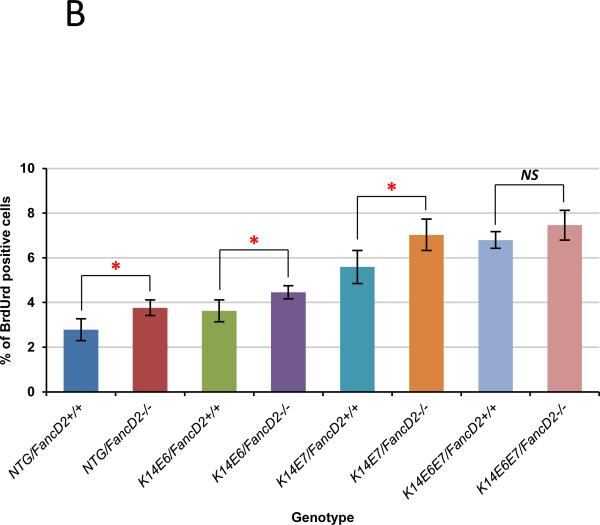
The observed neoplastic disease induced by HPV16 oncogene expression and/or the deficiency of the FA pathway led us to monitor if cell proliferation in the epithelium of the female lower reproductive tract was modulated by the FA pathway in the presence or absence of HPV oncogene expression. To score the frequency of cells supporting DNA synthesis in mice sufficient or deficient in the FA pathway, groups of mice (n ≥ 3) of the different genotypes described below were intraperitoneally injected with BrdUrd 1 hour before being sacrificed and sections of tissues subjected to BrdUrd-specific immunohistochemistry (Fig. 3A). BrdUrd-positive cells were quantified in cervical tissues from each genotype. In the cervical epithelium, on fancD2-sufficient background, both E6 and E7 independently and together (K14E6/FancD2+/+, K14E7/FancD2+/+, and K14E6E7/FancD2+/+) significantly increased the frequency of BrdUrd-positive cells compared to NTG/FancD2+/+ mice (Fig. 3B). E7 induced proliferation to a greater extent than E6 (K14E6/FancD2+/+ vs. K14E7/FancD2+/+; P=0.03 in Fig. 3B). In the context of the K14E6E7/FancD2+/+ mice, E6 and E7 showed additive effects on cell proliferation (K14E7/FancD2+/+ vs. K14E6E7/FancD2+/+; P=0.03 in Fig. 3B). Deficiency of fancD2 led to an increase in cell proliferation, regardless of HPV16 oncogene expression status. Except for K14E6E7 mice, the increased frequencies of BrdUrd-positive cells by fancD2 loss in NTG, K14E6, and K14E7 mice were statistically significant (P<0.05 in Fig. 3B). This increased DNA synthesis observed in fancD2 null mice is consistent with the knowledge that FancD2 regulates S-phase check point (31, 32) and the observations that deficiencies in the FA pathway lead to induced cell proliferation in other cell/tissue contexts (22, 23, 33). Our findings indicate that HPV oncogene expression and deficiency in the FA pathway are able to independently and additively induce cell proliferation in the epithelium of the cervix.
Figure 3. Cell proliferation modulated by HPV oncogenes and fancD2 deficiency.
A, to monitor the newly synthesized DNA at the cervical epithelia in each genotype of mice, mice were intraperitoneally injected with BrdUrd 1 hour before euthanized. Immunofluorescence with anti-BrdUrd (Red) was performed. DAPI (Blue) was used for a nuclear counterstaining. Scale bar, 50 μm. The white dot line indicates basal membrane. B, at least three mice from NTG, K14E6, K14E7, and K14E6E7 mice in the presence and absence of fancD2 expression were randomly selected and more than eight image frames of cells at the epithelia of cervix were quantified for each genotype of mice. The amount of BrdUrd-positive cells over total number of cells was plotted in each case (columns); bars, Standard deviation (SD). Asterisk (*) means significant difference (P<0.05). NS means no significant difference (P>0.05). In the epithelial layer of the cervical tissues, HPV oncogene expression significantly induced the number of BrdUrd-positive cells (NTG/FancD2+/+ vs. K14E6/FancD2+/+, K14E7/FancD2+/+, or K14E6E7/FancD2+/+; P<0.05). fancD2 deficiency statistically increased proliferation in NTG, K14E6, and K14E7 mice (NTG/FancD2+/+ vs. NTG/FancD2−/−, K14E6/FancD2+/+ vs. K14E6/FancD2−/−, and K14E7/FancD2+/+ vs. K14E7/FancD2−/−; P<0.05), not in K14E6E7 mice (K14E6E7/FancD2+/+ vs. K14E6E7/FancD2−/−; P=0.15). All statistical analyses were performed using a two-sided Wilcoxon Rank sum test.
FA pathway suppresses DNA damage caused by HPV16 E7 expression
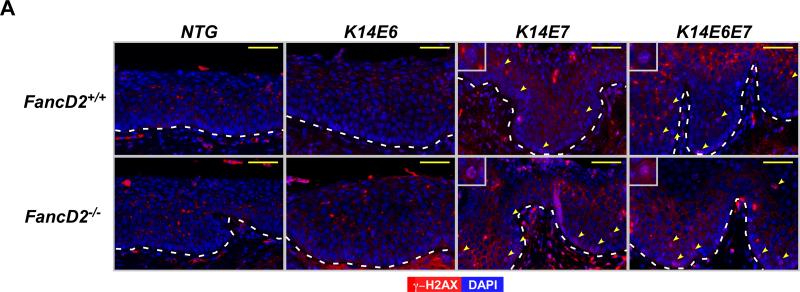
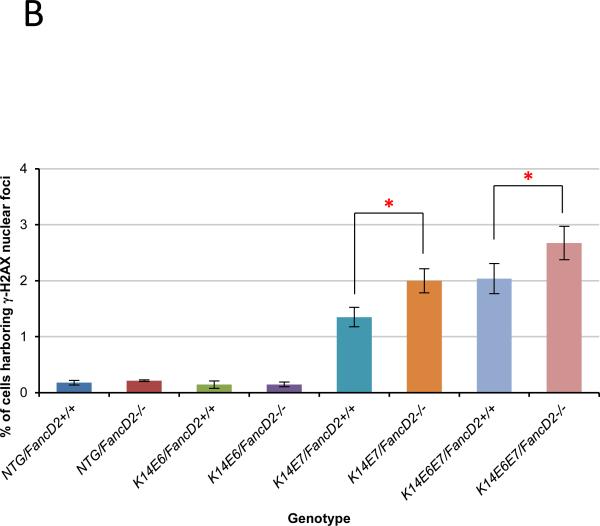
The hypothesis that HPV16 E7 causes DNA damage and that this leads to genomic instability, is supported by several in vitro observations: a) E7 increases single- and/or double-strand DNA breaks (20, 34, 35), b) E7 causes chromosomal abnormalities including losses or gains of specific alleles (35, 36) and c) E7 induces gross genetic changes as scored by karyotypic analyses for chromatid breaks, fusions or polyploidy (36). In the absence of fancD2 expression, E7 induced genomic instability was found to be further increased in an in vitro assay (21). Based on these prior findings, we decided to monitor the presence of DNA damage by quantifying the abundance of cells harboring γ-H2AX nuclear-foci, a surrogate marker for DNA double strand breaks (DSBs). Immunofluorescence for anti-γ-H2AX was performed on sections of the female lower reproductive tract from each genotype of mice (Fig. 4A). In NTG and K14E6 mice, there were few cells positive for γ-H2AX nuclear-foci in the epithelium of the cervix regardless of fancD2 gene status (Fig. 4A). In contrast, E7 expression led to a statistically significant increase in the frequency of the cell with γ-H2AX positive nuclear-foci in the cervical epithelia. This E7-driven DNA damage was significantly elevated by fancD2 deficiency in the cervical epithelial cells (K14E7/FancD2+/+ vs. K14E7/FancD2−/− and K14E6E7/FancD2+/+ vs. K14E6E7/FancD2−/−; P<0.05 in Fig. 4B). Although E6 expression was not sufficient to cause DNA damage, it caused a greater than additive effect on the DNA damage in the epithelial cells in K14E6E7 mice on fancD2-sufficient background (K14E7/FancD2+/+ vs. K14E6E7/FancD2+/+; P=0.03 in Fig. 4B). We used an alternative marker for DSBs (37), p53 binding protein 1 (53BP1), to indirectly measure the level of DNA damage in these same tissues. As observed with γ-H2AX foci staining, the number of 53BP1 nuclear foci-positive cells was increased by fancD2 loss only in the presence of E7 expression (Supplementary figure 2). These results support the hypothesis that the FA pathway suppresses primarily the capacity of E7 to induce DNA damage.
Figure 4. FancD2 deficiency induced E7-dependent DNA damage response via β-H2AX.
A, to detect DNA damage response, tissue sections from each group were stained for anti-γ-H2AX (Red) antibody. DAPI (Blue) is used for a nuclear counterstaining. Scale bar, 50 μm. γ-H2AX nuclear-foci positive cells are highlighted by yellow arrows in the images. Insets provide magnified views of cells with γ-H2AX positive nuclear-foci. The white dot line indicates basal membrane. B, at least three mice from NTG, K14E6, K14E7, and K14E6E7 mice in the presence and absence of fancD2 expression were randomly selected and more than eight image frames of cells at the epithelia of cervix were quantified for each mouse. The amount of γ-H2AX nuclear-foci positive cells over total number of cells was plotted in each case (columns); bar, Standard deviation (SD). In the epithelial layer of the cervical tissues, E7 expression and E6/E7 double expression significantly induced the number of γ-H2AX nuclear-foci positive cells on fancD2-sufficient background (NTG/FancD2 or K14E6/FancD2+/+ vs. K14E7/FancD2+/+ or K14E6E7/FancD2+/+; P<0.05). fancD2 deficiency statistically increased proliferation in K14E7 and K14E6E7 mice (K14E7/FancD2+/+ vs. K14E7/FancD2−/− and K14E6E7/FancD2+/+ vs. K14E6E7/FancD2−/−; P<0.05), not in NTG and K14E6 mice. Asterisk (*) means significant difference (P<0.05). NS means no significant difference (P>0.05). All statistical comparisons were performed using a two-sided Wilcoxon Rank sum test.
DNA damage was induced in mice deficient for pocket protein family members, pRb, p107, and p130 that are well-known targets of E7
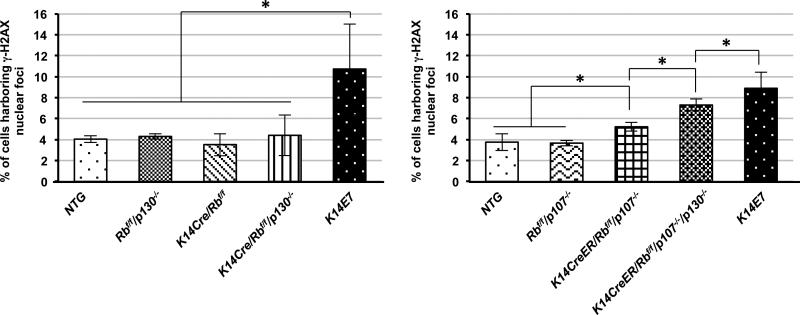
Our mouse model studies described above (Fig. 3 and 4) support the conclusion that increases in DNA damage response better correlate with the development of neoplastic disease including carcinomas in the cervical regions than do increases in cell proliferation. Because the increase in DNA damage response was observed exclusively with E7, not E6, we focused next on defining the mechanism by which E7 induces DNA damage responses. HPV16 E7 associates with more than a hundred cellular proteins (38) and deregulates multiple cellular processes (15). Among the E7 target proteins, pRb, and its related ‘pocket’ proteins, p107 and p130, are the most well-known target proteins of E7 and long considered the most relevant targets of E7 in the context of HPV-associated carcinogenesis (10). To test whether deficiency of these three pocket proteins recapitulates the observed E7-driven DNA damage in the cervical epithelium, we utilized our germ-line or conditional knockout mice in pRb, p107, or p130 protein for avoiding or decreasing morbidity of mice (11); K14Cre/pRbf/f, pRbf/f/p130−/−, and K14Cre/pRbf/f/p130−/− mice on the same FVB/129/C57 mixed genetic background and pRbf/f/p107−/−, K14CreER/pRbf/f/p107−/− and K14CreER/pRbf/f/p130f/f/p107−/− mice on the same CD1/129/C57 mixed genetic background. These mice were treated with 17β-estradiol for 6 weeks at 6-7 weeks of age. Immunofluorescence for anti-γ-H2AX was performed on sections of the female lower reproductive tract from each genotype of mice (data not shown). We quantified the abundance of cells harboring γ-H2AX nuclear-foci in the cervical epithelia of the each genotype of mice. Single knockout mice (K14Cre/pRb/f/f, pRbf/f/p130−/−, and pRbf/f/p107−/−) failed to increase the frequency of the cell harboring γ-H2AX nuclear-foci compared with NTG mice (Fig. 5). Combined loss of pRb and p107 significantly increased the number of γ-H2AX nuclear-foci positive cells (K14CreER/pRbf/f/p107−/− vs. NTG or pRbf/f/p107−/−; P=0.05 in Fig. 5). However, pRb and p130 double deletion did not cause a significant increase in the abundance of cells with γ-H2AX nuclear-foci (K14Cre/pRbf/f/p130−/− vs. NTG; P=0.66 in Fig. 5). Interestingly, our triple knockout mice (K14CreER/pRbf/f/p130f/f/p107−/−) significantly induced the number of γ-H2AX foci positive cells compared to NTG, p107, or K14CreER/pRbf/f/p107−/− (P<0.05 in Fig. 5). However, this increase in DNA damage in the triple knockout mice remained significantly lower than that observed in K14E7 mice (K14CreEr/pRbf/f/p130f/f/p107−/− vs. K14E7; P= 0.05 in Fig. 5). These findings indicate that E7's inactivation of pocket proteins contributes to but is not sufficient to account for all of its induction of DNA damage.
Figure 5. Deficiency of Pocket protein family members Increased DNA damage via γ-H2AX in the cervical epithelia.
A and B. At least three mice from each genotype were randomly selected and more than eight image frames of cells at the epithelia of cervix were quantified for each mouse. The amount of γ-H2AX nuclear-foci positive cells over total number of cells was plotted in each case (columns); bar, Standard deviation (SD). A. In the epithelial layer of the cervical tissues, K14Cre/pRbf/f, pRbf/f/p130−/−, and K14Cre/pRbf/f/p130−/− mice failed to increase the number of γ-H2AX nuclear-foci positive cells compared with NTG mice (P>0.05). E7 expression cause a significant increase in DNA damage compared with NTG, K14Cre/pRbf/f, pRbf/f/p130−/−, or K14Cre/pRbf/f/p130−/− mice (P<0.05). B. Single deficiency of p107 failed to increase DNA damage via γ-H2AX foci (pRbf/f/p107−/− vs. NTG; P>0.05). Double deletion of pRb and p107 significantly induced the number of cells harboring with γ-H2AX foci compared with single deletion of p107 (K14CreER/pRbf/f/p107−/− vs. pRbf/f/p107−/−; P<0.05). The frequency of γ-H2AX foci positive cells in the triple knockout mice was significantly higher than it in pRb and p107 double knockout mice, but was significantly lower than it in K14E7 mice. (K14CreER/pRbf/f/p130f/f/p107−/− vs. K14CreER/pRbf/f/p107−/−; P<0.05 and K14CreER/pRbf/f/p130f/f/p107−/− vs. K14E7; P<0.05). Asterisk (*) means significant difference (P<0.05). NS means no significant difference (P>0.05). All statistical comparisons were performed using a two-sided Wilcoxon Rank sum test. Note that In Fig. 4, E7 expression induced γ-H2AX in only ~1.5% of cells, but in the data presented in this figure, it induces it in ~10% of cells. This difference is solely attributed to the difference in genetic backgrounds between the mice used in the studies reported in these two figures, not due to differences in staining efficiency, because the same low % of cells with γ-H2AX positive foci was observed in sections of K14E7 transgenic mice taken from the study reported in Fig. 4, when they were stained alongside the samples from the experiment reported in this figure.
Discussion
The HPV etiology of cervical/vaginal cancer in the FA patient population has remained unclear due to a very small number and limited access to FA patients. In this study, we provide evidence that the incidence of HPV16-associated SCCs in the female lower reproductive tract is significantly accelerated in mice deficient for fancD2 (Fig. 1), and that this increased susceptibility is driven by the E7 oncogene, which clearly induces DNA damage responses in these tissues. These findings compare favorably with our prior study in which E7 transgenic mice were more susceptible to head/neck cancers (HNCs) on the same FA deficient background (23). An interesting difference between these two mouse model studies is that, in the HNC model, HNCs were induced by treatment with a mutagen, 4-NQO. This finding could easily be rationalized by the knowledge that E7 induces DNA damage responses, and thereby could increase the efficiency with which a mutagen causes mutations. But in the current study, estrogen is used as a co-carcinogen, and its role in inducing carcinogenesis within the lower reproductive tract is dependent upon its ability to induce cell proliferation through its nuclear receptor ERα (39), and therefore is not likely to reflect the argued mutagenic property of estrogen metabolites. A unifying explanation to explain the results of both studies is that E7 itself is indirectly promoting the accumulations of damaged DNA and/or preventing its repair, and this is primarily responsible for the increased susceptibility to cancer in FA-deficient mice, regardless of the mechanism of action of the cofactor used in helping to drive carcinogenesis at the different tissue sites. This hypothesis does not discount the role of HPV oncogenes E6 and E7 in modulating other biological processes including cell proliferation, differentiation and apoptosis; rather, it provides insight into the reason for the specific synergy between E7 and FA-deficiency.
HPV16 E6 augmentation of E7-induced DNA damage correlates with its synergistic role in HPV-associated carcinogenesis
By studying E6/E7 double transgenic mice as well as the two single HPV16 oncogene transgenic mice, this current study provided several new insights into our understanding the potential role of HPV oncogenes in causing cancers in FA patients. Most surprising among these insights was the observation that deficiency in the FA pathway did not increase cancer susceptibility in E6-transgenic mice (Fig. 1). A previous study showed that loss of fancD2 induced apoptosis mediated by p53, causing developmental abnormalities in vivo. This p53-dependent apoptosis was prevented by silencing p53 expression (40). Furthermore, heterozygosity in the p53 gene increased susceptibility of epithelial tumors in fancD2 knockout mice, with most of these tumors showing loss of heterozygosity (LOH) in p53 alleles. Presumably, the loss of p53 gave an advantage to cells deficient in the FA pathway by preventing p53-mediated apoptosis induced by DNA damage that would normally have been repaired by a functional FA pathway (41). Accumulation of the unrepaired DNA damage was predicted to contribute to tumor development in the absence of p53 and FancD2. In our study, E6 inactivation of p53 apparently was not sufficient to significantly increase the susceptibility of FA-deficient mice to develop cancers of the female lower reproductive tract (Table 1 and Fig. 1). One possible explanation is the fact that residual p53 activity is retained in the E6-expressing tissues in our mouse model (42). We also observed that E6 augmented the level of E7-driven DNA damage responses, though, alone, E6 failed to induce the level of DNA damage (Fig. 4). One hypothesis to explain this observation is that in our in vivo mouse model 1) E6 does not induce DNA damage, whereas E7 does; 2) the reduction in p53 activity in the presence of E6 leads to a further accumulation of E7-induced DNA damage, and 3) this increased accumulation of damaged DNA increases the propensity for tumors to arise. Such a hypothesis is supported by our observation that cancer incidence in the double transgenic mice was higher than E7 single transgenic mice (Fig. 1).
Pocket proteins as relevant E7 targets in the induction of DNA damage in the cervical epithelium
We discovered that the combined deletion of two or more pocket protein family members, cell-cycle regulators that are well-known targets of E7, induced DNA damage in vivo. This supports the hypothesis that endogenous DNA damage, which should be repaired before cells enter and/or pass through S phase, accumulates in cells not expressing pocket protein family members (43). More interestingly, in the double deletions of pRb/107 and pRb/p130, p107 was more potent in suppressing DNA damage than p130 in the cervical epithelium (Fig. 5). Each pocket protein family member regulates a subset of the family of E2F transcription factors (43), even though they all share a conserved overall structure, including sequence homology in a large C-terminal domain known to mediate their interaction with viral oncoproteins such as SV40 LT, Adenovirus E1A, and HPV E7 (44-47). We posit that in the stratified cervical epithelium, p107 and p130 play spatially different roles in regulating the cell cycle, and that this difference explains the differences in degrees of DNA damage arising in cells nulligenic for either of these two pocket proteins. Specifically, we previously learned that p107 and p130 are differentially expressed within the cervical epithelium; p107 is highly expressed in the basal and parabasal epithelial layers, whereas p130 is expressed mainly in the supra-parabasal cells (11). This difference in the location of expression may be one explanation for why we found differences in the levels of DNA damage in the stratified cervical epithelial layers of mice deficient for pRb/p107 vs. pRb/p130. Our findings are consistent with prior studies showing p107 and p130 are not simply replacements of pRb; rather, they possess specific tumor suppressing capabilities within different tissue contexts (48-51). Nevertheless, it is interesting to note that the triple deletion of pRb, p107, and p130 still failed to completely recapitulate E7's level of induction of DNA damage as scored by the presence of γ-H2AX nuclear-foci (Fig. 5). Interestingly this correlates with the insufficiency of the triple deletion of the pocket proteins to fully recapitulate the oncogenic potential of E7 in the same tissue (11). We conclude that E7's ability to target other cellular factors contributes to its ability to induce DNA damage. While we have not attempted to identify other target(s) of E7 that are relevant here, it is interesting to note that E7's inactivation of the tumor suppressor p21 also contributes to E7's oncogenic potential in the cervix (9).
Deficiency in the FA pathway and HPV infection
While the data from this study points to a selective influence of FA deficiency on promoting the oncogenicity of the HPV oncogene, E7, FA deficiency may promote other stages of HPV disease, specifically at the level of viral infection. In our current study we observed that fancD2 deficiency causes hyperplasia of the epithelia lining the female lower reproductive tract (Fig. 3B), similar to what we previously observed in the epithelia lining the head and neck (23). Both are sites of natural HPV infection. These in vivo findings corroborate in vitro studies identifying FancD2 as an S-phase regulator with post-translational modification via the ATM and ATR pathways (31, 32, 52). Importantly, cell cycle progression is critical and necessary for the establishment of HPV infections (53). Thus it is reasonable to hypothesize that the hyperplasia resulting from fancD2 deficiency not only contributes to tumorigenesis but also to the establishment of HPV infections. In support of this hypothesis, a recent study analyzing the presence of HPV-DNA in oral rinses found a very high prevalence of HPV infections in the oral cavity of FA patients (54). FA deficiency may also contribute to a later stage of the HPV infectious life cycle, the production of progeny virus. The HPV life cycle is tied to the differentiation of the host epithelium, wherein progeny viruses are produced in the terminally differentiating compartment (55). We previously reported that the HPV16 E7 reprograms suprabasal cells to support DNA synthesis, which is essential for the production of progeny virus (9). Recent studies argued that the FA pathway suppresses the ability of HPV to induce DNA synthesis in the suprabasal compartment (22, 56).
Taken together, we conclude that FA deficiency promotes HPV-associated carcinogenesis at multiple levels including two different steps in the infectious life cycle as well as in directly potentiating the oncogenic properties of the viral E7 oncogene.
Materials and Methods
Mice
K14E6 transgenic mice (57) and K14E7 transgenic mice (58) on the inbred FVB genetic background, were crossed to fancD2 gene knockout (FancD2−/−) mice (59) on the inbred 129 genetic background, to generate F1 mice, K14E6/FancD2+/− and K14E7/FancD2+/− on the same FVB/129 mixed background. All experimental mice (NTG/FancD2+/+, NTG/FancD2−/−, K14E6/FancD2+/+, K14E6/FancD2−/−, K14E7/FancD2+/+, K14E7/FancD2−/−, K14E6E7/FancD2+/+, and K14E6E7/FancD2−/− mice) were females generated by intercrossing F1 mice. K14Cre/pRbf/f, pRbf/f /p130−/−, and K14Cre/pRbf/f/p130−/− were generated on the same FVB/129/C57 mixed genetic background as described previously (11). pRbf/f/p107−/−, K14CreER/pRbf/f/p107−/− and K14CreER/pRbf/f/p130f/f/p107−/− were generated on the same CD1/129/C57 mixed genetic background as described previously (11). All mice were genotyped by PCR. The mice were housed in the Association for Assessment of Laboratory Animal Care-approved McArdle Laboratory Animal Care Unit. All experimental procedures were carried out in accordance with our animal protocol approved by the University of Wisconsin Institutional Animal Care and Use Committee.
Estrogen treatment and histopathological analyses of lower reproductive tract
Six to seven week old, virgin female mice were implanted with pellets delivering 17β-estradiol (0.05mg/60 days) for a period of 6 months to achieve a state of persistent estrus previously shown to be sufficient to induce cervical cancers in HPV16 oncogene-transgenic mice (24, 27). After 6 months treatment with 17β-estradiol, the lower reproductive tract was harvested and fixed according to previously described methods (24) and sectioned. Every tenth, 5 μm section was stained with hematoxylin and eosin (H&E) and histopathologically evaluated for the worst stage of neoplastic disease in the lower reproductive tract (cervix and vagina).
Immunofluorescence
Immunofluorescence was performed as described previously (24, 27). Antibodies used included anti-p16 (1:50 in 5% horse serum; M156, Santa Cruz Biotech.), anti-Mcm7 (1:200 in 5% horse serum; Neomarkers), anti-BrdUrd (1:50 in 5% horse serum; Calbiochem), and anti-γ-H2AX (1:100 in 5% horse serum; Millipore)
Quantifications of newly synthesized DNA and γ-H2AX nuclear-foci positive cells
To quantify the level of cell proliferation in the epithelium of the cervix, BrdUrd-positive cells were scored and divided by the total number of cells in each epithelial compartment to derive the percentage BrdUrd-positive cells. To evaluate DNA damage response, γ-H2AX nuclear-foci positive cells were quantified in the epithelial layers of the cervical epithelial compartment. For these quantifications, at least three mice of each genotype were randomly selected and eight or more 40X microscopic fields of the cervical tissues were scored per mouse sample.
Statistical analysis
Barnard's exact test was used to determine the significance of difference in cancer incidence between each genotype of mice. Wilcoxon rank sum test was used to determine the significance of difference in the severity of the disease in the lower reproductive tract as previously described (24). To determine the significance for BrdUrd-positive nuclei and γ-H2AX foci positive cells between each group of mice, Wilcoxon rank-sum test was used.
Supplementary Material
Acknowledgements
We thank Norman Drinkwater for advice on statistical analysis of the data reported in this study and Markus Grompe for the fancD2-null mouse strain. This study was supported by grants from the NIH (CA098428, CA022443).
Footnotes
Conflict of interest
The authors declared no conflict of interest.
Literature cited
- 1.Kutler DI, Singh B, Satagopan J, Batish SD, Berwick M, Giampietro PF, et al. A 20-year perspective on the International Fanconi Anemia Registry (IFAR). Blood. 2003 Feb 15;101(4):1249–56. doi: 10.1182/blood-2002-07-2170. [DOI] [PubMed] [Google Scholar]
- 2.Garner E, Smogorzewska A. Ubiquitylation and the Fanconi anemia pathway. FEBS Lett. [Research Support, N.I.H., Extramural Research Support, Non-U.S. Gov't Review] 2011 Sep 16;585(18):2853–60. doi: 10.1016/j.febslet.2011.04.078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Joenje H, Patel KJ. The emerging genetic and molecular basis of Fanconi anaemia. Nat Rev Genet. 2001 Jun;2(6):446–57. doi: 10.1038/35076590. [DOI] [PubMed] [Google Scholar]
- 4.Crossan GP, Patel KJ. The Fanconi anaemia pathway orchestrates incisions at sites of crosslinked DNA. J Pathol. [Research Support, Non-U.S. Gov't Review] 2012 Jan;226(2):326–37. doi: 10.1002/path.3002. [DOI] [PubMed] [Google Scholar]
- 5.Kennedy RD, D'Andrea AD. The Fanconi Anemia/BRCA pathway: new faces in the crowd. Genes Dev. 2005 Dec 15;19(24):2925–40. doi: 10.1101/gad.1370505. [DOI] [PubMed] [Google Scholar]
- 6.Knipscheer P, Raschle M, Smogorzewska A, Enoiu M, Ho TV, Scharer OD, et al. The Fanconi anemia pathway promotes replication-dependent DNA interstrand cross-link repair. Science. [Research Support, N.I.H., Extramural Research Support, Non-U.S. Gov't Research Support, U.S. Gov't, Non-P.H.S.] 2009 Dec 18;326(5960):1698–701. doi: 10.1126/science.1182372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schiffman M, Castle PE, Jeronimo J, Rodriguez AC, Wacholder S. Human papillomavirus and cervical cancer. Lancet. [Review] 2007 Sep 8;370(9590):890–907. doi: 10.1016/S0140-6736(07)61416-0. [DOI] [PubMed] [Google Scholar]
- 8.Watson M, Saraiya M, Ahmed F, Cardinez CJ, Reichman ME, Weir HK, et al. Using population-based cancer registry data to assess the burden of human papillomavirus-associated cancers in the United States: overview of methods. Cancer. [Research Support, U.S. Gov't, P.H.S.] 2008 Nov 15;113(10 Suppl):2841–54. doi: 10.1002/cncr.23758. [DOI] [PubMed] [Google Scholar]
- 9.Shin MK, Balsitis S, Brake T, Lambert PF. Human papillomavirus E7 oncoprotein overrides the tumor suppressor activity of p21Cip1 in cervical carcinogenesis. Cancer Res. 2009 Jul 15;69(14):5656–63. doi: 10.1158/0008-5472.CAN-08-3711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Munger K, Werness BA, Dyson N, Phelps WC, Harlow E, Howley PM. Complex formation of human papillomavirus E7 proteins with the retinoblastoma tumor suppressor gene product. Embo J. 1989 Dec 20;8(13):4099–105. doi: 10.1002/j.1460-2075.1989.tb08594.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shin MK, Sage J, Lambert PF. Inactivating all three rb family pocket proteins is insufficient to initiate cervical cancer. Cancer Res. 2012 Oct 15;72(20):5418–27. doi: 10.1158/0008-5472.CAN-12-2083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shin MK, Pitot HC, Lambert PF. Pocket proteins suppress head and neck cancer. Cancer Res. 2012 Mar 1;72(5):1280–9. doi: 10.1158/0008-5472.CAN-11-2833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Balsitis SJ, Sage J, Duensing S, Munger K, Jacks T, Lambert PF. Recapitulation of the effects of the human papillomavirus type 16 E7 oncogene on mouse epithelium by somatic Rb deletion and detection of pRb-independent effects of E7 in vivo. Mol Cell Biol. 2003 Dec;23(24):9094–103. doi: 10.1128/MCB.23.24.9094-9103.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Collins AS, Nakahara T, Do A, Lambert PF. Interactions with pocket proteins contribute to the role of human papillomavirus type 16 E7 in the papillomavirus life cycle. J Virol. 2005 Dec;79(23):14769–80. doi: 10.1128/JVI.79.23.14769-14780.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.McLaughlin-Drubin ME, Munger K. The human papillomavirus E7 oncoprotein. Virology. 2009 Feb 20;384(2):335–44. doi: 10.1016/j.virol.2008.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shai A, Brake T, Somoza C, Lambert PF. The human papillomavirus E6 oncogene dysregulates the cell cycle and contributes to cervical carcinogenesis through two independent activities. Cancer Res. 2007 Feb 15;67(4):1626–35. doi: 10.1158/0008-5472.CAN-06-3344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Genther SM, Sterling S, Duensing S, Munger K, Sattler C, Lambert PF. Quantitative role of the human papillomavirus type 16 E5 gene during the productive stage of the viral life cycle. J Virol. 2003 Mar;77(5):2832–42. doi: 10.1128/JVI.77.5.2832-2842.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang SS, Bratti MC, Rodriguez AC, Herrero R, Burk RD, Porras C, et al. Common variants in immune and DNA repair genes and risk for human papillomavirus persistence and progression to cervical cancer. The Journal of infectious diseases. [Research Support, N.I.H., Extramural Research Support, N.I.H., Intramural Research Support, Non-U.S. Gov't] 2009 Jan 1;199(1):20–30. doi: 10.1086/595563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Juko-Pecirep I, Ivansson EL, Gyllensten UB. Evaluation of Fanconi anaemia genes FANCA, FANCC and FANCL in cervical cancer susceptibility. Gynecologic oncology. [Research Support, Non-U.S. Gov't] 2011 Aug;122(2):377–81. doi: 10.1016/j.ygyno.2011.04.014. [DOI] [PubMed] [Google Scholar]
- 20.Duensing S, Munger K. The human papillomavirus type 16 E6 and E7 oncoproteins independently induce numerical and structural chromosome instability. Cancer Res. 2002 Dec 1;62(23):7075–82. [PubMed] [Google Scholar]
- 21.Spardy N, Duensing A, Charles D, Haines N, Nakahara T, Lambert PF, et al. The human papillomavirus type 16 E7 oncoprotein activates the Fanconi anemia (FA) pathway and causes accelerated chromosomal instability in FA cells. J Virol. 2007 Dec;81(23):13265–70. doi: 10.1128/JVI.01121-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hoskins EE, Morris TA, Higginbotham JM, Spardy N, Cha E, Kelly P, et al. Fanconi anemia deficiency stimulates HPV-associated hyperplastic growth in organotypic epithelial raft culture. Oncogene. 2009 Feb 5;28(5):674–85. doi: 10.1038/onc.2008.416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Park JW, Pitot HC, Strati K, Spardy N, Duensing S, Grompe M, et al. Deficiencies in the Fanconi anemia DNA damage response pathway increase sensitivity to HPV-associated head and neck cancer. Cancer Res. 2010 Dec 1;70(23):9959–68. doi: 10.1158/0008-5472.CAN-10-1291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Riley RR, Duensing S, Brake T, Munger K, Lambert PF, Arbeit JM. Dissection of human papillomavirus E6 and E7 function in transgenic mouse models of cervical carcinogenesis. Cancer Res. 2003 Aug 15;63(16):4862–71. [PubMed] [Google Scholar]
- 25.Alter BP, Greene MH, Velazquez I, Rosenberg PS. Cancer in Fanconi anemia. Blood. 2003 Mar 1;101(5):2072. doi: 10.1182/blood-2002-11-3597. [DOI] [PubMed] [Google Scholar]
- 26.Strati K, Pitot HC, Lambert PF. Identification of biomarkers that distinguish human papillomavirus (HPV)-positive versus HPV-negative head and neck cancers in a mouse model. Proc Natl Acad Sci U S A. 2006 Sep 19;103(38):14152–7. doi: 10.1073/pnas.0606698103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Brake T, Connor JP, Petereit DG, Lambert PF. Comparative analysis of cervical cancer in women and in a human papillomavirus-transgenic mouse model: identification of minichromosome maintenance protein 7 as an informative biomarker for human cervical cancer. Cancer Res. 2003 Dec 1;63(23):8173–80. [PubMed] [Google Scholar]
- 28.Pyeon D, Newton MA, Lambert PF, den Boon JA, Sengupta S, Marsit CJ, et al. Fundamental differences in cell cycle deregulation in human papillomavirus-positive and human papillomavirus-negative head/neck and cervical cancers. Cancer Res. 2007 May 15;67(10):4605–19. doi: 10.1158/0008-5472.CAN-06-3619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Begum S, Cao D, Gillison M, Zahurak M, Westra WH. Tissue distribution of human papillomavirus 16 DNA integration in patients with tonsillar carcinoma. Clin Cancer Res. 2005 Aug 15;11(16):5694–9. doi: 10.1158/1078-0432.CCR-05-0587. [DOI] [PubMed] [Google Scholar]
- 30.Gillison ML, Lowy DR. A causal role for human papillomavirus in head and neck cancer. Lancet. 2004 May 8;363(9420):1488–9. doi: 10.1016/S0140-6736(04)16194-1. [DOI] [PubMed] [Google Scholar]
- 31.Taniguchi T, Garcia-Higuera I, Andreassen PR, Gregory RC, Grompe M, D'Andrea AD. S-phase-specific interaction of the Fanconi anemia protein, FANCD2, with BRCA1 and RAD51. Blood. 2002 Oct 1;100(7):2414–20. doi: 10.1182/blood-2002-01-0278. [DOI] [PubMed] [Google Scholar]
- 32.Pichierri P, Rosselli F. The DNA crosslink-induced S-phase checkpoint depends on ATR-CHK1 and ATR-NBS1-FANCD2 pathways. Embo J. 2004 Mar 10;23(5):1178–87. doi: 10.1038/sj.emboj.7600113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhang QS, Marquez-Loza L, Eaton L, Duncan AW, Goldman DC, Anur P, et al. Fancd2−/− mice have hematopoietic defects that can be partially corrected by resveratrol. Blood. [Research Support, N.I.H., Extramural Research Support, U.S. Gov't, Non-P.H.S.] 2010 Dec 9;116(24):5140–8. doi: 10.1182/blood-2010-04-278226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Duensing S, Duensing A, Flores ER, Do A, Lambert PF, Munger K. Centrosome abnormalities and genomic instability by episomal expression of human papillomavirus type 16 in raft cultures of human keratinocytes. J Virol. 2001 Aug;75(16):7712–6. doi: 10.1128/JVI.75.16.7712-7716.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Duensing S, Munger K. Human papillomaviruses and centrosome duplication errors: modeling the origins of genomic instability. Oncogene. 2002 Sep 9;21(40):6241–8. doi: 10.1038/sj.onc.1205709. [DOI] [PubMed] [Google Scholar]
- 36.White AE, Livanos EM, Tlsty TD. Differential disruption of genomic integrity and cell cycle regulation in normal human fibroblasts by the HPV oncoproteins. Genes Dev. 1994 Mar 15;8(6):666–77. doi: 10.1101/gad.8.6.666. [DOI] [PubMed] [Google Scholar]
- 37.Huyen Y, Zgheib O, Ditullio RA, Jr., Gorgoulis VG, Zacharatos P, Petty TJ, et al. Methylated lysine 79 of histone H3 targets 53BP1 to DNA double-strand breaks. Nature. 2004 Nov 18;432(7015):406–11. doi: 10.1038/nature03114. [DOI] [PubMed] [Google Scholar]
- 38.Huh KW, DeMasi J, Ogawa H, Nakatani Y, Howley PM, Munger K. Association of the human papillomavirus type 16 E7 oncoprotein with the 600-kDa retinoblastoma protein-associated factor, p600. Proc Natl Acad Sci U S A. 2005 Aug 9;102(32):11492–7. doi: 10.1073/pnas.0505337102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chung SH, Wiedmeyer K, Shai A, Korach KS, Lambert PF. Requirement for estrogen receptor alpha in a mouse model for human papillomavirus-associated cervical cancer. Cancer Res. 2008 Dec 1;68(23):9928–34. doi: 10.1158/0008-5472.CAN-08-2051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Liu TX, Howlett NG, Deng M, Langenau DM, Hsu K, Rhodes J, et al. Knockdown of zebrafish Fancd2 causes developmental abnormalities via p53-dependent apoptosis. Dev Cell. 2003 Dec;5(6):903–14. doi: 10.1016/s1534-5807(03)00339-3. [DOI] [PubMed] [Google Scholar]
- 41.Houghtaling S, Granville L, Akkari Y, Torimaru Y, Olson S, Finegold M, et al. Heterozygosity for p53 (Trp53+/−) accelerates epithelial tumor formation in fanconi anemia complementation group D2 (Fancd2) knockout mice. Cancer Res. 2005 Jan 1;65(1):85–91. [PubMed] [Google Scholar]
- 42.Shai A, Pitot HC, Lambert PF. p53 Loss synergizes with estrogen and papillomaviral oncogenes to induce cervical and breast cancers. Cancer Res. 2008 Apr 15;68(8):2622–31. doi: 10.1158/0008-5472.CAN-07-5266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sun A, Bagella L, Tutton S, Romano G, Giordano A. From G0 to S phase: a view of the roles played by the retinoblastoma (Rb) family members in the Rb-E2F pathway. J Cell Biochem. 2007 Dec 15;102(6):1400–4. doi: 10.1002/jcb.21609. [DOI] [PubMed] [Google Scholar]
- 44.Mayol X, Grana X, Baldi A, Sang N, Hu Q, Giordano A. Cloning of a new member of the retinoblastoma gene family (pRb2) which binds to the E1A transforming domain. Oncogene. 1993 Sep;8(9):2561–6. [PubMed] [Google Scholar]
- 45.Hannon GJ, Demetrick D, Beach D. Isolation of the Rb-related p130 through its interaction with CDK2 and cyclins. Genes Dev. 1993 Dec;7(12A):2378–91. doi: 10.1101/gad.7.12a.2378. [DOI] [PubMed] [Google Scholar]
- 46.Ewen ME, Ludlow JW, Marsilio E, DeCaprio JA, Millikan RC, Cheng SH, et al. An N-terminal transformation-governing sequence of SV40 large T antigen contributes to the binding of both p110Rb and a second cellular protein, p120. Cell. 1989 Jul 28;58(2):257–67. doi: 10.1016/0092-8674(89)90840-4. [DOI] [PubMed] [Google Scholar]
- 47.Dyson N, Buchkovich K, Whyte P, Harlow E. The cellular 107K protein that binds to adenovirus E1A also associates with the large T antigens of SV40 and JC virus. Cell. 1989 Jul 28;58(2):249–55. doi: 10.1016/0092-8674(89)90839-8. [DOI] [PubMed] [Google Scholar]
- 48.Simpson DS, Mason-Richie NA, Gettler CA, Wikenheiser-Brokamp KA. Retinoblastoma family proteins have distinct functions in pulmonary epithelial cells in vivo critical for suppressing cell growth and tumorigenesis. Cancer Res. 2009 Nov 15;69(22):8733–41. doi: 10.1158/0008-5472.CAN-09-1359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Schaffer BE, Park KS, Yiu G, Conklin JF, Lin C, Burkhart DL, et al. Loss of p130 accelerates tumor development in a mouse model for human small-cell lung carcinoma. Cancer Res. 2010 May 15;70(10):3877–83. doi: 10.1158/0008-5472.CAN-09-4228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Robanus-Maandag E, Dekker M, van der Valk M, Carrozza ML, Jeanny JC, Dannenberg JH, et al. p107 is a suppressor of retinoblastoma development in pRb-deficient mice. Genes Dev. 1998 Jun 1;12(11):1599–609. doi: 10.1101/gad.12.11.1599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ho VM, Schaffer BE, Karnezis AN, Park KS, Sage J. The retinoblastoma gene Rb and its family member p130 suppress lung adenocarcinoma induced by oncogenic K-Ras. Oncogene. 2009 Mar 12;28(10):1393–9. doi: 10.1038/onc.2008.491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Taniguchi T, Garcia-Higuera I, Xu B, Andreassen PR, Gregory RC, Kim ST, et al. Convergence of the fanconi anemia and ataxia telangiectasia signaling pathways. Cell. 2002 May 17;109(4):459–72. doi: 10.1016/s0092-8674(02)00747-x. [DOI] [PubMed] [Google Scholar]
- 53.Pyeon D, Pearce SM, Lank SM, Ahlquist P, Lambert PF. Establishment of human papillomavirus infection requires cell cycle progression. PLoS pathogens. [Research Support, N.I.H., Extramural Research Support, Non-U.S. Gov't] 2009 Feb;5(2):e1000318. doi: 10.1371/journal.ppat.1000318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.de Araujo MR, Rubira-Bullen IR, Santos CF, Dionisio TJ, Bonfim CM, De Marco L, et al. High prevalence of oral human papillomavirus infection in Fanconi's anemia patients. Oral Dis. [Comparative Study Research Support, Non-U.S. Gov't] 2011 Sep;17(6):572–6. doi: 10.1111/j.1601-0825.2011.01803.x. [DOI] [PubMed] [Google Scholar]
- 55.Longworth MS, Laimins LA. Pathogenesis of human papillomaviruses in differentiating epithelia. Microbiol Mol Biol Rev. 2004 Jun;68(2):362–72. doi: 10.1128/MMBR.68.2.362-372.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hoskins EE, Morreale RJ, Werner SP, Higginbotham JM, Laimins LA, Lambert PF, et al. The Fanconi Anemia Pathway Limits Human Papillomavirus Replication. Journal of virology. 2012 May 23; doi: 10.1128/JVI.00408-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Song S, Pitot HC, Lambert PF. The human papillomavirus type 16 E6 gene alone is sufficient to induce carcinomas in transgenic animals. J Virol. 1999 Jul;73(7):5887–93. doi: 10.1128/jvi.73.7.5887-5893.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Herber R, Liem A, Pitot H, Lambert PF. Squamous epithelial hyperplasia and carcinoma in mice transgenic for the human papillomavirus type 16 E7 oncogene. J Virol. 1996 Mar;70(3):1873–81. doi: 10.1128/jvi.70.3.1873-1881.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Houghtaling S, Timmers C, Noll M, Finegold MJ, Jones SN, Meyn MS, et al. Epithelial cancer in Fanconi anemia complementation group D2 (Fancd2) knockout mice. Genes Dev. 2003 Aug 15;17(16):2021–35. doi: 10.1101/gad.1103403. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.